- 1Blood Diseases Institute, Affiliated Hospital of Xuzhou Medical University, Xuzhou Medical University, Xuzhou, China
- 2Department of Hematology, Affiliated Hospital of Xuzhou Medical University, Xuzhou, China
- 3Department of Hematology, The Third Affiliated Hospital of Soochow University, Changzhou, China
- 4Department of Neurosurgery, Institute of Nervous System Diseases, Xuzhou Medical University, Xuzhou, China
Acute myeloid leukemia (AML) requires refined risk stratification tools to drive decisions concerning effective therapeutic strategies. Here, genome-wide screening was carried out for identifying miRNA molecules capable of predicting treatment outcome in AML patients based on the TCGA dataset. We identified miR-340 as a prognostic factor for selecting treatment between chemotherapy and allogeneic transplantation (allo-HSCT). In multivariable analyses, low miR-340 expression independently predicted reduced OS (HR = 2.07, P = 0.004) and EFS (HR = 1.909, P = 0.01) independent of other well-known prognostic factors. Meanwhile, allo-HSCT overcome deleterious outcomes related to low miR-340. Cases administered allo-HSCT showed markedly improved OS (HR = 0.316, P < 0.0001) and EFS (HR = 0.391, P = 0.002) in comparison with those receiving chemotherapy in the low miR-340 group. Gene expression assessment revealed that elevated miR-340 amounts were negatively correlated with HOXA/HOXB cluster levels, as well as the amounts of the HOX cofactor MEIS1. Strikingly, in silico analysis pointing to HOXA10, HOXB2, and MEIS1 as miR-340 targets. The miR-340 expression may help identify cases requiring strategies for selecting the optimal therapeutic option between chemotherapy and allo-HCST. AML cases showing low miR-340 levels should be strongly considered for early allo-HSCT treatment.
Introduction
Acute myeloid leukemia (AML), the commonest acute leukemia type affecting adult individuals, results from a fast clonal expansion of cancerous myeloblasts (1, 2). AML has a complex and heterogeneous nature (3). Thus, it shows variable outcomes in affected individuals upon administration of current treatments. Cytotoxic chemotherapeutics are considered the mainstay therapeutic option for AML (4, 5). Meanwhile, allogeneic hematopoietic stem cell transplantation (allo-HSCT) provides potent antileukemic effects and potential cure in high-risk patients (6, 7). In AML cases, prognosis is associated with multiple intrinsic parameters such as cytogenetic and genetic modifications. For example, NPM1, CEBPA, FLT3, IDH1, IDH2, and TET2 mutations show associations with patient outcome, and are used for AML prognosis (8–10). Although leukemogenesis is mechanistically well-understood, most AML cases remain uncured. Indeed, no currently available classification system is completely accurate. This suggests an urgent need for identifying new prognostic markers to further ameliorate AML stratification and select rational treatment options.
MicroRNAs (miRNAs) specifically bind mRNA targets and reduce the expression of encoded proteins by translation inhibition (11). Dysregulated miRNAs in AML affects cell growth and hematopoietic differentiation (12, 13). Furthermore, aberrant expression of miRNAs is related to patient outcome (14). For instance, elevated miR-181a content predicts favorable outcome in AML with normal cytogenetic features (15). Patients expressing elevated miR-212 amounts tend to show longer survival in all cytogenetic subtypes (16). Meanwhile, overexpression of miR-3151 is related to decreased overall and disease-free survival in cytogenetically normal AML (17). Moreover, combination of miR-3151 with the associated host gene BAALC is more efficient in predicting worse outcome. However, most published reports did not distinguish the effects of chemotherapeutics and allo-HSCT on treatment outcome. It is generally accepted that a marker's value in AML prognosis depends on the therapeutic method administered. Thus, miRNAs could show distinct prognostic values between chemotherapy and allo-HSCT treatment.
Here, we performed genome-wide screening to identify miRNAs with significant prognostic values in AML cases administered chemotherapeutics. We identified miR-340 as a prognostic factor independently of reported clinico-molecular predictive factors in AML. In addition, whether all-HCST overcomes the adverse prognostic impact of miR-340 expression was assessed. To assess molecular mechanisms, the expression levels of genes and miRNAs throughout the genome were determined.
Methods
Patients
All the 162 cases assessed were newly diagnosed AML (non-M3 FAB subtypes) based on the WHO classification. RNA-Seq data for the above AML cases were provided by The Cancer Genome Atlas (Project ID: TCGA-LAML) (18). These AML cases were included in a single-institution tissue banking protocol that had approval from the human studies committee at Washington University (WU HSC #01-1014). All samples were collected from patients with previously untreated de novo AML. MicroRNA and mRNA levels were determined before treatment. The risk classification of AML is based on the NCCN recommendation which includes cytogenetics as well as gene mutational status. Patient treatment followed the NCCN guidelines. Among the 162 cases, 148 (91.4%) received standard induction chemotherapy (3 and 7 days of anthracycline and cytarabine, respectively); the remaining 14 patients received decitabine or azacitidine. Patients with unfavorable risk underwent allo-HSCT if medically suitable for transplantation and there was a matched donor. Some cases with intermediate risk were also administered allo-HSCT during the disease course. A total of 90 cases were solely administered chemotherapy, while 72 also underwent allo-HSCT. Clinical data, such as treatment approach and outcomes, can be found on the TCGA website (Supplementary Table 1).
Gene-Expression Profiling
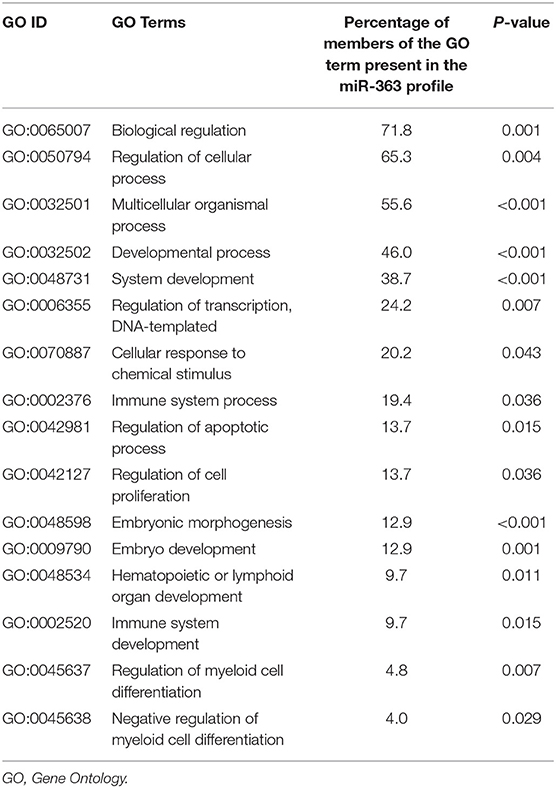
One hundred and fifty-five of the 162 cases were assessed for both miRNA and mRNA expression levels. Their specimens were employed to identify genes associated with miR-340 expression. In mRNA-seq, genes showing expression levels equal or inferior to a noise threshold of RPKM (Reads per kilobase per million mapped reads) in ≥75% of specimens were excluded. In miRNA-seq, RPM (Reads per million reads) was employed for normalization. Data underwent log2 transformation pre-analysis. The mRNA levels were compared between the high and low miRNA groups, with a univariate significance level at 0.005 to identify differentially expressed genes (DEGs). Then, hierarchical clustering using the multiple experiment viewer (MeV) 4.9.0 software was carried out to order gene rows; miRBase Targets V7 and Targetscan Release 7.1 were used for predicting microRNA targets. Gene Ontology enrichment was performed of genes associated with miR-340 using the Database for Annotation, Visualization, and Integrated Discovery (DAVID).
Statistical Analysis
Baseline features were compared between the high and low miRNA groups. Median levels of miR-340 were employed to identify patients with low and high miRNA levels. The miR-340 median values in the chemotherapy and allo-HSCT groups were 82.1 (13.2–227.5) and 75.0 (18.9–286.1), respectively. The Mann-Whitney U-test was employed to assess associations of two continuous variables. Fisher's exact test or the chi-square test was employed for categorical variables. Overall survival (OS) was defined as time between case enrolment and death, censoring live cases at final follow-up. Event-free survival (EFS) was defined as time between case enrolment and induction failure, recurrence, or death. Estimated OS and EFS distributions were determined by Kaplan-Meier curves, with the log-rank test carried out for comparing survival data.
The univariate Cox proportional hazards model was employed for evaluating associations of 340 expression with OS and EFS. The multivariate Cox proportional hazards model was used for identifying factors that affect survival. The factors assessed for model inclusion comprised miR-340 expression amounts; FLT3-ITD, NPM1, DNMT3A, RUNX1, TP53, TET2, MLL-PTD, IDH1/IDH2, and NRAS/KRAS mutation statuses; age; karyotype; and WBC involvement. Parameters with p = 0.10 in univariate analysis were further assessed by multivariate analysis (backward selection), in which p < 0.05 indicated significance. R v3.1.5, GraphPad Prism and SPSS were employed for data analysis, and P < 0.05 indicated statistically significant differences.
Results
Associations of miR-340 Levels With Clinico-Molecular Features
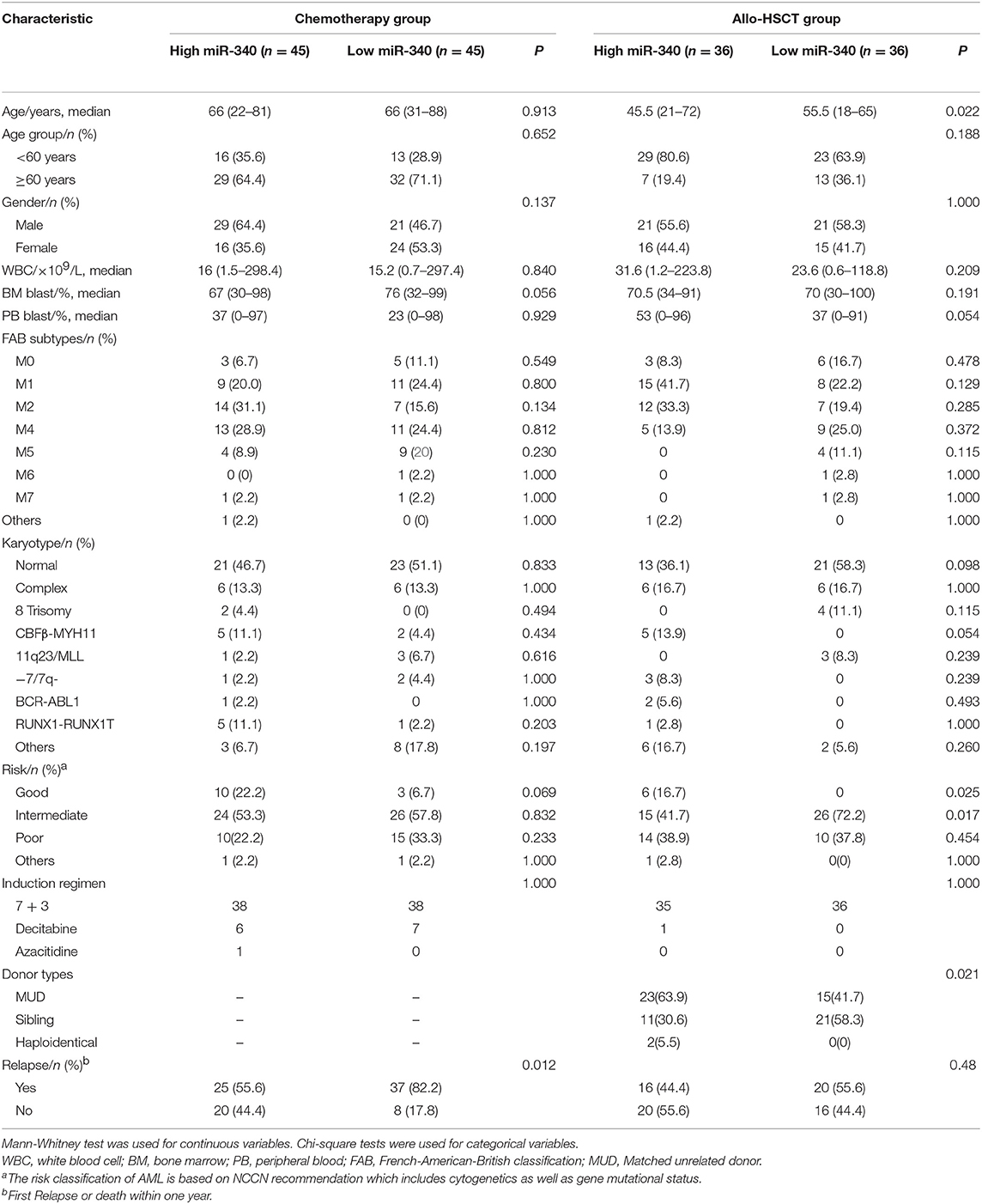
The chemotherapy and allo-HSCT groups were each subdivided into two according to median amounts of miRNAs, respectively. Tables 1, 2 summarize the associations of clinico-genetic features with miR-340 levels. In the chemotherapy group, cases with low miR-340 levels showed a higher rate of first relapse or death within 1 year after enrolment in comparison with those expressing high miR-340 amounts (82.2 vs. 55.6%, P = 0.012). There were no significant associations of other miR-340 expression distributions with clinical features. The above findings suggested that miR-340 expression may predict prognosis independently from known molecular features.
Prognostic Value of miR-340 in Cases Administered Chemotherapy or Allo-HSCT
To obtain prognostic markers for refining AML stratification, a genome-wide screen of miRNAs in AML patients was carried out; miR-340 was detected as a novel molecule predicting prognosis in AML cases treated by chemotherapy. In the chemotherapy group, low miR-340 expressers showed starkly reduced OS (P = 0.0005) and EFS (P = 0.0005) in comparison with high expressers (Figure 1A). Notably, 5 year OS rates were 35.6 and 5.4% in the high and low miR-340 expression groups, respectively. However, miR-340 levels were not associated with patient outcome in AML after allo-HCST (Figure 1B). The above findings indicated that low miR-340 was an unfavorable prognostic factor in AML cases administered chemotherapy.
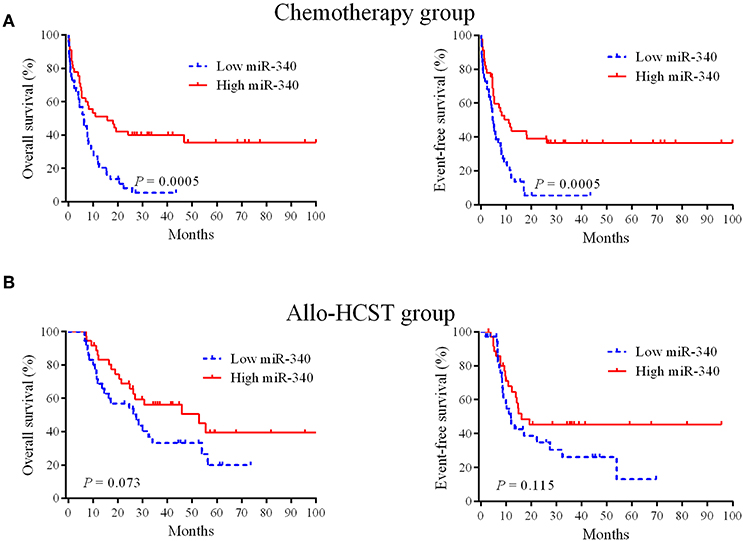
Figure 1. Kaplan-Meier survival curves based on miR-340 expression. (A) Cases highly expressing miR-340 showed markedly prolonged OS and EFS in the chemotherapy group (n = 90). (B) Effects of miR-340 levels on OS and EFS in cases administered allo-HSCT (n = 72).
MiR-340 Is Independently Associated With Clinical Outcome in Patients With AML
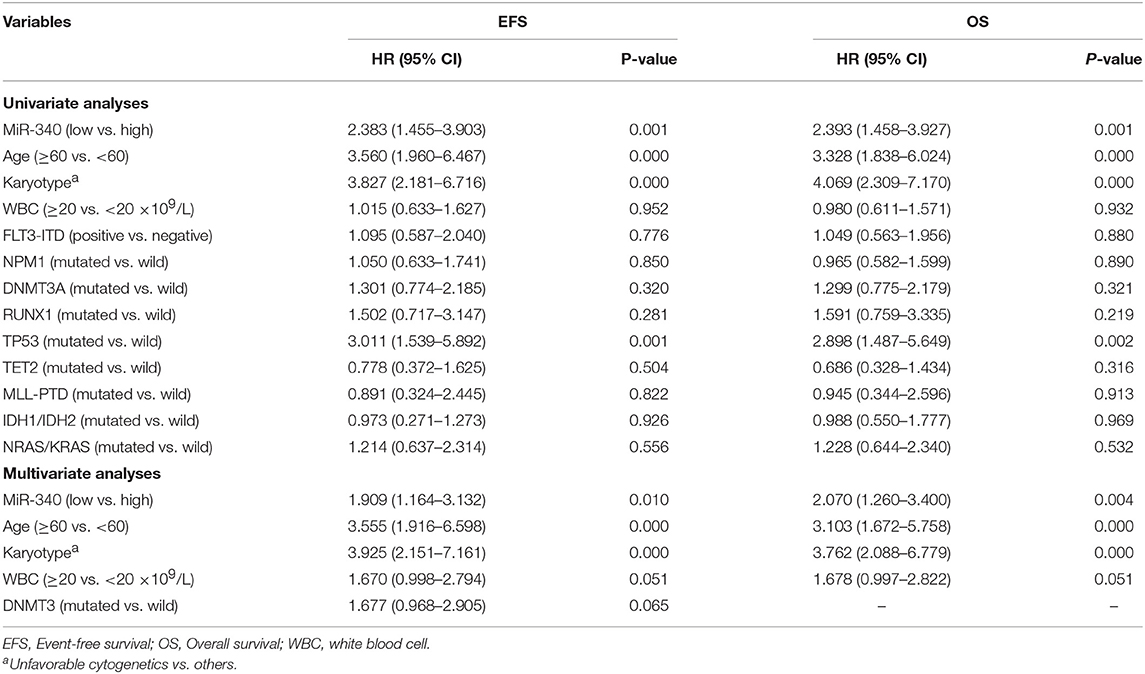
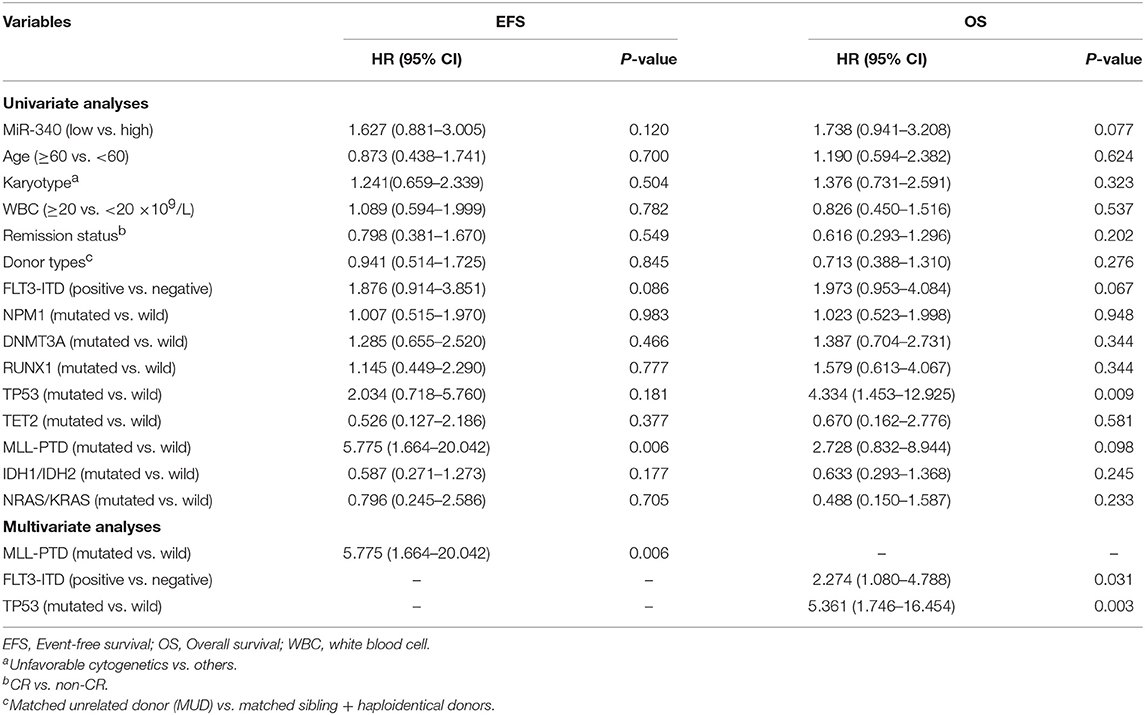
To assess whether miR-340 levels independently predict survival in AML, univariable and multivariate cox analyses were carried out. Univariate analysis (Table 3) revealed low miR-340 had a significant association with reduced OS (HR = 2.393, P = 0.001) and EFS (HR = 2.383, P = 0.001) in patients treated by chemotherapy. In multivariate analysis, miR-340 and many known prognostic factors were evaluated (Table 3). Low miR-340 expression was independently predictive of reduced OS (HR = 2.07, P = 0.004) and EFS (HR = 1.909, P = 0.01) upon adjustment for age (P < 0.0001) and karyotype risk (P < 0.0001). In patients administered allo-HSCT, univariate analysis revealed TP53-mutant cases displayed adverse OS (P = 0.009); however, the miR-340 expression status was not associated with OS and EFS (Table 4). Multivariable analysis showed TP53 and FLT3-ITD mutations were independently predictive of reduced OS (P = 0.003 and P = 0.031, respectively). Therefore, the miR-340 expression status did not remain a survival predictor after allo-HSCT.
Allo-HSCT Overcomes the Adverse Prognostic Effects of Low miR-340 Expression in AML
Whether allo-HSCT overcomes the unfavorable effects of low miR-340 expression was assessed. The 162 cases were assigned to two groups based on miR-340 levels. Then, each group was further divided into the chemotherapy and allo-HSCT groups. In cases with low miR-340 expression, significantly longer OS (HR = 0.316, 95% CI 0.167–0.459, P < 0.0001) and EFS (HR = 0.391, 95% CI 0.231–0.622, P = 0.002) were observed after allo-HSCT (Figure 2A). However, there were no differences in OS (P = 0.060) and EFS (P = 0.162) between the allo-HSCT and chemotherapy groups in high miR-340 expressers (Figure 2B). The above findings indicated miR-340 might constitute a novel molecule for detecting cases requiring strategies to select a rational therapeutic management.
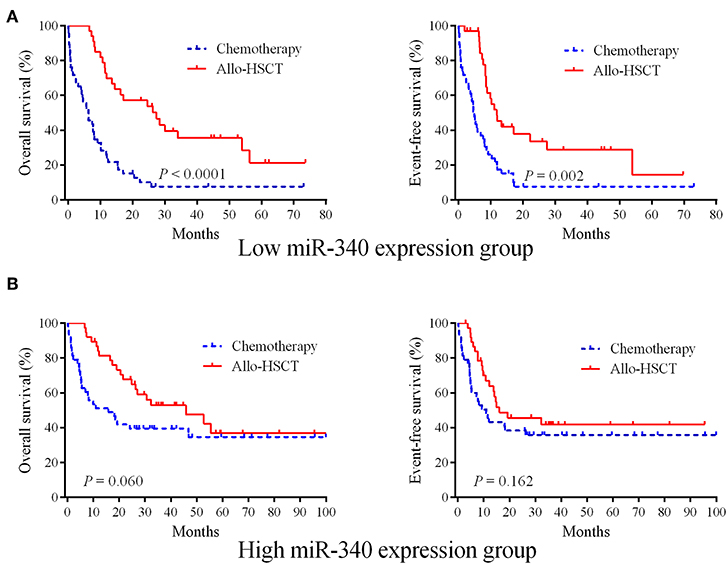
Figure 2. Allo-HSCT overcomes the adverse prognostic influence of low miR-340 expression in AML. (A) The 162 cases were divided into two groups according to median miR-340 levels. Kaplan-Meier survival curves for cases administered chemotherapy (n = 47) and allo-HSCT (n = 34), respectively, in the low miR-340 group. (B) Kaplan-Meier survival curves for cases administered chemotherapy (n = 43) and allo-HSCT (n = 8), respectively, in the high miR-340 group.
Biological Insights Into miR-340 Function in AML
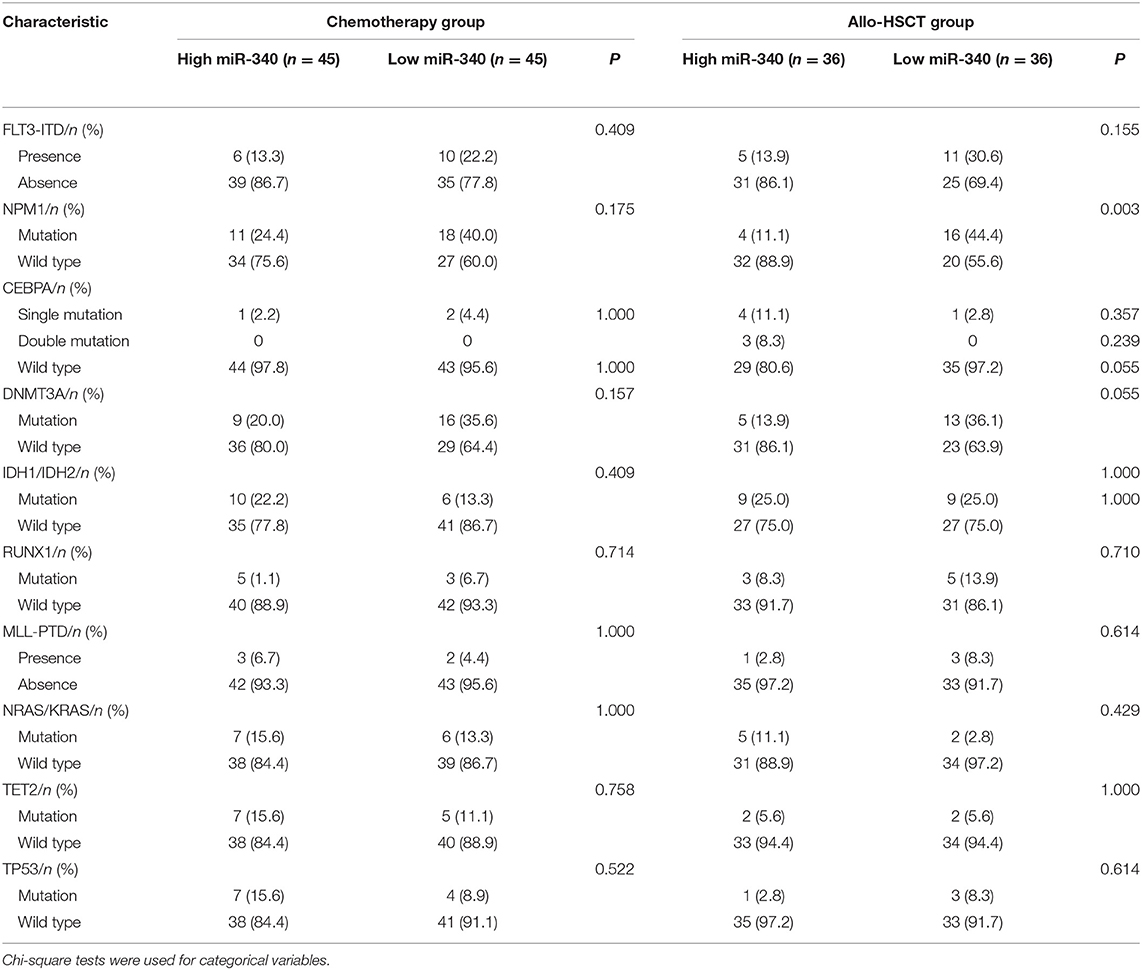
To gain insights into the biological functions of miR-340, gene expression signatures related to the miRNA expression were assessed in AML patients. As shown in Figure 3, the expression levels of 135 genes showed significant associations with miR-340 expression, with 61 and 74 positively and negatively correlated, respectively. In this study, miR-340 expression was negatively associated with HOXA/HOXB cluster and MEIS1 amounts. The latter genes are important in AML leukemogenesis and self-renewal features in leukemic stem cells (19). We further analyzed the biological significance of these genes by separating AML cases into two groups according to median miR-340 expression. In the low miR-340 expression group, the HOXA1 (P = 0.0112), HOXA2 (P = 0.019), HOXA3 (P < 0.0001), HOXA4 (P = 0.0005), HOXA5 (P < 0.0001), HOXA6 (P < 0.0001), HOXA7 (P < 0.0001), HOXA9 (P = 0.0004), HOXA10 (P < 0.0001), HOXB2 (P = 0.0026), HOXB3 (P < 0.0001), HOXB4 (P < 0.0001), HOXB6 (P < 0.0001), MEIS1 (P = 0.0001), and PRDM16 (P = 0.0002) genes were all expressed at higher levels compared with high miR-340 expression group. Notably, the histone H3K4 methyltransferase PRDM16, a gene that predicts adverse outcome in AML (20), was downregulated in the high miR-340 group. Strikingly, HOXA10, HOXB2, MEIS1 and PRDM16 were identified as miR-340 targets by in silico prediction. These findings corroborated clinical data for AML generated by miRNA analysis.
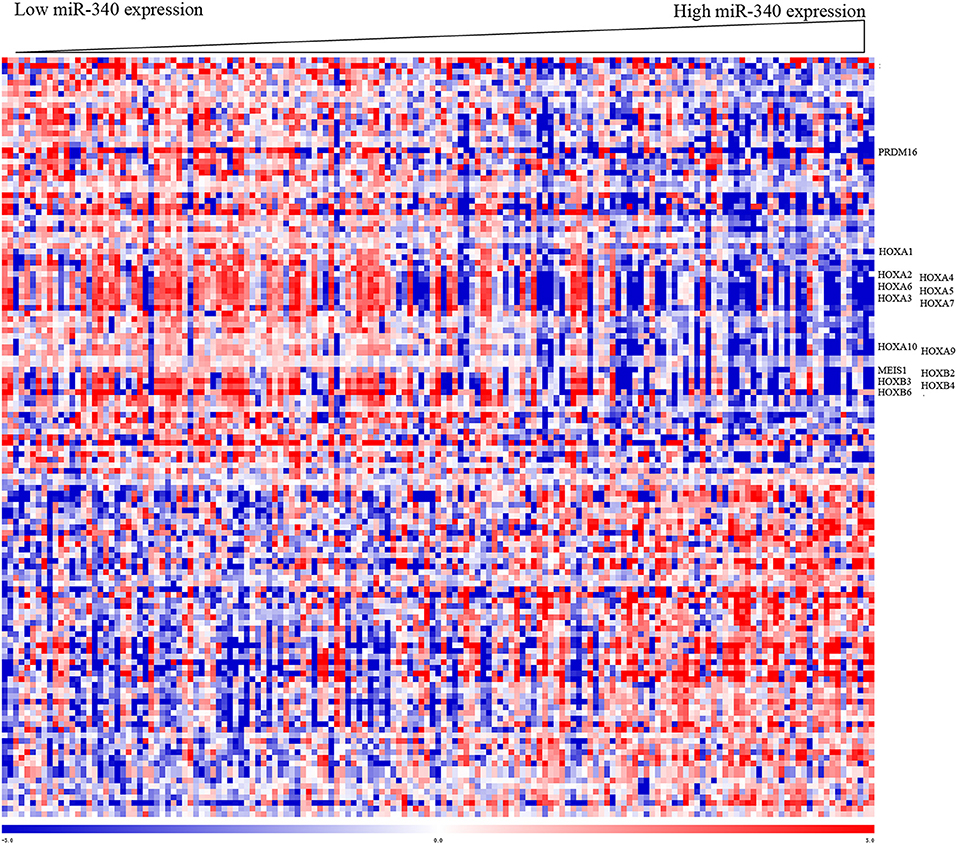
Figure 3. Heat map of the gene expression signature related to miR-340 expression in AML. Cases (columns) were ordered from left to right by increasing miR-340 levels. Genes (rows) were ordered by hierarchical cluster analysis. Blue and red reflect expression levels lower and above median values for the indicated genes, respectively; miR-340 associated genes are indicated.
Gene Ontology revealed that genes contributing to biological regulation, cellular and developmental processes, transcription regulation, immune system process, cell apoptosis and proliferation, myeloid cell differentiation, hematopoietic organ development, cellular response to chemical stimulus and embryonic morphogenesis were remarkably overrepresented among DEGs related to miR-340 expression (Table 5).
Discussion
Identifying new prognostic markers is of great importance in adult AML, because currently available molecular stratification approaches do not completely capture the heterogeneity of such cases (21–23). The significance of miRNAs as predictive biomarkers in AML patients remains mostly unclear. The present work demonstrated that miR-340 independently predicted prognosis of AML patients administered chemotherapy, providing an important tool for risk-stratifying AML cases. Of greater importance, allo-HSCT overcame miR-340-associated unfavorable outcomes.
Aberrant miRNA expression has a potential prognostic value in AML (24). High amounts of miR-155 (25), miR-196b (26), miR-362 (27), miR-363 (22), and miR-551b (28) show associations with adverse clinical outcomes. Conversely, elevated amounts of miR-9* (29), miR-34a (30), miR-181a (15), miR-193b (31), miR-212 (16) and miR-425 (14) predict favorable prognosis. As shown above, miR-340 expression independently predicted prognosis in a heterogeneous population of AML cases treated by chemotherapy. Low miR-340 amounts showed associations with reduced survival and risk of disease relapse or death. These findings suggest that miR-340 could enhance the prognostic value of currently known biomarkers in AML. Meanwhile, identifying low miR-340 expression as an adverse prognostic factor could guide intervention with synthetic miR-340 molecules. These findings indicate miR-340 functions independently to impact treatment outcome, and may drive leukemogenesis.
Routine therapeutic approaches after AML remission include conventional chemotherapy and allo-HCST (32). However, prognostic biomarkers efficiently guiding treatment selection are lacking. As demonstrated above, low miR-340 expressers treated by allo-HSCT had remarkably prolonged OS and EFS compared with the chemotherapy group. Notably, no benefit of allo-HSCT was found in cases highly expressing miR-340. The above results suggest that allo-HSCT as first line treatment might be of limited value in cases highly expressing miR-340. Thus, miR-340 levels might help identify individuals requiring strategies for selecting the most suitable therapeutic option between chemotherapy and allo-HCST. Low miR-340 expressing cases may be strongly considered for early allo-HSCT.
The tumor suppressor activity of miR-340 has been described in solid malignancies such as breast, colorectal and non-small cell lung cancers, as well as osteosarcoma and glioblastoma (33–35). However, miR-340's biological role in AML cells is largely undefined. As shown above, miR-340 levels were negatively correlated with HOXA/HOXB cluster and MEIS1 amounts. HOX genes are essential regulators of hematopoietic development and stem cell self-renewal (36). Abnormal overexpression of HOXA and MEIS1 represents a known hallmark of AML (19). Furthermore, artificial overexpression of HOXA7, HOXA9, or HOXA10 in combination with MEIS1 leads to rapid onset AML in animal models (37). High expression of HOXA9 in leukemia blasts predicts adverse outcome in AML. High expression levels of HOXA10 (38, 39) and MEIS1 (40) genes also predict poor outcome in AML. Notably, HOXA10, HOXB2, and MEIS1 were predicted in silico to be miR-340 targets in this study. Although they are potential therapeutic targets, developing chemical inhibitors of transcription factor remains challenging. Targeting these genes with synthetic miR-340 compounds may be a potential therapeutic strategy. Taken together, the miR-340 associated gene signature may confirm the notion that AML features miRNA expression changes. However, the mechanisms involved in miR-340 modulation and associated effects on patient outcome in AML after treatment deserve further attention.
Conclusions
MiR-340 amounts are independently associated with treatment outcome in a highly heterogeneous population of AML patients. MiR-340 amount determination may provide a powerful tool for the identification of a patient subset with adverse outcome in AML, and might help ameliorate stratification risk and inform therapeutic decision making for AML patients. Interestingly, allo-HSCT may overcome miR-340-associated deleterious effects in AML. Therefore, miR-340 expression analysis could help identify patients requiring complex approaches in selecting the most suitable therapeutic option between chemotherapy and allo-HCST.
Data Availability Statement
All datasets supporting the conclusions contained in the present report are included in the manuscript. All clinical data, including treatment approaches and outcomes, are publicly accessible from the TCGA website (https://cancergenome.nih.gov).
Author Contributions
MN, NZ, and RW carried out study design and computational analyses. TS, YF, YS, XL, KZ, SZ, and LX participated in statistical analysis. YY and KX contributed to study design and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Nos. 81670142, 81772658, 81870163, 81700179, and 81700199); the Jiangsu Provincial Key Research and Development Program (Nos. BE2017638 and BE2017636); the Natural Science Foundation of Jiangsu Province (Nos. BK20180104, BK20151153, and BK20160226), The Foundation of Jiangsu Province Six Talents Peak (No. 2017-WSN-120), Xuzhou Key Research and Application of Basic Research Projects (No. KC17158), Postgraduate Research and Practice Innovation Program of Jiangsu Province (Nos. SJCX18_0704 and SJKY19_2110), and the Natural Science Foundation of Jiangsu higher education institutions (No. 16KJA320003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.01058/full#supplementary-material
Supplementary Table 1. Clinical data.
Abbreviations
AML, acute myeloid leukemia; Allo-HSCT, allogeneic hematopoietic stem cell transplantation; TCGA, The Cancer Genome Atlas; OS, overall survival; EFS, event-free survival.
References
1. Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood. (2017) 129:1627–35. doi: 10.1182/blood-2016-10-696039
2. Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. (2016) 127:29–41. doi: 10.1182/blood-2015-07-604496
3. Ball M, List AF, Padron E. When clinical heterogeneity exceeds genetic heterogeneity: thinking outside the genomic box in chronic myelomonocytic leukemia. Blood. (2016) 128:2381–7. doi: 10.1182/blood-2016-07-692988
4. Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. (2017) 129:424–47. doi: 10.1182/blood-2016-08-733196
5. Niu M, Shen Y, Qi J, Liu X, Sang W, Wu Q, et al. Effects of realgar (As4S4) on degradation of PML-RARA harboring acquired arsenic-resistance mutations. Ann. Hematol. (2017) 96:1945–8. doi: 10.1007/s00277-017-3093-8
6. Grimm J, Bill M, Jentzsch M, Beinicke S, Hantschel J, Goldmann K, et al. Clinical impact of clonal hematopoiesis in acute myeloid leukemia patients receiving allogeneic transplantation. Bone Marrow Transplant. (2018) 54:1189–97. doi: 10.1038/s41409-018-0413-0
7. Ladetto M, Bottcher S, Kroger N, Pulsipher MA, Bader P. Methods and role of minimal residual disease after stem cell transplantation. Bone Marrow Transplant. (2019) 54:681–90. doi: 10.1038/s41409-018-0307-1
8. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. (2016) 374:2209–21. doi: 10.1056/NEJMoa1516192
9. Tawana K, Wang J, Renneville A, Bodor C, Hills R, Loveday C, et al. Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood. (2015) 126:1214–23. doi: 10.1182/blood-2015-05-647172
10. Ostronoff F, Othus M, Lazenby M, Estey E, Appelbaum FR, Evans A, et al. Prognostic significance of NPM1 mutations in the absence of FLT3-internal tandem duplication in older patients with acute myeloid leukemia: a SWOG and UK National Cancer Research Institute/Medical Research Council report. J Clin Oncol. (2015) 33:1157–64. doi: 10.1200/JCO.2014.58.0571
11. Yeh CH, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. (2016) 15:37. doi: 10.1186/s12943-016-0518-2
12. Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. (2017) 130:1290–301. doi: 10.1182/blood-2016-10-697698
13. Trino S, Lamorte D, Caivano A, Laurenzana I, Tagliaferri D, Falco G, et al. MicroRNAs as new biomarkers for diagnosis and prognosis, and as potential therapeutic targets in acute myeloid leukemia. Int J Mol Sci. (2018) 19:E460. doi: 10.3390/ijms19020460
14. Yang C, Shao T, Zhang H, Zhang N, Shi X, Liu X, et al. MiR-425 expression profiling in acute myeloid leukemia might guide the treatment choice between allogeneic transplantation and chemotherapy. J Trans. Med. (2018) 16:267. doi: 10.1186/s12967-018-1647-8
15. Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. (2010) 28:5257–64. doi: 10.1200/JCO.2010.29.2953
16. Sun SM, Rockova V, Bullinger L, Dijkstra MK, Dohner H, Lowenberg B, et al. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia. (2013) 27:100–6. doi: 10.1038/leu.2012.158
17. Eisfeld AK, Marcucci G, Maharry K, Schwind S, Radmacher MD, Nicolet D, et al. miR-3151 interplays with its host gene BAALC and independently affects outcome of patients with cytogenetically normal acute myeloid leukemia. Blood. (2012) 120:249–58. doi: 10.1182/blood-2012-02-408492
18. Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. New Engl J Med. (2013) 368:2059–74. doi: 10.1056/NEJMoa1301689
19. Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O'Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. (2015) 29:1279–89. doi: 10.1038/leu.2015.6
20. Matsuo H, Goyama S, Kamikubo Y, Adachi S. The subtype-specific features of EVI1 and PRDM16 in acute myeloid leukemia. Haematologica. (2015) 100:e116–7. doi: 10.3324/haematol.2015.124396
21. Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. (2014) 25:379–92. doi: 10.1016/j.ccr.2014.01.031
22. Zhang H, Zhang N, Wang R, Shao T, Feng Y, Yao Y, et al. High expression of miR-363 predicts poor prognosis and guides treatment selection in acute myeloid leukemia. J Trans Med. (2019) 17:106. doi: 10.1186/s12967-019-1858-7
23. Niu M, Feng Y, Zhang N, Shao T, Zhang H, Wang R, et al. High expression of miR-25 predicts favorable chemotherapy outcome in patients with acute myeloid leukemia. Cancer Cell Int. (2019) 19:122. doi: 10.1186/s12935-019-0843-9
24. Szymczyk A, Macheta A, Podhorecka M. Abnormal microRNA expression in the course of hematological malignancies. Cancer Manag Res. (2018) 10:4267–77. doi: 10.2147/CMAR.S174476
25. Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. (2013) 31:2086–93. doi: 10.1200/JCO.2012.45.6228
26. Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nature Commun. (2012) 3:688. doi: 10.1038/ncomms1681
27. Ma QL, Wang JH, Yang M, Wang HP, Jin J. MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J Trans Med. (2018) 16:68. doi: 10.1186/s12967-018-1445-3
28. de Leeuw DC, Verhagen HJ, Denkers F, Kavelaars FG, Valk PJ, Schuurhuis GJ, et al. MicroRNA-551b is highly expressed in hematopoietic stem cells and a biomarker for relapse and poor prognosis in acute myeloid leukemia. Leukemia. (2016) 30:742–6. doi: 10.1038/leu.2015.160
29. Nowek K, Sun SM, Dijkstra MK, Bullinger L, Dohner H, Erkeland SJ, et al. Expression of a passenger miR-9* predicts favorable outcome in adults with acute myeloid leukemia less than 60 years of age. Leukemia. (2016) 30:303–9. doi: 10.1038/leu.2015.282
30. Huang Y, Zou Y, Lin L, Ma X, Chen H. Identification of serum miR-34a as a potential biomarker in acute myeloid leukemia. Cancer Biomarkers. (2018) 22:799–805. doi: 10.3233/CBM-181381
31. Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, et al. Endogenous tumor suppressor microRNA-193b: therapeutic and prognostic value in acute myeloid leukemia. J Clin Oncol. (2018) 36:1007–16. doi: 10.1200/JCO.2017.75.2204
32. Cornelissen JJ, Versluis J, Passweg JR, van Putten WL, Manz MG, Maertens J, et al. Comparative therapeutic value of post-remission approaches in patients with acute myeloid leukemia aged 40-60 years. Leukemia. (2015) 29:1041–50. doi: 10.1038/leu.2014.332
33. Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J, Zhao JJ, et al. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer. (2011) 117:2842–52. doi: 10.1002/cncr.25860
34. Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. (2015) 34:3240–50. doi: 10.1038/onc.2014.267
35. Fiore D, Donnarumma E, Roscigno G, Iaboni M, Russo V, Affinito A, et al. miR-340 predicts glioblastoma survival and modulates key cancer hallmarks through down-regulation of NRAS. Oncotarget. (2016) 7:19531–47. doi: 10.18632/oncotarget.6968
36. Collins EM, Thompson A. HOX genes in normal, engineered and malignant hematopoiesis. Int J Dev Biol. (2018) 62:847–56. doi: 10.1387/ijdb.180206at
37. Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. (2007) 26:6766–76. doi: 10.1038/sj.onc.1210760
38. Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. (2010) 115:2910–8. doi: 10.1182/blood-2009-04-216606
39. Yao J, Fang LC, Yang ZL, Huang H, Li Y, Deng J, et al. Mixed lineage leukaemia histone methylases 1 collaborate with ERalpha to regulate HOXA10 expression in AML. Biosci Rep. (2014) 34:e00156. doi: 10.1042/BSR20140116
Keywords: miR-340, acute myeloid leukemia, clinical outcome, chemotherapy, allo-HCST
Citation: Niu M, Zhang N, Wang R, Shao T, Feng Y, Shen Y, Liu X, Zhao K, Zhu S, Xu L, Yao Y and Xu K (2019) MiR-340 Is a Biomarker for Selecting Treatment Between Chemotherapy and Allogeneic Transplantation in Acute Myeloid Leukemia. Front. Oncol. 9:1058. doi: 10.3389/fonc.2019.01058
Received: 11 July 2019; Accepted: 27 September 2019;
Published: 11 October 2019.
Edited by:
Basem M. William, The Ohio State University, United StatesReviewed by:
Apostolos Zaravinos, European University Cyprus, CyprusMichael Diamantidis, University Hospital of Larissa, Greece
Copyright © 2019 Niu, Zhang, Wang, Shao, Feng, Shen, Liu, Zhao, Zhu, Xu, Yao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yao Yao, eWFveTQxMiYjeDAwMDQwOzEyNi5jb20=; Kailin Xu, bGlobWRsJiN4MDAwNDA7MTYzLmNvbQ==
†These authors have contributed equally to this work
 Mingshan Niu1,2†
Mingshan Niu1,2† Kailin Xu
Kailin Xu