- Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
Over the last decade several components have been identified to be differentially expressed in subsets of mesodiencephalic dopaminergic (mdDA) neurons. These differences in molecular profile have been implied to be involved in the selective degeneration of the SNc neurons in Parkinson’s disease. The emergence and maintenance of individual subsets is dependent on different transcriptional programs already present during development. In addition to the influence of transcription factors, recent studies have led to the hypothesis that modifications of histones might also influence the developmental program of neurons. In this study we focus on the histone methyltransferase EZH2 and its role in the development and maintenance of mdDA neurons. We generated two different conditional knock out (cKO) mice; an En1Cre driven cKO, for deletion of Ezh2 in mdDA progenitors and a Pitx3Cre driven cKO, to study the effect of post-mitotic deletion of Ezh2 on mdDA neurons maturation and neuronal survival. During development Ezh2 was found to be important for the generation of the proper amount of TH+ neurons. The loss of neurons primarily affected a rostrolateral population, which is also reflected in the analysis of the subset marks, Ahd2 and Cck. In contrast to early genetic ablation, post-mitotic deletion of Ezh2 did not lead to major developmental defects at E14.5. However, in 6 months old animals Cck was found ectopically in the rostral domain of mdDA neurons and Ahd2 expression was reduced in more mediocaudal positioned cells. In addition, Pitx3Cre driven deletion of Ezh2 led to a progressive loss of TH+ cells in the VTA and these animals display reduced climbing behavior. Together, our data demonstrates that Ezh2 is important for the generation of mdDA neurons during development and that during adult stages Ezh2 is important for the preservation of proper neuronal subset identity and survival.
Introduction
The Substantia Nigra pars compacta (SNc) and the ventral tegmental area (VTA) are two neuronal sub-populations of the mesodiencephalic dopaminergic (mdDA) system, that can already be distinguished during development (Smits et al., 2006; Veenvliet et al., 2013). The projections of the SNc to the dorsolateral Striatum form the nigral-striatal pathway involved in the control of voluntary movement and body posture, while the VTA is involved in the control of emotion-related behavior by innervating the nucleus accumbens, the amygdala and the prefrontal cortex (Prakash and Wurst, 2006; Veenvliet and Smidt, 2014). Even though both groups of neurons use dopamine (DA) as their neurotransmitters, they are molecular distinct and depend on unique transcriptional programs for their development and survival (Veenvliet and Smidt, 2014). Both subsets arise from the same progenitor pool under the influence of differentially expressed transcription factors like Pitx3 and En1 (Veenvliet et al., 2013; Panman et al., 2014). Birth-dating experiments showed that neurons of the SNc are born first (Bayer et al., 1995; Bye et al., 2012) and the induction of the SNc phenotype has been shown to be dependent on a complex interplay of transcription factors to antagonize the VTA phenotype. After the establishment of the SNc phenotype it is hypothesized that the remaining DA neurons acquire a VTA phenotype by default (Veenvliet et al., 2013; Panman et al., 2014).
Recent studies have shown that developmental transitions are influenced by Enhancer of Zeste homolog 2 (EZH2) and polycomb repressive complex (PRC) 2 activity (Hirabayashi et al., 2009; Pereira et al., 2010). EZH2 functions as the methyltransferase of the PRC2 complex, which catalyzes the mono-di and tri-methylation of Histone 3 lysine 27 (H3K27) (Cao et al., 2002; Cao and Zhang, 2004b). Methylation of H3K27 negatively influences gene expression by promoting chromatin compaction and shows a highly dynamic profile during development (Cao et al., 2002; Francis et al., 2004; Margueron et al., 2008; Mohn et al., 2008; Ezhkova et al., 2009). Conditional removal of Ezh2 from cortical progenitors led to a global loss of H3K27 tri-methylation (H3K27me3) and shifted the balance between self-renewal and differentiation, in favor of differentiation (Pereira et al., 2010). A similar role for Ezh2 was found in neuronal progenitors (NPs) of the dorsal midbrain. Wnt1Cre driven deletion of Ezh2 led to reduced number of NPs in the dorsal midbrain, due to elevated cell cycle exit and differentiation (Zemke et al., 2015). In addition to a role in neuronal development, Ezh2 has also been linked to several neurodegenerative disorders (Li et al., 2013; Södersten et al., 2014; von Schimmelmann et al., 2016). L-Dopa was found to be capable of negatively influencing the binding of PRC2 to target genes in the Striatum of hemiparkinsonian mice, leading to a de-repression of PRC2 target genes and levodopa-induced dyskinesia (Södersten et al., 2014). In addition, post-mitotic deletion of Ezh2 in medium spiny neurons and cerebellar purkinje cells, in combination with an Ezh1 null mutant, led to the de-repression of PRC2 targets and a progressive and fatal degeneration of Ezh2 deficient neurons (von Schimmelmann et al., 2016).
The hallmark of Parkinson’s disease is the specific degeneration of neurons in the SNc, while neurons of the VTA remain largely unaffected (Braak et al., 2003). The specific vulnerability of the SNc neurons is in part thought to be caused by the molecular programming specifics of these neurons. Major progress has been made in unraveling the transcriptional programs involved in the specification and survival of the different subsets of the mdDA system, however, little is know about the influence of epigenetics on these processes. In this study we aimed to gain further insight into the role of Ezh2 in the formation and maintenance of mdDA neurons. To accomplish this we generated two different conditional knock out (cKO) mice; an En1Cre driven cKO (Kimmel et al., 2000) for deletion of Ezh2 in mdDA progenitors and a Pitx3Cre driven (Smidt et al., 2012) cKO to study the effects of post-mitotic deletion of Ezh2 on mdDA neuronal maturation and - survival. Importantly, deletion of Ezh2 from mdDA progenitors led to a general loss of H3K27me3 in the Cre-recombinant area, while H3K27me3 was still present in cells where Ezh2 was removed post-mitotically. Analysis of the amount of TH+ cells in developing En1Cre/+;Ezh2 L/L embryos showed that at E12.5 normal numbers of TH+ cells are generated, however, by E14.5 significantly fewer TH+ neurons are detected. The loss of neurons primarily affects the rostrolateral population, which is confirmed through analysis of the subset marks, Ahd2 and Cck. Expression of the rostrolateral mark, Ahd2, is significantly reduced in the En1Cre/Ezh2 cKO, while the expression level of the caudomedial mark, Cck, is not affected by the loss of Ezh2. In contrast to early genetic ablation, post-mitotic deletion of Ezh2 did not lead to major alterations in the expression of DA marks at E14.5. However, in 6 months old animals Cck was found ectopically in the rostral domain of mdDA neurons and Ahd2 expression was reduced in more mediocaudal positioned cells. Further analysis of Pitx3Cre/+; Ezh2 animals demonstrated a progressive loss of TH+ cells in the VTA and reduced climbing behavior in Pitx3Cre/+; Ezh2 L/L animals. Together, our data demonstrate that Ezh2 is important for the formation of the population of mdDA neurons during development and that during adult stages Ezh2 is important for the maintenance of the proper neuronal identity. In addition, our study confirms the initial suggestions that proper Ezh2 functioning is important for cellular survival, since in our mouse models mdDA neuronal survival is affected and leads to substantial losses.
Materials and Methods
Ethics Statement
All animal studies are performed in accordance with local animal welfare regulations, as this project has been approved by the animal experimental committee (Dier Ethische Commissie, Universiteit van Amsterdam; DEC-UvA), and international guidelines.
Animals
All lines were maintained on a C57BL/6J background (Charles River). Ezh2-floxed animals were generated by S. H. Orkin and a kind gift from F. Zilbermann (Friedrich Miescher Institute, Switzerland) and have been previously described (Shen et al., 2008). The En1Cre line has been generated by A. L. Joyner and was a kind gift from S. Blaess (Rheinische Friedrich-Wilhelms-Universität, Germany) (Kimmel et al., 2000). En1Cre/+ animals were crossed with En1Cre-ERT +/+; R R26RYFP/R26RYFP to obtain En1Cre/+; R26RYFP/R26RYFP (Kimmel et al., 2000). The Pitx3Cre line has been previously generated in our lab (Smidt et al., 2012). Ezh2 L/L animals were crossed with En1Cre/+ or Pitx3Cre/Cre animals to obtain En1Cre/+; Ezh2 L/+ or Pitx3Cre/Cre; Ezh2 L/+ animals. For the generation of embryos we crosses En1Cre/+; Ezh2 L/+ or Pitx3Cre/Cre; Ezh2 L/+ animals with Ezh2 L/+ animals. Embryos were isolated at embryonic day (E) 12.5, E14.5, considering the morning of plug formation as E0.5. Pregnant or adult mice were euthanized by CO2 asphyxiation and embryos or brains were collected in 1 × PBS and immediately frozen on dry-ice (fresh frozen) or fixed by immersion in 4% paraformaldehyde (PFA) for 4–8 h at 4°C. After PFA incubation, samples were cryoprotected O/N in 30% sucrose at 4°C. Embryos or brains were frozen on dry-ice and stored at -80°C. Cryosections were slices at 16 μm, mounted at Superfrost plus slides, air-dried and stored at -80°C until further use.
Genotyping
The genotyping for the Ezh2-flox allele was executed with 50–100 ng of genomic DNA together with forward primer 5′-ACCATGTGCTGAAACCAACAG-3′ and reverse primer 5′-TGACATGGGCCTCATAGTGAC-3′ resulting in a 395 bp product for the wildtype allele and a 361 bp product for floxed allele.
Genotyping of the En1Cre allele was performed with 50–100 ng of genomic DNA together with primer pair En1Cre 5UTR_F3 5′-CTTCGCTGAGGCTTCGCTTT-3′ and En1Cre Cre_R2 5′-AGTTTTTACTGCCAGACCGC-3′ resulting in a product at 240 bp for the Cre-allele.
Pitx3-Cre genotyping was done by two different PCR’s using 50–100 ng of genomic DNA for both reactions. The mutant allele was detected by using primer pair forward 5′-GCATGATTTCAGGGATGGAC and reverse 5′-ATGCTCCTGTCTGTGTGCAG, resulting in a product of 750 bp for a mutant allele, and no product in wild-type animals. To additionally detect the wildtype allele primers were designed around Pitx3 exon 2 and exon 3′ forward 5′-CAAGGGGCAGGAGCACA and reverse 5′-GTGAGGTTGGTCCACACCG, resulting in a product of 390 bp for the wildtype allele and no product for the mutant allele.
Genotyping for the R26R-YFP allele was performed using 3 primers Rosa_mutant primer 5′-AAGACCGCGAAGAGTTTGTC-3′, Rosa_wildtype primer 5′-GGAGCGGGAGAAATGGATATG-3′ and a Rosa_common primer 5′-AAAGTCGCTCTGAGTTGTTAT-3′ with 50–100 ng of genomic DNA. The PCR reaction gave a product at 320 bp for the mutant R26R-YFP allele and a product of 600 bp for the wildtype allele.
In situ Hybridization
In situ hybridization with digoxigenin (DIG)-labeled RNA probes was performed as described (Smits et al., 2003; Smidt et al., 2004). DIG-Labeled RNA probes for Th, Ahd2, Cck and Dat have been, respectively, described (Grima et al., 1985; Smits et al., 2003; Jacobs et al., 2007, 2011). The Calb1 probe was a 509 bp fragment directed against bp 196–704 of the Calbindin D 28K mRNA (NM_009788).
Immunohistochemistry
Fluorescent Immunohistochemistry
Cryosections were blocked with 4% HiFCS in 1× THZT [50 mM Tris–HCL, pH 7.6; 0.5M NaCl; 0.5% Triton X-100] and incubated with a primary antibody [Rb-TH (Pelfreeze 1:1000), Sh-TH (Millipore AB1542, 1:750), Rb-H3K27me3 (Millipore, 17–622 1:2000), Rb-Pitx3 (1:750, Smidt et al., 2000)] in 1× THZT overnight at room temperature. The following day the slides were washed and incubated for 2 hrs at room temperature with secondary Alexa Fluor antibody [anti-rabbit, anti-sheep (Invitrogen, 1:1000) in 1× TBS]. After immunostaining nuclei were staining with DAPI (1:3000) and embedded in FluorSave (Calbiogen) and analyzed with the use of a fluorescent microscope (Leica). All washes were done in TBS and double stainings were performed sequential, with immunostaining for TH being done first, followed by the staining for H3K27me3. The antibody against H3K27me3 requires antigen retrieval, which was executed as follows; slides were incubates 10 min in PFA after which they were submersed in 0.1 M citrate buffer pH 6.0 for 3 min at 1000 W followed by 9 min at 200 W. Slides were left to cool down, after which protocol was followed as described above.
Quantitative Analysis
Quantification of TH-expressing neurons 3 and 6 months old midbrain was performed in ImageJ as follows. Cells were counted in 10–12 matching coronal sections (3 months old; n = 3, 6 months old; n = 3). Cells were counted as TH+neurons when TH staining co-localized with nuclear DAPI staining. The separation of the SNc and VTA were made based on anatomical landmarks. Everything rostral of the supramammillary decussation was considered as SNc and distinction between the SNc and the VTA was made based on the tracts medial lemniscus, positioned between the SNc and VTA. Quantification of the TH-expressing cells in embryos was performed in ImageJ with particle analysis. TH (green) and DAPI (blue) images were turned into binary images using default settings, after which the binary DAPI image was used as an overlay on top of the TH image via image calculator ‘and.’ This generates a binary image of only the cells that are positive for both TH and DAPI, which were then counted by using the ‘analyze particles’ function of ImageJ. For E12.5 10–14 matching sagittal sections were analyzed and for E14.5 15–17 matching sagittal sections were analyzed. The separation between medial and lateral was made based on the retroflex fasciculus. All sections, including the section portraying the retroflex fasciculus were considered lateral. Statistical analysis was performed via a student’s t-test.
Quantitative PCR (qPCR)
RNA was isolated from dissected E14.5 midbrains of En1Cre/+; Ezh2+/+ and Ezh2 cKO embryos. RNA was isolated with TRIzol (Thermo Fisher Scientific) according to manufacturers protocol. Two midbrains were pooled for the Ezh2 cKO samples and a single midbrain was used per sample for the wildtype (Wildtype n = 4 and En1Cre/+; Ezh2 L/L n = 3). Relative expression levels were determined by using the QuantiTect SYBR green PCR LightCycler Kit (Qiagen) according to the manufacturers instructions. For each reaction 10 ng (dissected midbrain) of total RNA was used as input. Primers used for Th, Ahd2, Cck and were previously published (Jacobs et al., 2011). Primers for Pitx3 were designed as follows: forward 5′-CTTCCAGAGGAATCGCTACCCT and reverse 5′-CTGCGAAGCCACCTTTGCACA (product size 164 bp).
Behavioral Analysis
Climbing Test
Climbing behavior was assessed as described before (Protais et al., 1976; Smidt et al., 2004) during the dark (active) phase between 21.00 and 23.00. Animals were assessed twice, once at 3 months and once at 6 months. Animals were placed in the climbing cylinders and acclimatized for 30 min. All behavioral observations were done in a separate behavioral room to which the animals were transported 1 h prior to the experiment.
Open Field Test
The open field consisted of a plastic open rectangular box (54 cm × 37 cm × 33 cm) with bedding material on the bottom. Locomotor activity was monitored for 15 min using a fully automated observation system (EthoVision, Noldus Information Technology, Netherlands). The animals were tested twice, once at 3 months and once at 6 months of age. Measurements were performed during the dark (active) phase between 21.00 and 23.00 and in a separate behavioral room to which the animals were transferred 1 h prior to the experiment.
Results
H3K27me3 Is Lost in the Cre-Recombinant Area of En1Cre Driven Ezh2 cKOs
Previous studies have shown that the conditional deletion of Ezh2 in neuronal progenitors shift the balance between self-renewal and differentiation, in favor of differentiation (Pereira et al., 2010; Zemke et al., 2015; Feng et al., 2016). Conditional deletion of Ezh2 in cortical progenitors showed that an increased fraction of cortical progenitors leave the cell cycle at an earlier time-point, leading to a substantial thinner cortex (Pereira et al., 2010). Matching results were obtained for the dorsal midbrain, where the loss of Ezh2 leads to a reduced neuroepithelial thickness, by negatively affecting proliferation and canonical Wnt-signaling (Zemke et al., 2015). To study whether Ezh2 affects the differentiation of mdDA neurons we crossed Ezh2-floxed mice (Shen et al., 2008) with En1Cre animals (Kimmel et al., 2000), deleting Ezh2 from ventral midbrain progenitors from E10.5 onward (Sunmonu et al., 2009; Zemke et al., 2015). As described above, EZH2 is the methyltransferase of PRC2, which catalyzes the methylation of H3K27 (Cao and Zhang, 2004b; Margueron et al., 2008; Shen et al., 2008) and in previous studies in which Ezh2 is conditionally removed from neuronal progenitors a widespread loss of H3K27me3 is observed (Hirabayashi et al., 2009; Pereira et al., 2010; Zemke et al., 2015). To determine whether En1Cre driven deletion of Ezh2 affects the presence of H3K27me3 we performed immunohistochemistry experiments for H3K27Me3 at E14.5. The Cre-recombinant area was visualized using the R26R-YFP reporter allele (Srinivas et al., 2001) and in accordance with previous studies, H3K27me3 is lost in the Cre-recombinant area of En1Cre/+;Ezh2 L/L; R26R-YFP/R26R-YFP embryos [Figure 1 (1, right panel)], while the mark is still present in wildtypes [Figure 1 (1, left panel)]. Analysis of the periphery of the En1Cre expression domain demonstrated a mixture of H3K27me3 positive and negative cells in these Ezh2 cKO embryos [Figure 1 (2, right panel), arrowhead], suggesting that the recombination at the border regions is incomplete. Together, our data shows that the early removal of Ezh2 is sufficient to disturb general PRC2 functioning and ablates the tri-methylation of H3K27 in the Cre-recombinant area.
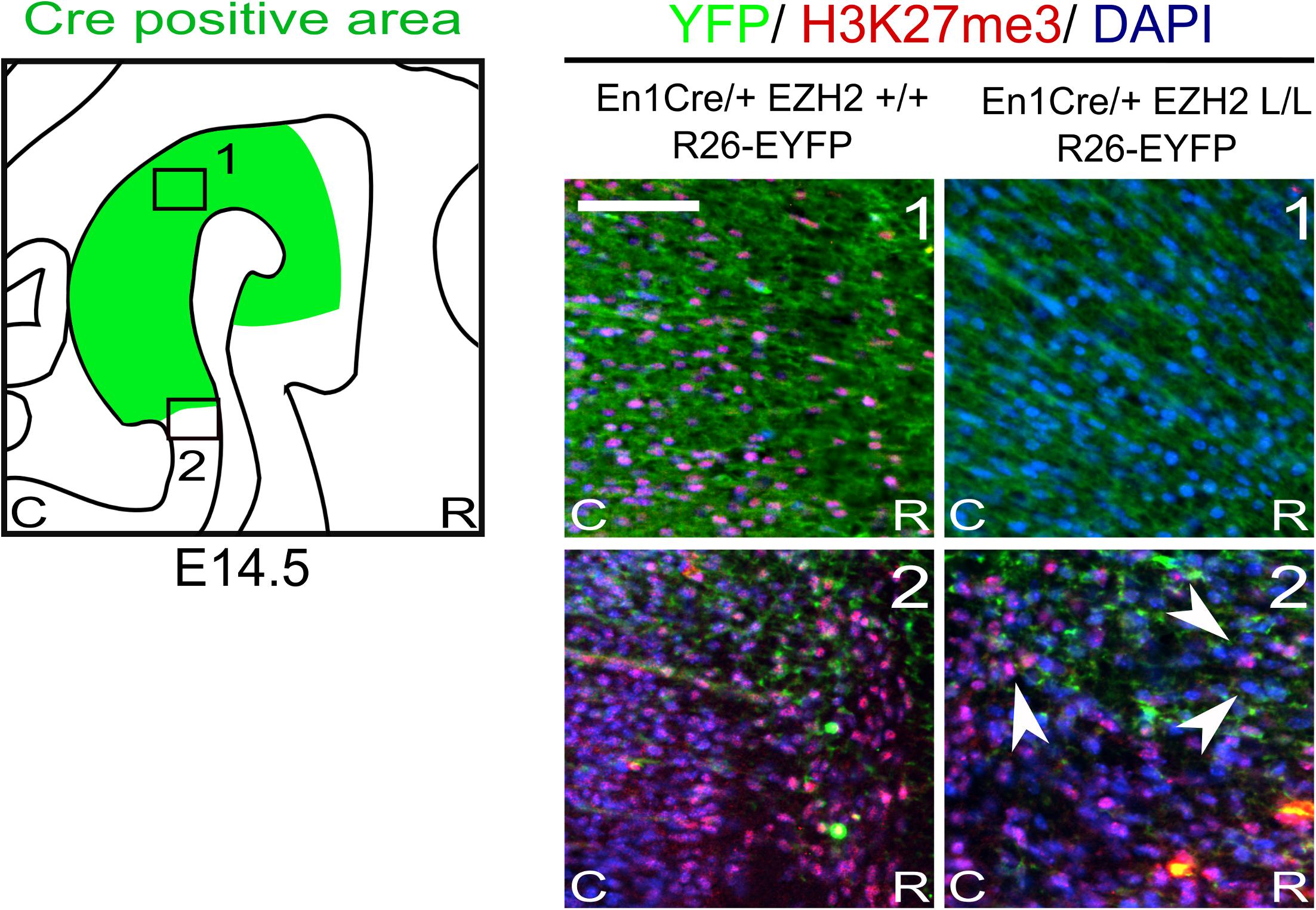
Figure 1. H3K27me3 is lost in the Cre-recombinant area of En1Cre driven Ezh2 cKO embryos. Analysis of the presence of H3K27me3 (red) in the Cre recombinant area, marked by YFP (green), in En1Cre/+; Ezh2+/+ and En1Cre/+; Ezh2 L/L E14.5 embryos. In En1Cre/+; Ezh2 L/L embryos H3K27me3 could not be detected in the midbrain at E14.5, while it was present in wildtype control embryos (1). Outside of the Cre-recombinant area H3K27me3 was present in both the wildtype and En1Cre/Ezh2 mutant (2). Co-localization with DAPI staining was used to confirm nuclear staining. Scale bars = 60 μM.
En1Cre/+; Ezh2 L/L Embryos Display a Disorganized mdDA Domain and a Reduction in TH Positive Neurons
mdDA progenitors develop at the ventricular zone of the ventral midbrain (Brodski et al., 2003; Placzek and Briscoe, 2005; Ono et al., 2007; Mesman et al., 2014). Around embryonic day (E) 10.5 the first progenitors exit the cell cycle to give rise to post-mitotic mdDA precursors that will start to express the rate-limiting enzyme of DA synthesis, Tyrosine Hydroxylase (TH) (Bayer et al., 1995). To study the effect of En1Cre driven deletion of Ezh2 on mdDA neurogenesis we quantified the amount of TH+ neurons at E12.5, at the peak of neurogenesis (Bayer et al., 1995; Bye et al., 2012), and E14.5, before the first wave of apoptosis commences (Bayer et al., 1995; Zhang et al., 2007; Bye et al., 2012), by performing immunohistochemistry for TH (Figure 2). Spatial analysis of the TH+ domain at E12.5 did not reveal any changes in the DA region between En1Cre/+; Ezh2 +/+ and En1Cre/+;Ezh2 L/L embryos. Also quantification of the amount of TH+ cells did not show any significant differences between the Ezh2 cKO and wildtype animals (Figure 2A). However, when examining the total number of TH+ neurons at E14.5 a loss of TH+ cells was found in the En1Cre/+; Ezh2 L/L midbrain (n = 3, P < 0.05, two-tailed) (Figure 2B).
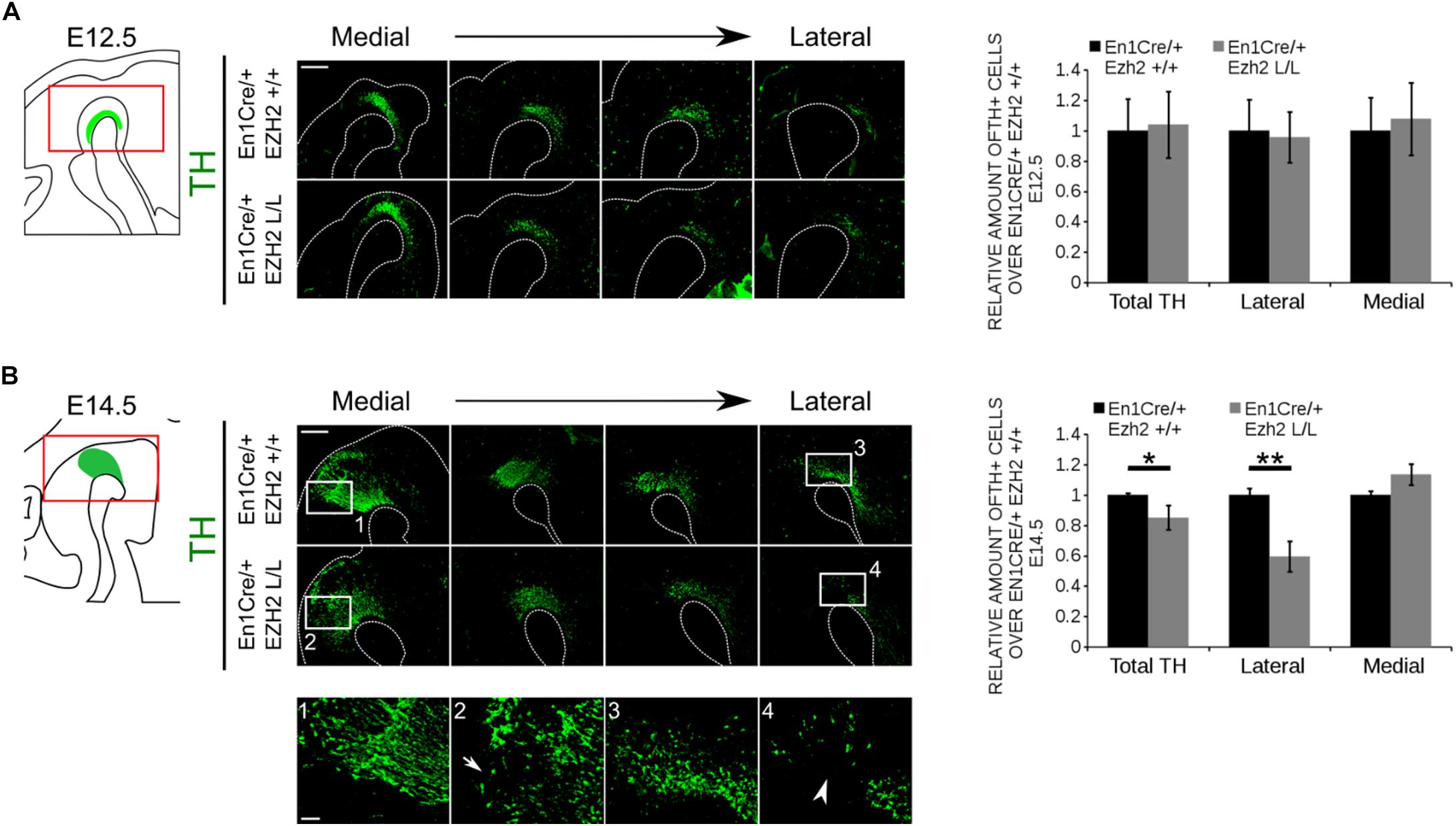
Figure 2. En1Cre driven deletion of Ezh2 affects the amount of TH+ cells at E14.5. Protein expression of TH was evaluated at E12.5 (A) and E14.5 (B). (A) Quantification of the amount of TH positive cells in sagittal midbrain sections at E12.5 demonstrated no significant changes in the number of TH+ cells between the En1Cre/+;Ezh2 +/+ (n = 3, black bar) and En1Cre/+; Ezh2 L/L (n = 3, gray bar) embryos. Wildtypes were set at 1. In addition TH+ neurons were normally distributed over the lateral and medial mdDA domain. Scale bars = 400 μM. (B) Quantification of the amount of TH+ in sagittal midbrain sections at E14.5. The total amount of TH+ neurons were reduced with ∼15% in the Ezh2 cKO embryos (n = 3, ∗P < 0.05, two-tailed). Subdivision into the lateral and medial population based on the location of the retroflex fasciculus demonstrates that in the lateral population ∼41% of the TH+ cells are lost in En1Cre/+; Ezh2 L/L embryos (4, white arrowhead) (n = 3, ∗∗P < 0.01, two-tailed), while an upward trend in the amount of TH+ cells in the medial domain is observed (2, arrow) (n = 3, P = 0.089, two-tailed). Wildtypes were set at 1. Scale bars = 400 μM and for 1–4 80 μM.
During neurogenesis post-mitotic mdDA cells migrate via radial and tangential migration to their final location to form different subsets of mdDA neurons (Shults et al., 1990; Kawano et al., 1995; Smits et al., 2013). While neurons destined to become the SNc are predominantly found in the rostrolateral population, the neurons that form the VTA are located caudomedial (Veenvliet et al., 2013). When examining the expression pattern of TH, we observed that mostly the rostrolateral population was affected in En1Cre/+; Ezh2 L/L animals [Figure 2B (4), arrowhead]. Quantification of the amount of cells found rostrolateral and caudomedial showed that ∼ 40% of the rostrolateral neurons were lost (n = 3, P < 0.01, two-tailed), while an upward trend in the number of TH+ neurons located caudomedial was found [Figure 2B (2)] (n = 3, p = 0.08, two-tailed), suggesting that, next to a loss, TH+ neurons may be dislocated. In agreement with sagittal sections, the most extensive loss of TH+ neurons in coronal sections was observed in rostral sections of En1Cre/Ezh2 mutant embryos (Supplementary Figure S1, arrowheads). Together, these data show that Ezh2 is important for the generation of the proper amount of TH+ neurons at the proper positions and that the loss of Ezh2 mainly affects the rostrolateral population of TH+ cells.
MdDA Subsets Are Differently Affected by En1Cre Driven Deletion of Ezh2
As described above, different mdDA subsets can already be distinguished during embryonic development based on their anatomical location (Smits et al., 2013; Veenvliet et al., 2013; La Manno et al., 2016). In addition to their location, each mdDA subset is characterized by a unique molecular code (Di Salvio et al., 2010; Di Giovannantonio et al., 2013; Veenvliet et al., 2013; Panman et al., 2014; La Manno et al., 2016), with Ahd2 and Cck as hallmarks for the rostrolateral and caudomedial populations, respectively (Jacobs et al., 2007; Veenvliet et al., 2013). Since En1Cre driven deletion of Ezh2 leads to a major loss of TH+ cells in the rostrolateral population, we aimed to confirm this by analyzing the expression of these subsets marks. The expression of Th, Cck and Ahd2 was analyzed using in situ hybridization and the levels were quantified by qPCR (Figure 3). In accordance with TH protein data, Th expression was affected in rostrolateral sections of E14.5 En1Cre/Ezh2 mutant midbrains (Figure 3A arrowhead). Noteworthy, the overall expression level was not significantly altered (Figure 3B). Analysis of two subset marks, Ahd2 and Cck, demonstrated that the overall expression level of the rostrolateral mark Ahd2 was reduced to ∼27% in En1Cre/+; Ezh2 L/L animals (n = 3) (Figure 3B) in comparison to wildtypes (n = 4) (P < 0.05, two-tailed) and spatial expression analysis showed that signal was lost in both lateral and medial sections of the midbrain of En1Cre driven Ezh2 cKO embryos (Figure 3A, arrowheads). In contrast, Cck expression was found to be extended rostrally in the medial-ventral-midbrain in En1Cre/+; Ezh2 L/L embryos (Figure 3A, arrows). Again, the overall expression level was comparable to wildtypes (Figure 3B).
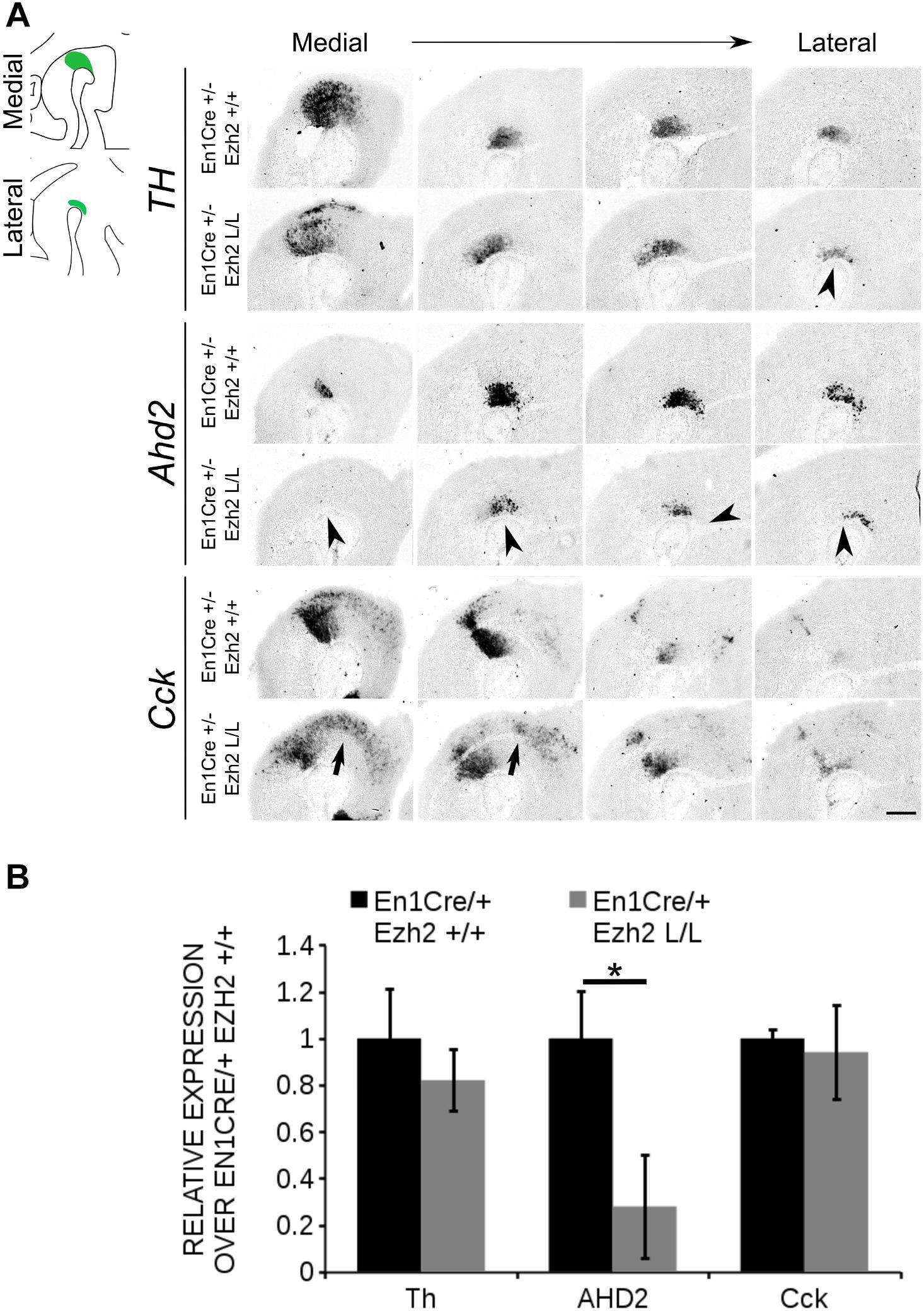
Figure 3. Ahd2 expression is reduced in En1Cre/+; Ezh2 L/L embryos at E14.5. (A) In situ hybridization for Th, Cck, and Ahd2 in sagittal E14.5 midbrain sections. Ahd2 expression is reduced in both lateral en medial sections (lowest panel, arrowheads), while Th expression is decreased only in the lateral sections (upper panel, arrowhead). Expression of Cck was found ectopically in the rostral midbrain (arrows). (B) Quantification of the expression levels of Th, Cck, and Ahd2 via qPCR showed that Th and Cck levels were not significantly altered in the En1Cre/+; Ezh2 L/L (gray bars), but levels of Ahd2 were reduced to ∼20% of the wildtype level (black bar) (n = 3, ∗P < 0.05, two-tailed). Wildtype levels were set at 1. Scale bars = 280 μM.
The phenotype observed in En1Cre/+; Ezh2 L/L embryos is partially reminiscent of defects observed in Pitx3 mutants (Jacobs et al., 2007, 2011), in which Ahd2 expression is lost and expression of Cck is up-regulated and expanded into the rostrolateral mdDA population (Jacobs et al., 2007, 2011). To verify whether the programming deficiency observed in En1Cre/+; Ezh2 L/L animals is not due to the possible lack of Pitx3 we performed immunohistochemistry for PITX3 at E14.5 and quantified the mRNA levels by means of qPCR (Figure 4). PITX3 was detected in lateral and medial midbrain sections of both En1Cre/+; Ezh2 +/+ and En1Cre/+; Ezh2 L/L embryos and mimicked the expression pattern of TH (Figure 4A). Quantification of the mRNA levels showed that the Pitx3 levels are not significantly different between En1Cre/+; Ezh2 L/L and control animals (Figure 4B), indicating that the observed phenotype in En1Cre driven Ezh2 cKOs is not due to any Pitx3 deficiency. In summary, our data show that a loss of Ezh2 affects the positioning of the Th population and differently affects mdDA subsets, leading to the loss of rostrolateral Ahd2 expression and extension of the Cck expression domain.
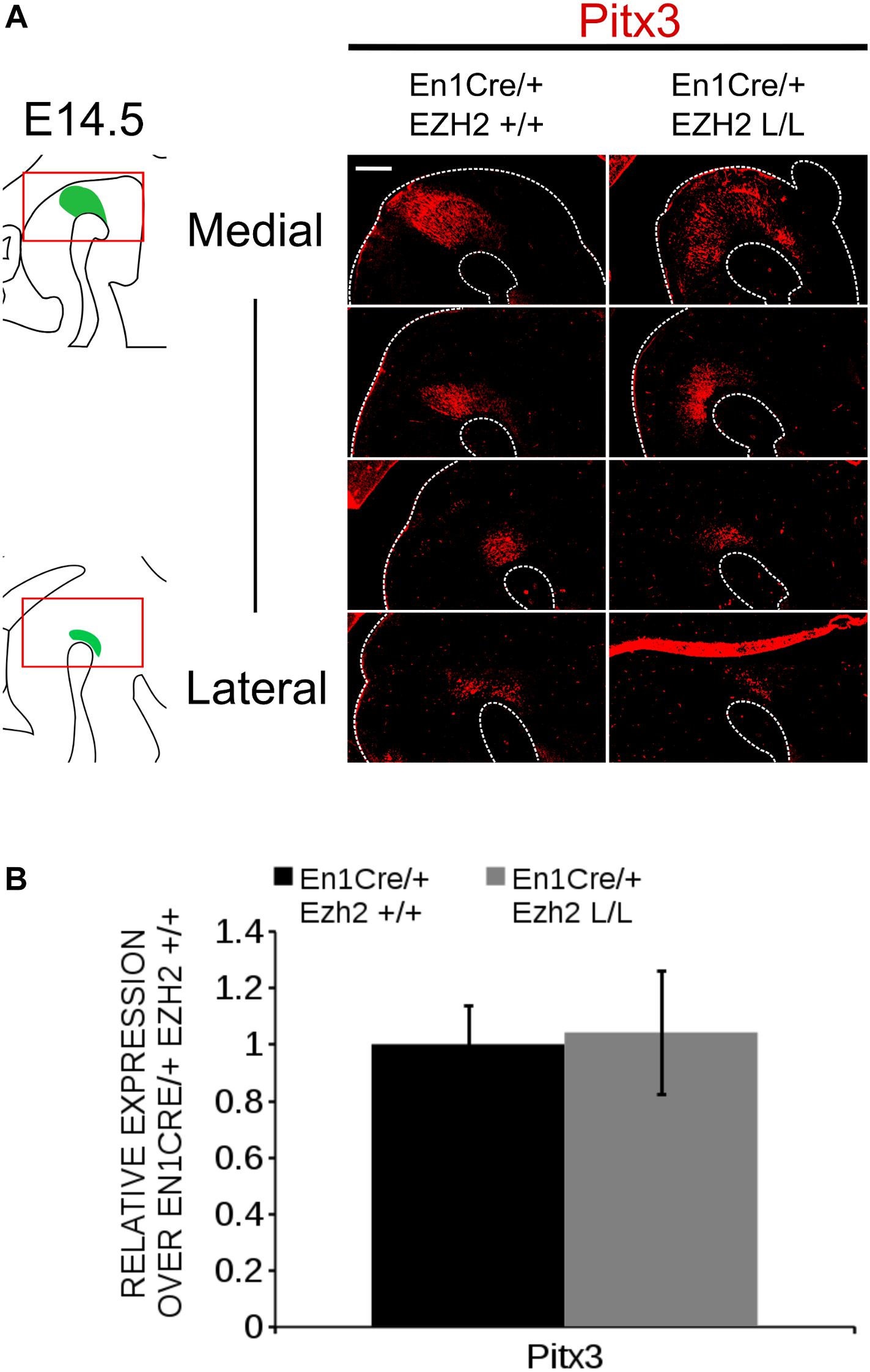
Figure 4. Pitx3 expression is not influenced by En1Cre driven deletion of Ezh2 at E14.5. (A) Analysis of the PITX3 protein expression in E14.5 En1Cre/+; EZh2 +/+ and En1Cre/+;Ezh2 L/L midbrains by means of immunohistochemistry. PITX3 expression can be detected in both the medial and lateral sections of the midbrain of Ezh2 cKO embryos, but less PITX3+ cells are present in the lateral sections of En1Cre/+; Ezh2 L/L embryos in comparison to wildtype littermates (white arrowheads). (B) Quantification of the mRNA levels by means of qPCR demonstrated not significant changes in expression levels between the En1Cre/+; Ezh2+/+ (black bar) and the Ezh2 cKO (gray bar) (n = 3). Wildtype levels where set at 1. Scale bars = 60 μM.
Post-mitotic Deletion of Ezh2 Affects the Expression of Subset Specific Factors Cck and Ahd2 in 6 Months Old Animals
Next to early developmental influences, we aimed to study the effect of Ezh2 deletion on mdDA neuronal maturation. However, similar to Wnt1Cre/+;Ezh2 mutants (Zemke et al., 2015), En1Cre driven deletion of Ezh2 leads to prenatal lethality. We therefore generated a second model, in which Ezh2 was deleted specifically in post-mitotic mdDA progenitors by crossing the Ezh2-floxed line (Shen et al., 2008) with Pitx3Cre animals (Smidt et al., 2012), to study mdDA neuronal maturation. To substantiate the initial phenotype found in En1Cre/Ezh2 mutants we examined the mdDA markers Th, Ahd2 and Cck in Pitx3/Ezh2 mutants. At E14.5 no differences in Th expression could be observed between Pitx3Cre/+;Ezh2 +/+ and Pitx3Cre/+; Ezh2 L/L embryos (Figure 5). Moreover, Ahd2 and Cck marked the default distinct mdDA sub-domains (Veenvliet et al., 2013) in the sagittal midbrain of both wildtype and Pitx3Cre/+; Ezh2 L/L embryos (Figure 5). After the analysis of the embryonic stage, we proceeded with examining the expression of Th, Cck and Ahd2 in the midbrain of 3 and 6 months old Pitx3Cre/+; Ezh2 L/L animals (Figure 6). Examination of the expression of Th demonstrated no clear expression pattern differences between Pitx3Cre/+; Ezh2+/+ and Pitx3Cre/+; Ezh2 L/L genotypes (Figure 6A). Assessment of the expression pattern of both Cck and Ahd2 revealed that post-mitotic deletion of Ezh2 influences the expression of both marks in a similar direction as early deletion of Ezh2 alters Cck and Ahd2 expression (Figures 2, 6B,C). In 6 months old Ezh2 cKO animals ectopic expression of Cck was found in the rostrolateral domain of mdDA neurons (Figure 6B, arrowheads), while Ahd2 expression was reduced in caudal areas (Figure 6C, arrow). Interestingly, the expression of Cck and Ahd2 was unaffected in 3 months old Pitx3Cre/+; Ezh2 L/L animals and Ahd2 was also normally expressed in the rostral domain in 6 months old mutants (Figures 6B,C). Next to these mdDA markers, we also included the expression pattern of the Dopamine transporter (Dat) in the adult midbrain (Figure 6D). During development Dat expression partially overlaps with the expression domains of both Ahd2 and Cck (Veenvliet et al., 2013) and in the adult midbrain, expression can be detected in both the VTA and the SNc (Figure 6D). Similar to Th, no considerable differences could be observed between Pitx3Cre/+; Ezh2+/+ and Pitx3Cre/+; Ezh2 L/L animals at both 3 and 6 months of age. Together these results suggest that post-mitotic deletion of Ezh2 does not influence the initial developmental programming of mdDA neurons, but that over time the expression of subset specific marks Cck and Ahd2 are altered in a corresponding manner as observed in En1Cre driven Ezh2 cKOs.
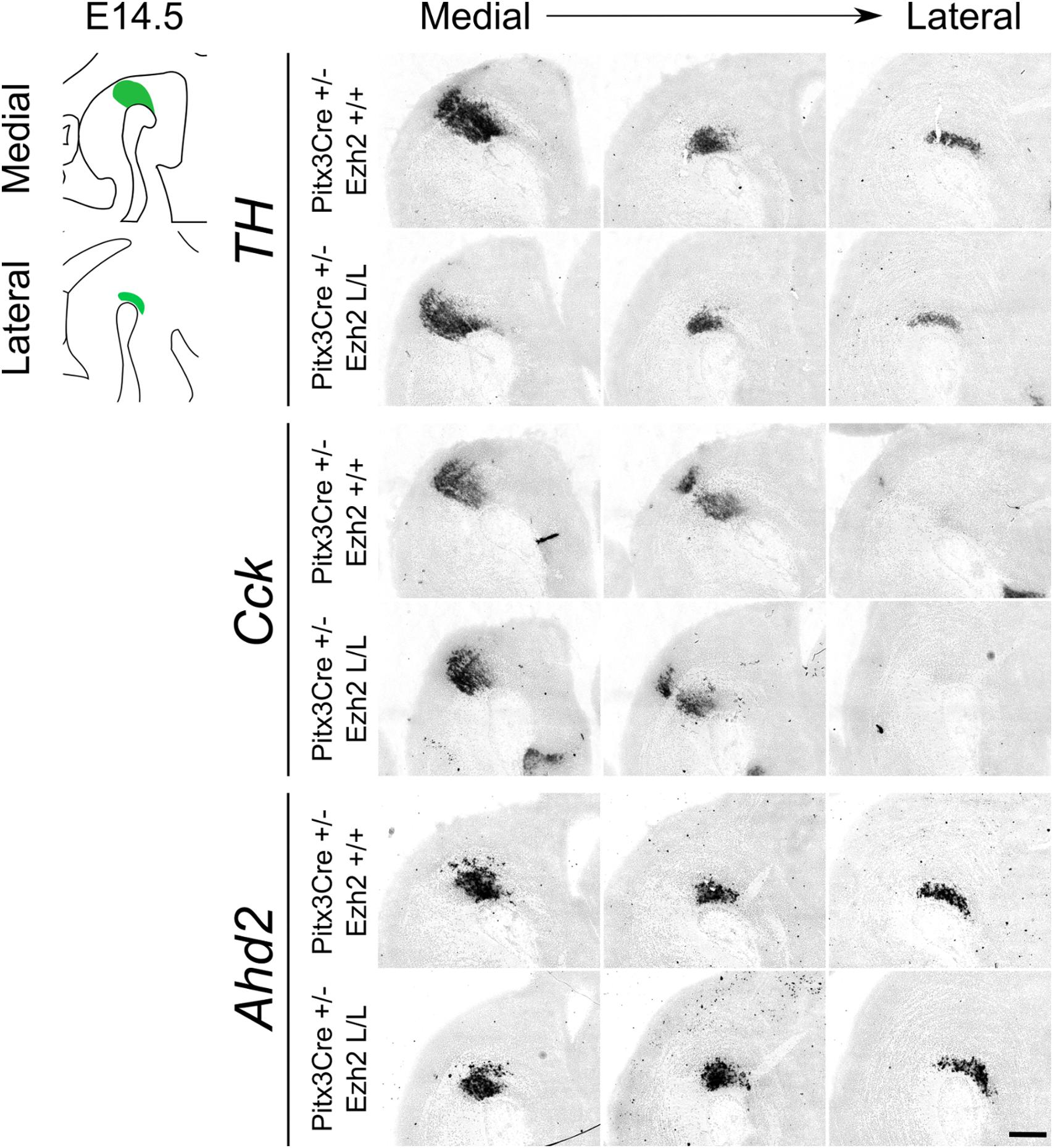
Figure 5. Post-mitotic deletion of Ezh2 does not change the expression pattern of Cck and Ahd2 at E14.5. Spatial expression of Th, Cck, and Ahd2 were analyzed in E14.5 sagittal midbrain sections of Pitx3Cre/+; Ezh2 +/+ and Pitx3Cre/+: Ezh2 L/L embryos with in situ hybridization. Th, Cck, and Ahd2 all show a normal spatial expression in both lateral and medial sections in the Ezh2 cKO in comparison to the wildtype sections. Scale bars = 300 μM.
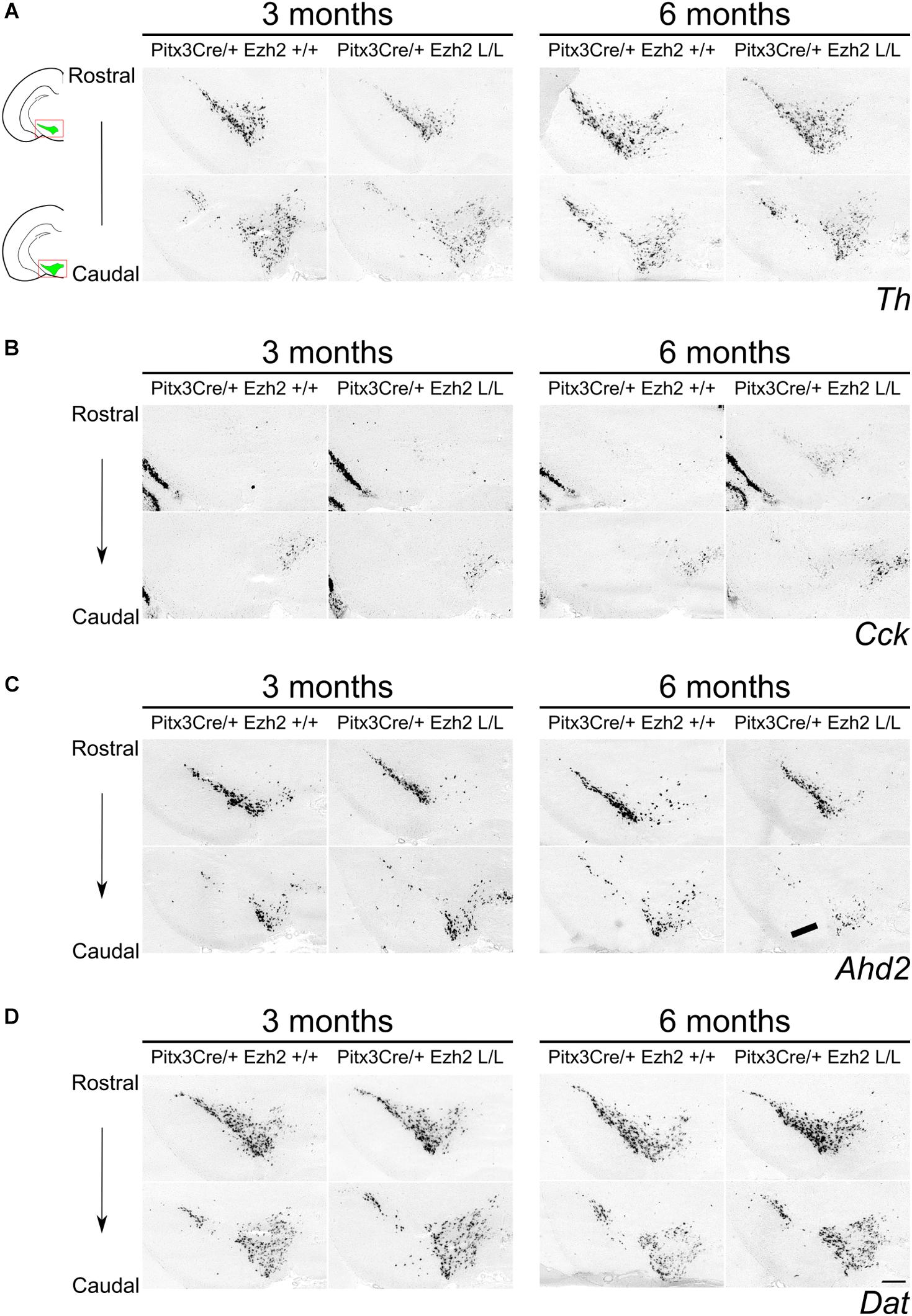
Figure 6. Expression of Cck and Ahd2 is altered in the midbrain of 6 months old Pitx3Cre/+; Ezh2 L/L animals. In situ hybridization of Th, Dat, Cck, and Ahd2 in 3 and 6 months old wildtype and Ezh2 cKO animals. (A) Analysis expression pattern of the general DA mark Th does not reveal considerable changes in expression between the Pitx3Cre/+; Ezh2+/+ and the Ezh2 cKO. (B) Cck is ectopically expressed in the rostral sections of the midbrain of 6 months old Pitx3Cre/+; Ezh2 L/L animals (arrowheads). This rostral expansion of Cck expression is not observed in 3 months old Ezh2 cKO animals (left panel). (C) Expression of Ahd2 is progressively reduced in the caudal midbrain of 6 months old Pitx3Cre/+; Ezh2 L/L animals in comparison to wildtype and 3 months old Ezh2 cKO animals (arrow), while expression in the rostral sections is maintained. (D) The expression pattern of Dat in Pitx3Cre/+;Ezh2 L/L adult midbrain is similar to wildtype littermates at both 3 and 6 months of age. Scale bars = 300 μM.
MdDA Neurons Are Lost in Aging Animals Lacking Ezh2
Previous studies have already shown that post-mitotic PRC2 deficiency can lead to delayed changes in expression of genes, including several important regulators of cell death (Sun et al., 2014; von Schimmelmann et al., 2016) and defective PRC2 activity has been associated with several degenerative diseases, including Huntington’s Disease (Seong et al., 2010; Li et al., 2013; Södersten et al., 2014; von Schimmelmann et al., 2016). We thus hypothesized that Pitx3Cre driven deletion of Ezh2 might affects the survival of mdDA neurons next to programming defects as observed above. To establish whether cells are lost in Pitx3/Ezh2 mutants we assessed the amount of TH+ neurons in the SNc and the VTA at two different ages, 3 and 6 months, by means of immunohistochemistry (Figure 7). Quantification of the amount of TH+ cells in the VTA and the SNc of 3 months old Pitx3Cre/+; Ezh2+/+ (n = 3) and Pitx3/+; Ezh2 L/L (n = 3) animals showed no significant differences in the amount of cells (Figures 7A,B). Interestingly, assessment of the amount of TH+ cells at 6 months of age showed a significant decrease in the amount of TH+ neurons in Pitx3Cre/Ezh2 mutants (∼ 33% loss, n = 3, P < 0.05, two-tailed) (Figure 7C). Separate quantification of the cells in the VTA and SNc (Figure 7A, right panel, white dashed lines) demonstrated that only in the VTA of Pitx3Cre/+; Ezh2 L/L animals a significant amount of TH+ cells is lost (∼32% loss, n = 3, P < 0.05, two-tailed) (Figure 7A, right panel, arrowhead, C), although a downward trend in the amount of TH+ in the SNc domain was detected (∼ 15% loss, n = 3, P = 0.07, two-tailed) (Figure 7C). The loss of cells in the VTA was confirmed by a loss of expression of Calbindin D28K (Calb1), a mark expressed by all cells of the VTA (Supplementary Figure S2, arrowhead). Taken together our results demonstrate that post-mitotic deletion of Ezh2 in mdDA neurons leads to a progressive loss of TH+ cells in the VTA, thereby supporting the argument that lower EZH2/PRC2 activity may lead to neurodegeneration in specific neuronal populations.
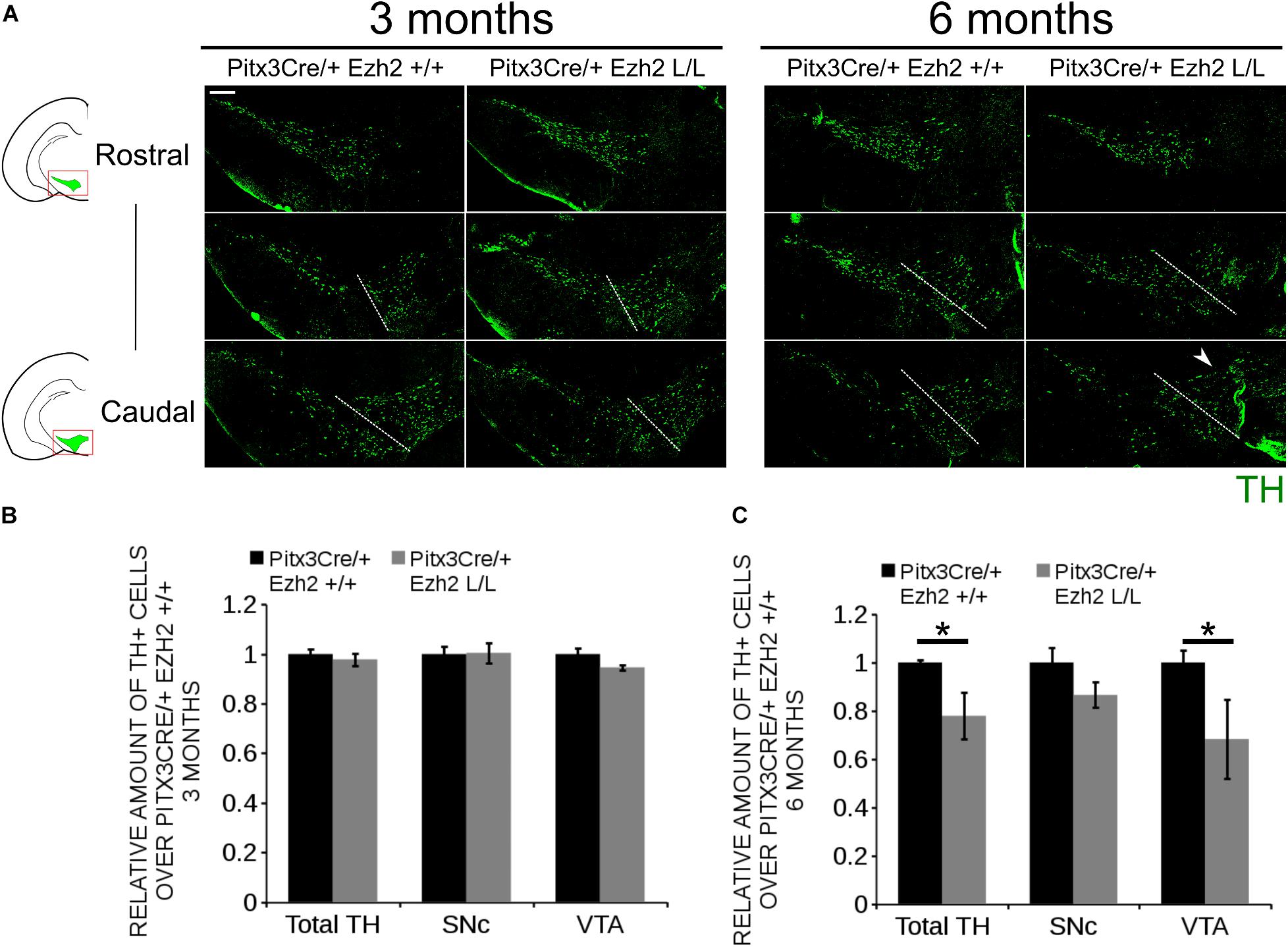
Figure 7. Progressive loss of TH+ cells in the VTA of 6 months old Pitx3/+: Ezh2 L/L animals. (A) Immunohistochemistry for TH in coronal sections of the midbrain of 3 and 6 months old Pitx3Cre/+; Ezh2 +/+ and Pitx3Cre/+; Ezh2 L/L animals. Loss of TH+ neurons was observed in the VTA of 6 months old Ezh2 cKO animals (white arrowhead). White dotted lines represent the border between what was considered SNc and VTA. (B) Quantification of the total amount of TH+ cells and the separation of the TH+ population into SNc and VTA TH+ neurons demonstrated no significant changes in either TH+ populations between 3 months old Pitx3Cre/+; Ezh2+/+ (black bars) and Pitx3Cre/+; Ezh2 L/L (gray bars) animals (n = 3). Wildtypes were set at 1. (C) Quantification of the total number of TH+ cells at 6 months of age showed a ∼22% reduction in the amount of TH+ cells in the Ezh2 cKO (n = 3, ∗P < 0.05, two-tailed). Separations into the SNc and the VTA showed that most cells are lost in the VTA (∼ 32% loss, n = 3, ∗P < 0.05, two-tailed) and down-ward trend is observed in the amount of TH+ cells in the SNc (n = 3, P = 0.07, two-tailed). Wildtypes were set at 1. Scale bars = 200 μM.
H3K27me3 Is Still Present in mdDA Neurons of 6 Months Old Pitx3Cre Driven Ezh2 cKOs
As described above, EZH2 functions as the methyltransferase of the PRC2 complex, which catalyzes the tri-methylation of H3K27 (Cao et al., 2002; Cao and Zhang, 2004a). The phenotype observed in Pitx3Cre/+; Ezh2 L/L animals suggest that PRC2 activity might be altered, which could affect the presence of H3K27me3 in mdDA neurons of these animals. To verify whether the phenotype of Pitx3Cre/+; Ezh2 L/L animals is a consequence of an overall loss of H3K27me3, we performed double immunohistochemistry for H3K27me3 and TH at 6 months of age (Figure 8). We analyzed the presence of H3K27me3 in TH+ neurons in both the SNc and the VTA. In the SNc H3K27me3 was present in TH+ neurons in Pitx3Cre/+; Ezh2 L/L animals in a comparable manner to wildtype littermates [Figure 8A (1–4)]. Similar results were obtained for the VTA, where H3K27me3 was also detected in TH+ cells of Pitx3Cre/Ezh2 cKOs [Figure 8B (1–4)]. Together our results show that post-mitotic deletion of Ezh2 alone is not sufficient to remove overall H3K27me3 marks.
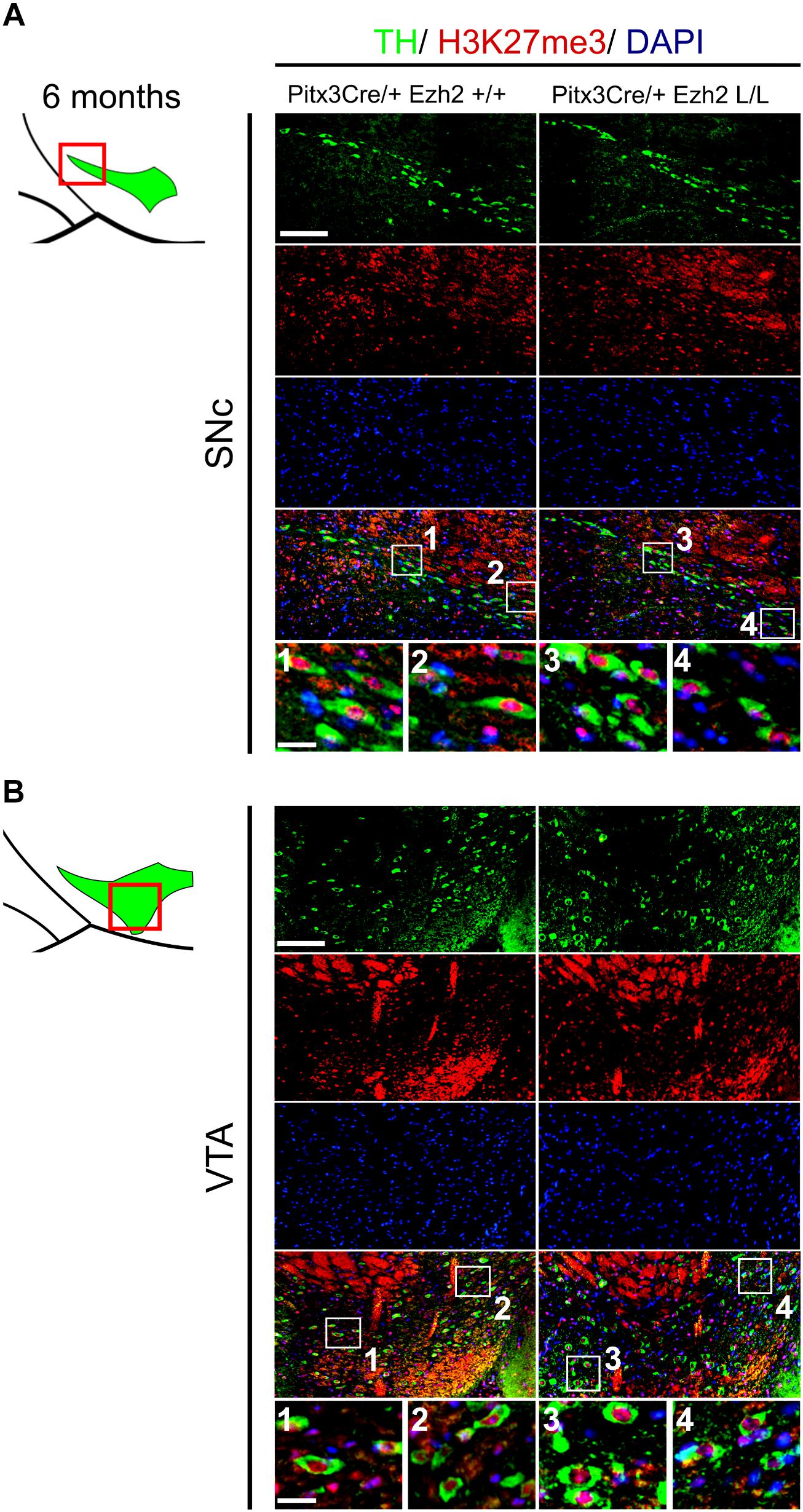
Figure 8. H3K27me3 can still be detected in the SNc and VTA of Ezh2 cKO animals at 6 months of age. (A,B) Immunohistochemistry for TH (green) and H3K27me3 (red) in coronal sections of 6 months old midbrains demonstrated that H3K27me3 is still present in the SNc (A, 3,4)and VTA (B 3,4) of Pitx3Cre/+; Ezh2 L/L animals. Co-localization of H3K27me3 and DAPI staining was used to determine nuclear localization of the H3K27me3 staining. Scale bars = 90 μM and for 1–4 30 μM.
Pitx3Cre/Ezh2 Mutants Display Behavioral Changes at 6 Months of Age
The different populations of mdDA neurons have characteristic projection areas and are involved in the regulation of different types of behavior (Lynd-Balta and Haber, 1994; Haber et al., 2000; Prakash and Wurst, 2006). In Pitx3Cre/+; Ezh2 L/L animals a loss of mdDA neurons is observed which might affect the dopaminergic output and alter the behavior of these animals. For this reason we assessed spontaneous locomotor activity in an open field test and climbing behavior of Pitx3Cre/+; Ezh2+/+ and Pitx3Cre/+; Ezh2 L/L animals (Figure 9). An age-dependent effect on the climbing behavior was observed (Figures 9A,B). Six months old Ezh2 mutant mice (n = 5) demonstrated a significantly lower climbing score than wildtype control animals (n = 5) (∼ 20% lower, P < 0.05, two-tailed) (Figure 9A), while in 3 months old Pitx3Cre/; Ezh2 L/L animals climbing behavior was not affected (n = 8) (Figure 9A). In contrast to the climbing behavior, spontaneous locomotor activity, assessed by distance walked and walking speed in an open field test, was not affected in both 3 months (n = 8) and 6 months (n = 5) old Pitx3Cre/+; Ezh2 L/L animals in comparison to wildtype littermates (Figures 9C,D). These data demonstrate that the loss of Ezh2 does not affect general locomotor activity, but specifically influences the more complex climbing behavior representing the loss of DA neurons in 6 months old Pitx3Cre/+; Ezh2 L/L animals.
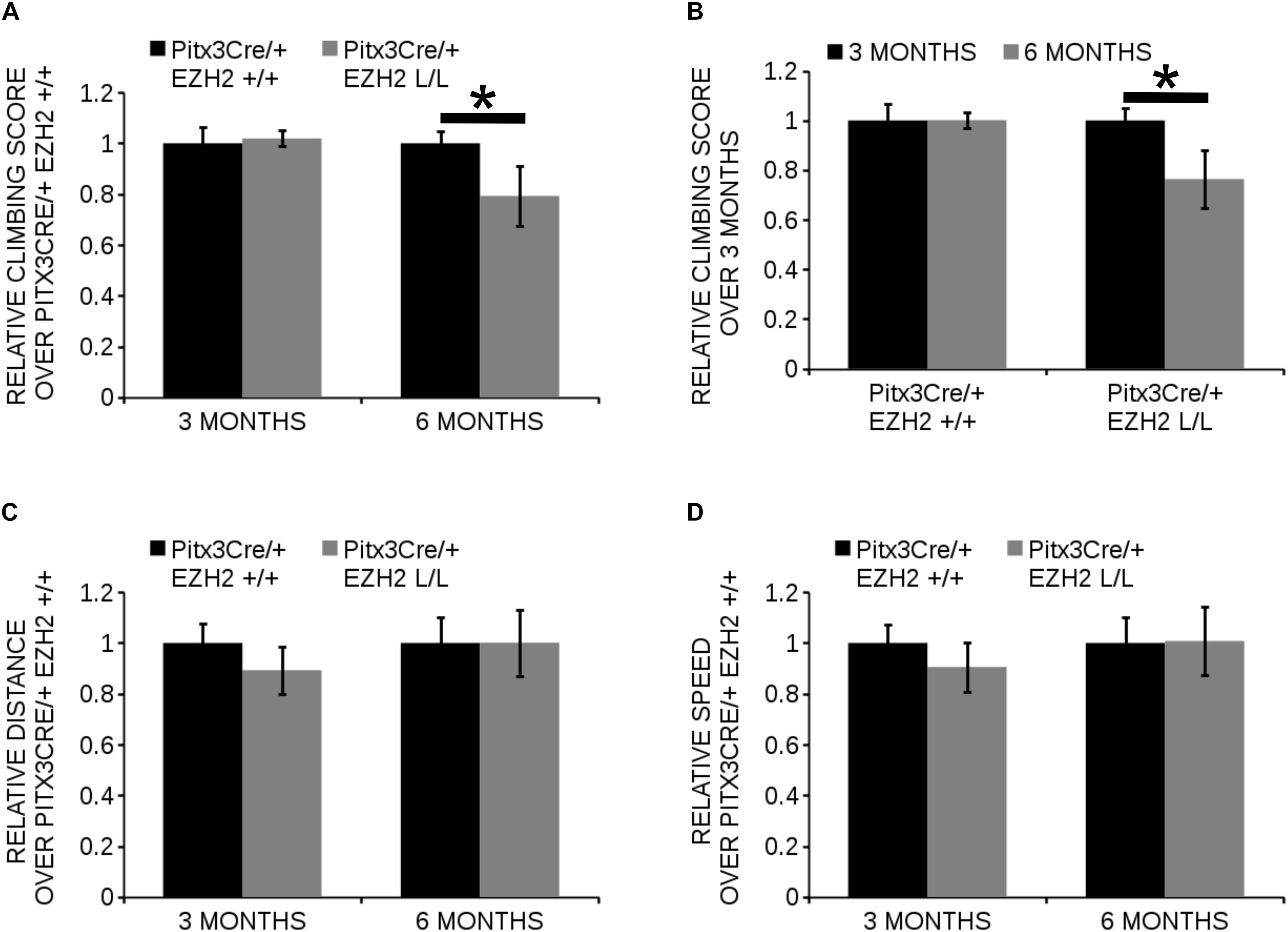
Figure 9. Pitx3Cre driven deletion of Ezh2 influences climbing behavior of 6 months old animals. (A,B) Evaluation of climbing behavior at 3 and 6 months of age. (A) Six months old Pitx3Cre/+;Ezh2 L/L (gray bars) animals have a significantly lower climbing score than wildtype littermates (black bars) (∼ 21% reduction, n = 5, ∗P < 0.05, two-tailed). This is not observed at 3 months of age (n = 8). (B) Climbing-scores progressively reduce in the Ezh2 cKO between 3 months (n = 8, black bars) and 6 months (n = 5, gray bars) (∼21% reduction, ∗P < 0.05, two-tailed), while Pitx3Cre/+;Ezh2+/+ animals show similar climbing behavior at 3 months (n = 8, black bars) and 6 months (n = 5, gray bars). (C,D) Locomotor activity was monitored in an automated set up and the distance walked (in cm) was not significantly altered at 3 months (n = 8) or 6 months (n = 5) in Pitx3Cre/+; Ezh2 L/L animals (C), nor was the speed (in cm/s) (D).
Discussion
Over the last decade several protein have been identified to be differentially expressed in the SNc and the VTA (Chung et al., 2005). These differences in molecular profile have been implied to partially cause the selective degeneration of the SNc neurons in Parkinson’s disease, while neurons of the VTA remain unaffected (Oliveira et al., 2017). Individual subsets are dependent on different transcriptional programs for their development and maintenance (Hwang et al., 2003; Smidt et al., 2004; Smits et al., 2013; Veenvliet et al., 2013; Panman et al., 2014). In addition to the influence of transcription factors, recent studies have led to the hypothesis that modifications of histones might also influence the developmental program of neurons (Pereira et al., 2010; Zemke et al., 2015; Feng et al., 2016). H3K27me3 is a modification associated with gene-silencing and it shows a highly dynamic profile during development (Mikkelsen et al., 2007; Mohn et al., 2008; Chou et al., 2011; Yu et al., 2011). The methylation of H3K27me3 is catalyzed by the PRC2 complex, of which EZH2 is the methyltransferase (Cao and Zhang, 2004b). Previous studies in which Ezh2 was conditionally ablated showed that Ezh2 is involved during several stages of neurodevelopment (Ezhkova et al., 2009; Pereira et al., 2010; Di Meglio et al., 2013; Zemke et al., 2015; Feng et al., 2016). In the current study we focused on the role of Ezh2 during the development, programming and maintenance of mdDA neurons. The conditional removal of Ezh2 in early mdDA progenitors resulted in reduced amounts of TH-expressing cells at E14.5. Previous studies showed that neurogenesis is initiated earlier at the expense of self-renewal in embryos where Ezh2 was conditionally removed from cortical neuroprogenitors (Pereira et al., 2010). Neurogenesis of mdDA neurons is initiated around E10.5 and ceases around E13.5, with the first TH+ neurons being detected around E11.5 (Bayer et al., 1995; Mesman et al., 2014). A shift in the balance between self-renewal and differentiation, at the expanse of self-renewal, might explain why less TH+ neurons are present at E14.5 in the Ezh2 cKO, however, this is not reflected by the amount of neurons present at E12.5, as the number of TH+ cells are not increased at this time point. Another possible reason for the reduced number of TH+ neurons might be that mdDA progenitors and mdDA neurons are produced less efficient due to reduced canonical WNT signaling (Tang et al., 2009). In Wnt1Cre/+; Ezh2 L/L embryos inhibitors of the WNT signaling pathway were up-regulated and less β-galactosidase-positive neural cells were detected in the dorsal midbrain (Zemke et al., 2015). In addition, analysis of Wnt1 expression in En1Cre/+; Ezh2 L/L embryos revealed reduced levels of Wnt1 at E12.5 (Wever et al., unpublished data), suggesting that WNT signaling is also affected in our model.
Interestingly, the loss of TH+ cells mostly affected the Ahd2+ population, which is generated first according to birth-dating studies (Bayer et al., 1995; Bye et al., 2012), while the later born caudomedial population of Cck+ cells remains largely unaffected. These results are in contrast with the results obtained by Pereira et al. where the earlier onset of cortical neurogenesis led to the loss of later born layer 2–4, due to a depletion of the neuroprogenitor pool (Pereira et al., 2010).
The loss of the Ahd2 population is also observed in Pitx3 null mutants (Jacobs et al., 2007), however, in En1Cre/+; Ezh2 L/L animals Pitx3 levels are not affected and PITX3 expression mimicked TH, suggesting that the loss of the Ahd2 positive population is not due to an effect of Ezh2 on Pitx3 expression. The impaired development of the rostrolateral population might be partially due to defective tangential migration. MdDA neurons are generated at the ventricular zone of the ventral midbrain and migrate to their final position first via radial migration followed by tangential migration (Kawano et al., 1995). The dislocated TH+ cells in the medial sections and the loss of rostrolateral cells, suggest that tangential migration might be affected in En1Cre/+; Ezh2 L/L embryos. Lateral migration is dependent on the interaction of mdDA neurons with tangential fibers (Kang et al., 2010). Reelin, an extracellular matrix glycoprotein, has been found to be essential for formation of these fibers, as genetic ablation of Reelin led to the loss of tangential fibers, while radial glial fibers were formed normally (Kang et al., 2010). A previous study demonstrated that genetic ablation of Ezh2 in cortical progenitors affected neuronal migration by influencing Reelin expression (Zhao et al., 2015). In addition EZH2 was found to be required to maintain the tangential migratory program of pontine neurons (Di Meglio et al., 2013), hinting toward a role for EZH2 in tangential migration.
Besides a role for Ezh2 in early development, we also demonstrated that Ezh2 is important for the preservation of neuronal identity and the survival of a subset of TH+ neurons. Post-mitotic deletion of Ezh2 leads to a progressive up-regulation of Cck in the rostral population of the mdDA system and a loss of Ahd2 expression in the more caudal population. Cck has been found to be a target of PRC2 during development and loss of Ezh2 in neuronal progenitors leads to a significant up-regulation of Cck expression (Bracken et al., 2006; Mikkelsen et al., 2007; Ku et al., 2008; Pereira et al., 2010). We thus theorize that the progressive appearance of Cck in the rostral population of 6 months old Pitx3/Ezh2 mutants is because H3K27me3 on the Cck promoter is not maintained over time, leading to the de-repression of the promoter. In contrast, the promoter of Ahd2 has not been associated with EZH2 binding and H3K27me3 (Mikkelsen et al., 2007), suggesting that the loss of Ahd2 in the caudal mdDA system is not due to a direct effect of EZH2 on Ahd2 expression. In addition to alterations in expression of subset marks, Pitx3Cre/Ezh2 mutants also display a progressive loss of TH+ cells in the VTA. The loss of TH+ cells was reflected by reduced expression of the VTA subset mark, Calb1. In contrast to Cck, Calb1 is expressed by all neurons of the VTA (La Manno et al., 2016; Brignani and Pasterkamp, 2017), suggesting that Pitx3Cre driven deletion of Ezh2 specifically affects the Calb+Cck- population of VTA neurons. Calb1 has been shown to promote resistance against neurodegeneration (German et al., 1992; McMahon et al., 1998) and the reduced levels of Calb1 in the more caudal region of the mdDA system might contribute the loss of TH+ neurons, however, the loss of Calb1 might also be a consequence of the initial loss of neurons. Next to a loss in DA neurons, Pitx3Cre/+; Ezh2 L/L also showed reduced climbing behavior without affecting general locomotor activity. It is hypothesized that climbing behavior requires different and more complex dopaminergic mechanisms than horizontal locomotor behavior (Cabib and Puglisi-Allegra, 1985; Moore and Axton, 1988; Usiello et al., 2000), including projections from the VTA to the Nucleus accumbens (Costall et al., 1983, 1985; Salamone, 1992), which might explain the specific reduction in climbing behavior. However, it needs to be noted that Pitx3 is also expressed in the muscles and lens and Pitx3Cre driven deletion of Ezh2 may also affect these systems. In addition, Pitx3Cre/+; Ezh2 L/L animals had a higher change of dying prematurely (∼35% higher).
Even though both En1/Ezh2 and Pitx3/Ezh2 mutants display a disturbance in Cck and Ahd2 expression and a loss of TH+ cells, the severity of the phenotype differs. The phenotype observed in En1Cre/+; Ezh2 L/L embryos is probably a consequence of an overall defect in PRC2 functioning and a global loss of H3K27me3 in the midbrain area. In contrast, the effects observed in Pitx3Cre driven Ezh2 cKOs can not be explained by a general lack of H3K27Me3, as the mark was still detected in the SNc and VTA of 6 months old Pitx3Cre/+; Ezh2 L/L animals. However, EZH2 has also been shown to have PRC2-independent activity and active transcription of genes marked by H3K27me3 has also been observed before, suggesting that changes in Ezh2 functioning might not be reflected by changes in H3K27me3 (Bracken et al., 2006; Mikkelsen et al., 2007; Mohn et al., 2008; Xu et al., 2012). In addition, local changes in H3K27me3 levels are not visualized by immunohistochemistry for H3K27me3.
Taken together, this study shows that next to a role in development, Ezh2 is also important for the maintenance and survival of cells of a small group of mdDA neurons. Interestingly, the loss of Ezh2 mainly affects the embryonic development of the rostrolateral population, destined to become the SNc, while cells of the VTA are lost when Ezh2 is removed post-mitotically. This suggests that Ezh2 has different functions during development and neuronal maintenance.
Author Contributions
IW performed the experiment and wrote the first draft of the manuscript. LvO and CW assisted in experiments. MS funded the study, provided resources, supervised IW, LvO, and CW, and revised the manuscript.
Funding
This work was sponsored by the NWO-ALW (Nederlandse Organisatie voor Wetenschappelijk Onderzoek-Aard en Levenswetenschappen) VICI grant (865.09.002) awarded to MS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Frederic Zilbermann of the Friedrich Miescher Institute for his generous gift of the Ezh2-floxed mouse line and Dr. Sandra Blaess for kindly providing us with the En1Cre; R26RYFP mouse strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2018.00491/full#supplementary-material
FIGURE S1 | TH expression is reduced in rostral sections of the embryonic midbrain of En1/Ezh2 mutants. Analysis of TH expression in coronal E14.5 midbrain sections by means of immunohistochemistry. Expression of TH is reduced in the rostral sections (white arrowheads). Scale bars = 200 μM.
FIGURE S2 | Pitx3Cre driven deletion of Ezh2 induces a progressive loss of Calb1 staining in the VTA. Analysis of the expression of Calb1 in coronal midbrain sections of 3 and 6 months old Pitx3Cre/+; Ezh2+/+ and Pitx3Cre/+; Ezh2 L/L animals by means of in situ hybridization. Expression of Calb1 is lost in the caudal VTA of 6 months old Ezh2 cKO animals (black arrowhead), while expression can still be detected in this location at 3 months (left panel). Scale bars = 300 μM.
References
Bayer, S. A., Wills, K. V., Triarhou, L. C., and Ghetti, B. (1995). Time of neuron origin and gradients of neurogenesis in midbrain dopaminergic neurons in the mouse. Exp. Brain Res. 105, 191–199. doi: 10.1007/BF00240955
Braak, H., Tredici, K. D., Rüb, U., Vos, R. A. I., de Steur, E. N. H. J., and Braak, E. (2003). Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211. doi: 10.1016/S0197-4580(02)00065-9
Bracken, A. P., Dietrich, N., Pasini, D., Hansen, K. H., and Helin, K. (2006). Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136. doi: 10.1101/gad.381706
Brignani, S., and Pasterkamp, R. J. (2017). Neuronal subset-specific migration and axonal wiring mechanisms in the developing midbrain dopamine system. Front. Neuroanat. 11:55. doi: 10.3389/fnana.2017.00055
Brodski, C., Weisenhorn, D. M. V., Signore, M., Sillaber, I., Oesterheld, M., Broccoli, V., et al. (2003). Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain–hindbrain organizer. J. Neurosci. 23, 4199–4207. doi: 10.1523/JNEUROSCI.23-10-04199.2003
Bye, C. R., Thompson, L. H., and Parish, C. L. (2012). Birth dating of midbrain dopamine neurons identifies A9 enriched tissue for transplantation into Parkinsonian mice. Exp. Neurol. 236, 58–68. doi: 10.1016/j.expneurol.2012.04.002
Cabib, S., and Puglisi-Allegra, S. (1985). Different effects of apomorphine on climbing behavior and locomotor activity in three strains of mice. Pharmacol. Biochem. Behav. 23, 555–557. doi: 10.1016/0091-3057(85)90418-6
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., et al. (2002). Role of histone H3 Lysine 27 methylation in polycomb-group silencing. Science 298, 1039–1043. doi: 10.1126/science.1076997
Cao, R., and Zhang, Y. (2004a). SUZ12 Is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15, 57–67. doi: 10.1016/j.molcel.2004.06.020
Cao, R., and Zhang, Y. (2004b). The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155–164.
Chou, R.-H., Yu, Y.-L., and Hung, M.-C. (2011). The roles of EZH2 in cell lineage commitment. Am. J. Transl. Res. 3, 243–250.
Chung, C. Y., Seo, H., Sonntag, K. C., Brooks, A., Lin, L., and Isacson, O. (2005). Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum. Mol. Genet. 14, 1709–1725. doi: 10.1093/hmg/ddi178
Costall, B., Eniojukan, J. F., and Naylor, R. J. (1983). The mesolimbic nucleus accumbens is critically involved with the mediation of the motor inhibitory and facilitatory effects of dopamine agonists on mouse spontaneous climbing behaviour. Eur. J. Pharmacol. 96, 201–210. doi: 10.1016/0014-2999(83)90309-6
Costall, B., Eniojukan, J. F., and Naylor, R. J. (1985). Dopamine agonist action in mesolimbic, cortical and extrapyramidal areas to modify spontaneous climbing behaviour of the mouse. Psychopharmacology 86, 452–457. doi: 10.1007/BF00427907
Di Giovannantonio, L. G., Di Salvio, M., Acampora, D., Prakash, N., Wurst, W., and Simeone, A. (2013). Otx2 selectively controls the neurogenesis of specific neuronal subtypes of the ventral tegmental area and compensates En1-dependent neuronal loss and MPTP vulnerability. Dev. Biol. 373, 176–183. doi: 10.1016/j.ydbio.2012.10.022
Di Meglio, T., Kratochwil, C. F., Vilain, N., Loche, A., Vitobello, A., Yonehara, K., et al. (2013). Ezh2 orchestrates topographic tangential migration and connectivity of mouse precerebellar neurons. Science 339, 204–207. doi: 10.1126/science.1229326
Di Salvio, M., Di Giovannantonio, L. G., Acampora, D., Prosperi, R., Omodei, D., Prakash, N., et al. (2010). Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat. Neurosci. 13, 1481–1488. doi: 10.1038/nn.2661
Ezhkova, E., Pasolli, H. A., Parker, J. S., Stokes, N., Su, I.-H. S. I., Hannon, G., et al. (2009). Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122–1135. doi: 10.1016/j.cell.2008.12.043
Feng, X., Juan, A. H., Wang, H. A., Ko, K. D., Zare, H., and Sartorelli, V. (2016). Polycomb Ezh2 controls the fate of GABAergic neurons in the embryonic cerebellum. Development 143, 1971–1980. doi: 10.1242/dev.132902
Francis, N. J., Kingston, R. E., and Woodcock, C. L. (2004). Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577. doi: 10.1126/science.1100576
German, D. C., Manaye, K. F., Sonsalla, P. K., and Brooks, B. A. (1992). Midbrain dopaminergic cell loss in parkinson’s disease and MPTP-induced parkinsonism: sparing of calbindin-D25k—containing cellsa. Ann. N. Y. Acad. Sci. 648, 42–62. doi: 10.1111/j.1749-6632.1992.tb24523.x
Grima, B., Lamouroux, A., Blanot, F., Biguet, N. F., and Mallet, J. (1985). Complete coding sequence of rat tyrosine hydroxylase mRNA. Proc. Natl. Acad. Sci. U.S.A. 82, 617–621. doi: 10.1073/pnas.82.2.617
Haber, S. N., Fudge, J. L., and McFarland, N. R. (2000). Striatonigrostriatal pathways in primates form an ascending spiral from the shell to the Dorsolateral striatum. J. Neurosci. 20, 2369–2382. doi: 10.1523/JNEUROSCI.20-06-02369.2000
Hirabayashi, Y., Suzki, N., Tsuboi, M., Endo, T. A., Toyoda, T., Shinga, J., et al. (2009). Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613. doi: 10.1016/j.neuron.2009.08.021
Hwang, D.-Y., Ardayfio, P., Kang, U. J., Semina, E. V., and Kim, K.-S. (2003). Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Mol. Brain Res. 114, 123–131. doi: 10.1016/S0169-328X(03)00162-1
Jacobs, F. M. J., Smits, S. M., Noorlander, C. W., von Oerthel, L., von Linden, A. J., van der, et al. (2007). Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development 134, 2673–2684. doi: 10.1242/dev.02865
Jacobs, F. M. J., Veenvliet, J. V., Almirza, W. H., Hoekstra, E. J., Oerthel, L., and von Linden, A. J. A. (2011). Retinoic acid-dependent and -independent gene-regulatory pathways of Pitx3 in meso-diencephalic dopaminergic neurons. Development 138, 5213–5222. doi: 10.1242/dev.071704
Kang, W.-Y., Kim, S.-S., Cho, S.-K., Kim, S., Suh-Kim, H., and Lee, Y.-D. (2010). Migratory defect of mesencephalic dopaminergic neurons in developing reeler mice. Anat. Cell Biol. 43, 241–251. doi: 10.5115/acb.2010.43.3.241
Kawano, H., Ohyama, K., Kawamura, K., and Nagatsu, I. (1995). Migration of dopaminergic neurons in the embryonic mesencephalon of mice. Brain Res. Dev. Brain Res. 86, 101–113. doi: 10.1016/0165-3806(95)00018-9
Kimmel, R. A., Turnbull, D. H., Blanquet, V., Wurst, W., Loomis, C. A., and Joyner, A. L. (2000). Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 14, 1377–1389.
Ku, M., Koche, R. P., Rheinbay, E., Mendenhall, E. M., Endoh, M., Mikkelsen, T. S., et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4:e1000242. doi: 10.1371/journal.pgen.1000242
La Manno, G., Gyllborg, D., Codeluppi, S., Nishimura, K., Salto, C., Zeisel, A., et al. (2016). Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566.e–580.e. doi: 10.1016/j.cell.2016.09.027
Li, J., Hart, R. P., Mallimo, E. M., Swerdel, M. R., Kusnecov, A. W., and Herrup, K. (2013). EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat. Neurosci. 16, 1745–1753. doi: 10.1038/nn.3564
Lynd-Balta, E., and Haber, S. N. (1994). The organization of midbrain projections to the striatum in the primate: sensorimotor-related striatum versus ventral striatum. Neuroscience 59, 625–640. doi: 10.1016/0306-4522(94)90182-1
Margueron, R., Li, G., Sarma, K., Blais, A., Zavadil, J., Woodcock, C. L., et al. (2008). Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518. doi: 10.1016/j.molcel.2008.11.004
McMahon, A., Wong, B. S., Iacopino, A. M., Ng, M. C., Chi, S., and German, D. C. (1998). Calbindin-D28k buffers intracellular calcium and promotes resistance to degeneration in PC12 cells. Mol. Brain Res. 54, 56–63. doi: 10.1016/S0169-328X(97)00305-7
Mesman, S., von Oerthel, L., and Smidt, M. P. (2014). Mesodiencephalic dopaminergic neuronal differentiation does not involve GLI2A-mediated SHH-signaling and is under the direct influence of canonical WNT signaling. PLoS One 9:e97926. doi: 10.1371/journal.pone.0097926
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. doi: 10.1038/nature06008
Mohn, F., Weber, M., Rebhan, M., Roloff, T. C., Richter, J., Stadler, M. B., et al. (2008). Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766. doi: 10.1016/j.molcel.2008.05.007
Moore, N. A., and Axton, M. S. (1988). Production of climbing behaviour in mice requires both D1 and D2 receptor activation. Psychopharmacology 94, 263–266. doi: 10.1007/BF00176857
Oliveira, M. A. P., Balling, R., Smidt, M. P., and Fleming, R. M. T. (2017). Embryonic development of selectively vulnerable neurons in Parkinson’s disease. NPJ Parkinsons Dis. 3:21. doi: 10.1038/s41531-017-0022-4
Ono, Y., Nakatani, T., Sakamoto, Y., Mizuhara, E., Minaki, Y., Kumai, M., et al. (2007). Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134, 3213–3225. doi: 10.1242/dev.02879
Panman, L., Papathanou, M., Laguna, A., Oosterveen, T., Volakakis, N., Acampora, D., et al. (2014). Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep. 8, 1018–1025. doi: 10.1016/j.celrep.2014.07.016
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M.-W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962. doi: 10.1073/pnas.1002530107
Placzek, M., and Briscoe, J. (2005). The floor plate: multiple cells, multiple signals. Nat. Rev. Neurosci. 6, 230–240. doi: 10.1038/nrn1628
Prakash, N., and Wurst, W. (2006). Development of dopaminergic neurons in the mammalian brain. Cell. Mol. Life Sci. 63, 187–206. doi: 10.1007/s00018-005-5387-6
Protais, P., Costentin, J., and Schwartz, J. C. (1976). Climbing behavior induced by apomorphine in mice: a simple test for the study of dopamine receptors in striatum. Psychopharmacology 50, 1–6. doi: 10.1007/BF00634146
Salamone, J. D. (1992). Complex motor and sensorimotor functions of striatal and accumbens dopamine: involvement in instrumental behavior processes. Psychopharmacology 107, 160–174. doi: 10.1007/BF02245133
Seong, I. S., Woda, J. M., Song, J.-J., Lloret, A., Abeyrathne, P. D., Woo, C. J., et al. (2010). Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 19, 573–583. doi: 10.1093/hmg/ddp524
Shen, X., Liu, Y., Hsu, Y.-J., Fujiwara, Y., Kim, J., Mao, X., et al. (2008). EZH1 mediates methylation on histone h3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502. doi: 10.1016/j.molcel.2008.10.016
Shults, C. W., Hashimoto, R., Brady, R. M., and Gage, F. H. (1990). Dopaminergic cells align along radial glia in the developing mesencephalon of the rat. Neuroscience 38, 427–436. doi: 10.1016/0306-4522(90)90039-7
Smidt, M. P., Asbreuk, C. H. J., Cox, J. J., Chen, H., Johnson, R. L., and Burbach, J. P. H. (2000). A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat. Neurosci. 3, 337–341. doi: 10.1038/73902
Smidt, M. P., Oerthel, L., von Hoekstra, E. J., Schellevis, R. D., and Hoekman, M. F. M. (2012). Spatial and temporal lineage analysis of a pitx3-driven cre-recombinase knock-in mouse model. PLoS One 7:e42641. doi: 10.1371/journal.pone.0042641
Smidt, M. P., Smits, S. M., Bouwmeester, H., Hamers, F. P. T., Linden, A. J. A., van der Hellemons, A. J. C. G. M., et al. (2004). Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development 131, 1145–1155. doi: 10.1242/dev.01022
Smits, S. M., Burbach, J. P. H., and Smidt, M. P. (2006). Developmental origin and fate of meso-diencephalic dopamine neurons. Prog. Neurobiol. 78, 1–16. doi: 10.1016/j.pneurobio.2005.12.003
Smits, S. M., Ponnio, T., Conneely, O. M., Burbach, J. P. H., and Smidt, M. P. (2003). Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur. J. Neurosci. 18, 1731–1738. doi: 10.1046/j.1460-9568.2003.02885.x
Smits, S. M., von Oerthel, L., Hoekstra, E. J., Burbach, J. P. H., and Smidt, M. P. (2013). Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons. PLoS One 8:e76037. doi: 10.1371/journal.pone.0076037
Södersten, E., Feyder, M., Lerdrup, M., Gomes, A.-L., Kryh, H., Spigolon, G., et al. (2014). Dopamine signaling leads to loss of polycomb repression and aberrant gene activation in experimental parkinsonism. PLoS Genet. 10:e1004574. doi: 10.1371/journal.pgen.1004574
Srinivas, S., Watanabe, T., Lin, C. S., William, C. M., Tanabe, Y., Jessell, T. M., et al. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4. doi: 10.1186/1471-213X-1-4
Sun, D., Luo, M., Jeong, M., Rodriguez, B., Xia, Z., Hannah, R., et al. (2014). Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14, 673–688. doi: 10.1016/j.stem.2014.03.002
Sunmonu, N. A., Chen, L., and Li, J. Y. H. (2009). Misexpression of Gbx2 throughout the mesencephalon by a conditional gain-of-function transgene leads to deletion of the midbrain and cerebellum in mice. Genesis 2000, 667–673. doi: 10.1002/dvg.20546
Tang, M., Miyamoto, Y., and Huang, E. J. (2009). Multiple roles of β-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Dev. Camb. Engl. 136, 2027–2038. doi: 10.1242/dev.034330
Usiello, A., Baik, J.-H., Rougé-Pont, F., Picetti, R., Dierich, A., LeMeur, M., et al. (2000). Distinct functions of the two isoforms of dopamine D2 receptors. Nature 408, 199–203. doi: 10.1038/35041572
Veenvliet, J. V., Santos, M. T., dos Kouwenhoven, W. M., von Oerthel, L., Lim, J. L., van der Linden, A. J., et al. (2013). Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 140, 4116–4116. doi: 10.1242/dev.094565
Veenvliet, J. V., and Smidt, M. P. (2014). Molecular mechanisms of dopaminergic subset specification: fundamental aspects and clinical perspectives. Cell. Mol. Life Sci. 71, 4703–4727. doi: 10.1007/s00018-014-1681-5
von Schimmelmann, M., Feinberg, P. A., Sullivan, J. M., Ku, S. M., Badimon, A., Duff, M. K., et al. (2016). Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 19, 1321–1330. doi: 10.1038/nn.4360
Xu, K., Wu, Z. J., Groner, A. C., He, H. H., Cai, C., Lis, R. T., et al. (2012). EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science 338, 1465–1469. doi: 10.1126/science.1227604
Yu, Y.-L., Chou, R.-H., Chen, L.-T., Shyu, W.-C., Hsieh, S.-C., Wu, C.-S., et al. (2011). EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1C-dependent calcium signaling. J. Biol. Chem. 286, 9657–9667. doi: 10.1074/jbc.M110.185124
Zemke, M., Draganova, K., Klug, A., Schöler, A., Zurkirchen, L., Gay, M. H.-P., et al. (2015). Loss of Ezh2 promotes a midbrain-to-forebrain identity switch by direct gene derepression and Wnt-dependent regulation. BMC Biol. 13:103. doi: 10.1186/s12915-015-0210-9
Zhang, J., Pho, V., Bonasera, S. J., Holzmann, J., Tang, A. T., Hellmuth, J., et al. (2007). Essential function of HIPK2 in TGFβ-dependent survival of midbrain dopamine neurons. Nat. Neurosci. 10, 77–86. doi: 10.1038/nn1816
Keywords: development, epigenetics, midbrain, dopamine, histone methylation
Citation: Wever I, von Oerthel L, Wagemans CMRJ and Smidt MP (2019) EZH2 Influences mdDA Neuronal Differentiation, Maintenance and Survival. Front. Mol. Neurosci. 11:491. doi: 10.3389/fnmol.2018.00491
Received: 12 October 2018; Accepted: 19 December 2018;
Published: 17 January 2019.
Edited by:
Björn Spittau, Universitätsmedizin Rostock, GermanyReviewed by:
Claude Brodski, Ben-Gurion University of the Negev, IsraelCarlos G. Dotti, Spanish National Research Council (CSIC), Spain
Copyright © 2019 Wever, von Oerthel, Wagemans and Smidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marten P. Smidt, bS5wLnNtaWR0QHV2YS5ubA==
 Iris Wever
Iris Wever Lars von Oerthel
Lars von Oerthel Cindy M. R. J. Wagemans
Cindy M. R. J. Wagemans Marten P. Smidt
Marten P. Smidt