- 1Genetics Laboratory, Department of Neuroimaging, Department of Psychiatry, Nankai University Affiliated Anding Hospital, Tianjin Anding Hospital, Tianjin, China
- 2Psychiatric Genetic Laboratory, Department of Psychiatry, Jining Medical University, Jining, China
- 3Department of Psychiatric Genetics, Tianjin Medical University, Tianjin, China
- 4Department of Psychiatry, Wenzhou Seventh People’s Hospital, Wenzhou, China
- 5Department of Neuroimaging Laboratory, Qilu Hospital of Shandong University, Jinan, China
- 6Department of Pharmacy, The First Hospital of Hebei Medical University, Shijiazhuang, China
- 7GHM Institute of CNS Regeneration, Jinan University, Guangzhou, China
Schizophrenia (SCZ) is a common psychiatric disorder with polygenetic pathogenesis. Among the many identified candidate genes and loci, the group of tumour suppressor genes has drawn our interest. In this mini-review article, we describe evidence of a correlation between major tumour suppressor genes and SCZ development. Genetic mutations ranging from single nucleotide polymorphisms to large structural alterations have been found in tumour-related genes in patients with SCZ. Epigenetic mechanisms, including DNA methylation/acetylation and microRNA regulation of tumour suppressor genes, have also been implicated in SCZ. Beyond genetic correlations, we hope to establish causal relationships between tumour suppressor gene function and SCZ risk. Accumulating evidence shows that tumour suppressor genes may mediate cell survival and neural development, both of which contribute to SCZ aetiology. Moreover, converging intracellular signalling pathways indicate a role of tumour suppressor genes in SCZ pathogenesis. Tumour suppressor gene function may mediate a direct link between neural development and function and psychiatric disorders, including SCZ. A deeper understanding of how neural cell development is affected by tumour suppressors may lead to improved anti-psychotic drugs.
Genetic Factors of Schizophrenia
Schizophrenia (SCZ) has a worldwide prevalence of approximately 1% (Leucht et al., 2007). The clinical symptoms of SCZ include delusions, hallucinations, social deficits, and emotional and cognitive impairments (Sass and Parnas, 2003). SCZ has become a major health burden as it affects patients’ well-being as well as the welfare of their families and society at large (Millier et al., 2014). The disease typically has an early onset around adolescence (Häfner et al., 1993), raising the possibility of genetic factors contributing to its pathogenesis. In addition, classical twin studies of SCZ have revealed a high heritability (Sullivan et al., 2003). However, early efforts to identify candidate SCZ genes by linkage analysis did not produce consistent results (Shi et al., 2008), suggesting that SCZ aetiology could not be attributed to a single gene mutation. We now understand that genetic contributions to SCZ are inherently complex and include structural variations and single-nucleotide variations (Chen et al., 2015). Modern genetic techniques, especially genome-wide association studies and next-generation DNA sequencing approaches, have significantly accelerated the identification of SCZ risk genes and loci. Perhaps unsurprisingly, these studies, using data from very large patient populations, have found that multiple gene variants are significantly associated with SCZ, further supporting the polygenic and heterogeneous nature of SCZ aetiology.
At present, the principal candidate loci for SCZ include the Disrupted in SCZ-1 gene, which regulates neurodevelopment (Miyoshi et al., 2003), and the major histocompatibility complex genes (Purcell et al., 2009), suggesting immune system involvement in SCZ pathology. Other major risk genes include those encoding transcription factors and synaptic proteins such as voltage-gated calcium channels (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). In addition, a group of tumour suppressor genes have also been associated with SCZ (Catts and Catts, 2000). Their possible involvement is bolstered by a recently published transcriptomic meta-analysis that demonstrated an inverse relationship between psychiatric disorders and cancer risk, and identified three major signalling cascades including the p53, Wnt, and peptidylprolyl cis/trans isomerase NIMA-interacting 1 pathways that might mediate this relationship (Ibáñez et al., 2014). In this review article, we will revisit the evidence for the association between SCZ and tumour suppressor genes. Efforts to understand this relationship may result in the identification of novel biomarkers for early SCZ diagnosis and aid in the exploration of SCZ pathology.
The Correlation Between Tumour Suppressor Genes and SCZ
Epidemiological Studies of Tumour Susceptibility in Patients With SCZ
The first argument for the relationship between SCZ and tumour suppressor genes is based on epidemiological surveys indicating reduced cancer risk in schizophrenics and their relatives. For example, a well-designed study found a significantly lower risk of cancer in patients with familial aggregated SCZ and their biological parents when compared to that in the general population (Gal et al., 2012). Similar results were obtained in sibling studies reporting a reduced cancer risk in the affected siblings of patients with SCZ (Levav et al., 2007). We speculate that the upregulation of tumour suppressor gene expression may confer susceptibility to SCZ. However, Lichtermann et al. (2001) provided contradictory evidence by showing that patients with SCZ had a higher risk of lung or pharyngeal cancer than their unaffected relatives, whose risk was lower than that in the general population. The authors argued that the elevated cancer risk in patients with SCZ was likely attributable to their unhealthy lifestyles, whereas the lower-than-normal risk in unaffected siblings might reflect the protective effects of increased tumour suppressor gene expression uncompromised by unhealthy lifestyle changes. In any case, this evidence is consistent with an interaction between SCZ and tumour suppressor gene function.
Gene Association Studies Linking TP53 and Wnt Pathway Genes to SCZ
The earliest evidence correlating SCZ with a tumour suppressor gene was the finding that SCZ was associated with mutations in the TP53 gene (Catts and Catts, 2000). This gene encodes the well-established tumour suppressor protein p53 (Jiang et al., 2011), and TP53 mutations are frequently observed in various human cancers (Levine and Oren, 2009). The role of p53 in SCZ pathogenesis was further supported by three separate lines of evidence: (1) two new SCZ candidate genes were found on human chromosome 6q21, which was previously shown to contain a tumour suppressor gene (Morelli et al., 2000), and an SCZ-associated gene locus containing the common fragile site FRA6F was observed in various human leukaemias (Morelli et al., 2002); (2) increased apoptosis was reported to result in neurodevelopmental abnormalities, including SCZ (Sanders et al., 2013); and (3) p53 was reported to induce cellular apoptosis to prevent malignant transformation and tumour development (Vousden and Prives, 2009).
The involvement of multiple tumour suppressor genes in SCZ indicates that certain cellular mechanisms may regulate both tumourigenesis and neural function. Among such putative mechanisms, Wnt signalling is widely reported to be involved in SCZ pathogenesis (Peng et al., 2014). In fact, Wnt signalling is a pleiotropic pathway mediating nearly every aspect of cell growth, including tumorigenesis. For example, Wnt1 was identified as an oncogene (Nusse et al., 1984). It is not unexpected that the Wnt pathway can mediate SCZ by modulating neurodevelopment. In the canonical Wnt pathway, Akt kinase, which is a glycogen synthase kinase 3β inhibitor, and β-catenin are the major downstream effector proteins. An early study reported decreased β-catenin expression in the hippocampal regions of patients with SCZ (Cotter et al., 1998). More compelling evidence was recently obtained by demonstrating abnormal Wnt signalling in human-induced pluripotent stem cells from patients with SCZ during differentiation into neural progenitor cells (Topol et al., 2015). In addition, frizzled protein 7, which is a Wnt receptor, was recently found to be upregulated in patients with SCZ (Hoseth et al., 2018). The second tumour suppressor gene product of the canonical Wnt/β-catenin pathway to be associated with SCZ is adenomatous polyposis coli (APC). In an animal study using the N-methyl-D-aspartate receptor antagonist MK-801 to induce SCZ-like behaviours, APC gene expression in the prefrontal cortex and ventral tegmental area was associated with SCZ symptoms (Yu et al., 2011). Furthermore, a systematic study using the transmission disequilibrium test identified three single nucleotide polymorphisms (SNPs) of the APC gene that are correlated with SCZ (Cui et al., 2005). Taken together, these gene associations suggest a possible link between the Wnt signalling pathway and SCZ.
Other Candidate Tumour Suppressor Genes Associated With SCZ
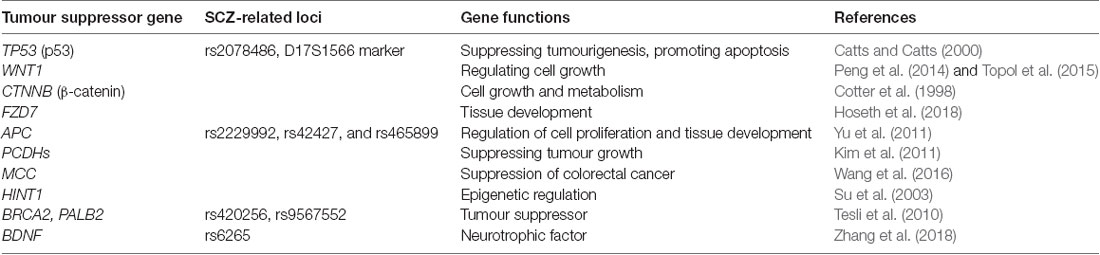
Other tumour suppressor genes have also been associated with SCZ. For example, transforming growth factor-beta type II serine/threonine kinase receptor on chromosome 3p22 was shown to be transcriptionally upregulated in patients with SCZ, and its transcription was normalised after antipsychotic treatment (Numata et al., 2008). Protocadherins have also been associated with SCZ and tumour suppressor functions (Kim et al., 2011). Similar to lung cancer, the prevalence of colorectal cancer is reported to be lower in SCZ cohorts than in unaffected individuals (Catts et al., 2008). In a detailed study, allele-specific expression of the mutated in colorectal cancer gene at the rs2227948 and rs2227947 loci was found to be significantly different between patients with SCZ and healthy individuals (Wang et al., 2016), suggesting that mutated in colorectal cancer, a potential tumour suppressor gene, might be involved in SCZ. The tumour suppressor gene histidine triad nucleotide-binding protein 1 is down-regulated in the prefrontal cortex of patients with SCZ (Elashoff et al., 2007). In human patients with SCZ, histidine triad nucleotide-binding protein 1 is associated with acute behavioural changes (Su et al., 2003), whereas histidine triad nucleotide-binding protein 1 knockout mice show elevated anxiety- and depression-like behaviours (Sun et al., 2017). We have summarised the major tumour suppressor genes associated with SCZ described to date in Table 1.
Genetics of Tumour Suppressors and SCZ Susceptibility
Single gene mutations can present in multiple forms, from SNPs to whole-gene duplications or inversions/deletions (indels). Unique phenotypes are associated with different mutant alleles and loci. In genetic studies of SCZ, a number of different candidate loci have been identified. The relationship between p53 polymorphisms and SCZ was reported in an early study showing that patients carrying specific alleles had reduced vulnerability to lung cancer (Park et al., 2004). Specifically, whereas the rs1042522 locus (containing a Pro72Arg mutation and a 16-bp insertion) was not significantly associated with an increased risk of SCZ (Papiol et al., 2004), rs2078486 and other SNP loci in the TP53 gene appeared to significantly increase or decrease SCZ susceptibility (Yang et al., 2004). In a subsequent study, transmission disequilibrium test-based analyses consistently linked specific p53 polymorphisms, including CAA indels and a 16-bp indel, to SCZ pathogenesis in independent patient cohorts (Ni et al., 2005). Moreover, a study of an isolated Spanish population revealed structural variations in TP53 at the D17S1566 marker that conferred a high risk of SCZ (Tabarés-Seisdedos et al., 2008). A case-control study further elaborated the relationship between the TP53 codon 72 polymorphism and SCZ risk (Lung et al., 2009), whereas TP53 alleles carrying BstUI (exon 4) and MspI (intron 6) restriction sites were shown to confer greater susceptibility to lung cancer in Turkish patients with SCZ (Ozbey et al., 2011). Taken together, these studies demonstrate the existence of multiple TP53 loci associated with increased susceptibility to SCZ.
Additional candidate genes and loci associated with SCZ continue to be identified. A genomic study focusing on human chromosome 8p has identified dozens of risk loci, encoding both protein and microRNA in each region, including various tumour suppressor genes with structural variations, such as copy number variants, microdeletions, and microduplications, all of which contribute to SCZ (Tabarés-Seisdedos and Rubenstein, 2009). The breast cancer 2 and partner and localiser of breast cancer 2 genes, two commonly known risk genes for breast cancer, have been reported to be associated with SCZ at the rs420256 and rs9567552 loci, respectively (Tesli et al., 2010). Polymorphisms of glutathione peroxidase, in which higher numbers of GAG repeats can increase the risk of SCZ, are associated with prostate and colon cancer development (Zmorzyński et al., 2015). The genome-wide association study approach has revealed other candidate loci conferring susceptibility to SCZ. The gene encoding the epigenetic reader bromodomain-containing protein 4 is a major risk locus in both breast cancer and SCZ (Zuber et al., 2017). A study of bromodomain-containing protein 4 expression revealed that the rs138880 allele was associated with SCZ (Dyrvig et al., 2017). Moreover, analysis of X-ray repair cross-complementing 4 polymorphisms revealed protective effects of the rs6452536 and rs35268 loci against SCZ and colorectal cancer in a Chinese population (Wang et al., 2010). A genome-wide association study of oral squamous cell carcinoma identified a novel SNP at the X-ray repair cross-complementing 4 rs1412115 locus, in which an A > G substitution increases the risk of oral squamous cell carcinoma and is also related to SCZ (Ma et al., 2015). Opposing evidence also exists, as another study of cervical squamous cell carcinoma revealed correlations between TATC or CAA indel polymorphisms and increased risk of both cancer and SCZ (Shi et al., 2012). Nonetheless, the convergence of genetic risk for multiple tumour types and SCZ strongly indicates common underlying cellular mechanisms. In examining the Wnt signalling pathway, the gene encoding a major Wnt receptor, frizzled-3, was found to have a number of SNPs associated with SCZ pathogenesis (Katsu et al., 2003). Similar patterns for SCZ-related SNPs have been found in a downstream Wnt effector gene, Dickkopf 4, in a Chinese population (Proitsi et al., 2008). In addition, three SNPs in APC exons that are associated with SCZ, including rs2229992, rs42427, and rs465899 have been identified (Chambers and Perrone-Bizzozero, 2004). The gene encoding brain-derived neurotropic factor, which is indirectly related to the Wnt pathway, has also been implicated in suppressing tumour pathogenesis (Cao et al., 2010), and its rs6265 SNP genotype has recently been found to affect SCZ pathogenesis (Zhang et al., 2018). The major tumour suppressor genes and loci related to SCZ are summarised in Table 1.
Epigenetic Regulation of Tumour Suppressor Genes in SCZ
In addition to the direct regulation of protein expression and function by variants in protein-coding regions, epigenetic mechanisms play an important role in SCZ onset by regulating protein expression without altering the gene sequence. Evidence is continuously growing for the role of epigenetics in SCZ occurrence (Dempster et al., 2013). Mutations in both coding and non-coding regions can affect gene expression levels, thus affecting biological function. In a study investigating the correlation between SCZ risk and variants of the deleted in colorectal cancer gene, the SNP locus rs2270954 was found to reside in the 3’ untranslated region, thus presumably mediating gene function without affecting the protein sequence (Grant et al., 2012). Recent studies have shown that the correlation between SCZ and tumorigenesis may be regulated epigenetically through the involvement of microRNAs (Rizos et al., 2016). For example, miR-183, previously shown to mediate the expression of multiple tumour suppressor genes, was upregulated in a cohort of patients with SCZ (Rizos et al., 2012). Similarly, miR-137 may play a dual role in brain tumour suppression and neural development underlying psychiatric disease, including SCZ (Mahmoudi and Cairns, 2017). Another recently identified non-coding RNA, miR-193a-3p, is a circulatory marker for various tumours and has also been identified in patients with SCZ (Grossi et al., 2017). These studies have demonstrated that both genetic and epigenetic regulation of certain tumour suppressor genes contributes to the risk of SCZ.
Cellular and Molecular Mechanisms of Tumour Suppressor Gene Involvement in SCZ
Although the abovementioned evidence highlights the correlation between tumour suppressor genes and SCZ susceptibility, we still lack evidence of a causal relationship. One possible hypothesis is that tumour suppressor genes participate in SCZ pathogenesis through neurodevelopment or neural functions. The first piece of supporting evidence for the above hypothesis was reported for the p53 gene, whose over-expression was shown to lead to excessive neuronal death and impaired neural function (Hughes et al., 1997). In patients with SCZ, p53 activation can enhance apoptosis in dermal fibroblasts (Catts et al., 2006) and stem-cell-derived fibroblasts from medication-free individuals (Gassó et al., 2014). Therefore, p53 hyper-activation may enhance tumour surveillance at the cost of higher neuronal apoptosis, which impairs psychiatric function. In addition, transcriptional regulation may help to explain the potentially causal relationship between tumour suppressor gene activity and SCZ. Dysbindin-1, the product of the SCZ risk gene dystrobrevin binding protein 1, increases TP53 gene expression and leads to excessive neurite outgrowth (Ma et al., 2011). The above developmental effects of p53 highlight possible routes by which tumour suppressor- and SCZ-associated genes may interact to affect SCZ risk.
Other tumour suppressor genes may also contribute to SCZ pathogenesis due to their effects on neural development, including axon guidance and dendritic arborisation. They include SCZ risk genes such as disrupted in SCZ-1 (Miyoshi et al., 2003; Mackie et al., 2007). Wnt signalling is associated with SCZ pathogenesis and regulates brain development during both embryonic and postnatal stages (Inestrosa et al., 2012). Specifically, canonical and non-canonical Wnt pathways mediate axon guidance and dendritic arborisation, indicating possible roles in SCZ development. Aberrant Wnt signalling and decreased neurite numbers were observed in reprogrammed neurons derived from human-induced pluripotent stem cells in patients with SCZ (Brennand et al., 2011). Given that Wnt activation can facilitate tumour growth, its deactivating mutations may simultaneously reduce tumorigenesis and impair neuronal development, probably increasing SCZ risk. In addition to direct regulation of neural development, the Wnt pathway may be modified epigenetically, as the tumour-suppressing miR-137 can simultaneously modulate Akt pathway activity (Thomas et al., 2017) and the expression of brain-derived neurotrophic factor (Hill et al., 2014), highlighting its dual effects on tumour growth and SCZ risk. Downstream of the Wnt pathway, converging molecular pathways further orchestrate tumour cell surveillance and neurodevelopment. For example, heterozygous knockout of the tumour suppressor gene APC leads to development of age-dependent working memory deficits and hypoactivity (Koshimizu et al., 2011). A mutation in the mouse APC gene has also been reported to result in abnormal dendritic spine formation and long-term potentiation in hippocampal neurons, and is associated with impaired social interaction (Onouchi et al., 2014). Although some of the above findings are inconsistent with our proposed model, they still suggest the involvement of tumour-related genes in neural function.
SCZ pathogenesis is inherently a polygenic event involving interactions among multiple risk genes. A recent association study reported that the interaction between the SNP loci of two tumour suppressor genes, thioredoxin interacting protein and AF1q, contributed to SCZ susceptibility (Su et al., 2017). The molecular mechanisms underlying such gene interactions mainly resides in epigenetic or transcriptional regulation. For example, Wnt activation can epigenetically enhance the expression of transcription factor 4, which is a transcription factor for SCZ risk genes (Hennig et al., 2017). Interestingly, transcription factor 4 mediates apoptosis and epithelial-mesenchymal transition genes in addition to neurodevelopmental factors (Forrest et al., 2013), making it a potent “dual player” candidate for SCZ and cancer. Upstream of Wnt signalling, disrupted in SCZ-1 can stimulate the transcriptional activity of β-catenin (Boccitto et al., 2016) and upregulate Wnt activity. Outside the Wnt pathway, other tumour suppressor gene may also interact with SCZ risk genes, such as protein phosphatase 2A (Palanichamy et al., 2017).
In addition to converging molecular pathways and transcriptional regulation between tumour suppressor and SCZ risk genes, shared epigenetic regulation of these two groups of genes may confer SCZ and cancer phenotypes. For example, age-related DNA methylation can elevate the risk of both SCZ and cancer (Jenkins et al., 2014). Histone deacetylase 2 regulates metabotropic glutamate receptor 2 promoter activity (Kurita et al., 2012) in addition to p53 expression levels (Wagner et al., 2014). Therefore, epigenetic regulation of DNA represents another mechanism linking cancer risk and SCZ, although not all studies have reported results consistent with our hypothesis of an inverse correlation between cancer and SCZ risk. As an additional layer of gene function regulation, post-translational modification of tumour gene protein products has also been implicated in SCZ. One study reported that a novel tumour suppressor gene, zinc fingers DHHC-type 14, encoded an enzyme for S-acylation that is implicated in SCZ (Greaves and Chamberlain, 2014). Taken together, the separate lines of evidence discussed above support the existence of genetic and epigenetic regulatory networks underlying SCZ pathogenesis and tumorigenesis.
Conclusion and Future Perspectives
We have summarised the recent major progress in understanding the roles of tumour suppressor genes in SCZ pathogenesis and discussed potential underlying cellular and molecular mechanisms. Tumour suppressor genes have been associated with neurodevelopmental disorders other than SCZ, such as autism (Crespi, 2011). These findings reinforce the general concept that abnormalities in neurodevelopment can lead to a spectrum of behavioural deficits that spans autism, SCZ, and other syndromes. Research on the role of tumour suppressor genes in SCZ has important implications for drug development. Typical antipsychotics such as haloperidol can cause neuronal apoptosis via p53 activation, whilst atypical drugs can have the opposite effect of neuroprotection (Nandra and Agius, 2012). We therefore expect that atypical antipsychotic drugs should have better efficacy and fewer adverse effects in SCZ cases with tumour suppressor gene hyperactivation. In reviewing associations of SCZ with the Wnt pathway, we hypothesise that drugs targeting this pathway may be effective antipsychotics (Freyberg et al., 2010; Singh, 2013). The feasibility of correcting epigenetic abnormalities in vivo warrants more research and development as a potential pharmacological approach for SCZ. Histone deacetylase 2 is a critical mediator of atypical antipsychotic drug action (Kurita et al., 2012) and the tumorigenic activity of p53 (Wagner et al., 2014). While anti-tumour drugs targeting specific cellular and molecular pathways are being rapidly developed, antipsychotic drugs are meeting hurdles in the traditional path. Therefore, screening anti-tumour drugs for potential neural modulating properties may provide promising fast-track candidates for SCZ treatment. Conversely, recent studies have suggested that anti-psychotic drugs might be effective in decreasing cancer risk (Shi et al., 2015), which may partially explain the lower cancer incidence observed in patients with SCZ (Raviv et al., 2014; Xu et al., 2017). However, the anti-tumorigenic effect of anti-psychotic drugs is unlikely to fully explain those surveys. Rather, we hypothesise that tumour suppressor genes may protect patients with SCZ from cancer onset. This “double-edged sword” model (Figure 1) reflects the intricate balance between preventing dysregulated cell growth and protecting against abnormal neural development, in which a higher risk of tumour onset is traded for a lower incidence of SCZ, and vice versa. In summary, tumour suppressor genes and SCZ are highly correlated genetically, molecularly, and potentially pharmacologically. Further investigation is required to achieve a better, clinically translatable understanding of the cellular mechanisms of tumour suppressor gene involvement in neurodevelopment and behaviour.
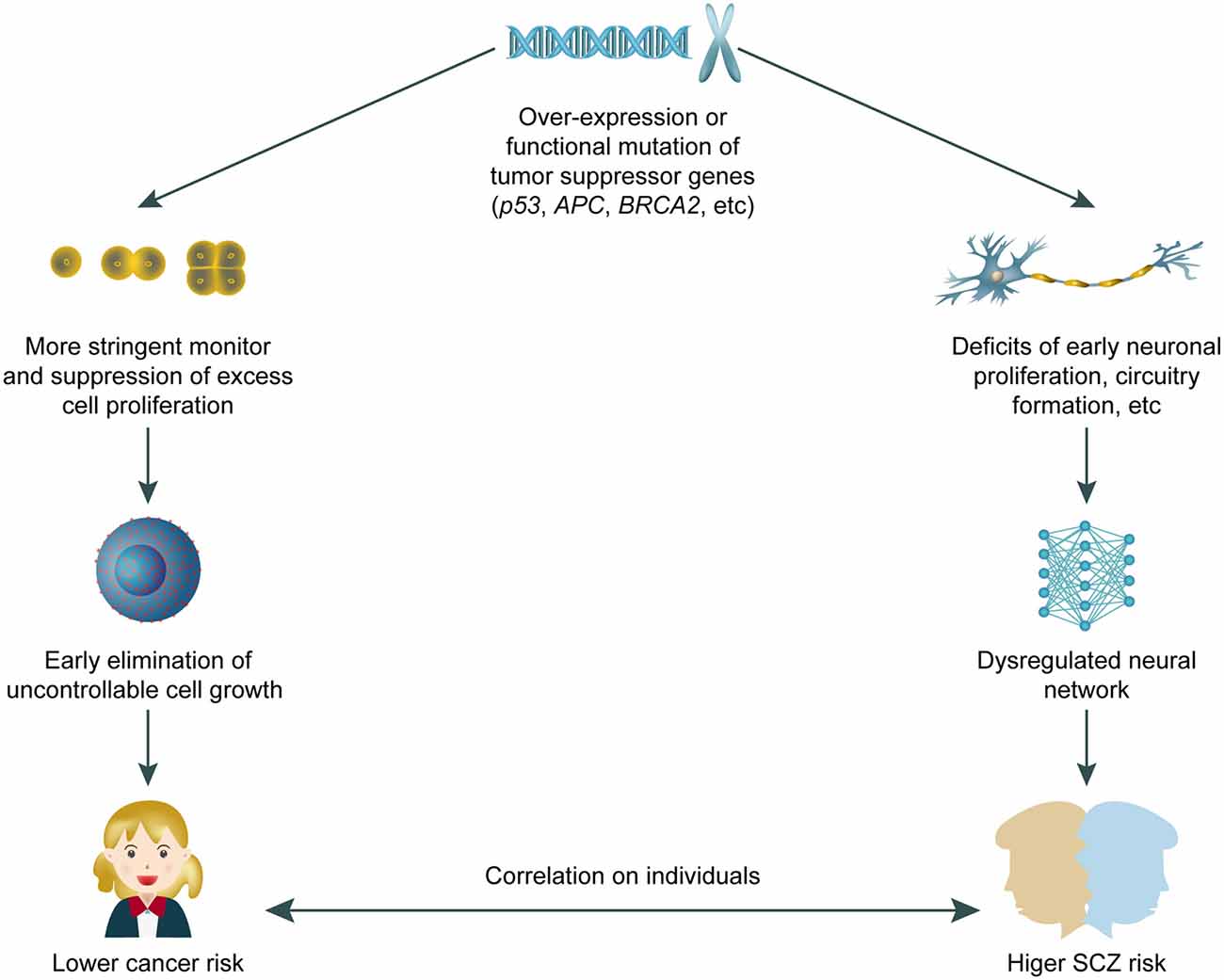
Figure 1. A working model for “double edged” effects of tumour suppressor genes on both cancer and schizophrenia (SCZ) risk. The over-expression or gain-of-function mutation of tumour suppressor genes confer dual effects including closer monitoring of unwanted cell proliferation, plus potential interruption on neural development that leads to higher SCZ risk in adults. The correlation and potentially “trade-off” between SCZ and cancer risk requires further study.
Hypothesis
Based on our current knowledge of tumour suppressor gene involvement in SCZ pathogenesis, we propose a working model in which at least certain SCZ cases can be attributed to functional hyperactivation of tumour suppressor genes, resulting in a concomitant reduction in susceptibility to cancers. Because tumour suppressor genes can inhibit cell proliferation and growth, they can also affect normal development or activity of neural tissues, thus leading to SCZ.
Author Contributions
CZhuo: study idea and design. HT, SL, and JL: systematic review, extraction, and analysis. FJ, CC, CL, MC, CZhou, LZ, and CZhuo: drafting of the manuscript. CZhuo, XS, JL, DW, and LZ: critical revision of the manuscript for important intellectual content. CZhuo, CC, CL, JL, LZ, and DW: access to data and had full access to all the data in the study and take the responsibility for the integrity of the data and the accuracy of the data analysis. All authors have reviewed the manuscript.
Funding
This work was supported by grants from the Tianjin Health Bureau Foundation (2014KR02 to CZhuo), National Natural Science Foundation of China (81871052 to CZhuo), and the Key Projects of the Natural Science Foundation of Tianjin, China (17JCZDJC35700 to CZhuo).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LC and the handling Editor declared their shared affiliation.
Acknowledgments
We would like to thank Prof. Fernando Gose at the Department of Psychiatry and Behavioural Sciences, Johns Hopkins School of Medicine, for his invaluable advice on the methodologies reviewed in this study and the revision of this manuscript.
References
Boccitto, M., Doshi, S., Newton, I. P., Nathke, I., Neve, R., Dong, F., et al. (2016). Opposing actions of the synapse-associated protein of 97-kDa molecular weight (SAP97) and Disrupted in Schizophrenia 1 (DISC1) on Wnt/β-catenin signaling. Neuroscience 326, 22–30. doi: 10.1016/j.neuroscience.2016.03.048
Brennand, K. J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., et al. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225. doi: 10.1038/nature09915
Cao, L., Liu, X., Lin, E. J., Wang, C., Choi, E. Y., Riban, V., et al. (2010). Environmental and genetic activation of a brain-adipocyte BDNF/leptin axis causes cancer remission and inhibition. Cell 142, 52–64. doi: 10.1016/j.cell.2010.05.029
Catts, V. S., and Catts, S. V. (2000). Apoptosis and schizophrenia: is the tumour suppressor gene, p53, a candidate susceptibility gene? Schizophr. Res. 41, 405–415. doi: 10.1016/s0920-9964(99)00077-8
Catts, V. S., Catts, S. V., McGrath, J. J., Feron, F., McLean, D., Coulson, E. J., et al. (2006). Apoptosis and schizophrenia: a pilot study based on dermal fibroblast cell lines. Schizophr. Res. 84, 20–28. doi: 10.1016/j.schres.2006.03.016
Catts, V. S., Catts, S. V., O’Toole, B. I., and Frost, A. D. (2008). Cancer incidence in patients with schizophrenia and their first-degree relatives—a meta-analysis. Acta Psychiatr. Scand. 117, 323–336. doi: 10.1111/j.1600-0447.2008.01163.x
Chambers, J. S., and Perrone-Bizzozero, N. I. (2004). Altered myelination of the hippocampal formation in subjects with schizophrenia and bipolar disorder. Neurochem. Res. 29, 2293–2302. doi: 10.1007/s11064-004-7039-x
Chen, J., Cao, F., Liu, L., Wang, L., and Chen, X. (2015). Genetic studies of schizophrenia: an update. Neurosci. Bull. 31, 87–98. doi: 10.1007/s12264-014-1494-4
Cotter, D., Kerwin, R., al-Sarraji, S., Brion, J. P., Chadwich, A., Lovestone, S., et al. (1998). Abnormalities of Wnt signalling in schizophrenia—evidence for neurodevelopmental abnormality. Neuroreport 9, 1379–1383. doi: 10.1097/00001756-199805110-00024
Cui, D. H., Jiang, K. D., Jiang, S. D., Xu, Y. F., and Yao, H. (2005). The tumor suppressor adenomatous polyposis coli gene is associated with susceptibility to schizophrenia. Mol. Psychiatry 10, 669–677. doi: 10.1038/sj.mp.4001653
Dempster, E., Viana, J., Pidsley, R., and Mill, J. (2013). Epigenetic studies of schizophrenia: progress, predicaments, and promises for the future. Schizophr. Bull. 39, 11–16. doi: 10.1093/schbul/sbs139
Dyrvig, M., Qvist, P., Lichota, J., Larsen, K., Nyegaard, M., Børglum, A. D., et al. (2017). DNA methylation analysis of BRD1 promoter regions and the schizophrenia rs138880 risk allele. PLoS One 12:e0170121. doi: 10.1371/journal.pone.0170121
Elashoff, M., Higgs, B. W., Yolken, R. H., Knable, M. B., Weis, S., Webster, M. J., et al. (2007). Meta-analysis of 12 genomic studies in bipolar disorder. J. Mol. Neurosci. 31, 221–243.
Forrest, M. P., Waite, A. J., Martin-Rendon, E., and Blake, D. J. (2013). Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One 8:e73169. doi: 10.1371/journal.pone.0073169
Freyberg, Z., Ferrando, S. J., and Javitch, J. A. (2010). Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am. J. Psychiatry 167, 388–396. doi: 10.1176/appi.ajp.2009.08121873
Gal, G., Goral, A., Murad, H., Gross, R., Pugachova, I., Barchana, M., et al. (2012). Cancer in parents of persons with schizophrenia: is there a genetic protection? Schizophr. Res. 139, 189–193. doi: 10.1016/j.schres.2012.04.018
Gassó, P., Mas, S., Molina, O., Lafuente, A., Bernardo, M., and Parellada, E. (2014). Increased susceptibility to apoptosis in cultured fibroblasts from antipsychotic-naive first-episode schizophrenia patients. J. Psychiatr. Res. 48, 94–101. doi: 10.1016/j.jpsychires.2013.09.017
Grant, A., Fathalli, F., Rouleau, G., Joober, R., and Flores, C. (2012). Association between schizophrenia and genetic variation in DCC: a case-control study. Schizophr. Res. 137, 26–31. doi: 10.1016/j.schres.2012.02.023
Greaves, J., and Chamberlain, L. H. (2014). New links between S-acylation and cancer. J. Pathol. 233, 4–6. doi: 10.1002/path.4339
Grossi, I., Salvi, A., Abeni, E., Marchina, E., and De Petro, G. (2017). Biological function of MicroRNA193a-3p in health and disease. Int. J. Genomics 2017:5913195. doi: 10.1155/2017/5913195
Häfner, H., Maurer, K., Löffler, W., and Riecher-Rössler, A. (1993). The influence of age and sex on the onset and early course of schizophrenia. Br. J. Psychiatry 162, 80–86. doi: 10.1192/bjp.162.1.80
Hennig, K. M., Fass, D. M., Zhao, W. N., Sheridan, S. D., Fu, T., Erdin, S., et al. (2017). WNT/β-catenin pathway and epigenetic mechanisms regulate the pitt-hopkins syndrome and schizophrenia risk gene TCF4. Mol. Neuropsychiatry 3, 53–71. doi: 10.1159/000475666
Hill, M. J., Donocik, J. G., Nuamah, R. A., Mein, C. A., Sainz-Fuertes, R., and Bray, N. J. (2014). Transcriptional consequences of schizophrenia candidate miR-137 manipulation in human neural progenitor cells. Schizophr. Res. 153, 225–230. doi: 10.1016/j.schres.2014.01.034
Hoseth, E. Z., Krull, F., Dieset, I., Mørch, R. H., Hope, S., Gardsjord, E. S., et al. (2018). Exploring the Wnt signaling pathway in schizophrenia and bipolar disorder. Transl. Psychiatry 8:55. doi: 10.1038/s41398-018-0102-1
Hughes, P. E., Alexi, T., and Schreiber, S. S. (1997). A role for the tumour suppressor gene p53 in regulating neuronal apoptosis. Neuroreport 8, v–xii.
Ibáñez, K., Boullosa, C., Tabarés-Seisdedos, R., Baudot, A., and Valencia, A. (2014). Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 10:e1004173. doi: 10.1371/journal.pgen.1004173
Inestrosa, N. C., Montecinos-Oliva, C., and Fuenzalida, M. (2012). Wnt signaling: role in Alzheimer disease and schizophrenia. J. Neuroimmune Pharmacol. 7, 788–807. doi: 10.1007/s11481-012-9417-5
Jenkins, T. G., Aston, K. I., Pflueger, C., Cairns, B. R., and Carrell, D. T. (2014). Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet. 10:e1004458. doi: 10.1371/journal.pgen.1004458
Jiang, D., Brady, C. A., Johnson, T. M., Lee, E. Y., Park, E. J., Scott, M. P., et al. (2011). Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc. Natl. Acad. Sci. U S A 108, 17123–17128. doi: 10.1073/pnas.1111245108
Katsu, T., Ujike, H., Nakano, T., Tanaka, Y., Nomura, A., Nakata, K., et al. (2003). The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci. Lett. 353, 53–56. doi: 10.1016/s0304-3940(03)01110-8
Kim, S. Y., Yasuda, S., Tanaka, H., Yamagata, K., and Kim, H. (2011). Non-clustered protocadherin. Cell Adh. Migr. 5, 97–105. doi: 10.4161/cam.5.2.14374
Koshimizu, H., Fukui, Y., Takao, K., Ohira, K., Tanda, K., Nakanishi, K., et al. (2011). Adenomatous polyposis coli heterozygous knockout mice display hypoactivity and age-dependent working memory deficits. Front. Behav. Neurosci. 5:85. doi: 10.3389/fnbeh.2011.00085
Kurita, M., Holloway, T., García-Bea, A., Kozlenkov, A., Friedman, A. K., Moreno, J. L., et al. (2012). HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat. Neurosci. 15, 1245–1254. doi: 10.1038/nn.3181
Leucht, S., Burkard, T., Henderson, J., Maj, M., and Sartorius, N. (2007). Physical illness and schizophrenia: a review of the literature. Acta Psychiatr. Scand. 116, 317–333. doi: 10.1111/j.1600-0447.2007.01095.x
Levav, I., Lipshitz, I., Novikov, I., Pugachova, I., Kohn, R., Barchana, M., et al. (2007). Cancer risk among parents and siblings of patients with schizophrenia. Br. J. Psychiatry 190, 156–161. doi: 10.1192/bjp.bp.106.024943
Levine, A. J., and Oren, M. (2009). The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758. doi: 10.1038/nrc2723
Lichtermann, D., Ekelund, J., Pukkala, E., Tanskanen, A., and Lönnqvist, J. (2001). Incidence of cancer among persons with schizophrenia and their relatives. Arch. Gen. Psychiatry 58, 573–578. doi: 10.1001/archpsyc.58.6.573
Lung, F. W., Shu, B. C., Kao, W. T., Chen, C. N., Ku, Y. C., and Tzeng, D. S. (2009). Association of DRD4 uVNTR and TP53 codon 72 polymorphisms with schizophrenia: a case-control study. BMC Med. Genet. 10:147. doi: 10.1186/1471-2350-10-147
Ma, L., Chen, J., Song, X., Yuan, H., Wang, Y., Wu, Y., et al. (2015). Evidence that the genetic polymorphism rs1412115 on chromosome 10 is associated with risk for oral squamous cell carcinoma. Gene 560, 137–139. doi: 10.1016/j.gene.2015.01.049
Ma, X., Fei, E., Fu, C., Ren, H., and Wang, G. (2011). Dysbindin-1, a schizophrenia-related protein, facilitates neurite outgrowth by promoting the transcriptional activity of p53. Mol. Psychiatry 16, 1105–1116. doi: 10.1038/mp.2011.43
Mackie, S., Millar, J. K., and Porteous, D. J. (2007). Role of DISC1 in neural development and schizophrenia. Curr. Opin. Neurobiol. 17, 95–102. doi: 10.1016/j.conb.2007.01.007
Mahmoudi, E., and Cairns, M. J. (2017). MiR-137: an important player in neural development and neoplastic transformation. Mol. Psychiatry 22, 44–55. doi: 10.1038/mp.2016.150
Millier, A., Schmidt, U., Angermeyer, M. C., Chauhan, D., Murthy, V., Toumi, M., et al. (2014). Humanistic burden in schizophrenia: a literature review. J. Psychiatr. Res. 54, 85–93. doi: 10.1016/j.jpsychires.2014.03.021
Miyoshi, K., Honda, A., Baba, K., Taniguchi, M., Oono, K., Fujita, T., et al. (2003). Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol. Psychiatry 8, 685–694. doi: 10.1038/sj.mp.4001352
Morelli, C., Karayianni, E., Magnanini, C., Mungall, A. J., Thorland, E., Negrini, M., et al. (2002). Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene 21, 7266–7276. doi: 10.1038/sj.onc.1205573
Morelli, C., Magnanini, C., Mungall, A. J., Negrini, M., and Barbanti-Brodano, G. (2000). Cloning and characterization of two overlapping genes in a subregion at 6q21 involved in replicative senescence and schizophrenia. Gene 252, 217–225. doi: 10.1016/s0378-1119(00)00231-6
Nandra, K. S., and Agius, M. (2012). The differences between typical and atypical antipsychotics: the effects on neurogenesis. Psychiatr. Danub. 24, S95–S99.
Ni, X., Trakalo, J., Valente, J., Azevedo, M. H., Pato, M. T., Pato, C. N., et al. (2005). Human p53 tumor suppressor gene (TP53) and schizophrenia: case-control and family studies. Neurosci. Lett. 388, 173–178. doi: 10.1016/j.neulet.2005.06.050
Numata, S., Ueno, S., Iga, J., Yamauchi, K., Hongwei, S., Hashimoto, R., et al. (2008). TGFBR2 gene expression and genetic association with schizophrenia. J. Psychiatr. Res. 42, 425–432. doi: 10.1016/j.jpsychires.2007.04.002
Nusse, R., van Ooyen, A., Cox, D., Fung, Y. K., and Varmus, H. (1984). Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature 307, 131–136. doi: 10.1038/307131a0
Onouchi, T., Kobayashi, K., Sakai, K., Shimomura, A., Smits, R., Sumi-Ichinose, C., et al. (2014). Targeted deletion of the C-terminus of the mouse adenomatous polyposis coli tumor suppressor results in neurologic phenotypes related to schizophrenia. Mol. Brain 7:21. doi: 10.1186/1756-6606-7-21
Ozbey, U., Yüce, H., Namli, M., and Elkiran, T. (2011). Investigation of differences in P53 gene polymorphisms between schizophrenia and lung cancer patients in the turkish Population. Genet. Res. Int. 2011:483851. doi: 10.4061/2011/483851
Palanichamy, K., Kanji, S., Gordon, N., Thirumoorthy, K., Jacob, J. R., Litzenberg, K. T., et al. (2017). NNMT silencing activates tumor suppressor PP2A, inactivates oncogenic STKs and inhibits tumor forming ability. Clin. Cancer Res. 23, 2325–2334. doi: 10.1158/1078-0432.ccr-16-1323
Papiol, S., Arias, B., Barrantes-Vidal, N., Guitart, M., Salgado, P., Catalán, R., et al. (2004). Analysis of polymorphisms at the tumor suppressor gene p53 (TP53) in contributing to the risk for schizophrenia and its associated neurocognitive deficits. Neurosci. Lett. 363, 78–80. doi: 10.1016/s0304-3940(04)00361-1
Park, J. K., Lee, H. J., Kim, J. W., Park, Y. H., Lee, S. S., Chang, H. I., et al. (2004). Differences in p53 gene polymorphisms between Korean schizophrenia and lung cancer patients. Schizophr. Res. 67, 71–74. doi: 10.1016/s0920-9964(03)00155-5
Peng, Y., Xu, Y., and Cui, D. (2014). Wnt signaling pathway in schizophrenia. CNS Neurol. Disord. Drug Targets 13, 755–764. doi: 10.2174/1871527312666131223113521
Proitsi, P., Li, T., Hamilton, G., Di Forti, M., Collier, D., Killick, R., et al. (2008). Positional pathway screen of wnt signaling genes in schizophrenia: association with DKK4. Biol. Psychiatry 63, 13–16. doi: 10.1016/j.biopsych.2007.03.014
Purcell, S. M., Wray, N. R., Stone, J. L., Visscher, P. M., O’Donovan, M. C., Sullivan, P. F., et al. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. doi: 10.1038/nature08185
Raviv, G., Laufer, M., Baruch, Y., and Barak, Y. (2014). Risk of prostate cancer in patients with schizophrenia. Compr. Psychiatry 55, 1639–1642. doi: 10.1016/j.comppsych.2014.05.007
Rizos, E., Siafakas, N., Koumarianou, A., Katsantoni, E., Filippopoulou, A., Ntounas, P., et al. (2012). miR-183 as a molecular and protective biomarker for cancer in schizophrenic subjects. Oncol. Rep. 28, 2200–2204. doi: 10.3892/or.2012.2052
Rizos, E., Siafakas, N., Skourti, E., Papageorgiou, C., Tsoporis, J., Parker, T. H., et al. (2016). miRNAs and their role in the correlation between schizophrenia and cancer (Review). Mol. Med. Rep. 14, 4942–4946. doi: 10.3892/mmr.2016.5853
Sanders, A. R., Göring, H. H., Duan, J., Drigalenko, E. I., Moy, W., Freda, J., et al. (2013). Transcriptome study of differential expression in schizophrenia. Hum. Mol. Genet. 22, 5001–5014. doi: 10.1093/hmg/ddt350
Sass, L. A., and Parnas, J. (2003). Schizophrenia, consciousness, and the self. Schizophr. Bull. 29, 427–444. doi: 10.1093/oxfordjournals.schbul.a007017
Schizophrenia Working Group of the Psychiatric Genomics Consortium. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. doi: 10.1038/nature13595
Shi, J., Gershon, E. S., and Liu, C. (2008). Genetic associations with schizophrenia: meta-analyses of 12 candidate genes. Schizophr. Res. 104, 96–107. doi: 10.1016/j.schres.2008.06.016
Shi, X. N., Li, H., Yao, H., Liu, X., Li, L., Leung, K. S., et al. (2015). In silico identification and in vitro and in vivo validation of anti-psychotic drug fluspirilene as a potential CDK2 inhibitor and a candidate anti-cancer drug. PLoS One 10:e0132072. doi: 10.1371/journal.pone.0132072
Shi, S., Zhou, B., Wang, Y., Chen, Y., Zhang, K., Wang, K., et al. (2012). Genetic variation in RTN4 3’-UTR and susceptibility to cervical squamous cell carcinoma. DNA Cell Biol. 31, 1088–1094. doi: 10.1089/dna.2011.1548
Singh, K. K. (2013). An emerging role for Wnt and GSK3 signaling pathways in schizophrenia. Clin. Genet. 83, 511–517. doi: 10.1111/cge.12111
Su, Y., Ding, W., Xing, M., Qi, D., Li, Z., and Cui, D. (2017). The interaction of TXNIP and AFq1 genes increases the susceptibility of schizophrenia. Mol. Neurobiol. 54, 4806–4812. doi: 10.1007/s12035-016-9954-7
Su, T., Suzui, M., Wang, L., Lin, C. S., Xing, W. Q., and Weinstein, I. B. (2003). Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc. Natl. Acad. Sci. U S A 100, 7824–7829. doi: 10.1073/pnas.1332160100
Sullivan, P. F., Kendler, K. S., and Neale, M. C. (2003). Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192. doi: 10.1001/archpsyc.60.12.1187
Sun, L., Liu, P., Liu, F., Zhou, Y., Chu, Z., Li, Y., et al. (2017). Effects of Hint1 deficiency on emotional-like behaviors in mice under chronic immobilization stress. Brain Behav. 7:e00831. doi: 10.1002/brb3.831
Tabarés-Seisdedos, R., Mata, I., Escámez, T., Vieta, E., López-Ilundain, J. M., Salazar, J., et al. (2008). Evidence for association between structural variants in lissencephaly-related genes and executive deficits in schizophrenia or bipolar patients from a Spanish isolate population. Psychiatr. Genet. 18, 313–317. doi: 10.1097/ypg.0b013e3283118725
Tabarés-Seisdedos, R., and Rubenstein, J. L. (2009). Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol. Psychiatry 14, 563–589. doi: 10.1038/mp.2009.2
Tesli, M., Athanasiu, L., Mattingsdal, M., Kähler, A. K., Gustafsson, O., Andreassen, B. K., et al. (2010). Association analysis of PALB2 and BRCA2 in bipolar disorder and schizophrenia in a scandinavian case-control sample. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B, 1276–1282. doi: 10.1002/ajmg.b.31098
Thomas, K. T., Anderson, B. R., Shah, N., Zimmer, S. E., Hawkins, D., Valdez, A. N., et al. (2017). Inhibition of the schizophrenia-associated MicroRNA miR-137 disrupts nrg1α neurodevelopmental signal transduction. Cell Rep. 20, 1–12. doi: 10.1016/j.celrep.2017.06.038
Topol, A., Zhu, S., Tran, N., Simone, A., Fang, G., and Brennand, K. J. (2015). Altered WNT signaling in human induced pluripotent stem cell neural progenitor cells derived from four schizophrenia patients. Biol. Psychiatry 78, e29–e34. doi: 10.1016/j.biopsych.2014.12.028
Vousden, K. H., and Prives, C. (2009). Blinded by the light: the growing complexity of p53. Cell 137, 413–431. doi: 10.1016/j.cell.2009.04.037
Wagner, T., Brand, P., Heinzel, T., and Krämer, O. H. (2014). Histone deacetylase 2 controls p53 and is a critical factor in tumorigenesis. Biochim. Biophys. Acta 1846, 524–538. doi: 10.1016/j.bbcan.2014.07.010
Wang, Y., Cao, Y., Huang, X., Yu, T., Wei, Z., McGrath, J., et al. (2016). Allele-specific expression of mutated in colorectal cancer (MCC) gene and alternative susceptibility to colorectal cancer in schizophrenia. Sci. Rep. 6:26688. doi: 10.1038/srep26688
Wang, Y., Wang, L., Li, X., Liu, B., Zhao, Q., Chen, P., et al. (2010). Polymorphisms of XRCC4 are involved in reduced colorectal cancer risk in Chinese schizophrenia patients. BMC Cancer 10:523. doi: 10.1186/1471-2407-10-523
Xu, D., Chen, G., Kong, L., Zhang, W., Hu, L., Chen, C., et al. (2017). Lower risk of liver cancer in patients with schizophrenia: a systematic review and meta-analysis of cohort studies. Oncotarget 8, 102328–102335. doi: 10.18632/oncotarget.21679
Yang, Y., Xiao, Z., Chen, W., Sang, H., Guan, Y., Peng, Y., et al. (2004). Tumor suppressor gene TP53 is genetically associated with schizophrenia in the Chinese population. Neurosci. Lett. 369, 126–131. doi: 10.1016/j.neulet.2004.07.068
Yu, J., Qi, D., Xing, M., Li, R., Jiang, K., Peng, Y., et al. (2011). MK-801 induces schizophrenic behaviors through downregulating Wnt signaling pathways in male mice. Brain Res. 1385, 281–292. doi: 10.1016/j.brainres.2011.02.039
Zhang, Y., Fang, X., Fan, W., Tang, W., Cai, J., Song, L., et al. (2018). Interaction between BDNF and TNF-α genes in schizophrenia. Psychoneuroendocrinology 89, 1–6. doi: 10.1016/j.psyneuen.2017.12.024
Zmorzyński, S., Świderska-Kołacz, G., Koczkodaj, D., and Filip, A. A. (2015). Significance of polymorphisms and expression of enzyme-encoding genes related to glutathione in hematopoietic cancers and solid tumors. Biomed. Res. Int. 2015:853573. doi: 10.1155/2015/853573
Keywords: schizophrenia risk gene, tumour suppressor gene, neurodevelopment, Wnt pathway, molecular targeting drugs
Citation: Zhuo C, Wang D, Zhou C, Chen C, Li J, Tian H, Li S, Ji F, Liu C, Chen M and Zhang L (2019) Double-Edged Sword of Tumour Suppressor Genes in Schizophrenia. Front. Mol. Neurosci. 12:1. doi: 10.3389/fnmol.2019.00001
Received: 07 June 2018; Accepted: 07 January 2019;
Published: 12 February 2019.
Edited by:
Margaret M. DeAngelis, University of Utah, United StatesReviewed by:
Ming Li, Kunming Institute of Zoology, ChinaLara Sabrina Carroll, University of Utah, United States
Copyright © 2019 Zhuo, Wang, Zhou, Chen, Li, Tian, Li, Ji, Liu, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanjun Zhuo, chuanjunzhuotjmh@163.com
† These authors have contributed equally to this work
 Chuanjun Zhuo
Chuanjun Zhuo Dawei Wang
Dawei Wang Chunhua Zhou6†
Chunhua Zhou6† Li Zhang
Li Zhang