- 1Department of Pathology, Dunedin School of Medicine, University of Otago, Dunedin, New Zealand
- 2Maurice Wilkins Centre for Molecular Biodiscovery, Auckland, New Zealand
The long non-coding RNA (LncRNA) antisense RNA in the INK4 locus (ANRIL) is overexpressed in several cancers including breast cancer. To better understand the role of ANRIL in breast cancer this study investigated where ANRIL was expressed in breast tumors using in situ hybridization by RNAscope. Additional RNAscope assays for IL6, CCL2, and POSTN were used to establish whether ANRIL correlated with increased tumor promoting cytokines. Breast tumors with ANRIL over expressed from real-time quantitative (RT-q) PCR assays were selected for analysis using RNAscope. All tumors showed ANRIL expression in malignant cells, but amongst tumors ANRIL showed different subcellular locations with 56% of tumors with ANRIL only in the nucleus, 16% with ANRIL only in the cytoplasm and 28% with ANRIL in both the nucleus and cytoplasm. Cases with nuclear ANRIL were positively correlated with POSTN expression in malignant cells (ρ = 0.57, P = 0.0086), and no correlation was found between ANRIL and IL6 or CCL2. Reduced POSTN was also found using siRNA to ANRIL in MDA-MB-231 and MCF7 breast cancer cells. These data indicate that ANRIL is expressed in malignant breast cells, and suggest its subcellular location may indicate its function in cancer progression.
Introduction
Given that the progression of breast cancer is associated with poor prognosis, a better understanding of breast cancer progression will better aid targeted patient management and treatment (1). Long non-coding RNAs (LncRNAs) are generally defined as RNA molecules longer than 200 nucleotides with no coding ability (2). They have been proposed to function in multiple cancer pathways including promoting proliferation, migration, invasion, inflammation, and metastasis (2, 3). One such regulatory LncRNA, ANRIL, spans over 126 kbp of genomic sequence and three tumor suppressor genes encoded at 9p21 (4–6). In inflammatory conditions including cardiovascular disease and ulcerative colitis ANRIL expression correlates with increased expression of three pro-inflammatory cytokines IL6, CCL2, and POSTN (4, 7–10).
ANRIL is also over expressed in many cancers including breast (11–16), prostate (5), colorectal (17), gastric (18), and brain (19). Amongst breast cancers multiple studies have found elevated ANRIL in triple negative tumors (12, 14–16). Amongst this subtype, ANRIL was elevated in 25–40% of cases, associated with a poorer prognosis, and was included as part of a three non-coding RNA signature proposed for distinguishing triple negative breast cancer from other cancer types (15). Overexpression of ANRIL does occur in some estrogen receptor (ER) and progesterone receptor (PR) positive breast cancers. These include tumors from women that carried the minor allele of the rs11515 polymorphism in the 3′ UTR of cyclin dependent kinase inhibitor 2A (CDKN2A) that encodes both the p16INK4a and p14ARF tumor suppressors (13). In addition to increased ANRIL, breast tumors from those with the rs11515 minor allele had reduced expression of p16INK4a offering one explanation of how ANRIL could be contributing to breast cancer progression. A role for ANRIL in promoting breast cancer proliferation was also found when ANRIL was knocked down in breast cancer cell lines and tumor xenografts (16).
Despite ANRIL being well-studied as a biomarker in breast cancer, it is uncertain if ANRIL is expressed within tumors or other cells that are part of the breast tumor microenvironment. The subcellular localization of ANRIL and whether it correlates with IL6, CCL2, and POSTN expression, found in other conditions, in breast cancer is also unknown (4, 7–10). To investigate this, we used RNAscope to examine the cell type and the subcellular location containing ANRIL in breast tumors. Whether ANRIL expression correlates with IL6, CCL2, and POSTN in breast tumors was also investigated to provide evidence of how ANRIL may contribute to inflammation in breast cancer (20–22).
Materials and Methods
Study Cohort
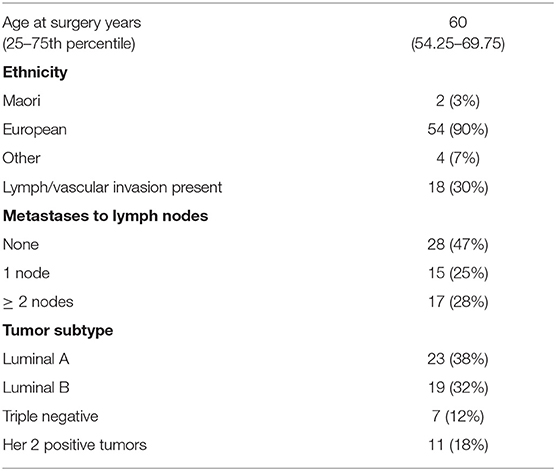
The study cohort consisted of 60 women diagnosed with primary breast cancer who underwent surgery at Dunedin hospital, New Zealand. Clinicopathological data for the cohort is provided in Table 1. Blocks of formalin fixed paraffin embedded material (FFPE) from breast tumor and separate blocks of normal tissue adjacent to tumor tissue were available. Ethical approval was obtained from the Lower South Regional Ethics Committee, Ministry of Health, New Zealand (Ethics Committee Approval LRS/10/09/035) and all participating women gave written and informed consent for inclusion in the study. Standard procedures including culturally appropriate tissue handling and disposal protocols were followed. In addition to the breast cohort, commercially available RNA (Takara Bio, Shiga, Japan) supplied on dry ice and stored at −80°C before use from a variety of tumors and normal tissues were also quantified for ANRIL expression. Breast cancers were separated into molecular subtypes (Luminal A, Luminal B, Her 2 positive and triple negative breast cancer) using immunohistochemical surrogates for molecular classifications (23). The luminal A group were classified as ER and PR (≥20%) positive, Her 2 negative and low for Ki67 (<14%); Luminal B were classified as ER and PR (≥20%) positive and high for Ki67 (≥14%), or ER positive with low PR positive cells (<20%); Her 2 positive were all Her 2 positive tumors; and the triple negative breast cancer were classified as all tumors that were negative for ER, PR, and Her 2.
RNA Extraction
Sections (10 sections at 10 μm) from FFPE breast tumor and normal tissue adjacent to the tumor tissue blocks were subjected to RNA extraction using the RecoverAll™ Total Nucleic Acid Isolation Kit (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Eluted RNA was subject to further purification using the RNA Clean-Up and Concentration Kit™ (Norgen Biotek, Thorold, ON, Canada) and incubated at 70°C for 20 min immediately prior to cDNA synthesis (24). High Capacity RNA-to-cDNA Kit (ThermoFisher Scientific, Waltham, MA, USA) was used to convert total RNA to cDNA according to the manufacturer's protocol.
Quantification of ANRIL Using TaqMan Assays
Quantification of ANRIL (assay ID Hs01390879_m1) was performed using the Applied Biosystems TaqMan Non-Coding RNA Assay (ThermoFisher Scientific, Waltham, MA, USA) with hypoxanthine phosphoribosyltransferase (HPRT) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) assays (IDs Hs99999909_m1 and Hs03929097_g, respectively) included as reference genes. Gene expression was quantified in triplicate from single cDNA preparations using the LightCycler 480 (LC480) qPCR machine (Roche, Basel, Switzerland). Relative gene expression was calculated using double delta Ct (ΔΔCT) method and qbase+ software (25).
RNAscope in situ Hybridization
RNAscope 2.5 HD Assay-BROWN manual kit™ (Advanced Cell Diagnostics, Newark, CA, USA) was used to measure ANRIL (Hs-CDKN2B-AS1), IL-6 (Hs-IL6), CCL2 (Hs-CCL2), and POSTN (Hs-POSTN) on FFPE tissue sections. Positive (Ubiquitin C, UBC) and negative control (the bacterial gene DapB) probes were performed on serial tissue sections. Tissue blocks were sectioned at 4 μm thickness and mounted onto Leica BOND™ Plus Slides (Leica Biosystems, Wetzlar, Germany). The assays were performed using the manufacturer's protocol. Stained slides were scanned into the Aperio Scanscope CS digital pathology system slide scanner under 400x magnification (Leica Biosystems, Wetzlar, Germany). The number of dots per cell and the number of positive cells were calculated to provide a score to evaluate the amount of staining present. Expression of ANRIL was scored 0–3 as outlined below. A score of 0 (no staining or <1 dot in a total of ten cells), 1 (1–3 dots per cell with <20% of cells per high powered field (hpf, at x400 magnification) positive), 2 (4–9 dots per cell in at least 20% of cells positive or >20% of cells per hpf with at least 1 dot per cell), 3 (>10 dots per cell and at least 20% of cells positive per hpf or >40% of cells with at least 1 dot per cell). For the correlation between ANRIL and POSTN the average amount of dots per 200 malignant cells were calculated from the average of three fields with 200 cells per field counted. Small clusters of staining were considered to contain ten dots and large clusters 20 dots.
RNAscope was also performed on MCF7 cells treated with siRNA toward ANRIL or a control siRNA. Cells from 6 well-plates were combined and made into a cell clot using HistoGelTM specimen processing gel (Thermo Fisher Scientific, Massachusetts, USA). Cell clots were fixed in 10% neutral buffered formaldehyde overnight, and processed into paraffin wax. Cell clot sections (5 μm) were subjected to RNAscope for ANRIL, POSTN, FBXO18, TERC, PPIB, UBC, and DapB according to the manufacturer's instructions for cells. Cells were imaged using a (Zeiss LSM510; Carl Zeiss, Thornwood, NY, USA), and images were analyzed by Zeiss LSM Image Examiner software version 3.2.0.115 (Carl Zeiss), or imaged using the Aperio Scanscope CS digital pathology system.
Cell Culture
MCF7 and MDA-MB-231 cell lines (American Type Culture Collection, ATCC) were cultured in Dulbecco's Modified Eagle Medium (DMEM) from Life Technologies (Gibco BRL, Grand Island, NY, USA) and supplemented with 10% fetal bovine serum (FBS) in a 37°C humidified incubator at 5% CO2.
siRNA
Cells were transfected with Lincode Human CDKN2B-AS1 (R-188105-00-0005) siRNA; Dharmacon, Chicago, IL, USA) using Lipofectamine 2000 (Invitrogen, Burlington, ON, Canada). ON-Target plus Non-targeting Pool (D-001810-10-05) was used as a negative control as suggested by the manufacturer. siRNAs were transfected at a final concentration of 5 or 10 nM using Lipofectamine RNAiMax (Invitrogen). The siRNA and RNAiMax were combined in additional culture media and the resulting lipoplexes were added to the wells with a total of 1 × 104 cells/2 mL for overnight transfection. After 24 h the cultured media was replaced with fresh media and finally harvested at 48 h in total. Each experiment was performed at least in triplicate. ANRIL knockdown was verified by real-time RT-PCR using GAPDH and HPRT as reference genes and tested for ANRIL and POSTN expression as detailed elsewhere.
Cytotoxicity Assay
Cells were seeded at a concentration of 1 × 105 cells/mL into a 96-well plate and assayed using the Vybrant MTT Cell Proliferation Assay kit (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer's protocol for adherent cells.
Statistical Analysis
Student's t test (two tailed) was used to compare two groups, one- way ANOVA for multiple comparisons was used to compare more than two groups, and Pairwise Spearman correlation was performed between ANRIL and POSTN expression using GraphPad Prism7 software (California, USA). A P value < 0.05 was considered statistically significant.
Results
ANRIL Is Expressed Within Malignant Cells in Different Subcellular Localizations in Breast Tumors
To determine where ANRIL was located in breast tumors we first determined the expression of ANRIL in 60 breast tumors, and other tissues from a commercially available source using RT-qPCR. Expression of ANRIL was found in all tissues in varying amounts being lowest in lung cancer, kidney, lymph node, and smooth muscle and highest in spleen, thymus, pituitary gland, stomach cancer and leukemia (Figures 1A,B). The presence of ANRIL mRNA in a wide variety of tissues including hemopoietic tissues suggested ANRIL could be expressed in both tumor cells and the tumor microenvironment. In breast tumors ANRIL was increased in RNA extracted from tumor tissue compared to the patient-matched normal tissue adjacent to the tumor ANRIL (Figure 1C, P = 0.0037). An analysis of ANRIL based on breast tumor subtypes compared to the normal associated tissues showed ANRIL was increased in Luminal A (Figure 1D, P = 0.0017) and Luminal B (Figure 1D, P = 0.0008) tumors compared to the normal associated tissue. Although the highest average expression of ANRIL was found in triple negative tumors, there was no significant difference in ANRIL when compared to the normal associated tissue. A comparison of ANRIL amongst tumor tissues showed that ANRIL was increased in triple negative tumors compared to Luminal A (P = 0.011) and B (P = 0.027) with no significance found compared to Her 2 positive tumors (P = 0.35).
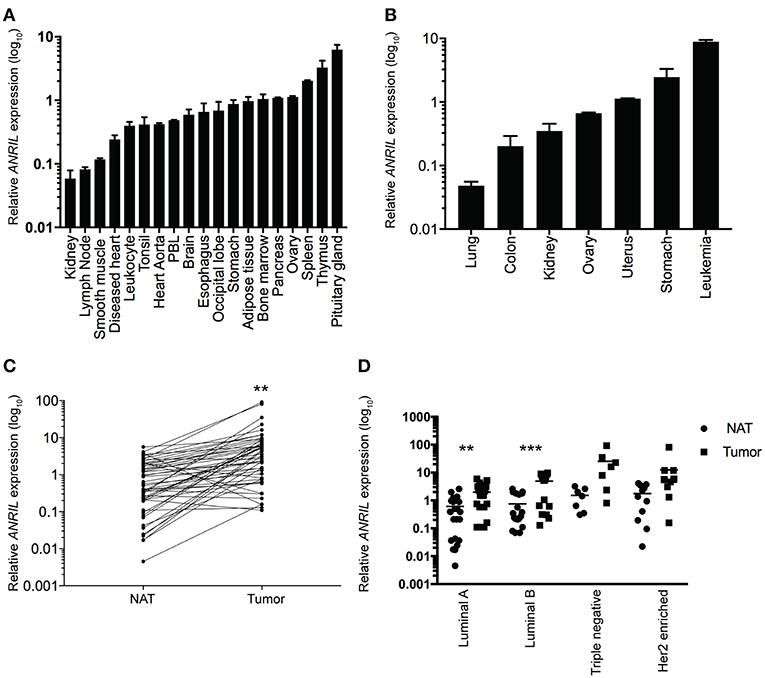
Figure 1. Relative ANRIL expression levels in various normal and cancer tissues including primary breast cancer. (A) RT-qPCR results from normal commercially available tissues showed a range of ANRIL expression levels suggesting ANRIL could be expressed as part of the tumor micro environment. (B) RT-qPCR results from cancerous commercially available tissues showed a range of ANRIL expression levels. Data represented as mean ± SEM for four replicates of each sample. (C) Line plot showing higher ANRIL levels in breast cancer tissue compared to the matched normal tissue adjacent to the tumor (NAT) from 60 breast cancer patients measured from RT-qPCR assays. ANRIL in tumors was significantly higher than NAT (**P < 0.01). (D) Profile of ANRIL expression amongst different tumor subtypes showed in Luminal A and Luminal B tumors ANRIL was increased in tumors compared to NAT (***P < 0.001). Data represented as mean ± SD for three replicates of each sample. All RT-qPCR normalized to GAPDH and HPRT.
To determine the cell type responsible for ANRIL expression in breast tumors, 25 cases with sufficient tissue available and those with ANRIL expressed >2.5-fold compared to the mean expression of the house keeping genes were selected and subjected to analyses using RNAscope. Cases included 5 Luminal A, 8 Luminal B, 4 triple negative, and 8 Her 2 positive tumors. Tissue sections were first assessed using a positive control probe for UBC to check for sufficient RNA quality and sequential sections stained for ANRIL and a negative control probe. All cases showed strong staining for UBC and variable amounts of ANRIL staining that was absent in the negative control. All cases showed the majority of ANRIL expression in malignant epithelial cells with no expression identified in normal mammary epithelium in the same section. In fifteen (60%) cases, scattered non-malignant cells were also positive for ANRIL and included endothelial cells (1–9 dots in positive cells and up to three cells positive focally) and other stromal cells (3–9 dots positive in individual cells). The RNAscope assay revealed different cellular localization for ANRIL (Figure 2) in malignant cells with staining in the nucleus (Figure 2A) and cytoplasm (Figure 2B). The majority of cases had ANRIL only in the nucleus (n = 14, 56% and Figure 2A top panel), 28% of cases had staining in the nucleus and outside of the nucleus (n = 7), and 16% of cases had ANRIL staining outside of the nucleus (n = 4, and Figure 2C). Although the number of tumors in individual tumor subtypes were limited. The majority of Luminal A and Her 2 and all triple negative breast tumors had ANRIL only in the nucleus. Luminal B tumors showed more of a mixed location for ANRIL with one third of tumors with ANRIL in the nucleus, one third with ANRIL outside the nucleus and one third with ANRIL in both the nucleus and outside the nucleus.
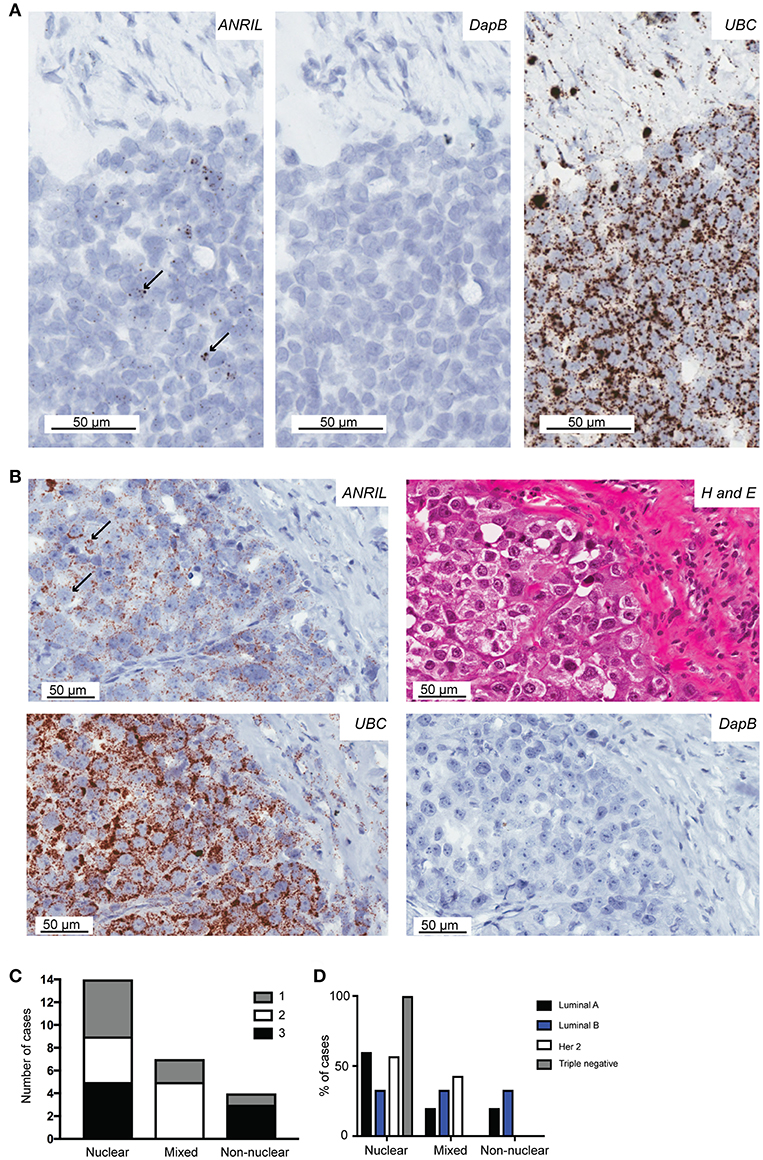
Figure 2. The localization of ANRIL in breast tumors using RNAscope in situ hybridization. Using RNAscope different subcellular localizations of ANRIL were revealed in breast tumors. (A) Left panel, Nuclear ANRIL (brown dots) in malignant cells; Middle panel, the negative control DapB; and Right panel, the positive control UBC. (B) Top left panel, Non-nuclear ANRIL (brown dots) in malignant cells; Top right, hematoxylin and eosin staining showing cellular morphology confirming presence of malignant cells; Bottom left, the positive control UBC; Bottom right, the negative control DapB. Magnification x400. (C) Summary of the ANRIL RNAscope assay results across all breast tumors analyze including the subcellular localization of the staining. The amount of staining was semi quantified using a score based on the number of positive dots and the number of positive cells (0 lowest-3 highest staining). (D) Summary of the ANRIL location results across all breast tumor subtypes.
Overall these findings suggest ANRIL was largely present in malignant cells and its presence inside and outside of the nucleus suggests ANRIL is heterogeneously located amongst breast tumors.
ANRIL Correlated With POSTN in Malignant Breast Cells
To determine if malignant cells that expressed ANRIL also expressed CCL2, IL6 or POSTN, the RNAscope analyses were extended to include each cytokine and the areas with ANRIL staining were correlated with the other markers. Only one tumor had IL6 staining in malignant cells with ANRIL staining and no cases showed CCL2 staining in cells with ANRIL. Areas with ANRIL positivity in malignant cells also showed POSTN staining (Figure 3A). An analysis including all ANRIL positive tumors found no correlation between ANRIL and POSTN (Figure 3B, left panel); however, a positive correlation was found when only tumors with nuclear staining were included (Figure 3B, right panel). These results suggest in breast tumors nuclear ANRIL is correlated with POSTN expression.
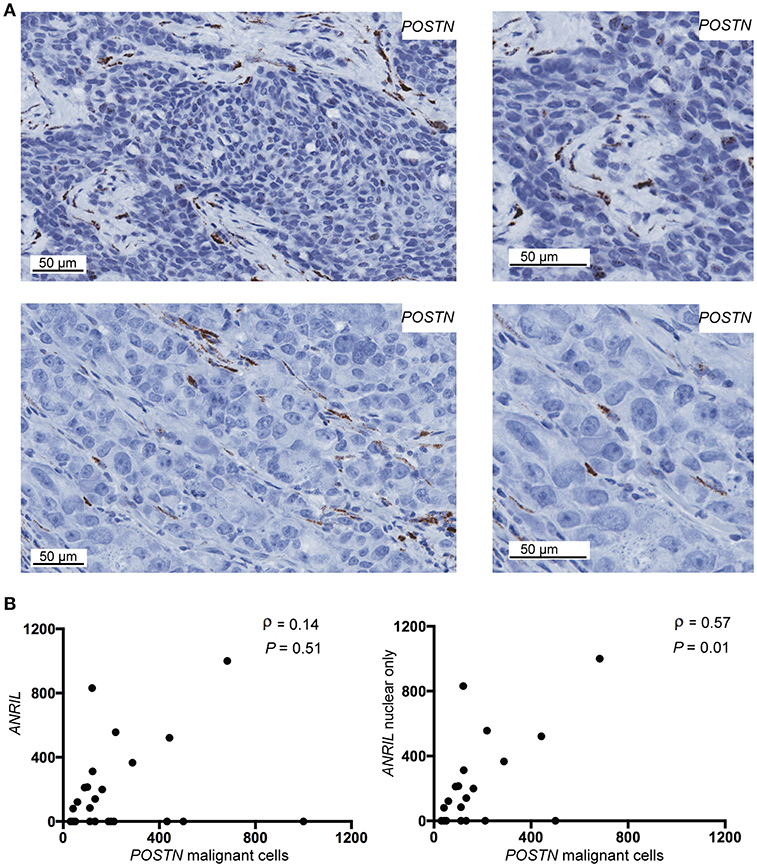
Figure 3. The correlation between POSTN and ANRIL in breast tumors using RNAscope in situ hybridization. (A) Using RNAscope POSTN was expressed in the malignant and stroma (top panel) or the stroma only (bottom panel) in breast tumors. Images at the same magnification but enlarged are shown to the right to better illustrate positively stained cells. (B) Scatter plot illustrating ANRIL and POSTN expression in malignant cells. Spearman's correlation coefficient (ρ) between ANRIL and POSTN revealed no association when all ANRIL expressing tumors were included in the analysis (left panel), but a significant correlation when cytoplasmic only expressing tumors were excluded (right panel).
ANRIL Correlated With POSTN in Breast Cell Lines
To test the correlation between ANRIL and POSTN found in breast tumors siRNA was used to knockdown ANRIL levels in two breast cancer cell lines and the effect on POSTN expression measured using RT-qPCR. Two cell lines were selected MDA-MB-231 and MCF7 cells that expressed ANRIL (26) In both cell lines ANRIL was knocked down relative to cells treated with the negative control siRNA (Figure 4A). Cells with reduced ANRIL also had reduced POSTN expression (Figure 4B). The MTT assay showed no significance difference between MCF7 cells treated with siANRIL and those treated with the control siRNA (Figure 4C). MCF7 cells treated with siRNA for ANRIL and the negative control siRNA were also investigated using RNAscope. There was no difference in expression of genes not expected to be affected by ANRIL (PPIB, FBXO18 and TERC) in MCF7 cells treated with the siANRIL compared to those treated with the control siRNA (Figure 4D). However, RNAscope for ANRIL and POSTN showed that although ANRIL and POSTN expression were low in cells treated with the negative control siRNA it was further reduced in cells treated with siRNA for ANRIL (Figure 5), consistent with the findings using RT-qPCR.
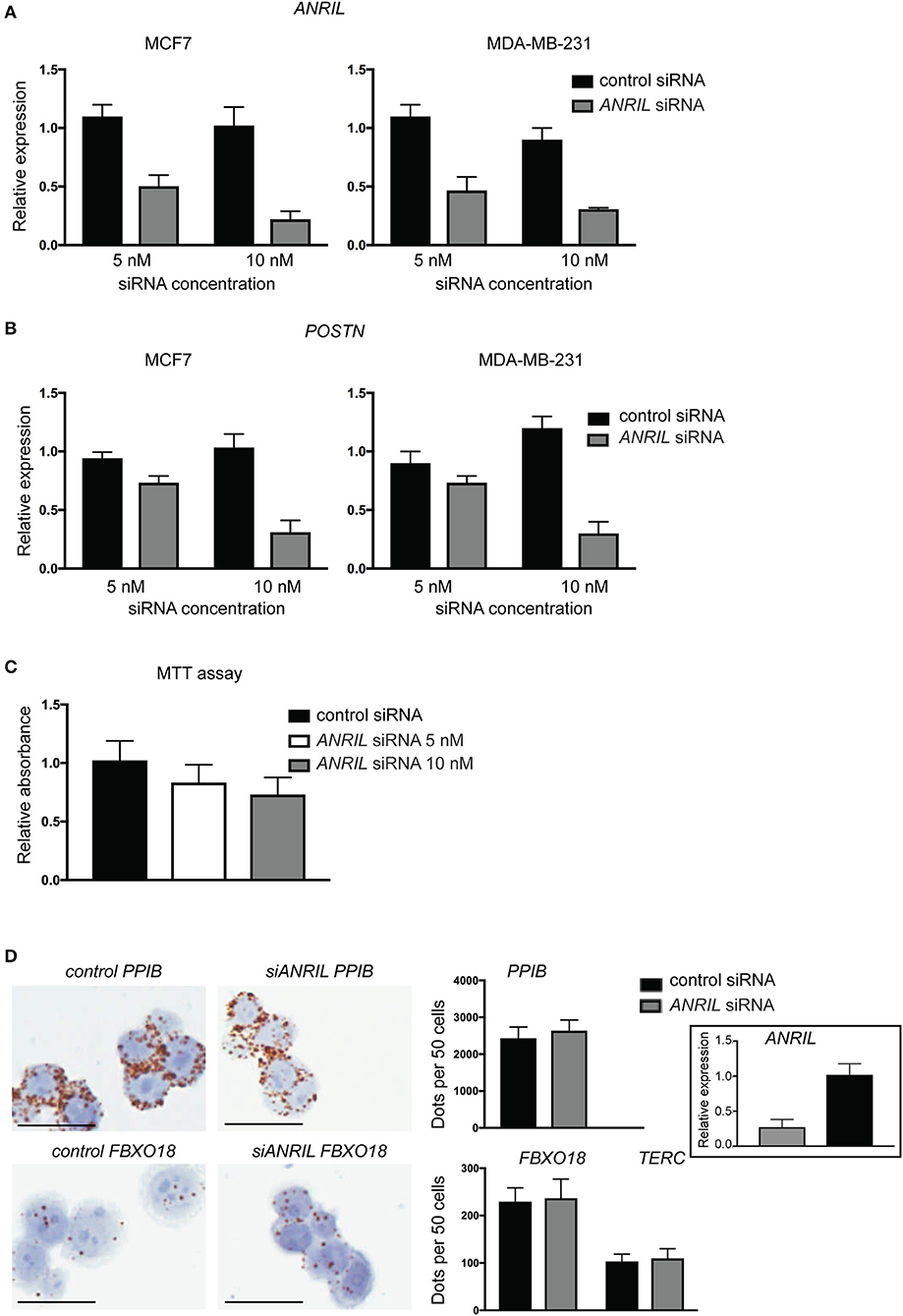
Figure 4. siRNA toward ANRIL in breast cancer cell lines is associated with reduced POSTN expression using RT-qPCR. (A) Breast cancer cell lines MCF7 and MDA-MB-231 were treated with siRNAs toward ANRIL or a negative control at two different concentrations. (B) Cells treated with siRNA toward ANRIL or the negative control were also measured for POSTN. All RT-qPCRs were normalized to GAPDH and HPRT and results expressed relative to expression found in untreated cells. (C) The MTT assay was performed on MCF7 cells to identify if siANRIL affected cell proliferation; no significance differences were found. (D) MCF7 cells were treated with siRNAs toward ANRIL or a negative control siRNA (control siRNA) at 10 nM, cells harvested, fixed, and processed into paraffin wax. Cell sections were subjected to RNAscope for genes not expected to be affected by siANRIL (PPIB, FBXO18, and TERC). No difference in positive staining (brown dots) was found in cells treated with the control siRNA compared to those treated with siANRIL. Scale bar, 25 μm taken at 400x magnification. Data represented as mean ± SD for at least three replicates of each sample. Boxed insert siRNA for PPIB and FBXO18 experiments.
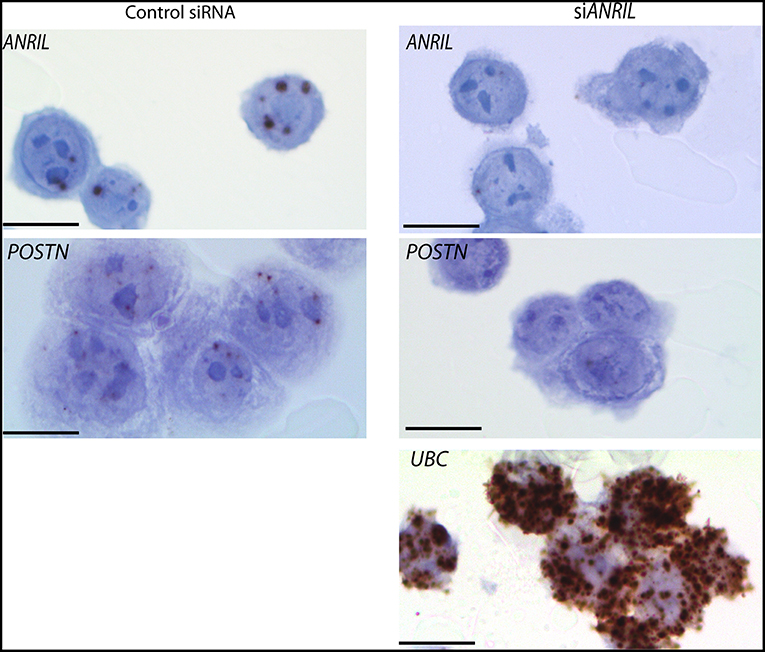
Figure 5. Reduced POSTN in MCF7 cells treated with siRNA toward ANRIL using RNAscope. The breast cancer cell line MCF7 was treated with siRNAs toward ANRIL or a negative control siRNA (control siRNA) at 10 nM, cells harvested, fixed, and processed into paraffin wax. Cell sections were subjected to RNAscope for ANRIL, POSTN, and UBC. Increased positive staining (brown dots) for ANRIL and POSTN was found in cells treated with the control siRNA. Scale bar, 20 μm taken at 1000x magnification under oil.
Discussion
ANRIL is a LncRNA recently found to be elevated in a range of cancers including breast cancer. In this study, we applied an in situ analysis and identified ANRIL largely localized to malignant cells with heterogeneity in its subcellular location. The correlation between POSTN and ANRIL was found when only tumors with nuclear ANRIL were included, suggesting the subcellular localization of ANRIL will be important in determining the potential effects of ANRIL toward cancer progression.
Further support toward ANRIL being a potential biomarker for breast cancer was evident with elevated ANRIL in the tumor compared to the normal adjacent tissue. Elevated ANRIL is most commonly found in triple negative breast tumors (12, 14, 16). The finding of elevated ANRIL in the tumor when compared to normal adjacent tissue from the same donor irrespective of the subtype suggests ANRIL may be more universally increased in breast cancer, but the highest levels occur in triple negative tumors. Finding ANRIL predominately in malignant cells provides further support toward ANRIL being a biomarker in breast cancer.
ANRIL may contribute to the tumor microenvironment by supplying a protein that remodels the extracellular matrix, promoting metastasis (27). A correlation between ANRIL and POSTN has been found in an investigation of genetic risk for cardiovascular disease (10). Periostin induces many other cancer promoting abilities when administered to various cell lines (27). The finding of nuclear but not cytoplasmic ANRIL associated with POSTN is consistent with different functions of ANRIL. One of the best-known mechanisms for ANRIL is gene silencing via epigenetic regulation involving recruiting and binding to the components of polycomb group of transcriptional repressor proteins (PcG), namely, PRC1 and PRC2 (14). Using chromatin fractionation, although not exclusively for ANRIL Ray et al. (28) found a number of lncRNAs involved in PcG biology to be primarily localized to the nucleus (28), and in a second study nuclear extracts of prostate cancer cells showed ANRIL association with CBX7 protein a member of the PRC1 complex (5). The tumor microenvironment may increase ANRIL. In fibroblast and osteosarcoma cell lines, ANRIL levels rise in response to a variety of DNA damaging agents via an ATM-induced E2F1 transcription-mediated pathway (29) and ANRIL contributes to pro-inflammatory cytokine expression by binding to YingYang-1 (YY1) in response to NFκB signaling (8). POSTN is regulated by YY1, so it is possible that periostin may be an additional cytokine to IL6/8 regulated by the YY1/ANRIL complex (30). Considering periostin is commonly produced by stromal cells the relevance of the contribution from malignant cells is unclear. In breast cancer POSTN was elevated in malignant cells and associated with poorer patient progression free and overall survival (31). In a second study POSTN was associated with a poorer prognosis in triple negative tumors (32). However, in other studies POSTN in stromal cells was thought to be important in breast cancer progression (32, 33).
The subcellular localization of ANRIL between breast tumors was heterogeneous. In an extensive survey of 61 LncRNA that included ANRIL in three cell lines revealed that individual LcRNAs showed cell-to-cell variability further extending the benefits of visualization LcRNAs directly (34). In cancer cell lines ANRIL was located in the nucleus with one or two large foci (34, 35) or in both the nucleus or cytoplasmic fraction (36). In non-transformed cells human umbilical vein endothelial cells (HUVEC) and normal lung tissue ANRIL was present in the nucleus and cytoplasm including perinuclear cytoplasmic staining (37, 38). Different isoforms of ANRIL exist, linear (exons 1–3, 15–20) or circular (exons 4–14) forms (39). In melanoma cell lines, linear isoforms of ANRIL were specifically found to be enriched in the nucleus and circular isoforms enriched in the cytoplasmic (36). The ANRIL probe designed by RNAscope spans from base pairs 9-1252 (exons 1–6) spanning exons in both the linear and cytoplasmic isoforms, thus the different subcellular localization found in breast tumors may reflect different isoforms being present.
Conclusion
This study highlights that ANRIL exists in different subcellular locations in breast tumors, which may affect its functionality, for example whether nuclear ANRIL is associated with increased POSTN expression in malignant epithelial cells.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
Ethics Committee Approval LRS/10/09/035.
Author Contributions
PM-M performed the RNAscope and qPCR analyzes, analyzed and interpreted the data, and co-wrote the manuscript. HC designed some of the analyses, interpreted the data, and edited the manuscript. NH designed the study, analyzed and interpreted the data, edited the manuscript, and obtained the ethical approval. TS conceived the study, analyzed and interpreted the data, obtained the funding, and co-wrote the manuscript.
Funding
HS and JC Anderson Charitable Trust.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Drs. Michael Landmann and Simone Petrich for contribution of cases to the study. Ms. Sankalita Ray is thanked for her assistance with siRNA experiments. Ms. Vicky Phillips, Ms. Amanda Fisher (Histology OMNI unit, University of Otago) for technical assistance on the project. Drs. Janice Royds, Anna Pilbow, and Margaret Currie and Ms. Helen Morrin for insightful discussion that aided the inception of this study.
References
1. Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. (2011) 11:735–48. doi: 10.1038/nrc3125
2. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
3. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. (2014) 1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012
4. Aguilo F, Cecilia SD, Walsh MJ. Long non-coding RNA ANRIL and polycomb in human cancers and cardiovascular disease. Curr Top Microbiol Immunol. (2016) 394:29–39. doi: 10.1007/82_2015_455
5. Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. (2010) 38:662–74. doi: 10.1016/j.molcel.2010.03.021
6. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. (2007) 67:3963–9. doi: 10.1158/0008-5472.CAN-06-2004
7. Qiao C, Yang L, Wan J, Liu X, Pang C, You W, et al. Long noncoding RNA ANRIL contributes to the development of ulcerative colitis by miR-323b-5p/TLR4/MyD88/NF-kappaB pathway. Biochem Biophys Res Commun. (2018) 508:217–24. doi: 10.1016/j.bbrc.2018.11.100
8. Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, et al. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. (2016) 13:98–108. doi: 10.1080/15476286.2015.1122164
9. Zhang B, Wang D, Ji TF, Shi L, Yu JL. Overexpression of lncRNA ANRIL up-regulates VEGF expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating NF-kappaB signaling pathway in a rat model. Oncotarget. (2017) 8:17347–59. doi: 10.18632/oncotarget.14468
10. Pilbrow AP, Folkersen L, Pearson JF, Brown CM, McNoe L, Wang NM, et al. The chromosome 9p21.3 coronary heart disease risk allele is associated with altered gene expression in normal heart and vascular tissues. PLoS ONE. (2012) 7:e39574. doi: 10.1371/journal.pone.0039574
11. Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. (2010) 42:504–7. doi: 10.1038/ng.586
12. Iranpour M, Soudyab M, Geranpayeh L, Mirfakhraie R, Azargashb E, Movafagh A, et al. Expression analysis of four long noncoding RNAs in breast cancer. Tumour Biol. (2015) 37:2933–40. doi: 10.1007/s13277-015-4135-2
13. Royds JA, Pilbrow AP, Ahn A, Morrin HR, Frampton C, Russell IA, et al. The rs11515 polymorphism is more frequent and associated with aggressive breast tumors with increased ANRIL and decreased p16 (INK4a) expression. Front Oncol. (2015) 5:306. doi: 10.3389/fonc.2015.00306
14. Meseure D, Vacher S, Drak Alsibai K, Nicolas A, Chemlali W, Caly M, et al. Expression of ANRIL - polycomb complexes - CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. (2016) 14:623–33. doi: 10.1158/1541-7786.MCR-15-0418
15. Liu M L., -Xing Q, Liu JY. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine. (2017) 96:e6222. doi: 10.1097/MD.0000000000006222
16. Xu ST, Xu JH, Zheng ZR, Zhao QQ, Zeng XS, Cheng SX, et al. Long non-coding RNA ANRIL promotes carcinogenesis via sponging miR-199a in triple-negative breast cancer. Biomed Pharmacother. (2017) 96:14–21. doi: 10.1016/j.biopha.2017.09.107
17. Zhang Z, Feng L, Liu P, Duan W. ANRIL promotes chemoresistance via disturbing expression of ABCC1 by regulating the expression of Let-7a in colorectal cancer. Biosci Rep. (2018) 38:BSR20180620. doi: 10.1042/BSR20180620
18. Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. (2014) 5:2276–92. doi: 10.18632/oncotarget.1902
19. Paul Y, Thomas S, Patil V, Kumar N, Mondal B, Hegde AS, et al. Genetic landscape of long noncoding RNA (lncRNAs) in glioblastoma: identification of complex lncRNA regulatory networks and clinically relevant lncRNAs in glioblastoma. Oncotarget. (2018) 9:29548–64. doi: 10.18632/oncotarget.25434
20. Tsuyada A, Chow A, Wu J, Somlo G, Chu P, Loera S, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. (2012) 72:2768–79. doi: 10.1158/0008-5472.CAN-11-3567
21. Kumari N, Dwarakanath BS, Das A, Bhatt AN. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. (2016) 37:11553–72. doi: 10.1007/s13277-016-5098-7
22. Gonzalez-Gonzalez L, Alonso J. Periostin: a matricellular protein with multiple functions in cancer development and progression. Front Oncol. (2018) 8:225. doi: 10.3389/fonc.2018.00225
23. Tang P, Tse GM. Immunohistochemical surrogates for molecular classification of breast carcinoma: a 2015 update. Arch Pathol Lab Med. (2016) 140:806–14. doi: 10.5858/arpa.2015-0133-RA
24. Li J, Smyth P, Cahill S, Denning K, Flavin R, Aherne S, et al. Improved RNA quality and TaqMan(®)Pre-amplification method (PreAmp) to enhance expression analysis from formalin fixed paraffin embedded (FFPE) materials. BMC Biotechnol. (2008) 8:10. doi: 10.1186/1472-6750-8-10
25. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protocols. (2008) 3:1101–8. doi: 10.1038/nprot.2008.73
26. Lee J, Jung JH, Chae YS, Park HY, Kim WW, Lee SJ, et al. Long noncoding RNA snaR regulates proliferation, migration and invasion of triple-negative breast cancer cells. Anticancer Res. (2016) 36:6289–95. doi: 10.21873/anticanres.11224
27. González-González L, Alonso J. Periostin: a matricellular protein with multiple functions in cancer development and progression. Front. Oncol. (2018) 8:225.
28. Ray MK, Wiskow O, King MJ, Ismail N, Ergun A, Wang Y, et al. CAT7 and cat7l Long Non-coding RNAs tune polycomb repressive complex 1 function during human and zebrafish development. J Biol Chem. (2016) 291:19558–72. doi: 10.1074/jbc.M116.730853
29. Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, et al. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. (2013) 25:1086–95. doi: 10.1016/j.cellsig.2013.02.006
30. Romeo F, Falbo L, Di Sanzo M, Misaggi R, Faniello MC, Barni T, et al. Negative transcriptional regulation of the human periostin gene by YingYang-1 transcription factor. Gene. (2011) 487:129–34. doi: 10.1016/j.gene.2011.07.025
31. Kim GE, Lee JS, Park MH, Yoon JH. Epithelial periostin expression is correlated with poor survival in patients with invasive breast carcinoma. PLoS ONE. (2017) 12:e0187635. doi: 10.1371/journal.pone.0187635
32. Lambert AW, Wong CK, Ozturk S, Papageorgis P, Raghunathan R, Alekseyev Y, et al. Tumor cell-derived periostin regulates cytokines that maintain breast cancer stem cells. Mol Cancer Res. (2016) 14:103–13. doi: 10.1158/1541-7786.MCR-15-0079
33. Ratajczak-Wielgomas K, Grzegrzolka J, Piotrowska A, Matkowski R, Wojnar A, Rys J, et al. Expression of periostin in breast cancer cells. Int J Oncol. (2017) 51:1300–10. doi: 10.3892/ijo.2017.4109
34. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. (2015) 16:20. doi: 10.1186/s13059-015-0586-4
35. Shinjo K, Yamashita Y, Yamamoto E, Akatsuka S, Uno N, Kamiya A, et al. Expression of chromobox homolog 7 (CBX7) is associated with poor prognosis in ovarian clear cell adenocarcinoma via TRAIL-induced apoptotic pathway regulation. Int J Cancer. (2014) 135:308–18. doi: 10.1002/ijc.28692
36. Sarkar D, Oghabian A, Bodiyabadu PK, Joseph WR, Leung EY, Finlay GJ, et al. Multiple isoforms of ANRIL in melanoma cells: structural complexity suggests variations in processing. Int J Mol Sci. (2017) 18:1378. doi: 10.3390/ijms18071378
37. Thomas AA, Feng B, Chakrabarti S. ANRIL: a regulator of VEGF in diabetic retinopathy. Invest Ophthalmol Visual Sci. (2017) 58:470–80. doi: 10.1167/iovs.16-20569
38. Kang YH, Kim D, Jin EJ. Down-regulation of phospholipase D stimulates death of lung cancer cells involving up-regulation of the long ncRNA ANRIL. Anticancer Res. (2015) 35:2795–803.
Keywords: LncRNA, ANRIL, breast cancer, periostin, CCL2, IL-6, RNAscope
Citation: Mehta-Mujoo PM, Cunliffe HE, Hung NA and Slatter TL (2019) Long Non-coding RNA ANRIL in the Nucleus Associates With Periostin Expression in Breast Cancer. Front. Oncol. 9:885. doi: 10.3389/fonc.2019.00885
Received: 18 March 2019; Accepted: 27 August 2019;
Published: 11 September 2019.
Edited by:
Ivan Garcia-Bassets, University of California, San Diego, United StatesReviewed by:
Chunru Lin, University of Texas MD Anderson Cancer Center, United StatesWenbo Li, University of Texas, United States
Qidong Hu, National University of Singapore, Singapore
Copyright © 2019 Mehta-Mujoo, Cunliffe, Hung and Slatter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tania L. Slatter, dGFuaWEuc2xhdHRlckBvdGFnby5hYy5ueg==
 Paulomi M. Mehta-Mujoo
Paulomi M. Mehta-Mujoo Heather E. Cunliffe
Heather E. Cunliffe Noelyn A. Hung
Noelyn A. Hung Tania L. Slatter
Tania L. Slatter