- Department of Biochemistry and Molecular Biology, Central University of Kerala, Kasaragod, India
Generally, changes in the metabolic status of cells under conditions like hypoxia and accumulation of lactate can be sensed by various sensing mechanisms, leading to modulation of a number of signal transduction pathways and transcription factors. Several of the proangiogenic cytokines like VEGF, FGF, PDGF, TGF-β, Ang-2, ILs, etc. are secreted by cancer cells, under hypoxic microenvironment. These cytokines bind to their receptors on the endothelial cells and activates a number of signaling pathways including Akt/PIP3, Src, p38/MAPK, Smad2/3, etc., which ultimately results in the proliferation and migration of endothelial cells. Transcription factors that are activated in response to the metabolic status of tumors include HIFs, NF-κb, p53, El-2, and FOXO. Many of these transcription factors has been reported to be regulated by a class of histone deacetylase called sirtuins. Sirtuins are NAD+ dependent histone deacetylases that play pivotal role in the regulation of tumor cell metabolism, proliferation, migration and angiogenesis. The major function of sirtuins include, deacetylation of histones as well as some non-histone proteins like NF-κB, FOXOs, PPAR⋎, PGC1-α, enzymes like acetyl coenzymeA and structural proteins like α tubulin. In the cell, sirtuins are generally considered as the redox sensors and their activities are dependent on the metabolic status of the cell. Understanding the intricate regulatory mechanisms adopted by sirtuins, is crucial in devising effective therapeutic strategies against angiogenesis, metastasis and tumor progression. Keeping this in mind, the present review focuses on the role of sirtuins in the process of tumor angiogenesis and the regulatory mechanisms employed by them.
Introduction
Angiogenesis, the process of formation of new blood vessels from pre-existing ones, is essential for the normal growth, development and wound healing. Apart from this, angiogenesis is also inevitable for tumor growth and metastasis (1–5). The expression and secretion of various modulators of angiogenesis is regulated by microenvironmental factors like hypoxia and accumulation of different metabolites (6–8). Under conditions like hypoxia, a number of signal transduction pathways and transcription factors like PPARα, PGC-1α, AMPK, FOXOs, etc. gets activated (9, 10). The expression and activation of these transcription factors has been reported to be regulated by a class of histone deacetylase called sirtuins or SIRT (11, 12). Sirtuins are NAD+ dependent histone deacetylases that play a vital role in the regulation of metabolism, aging, oncogenesis, angiogenesis and cancer progression (13, 14). It has been reported that SIRT1 can function as a redox sensor, and its activity might be dependent on the overall metabolic status of the cell (15), since it has been shown to regulate the stabilization of transcription factors such as HIF1α under hypoxic conditions (16). Therefore, understanding the regulatory mechanisms employed by sirtuins to modulate tumor angiogenesis is essential for developing effective anti-cancer and anti-angiogenic therapeutic strategies.
Sirtuins: Mechanism of Action and Classification
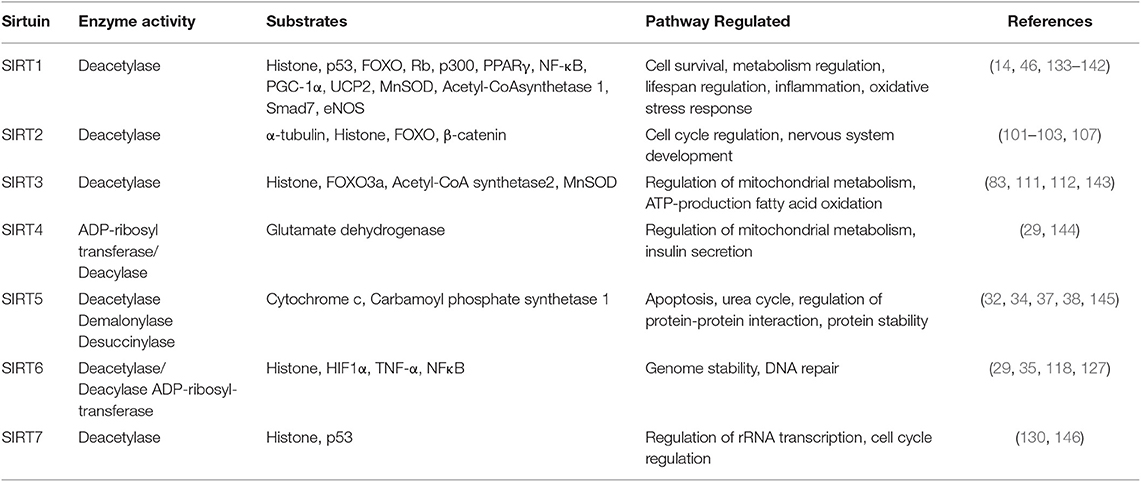
In mammals, seven homologs of sirtuins, i.e., SIRT1–SIRT7 (17, 18) which were initially described as class III HDACs (Histone deacetylases), are now known as class III KDACS (Lysine deacetylases) (19). The proposed mechanism of Sirtuin deacetylation is reported to be ADPR-peptidyl-imidate (20) where, Sirtuins catalyze NAD+-dependent deacetylation of acetyl lysine, producing nicotinamide, deacetylated lysine, and 2′-O-acetyl-ADP-ribose (21). The major function of sirtuins involve, removal of acetyl groups from the acetyl lysine-modified proteins (22, 23). The reaction gets initiated when, NAD+ binds to the catalytic site of sirtuin, with the C1 of NAD+ getting placed at the channel junction that, houses the acetyl lysine (24). To understand how increasing levels of NAD+ affects sirtuin activity, NAD+ synthesis was enhanced by supplementing different precursors for NAD+ like nicotinic acid to Preiss-Handler pathway and the result showed increased activation of sirtuins and other enzymes which are NAD+ dependent (25). Cellular [NAD+]/[NADH] ratio is reported to control deacetylase activity of the sirtuins where, NAD+ works as activator, and both nicotinamide and reduced nicotinamide adenine dinucleotide (NADH) acts as inhibitors (26, 27). Sirtuins (SIRT1-3, 5, and 7) catalyze deacetylation reaction on lysine residues of target proteins (28) whereas, SIRT4 and SIRT6 catalyze ADP-ribosylation reaction, by transferring ribosyl moiety to the substrates (29). Sirtuins carry out transcriptional repression where acetylated histones H1, H3, and H4 act as substrates (30). In addition, a number of non-histone proteins like nuclear factor-κB (NFκB), forkhead box type O transcription factors (FOXO), peroxisome proliferator-activated receptor ⋎ (PPAR⋎), coactivator 1α (PGC-1α), enzymes like acetyl coenzyme A (CoA) synthetase 2 (AceCS2), and structural proteins, such as α-tubulin are also deacetylated by sirtuins (29). In contrast to other KDACs, whose only function include deacetylation (31), sirtuins can also remove other groups like glutaryl (32), crotonyl (33), succinyl (34), palmitoyl (35), and myristoyl (36) groups. SIRT1-3 is reported to deacylate hydrophobic (butyryl group) and SIRT5, acidic acyl group (Malonyl group) in histones (37, 38). Also, some non-histone proteins like IDH2, MnSOD and TNFα have been reported to be deacylated by sirtuins (39). It is observed that SIRT4 has both, deacylase activity in leucine metabolism and lipoamidase activity in decarboxylation of pyruvate, to generate acetyl CoA (40, 41). The intracellular distribution of sirtuins differs. While SIRT1, 6 and 7 are located within the nucleus, SIRT2 is located in the cytoplasm and SIRT 3, 4, and 5 are located within mitochondria (42). In addition, SIRT1 and SIRT3 are known to shuttle to cytoplasm and nucleus, respectively (43, 44). These findings establish sirtuins as important players in epigenetic gene regulations.
Sirtuins in Endothelial Cell Functions
Different classes of sirtuins have been widely studied in endothelial cell growth and maintenance (45–47). While, blocking the function of SIRT1 reduced endothelial sprout formation, migration and the assembly of primitive vascular network (14) it was observed that, knockdown of SIRT1 altered the levels of sprouting angiogenesis due to reduction of MMP14 expression (48, 49). Potente et al. reported that SIRT1 deacetylates FOXO1, a negative regulator of angiogenesis, as SIRT1- deficient ECs showed abnormal angiogenic behavior due to FOXO1 activity (14). Nutrient deprivation and cellular energy shortage increase the levels of NAD+ and thus the expression and activation of sirtuins (50). It was observed that, endothelial tip cells employ anaerobic glycolysis for generating ATP (Warburg effect) rather than oxidative phosphorylation (51). In addition to promoting endothelial cell proliferation and angiogenesis, this makes ECs more resistant to hypoxia too (52, 53). Recent studies have established that SIRT1 modulates tip and stalk behavior through deacetylation of intracellular domain (NICD) of NOTCH1 in tumor associated endothelial cells (54). Interestingly, sirtuins also regulate, endothelial homeostasis by modulating the endothelial nitric oxide synthase (eNOS) (55). Recent studies reveal that endothelial SIRT1 deficiency, causes fibrosis due to aberrant secretion of ligands of Wnt and Notch pathways, as well as proteolytic fragments of glycocalyx core protein (56). Some studies also reported that SIRT1 can mediate transcriptional repression in association with Hey2 and Hes1 during vascular development (57, 58). In ECs, over expression of SIRT1 prevents cellular senescence, enhances vasodilatory responses, and alleviates aging-induced vascular impairment (59, 60) whereas, SIRT1 deficiency results in reduced migration in response to chemoattractant (14). In addition to SIRT1, SIRT2 is reported to regulate the survival and energy metabolism of ECs. Studies by Zhang et al. demonstrated that SIRT2 inhibition reduce the survival rate of PIEC cells as it causes mitochondrial depolarization (61). SIRT2 is also reported to promote Ang II-induced cytoskeletal remodeling in ECs (62). In addition, SIRT2 knock down studies revealed altered expression of migration associated genes like CALD1 (caldesmon) and CNN2 (calponin) (63, 64). SIRT3 was observed to increase survival of ECs especially during hypoxia through elevating the levels of deacetylation of FOXO3 (65). Recent studies also revealed that in SIRT3 deficient endothelial cells the expression of PFKFB3 was downregulated causing attenuation of glycolysis and angiogenesis (66). SIRT4 however appears to inhibit mononuclear cell adhesion to pulmonary microvascular endothelial cells through repression of E-selectin and VCAM-1 (67). While, angiogenic capacity of endothelial progenitor cells were significantly reduced due to down regulation of CXCR4/JAK2/SIRT5 signaling (68), it was observed that, SIRT6 protects endothelial cells from DNA damage and telomere dysfunction (62, 69). Though various members of the SIRT family has been implicated in the regulation of EC biology, the role of SIRT-1 is the most widely studied ones.
Sirtuins in Tumor Angiogenesis and their Regulatory Strategies
Expression pattern of sirtuins varies in different types of cancers. While, SIRT1, 4, 5, and 7 have been reported to be upregulated in certain cancers (70–72), the same SIRT1as well as SIRT2 and SIRT6 is shown to be downregulated in breast cancer, hepatic cell carcinoma (73), gliomas, gastric carcinomas (74, 75) and colon adenocarcinoma (76). SIRT1 mainly, mediates heterochromatin formation by deacetylation of histone H1 K26, histone H3 K9 and histone H4 K16, thereby causing deacetylation of non-histone proteins, like transcription factors (E2F1, p53, FOXO, BCL6, p53, Rb), DNA repair proteins and signaling factors (77). SIRT1 mediates regulation of gene expression in response to metabolic status by modulating FOXOs (78). Such a deacetylation of FOXOs by SIRT1 alters various signaling pathways, inhibit apoptosis and regulates mechanisms involved in oxidative stress (79, 80). In general, p53 negatively regulates angiogenesis either, by increasing the production of anti-angiogenic factors or inhibiting pro-angiogenic factors (81). SIRT1 has been reported to regulate neovascularization, through reducing the transcriptional activity of p53 by deacetylation of lysine (45, 46, 82). Apart from SIRT1, SIRT3 and SIRT7 has also been reported to deacetylate p53 thus, negating p53 activity (83, 84). SIRT1 also deacetylates other transcription factors like p73, E2F1, SMAD 7, NFKB and modulate apoptosis and inflammatory responses (45, 46, 85). It was observed that resveratrol, a SIRT1 activator reduced total VEGFR2 expression and inhibited phosphorylation of VEGFR2 by VEGF (86). Also, SIRT1 negatively modulates Delta-like ligand 4 (DLL4)/Notch pathway, inactivates elongation factor2 through activation of ELF2kinase and ultimately inhibits the proliferation and migration of vascular endothelial cells (87, 88). It is reported that SIRT1 deacetylation at K14 and K20 of PH domain is necessary for binding of Akt to PIP3 and further activation during tumor angiogenesis (89, 90). Several reports suggest that, SIRT1 deacetylate eNOS, stimulate its activity and enhance NO production and tumor angiogenesis (91, 92). Also, FOXO1 and FOXO3 have been reported to repress eNOS, suggesting a link between SIRT1, FOXO and eNOS (93). Increase in SIRT1 deacetylase activity and a consecutive HIF2α activity in ECs, results in acidification and reprogramming toward glutamine metabolism during induction of angiogenesis (94, 95). Studies by Kunhiraman et al. and Edatt et al. reveal that glycolytic inhibition using 2-DG at a sublethal concentration increased the expression and activity of SIRT1, causing reduced expression of angiogenesis associated genes like VEGF and MMP9 (96, 97). Contrary to these reports, Portmann et al., Li et al., and Suzuki et al., report that SIRT1 and VEGF expression is positively correlated during hypoxia induced angiogenesis in breast cancer and lung cancer (98–100). It therefore appears that, the SIRT-1 mediated regulation of angiogenesis and factors regulating it, is largely context dependent.
Like SIRT1, SIRT2 also deacetylate proteins like α-tubulin and histones, being co-localized with tubulin (101–103). Hu et al., demonstrated that SIRT2 knockdown prevented STAT3 phosphorylation and translocation to nucleus, thus decreasing the secretion of VEGF (104, 105). In addition, SIRT2 is reported to directly interact with β-catenin thereby altering the expression of genes like MMPs during tumor angiogenesis (106, 107). Also, it is observed that, SerRS (seryl-tRNA synthetase) plays tumor suppressor and anti-angiogenic role by collaborating with SIRT2 to antagonize c-Myc, a known angiogenic and oncogenic gene (108). Another class of sirtuins, SIRT3 is reported to mediate deacetylation of histones, regulate the stability of tubulin polymers (44, 109, 110), mediate induction of uncoupling protein−1 and regulate Acetyl-CoA synthetase activity (111, 112). Contrary to SIRT1 and SIRT2, SIRT3 is reported to have an opposing effect on angiogenesis, as loss of SIRT3 in human breast cancers, resulted in the upregulation of HIF-1α target genes like VEGF and genes involved in glycolysis (113, 114). Interestingly, it was observed that, SIRT3 overexpression reduced angiogenesis by negatively regulating ROS production, glycolysis as well as HIF-1α stabilization, ultimately resulting in a negative regulation of Warburg effect (115). SIRT4, 5, and 6 has been majorly reported to carry out ADP-ribosylation, desuccinylation and demalonylation rather that deacetylation (34, 116–118). ADP-ribosylation, regulate the activity of glutamate dehydrogenase and PARP (119, 120). Studies from our lab and others has demonstrated that PARPs can regulate the VEGF/VEGFR2 signaling circuit by either transactivation of VEGFR2 or poly ADP ribosylating VEGF to reduce its activity (7, 96, 121). Desuccinylation by SIRT5 suppresses the activities of pyruvate dehydrogenase complex and succinate dehydrogenase (117) leading to the accumulation of succinate and mitochondrial reactive oxygen species, thereby activating HIF1α (122). SIRT5 can also cause desuccinylation and negative regulation of S100A10, a protein that regulate invasion and motility (123). Generally, NAD+ levels influence the secretion of various cytokines by inflammatory cells (124). It was found that SIRT6 over expression in pancreatic cancer cells increased TNFα and IL8 production through ADP-ribosylation mediated Ca2+ responses (125) and elevated levels of IL8 led to local inflammation, angiogenesis, and EMT (126). Interestingly, Kawahara et al. demonstrated SIRT6 interaction with RELA subunit of NFκB to regulate the expression of its target genes involved in tumor progression through deacetylation of promoter region (127, 128). SIRT6 is also reported as a corepressor of HIF1α by deacetylating H3K9 causing downregulation of the expression of genes involved in energy metabolism (129). Furthermore, it is observed that SIRT7 can inhibit HIF1α through a mechanism that is independent of its catalytic activity and regulate the expression of downstream genes like VEGF A and erythropoeitin (130). Also, downregulation of SIRT7 during breast cancer lung metastasis, caused activation of TGFβ signaling pathway and angiogenesis (131). Contrary to this, SIRT7 has been reported to promote angiogenic response by modulating endothelial cell function and VEGF like growth factor expression in mice (132). Altogether these contradicting roles played by sirtuins in tumorigenesis and angiogenesis, highlights the epigenetic regulations involved and unravels the therapeutic potential of sirtuin modulators in treatment of tumor progression by targeting tumor angiogenesis (Table 1).
Post Transcriptional Regulation of Sirtuins: Importance of miRNAs in Tumor Angiogenesis
Along with cytokine and transcriptional factor mediated regulation, precise and effective post transcriptional level regulation are also employed by sirtuins through RNA binding proteins (RBPs) and small non-coding RNA molecules. Micro RNAs are a group of small non-coding RNAs, known as the micro regulators of gene expression. For e.g., miR-34a has been reported to retard endothelial progenitor cell (EPC) mediated angiogenesis by targeting SIRT1 and thereby elevating the levels of acetylated FOXO1, leading to endothelial cell (EC) senescence and cell cycle arrest (147, 148). Similarly, miR-217 has been reported to induce senescence of ECs by modulating the levels of acetylated FOXO1 in a SIRT1 dependent mechanism (149). However, miR-217 has also been reported to promote angiogenesis of Human cytomegalovirus infected endothelial cells by inhibiting SIRT1 and FOXO3A (150). Further, report from our group suggests that miR-106a regulates the expression of MMP9 during cell migration by directly targeting SIRT1 mRNA (151). Our group has also reported that the horizontal transfer of miR-23a from tumor cell colonies can induces angiogenesis by targeting SIRT1 in the recipient endothelial cells (152). Further, miR-212 has been reported to exhibit anti-angiogenic properties, by targeting SIRT1 and Gab1 in endothelial cells (153). SIRT1 has also been reported to inhibit the anti-angiogenic- Notch signaling pathway (54). In addition, TGFβ mediated suppression of SIRT1 expression leading to the activation of Notch signaling pathway in ECs was reported to be depended partly on miR-212 (153). Further, a key micro regulator of angiogenesis and hypoxia responses, miR-138-P_5P has been reported to target SIRT1 (154–156).
Conclusion: Future Perspectives and Novel Therapeutic Approaches
During the past decade, sirtuins have emerged as critical regulators of endothelial cell behavior and have been directly linked to tumor angiogenesis through multiple signaling pathways and cross-talks (Figure 1). Lack of long-term therapeutic efficacy of current anti-angiogenic strategies requisite for novel angiogenesis inhibitors targeting sirtuins (157, 158). Recent discoveries suggest that employing sirtuin isoform specific modulators is a potent anti-angiogenic strategy. Endothelial microparticles enriched with Sirt6 mRNA induces EC angiogenesis, increases eNOS phosphorylation and prevents release of inflammatory chemokines in diabetic patients (159). Novel approaches like employing various metal (160–162) and inorganic NPs (163–165) have been reported to modulate angiogenesis. Many studies revealed that the shape, size and surface charge of the nanoparticles plays a crucial role in their angiogenic behavior (166, 167). Recently our group has reported that carbon-based nanoparticles (carbon quantum dots) with size <6 nm, inhibit angiogenic process and significantly reduce the expression level of VEGF, VEGFR2, and FGF (168). SirtuinsNano-particle based phytochemicals are reported to regulate sirtuins in cardioprotective treatment strategies (169). So far, no reports are available on the direct correlation with nano particles targeting sirtuins in tumor angiogenesis. Mechanistic studies are under progress on the development of NPs targeting sirtuins and further, tumor angiogenesis. Future studies that unveil the role of potent sirtuin modulators like CQDs at the crossroads of tumor angiogenesis will provide insights for designing novel anti-angiogenic therapies targeting sirtuin.
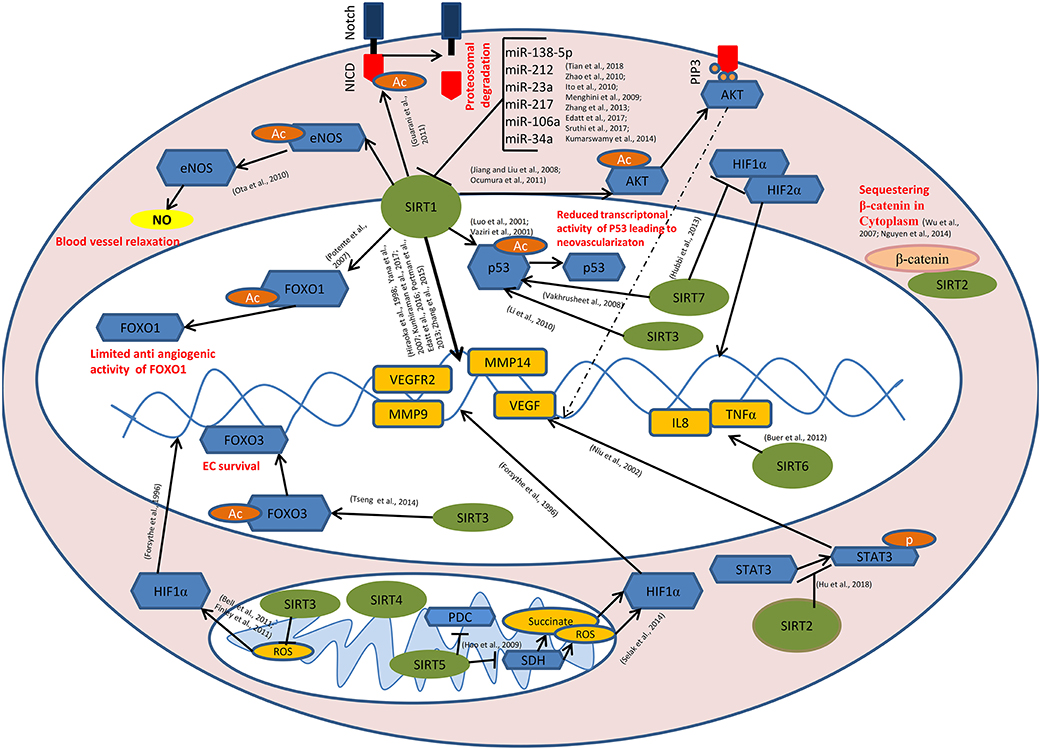
Figure 1. Role of sirtuins in Tumor Angiogenesis: SIRT1 mediates deacetylation of FOXO1, p53, AKT, eNOS, and the intra cellular domain of the Notch protein (NICD) leading to the reduced anti-angiogenic activity of FOXO1, reduced transcriptional activity of p53, induction of AKT signaling causing the transcriptional activation of pro angiogenic genes, enhanced endothelial NO production causing blood vessel relaxation and disassembly followed by the proteasomal degradation of Notch protein respectively. SIRT1 also modulates the expression of VEGF, VEGFR2, MMP9, MMP14, etc. directly by its histone deacetylase activity. miR-34a, miR-106a, miR-217, miR-23a, miR-212, and miR-138-5p targets SIRT1 at post transcriptional level. SIRT3 and SIRT7 catalyze the deacetylation of p53. SIRT7 inhibits HIF-1α stabilization and hence its nuclear translocation. Binding of SIRT2 with β-catenin leads to the sequestration of β-catenin in the cytoplasm, causing modulation in the expression of β-catenin responsive genes including MMPs. SIRT6 mediates the transcriptional activation of IL8 and TNFα which, in turn positively modulates tumor angiogenesis. SIRT2 inhibits STAT3 phosphorylation and its nuclear translocation. SIRT5 inhibits pyruvate dehydrogenase complex (PDC) and succinate dehydrogenase (SDH) causing the accumulation of succinate and reactive oxygen species (ROS) in the mitochondria, leading to HIF-1α activation. SIRT3 negatively regulates mitochondrial ROS production and hence HIF-1α stabilization. SIRT3 mediates deacetylation of FOXO3, thereby promoting endothelial cell (EC) survival under hypoxia.  - Sirtuins,
- Sirtuins,  - transcription factors/enzymes/signaling molecules,
- transcription factors/enzymes/signaling molecules,  - downstream genes,
- downstream genes,  - acetyl(Ac)/phosphate(p) group,
- acetyl(Ac)/phosphate(p) group,  - β-catenin,
- β-catenin,  - Nitric oxide,
- Nitric oxide,  - Succinate/reactive oxygen species (ROS).
- Succinate/reactive oxygen species (ROS).
Author Contributions
LE and VK contributed to conception and manuscript writing. VR, GR, and SS searched the literature. AP collected data and designed the scheme for the regulation of tumor angiogenesis by sirtuins. LE and VK participated in its coordination and modification. All the authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial assistance in the form of fellowships to LE, AP, GR, VR, and SS, received from DST-SERB, DBT, and KSCSTE is gratefully acknowledged.
References
1. Stephenson JA, Goddard JC, Taan OA, Dennison AR, Morgan B. Tumour angiogenesis: a growth area—from John Hunter to Judah Folkman and beyond. J Cancer Res. (2013) 2013:895019. doi: 10.1155/2013/895019
2. Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. (2000) 407:249–57. doi: 10.1038/35025220
3. Folkman J. Tumor angiogenesis: from bench to bedside. In: Marmé D, Fusenig N, editors. Tumor Angiogenesis. Berlin; Heidelberg: Springer (2008). p. 3–28.
4. Hall AP. The role of angiogenesis in cancer. Comp Clin Path. (2005) 13:95–9. doi: 10.1007/s00580-004-0533-3
5. Hunter J. (1861). Essays and Observations on Natural History, Anatomy, Physiology, and Geology. London: Van Voorst.
6. Ohno-Matsui K, Hirose A, Yamamoto S, Saikia J, Okamoto N, Gehlbach P, et al. Inducible expression of vascular endothelial growth factor in adult mice causes severe proliferative retinopathy and retinal detachment. Am J Pathol. (2002) 160:711–9. doi: 10.1016/S0002-9440(10)64891-2
7. Kumar VBS, Viji RI, Kiran MS, Sudhakaran PR. Endothelial cell response to lactate: implication of PAR modification of VEGF. J Cell Physiol. (2007) 211:477–85. doi: 10.1002/jcp.20955
8. Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. (1996) 16:4604–13. doi: 10.1128/MCB.16.9.4604
9. Kumar VBS, Binu S, Soumya SJ, Haritha K, Sudhakaran PR. Regulation of vascular endothelial growth factor by metabolic context of the cell. Glycoconjugate J. (2014) 31–7, 427–434. doi: 10.1007/s10719-014-9547-5
10. Jensen KS, Binderup T, Jensen KT, Therkelsen I, Borup R, Nilsson E, et al. FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. EMBO J. (2011) 30:4554–70. doi: 10.1038/emboj.2011.323
11. Maher JC, Wangpaichitr M, Savaraj N, Kurtoglu M, Lampidis TJ. Hypoxia-inducible factor-1 confers resistance to the glycolytic inhibitor 2-deoxy-D-glucose. Mol Cancer Ther. (2007) 6:732–41. doi: 10.1158/1535-7163.MCT-06-0407
12. Balaiya S, Ferguson LR, Chalam KV. Evaluation of sirtuin role in neuroprotection of retinal ganglion cells in hypoxia. Investig Ophthalmol Vis Sci. (2012) 53:4315–22. doi: 10.1167/iovs.11-9259
13. Balcerczyk A, Pirola L. Therapeutic potential of activators and inhibitors of sirtuins. Biofactors. (2010) 36:383–93. doi: 10.1002/biof.112
14. Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. (2007) 21:2644–58. doi: 10.1101/gad.435107
15. Aguilar AL, Ranjit S, Stringari C, OrozcoSR, Gratton E, Sassone CP. Spatial dynamics of SIRT1 and the subnuclear distribution of NADH species. Proc Natl Acad Sci USA. (2016) 113:12715–20. doi: 10.1073/pnas.1609227113
16. Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. (2009) 104:879–86. doi: 10.1161/CIRCRESAHA.108.193102
17. Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. (1999) 260:273–9. doi: 10.1006/bbrc.1999.0897
18. Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. (2000) 273:793–8. doi: 10.1006/bbrc.2000.3000
19. Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. (2003) 370:737–49. doi: 10.1042/bj20021321
20. Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. (2006) 75:435–65. doi: 10.1146/annurev.biochem.74.082803.133500
21. Jackson MD, Denu JM. Structural identification of 2′-and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β-NAD+-dependent histone/protein deacetylases. J Biol Chem. (2002) 277:18535–44. doi: 10.1074/jbc.M200671200
22. Imai SI, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. (2000) 403:795–800. doi: 10.1038/35001622
23. Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. (2000) 97:5807–11. doi: 10.1073/pnas.110148297
24. Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog–NAD complex. Cell. (2001) 105:269–79. doi: 10.1016/S0092-8674(01)00317-8
25. Frothingham R, Meeker OCWA, Talbot EA, George JW, Kreuzer KN. Identification, cloning, and expression of the Escherichia coli pyrazinamidase and nicotinamidase gene, pncA. Antimicrob Agents Chemother. (1996) 40:1426–31. doi: 10.1128/AAC.40.6.1426
26. Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. (2003) 423:181–5. doi: 10.1038/nature01578
27. Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. (2000) 289:2126–8. doi: 10.1126/science.289.5487.2126
28. Vaquero A. The conserved role of sirtuins in chromatin regulation. Int J Dev Biol. (2009) 53–3, 303–322. doi: 10.1387/ijdb.082675av
29. Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. (2007) 21:1745–55. doi: 10.1210/me.2007-0079
30. Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. (2005) 121:515–27. doi: 10.1016/j.cell.2005.03.035
31. Madsen AS, Olsen CA. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew Chem. (2012) 124:9217–21. doi: 10.1002/ange.201203754
32. Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. (2014) 19:605–17. doi: 10.1016/j.cmet.2014.03.014
33. Bao X, Wang Y, Li X, Li X, Liu Z, Yang T, et al. Identification of ‘erasers' for lysine crotonylated histone marks using a chemical proteomics approach. Elife. (2014) 3:e02999. doi: 10.7554/eLife.02999
34. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. (2011) 334:806–9. doi: 10.1126/science.1207861
35. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. (2013) 496:110–3. doi: 10.1038/nature12038
36. Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. (2013) 288:31350–6. doi: 10.1074/jbc.C113.511261
37. Feldman JL, Kristin E, Dittenhafer-Reed KN, Thelen JN, Ito A, Yoshida M, et al. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry. (2015) 54:3037–50. doi: 10.1021/acs.biochem.5b00150
38. Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. (2011) 10:M111.012658. doi: 10.1074/mcp.M111.012658
39. Chen B, Zang W, Wang J, Huang Y, He Y, Yan L, et al. The chemical biology of sirtuins. Chem Soc Rev. (2015) 44:5246–64. doi: 10.1039/C4CS00373J
40. Anderson KA, Huynh FK, Fisher WK, Stuart JD, Peterson BS, Douros JD, et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. (2017) 25:838–55. doi: 10.1016/j.cmet.2017.03.003
41. Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. (2014) 159:1615–25. doi: 10.1016/j.cell.2014.11.046
42. Jing H, Lin H. Sirtuins in epigenetic regulation. Chem Rev. (2015) 115:2350–75. doi: 10.1021/cr500457h
43. Jin Q, Yan T, Ge X, Sun C, Shi X, Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J Cell Physiol. (2007) 213:88–97. doi: 10.1002/jcp.21091
44. Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. (2007) 21:920–8. doi: 10.1101/gad.1527307
45. Vaziri H, Dessain SK, Eaton EN, Imai SI, Frye RA, Pandita TK, et al. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. (2001) 107:149–59. doi: 10.1016/S0092-8674(01)00527-X
46. Luo J, Nikolaev AY, Imai SI, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. (2001) 107:137–48. doi: 10.1016/S0092-8674(01)00524-4
47. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. (2004) 303:2011–5. doi: 10.1126/science.1094637
48. Hiraoka N, Allen E, Apel IJ, Gyetko MR, Weiss SJ. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. (1998) 95:365–77. doi: 10.1016/S0092-8674(00)81768-7
49. Yana I, Sagara H, Takaki S, Takatsu K, Nakamura K, Nakao K, et al. Crosstalk between neovessels and mural cells directs the site-specific expression of MT1-MMP to endothelial tip cells. J Cell Sci. (2007) 120:1607–14. doi: 10.1242/jcs.000679
50. Cantó C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, et al. The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. (2012) 15:838–47. doi: 10.1016/j.cmet.2012.04.022
51. Culic O, Gruwel ML, Schrader J. Energy turnover of vascular endothelial cells. Am J Physiol Cell Physiol. (1997) 273:205–13. doi: 10.1152/ajpcell.1997.273.1.C205
52. De BK, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. (2013) 154:651–63. doi: 10.1016/j.cell.2013.06.037
53. Fitzgerald GA, Soro AI, De Bock K. The Warburg effect in endothelial cells and its potential as an anti-angiogenic target in cancer. Front Cell Dev Biol. (2018) 6:100–17. doi: 10.3389/fcell.2018.00100
54. Guarani V, Deflorian G, Franco CA, Krüger M, Phng LK, Bentley K, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. (2011) 473:234–8. doi: 10.1038/nature09917
55. Wallerath T, Li H, Ambrust UG, Schwarz PM, Förstermann U. A blend of polyphenolic compounds explains the stimulatory effect of red wine on human endothelial NO synthase. Nitric Oxide. (2005) 12:97–104. doi: 10.1016/j.niox.2004.12.004
56. Lipphardt M, Dihazi H, Müller GA, Goligorsky M. Fibrogenic secretome of sirtuin 1-deficient endothelial cells: Wnt, notch and glycocalyx rheostat. Front Physiol. (2018) 9:1325. doi: 10.3389/fphys.2018.01325
57. Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1-and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. (2003) 301:250–7. doi: 10.1016/S0006-291X(02)03020-6
58. Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. (2004) 18:901–11. doi: 10.1101/gad.291004
59. Guo S, Long M, Li X, Zhu S, Zhang M, Yang Z. Curcumin activates autophagy and attenuates oxidative damage in EA. hy926 cells via the Akt/mTOR pathway. Mol Med Rep. (2016) 13:2187–93. doi: 10.3892/mmr.2016.4796
60. Guo L, Zhou SR, Wei XB, Liu Y, Chang XX, Liu Y, et al. Acetylation of mitochondrial trifunctional protein α-subunit enhances its stability to promote fatty acid oxidation and is decreased in nonalcoholic fatty liver disease. Mol Cell Biol. (2016) 36:2553–67. doi: 10.1128/MCB.00227-16
61. Zhang J, Wang C, Nie H, Wu D, Ying W. SIRT2 plays a significant role in maintaining the survival and energy metabolism of PIEC endothelial cells. Int J Physiol Pathophysiol Pharmacol. (2016) 8:120.
62. D'Onofrio N, Servillo L, Giovane A, Casale R, Vitiello M, Marfella R, et al. Ergothioneine oxidation in the protection against high-glucose induced endothelial senescence: involvement of SIRT1 and SIRT6. Free Radic Biol Med. (2016) 96:211–22. doi: 10.1016/j.freeradbiomed.2016.04.013
63. Liu J, Wu X, Wang X, Zhang Y, Bu P, Zhang Q, et al. Global gene expression profiling reveals functional importance of Sirt2 in endothelial cells under oxidative stress. Int J Mol Sci. (2013) 14:5633–49. doi: 10.3390/ijms14035633
64. Yang D, Lee J, Yang X, Komarova Y. SIRT2 stabilizes endothelial adherens junctions. FASEB J. (2019) 33:682–8.
65. Tseng AHH, Wu LH, Shieh SS, Wang DL. SIRT3 interactions with FOXO3 acetylation, phosphorylation and ubiquitinylation mediate endothelial cell responses to hypoxia. Biochem J. (2014) 464:157–68. doi: 10.1042/BJ20140213
66. He X, Zeng H, Chen ST, Roman RJ, Aschner JL, Didion S, et al. Endothelial specific SIRT3 deletion impairs glycolysis and angiogenesis and causes diastolic dysfunction. J Mol Cell Cardiol. (2017) 112:104–13. doi: 10.1016/j.yjmcc.2017.09.007
67. Chen Y, Wang H, Luo G, Dai X. SIRT4 inhibits cigarette smoke extracts-induced mononuclear cell adhesion to human pulmonary microvascular endothelial cells via regulating NF-κB activity. Toxicol Lett. (2014) 226:320–7. doi: 10.1016/j.toxlet.2014.02.022
68. Yu BB, Zhi H, Zhang XY, Liang JW, He J, Su C, et al. Mitochondrial dysfunction-mediated decline in angiogenic capacity of endothelial progenitor cells is associated with capillary rarefaction in patients with hypertension via downregulation of CXCR4/JAK2/SIRT5 signaling. EBioMedicine. (2019) 42:64–75. doi: 10.1016/j.ebiom.2019.03.031
69. Cardus A, Uryga AK, Walters G, Erusalimsky JD. SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovasc Res. (2012) 97:571–9. doi: 10.1093/cvr/cvs352
70. Stünkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Tellez MS, et al. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J Healthcare Nutr Technol. (2007) 2:1360–8. doi: 10.1002/biot.200700087
71. Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. (2005) 19:1751–9. doi: 10.1038/sj.leu.2403910
72. Kozako T. New strategy of adult T-cell leukemia treatment targeted for anti-tumor immunity and a longevity gene-encoded protein. Yakugaku Zasshi. (2011) 131:1061–72. doi: 10.1248/yakushi.131.1061
73. Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. (2008) 14:312–23. doi: 10.1016/j.ccr.2008.09.001
74. Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. (2007) 26:945–57. doi: 10.1038/sj.onc.1209857
75. Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA. (2005) 102:16013–8. doi: 10.1073/pnas.0500090102
76. Sebastián C, Zwaans BMM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. (2012) 151:1185–99. doi: 10.1016/j.cell.2012.10.047
77. Alcaín FJ, Villalba JM. Sirtuin inhibitors. Expert Opin Ther Pat. (2009) 19:283–94. doi: 10.1517/13543770902755111
78. Cantó C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. (2009) 20:325–31. doi: 10.1016/j.tem.2009.03.008
79. Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. (2007) 35:6984–94. doi: 10.1093/nar/gkm703
80. Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. (2004) 101:10042–7. doi: 10.1073/pnas.0400593101
81. Teodoro JG, Evans SK, Green MR. Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J Mol Med. (2007) 85:1175–86. doi: 10.1007/s00109-007-0221-2
82. Lynch CJ, Shah ZH, Allison SJ, Ahmed SF, Ford J, Warnock LJ, et al. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PLoS ONE. (2010) 5:e13502. doi: 10.1371/journal.pone.0013502
83. Li SD, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS ONE. (2010) 5:e10486. doi: 10.1371/journal.pone.0010486
84. Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. (2008) 102:703–10. doi: 10.1161/CIRCRESAHA.107.164558
85. Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. (2003) 100:10794–9. doi: 10.1073/pnas.1934713100
86. Zhang H, He S, Spee C, Ishikawa K, Hinton DR. SIRT1 mediated inhibition of VEGF/VEGFR2 signaling by Resveratrol and its relevance to choroidal neovascularization. Cytokine. (2015) 76:549–52. doi: 10.1016/j.cyto.2015.06.019
87. Khan AA, Dace DS, Ryazanov AG, Kelly J, Apte RS. Resveratrol regulates pathologic angiogenesis by a eukaryotic elongation factor-2 kinase-regulated pathway. Am J Pathol. (2010) 177:481–92. doi: 10.2353/ajpath.2010.090836
88. Xie M, Liu M, He CS. SIRT1 regulates endothelial Notch signaling in lung cancer. PLoS ONE. (2012) 7:e45331. doi: 10.1371/journal.pone.0045331
89. Jiang BH, Liu LZ. AKT signaling in regulating angiogenesis. Curr Cancer Drug Targets. (2008) 8:19–26. doi: 10.2174/156800908783497122
90. Okumura N, Yoshida H, Kitagishi Y, Murakami M, Nishimura Y, Matsuda S. PI3K/AKT/PTEN signaling as a molecular target in leukemia angiogenesis. Adv Hematol. (2012) 2012:843085. doi: 10.1155/2012/843085
91. Ota H, Eto M, Ogawa S, Iijima K, Akishita M, Ouchi Y. SIRT1/eNOS axis as a potential target against vascular senescence, dysfunction and atherosclerosis. J Atheroscler Thromb. (2010) 17:431–5. doi: 10.5551/jat.3525
92. Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci USA. (2007) 104:14855–60. doi: 10.1073/pnas.0704329104
93. Potente M, Urbich C, Sasaki KI, Hofmann WK, Heeschen C, Aicher A, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. (2005) 115:2382–92. doi: 10.1172/JCI23126
94. Fukumura D, Xu L, Chen Y, Gohongi T, Seed B, Jain RK. Hypoxia and acidosis independently up-regulate vascular endothelial growth factor transcription in brain tumors in vivo. Cancer Res. (2001) 61:6020–4.
95. Corbet C, Draoui N, Polet F, Pinto A, Drozak X, Riant O, et al. The SIRT1/HIF2α axis drives reductive glutamine metabolism under chronic acidosis and alters tumor response to therapy. Cancer Res. (2014) 74:5507–19. doi: 10.1158/0008-5472.CAN-14-0705
96. Kunhiraman H, Edatt L, Thekkeveedu S, Poyyakkara A, Raveendran V, Kiran MS, et al. 2-Deoxy Glucose modulates expression and biological activity of VEGF in a SIRT-1 dependent mechanism. J Cell Biochem. (2017) 118:252–62. doi: 10.1002/jcb.25629
97. Edatt L, Haritha K, Sruthi TV, Aswini P, Sameer Kumar VB. 2-Deoxy glucose regulate MMP-9 in a SIRT-1 dependent and NFkB independent mechanism. Mol Cell Biochem. (2016) 423–2, 197–206. doi: 10.1007/s11010-016-2837-4
98. Portmann S, Fahrner R, Lechleiter A, Keogh A, Overney S, Laemmle A, et al. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. (2013) 12:499–508. doi: 10.1158/1535-7163.MCT-12-0700
99. Li C, Wang L, Zheng L, Zhan X, Xu B, Jiang J, et al. SIRT1 expression is associated with poor prognosis of lung adenocarcinoma. Onco Targets Ther. (2015) 8:977. doi: 10.2147/OTT.S82378
100. Suzuki K, Hayashi R, Ichikawa T, Imanishi S, Yamada T, Inomata M, et al. SRT1720, a SIRT1 activator, promotes tumor cell migration, and lung metastasis of breast cancer in mice. Oncol Rep. (2012) 27:1726–32. doi: 10.3892/or.2012.1750
101. Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. (2003) 23:3173–85. doi: 10.1128/MCB.23.9.3173-3185.2003
102. North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. (2003) 11:437–44. doi: 10.1016/S1097-2765(03)00038-8
103. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. (2005) 16:4623–35. doi: 10.1091/mbc.e05-01-0033
104. Hu F, Sun X, Li G, Wu Q, Chen Y, Yang X, et al. Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis. (2018) 10:9–23. doi: 10.1038/s41419-018-1260-z
105. Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. (2002) 21:2000–8. doi: 10.1038/sj.onc.1205260
106. Wu B, Crampton SP, Hughes CCW. Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity. (2007) 26:227–39. doi: 10.1016/j.immuni.2006.12.007
107. Nguyen P, Lee S, Leins DL, Trepel J, Smart DDK. SIRT2 interacts with β-catenin to inhibit Wnt signaling output in response to radiation-induced stress. Mol Cancer Res. (2014) 12:1244–53. doi: 10.1158/1541-7786.MCR-14-0223-T
108. Shi Y, Xu X, Zhang Q, Fu G, Mo Z, Wang GS, et al. tRNA synthetase counteracts c-Myc to develop functional vasculature. Elife. (2014) 3:e02349. doi: 10.7554/eLife.02349
109. Khalek WA, Cortade F, Ollendorff V, Lapasset L, Tintignac L, Chabi B, et al. SIRT3, a mitochondrial NAD+-dependent deacetylase, is involved in the regulation of myoblast differentiation. PLoS ONE. (2014) 9:e114388. doi: 10.1371/journal.pone.0114388
110. Wu K, Wang L, Chen Y, Pirooznia M, Singh K, Wälde S, et al. GCN5L1 interacts with αTAT1 and RanBP2 to regulate hepatic α-tubulin acetylation and lysosome trafficking. J Cell Sci. (2018) 131:jcs221036. doi: 10.1242/jcs.221036
111. Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. (2006) 103:10224–9. doi: 10.1073/pnas.0603968103
112. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. (2005) 280:13560–7. doi: 10.1074/jbc.M414670200
113. Bell EL, Emerling BM, Ricoult SJH, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. (2011) 30:2986–96. doi: 10.1038/onc.2011.37
114. Finley LWS, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. (2011) 19:416–28. doi: 10.1016/j.ccr.2011.02.014
115. Schumacker PT. SIRT3 controls cancer metabolic reprogramming by regulating ROS and HIF. Cancer Cell. (2011) 19:299–300. doi: 10.1016/j.ccr.2011.03.001
116. Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. (2005) 280:21313–20. doi: 10.1074/jbc.M413296200
117. Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. (2009) 325:1139–42. doi: 10.1126/science.1175689
118. Sauve AA. Sirtuin chemical mechanisms. Biochim Biophys Acta. (2010) 1804:1591–603. doi: 10.1016/j.bbapap.2010.01.021
119. Herrero YA, Bakhit SMA, Franke P, Weise C, Schweiger M, Jorcke D, et al. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. (2001) 20:2404–12. doi: 10.1093/emboj/20.10.2404
120. Cantó C, Sauve AA, Bai P. Crosstalk between poly (ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med. (2013) 34:1168–201. doi: 10.1016/j.mam.2013.01.004
121. Mathews MT, Berk BC. PARP-1 inhibition prevents oxidative and nitrosative stress–induced endothelial cell death via transactivation of the VEGF receptor 2. Arterioscler Thromb Vasc Biol. (2008) 28:711–7. doi: 10.1161/ATVBAHA.107.156406
122. Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson GD, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. (2005) 7:77–85. doi: 10.1016/j.ccr.2004.11.022
123. Wang C, Zhang C, Li X, Shen J, Xu Y, Shi H, et al. CPT 1A-mediated succinylation of S100A10 increases human gastric cancer invasion. J Cell Mol Med. (2019) 23:293–305. doi: 10.1111/jcmm.13920
124. Van GF, Gallí M, Gueydan C, Kruys V, Prevot PP, Bedalov A, et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med. (2009) 15:206–10. doi: 10.1038/nm.1906
125. Bauer I, Grozio A, Lasigliè D, Basile G, Sturla L, Magnone M, et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. (2012) 287:40924–37. doi: 10.1074/jbc.M112.405837
126. Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C. IL-8 signaling plays a critical role in the epithelial–mesenchymal transition of human carcinoma cells. Cancer Res. (2011) 71:5296–306. doi: 10.1158/0008-5472.CAN-11-0156
127. Kawahara TLA, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. (2009) 136:62–74. doi: 10.1016/j.cell.2008.10.052
128. Basseres DS, Baldwin AS. Nuclear factor-κB and inhibitor of κB kinase pathways in oncogenic initiation and progression. Oncogene. (2006) 25:6817–30. doi: 10.1038/sj.onc.1209942
129. Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell. (2010) 140:280–93. doi: 10.1016/j.cell.2009.12.041
130. Hubbi ME, Hu H, Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Biol Chem. (2013) 288:20768–75. doi: 10.1074/jbc.M113.476903
131. Tang X, Shi L, Xie N, Liu Z, Qian M, Meng F, et al. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat Commun. (2017) 8:318. doi: 10.1038/s41467-017-00396-9
132. Rokutanda T, Izumiya Y, Araki S, Hanatani S, Bober E, Braun T, et al. Sirt7 regulates endothelial cell functions and promotes angiogenic response in mice model of hindlimb ischemia. Circulation. (2012) 126:A10980.
133. Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. (2005) 280:16456–60. doi: 10.1074/jbc.M501485200
134. Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. (2004) 23:2369–80. doi: 10.1038/sj.emboj.7600244
135. Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. (2003) 12:51–62. doi: 10.1016/S1097-2765(03)00226-0
136. Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. (2006) 4:e31. doi: 10.1371/journal.pbio.0040031
137. Picard F, Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado D. O. R., et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR. Nature. (2004) 429:771–6. doi: 10.1038/nature02583
138. Zhang W, Huang Q, Zeng Z, Wu J, Zhang Y, Chen Z. Sirt1 inhibits oxidative stress in vascular endothelial cells. Oxid Med Cell Longev. (2017) 2017:7543973. doi: 10.1155/2017/7543973
139. Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. (2005) 310:314–7. doi: 10.1126/science.1117728
140. Sahar S, Masubuchi S, Mahan KE, Vollmer S, Galla L, Ceglia N, et al. Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J Biol Chem/. (2014) 289:6091–7. doi: 10.1074/jbc.M113.537191
141. Zerr P, Zerr KP, Huang J, Tomcik M, Sumova B, Distler O, et al. Sirt1 regulates canonical TGF-β signalling to control fibroblast activation and tissue fibrosis. Ann Rheum Dis. (2016) 75:226–33. doi: 10.1136/annrheumdis-2014-205740
142. Patel VB, Preedy VR. (Eds.). Handbook of Nutrition, Diet, and Epigenetics. Cham: Springer (2019).
143. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. (2010) 464:121–5. doi: 10.1038/nature08778
144. Han Y, Zhou S, Coetzee S, Chen A. SIRT4 and its roles in energy and redox metabolism in health, disease and during exercise. Front Physiol. (2019) 10:1006. doi: 10.3389/fphys.2019.01006
145. Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CFW, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. (2008) 382:790–801. doi: 10.1016/j.jmb.2008.07.048
146. Barber MF, Michishita KE, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. (2012) 487:114–8. doi: 10.1038/nature11043
147. Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. American J Physiol Endocrinol Metab. (2010) 299:E110–16. doi: 10.1152/ajpendo.00192.2010
148. Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. (2010) 398:735–40. doi: 10.1016/j.bbrc.2010.07.012
149. Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator. Circulation. (2009) 120:1524–32. doi: 10.1161/CIRCULATIONAHA.109.864629
150. Zhang S, Liu L, Wang R, Tuo H, Guo Y, Yi L, et al. MicroRNA-217 promotes angiogenesis of human cytomegalovirus-infected endothelial cells through downregulation of SIRT1 and FOXO3A. PLoS ONE. (2013) 8:e83620. doi: 10.1371/journal.pone.0083620
151. Edatt L, Maurya AK, Raji G, Kunhiraman H, Kumar VBS. MicroRNA106a regulates matrix metalloprotease 9 in a sirtuin-1 dependent mechanism. J Cell Physiol. (2018) 233:238–48. doi: 10.1002/jcp.25870
152. Sruthi TV, Edatt L, Raji GR, Kunhiraman H, Shankar SS, Shankar V, et al. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J Cell Physiol. (2018) 233:3498–514. doi: 10.1002/jcp.26202
153. Kumarswamy R, Volkmann I, Beermann J, Napp LC, Jabs O, Bhayadia R, et al. Vascular importance of the miR-212/132 cluster. Eur Heart J. (2014) 35:3224–31. doi: 10.1093/eurheartj/ehu344
154. Zhang Y, Du X, Li W, Sang H, Qian A, Sun L, et al. Resveratrol improves endothelial progenitor cell function through miR-138 by targeting focal adhesion kinase (FAK) and promotes thrombus resolution in vivo. Med Sci Monit. (2018) 24:951. doi: 10.12659/MSM.906116
155. Zheng Z, Zhao B. Astragalus polysaccharide protects hypoxia-induced injury by up-regulation of miR-138 in rat neural stem cells. Biomed Pharmacother. (2018) 102:295–301. doi: 10.1016/j.biopha.2018.03.040
156. Tian F, Yuan C, Yue H. MiR-138/SIRT1 axis is implicated in impaired learning and memory abilities of cerebral ischemia/reperfusion injured rats. Exp Cell Res. (2018) 367:232–40. doi: 10.1016/j.yexcr.2018.03.042
157. Vernucci E, Tomino C, Molinari F, Limongi D, Aventaggiato M, Sansone L, et al. Mitophagy and oxidative stress in cancer and aging: focus on sirtuins and nanomaterials. Oxid Med Cell Longev. (2019) 2019:6387357. doi: 10.1155/2019/6387357
158. Christovam AC, Theodoro V, Mendonça FAS, Esquisatto MAM, Santos GMT, Amaral MEC. Activators of SIRT1 in wound repair: an animal model study. Arch Dermatol Res. (2019) 311:193–201. doi: 10.1007/s00403-019-01901-4
159. Jing T, Ya SK, Xue JW, Han JH, Yan L, Yi AY, et al. Sirt6 mRNA-incorporated endothelial microparticles (EMPs) attenuates DM patient-derived EMP-induced endothelial dysfunction. Oncotarget. (2017) 8:114300–13. doi: 10.18632/oncotarget.23259
160. Arvizo RR, Saha S, Wang E, Robertson JD, Bhattacharya R, Mukherjee P. Inhibition of tumor growth and metastasis by a self-therapeutic nanoparticle. Proc Natl Acad Sci USA. (2013) 110:6700–5. doi: 10.1073/pnas.1214547110
161. Yang T, Yao Q, Cao F, Liu Q, Liu B, Wang XH. Silver nanoparticles inhibit the function of hypoxia-inducible factor-1 and target genes: insight into the cytotoxicity and antiangiogenesis. Int J Nanomed. (2016) 11:6679. doi: 10.2147/IJN.S109695
162. Li W, Zhao X, Du B, Li X, Liu S, Yang XY, et al. Gold nanoparticle–mediated targeted delivery of recombinant human endostatin normalizes tumour vasculature and improves cancer therapy. Sci Rep. (2016) 6:30619. doi: 10.1038/srep30619
163. Xu Y, Wen Z, Xu Z. Chitosan nanoparticles inhibit the growth of human hepatocellular carcinoma xenografts through an antiangiogenic mechanism. Anticancer Res. (2009) 29:5103–9.
164. Jin H, Pi J, Yang F, Wu C, Cheng X, Bai H, et al. Ursolic acid-loaded chitosan nanoparticles induce potent anti-angiogenesis in tumor. Appl Microbiol Biotechnol. (2016) 100:6643–52. doi: 10.1007/s00253-016-7360-8
165. Hu H, You Y, He L, Chen T. The rational design of NAMI-A-loaded mesoporous silica nanoparticles as antiangiogenic nanosystems. J Mater Chem B. (2015) 3:6338–46. doi: 10.1039/C5TB00612K
166. Arvizo RR, Rana S, Miranda OR, Bhattacharya R, Rotello VM, Mukherjee P. Mechanism of anti-angiogenic property of gold nanoparticles: role of nanoparticle size and surface charge. Nanomed Nanotechnol Biol Med. (2011) 7:580–7. doi: 10.1016/j.nano.2011.01.011
167. Shi X, Zhou K, Huang F, Wang C. Interaction of hydroxyapatite nanoparticles with endothelial cells: internalization and inhibition of angiogenesis in vitro through the PI3K/Akt pathway. Int J Nanomed. (2017) 12:5781–95. doi: 10.2147/IJN.S140179
168. Shereema TM, Sruthi TV, Kumar VBS, Rao TP, Shankar SS. Angiogenic profiling of synthesized carbon quantum dots. Biochemistry. (2015) 54:6352–6. doi: 10.1021/acs.biochem.5b00781
Keywords: sirtuins, tumor angiogenesis, metabolism, histone deacetylases, endothelial cells, signaling pathways
Citation: Edatt L, Poyyakkara A, Raji GR, Ramachandran V, Shankar SS and Kumar VBS (2020) Role of Sirtuins in Tumor Angiogenesis. Front. Oncol. 9:1516. doi: 10.3389/fonc.2019.01516
Received: 01 November 2019; Accepted: 16 December 2019;
Published: 17 January 2020.
Edited by:
Stefano Falone, University of L'Aquila, ItalyReviewed by:
Vincenzo Carafa, University of Campania Luigi Vanvitelli, ItalySimona Delle Monache, University of L'Aquila, Italy
Dante Rotili, Sapienza University of Rome, Italy
Copyright © 2020 Edatt, Poyyakkara, Raji, Ramachandran, Shankar and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: V. B. Sameer Kumar, c2FtZWVya3VtYXJ2YkBnbWFpbC5jb20=; c2t1bWFydmJAY3VrZXJhbGEuYWMuaW4=
 Lincy Edatt
Lincy Edatt Aswini Poyyakkara
Aswini Poyyakkara Grace R. Raji
Grace R. Raji Vishnu Ramachandran
Vishnu Ramachandran S. Sharath Shankar
S. Sharath Shankar V. B. Sameer Kumar
V. B. Sameer Kumar