- 1Developmental Biology, Institute Biology 1, Faculty of Biology, Albert Ludwig University of Freiburg, Freiburg im Breisgau, Germany
- 2BIOSS Centre for Biological Signalling Studies, University of Freiburg, Freiburg im Breisgau, Germany
- 3CIBSS – Centre for Integrative Biological Signalling Studies, University of Freiburg, Freiburg im Breisgau, Germany
- 4Department of Neurobiology & Anatomy, University of Utah, Salt Lake City, UT, United States
The hypothalamus is characterized by great neuronal diversity, with many neuropeptides and other neuromodulators being expressed within its multiple anatomical domains. The regulatory networks directing hypothalamic development have been studied in detail, but, for many neuron types, control of differentiation is still not understood. The highly conserved Brain-specific homeobox (Bsx) transcription factor has previously been described in regulating Agrp and Npy expression in the hypothalamic arcuate nucleus (ARC) in mice. While Bsx is expressed in many more subregions of both tuberal and mamillary hypothalamus, the functions therein are not known. Using genetic analyses in zebrafish, we show that most bsx expression domains are dependent on Nkx2.1 and Nkx2.4 homeodomain transcription factors, while a subset depends on Otp. We show that the anatomical pattern of the ventral forebrain appears normal in bsx mutants, but that Bsx is necessary for the expression of many neuropeptide encoding genes, including agrp, penka, vip, trh, npb, and nts, in distinct hypothalamic anatomical domains. We also found Bsx to be critical for normal expression of two Crh family members, crhb and uts1, as well as crhbp, in the hypothalamus and the telencephalic septal region. Furthermore, we demonstrate a crucial role for Bsx in serotonergic, histaminergic and nitrergic neuron development in the hypothalamus. We conclude that Bsx is critical for the terminal differentiation of multiple neuromodulatory cell types in the forebrain.
Introduction
Together with the pituitary gland, the hypothalamus represents the main neuroendocrine center of the body, regulating core physiological parameters such as growth, body temperature, fluid balance, feeding and energy expenditure, stress and arousal, fatigue and sleep, maternal bonding, and lactation. This functional diversity is reflected by the cellular heterogeneity of the hypothalamus, which consists of multiple distinct nuclei harboring a great variety of cell types (Bedont et al., 2015). Beyond truly neuroendocrine cells secreting neurohormones, a great variety of neuromodulators are expressed in the hypothalamus (Romanov et al., 2019). These neuromodulators influence the response behavior of numerous neural circuits, both within and beyond the hypothalamus (Lee and Dan, 2012; Nadim and Bucher, 2014). Biochemically, neuromodulators range from small molecules, such as nitric oxide or monoamines, to peptides, with more than 100 different neuropeptides being known in the human brain (Wang et al., 2015; Russo, 2017).
Thus, the hypothalamus represents an excellent model to study how extraordinary cellular heterogeneity arises from a limited number of progenitor cells. Previous work already suggested that multiple signaling pathways and a highly combinatorial network of transcription factors are required to generate the great diversity of neuromodulatory cell types in the hypothalamus (reviewed in Bedont et al., 2015; Xie and Dorsky, 2017; Alvarez-Bolado, 2019). Deciphering these signaling pathways and transcription factors will deepen our understanding of how this complex brain structure develops, and ultimately may facilitate the establishment of in vitro differentiation protocols for use in regenerative approaches to treat neuroendocrine disorders (Suga, 2019). Extensive evolutionary conservation of hypothalamus anatomy, development, and function facilitates comparisons between different animal models from fish to mammals (Löhr and Hammerschmidt, 2011; Machluf et al., 2011; Dominguez et al., 2015; Puelles and Rubenstein, 2015; Santos-Duran et al., 2015; Xie and Dorsky, 2017; Alié et al., 2018; Schredelseker and Driever, 2020).
The highly conserved Brain-specific Homeobox (Bsx) transcription factor has first been reported in Drosophila (Jones and McGinnis, 1993). Bsx expression in several subregions of the hypothalamus, the pineal gland, and the telencephalic septum (TelSep) has been described for mice (Cremona et al., 2004) and zebrafish (Schredelseker and Driever, 2018). While recently Bsx functions in the development of the epithalamus have been elucidated (D’Autilia et al., 2010; Schredelseker and Driever, 2018; Mano et al., 2019), no data exist on the role of Bsx in other forebrain regions of teleosts.
In the mouse ARC, Bsx has been shown to be coexpressed with Agouti-related neuropeptide (Agrp) and Neuropeptide Y (Npy) (Sakkou et al., 2007). Moreover, Agrp and Npy expression has been found to be strongly reduced during mouse embryonic development in Bsx mutants (Sakkou et al., 2007). Follow-up studies showed that upon activation by Ghrelin (Nogueiras et al., 2008) Bsx directly binds the promoter regions of Agrp and Npy (Lee et al., 2013). Bsx has been discussed nearly exclusively as a regulator of orexigenic peptide expression in the ARC (Burbridge et al., 2016; Alvarez-Bolado, 2019). Bsx functions beyond the regulation of orexigenic factors in cells of the melanocortin system have been scarcely explored, but lactation deficiencies in bsx mutant mice have been reported (McArthur and Ohtoshi, 2007). Given the much broader expression domains of Bsx, however, we hypothesized that Bsx has additional functions in the development of other cell types in the hypothalamus. Here, we assessed Bsx functions in the secondary prosencephalon with a focus on the bHyp, where bsx is broadly expressed in domains that we recently characterized in detail (Schredelseker and Driever, 2020). We identified transcription factors that regulate the expression of bsx in the hypothalamus. In bsx mutant embryos, we found patterning in the secondary prosencephalon to be normal. To identify Bsx roles in neuronal differentiation, we focused on peptidergic and aminergic neuromodulators. Comparing wildtype and bsx mutant zebrafish embryos, we analyzed the expression of genes encoding zebrafish homologs of the neuropeptides assessed by Díaz et al. (2014), with the exception of oxytocin (oxt), which in the embryonic hypothalamus of both zebrafish and mouse is only expressed in alar regions (Eaton and Glasgow, 2007; Díaz et al., 2014), where bsx is not expressed. We extended our analysis to additional markers for peptidergic, nitrergic and monoaminergic neurons. For 13 of the 26 markers analyzed, we detected absent or strongly reduced expression in defined bHyp subregions of bsx mutant embryos, demonstrating that Bsx exerts functions beyond the specification of orexigenic neurons in the ARC. In addition, we found Bsx to be required for uts1 expression in the TelSep.
Notably, we found that Bsx functions are not restricted to a single hypothalamic nucleus, and that Bsx is also not selectively required for expression of a particular gene specific to a certain neuromodulatory cell type. Instead, Bsx appears crucial for expression of multiple genes in distinct clusters distributed over several distinct hypothalamic areas, while the same genes are expressed independently of Bsx in other areas. This supports the idea that the development of the numerous neuromodulatory cell types in the hypothalamus is controlled by transcription factors in a highly combinatorial manner. By demonstrating that Bsx is a determinant of a surprisingly large number of hypothalamic and septal neuromodulatory cell populations, we propose to replace the notion of Bsx as a transcriptional regulator in a single neuron type by a model that presents Bsx as a major developmental factor in many neuromodulatory cell types within and beyond the hypothalamus. Bsx is a crucial component of a so far not fully understood complex combinatorial code for neuromodulatory neuron differentiation.
Results
bsx Expression Is Differentially Regulated by Homeobox Transcription Factors in Different Regions of the Secondary Prosencephalon
While bsx expression domains in the bHyp have recently been characterized in detail (Schredelseker and Driever, 2020), no data exist on the upstream regulation of bsx expression in the hypothalamus. Severe hypoplasia and deformities in the bHyp were described for Nkx2.1 mutant mice (Kimura et al., 1996) and critical functions of Nkx-homeodomain factors Nkx2.1, Nkx2.4a, and Nkx2.4b were revealed in zebrafish hypothalamus development (Manoli and Driever, 2014). To asses if hypothalamic bsx expression depends on the activity of early acting Nkx-homeodomain transcription factors, we used TALENs to generate loss-of-function alleles for nkx2.1, nkx2.4a and nkx2.4b (Supplementary Figure S1). We analyzed bsx expression in single and compound mutants by in situ hybridization, and found it to be lost in nkx2.1, nkx2.4a, and nkx2.4b triple mutants (here abbreviated nkx2.1/2.4a/b) in all hypothalamic areas of embryos 4 dpf (Figures 1a–d). However, expression of bsx is preserved in nkx2.1/2.4a/b triple mutants in two ventral forebrain areas. The first one might correspond to an expression domain which in wildtype embryos is located at the border between the alar and basal plate at the rostrocaudal level of the border between diencephalon and secondary prosencephalon (Figures 1a–d, arrow), where bsx is expressed in close proximity to the th expressing dopaminergic diencephalic cluster 2 (DC2) cells in the “ventral posterior tuberculum” [“PTv”; Figures 2c–c′′ and (Schredelseker and Driever, 2020)]. However, this putative “PTv” domain seem to be reduced in all triple mutants analyzed, suggesting that also this domain is not fully independent of Nkx2.1/2.4a/b factors. Second, bsx expression in the TelSep region, which in mice is fused at the midline upon Nkx2.1 loss (Kimura et al., 1996), is normal in nkx2.1/2.4a/b triple mutants (Figures 1a–d, arrowhead).
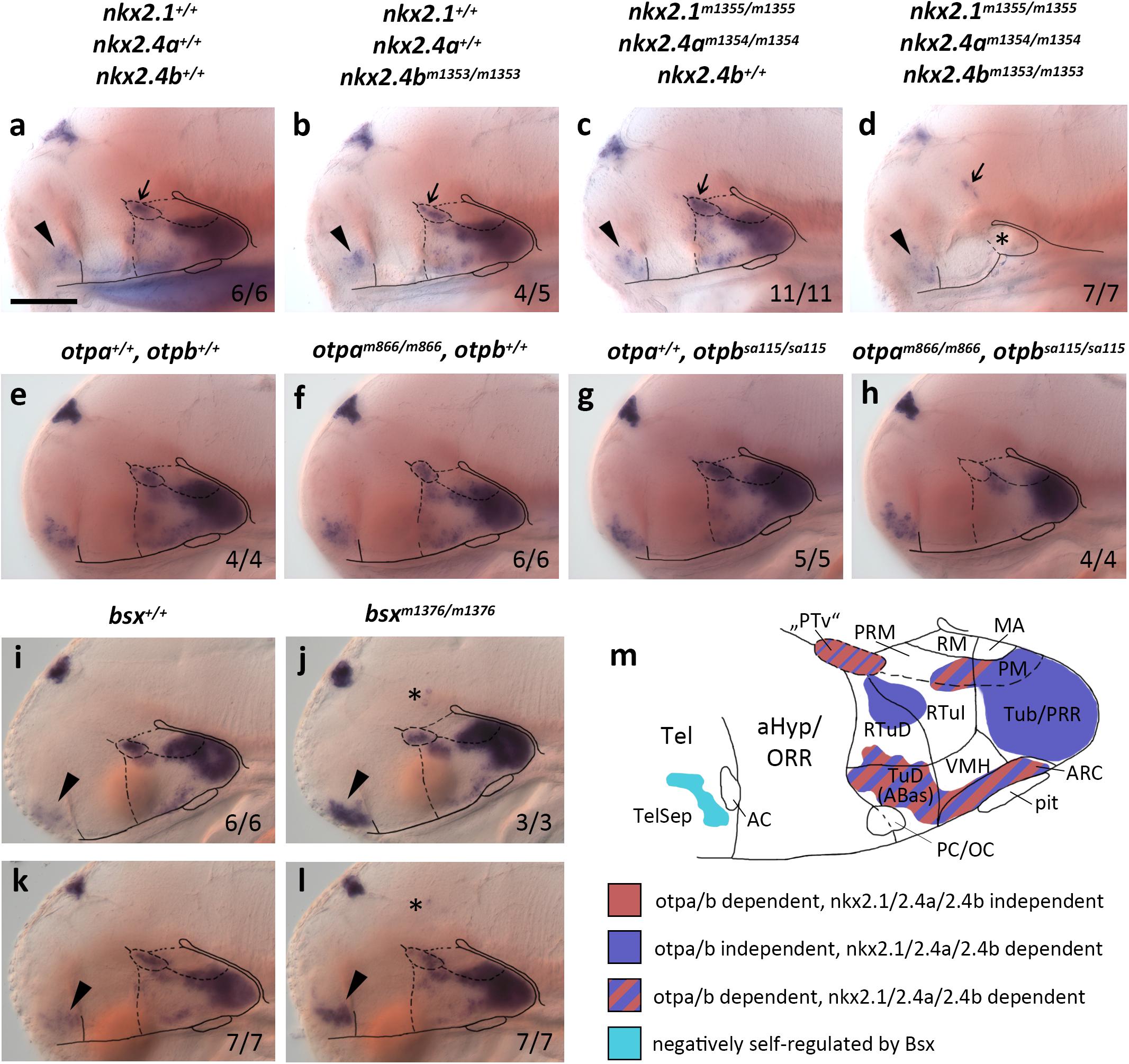
Figure 1. bsx expression depends on Nkx2.1, Nkx2.4, and Otp transcription factors. (a–l) Lateral views of wildtype and mutant (genotypes as indicated above panels) 4 dpf (a–d), 3 dpf (e–h,k,l) or 2 dpf (i,j) embryos, stained by in situ hybridization for bsx expression. All images are minimum intensity projections of 50 brightfield focal planes (distance 1 μm). Anterior is to the left. Scale bar (a–l): 100 μm. The ratio of observed phenotype and total analyzed embryos per genotype [n (phenotype)/n (total)] is indicated in right bottom corner of each panel. Arrowheads indicates bsx expression in the telencephalic septal region; small arrows indicate bsx expression which might correspond to “PTv” region; asterisk in (d) indicates tissue which might correspond to residual basal hypothalamus (bHyp) but which was not confirmed through marker gene expression in nkx2.1, nkx2.4a, nkx2.4b triple mutants; asterisks in (j,l) indicate ectopic bsx expressing cells. (m) Schematic drawing of a lateral view of bsx expressing forebrain regions of a zebrafish embryo 3–4 dpf illustrating which of the bsx expression domains depend on Otp or Nkx2.1 and Nkx2.4 transcription factors as indicated by color code. Anterior at left. For abbreviations see list.
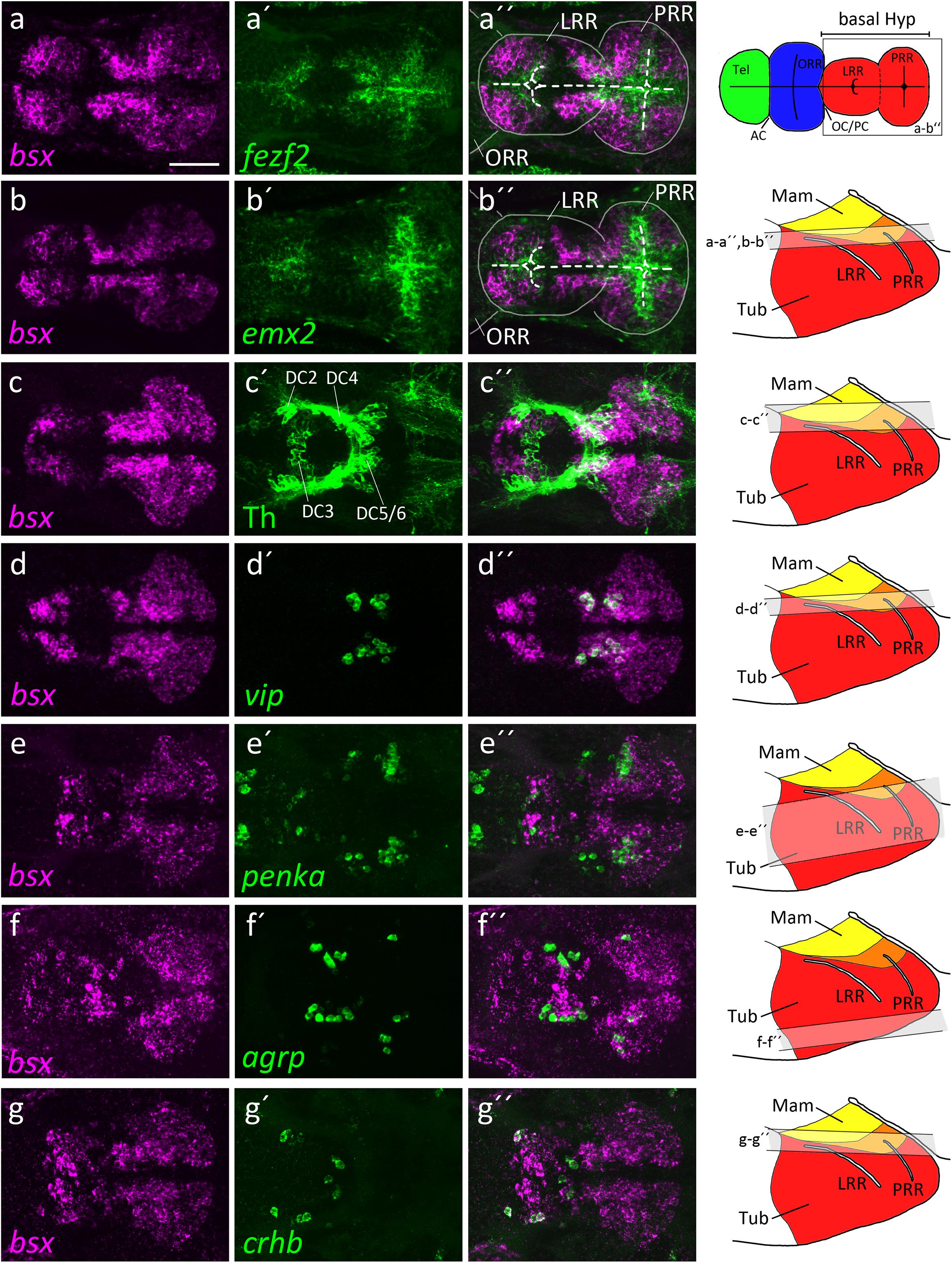
Figure 2. bsx expression within the bHyp in relation to expression of neural progenitor markers and neuropeptidergic genes. Dorsal view of confocal sections of zebrafish embryos at 2 dpf (a–b′′), 3 dpf (c–e′′) or 4 dpf (f–g′′) after double-fluorescent whole-mount in situ hybridization using probes as indicated. Maximum intensity Z-projections of 30 (a–b′′,d–d′′), 40 (f–g′′), 50 (c–c′′), or 70 (e–e′′) single confocal planes (1 μm steps) are shown. Schematics on the right show lateral view of the bHyp for 2–4 dpf embryos indicating which planes were selected for Z-projections in the panels indicated. Expression of bsx is detected further away from the ventricle [white dashed line in (a–b′′)] than expression of neuronal progenitor markers (a–b′′). bsx expression colocalizes in neurons expressing Th [anti-Th immunostain; (c–c′′)] or neuropeptidergic transcripts (d–g′′). Scale bar in (a) for all images: 50 μm. Schematics in the top right represents a model of the zebrafish forebrain highlighting the ventricular recesses. Anterior at left. For abbreviations see list.
Single nkx2.4b mutants or nkx2.1, nkx2.4a double mutants show reduced expression of bsx in the bHyp (Figures 1b,c), which is, however, not nearly as severe as in the triple mutants (Figure 1d). These data are consistent with previous studies showing partial redundancy of Nkx2.1 and Nkx2.4 transcription factors in hypothalamus patterning (Manoli and Driever, 2014). As expected, bsx expression in the pineal complex is unaffected in nkx2.1/2.4a/b triple mutants (Figures 1a–d).
We recently showed that bsx is coexpressed with orthopedia homeobox a (otpa) in mamillary regions as well as in the terminal tuberal hypothalamus (Schredelseker and Driever, 2020). The two paralogs otpa and otpb function partially redundantly in the ventral forebrain (Ryu et al., 2007; Fernandes et al., 2013). For Otp mutant mice, reduced Bsx expression has been reported only for the ARC (Lee et al., 2018). In zebrafish embryos, however, we found bsx expression to be lost in otpa/otpb double mutants not only in the ARC but also in all other regions in which bsx colocalizes with otpa (3 dpf; Figures 1e–h and Supplementary Figure S2). We concluded that in all bsx+/otpa+ regions, i.e., the “PTv”, parts of the Mam as well as the ARC and the dorsotuberal/anterobasal (TuD/ABas) region, bsx expression is strictly dependent on Otp transcription factors. To investigate whether Otpa is also sufficient to induce ectopic bsx expression, we assessed Tg(hsp70l:otpa-ires-egfp-caax) embryos, which, upon heat shock, express otpa broadly throughout the brain (Supplementary Figures S3A,B). However, bsx expression was normal in embryos 2 dpf even after three waves of otpa overexpression induced by heat shocks performed at 12, 24, and 30 hpf (Supplementary Figures S3C,D), indicating that Otpa is not sufficient to activate ectopic bsx expression.
We further assessed bsx expression in bsx mutants (Schredelseker and Driever, 2018) to investigate potential autoregulative Bsx functions. We found bsx to be expressed normally in the pineal complex and hypothalamus but to be upregulated in the TelSep area in bsx mutant embryos (2 dpf: Figures 1i,j; 3 dpf: Figures 1k,l), suggesting negative autoregulation in this area. We also found few dispersed bsx expressing cells around the di-mesencephalic boundary in all bsx mutant embryos (Figures 1j,l, asterisk) but never in wildtypes. We summarized the upstream regulation of bsx in different domains of the forebrain in Figure 1m.
bsx Is Expressed in Differentiating Neurons Rather Than in Neural Stem Cells
The hypothalamus, like many brain regions, displays a pronounced centrifugal ventricular to mantle gradient of neurogenesis and cell maturation (Affaticati et al., 2015; Alvarez-Bolado, 2019). By double-fluorescent in situ hybridization, we compared bsx expression along the apical to basal axis of the hypothalamic neuroepithelium with expression of genes characteristic for proliferating neural stem cells or postmitotic differentiated neurons. Expression of the neural stem and radial glia markers emx2 and fezf2 (Cecchi, 2002; Yang et al., 2012; Alvarez-Bolado, 2019) are restricted to areas close to the ventricle, while bsx expression was found to be located further away from the ventricle in mantle zones (Figures 2a–b′′).
We then compared bsx expression to markers of differentiated neurons. We detected bsx expression in dopaminergic DC5 and 6 cells in the mamillary hypothalamus (Figures 2c–c′′). In the mamillary hypothalamus, we also found cells expressing both bsx and vasoactive intestinal peptide (vip) (Figures 2d–d′′). The location of those vip+ cells within the otpa+ positive parts of the mamillary hypothalamus has been shown by others (Wolf and Ryu, 2013). We recently found medial regions of this brain area to correspond to mamillary hypothalamus, while more lateral regions express markers characteristic for the tuberal hypothalamus (Schredelseker and Driever, 2020). Within the tuberal hypothalamus, lateral to the vip expressing cells, as well as in the TuD/ABas region, we found cells expressing both bsx and the endogenous opioid preproprotein proenkephalin a (penka) (Figures 2e–e′′). In an adjacent region of the terminal tuberal hypothalamus, corresponding to the ARC region (Schredelseker and Driever, 2020), we also detected agrp cells within the bsx expressing territories (Figures 2f–f′′). Notably, in the border region between alar and basal plate at the rostrocaudal level of the border between hypothalamus and diencephalon, in which also the th expressing dopaminergic DC2 cluster is located, we found crhb cells to express bsx (Figures 2g–g′′). Taken together, these data suggest that bsx is expressed in mantle postmitotic differentiating neurons rather than in ventricular neural stem or progenitor cells.
Forebrain Patterning Is Normal in bsx Mutants
To investigate a potential role of Bsx in hypothalamus patterning, we compared bsx mutants with wildtype siblings for gene expression patterns which define specific progenitor domains in the embryonic hypothalamus. sonic hedgehog a (shha) expression in the secondary prosencephalon marks the alar-basal boundary [(Puelles et al., 2012); Figure 3a, red dashed line; for regional organization of the hypothalamus see Figure 1m]. Expression of shha being unaffected in bsx mutants suggests a normal course of the alar-basal boundary in bsx mutants (Figure 3a). Expression of pax6a and pax7a is unaffected in bsx mutants compared to wildtype (Figures 3b,c, arrowhead), indicating regular formation of alar and basal parts of prosomer 3, respectively. Unchanged expression of pax7a in the pituitary further indicates proper formation of the pituitary intermediate lobe (Figure 3c, arrow). pax6a in the telencephalon and in the progenitor cells lining the optic recess (Figure 3b, arrow) is also expressed identically to wildtype siblings in bsx mutants.
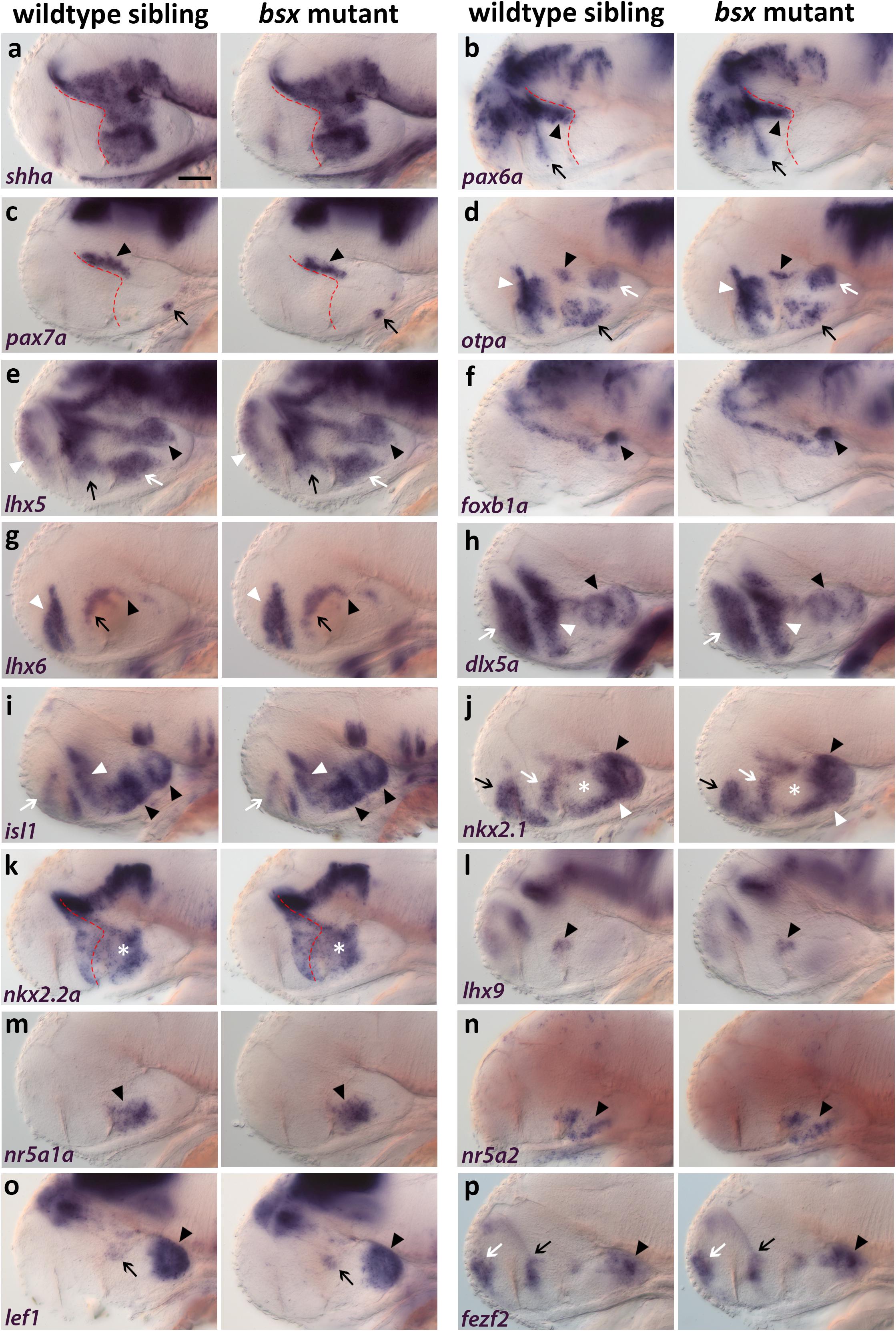
Figure 3. Forebrain patterning is normal in bsx mutants. (a–p) Lateral views of 2 dpf zebrafish embryo heads stained by in situ hybridization using probes as indicated. No differences in expression of any marker was observed between wildtype and bsx mutant embryos. All images are minimum intensity projections of 40 brightfield focal planes (distance 1 μm). Anterior is to the left. Scale bar in (a) for all images: 50 μm. Anterior at left. Number n of embryos analyzed (wildtype) = 3 (a,e,h,i); 5 (b,m); 6 (c,p); 10 (d); 9 (f,n); 14 (g); 8 (j,k,o); 12 (l). n (bsx mutants) = 3 (a); 11 (b,g); 4 (c,j); 12 (d); 5 (e,l,o); 9 (f,n,p); 8 (h,i); 10 (k); 6 (m).
otpa expression is normal in the “PTv” (Figure 3d, black arrowhead), perimamillary (PM) region (Figure 3d, white arrow), TuD/ABas area (Figure 3d, black arrow) as well as in the aHyp/ORR (Figure 3d, white arrowhead). Similarly, lhx5 expression is unchanged in mamillary regions (Figure 3e, black arrowhead), TuD/ABas (Figure 3e, white arrow), aHyp/ORR (Figure 3e, black arrow) and the telencephalon (Figure 3e, white arrowhead). Normal foxb1a expression indicates the Mam to develop properly (Figure 3f, black arrowhead). lhx6 expression is unchanged in the pallidum (Figure 3g, white arrowhead), in the aHyp/ORR (Figure 3g, black arrow), as well as in the bHyp (Figure 3g, black arrowhead) where in mouse it has been demonstrated to selectively mark RTuV/TuV (Puelles et al., 2012).
Also other tuberal regions appear to be well developed in bsx mutants, as judged from unaltered expression of dlx5a (Figure 3h, black arrowhead), which marks all tuberal regions except the core region of the ventromedial hypothalamus (VMH) (Puelles et al., 2012; Morales-Delgado et al., 2014), and of isl1 (Figure 3i, black arrowheads), which marks all tuberal regions (Puelles et al., 2012). Expression of dlx5a and isl1 is also unchanged in the telencephalon (Figures 3h,i, white arrow) and aHyp/ORR (Figures 3h,i, white arrowhead). nkx2.1 expression is normal in both mamillary and tuberal regions (Figure 3j, black and white arrowheads, respectively) as well as in the telencephalon and aHyp/ORR (Figure 3j, black and white arrow, respectively). The gap in nkx2.1 expression in the intermediate and dorsal retrotuberal (RTuI/RTuD) regions (Figure 3j, white asterisk) was observed previously (Rohr et al., 2001; Manoli and Driever, 2014), and is also unchanged in bsx mutants. In this nkx2.1 negative territory, nkx2.2a is expressed (Figure 3k, white asterisk). nkx2.2a has been described to be expressed in both the liminal and subliminal band along the alar-basal boundary (Puelles et al., 2012; Figure 3k, red dashed line), and is expressed identically to wildtype in bsx mutants. lhx9 expression indicates normal development of the RTuD/PBas region (Puelles et al., 2012; Figure 3l, black arrowhead). nr5a1a and nr5a2 are expressed in equal patterns in wildtype and bsx mutant embryos, suggesting proper formation of TuD/ABas, VMH and the ARC region (Kurrasch et al., 2007; Bedont et al., 2015; Xie and Dorsky, 2017) (Figures 3m,n, black arrowheads). The PRR also appears to develop orderly, as observed through lef1 and fezf2 expression (Levkowitz et al., 2003; Wang et al., 2012; Figures 3o,p, black arrowheads). A small expression domain of lef1 in the RTu region (Figure 3o, black arrow), which has previously been described in mice (Ferran et al., 2015), also appears to be unchanged in bsx mutants. fezf2 expression is normal along the optic recess and in the telencephalon (Figure 3p, black and white arrow, respectively). In conclusion, we could not detect any difference in patterning of the secondary prosencephalon between wildtypes and bsx mutants.
Expression of agrp and penka, but Not pomca or npy, in Terminal Regions of the Tuberal Hypothalamus Depends on Bsx
In the mouse ARC, Bsx has been shown to regulate expression of Agrp and Npy, but not Pomc (Sakkou et al., 2007). We found agrp expression in the putative ARC area to be lost in bsx mutants at 3 dpf (Figures 4a–d, arrowheads) and 4 dpf (Supplementary Figures S4A–D, arrowheads), while pomca is expressed normally at both 3 dpf (Supplementary Figure S8I) and 4 dpf (Supplementary Figure S9I), indicating that Bsx functions are conserved in zebrafish. We observed a few dispersed agrp expressing cells in peduncular RTu regions which are unaffected by loss of Bsx (Figures 4a–d). We do not know whether these cells represent a distinct cluster which remains anatomically and potentially functionally disjunct from the ARC population, whether those cells are migratory, and/or whether those cells express agrp only transiently during embryonic development. Notably, npy is not expressed in the ARC area in zebrafish larvae, and we could not detect altered npy expression in any brain region of bsx mutant embryos at 3 dpf (Supplementary Figure S8H) or 4 dpf (Supplementary Figure S9H).
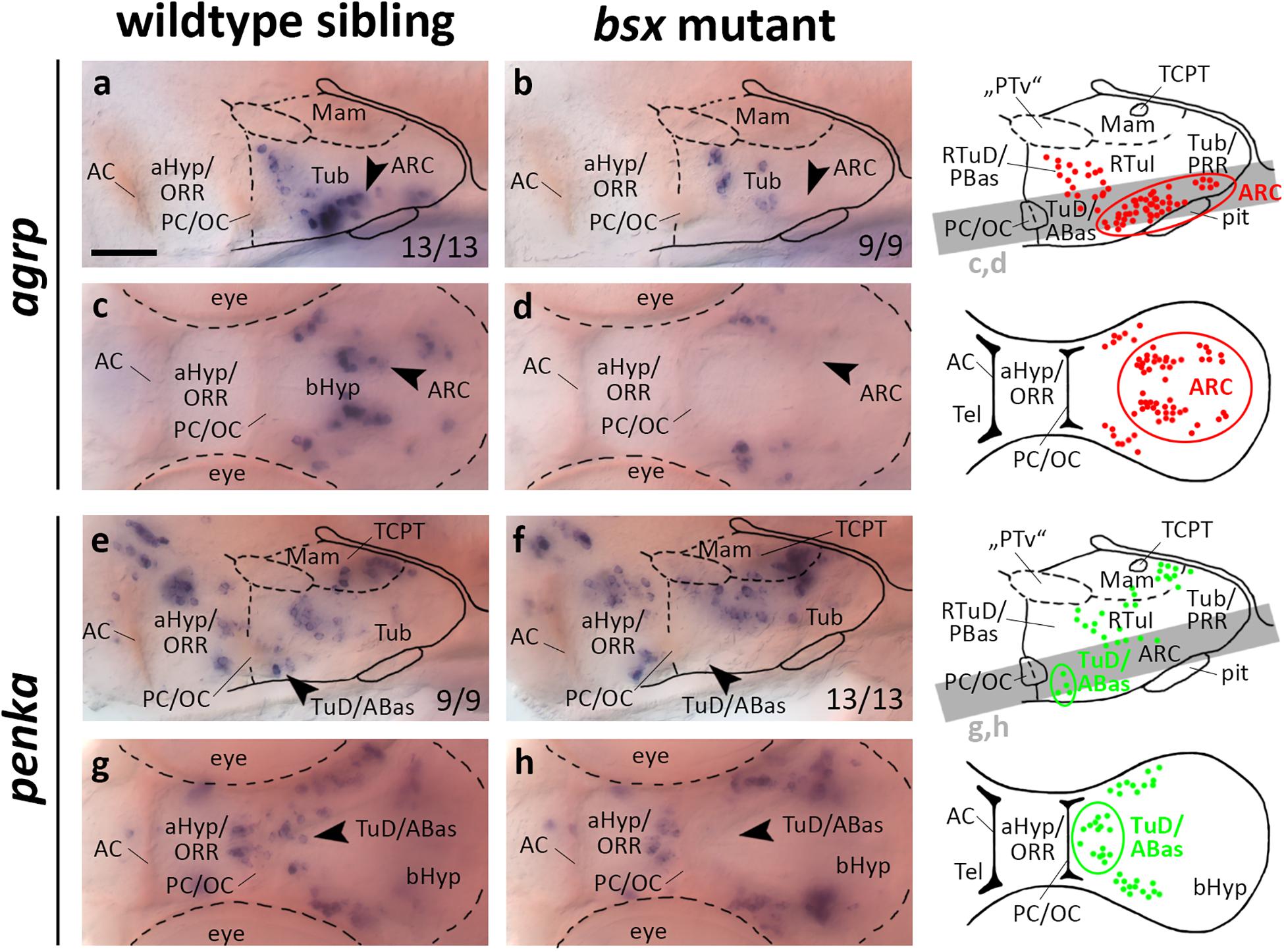
Figure 4. agrp and penka expression domains in the terminal tuberal hypothalamus are lost in bsx mutant embryos. Lateral (a,b,e,f) and dorsal views (c,d,g,h) of the ventral forebrain in 3 dpf embryos stained by in situ hybridization using probes as indicated. All images are minimum intensity projections of 40 brightfield focal planes (distance 1 μm). Scale bar in (a) for all images: 50 μm. Anterior at left. Schematics to the right show location of agrp (red) or penka (green) expressing cells as observed by stainings and indicate the focal planes which are shown in dorsal view pictures and schematics. Cells with Bsx-dependent expression are circled. Numbers n (phenotype shown)/n (total analyzed) as indicated.
Given that bsx, in mice and in teleosts, is expressed not only in the ARC, but also in multiple other regions of the bHyp (Cremona et al., 2004; Schredelseker and Driever, 2020; Figures 1, 2), we analyzed several other neuropeptidergic cell populations therein. Since we demonstrated several clusters of penka expressing neurons to be located within the bsx expression domain in both the LRR and PRR (Figures 2e–e′′), we analyzed penka expression in bsx mutant embryos. We found penka expression in one cell cluster within the LRR to be lost in bsx mutants at 3 dpf (Figures 4e–h, arrowheads) and 4 dpf (Supplementary Figures S4E–H, arrowheads). In prosomeric anatomical terms this cluster is located in the terminal TuD/ABas region (Schredelseker and Driever, 2020). We concluded that in addition to the role of Bsx on agrp neuron differentiation, which has been shown previously in mice (Sakkou et al., 2007), Bsx also functions in differentiation of penka neurons in the terminal tuberal hypothalamus in zebrafish.
Expression of vip, npb, and trh in Subregions of the Mamillary Region Is Dependent on Bsx
In mice, vip expressing cells originate in the aHyp (Díaz et al., 2014) and play a crucial role in circadian rhythm regulation (Maywood et al., 2006). In contrast, during zebrafish development vip is expressed not only in the aHyp/ORR but also in mamillary regions of the bHyp, where vip expression is dependent on Otp transcription factors (Wolf and Ryu, 2013). We found the mamillary vip cluster to be absent in bsx mutants at 3 dpf (Figures 5a–d, arrowheads) and 4 dpf (Supplementary Figures S5A–D, arrowheads). Taken together with the observation that bsx expression in the Mam is reduced in otpa/otpb double mutants (Figures 1e–h), we hypothesized that Bsx acts downstream of Otp factors in the differentiation of vip neurons.
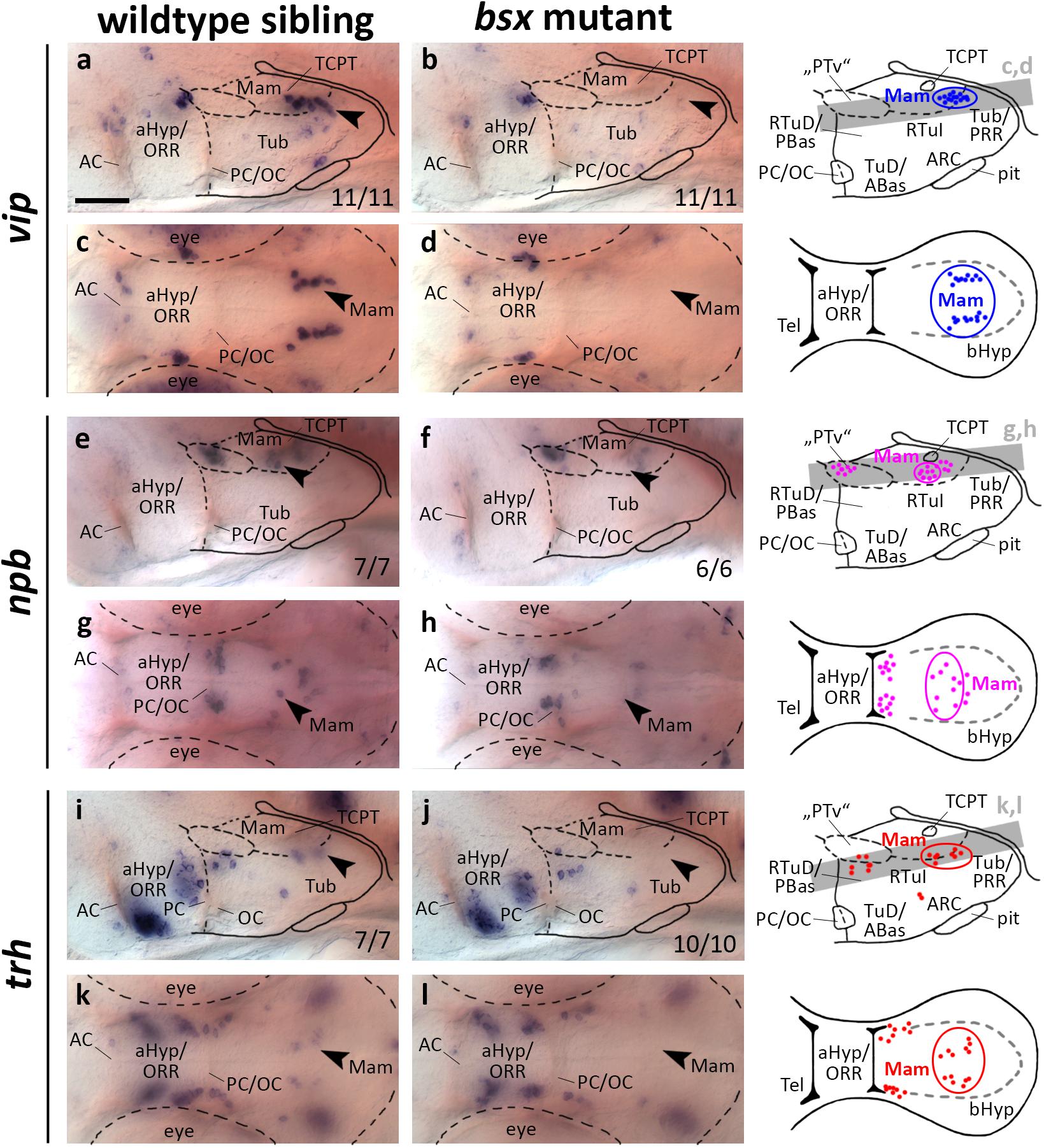
Figure 5. vip, npb, and trh expression domains in mamillary hypothalamic regions are lost in bsx mutant embryos. Lateral (a,b,e,f,i,j) and dorsal views (c,d,g,h,k,l) of the ventral forebrain in 3 dpf embryos stained by in situ hybridization using probes as indicated. All images are minimum intensity projections of 40 brightfield focal planes (distance 1 μm). Scale bar in (a) for all images: 50 μm. Anterior at left. Schematics to the right show location of vip (blue), npb (magenta), or trh (red) expressing cells as observed by stainings and indicate the focal planes which are shown in dorsal view pictures and schematics. Cells with Bsx-dependent expression are circled. Numbers n (phenotype shown)/n (total analyzed) as indicated.
In a neighboring domain within the Mam, in close proximity to the tract of the commissure of the caudal tuberculum (TCPT; Wilson et al., 1990), we found expression of npb to be strongly reduced upon Bsx loss both at 3 dpf (Figures 5e–h, arrowheads) and 4 dpf (Supplementary Figures S5E–H, arrowheads). In mouse, Npb is expressed in multiple nuclei within the telencephalon, aHyp and tuberal hypothalamus (Takenoya et al., 2013), and has been shown to be involved in pain processing, circadian rhythm, sleep/wake regulation, and feeding behavior (Dvorakova, 2018). To our knowledge, thus far, no reports exist on npb expression in mamillary regions, or on npb functions in zebrafish.
Thyrotropin-releasing hormone (Trh) neurons have been implicated in energy homeostasis, arousal and cold response (Hollenberg, 2008; Hara et al., 2009; Zhang et al., 2018). We found a cluster of trh expressing cells that is absent in the Mam of bsx mutants at 3 dpf (Figures 5i–l, arrowheads) and 4 dpf (Supplementary Figures S5I–L, arrowheads). We conclude that Bsx is essential for differentiation of neuropeptidergic cell clusters not only within the developing ARC but also in multiple other regions within the bHyp.
Expression of hcrt, cart4, sst1.1, pdyn, galn, avp, pmch, pmchl, npvf, and nmu Is Normal in bsx Mutant Embryos
Of the 16 neuropeptide encoding genes analyzed by Díaz et al. (2014), for 11 expression has been reported to originate in the bHyp in mouse. Except for growth hormone-related hormone (ghrh), which we could not detect in the zebrafish hypothalamus at 3 or 4 dpf (data not shown), and oxytocin, which in zebrafish larvae as in mice is restricted to aHyp regions (Unger and Glasgow, 2003; Díaz et al., 2014), we found zebrafish homologs for all other genes to be expressed in at least few cells within the bHyp. In addition to npy and pomca which we previously mentioned, we detected the expression of hypocretin (hcrt), cocaine- and amphetamine-regulated transcript 4 (cart4), somatostatin 1.1 (sst1.1), prodynorphin (pdyn), galanin (galn), arginine vasopressin (avp), pro-melanin-concentrating hormone (pmch), pro-melanin-concentrating hormone, like (pmchl), neuropeptide VF precursor (npvf), and neuromedin U (nmu) in bsx mutants to be similar to wildtype both at 3 dpf (Supplementary Figure S8) and 4 dpf (Supplementary Figure S9). Therefore, many neuropeptide encoding genes are normally expressed in bsx mutants, and Bsx appears to be specifically required only for expression of a defined group of neuropeptide precursor genes.
Bsx Is Required for Normal Development of the Crh System and for uts1 Expression in the Telencephalic Septal Region
In teleosts, four genes encode the corticotropin-releasing hormone (Crh) family of ligands: crha, crhb, uts1, and urocortin 3 like (Cardoso et al., 2016). Their binding to the Crh receptors is additionally regulated through presence and concentration of a Crh binding protein (Crhbp) (Ketchesin et al., 2017). The Crh system is a core component of the stress response acting through the hypothalamic-pituitary-adrenal axis (Denver, 2009), but has also been implicated in energy homeostasis and appetite regulation (Matsuda, 2013).
In the embryonic zebrafish brain crhb expressing cells in the “PTv” region have been found not only to be located in close proximity to the th expressing DC2 dopaminergic neurons, but also to be expressed under control of shared transcriptional regulation (Löhr et al., 2009). For instance, “PTv” crhb neurons also depend on the Otp transcription factors (Löhr et al., 2009). When we assessed crhb expression in the “PTv” region in bsx mutants, we found this cell cluster to be absent (Figures 6a–d, arrowheads, and Supplementary Figures S6A–D, arrowheads). Taken together with our observation that bsx expression in this region is dependent on Otp (Figures 1e–h), we conclude that Otp may contribute to differentiation of crhb neurons in the “PTv” area by activation of bsx expression, or that both factors may act in a combinatorial manner in crhb differentiation.
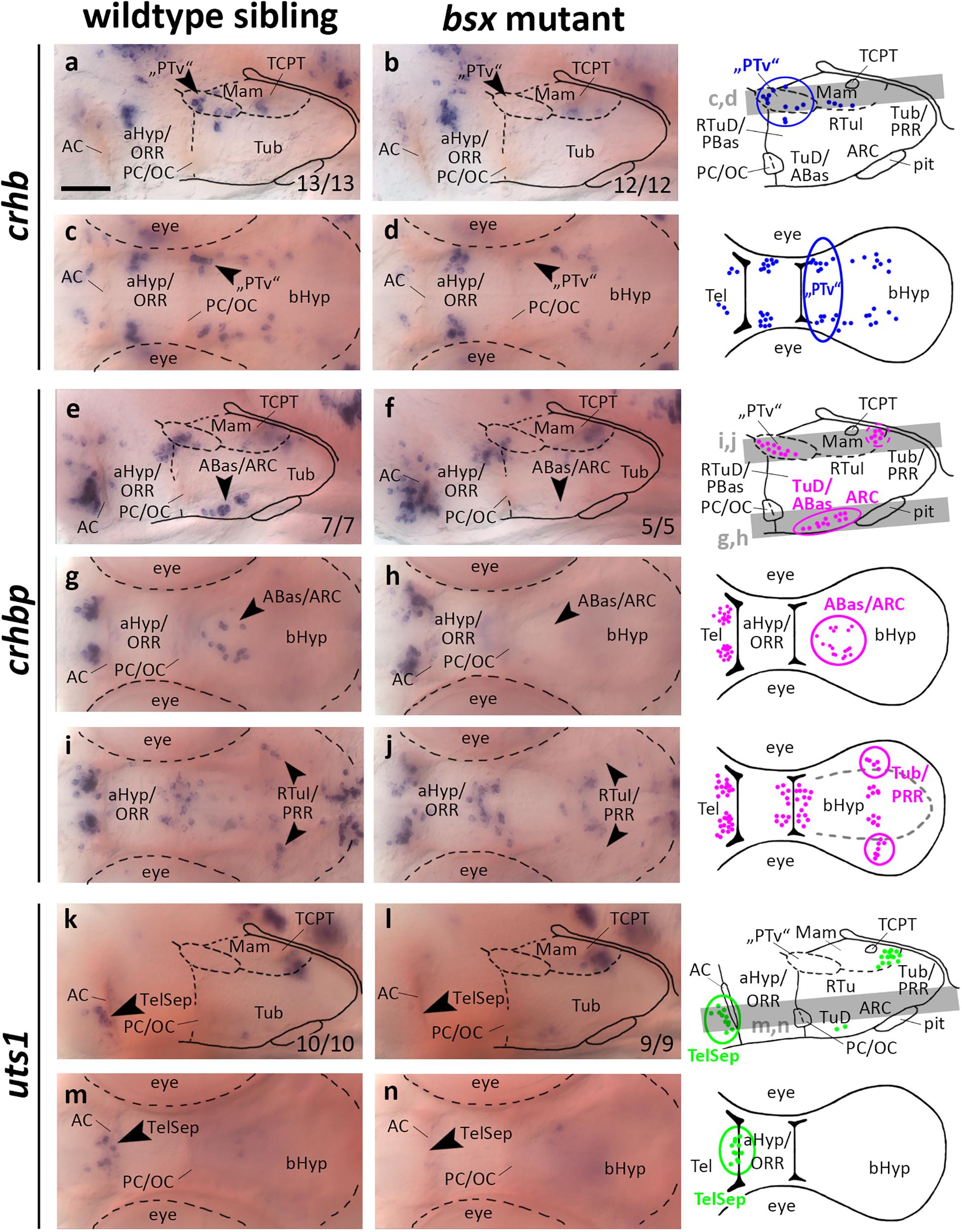
Figure 6. crhb, crhbp and uts1 expression domains in the secondary prosencephalon are lost in bsx mutant embryos. Lateral (a,b,e,f,k,l) and dorsal views (c,d,g–j,m,n) of the ventral forebrain in 3 dpf embryos stained by in situ hybridization using probes as indicated. All images are minimum intensity projections of 40 brightfield focal planes (distance 1 μm). Scale bar in (a) for all images: 50 μm. Anterior at left. Schematics to the right show location of crhb (blue), crhbp (magenta), or uts1 (green) expressing cells as observed by stainings and indicate the focal planes which are shown in dorsal view pictures and schematics. Cells with Bsx-dependent expression are circled. Numbers n (phenotype shown)/n (total analyzed) as indicated.
crhbp in the embryonic zebrafish is expressed in the telencephalon, aHyp/ORR, “PTv” area, and the border region between tuberal and mamillary regions of the bHyp (Figures 6e–j and Supplementary Figures S6E–H, arrowheads). Additionally, a cluster of crhbp expressing cells was found in the terminal tuberal hypothalamus reaching from the TuD/ABas region to the ARC (Figures 6e–h, black arrowheads). This cluster was not detected in bsx mutants (Figures 6f,h, arrowheads, and Supplementary Figures S6E–H, arrowheads), indicating a differentiation defect of those neurons upon loss of Bsx function. In the border region between mamillary and tuberal regions we found a far laterally located cluster of crhbp expressing cells to be absent in bsx mutants (Figures 6i,j, arrowheads), while more medially located crhbp cells appear to be unaffected. Based on our previous studies (Schredelseker and Driever, 2020), within the PRR we associated the Bsx-dependent lateral crhbp cells with tuberal regions and the Bsx-independent cells with mamillary regions.
urotensin 1 (uts1) expression in the zebrafish Mam has been demonstrated to be dependent on Otp transcription factors (Wolf and Ryu, 2013). A member of the mammalian homologs to Urotensin, Urocortin 1, has been detected in the lateral telencephalic septum (Kozicz et al., 1998), which is of pallidal origin in mice (Medina and Abellán, 2012). We detected a few uts1 expressing cells adjacent to the AC in an area hypothesized to be homologous to the mammalian TelSep region (Schredelseker and Driever, 2020; Figure 6k, arrowhead). We could not detect any uts1 expressing neurons in this region in bsx mutant embryos 3 dpf (Figures 6k–n, arrowheads) or 4 dpf (Supplementary Figures S6I–L, arrowheads). Since we have demonstrated the pallidum to develop normally as inferred from normal expression of nkx2.1 and lhx6 (Figure 3), we conclude that Bsx is critical for differentiation of uts1 neurons, but not for patterning of the pallidal domain.
Bsx Functions in Differentiation of Monoaminergic Cells in the Posterior Recess Region
For the PRR we recently proposed homology of ventral and dorsolateral parts to tuberal regions, and of dorsomedial parts to mamillary regions, based on gene expression data (Schredelseker and Driever, 2020). Still, the phylogenetic relationship of the PRR remains particularly elusive as most phyla lack a posterior hypothalamic recess (Vernier, 2017; Yamamoto et al., 2017). Cell types within the teleost-specific PRR are well characterized and include many monoaminergic cerebrospinal fluid contacting (CSF-c) cells (Eriksson et al., 1998; Filippi et al., 2010; Xavier et al., 2017; Xie et al., 2017).
Since bsx is expressed in the PRR, we aimed to assess the differentiation of monoaminergic cells therein. We made use of vmat2 as a marker for all monoaminergic cell types, of hdc as a specific marker for histaminergic cells, and of th as well as its paralog th2 to label dopaminergic cells. Since tryptophan hydroxylase expression is low in hypothalamic serotonergic cells (Bellipanni et al., 2002), we made use of an antibody recognizing a 5-HT/paraformaldehyde conjugate (Bosco et al., 2013). We found the expression of vmat2 to be strongly reduced in the PRR (Figures 7a–d, arrowheads, and Supplementary Figures S7A–D, arrowheads), indicating that monoaminergic cells develop abnormally in bsx mutants. The presence of hdc expressing neurons in the PRR supports our hypothesis that parts of the PRR are homologous to the RTuV/TuV territory as defined by the prosomeric model, in which RTuV/TuV has been proposed to be the only source of histaminergic neurons in the brain (Puelles et al., 2012). We detected no hdc expressing cells in bsx mutants 3 dpf (Figures 7e–h, arrowheads) and 4 dpf (Supplementary Figures S7E–H, arrowheads), and concluded that all histaminergic cells present in the brain at these embryonic stages are strictly Bsx-dependent. We found both the expression of th1 (DC7; Supplementary Figures S10A–H) and th2 (Supplementary Figures S10I–L) to be normal in the bsx mutant PRR, suggesting dopaminergic cells to develop in a Bsx-independent manner. The th expressing cell clusters DC2, 4, 5, and 6 have previously been shown to be dependent on Otp (Ryu et al., 2007). Since we found multiple Otp-dependent expression domains of neuropeptidergic genes also to be affected in bsx mutants, we were surprised to find th expression in these clusters to be unaffected by Bsx loss (Supplementary Figures S10A–H). This indicates that Otp transcription factors function independently of Bsx in dopaminergic differentiation.
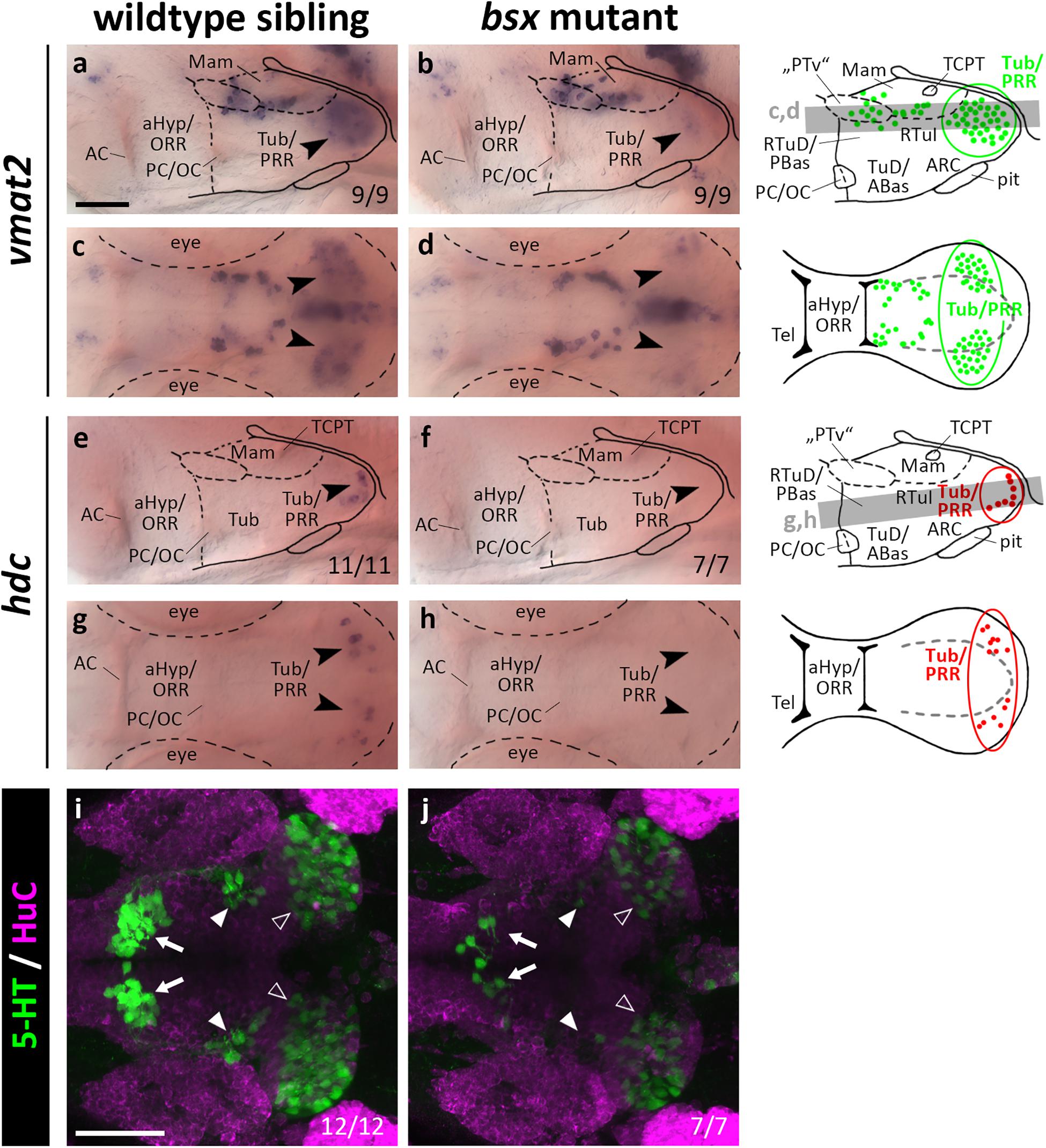
Figure 7. vmat2 expression, hdc expression and 5-HT immunoreactivity in the posterior recess region are reduced or absent in bsx mutant embryos. Lateral (a,b,e,f) and dorsal views (c,d,g,h) of the ventral forebrain in 3 dpf embryos stained by in situ hybridization using probes as indicated. (i,j) Dorsal view of the ventral forebrain in 3 dpf embryos immunostained using 5-HT (green) and HuC antibody (magenta). Images show minimum intensity projections of 40 brightfield focal planes (a–h; 1 μm steps) or maximum intensity projections of 40 confocal planes (i,j; 1 μm steps). Scale bars in (a) for (a–h) and in (i) for (i) and (j): 50 μm. Anterior at left. Schematics to the right show location of vmat2 (green) and hdc (red) expressing cells as observed in stainings and indicate the focal planes which are shown in dorsal view pictures and schematics. Cells with Bsx-dependent expression are circled. Numbers n (phenotype shown)/n (total analyzed) as indicated.
In bsx mutants, 5-HT immunoreactivity is strongly reduced not only in the PRR (Figures 7i,j, white outline arrowheads) but also in the other two hypothalamic serotonergic cell clusters (Figures 7i,j, filled white arrowheads and arrows). We conclude that Bsx is crucial for the terminal differentiation of histaminergic and serotonergic, but not dopaminergic cells in the hypothalamus.
Bsx Is Required for Normal nts and nos1 Expression in the Posterior Recess Region
Neurotensin attracted much attention as an important neuromodulator of multiple, and most notably dopaminergic, neuronal circuits (Dobner and Carraway, 2013). We detected nts expression in the PRR of wildtype but not bsx mutant embryos 3 dpf (Figures 8a–d, arrowheads). At 4 dpf we detected few nts expressing cells in the PRR of bsx mutants, while wildtype embryos at that stage showed a stronger signal (Supplementary Figures S11A–D, arrowheads).
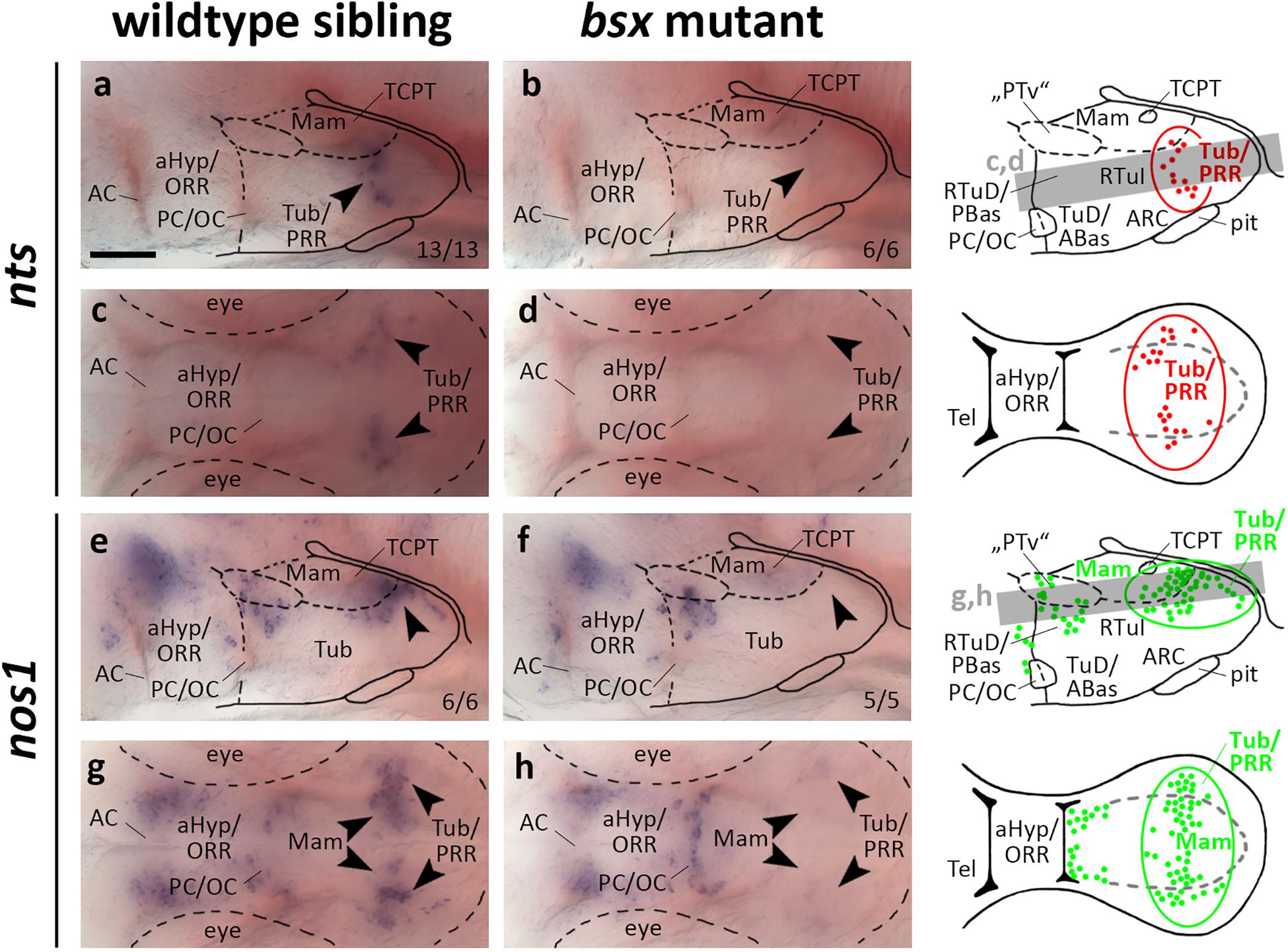
Figure 8. nts and nos1 expression in the posterior recess region is lost in bsx mutant embryos. Lateral (a,b,e,f) and dorsal views (c,d,g,h) of the ventral forebrain in 3 dpf embryos stained by in situ hybridization using probes as indicated. All images are minimum intensity projections of 40 brightfield focal planes (distance 1 μm). Scale bar in (a) for all images: 50 μm. Anterior at left. Schematics to the right show location of nts (red) or nos1 (green) expressing cells as observed in stainings and indicate the focal planes which are shown in dorsal view pictures and schematics. Cells with Bsx-dependent expression are circled. Numbers n (phenotype shown)/n (total analyzed) as indicated.
nos1 has been demonstrated to be coexpressed in th expressing neurons in Xenopus (Lopez et al., 2005) and zebrafish embryos (Poon et al., 2003). Functionally, hypothalamic nos1 has been implicated in energy homeostasis and feeding regulation as well as fertility and reproduction (Leshan et al., 2012; Sutton et al., 2014; Chachlaki et al., 2017). We detected widespread expression of nos1 in the PRR reaching from medial mamillary regions to lateral Tub regions, which, however, is absent in bsx mutants at 3 dpf (Figures 8e–h, arrowheads). At 4 dpf we found few nos1 expressing cells in the more laterally located tuberal regions of bsx mutants while the more medial cells in mamillary regions remained absent (Supplementary Figures S11E–H, arrowheads). Another nos1 expression domain, consisting of cells spread along the “PTv” and RTuD/PBas region, was unaffected by Bsx loss both at 3 dpf (Figures 8e–h, arrowheads) and 4 dpf (Supplementary Figures S11E–H, arrowheads), indicating that nos1 expression in the PRR, but not the “PTv” or RTuD/PBas region, strictly depends on Bsx. We conclude that Bsx functions in the differentiation of the nitric oxide and neurotensin neuromodulatory systems in the PRR.
Discussion
In wildtype and bsx mutant embryos, we analyzed the expression of different monoamine pathway components and multiple neuropeptidergic precursor genes, for many of which the expression in the zebrafish forebrain has not previously been described in detail. We demonstrate that Bsx is necessary not only for normal expression of a surprisingly large number of peptidergic neuromodulators, but also for the expression of enzymes involved in the synthesis of small neuromodulatory molecules, such as histamine and nitric oxide, and for normal serotonin levels in the hypothalamus (Figure 9). Even though the patterning and gross anatomical structure of the secondary prosencephalon is unchanged in bsx mutants, they do not develop the full complement of neuromodulatory molecules in the forebrain, which might have strong implications on physiology and behavior of animals devoid of functional Bsx.
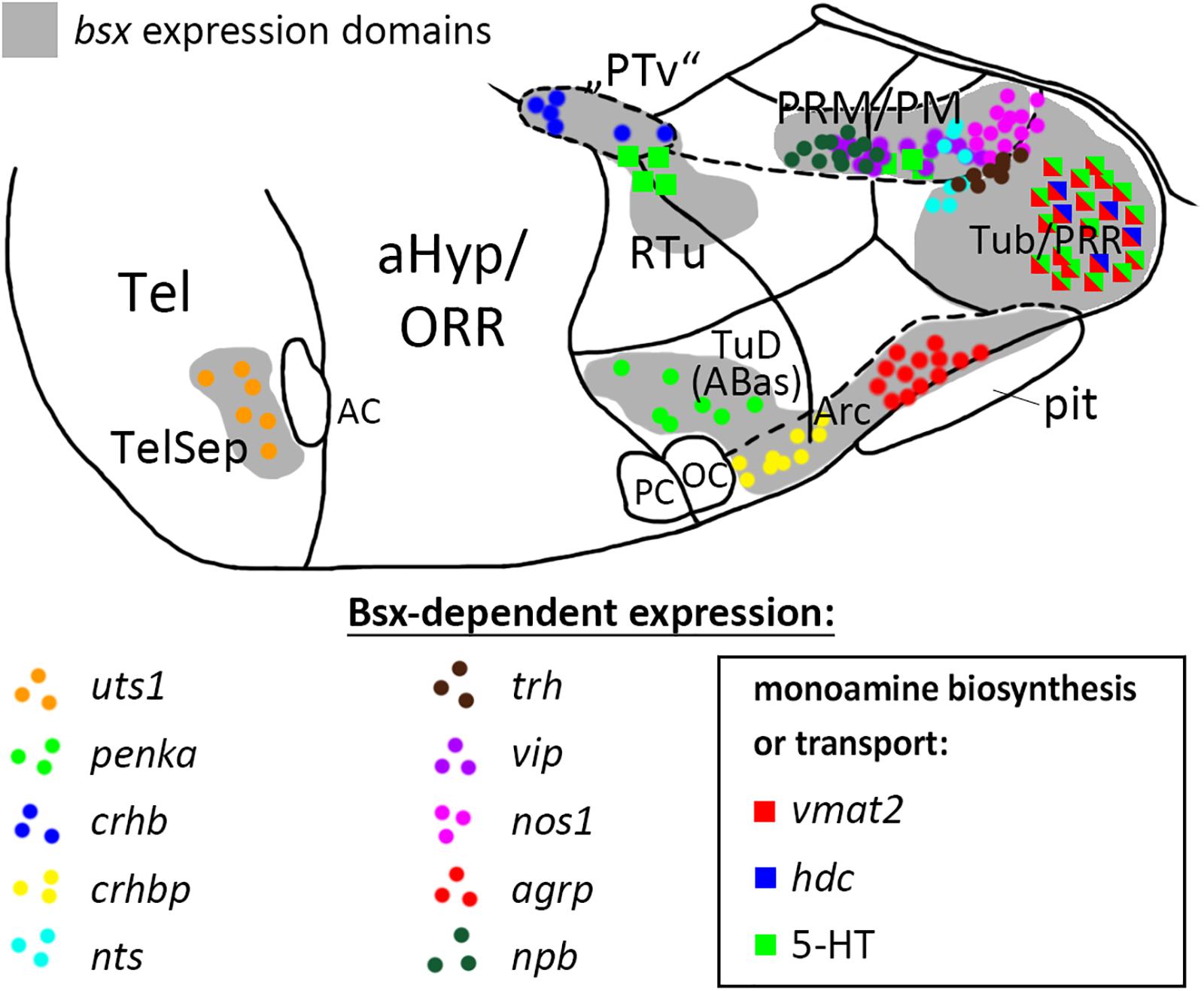
Figure 9. Distribution of Bsx-dependent neuronal cell types. Schematic representation of a sagittal view of the ventral forebrain in 3–4 dpf embryos showing bsx expression domains in gray. Only Bsx-dependent neuronal cells are shown and color coded as indicated. The number of symbols reflects the spatial extend of the affected domain but not cell numbers For abbreviations see list. Anterior at left.
Our data reveal that all bsx expression in the basal prosencephalon depends on combined activity of nkx2.1 and nkx2.4a/b genes in zebrafish. Expression of these three genes is maintained during all phases of bsx expression studied here (Manoli and Driever, 2014), thus it is possible that Nkx2.1 and 2.4 transcription factors may directly control bsx expression. However, embryos devoid of nkx2.1 and nkx2.4a/b develop severe patterning abnormalities in the hypothalamus with lack of most or all hypothalamic regions in which bsx will be expressed (Manoli and Driever, 2014). Therefore, also indirect regulation is possible. Downregulation of bsx expression in the ARC upon loss of the Otp transcription factor has recently been shown in mice (Lee et al., 2018), and our data reveal conserved Otp regulation of bsx in teleosts. It is unclear how well conserved this gene regulatory network is beyond the ARC, as for Otp knock-out mice no reports exist on Bsx expression in those areas of the mouse bHyp for which we demonstrate dependency on Otp factors in zebrafish. Other functions of Otp, in contrast, appear to be Bsx-independent, since expression of th and avp is reduced in otp mutants (Ryu et al., 2007; Fernandes et al., 2013), but not in bsx mutants.
We recently demonstrated prosomeric organization of the bHyp to be highly conserved in mammalian and teleost embryos (Schredelseker and Driever, 2020). The homology relations of hypothalamic subregions that we proposed in this model are also supported by the expression domains of many neuropeptidergic and aminergic genes that were assessed in the present study. This combined information may also help to understand additional anatomical features. For instance, the location of commissures, visible in stained embryos by mere tissue contrast, is in line with our anatomical analysis and shall be illustrated by one example. We proposed large parts of what was previously suggested to belong to the “PTv” (thus being a basal part of prosomer 3), to actually correspond to mamillary hypothalamic regions (Schredelseker and Driever, 2020). We should thus reconsider the anatomical status (and the name) of the TCPT (Wilson et al., 1990), and, based on its location, hypothesize this commissure to correspond to the retromamillary commissure in mouse (Puelles et al., 2012).
The high degree of conservation in both protein structure and expression pattern of Bsx suggests that many of the herein shown Bsx functions in neuronal differentiation might be conserved in other phyla. However, in demonstrating the loss of monoaminergic markers in the PRR, we also describe Bsx functions which might have undergone substantial evolutionary divergence. The special phylogenetic status of the PRR has been discussed previously (Vernier, 2017; Xavier et al., 2017; Schredelseker and Driever, 2020). The dependence of the monoaminergic CSF-c cells that densely populate the PRR on Bsx might help elucidating the evolutionary relationship of these neurons across vertebrates.
With respect to neuropeptides, we find conservation of agrp regulation by Bsx in zebrafish, as reported for mice (Sakkou et al., 2007). As of yet, no reports exist on Bsx functions in other subregions of the developing hypothalamus beyond the ARC. Here, we present Bsx functions in the differentiation of monoaminergic and neuropeptidergic cell types within several subregions of both, the tuberal and mamillary hypothalamus, and the telencephalic septum (summarized in Figure 9, only cell types affected in bsx mutants are shown). It should be noted that for all regions in which we found a neuronal differentiation gene not to be expressed in bsx mutants, we found other marker genes to be unaffected by Bsx loss. Moreover, for each expressed neuronal differentiation gene affected by Bsx loss in specific regions, we found expression of the same neuronal differentiation gene being not regulated by Bsx in other regions. Together, these findings support the conclusion that the differentiation program for the hypothalamus is regulated in a highly combinatorial manner, and that Bsx functions are context-dependent, rather than Bsx being a specific determinant for a defined cell type. In the case of penka, pmchl, avp, vip and npb, we observed, in some embryos, ectopic expression and/or a seemingly denser in situ hybridization staining intensity in certain Bsx-independent clusters, which, however, we did not quantify. If confirmed in future studies, this would indicate that Bsx regulates fate switches in the differentiation programs of neuromodulatory cells in the hypothalamus, such that loss of one cell type may correspond to more cells of another type. Bsx function appears to be limited to differentiation rather than patterning mechanisms, as the gross anatomy of the bHyp is unchanged in bsx mutants. Notably, Jones and McGinnis previously hypothesized in 1993 that the Drosophila bsx homolog bsh “is not required for the actual morphology of brain cells, the axonal pathways, etc., but instead regulates expression of neurotransmitters, receptors, channels, synaptic specializations, or other effector molecules that are critical to the physiologic function of particular neural cells” (Jones and McGinnis, 1993).
We were surprised by the strong effects of Bsx loss on histaminergic and serotonergic differentiation in the hypothalamus. Two different Ets domain transcription factors, Pet1 and Etv5b, specify Raphe nucleus and hypothalamic serotonergic neurons, respectively (Hendricks et al., 2003; Bosco et al., 2013). Here, we show that Bsx is an additional component of the transcriptional network that specifies serotonergic differentiation in the hypothalamus, but not the Raphe nucleus. Since in mammals serotonergic somata became restricted to the Raphe, functional characterization of the hypothalamic serotonergic system is sparse. Recent work suggests that the activity of hypothalamic serotonergic cells correlates with hunger states in zebrafish (Wee et al., 2019). The histaminergic system has been well described in zebrafish, but transcriptional regulators of its development have not been reported so far (Chen et al., 2016).
While bsx is also expressed in a subset of dopaminergic neurons in the mamillary hypothalamus, the expression of the dopaminergic marker genes th and th2 is normal in bsx mutants. DC2, 4, 5, and 6 dopaminergic cells and adjacent neuropeptidergic cell types, including crhb expressing neurons, are specified by a shared transcriptional network (Löhr et al., 2009). So far, no transcription factors specific for only one of those two lineages have been found. In demonstrating that th expression is unaffected by Bsx loss, while adjacent crhb expression is lost, we identify Bsx as a factor that separates those lineages. A model has been put forward that neurons, particularly in the hypothalamus, may frequently express more than one neurotransmitter type, including neuropeptide precursor peptides (Romanov et al., 2019). Therefore, it is tempting to speculate that Bsx may be required in dopaminergic neurons to establish the full complement of neuromodulatory factors which they might express in addition to th.
It has recently been suggested that the functional homolog of the crhb expressing neurons in the mammalian PVN, which stimulate corticotropes in the anterior pituitary, are located in the neurosecretory preoptic area (Herget et al., 2014), a well characterized region densely populated by various neuropeptidergic cell types within the aHyp/ORR. In contrast, the functions of the Bsx-dependent clusters expressing crhb, crhbp and uts1 in the “PTv,” PRR and TelSep region, respectively, are poorly understood. uts1 was the only gene for which we found TelSep expression to be Bsx-dependent, providing a first description of a Bsx function in the telencephalon. However, we also found several other genes encoding neuropeptidergic precursors to be expressed in this region, suggesting that the TelSep might act as a neuromodulatory center in zebrafish.
Even though a function in feeding regulation has been suggested for many of the analyzed neuropeptides, they are known to exert multiple functions beyond energy homeostasis, ranging from stress response behavior to the regulation of sleep and arousal. While so far phenotypes of Bsx knock-out animals, such as high body fat and reduced locomotion (Sakkou et al., 2007), were discussed only in the light of Bsx as a regulator of Agrp and Npy expression, alternative explanations should be considered given the breadth of neuropeptides affected.
While Bsx in mouse has been shown to directly bind to the promoters of the Agrp and Npy genes, we do not know whether this is the case in zebrafish for agrp, npy, or any of the other genes for which we found expression to depend on Bsx. It might well be that expression of some of the affected genes in the expected areas was not detected due to a failure of precursors or neurons to migrate or survive in bsx mutants. Such roles have recently been described for the Otp transcription factor, which shall be used as an example to illustrate the highly combinatorial action of transcription factors in hypothalamus development. In Otp knock-out mice, Trh, Crh, Avp, and Oxt and Sst expression is lost in specific subregions of the aHyp (Acampora et al., 1999). In the ARC of Otp mutant mice, Agrp, Npy and Sst expression is absent while Ghrh and Pomc expression is normal (Acampora et al., 1999; Lee et al., 2018). A careful re-examination of Otp mutant mice revealed that all Ghrh expressing cells in the bHyp which originate in alar territories, but migrate to the VMH region, are absent upon Otp loss, while the intrinsically basal Ghrh expression in the ARC is unaffected (Morales-Delgado et al., 2014). Unfortunately, we could not analyze ghrh expression in bsx mutant zebrafish since we could not detect any signal at the embryonic stages analyzed when using a ghrh probe.
Since serotonin, Vip and Trh have been implicated in lactation in mammals (Kato et al., 1978; Crowley, 2014), the defects in these systems might contribute to the nursing defects which were described for Bsx mutant female mice (McArthur and Ohtoshi, 2007). The hypothalamic nitric oxide system has been linked to ovarian cyclicity and the onset of puberty (Chachlaki et al., 2017) and, notably, BSX was identified as a locus for age at menarche in GWAS (Elks et al., 2010; Demerath et al., 2013). Moreover, our data might help to reevaluate endocrine features of patients suffering from Jacobsen syndrome, a disease in which chromosome 11 deletions which encompass the BSX locus, were implicated (Haghi et al., 2004; Coldren et al., 2009).
Materials and Methods
Zebrafish Strains, Maintenance and Heat Shock Treatment
Zebrafish embryos of the ABTL strain were obtained through natural breeding, incubated in constant darkness at 28°C, and staged according to Kimmel et al. (1995). The following transgenic or mutant lines were used: Tg(hsp70l:otpa-IRES-gfp-caax)m1178 (generated by Heiko Löhr, Driever Lab), bsxm1376 (Schredelseker and Driever, 2018), otpam866 (Ryu et al., 2007), otpbsa115 (Fernandes et al., 2012), nkx2.1m1355, nkx2.4am1354, nkx2.4bm1353 (this study). All mutant embryos were generated by incrossing heterozygous parent animals. Heat shock was performed by keeping petri dishes with zebrafish embryos in a 39°C water bath for 1 h. Heat shock transgenic embryos were identified through GFP signal by brightfield fluorescent microscopy. All experiments were carried out in accordance with the German Animal Welfare Act.
Generation of the Heat Shock-Inducible otpa Overexpressing Line
The open reading frame of the otpa gene (NCBI RefSeq NM_001128703.1) was PCR-amplified from a pCS2-otpa plasmid (Ryu et al., 2007) as a template with the following primers containing attB1/2 sites (in bold):
5′GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGC TTTCGCATGCCGACCTGCTG3′,
5′GGGGACCACTTTGTACAAGAAAGCTGGGTATTAGG TGAAGCTCATGGACACTGTG3′
(start and stop codons underlined). PCR-products were combined with a pDONR221 (Kwan et al., 2007) donor vector in a Gateway BP recombination reaction (Thermo Fisher) to generate the middle entry clone pME-otpa which was then recombined with a p5E-hsp70l and p3E-IRES-egfp-caax-pA onto a pDestTol2CG2 [for all three plasmids see Kwan et al. (2007)] in a Gateway LR reaction (Thermo Fisher). The resulting hsp70l-otpa-IRES-egfp-caax-pA-CG2 construct containing Tol2 recombination sites was injected into single-cell-staged embryos together with transposase mRNA, which was transcribed in vitro from pCS2FA-transposase (Kwan et al., 2007) using the mMessage mMachine Sp6 Transcription Kit (Thermo Fisher). The cmlc2:gfp cassette (CG2) present on the construct allowed easy screening for germline line transgenic animals through GFP signal in the heart.
Targeted Mutagenesis of nkx2.1 and nkx2.4a/b Alleles
TALEN target sites were identified for Exon 1 of nkx2.1 (NCBI Gene ID: 81883), nkx2.4a (NCBI Gene ID: 562300) or nkx2.4b (NCBI Gene ID: 58112) gene using the Mojo Hand design tool, Version 1.5 (Neff et al., 2013). The following TALEN plasmids were assembled using the Golden Gate TALEN and TAL Effector Kit 2.0 onto an RCI-Script-GoldyTALEN backbone (addgene.org), (Cermak et al., 2011): nkx2.1 left – GGAGTTTCCCAGCTGTC; nkx2.1 right – GGGCTACTGTAACGGG (binding strand: CCC GTTACAGTAGCCC); nkx2.4a left: GGAAACCTCACATCTCC; nkx2.4a right – GCAACCTCAGGTGTCCC (binding strand: GGGACACCTGAGGTTGC); nkx2.4b left – GAGAAACGGCGCCACGG; nkx2.4b right – GCTCGAA CCCGGAGCCG (binding strand: CGGCTCCGGGT TCGAGC). TALEN assembly products were verified by DNA sequencing and restriction digest with BamHI (NEB) and BsaI (NEB). Purified products were used for in vitro transcription of mRNA using the mMessage mMachine T3 Transcription Kit (Thermo Fisher Scientific). For nkx2.4a or nkx2.4b 200-300 pg of each TALEN mRNA were injected into wildtype one-cell stage embryos. Efficiency of TALEN-mediated indel generation was assayed through PCR and restriction enzyme digest on pooled injected embryos 3 dpf. Potential mutant alleles of F1 animals were sequenced and frameshift indels identified. nkx2.4am1354, nkx2.4bm1353 double heterozygous F2 animals were raised and identified through genotyping. Since nkx2.4a and nkx2.1 are both located on chromosome 17, we introduced the nkx2.1 mutation de novo in the nkx2.4am1354, nkx2.4bm1353 fish. 200–300 pg nkx2.1 TALEN mRNA was injected into one cell stage embryos from crosses of nkx2.4am1354, nkx2.4bm1353 fish crossed with ABTL fish. A new mutant nkx2.4bm1353 allele with a small deletion generating a frameshift was identified by sequencing in F2 animals. Triple heterozygous nkx2.1m1355, nkx2.4am1354, nkx2.4bm1353 animals were genotyped and crossed to generate nkx2.1, nkx2.4a, nkx2.4b triple homozygous embryos.
Genotyping
All breeding adult animals and experimental embryos were genotyped through PCR, followed by restriction digest. Fin or tail biopsies were lysed in 50 mM NaOH for 45 min at 95°C, then neutralized with 1/10 volume of 1M Tris-HCl pH 7.5 before being used as template. Primer sequences, amplicon length, restriction enzymes and length of digestion products are given in Supplementary Table S2.
Cloning of cDNA Fragments Used as Templates for in situ Hybridization
Gene fragments were PCR amplified from cDNA and cloned into pCRII-TOPO (Invitrogen) as has been described (Schredelseker and Driever, 2018) using primer sequences shown in Supplementary Table S1. Plasmids were linearized with restriction enzymes (Supplementary Table S1) and used as template for in vitro transcription of digoxigenin (DIG)-labeled probe using T3, T7, or SP6 RNA polymerase [Supplementary Table S1; Schredelseker and Driever (2018)].
In situ Hybridization and Immunohistochemistry
Gene IDs for all antisense probes generated are provided in Table 1. Whole-mount DIG-labeled RNA in situ hybridization based on alkaline phosphatase reaction was carried out as previously described (Schredelseker and Driever, 2018). Tyramide signal amplification (TSA) fluorescent in situ hybridization (FISH) was carried out as described previously (Ronneberger et al., 2012). For combined FISH/fluorescent immunohistochemistry (FIHC), after detection of a single DIG-labeled probe through TSA reaction using Alexa Fluor 488, peroxidase activity was inactivated through extensive washing in TNT [100 mM Tris-HCl pH7.5, 150 mM NaCl, 0.5% Tween20] and incubation in 1% H2O2/TNT for 30 min at room temperature (RT). Embryos were then washed in PBTD [1% DMSO/PBST (0.1% Tween20 in PBS)] and blocked for 2 h in Blocking Solution [5% goat serum (Sigma-Aldrich #G9023), 1% blocking reagent (Roche, #1096176), 1% BSA (Sigma-Aldrich #A6003) in PBTD] at RT. Embryos were incubated with primary rabbit anti 5-HT antibody (Sigma-Aldrich #S5545; 1:200), primary mouse HuC/HuD antibody (16A11, Thermo Fisher Scientific #A-21271; 1:500) or preabsorbed primary rabbit anti-Th antibody (Ryu et al., 2007; 1:500) 1:500 in Blocking Solution over night at 4°C. Embryos were washed all day in PBTD at RT, then incubated with secondary goat anti-rabbit IgG Alexa 488 or 546 antibody (Thermo Fisher Scientific, #A-11008 or #A-11035; 1:1000) and secondary goat anti-mouse Alexa 555 antibody (Thermo Fisher Scientific, #A-21425; 1:1000) in PBTD over night at 4°C. Embryos were washed several times in PBTD and PBST, then transferred to 80% glycerol/PBST and stored at 4°C until imaged. All mutant embryos came from incrosses of heterozygous parent animals and were stained in the same tube and/or wells together with their wildtype siblings, and, after the staining procedure, but before imaging, were genotyped based on tail clip DNA. Heat shock transgenic embryos, identified by GFP signal, were presorted and stained in tubes and/or wells separate from their wildtype siblings, but the staining procedure was performed in parallel, with all solutions being applied at the same time and from the same stock.
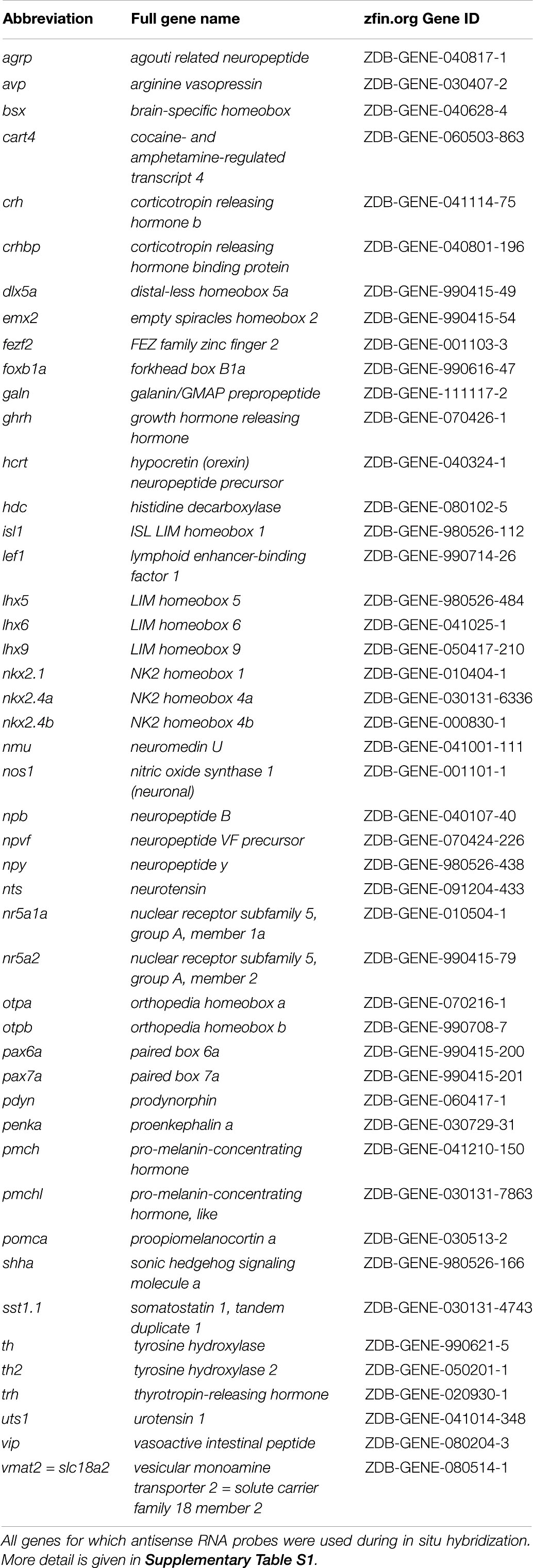
Table 1. Abbreviations, full names, and zfin.org Gene ID of genes in this study.
For all previously unpublished probes we performed in silico alignment analysis using the BLASTN discontiguous megablast algorithm on the Danio rerio nucleotide collection (taxid: 7955) with default parameters and for most probe sequences found no hits except the specific transcript which we aimed to target. There are, however, three exceptions. Our nos1 probe has some similarity to a nos2a transcript (XM_005165296.4). However, nos2a was found not to be expressed in the larval zebrafish until 5 dpf (Thisse and Thisse, 2005). For nr5a1a we cannot exclude hybridization of our probe to nr5a1b (NM_212834.1) or nr5a2 (NM_001313729.1). Since we used nr5a1a staining only to demonstrate that forebrain patterning is normal, we concluded potential hybridization to those other genes to be rather inconsequential for our statement. Our vmat2 probe also has some similarity to a vmat1 transcript (XM_021478484.1), which, however, is not expressed in the larval zebrafish brain (Puttonen et al., 2017). We also found the expression patterns generated by all in situ hybridization probe sequences cloned in this study to match the expression patterns of the same gene as has been published previously (for references see Supplementary Table S1).
Imaging and Figure Preparation
Alkaline phosphatase blue in situ hybridization stained embryos were mounted for brightfield microscopy in 80% glycerol, 20% PBST, 1 mM EDTA. Images were obtained using an AxioCam CC1 on an AxioPlan2 microscope with a PLAN-NEOFLUAR 20×/0.5 or 10×/0.3 objective and DIC optics using the AxioVs40 Software (all Zeiss). From image stacks (1 μm step size), minimum intensity projections of z-planes as indicated in the figure legends were generated using ImageJ. Embryos stained by fluorescent in situ hybridization and immunohistochemistry were recorded using a Zeiss LSM 510 or LSM 880. All figures were assembled using Adobe Photoshop CS4 or CS6. When linear adjustment of levels was made, histograms were clipped to the same values for experimental and control images.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Regierungspräsidium Freiburg, Freiburg, Germany.
Author Contributions
TS conceptualized and designed the study and performed the experiments, assembled the figures, and wrote the first draft of the manuscript. FV generated the nkx2.1 and nkx2.4a/b mutant lines. RD communicated unpublished data and suggested hypothalamic markers. WD contributed to design, supervision and editing, and provided project administration and funding acquisition.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC-2189 – Project ID: 390939984 [Gefördert durch die Deutsche Forschungsgemeinschaft (DFG) im Rahmen der Exzellenzstrategie des Bundes und der Länder – EXC-2189 – Projektnummer 390939984] and the Excellence Initiative of the German Federal and State Governments (BIOSS - EXC 294).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many thanks go to Nadja Bischoff, Daniel Armbruster, Daniela Reuter-Schmitt, and Dörte Meyer for technical help with the staining procedures or genotyping. Many thanks go to Eva Carl for cloning the fezf2 plasmid, to Jochen Holzschuh for cloning the vmat2 plasmid and to Pooja Sant for cloning the npb plasmid used for probe synthesis. Many thanks go to Heiko Löhr for generating the Tg(hsp70l:otpa-IRES-gfp-caax)m1178 line and for discussions. Many thanks go to Matthias Hammerschmidt for discussions and for providing the agrp, pomca, hcrt, nr5a2 and npy plasmids. Many thanks go to Sabine Götter for excellent fish care and to the Life Imaging Center Freiburg for technical support with confocal microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2020.00525/full#supplementary-material
Abbreviations
5-HT, 5-hydroxytryptamine/serotonin; ABas, anterobasal area; AC, anterior commissure; aHyp, alar hypothalamus; ARC, arcuate nucleus; bHyp, basal hypothalamus; CSF-c, cerebrospinal fluid-contacting; DC, diencephalic cluster; dpf, days post fertilization; GWAS, genome-wide association study; Hpf, hours post fertilization; LRR, lateral recess region; Mam, mamillary region; OC, optic commissure; ORR, optic recess region; PBas, posterobasal area; PC, posterior commissure; Pit, pituitary; PM, perimamillary domain; PRM, periretromamillary domain; PRR, posterior recess region; “PTv”, posterior tuberculum, ventral part; PVN, paraventricular nucleus; RM, retromamillary domain; RTu, retrotuberal domain; RTuD, retrotuberal domain, dorsal part; RTuI, retrotuberal domain, intermediate part; RTuV, retrotuberal domain, ventral part; TALEN, transcription activator-like effector nuclease; TCPT, tract of the commissure of the posterior tuberculum; TelSep, telencephalic septum; Tub, tuberal region; TuD, tuberal domain, dorsal part; TuI, tuberal domain, intermediate part; TuV, tuberal domain, ventral part; VMH, ventromedial hypothalamic nucleus region.
References
Acampora, D., Postiglione, M. P., Avantaggiato, V., Di Bonito, M., Vaccarino, F. M., Michaud, J., et al. (1999). Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the Orthopedia gene. Genes Dev. 13, 2787–2800. doi: 10.1101/gad.13.21.2787
Affaticati, P., Yamamoto, K., Rizzi, B., Bureau, C., Peyrieras, N., Pasqualini, C., et al. (2015). Identification of the optic recess region as a morphogenetic entity in the zebrafish forebrain. Sci. Rep. 5:8738. doi: 10.1038/srep08738
Alié, A., Devos, L., Torres-Paz, J., Prunier, L., Boulet, F., Blin, M., et al. (2018). Developmental evolution of the forebrain in cavefish, from natural variations in neuropeptides to behavior. eLife 7:e32808. doi: 10.7554/eLife.32808
Alvarez-Bolado, G. (2019). Development of neuroendocrine neurons in the mammalian hypothalamus. Cell Tissue Res. 375, 23–39. doi: 10.1007/s00441-018-2859-1
Bedont, J. L., Newman, E. A., and Blackshaw, S. (2015). Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip. Rev. Dev. Biol. 4, 445–468. doi: 10.1002/wdev.187
Bellipanni, G., Rink, E., and Bally-Cuif, L. (2002). Cloning of two tryptophan hydroxylase genes expressed in the diencephalon of the developing zebrafish brain. Mech. Dev. 119, (Suppl. 1), S215–S220. doi: 10.1016/s0925-4773(03)00119-9
Bosco, A., Bureau, C., Affaticati, P., Gaspar, P., Bally-Cuif, L., and Lillesaar, C. (2013). Development of hypothalamic serotoninergic neurons requires Fgf signalling via the ETS-domain transcription factor Etv5b. Development 140, 372–384. doi: 10.1242/dev.089094
Burbridge, S., Stewart, I., and Placzek, M. (2016). Development of the neuroendocrine hypothalamus. Compr. Physiol. 6, 623–643. doi: 10.1002/cphy.c150023
Cardoso, J. C. R., Bergqvist, C. A., Félix, R. C., and Larhammar, D. (2016). Corticotropin-releasing hormone family evolution: five ancestral genes remain in some lineages. J. Mol. Endocrinol. 57, 73–86. doi: 10.1530/JME-16-0051
Cecchi, C. (2002). Emx2: a gene responsible for cortical development, regionalization and area specification. Gene 291, 1–9. doi: 10.1016/s0378-1119(02)00623-6
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39:e82. doi: 10.1093/nar/gkr218
Chachlaki, K., Garthwaite, J., and Prevot, V. (2017). The gentle art of saying NO: how nitric oxide gets things done in the hypothalamus. Nat. Rev. Endocrinol. 13, 521–535. doi: 10.1038/nrendo.2017.69
Chen, Y.-C., Semenova, S., Rozov, S., Sundvik, M., Bonkowsky, J. L., and Panula, P. (2016). A novel developmental role for dopaminergic signaling to specify hypothalamic neurotransmitter identity. J. Biol. Chem. 291, 21880–21892. doi: 10.1074/jbc.M115.697466
Coldren, C. D., Lai, Z., Shragg, P., Rossi, E., Glidewell, S. C., Zuffardi, O., et al. (2009). Chromosomal microarray mapping suggests a role for BSX and Neurogranin in neurocognitive and behavioral defects in the 11q terminal deletion disorder (Jacobsen syndrome). Neurogenetics 10, 89–95. doi: 10.1007/s10048-008-0157-x
Cremona, M., Colombo, E., Andreazzoli, M., Cossu, G., and Broccoli, V. (2004). Bsx, an evolutionary conserved Brain Specific homeoboX gene expressed in the septum, epiphysis, mammillary bodies and arcuate nucleus. Gene Expr. Patterns 4, 47–51. doi: 10.1016/s1567-133x(03)00151-0
Crowley, W. R. (2014). Neuroendocrine regulation of lactation and milk production. Compr. Physiol. 5, 255–291. doi: 10.1002/cphy.c140029
D’Autilia, S., Broccoli, V., Barsacchi, G., and Andreazzoli, M. (2010). Xenopus Bsx links daily cell cycle rhythms and pineal photoreceptor fate. Proc. Natl. Acad. Sci. U.S.A. 107, 6352–6357. doi: 10.1073/pnas.1000854107
Demerath, E. W., Liu, C.-T., Franceschini, N., Chen, G., Palmer, J. R., Smith, E. N., et al. (2013). Genome-wide association study of age at menarche in African-American women. Hum. Mol. Genet. 22, 3329–3346. doi: 10.1093/hmg/ddt181
Denver, R. J. (2009). Structural and functional evolution of vertebrate neuroendocrine stress systems. Ann. N. Y. Acad. Sci. 1163, 1–16. doi: 10.1111/j.1749-6632.2009.04433.x
Díaz, C., Morales-Delgado, N., and Puelles, L. (2014). Ontogenesis of peptidergic neurons within the genoarchitectonic map of the mouse hypothalamus. Front. Neuroanat. 8:162. doi: 10.3389/fnana.2014.00162
Dobner, P. R., and Carraway, R. E. (2013). “Chapter 117 - Neurotensin/Neuromedin N,” in Handbook of Biologically Active Peptides, 2nd Edn, ed. A. J. Kastin, (Boston, MA: Academic Press), 875–882.
Dominguez, L., Gonzalez, A., and Moreno, N. (2015). Patterns of hypothalamic regionalization in amphibians and reptiles: common traits revealed by a genoarchitectonic approach. Front. Neuroanat. 9:3. doi: 10.3389/fnana.2015.00003
Dvorakova, C. M. (2018). Distribution and function of neuropeptides W/B signaling system. Front Physiol 9:981. doi: 10.3389/fphys.2018.00981
Eaton, J. L., and Glasgow, E. (2007). Zebrafish orthopedia (otp) is required for isotocin cell development. Dev. Genes Evol. 217, 149–158. doi: 10.1007/s00427-006-0123-2
Elks, C. E., Perry, J. R. B., Sulem, P., Chasman, D. I., Franceschini, N., He, C., et al. (2010). Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 42, 1077–1085. doi: 10.1038/ng.714
Eriksson, K. S., Peitsaro, N., Karlstedt, K., Kaslin, J., and Panula, P. (1998). Development of the histaminergic neurons and expression of histidine decarboxylase mRNA in the zebrafish brain in the absence of all peripheral histaminergic systems. Eur. J. Neurosci. 10, 3799–3812. doi: 10.1046/j.1460-9568.1998.00394.x
Fernandes, A. M., Beddows, E., Filippi, A., and Driever, W. (2013). Orthopedia transcription factor otpa and otpb paralogous genes function during dopaminergic and neuroendocrine cell specification in larval Zebrafish. PLoS One 8:e75002. doi: 10.1371/journal.pone.0075002
Fernandes, A. M., Fero, K., Arrenberg, A. B., Bergeron, S. A., Driever, W., and Burgess, H. A. (2012). Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr. Biol. 22, 2042–2047. doi: 10.1016/j.cub.2012.08.016
Ferran, J. L., Puelles, L., and Rubenstein, J. L. (2015). Molecular codes defining rostrocaudal domains in the embryonic mouse hypothalamus. Front. Neuroanat. 9:46. doi: 10.3389/fnana.2015.00046
Filippi, A., Mahler, J., Schweitzer, J., and Driever, W. (2010). Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J. Comp. Neurol. 518, 423–438. doi: 10.1002/cne.22213
Haghi, M., Dewan, A., Jones, K. L., Reitz, R., Jones, C., and Grossfeld, P. (2004). Endocrine abnormalities in patients with Jacobsen (11q-) syndrome. Am. J. Med. Genet. Part A 129A, 62–63. doi: 10.1002/ajmg.a.30248
Hara, J., Gerashchenko, D., Wisor, J. P., Sakurai, T., Xie, X., and Kilduff, T. S. (2009). Thyrotropin-releasing hormone increases behavioral arousal through modulation of hypocretin/orexin neurons. J. Neurosci. 29, 3705–3714. doi: 10.1523/JNEUROSCI.0431-09.2009
Hendricks, T. J., Fyodorov, D. V., Wegman, L. J., Lelutiu, N. B., Pehek, E. A., Yamamoto, B., et al. (2003). Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron 37, 233–247. doi: 10.1016/s0896-6273(02)01167-4
Herget, U., Wolf, A., Wullimann, M. F., and Ryu, S. (2014). Molecular neuroanatomy and chemoarchitecture of the neurosecretory preoptic-hypothalamic area in zebrafish larvae. J. Comp. Neurol. 522, 1542–1564. doi: 10.1002/cne.23480
Hollenberg, A. N. (2008). The role of the thyrotropin-releasing hormone (TRH) neuron as a metabolic sensor. Thyroid 18, 131–139. doi: 10.1089/thy.2007.0251
Jones, B., and McGinnis, W. (1993). A new Drosophila homeobox gene, bsh, is expressed in a subset of brain cells during embryogenesis. Development 117, 793–806.
Kato, Y., Iwasaki, Y., Iwasaki, J., Abe, H., Yanajhara, N., and Imura, H. (1978). Prolactin release by vasoactive intestinal polypeptide in rats. Endocrinology 103, 554–558. doi: 10.1210/endo-103-2-554
Ketchesin, K. D., Stinnett, G. S., and Seasholtz, A. F. (2017). Corticotropin-releasing hormone-binding protein and stress: from invertebrates to humans. Stress 20, 449–464. doi: 10.1080/10253890.2017.1322575
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kimura, S., Hara, Y., Pineau, T., Fernandez-Salguero, P., Fox, C. H., Ward, J. M., et al. (1996). The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69.
Kozicz, T., Yanaihara, H., and Arimura, A. (1998). Distribution of urocortin-like immunoreactivity in the central nervous system of the rat. J. Comp. Neurol. 391, 1–10. doi: 10.1002/(sici)1096-9861(19980202)391:1<1::aid-cne1>3.0.co;2-6
Kurrasch, D. M., Cheung, C. C., Lee, F. Y., Tran, P. V., Hata, K., and Ingraham, H. A. (2007). The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J. Neurosci. 27, 13624–13634. doi: 10.1523/JNEUROSCI.2858-07.2007
Kwan, K. M., Fujimoto, E., Grabher, C., Mangum, B. D., Hardy, M. E., Campbell, D. S., et al. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099. doi: 10.1002/dvdy.21343
Lee, B., Kim, J., An, T., Kim, S., Patel, E. M., Raber, J., et al. (2018). Dlx1/2 and Otp coordinate the production of hypothalamic GHRH- and AgRP-neurons. Nat. Commun. 9:2026. doi: 10.1038/s41467-018-04377-4
Lee, B., Kim, S.-G., Kim, J., Choi, K. Y., Lee, S., Lee, S.-K., et al. (2013). Brain-specific homeobox factor as a target selector for glucocorticoid receptor in energy balance. Mol. Cell. Biol. 33, 2650–2658. doi: 10.1128/MCB.00094-13
Lee, S.-H., and Dan, Y. (2012). Neuromodulation of Brain States. Neuron 76, 209–222. doi: 10.1016/j.neuron.2012.09.012
Leshan, R. L., Greenwald-Yarnell, M., Patterson, C. M., Gonzalez, I. E., and Myers, M. G. Jr. (2012). Leptin action through hypothalamic nitric oxide synthase-1-expressing neurons controls energy balance. Nat. Med. 18, 820–823. doi: 10.1038/nm.2724
Levkowitz, G., Zeller, J., Sirotkin, H. I., French, D., Schilbach, S., Hashimoto, H., et al. (2003). Zinc finger protein too few controls the development of monoaminergic neurons. Nat. Neurosci. 6, 28–33. doi: 10.1038/nn979
Löhr, H., and Hammerschmidt, M. (2011). Zebrafish in endocrine systems: recent advances and implications for human disease. Annu. Rev. Physiol. 73, 183–211. doi: 10.1146/annurev-physiol-012110-142320
Löhr, H., Ryu, S., and Driever, W. (2009). Zebrafish diencephalic A11-related dopaminergic neurons share a conserved transcriptional network with neuroendocrine cell lineages. Development 136, 1007–1017. doi: 10.1242/dev.033878
Lopez, J. M., Moreno, N., Morona, R., Munoz, M., and Gonzalez, A. (2005). Colocalization of nitric oxide synthase and monoamines in neurons of the amphibian brain. Brain Res. Bull. 66, 555–559. doi: 10.1016/j.brainresbull.2005.05.008
Machluf, Y., Gutnick, A., and Levkowitz, G. (2011). Development of the zebrafish hypothalamus. Ann. N. Y. Acad. Sci. 1220, 93–105. doi: 10.1111/j.1749-6632.2010.05945.x
Mano, H., Asaoka, Y., Kojima, D., and Fukada, Y. (2019). Brain-specific homeobox Bsx specifies identity of pineal gland between serially homologous photoreceptive organs in zebrafish. Commun. Biol. 2:364. doi: 10.1038/s42003-019-0613-1
Manoli, M., and Driever, W. (2014). nkx2.1 and nkx2.4 genes function partially redundant during development of the zebrafish hypothalamus, preoptic region, and pallidum. Front. Neuroanat. 8:145. doi: 10.3389/fnana.2014.00145
Matsuda, K. (2013). Regulation of feeding behavior and psychomotor activity by corticotropin-releasing hormone (CRH) in fish. Front Neurosci 7:91. doi: 10.3389/fnins.2013.00091
Maywood, E. S., Reddy, A. B., Wong, G. K., O’Neill, J. S., O’Brien, J. A., McMahon, D. G., et al. (2006). Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605. doi: 10.1016/j.cub.2006.02.023
McArthur, T., and Ohtoshi, A. (2007). A brain-specific homeobox gene, Bsx, is essential for proper postnatal growth and nursing. Mol. Cell. Biol. 27, 5120–5127. doi: 10.1128/MCB.00215-07
Medina, L., and Abellán, A. (2012). “Chapter 7 - Subpallial Structures,” in The Mouse Nervous System, eds C. Watson, G. Paxinos, and L. Puelles, (San Diego, CA: Academic Press), 173–220.
Morales-Delgado, N., Castro-Robles, B., Ferrán, J. L., Martinez-de-la-Torre, M., Puelles, L., and Díaz, C. (2014). Regionalized differentiation of CRH, TRH, and GHRH peptidergic neurons in the mouse hypothalamus. Brain Struct. Funct. 219, 1083–1111. doi: 10.1007/s00429-013-0554-2
Nadim, F., and Bucher, D. (2014). Neuromodulation of neurons and synapses. Curr. Opin. Neurobiol. 29, 48–56. doi: 10.1016/j.conb.2014.05.003
Neff, K. L., Argue, D. P., Ma, A. C., Lee, H. B., Clark, K. J., and Ekker, S. C. (2013). Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics 14:1. doi: 10.1186/1471-2105-14-1
Nogueiras, R., López, M., Lage, R., Perez-Tilve, D., Pfluger, P., Mendieta-Zerón, H., et al. (2008). Bsx, a novel hypothalamic factor linking feeding with locomotor activity, is regulated by energy availability. Endocrinology 149, 3009–3015. doi: 10.1210/en.2007-1684
Poon, K. L., Richardson, M., Lam, C. S., Khoo, H. E., and Korzh, V. (2003). Expression pattern of neuronal nitric oxide synthase in embryonic zebrafish. Gene Expr. Patterns 3, 463–466. doi: 10.1016/s1567-133x(03)00063-2
Puelles, L., Martinez-de-la-Torre, M., Bardet, S., and Rubenstein, J. L. R. (2012). “Chapter 8 - Hypothalamus,” in The Mouse Nervous System, eds C. Watson, G. Paxinos, and L. Puelles, (San Diego, CA: Academic Press), 221–312.
Puelles, L., and Rubenstein, J. L. (2015). A new scenario of hypothalamic organization: rationale of new hypotheses introduced in the updated prosomeric model. Front. Neuroanat. 9:27. doi: 10.3389/fnana.2015.00027
Puttonen, H. A. J., Semenova, S., Sundvik, M., and Panula, P. (2017). Storage of neural histamine and histaminergic neurotransmission is VMAT2 dependent in the zebrafish. Sci. Rep. 7:3060. doi: 10.1038/s41598-017-02981-w
Rohr, K. B., Barth, K. A., Varga, Z. M., and Wilson, S. W. (2001). The nodal pathway acts upstream of hedgehog signaling to specify ventral telencephalic identity. Neuron 29, 341–351. doi: 10.1016/s0896-6273(01)00210-0
Romanov, R. A., Alpár, A., Hökfelt, T., and Harkany, T. (2019). Unified classification of molecular, network, and endocrine features of hypothalamic neurons. Annu. Rev. Neurosci. 9, 1–26. doi: 10.1146/annurev-neuro-070918-050414
Ronneberger, O., Liu, K., Rath, M., Rueβ, D., Mueller, T., Skibbe, H., et al. (2012). ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat. Methods 9, 735–742. doi: 10.1038/nmeth.2076
Russo, A. F. (2017). Overview of neuropeptides: Awakening the senses? Headache 57, 37–46. doi: 10.1111/head.13084
Ryu, S., Mahler, J., Acampora, D., Holzschuh, J., Erhardt, S., Omodei, D., et al. (2007). Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr. Biol. 17, 873–880. doi: 10.1016/j.cub.2007.04.003
Sakkou, M., Wiedmer, P., Anlag, K., Hamm, A., Seuntjens, E., Ettwiller, L., et al. (2007). A role for brain-specific homeobox factor Bsx in the control of hyperphagia and locomotory behavior. Cell Metab. 5, 450–463. doi: 10.1016/j.cmet.2007.05.007
Santos-Duran, G. N., Menuet, A., Lagadec, R., Mayeur, H., Ferreiro-Galve, S., Mazan, S., et al. (2015). Prosomeric organization of the hypothalamus in an elasmobranch, the catshark Scyliorhinus canicula. Front. Neuroanat. 9:37. doi: 10.3389/fnana.2015.00037
Schredelseker, T., and Driever, W. (2018). Bsx controls pineal complex development. Development 145:dev163477. doi: 10.1242/dev.163477
Schredelseker, T., and Driever, W. (2020). Conserved genoarchitecture of the basal hypothalamus in Zebrafish embryos. Front. Neuroanat. 14:3. doi: 10.3389/fnana.2020.00003
Suga, H. (2019). Application of pluripotent stem cells for treatment of human neuroendocrine disorders. Cell Tissue Res. 375, 267–278. doi: 10.1007/s00441-018-2880-4
Sutton, A. K., Pei, H., Burnett, K. H., Myers, M. G. Jr., Rhodes, C. J., and Olson, D. P. (2014). Control of food intake and energy expenditure by Nos1 neurons of the paraventricular hypothalamus. J. Neurosci. 34, 15306–15318. doi: 10.1523/JNEUROSCI.0226-14.2014
Takenoya, F., Kageyama, H., Kang, K. S., and Shioda, S. (2013). “Chapter 115 - Neuropeptide B/W,” in Handbook of Biologically Active Peptides, Second Edn, ed. A. J. Kastin, (Boston, MA: Academic Press), 863–868.
Thisse, C., and Thisse, B. (2005). High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. Available online at at: https://zfin.org/ZDB-PUB-051025-1 (accessed May 12, 2020).
Unger, J. L., and Glasgow, E. (2003). Expression of isotocin-neurophysin mRNA in developing zebrafish. Gene Expr. Patterns 3, 105–108. doi: 10.1016/s1567-133x(02)00064-9
Vernier, P. (2017). “1.04 - The brains of teleost fishes,” in Evolution of Nervous Systems (Second Edition), ed. J. H. Kaas, (Oxford: Academic Press), 59–75.
Wang, X., Kopinke, D., Lin, J., McPherson, A. D., Duncan, R. N., Otsuna, H., et al. (2012). Wnt signaling regulates postembryonic hypothalamic progenitor differentiation. Dev. Cell 23, 624–636. doi: 10.1016/j.devcel.2012.07.012
Wang, Y., Wang, M., Yin, S., Jang, R., Wang, J., Xue, Z., et al. (2015). NeuroPep: a comprehensive resource of neuropeptides. Database 2015:bav038. doi: 10.1093/database/bav038
Wee, C. L., Song, E. Y., Johnson, R. E., Ailani, D., Randlett, O., Kim, J.-Y., et al. (2019). A bidirectional network for appetite control in larval zebrafish. eLife 8:e43775. doi: 10.7554/eLife.43775
Wilson, S. W., Ross, L. S., Parrett, T., and Easter, S. S. (1990). The development of a simple scaffold of axon tracts in the brain of the embryonic zebrafish, Brachydanio rerio. Development 108, 121–145.
Wolf, A., and Ryu, S. (2013). Specification of posterior hypothalamic neurons requires coordinated activities of Fezf2, Otp, Sim1a and Foxb1.2. Development 140, 1762–1773. doi: 10.1242/dev.085357
Xavier, A. L., Fontaine, R., Bloch, S., Affaticati, P., Jenett, A., Demarque, M., et al. (2017). Comparative analysis of monoaminergic cerebrospinal fluid-contacting cells in Osteichthyes (bony vertebrates). J. Comp. Neurol. 525, 2265–2283. doi: 10.1002/cne.24204
Xie, Y., and Dorsky, R. I. (2017). Development of the hypothalamus: conservation, modification and innovation. Development 144, 1588–1599. doi: 10.1242/dev.139055
Xie, Y., Kaufmann, D., Moulton, M. J., Panahi, S., Gaynes, J. A., Watters, H. N., et al. (2017). Lef1-dependent hypothalamic neurogenesis inhibits anxiety. PLoS Biol. 15:e2002257. doi: 10.1371/journal.pbio.2002257
Yamamoto, K., Bloch, S., and Vernier, P. (2017). New perspective on the regionalization of the anterior forebrain in Osteichthyes. Dev. Growth Differ. 59, 175–187. doi: 10.1111/dgd.12348
Yang, N., Dong, Z., and Guo, S. (2012). Fezf2 regulates multilineage neuronal differentiation through activating basic helix-loop-helix and homeodomain genes in the zebrafish ventral forebrain. J. Neurosci. 32, 10940–10948. doi: 10.1523/JNEUROSCI.2216-12.2012
Keywords: neuromodulators, evolutionary conservation, brain-specific homeobox (bsx), zebrafish, basal hypothalamus, neuropeptides, aminergic neurons, nitrergic neurons
Citation: Schredelseker T, Veit F, Dorsky RI and Driever W (2020) Bsx Is Essential for Differentiation of Multiple Neuromodulatory Cell Populations in the Secondary Prosencephalon. Front. Neurosci. 14:525. doi: 10.3389/fnins.2020.00525
Received: 29 November 2019; Accepted: 28 April 2020;
Published: 03 June 2020.
Edited by:
Jackson Cioni Bittencourt, University of São Paulo, BrazilReviewed by:
David Deitcher, Cornell University, United StatesGirardet Clémence, Sorbonne Université, France
Alex C. Manhães, Universidade Federal do Estado do Rio de Janeiro, Brazil
Copyright © 2020 Schredelseker, Veit, Dorsky and Driever. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Theresa Schredelseker, c2NocmVkZXRAdGNkLmll; Wolfgang Driever, ZHJpZXZlckBiaW9sb2dpZS51bmktZnJlaWJ1cmcuZGU=
 Theresa Schredelseker
Theresa Schredelseker Florian Veit
Florian Veit Richard I. Dorsky
Richard I. Dorsky Wolfgang Driever
Wolfgang Driever