- 1Department of Epidemiology, School of Public Health, China Medical University, Shenyang, China
- 2Key Laboratory of Cancer Etiology and Prevention, Liaoning Provincial Department of Education, China Medical University, Liaoning, China
Background: Recent studies have investigated the relationships between PLCE1 polymorphisms and cancer susceptibility. However, some findings lack consistency.
Objectives: In the current study, we conducted a meta-analysis to more accurately evaluate the relationships between PLCE1 (rs2274223, rs3765524, rs753724, rs11187842, and rs7922612) single nucleotide polymorphisms (SNPs) and risk for different types of cancer.
Methods: We performed a comprehensive search strategy in PubMed, Web of Science, Medline, EMbase, and Scopus for articles available until 19 March 2018. A total of 54 case-control studies comprising 17,955 cases and 20,400 controls were included in the current meta-analysis, which together comprised a total of 32 publications. The pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate relationships between the PLCE1 polymorphisms and cancer susceptibility. All statistical analyses were performed using Stata 11 software.
Results: Results of the meta-analysis demonstrated that the rs2274223 polymorphism showed a significant correlation with increased overall cancer susceptibility (AG vs. AA: OR 1.168, 95% CI 1.084–1.259; GG vs. AA: OR 1.351, 95% CI 1.163–1.570; AG+GG vs. AA: OR 1.193, 95% CI 1.103–1.290; GG vs. AA+AG: OR 1.262, 95% CI 1.102–1.446; G vs. A: OR 1.163, 95% CI 1.089–1.242). Results of subgroup analysis showed that the rs2274223 polymorphism was associated with higher risk for esophageal cancer and gastric cancer relative to colorectal cancer and head and neck cancer. In addition, the rs2274223 polymorphism was found to be associated with increased cancer risk, especially among the subgroups comprising Asians, studies with population-based controls, studies employing the TaqMan genotyping method, and studies consistent with Hardy-Weinberg equilibrium (HWE). The association between the rs3765524 polymorphism and reduced overall cancer risk was detected in one specific genetic model (CT vs. CC: OR 0.681, 95% CI 0.523–0.886). Results of subgroup analysis showed that the rs3765524 polymorphism was associated with cancer risk in a specific genetic model among the subgroups of colorectal cancer, esophageal cancer, Asians, studies with population-based controls, and studies consistent with HWE. However, relationships among the PLCE1 rs753724, rs11187842, and rs7922612 polymorphisms and tumor risk were not identified.
Conclusions: Results of the current meta-analysis suggested that PLCE1 (rs2274223, rs3765524) polymorphisms are associated with cancer susceptibility.
Introduction
Cancer has become a major threat to public health worldwide (1). In 2018, there is a predicted 1,735,350 new cancer cases, which are equivalent to over 4,700 new cancer diagnoses each day in the United States, which correspond to an expected 609,640 cancer deaths (2). In addition, cancer has become the leading cause of death in China (3). Therefore, there is an urgent need to investigate cancer, identify relevant biomarkers, and develop strategies for active prevention and early diagnosis and treatment. Cancer is well-established to be the result of a combination of genetic and environmental factors. In the last few decades, extensive experimental and epidemiological findings demonstrated the close association between genetic alterations and tumor risk (4). Single nucleotide polymorphisms (SNPs), the most common form of gene alteration in the human genome, refers to single-nucleotide variations with distribution frequencies that are >1% in the population.
Phospholipase C epsilon1 (PLCE1), which is located on chromosome 10q23, is a member of the phospholipase C protein family (5). In 2010, the results of genome-wide association studies indicated that PLCE1 is associated with cancer risk (6, 7). Since then, multiple researchers investigated the relationship between PLCE1 polymorphisms and cancer risk. Cui et al. (8) explored the association between PLCE1 polymorphisms and risk for esophageal squamous cell carcinoma. Li (9), Zhang (10) and other authors investigated the relationship between PLCE1 polymorphisms and colorectal cancer risk. Yuan (11), Malik (12) and other authors investigated the association between PLCE1 polymorphisms and gastric carcinoma risk. Sharma (13) and other authors showed that PLCE1 polymorphisms were associated with susceptibility to developing gall bladder cancer. Among all studies that investigated PLCE1 polymorphisms and cancer susceptibility, the SNPs rs2274223, rs3765524, rs753724, rs11187842, and rs7922612 were five of the most extensively studied polymorphic loci. However, we noted significant differences in the results, sample size, race, or selection of controls among the different studies. In addition, the latest meta-analysis on the relationship between the rs2274223 polymorphism and the overall cancer risk was published in 2015 (14). Furthermore, to the best of our knowledge, no studies conducted meta-analysis of the association of rs3765524, rs753724, rs11187842, and rs7922612 polymorphisms with overall cancer risk. Therefore, in the present study, we summarized all currently qualified case-control studies to obtain a more accurate understanding of the relationship between the PLCE1 polymorphism rs2274223 and overall cancer risk [(15) studies were added to the current meta-analysis from the meta-analysis published in 2015 (14)]. And we firstly performed a meta-analysis of the association between the rs3765524, rs753724, rs11187842, and rs7922612 polymorphisms and cancer risk in the overall population.
Methods
Literature Search
We carried a comprehensive search strategy to retrieve qualified publications from PubMed and Web of Science until 19 March 2018. The search queries comprised a combination of the Medical Subject Headings (MeSH) and the following keywords: (rs2274223 OR rs3765524 OR rs753724 OR rs11187842 OR rs7922612) OR (PLCE1 OR PLCE OR PPLC OR NPHS3) and (cancer OR tumor OR carcinoma OR neoplasm OR malignancy). In addition, we searched literatures from Medline, EMbase, and Scopus, as complementary data. The references of qualified articles or other reviews were additionally searched. For publications with no available original data, we contacted the authors to ensure that data from all qualified literatures were included in the current meta-analysis. The authors of three out of six publications responded.
Inclusion Criteria and Exclusion Criteria
The inclusion criteria for qualified literatures were as follows: (a) The studies evaluated the associations between PLCE1 polymorphisms (rs2274223 or rs3765524 or rs753724 or rs11187842 or rs7922612) and cancer risk. (b) The study had available genotyping data required for the calculation of the odds ratios (ORs) with 95% confidence intervals (95% CIs). (c) The studies were case-control studies. (d) Studies were complete original articles. Exclusion criteria of qualified literatures were as follows: (a) Articles did not estimate the relationships between the PLCE1 (rs2274223, rs3765524, rs753724, rs11187842, or rs7922612) polymorphisms and cancer susceptibility. (b) The article was a repeated publication. (d) Primary data were missing and were not obtained after contacting the authors. (e) The subjects were not human. Two researchers independently retrieved the literature. In the case of different views in the selected literature, the two researchers discussed to reach an agreement or the decision was made by an independent researcher (Xuelian Li).
Reporting Items
Two investigators independently gathered data from each selected article, including the first author, year, country, ethnicity, tumor type, genotyping methods, the source of control, number of cases and controls, and the P-values of the HWE test of the controls. In the case of different views, the two researchers reached an agreement through discussion.
Quality Score Assessment
All qualified literatures were individually assessed by two researchers based on the Newcastle-Ottawa scale (NOS) (16). The assessment results indicated that all selected literatures were of relatively high quality (all NOS scores were ≥6). In addition, the two researchers assessed the quality of the studies using the STREGA (strengthening report of genetic association studies) quality score system (15). All STREGA scores were >12, which indicated that the quality of the studies was moderate-high or high.
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) was examined by performing a Chi-square test in the controls. Heterogeneity was evaluated by conducting Q-test and I2-test. In addition, the pooled ORs with 95% CIs were calculated based on the random effects model when heterogeneity was significant (I2 > 50%) (17). Otherwise, pooled ORs with 95% CIs were calculated according to the fixed-effects model (18). The pooled ORs with 95% CIs were used to evaluate relationships between the PLCE1 polymorphisms (rs2274223, rs3765524, rs753724, rs11187842, and rs7922612) and cancer susceptibility. To investigate the potential sources of heterogeneity across different studies, stratification and meta-regression analyses were conducted. Sensitivity analyses were carried out to evaluate the stability of the results. The effect of publication bias was evaluated using Begg's funnel plot (19) and Egger's test (20). All the above analyses were performed using Stata 11 software. P < 0.05 was considered statistically significant.
Results
Study Characteristics
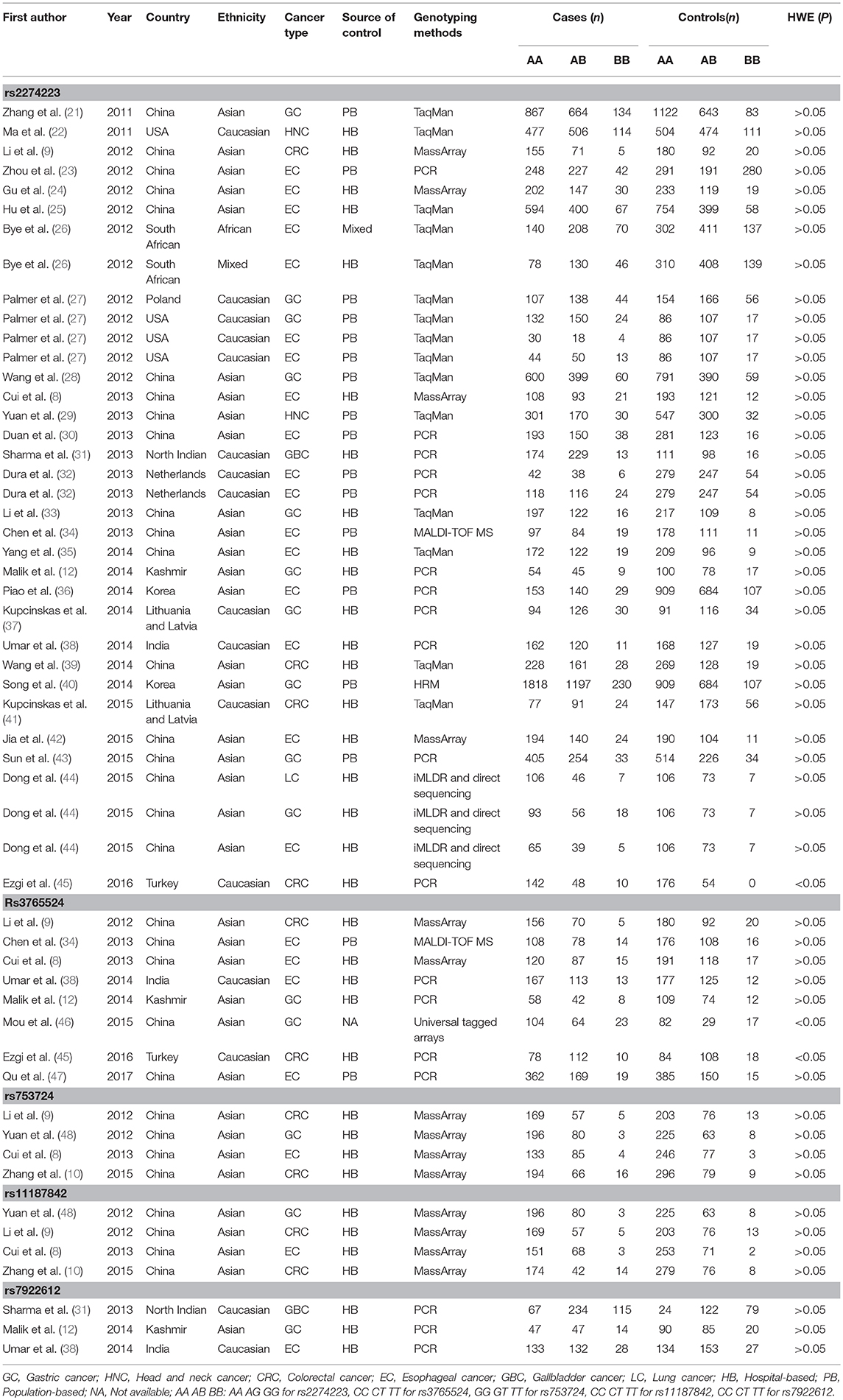
A total of 32 literatures were eventually included based on the above described comprehensive search strategy. The workflow of the enrollment in the meta-analysis is presented in Figure 1. A total of 54 case-control studies comprising 17,955 cases and 20,400 controls were included in the 32 publications. Five target SNPs were investigated in the current meta-analysis. The main characteristics of the 54 case-control studies and the genotype distribution information of the five polymorphisms are summarized in Table 1. The rs2274223, rs3765524, rs753724, and rs11187842, rs7922612 polymorphisms were involved in 35 (8, 9, 12, 21–45), eight (8, 9, 12, 34, 38, 45–47), four (8–10, 48), four (8–48), and three studies (12, 31, 38), respectively. The different cancer types investigated included gastric cancer, head and neck cancer, colorectal cancer, esophageal cancer, gall bladder cancer, and lung cancer. Of all case-control studies, only genotype frequencies of three studies among the controls were not consistent with HWE (45, 46). A total of 36 studies involved Asians; 16 studies involved Caucasians; one study involved Africans; and one study involved individuals of mixed ancestry. Meanwhile, 35 studies were hospital-based, and 17 studies were population-based. All studies were case-control studies.
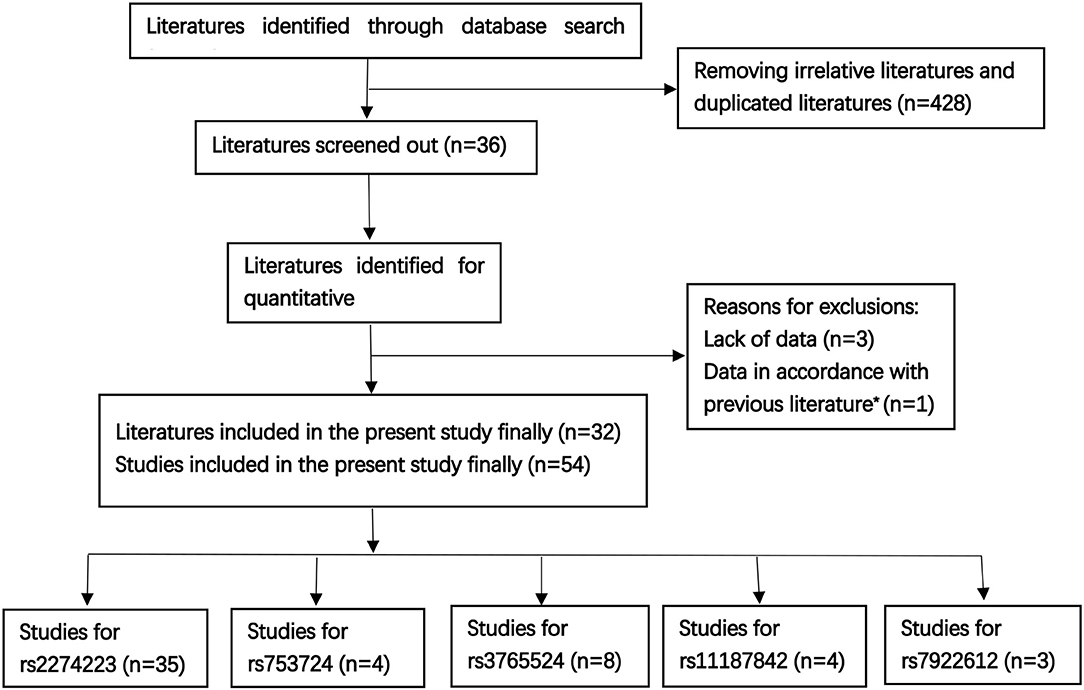
Figure 1. The workflow of the enrollment [Literatures sources: PubMed n = 112, Web of Science n = 201, other literatures were supplemented by Medline, EMbase, and Scopus, as well as the references of qualified literatures or other reviews; previous literature* (13)].
Meta-Analysis of the Relationship Between the PLCE1 rs2274223 Polymorphism and Cancer Risk
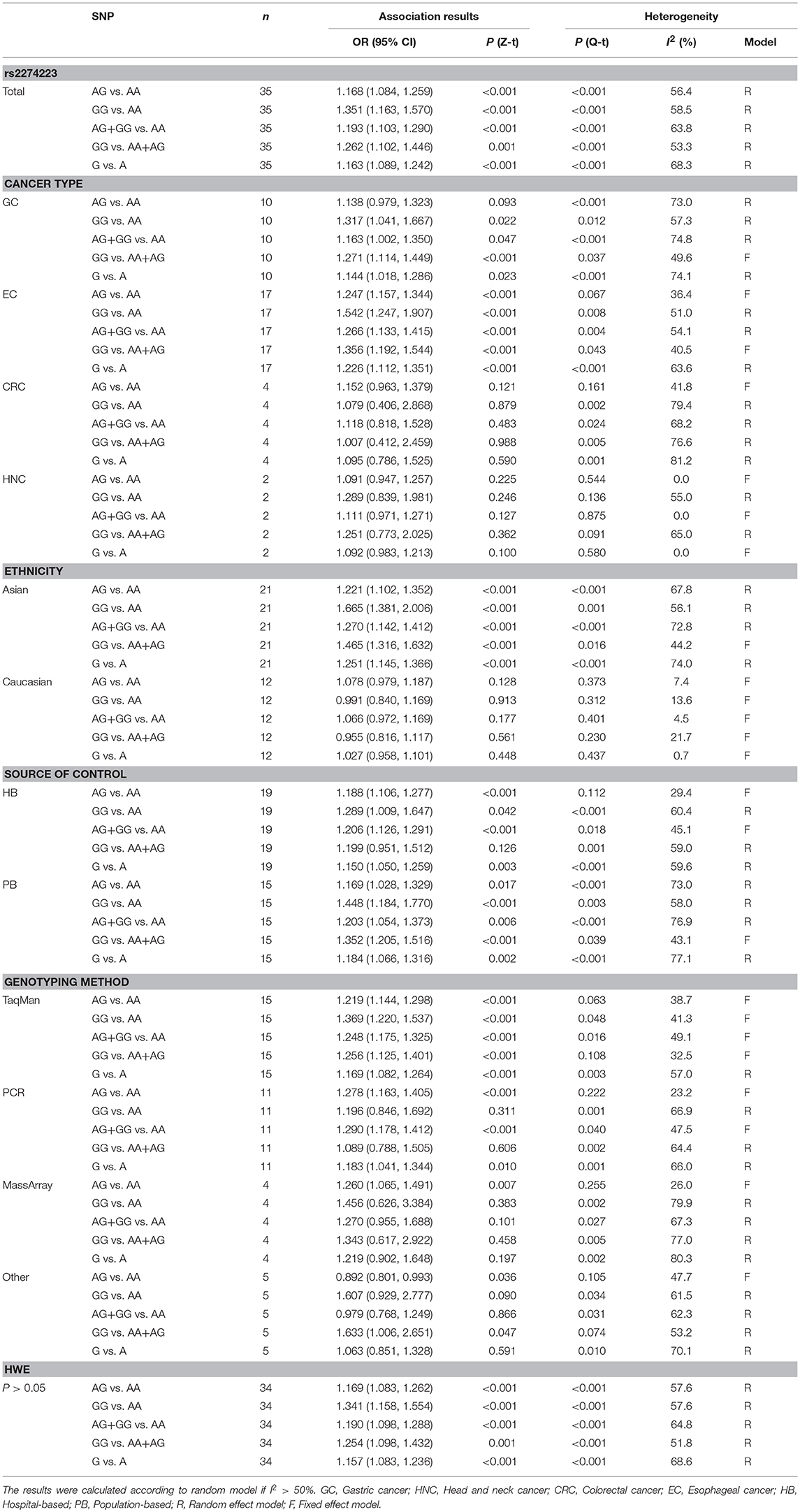
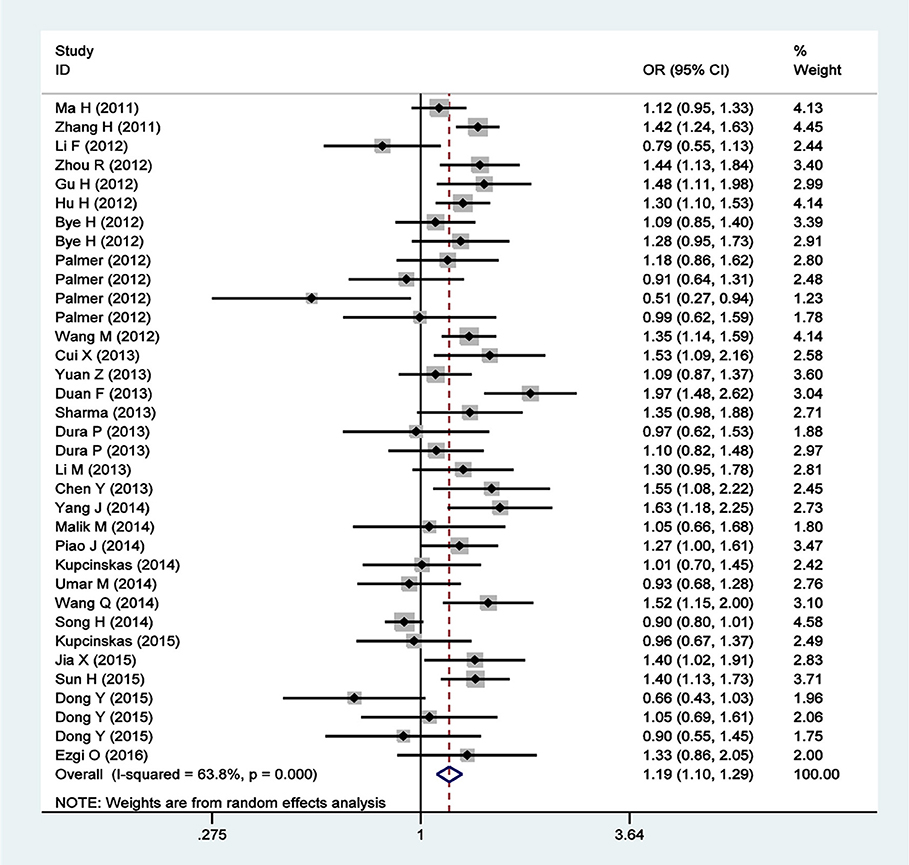
A total of 35 qualified case-control studies were included in this meta-analysis, which assessed the relationship between the PLCE1 rs2274223 polymorphism and cancer susceptibility. We evaluated heterogeneity and selected the random effects model or the fixed-effects model based on the results of Q-test and I2-values. Results of the meta-analysis of the relationship between the PLCE1 rs2274223 polymorphism and cancer risk are shown in Table 2 and Figure 2. Results showed a correlation between the rs2274223 polymorphism with significantly increased overall cancer susceptibility in all genetic models [AG vs. AA: OR 1.168, 95% CI 1.084–1.259 (P < 0.001); GG vs. AA: OR 1.351, 95% CI 1.163–1.570 (P < 0.001); AG+GG vs. AA: OR 1.193, 95% CI 1.103–1.290 (P < 0.001); GG vs. AA+AG: OR 1.262, 95% CI 1.102–1.446 (P = 0.001); G vs. A: OR 1.163, 95% CI 1.089–1.242 (P < 0.001)]. To further study the association between rs2274223 polymorphism and cancer risk, we carried out stratified analyses according to cancer type, ethnicity, the source of control, genotyping methods, and HWE. The results of subgroup analyses are also shown in Table 2. Results according to the cancer type indicated that the rs2274223 polymorphism was associated with a higher risk of gastric cancer in four genetic models [GG vs. AA: OR 1.317, 95% CI 1.041–1.667 (P = 0.022); AG+GG vs. AA: OR 1.163, 95% CI 1.002–1.350 (P = 0.047); GG vs. AA+AG: OR 1.271, 95% CI 1.114–1.449 (P < 0.001); G vs. A: OR 1.144, 95% CI 1.018–1.286 (P = 0.023)]. Meanwhile, the rs2274223 polymorphism was related to a significantly increased risk of esophageal cancer in all genetic models [AG vs. AA: OR 1.247, 95% CI 1.157–1.344 (P < 0.001); GG vs. AA: OR 1.542, 95% CI 1.247–1.907 (P < 0.001); AG+GG vs. AA: OR 1.266, 95% CI 1.133–1.415 (P < 0.001); GG vs. AA+AG: OR 1.356, 95% CI 1.192–1.544 (P < 0.001); G vs. A: OR 1.226, 95% CI 1.112–1.351 (P < 0.001)]. However, we found no statistically significant associations between the rs2274223 polymorphism and risks of head and neck cancer and colorectal cancer. The results of subgroup analyses according to ethnicity indicated that the rs2274223 polymorphism increased cancer susceptibility in Asians [AG vs. AA: OR 1.221, 95% CI 1.102–1.352 (P < 0.001); GG vs. AA: OR 1.665, 95% CI 1.381–2.006 (P < 0.001); AG+GG vs. AA: OR 1.270, 95% CI 1.142–1.412 (P < 0.001); GG vs. AA+AG: OR 1.465, 95% CI 1.316–1.632 (P < 0.001); G vs. A: OR 1.251, 95% CI 1.145–1.366 (P < 0.001)]. However, the association between rs2274223 polymorphism and cancer risk was not identified in Caucasians. The results of subgroup analyses based on the source of control showed that the rs2274223 polymorphism was associated with significantly increased risk of tumor in hospital-based subgroup [AG vs. AA: OR 1.188, 95% CI 1.106–1.277 (P < 0.001); GG vs. AA: OR 1.289, 95% CI 1.009–1.647 (P = 0.042); AG+GG vs. AA: OR 1.206, 95% CI 1.126–1.291 (P < 0.001); G vs. A: OR 1.150, 95% CI 1.050–1.259 (P = 0.003)]. Meanwhile, the statistically significant relationship between the rs2274223 polymorphism and cancer susceptibility was also detected in the population-based subgroup [AG vs. AA: OR 1.169, 95% CI 1.028–1.329 (P = 0.017); GG vs. AA: OR 1.448, 95% CI 1.184–1.770 (P < 0.001); AG+GG vs. AA: OR 1.203, 95% CI 1.054–1.373 (P = 0.006); GG vs. AA+AG: OR 1.352, 95% CI 1.205–1.516 (P < 0.001); G vs. A: OR 1.184, 95% CI 1.066–1.316 (P = 0.002)]. The results of subgroup analyses based on genotyping methods indicated that rs2274223 polymorphism might increase tumor risk in all genetic models in TaqMan subgroup. In addition, the rs2274223 polymorphism was related to a significantly higher risk of tumor in three genetic models in the PCR subgroup [AG vs. AA: OR 1.278, 95% CI 1.163–1.405 (P < 0.001); AG+GG vs. AA: OR 1.290, 95% CI 1.178–1.412 (P < 0.001); G vs. A: OR 1.183, 95% CI 1.041–1.344 (P = 0.010)]. However, the rs2274223 polymorphism was associated with tumor risk in a few genetic models in MassArray subgroup and other subgroup. Out of the 35 qualified case-control studies, only one study did not satisfy the HWE. After removing this study, the statistically significant association between rs2274223 polymorphism and cancer risk still existed [AG vs. AA: OR 1.169, 95% CI 1.083–1.262 (P < 0.001); GG vs. AA: OR 1.341, 95% CI 1.158–1.554 (P < 0.001); AG+GG vs. AA: OR 1.190, 95% CI 1.098–1.288 (P < 0.001); GG vs. AA+AG: OR 1.254, 95% CI 1.098–1.432 (P = 0.001); G vs. A: OR 1.157, 95% CI 1.083–1.236 (P < 0.001)].
Meta-Analysis of the Association Between PLCE1 rs3765524 Polymorphism and Cancer Susceptibility
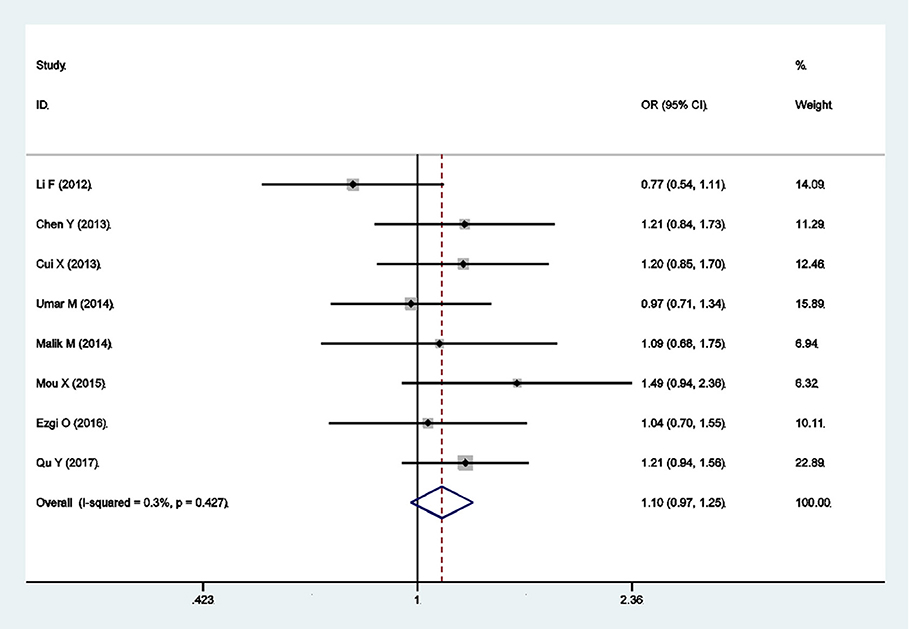
There were eight qualified case-control studies in this meta-analysis, which assessed the relationship between the PLCE1 rs3765524 polymorphism and cancer susceptibility. The results of meta-analysis on the relationship between the PLCE1 rs3765524 polymorphism and cancer risk are summarized in Table 3 and Figure 3. The association between the rs3765524 polymorphism and overall cancer risk was identified in one genetic model [CT vs. CC: OR 0.681, 95% CI 0.523–0.886 (P = 0.004)]. The results of subgroup analyses are shown in Table 3. The results of subgroup analyses based on cancer type showed that the rs3765524 polymorphism was associated with risk of esophageal cancer in two genetic models [CT vs. CC: OR 0.611, 95% CI 0.515–0.726 (P < 0.001); T vs. C: OR 1.154, 95% CI 1.014–1.313 (P = 0.029)]. In addition, the rs3765524 polymorphism was associated with colorectal cancer susceptibility in the specific genetic models [TT vs. CC: OR 0.431, 95% CI 0.229–0.811 (P = 0.009); TT vs. CT+CC: OR 0.429, 95% CI 0.232–0.794 (P = 0.007)]. However, the observed relationship between rs3765524 polymorphism and risk of gastric cancer was not statistically significant. Subgroup analyses according to ethnicity identified an association between the rs3765524 polymorphism and cancer risk in Asians [CT vs. CC: OR 0.579, 95% CI 0.492–0.680 (P < 0.001)]. However, the association was not statistically significant in the Caucasian population. The results of stratified analyses based on the source of controls showed that the CT genotype of rs3765524 decreased cancer susceptibility in the population-based subgroup relative to CC genotype [CT vs. CC: OR 0.568, 95% CI 0.371–0.870 (P = 0.009)]. However, the results of stratified analyses were not statistically significant in the hospital-based subgroup. The results of subgroup analyses based on genotyping method indicated that the rs3765524 polymorphism is not associated with tumor risk in each subgroup. Finally, we carried out subgroup analyses based on HWE. In the subgroup whose genotype frequencies among controls was consistent with HWE, the rs3765524 polymorphism was associated with cancer risk in only one genetic model [CT vs. CC: OR 0.594, 95% CI 0.511–0.691 (P < 0.001)]. However, the results were not statistically significant in the subgroup whose genotype frequencies among controls were not consistent with HWE.
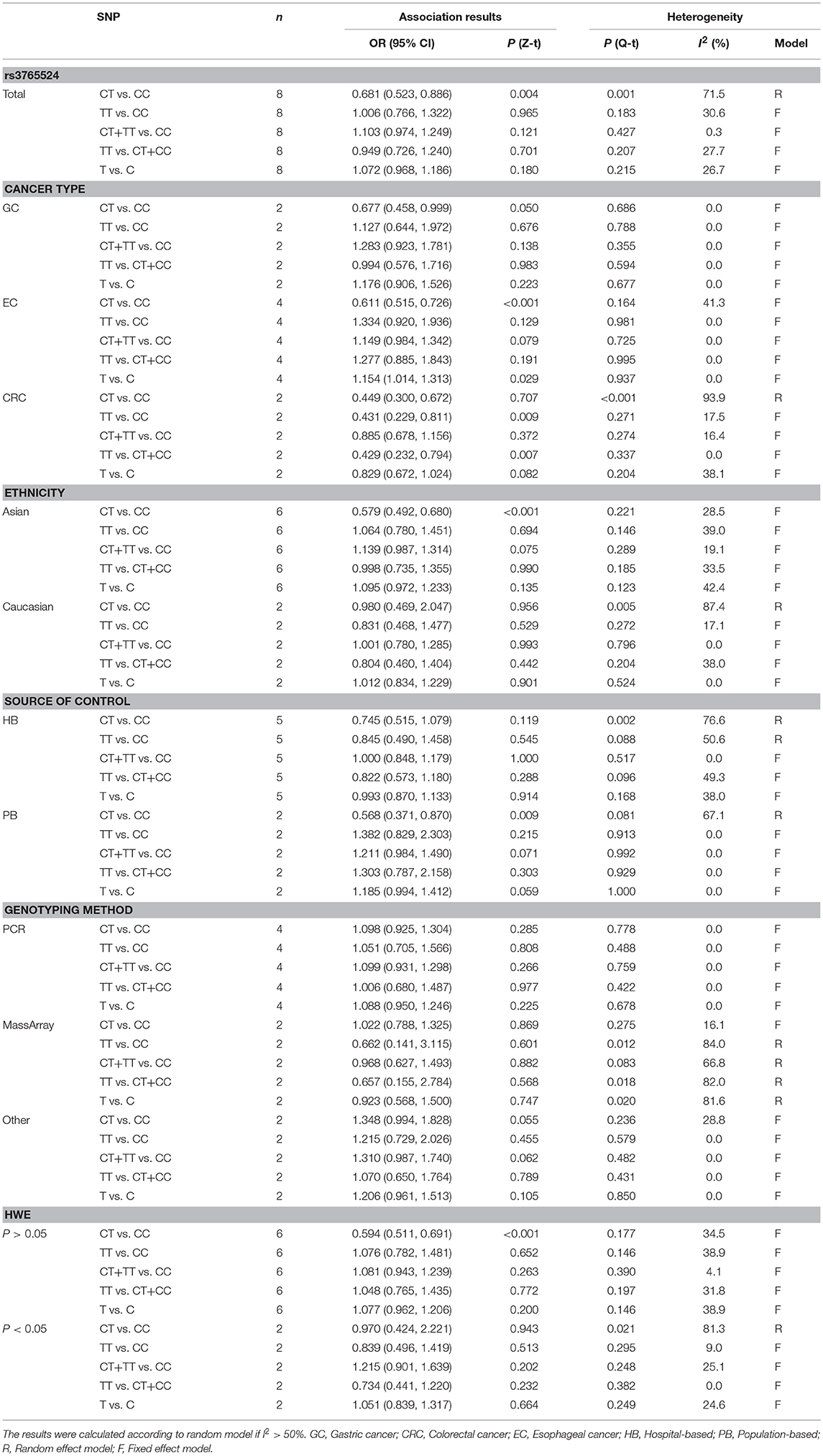
Table 3. Meta-analysis of the association between PLCE1 rs3765524 polymorphism and cancer susceptibility.
Meta-Analysis of the Relationship Between the PLCE1 rs753724, rs11187842, and rs7922612 Polymorphisms and Cancer Risk
The rs753724, rs11187842, and rs7922612 polymorphisms were involved in four, four, and three case-control studies, respectively. The results of meta-analysis on the association between PLCE1 (rs753724, rs11187842, and rs7922612) polymorphisms and cancer risk are summarized in Table 4 and Figure S1. Results indicated no significant relationship between the PLCE1 rs753724, rs11187842, and rs7922612 polymorphisms and cancer risk. In addition, further subgroup analysis did not identify statistically significant relationships in any genetic model.
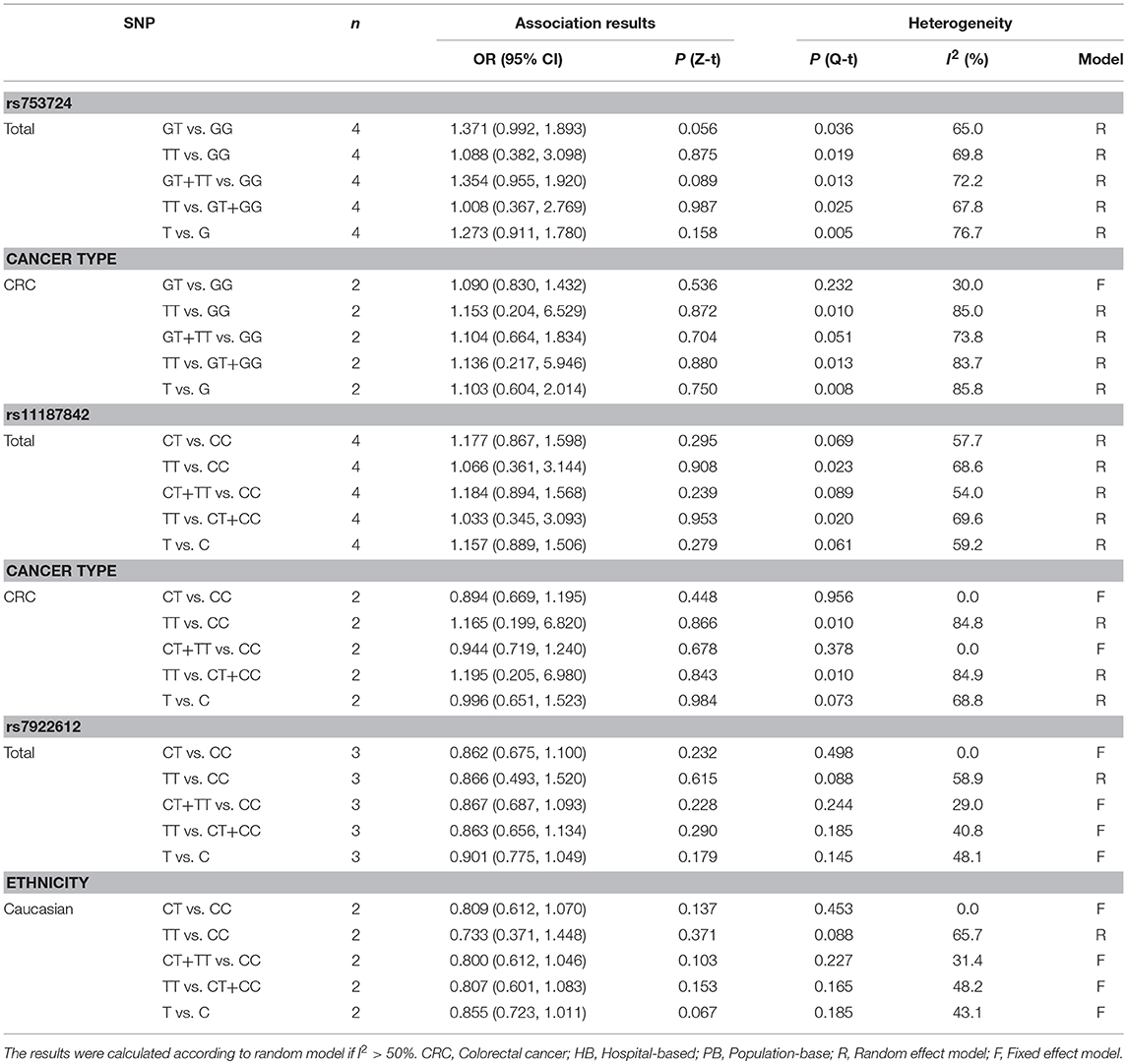
Table 4. Meta-analysis of the relationship between PLCE1 rs753724, rs11187842, rs7922612 polymorphisms and cancer risk.
Heterogeneity, Sensitivity Analysis, and Publication Bias
Substantial heterogeneities were identified in our meta-analysis. For example, we observed significant heterogeneity in the overall analysis for rs2274223 (I2 > 50%). Therefore, we conducted meta-regression analyses to investigate the source of heterogeneity for rs2274223. Results suggested that ethnicity is the likely source of heterogeneity for rs2274223 in the three genetic models (GG vs. AA: P = 0.009; GG vs. AA+AG: P = 0.009; G vs. A: P = 0.048). The genotyping method is a possible source of heterogeneity for rs2274223 in one genetic model (AG vs. AA: P = 0.020) (Table S1). The results of stratified analyses for rs2274223 were basically consistent with results of meta-regression. However, identifying the source of heterogeneity for rs3765524, rs753724, rs11187842, and rs7922612 was difficult based on stratified analyses. Further, results of sensitivity analysis suggested that the results of meta-analysis were not influenced by any single study in all genetic models for all five polymorphisms, which indicated that our analysis was robust and stable (Figure 4). Next, publication bias was evaluated by Egger's test and funnel plot. (Table S2 and Figure S2). The results of Egger's test showed that all P-values were >0.05 and that the funnel plots were relatively symmetrical, indicating no publication bias was detected in the current analysis.
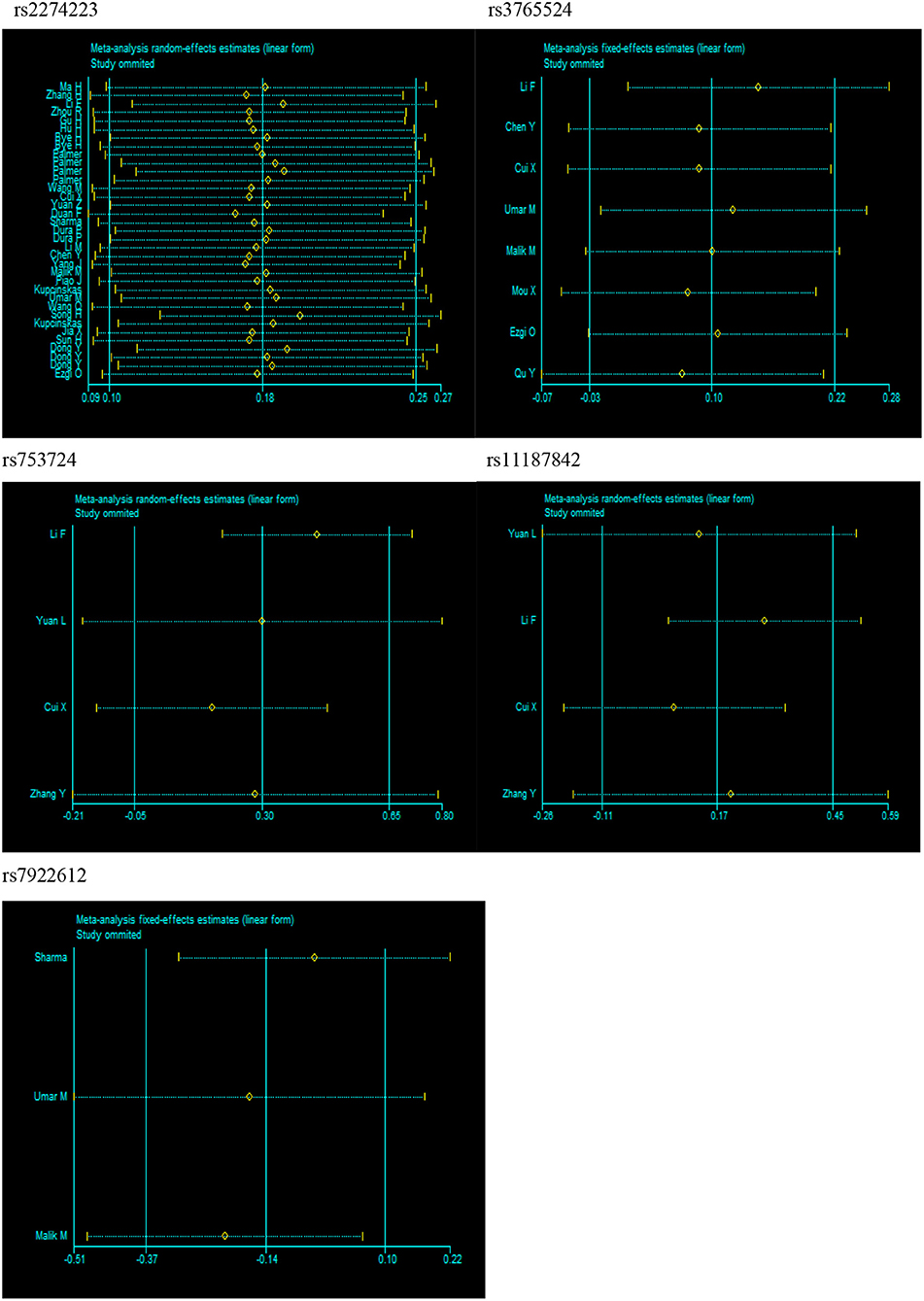
Figure 4. Sensitivity analysis for PLCE1 polymorphisms and cancer risk in dominant model (rs2274223: AG+GG vs. AA; rs3765524: CT+TT vs. CC; rs753724: GT+TT vs. GG; rs11187842: CT+TT vs. CC; rs7922612: CT+TT vs. CC).
Discussion
Tumor pathogenesis involves both genetic and environmental factors. As the effects of genetic mutations on cancer continued to be revealed, many authors have focused on the associations between SNPs and cancer susceptibility. PLCE1 is one of the members of the phospholipase C protein family, which can interact with Ras and participate in cellular signal transduction, produce secondary messengers by hydrolyzing phosphatidylinositol-4, 5-bisphosphate, and regulate cell growth, differentiation, apoptosis, and angiogenesis. (49–53) The role of PLCE1 in cancer remains controversial. The studies of Wang et al. (54, 55) demonstrated that PLCE1 plays a tumor suppressor role in colorectal carcinoma. However, some studies indicated that PLCE1 acts as an oncogene in numerous cancers, such as non-small cell lung cancer (56) and head and neck cancer (57). Recent years have witnessed an increasing number of studies that investigate PLCE1 polymorphisms and cancer susceptibility. Likewise, several meta-analyses assessed the association between PLCE1 polymorphisms and cancer risk. However, most of these studies focused on the relationship between PLCE1 polymorphisms and digestive tract cancer rather than the overall tumor risk.
Our current findings showed that the rs2274223 polymorphism was associated with overall tumor susceptibility in five genetic models, consistent with the results reported by Xue et al. (14). However, the current results were slightly different from those reported by Umar (58), in which the rs2274223 polymorphism showed no significant association with overall cancer susceptibility in one specific genetic model (GG vs. AG+AA). Further stratified analysis revealed that the rs2274223 polymorphism was associated with gastric cancer and esophageal cancer susceptibility, but not with other types of cancer. The above findings were consistent with those reported by Umar (58), but slightly different from the findings of Xue et al. (14), which suggested that rs2274223 polymorphism was not associated with susceptibility to gastric cancer. The results based on the esophageal cancer subgroup were consistent with the results of Wang et al. (59) and Guo et al. (60). Moreover, the results of the stratified analysis indicated that the rs2274223 polymorphism was associated with cancer susceptibility in Asians but not in Caucasians, consistent with the findings of Umar et al. (58). Results of subgroup analysis according to the source of controls identified a relationship between rs2274223 polymorphism and tumor risk regardless of whether controls were obtained from a hospital or a population and were also consistent with the findings of Umar et al. (58). For rs3765524, the results of the present meta-analysis showed that the association between the rs3765524 polymorphism and overall cancer risk was identified in only one genetic model (CT vs. CC). The results of stratified analysis indicated that the rs3765524 polymorphism was associated with colorectal cancer and esophageal cancer susceptibility but not with the other types of cancer. The above findings were distinct from those of Mocellin et al. (61), which identified an association between the rs3765524 polymorphism and gastric cancer susceptibility. Finally, our results of both the total cancer analysis or subgroup analysis indicated that the rs753724, rs11187842, and rs7922612 polymorphisms were not related to tumor risk. The consistencies between the current and previous meta-analyses might be because some of the literatures included in meta-analyses were the same. Meanwhile, the inconsistencies between the current and previous meta-analyses could be attributed to differences in inclusion criteria. For example, the present meta-analysis specifically required that the qualified studies were case-control studies, which was different from the meta-analysis of Mocellin et al. (61).
Some limitations still existed in the present analysis, though the analysis was performed carefully. First, relatively few qualified studies were included for investigating rs753724, rs11187842, and rs7922612, and some subgroups included in the stratified analysis had low sample sizes, which might have affected statistical results. Second, unified adjustment about confounders could not be carried out in our analysis because the original data were not obtained. Third, ICD-O codes of cancers from qualified studies were not obtained, and differences in cancers included in the studies might lead to biases. Finally, unpublished materials were not obtained, which might have caused publication bias, although publication bias was not detected based on Begg's funnel plots and Egger's test in this meta-analysis.
Conclusions
Our findings indicated that the PLCE1 rs2274223 polymorphism is significantly associated with cancer susceptibility in the overall population. On the other hand, the PLCE1 rs753724, rs11187842, and rs7922612 polymorphisms showed no significant associations with cancer risk. In addition, the results suggested that the PLCE1 rs3765524 polymorphism is associated with overall cancer risk under the heterozygote model (CT vs. CC).
Author Contributions
XiaL and MJ collected the data. XiaL analyzed the data and wrote the paper. WT, XueL, and BZ read and revised the paper. All the authors supported the submission of this manuscript.
Funding
This study is supported by grant no.81502878 from National Natural Science Foundation of China and no.201501017 from the Doctoral Scientific Research Foundation of Liaoning Province.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the participants in the present study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2018.00613/full#supplementary-material
References
1. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, et al. The global burden of cancer 2013. JAMA Oncol. (2015) 1:505–27. doi: 10.1001/jamaoncol.2015.0735
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
4. Dixon K, Kopras E. Genetic alterations and DNA repair in human carcinogenesis. Sem Cancer Biol. (2004) 14:441–8. doi: 10.1016/j.semcancer.2004.06.007
5. Wing MR, Bourdon DM, Harden TK. PLC-epsilon: a shared effector protein in Ras-, Rho-, and G alpha beta gamma-mediated signaling. Mol Interv. (2003) 3:273–80. doi: 10.1124/mi.3.5.273
6. Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. (2010) 42:764–7. doi: 10.1038/ng.649
7. Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. (2010) 42:759–63. doi: 10.1038/ng.648
8. Cui XB, Chen YZ, Pang XL, Liu W, Hu JM, Li SG, et al. Multiple polymorphisms within the PLCE1 are associated with esophageal cancer via promoting the gene expression in a Chinese Kazakh population. Gene (2013) 530:315–22. doi: 10.1016/j.gene.2013.08.057
9. Li FX, Yang XX, He XQ, Hu NY, Wu YS, Li M. Association of 10q23 with colorectal cancer in a Chinese population. Mol Biol Rep. (2012) 39:9557–62. doi: 10.1007/s11033-012-1820-8
10. Zhang Y, Gong Y, Du S, Yan M, Geng T, Feng T, et al. The association between phospholipase C epsilon gene (PLCE1) polymorphisms and colorectal cancer risk in a Chinese Han population: a case-control study. Int J Clin Exp Med. (2015) 8:19360–6.
11. Yuan J, Li Y, Tian T, Li N, Zhu Y, Zou J, et al. Risk prediction for early-onset gastric carcinoma: a case-control study of polygenic gastric cancer in Han Chinese with hereditary background. Oncotarget (2016) 7:33608–15. doi: 10.18632/oncotarget.9025
12. Malik MA, Srivastava P, Zargar SA, Mittal B. Phospholipase C epsilon 1 (PLCE1) haplotypes are associated with increased risk of gastric cancer in Kashmir Valley. Saudi J Gastroenterol. (2014) 20:371–7. doi: 10.4103/1319-3767.145330
13. Sharma KL, Rai R, Srivastava A, Sharma A, Misra S, Kumar A, et al. A multigenic approach to evaluate genetic variants of PLCE1, LXRs, MMPs, TIMP, and CYP genes in gallbladder cancer predisposition. Tumour Biol. (2014) 35:8597–606. doi: 10.1007/s13277-014-2094-7
14. Xue W, Zhu M, Wang Y, He J, Zheng L. Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: proof from a meta-analysis. Sci Rep. (2015) 5:7986. doi: 10.1038/srep07986
15. Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. STrengthening the REporting of Genetic Association studies (STREGA)–an extension of the STROBE statement. Eur J Clin Invest. (2009) 39:247–66. doi: 10.1111/j.1365-2362.2009.02125.x
16. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
17. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials (1986) 7:177–88. doi: 10.1016/0197-2456(86)90046-2
18. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. (1959) 22:719–48.
19. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics (1994) 50:1088–101. doi: 10.2307/2533446
20. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629
21. Zhang H, Jin G, Li H, Ren C, Ding Y, Zhang Q, et al. Genetic variants at 1q22 and 10q23 reproducibly associated with gastric cancer susceptibility in a Chinese population. Carcinogenesis (2011) 32:848–52. doi: 10.1093/carcin/bgr051
22. Ma H, Wang LE, Liu Z, Sturgis EM, Wei Q. Association between novel PLCE1 variants identified in published esophageal cancer genome-wide association studies and risk of squamous cell carcinoma of the head and neck. BMC Cancer (2011) 11:258. doi: 10.1186/1471-2407-11-258
23. Zhou RM, Li Y, Wang N, Liu BC, Chen ZF, Zuo LF. PLC-epsilon1 gene polymorphisms significantly enhance the risk of esophageal squamous cell carcinoma in individuals with a family history of upper gastrointestinal cancers. Arch Med Res. (2012) 43:578–84. doi: 10.1016/j.arcmed.2012.09.006
24. Gu H, Ding G, Zhang W, Liu C, Chen Y, Chen S, et al. Replication study of PLCE1 and C20orf54 polymorphism and risk of esophageal cancer in a Chinese population. Mol Biol Rep. (2012) 39:9105–11. doi: 10.1007/s11033-012-1782-x
25. Hu H, Yang J, Sun Y, Yang Y, Qian J, Jin L, et al. Putatively functional PLCE1 variants and susceptibility to esophageal squamous cell carcinoma (ESCC): a case-control study in eastern Chinese populations. Ann Surg Oncol. (2012) 19:2403–10. doi: 10.1245/s10434-011-2160-y
26. Bye H, Prescott NJ, Lewis CM, Matejcic M, Moodley L, Robertson B, et al. Distinct genetic association at the PLCE1 locus with oesophageal squamous cell carcinoma in the South African population. Carcinogenesis (2012) 33:2155–61. doi: 10.1093/carcin/bgs262
27. Palmer AJ, Lochhead P, Hold GL, Rabkin CS, Chow WH, Lissowska J, et al. Genetic variation in C20orf54, PLCE1 and MUC1 and the risk of upper gastrointestinal cancers in Caucasian populations. Eur J Cancer Prev. (2012) 21:541–4. doi: 10.1097/CEJ.0b013e3283529b79
28. Wang M, Zhang R, He J, Qiu L, Li J, Wang Y, et al. Potentially functional variants of PLCE1 identified by GWASs contribute to gastric adenocarcinoma susceptibility in an eastern Chinese population. PLoS ONE (2012) 7:e31932. doi: 10.1371/journal.pone.0031932
29. Yuan Z, Yuan H, Ma H, Chu M, Wang Y, Hu Z, et al. Genetic variants at 10q23 are associated with risk of head and neck cancer in a Chinese population. Oral Oncol. (2013) 49:332–5. doi: 10.1016/j.oraloncology.2012.10.010
30. Duan F, Xie W, Cui L, Wang P, Song C, Qu H, et al. Novel functional variants locus in PLCE1 and susceptibility to esophageal squamous cell carcinoma: based on published genome-wide association studies in a central Chinese population. Cancer Epidemiol. (2013) 37:647–52. doi: 10.1016/j.canep.2013.04.009
31. Sharma KL, Umar M, Pandey M, Misra S, Kumar A, Kumar V, et al. Association of potentially functional genetic variants of PLCE1 with gallbladder cancer susceptibility in north Indian population. J Gastrointest Cancer (2013) 44:436–43. doi: 10.1007/s12029-013-9537-z
32. Dura P, Bregitha CV, te Morsche RH, Roelofs HM, Kristinsson JO, Wobbes T, et al. GWAS-uncovered SNPs in PLCE1 and RFT2 genes are not implicated in Dutch esophageal adenocarcinoma and squamous cell carcinoma etiology. Eur J Cancer Prevent. (2013) 22:417–9. doi: 10.1097/CEJ.0b013e32835c7f53
33. Li M, Huang L, Qiu H, Fu Q, Li W, Yu Q, et al. Helicobacter pylori infection synergizes with three inflammation-related genetic variants in the GWASs to increase risk of gastric cancer in a Chinese population. PLoS ONE (2013) 8:e74976. doi: 10.1371/journal.pone.0074976
34. Chen YZ, Cui XB, Pang XL, Li L, Hu JM, Liu CX, et al. [Relationship between rs2274223 and rs3765524 polymorphisms of PLCE1 and risk of esophageal squamous cell carcinoma in a Kazakh Chinese population]. Chin J Pathol. (2013) 42:795–800.
35. Yang J, Wu H, Wei S, Xiong H, Fu X, Qi Z, et al. HPV seropositivity joints with susceptibility loci identified in GWASs at apoptosis associated genes to increase the risk of Esophageal Squamous Cell Carcinoma (ESCC). BMC Cancer (2014) 14:501. doi: 10.1186/1471-2407-14-501
36. Piao JM, Shin MH, Kim HN, Song HR, Kweon SS, Choi JS, et al. Replication of results of genome-wide association studies on esophageal squamous cell carcinoma susceptibility loci in a Korean population. Dis Esophagus (2014) 27:798–801. doi: 10.1111/dote.12155
37. Kupcinskas J, Wex T, Link A, Bartuseviciute R, Dedelaite M, Kevalaite G, et al. PSCA and MUC1 gene polymorphisms are associated with gastric cancer and pre-malignant gastric conditions [corrected]. Anticancer Res. (2014) 34:7167–75.
38. Umar M, Upadhyay R, Kumar S, Ghoshal UC, Mittal B. Role of novel and GWAS originated PLCE1 genetic variants in susceptibility and prognosis of esophageal cancer patients in northern Indian population. Tumour Biol. (2014) 35:11667–76. doi: 10.1007/s13277-014-2458-z
39. Wang Q, Chen P, Chen D, Liu F, Pan W. Association between phospholipase C epsilon gene (PLCE1) polymorphism and colorectal cancer risk in a Chinese population. J Int Med Res. (2014) 42:270–81. doi: 10.1177/0300060513492484
40. Song HR, Kim HN, Kweon SS, Choi JS, Shim HJ, Cho SH, et al. Common genetic variants at 1q22 and 10q23 and gastric cancer susceptibility in a Korean population. Tumour Biol. (2014) 35:3133–7. doi: 10.1007/s13277-013-1409-4
41. Kupcinskas J, Gyvyte U, Bruzaite I, Leja M, Kupcinskaite-Noreikiene R, Pauzas H, et al. Common genetic variants of PSCA, MUC1 and PLCE1 genes are not associated with colorectal cancer. Asian Pac J Cancer Prev. (2015) 16:6027–32. doi: 10.7314/APJCP.2015.16.14.6027
42. Jia X, Liu P, Zhang M, Feng T, Tang H, Tang Z, et al. Genetic variants at 6p21, 10q23, 16q21 and 22q12 are associated with esophageal cancer risk in a Chinese Han population. Int J Clin Exp Med. (2015) 8:19381–7.
43. Sun H, Wu X, Wu F, Li Y, Yu Z, Chen X, et al. Associations of genetic variants in the PSCA, MUC1 and PLCE1 genes with stomach cancer susceptibility in a Chinese population. PLoS ONE (2015) 10:e0117576. doi: 10.1371/journal.pone.0117576
44. Dong Y, Chen J, Chen Z, Tian C, Lu H, Ruan J, et al. Evaluating the association of eight polymorphisms with cancer susceptibility in a Han Chinese population. PLoS ONE (2015) 10:e0132797. doi: 10.1371/journal.pone.0132797
45. Ezgi O, Merve A, Hakan YT, Gul O. Genetic variations in Phospholipase C-epsilon 1 (PLCE1) and susceptibility to colorectal cancer risk. Biochem Genet. (2016) 54:826–9. doi: 10.1007/s10528-016-9759-4
46. Mou X, Li T, Wang J, Ali Z, Zhang Y, Chen Z, et al. Genetic variation of BCL2 (rs2279115), NEIL2 (rs804270), LTA (rs909253), PSCA (rs2294008) and PLCE1 (rs3765524, rs10509670) genes and their correlation to gastric cancer risk based on universal tagged arrays and Fe$sub$3$/sub$O$sub$4$/sub$ magnetic nanoparticles. J Biomed Nanotechnol. (2015) 11:2057–66. doi: 10.1166/jbn.2015.2113
47. Qu Y, Zhang S, Cui L, Wang K, Song C, Wang P, et al. Two novel polymorphisms in PLCE1 are associated with the susceptibility to esophageal squamous cell carcinoma in Chinese population. Dis Esophagus (2017) 30:1–7. doi: 10.1111/dote.12463
48. Yuan LJ, Jin TB, Yin JK, Du XL, Wang Q, Dong R, et al. Polymorphisms of tumor-related genes IL-10, PSCA, MTRR and NOC3L are associated with the risk of gastric cancer in the Chinese Han population. Cancer Epidemiol. (2012) 36:e366–72. doi: 10.1016/j.canep.2012.05.016
49. Bunney TD, Harris R, Gandarillas NL, Josephs MB, Roe SM, Sorli SC, et al. Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol cell (2006) 21:495–507. doi: 10.1016/j.molcel.2006.01.008
50. Zhang Y, Wang R, Zhu L, Zhang S, Yuan H, Jiang H. Meta-analysis of phospholipase C epsilon 1 polymorphism and cancer risk. Cancer Biomark. (2013) 13:483–9. doi: 10.3233/CBM-130388
51. Luo XP. Phospholipase C epsilon-1 inhibits p53 expression in lung cancer. Cell bIochem Funct. (2014) 32:294–8. doi: 10.1002/cbf.3015
52. Bunney TD, Baxendale RW, Katan M. Regulatory links between PLC enzymes and Ras superfamily GTPases: signalling via PLCepsilon. Adv Enzyme Regul. (2009) 49:54–8. doi: 10.1016/j.advenzreg.2009.01.004
53. Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. (2006) 281:14026–40. doi: 10.1074/jbc.M507734200
54. Wang X, Zbou C, Qiu G, Fan J, Tang H, Peng Z. Screening of new tumor suppressor genes in sporadic colorectal cancer patients. Hepato-Gastroenterology (2008) 55:2039–44.
55. Wang X, Zhou C, Qiu G, Yang Y, Yan D, Xing T, et al. Phospholipase C epsilon plays a suppressive role in incidence of colorectal cancer. Med Oncol. (2012) 29:1051–8. doi: 10.1007/s12032-011-9981-1
56. Chen G, Hu J, Huang Z, Yang L, Chen M. MicroRNA-1976 functions as a tumor suppressor and serves as a prognostic indicator in non-small cell lung cancer by directly targeting PLCE1. Biochem Biophys Res Commun. (2016) 473:1144–51. doi: 10.1016/j.bbrc.2016.04.030
57. Stolzel F, Pfirrmann M, Aulitzky WE, Kaufmann M, Bodenstein H, Bornhauser M, et al. Risk stratification using a new prognostic score for patients with secondary acute myeloid leukemia: results of the prospective AML96 trial. Leukemia (2011) 25:420–8. doi: 10.1038/leu.2010.279
58. Umar M, Upadhyay R, Mittal B. PLCE1 rs2274223 A>G polymorphism and cancer risk: a meta-analysis. Tumour Biol. (2013) 34:3537–44. doi: 10.1007/s13277-013-0932-7
59. Wang J, Lin L, Wang HQ, Chen N. PLCE1 rs2274223 polymorphism contributes to risk of esophageal cancer: evidence based on a meta-analysis. Tumour Biol. (2014) 35:6925–31. doi: 10.1007/s13277-014-1914-0
60. Guo LY, Yang N, Hu D, Zhao X, Feng B, Zhang Y, et al. PLCE1 rs2274223 polymorphism and susceptibility to esophageal cancer: a meta-analysis. Asian Pac J Cancer Prev. (2014) 15:9107–12. doi: 10.7314/APJCP.2014.15.21.9107
Keywords: PLCE1, cancer, polymorphism, susceptibility, meta-analysis
Citation: Li X, Li X, Jiang M, Tian W and Zhou B (2018) Single Nucleotide Polymorphisms in PLCE1 for Cancer Risk of Different Types: A Meta-Analysis. Front. Oncol. 8:613. doi: 10.3389/fonc.2018.00613
Received: 14 September 2018; Accepted: 29 November 2018;
Published: 11 December 2018.
Edited by:
Jianguang Ji, Lund University, SwedenReviewed by:
Martha Jane Felini, University of North Texas Health Science Center, United StatesGuangwen Cao, Second Military Medical University, China
Ni Li, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2018 Li, Li, Jiang, Tian and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baosen Zhou, YnN6aG91QGNtdS5lZHUuY24=
 Xiaoying Li
Xiaoying Li Xuelian Li
Xuelian Li Min Jiang
Min Jiang Wen Tian1,2
Wen Tian1,2 Baosen Zhou
Baosen Zhou