- 1Immunopathology Laboratory, School of Pharmacy, University of Camerino, Camerino, Italy
- 2Immunopathology Laboratory, School of Biosciences, Biotechnology and Veterinary Medicine, University of Camerino, Camerino, Italy
- 3Oncology Unit, Macerata Hospital, Macerata, Italy
Bladder cancer (BC) is ones of the most common cancer worldwide. It is classified in muscle invasive (MIBC) and muscle non-invasive (NMIBC) BC. NMIBCs frequently recur and progress to MIBCs with a reduced survival rate and frequent distant metastasis. BC detection require unpleasant and expensive cystoscopy and biopsy, which are often accompanied by several adverse effects. Thus, there is an urgent need to develop novel diagnostic methods for initial detection and surveillance in both MIBCs and NMIBCs. Multiple urine-based tests approved by FDA for BC detection and surveillance are commercially available. However, at present, sensitivity, specificity and diagnostic accuracy of these urine-based assays are still suboptimal and, in the attend to improve them, novel molecular markers as well as multiple-assays must to be translated in clinic. Now there are growing evidence toward the use of minimally invasive “liquid biopsy” to identify biomarkers in urologic malignancy. DNA- and RNA-based markers in body fluids such as blood and urine are promising potential markers in diagnostic, prognostic, predictive and monitoring urological malignancies. Thus, circulating cell-free DNA, DNA methylation and mutations, circulating tumor cells, miRNA, IncRNA and mRNAs, cell-free proteins and peptides, and exosomes have been assessed in urine specimens. However, proteomic and genomic data must to be validated in well-designed multicenter clinical studies, before to be employed in clinic oncology.
Introduction
Bladder cancer (BC) represents the 9th and 4th most common cancer worldwide and in men in the USA, respectively (1, 2). Its main histological type is urothelial carcinoma (UC). About 70–80% of BC is diagnosed as non-muscle invasive BC (NMIBC) and 20–30% as muscle invasive (MIBC). Because 10–30% of patients with NMIBC progress to invasive disease (3–8), early diagnosis and early detection of recurrence are very important. BC diagnosis requires cystoscopy and biopsy, which are unpleasant and costly procedures (9). It is necessary to develop new diagnostic methods less invasive and expensive for BC diagnosis and surveillance. The Food and Drug Administration (FDA) has approved the use of multiple urine-based tests that are commercially available. However, none of these tests has been routinely used and incorporated in the American Urological Association or in the European Association of Urology clinical guidelines for BC treatment (10). In this mini-review we discuss the clinical implementation by the use of novel molecular approaches and liquid biopsy in BC.
At present, the gold standard methods for BC diagnosis are urine cytology and cystoscopy. Cytopathology of urine specimens is the widely used non-invasive test for detection and surveillance of BC (11–13). Cytology is very specific (about 86%), but it is low sensitive (48%) limiting its use in low-grade BC (14–16). Diagnostic accuracy of urinary cytology is subjective, depending on cytopathologist expertise (17). Thus, new molecular-based urinary tests for reducing or substituting, the endoscopy frequency in BC recurrence patients, are required (18, 19).
Advanced technology utilizes patients' urine as samples instead of primary BC tissues to identify novel predictive biomarkers. At present, the major problem is to translate the extensive proteomic and genomic data in clinical practice and to validate the expression of these biomarkers in well-designed multicenter clinical studies (20).
Proteomic and Peptidomic Analysis
Proteomic analyses have opened a new horizon for cancer biomarker discovery (21). At present, seven tests are available: FDA approved six on seven of these tests, and the last one is in agree with the Clinical Laboratory Improvement Act standards. NMP22, NMP22 BladderChek, and UroVysion have FDA approval for BC diagnosis and surveillance; immunocytology (uCyt+), BTA-TRAK, and BTA-STAT have been approved only for surveillance (22–26).
In order to improve sensitivity, specificity and diagnostic accuracy in BC diagnosis, novel protein markers, waiting to be approved, are used experimentally. BCLA-1 and BCLA-4 are nuclear matrix proteins specifically targeting BC tissues, with no interference with infection, smoking, catheterization or cystitis (27). In patients with hematuria, aurora A kinase (AURKA) discriminates between low-grade BC vs. normal patients (28). The Aura Tek FDP Test™ in urine can detect BC recurrence (29). The activated leukocyte cell adhesion molecule (ALCAM), a cell adhesion molecule (30), positively correlates with tumor stage and overall survival (OS), after adjusting for patients, clinical features and Bacillus Calmette-Guerin treatment (31). Nicotinamide N-methyltransferase is high in BC patients and correlate with histological grade (32). Apurinic/apyrimidinic endonuclease 1/redox factor-1 (APE/Ref-1) levels are higher in BC, respect to non-BC, and correlate with tumor grade and stage; moreover it is high also in patients with recurrence history of BC (33). The cytokeratin-20 (CK20) urine RT-PCR assay shows 78–87% sensitivity and 56–80% specificity for urothelial BC detection, with improved diagnostic accuracy in tumor progression (34) but it has poor performance for low-grade tumors. Higher levels of CK8 and CK18 was detected in the urine by UBC Rapid test in high- vs low-grade BC (35).
As multiple markers for BC detection, increased urinary levels of apolipoprotein A1, A2, B, C2, C3, E (APOA1, APOA2, APOB, APOC2, APOC3, APOE) were found in BC relative to healthy controls (36, 37). A signature of 4 urinary fragments of uromodulin, collagen α-1 (I), collagen α-1 (III), and membrane-associated progesterone receptor component 1 seems to discriminate MIBCs from NMIBCs (38). Other panel employs IL-8, MMP-9/10, ANG, APOE, SDC-1, α1AT, PAI-1, VEGFA, and CA9 to diagnose BC starting from urine samples (39). The advantage of these multi-urinary protein biomarkers was evident in high- and low-grade and high- and low-stage disease (39). The combination of urinary markers such as midkine (MDK) and synuclein G or MDK, ZAG2 and CEACAM1 (40), angiogenin and clusterin (41) evaluated by immunoassay and urine cytology increases the sensitivity and specificity in NMIBC diagnosis (40). Increased CK20 and Insulin Like Growth Factor II (IGFII) levels were detected in the urine sediments of NMIBC patients compared to controls (42). Increased levels of urinary HAI-1 and Epcam evaluated by ELISA, are prognostic biomarkers in high-risk NMIBC patients (43). Urinary survivin evaluated by chemiluminescence enzyme immunoassay correlates with tumor stage, lymph node and distant metastases and represents a potential marker for preliminary BC diagnosis (44). Snail overexpression represents an independent prognostic factor for tumor recurrence in NMIBC (45). Finally, specific glycoproteins were identified by glycan-affinity glycoproteomics nanoplatforms in the urine of low- and high-grade NMIBC; among these, increased urinary CD44 levels were evidenced in high-grade MIBC (46).
Urinary metabolomics signature could also be useful in early BC. By ultra-performance liquid chromatography time and mass spectrometry, imidazole-acetic acid was evidenced in BC (47). Moreover, acid trehalose, nicotinuric acid, AspAspGlyTrp peptide were upregulated; inosinic acid, ureidosuccinic acid and GlyCysAlaLys peptide were downregulated in BC, but not in normal cohort (48). A metabolite panel with indolylacryloylglycine, N2-galacturonyl-L-lysine and aspartyl-glutamate permits to discriminate high- vs. low-grade BC (49). In addition, the alteration of phenylalanine, arginine, proline and tryptophan metabolisms was evidenced by UPLC-MS in NMBIC (50).
Circulating Tumor and Cell-Free DNA
Tumors release DNA fragments into circulation, called circulating tumor DNA (ctDNA) containing tumor-specific mutations, variations of copy number and alterations in DNA methylation status. This ctDNA reflects the heterogeneity of tumor subclones. In BC patients, ctDNA is detectable in over 70% of urine samples (51) and it allows to discriminate between BC patients and control subjects (52). CtDNA measures about 180 and 200 base pairs. It is easily accessible, but it is rapidly cleared from circulation following systemic therapy (53). PCR-based approaches, and more recently, digital-PCR and genome sequencing, represent the methods of choice for cell-free DNA (cfDNA) analysis.
DNA Methylation
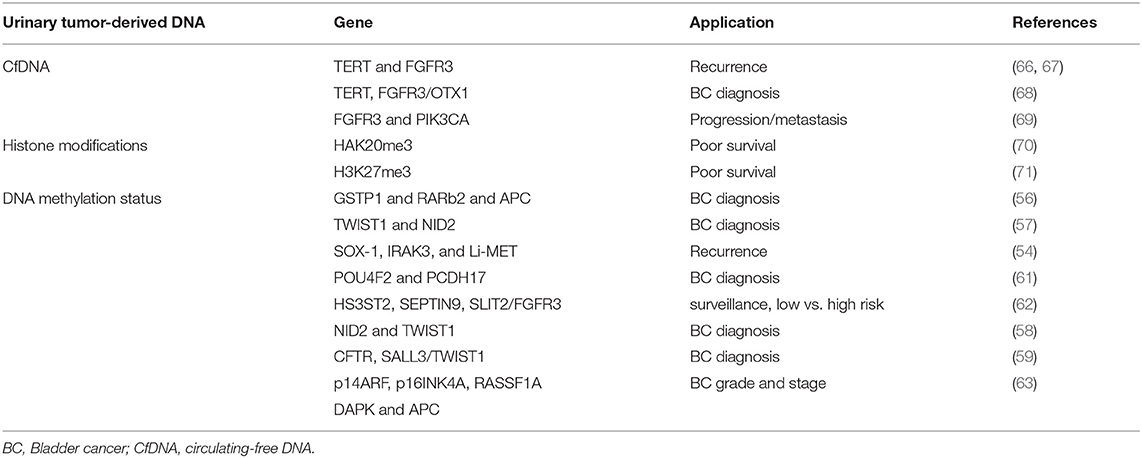
The methylation status of tumor-related genes represents a very important epigenetic alteration affecting cancer initiation and progression. Hyper- and hypo-methylated regions are identified in BC and in premalignant lesions. Alterations in DNA methylation status are chemically stable, develop early during tumorigenesis and can be assessed in circulating cfDNA fragments and in cells shed into the urine (54). A significant prevalence of methylated genes, for example APC and cyclin D2, was found in the urine from malignant vs. benign cases (55). Hyper-methylation in GSTP1 and RARβ2 and APC genes has been identified in the urine from BC patients (56). The evaluation of Twist Family BHLH Transcription Factor 1 (TWIST1) and NID2 genes methylation status in urine permits to differentiate primary BC patients from controls with 90% sensitivity and 93% specificity (57). In addition, the evaluation of the methylation status of NID2 and TWIST1 or CFTR, SALL3 and TWIST1 genes in urinary cells in combination with cytology, has been found to increase sensitivity and high negative predictive value in BC patients (58, 59). The analysis of 1,370 loci specific DNA methylation patterns seem to permit to distinguish NMIBC from MIBC (60). Sun and coworkers demonstrated higher recurrence predictivity than urine cytology and cystoscopy (80 vs. 35 vs. 15%) by using SOX-1, IRAK3, and Li-MET genes methylation status from urine sediments of BC patients (54). POU4F2 and PCDH17 methylation levels in urine distinguish BC from normal controls with 90% sensitivity and 94% specificity (61). Promoter hyper-methylation of HS3ST2, SEPTIN9 and SLIT2 genes combined with FGFR3 mutation showed 97.6% sensitivity and 84.8% specificity for diagnosis, surveillance and risk stratification in low- or high-risk NMIBC patients (62). Finally, the methylation status of p14ARF, p16INK4A, RASSF1A, DAPK, and APC tumor suppressor genes has been found to correlate with BC grade and stage (63).
Altogether, although promising results were obtained, accuracy of urinary methylated DNA is variable and results still await validation studies and complementary markers for clinical implementation (64, 65). In this regard, the recent introduction of the methylation-sensitive High Resolution Melting and Methylated CpG Island Recovery methods could further increases the sensitivity for the detection of methylome in BC urine (Table 1) (72, 73).
cfDNA, Mutation and Microsatellite Alterations
Since tumor-derived DNA can be released into circulation and mutations in cfDNA can be detected in various biological fluids, their use as non-invasive cancer biomarkers has been proposed. Urinary TERT promoter mutations, that occur early in urothelial neoplasia, FGFR3 mutation and telomere length correlate with high-risk BC recurrence (66, 67). TERT, evaluated by telomeric repeat amplification protocol, in combination with FGF3 and OTX1 shows high sensitivity in NMIBCs as well as in pT1 tumors and in high-grade BC (68). In addition, increased FGFR3 and PIK3CA mutated DNA levels in urine has been found to be indicative of progression and metastasis in NMIBC (69). Microsatellite analysis in circulating DNA of BC patients targets highly polymorphic, short tandem repeats. Loss of heterozygosity (LOH) analysis is more sensitive than urine cytology (97 vs. 79%), particularly for low-grade BC diagnosis. It also significantly improves the detection of low-grade and low-stage BC, with 95% sensitivity for G1-G2 grades and 100% for pTis and pTa tumors (Table 1) (74).
Histone Tail Modifications
The levels of histone methylation are lower in advanced tumors respect to controls and correlated to poor survival. Thus, increased levels of HAK20me3 were evidenced in a MIBC subset (70); furthermore high H3K27me3 levels correlate with worse survival after cystectomy in pT1-3 and pN- BC patients (71). H2AFX1 gene methylation was detected in paraffin-embedded BC and its expression correlated with increased recurrence rates (Table 1) (75).
Urinary Tumor RNA
Several RNA classes, messenger RNAs (mRNAs), microRNAs (miRs) and long non-coding RNAs (lncRNAs), have been recognized as potential non-invasive cancer biomarkers (76). Altered levels of circulating RNAs in cancer, which returned to normal following surgery have been reported (77), suggesting release of RNA molecules from tumors.
miRNAs (miRNAs)
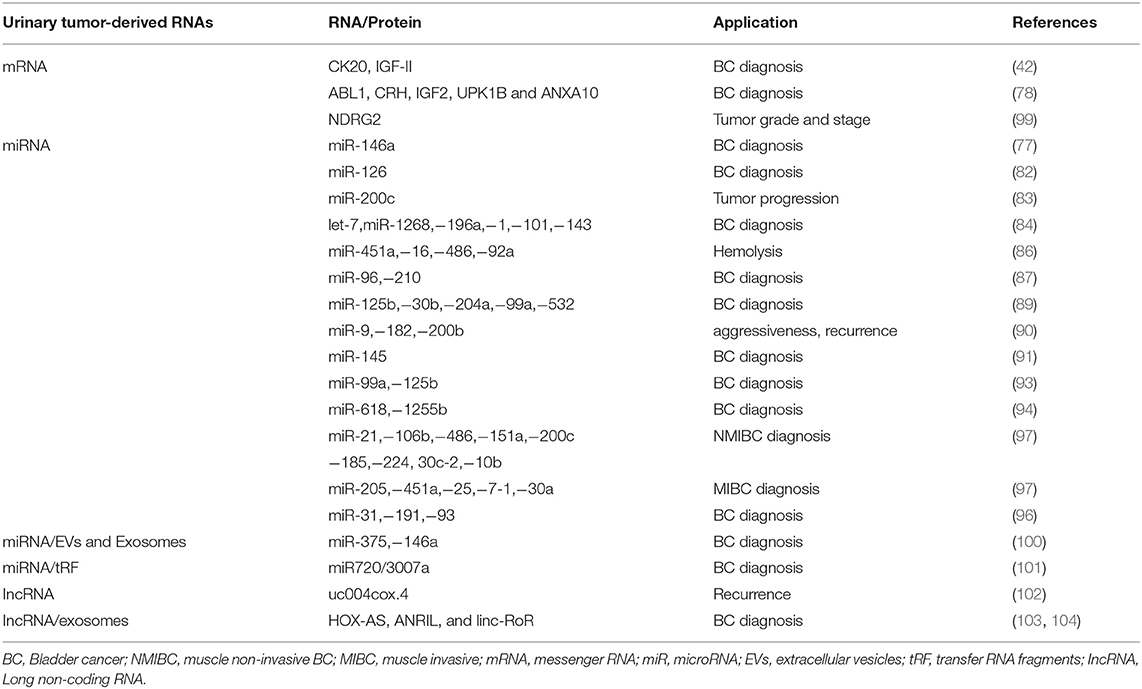
miRNAs are short (21–23 nucleotides length) non-coding RNAs regulating gene expression by pairing to the 3′untranslated region (UTR) of their target mRNA. Several miRNAs have been found to play an important role in tumorigenesis, progression and metastasis of cancer cells (78, 79). Urine seems to be a good source for miRNA detection for its content of cell-free nucleic acid in supernatant or sediments (80). However, the diagnostic significance in the detection of miRs in urine as respect to blood of BC patients is controversial (81). MiR-126 urinary levels were found to be enhanced in BC compared to healthy controls (82). Urine miR-146a-5p is significantly increased in high-grade BC (77). Low miR-200c expression correlates with tumor progression in NMIBCs (83). Chen et al. detected 74 miRNAs, of which 33 upregulated and 41 downregulated in BC compared to healthy patients (84). The most interesting are let-7miR, mir-1268, miR-196a, miR-1, miR-100, miR-101, and miR-143 (84). MiR-200 was identified as epithelial–mesenchymal transition regulator in BC cells by targeting Zinc Finger E-Box Binding Homeobox 1 (ZEB1), ZEB2 and Epidermal growth factor receptor (EGFR) (85). Some miRNAs have been associated with hemolysis including miR-451a, miR-16, miR-486-5p, and miR-92a (86). Eissa et al. by screening BC patients with negative cystoscopy, identified miR-96 and miR-210 in BC (87). Sapre et al., by using a panel of 12 miRNA, reduced the cystoscopy rates by 30% by increasing sensitivity and specificity (88). MiR-125b, miR-30b, miR-204, miR-99a, and miR-532-3p were downregulated in BC patient's urine supernatant, with miR-125 levels (95.7% specificity, 59.3% sensitivity) (89). MiR-9, miR-182 and miR-200b correlated with MIBC aggressiveness, recurrence-free and OS (90). MiR-145 distinguishes NMIBCs from non-BCs (91). MiR-144-5p inhibited BC proliferation, affecting CCNE1, CCNE2, CDC25A, PKMYT1 target genes (92). Cell-free urinary miR-99a and miRNA-125b were found to be downregulated in the urine supernatants of BC patients (sensitivity 86.7%; specificity 81.1%) (93). Urinary levels of miR-618 and miR-1255b-5p in MIBC patients were increased in comparison to controls (94). Multiple miRNA assay shows higher diagnostic performance than single RNA assay (95). By whole genome analysis increased miR-31-5p, miR-191-5p and miR-93-5p levels were identified in the urine of BC patients as compared to controls (96).
Recently, a miRNA profile, identified in urine by next-generation sequencing (NGS) analysis, has been capable to stratify different BC subtypes (97). In NMIBC G1/G2 patients a miR-205-5p upregulation compared to controls was observed. Among NMIBC G3, upregulation of miR-21-5p, miR-106b-3p, mir-486-5p, miR-151a-3p, miR-200c-3p, miR-185-5p, miR-185-5p and miR-224-5p and downregulation of miR-30c-2-5p and miR-10b-5p were observed. In MIBCs, miR-205-5p, miR-451a, miR-25-3p and miR-7-1-5p were upregulated, while miR-30a-5p was downregulated compared to controls (97). The application of NGS have increased the diagnostic accuracy. However results obtained in NGS were only partially overlapping with that obtained by qRT-PCR (98) (Table 2).
Long Non Coding RNAs (IncRNAs)
Long non coding RNAs (lncRNAs) regulate gene expression or epigenetic levels. Several findings show lncRNA changes in cancers suggesting a role in the promotion of tumor development and progression (105, 106). The use of lncRNAs as non-invasive BC marker has recently interested (107). Circulating urothelial carcinoma antigen 1 (UCA1) levels in urinary sediments represents a potential diagnostic marker for UC, with 81% sensitivity and 92% specificity (108). Du et al. describe high uc004cox.4 IncRNA level association with poor recurrence-free survival in NMIBCs (102). The retrotrasposome, long interspaced element-1 (LINE-1) has been found to be hypo-methylated and its expression was associated with long recurrence-free and tumor specific survival in BC (109) (Table 2).
Messenger RNAs (mRNAs)
Circulating messenger RNAs (mRNAs) were detected in cancer patients, although the majority of circulating mRNAs are degraded by RNases (110). Given their role in intracellular protein translation, their presence reflects the status of intracellular processes and they are potential cancer biomarkers. Urine Ubiquitin Conjugating Enzyme E2 C (UBE2C) mRNA levels were higher in BC patients, compared to normal and hematuria specimens (111). The expression of isoleucine glutamine motif-containing GTAase-activating proteins (IQGAP3) mRNA in urine was found higher in BC than in controls (112). Further analysis of IQGAP3, with respect to tumor invasiveness and grade also yielded a high diagnostic accuracy, suggesting that IQGAP3 can be used to discriminate BC from non-BC patients with hematuria (112).
In regard to mRNAs extracted by exfoliated urinary cells, the Xpert BC Monitor measuring ABL1, corticotropin releasing hormone (CRH), IGF2, uroplakin 1B (UPK1B), annexin A10 (ANXA10) mRNAs by RT-PCR, increased the overall sensitivity over urinary cytology in low-grade and pTa disease (113).
In addition, the presence of carbonic anhydrase 9 (CAIX) splice variant mRNA in the urine, increased the diagnostic performance for BC (90% sensitivity and 72% specificity) (114). The downregulation of N-Myc downstream-regulated gene 2 (NDRG2) mRNA levels in the urine of BC patients correlated with tumor grade and stage (99) (Table 2).
Transfer RNA Fragments (tRFs)
Elevated levels of transfer RNA fragments (tRF) are found in cancer (115). tRF are 14-32 base long single-stranded RNA derived from mature o precursor tRNA. They are grouped into 3 classes (tRF-1, −3, and −5) and, depending of their cleavage site within a mature RNA, they are further divided in 5 subclasses. The first identified tRF in NMIBCs was miR720/3007a (101) (Table 2).
Extracellular Vesicles (EVs) and Exosomes
Extracellular Vesicles (EVs) enrichment was found in BC patient urine. EVs, analyzed by MS based proteomics, demonstrated specific protein and miRNAs pattern in BC patients (116). By using a microarray platform and RT-PCR analysis, miR-375, and miR146a have been found to specifically identify high-grade and low-grade BC, respectively (100). The application of nanowires anchored into a microfluidic substrate will enable the efficiency of EV collection, thus permitting to identify EV harboring miRNAs (117).
Exosomes are membrane vesicles secreted in nearly all body fluids at elevated levels in cancer patients relative to healthy subjects (118, 119). They realize intercellular communication through transferring distinct biologically active molecules (RNAs, DNA, and proteins), thus influencing the therapeutic responses. The HOX transcript antisense RNA (HOTAIR) together with other IncRNA, such as HOX-AS-2, ANRIL, and linc-RoR, were augmented in urinary exosomes from high-grade MIBC patients (103). Loss of HOTAIR expression in BC cells alters the expression of SNA1, TWIST1, ZEB1, ZO1, MMP-1, Laminin Subunit Beta 3 (LAMB3), and Laminin Subunit Gamma 2 (LAMC2) epithelial-to mesenchymal transition genes. Moreover, the tumor-associated calcium-signal transducer 2 (TACSTD2) was found in BC exosomes by proteomic analysis (104). EVs can also promote BC progression by delivering the protein EGF-like repeat and discoidin I-like domain-containing protein-3 (120).
Exosomes in urine also contain miRNAs, in particular miR-1224-3p, miR-135b, and miR15b; in particular, miR-126/miR-152 ratio correlated with positive BC diagnosis (121) (Table 2).
Although EVs and exosomes represent an interesting source of cancer biomarkers, the lack of accurate isolation and detection methods affects their utilization in practice. In the next future, the development of sensitive capture platforms for exosomes, likely increases their introduction into clinic.
Urinary Microbiome
Dysbiosis of urinary microbiome has been suggested to be involved in bladder tumorigenesis. Recently, Wu et al. by analyzing DNA extracted by urine pellets, observed specific enrichment of Acinetobacter, Anaerococcus, and Sphingobacterium in BC cohort as respect to controls (122). Moreover, the increase of Herbaspirillum, Porphyrobacter, and Bacteroides in high-risk BC patients suggested that these genera may represent new potential biomarkers (122).
Conclusions and Perspectives
We provide the state of art into the use of urinary biomarkers as tool to aid diagnosis of BC. Urine cytology, utilized for decades, shows poor sensitivity, particularly for low-grade tumors. The addition of immunoassay and FISH analysis has provided an additional diagnostic armamentarium to determine which patients may need further evaluation. At present, there are growing evidence toward the use of “Liquid Biopsy” to identify urinary biomarkers such as circulating cell-free DNA, DNA methylation, miRNA, cell-free proteins/peptides and exosomes, useful for discriminating NMIBC from MIBC (123). The potential introduction of “smart toilets” working with a more advanced “nano-sensor” able to detect RNA and proteins in urine is close to reality, more that we think (124). However, now in clinical reality, there is an urgent need to validate the recently discovered extensive proteomic and genomic, epigenomic, transcriptomic and metabolomic data as urinary biomarkers in well-designed multicenter clinical studies (125, 126).
Author Contributions
GS, MM conception and design. GS drafting the manuscript. CA and NB critical revision of the manuscript.
Funding
The authors are grateful to AIRC IG 2014 and Post-doctoral Fellowships 2018 Fondazione Veronesi.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. (2015) 65:5–29. doi: 10.3322/caac.21254
2. van Rhijn BW, Burger M, Lotan Y, Solsona E, Stief CG, Sylvester RJ, et al. Recurrence and progression of disease in non-muscle-invasive bladder cancer: from epidemiology to treatment strategy. Eur Urol. (2009) 56:430–42. doi: 10.1016/j.eururo.2009.06.028
3. Prout GR, Barton BA, Griffin PP, Friedell GH. Treated history of noninvasive grade 1 transitional cell carcinoma. J Urol. (1992) 148:1413–9. doi: 10.1016/S0022-5347(17)36924-0
4. Herr HW. Tumor progression and survival of patients with high grade, noninvasive papillary (TaG3) bladder tumors: 15-year outcome. J Urol. (2000) 163:60–2. doi: 10.1016/s0022-5347(05)67972-4
5. Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffiuoux C, Denis L, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. (2006) 49:466–7. doi: 10.1016/j.eururo.2005.12.031
6. Johnson MI, Merrilees D, Robson WA, Lennon T, Masters J, Orr KE, et al. Oral ciprofloxacin or trimethoprim reduces bacteriuria after flexible cystoscopy. BJU Int. (2007) 100:826–9. doi: 10.1111/j.1464-410X.2007.07093.x
7. Soloway MS. Bladder cancer: lack of progress in bladder cancer—what are the obstacles? Nat Rev Urol. (2013) 10:5–6. doi: 10.1038/nrurol.2012.219
8. Türkölmez K, Tokgöz H, Reşorlu B, Köse K, Bedük Y. Muscle-invasive bladder cancer: predictive factors and prognostic difference between primary and progressive tumors. Urology (2007) 70:477–81. doi: 10.1016/j.urology.2007.05.008
9. Burke DM, Shackley DC, O'Reilly PH. The community-based morbidity of flexible cystoscopy. BJU Int. (2002) 89:347–9. doi: 10.1046/j.1464-4096.2001.01899.x
10. Zuiverloon TCM, de Jong FC, Theodorescu D. Clinical decision making in surveillance of non-muscle-invasive bladder cancer: the evolving roles of urinary cytology and molecular markers. Oncology (Williston Park) (2017) 31:855–62.
11. Têtu B. Diagnosis of urothelial carcinoma from urine. Mod Pathol. (2009) 22 (Suppl 2):S53–9. doi: 10.1038/modpathol.2008.193
12. Sapre N, Hong MK, Huang JG, Pedersen J, Ryan A, Anderson P, et al. Bladder cancer biorepositories in the “-omics” era: integrating quality tissue specimens with comprehensive clinical annotation. Biopreserv Biobank (2013) 11:166–72. doi: 10.1089/bio.2012.0062
13. Yafi FA, Brimo F, Steinberg J, Aprikian AG, Tanguay S, Kassouf W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol. (2015) 33:66.e25–31. doi: 10.1016/j.urolonc.2014.06.008
14. Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology (2003) 61:109–18. doi: 10.1016/S0090-4295(02)02136-2
15. Simon MA, Lokeshwar VB, Soloway MS. Current bladder cancer tests: unnecessary or beneficial? Crit Rev Oncol Hematol. (2003) 47:91–107. doi: 10.1016/S1040-8428(03)00074-X
16. van der Aa MN, Steyerberg EW, Sen EF, Zwarthoff EC, Kirkels WJ, van der Kwast TH, et al. Patients' perceived burden of cystoscopic and urinary surveillance of bladder cancer: a randomized comparison. BJU Int. (2008) 101:1106–10. doi: 10.1111/j.1464-410X.2007.07224.x
17. Shariat SF, Karam JA, Lotan Y, Karakiewizc PI. Critical evaluation of urinary markers for bladder cancer detection and monitoring. Rev Urol. (2008) 10:120–35.
18. Lokeshwar VB, Habuchi T, Grossman HB, Murphy WM, Hautmann SH, Hemstreet GP, et al. Bladder tumor markers beyond cytology: international Consensus Panel on bladder tumor markers. Urology (2005) 66 (6 Suppl 1):35–63. doi: 10.1016/j.urology.2005.08.064
19. Owens CL, VandenBussche CJ, Burroughs FH, Rosenthal DL. A review of reporting systems and terminology for urine cytology. Cancer Cytopathol. (2013) 121:9–14. doi: 10.1002/cncy.21253
20. Ralla B, Stephan C, Meller S, Dietrich D, Kristiansen G, Jung K. Nucleic acid-based biomarkers in body fluids of patients with urologic malignancies. Crit Rev Clin Lab Sci. (2014) 51:200–31. doi: 10.3109/10408363.2014.914888
21. Di Meo A, Pasic MD, Yousef GM. Proteomics and peptidomics: moving toward precision medicine in urological malignancies. Oncotarget (2016) 7:52460–74. doi: 10.18632/oncotarget.8931
22. Kim WT, Cho NH, Ham WS, Lee JS, Ju HJ, Kwon YU, et al. Comparison of the efficacy of urine cytology, Nuclear Matrix Protein 22 (NMP22), and Fluorescence in Situ Hybridization (FISH) for the diagnosis of bladder cancer. Korean J Urol. (2009) 50:6–11. doi: 10.4111/kju.2009.50.1.6
23. Hajdinjak T. UroVysion FISH test for detecting urothelial cancers: meta-analysis of diagnostic accuracy and comparison with urinary cytology testing. Urol Oncol. (2008) 26:646–51. doi: 10.1016/j.urolonc.2007.06.002
24. Horstmann M, Patschan O, Hennenlotter J, Senger E, Feil G, Stenzl A. Combinations of urine-based tumour markers in bladder cancer surveillance. Scand J Urol Nephrol. (2009) 43:461–6. doi: 10.3109/00365590903296837
25. Todenhöfer T, Hennenlotter J, Esser M, Mohrhardt S, Tews V, Aufderklamm S, et al. Combined application of cytology and molecular urine markers to improve the detection of urothelial carcinoma. Cancer Cytopathol. (2013) 121:252–60. doi: 10.1002/cncy.21247
26. He H, Han C, Hao L, Zang G. ImmunoCyt test compared to cytology in the diagnosis of bladder cancer: a meta-analysis. Oncol Lett. (2016) 12:83–8. doi: 10.3892/ol.2016.4556
27. Deininger S, Hennenlotter J, Rausch S, Docktor K, Neumann E, da Costa IA, et al. No influence of smoking status on the performance of urine markers for the detection of bladder cancer. J Cancer Res Clin Oncol. (2018) 144:1367–73. doi: 10.1007/s00432-018-2639-z
28. de Martino M, Shariat SF, Hofbauer SL, Lucca I, Taus C, Wiener HG, et al. Aurora A Kinase as a diagnostic urinary marker for urothelial bladder cancer. World J Urol. (2015) 33:105–10. doi: 10.1007/s00345-014-1267-8
29. Siemens DR, Morales A, Johnston B, Emerson L. A comparative analysis of rapid urine tests for the diagnosis of upper urinary tract malignancy. Can J Urol. (2003) 10:1754–8.
30. Rosso O, Piazza T, Bongarzone I, Rossello A, Mezzanzanica D, Canevari S, et al. The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res. (2007) 5:1246–53. doi: 10.1158/1541-7786.MCR-07-0060
31. Egloff SA, Du L, Loomans HA, Starchenko A, Su PF, Ketova T, et al. Shed urinary ALCAM is an independent prognostic biomarker of three-year overall survival after cystectomy in patients with bladder cancer. Oncotarget (2017) 8:722–41. doi: 10.18632/oncotarget.13546
32. Pozzi V, Di Ruscio G, Sartini D, Campagna R, Seta R, Fulvi P, et al. Clinical performance and utility of a NNMT-based urine test for bladder cancer. Int J Biol Mark. (2018) 33:94–101. doi: 10.5301/ijbm.5000311
33. Choi S, Shin JH, Lee YR, Joo HK, Song KH, Na YG, et al. Urinary APE1/Ref-1: A Potential bladder cancer biomarker. Dis Mark. (2016) 2016:7276502. doi: 10.1155/2016/7276502
34. Mi Y, Zhao Y, Shi F, Zhang M, Wang C, Liu X. Diagnostic accuracy of urine cytokeratin 20 for bladder cancer: a meta-analysis. Asia Pac J Clin Oncol. (2018) doi: 10.1111/ajco.13024. [Epub ahead of print].
35. Ecke TH, Weiß S, Stephan C, Hallmann S, Barski D, Otto T, et al. UBC® Rapid Test for detection of carcinoma in situ for bladder cancer. Tumor Biol. (2017) 39:1010428317701624. doi: 10.1177/1010428317701624
36. Chen YT, Chen CL, Chen HW, Chung T, Wu CC, Chen CD, et al. Discovery of novel bladder cancer biomarkers by comparative urine proteomics using iTRAQ technology. J Proteome Res. (2010) 5:5803–15. doi: 10.1021/pr100576x
37. Chen YT, Chen HW, Domanski D, Smith DS, Liang KH, Wu CC, et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J Proteomics (2012) 75:3529–45. doi: 10.1016/j.jprot.2011.12.031
38. Schiffer E, Vlahou A, Petrolekas A, Stravodimos K, Tauber R, Geschwend JE, et al. Prediction of muscle-invasive bladder cancer using urinary proteomics. Clin Cancer Res. (2009) 15:4935–43. doi: 10.1158/1078-0432.CCR-09-0226
39. Masuda N, Ogawa O, Park M, Liu AY, Goodison S, Dai Y, et al. Meta-analysis of a 10-plex urine-based biomarker assay for the detection of bladder cancer. Oncotarget (2018) 9:7101–11. doi: 10.18632/oncotarget.23872
40. Soukup V, Kalousová M, Capoun O, Sobotka R, Breyl Z, Pešl M, et al. Panel of urinary diagnostic markers for non-invasive detection of primary and recurrent urothelial urinary bladder carcinoma. Urol Int. (2015) 95:56–64. doi: 10.1159/000368166
41. Shabayek MI, Sayed OM, Attaia HA, Awida HA, Abozeed H. Diagnostic evaluation of urinary angiogenin (ANG) and clusterin (CLU) as biomarker for bladder cancer. Pathol Oncol Res. (2014) 20:859–66. doi: 10.1007/s12253-014-9765-y
42. Salomo K, Huebner D, Boehme MU, Herr A, Brabetz W, Heberling U, et al. Urinary transcript quantitation of CK20 and IGF2 for the non-invasive bladder cancer detection. J Cancer Res Clin Oncol. (2017) 143:1757–69. doi: 10.1007/s00432-017-2433-3
43. Snell KIE, Ward DG, Gordon NS, Goldsmith JC, Sutton AJ, Patel P, et al. Exploring the roles of urinary HAI-1, EpCAM & EGFR in bladder cancer prognosis & risk stratification. Oncotarget (2018) 9:25244–53. doi: 10.18632/oncotarget.25397
44. Yang Y, Xu J, Zhang Q. Detection of urinary surviving using a magnetic paricles-based chemiluminescence immunoassay for the preliminary diagnosis of bladder cancer and renal cell carcinoma combined with LAPTM4B. Oncol Lett. (2018) 15:7923–33. doi: 10.3892/ol.2018.8317
45. Santi R, Cai T, Nobili S, Galli IC, Amorosi A, Comperat E, et al. Snail immunohistochemical overexpression correlates to recurrence risk in non-muscle invasive bladder cancer: results from a longitudinal cohort study. Virchows Arch. (2018) 472:605–13. doi: 10.1007/s00428-018-2310-8
46. Azevedo R, Soares J, Gaiteiro C, Peixoto A, Lima L, Ferreira D, et al. Glycan affinity magnetic nanoplatforms for urinary glycobiomarkers discovery in bladder cancer. Talanta (2018) 184:347–55. doi: 10.1016/j.talanta.2018.03.028
47. Shao CH, Chen CL, Lin JY, Chen CJ, Fu SH, Chen YT, et al. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget (2017) 8:38802–10. doi: 10.18632/oncotarget.16393
48. Shen C, Sun Z, Chen D, Su X, Jiang J, Li G, et al. Developing urinary metabolomic signatures as early bladder cancer diagnostic markers. OMICS (2015) 19:1–11. doi: 10.1089/omi.2014.0116
49. Liu X, Cheng X, Liu X, He L, Zhang W, Wang Y, et al. Investigation of urinary metabolic variations and the application in bladder cancer biomarker discovery. Int J Cancer (2018) 143:408–18. doi: 10.1002/ijc.31323
50. Loras A, Trassierra M, Sanjuan-Herráez D, Martínez-Bisbal MC, Castell JV, Quintás G, et al. Bladder cancer recurrence surveillance by urine metabolomics analysis. Sci Rep. (2018) 8:9172. doi: 10.1038/s41598-018-27538-3
51. Goessl C, Müller M, Straub B, Miller K. DNA alterations in body fluids as molecular tumor markers for urological malignancies. Eur Urol. (2002) 41:668–76. doi: 10.1016/S0302-2838(02)00126-4
52. Brisuda A, Pazourkova E, Soukup V, Horinek A, Hrbáček J, Capoun O, et al. Urinary cell-free DNA quantification as non-invasive biomarker in patients with bladder cancer. Urol Int. (2016) 96:25–31. doi: 10.1159/000438828
53. Qin Z, Ljubimov VA, Zhou C, Tong Y, Liang J. Cell-free circulating tumor DNA in cancer. Chin J Cancer (2016) 35:36. doi: 10.1186/s40880-016-0092-4
54. Su SF, de Castro Abreu AL, Chihara Y, Tsai Y, Andreu-Vieyra C, Daneshmand S, et al. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin Cancer Res. (2014) 20:1978–89. doi: 10.1158/1078-0432.CCR-13-2637
55. Pu RT, Laitala LE, Clark DP. Methylation profiling of urothelial carcinoma in bladder biopsy and urine. Acta Cytol. (2006) 50:499–506. doi: 10.1159/000326003
56. Hauser S, Kogej M, Fechner G, VON Pezold J, Vorreuther R, Lümmen G, et al. Serum DNA hypermethylation in patients with bladder cancer: results of a prospective multicenter study. Anticancer Res. (2013) 33:779–84.
57. Renard I, Joniau S, van Cleynenbreugel B, Collette C, Naômé C, Vlassenbroeck I, et al. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur Urol. (2010) 58:96–104. doi: 10.1016/j.eururo.2009.07.041
58. Fantony JJ, Longo TA, Gopalakrishna A, Owusu R, Lance RS, Foo WC, et al. Urinary NID2 and TWIST1 methylation to augment conventional urine cytology for the detection of bladder cancer. Cancer Biomark. (2017) 18:381–7. doi: 10.3233/CBM-160261
59. van der Heijden AG, Mengual L, Ingelmo-Torres M, Lozano JJ, van Rijt-van de Westerlo CCM, Baixauli M, et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin Epigenetics. (2018) 10:71. doi: 10.1186/s13148-018-0496-x
60. Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. (2010) 70:8169–78. doi: 10.1158/0008-5472.CAN-10-1335
61. Wang Y, Yu Y, Ye R, Zhang D, Li Q, An D, et al. An epigenetic biomarker combination of PCDH17 and POU4F2 detects bladder cancer accurately by methylation analyses of urine sediment DNA in Han Chinese. Oncotarget (2016) 7:2754–64. doi: 10.18632/oncotarget.6666
62. Roperch JP, Grandchamp B, Desgrandchamps F, Mongiat-Artus P, Ravery V, Ouzaid I, et al. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer (2016) 16:704. doi: 10.1186/s12885-016-2748-5
63. Pietrusinski M, Kępczyński Ƚ, Jędrzejczyk A, Borkowska E, Traczyk-Borszyńska M, et al. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark. 2017:18:47–59. doi: 10.3233/CBM-160673
64. Peng M, Chen C, Hulbert A, Brock MV, Yu F. Non-blood circulating tumor DNA detection in cancer. Oncotarget (2017) 8:69162–73. doi: 10.18632/oncotarget.19942
65. Bosschieter J, Lutz C, Segerink LI, Vis AN, Zwarthoff EC, A van Moorselaar RJ, et al. The diagnostic accuracy of methylation markers in urine for the detection of bladder cancer: a systematic review. Epigenomics (2018) 10:673–87. doi: 10.2217/epi-2017-0156
66. Kinde I, Munari E, Faraj SF, Hruban RH, Schoenberg M, Bivalacqua T, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. (2013) 73:7162–7. doi: 10.1158/0008-5472.CAN-13-2498
67. Hosen I, Rachakonda PS, Heidenreich B, de Verdier PJ, Ryk C, Steineck G, et al. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. Int J Cancer (2015) 137:1621–9. doi: 10.1002/ijc.29526
68. Beukers W, van der Keur KA, Kandimalla R, Vergouwe Y, Steyerberg EW, Boormans JL, et al. FGFR3, TERT and OTX1 as a urinary biomarker combination for surveillance of patients with bladder cancer in a large prospective multicenter study. J Urol. (2017) 197:1410–8. doi: 10.1016/j.juro.2016.12.096
69. Christensen E, Birkenkamp-Demtröder K, Nordentoft I, Høyer S, van der Keur K, van Kessel K, et al. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur Urol. (2017) 71:961–9. doi: 10.1016/j.eururo.2016.12.016
70. Schneider AC, Heukamp LC, Rogenhofer S, Fechner G, Bastian PJ, von Ruecker A, et al. Global histone H4K20 trimethylation predicts cancer-specific survival in patients with muscle-invasive bladder cancer. BJU Int. (2011) 108:E290–6. doi: 10.1111/j.1464-410X.2011.10203.x
71. Liu J, Li Y, Liao Y, Mai S, Zhang Z, Liu Z, et al. High expression of H3K27me3 is an independent predictor of worse outcome in patients with urothelial carcinoma of bladder treated with radical cystectomy. Biomed Res Int. (2013) 2013:390482. doi: 10.1155/2013/390482
72. Hussmann D, Hansen LL. Methylation-Sensitive High Resolution Melting (MS-HRM). Methods Mol Biol. (2018) 1708:551–71. doi: 10.1007/978-1-4939-7481-8_28
73. Tommasi S, Besaratinia A. A versatile assay for detection of aberrant DNA methylation in bladder cancer. Methods Mol Biol. (2018) 1655:29–41. doi: 10.1007/978-1-4939-7234-0_3
74. Seripa D, Parrella P, Gallucci M, Gravina C, Papa S, Fortunato P, et al. Sensitive detection of transitional cell carcinoma of the bladder by microsatellite analysis of cells exfoliated in urine. Int J Cancer (2001) 95:364–9. doi: 10.1002/1097-0215(20011120)95:6<364::AID-IJC1064>3.0.CO;2-V
75. García-Baquero R, Puerta P, Beltran M, Alvarez-Mújica M, Alvarez-Ossorio JL, Sánchez-Carbayo M. Methylation of tumor suppressor genes in a novel panel predicts clinical outcome in paraffin-embedded bladder tumors. Tumour Biol. (2014) 35:5777–86. doi: 10.1007/s13277-014-1767-6
76. Bryzgunova OE, Laktionov PP. Extracellular nucleic acids in urine: sources, structure, diagnostic potential. Actanaturae (2015) 7:48–54.
77. Sasaki H, Yoshiike M, Nozawa S, Usuba W, Katsuoka Y, Aida K, et al. expression level of urinary microrna-146a-5p is increased in patients with bladder cancer and decreased in those after transurethral resection. Clin Genitourin Cancer (2016) 14:e493–9. doi: 10.1016/j.clgc.2016.04.002
78. Sethi S, Sethi S, Bluth MH. Clinical Implication of micrornas in molecular pathology: an update for 2018. Clin Lab Med. (2018) 38:237–51. doi: 10.1016/j.cll.2018.02.003
79. Liu X, Wu Y, Wu Q, Wang Q, Yang Z, Li L. MicroRNAs in biofluids are novel tools for bladder cancer screening. Oncotarget (2017) 8:32370–9. doi: 10.18632/oncotarget.16026
80. Fuessel S, Lohse-Fischer A, Vu Van D, Salomo K, Erdmann K, Wirth MP. quantification of micrornas in urine-derived specimens. Methods Mol Biol. (2018) 1655:201–26. doi: 10.1007/978-1-4939-7234-0_16
81. Xiao S, Wang J, Xiao N. MicroRNAs as noninvasive biomarkers in bladder cancer detection: a diagnostic meta-analysis based on qRT-PCR data. Int J Biol Markers (2016) 31:e276–85. doi: 10.5301/jbm.5000199
82. Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. (2010) 28:655–61. doi: 10.1016/j.urolonc.2009.01.027
83. Wiklund ED, Gao S, Hulf T, Sibbritt T, Nair S, Costea DE, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE (2011) 6:e27840. doi: 10.1371/journal.pone.0027840
84. Chen YH, Wang SQ, Wu XL, Shen M, Chen ZG, Chen XG, et al. Characterization of microRNAs expression profiling in one group of Chinese urothelial cell carcinoma identified by Solexa sequencing. Urol Oncol. (2013) 31:219–27. doi: 10.1016/j.urolonc.2010.11.007
85. Braicu C, Cojocneanu-Petric R, Chira S, Truta A, Floares A, Petrut B, et al. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomed. (2015) 10:791–800. doi: 10.2147/IJN.S72904
86. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res. (2012) 5:492–7. doi: 10.1158/1940-6207.CAPR-11-0370
87. Eissa S, Matboli M, Essawy NO, Kotb YM. Integrative functional genetic-epigenetic approach for selecting genes as urine biomarkers for bladder cancer diagnosis. Tumour Biol. (2015) 36:9545–52. doi: 10.1007/s13277-015-3722-6
88. Sapre N, Macintyre G, Clarkson M, Naeem H, Cmero M, Kowalczyk A, et al. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br J Cancer (2016) 114:454–62. doi: 10.1038/bjc.2015.472
89. Pospisilova S, Pazourkova E, Horinek A, Brisuda A, Svobodova I, Soukup V, et al. MicroRNAs in urine supernatant as potential non-invasive markers for bladder cancer detection. Neoplasma (2016) 63:799–808. doi: 10.4149/neo_2016_518
90. Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. MicroRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer (2013) 132:2479–91. doi: 10.1002/ijc.27949
91. Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. (2012) 41:1871–8. doi: 10.3892/ijo.2012.1622
92. Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer (2015) 113:282–9. doi: 10.1038/bjc.2015.195
93. Zhang DZ, Lau KM, Chan ES, Wang G, Szeto CC, Wong K, et al. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PLoS ONE (2014) 9:e100793. doi: 10.1371/journal.pone.0100793
94. Tölle A, Jung M, Rabenhorst S, Kilic E, Jung K, Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep. (2013) 30:1949–56. doi: 10.3892/or.2013.2621
95. Chen L, Cui Z, Liu Y, Bai Y, Lan F. MicroRNAs as biomarkers for the diagnostics of bladder cancer: a meta-analysis. Clin Lab. (2015) 61:1101–8.
96. Juracek J, Peltanova B, Dolezel J, Fedorko M, Pacik D, Radova L, et al. Genome-wide identification of urinary cell-free microRNAs for non-invasive detection of bladder cancer. J Cell Mol Med. (2018) 22:2033–8. doi: 10.1111/jcmm.13487
97. Pardini B, Cordero F, Naccarati A, Viberti C, Birolo G, Oderda M, et al. MicroRNA profiles in urine by next-generation sequencing can stratify bladder cancer subtypes. Oncotarget (2018) 9:20658–69. doi: 10.18632/oncotarget.25057
98. Matullo G, Naccarati A, Pardini B. MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids. Int J Cancer (2016) 138:2334–45. doi: 10.1002/ijc.2989
99. Zhang M, Ren B, Li Z, Niu W, Wang Y. Expression of N-Myc Downstream-Regulated Gene 2 in bladder cancer and its potential utility as a urinary diagnostic biomarkers. Med Sci Monit. (2017) 23:4644–9. doi: 10.12659/MSM.901610
100. Andreu Z, Otta Oshiro R, Redruello A, López-Martín S, Gutiérrez-Vázquez C, Morato E, et al. Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur J Pharm Sci. (2017) 98:70–9. doi: 10.1016/j.ejps.2016.10.008
101. Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer (2015) 14:194. doi: 10.1186/s12943-015-0466-2
102. Du L, Duan W, Jiang X, Zhao L, Li J, Wang R, et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. (2018) 22:2838–45. doi: 10.1111/jcmm.13578
103. Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A, et al. Expression of the long non-coding rna hotair correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE (2016) 11:e0147236. doi: 10.1371/journal.pone.0147236
104. Chen CL, Lai YF, Tang P, Chien KY, Yu JS, Tsai CH et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res. (2012) 11:5611–29. doi: 10.1021/pr3008732
105. Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. (2014) 65:1140–51. doi: 10.1016/j.eururo.2013.12.003
106. Terracciano D, Ferro M, Terreri S, Lucarelli G, D'Elia C, Musi G, et al. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl Res. (2017) 184:108–17. doi: 10.1016/j.trsl.2017.03.005
107. Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. (2014) 281:1750–8. doi: 10.1111/febs.12737
108. Peter S, Borkowska E, Drayton RM, Rakhit CP, Noon A, Chen W, et al. Identification of differentially expressed long noncoding RNAs in bladder cancer. Clin Cancer Res. (2014) 20:5311–21. doi: 10.1158/1078-0432.CCR-14-0706
109. Neuhausen A, Florl AR, Grimm MO, Schulz WA. DNA methylation alterations in urothelial carcinoma. Cancer Biol Ther. (2006) 5:993–1001. doi: 10.4161/cbt.5.8.2885
110. Deligezer U, Erten N, Akisik EE, Dalay N. Circulating fragmented nucleosomal DNA and caspase-3 mRNA in patients with lymphoma and myeloma. Exp Mol Pathol. (2006) 80:72–6. doi: 10.1016/j.yexmp.2005.05.001
111. Kim WT, Jeong P, Yan C, Kim YH, Lee IS, Kang HW, et al. UBE2C cell-free RNA in urine can discriminate between bladder cancer and hematuria. Oncotarget (2016) 7:58193–202. doi: 10.18632/oncotarget.11277
112. Kim WT, Kim YH, Jeong P, Seo SP, Kang HW, Kim YJ, et al. Urinary cell-free nucleic acid IQGAP3: a new non-invasive diagnostic marker for bladder cancer. Oncotarget (2018) 9:14354–65. doi: 10.18632/oncotarget
113. Pichler R, Fritz J, Tulchiner G, Klinglmair G, Soleiman A, Horninger W, et al. Increased accuracy of a novel mRNA-based urine test for bladder cancer surveillance. BJU Int. (2018) 121:29–37. doi: 10.1111/bju.14019
114. Malentacchi F, Vinci S, Melina AD, Kuncova J, Villari D, Nesi G, et al. Urinary carbonic anhydrase IX splicing messenger RNA variants in urogenital cancers. Urol Oncol. (2016) 34:292.e9–292.e16. doi: 10.1016/j.urolonc.2016.02.01
115. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA (2010) 16:673–95. doi: 10.1261/rna.2000810
116. Lee J, McKinney KQ, Pavlopoulos AJ, Niu M, Kang JW, Oh JW, et al. Altered proteome of extracellular vesicles derived from bladder cancer patients urine. Mol Cells (2018) 41:179–87. doi: 10.14348/molcells.2018.2110
117. Yasui T, Yanagida T, Ito S, Konakade Y, Takeshita D, Naganawa T, et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci Adv. (2017) 3:e1701133. doi: 10.1126/sciadv.1701133
118. Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget (2015) 6:37151–68. doi: 10.18632/oncotarget.6022
119. Rosell R, Wei J, Taron M. Circulating MicroRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer (2009) 10:8–9. doi: 10.3816/CLC.2009.n.001
120. Beckham CJ, Olsen J, Yin PN, Wu CH, Ting HJ, Hagen FK, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol. (2014) 192:583–92. doi: 10.1016/j.juro.2014.02.035
121. Huang X, Liang M, Dittmar R, Wang L. Extracellular microRNAs in urologic malignancies: chances and challenges. Int J Mol Sci. (2013) 14:14785–99. doi: 10.3390/ijms140714785
122. Wu P, Zhang G, Zhao J, Chen J, Chen Y, Huang W, et al. Profiling the urinary microbiota in male patients with bladder cancer in China. Front Cell Infect Microbiol. (2018) 8:167. doi: 10.3389/fcimb.2018.00167
123. Ward DG, Bryan RT. Liquid biopsies for bladder cancer. Transl Androl Urol. (2017) 6:331–5. doi: 10.21037/tau.2017.03.08
125. Piao XM, Byun YJ, Kim WJ, Kim J. Unmasking molecular profiles of bladder cancer. Invest Clin Urol. (2018) 59:72–82. doi: 10.4111/icu.2018.59.2.72
Keywords: urinary biomarkers, bladder cancer, liquid biopsy, microRNA, exosomes
Citation: Santoni G, Morelli MB, Amantini C and Battelli N (2018) Urinary Markers in Bladder Cancer: An Update. Front. Oncol. 8:362. doi: 10.3389/fonc.2018.00362
Received: 29 June 2018; Accepted: 16 August 2018;
Published: 07 September 2018.
Edited by:
Rodolfo Montironi, Università Politecnica delle Marche, ItalyReviewed by:
Simona Di Francesco, Independent researcher, Chieti, ItalyRiccardo Autorino, Virginia Commonwealth University, United States
Copyright © 2018 Santoni, Morelli, Amantini and Battelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giorgio Santoni, Z2lvcmdpby5zYW50b25pQHVuaWNhbS5pdA==
 Giorgio Santoni
Giorgio Santoni Maria B. Morelli
Maria B. Morelli Consuelo Amantini
Consuelo Amantini Nicola Battelli3
Nicola Battelli3