
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 15 October 2018
Sec. Gastrointestinal Cancers
Volume 8 - 2018 | https://doi.org/10.3389/fonc.2018.00371
Colorectal cancer (CRC) is the third most common cancer worldwide and its pathogenesis has been extensively explored over the past decades. Recently, microorganisms in the gastrointestinal tract have emerged as potential etiological agents. In particular, a direct proportional association between Fusobacterium and CRC has been described. Since then, the functional impact of Fusobacterium in CRC development has been studied using various mouse models. Although some epidemiologic studies did not establish an obvious relationship between Fusobacterium and CRC, numerous pathogenic mechanisms leading to the disease have been described. For instance, Fusobacterium can activate the E-cadherin/β-catenin signaling pathway and is associated with particular epigenetic phenotype, such as microsatellite instability (MSI) and hypermethylation, via its strong adhesive and invasive abilities resulting in malignant transformation of epithelial cells. Also, Fusobacterium could alter the tumor microenvironment (TME) significantly by myeloid-derived suppressor cells (MDSCs), tumor associated macrophages (TAMs), and tumor associated neutrophils (TANs) recruitment and local immune suppression. Herein, we provide an in-depth review of the relationship between Fusobacterium and colorectal cancer. In light of the emergence of microbiome-based therapeutics, potential therapies and preventive strategies for colorectal cancer related to Fusobacterium are also discussed.
Commensal bacteria in the colon might play a significant role in the maintenance of health (1–4). Intestinal microbiota promotes the maturation of human immune system and maintenance of natural barrier integrity (5). Bacterial dysbiosis in the gut has been associated with numerous human diseases, including obesity (6, 7), intestinal diseases (8, 9), cardiovascular diseases (10), autism (11), malignancies (12–14) and others. Garrett et al. identified that colitis could be transferred from T-bet−/− × Rag2−/− ulcerative colitis (TRUC) mice to wild type mice with particular intestinal bacteria (Proteus mirabilis and Klebsiella pneumoniae) transplantation (15). The metabolic phenotypes and adiposity status could also be modulated by cultured gut microbiota from human and coordinate diets in mice (6). This finding denotes the importance of gut microbiota symbiosis and the dysfunctional proportion could exacerbate human diseases, since both normal and pathogenic flora are important in the regulation of homeostasis (16). Particularly, colonic dysbiosis has been associated with CRC (17) (12, 18) (19) (20). Among the multitudinous genera, Fusobacterium stood out as being oftentimes increased in CRC (14, 21) (Table 1). As in the case of Helicobacter Pylori correlation with gastric cancer, Fusobacterium may be an essential microbial carcinogen that fuels the initiation and development of CRC (21, 22). Fusobacterium is a genus of gram-negative anaerobic bacteria. It may act as a main anchor of biofilms that can induce periodontitis (23, 24), vaginitis (25) and other infections (26). Fusobacterium was considered as part of the normal flora of the oropharynx formerly, but lately its pathogenic role especially as a driver of periodontitis (27) and its association with intestinal diseases has been demonstrated. Although it is still unclear whether Fusobacterium is the passenger or driver of CRC, many studies have concluded that Fusobacterium is a novel risk factor for CRC development and progression, as well as a determinant affecting patient survival outcomes (13, 28, 29). Kostic et al. (14) illustrated that in colorectal adenoma, an early event in CRC development, Fusobacterium is found to be enriched in comparison with surrounding normal tissue suggesting an essential role of Fusobacterium in the early onset of CRC. Moreover, a recent retrospective study with 13,096 adult patients suggested that those presented with Fusobacterium nucleatum (one of the species of Fusobacterium) have increased risk of CRC (30). Additionally, significantly larger proportions of Fusobacterium has been detected from feces of adenoma and CRC patients in comparison to healthy controls, which further confirms Kostic's finding. Distal metastasis of colonic cancer was also found to be colonized with Fusobacterium and other assembled microbes. Investigators have also shown that tumor proliferation and cancer growth could be reduced via decreasing the load of Fusobacterium by antibiotic treatment (metronidazole) (31). Moreover, Fusobacterium is associated with certain epigenetic phenotypes of CRC –high degrees of microsatellite instability (MSI) and CpG island methylation phenotype (CIMP) (32, 33), which could provide promising opportunities to develop diagnostic tools or treatment biomarkers for CRC.
Fusobacterium is a cylindrical shaped, gram-negative, non-spore-forming, strictly anaerobic genus. Although Fusobacterium is part of the normal microbiome, recent findings indicated that increased Fusobacterium levels have been detected in various inflammatory (39–41) and cancer samples (33). There are 14 species in Fusobacterium, such as F. necrophorum (inhabitant of the alimentary tract and being responsible for Lemierre' syndrome) and F. varium (found in the ulcerative colitis). Among them, F. nucleatum is one of the key pathogens which plays a role in oral plaque formation, due to its adhesive ability, serving as a bridge organism during colonization and biofilm formation (42). Although several studies suggest that Fusobacterium strains might vary in their virulence potential, it is has been speculated that some Fusobacterium strains can acquire genes through horizontal transfer and obtain increased virulence potential from different species and strains (43–45). Regardless of the mechanism in which Fusobacterium attains its virulence, evidence points to the positive correlation of this Fusobacterium toward CRC malignancy.
Fusobacterium is an invasive organism. Fusobacterium invades host with the aid of a surface adhesion molecule called FadA, which is abundantly expressed on Fusobacterium (46). Rubinstein and his colleagues suggested that FadA, a membrane protein, encoded by Fusobacterium binds to E-cadherin on the epithelial cell surface and leads to E-cadherin phosphorylation and internalization. This, in turn, activates β-catenin signaling pathways (22) and consequently leads to inflammation and tumorigenesis gene transcription (such as NF-κB, Myc ad Cyclin D1, lymphoid enhance factor/T cell factor). Furthermore, they detected higher FadA expression levels in CRC tissue compared with normal tissue. Consistently, the expression of oncogenes, inflammatory genes and Wnt genes are increased in CRC cells under modulation of purified FadA. Further experiments with purified FadA or F. nucleatum could only stimulated cell lines with APC or β-catenin mutations but not non-cancerous HEK-293 cells, indicated that oncogenesis promoted by Fusobacterium was secondary to these significant mutation events. As an invasive organism that survives inside host cells, Fusobacterium is also capable of releasing RNA into the host cell cytoplasm that is detected by cytosolic retinoic acid-inducible gene 1 (RIG-1), triggering activation of NF-κB and activating inflammatory genes and oncogenes (47, 48). Furthermore, FadA binds to vascular endothelial cadherin (VE-cadherin), a cell-cell junction molecule identified as the endothelial receptor for FadA. FadA binding causes VE-cadherin to relocate and increases the endothelial permeability, which then facilitates Fusobacterium and other bacteria species to penetrate into the blood stream (49). Fusobacterium also has a strong ability to induce co-aggregation and shuttle unrelated microbes [in particular Streptococcus and Campylobacter (50)] into host cell via Fap2, a large membrane protein present in Fusobacterium The shuttled microbes are known to have weak binding ability to host cells or they are non-invasive (51, 52). Being shuttled into the host cells by Fusobacterium, the toxic effects on endothelial cells become multifold.
Normally, the interaction between intestinal microbiota and the host contributes to the maintenance of homeostasis and normal mucosal immunity. Abnormalities on either side will disrupt this balance and brings about disease and malignancy. It has already been established that lesions on the intestinal mucosa impair barrier function (53). In tumor microenvironment (TME), the pH value of the tumor tissue is around 6.5-6.9, which is slightly lower than the normal physicological pH of 7.4 (51, 52), This slight change of pH can significantly affect the refined composition of microbial community in TME (54). In APC gene deleted mouse models (CRC animal models), significant defective intestinal barrier function at tumor sites has been described and therefore concluded that changes in the local environment and a deficient barrier could provide favorable condition for Fusobacterium to reproduce and cause further mucosal injury (55). Hence, it is hypothesized that the growth of Fusobacterium, normally a commensal at low abundance, is increased with disruption of intestinal homeostasis and in turn accelerates tumorigenesis, consequently forming a vicious positive feedback cycle.
However, as only one of the estimated 103 microbial species in the colon (56), Fusobacterium cannot be studied in isolation and some studies have identified other bacterial species that may also contribute to colorectal carcinogenesis and have interaction with Fusobacterium. These bacteria include and Escherichia coli (E. coli) (20), Bacteroides fragilis (B. fragilis) (57), H. pylori (58), Enterococcus faecalis (E.faecalis) (59), Streptococcus bovis (S. bovis) (60) and Clostridium septicum (C. septicum) (61). Moreover, as an oral commensal and a periodontal pathogen, Fusobacterium also interacts with candida albicans within the oral cavity (62). As evidenced by other oral community members (such as members of the Porphyromonas) that has been found on colonic tumors (63), Fusobacterium may also play a role in the link between oral and intestinal microbiota. Fusobacterium has defense ability against human neutrophilic peptide-1 (64) and is one of the most important anaerobes that promote the formation of dental biofilms (24, 65). The polymicrobial nature of oral biofilms and the asaccharolytic metabolism of many of these species allows them to survive comfortably in the microenvironment of colonic lesions (66). Consequently, co-occurrence patterns of Fusobacterium with other microbiota in the oral cavity may correlate with localized intestinal dysbiosis in the setting of colorectal carcinogenesis.
Sporadic colorectal cancer and colitis-associated colorectal cancer are predominant classes of CRC. Sporadic colorectal cancer develops in five stages—polyp, early adenoma, late adenoma, carcinoma and invasion; while colitis-associated cancer develops in six stages—colitis, indefinite dysplasia, low-grade dysplasia, high-grade dysplasia, carcinoma and invasion (67). In both cases, Fusobacterium was found to be an important associated factor in their development (34, 68, 69). Oncogenesis caused by Fusobacterium is closely related to the inflammatory state of the tumor. However, the crosstalk between each inflammatory component of the tumor microenvironment is very complicated. For example, the abundance of tumor infiltrating lymphocytes (TILs) correlate with improved clinical outcomes in colorectal patients (70–72), while the presence of myeloid derived suppressive cells (MDSCs) (73) and regulatory T cells (Tregs) (74) indicate immune inhibitory status and are associated with poor prognosis. Hereby, the detailed mechanism of the interaction of Fusobacterium with each TME component in inflammatory and malignancy state is discussed as follows (Figure 1).
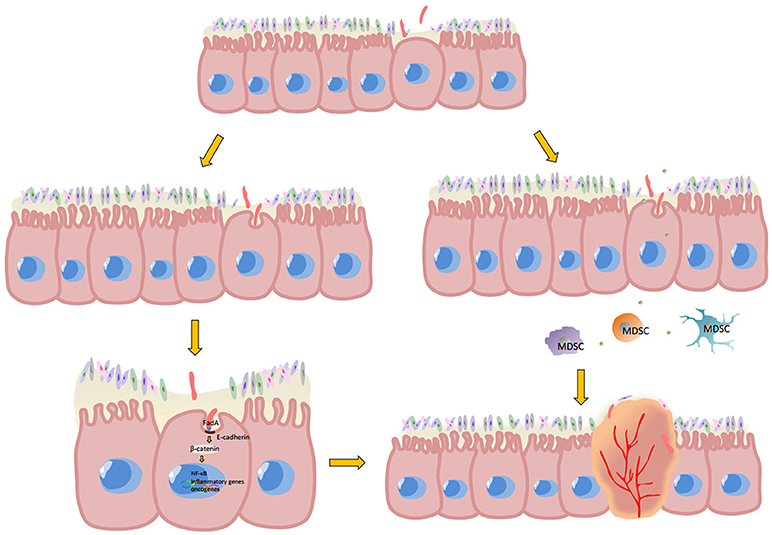
Figure 1. Potential mechanism of Fusobacterium-associated CRC. Mutant epithelial cells cause local intestinal barrier impairment, which gives Fusobacterium the opportunity for adherence and subsequent invasion into epithelial cells. Once FadA, a membrane protein of Fusobacterium, combines with E-cadherin and is internalized by epithelial cells, the β-catenin signaling pathway is activated. Phosphorylated β-catenin would enter the cell nucleus from cytoplasm and promote NF-κB genes, pro-inflammatory genes and the expression of many other oncogenes. Moreover, the microenvironment of malignancies is anoxic and acidic, which would be more suitable for Fusobacterium reproduction compared to other bacteria. The metabolites of aggregated Fusobacterium. Then recruit MDSCs, in turn suppressing anti-tumor immunity and promote CRC carcinogenesis.
Kostic et al. (21) found that CD11b+ myeloid derived suppressive cells (MDSCs), including granulocytes/tumor associated neutrophils (TANs) and macrophages/tumor associated macrophages (TAMs), are more abundant in tumor tissues than normal tissue of Fusobacterium-fed APC Min mice. MDSCs are a heterogeneous group of immune cells differentiating from the myeloid lineage (21, 75). MDSCs are highly abundant in some pathological situations such as chronic infections and cancer. Due to their immunosuppressive activities and interaction with other immune cell types, tumor tissue with high infiltration of MDSCs may predict poor prognosis and drug resistance (76, 77). TANs and TAMs, the special subset of MDSCs, also contribute to the inhibition of anti-tumor immunity and result in tumor progression and metastasis, which has been verified in considerable experimental data from clinical and pre-clinical studies (48, 56).
In mouse models, MDSCs express both myeloid lineage differentiation antigen Gr1 (granulocyte differentiation antigen1) and CD11b and are further classified into two subtypes—monocytic and granulocytic (78–80). F. nucleatum increases the production of reactive oxygen species (ROS) (81) and inflammatory cytokines such as IL-10 in CRC (34), possibly by recruiting MDSCs. ROS normally serve as second messengers to regulate many of the intracellular signaling cascades that govern multiple cellular activities (81), which causes DNA damage and other cell injuries. It is well documented that inflammatory oxidative stress can lead to p53 mutation, thus promoting oncogenesis (82). Oxidative stress also contributes to aberrant hypermethylation (83), which in turn causes tumor-suppressive gene inactivation and carcinogenesis (84, 85). Hypermethylation could also occur in promoter regions of mismatch repair protein (MLH1) gene and result in microsatellite instability (MSI) (86–88), which was recognized as an early molecular phenomenon in CRC. Moreover, MDSCs can disrupt the anti-tumor abilities of the immune system by secreting arginase-1 (89), which suppresses T cell activity and induces tumor intolerance (90, 91).
MDSCs also interact with other immune cell types, such as an anti-tumor subtype of dendritic cells (CD103+ DCs). Mice treated with Fusobacterium were found to have more CD103+ DCs within tumors compared with control groups (21). These cells regulate immune responses by promoting the activation of Foxp3+ regulatory T cells, a CD4+ T cell subset that inhibits cytotoxic and effector T cells, and results in restrained antitumor immunity (92). In addition, immunohistochemical analysis indicate that a higher amount of Fusobacterium were associated with a lower density of tumor inhibiting CD3+ T cells in tumor tissue, thereby enabling tumor to escape immune surveillance (37). Additionally, Fusobacterium could suppress immune function by arresting T cells in the Mid-G1 phase of the cell cycle. Shenker and Datar discovered that Fusobacterium suspended T cell proliferation at Mid-G1 phase by regulating different cyclin levels via an immunosuppressive protein, F. nucleatum immunosuppressive protein (FIP) (93). They also found that proliferating cell nuclear antigen (PCNA) was significantly decreased in expression. PCNA is a DNA clamp that acts as a processivity factor for DNA polymerase δ and accelerates cell transition through early to mid-G1. With decreased proliferative ability, T cell would fail to function or attack cancer cells thus progressing to an immunosuppressive phase.
Fap2, an outer membrane protein of Fusobacterium, is also involved in tumor suppression activity. Kaplan found that Fap2, together with adhesin RadD can cause lymphocyte death by direct contact with target lymphocytes. Either Fap2 or RadD alone could induce lymphocyte death, though at a much lower level. Chamutal Gur also discovered that Fap2 could bind to TIGIT, an inhibitory receptor presents on all human NK cells and various T cells, protecting tumors from immune attack by inhibiting effective immune cell activity (94).
Recent findings support the claim that Fusobacterium could enhance the development from being in inflammatory state to malignancy. This finding was first reported by Kostic (21) and colleagues when APCMin/+ mice were introduced to human isolates of Fusobacterium nucleatum. Higher level of inflammation and more colonic tumors were found in the F. nucleatum group compared to control. However the colitis mice model (IL-10−/− and Rag2−/−/T-bet−/−) treated with F. nucleatum did not develop colonic tumors. This may suggest that Fusobacterium induce oncogenesis downstream of the APC pathway and the tumorigenesis does not depend on pre-existing colitis condition because the colitis mice did not develop colon tumors after F. nucleatum introduction. They also revealed the expression signature of inflammation: the higher expression of COX-2, IL-1 β, IL-6, IL-8, TNF, and MMP3 by human and mouse cell line while co-culturing with Fusobacterium suggests an NF-κB -driven pro-inflammatory response. Rubinstein et al. further indicated that the elevated expression of inflammatory genes such as NF-κB, IL-6, IL-8 and IL-18 were correlated with FadA level in CRC tissues, which is consistent with the former study. Additionally, Fusobacterium also binds to Toll-like receptor 4 (TLR4) of epithelial cells and activate the TLR4/MYD88/NF-κB signaling pathway. NF-κB p50/p65 separated with phosphorylated IκB and bound to DNA, followed with microRNA-21 transcription. microR-21 could inhibit RAS p21 GTPase activating protein (RASA1) and therefore activate the RAS/RAF/MEK/ERK signaling pathway (68). Thus, in the tumor-initiation period, Fusobacterium exerts its tumor-promoting action through the augmentation of local inflammation. Furthermore, some indicative molecules in human colon samples, such as COX-2, IL-1β, IL-6, IL-8, TNF-α, and MMP3, are suggestive of activation of NK-κB-driven inflammation (21, 55, 95). Among these expression signatures, IL-8, TNF-α and other chemokines could recruit neutrophils and macrophages, which synthesize nitric oxide (NO) and cause oxidative stress to epithelial and stromal cells. This results in DNA damage and consequently activation of p53 transcription which in turn suppresses tumorigenesis by inducing G1-S arrest, DNA repair and cell apoptosis. Moreover, p53 overexpression also leads to TP53 mutation, which is a key event during CRC development. Additionally, chronic inflammation and ROS production cause many other mutations (such as CHD7 and CHD8, members of the chromodomain helicase/ATP-dependent chromatin remodeling family (33, 96) and genomic instability), all of which would accelerate CRC development.
Intriguingly, high load of Fusobacterium was recently reported to be associated with a specific epigenetic phenotype of CRC. Fusobacterium detected in colorectal cancer tissues was related to CpG island methylator phenotype (CIMP) status, high MSI and MLH1 hypermethylation (33) and up-regulating expression of microRNA-21. Interestingly, these specific molecular features of colorectal cancer occur mostly in the ascending colon (97, 98), which is also the most common colonization site of Fusobacterium in the GI tract. This might indicate some association between Fusobacterium biogeography and the colonic mucosal microenvironment.
CIMP is characterized by simultaneous hypermethylation of numerous CpG islands surrounding the promoter regions of several genes. The high level of methylation of CpG island indicates chronic inflammation and an aggravated immune response (99). Microsatellite instability is the somatic accumulation of length variations in repetitive DNA sequences (33). It has been established that defects in the DNA mismatch repair (MMR) pathway lead to disincorporation, insertions and deletions in microsatellite in this repetitive DNA sequences. MSI is frequently observed in both hereditary and sporadic CRC (100). Inflammatory state and reactive oxygen stress produced by Fusobacterium may contribute to epigenetic silencing of the MMR protein MLH1 and reduction of its enzymatic activity, which leads to MSI CRC (101).
microRNAs, a small non-coding RNA functioning in RNA silence and regulating post-transcriptional gene expression, rapidly emerging as promising diagnostic and therapeutic targets, may be involved in the progression of cancer as well (102). Studies suggest that Fusobacterium might raise the level of microRNA-21 in tumor cells via epigenetic regulation during macrophage inflammatory response (68). microRNA-21 in turn increases the levels of IL-10 and PGE2 (prostaglandin E2), which inhibit antitumor immunity mediated by T cells in the TME (103, 104), and therefore high level of microRNA-21 usually indicates worse clinical outcomes (105). Nevertheless, further studies are needed in order to pinpoint the relationship between Fusobacterium and microRNAs and its significance.
Numerous studies have found that enrichment of Fusobacterium is related to worse clinical outcome in CRC patients (96). Analogous to Helicobacter Pylori in the setting of gastric cancer, Fusobacterium may be an essential pathogen that fuels the initiation and development of CRC. Research has suggested that reducing the abundance of Fusobacterium may help patients with intestinal diseases such as IBD to recover (106). Therefore, therapies that specifically target Fusobacterium or their mechanism of action could be developed for the prevention or treatment of CRC.
As we discussed early, FadA, a membrane protein of Fusobacterium, induces adhesion and invasion, and is the originator of epithelial injury. In FadA-E cadherin pathway, FadA gene copy number has been found to have a direct correlation with either healthy, pre-cancerous or CRC states (22), suggesting that FadA may be a promising biomarker in CRC diagnosis. Targeting FadA blockade development is also promising by inhibiting its adhesive and invasive abilities. As E-cadherin internalization via clathrin is a key process during FadA/E-cadherin/β-catenin pathway, a clathrin inhibitor (Pitstop2) could block the pro-carcinogenesis pathway (22). Similarly, Fap2, which ferries other bacteria into the host cell, is also a membrane protein of Fusobacterium The hemagglutination and coaggregation function of Fusobacterium can be inhibited by galactose (52). This indicates that membrane blockers may suppress key pathogenic features of Fusobacterium, providing a further target in the early stages of Fusobacterium pathogenesis. However, the possibility should be taken into consideration that abundance of FadA might only reflect Fusobacterium infection status at a given time point. Since E. coli and Streptococcus gallyliticus (107) and many other microorganisms could also be significant pathogenesis factors and promote oncogenesis via different mechanisms, FadA alone may not be sufficient. Moreover, Fusobacterium is a heterogeneous genus and further gene and protein differences between low-virulence strains and high-virulence strains will need to be probed by genome and proteome analysis (108). Such approaches may allow for characterizing relevant species at a much more granular level and identifying those equipped with highly oncogenic motifs.
Studies have found that some F. nucleatum strains may acquire genes through horizontal transfer, indicating the close connection between Fusobacterium and other gut microbes (44, 45). This can be seen in the therapeutic effects of microecologic products (109), such as probiotics and prebiotics, as they reduce the overall virulence genera through regulating the whole composition of microbes and cut down the possibility for Fusobacterium to obtain virulence genes. For example, the amount of Fusobacterium was reduced in fecal samples and the microbial diversity increased following probiotic intervention in CRC patients, suggesting that microecologic products would benefit CRC patient for suppressing CRC-associated genera (110). Though limited literature has proved a direct relationship between Fusobacterium colonized in gingival sulcus and colorectal cancer tumorigenesis, there might be some connections between oral flora and gut flora, as well as connections between oral infectious diseases and intestinal diseases (such as CRC). Combined detection of fecal and oral microbes may also enhance prediction of adenomas or CRC in addition to other risk factors (111). It is possible that altering intestinal Fusobacterium abundance could be approached by targeting oral Fusobacterium, however further studies are needed for confirmation.
As stated earlier, Fusobacterium has a complex influence on the tumor microenvironment. Either chemotherapy or targeted therapies for CRC face the serious dilemma of drug resistance (112, 113). Compared with other tumors, CRC uniquely occurs in an intricate microenvironment owing to the co-existence of the huge diversity of microorganisms within the gut microbiome. It is now understood that drug resistance is closely related to the tumor microenvironment (114, 115). In this sense, the microenvironment regulated by Fusobacterium might account for drug resistance in CRC. On the one hand, Fusobacterium nucleatum has been found to promote chemoresistance to colorectal cancer by modulating autophagy (116). Via binding to TLR4 on colorectal cancer cells, Fusobacterium nucleatum activated the TLR4/MYD88 innate immune signaling pathway and miRNA-18a and miRNA-4802 were downregulated sequentially, which resulted in ULK1 and ATK7 expression. Both ULK1 and ATG7 are important elements of autophagy, the downregulation of which could decrease CRC cell apoptosis induced by chemotherapy agents. On the other hand, Fusobacterium might contribute to anti-VEGF pathway agents, such as bevacizumab. Fusobacterium promotes inflammation and cause aberrant extracellular matrix (ECM) formation, which accelerate VEGF synthesis and secretion. Many of the Fusobacterium –associated genes, including IL-6 and IL-8, also play significant roles in alternative pro-angiogenesis pathways (117). Besides, MDSCs recruited by Fusobacterium are also associated with drug resistance (118, 119). With overactive inflammation-tumorigenesis sequences and a local immunosuppressive state, it is more likely for patients to undergo drug resistance to immune therapies. Facing treatment failure, strategies to lower the abundance of Fusobacterium may be helpful to reduce drug resistance.
Some studies implicate Fusobacterium in certain epigenetic changes of CRC, such as CpG island methylation phenotype (CIMP), MSI and microRNA expression. However, further exploration is needed to uncover mechanism of how Fusobacterium impacts on CRC epigenetic changes. Particularly, CRC tissues enriched with Fusobacterium may be associated with MSI (96). Interestingly, high-abundance of Fusobacterium suggests poor prognosis of CRC while MSI forecasts favorable prognosis. One possible explanation is that MSI is not a predominant factor in the interaction between Fusobacterium and host cells, and other factors should also be taken into consideration. Recently, MSI is regarded as a strong biomarker for PD-1 blockade (120). It would be a possible explanation that CRC cells with frameshift mutations in the absence of normal mismatch repair function produce a mutation-associated neoantigen, which may activate anti-tumor immunity and enhance the effect of PD-L1 blockade. Some studies have indicated the relationship between gut microbiota and checkpoint immunotherapy recently (121–124), but whether Fusobacterium impact on checkpoint inhibitors still remains unclear. Further studies are needed to clarify whether Fusobacterium influences mismatch repair signaling pathways and alters the patient's immune response.
Colorectal cancer is one of the most common cancers worldwide and an understanding of its relationship with the gut microbiota is beginning to emerge. Interacting with numerous microbes, the initiation and development of colorectal cancer may be fueled by pathogens or inhibited by probiotics. Researchers have demonstrated the correlation between Fusobacterium with colonic adenomas and colorectal cancer. Whether this is a cause or just an association with CRC remains unclear, although some potential mechanisms have been elucidated. While FadA-induced cell invasion and subsequent signaling pathway activation is one of the most accepted mechanisms, nevertheless, based on the evidence so far, screening or risk stratification strategies utilizing Fusobacterium should be developed. In accordance with the broad public acceptance of probiotics and prebiotics, microbiome-targeted therapies for CRC are a promising avenue for future development. Notably, fecal microbiota transplantation is now considered standard of care for recurrent Clostridium difficile infection, with promising results in other conditions reported (125, 126). However, not all of the study results are in agreement with a “causal relationship” conclusion: Dejea et al. found that no consistent bacterial genus associated with tumors by high-throughput sequencing in 30 CRC and 6 adenoma human samples (127). In another paper, Dejea and colleagues studied patchy biofilms in colonic mucosa of patients with an APC gene mutations (128). Fusobacterium was not the predominant component of the biofilms while Escherichia coli and Bacteroides fragilis were prominently associated. Moreover, although Fusobacterium was more abundant in CRC biopsies and fecal samples than normal control group (129), the elevated level was only manifested in 20–30% of CRC patients and has not been consistent. In a cohort study of 137217 adults (29) to explore the association between diets with CRC prevalence, a stronger relationship between diet and CRC was found in F. nucleatum enriched individuals than subgroups without F. nucleatum detection. This finding also suggests that Fusobacterium may play different roles in different subtypes of CRC. Additionally, the interaction between the microbiota and carcinogenesis in the colon is sophisticated. Studies have also found associations with other microorganisms triggering neoplasia as well as associated mechanisms. For instance, Enterotoxigenic Bacteoides fragilis promotes Th17 development by Tregs, limiting the availability of IL2 in the local microenvironment (130) while Escherichia coli releases colibactin, a genomic product of polyketide synthase island, which is carcinogenic and promotes CRC development (20).
In conclusion, how Fusobacterium impacts CRC warrants large-scale cohort studies and laboratory experiments. Some key studies suggest that Fusobacterium could activate the β-catenin signaling pathway to promote oncogene transcription and alter the tumor microenvironment to induce immune suppression, while others suggested that Fusobacterium is associated with epigenetic changes of malignant epithelial cells. Potential microbiome therapies in colorectal cancer are still some time away but are likely to emerge as microbiome research continues to expand into oncological research. It is our hope that this review provides new insights for further CRC and Fusobacterium research.
ZZ conceived and wrote the paper. HH, HY, and JC reviewed and edited the manuscript. All authors read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I would like to express my gratitude to Prof. Phei Er Saw who helped in editing and review during the writing of this manuscript. This work was supported by grants from Natural Science Foundation of China (81672738, 81730077, 81372819, 81572596, U1601223), National Key R&D Program of China (2016YFC1302301), Science and Technology Program of Guangzhou (201704020095), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S252), Guangzhou Science and Technology Bureau (2014J4100170, 201704020131), Grants from Natural Science Foundation of Guangdong Province (2017A030313828), the Fundamental Research Funds for the Central Universities (17ykjc14).
MSI, microsatellite instability; MDSCs, myeloid-derived suppressive cells; TAMs, tumor associated macrophages; TANs, tumor associated neutrophils; TRUC mice, T-bet−/− × Rag2−/− ulcerative colitis mice; CRC, colorectal cancer; IBD, inflammatory bowel disease; NF-κB, Nuclear factor kappa-light-chain-enhancer of activated B cells; F. nucleatum, Fusobacterium nucleatum; APC, adenomatous polyposis coli; RIG-1, retinoic acid inducible gene I; Tregs, regulatory T cells; Gr1, granulocyte differentiation antigen 1; CD, cluster of differentiation; ROS, reactive oxygen species; MLH1, human mutL homolog 1; FIP, Fusobacterium nucleatum immunosuppressive protein; PCNA, proliferating cell nuclear antigen; TIGIT, T cell immunoreceptor with Ig and ITIM domains; NK cell, natural killer cell; COX2, cyclo-oxygenase-2; IL, interleukin; TNF, tumor necrosis factor; MMP3, matrix metalloproteinase-3; TLR, toll-like receptor; RASA1, RASp21 GTPase activating protein; RAS, a class of small G-protein; RAF, rapidly accelerated fibrosarcoma; MEK, mitogen-activated protein kinase-ERK kinase; ERK, extracellular regulated mitogen-activated protein kinase; TP53, tumor protein p53; CHD, chromodomain helicase DNA binding protein; CIMP, CpG island methylator phenotype; GI tract, gastrointestinal tract; MMR, mismatch repair; PGE2, prostaglandin E2; MYD88, myeloid differentiation primary response gene 88; ULK1, unc-51 like autophagy activating kinase 1; ATG7, autophagy-related protein 7; ECM, extracellular matrix; VEGF, vascular endothelial growth factor; PD-1, programmed cell death 1; Th17 cells, T helper 17 cells.
1. Dickson RP. The microbiome and critical illness. Lancet Respir Med. (2016) 4:59–72. doi: 10.1016/S2213-2600(15)00427-0
2. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535:65–74. doi: 10.1038/nature18847
3. Cresci GA, Bawden E. Gut microbiome: what we do and don't know. Nutr Clin Pract. (2015) 30:734–46. doi: 10.1177/0884533615609899
4. Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell (2014) 156:408–11. doi: 10.1016/j.cell.2014.01.025
5. C. Human Microbiome Project. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486:207–14. doi: 10.1038/nature11234
6. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. doi: 10.1126/science.1241214
7. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature (2009) 457:480–4. doi: 10.1038/nature07540
8. Roy U, Galvez EJC, Iljazovic A, Lesker TR, Blazejewski AJ, Pils MC, et al. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep. (2017) 21:994–1008. doi: 10.1016/j.celrep.2017.09.097
9. Jacobs JP, Lin L, Goudarzi M, Ruegger P, McGovern DP, Fornace AJ Jr, et al. Microbial, metabolomic, and immunologic dynamics in a relapsing genetic mouse model of colitis induced by T-synthase deficiency. Gut Microbes. (2017) 8:1–16. doi: 10.1080/19490976.2016.1257469
10. Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. (2017) 8:845. doi: 10.1038/s41467-017-00900-1
11. Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome (2017) 5:10. doi: 10.1186/s40168-016-0225-7
12. Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut (2016) 65:330–9. doi: 10.1136/gutjnl-2015-309990
13. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. (2012) 22:299–306. doi: 10.1101/gr.126516.111
14. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. (2012) 22:292–8. doi: 10.1101/gr.126573.111
15. Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. (2010) 8:292–300. doi: 10.1016/j.chom.2010.08.004
16. Kataoka K. The intestinal microbiota and its role in human health and disease. J Med Invest. (2016) 63:27–37. doi: 10.2152/jmi.63.27
17. Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, et al. Towards the human colorectal cancer microbiome. PLoS ONE (2011) 6:e20447. doi: 10.1371/journal.pone.0020447
18. Scanlan PD, Shanahan F, Clune Y, Collins JK, O'Sullivan GC, O'Riordan M, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. (2008) 10:789–98. doi: 10.1111/j.1462-2920.2007.01503.x
19. Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. (2013) 98:111–20. doi: 10.3945/ajcn.112.056689
20. Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (2012) 338:120–3. doi: 10.1126/science.1224820
21. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. (2013) 14:207–15. doi: 10.1016/j.chom.2013.07.007
22. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. (2013) 14:195–206. doi: 10.1016/j.chom.2013.07.012
23. Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. (2010) 8:471–80. doi: 10.1038/nrmicro2381
24. Okuda T, Kokubu E, Kawana T, Saito A, Okuda K, Ishihara K. Synergy in biofilm formation between Fusobacterium nucleatum and Prevotella species. Anaerobe (2012) 18:110–6. doi: 10.1016/j.anaerobe.2011.09.003
25. Machado A, Cerca N. Influence of biofilm formation by gardnerella vaginalis and other anaerobes on bacterial vaginosis. J Infect Dis. (2015) 212:1856–61. doi: 10.1093/infdis/jiv338
26. Sanmillan JL, Pelegrin I, Rodriguez D, Ardanuy C, Cabellos C. Primary lumbar epidural abscess without spondylodiscitis caused by Fusobacterium necrophorum diagnosed by 16S rRNA PCR. Anaerobe (2013) 23:45–7. doi: 10.1016/j.anaerobe.2013.06.014
27. Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. (2012) 6:1176–85. doi: 10.1038/ismej.2011.191
28. Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. (2014) 33:1381–90. doi: 10.1007/s10096-014-2081-3
29. Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. (2017) 3:921–7. doi: 10.1001/jamaoncol.2016.6374
30. Kwong TNY, Wang X, Nakatsu G, Chow TC, Tipoe T, Dai RZW, et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology (2018) 155:383–90.e8. doi: 10.1053/j.gastro.2018.04.028
31. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (2017) 358:1443–8. doi: 10.1126/science.aal5240
32. Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget (2015) 6:7209–20. doi: 10.18632/oncotarget.3109
33. Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. (2014) 74:1311–8. doi: 10.1158/0008-5472.CAN-13-1865
34. McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS ONE (2013) 8:e53653. doi: 10.1371/journal.pone.0053653
35. Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut (2017) 66:1441–8. doi: 10.1136/gutjnl-2016-312766
36. Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer (2015) 137:1258–68. doi: 10.1002/ijc.29488
37. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. (2015) 1:653–61. doi: 10.1001/jamaoncol.2015.1377
38. Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, et al Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. (2016) 6:33440. doi: 10.1038/srep33440
39. Sekizuka T, Ogasawara Y, Ohkusa T, Kuroda M. Characterization of Fusobacterium varium Fv113-g1 isolated from a patient with ulcerative colitis based on complete genome sequence and transcriptome analysis. PLoS ONE (2017) 12:e0189319. doi: 10.1371/journal.pone.0189319
40. Ohkusa T, Sato N, Ogihara T, Morita K, Ogawa M, Okayasu I. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. (2002) 17:849–53. doi: 10.1046/j.1440-1746.2002.02834.x
41. Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut (2003) 52:79–83. doi: 10.1136/gut.52.1.79
42. Kolenbrander PE, Palmer RJ Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000 (2006) 42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x
43. Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes (2011) 2:294–8. doi: 10.4161/gmic.2.5.18603
44. Ang MY, Dutta A, Wee WY, Dymock D, Paterson IC, Choo SW. Comparative Genome Analysis of Fusobacterium nucleatum. Genome Biol Evol. (2016) 8:2928–38. doi: 10.1093/gbe/evw199
45. Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, et al. Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable Fusobacterium. PLoS ONE (2007) 2:e659. doi: 10.1371/journal.pone.0000659
46. Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. (2005) 187:5330–40. doi: 10.1128/JB.187.15.5330-5340.2005
47. Lee P, Tan KS. Fusobacterium nucleatum activates the immune response through retinoic acid-inducible gene I. J Dent Res. (2014) 93:162–8. doi: 10.1177/0022034513516346
48. Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. (2014) 162:54–61. doi: 10.1016/j.imlet.2014.05.014
49. Fardini Y, Wang X, Temoin S, Nithianantham S, Lee D, Shoham M, et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. (2011) 82:1468–80. doi: 10.1111/j.1365-2958.2011.07905.x
50. Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome (2013) 1:16. doi: 10.1186/2049-2618-1-16
51. Edwards AM, Grossman TJ, Rudney JD. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun. (2006) 74:654–62. doi: 10.1128/IAI.74.1.654-662.2006
52. Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW. Bachrach G, Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. (2015) 83:1104–13. doi: 10.1128/IAI.02838-14
53. Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. (2006) 169:1901–9. doi: 10.2353/ajpath.2006.060681
54. Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon, Appl Environ Microbiol. (2005) 71:3692–700. doi: 10.1128/AEM.71.7.3692-3700.2005
55. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature (2012) 491:254–8. doi: 10.1038/nature11465
56. Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. (2006) 21:517–23. doi: 10.1016/j.tree.2006.06.013
57. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. (2012) 6:320–9. doi: 10.1038/ismej.2011.109
58. Grahn N, Hmani-Aifa M, Fransen K, Soderkvist P, Monstein HJ. Molecular identification of Helicobacter DNA present in human colorectal adenocarcinomas by 16S rDNA PCR amplification and pyrosequencing analysis, J Med Microbiol. (2005) 54:1031–5. doi: 10.1099/jmm.0.46122-0
59. Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. (2008) 23:1298–303. doi: 10.1111/j.1440-1746.2008.05490.x
60. Krishnan S, Eslick GD. Streptococcus bovis infection and colorectal neoplasia: a meta-analysis. Colorectal Dis. (2014) 16:672–80. doi: 10.1111/codi.12662
61. Mirza NN, McCloud JM, Cheetham MJ. Clostridium septicum sepsis and colorectal cancer-a reminder. World J Surg Oncol. (2009) 7:73. doi: 10.1186/1477-7819-7-73
62. Bor B, Cen L, Agnello M, Shi W, He X. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci Rep. (2016) 6:27956. doi: 10.1038/srep27956
63. Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, et al. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS ONE (2016) 11:e0155362. doi: 10.1371/journal.pone.0155362
64. Keskin M, Kononen E, Soderling E, Isik G, Firatli E, Uitto VJ, et al. Increased proliferation and decreased membrane permeability as defense mechanisms of Fusobacterium nucleatum against human neutrophilic peptide-1. Anaerobe (2014) 30:35–40. doi: 10.1016/j.anaerobe.2014.08.001
65. Kouidhi B, Al Qurashi YM, Chaieb K. Drug resistance of bacterial dental biofilm and the potential use of natural compounds as alternative for prevention and treatment. Microb Pathog. (2015) 80:39–49. doi: 10.1016/j.micpath.2015.02.007
66. Flynn KJ, Baxter NT, Schloss PD. Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere (2016) 1: doi: 10.1128/mSphere.00102-16
67. West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. (2015) 15:615–29. doi: 10.1038/nri3896
68. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and Up-regulating expression of microRNA-21. Gastroenterology (2017) 152:851–66 e24. doi: 10.1053/j.gastro.2016.11.018
69. Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. (2011) 17:1971–8. doi: 10.1002/ibd.21606
70. Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. (2005) 102:18538–43. doi: 10.1073/pnas.0509182102
71. Leffers N, Gooden MJ, de Jong RA, Hoogeboom BN, ten Hoor KA, Hollema H, et al Prognostic significance of tumor-infiltrating T-lymphocytes in primary and metastatic lesions of advanced stage ovarian cancer. Cancer Immunol Immunother. (2009) 58:449–59. doi: 10.1007/s00262-008-0583-5
72. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science (2006) 313:1960–4. doi: 10.1126/science.1129139
73. Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin Cancer Res. (2005) 11:6713–21. doi: 10.1158/1078-0432.CCR-05-0883
74. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell (2008) 133:775–87. doi: 10.1016/j.cell.2008.05.009
75. Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. (2007) 25:2546–53. doi: 10.1200/JCO.2006.08.5829
76. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. (2012) 12:253–68. doi: 10.1038/nri3175
77. Coussens LM, Pollard JW. Leukocytes in mammary development and cancer. Cold Spring Harb Perspect Biol. (2011) 3:doi: 10.1101/cshperspect.a003285
78. Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. (2008) 181:5791–802. doi: 10.4049/jimmunol.181.8.5791
79. Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med. (2006) 203:2691–702. doi: 10.1084/jem.20061104
80. Poschke I, Kiessling R. On the armament and appearances of human myeloid-derived suppressor cells. Clin Immunol. (2012) 144:250–68. doi: 10.1016/j.clim.2012.06.003
81. Wang LS, Kuo CT, Huang YW, Stoner GD, Lechner JF. Gene-diet interactions on colorectal cancer risk. Curr Nutr Rep. (2012) 1:132–41. doi: 10.1007/s13668-012-0023-1
82. Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis (2010) 31:37–49. doi: 10.1093/carcin/bgp272
83. Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, et al. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology (2008) 135:2128–40, 2140 e1-8. doi: 10.1053/j.gastro.2008.07.027
84. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. (2007) 8:286–98. doi: 10.1038/nrg2005
85. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. (2002) 3:415–28. doi: 10.1038/nrg816
86. Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. (1997) 57:808–11.
87. Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. (1998) 95:6870–5.
88. Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. (2001) 61:4689–92.
89. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. (2009) 9:162–74. doi: 10.1038/nri2506
90. Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. (2005) 5:641–54. doi: 10.1038/nri1668
91. Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. (2003) 24:302–6. doi: 10.1016/S1471-4906(03)00132-7
92. Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. (2007) 204:1757–64. doi: 10.1084/jem.20070590
93. Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. (1995) 63:4830–6.
94. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42:344–55. doi: 10.1016/j.immuni.2015.01.010
95. Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K, Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. (2011) 79:2597–607. doi: 10.1128/IAI.05118-11
96. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut (2016) 65:1973–80. doi: 10.1136/gutjnl-2015-310101
97. Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut (2012) 61:847–54. doi: 10.1136/gutjnl-2011-300865
98. Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology (2015) 148:77–87 e2. doi: 10.1053/j.gastro.2014.09.038
99. Leung A, Tsoi H, Yu J. Fusobacterium and Escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol. (2015) 9:651–7. doi: 10.1586/17474124.2015.1001745
100. Westdorp H, Fennemann FL, Weren RD, Bisseling TM, Ligtenberg MJ, Figdor CG, et al. Opportunities for immunotherapy in microsatellite instable colorectal cancer. Cancer Immunol Immunother. (2016) 65:1249–59. doi: 10.1007/s00262-016-1832-7
101. Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. (2016) 22:557–66. doi: 10.3748/wjg.v22.i2.557
102. Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. (2014) 455:43–57. doi: 10.1016/j.bbrc.2014.08.001
103. Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. (2010) 11:141–7. doi: 10.1038/ni.1828
104. Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci Signal (2011) 4:ra75. doi: 10.1126/scisignal.2001868
105. Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA (2008) 299:425–36. doi: 10.1001/jama.299.4.425
106. Nomura T, Ohkusa T, Okayasu I, Yoshida T, Sakamoto M, Hayashi H, et al. Mucosa-associated bacteria in ulcerative colitis before and after antibiotic combination therapy. Aliment Pharmacol Ther. (2005) 21:1017–27. doi: 10.1111/j.1365-2036.2005.02428.x
107. Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes (2010) 1:138–47. doi: 10.4161/gmic.1.3.12360
108. Manson McGuire, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, et al. Evolution of invasion in a diverse set of Fusobacterium species. MBio (2014) 5:e01864. doi: 10.1128/mBio.01864-14
109. Lee JY, Chu SH, Jeon JY, Lee MK, Park JH, Lee DC, et al. Effects of 12 weeks of probiotic supplementation on quality of life in colorectal cancer survivors: a double-blind, randomized, placebo-controlled trial. Dig Liver Dis. (2014) 46:1126–32. doi: 10.1016/j.dld.2014.09.004
110. Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgard L, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. (2017) 4:e000145. doi: 10.1136/bmjgast-2017-000145
111. Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut (2017) 67:1454–63. doi: 10.1136/gutjnl-2017-314814
112. Temraz S, Mukherji D, Alameddine R, Shamseddine A. Methods of overcoming treatment resistance in colorectal cancer. Crit Rev Oncol Hematol. (2014) 89:217–30. doi: 10.1016/j.critrevonc.2013.08.015
113. Seow HF, Yip WK, Fifis T. Advances in targeted and immunobased therapies for colorectal cancer in the genomic era. Onco Targets Ther. (2016) 9:1899–920. doi: 10.2147/OTT.S95101
114. Sun Y. Tumor microenvironment and cancer therapy resistance. Cancer Lett. (2016) 380:205–15. doi: 10.1016/j.canlet.2015.07.044
115. Borriello L, DeClerck YA. [Tumor microenvironment and therapeutic resistance process]. Med Sci. (2014) 30:445–51. doi: 10.1051/medsci/20143004021
116. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170:548–63 e16. doi: 10.1016/j.cell.2017.07.008
117. Ulivi P, Marisi G, Passardi A. Relationship between hypoxia and response to antiangiogenic therapy in metastatic colorectal cancer. Oncotarget (2016) 7:46678–91. doi: 10.18632/oncotarget.8712
118. Romano A, Parrinello NL, La Cava P, Tibullo D, Giallongo C, Camiolo G, et al. PMN-MDSC and arginase are increased in myeloma and may contribute to resistance to therapy. Expert Rev Mol Diagn. (2018) 18:675–83. doi: 10.1080/14737159.2018.1470929
119. Feng PH, Yu CT, Chen KY, Luo CS, Wu SM, Liu CY, et al S100A9(+) MDSC and TAM-mediated EGFR-TKI resistance in lung adenocarcinoma: the role of RELB. Oncotarget (2018) 9:7631–43. doi: 10.18632/oncotarget.24146
120. Diaz LA Jr, Le DT. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med. (2015) 373:1979. doi: 10.1056/NEJMc1510353
121. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350:1084–9. doi: 10.1126/science.aac4255
122. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359:104–8. doi: 10.1126/science.aao3290
123. West NR, Powrie F. Immunotherapy not working? check your microbiota. Cancer Cell (2015) 28:687–9. doi: 10.1016/j.ccell.2015.11.010
124. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359:97–103. doi: 10.1126/science.aan4236
125. Rossen NG, MacDonald JK, de Vries EM, D'Haens GR, de Vos WM, Zoetendal EG, et al. Fecal microbiota transplantation as novel therapy in gastroenterology: a systematic review. World J Gastroenterol. (2015) 21:5359–71. doi: 10.3748/wjg.v21.i17.5359
126. Borody T, Fischer M, Mitchell S, Campbell J. Fecal microbiota transplantation in gastrointestinal disease: 2015 update and the road ahead. Expert Rev Gastroenterol Hepatol. (2015) 9:1379–91. doi: 10.1586/17474124.2015.1086267
127. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA. (2014) 111:18321–6. doi: 10.1073/pnas.1406199111
128. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science (2018) 359:592–7. doi: 10.1126/science.aah3648
129. Flemer B, Herlihy M, O'Riordain M, Shanahan F, O'Toole PW. Tumour-associated and non-tumour-associated microbiota:Addendum. Gut Microbes (2018) 1–5. doi: 10.1080/19490976.2018.1435246
Keywords: Fusobacterium, colorectal cancer, tumor microenvironment, epigenetic changes, tumor immunity
Citation: Zhou Z, Chen J, Yao H and Hu H (2018) Fusobacterium and Colorectal Cancer. Front. Oncol. 8:371. doi: 10.3389/fonc.2018.00371
Received: 06 March 2018; Accepted: 21 August 2018;
Published: 15 October 2018.
Edited by:
Elizabeth J. Ryan, St. Vincent's University Hospital, IrelandReviewed by:
Angel Lanas, Universidad de Zaragoza, SpainCopyright © 2018 Zhou, Chen, Yao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Herui Yao, eWFvaGVydWlAMTYzLmNvbQ==
Hai Hu, aHVoYWlAbWFpbC5zeXN1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.