- 1Department of Emergency Medicine, Shengjing Hospital of China Medical University, Shenyang, China
- 2Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, China
Motor neuron disease (MND) is a kind of neurodegenerative disease that selectively invades spinal cord anterior horn cells, brainstem motor neurons, cortical pyramidal cells and the pyramidal tract. The main clinical features are the symptoms and signs of impaired upper and lower motor neurons, manifested as muscle weakness, atrophy and pyramidal tract signs. Histopathology has shown the disappearance of pyramidal cells in the motor cortex, loss of motor neurons in the anterior horn of the spinal cord and brainstem, and degeneration of the corticospinal tract. Due to the lack of effective treatment methods, the prognosis is generally poor, so it is of great significance to confirm the diagnosis early by various means. However, the current diagnosis of MND mainly relies on the combination of clinical manifestations and neurophysiological examinations, lacking effective means of early diagnosis. Circulating microRNA (CmiRNA) is a kind of stable miRNA molecule in serum, plasma and other body fluids, which has the characteristics of distinct differential expression, sensitive detection and convenient sample collection. As a possible new biomarker of MND, CmiRNA can not only reveal the pathophysiological process of MND, but also monitor disease progression and response to drug therapy. With the development of miRNA detection technology, more and more CmiRNAs as biomarkers with potential diagnostic value have been investigated. In this review, we explored the possibility of circulating samples as different sources of biomarkers for the diagnosis of MND, analyzing the progress of CmiRNA detection techniques, and presenting potential diagnostic MND biomarkers that have been reported.
Introduction
Motor neuron disease (MND) is a kind of neurodegenerative disease that selectively invades spinal cord anterior horn cells, brainstem motor neurons, cortical pyramidal cells and the pyramidal tract, which was first reported by Charcot in 1869. The main clinical features are the symptoms and signs of impaired upper and lower motor neurons, manifested as muscle weakness, atrophy and pyramidal tract signs (Luo et al., 2019; Ragagnin et al., 2019). Histopathology has shown the disappearance of pyramidal cells in the motor cortex, loss of motor neurons in the anterior horn of the spinal cord and brainstem, and degeneration of the corticospinal tract (Statland et al., 2015; Taylor et al., 2016). At present, the pathogenesis of the disease is still unclear. Different theories have been put forward in research in biochemistry, molecular biology, cell biology and other basic medical fields (Bartoletti et al., 2019; Lai and Ichida, 2019; Yang Y. et al., 2019). The incidence and prevalence of MND vary with age, gender, race and region, and are related to genetic factors, occupational characteristics and special material contact history (Veldink, 2017; Tank et al., 2018). MND is mainly divided into four types: amyotrophic lateral sclerosis (ALS), progressive muscular atrophy (PMA), progressive bulbar palsy (PBP) and primary lateral sclerosis (PLS) (Statland et al., 2015; Goncalves et al., 2016). Due to the lack of effective treatment and poor prognosis of MND, it is of great significance to diagnose MND as early as possible by various means (Westeneng et al., 2018; Liu et al., 2019; Sharrad and Schultz, 2019). There is continuous research and development on various biomarkers of the disease. The levels of kynurenine pathway metabolites (KPMs) are known to be dysregulated in the serum, cerebrospinal fluid (CSF), and tissue of ALS patients (Chen et al., 2010), and Tan and Guillemin suggested that KPMs could be a potential sensitive and specific diagnostic biomarker for ALS patients (Tan and Guillemin, 2019). Neurofilaments (Nfs) are neuron-specific cytoskeletal proteins, and it was reported that increased levels in biological fluids are proportionally associated with degree of axonal damage, representing potential biomarkers in MND (Gaiottino et al., 2013). Gagliardi et al. (2019) believed that Nfs may be a promising biomarker for diagnosing MND, predicting disease progression, and reflecting response to pharmacological intervention. Pathological 43-kDa transactive responsive DNA-binding protein (TDP-43) has been validated as the major disease protein in ALS (Neumann et al., 2006). Geser et al. (2011) concluded that TDP-43 was also an MND/LMN or PMA proteinopathy similar to sporadic ALS, which might reflect MND progression. However, limited by low sensitivity and specificity, these markers are not suitable as a routine test for clinical examination.
Circulating miRNAs (CmiRNAs) are a class of miRNAs found in body fluids, mainly found in blood, urine, saliva, tears, milk, or amniotic fluid, which are mainly produced by the efflux of miRNAs from tissue or cells into biofluids (Jin and Xing, 2017). CmiRNAs in peripheral biofluids have been extensively investigated as biomarkers for early diagnosis and monitoring disease progression, such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) (Hajjari et al., 2017; Li et al., 2017; Katsu et al., 2019). miRNAs are small non-protein-encoding RNAs, usually between 19 and 25 bases in length, that can bind to RISC to regulate mRNA expression. Dysregulated miRNAs have been reported associated with the pathogenesis of MND, permitting CmiRNAs to be biomarkers in biofluids for the diagnosis and progression of MND (De Paola et al., 2019; Katsu et al., 2019). This review summarizes the progress of research on the sources and functions of CmiRNAs, the potential of diagnostic markers, and the clinical application prospects, together with problems in MND-associated miRNA biomarkers.
Source, Existence, Extraction, and Detection of miRNAs and Their Biomarker Potential
Under normal conditions, CmiRNAs are mainly derived from the active secretion of cells, the release of cells that are apoptotic or necrotic, and the lysis of circulating cells. In the disease state, some CmiRNAs may come from diseased tissue cells, such as tumor cells (Matamala et al., 2018). miRNAs may be passively leaked from a variety of damaged tissues or cells, like other substances. For example, miRNA-206 is specifically expressed in muscle tissue, and it can be detected in serum after involvement of muscle tissue in ALS patients (Valsecchi et al., 2015; Pegoraro et al., 2017). There are two forms of active secretion of miRNA: one is that the free miRNA is directly secreted by cells, and the other is that miRNA is first selectively wrapped in membrane structures such as exosomes and minute bodies, and then enters the humoral circulation from endocrinology to exocytosis in the wrapped form (Iguchi et al., 2010; Ogawa et al., 2010; Burgos et al., 2014; Srivastava et al., 2018).
The current methods of extracting miRNAs from biofluids are varied in different studies. Commonly used methods of miRNA extraction from biofluids include Trizol reagent and miRNA kits. Due to the small size and low expression of CmiRNAs fragments, quantitative detection is limited.
Despite this, a number of effective CmiRNAs detection methods have been developed in recent years, including not only traditional cDNA library cloning, Northern blot and qRT-PCR, but also newly developed gene chip technology, high-throughput sequencing and nanostring nCounter (Morozova and Marra, 2008; Roberts et al., 2014; Ricci et al., 2018). With the development of molecular biology technology, the detection methods for CmiRNAs have been improved and developed, the new technologies enable the detection of CmiRNAs with greater sensitivity and precision, providing technical support for CmiRNAs to become diagnostic biomarkers (Figure 1).
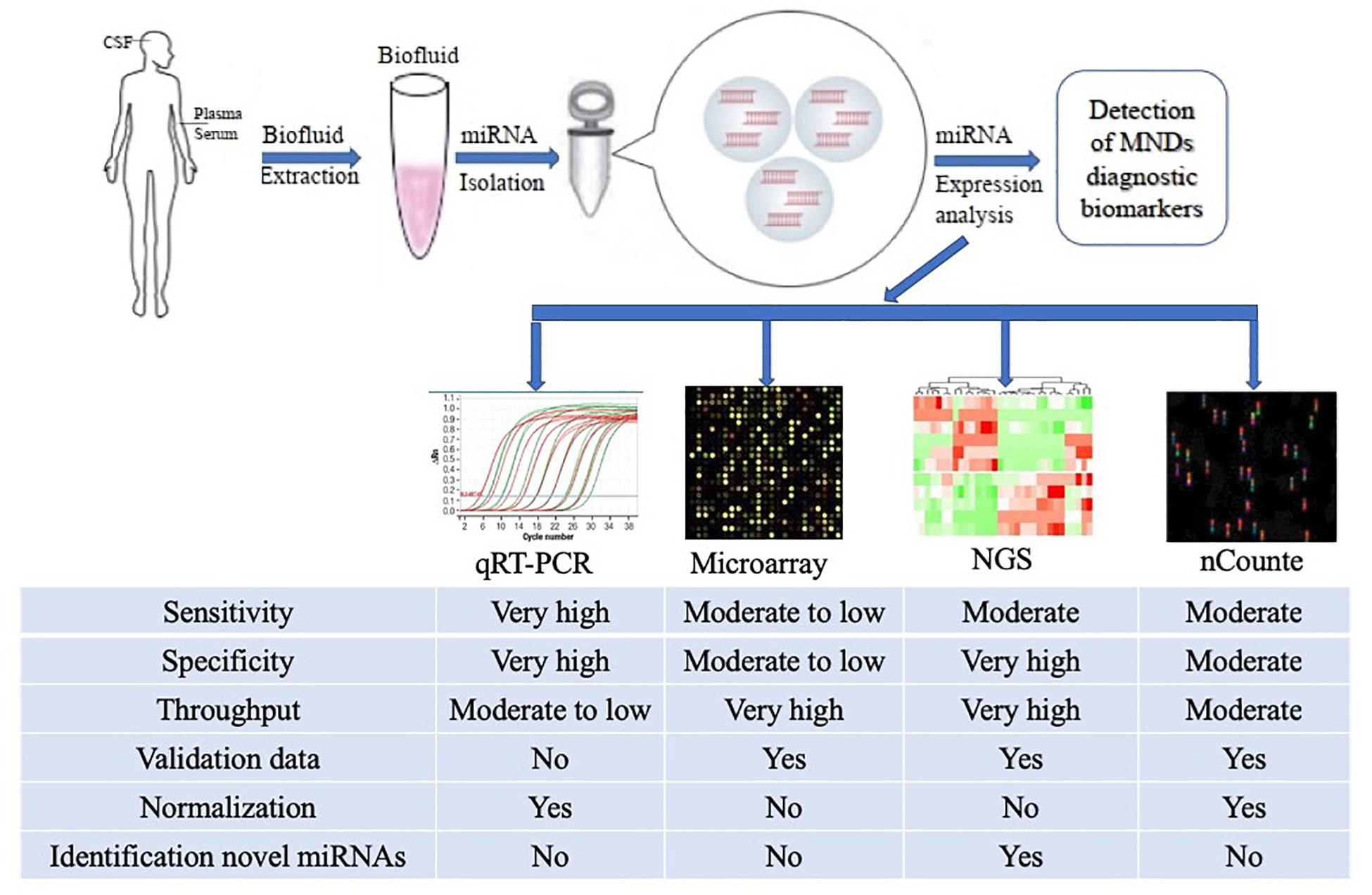
Figure 1. Methodological aspects to consider in the study of circulating miRNAs as biomarkers for ALS and other MNDs. The comparison among the common characteristics of miRNA detection platforms are summarized in the figure. qRT-PCR, quantitative Real-Time Polymerase Chain Reaction; NGS, Next Generation Sequencing.
CmiRNAs are not present in free form but are bound to specific proteins or encapsulated in active vesicles in the form of complexes in biofluids, thus providing the ability to resist RNase degradation and stable storage (Vickers et al., 2011; Zetterberg and Burnham, 2019). CmiRNAs have been repeatedly demonstrated to show substantial expression differences under different pathological conditions, which are closely related to the pathological state of individuals. Yang Q. et al. (2019) suggested that the serum level of miRNA-133b was significantly downregulated in AD patients, and that it may serve as a novel diagnostic biomarker for AD. miRNA-30c-5p has been reported to be significantly upregulated in multiple system atrophy (MSA) patients, and Vallelunga et al. (2019) considered that serum miRNA-30-5p could be a biomarker for the diagnosis and progression of MSA. Regarding CmiRNAs as diagnostic markers for MND diagnosis, relatively few studies have been published, but some breakthroughs have been made. In the next section, we review the findings of the most significant studies investigating the underlying role of CmiRNAs as diagnostic biomarkers in MND (Table 1).
Circulating miRNAs as Biomarkers in ALS
Amyotrophic lateral sclerosis is a progressive paralytic disorder characterized by degenerative changes in brain and spinal motor neurons, usually with insidious local weakness and then rapid progression to the paralysis of most muscles, including the diaphragm (Van Zundert and Brown, 2017). According to the global ALS epidemiological survey, the number of new ALS patients per year is 1–2 per 100,000 people, and incidence and prevalence increase with age. About 10% of ALS cases are familial, and the rest are sporadic (Doi et al., 2014; Talbott et al., 2016). Abnormal protein aggregation is found in the cytoplasm of motor cells in most patients with ALS, mainly TAR DNA-binding protein 43 (TDP 43). Since the superoxide dismutase 1 (SOD1) gene was shown to be associated with ALS in 1993, at least 25 related genes have been identified to date, including TDP 43 and C9orf72 (Neumann et al., 2006; Therrien et al., 2016).
Serum levels of miRNA in ALS patients were analyzed in four studies by using either qRT-PCR or microarray. Raheja et al. enrolled 23 ALS patients (3 familial and 20 sporadic) and 30 healthy controls; they identified 13 differentially expressed miRNAs, including 7 upregulated (miRNA-1, miRNA-19a-3p, miRNA-133a-3p, miRNA-133b, miRNA-192-3p, miRNA-192-5p, and miRNA-144-5p) and 6 downregulated (let-7d-3p, miRNA-139-5p, miRNA-320a, miRNA-320b, miRNA-320c, and miRNA-425-5p) miRNAs in ALS patients compared to healthy controls using qRT-PCR. By logistic regression and ROC curve analysis, the 7 up-regulated miRNAs and 6 down-regulated miRNAs had ideal diagnostic value to distinguish between ALS and healthy controls (Raheja et al., 2018). In a study by Matamala et al. (2018) the differential expression of miRNAs was first detected in the serum of SOD1 transgenic mice by using next-generation sequencing. Further verification of differential miRNAs in ALS patients’ serum by qRT-PCR revealed that there was substantial dysregulated expression of miRNA-142-3p and miRNA-1249-3p. In addition, the authors found a negative correlation between miRNA-142-3p and revised ALS functional rating scale and suggested that the serum level of miRNA-142-3p could be a potential biomarker for ALS severity (Matamala et al., 2018). In a qRT-PCR analysis of 18 sporadic ALS (sALS) patients and 16 healthy controls, Freischmidt et al. (2015) observed 2 significantly downregulated serum miRNAs (miRNA-1234-3p and miRNA-1825), and demonstrated that there was dysregulation of miRNA-1825 in both sALS and familial ALS (fALS) patients, and that significant downregulation of miRNA-1234-3p was restricted to patients with sALS. Another study including a larger number of sALS patients in validation cohorts distinguished a set of 3 CmiRNAs (miRNA-143-3p, miRNA-206, and miRNA-374b-5p) in serum. The levels of miRNA-143-3p and miRNA-206 were significantly increased, while the levels of miRNA-374b-5p were significantly decreased, which was not affected by Riluzole treatment. In a further analysis of the clinical significance of three candidate CmiRNAs by subgroup, Waller et al. determined that miRNA-143-3p was associated with lower limb onset of ALS patients (Waller et al., 2017a).
A number of studies have investigated plasma levels of miRNAs using high-throughput next-generation sequencing. In a study of dysregulated miRNAs associated with sALS, which included 56 ALS patients and 20 healthy controls, Liguori et al. (2018) identified 38 downregulated miRNAs in sALS patients. The authors also observed that the expression levels of miRNA-130a-3p, miRNA-151b, and miRNA-221-3p were positively correlated with sALS progression. These authors further suggested that these CmiRNAs could serve not only as biomarkers for diagnosis but also for monitoring disease progression (Liguori et al., 2018). In another study comparing plasma miRNAs between 16 sALS patients and 10 healthy controls using microarray analysis, validated by qPCR, Takahashi et al. (2015) found two differentially expressed miRNAs (downregulated miRNA-4299 and upregulated miRNA-4649-5p), regardless of other clinical features, concluding that these two plasma CmiRNAs had the potential to be ALS diagnosis biomarkers. By using microarray, Katsu et al. (2019) identified 30 differentially expressed miRNAs in ALS plasma, including 13 upregulated and 17 downregulated miRNAs, and they further investigated the relationship between these miRNAs and ALS, and suggested that four miRNAs (miRNA-24-3p, miRNA-1268a, miRNA-3911, and miRNA-4646-5) were enriched in synaptic vesicle exocytosis and processes. Detection of these biomarkers helps get a better understanding of ALS pathophysiology and could be used for early diagnosis (Katsu et al., 2019).
There are few studies on differential miRNA expression in CSF samples from ALS patients. Using qRT-PCR, Benigni et al. (2016) identified eight significantly deregulated miRNAs in CSF samples from 24 ALS patients, where statistical analysis results showed that upregulated miRNA-181a-5p and downregulated miRNA-15b-5p and miRNA-21-5p had the highest diagnostic accuracy, confirming the application potential as ALS diagnostic biomarkers. Eleven dysregulated miRNAs were detected by qRT-PCR in the CSF of sALS patients associated with neural and glial activity: 7 miRNAs (miRNA-124-3p, miRNA-126-5p, miRNA-127-3p, miRNA-23b-3p, miRNA-27b-3p, miRNA-9-5p, and miRNA-99b-5p) were upregulated and 4 miRNAs (let-7f-5p, miRNA-150-5p, miRNA-142-5p, and miRNA-378a-3p) downregulated (Waller et al., 2017b).
miRNA expression in the leukocytes of sALS patients was also addressed in one study. By using microarray technology in leukocytes obtained from 5 sALS patients, Chen et al. identified 11 differentially expressed miRNAs, including 4 upregulated and 7 downregulated miRNAs. The results of four of seven miRNAs (miRNA-183, miRNA-193b, miRNA-451, and miRNA-3935) were validated by qRT-PCR, and the authors showed that all these four miRNAs showed high diagnostic accuracy for ALS (Chen et al., 2016).
miRNA-338-3p is a well-studied CmiRNA in the previous studies. De Felice et al. (2014) demonstrated that miRNA -338-3p was upregulated in peripheral leukocytes, serum, and CSF from sALS patients. Their results further showed signs of miRNA-338-3p being localized in the gray matter of spinal cord tissues from sALS patients, and the authors considered the miRNA to be an effective biomarker in the early diagnosis of sALS (De Felice et al., 2014).
In addition to microarray analysis and qRT PCR validation, most CmiRNA studies included the optimal cut-off value for ROC analysis (Takahashi et al., 2015; Liguori et al., 2018; Matamala et al., 2018; Raheja et al., 2018). In most studies, CmiRNAs have been used as biomarkers to distinguish ALS from healthy controls, while a few studies have used them to distinguish patients with fALS from those with sALS. No matter what the purpose of the studies, we found that most results suggested that CmiRNAs, as biomarkers, have high sensitivity (up to 94%) and specificity (up to 87%), indicating that CmiRNAs have great potential as biomarkers in clinical practice (Benigni et al., 2016; Verde et al., 2019).
Although the specimens are from different sources, it is not difficult to find that CmiRNAs have proven their clinical value in many aspects based on previous researches. miRNA-24-3p, miRNA-1268a, miRNA-3911, miRNA-4646-5, miRNA-4299, and miRNA-4649-5p in plasma could be potential CmiRNAs for ALS diagnosis, miRNA-181a-5p, miRNA-15b-5p, and miRNA-21-5p in CSF also had high diagnostic accuracy for ALS; miRNA-1234-3p, miRNA-143-3p, miRNA-206, and miRNA-374b-5p in serum were restricted to patients with sALS in distinguishing fALS. Serum miRNA-142-3p could be a potential biomarker for ALS severity; miRNA-130a-3p, miRNA-151b, and miRNA-221-3p in plasma were positively correlated with sALS progression. At the same time, we cannot deny that there are few repetitive CmiRNAs in previous studies, which may be related to the involvement of multiple genes associated with genetic ALS and have different pathological pathways related to RNA metabolism. All this reflects the complexity of ALS diagnosis, and future studies with larger patient numbers and a higher methodological standardization might promote the clinical application of CmiRNAs as diagnostic biomarkers.
CmiRNAs as Biomarkers in Other MNDs
SMA is a single gene autosomal recessive genetic disease, which is mainly caused by homozygous deletion of SMN1 gene. It is characterized by degeneration of spinal cord anterior horn cells and muscle atrophy and weakness (Lefebvre et al., 1995; Faravelli et al., 2015). Recent studies have confirmed a critical role of miRNAs in the pathogenesis of SMA. In addition, dysregulated motor neuron−specific miRNAs are not only involved in the SMA motor neuron phenotype but may also be biomarkers and therapeutic targets (Haramati et al., 2010; Kye and Goncalves Ido, 2014; Magri et al., 2018). It was reported that miRNA-146a was significantly upregulated in the SMA spinal cord mouse model, where Sison et al. (2017) investigated the upregulated miRNA-146a-induced motor neuron loss in vitro and suggested astrocyte-produced miRNA-146a may be a contributing factor in SMA pathology. Kye et al. (2014) observed an upregulated expression of miRNA-183 in SMA knockdown in cortical neurons and suggested that miRNA-183 regulates axonal local translation via the mTOR pathway. Catapano et al. (2016) examined the expression changes of miRNA-9, -132, and -206 in SMA mouse tissues and patient serum. The authors found that the expression of miRNA-9 and miRNA-132 was significantly decreased in the spinal cord of SMA mice, but the expression in muscle was significantly increased, and that the expression of miRNA-206 was increased both in spinal cord and muscle tissue. These three miRNAs were also differentially expressed in the serum of SMA mice, and this change was prior to that in spinal cord and muscle tissue. miRNAs were detected by qRT-PCR in the serum of SMA patients. The expression levels of miRNA-9 and miRNA-132 were significantly increased in the serum of patients, but there was no significant change in miRNA-206 levels, suggesting that miRNA-9 and miRNA-132 can serve as non-invasive biomarkers (Catapano et al., 2016). Despite limited current researches, motor neuron-specific miRNAs may have great potential in the diagnosis of SMA. For example, miRNA-9 and miRNA-132 in serum, miRNA-206 in muscle tissue could be used to diagnose SMA. However, more clinical studies are needed to further validate the diagnostic efficacy of these CmiRNAs.
Advantages and Problems of CmiRNAs as Biomarkers
At present, biomarkers in biofluid are mainly based on the change in expression level of some specific proteins, such as β-amyloid (Aβ) and tau protein in peripheral blood, for diagnosis of AD (McDade and Bateman, 2017), and detection of α-synuclein for diagnosis of PD and monitoring disease progression as well (Atik et al., 2016). However, the complexity of protein composition, the variation caused by post-transcriptional modification, the low level of protein biomarkers, the stability of samples and the sensitivity of assays restrict the further search for new protein biomarkers to some extent.
In clinical practice, miRNAs have been identified in blood, urine, saliva and other easily available biofluids. These CmiRNAs show extraordinary stability and anti-degradation ability, showing the basic characteristics of miRNAs in biofluid as ideal for disease biomarkers (Mushtaq et al., 2016). First, some of the CmiRNAs are released into membrane-bound vesicles by the cells, which can protect them from RNase degradation. These CmiRNAs are transported by vesicles, which come from the release of multivesicular bodies and exosomes (Orozco and Lewis, 2010). Secondly, 90% of miRNAs in blood are in the form of a nucleic acid-protein complex, because most of the blood CmiRNAs are associated with argonaute2 (ago2) protein. Ago2 is an effective component of miRNA induced silent complex (miRISC). It can directly bind to miRNAs and mediate the post-transcriptional regulation of mRNA. The reason why they can exist outside the cell is the high stability of Ago2-miRNA protein complexes (Bose and Bhattacharyya, 2018). In addition, CmiRNAs can also bind to non-vesicular lipoproteins and RNA-binding proteins. Protein-binding miRNA is a signal molecule which is actively secreted by cells and functions in intercellular communication. Therefore, CmiRNAs analysis is mainly to detect the level of miRNA-protein complexes, rather than vesicles (Wang et al., 2010; Vickers et al., 2011).
However, there are still many problems to be solved for the use of CmiRNAs as disease biomarkers. (1) Current research specimens are small in quantity, and systematic large-scale collaborative studies are lacking; at the same time, due to differences in detection methods and assessment standards, some research results and conclusions lack consistency. (2) The low efficiency of extracting miRNAs and the limited sensitivity and specificity of CmiRNA detection, which limits the application of miRNAs as disease biomarkers in clinical practice. In addition, the expression levels of miRNAs may change in a variety of diseases, such as miRNA-9, which can be differentially expressed in Huntington’s disease, SMA and PD (Catapano et al., 2016; Chang et al., 2017). (3) Studies have also found that serum and plasma miRNA expression profiles are not completely consistent. The mechanism of production and transport of serum or plasma miRNAs is still unclear (Wang et al., 2012).
Conclusion
Emerging studies have shown that CmiRNAs can serve as potential biomarkers for neurodegenerative diseases and can be used in clinical diagnosis and monitoring disease progression and response to treatment measures. On the basis of the review of previous studies on the use of CmiRNAs as biomarkers of MND, we found that although the candidate CmiRNAs have very low reproducibility, it is not difficult to see after statistical analysis of the data obtained that CmiRNAs as diagnostic biomarkers have very ideal sensitivity and specificity. In addition, CmiRNAs can be easily detected and differentially expressed in a variety of body fluids, and can thus be promising diagnostic biomarkers for MND. To further apply CmiRNAs as biomarkers in clinical practice as soon as possible, it is necessary to ensure strict standardized test methods, as well as research and validation cohorts from multiple centers, and require selected patients to prioritize genetic diagnosis to improve the accuracy of the diagnosis.
Author Contributions
LW wrote the manuscript primarily. LW and LZ produced the figure. LZ contributed to the editing of this review. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Atik, A., Stewart, T., and Zhang, J. (2016). Alpha-synuclein as a biomarker for Parkinson’s disease. Brain Pathol. 26, 410–418. doi: 10.1111/bpa.12370
Bartoletti, M., Bosco, D. A., Da Cruz, S., Lagier-Tourenne, C., Liachko, N., Markmiller, S., et al. (2019). Phenotypic suppression of ALS/FTD-associated neurodegeneration highlights mechanisms of dysfunction. J. Neurosci. 39, 8217–8224. doi: 10.1523/JNEUROSCI.1159-19.2019
Benigni, M., Ricci, C., Jones, A. R., Giannini, F., Al-Chalabi, A., and Battistini, S. (2016). Identification of miRNAs as potential biomarkers in cerebrospinal fluid from amyotrophic lateral sclerosis patients. Neuromol. Med. 18, 551–560.
Bose, M., and Bhattacharyya, S. N. (2018). Target mRNA-driven biogenesis of cognate MicroRNAs in vitro. Methods Mol. Biol. 1733, 27–39. doi: 10.1007/978-1-4939-7601-0_2
Burgos, K., Malenica, I., Metpally, R., Courtright, A., Rakela, B., Beach, T., et al. (2014). Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 9:e94839. doi: 10.1371/journal.pone.0094839
Catapano, F., Zaharieva, I., Scoto, M., Marrosu, E., Morgan, J., Muntoni, F., et al. (2016). Altered levels of MicroRNA-9, -206, and -132 in spinal muscular atrophy and their response to antisense oligonucleotide therapy. Mol. Ther. Nucleic Acids 5:e331. doi: 10.1038/mtna.2016.47
Chang, K. H., Wu, Y. R., and Chen, C. M. (2017). Down-regulation of miR-9∗ in the peripheral leukocytes of Huntington’s disease patients. Orphanet. J. Rare Dis. 12:185. doi: 10.1186/s13023-017-0742-x
Chen, Y., Stankovic, R., Cullen, K. M., Meininger, V., Garner, B., Coggan, S., et al. (2010). The kynurenine pathway and inflammation in amyotrophic lateral sclerosis. Neurotox Res. 18, 132–142. doi: 10.1007/s12640-009-9129-7
Chen, Y., Wei, Q., Chen, X., Li, C., Cao, B., Ou, R., et al. (2016). Aberration of miRNAs expression in leukocytes from sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 9:69. doi: 10.3389/fnmol.2016.00069
De Felice, B., Annunziata, A., Fiorentino, G., Borra, M., Biffali, E., Coppola, C., et al. (2014). miR-338-3p is over-expressed in blood, CFS, serum and spinal cord from sporadic amyotrophic lateral sclerosis patients. Neurogenetics 15, 243–253. doi: 10.1007/s10048-014-0420-2
De Paola, E., Verdile, V., and Paronetto, M. P. (2019). Dysregulation of microRNA metabolism in motor neuron diseases: novel biomarkers and potential therapeutics. Noncoding RNA Res. 4, 15–22. doi: 10.1016/j.ncrna.2018.12.001
Doi, Y., Atsuta, N., Sobue, G., Morita, M., and Nakano, I. (2014). Prevalence and incidence of amyotrophic lateral sclerosis in Japan. J. Epidemiol. 24, 494–499.
Faravelli, I., Nizzardo, M., Comi, G. P., and Corti, S. (2015). Spinal muscular atrophy–recent therapeutic advances for an old challenge. Nat. Rev. Neurol. 11, 351–359. doi: 10.1038/nrneurol.2015.77
Freischmidt, A., Muller, K., Zondler, L., Weydt, P., Mayer, B., Von Arnim, C. A., et al. (2015). Serum microRNAs in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 36, e2615–e2620.
Gagliardi, D., Meneri, M., Saccomanno, D., Bresolin, N., Comi, G. P., and Corti, S. (2019). Diagnostic and prognostic role of blood and cerebrospinal fluid and blood neurofilaments in amyotrophic lateral sclerosis: a review of the literature. Int. J. Mol. Sci. 20:4152. doi: 10.3390/ijms20174152
Gaiottino, J., Norgren, N., Dobson, R., Topping, J., Nissim, A., Malaspina, A., et al. (2013). Increased neurofilament light chain blood levels in neurodegenerative neurological diseases. PLoS One 8:e75091. doi: 10.1371/journal.pone.0075091
Geser, F., Stein, B., Partain, M., Elman, L. B., Mccluskey, L. F., Xie, S. X., et al. (2011). Motor neuron disease clinically limited to the lower motor neuron is a diffuse TDP-43 proteinopathy. Acta Neuropathol. 121, 509–517.
Goncalves, I. D., Rehorst, W. A., and Kye, M. J. (2016). Dysregulation of RNA mediated gene expression in motor neuron diseases. CNS Neurol. Disord. Drug Targets 15, 887–895.
Hajjari, S. N., Mehdizadeh, M., Sadigh-Eteghad, S., Shanehbandi, D., Teimourian, S., and Baradaran, B. (2017). Secretases-related miRNAs in Alzheimer’s disease: new approach for biomarker discovery. Neurol. Sci. 38, 1921–1926.
Haramati, S., Chapnik, E., Sztainberg, Y., Eilam, R., Zwang, R., Gershoni, N., et al. (2010). miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. U.S.A. 107, 13111–13116.
Iguchi, H., Kosaka, N., and Ochiya, T. (2010). Secretory microRNAs as a versatile communication tool. Commun. Integr. Biol. 3, 478–481.
Jin, F., and Xing, J. (2017). Circulating pro-angiogenic and anti-angiogenic microRNA expressions in patients with acute ischemic stroke and their association with disease severity. Neurol. Sci. 38, 2015–2023.
Katsu, M., Hama, Y., Utsumi, J., Takashina, K., Yasumatsu, H., Mori, F., et al. (2019). MicroRNA expression profiles of neuron-derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci. Lett. 708:134176.
Kye, M. J., and Goncalves Ido, C. (2014). The role of miRNA in motor neuron disease. Front. Cell Neurosci. 8:15. doi: 10.3389/fncel.2014.00015
Kye, M. J., Niederst, E. D., Wertz, M. H., Goncalves Ido, C., Akten, B., Dover, K. Z., et al. (2014). SMN regulates axonal local translation via miR-183/mTOR pathway. Hum. Mol. Genet. 23, 6318–6331.
Lai, J. D., and Ichida, J. K. (2019). C9ORF72 protein function and immune dysregulation in amyotrophic lateral sclerosis. Neurosci. Lett. 713:134523. doi: 10.1016/j.neulet.2019.134523
Lefebvre, S., Burglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., et al. (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165.
Li, N., Pan, X., Zhang, J., Ma, A., Yang, S., Ma, J., et al. (2017). Plasma levels of miR-137 and miR-124 are associated with Parkinson’s disease but not with Parkinson’s disease with depression. Neurol. Sci. 38, 761–767. doi: 10.1007/s10072-017-2841-9
Liguori, M., Nuzziello, N., Introna, A., Consiglio, A., Licciulli, F., D’errico, E., et al. (2018). Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 11:288. doi: 10.3389/fnmol.2018.00288
Liu, J., Zhang, X., Ding, X., Song, M., and Sui, K. (2019). Analysis of clinical and electrophysiological characteristics of 150 patients with amyotrophic lateral sclerosis in China. Neurol. Sci. 40, 363–369. doi: 10.1007/s10072-018-3633-6
Luo, L., Song, Z., Li, X., Huiwang, Zeng, Y., and Qinwang, et al. (2019). Efficacy and safety of edaravone in treatment of amyotrophic lateral sclerosis-a systematic review and meta-analysis. Neurol. Sci. 40, 235–241. doi: 10.1007/s10072-018-3653-2
Magri, F., Vanoli, F., and Corti, S. (2018). miRNA in spinal muscular atrophy pathogenesis and therapy. J. Cell Mol. Med. 22, 755–767. doi: 10.1111/jcmm.13450
Matamala, J. M., Arias-Carrasco, R., Sanchez, C., Uhrig, M., Bargsted, L., Matus, S., et al. (2018). Genome-wide circulating microRNA expression profiling reveals potential biomarkers for amyotrophic lateral sclerosis. Neurobiol. Aging 64, 123–138. doi: 10.1016/j.neurobiolaging.2017.12.020
Morozova, O., and Marra, M. A. (2008). Applications of next-generation sequencing technologies in functional genomics. Genomics 92, 255–264. doi: 10.1016/j.ygeno.2008.07.001
Mushtaq, G., Greig, N. H., Anwar, F., Zamzami, M. A., Choudhry, H., Shaik, M. M., et al. (2016). miRNAs as circulating biomarkers for Alzheimer’s disease and Parkinson’s disease. Med. Chem. 12, 217–225.
Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133.
Ogawa, R., Tanaka, C., Sato, M., Nagasaki, H., Sugimura, K., Okumura, K., et al. (2010). Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem. Biophys. Res. Commun. 398, 723–729. doi: 10.1016/j.bbrc.2010.07.008
Orozco, A. F., and Lewis, D. E. (2010). Flow cytometric analysis of circulating microparticles in plasma. Cytometry A 77, 502–514. doi: 10.1002/cyto.a.20886
Pegoraro, V., Merico, A., and Angelini, C. (2017). Micro-RNAs in ALS muscle: differences in gender, age at onset and disease duration. J. Neurol. Sci. 380, 58–63. doi: 10.1016/j.jns.2017.07.008
Ragagnin, A. M. G., Shadfar, S., Vidal, M., Jamali, M. S., and Atkin, J. D. (2019). Motor neuron susceptibility in ALS/FTD. Front. Neurosci. 13:532. doi: 10.3389/fnins.2019.00532
Raheja, R., Regev, K., Healy, B. C., Mazzola, M. A., Beynon, V., Von Glehn, F., et al. (2018). Correlating serum micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve 58, 261–269. doi: 10.1002/mus.26106
Ricci, C., Marzocchi, C., and Battistini, S. (2018). MicroRNAs as biomarkers in amyotrophic lateral sclerosis. Cells 7:219.
Roberts, T. C., Coenen-Stass, A. M., and Wood, M. J. (2014). Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One 9:e89237. doi: 10.1371/journal.pone.0089237
Sharrad, D. F., and Schultz, D. W. (2019). Factors contributing to delays in the diagnosis of motor neuron disease - A south Australian study. J. Neurol. Sci. 407:116540. doi: 10.1016/j.jns.2019.116540
Sison, S. L., Patitucci, T. N., Seminary, E. R., Villalon, E., Lorson, C. L., and Ebert, A. D. (2017). Astrocyte-produced miR-146a as a mediator of motor neuron loss in spinal muscular atrophy. Hum. Mol. Genet. 26, 3409–3420. doi: 10.1093/hmg/ddx230
Srivastava, A., Moxley, K., Ruskin, R., Dhanasekaran, D. N., Zhao, Y. D., and Ramesh, R. (2018). A non-invasive liquid biopsy screening of urine-derived exosomes for miRNAs as biomarkers in endometrial cancer patients. AAPS J. 20:82. doi: 10.1208/s12248-018-0220-y
Statland, J. M., Barohn, R. J., Mcvey, A. L., Katz, J. S., and Dimachkie, M. M. (2015). Patterns of weakness, classification of motor neuron disease, and clinical diagnosis of sporadic amyotrophic lateral sclerosis. Neurol. Clin. 33, 735–748. doi: 10.1016/j.ncl.2015.07.006
Takahashi, I., Hama, Y., Matsushima, M., Hirotani, M., Kano, T., Hohzen, H., et al. (2015). Identification of plasma microRNAs as a biomarker of sporadic amyotrophic lateral sclerosis. Mol. Brain 8:67. doi: 10.1186/s13041-015-0161-7
Talbott, E. O., Malek, A. M., and Lacomis, D. (2016). The epidemiology of amyotrophic lateral sclerosis. Handb. Clin. Neurol. 138, 225–238. doi: 10.1016/B978-0-12-802973-2.00013-6
Tan, V. X., and Guillemin, G. J. (2019). Kynurenine pathway metabolites as biomarkers for amyotrophic lateral sclerosis. Front. Neurosci. 13:1013. doi: 10.3389/fnins.2019.01013
Tank, E. M., Figueroa-Romero, C., Hinder, L. M., Bedi, K., Archbold, H. C., Li, X., et al. (2018). Abnormal RNA stability in amyotrophic lateral sclerosis. Nat. Commun. 9:2845. doi: 10.1038/s41467-018-05049-z
Taylor, J. P., Brown, R. H. Jr., and Cleveland, D. W. (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. doi: 10.1038/nature20413
Therrien, M., Dion, P. A., and Rouleau, G. A. (2016). ALS: recent developments from genetics studies. Curr. Neurol. Neurosci. Rep. 16:59. doi: 10.1007/s11910-016-0658-1
Vallelunga, A., Iannitti, T., Dati, G., Capece, S., Maugeri, M., Tocci, E., et al. (2019). Serum miR-30c-5p is a potential biomarker for multiple system atrophy. Mol. Biol. Rep. 46, 1661–1666. doi: 10.1007/s11033-019-04614-z
Valsecchi, V., Boido, M., De Amicis, E., Piras, A., and Vercelli, A. (2015). Expression of muscle-specific MiRNA 206 in the progression of disease in a murine SMA model. PLoS One 10:e0128560. doi: 10.1371/journal.pone.0128560
Van Zundert, B., and Brown, R. H. Jr. (2017). Silencing strategies for therapy of SOD1-mediated ALS. Neurosci. Lett. 636, 32–39. doi: 10.1016/j.neulet.2016.07.059
Veldink, J. H. (2017). ALS genetic epidemiology ‘How simplex is the genetic epidemiology of ALS?’. J. Neurol. Neurosurg. Psychiatry 88:537.
Verde, F., Silani, V., and Otto, M. (2019). Neurochemical biomarkers in amyotrophic lateral sclerosis. Curr. Opin. Neurol. 32, 747–757. doi: 10.1097/WCO.0000000000000744
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433. doi: 10.1038/ncb2210
Waller, R., Goodall, E. F., Milo, M., Cooper-Knock, J., Da Costa, M., Hobson, E., et al. (2017a). Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 55, 123–131. doi: 10.1016/j.neurobiolaging.2017.03.027
Waller, R., Wyles, M., Heath, P. R., Kazoka, M., Wollff, H., Shaw, P. J., et al. (2017b). Small RNA sequencing of sporadic amyotrophic lateral sclerosis cerebrospinal fluid reveals differentially expressed miRNAs related to neural and glial activity. Front. Neurosci. 11:731. doi: 10.3389/fnins.2017.00731
Wang, K., Yuan, Y., Cho, J. H., Mcclarty, S., Baxter, D., and Galas, D. J. (2012). Comparing the MicroRNA spectrum between serum and plasma. PLoS One 7:e41561. doi: 10.1371/journal.pone.0041561
Wang, K., Zhang, S., Weber, J., Baxter, D., and Galas, D. J. (2010). Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38, 7248–7259. doi: 10.1093/nar/gkq601
Westeneng, H. J., Debray, T. P. A., Visser, A. E., Van Eijk, R. P. A., Rooney, J. P. K., Calvo, A., et al. (2018). Prognosis for patients with amyotrophic lateral sclerosis: development and validation of a personalised prediction model. Lancet Neurol. 17, 423–433. doi: 10.1016/S1474-4422(18)30089-9
Yang, Q., Zhao, Q., and Yin, Y. (2019). miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 18, 2711–2718. doi: 10.3892/etm.2019.7855
Yang, Y., Halliday, G. M., Kiernan, M. C., and Tan, R. H. (2019). TDP-43 levels in the brain tissue of ALS cases with and without C9ORF72 or ATXN2 gene expansions. Neurology 93, e1748–e1755. doi: 10.1212/WNL.0000000000008439
Keywords: circulating microRNA, biomarker, motor neuron disease, neurodegeneration, blood, cerebral spinal fluid
Citation: Wang L and Zhang L (2020) Circulating MicroRNAs as Diagnostic Biomarkers for Motor Neuron Disease. Front. Neurosci. 14:354. doi: 10.3389/fnins.2020.00354
Received: 21 December 2019; Accepted: 24 March 2020;
Published: 16 April 2020.
Edited by:
Francesca Trojsi, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Maria Shadrina, Institute of Molecular Genetics (RAS), RussiaLucia Scisciola, University of Campania Luigi Vanvitelli, Italy
Valeria Valsecchi, University of Naples Federico II, Italy
Copyright © 2020 Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Zhang, Y211X2xpanVhbnpoYW5nQDE2My5jb20=
 Lin Wang
Lin Wang Lijuan Zhang
Lijuan Zhang