- 1Stem Cell and Neurogenesis Unit, Division of Neuroscience, San Raffaele Scientific Institute, Milan, Italy
- 2CNR Institute of Neuroscience, Milan, Italy
In recent years, the need to derive sources of specialized cell types to be employed for cell replacement therapies and modeling studies has triggered a fast acceleration of novel cell reprogramming methods. In particular, in neuroscience, a number of protocols for the efficient differentiation of somatic or pluripotent stem cells have been established to obtain a renewable source of different neuronal cell types. Alternatively, several neuronal populations have been generated through direct reprogramming/transdifferentiation, which concerns the conversion of fully differentiated somatic cells into induced neurons. This is achieved through the forced expression of selected transcription factors (TFs) in the donor cell population. The reprogramming cocktail is chosen after an accurate screening process involving lists of TFs enriched into desired cell lineages. In some instances, this type of studies has revealed the crucial role of TFs whose function in the differentiation of a given specific cell type had been neglected or underestimated. Herein, we will speculate on how the in vitro studies have served to better understand physiological mechanisms of neuronal development in vivo.
Introduction
Over the years, crucial extrinsic and intrinsic mechanisms regulating the acquisition of cell fate during neural development have been elucidated. Gradients of morphogens secreted by organizer centers instruct neural progenitor cells (NPCs) to activate the expression of transcription factor (TF) cascades that guide cells through every single step of the fate acquisition process. Genetic studies in vitro and in vivo, essentially based on the gain- and loss-of-function experiments, revealed that large arrays of TF cascades are indeed responsible for the specification of different neuronal subtypes.
This mechanistic knowledge was critical for the field of cell reprogramming to emerge. Indeed, the possibility to convert a cell type into another has been strictly dependent on seminal findings accumulated over the last 30 years in neurodevelopmental biology.
Back in the 1950s, it was not yet clear whether all cells belonging to the same organism contained the same set of genes. On this line, Weismann had suggested that genes whose function was no longer required might be lost or permanently inactivated in a specific cell type, seeding the concept that cell fate acquisition is an irreversible process being associated with loss of genetic material. This concept, well represented by the famous Waddington’s landscape (Waddington, 1957) was later challenged by Gurdon’s work. He performed pioneer experiments of somatic nuclei transfer in Xenopus oocytes during his Ph.D. studies, providing the first evidence for the preservation of genome integrity after cellular differentiation (Gurdon, 1962).
Up to date, it is consolidated the concept that epigenetic-mediated gene silencing, rather than gene loss, accompanies cell fate acquisition. This evidence opened a crack toward the plasticity of cell identity and the possibility of altering the fate of a differentiated cell.
In 1988, MyoD ectopic expression in mouse embryonic fibroblasts (MEFs) was revealed sufficient to convert them into muscle cells (Tapscott et al., 1988). Two decades later the breakthrough from the Yamanaka’s group showed that somatic cells can be reverted to a pluripotent state forcing the expression of the four factors Oct4, Sox2, Klf4, and c-Myc (Takahashi and Yamanaka, 2006), that are mediating global chromatin remodeling allowing for the expression of the pluripotency gene machinery (Takahashi and Yamanaka, 2006; Boissart et al., 2012). First successful conversion of MEFs into functional induced neurons (iNs) was described a few years later through Ascl1, Brn2, and Myt1l misexpression (Vierbuchen et al., 2010). After this study, many others attempted to modify or enrich this TF combination to induce MEF differentiation toward specific neuronal subtypes localized to defined areas of the brain (reviewed in Masserdotti et al., 2016).
All these works highlighted the ability of accurately selected cocktail of TFs to alter the fate of fully differentiated cells and to obtain functional neuronal cells.
To define a cell reprogramming gene cocktail, the TFs to be tested in the screening are chosen for their capability to impose that specific neuronal fate (master regulator genes) or among genes enriched in the target cell population, but not necessarily with their functions already addressed. Very recently, unbiased screenings of TFs for neuronal conversion have been also performed with very informative results (Liu et al., 2018; Tsunemoto et al., 2018). Once the candidate TF list is selected, they are delivered in donor cells according to a “narrow down” or an “add one” strategy. Generally, TFs are delivered and expressed all simultaneously in the donor cells although they control different phases of the cell fate acquisition process. In other cases, genetic tricks (i.e., mix of constitutive promoter guided- and inducible promoter guided-TFs whose expression can be turned off at a defined time) are employed to allow sequential expression of TFs required in different phases of differentiation in a manner that tries to recapitulate the expression timing observed during in vivo development (Au et al., 2013; Colasante et al., 2015). Finally, in an even more sophisticated experimental setting, the endogenous loci of the desired TFs can be activated using the CRISPR/Cas9 system (Black et al., 2016; Liu et al., 2018). In all these cases, the final output of these studies can meet the initial expectations, but unpredicted results have not rarely been reported. In fact, in some instances, new features for the mechanisms of action of TFs have been emerging. In others, TFs whose role was not considered determining for a specific neuronal fate acquisition during in vivo development, have come out as pivotal in the neuronal specification during direct cell reprogramming. Even more surprisingly is the identification of TFs not related to neuronal development that are able to impose a neuronal identity when overexpressed in heterologous cells.
This predictive value of the direct cell reprogramming methodology can be likely explained by the fact that during this process selected TFs are forced to operate in donor cell populations that are very distant from the target neuronal cells. This is the case for the fibroblast-to-neuron conversion, as fibroblasts have a mesodermic origin in the embryo contrary to the ectoderm-derived neurons. According to this different ontogeny, fibroblasts present both divergent global gene expression profiles and chromatin states compared to neurons. In this “unfavorable environment,” some neuronal TFs unexpectedly revealed to have a pioneer function being able to “open up” the chromatin and activate genes that are silenced in donor cells. Conversely, in vivo, their function might be facilitated by other TFs expressed earlier in the transcriptional cascades or their function might be hidden by complex gene regulation networks. With its ability to directly challenge TFs, the direct neuronal reprogramming provides a unique experimental system where to better appreciate their role in a relatively simple in vitro assay with a clear phenotypic analysis outcome.
New Insights Into the Roles of the Proneural TFs
Deepening Our Understanding of Classical Proneural TFs: Ascl1 and Neurog2
Textbook developmental biology studies revealed that Achaete-scute homolog 1 (Ascl1) and Neurogenin2 (Neurog2) are the prominent pro-neural factors in charge of the neuronal identity specification in the nervous system (Horton et al., 1999; Bertrand et al., 2002; Parras et al., 2002; Mattar et al., 2004; Schuurmans et al., 2004; Britz et al., 2006; Poitras et al., 2007; Kovach et al., 2013). These two TFs are expressed in a complementary manner in the telencephalon: Neurog2 is expressed in dorsal progenitors and instruct them to generate glutamatergic neurons, whereas Ascl1 is expressed in ventral progenitor cells contributing to the acquisition of GABAergic fate.
With this well-established background, it seemed pretty consistent that the forced expression of Ascl1 was shown essential to obtain neurons from both murine and human fibroblasts (Vierbuchen et al., 2010; Caiazzo et al., 2011; Kim et al., 2011; Pfisterer et al., 2011; Torper et al., 2013; Colasante et al., 2015; Table 1). The relevant role of Ascl1 has also been demonstrated in the reprogramming of other cell types that are more plastic than terminally differentiated fibroblasts or more closely related to neurons. Indeed, Ascl1 alone can guide the conversion of murine or human embryonic stem cells (ESCs) (Chanda et al., 2014) and astrocytes (Berninger et al., 2007; Heinrich et al., 2010; Liu et al., 2015; Masserdotti et al., 2015; Chouchane et al., 2017) into neurons (Table 1).
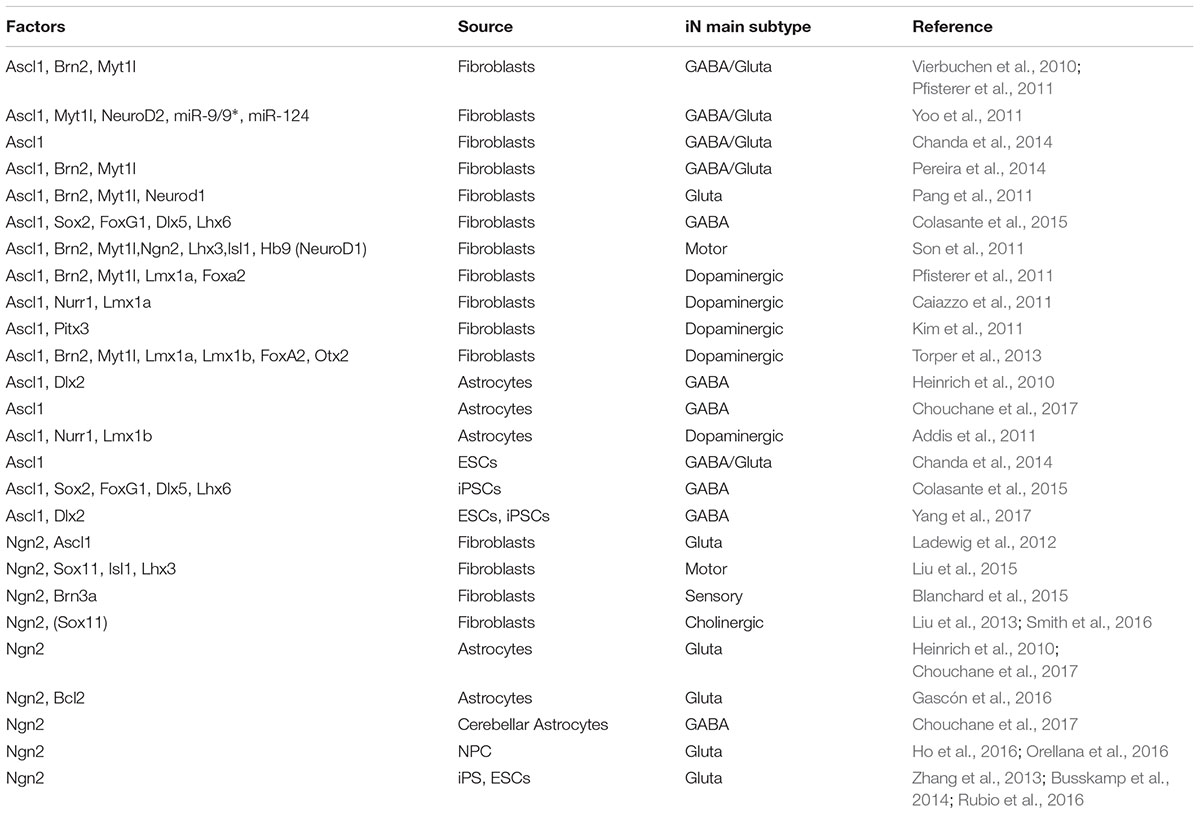
Table 1. Summary of the TF combinations that include Ascl1 or Ngn2 to directly reprogram somatic or pluripotent cells into specific iN subtypes.
Very soon in the field, an amazing difference in the ability in fibroblast-to-neuron conversion was observed between Ascl1 and its glutamatergic alter ego, Neurog2. Indeed, several studies indicated that the induction of Neurog2 cannot reprogram fibroblasts efficiently while it can generate neurons when overexpressed in ESCs, induced pluripotent stem cells (iPSCs), NPCs and astrocytes (Berninger et al., 2007; Heinrich et al., 2010; Vierbuchen et al., 2010; Liu et al., 2013; Zhang et al., 2013; Busskamp et al., 2014; Chanda et al., 2014; Masserdotti et al., 2015; Ho et al., 2016; Orellana et al., 2016; Rubio et al., 2016; Table 1). As expected, in most of these cases the neurons acquired a glutamatergic identity. Only when it is induced in murine astrocytes of cerebellar origin, Neurog2 promotes the generation of GABAergic neurons in according to its role during embryo development where it drives the differentiation of GABAergic Purkinje cells (Florio et al., 2012; Chouchane et al., 2017).
The poor efficiency of Neurog2 in the fibroblast-to-neuron conversion can be raised dramatically when Neurog2 is expressed together with other transcriptional factors and/or in the presence of small molecules in the media (Son et al., 2011; Liu et al., 2012, 2013, 2015, 2016; Aravantinou-Fatorou et al., 2015; Blanchard et al., 2015).
We tried to clarify this intriguing difference between Ascl1 and Neurog2 in reprogramming efficiency of fibroblasts comparing side by side their direct molecular targets. To this aim, we took advantage of chromatin immunoprecipitation-sequencing (ChIP-seq) data already available in the literature. Smith et al. (2016) transduced MRC-5 human fetal fibroblasts with lentiviruses expressing either ASCL1, NEUROG2, or NEUROG2 together with the small molecules forskolin and dorsomorphin to increase the reprogramming efficiency. Then, ChIP-seq analyses were performed at 2.5, 3, and 4 days after the infection for each condition and we cross-referenced the datasets merging all time points together (Figure 1). Focusing our attention on the conditions where (i) ASCL1 or (ii) NEUROG2 were induced, we found that ASCL1 and NEUROG2 share a consistent number of direct targets (1863) although maintaining many other exclusive (319+729 for ASCL1 and 811+5965 for NEUROG2) (Figure 1A).
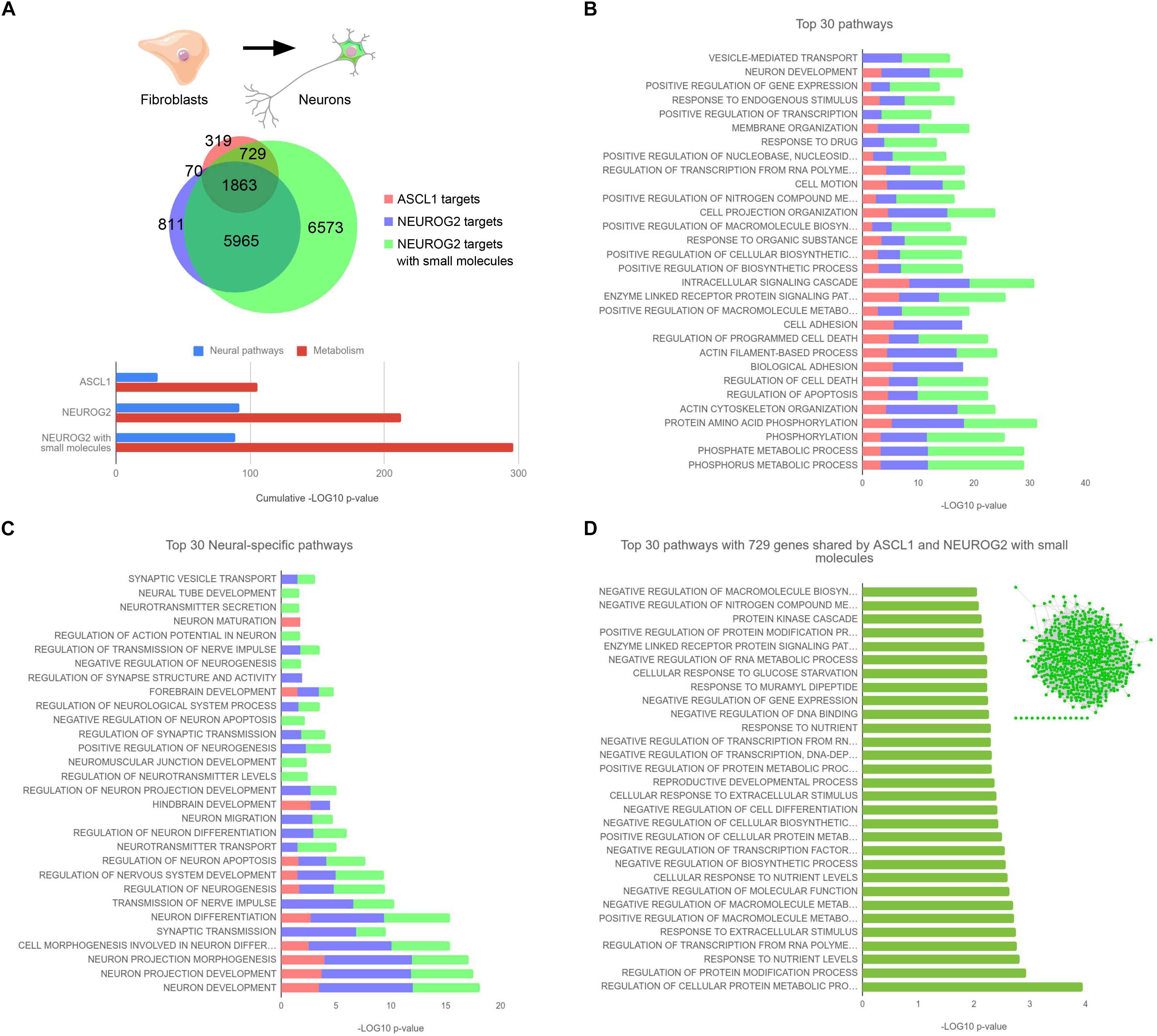
Figure 1. (A) Area-proportional Venn diagram depicting the total number of ASCL1 and NEUROG2 targets. The proportion of shared targets for the 3 experimental conditions (ASCL1, NEUROG2, and NEUROG2 with small molecules) are indicated. Bottom, result summary showing the cumulative enrichment of neural or metabolic pathways in the 3 conditions. (B) Top 30 pathways enriched in the 3 experimental conditions. (C) Top 30 neural-specific pathways in the 3 experimental conditions. (D) Top 30 pathways of the 729 genes shared by ASCL1 and NEUROG2 with small molecules. Inset: gene network generated with the GeneMania Cytoscape plugin, built in accordance with Physical interactions, Co-expression, Co-localization, corresponding Pathway, and Genetic interactions showing one single molecular hub of 715 genes, with 636 edges and a clustering coefficient of 0.135. ChIP-seq peaks were downloaded from NCBI GEO (GSE43916, GSE63621, and GSE75912) and annotated with annotatePeaks (Heinz et al., 2010). Functional enrichment analysis was performed with DAVID (Huang da et al., 2009). Filtering, statistics, and plotting were performed within the R environment.
Surprisingly, NEUROG2 targets are about nine times more abundant than those of ASCL1. Considering the difference in the reprogramming efficiency of these two TFs, we hypothesized that ASCL1 might be more efficient in the neuronal program activation by binding mainly neural genes among its targets. However, when we analyzed more deeply the top 30 GO (gene ontology) pathways targeted by ASCL1, we realized that most of them correlate with the activation of non-neural specific genes, i.e., GO related to alterations in intracellular pathways and metabolic changes (Figure 1B). Very few of them were instead related to neural differentiation program, for example, neuronal development (GO 0048666) and alterations in the cytoskeleton and plasma membrane (GO 0016044, GO 0007155, GO 0030029, and GO 0030036). Interestingly, the top 30 targets of NEUROG2 were classified in GO categories similar to the ones observed in the case of ASCL1. When we focused on the Top 30 neural-specific pathways, it clearly emerged that NEUROG2 exhibited even a higher enrichment of genes promoting neuralization in comparison to ASCL1 (Figure 1C), not confirming our initial hypothesis.
To add more information to this picture, we analyzed also ChIP-seq datasets of NEUROG2 in presence of small molecules: in this condition, the access of NEUROG2 to chromatin is even more enhanced likely due to the chromatin remodeling mediated by the small molecules possibly through the activation of SOX4 (Smith et al., 2016). In particular, in the presence of forskolin and dorsomorphin, NEUROG2 is able to bind to 729 novel genes that in the previous comparison were exclusive targets of ASCL1 (Figure 1A). Supposing that they contain the key genes responsible for the success of the reprogramming, we analyzed them more accurately. Again, we observed that these shared target genes do not belong mainly to neural categories (Figure 1D) suggesting that the activation of non-neural pathways, more than a prompt neuralization, is essential for the neuronal reprogramming in a “non-neuronal” context. Interestingly, we observe that most of these genes (715 out of 729) were either co-expressed, physically interacting, or belonging to the same molecular pathway, thus giving rise to a unique molecular network (Figure 1D, inset). This indicates that the activation of key regulatory genes that are connected to each other might guide the efficient conversion of fibroblasts into neurons. We also observed that 6573 targets are exclusive to the condition where the fibroblasts were treated with NEUROG2 and the small molecules (Figure 1A). Since NEUROG2 in the presence of small molecules generates cholinergic neurons, differently from ASCL1, we believe that these genes or at least part of them might be involved in promoting the cholinergic fate. Alternatively, we cannot exclude that those targets themselves might be responsible for the acquired success of NEUROG2 in the neuronal conversion process. In this case, the scenario would be different and would suggest that NEUROG2 plus small molecules and ASCL1 activate complementary gene regulatory networks that can independently reprogram fibroblasts into neurons. Further analysis is warranted to address this hypothesis.
Importantly, the prompt and massive neuralizing action of NEUROG2 may be beneficial for the reprogramming of cells that are more closely related to neurons (such as astrocytes) or more prone to differentiate into neurons (such as ESCs, iPSCs, or NPCs). Indeed, in agreement with this speculation and assuming that the occupancy profiles would be similar using other cells, it has been reported that NEUROG2, when overexpressed in human ESCs, generates mature neurons faster than ASCL1 (Chanda et al., 2014).
In synthesis, direct reprogramming enriched the classic knowledge on proneural genes highlighting different dynamics and kinetics in neural conversion mediated by Ascl1 and Neurog2 (summarized in Figure 1A, bottom). Those differences can underlie their different neuronal reprogramming efficiencies in different cellular lineages.
The Emerging Function of the Previously Overlooked Proneural TF Myt1l
In other studies, direct cell reprogramming uncovered the importance during neuronal differentiation of TFs whose roles remained overlooked or underestimated during embryonic development. This is the case of Myt1l, encoding for a member of the zinc finger superfamily of TFs. Little work was performed in the past to understand the role of Myt1l in neural development until it emerged as an important factor in direct reprogramming (Vierbuchen et al., 2010). Since Myt1l is specifically expressed in neurons (Kim et al., 1997; Matsushita et al., 2014) it was selected together with other 18 genes as candidates to achieve fibroblast-to-neuron conversion. After careful screening, the authors defined that the minimal cocktail of 3TF necessary to reprogram fibroblasts included Myt1l indicating its importance, previously neglected, in neuronal development. Since then, Myt1l has been used in numerous protocols to reprogram non-neuronal cells into neurons (Ambasudhan et al., 2011; Marro et al., 2011; Pang et al., 2011; Pfisterer et al., 2011; Son et al., 2011; Yoo et al., 2011; Torper et al., 2013; Victor et al., 2014; Wainger et al., 2015). The experimental evidence indicate that Myt1l alone is not sufficient to obtain neurons, but it improves instead the efficiency of conversion and the morphology of the neurons when used together with Ascl1 or miRNA (for example, in MEF (Vierbuchen et al., 2010), in human ES (Pang et al., 2011), in human fibroblasts (Victor et al., 2014). The discovery of this important role for Myt1l sparked the scientific community interest to better determine its mechanism of action during reprogramming but also in vivo during development. Studies in the reprogramming context have demonstrated that Myt1l is not a pioneer factor since it binds mainly open and active chromatin (Wapinski et al., 2013; Mall et al., 2017). In contrast, Myt1l acts as a transcriptional repressor that downregulates different cascade of non-neuronal genes, such as Notch and Wnt pathway, to promote neurogenesis (Mall et al., 2017). Importantly, Notch repression has been also confirmed in a physiological context for both Myt1l and Myt1, a gene highly homologous to Myt1l (Vasconcelos et al., 2016; Mall et al., 2017). Other data indicate the importance of Myt1l in neuronal development: Myt1l overexpression in NSCs and in vivo increases neuronal differentiation (Mall et al., 2017) and mutations in Myt1l have been associated with intellectual disability, schizophrenia and autism (Li et al., 2012; De Rubeis et al., 2014; De Rocker et al., 2015).
Defining Gene Networks Responsible for Neuronal Subtype Specification
Direct reprogramming assays have further contributed to better tracking the functional interactions among TFs during the commitment of specific neuronal subtypes.
The discovery that Ascl1 alone or with Brn2 and Myt1l is able to generate glutamatergic iN (Vierbuchen et al., 2010; Chanda et al., 2014) and that, associated with other TFs, it seems necessary to generate every type of neurons (Caiazzo et al., 2011; Kim et al., 2011; Pfisterer et al., 2011; Torper et al., 2013; Colasante et al., 2015), resulted in direct contradiction with the undebated role of Ascl1 during development in the specification of GABAergic neurons. Indeed, Ascl1 was clearly described as an activator of Dlx1/2 (Casarosa et al., 1999; Yun et al., 2002) and its ectopic expression in cortical ventricular zone (VZ) is sufficient to upregulate Dlx1/2 (Fode et al., 2000), which in turn activates GAD65/67 expression (Stuhmer et al., 2002).
Five years later the first iN derivation, Colasante et al. (2015) suggested an answer to this conundrum revealing that Ascl1 is effective in activating the GABAergic reporter GAD67-GFP during MEF to neuron conversion only if combined with either Sox2 or Foxg1. Although their role in regulating the competence of telencephalic progenitors to adopt subpallial fates had been recently proposed (Manuel et al., 2010; Ferri et al., 2013), Colasante and colleagues, going deeper on the mechanism, showed that Ascl1, Sox2, and Foxg1 strictly cooperate in determining the GABAergic fate. They showed that SOX2 and ASCL1- but not FOXG1-interact to bind and activate Dlx1/2 enhancer, but the binding is allowed only when FOXG1 is also expressed. They hypothesized a chromatin pioneer role for FOXG1 that similarly to other forkhead-box TFs (Watts et al., 2011; Iwafuchi-Doi and Zaret, 2014), might open the repressed chromatin to enable SOX2 and ASCL1 binding to the Dlx1/2 locus.
Interestingly, cooperation between these three factors for the determination of a GABAergic fate was confirmed also in vivo. Cortical VZ cells express already Sox2 and Foxg1, for this reason, it is sufficient to express Ascl1 to allow a fate switch from glutamatergic to GABAergic (Fode et al., 2000). Conversely, the silencing of either Foxg1 or Sox2 in cortical VZ is sufficient to abolish the ability of Ascl1 to induce a GABAergic neuronal fate when overexpressed in the same compartment. In accordance with this report, astrocytes over-expressing Ascl1 can be converted in GABAergic neurons as they already express Sox2 and FoxG1 endogenously (Heinrich et al., 2010; Zhang et al., 2013; Masserdotti et al., 2015). The direct link between FOXG1 and GABAergic fate emerged also when Mariani et al. (2015) observed that overexpression of the TF FOXG1 is responsible for the overproduction of GABAergic neurons in brain organoids modeling of autism spectrum disorders.
Unexpected TFs Regulating Neuronal Reprogramming
Some recent works in the reprogramming field have contributed to identify TFs not previously related to neurogenesis that were unexpectedly able to generate differentiated neurons. The large majority of them has emerged by carrying out unbiased screenings of TFs able to produce neurons (Liu et al., 2018; Tsunemoto et al., 2018). In both these studies, the screening is based on systematic combinatorial strategies that are not exclusively relying on testing TFs differentially expressed between starting cells and desired target cells. Tsunemoto et al. (2018) tested 598 pairs of TFs cloned in doxycycline-inducible lentiviruses. They infected MEFs and observed which TF pair generated neurons that were functional. Surprisingly some of the identified TFs that could convert MEFs into neurons have never been related to the generation of neurons before, such as: OCT4, a well-known factor for cell pluripotency; Myf5, known mostly as a regulator of myogenesis; and Pit1, an anterior pituitary-specific TF. The screening also identified Ptf1a (Pancreas TF-1a), a TF that has been recently shown to generate NSCs when overexpressed in MEFs (Xiao et al., 2018).
Liu et al. (2018) performed an activatory CRISPR screening in mouse ESCs to systematically identify regulators of neuronal-fate specification. Among the top 20 TFs or DNA binding proteins most efficient in neuronal conversion, some of them (Ezh2, Suz12, Maz, Nr3c1, and Sin3b) were not preferentially enriched in neural cells as no differential expression was observed between the obtained neurons and mESCs. Ezh2 and Suz12 are two Polycomb-group proteins that act as global epigenetic regulators (Margueron and Reinberg, 2011) and can promote neuronal differentiation. Transcriptomic analyses suggested that Ezh2, in particular, acts mainly by inhibiting alternative endodermal and mesodermal lineages. Regarding the TFs not related with the neuronal identity the specific mechanism of action is yet to be defined. One plausible possibility is that, when expressed at supra-physiological levels, they are able to activate overlapping genetic pathways by ectopic binding to the same transcriptional targets through their sequence homology with their related neurogenic factors. Alternatively, they may have a yet unknown role during only a short window of time along the whole process of neuronal differentiation. Additional work is needed to fully investigate these alternative scenarios.
Conclusion
Direct cell reprogramming proved to be informative in defining the dynamics and specific roles for several TFs and clarifying their molecular functions in yet unexplored gene networks. More unbiased screenings of TFs for neuronal conversion should be pursued (Liu et al., 2018; Tsunemoto et al., 2018), since they can help to better define these networks identifying remaining TFs not yet related to neuronal development but that can facilitate cell conversion. Finally, cell reprogramming studies generated a renewed interest in better deciphering TF functions during development, stimulating new studies in vivo. In overall, direct cell reprogramming can be considered as the first stage where newly TFs or unprecedented functions of well-known TFs can make their debut.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
LM performed computational analysis. GC, AR, and VB wrote the manuscript.
Funding
This work was supported by the European Research Council (AdERC #340527).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Alessandro Sessa for critical reading of the manuscript.
References
Addis, R. C., Hsu, F. C., Wright, R. L., Dichter, M. A., Coulter, D. A., and Gearhart, J. D. (2011). Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One 6:e28719. doi: 10.1371/journal.pone.0028719
Ambasudhan, R., Talantova, M., Coleman, R., Yuan, X., Zhu, S., Lipton, S. A., et al. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118. doi: 10.1016/j.stem.2011.07.002
Aravantinou-Fatorou, K., Ortega, F., Chroni-Tzartou, D., Antoniou, N., Poulopoulou, C., Politis, P. K., et al. (2015). CEND1 and NEUROGENIN2 reprogram mouse astrocytes and embryonic fibroblasts to induced neural precursors and differentiated neurons. Stem Cell Rep. 5, 405–418. doi: 10.1016/j.stemcr.2015.07.012
Au, E., Ahmed, T., Karayannis, T., Biswas, S., Gan, L., and Fishell, G. (2013). A modular gain-of-function approach to generate cortical interneuron subtypes from ES cells. Neuron 80, 1145–1158. doi: 10.1016/j.neuron.2013.09.022
Berninger, B., Costa, M. R., Koch, U., Schroeder, T., Sutor, B., Grothe, B., et al. (2007). Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 27, 8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007
Bertrand, N., Castro, D. S., and Guillemot, F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. doi: 10.1038/nrn874
Black, J. B., Adler, A. F., Wang, H. G., D’Ippolito, A. M., Hutchinson, H. A., Reddy, T. E., et al. (2016). Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell. 19, 406–414. doi: 10.1016/j.stem.2016.07.001
Blanchard, J. W., Eade, K. T., Szucs, A., Lo Sardo, V., Tsunemoto, R. K., Williams, D., et al. (2015). Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 18, 25–35. doi: 10.1038/nn.3887
Boissart, C., Nissan, X., Giraud-Triboult, K., Peschanski, M., and Benchoua, A. (2012). miR-125 potentiates early neural specification of human embryonic stem cells. Development 139, 1247–1257. doi: 10.1242/dev.073627
Britz, O., Mattar, P., Nguyen, L., Langevin, L. M., Zimmer, C., Alam, S., et al. (2006). A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex 16(Suppl. 1), i138–i151. doi: 10.1093/cercor/bhj168
Busskamp, V., Lewis, N. E., Guye, P., Ng, A. H., Shipman, S. L., Byrne, S. M., et al. (2014). Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 10:760. doi: 10.15252/msb.20145508
Caiazzo, M., Dell’Anno, M. T., Dvoretskova, E., Lazarevic, D., Taverna, S., Leo, D., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227. doi: 10.1038/nature10284
Casarosa, S., Fode, C., and Guillemot, F. (1999). Mash1 regulates neurogenesis in the ventral telencephalon. Development 126, 525–534.
Chanda, S., Ang, C. E., Davila, J., Pak, C., Mall, M., Lee, Q. Y., et al. (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3, 282–296. doi: 10.1016/j.stemcr.2014.05.020
Chouchane, M., Melo de Farias, A. R., Moura, D. M. S., Hilscher, M. M., Schroeder, T., Leao, R. N., et al. (2017). Lineage reprogramming of astroglial cells from different origins into distinct neuronal subtypes. Stem Cell Rep. 9, 162–176. doi: 10.1016/j.stemcr.2017.05.009
Colasante, G., Lignani, G., Rubio, A., Medrihan, L., Yekhlef, L., Sessa, A., et al. (2015). Rapid conversion of fibroblasts into functional forebrain gabaergic interneurons by direct genetic reprogramming. Cell Stem Cell 17, 719–734. doi: 10.1016/j.stem.2015.09.002
De Rocker, N., Vergult, S., Koolen, D., Jacobs, E., Hoischen, A., and Zeesman, S. (2015). Refinement of the critical 2p25.3 deletion region: the role of MYT1L in intellectual disability and obesity. Genet. Med. 17, 460–466. doi: 10.1038/gim.2014.124
De Rubeis, S., He, X., Goldberg, A. P., Poultney, C. S., Samocha, K., Cicek, A. E., et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. doi: 10.1038/nature13772
Ferri, A., Favaro, R., Beccari, L., Bertolini, J., Mercurio, S., Nieto-Lopez, F., et al. (2013). Sox2 is required for embryonic development of the ventral telencephalon through the activation of the ventral determinants Nkx2.1 and Shh. Development 140, 1250–1261. doi: 10.1242/dev.073411
Florio, M., Leto, K., Muzio, L., Tinterri, A., Badaloni, A., Croci, L., et al. (2012). Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development 139, 2308–2320. doi: 10.1242/dev.075861
Fode, C., Ma, Q., Casarosa, S., Ang, S. L., Anderson, D. J., and Guillemot, F. (2000). A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14, 67–80.
Gascón, S., Murenu, E., Masserdotti, G., Ortega, F., Russo, G. L., Petrik, D., et al. (2016). Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell 18, 396–409. doi: 10.1016/j.stem.2015.12.003
Gurdon, J. B. (1962). Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 4, 256–273. doi: 10.1016/0012-1606(62)90043-X
Heinrich, C., Blum, R., Gascon, S., Masserdotti, G., Tripathi, P., Sanchez, R., et al. (2010). Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 8:e1000373. doi: 10.1371/journal.pbio.1000373
Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y. C., Laslo, P., et al. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–589. doi: 10.1016/j.molcel.2010.05.004
Ho, S. M., Hartley, B. J., Tcw, J., Beaumont, M., Stafford, K., Slesinger, P. A., et al. (2016). Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods 101, 113–124. doi: 10.1016/j.ymeth.2015.11.019
Horton, S., Meredith, A., Richardson, J. A., and Johnson, J. E. (1999). Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol. Cell. Neurosci. 14, 355–369. doi: 10.1006/mcne.1999.0791
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Iwafuchi-Doi, M., and Zaret, K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692. doi: 10.1101/gad.253443.114
Kim, J., Efe, J. A., Zhu, S., Talantova, M., Yuan, X., Wang, S., et al. (2011). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843. doi: 10.1073/pnas.1103113108
Kim, J. G., Armstrong, R. C., v Agoston, D., Robinsky, A., Wiese, C., Nagle, J., et al. (1997). Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J. Neurosci. Res. 50, 272–290. doi: 10.1002/(SICI)1097-4547(19971015)50:2<272::AID-JNR16>3.0.CO;2-A
Kovach, C., Dixit, R., Li, S., Mattar, P., Wilkinson, G., Elsen, G. E., et al. (2013). Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb. Cortex 23, 1884–1900. doi: 10.1093/cercor/bhs176
Ladewig, J., Mertens, J., Kesavan, J., Doerr, J., Poppe, D., Glaue, F., et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9, 575–578. doi: 10.1038/nmeth.1972
Li, W., Wang, X., Zhao, J., Lin, J., Song, X. Q., Yang, Y., et al. (2012). Association study of myelin transcription factor 1-like polymorphisms with schizophrenia in Han Chinese population. Genes Brain Behav. 11, 87–93. doi: 10.1111/j.1601-183X.2011.00734.x
Liu, M. L., Zang, T., and Zhang, C. L. (2016). Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Rep. 14, 115–128. doi: 10.1016/j.celrep.2015.12.018
Liu, M. L., Zang, T., Zou, Y., Chang, J. C., Gibson, J. R., Huber, K. M., et al. (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4:2183. doi: 10.1038/ncomms3183
Liu, X., Li, F., Stubblefield, E. A., Blanchard, B., Richards, T. L., Larson, G. A., et al. (2012). Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 22, 321–332. doi: 10.1038/cr.2011.181
Liu, Y., Miao, Q., Yuan, J., Han, S., Zhang, P., Li, S., et al. (2015). Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J. Neurosci. 35, 9336–9355. doi: 10.1523/JNEUROSCI.3975-14.2015
Liu, Y., Yu, C., Daley, T. P., Wang, F., Cao, W. S., Bhate, S., et al. (2018). CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 23, 758–771.e8. doi: 10.1016/j.stem.2018.09.003
Mall, M., Kareta, M. S., Chanda, S., Ahlenius, H., Perotti, N., Zhou, B., et al. (2017). Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544, 245–249. doi: 10.1038/nature21722
Manuel, M., Martynoga, B., Yu, T., West, J. D., Mason, J. O., and Price, D. J. (2010). The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development 137, 487–497. doi: 10.1242/dev.039800
Margueron, R., and Reinberg, D. (2011). The polycomb complex PRC2 and its mark in life. Nature 469, 343–349. doi: 10.1038/nature09784
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-dependent dysregulation of GABA/Glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390. doi: 10.1016/j.cell.2015.06.034
Marro, S., Pang, Z. P., Yang, N., Tsai, M. C., Qu, K., Chang, H. Y., et al. (2011). Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9, 374–382. doi: 10.1016/j.stem.2011.09.002
Masserdotti, G., Gascon, S., and Gotz, M. (2016). Direct neuronal reprogramming: learning from and for development. Development 143, 2494–2510. doi: 10.1242/dev.092163
Masserdotti, G., Gillotin, S., Sutor, B., Drechsel, D., Irmler, M., Jorgensen, H. F., et al. (2015). Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17, 74–88. doi: 10.1016/j.stem.2015.05.014
Matsushita, F., Kameyama, T., Kadokawa, Y., and Marunouchi, T. (2014). Spatiotemporal expression pattern of Myt/NZF family zinc finger transcription factors during mouse nervous system development. Dev. Dyn. 243, 588–600. doi: 10.1002/dvdy.24091
Mattar, P., Britz, O., Johannes, C., Nieto, M., Ma, L., Rebeyka, A., et al. (2004). A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev. Biol. 273, 373–389. doi: 10.1016/j.ydbio.2004.06.013
Orellana, D. I., Santambrogio, P., Rubio, A., Yekhlef, L., Cancellieri, C., Dusi, S., et al. (2016). Coenzyme a corrects pathological defects in human neurons of PANK2-associated neurodegeneration. EMBO Mol. Med. 8, 1197–1211. doi: 10.15252/emmm.201606391
Pang, Z. P., Yang, N., Vierbuchen, T., Ostermeier, A., Fuentes, D. R., Yang, T. Q., et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223. doi: 10.1038/nature10202
Parras, C. M., Schuurmans, C., Scardigli, R., Kim, J., Anderson, D. J., and Guillemot, F. (2002). Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16, 324–338. doi: 10.1101/gad.940902
Pereira, M., Pfisterer, U., Rylander, D., Torper, O., Lau, S., Lundblad, M., et al. (2014). Highly efficient generation of induced neurons from human fibroblasts that survive transplantation into the adult rat brain. Sci. Rep. 4:6330. doi: 10.1038/srep06330
Pfisterer, U., Kirkeby, A., Torper, O., Wood, J., Nelander, J., Dufour, A., et al. (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 108, 10343–10348. doi: 10.1073/pnas.1105135108
Poitras, L., Ghanem, N., Hatch, G., and Ekker, M. (2007). The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development 134, 1755–1765. doi: 10.1242/dev.02845
Rubio, A., Luoni, M., Giannelli, S. G., Radice, I., Iannielli, A., Cancellieri, C., et al. (2016). Rapid and efficient CRISPR/Cas9 gene inactivation in human neurons during human pluripotent stem cell differentiation and direct reprogramming. Sci. Rep. 6:37540. doi: 10.1038/srep37540
Schuurmans, C., Armant, O., Nieto, M., Stenman, J. M., Britz, O., Klenin, N., et al. (2004). Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 23, 2892–2902. doi: 10.1038/sj.emboj.7600278
Smith, D. K., Yang, J., Liu, M. L., and Zhang, C. L. (2016). Small molecules modulate chromatin accessibility to promote neurog2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep. 7, 955–969. doi: 10.1016/j.stemcr.2016.09.013
Son, E. Y., Ichida, J. K., Wainger, B. J., Toma, J. S., Rafuse, V. F., Woolf, C. J., et al. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218. doi: 10.1016/j.stem.2011.07.014
Stuhmer, T., Puelles, L., Ekker, M., and Rubenstein, J. L. (2002). Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb. Cortex 12, 75–85. doi: 10.1093/cercor/12.1.75
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Tapscott, S. J., Davis, R. L., Thayer, M. J., Cheng, P. F., Weintraub, H., and Lassar, A. B. (1988). MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411. doi: 10.1126/science.3175662
Torper, O., Pfisterer, U., Wolf, D. A., Pereira, M., Lau, S., Jakobsson, J., et al. (2013). Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 7038–7043. doi: 10.1073/pnas.1303829110
Tsunemoto, R., Lee, S., Szucs, A., Chubukov, P., Sokolova, I., Blanchard, J. W., et al. (2018). Diverse reprogramming codes for neuronal identity. Nature 557, 375–380. doi: 10.1038/s41586-018-0103-5
Vasconcelos, F. F., Sessa, A., Laranjeira, C., Raposo, A., Teixeira, V., Hagey, D. W., et al. (2016). MyT1 counteracts the neural progenitor program to promote vertebrate neurogenesis. Cell Rep. 17, 469–483. doi: 10.1016/j.celrep.2016.09.024
Victor, M. B., Richner, M., Hermanstyne, T. O., Ransdell, J. L., Sobieski, C., Deng, P. Y., et al. (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84, 311–323. doi: 10.1016/j.neuron.2014.10.016
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Sudhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. doi: 10.1038/nature08797
Waddington, C. H. (1957). The Strategy of the Genes; A Discussion of Some Aspects of Theoretical Biology. London: Allen & Unwin.
Wainger, B. J., Buttermore, E. D., Oliveira, J. T., Mellin, C., Lee, S., Saber, W. A., et al. (2015). Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 18, 17–24. doi: 10.1038/nn.3886
Wapinski, O. L., Vierbuchen, T., Qu, K., Lee, Q. Y., Chanda, S., Fuentes, D. R., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. doi: 10.1016/j.cell.2013.09.028
Watts, J. A., Zhang, C., Klein-Szanto, A. J., Kormish, J. D., Fu, J., Zhang, M. Q., et al. (2011). Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 7:e1002277. doi: 10.1371/journal.pgen.1002277
Xiao, D., Liu, X., Zhang, M., Zou, M., Deng, Q., Sun, D., et al. (2018). Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun. 9:2865. doi: 10.1038/s41467-018-05209-1
Yang, N., Chanda, S., Marro, S., Ng, Y. H., Janas, J. A., Haag, D., et al. (2017). Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 14, 621–628. doi: 10.1038/nmeth.4291
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. doi: 10.1038/nature10323
Yun, K., Fischman, S., Johnson, J., Hrabe de Angelis, M., Weinmaster, G., and Rubenstein, J. L. (2002). Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development 129, 5029–5040.
Keywords: stem cells, cell reprogramming, neuronal differentiation, brain development, transcription factor
Citation: Colasante G, Rubio A, Massimino L and Broccoli V (2019) Direct Neuronal Reprogramming Reveals Unknown Functions for Known Transcription Factors. Front. Neurosci. 13:283. doi: 10.3389/fnins.2019.00283
Received: 12 November 2018; Accepted: 11 March 2019;
Published: 26 March 2019.
Edited by:
Annalisa Buffo, University of Turin, ItalyReviewed by:
Mengqing Xiang, Sun Yat-sen University, ChinaEumorphia Remboutsika, National and Kapodistrian University of Athens Medical School, Greece
Copyright © 2019 Colasante, Rubio, Massimino and Broccoli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vania Broccoli, YnJvY2NvbGkudmFuaWFAaHNyLml0
†These authors share first authorship
 Gaia Colasante
Gaia Colasante Alicia Rubio
Alicia Rubio Luca Massimino
Luca Massimino Vania Broccoli
Vania Broccoli