
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 18 September 2019
Sec. Cancer Molecular Targets and Therapeutics
Volume 9 - 2019 | https://doi.org/10.3389/fonc.2019.00901
 Suraksha Tamang1†
Suraksha Tamang1† Varnali Acharya1†
Varnali Acharya1† Deepronil Roy1
Deepronil Roy1 Rinka Sharma1
Rinka Sharma1 Apeksha Aryaa1
Apeksha Aryaa1 Uttam Sharma1
Uttam Sharma1 Akanksha Khandelwal2
Akanksha Khandelwal2 Hridayesh Prakash3
Hridayesh Prakash3 Karen M. Vasquez4
Karen M. Vasquez4 Aklank Jain1*
Aklank Jain1*Limitations in current diagnostic procedures warrant identification of new methodologies to improve diagnoses of cancer patients. In this context, long non-coding RNAs (lncRNAs) have emerged as stable biomarkers which are expressed abundantly in tumors. Importantly, these can be detected at all stages of tumor development, and thus may provide potential biomarkers and/or therapeutic targets. Recently, we suggested that aberrant levels of lncRNAs can be used to determine the invasive and metastatic potential of tumor cells. Further, direct correlations of lncRNAs with cancer-derived inflammation, metastasis, epithelial-to-mesenchymal transition, and other hallmarks of cancer indicate their potential as biomarkers and targets for cancer. Thus, in this review we have discussed the importance of small nucleolar RNA host gene 12 (SNHG12), a lncRNA, as a potential biomarker for a variety of cancers. A meta-analysis of a large cohort of cancer patients revealed that SNHG12 may also serve as a potential target for cancer-directed interventions due to its involvement in unfolded protein responses, which many tumor cells exploit to both evade immune-mediated attack and enhance the polarization of effector immune cells (e.g., macrophages and T cells). Thus, we propose that SNHG12 may serve as both a biomarker and a druggable therapeutic target with promising clinical potential.
Although excellent progress has been made in the management of cancer over the past decade, unfortunately cancer-related deaths have not significantly decreased worldwide (1). Potential reasons for this may include the invasive nature of tumor cells, the heterogeneous steps involved in cancer initiation, progression, and metastasis, including the interlinking of various signaling molecules and pathways, resulting in poor detection and treatment of cancer. In this regard, regulatory molecules such as long non-coding RNAs (lncRNAs), micro RNAs (miRNAs), piwiRNAs (piRNAs), and other non-coding RNAs (ncRNAs) comprise ~90% of the human genome. Multiple studies have demonstrated that lncRNAs are indeed functional elements involved in a wide range of physiological and pathophysiological processes, and can act as oncogenes or tumor suppressor genes (2–6).
One such lncRNA, small nucleolar host gene 12 (SNHG12), also known as LNC04080, is an lncRNA located at the p35.3 region on chromosome 1. It is ~1,867 bases long (7) and encodes four small nucleolar RNAs (SNORA66, SNORA61, SNORA16A, and SNORD99) from its spliced introns (8). Studies have implicated SNHG12 in a number of cancers, such as breast (9), gastric (10), osteosarcoma (11), and glioma (12) and other cancer types detailed in this review. The altered expression of SNHG12 has been correlated with the viability, proliferation, metastasis, and invasion of tumor cells, impacting the prognosis and survival of cancer patients.
SNHG12 also acts as a competitive endogenous RNA (ceRNA) by harboring multiple miRNA binding sites, thereby regulating their downstream targets by “sponging” those miRNAs. Recent research describes the emerging role of ceRNAs in cancer etiology where various ncRNA molecules including lncRNAs, miRNAs, pseudogenes, and circular RNAs (circRNAs) share common miRNA response elements (MREs), thereby mutually regulating each other through cellular processes via complex RNA networks (13). MREs have been found in the 5'UTRs, coding sequences, and 3'UTRs of genes (14, 15). It has been reported that SNHG12 lncRNA has a higher density of MREs for target miRNAs, thereby increasing the likelihood of sharing and titrating miRNAs and preventing them from binding to other transcripts (16). According to various models, this miRNA-mediated lncRNA and mRNA crosstalk can be unidirectional, where changes in the levels of one transcript (ceRNA) can derepress the expression of the target transcript by miRNA sponging, as well as bidirectional, where both transcripts compete for the binding of miRNAs and regulate each other (17–19) as shown in Figure 1. Collectively, it has been suggested that optimal miRNA-mediated crosstalk depends on the relative abundance of participating molecules, i.e., lncRNAs and miRNAs, and maximum sponging activity is typically observed at equimolar concentrations of miRNAs and their target lncRNAs (Figure 1, middle panel). Examples of lncRNAs acting as ceRNAs for mRNA and miRNA have been reported in several publications (21, 22). For instance, highly up-regulated in liver cancer (HULC) and papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) transcripts contain miRNA-binding sites that inhibit the expression of the miRNAs and regulate downstream targets that aid in the progression of hepatocellular carcinoma (23) and thyroid cancer (24), respectively.
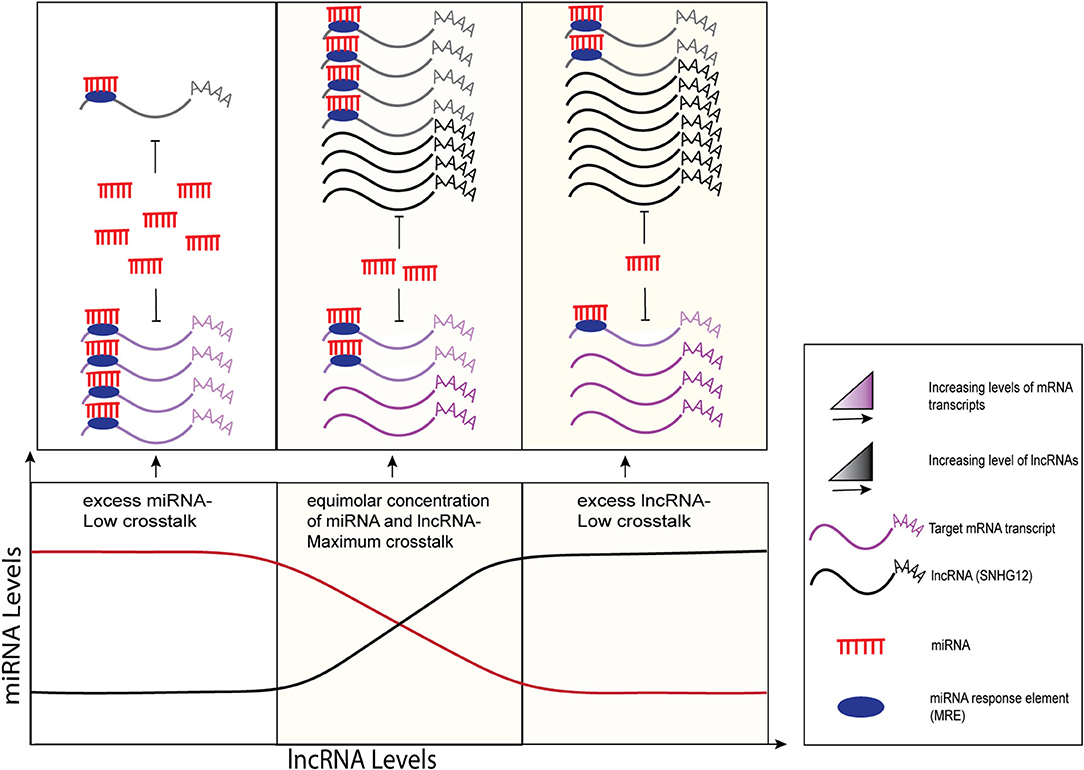
Figure 1. Graphical representation of sponging mechanisms of lncRNAs and miRNAs. MiRNA-mediated crosstalk depends on the relative abundance of its target transcripts. LncRNAs (black) regulate the expression levels of their target mRNA transcripts (purple) by competing for common miRNAs (red) via the MREs (filled dark blue ovals). Minimal crosstalk is observed when miRNAs are in excess (left side) or in limited numbers (right side) as compared to lncRNAs. At equimolar concentrations, maximum crosstalk is observed where lncRNAs sponge miRNAs, preventing them from binding to and repressing their target mRNAs. The figure modified from Ala et al. (17) and Tan et al. (20).
In this review, we discuss the proposed mechanistic roles of SNHG12 in the pathogenesis of several human cancers, including the regulatory molecules of SNHG12 in modulating several hallmarks of cancer. These include interactions that: (1) sustain proliferative signals via inhibition of miR-195 (12), miR-181a (25), miR-320 (10), miR-101-3p (26), and up-regulation of Gelsolin (12), Mitogen-Activated Protein Kinase 1/Mitogen-Activated Protein Kinase Kinase1 (MAPK1/MAP2K1) (25), phosphorylated Extracellular Signal-Related Kinase (pERK) (10), phosphorylated Protein Kinase B (pAkt) (10), Cyclin D1 (11, 27), and Forkhead Box P1 (FOXp1) (26); (2) enable replicative immortality via inhibition of Large Tumor Supressor Kinase 2 (LATS2) (28); (3) resist cell death via inhibition of miR-320a (29), Caspase 3 (30), miR-138 (31), miR-218 (32), miR-199a-5p (8), miR-424-5p (33), and up-regulation of MCL1 Apoptosis Regulator, BCL2 family member (Mcl-1) (29), Caspase 9 (32), Mixed Lineage Protein Kinase 3 (MLK3) (8), NF-KB inhibitor alpha (IKBα) (8), NFKB inhibitor beta (IKBβ) (8); (4) activate invasion and metastasis via inhibition of miR-101-3p (26), miR-424-5p (33), miR-125b (34), miR-195-5p (11), E-Cadherin (35), miR-218 (32), and upregulation of Forkhead Box P1 (FOXP1) (26), Matrix Metallopeptidase 3 (MMP13) (9), Matrix Metallopeptidase 2 (MMP2) (27), Signal Transducer and Activator of Transcription (STAT3) (34), Notch2 (11), Vimentin (35), N-Cadherin (35), Zinc Finger E-Box Binding Homeobox (ZEB2) (32), Matrix Metallopeptidase 9 (MMP9) (32), and Slug (32); (5) induce angiogenesis via targeting Histone Deacetylase 10 (HDAC10) (28); and (6) promote inflammation via targeting the Advanced Glycosylation End-Product Specific Receptor (AGER) (28). The diagrammatic representation of the impact of SNHG12 in these mechanisms and hallmarks of cancer are shown in Figure 2. We have highlighted the target molecules of SNHG12, which may serve as potential diagnostic and prognostic biomarkers, and/or as targets for novel strategies for cancer therapy.
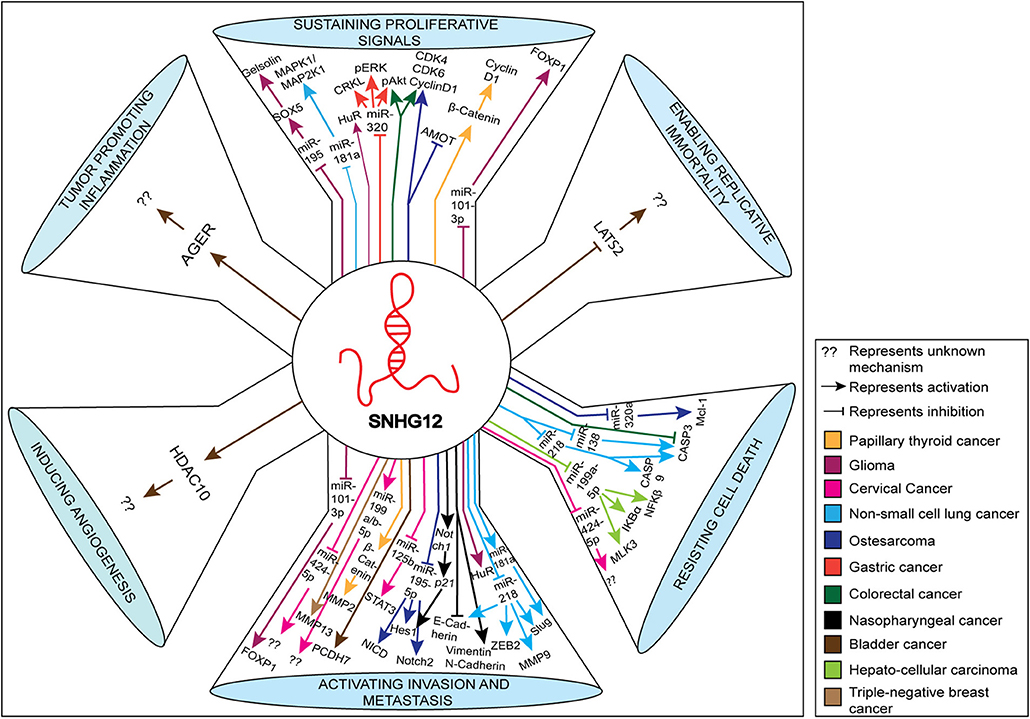
Figure 2. Schematic representation of various molecules and pathways targeted by SNHG12 in cancer. The various pathways shown represent hallmarks of cancer. The up-regulated and down-regulated molecules/pathways are depicted by up and down arrows, respectively.
SNHG12 expression was evaluated by two different research groups in over 100 GC patients combined, and they found an ~1.6-fold up-regulation of SNHG12 in GC tissues compared to adjacent normal tissues (10, 36). In both the studies, SNHG12 expression was positively correlated with tumor size, stage, lymph node metastasis, and distant metastasis; whereas, no correlation was found with other clinicopathological characteristics such as patient age and gender. Moreover, its high expression was negatively correlated with patient survival time and disease-free survival (10, 36). Similarly, the up-regulation of SNHG12 (up to ~15-fold) was observed in the GC cell lines SGC-7901, AGS, and BGC-823, compared to normal gastric epithelial (GES-1) cells, as outlined in Table 1. In addition, siRNA knockdown of SNHG12 in GC cells led to reduced proliferation and colony formation when assessed by CCK-8 staining and colony formation assays. Subsequently, both Zhang et al. (10) and Yang et al. (36) identified miR-199a/b-5p and miR-320 as potential target molecules of SNGH12 by using microRNAcode and LNCipedia database prediction analyses (Figure 2) and through dual-luciferase reporter assays they confirmed miR-199a/b-5p and miR-320 as its direct downstream targets (10, 36). Moreover, knockdown of SNHG12 in GC cell lines significantly up-regulated the levels of both the miRNAs showing a negative correlation between the two and reversing the effect of SNHG12 in cancer progression. Knockdown of SNHG12 also led to down-regulation of CRK Like Proto-Oncogene, Adaptor Protein (CRKL), p-AKT, and p-ERK proteins. Consistent with this, CRKL was previously reported to be a target of miR-320 in GC cells (52) (Figure 2). Importantly, miR-320-mediated CRKL inhibition was found to enhance cell proliferation, invasion, and metastasis of GC cells via up-regulation of the ERK and AKT pathways (52). It was suggested that SNHG12 up-regulated CRKL expression, thereby enhancing AKT and ERK signaling, thus promoting GC progression via sponging of miR-320 (10) (Figure 2). Thus, these studies implicated SNHG12 in GC by sponging miR-320 and miR-199a/b-5p, thereby regulating oncogenic pathways.
In another study, low plasma levels of SNHG12 were reported in 10 pre-operative GC patients compared to their post-operative samples and 10 healthy controls (53), which is in contrast to its expression in cancer tissue samples. Thus, additional studies are needed to understand the mechanistic roles of SNHG12 in GC to better define its potential as a candidate biomarker to facilitate early diagnosis of GC, and/or as a potential novel therapeutic target for the treatment of GC to overcome its high mortality rates (1).
Several studies have documented significantly up-regulated expression of SNHG12 in NSCLC tissue samples as well as NSCLC cell lines compared to normal control samples. Zhu et al. (54) observed an ~27-fold higher expression of SNHG12 in 10 lung adenocarcinoma (LAD) tissues compared to adjacent non-cancerous tissues through qRT-PCR experiments. Similarly, two additional studies observed an ~2.3-fold and ~1.3-fold up-regulation of SNHG12 in 22 and 40 NSCLC tumor samples compared to adjacent normal tissues, respectively (25, 32). Importantly, higher expression of SNHG12 was found to be negatively associated with the overall survival rate of NSCLC patients (32). Additionally, NSCLC cell lines (PC9, H1299, and A549) were examined for SNHG12 expression, where an ~2.5-fold up-regulation in PC9 cells, up to ~2.7-fold up-regulation in H1299 cells (25, 31) and as much as an ~3-fold up-regulation in A549 cells (25, 31) was observed compared to normal human bronchial epithelial (16-HBE) cells (Table 1).
Studies have shown that SNHG12, like other lncRNAs, acts as an endogenous sponge that competes for several miRNAs including hsa-miR-181b-5p, hsa-miR-181a-5p, hsa-miR-16-5p, hsa-miR-15a-5p, hsa-miR-195-5p, hsa-miR-497-5p, hsa-miR-181c-5p, hsa-miR-181d-5p, hsa-miR-15b-5p, hsa-miR-138, and hsa-miR-218 in NSCLC (31). It was found that alterations in miR-181a, a tumor suppressor miRNA (32, 37, 55), expression levels affected its downstream targets, such as MAPK and Slug (Figure 2), facilitating the development of NSCLC (38, 39, 56, 57). Likewise, over-expression of miR-138 suppressed cell proliferation and colony formation by increasing the apoptotic rate of NSCLC cells via up-regulated Caspase-3 activity, which suppressed NSCLC progression (37). Furthermore, altered expression of miR-218 was found to be associated with increased tumorigenesis by increasing the epithelial-to-mesenchymal transition in NSCLC by targeting Matrix Metallopeptidase 9 (MMP-9), Vimentin, Slug, Zinc Finger E-Box Binding Homeobox 2 (ZEB2), and E-cadherin proteins (Figure 2) (32). In addition, miR-218 up-regulation positively correlated with the expression levels of cleaved Caspase-3 and Caspase-9, thereby suppressing cell viability and cancer progression by triggering apoptosis. Indeed, dual-luciferase reporter assays confirmed the transcriptional regulation of SNHG12 with hsa-miR-181a, hsa-miR-138, and hsa-miR-218 (25, 31, 32). At the cellular level, knockdown of SNHG12 resulted in a significant decrease in cell proliferation, colony formation, and invasive potential of cells, but increased apoptosis by up-regulating the expression of the target miRNAs. Overall, these data revealed that SNHG12 negatively regulated the expression of its target miRNAs in NSCLC (32).
These composite results support the conclusion that SNHG12 is up-regulated in NSCLC where it plays a direct role in the modulation of cancer progression through down-regulating the expression of tumor-suppressor miRNAs, such as miR-138 and miR-181a, and it is also involved in NSCLC drug resistance. Thus, SNHG12 may serve as a potential diagnostic biomarker, and therapeutic target in the treatment of NSCLC, including patients that have developed resistance to therapy.
Wang et al. (9) have identified numerous differentially expressed lncRNAs through RNA sequencing in TNBC compared to non-tumorous breast tissues, and found an increased expression (~2.2-fold) of SNHG12 in TNBC (Table 1). Further, SNHG12 up-regulation was positively correlated with advanced tumor stage and size, while it negatively correlated with patient survival, thus demonstrating its clinical significance. Mechanistic investigation revealed that SNHG12 was a direct transcriptional target of c-MYC. Using the Multiple EM for Motif Elicitation tool for discovering motifs (MEME), the authors identified two putative c-MYC-binding motifs at 1,564 bp and 780 bp upstream of the SNHG12 transcription start site. This was confirmed when transfection of a c-MYC-containing plasmid increased SNHG12 levels in MDA-MB-231 and BT-549 TNBC cell lines, enhancing their viability and colony-forming capacity, increasing cell proliferation, and inhibiting apoptosis (9). Over-expression of SNHG12 also significantly increased the expression of MMP13 (Table 1; Figure 2), while SNHG12 knockdown constrained its expression.
In conclusion, SNHG12 was up-regulated in TNBC and promoted proliferation, while inhibiting apoptosis. These results implicate SNHG12 in TNBC, suggesting that it may provide a target for the treatment and diagnosis of TNBC.
Lan et al. (8) measured SNHG12 levels in 48 human HCCs and their adjacent normal tissues, where 24 HCCs showed significant up-regulation (by ~3.7-fold) in cancerous tissues (Table 1). SNHG12 up-regulation was positively correlated with tumor size, vascular invasion, and tumor node metastasis (TNM) stage, while it was negatively correlated with patient survival, demonstrating its clinical significance. However, while SNHG12 knockdown by siRNA reduced cell viability, colony formation, and inhibited cell cycle progression, its depletion increased apoptosis of SK-Hep1 and HCCLM9 cells lines in vitro (8).
The suspected crosstalk between SNHG12 and miR-199a/b-5p was analyzed by miRcode software (http://www.mircode.org/), and was confirmed by a decreased luciferase activity of a pMir-reporter-SNHG12-WT when transfected with a miR-199a/b-5p mimic in HCC cell lines. Furthermore, loss-of-function assays revealed that there was an inverse correlation between the expression of miR-199a-5p and SNHG12 (Figure 2) (40, 58). When miRNAs exert silencing functions on target protein-coding genes, RISC formation can occur, allowing binding to the target via Ago2. To confirm the speculation that Ago2 could directly bind to both SNHG12 and miR-199a/b-5p, RIP and qRT-PCR analyses were conducted. The results demonstrated that SNHG12 and miR-199a/b-5p were enriched in the Ago2-containing pellet (8). Results of chromatin isolation by RNA purification assays supported these findings by demonstrating that SNHG12 co-immunoprecipitated with miR-199a/b-5p, confirming their interaction (8). The decrease in miR-199a/b-5p expression was found to be associated with increased tumorigenesis, at least in part by regulation of the mitogen-activated protein kinase 3 (MLK3)/IkB-α/NF-kB pathway (Figure 2) (59). Indeed, immunoblot analysis indicate that elevated expression of miR-199a-5p and knockdown of SNHG12 suppressed the expression of MLK3 and phosphorylation of IkB-α and NF-kB in SK-Hep1 cells (Figure 2). In contrast, the phosphorylation of ERK1/2, a key regulator of MAPK signaling pathways, was not affected (8). These results suggested that SNGH12 induced tumorigenesis in HCC by regulating the MLK3/IkB-α/NF-kB pathway (Figure 2).
Wang et al. (30) measured SNHG12 levels in 60 human CRCs and their adjacent normal tissues, which showed significant up-regulation (by ~2.5-fold) in the cancerous tissues over the normal tissues (Table 1) (60). SNHG12 up-regulation was positively correlated with advanced tumor stage and size, while it negatively correlated with patient survival (60). Similarly, levels of SNHG12 were significantly up-regulated (~4.7-fold) in the HT29 CRC cell line compared to the normal human colonic epithelial cell line (HcoEpiC) (60). Consistent with these observations, over-expression of SNHG12 in SW480 and HT29 cell lines increased cell viability, colony-forming capacity, cell proliferation, and inhibited apoptosis (60). In addition, immunoblot analysis confirmed that SNHG12 increased protein levels of CDK4, CDK6, Cyclin D1 (CCND1), and decreased Caspase-3 levels (Figure 2). The authors also reported that the over-expression of SNHG12 significantly up-regulated (~3-fold) the expression of pAKT (Figure 2), while SNHG12 depletion constrained its expression (60).
In conclusion, these results suggested that SNHG12 promoted cell growth and inhibited cell apoptosis in CRC cells. Thus, it may serve as a potential diagnostic biomarker and therapeutic target and for the detection and treatment of CRC.
Jiang et al. (28) evaluated data on expression of lncRNAs in bladder cancer specimens from The Cancer Genome Atlas (TCGA) and GSE89006 datasets and found the expression of SNHG12 to be up-regulated as compared to the normal adjacent tissues by ~1.1 and ~1.2-fold, respectively (Table 1). The higher expression of SNHG12 was found to be associated with relatively shorter recurrence-free survival time (RFS). Moreover, RNA sequencing data from the Cancer Cell Line Encyclopedia (CCLE) revealed higher expression of SNHG12 in the bladder cancer cell lines SW780 and UMUC3, compared to normal control cell lines. Depletion of SNHG12 in SW780 and UMUC3 cell lines by siRNA resulted in impaired cellular proliferation, colony formation, and invasion potential of bladder cancer cell lines (28). Furthermore, it was observed that siRNA-mediated knockdown of SNGH12 in SW780 bladder cancer cells led to a ~9% increase in the apoptotic rate of the SW780 cells compared to control cells. SNGH12 knockdown also impaired the invasive capacity of SW780 cells compared to control cells (28). These data suggested a role for SNGH12 in bladder cancer progression and invasion. The TCGA and Gene Ontology (GO) analyses revealed that SNHG12 expression was positively associated with protein-coding genes such as Histone Deacetylase 10 (HDAC10) and Advanced Glycosylation End-product Specific Receptor (AGER), and negatively associated with other protein-coding genes such as Protocadherin 7 (PCDH7) and Large Tumor-Suppressor Kinase 2 (LATS2) (Figure 2), which are involved in biological pathways that assist in regulating cancer cell growth and proliferation.
These results provided evidence that SNHG12 up-regulation, which associated positively with the expression of certain oncogenes and inversely with tumor-suppressor genes, plays an essential role in the growth and proliferation of bladder cancer. The increased expression of SNHG12 also correlated with shorter survival time of patients, suggesting that it may be utilized as a potential biomarker for the improved prognosis, and as a target for the treatment of bladder cancer.
Ding et al. (27) examined 42 PTC tissues and found SNHG12 to be up-regulated by ~3.8-fold in the PTC tissues compared to normal adjacent tissue samples. Similarly, in the PTC cell lines K1, TPC-1, and BCPAP, the expression of SNHG12 was found to be up-regulated by ~3, ~5, and ~6-fold over controls, respectively (Table 1) (27). Decreased SNHG12 expression by SNHG12 interference sequences in the cancer cell lines, TPC-1, and BCPAP, resulted in decreased proliferative capacity, an increased apoptotic rate (~28% increase in K1 and >28% in TPC-1), and impaired metastatic and migration capacities, implicating SNHG12 in tumor cell proliferation. Further, the results of immunoblotting revealed that SNHG12 promoted PTC tissue proliferation and metastasis, in part, through Wnt/β-catenin signaling by up-regulating β-catenin and the downstream genes, Matrix Metalloproteinase 2 (MMP-2) and CCND1 (Figure 2). Similar results were found in in vivo experiments performed in BALB/c male nude mice, which demonstrated that knockdown of SNHG12 in the experimental group led to decreased numbers of metastatic tumors in the lungs of the mice carrying BCPAP cell xenografts compared with the control group (27).
Based on the findings mentioned above, it can be inferred that SNHG12 plays an important role in the proliferation and metastasis of papillary thyroid cancer tissues and cells by regulating the Wnt/β-catenin signaling pathway. Thus, it adds to the list of tumor types in which SNHG12 may serve as an effective tool in the diagnosis and treatment of cancer.
The expression of SNHG12 was examined by three different research groups in 33, 39, and 79 glioma tissue samples compared to adjacent normal tissues and were found to be up-regulated by ~3.3, ~5.5, and ~2.2-fold, respectively (Table 1) (12, 26, 41). The over-expression of SNHG12 was found to be associated with low overall survival and clinicopathological characteristics of patients such as age, WHO grade, and Karnofsky Performance Score (KPS) (26). Moreover, over-expression of SNHG12 was found to be associated with genotypes having mutations in the telomerase reverse transcriptase (TERT) gene and the isocitrate dehydrogenase ½ (IDH1/2) genes, along with 1p/19q co-deletion, and methylation of O-6-Methylguanine-DNA Methyltransferase (MGMT) (42). Additionally, RNA-binding proteins Tar DNA Binding Protein (TDP-43) and Hu-Antigen R (HuR) were reported to play oncogenic roles in the progression of glioma by binding to and stabilizing SNHG12 (12, 41). In three independent studies SNHG12 was found to be over-expressed by ~5.2, ~3.8, and ~11-fold in the U87 glioma cell line, and ~5.3, ~4.2, and ~13-fold in the U251 glioma cell line (Table 1) (12, 26, 41). Knockdown of SNHG12 in the U87 and U251 glioma cells resulted in decreased cell numbers, impaired proliferation, migration, and invasion, but increased the apoptotic rate in these cells compared to control human astrocytes (12, 26, 41). Similar results were obtained from in vivo experiments conducted in athymic BALB/c nude mice, where in one study, lentivirus-mediated transfection with sh-RNA against SNHG12 decreased tumor weight and volume of human U87 glioma cell xenografts in mice (26).
As previously mentioned, SNHG12 was found to act as ceRNA capable of sponging several miRNAs that serve as tumor suppressors in various cancers (11, 33, 61). MiR-101-3p and miR-195 were found to be putative targets of SNHG12, which inhibited the progression of glioma by down-regulating their target genes, Forkhead Box Protein P1 (FOXP1), and Sry-Box 5 (SOX5) (12, 26). A negative correlation was found between SNHG12 and its target miRNAs (miR-101-3p and miR-195) by qRT-PCR and immunoblot analyses, which demonstrated upregulation of the downstream targets (12, 26). Further, luciferase-reporter and RIP assays confirmed the direct interaction of SNHG12 with the above-mentioned miRNA targets, as well as their involvement in the formation of RISC complexes. FOXP1, a transcriptional factor in the FOXP subfamily of the FOX family, impacts the cell cycle, cell proliferation, differentiation, apoptosis, and has played an oncogenic role in various B-cell lymphomas (26). Similarly, SOX5, a transcription factor of the group D of sex-determining region (Sry)-related transcription factors (SOXD) protein family, acts as a transcriptional regulator and has been implicated in tumor progression. In addition, SOX5 was found to regulate the malignant progression of glioma cells via direct binding to the promoter of its downstream target gene Gelsolin, and the promoter of SNHG12. Consistent with these reports, knockdown of FOXP and SOX5 hindered cell proliferation, invasion, and migration potential in U87 and U251 cell lines, suggesting their oncogenic function in glioma cells.
Taken together, these findings suggest that SNHG12 acts as a key player in the progression of glioma by its involvement in the miR-101-3p/FOXP1 axis, and the miR-195/SOX5 axis, which are stabilized by the binding of the RNA-binding proteins, HuR and TDP43, respectively. Thus, SNHG12 may serve as a potential biomarker/target for glioma.
Dong et al. (34) measured the expression of SNHG12 in 76 paired cervical cancer tissues and found that it was up-regulated (by ~1.8-fold) compared to adjacent normal tissues. Increased levels of SNHG12 were found to be associated with clinicopathological manifestations such as lymph node metastasis, and International Federation of Gynecology and Obstetrics (FIGO) stage, where high-grade tumors (stage II) showed increased expression compared to low-grade tumors (stage I). High SNHG12 levels were also found to be negatively correlated with the overall survival of patients (34). Similarly, five different cervical cell lines showed increased levels of SNHG12 in two separate studies compared to the normal control cell lines viz. ~6-fold in CaSKi cells and ~5-fold in SiHa cells as compared to the control NC104 cells (34), and ~3.5-fold in HeLa cells and ~3.3-fold in SiHa cells as compared to the immortalized cervical epithelial cell line, ICE (33) (Table 1).
In addition to these observations, it was found that SNHG12 promoted cervical cell motility and aided in their proliferation and migration by regulating downstream targets via sponging of miR-424-5p and miR-125b, as outlined in Figure 2 (33, 34). According to previous reports, miR-424-5p functions as a tumor suppressor by enhancing apoptosis of cancer cells and blocking the G1/S transition (43). Similarly, miR-125b can also inhibit tumor growth by suppressing the expression of the anti-apoptotic gene, BCL-2, consequently enhancing apoptosis (44). These effects were apparent when shRNA-mediated knockdown of SNHG12 impeded cell metastasis in HeLa, SiHa, and CaSki cervical cell lines, corresponding to up-regulated levels of miR-125b and miR-424-5p (33). In an in vivo study, knockdown of SNHG12 resulted in smaller tumor volumes and reduced tumor weights in CaSki cervical cancer cell-derived human xenografts in nude mice compared to the control mice (34). Further analyses implied that the signal transducer and activator of transcription (STAT3) gene acted as a direct downstream target of miR-125b (Figure 2) (42); STAT3 is known to function both as a transcription factor and an oncogene (62). It was found that phosphorylated STAT3 elevated the levels of the anti-apoptotic genes BCL-2 like 1 (BCL-XL), Survivin, and Myeloid Cell Leukemia Sequence 1 (MCL-1) in endometrial and cervical cancer tissues relative to normal tissue samples, aiding in their progression (63). Consistent with these reports, qRT-PCR results and immunoblot analyses revealed that miR-125b mimicked repressed STAT3 expression in cervical cancer cell lines (HeLa and SiHa), and that STAT3 knockdown was associated with decreased cell migration and proliferation.
Taken together, these data suggested that SNHG12 plays an important role in promoting tumorigenesis and metastasis in cervical cancer via sponging of miRNAs, such as miR-125p and miR-424-5b. Moreover, since STAT3 is required by cells to regulate various biological processes, modulation of the components of the SNHG12/miR-125b/STAT3 signaling axis may reduce the progression of cervical cancer.
Liu et al. (35) evaluated the expression of SNHG12 in 129 human NPC tissue samples and found it to be up-regulated by ~1.5-fold compared to adjacent normal tissues using qRT-PCR (Table 1) (35). Up-regulated levels of SNHG12 correlated with clinical stage and grade, with no significant correlation with other clinical or pathological characteristics. Moreover, using Kaplan-Meier curves, the authors showed that high SNHG12 expression was correlated with shorter overall survival times (35). Likewise, the evaluation of SNHG12 expression in five different NPC cell lines revealed its up-regulation in CNE1 cells and SUNE1 cells by ~10-fold and ~7.7-fold, respectively, as compared to the control cell line, NP69 (Table 1). Moreover, immunoblot analysis revealed that SNHG12 over-expression was associated with increased levels of protein expression, such as Notch1 and its downstream target proteins p21 and Hes family BHLH transcription factor 1 (Hes1) in NPC cell lines (Figure 2) (35). Following these observations, the authors found that SNHG12 knockdown by siRNA increased the apoptotic rate, and decreased the proliferative and metastatic potentials of the NPC cell lines (CNE1 and SUNE1), presumably by inhibiting Notch signaling. Thus, a positive correlation was established between the expression of SNHG12 and the resultant activation of the Notch signaling pathway (35). In summary, these results provide evidence that SNHG12 may serve as a biomarker for early detection of NPC and/or as a target for more effective treatment strategies for NPC patients.
Osteosarcoma is the most common primary malignant bone tumor and a major cause of cancer-associated fatality rates in children and adolescents (64). The annual incidence is ~3.4 cases per million people with a peak incidence occurring in late adolescence (45). However, the 5-year survival rates in localized and metastatic osteosarcoma have stagnated since the 1980s (65), presumably due to the lack of advances in chemotherapeutic treatment strategies that can prevent the chemoresistance that often occurs with the traditional therapies. This highlights the urgent need to formulate new therapeutic strategies, which may be provided by identifying potential molecular targets involved in the development of osteosarcoma.
The expression of SNHG12 was examined by three different research groups in 20, 31, and 32 doxorubicin-resistant (DXR) osteosarcoma tissue samples and it was found to be up-regulated by ~3.4-fold, ~2.9-fold, and ~2.4-fold, respectively, as shown in Table 1 (11, 29, 46). Moreover, SNHG12 abundance was associated with various clinical parameters, including advanced tumor grade, tumor size, lymph node metastasis, and Enneking staging, while its over-expression was negatively associated with overall survival rate. In addition, SNHG12 expression in different osteosarcoma cell lines was found to be up-regulated by ~6.2-fold in 143B cells, up to ~3.9-fold in MG63 cells, ~5.4-fold in MG63/DXR cells, and up to ~5.6-fold in U2OS cells compared to normal human osteoblast cell lines (11, 29) (Table 1).
It is known that SNHG12 can act as a ceRNA for various miRNAs, including miR-125b (33), miR-199a/b-5p (8), and miR-320 (52). Further analysis identified and confirmed miR-320a and miR-195-5p as direct downstream targets of SNHG12 that regulated the expression of the MCL-1 and NOTCH2 genes, respectively, via sponging (Figure 2). Notch 2 is an important receptor in Notch signaling and has been shown to regulate metastasis and tumorigenesis of osteosarcoma (11). Immunoblot analysis revealed that over-expression of Notch 2 in osteosarcoma cell lines up-regulated protein levels of Notch intracellular domain (NICD) and Hes1, which are involved in Notch signaling (Figure 2). SiRNA-mediated knockdown of MCL-1 promoted apoptosis of osteosarcoma cells by increasing their sensitivity toward the anti-cancer drug, doxorubicin. MCL-1 is an anti-apoptotic member of the Bcl-2 protein family and a key factor in conferring resistance to some cancer types to conventional chemotherapy (29). Consistent with the above findings, it was shown that SNHG12 depletion resulted in over-expression of miRNA-320a and reduced the invasiveness and proliferative capacity of the osteosarcoma cell lines, 143B, U2OS, MG-63, and MG-63/DXR (11, 29, 46). In addition, depletion of SNHG12 increased the susceptibility of U2OS and MG-63 osteosarcoma cells to apoptosis and doxorubicin sensitivity, presumably by down-regulating Notch 2 and MCL-1 (11, 29). Moreover, immunoblot analysis showed that the levels of cell cycle progression-related proteins (CDK4, CDK6, and CCND1) were increased (4) in si-SNHG12 transfected U2OS and 143B cell lines as shown in Figure 2.
In a study by Ruan et al. (46), it was reported that SNHG12 acted as an oncogene by regulating the expression of the Angiomotin (AMOT) gene (Figure 2). The AMOT gene directly regulates Rac1-GTPase, and enhances cell migration and stabilizes new blood vessels by activating the ERK pathway (66, 67). Consistent with these reports, combined results of qRT-PCR and immunoblots revealed that the over-expression of SNHG12 correlated with increased AMOT gene expression, which further promoted the proliferation and migration capacity of osteosarcoma cells.
These findings suggest that SNHG12 is involved in the etiology of osteosarcoma via the miR-320a/MCL-1 axis, the miR-195-5p/Notch2 axis, and AMOT gene up-regulation, and thus might serve as a potential biomarker/target for the diagnosis and/or treatment of osteosarcoma.
Apart from the cancer types discussed above, very recently SNHG12 has also been implicated in several other cancers (Table 1); for example, prostate cancer, ovarian cancer, renal cell carcinoma, and lymphoma, discussed below.
Two independent studies have shown that SNHG12 levels were up-regulated (~1.8-fold) in paired human prostate cancer (PCa) tissue samples compared to normal adjacent tissue, and (~2.3-fold) in the DU145 prostate cancer cell line compared to normal prostate cells (47, 48). This upregulation was found to be positively correlated with positive lymph nodes, Gleason score, and advanced residual tumor grade along with poorer recurrence-free survival (47, 48). Moreover, high SNHG12 levels were also correlated with low levels of miR-133b (47) and miR-195 (48), which are known to be involved in regulating proliferation, migration, invasion, and the cell cycle of PCa by enhancing the p53 (47) and Wnt/β-catenin signaling pathways (48). Thus, suggesting that SNHG12 may have potential as a prognostic as well as a diagnostic biomarker for PCa.
Very recently it was demonstrated that SNHG12 was aberrantly up-regulated (~1.75-fold) in renal cell carcinoma cells compared to control samples (49). Corresponding to this upregulation, the high expression of SNHG12 was correlated with poor prognosis (49). Furthermore, decreased SNHG12 levels inhibited cell viability and anchorage-independent growth, and induced apoptosis. However, these SNHG12 silencing-repressive effects were partially reversed by increased miR-199a-5p expression. Corresponding to these results, further analysis revealed that SNHG12 promoted renal cell carcinoma by modulating HIF1α expression via the sponging of miR-199a-5p (49).
Sun and Fan (50) reported that SNHG12 was up-regulated (~1.7-fold) in ovarian cancer (OC) tissues as compared to the normal adjacent tissues, and a similar expression was observed in OC cell lines as compared to a normal ovarian epithelial cell line (50). Moreover, SNHG12 abundance was found to be associated with increased metastasis, high-grade tumor (stage III-IV), low overall survival rate, and poor prognosis. Bioinformatics prediction identified SOX4 as a target of miR-129 which in turn served as a target for SNHG12 (50). Similar to previous results, SNHG12 overexpression enhanced the proliferative and migratory capacity of cells by inducing the expression of SOX4 via repressing the expression of its target miRNA (miR-129) (50).
Zhu et al. (51) studied the regulatory mechanism of SNHG12 in conferring multidrug resistance (MDR) to Natural Killer/T-cell lymphoma (NKTCL). It was observed that SNHG12 expression was increased (~1.3-fold) in NKTCL tissues of stages 3/4 as compared to the NKTCL tissues of stages 1/2, and reactive hyperplasia of lymph node (RHLN) was positively associated with clinical grading (stage 3/4) (51). C-Myc was identified as a direct transcriptional regulator of SNHG12 where its overexpression aided in the proliferation of cells, increased MDR-associated protein levels (P-gp), and inhibited sensitivity to cisplatin (CCDP) by binding to the promoter sequence of SNHG12 thereby promoting its expression (51). Since c-Myc induced upregulation of SNHG12, promoted NKTCL, and inhibited its drug sensitivity, SNHG12 may act as a therapeutic target and novel biomarker for CCDP-resistant NKTCL patients.
SNHG12 has been reported to play an important role in MDR in tumor cells. For example, Wang et al. (25) performed qRT-PCR analyses in the NSCLC drug-resistant cell lines, A549/DDP, A549/PTX, and PC9/AB2, and found an ~5-fold, ~4.8-fold, and ~5.5-fold higher expression of SNHG12, respectively, and an ~2.5-fold lower expression of miR-181a compared to their parental cell lines A549 and PC9 (25) (Table 1). Moreover, silencing of SNHG12 using siRNA increased the expression of miR-181a, which correlated with improved sensitivity of the resistant cell lines A549/DDP, A549/PTX, and PC9/AB2, to cisplatin, paclitaxel, and gefitinib, respectively. Similar results were observed in a separate study with increased sensitivity of A549/DDP human tumor xenografts in athymic mice to cisplatin treatment (25). Evidence suggested that silencing of SNHG12 suppressed the expression of MAPK1 and MAP2K1, and upregulated miR-181a further contributing to the inhibition of the MAPK/Slug pathway. Thus, it is possible that SNHG12 may also influence the unfolded protein response (UPR), which is stabilized in a large variety of cancers in a MAPK-dependent manner (68, 69). Many tumor cells exploit UPR via MAPK for the polarization of tumor-associated macrophages and T cells and enhanced angiogenesis (70). p38 MAPK is an example of a protein in the MAPK family that is potentially involved in vascular endothelial growth factor (VEGF)-mediated angiogenesis (70) and functional exhaustion of CD4 and CD8+ T cells. Our recent study has suggested that intratumoral inhibition of p38 MAPK influenced M1 re-tuning of tumor-induced macrophages and decreased the levels of VEGF and TGF-beta in neuroendocrine tumors of the pancreas (71). Thus, from this point of view, it is likely that inhibition of SNHG12 may also influence angiogenesis and depolarization of tumor-primed refractory immune cells contributing to immune restoration, though further studies are warranted.
Based on the literature discussed above, it is evident that pharmacological targeting of SNHG12 may serve as a potential alternative strategy for the diagnosis and treatment of a variety of cancers. It is anticipated that inhibiting SNHG12 may also influence the unfolded protein response, which is at the interface with MAPK signaling, contributing to stress, and angiogenic responses. It is also anticipated that targeting SNHG12 may improve cancer-directed interventions by mitigating UPR and MAPK signaling in both tumors as well as non-tumorous components of tumor-micro milieu. In this context, it would be worthwhile to investigate the extent to which inhibiting SNHG12 might also augment spontaneous effector immune responses and attenuate resistance and relapse in cancer patients. Further, SNHG12 profiling may provide a useful component of personalized medicine for future cancer-directed interventions. Thus, we propose SNHG12 as a potential candidate for cancer-directed interventions and that targeting SNHG12 may allow advances in the diagnoses, prognosis, and/or treatment of a variety of cancer types.
ST, VA, DR, RS, AA, and AJ conceived the idea. ST, VA, KV, HP, and AJ wrote the majority of the manuscript. ST, VA, DR, US, and AK composed the figures and table. Critical revisions were made by ST, VA, US, AK, HP, KV, and AJ. All authors read and approved the final manuscript.
AJ would like to acknowledge support from the Department of Biotechnology, India for providing grant (6242-P30/RGCB/PMD/DBT/AKJN/2015), the Indian Council of Medical Research (5/13/81/2013-NCD-III), and the DST-SERB for Indo-Russian grant (INT/RUS/RFBR/P-311). This work was also supported by NIH/NCI grant (CA093729) to KV.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Khandelwal A, Bacolla A, Vasquez KM, Jain A. Long non-coding RNA: a new paradigm for lung cancer. Mol Carcinog. (2015) 54:1235–51. doi: 10.1002/mc.22362
3. Khandelwal A, Malhotra A, Jain M, Vasquez KM, Jain A. The emerging role of long non-coding RNA in gallbladder cancer pathogenesis. Biochimie. (2017) 132:152–60. doi: 10.1016/j.biochi.2016.11.007
4. Malhotra A, Jain M, Prakash H, Vasquez KM, Jain A. The regulatory roles of long non-coding RNAs in the development of chemoresistance in breast cancer. Oncotarget. (2017) 8:110671–84. doi: 10.18632/oncotarget.22577
5. Saluja R, Kumar A, Jain M, Goel SK, Jain A. Role of sphingosine-1-phosphate in mast cell functions and asthma and its regulation by non-coding RNA. Front Immunol. (2017) 8:587. doi: 10.3389/fimmu.2017.00587
6. Malhotra A, Sharma U, Puhan S, Chandra Bandari N, Kharb A, Arifa PP, et al. Stabilization of miRNAs in esophageal cancer contributes to radioresistance and limits efficacy of therapy. Biochimie. (2019) 156:148–57. doi: 10.1016/j.biochi.2018.10.006
7. Zhai W, Li X, Wu S, Zhang Y, Pang H, Chen W. Microarray expression profile of lncRNAs and the upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int J Oncol. (2015) 46:2125–37. doi: 10.3892/ijo.2015.2897
8. Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. (2017) 36:11. doi: 10.1186/s13046-016-0486-9
9. Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. (2017) 9:533–45.
10. Zhang H, Lu W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR320. Mol Med Rep. (2018) 17:2743–9. doi: 10.3892/mmr.2017.8143
11. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. (2018) 495:1822–32. doi: 10.1016/j.bbrc.2017.12.047
12. Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang Z, et al. Inhibition of TDP43-mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol Ther Nucleic Acids. (2018) 10:142–58. doi: 10.1016/j.omtn.2017.12.001
13. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. (2015) 52:710–8. doi: 10.1136/jmedgenet-2015-103334
14. Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. (2014) 4:6088. doi: 10.1038/srep06088
15. Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. (2018) 645:157–62. doi: 10.1016/j.gene.2017.12.036
16. Tan JY, Marques AC. miRNA-mediated crosstalk between transcripts: The missing “linc”? Bioessays. (2016) 38:295–301. doi: 10.1002/bies.201500148
17. Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Leopold V, et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA. (2013) 110:7154–9. doi: 10.1073/pnas.1222509110
18. Bosia C, Pagnani A, Zecchina R. Modelling competing endogenous RNA networks. PLoS ONE. (2013) 8:e66609. doi: 10.1371/journal.pone.0066609
19. Figliuzzi M, Marinari E, De Martino A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys J. (2013) 104:1203–13. doi: 10.1016/j.bpj.2013.01.012
20. Tan JY, Sirey T, Honti F, Graham B, Piovesan A, Merkenschlager M, et al. Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. (2015) 25:655–66. doi: 10.1101/gr.181974.114
21. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. doi: 10.1038/nature12986
22. Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. (2016) 32:211–24. doi: 10.1016/j.tig.2016.02.001
23. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. (2010) 38:5366–83. doi: 10.1093/nar/gkq285
24. Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. (2013) 5:1143–6. doi: 10.3892/etm.2013.933
25. Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. (2017) 8:84086–101. doi: 10.18632/oncotarget.20475
26. Sun Y, Liu J, Chu L, Yang W, Liu H, Li C, et al. Long noncoding RNA SNHG12 facilitates the tumorigenesis of glioma through miR-101-3p/FOXP1 axis. Gene. (2018) 676:315–21. doi: 10.1016/j.gene.2018.08.034
27. Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/beta-catenin signaling pathway. Cancer Biomark. (2018) 22:217–26. doi: 10.3233/CBM-170777
28. Jiang B, Hailong S, Yuan J, Zhao H, Xia W, Zha Z, et al. Identification of oncogenic long noncoding RNA SNHG12 and DUXAP8 in human bladder cancer through a comprehensive profiling analysis. Biomed Pharmacother. (2018) 108:500–7. doi: 10.1016/j.biopha.2018.09.025
29. Zhou B, Li L, Li Y, Sun H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. (2018) 106:850–7. doi: 10.1016/j.biopha.2018.07.003
30. Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. (2017) 50:e6079. doi: 10.1590/1414-431x20176079
31. Wang X, Qi G, Zhang J, Wu J, Zhou N, Li L, et al. Knockdown of long noncoding RNA small nucleolar RNA host gene 12 inhibits cell growth and induces apoptosis by upregulating miR-138 in nonsmall cell lung cancer. DNA Cell Biol. (2017) 36:892–900. doi: 10.1089/dna.2017.3830
32. Wang Y, Liang S, Yu Y, Shi Y, Zheng H. Knockdown of SNHG12 suppresses tumor metastasis and epithelial-mesenchymal transition via the Slug/ZEB2 signaling pathway by targeting miR-218 in NSCLC. Oncol Lett. (2019) 17:2356–64. doi: 10.3892/ol.2018.9880
33. Jin XJ, Chen XJ, Zhang ZF, Hu WS, Ou RY, Li S, et al. Long noncoding RNA SNHG12 promotes the progression of cervical cancer via modulating miR-125b/STAT3 axis. J Cell Physiol. (2019) 234:6624–32. doi: 10.1002/jcp.27403
34. Dong J, Wang Q, Li L, Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol Biochem. (2018) 45:2086–94. doi: 10.1159/000488045
35. Liu ZB, Tang C, Jin X, Liu SH, Pi W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomark. (2018) 23:603–13. doi: 10.3233/CBM-181873
36. Yang BF, Cai W, Chen B. LncRNA SNHG12 regulated the proliferation of gastric carcinoma cell BGC-823 by targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci. (2018) 22:1297–306. doi: 10.26355/eurrev_201803_14471
37. Han LP, Fu T, Lin Y, Miao JL, Jiang QF. MicroRNA-138 negatively regulates non-small cell lung cancer cells through the interaction with cyclin D3. Tumour Biol. (2016) 37:291–8. doi: 10.1007/s13277-015-3757-8
38. Ma Z, Qiu X, Wang D, Li Y, Zhang B, Yuan T, et al. MiR-181a-5p inhibits cell proliferation and migration by targeting Kras in non-small cell lung cancer A549 cells. Acta Biochim Biophys Sin. (2015) 47:630–8. doi: 10.1093/abbs/gmv054
39. Shi Q, Zhou Z, Ye N, Chen Q, Zheng X, Fang M. MiR-181a inhibits non-small cell lung cancer cell proliferation by targeting CDK1. Cancer Biomark. (2017) 20:539–46. doi: 10.3233/CBM-170350
40. Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. (2011) 19:232–43. doi: 10.1016/j.ccr.2011.01.001
41. Lei W, Wang ZL, Feng HJ, Lin XD, Li CZ, Fan D. Long non-coding RNA SNHG12promotes the proliferation and migration of glioma cells by binding to HuR. Int J Oncol. (2018) 53:1374–84. doi: 10.3892/ijo.2018.4478
42. Yin H, Sun Y, Wang X, Park J, Zhang Y, Li M, et al. Progress on the relationship between miR-125 family and tumorigenesis. Exp Cell Res. (2015) 339:252–60. doi: 10.1016/j.yexcr.2015.09.015
43. Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. (2013) 32:976–87. doi: 10.1038/onc.2012.121
44. Cui F, Li X, Zhu X, Huang L, Huang Y, Mao C, et al. MiR-125b inhibits tumor growth and promotes apoptosis of cervical cancer cells by targeting phosphoinositide 3-kinase catalytic subunit delta. Cell Physiol Biochem. (2012) 30:1310–8. doi: 10.1159/000343320
45. Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. (2009) 125:229–34. doi: 10.1002/ijc.24320
46. Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. (2016) 37:4065–73. doi: 10.1007/s13277-015-4256-7
47. Cheng G, Song Z, Liu Y, Xiao H, Ruan H, Cao Q, et al. Long noncoding RNA SNHG12 indicates the prognosis of prostate cancer and accelerates tumorigenesis via sponging miR-133b. J Cell Physiol. (2019). doi: 10.1002/jcp.29039. [Epub ahead of print].
48. Song J, Wu X, Ma R, Miao L, Xiong L, Zhao W. Long noncoding RNA SNHG12 promotes cell proliferation and activates Wnt/beta-catenin signaling in prostate cancer through sponging microRNA-195. J Cell Biochem. (2019) 120:13066–75. doi: 10.1002/jcb.28578
49. Chen Q, Zhou W, Du SQ, Gong DX, Li J, Bi JB, et al. Overexpression of SNHG12 regulates the viability and invasion of renal cell carcinoma cells through modulation of HIF1alpha. Cancer Cell Int. (2019) 19:128. doi: 10.1186/s12935-019-0782-5
50. Sun D, Fan XH. LncRNA SNHG12 accelerates the progression of ovarian cancer via absorbing miRNA-129 to upregulate SOX4. Eur Rev Med Pharmacol Sci. (2019) 23:2345–52. doi: 10.26355/eurrev_201903_17378
51. Zhu L, Zhang X, Fu X, Li Z, Sun Z, Wu J, et al. c-Myc mediated upregulation of long noncoding RNA SNHG12 regulates proliferation and drug sensitivity in natural killer/T-cell lymphoma. J Cell Biochem. (2019) 120:12628–37. doi: 10.1002/jcb.28529
52. Zhao Y, Dong Q, Wang E. MicroRNA-320 inhibits invasion and induces apoptosis by targeting CRKL and inhibiting ERK and AKT signaling in gastric cancer cells. Onco Targets Ther. (2017) 10:1049–58. doi: 10.2147/OTT.S123324
53. Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. (2018) 16:4689–98. doi: 10.3892/ol.2018.9199
54. Zhu TG, Xiao X, Wei Q, Yue M, Zhang LX. Revealing potential long non-coding RNA biomarkers in lung adenocarcinoma using long non-coding RNA-mediated competitive endogenous RNA network. Braz J Med Biol Res. (2017) 50:e6297. doi: 10.1590/1414-431x20176297
55. Braicu C, Gulei D, Cojocneanu R, Raduly L, Jurj A, Knutsen E, et al. miR-181a/b therapy in lung cancer: reality or myth? Mol Oncol. (2019) 13:9–25. doi: 10.1002/1878-0261.12420
56. He S, Zeng S, Zhou ZW, He ZX, Zhou SF. Hsa-microRNA-181a is a regulator of a number of cancer genes and a biomarker for endometrial carcinoma in patients: a bioinformatic and clinical study and the therapeutic implication. Drug Des Devel Ther. (2015) 9:1103–75. doi: 10.2147/DDDT.S73551
57. Li W, Qiu X, Jiang H, Han Y, Wei D, Liu J. Downregulation of miR-181a protects mice from LPS-induced acute lung injury by targeting Bcl-2. Biomed Pharmacother. (2016) 84:1375–82. doi: 10.1016/j.biopha.2016.10.065
58. Li SQ, Wang ZH, Mi XG, Liu L, Tan Y. MiR-199a/b-3p suppresses migration and invasion of breast cancer cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life. (2015) 67:768–77. doi: 10.1002/iub.1433
59. Song T, Zhang X, Yang G, Song Y, Cai W. Decrement of miR-199a-5p contributes to the tumorigenesis of bladder urothelial carcinoma by regulating MLK3/NF-kappaB pathway. Am J Transl Res. (2015) 7:2786–94.
60. Ouchen Wang FY, Liu Y, Lv L, Ma R, Chen C, Wang J, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Trans Res. (2017) 9:533.
61. Wang Y, Zhang X, Zou C, Kung HF, Lin MC, Dress A, et al. miR-195 inhibits tumor growth and angiogenesis through modulating IRS1 in breast cancer. Biomed Pharmacother. (2016) 80:95–101. doi: 10.1016/j.biopha.2016.03.007
62. Yuan J, Zhang F, Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep. (2015) 5:17663. doi: 10.1038/srep17663
63. Chen CL, Hsieh FC, Lieblein JC, Brown J, Chan C, Wallace JA, et al. Stat3 activation in human endometrial and cervical cancers. Br J Cancer. (2007) 96:591–9. doi: 10.1038/sj.bjc.6603597
64. Ando K, Heymann MF, Stresing V, Mori K, Redini F, Heymann D. Current therapeutic strategies and novel approaches in osteosarcoma. Cancers. (2013) 5:591–616. doi: 10.3390/cancers5020591
65. Lee CG, Dela Cruz CS, Ma B, Ahangari F, Zhou Y, Halaban R, et al. Chitinase-like proteins in lung injury, repair, and metastasis. Proc Am Thorac Soc. (2012) 9:57–61. doi: 10.1513/pats.201112-056MS
66. Levchenko T, Bratt A, Arbiser JL, Holmgren L. Angiomotin expression promotes hemangioendothelioma invasion. Oncogene. (2004) 23:1469–73. doi: 10.1038/sj.onc.1207264
67. Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. (2011) 19:527–40. doi: 10.1016/j.ccr.2011.02.017
68. Darling NJ, Cook SJ. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim Biophys Acta. (2014) 1843:2150–63. doi: 10.1016/j.bbamcr.2014.01.009
69. Yang F, Tang XY, Liu H, Jiang ZW. Inhibition of mitogen-activated protein kinase signaling pathway sensitizes breast cancer cells to endoplasmic reticulum stress-induced apoptosis. Oncol Rep. (2016) 35:2113–20. doi: 10.3892/or.2016.4580
70. Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat Rev Immunol. (2016) 16:469–84. doi: 10.1038/nri.2016.62
Keywords: long non-coding RNA, lncRNA, SNHG12, miRNA, cancer, microRNA, small nucleolar host gene 12
Citation: Tamang S, Acharya V, Roy D, Sharma R, Aryaa A, Sharma U, Khandelwal A, Prakash H, Vasquez KM and Jain A (2019) SNHG12: An LncRNA as a Potential Therapeutic Target and Biomarker for Human Cancer. Front. Oncol. 9:901. doi: 10.3389/fonc.2019.00901
Received: 04 May 2019; Accepted: 30 August 2019;
Published: 18 September 2019.
Edited by:
Kuzhuvelil B. Harikumar, Rajiv Gandhi Centre for Biotechnology, IndiaCopyright © 2019 Tamang, Acharya, Roy, Sharma, Aryaa, Sharma, Khandelwal, Prakash, Vasquez and Jain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aklank Jain, YWtsYW5rLmphaW5AY3VwLmVkdS5pbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.