
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 15 November 2019
Sec. Cancer Molecular Targets and Therapeutics
Volume 9 - 2019 | https://doi.org/10.3389/fonc.2019.01225
This article is part of the Research TopicThe Role of ncRNAs in Solid Tumors Prognosis: From Laboratory To Clinical UtilityView all 49 articles
Long noncoding RNAs (lncRNAs) are emerging as important regulators of numerous biological processes, especially in cancer development. Aberrantly expressed and specifically located in tumor cells, they exert distinct functions in different cancers via regulating multiple downstream targets such as chromatins, RNAs, and proteins. Differentiation antagonizing non-protein coding RNA (DANCR) is a cytoplasmic lncRNA that generally works as a tumor promoter. Mechanically, DANCR promotes the functions of vital components in the oncogene network by sponging their corresponding microRNAs or by interacting with various regulating proteins. DANCR's distinct expression in tumor cells and collective involvement in pro-tumor pathways make it a promising therapeutic target for broad cancer treatment. Herein, we summarize the functions and molecular mechanism of DANCR in human cancers. Furthermore, we introduce the use of CRISPR/Cas9, antisense oligonucleotides and small interfering RNAs as well as viral, lipid, or exosomal vectors for onco-lncRNA targeted treatment. Conclusively, DANCR is a considerable promoter of cancers with a bright prospect in targeted therapy.
Approximately 75% of the human genome can be transcribed into RNAs, yet no more than 2% encodes proteins (1). Non-coding RNAs occupy a large part of the human transcriptome and long non-coding RNAs (lncRNAs) [transcripts over 200 nt in length and lack the potential of coding protein (2)] are the major components (Figure 1A) (3). Extensive research on lncRNAs has been conducted for several decades and considerable progress has been achieved in this field. H19 is the first recognized lncRNA that can be transcribed from both protein coding and non-coding DNA (4). Its deficit in open reading frame and inactive interaction with ribosomes distinguish it from normal protein-coding mRNAs and extend human understanding of RNA transcriptome (5). X-inactive specific transcript (Xist) is one of the most well-known lncRNAs which is associated with the silencing of X chromosome (6). With the technical improvement of gene sequencing technology (7), the past 30 years have led to the discovery of a wide spectrum of evolutionally conserved and tissue-specific lncRNAs. They are located in cell nucleus and cytoplasm and play an important role in pathophysiological processes (8). Their various functions (promote tumor proliferation, motility, viability, and angiogenesis) in cancer regulation becomes a research focus in recent years (9) (Figure 1B). Mechanically, by interacting with proteins and RNAs, lncRNAs can regulate the transcription and post-transcription of RNAs or mediate the modification of proteins to adjust the cellular signaling pathways. LncRNAs have also been involved in the clinical cancer treatment. For example, prostate cancer antigen 3 has been approved by the Food and Drug Administration (FDA) as a biomarker in urine for prostate cancer diagnosis; it displays a better diagnostic effect compared with traditional serum prostate-specific antigen testing (10). Some lncRNAs, such as lncRNA Hox Transcript Antisense Intergenic RNA (HOTAIR) in breast cancer, are proven to enhance cells' resistance to certain drugs, which can guide drug selection in cancer chemotherapy (11). Onco-lncRNA targeted therapy is a novel and promising therapeutic method with collective efficacy, high specificity and few side effects (12). Many synthetic therapeutic products have been constructed and pre-clinical experiments have been conducted to promote the clinical application of lncRNA-targeted therapy.
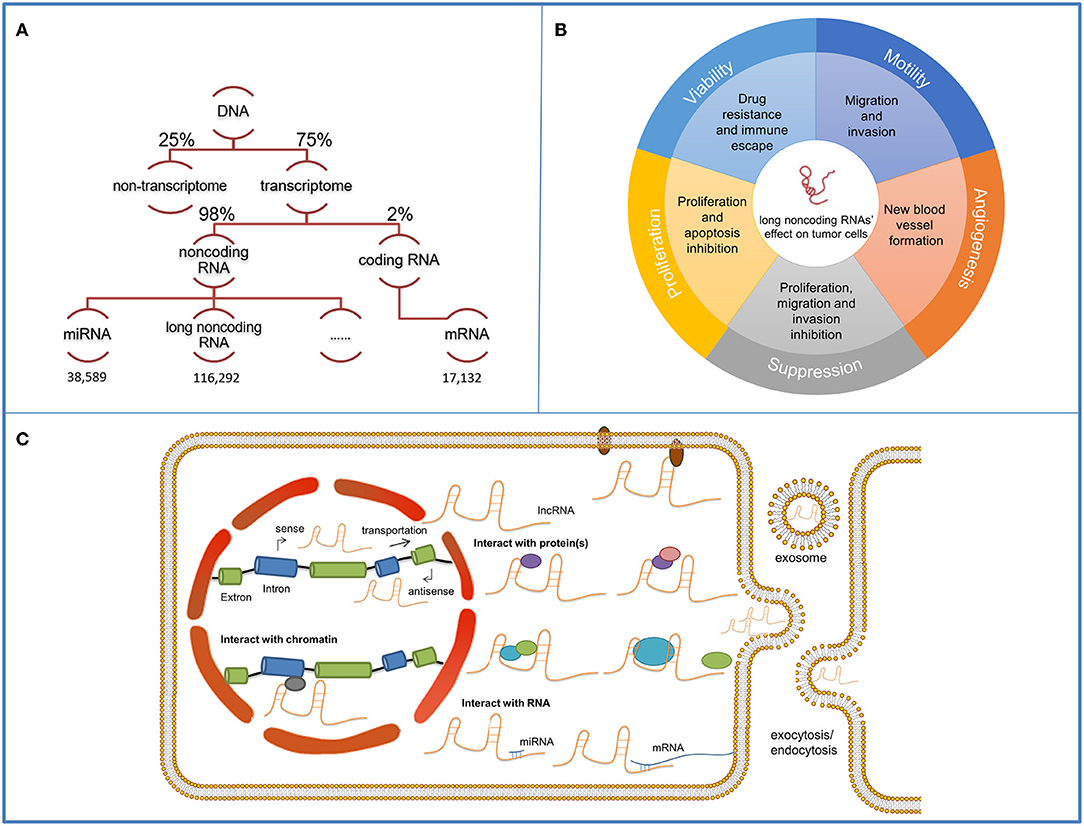
Figure 1. Functions and mechanisms of long non-coding RNAs (lncRNAs). The schematic illustrates lncRNAs and their functions and mechanisms in tumor cells. (A) Among the 75% actively transcribed sequences in the human genome, 98% are non-coding sequences where long non-coding sequences occupy the majority. The quantity of miRNAs, lncRNAs and miRNAs mapped on the schematic are obtained from the respective databases. (B) LncRNAs' functions on cancer development contain viability, motility, angiogenesis, proliferation promotion, and suppression. (C) LncRNAs have distinct working compartments. They can assemble with nucleus proteins and regulate transcription by interacting with the histone proteins or the promoter regions. They can also transmit to the cytoplasm and participate in signaling transmission by mediating the modification of components in cellular pathways. They can be contained in exosomes and delivered for intercellular communication. Mechanically, they can interact with protein/protein complex and form protein-RNA complex, serve as a scaffold and promote the assembling of proteins or complementary bind to RNAs to exert their functions.
Due to the overwhelming quantity of lncRNAs in the human genome and the lack of methods for predicting their functions, massive work is still needed to better understand the lncRNAs. LncRNA differentiation antagonizing non-protein coding RNA (DANCR) can serve as a paradigm for lncRNA investigation. DANCR is a typical oncogenic lncRNA which exerts multiple functions in cancer development. It has also demonstrated its value in cancer therapy as a promising target (12).
This article summarized the functional properties and mechanisms of DANCR in tumor progression. The general targeted therapies against onco-lncRNAs were introduced and the specific methods to target DANCR were discussed. This article can provide referential value for latter onco-lncRNA investigations and insights into the clinical treatment against cancers caused by aberrant expression and regulation of lncRNAs.
DANCR also named as Anti-Differentiation Non-coding RNA (ANCR), Small nucleolar RNA host gene protein 13(SNHG13), upregulating transcripts 2 (AGU2) is an oncogenic lncRNA located on chromosome 4 (13, 14). It was first identified as a transcript of unknown function whose expression is dramatically upregulated during adipogenesis of pluripotent human mesenchymal stem cells (hMSCs) (13). Later it was recognized as an anti-differentiation non-coding RNA as it promoted epidermal progenitor cell maintenance and prevented differentiation in osteoblasts (15, 16). Extensive data have shown that DANCR correlates with the prognosis of various human cancers, and exerts diverse oncogenic functions via multiple downstream targets. It will serve as a novel target for targeted therapy.
The discovery that DANCR over-expressed in colorectal cancer promoted tumor progression and predicted poor prognosis introduced the study of DANCR into the field of cancer (17). DANCR can serve as a novel biomarker of CRC progression, and its high expression is correlated with TNM stage, histologic grade, and lymph node metastasis (17). In addition, a recent meta-analysis and systemic review illustrated that DANCR expression is of significant prognostic value in various cancers (18). It is discovered that high lncRNA DANCR expression is associated with shorter overall survival and disease-free survival, predicting worse prognosis (18).
In the wake of the deepening of the research, DANCR was found to be aberrantly expressed in numerous cancers and exerted multiple regulating functions. The underlying mechanisms were also being constantly elucidated.
DANCR is a typical oncogenic lncRNA, overexpressed in various tumor cells. It can regulate the progression of numerous cancers by modulating the expression or activity of downstream targets and consequently promoting hallmarks of cancer including cell proliferation, cell motility, tumor angiogenesis, and cell viability.
In hepatocellular carcinoma, it is observed that down-regulated DANCR expression can boost cell apoptosis and cell cycle block in G1 which underlies that DANCR can promote tumor cell proliferation by inhibiting cell apoptosis and facilitating growth inhibition evasion (19). DANCR overexpressed in endometrial carcinoma can alleviate the inhibitory effect of miR-214 on target mRNAs and inhibit cell apoptosis and promote proliferation (20). In MYC-induced lymphoma, DANCR limits the transcription of Cell-cycle inhibitor p21 (CDKN1A) which can cause cell cycle arrest by restricting the activity of cyclin-dependent kinases. Inhibition of CDKN1A leads to accelerated G1/S shift and promote the proliferation of lymphoma cells (21, 22).
Cell motility refers to the metastasis and invasion properties of cells and malignant cells to vital organs and it is the major cause of cancer deaths (23). DANCR can promote tumor cell motility in various ways.
DANCR promotes the motility of nasopharyngeal carcinoma cells by stabilizing hypoxia inducible factor-1α, the subunit of Hypoxia inducible factor 1 (HIF-1) which are transcription factors that can activate several hypoxia-responsive genes in response to hypoxia microenvironment and promote the metastasis and invasion of cancer cells (24, 25). Matrix Metallopeptidase 16(MMP16) is one of the extracellular matrix (ECM) enzymes which can decompose the ECM physical barriers and promote the metastasis of cells (26). MMP16 can activate MMP2 for collagen degradation (27). DANCR can upregulate MMP16 protein and lead to enhanced motility of pancreatic cancer cells (28). Another promoting factor of carcinoma metastasis is Epithelial-mesenchymal transition (EMT), during which epithelial cells lose their epithelial characteristics and cell-cell contact, thus increasing their invasive potential and the migratory capacity and invasive potential of tumor cells (29). DANCR can modulate the EMT progression by upregulating Rho-associated coiled-coil containing protein kinase 1 (ROCK1) and promote metastasis in cervical cancer (30).
Tumor angiogenesis is one of the hallmarks of cancer which provide essential nutrients and oxygen to proliferating cancer cells (31). DANCR can increase vascular endothelial growth factor A (VEGF-A) expression, mediate vascular permeability as well as the multiplication and motion of endothelial cells and promote the neovascularization in ovarian cancer cell (32, 33).
The innate or acquired chemo-resistance of tumor cells is a main problem in the clinical cancer therapy. DANCR promotes the docetaxel resistance in prostate cancer cells by mediating the upregulation of multidrug resistance associated protein (MDR) and p-glycoprotein (P-gp), which bump many chemotherapeutic drugs out of the cells and limit the intracellular drug dosage to sub-therapeutic levels (34, 35). DANCR can also reduce the sensitivity of glioma cells to cisplatin through PI3K/Akt signaling pathway activation and mediate the recruitment of NF-κB protein to drug resistance genes for their enhanced transcription (36).
Generally, DANCR exerts multiple regulatory functions on tumor progression of numerous cancers, revealing its role as an important pro-tumor regulator (Table 1). And notably, DANCR, an oncogenic lncRNA in most cases, suppresses proliferation and motility and promotes apoptosis in renal carcinoma cells (RCCs), its mechanism remains unclear (61).
LncRNAs perform their multiple regulatory functions on tumors by interacting with numerous downstream targets (Figure 1C). The downstream targets include proteins and RNAs. LncRNAs can bind with histone-regulating proteins [such as Enhancer of zeste homolog 2 (EZH2), histone deacetylase (HDAC)], which catalyze the histone protein in chromatins, and the promoter-binding proteins, which can bind to the promoters or enhancers of certain genes, and guide the silencing of certain genes (34, 62, 63). They can interact with cytoplasm proteins, mRNAs and miRNAs and thus modulate the cells via stabilizing, regulating and modifying the components within signaling pathways (64). LncRNAs can also bind with integrated proteins and mediate the transmission of the signaling received from the extracellular environment (64). DANCR, as a typical lncRNA, exerts its regulatory functions mainly via protein-DANCR and miRNA-DANCR interaction (Table 2). Herein, we review some common targets of DANCR which are discovered to interact with DANCR in multiple cancers and further discuss the role of DANCR in particular cancers to better understand the regulatory mechanism of DANCR.
EZH2 is a common target protein of DANCR. Mechanically, EZH2 enables the silencing of gene expression by catalyzing histone H3 trimethylation at lysine 27 (H3K27me3) in target gene promoters (73). DANCR can bind with EZH2, guide it to silence the specific gene and thus regulate the cells. EZH2-DANCR interaction was first discovered in human fetal osteoblastic cell where the lncRNA-protein complex inhibited Runt-related transcription factor-2 expression and pursuant osteoblast differentiation (16). EZH2-DANCR mechanism can also be found in many cancers: In non-small-cell lung cancer cells, it promotes cell proliferation, migration, and invasion by inhibiting p21 expression (22). In gastric cancer cells it enhances cell migration and invasion through suppression of lncRNA-LET (Low Expression in Tumor) (72). In prostate cancer cells it boosts cell invasion by epigenetically silencing expression of tissue inhibitors of metalloproteinase 2/3 (58). In cholangiocarcinoma it regulates proliferation and migration by epigenetically silencing fructose-1,6-bisphosphatase 1. By silencing the tumor inhibitory gene via EZH2 recruitment, DANCR manages to promote cancer progression (45).
MicroRNAs are small regulating RNAs (−22 nt in length) derived from the short hairpin regions of RNA transcripts and they induce cleavage and suppression of translation via complementary binding to target RNAs (74). DANCR can acts as competitive endogenous RNA (ceRNA) and competitively bind tumor suppressive miRNAs to restore the functions of target oncogenic mRNAs. miR-216a is a common tumor suppressive miRNA and downstream target of DANCR. In lung cancer, DANCR induces cell proliferation and colony formation via sequestering miR-216a-5p (75). In breast cancer, DANCR targets miR-216a-5p, increases Nanog, SOX2, and OCT4 and promotes cell proliferation and invasion (42). In glioma, DANCR can restore LGR5 (Leucine-rich repeat-containing G protein-coupled receptor 5), PI3K, AKT, and p-AKT accumulation reduced by miR-216a, facilitating proliferation, migration, invasion, and angiogenesis and inhibited apoptosis (65). The massive targets of miR-216a complicate the functions of DANCR in cancer cells. In some cases, a novel DANCR-mRNA-miRNA interacting pathway is observed. For instance, DANCR competitively sponges the 3′UTR of transforming growth factor beta receptor 1 (TGFBR1) mRNA in cervical cancer cells, which blocks miR-665 from binding to and degrading TGFBR1 and promotes the TGFBR1-mediated oncogenic ERK (extracellular regulated protein kinases)/SMAD pathway (48).
In this section, the role of DANCR in particular cancers is discussed so as to clarify the molecular mechanisms of DANCR in a more specific way and instruct the therapy of DANCR aberrantly-expressed cancers (Figure 2).
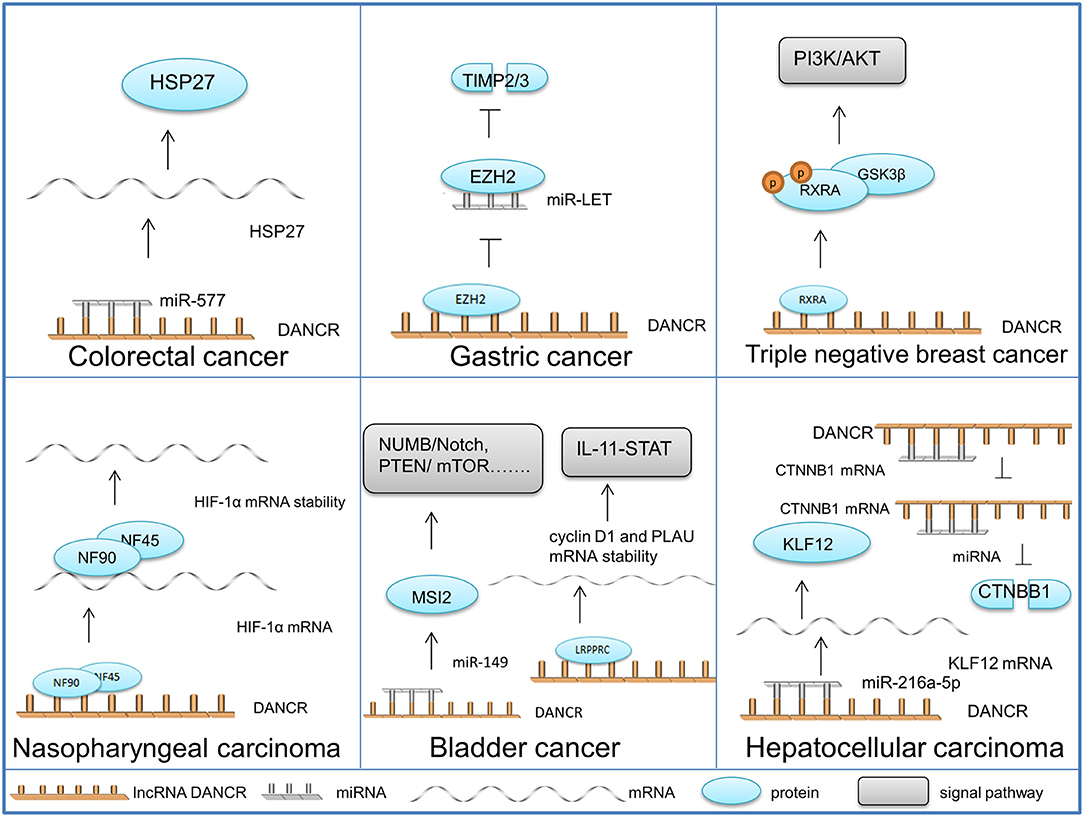
Figure 2. Functional properties and molecular mechanisms of DANCR in different cancers. The schematic illustrates the mechanisms of DANCR in different cancers.
DANCR acts as a miRNA sponge in colorectal cancer (CRC) cells, which is the most common role that DANCR plays in cancer development. Harboring the same miR-577 binding site, DANCR and heat shock protein 27 (HSP27) mRNA competitively bind to miR-577. The overexpression of DANCR in CRC cells alleviates the degradation impact of miR-577 on HSP27, which contributes to cell proliferation and metastasis (50).
DANCR activated by SALL-4 (Sal-like protein 4) (53) mediates the EZH2-HDAC3 (histone deacetylase 3) complex to regulate the transcription of lncRNA-LET. It binds to the protein complex and directs it to the promoter of LET (Low Expression in Tumor), thereby silencing the tumor suppressive RNA and promoting tumor metastasis (72).
HIF-1α, generally known as the core regulator of the hypoxic response, has been validated as a critical positive regulator of nasopharyngeal carcinoma (NPC) cell metastasis. DANCR can combine the NF90 (nuclear factor 90)/NF45 (nuclear factor 45) complex with its AU-rich elements (AREs) in 3′ region, which stabilizes HIF-1α mRNA and upgrades the motility of NPC cells (25).
In triple-negative breast cancer (TNBC), DANCR functions by mediating protein assembly and modification. DANCR can bind with the phosphorylation site of RXRA, recurring glycogen synthase kinase 3 beta (GSK3β) to sponge RXRA and promote RXRA ser phosphorylation. The structural change in RXRA suppresses its interaction with the promoter of phosphatidylinositol-4,5-biophosphate 3-kinase catalytic subunit alpha (PIK3CA), thereby enhancing PIK3CA in both mRNA and protein levels. Consequently, protein PIK3CA activates the P13K/AKT pathway, leading to the proliferation and tumor growth of TNBC (71).
Bladder cancer (BC) whose proper diagnosis and efficient treatment are current challenges in urology, is the most frequent malignancy of the urinary tract (76). The following two mechanisms of DANCR are discovered in BC cells: DANCR can combine with the miRNA-149 in BC cell cytoplasm, positively regulating the expression of Musashi RNA binding protein 2 (MSI2). Elevated MSI2 protein enhances the transcription and translation of the essential components in the oncogenic signaling pathways, such as NUMB/Notch, PTEN (Phosphatase and tensin homolog)/mTOR, TGF-β (transforming growth factor-β)/SMAD3, MYC, and cMET (77). DANCR can also direct a leucine-rich pentatricopeptide repeat containing (LRPPRC) to stabilize mRNA, thereby increasing cyclin D1 and PLAU (Plasminogen Activator, Urokinase) expression levels and activating IL-11 (Interleukin 11)-STAT3 (Signal Transducer and Activator of Transcription 3) signaling. Through the mechanisms above, DANCR facilitates the proliferation and invasion of BC cells (51).
Similar to CRC, oncogenic DANCR within hepatocellular carcinoma (HC) has also been found to have multiple regulating targets. KLF12 (Krueppel-like factor 12) can be positively regulated by DANCR through miR-216a-5p sponging, leading to malignant progression (19). DANCR can also competitively bind to the 3′UTR of Catenin beta-1 (CTNNB1) to block miRNA-mediated CTNNB1 suppression, leading to the initiation and progression of HC (78).
We can conclude that DANCR can regulate the signaling pathways by alleviating miRNA's inhibitory functions on target RNA, combining with proteins and guiding them to modulate the expression or the stability of RNAs or facilitating the integration and subsequent modifying of proteins. Moreover, DANCR can manipulate more than one target in cancer and join in multiple pathways. As we can see, due to the diverse mechanisms of DANCR and limited research on it, it is hard to foresee the unknown mechanisms and functions of DANCR in cancers. However, lncRNA's collective involvement in the pathways of numerous cancers does make it a promising target for cancer therapy.
The potency of lncRNA-based cancer therapy has been pointed out in previous years. Targeting lncRNAs for cancer treatment is a novel and promising therapy due to their collective therapeutic efficacy, high specificity, and few side effects (12). The possible molecular drugs against lncRNAs and nanoparticle vectors for their delivery are as follows (Figure 3).
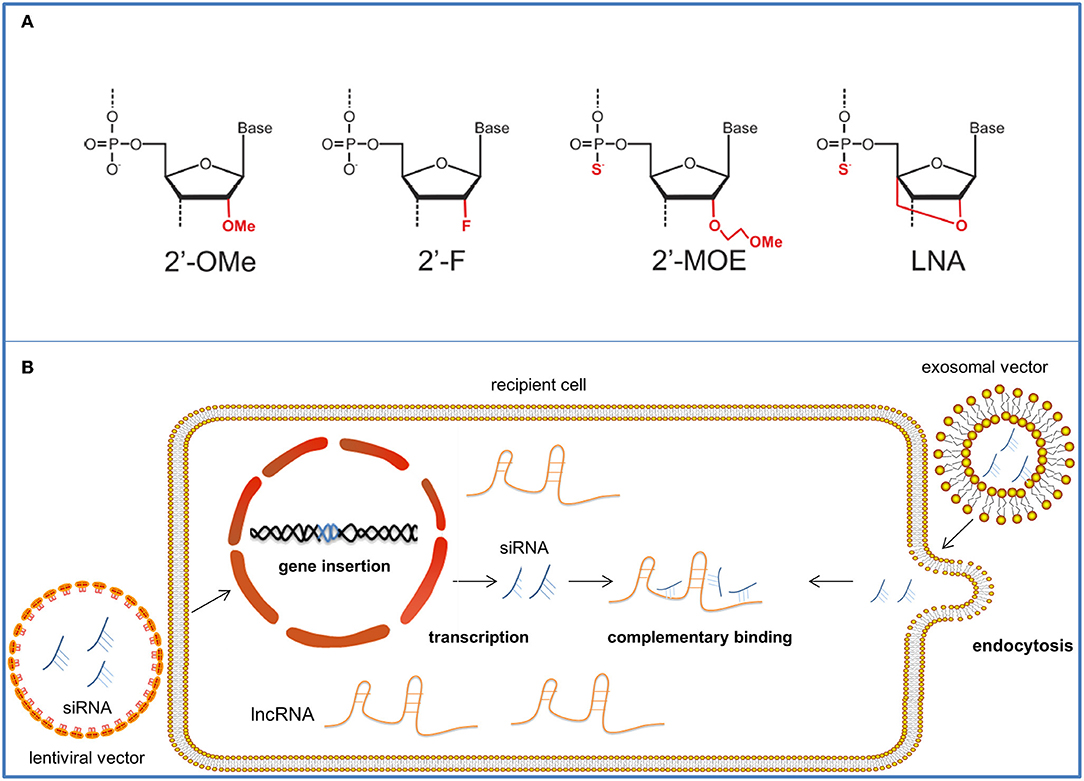
Figure 3. The therapeutic approaches targeting onco-lncRNA—DANCR. The schematic illustrates several targeting methods against onco-lncRNAs by using LNAs or siRNAs for lncRNA silencing and degradation. (A) Antisense oligonucleotides can be chemically modified to have strengthened affinity and stability. The figure shows four common modifications of ASOs. (B) SiRNAs are packaged in nanoparticle vectors for improved targeting. Lentiviral vectors can reverse and insert the siRNA sequence into the genome of target cells and achieve stable transfection. After transcription, siRNAs complementarily bind the target onco-lncRNAs and suppress their tumor promoter functions. Exosomal nanovectors can directly deliver the siRNAs to the target cell cytoplasm for onco-lncRNA inhibition through endocytosis.
The CRISPR/Cas9 technology can be used to target the endogenous loci of lncRNAs and restrict their transcription (79). The CRISPR system discovered in many bacteria as an immune mechanism for invading nucleic acid elimination can produce CRISPR RNAs (crRNAs) with certain “protospacer” regions complementary to the foreign DNA site (80). The crRNAs together with transactivating CRISPR RNAs (tracrRNAs) form the RNA pairs which combine and direct Cas9 nuclease to cleave complementary target-DNA sequences and cause DNA double-stranded break (DSB) (80). The CRISPR/cas9 system can be applied for target DNA cut and gene knockout via a designed single-guide RNA (sgRNA) complementary to the target gene (81). Progress has been made in the field: Cas9 nickases can cut one strand rather than both strands of the target DNA site (82). Catalytically inactive Cas9 (dCas9) enables the transcription regulation of target genes which may partly relieve the safety concern about irreversible frameshift disruptions caused by DNA cut (83). The dCas9 protein can be recruited to the coding region of genes and sterically block their transcription (84). Moreover, dCas9 can be fused to protein domains capable of recruiting repressive chromatin-modifying complexes, and lead to the silencing of particular genes determined by the engineered sgRNA (84). Recently, by targeting Erb-B2 Receptor Tyrosine Kinase 2(HER2) gene, CRISPR-Cas9 has managed to inhibit cell proliferation and tumorigenicity in HER2-positive breast cancer cells, demonstrating its potency in clinical application (85). The CRISPR/Cas9 technology has also been experimented for targeting oncogenic lncRNAs and further improvement is in demand for better efficacy (86).
Antisense oligonucleotides (ASOs) can be used in lncRNA-targeted therapy. They are synthetic and modified oligonucleotide single strands that can complementarily bind to certain sequences of their target RNAs. ASOs exert their inhibiting effect mainly by recruiting ribonuclease H (Rnase H) to degrade the targets. They comprise complementary DNA single strands to form DNA/RNA duplexes for Rnase H recognition and later cleavage (87). Vulnerable to various existing ribonucleases (88), ASOs are unstable in vivo. Combined with their relatively high charge and hydrophilicity (89), all these characteristics make crossing membranes of targeted cells difficult. Similar to other RNA therapeutic products, ASOs must be chemically modified to improve their pharmacological properties. Phosphorylation is the most commonly employed modification method (88) that involves substituting the phosphodiester backbones of the oligonucleotide strands for phosphonothioate (Ps) linkages (87). In this way, nucleotides become more hydrophobic and have higher adhesion to plasma proteins, enabling them to enter the targeted cells (68). Additionally, their resistance against nucleuses is advanced, making the products more stable (90). However, phosphorylation can reduce ASO's affinity to RNA and increase the likelihood of non-specific protein binding (90), which is harmful to regulation. In addition to phosphorylation, other widely used modifications include 2′ sugar modifications [such as 2′-O-methyl, 2′-fluoro (2′-F) and 2′- O-(2-methoxyethyl)(2′-MOE)],which all result in high affinity to RNA along with their diminished or disappeared ability to attract Rnase H (91). Uniformly modified ASOs are ASOs that completely lose their ability to recruit Rnase H for degradation, and they can sterically block certain domains to inhibit further interaction (92). Among all these synthetic products, locked nucleic acid (LNA), which is produced by changing the 20-hydroxy (20-OH) for 20,40-O-methylene bridge, exhibits the best affinity to RNAs (93). LNA can be used as the flank of GAPMER, which is a widely-used artificial complex for RNA targeting (88). The center of GAPMER is a DNA monomer which can enable Rnase H cleavage of the targeting mRNA, while LNA flanks increase its affinity to RNA and resistance to nuclease (69). Both miRNA affinity and nucleotide stability are achieved in this modification. Nusinersen, a splicing switching ASO, is a successful application of ASO in clinical treatment which was approved by the FDA in 2016 and became the first drug to treat spinal muscular atrophy (94). And to achieve better pharmaceutical effect, considerable work has to be done to achieve specific transportation (95).
We can also use small interfering RNAs (siRNAs) to target lncRNAs. In 2018 patisiran became the first approved RNAi drug to remedy hereditary transthyretin amyloidosis (70). SiRNAs are usually chemically synthesized or evolved from DNA-based small hairpin RNA (shRNA) (96). And compared to chemically synthesized siRNAs, effective shRNA expression DNA sequence could retain gene silencing for a longer period (97). Mechanically, siRNAs induce gene silencing by complementing with the target RNA and forming an RNA-induced silencing complex to splice and degrade the target (98). SiRNAs can be chemically modified to improve stability and prevent nuclease degradation. For example, boranophosphate siRNAs were 10 times more nuclease resistant than unmodified siRNAs with advanced silencing efficacy (99). Incorporation of a modest number of LNA modifications significantly prolongs the half-life of serum siRNAs (100). SiRNAs can also be packaged in synthetic carriers to enter the cell membrane (101).
Viral vectors were one of the earliest engineered nanoscale delivery systems used for transporting RNA products to targeted tissues. Viral vectors are known for their efficient transfer of codes to the cell interior and capability for escaping immunosurveillance by infected cells (102). In tumor treatment, lentiviral vectors have an advantage over other viral vectors. As a retroviral vector, the lentiviral vector avoids the drawback of transient transgene expression in the case of AdVs (102); reagent RNAs can be reversely transcribed into DNA with the help of reverse transcriptase and inserted into the genes of the targeted cells, which enables the RNA reagent to exert continual therapeutic effects on the fast-growing cell lines. It is less likely to cause an intense inflammatory response and can be processed in a relatively simple way (103). Moreover, LVs, with a maximum capacity of 8 kb nucleic strands, boast a relatively high capacity compared to other viral vectors. Although problems such as possible insertional mutagenesis (104), comparatively high immunogenicity and difficult large-scale production persist (102), the utilization of siRNA packaged in modified LVs for the modulation of onco-lncRNAs and their molecular targets will be a promising treatment for cancers.
Compared to viral vectors, non-viral vectors are of lower transfection efficiency (105). However, the low immunogenicity and advanced safety, relative convenient and inexpensive constructing process for large scale production make the latter one attractive in clinical therapy (102). Notably, owing to the hydrophobic nature of anchored lipids, lipid based vectors prevail over other non-lipid vectors for the extremely high transfection efficiency (106). Moreover, liposomes products comprise 70% of submissions to the FDA, which also proves the safety and effectiveness of liposomes (107). Generally, the liposome is composed of neutral or cationic amphiphilic lipid units. Neutral or positively charged polar group(s) makes up the hydrophilic “head” and a hydrophobic “tail” comprise fatty acid(s) (102). Compared to neutral ones, the cationic liposomes can integrate with negatively charged RNA molecules to form electrostatic lipoplexes, which can wrap and protect RNA from degradation by serum nucleases and promote endocytosis for cellular uptake (108). Better pharmacological properties can be achieved through liposome modulation. Conjugating a poly-(ethylene glycol) (PEG)-lipid within the lipid bilayer can extend circulating period and reduce the uptake by phagocytes (109, 110).Specific ligands and antibodies can be added to the lipid for enhanced delivery specificity (111). For example, the conjugation of transferrin to nanoparticles is demonstrated to be an efficient cancer targeted strategy due to extremely higher expression level of transferrin receptor on tumor cell surface (112).
Notably, exosomes can also play a part in cancer treatment by working as a novel and practical biological vector. Exosomes are vesicles released by cells into serums containing proteins, lipids and nucleic acids (113). They are involved in extracellular communication and mediate tumor progression via the contents within. As a biological vector, exosomes are less likely to activate the immune response and are highly stable in vivo (114), thereby prolonging their circulation duration. Their small sizes (30–150 nm) also enable them to function inside dense tissues such as osteoblasts. The general construction progress comprises five steps (114): (a) find a suitable parental cell for exosome vector production. For example, immature dendritic cells can produce exosomes that are deficient in T-cell activating factors so that they can cause minimal immune reaction (115). HEK293T cells are credited for high transferring efficacy as they can produce exosomes in large quantities. The produced exosomes can easily diffuse with targeted cells and release the inner therapeutic contents (116); (b) transfect the parental cells with plasmid containing the gene code of ligand proteins which can bind the receptor on targeted cells. In this way, exosomes are engineered with the desired ligands on their surface and they can specifically target the recipient cells. In previous practice, HEK293T cells were transfected with pDisplay encoding GE11, a ligand matching the receptors on recipient breast cancer cells, for improved targeting efficacy (117); (c) isolate the exosomes by ultracentrifugation or use of commercial kit, etc.; (d) package the therapeutic reagents into exosome vectors via electroporation; (e) inject the exosome vector into human internal environment, and the exosome can circulate in vivo and find its way to the target cells.
Alvarez-Erviti et al. pioneered the practice of applying engineered exosomes to deliver siRNA. They build neuronal cell-targeted exosomes and use them to pass through the blood-brain barrier and treat Alzheimer's disease (118). A recent trial using exosome vector delivering siRNA was conducted in HER2 positive breast cancer cells and BC cells (119). Although methods of exosome separation and exosomal carrier construction need considerable improvement, all these successful practices remark a bright prospect for therapeutic exosome vector.
To date, researches on targeting DANCR for cancer therapy remains limited. A previous study presented that the relative enrichment of the enzymes responsible for RNA degradation vary between cellular compartments, so the location of lncRNA can impact the suppressing efficacy of the molecular drugs on it. Comparatively, ASO is more capable of clearing the nuclear lncRNAs while RNAi have a better suppressive effect on lncRNAs in cytoplasm (120). Referring to this, the RNAi therapy is more suitable for the cytoplasmic oncogenic lncRNA DANCR (120). Moreover, being effectively suppressed by all 28 RNAi regents tested in the experiment further demonstrated that DANCR can be an ideal therapeutic target (120). Researchers should work on the construction of superior vector of the RNAi regents for better targeting effect. Remarkable progress has been made by Vaidya et al. who successfully constructed a non-viral nanoparticle carrier containing siDANCR and proved its repressive effect on the invasion and proliferation of TNBC cells via null mice injection (12). Overall, DANCR targeted therapy is of great promise and must be investigated further.
The review has shown the vital research value of DANCR. DANCR is also a critical oncogenic regulator which presents an increasingly important status in cancer study. It can regulate hallmarks of various cancers, indicate their progression and clinical outcomes and serve as a novel target for cancer targeted treatment. Researches on DANCR remain limited and there is an urgent need for further study on this critical onco-lncRNA.
The recent progress on RNA interaction identification method includes the refined variants of immunoprecipitation techniques (such as PAR-CLIP, HITS-CLIP Maps, iCLIP, hiCLIP, CLASH etc.) and new high-throughput RNA interactome analysis methods [such as Psoralen analysis of RNA interactions and structures (PARIS), sequencing of psoralen-crosslinked, ligated, and selected hybrids (SPLASH), ligation of interacting RNA followed by high-throughput sequencing (LIGR-seq), and MARIO] (121). Without any form of crosslinking, proximity proteomics is a novel method for RNA-protein interactions studies (122). Wide application of these techniques and further development of the new ones in the late future may bring forward a new impetus for the understanding of the diverse and complicated regulatory mechanisms of lncRNA in cancers. Also, advanced techniques are in demand for the lncRNA targeted therapy. Improved targeting methods and drug vectors are needed to reduce untoward effect and improve the efficacy and specificity of the therapy.
S-JJ conceived, wrote the manuscript, and completed the figures and tables. M-ZJ revised the manuscript. W-LJ and B-RX conceived, organized, and edited the text.
This work was supported by China Postdoctoral Science Foundation (No. 2019T120281), Natural Science Foundation of Heilongjiang (No. H2017049), and Applied Technology Research and Development Project of Harbin (2017RAQXJ231). This work was in part supported by the National Natural Science Foundation of China (No. 81872430) and the National Key Research and Development Program of China (No. 2017FYA0205302).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We apologize to those colleagues whose important work could not be cited due to space constraints.
1. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
3. Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. (2018) 48:8–15. doi: 10.1016/j.gde.2017.10.004
4. Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS. Long non-coding RNA: its evolutionary relics and biological implications in mammals: a review. J Anim Sci Technol. (2018) 60:25. doi: 10.1186/s40781-018-0183-7
5. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. (1990) 10:28–36. doi: 10.1128/MCB.10.1.28
6. Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. (1992) 71:515–26. doi: 10.1016/0092-8674(92)90519-I
7. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. (2018) 19:535–48. doi: 10.1038/s41576-018-0017-y
8. Carlevaro-Fita J, Johnson R. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol Cell. (2019) 73:869–83. doi: 10.1016/j.molcel.2019.02.008
9. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
10. Wei JT, Ziding F, Partin AW, Elissa B, Ian T, Lori S, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol. (2014) 32:4066–72. doi: 10.1200/JCO.2013.52.8505
11. Deng H, Zhang J, Shi JJ, Guo ZD, He CR, Ding L, et al. Role of long non-coding RNA in tumor drug resistance. Tumor Biol. (2016) 37:11623–31. doi: 10.1007/s13277-016-5125-8
12. Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X, Jiang H, et al. Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA facilitates effective triple-negative breast cancer therapy. Bioconjug Chem. (2019) 30:907–19. doi: 10.1021/acs.bioconjchem.9b00028
13. Kikuchi K, Fukuda M, Ito T, Inoue M, Yokoi T, Chiku S, et al. Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. (2009) 37:4987–5000. doi: 10.1093/nar/gkp426
14. Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. (2012) 26:338–43. doi: 10.1101/gad.182121.111
15. Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. (2015) 32:693–706. doi: 10.1016/j.devcel.2015.01.028
16. Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. (2013) 432:612–7. doi: 10.1016/j.bbrc.2013.02.036
17. Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. (2015) 8:11480–4.
18. Liu W, Wang QP, Guo J. Prognostic significance of long non-coding RNA DANCR expression in human cancers: a systematic review and meta-analysis. Biosci Rep. (2019) 39:BSR20190608. doi: 10.1042/BSR20190608
19. Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, et al. DANCR contributed to hepatocellular carcinoma malignancy via sponging miR-216a-5p and modulating KLF12. J Cell Physiol. (2019) 234:9408–16. doi: 10.1002/jcp.27625
20. Sun J, Gao S, Lu C. Knockdown of differentiation antagonizing non-protein coding RNA exerts anti-tumor effect by up-regulating miR-214 in endometrial carcinoma. Mol Cell Biochem. (2019) 460:9–15. doi: 10.1007/s11010-019-03565-0
21. Dutto I, Tillhon M, Cazzalini O, Stivala LA, Prosperi E. Biology of the cell cycle inhibitor p21(CDKN1A): molecular mechanisms and relevance in chemical toxicology. Arch Toxicol. (2015) 89:155–78. doi: 10.1007/s00204-014-1430-4
22. Guo L, Gu J, Hou S, Liu D, Zhou M, Hua T, et al. Long non-coding RNA DANCR promotes the progression of non-small-cell lung cancer by inhibiting p21 expression. Onco Targets Ther. (2019) 12:135–46. doi: 10.2147/OTT.S186607
23. Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. (2005) 5:591–602. doi: 10.1038/nrc1670
24. Kitajima Y, Ide T, Ohtsuka T, Miyazaki K. Induction of hepatocyte growth factor activator gene expression under hypoxia activates the hepatocyte growth factor/c-Met system via hypoxia inducible factor-1 in pancreatic cancer. Cancer Sci. (2008) 99:1341–7. doi: 10.1111/j.1349-7006.2008.00828.x
25. Wen X, Liu X, Mao YP, Yang XJ, Wang YQ, Zhang PP, et al. Long non-coding RNA DANCR stabilizes HIF-1alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. (2018) 8:5676–89. doi: 10.7150/thno.28538
26. Jablonska-Trypuc A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhibit Med Chem. (2016) 31:177–83. doi: 10.3109/14756366.2016.1161620
27. Jalali S, Singh S, Agnihotri S, Wataya T, Salehi F, Alkins R, et al. A role for matrix remodelling proteins in invasive and malignant meningiomas. Neuropathol Appl Neurobiol. (2015) 41:e16–28. doi: 10.1111/nan.12166
28. Luo Y, Wang Q, Teng L, Zhang J, Song J, Bo W, et al. LncRNA DANCR promotes proliferation and metastasis in pancreatic cancer by regulating miRNA-33b. FEBS Open Biol. (2019). doi: 10.1002/2211-5463.12732
29. Azab AK, Hu J, Quang P, Azab F, Pitsillides C, Awwad R, et al. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood. (2012) 119:5782–94. doi: 10.1182/blood-2011-09-380410
30. Liang H, Zhang C, Guan H, Liu J, Cui Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. (2019) 234:7266–78. doi: 10.1002/jcp.27484
31. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
32. Girling JE, Rogers PA. Regulation of endometrial vascular remodelling: role of the vascular endothelial growth factor family and the angiopoietin-TIE signalling system. Reproduction. (2009) 138:883–93. doi: 10.1530/REP-09-0147
33. Lin X, Yang F, Qi X, Li Q, Wang D, Yi T, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog. (2019). doi: 10.1002/mc.23117
34. Ma Y, Fan B, Ren Z, Liu B, Wang Y. Long noncoding RNA DANCR contributes to docetaxel resistance in prostate cancer through targeting the miR-34a-5p/JAG1 pathway. Onco Targets Ther. (2019) 12:5485–97. doi: 10.2147/OTT.S197009
35. Yuan Y, Cai T, Xia X, Zhang R, Chiba P, Cai Y. Nanoparticle delivery of anticancer drugs overcomes multidrug resistance in breast cancer. Drug Deliv. (2016) 23:3350–7. doi: 10.1080/10717544.2016.1178825
36. Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int. (2018) 118:233–41. doi: 10.1016/j.neuint.2018.03.011
37. Yang M, Wei W. Long non-coding RNAs in retinoblastoma. Pathol Res Pract. (2019) 215:152435. doi: 10.1016/j.prp.2019.152435
38. Zhang X, Yang J, Bian Z, Shi D, Cao Z. Long noncoding RNA DANCR promotes nasopharyngeal carcinoma progression by interacting with STAT3, enhancing IL-6/JAK1/STAT3 signaling. Biomed Pharmacother. (2019) 113:108713. doi: 10.1016/j.biopha.2019.108713
39. Zhang C, Wang L, Yang J, Fu Y, Li H, Xie L, et al. MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1. Eur J Pharmacol. (2019) 861:172590. doi: 10.1016/j.ejphar.2019.172590
40. Bai Y, Zhang G, Chu H, Li P, Li J. The positive feedback loop of lncRNA DANCR/miR-138/Sox4 facilitates malignancy in non-small cell lung cancer. Am J Cancer Res. (2019) 9:270–84.
41. Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. (2018) 103:94–100. doi: 10.1016/j.biopha.2018.03.053
42. Tao W, Wang C, Zhu B, Zhang G, Pang D. LncRNA DANCR contributes to tumor progression via targetting miR-216a-5p in breast cancer: lncRNA DANCR contributes to tumor progression. Biosci Rep. (2019) 39:BSR20181618. doi: 10.1042/BSR20181618
43. Sha S, Yuan D, Liu Y, Han B, Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. (2017) 6:1310–6. doi: 10.1242/bio.023135
44. Ma X, Wang X, Yang C, Wang Z, Han B, Wu L, et al. DANCR acts as a diagnostic biomarker and promotes tumor growth and metastasis in hepatocellular carcinoma. Anticancer Res. (2016) 36:6389–98. doi: 10.21873/anticanres.11236
45. Wang N, Zhang C, Wang W, Liu J, Yu Y, Li Y, et al. Long noncoding RNA DANCR regulates proliferation and migration by epigenetically silencing FBP1 in tumorigenesis of cholangiocarcinoma. Cell Death Dis. (2019) 10:585. doi: 10.1038/s41419-019-1810-z
46. Chen L, Liu J, Tang T, Zhang YC, Liu MZ, Xu LY, et al. lncRNA differentiation antagonizing nonprotein coding RNA overexpression accelerates progression and indicates poor prognosis in pancreatic ductal adenocarcinoma. Onco Targets Ther. (2018) 11:7955–65. doi: 10.2147/OTT.S167065
47. Yao Z, Chen Q, Ni Z, Zhou L, Wang Y, Yang Y, et al. Long non-coding RNA differentiation antagonizing nonprotein coding RNA (DANCR) promotes proliferation and invasion of pancreatic cancer by sponging miR-214-5p to regulate E2F2 expression. Med Sci Monit. (2019) 25:4544–52. doi: 10.12659/MSM.916960
48. Cao L, Jin H, Zheng Y, Mao Y, Fu Z, Li X, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. (2019) 110:913–25. doi: 10.1111/cas.13921
49. Gao YQ, Cheng HY, Liu KF. Long non-coding RNA DANCR upregulates IGF2 expression and promotes ovarian cancer progression. Eur Rev Med Pharmacol Sci. (2019) 23:3621–6. doi: 10.26355/eurrev_201905_17785
50. Wang Y, Lu Z, Wang N, Feng J, Zhang J, Luan L, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. (2018) 50:57. doi: 10.1038/s12276-018-0082-5
51. Chen Z, Chen X, Xie R, Huang M, Dong W, Han J, et al. DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing IL-11-STAT3 signaling and CCND1 expression. Mol Ther. (2019) 27:326–41. doi: 10.1016/j.ymthe.2018.12.015
52. Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335–5p and miR-1972 in osteosarcoma. Mol Cancer. (2018) 17:89. doi: 10.1186/s12943-018-0837-6
53. Pan L, Liang W, Gu J, Zang X, Huang Z, Shi H, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. (2018) 9:1915–30. doi: 10.18632/oncotarget.23019
54. Xu YD, Shang J, Li M, Zhang YY. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur Rev Med Pharmacol Sci. (2019) 23:2794–802. doi: 10.26355/eurrev_201904_17554
55. Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/beta-catenin signaling. Biomed Pharmacother. (2018) 102:602–7. doi: 10.1016/j.biopha.2018.03.116
56. Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. (2018) 38:BSR20171664. doi: 10.1042/BSR20171664
57. Lu Y, Hu Z, Mangala LS, Stine ZE, Hu X, Jiang D, et al. MYC targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levels. Cancer Res. (2018) 78:64–74. doi: 10.1158/0008-5472.CAN-17-0815
58. Jia J, Li F, Tang XS, Xu S, Gao Y, Shi Q, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. (2016) 7:37868–81. doi: 10.18632/oncotarget.9350
59. Zhao HF, Zhang ZC, Shi BK, Jiang XZ. DANCR sponges miR-135a to regulate paclitaxel sensitivity in prostate cancer. Eur Rev Med Pharmacol Sci. (2019) 23:6849–57. doi: 10.26355/eurrev_201908_18724
60. Zhang K, Lv J, Peng X, Liu J, Li C, Li J, et al. Down-regulation of DANCR acts as a potential biomarker for papillary thyroid cancer diagnosis. Biosci Rep. (2019) 39:BSR20181616. doi: 10.1042/BSR20181616
61. Jin L, Fu H, Quan J, Pan X, He T, Hu J, et al. Overexpression of long non-coding RNA differentiation antagonizing non-protein coding RNA inhibits the proliferation, migration and invasion and promotes apoptosis of renal cell carcinoma. Mol Med Rep. (2017) 16:4463–8. doi: 10.3892/mmr.2017.7135
63. Tang Q, Hann SS. HOTAIR: an oncogenic long non-coding RNA in human cancer. Cell Physiol Biochem. (2018) 47:893–913. doi: 10.1159/000490131
64. Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: a new regulator in cancer signaling. Cancer Lett. (2018) 419:27–9. doi: 10.1016/j.canlet.2018.01.008
65. Wang W, Li Y, Ma Q, Yan H, Su W. Differentiation antagonizing non-protein coding RNA modulates the proliferation, migration, and angiogenesis of glioma cells by targeting the miR-216a/LGR5 axis and the PI3K/AKT signaling pathway. Onco Targets Ther. (2019) 12:2439–49. doi: 10.2147/OTT.S196851
66. Guo D, Li Y, Chen Y, Zhang D, Wang X, Lu G, et al. DANCR promotes HCC progression and regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1 pathway. Cell Prolif. (2019) 52:e12628. doi: 10.1111/cpr.12628
67. Tang Y, Cao G, Zhao G, Wang C, Qin Q. LncRNA differentiation antagonizing non-protein coding RNA promotes proliferation and invasion through regulating miR-135a/NLRP37 axis in pancreatic cancer. Invest New Drugs. (2019). doi: 10.1007/s10637-019-00798-0
68. Geary RS. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin Drug Metab Toxicol. (2009) 5:381–91. doi: 10.1517/17425250902877680
69. Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. (2002) 30:1911–8. doi: 10.1093/nar/30.9.1911
70. Yang J. Patisiran for the treatment of hereditary transthyretin-mediated amyloidosis. Expert Rev Clin Pharmacol. (2019) 12:95–9. doi: 10.1080/17512433.2019.1567326
71. Tang J, Zhong G, Zhang H, Yu B, Wei F, Luo L, et al. LncRNA DANCR upregulates PI3K/AKT signaling through activating serine phosphorylation of RXRA. Cell Death Dis. (2018) 9:1167. doi: 10.1038/s41419-018-1220-7
72. Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. (2017) 37:BSR20171070. doi: 10.1042/BSR20171070
73. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. (2002) 298:1039–43. doi: 10.1126/science.1076997
74. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
75. Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX, Zhao XJ, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. (2018) 25:1073274818769849. doi: 10.1177/1073274818769849
76. Kutwin P, Konecki T, Cichocki M, Falkowski P, Jablonowski Z. Photodynamic diagnosis and narrow-band imaging in the management of bladder cancer: a review. Photomed Laser Surg. (2017) 35:459–64. doi: 10.1089/pho.2016.4217
77. Zhan Y, Chen Z, Li Y, He A, He S, Gong Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. (2018) 37:273. doi: 10.1186/s13046-018-0921-1
78. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. (2016) 63:499–511. doi: 10.1002/hep.27893
79. Elling R, Chan J, Fitzgerald KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur J Immunol. (2016) 46:504–12. doi: 10.1002/eji.201444558
80. Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. (2014) 32:347–55. doi: 10.1038/nbt.2842
81. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. (2012) 337:816–21. doi: 10.1126/science.1225829
82. Gao Y, Wu H, Wang Y, Liu X, Chen L, Li Q, et al. Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol. (2017) 18:13. doi: 10.1186/s13059-016-1144-4
83. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. (2014) 159:647–61. doi: 10.1016/j.cell.2014.09.029
84. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. (2013) 154:442–51. doi: 10.1016/j.cell.2013.06.044
85. Wang H, Sun W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. (2017) 385:137–43. doi: 10.1016/j.canlet.2016.10.033
86. Goyal A, Myacheva K, Gross M, Klingenberg M, Duran Arque B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. (2017) 45:e12. doi: 10.1093/nar/gkw883
87. Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. (2019) 18:421–46. doi: 10.1038/s41573-019-0017-4
88. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. (2018) 24:257–77. doi: 10.1016/j.molmed.2018.01.001
89. Bass BL. Double-stranded RNA as a template for gene silencing. Cell. (2000) 101:235–8. doi: 10.1016/S0092-8674(02)71133-1
90. Valerio LG Jr. Tenth anniversary of expert opinion on drug metabolism & toxicology. Expert Opin Drug Metab Toxicol. (2014) 10:767–8. doi: 10.1517/17425255.2014.920007
91. Monia BP, Lesnik EA, Gonzalez C, Lima WF, Mcgee D, Guinosso CJ, et al. Evaluation of 2'-modified oligonucleotides containing 2'-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. (1993) 268:14514.
92. Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. (2011) 478:123–6. doi: 10.1038/nature10485
93. Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. (2008) 452:896–9. doi: 10.1038/nature06783
94. Delivering the promise of RNA therapeutics. Nat Med. (2019) 25:1321. doi: 10.1038/s41591-019-0580-6
95. Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. (2010) 38:e3. doi: 10.1093/nar/gkp841
96. Nie L, Das Thakur M, Wang Y, Su Q, Zhao Y, Feng Y. Regulation of U6 promoter activity by transcriptional interference in viral vector-based RNAi. Genomics Proteomics Bioinformatics. (2010) 8:170–9. doi: 10.1016/S1672-0229(10)60019-8
97. Liao Y, Tang L. Inducible RNAi system and its application in novel therapeutics. Crit Rev Biotechnol. (2016) 36:630–8. doi: 10.3109/07388551.2014.1003030
98. Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. (2002) 110:563–74. doi: 10.1016/S0092-8674(02)00908-X
99. Hall AH, Wan J, Shaughnessy EE, Ramsay Shaw B, Alexander KA. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. (2004) 32:5991–6000. doi: 10.1093/nar/gkh936
100. Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. (2005) 33:439–47. doi: 10.1093/nar/gki193
101. Turner JJ, Jones SW, Moschos SA, Lindsay MA, Gait MJ. MALDI-TOF mass spectral analysis of siRNA degradation in serum confirms an RNAse A-like activity. Mol Biosyst. (2007) 3:43–50. doi: 10.1039/B611612D
102. Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. (2010) 17:295–304. doi: 10.1038/gt.2009.148
103. Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: basic to translational. Biochem J. (2012) 443:603–18. doi: 10.1042/BJ20120146
104. Matrai J, Cantore A, Bartholomae CC, Annoni A, Wang W, Acosta-Sanchez A, et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. (2011) 53:1696–707. doi: 10.1002/hep.24230
105. Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. (2017) 474:4219–51. doi: 10.1042/BCJ20170079
106. Alshamsan A, Haddadi A, Incani V, Samuel J, Lavasanifar A, Uludag H. Formulation and delivery of siRNA by oleic acid and stearic acid modified polyethylenimine. Mol Pharm. (2009) 6:121–33. doi: 10.1021/mp8000815
107. D'Mello SR, Cruz CN, Chen ML, Kapoor M, Lee SL, Tyner KM. The evolving landscape of drug products containing nanomaterials in the United States. Nat Nanotechnol. (2017) 12:523–9. doi: 10.1038/nnano.2017.67
108. Pathak K, Keshri L, Shah M. Lipid nanocarriers: influence of lipids on product development and pharmacokinetics. Crit Rev Ther Drug Carrier Syst. (2011) 28:357–93. doi: 10.1615/CritRevTherDrugCarrierSyst.v28.i4.20
109. Fang C, Shi B, Pei YY, Hong MH, Wu J, Chen HZ. In vivo tumor targeting of tumor necrosis factor-alpha-loaded stealth nanoparticles: effect of MePEG molecular weight and particle size. Eur J Pharm Sci. (2006) 27:27–36. doi: 10.1016/j.ejps.2005.08.002
110. Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomed. (2006) 1:297–315.
111. Perche F, Torchilin VP. Recent trends in multifunctional liposomal nanocarriers for enhanced tumor targeting. J Drug Deliv. (2013) 2013:705265. doi: 10.1155/2013/705265
112. Ulbrich K, Hekmatara T, Herbert E, Kreuter J. Transferrin- and transferrin-receptor-antibody-modified nanoparticles enable drug delivery across the blood-brain barrier (BBB). Eur J Pharm Biopharm. (2009) 71:251–6. doi: 10.1016/j.ejpb.2008.08.021
113. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. (2015) 219:278–94. doi: 10.1016/j.jconrel.2015.06.029
114. Xie X, Wu H, Li M, Chen X, Xu X, Ni W, et al. Progress in the application of exosomes as therapeutic vectors in tumor-targeted therapy. Cytotherapy. (2019) 21:509–24. doi: 10.1016/j.jcyt.2019.01.001
115. Quah BJ, O'Neill HC. The immunogenicity of dendritic cell-derived exosomes. Blood Cell Mol Dis. (2005) 35:94–110. doi: 10.1016/j.bcmd.2005.05.002
116. Li J, Chen X, Yi J, Liu Y, Li D, Wang J, et al. Identification and characterization of 293T cell-derived exosomes by profiling the protein, mRNA and microRNA components. PLoS ONE. (2016) 11:e0163043. doi: 10.1371/journal.pone.0163043
117. Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. (2013) 21:185–91. doi: 10.1038/mt.2012.180
118. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. (2011) 29:341–5. doi: 10.1038/nbt.1807
119. Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol. (2019) 187:352–64. doi: 10.1007/s12010-018-2813-4
120. Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. (2016) 44:863–77. doi: 10.1093/nar/gkv1206
121. Nguyen TC, Zaleta-Rivera K, Huang X, Dai X, Zhong S. RNA, action through interactions. Trends Genet. (2018) 34:867–82. doi: 10.1016/j.tig.2018.08.001
Keywords: long non-coding RNA, cancer, DANCR, mechanism, therapy
Citation: Jin S-J, Jin M-Z, Xia B-R and Jin W-L (2019) Long Non-coding RNA DANCR as an Emerging Therapeutic Target in Human Cancers. Front. Oncol. 9:1225. doi: 10.3389/fonc.2019.01225
Received: 10 July 2019; Accepted: 28 October 2019;
Published: 15 November 2019.
Edited by:
Zhaohui Huang, Affiliated Hospital of Jiangnan University, ChinaReviewed by:
Vijay Pandey, Tsinghua-Berkeley Shenzhen Institute, ChinaCopyright © 2019 Jin, Jin, Xia and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bai-Rong Xia, eGlhYmFpcm9uZzk5OTlAMTI2LmNvbQ==; Wei-Lin Jin, d2VpbGluamluQHlhaG9vLmNvbQ==; d2VpbGluamluQHNqdHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.