- Laboratory of Integrative Brain Sciences, Department of Biology and Center for Medical Life Science, Waseda University, Shinjuku, Japan
Gonadotropin-inhibitory hormone (GnIH) is a hypothalamic neuropeptide that was found in the brain of Japanese quail when investigating the existence of RFamide peptides in birds. GnIH was named because it decreased gonadotropin release from cultured anterior pituitary, which was located in the hypothalamo-hypophysial system. GnIH and GnIH precursor gene related peptides have a characteristic C-terminal LPXRFamide (X = L or Q) motif that is conserved in jawed vertebrates. Orthologous peptides to GnIH are also named RFamide related peptide or LPXRFamide peptide from their structure. A G-protein coupled receptor GPR147 is the primary receptor for GnIH. Similarity-based clustering of neuropeptide precursors in metazoan species indicates that GnIH precursor of vertebrates is evolutionarily related to FMRFamide precursor of mollusk and nematode. FMRFamide peptide is the first RFamide peptide that was identified from the ganglia of the venus clam. In order to infer the evolutionary history of the GnIH-GnIH receptor system we investigate the structural similarities between GnIH and its receptor and well-studied nematode Caenorhabditis elegans (C. elegans) FMRFamide-like peptides (FLPs) and their receptors. We also compare the functions of FLPs of nematode with GnIH of chordates. A multiple sequence alignment and phylogenetic analyses of GnIH, neuropeptide FF (NPFF), a paralogous peptide of GnIH, and FLP precursors have shown that GnIH and NPFF precursors belong to different clades and some FLP precursors have structural similarities to either precursor. The peptide coding regions of FLP precursors in the same clade align well with those of GnIH or NPFF precursors. Alignment of GnIH (LPXRFa) peptides of chordates and FLPs of C. elegans grouped the peptides into five groups according to the last C-terminal amino acid sequences, which were MRFa, LRFa, VRFa, IRFa, and PQRFa. Phylogenetic analysis of receptors suggested that GPR147 has evolutionary relationships with FLP receptors, which regulate reproduction, aggression, locomotion, and feeding. GnIH and some FLPs mediate the effect of stress on reproduction and behavior, which may also be a conserved property of these peptide systems. Future studies are needed to investigate the mechanism of how neuropeptide precursor genes are mutated to evolve new neuropeptides and their inheritance.
Introduction
A hypothalamic neuropeptide gonadotropin-inhibitory hormone (GnIH) was discovered in the Japanese quail (Coturnix japonica) brain, while studying the existence of Arg-Phe-NH2 (RFamide) peptides in birds, which have a characteristic C-terminal RFamide sequence (Tsutsui et al., 2000). Phe-Met-Arg-Phe-NH2 (FMRFamide) is the first RFamide peptide that was identified in the venus clam Macrocallista nimbosa ganglia, which has a cardioexcitatory function (Price and Greenberg, 1977). Since then, multiple RFamide peptides acting as hormones, neuromodulators and neurotransmitters have been found in cnidarians, nematodes, annelids, mollusks, and arthropods. Multiple immunohistochemical studies using antibodies against RFamide peptides of invertebrates suggested the presence of RFamide peptides in the central nervous system of vertebrates.
Tsutsui et al. (2000) have successfully isolated a peptide from 500 Japanese quail brains using high-performance liquid chromatography combined with a competitive enzyme-linked immunosorbent assay with an antibody against Arg-Phe-NH2. The C-terminal structure of the isolated peptide SIKPSAYLPLRFamide (Table 1) was found to be identical to the chicken LPLRFamide that was reported as the first RFamide peptide isolated in vertebrates (Dockray et al., 1983). However, the previously reported chicken LPLRFamide peptide can be the fragment of the chicken GnIH peptide that was identified to have a sequence of SIRPSAYLPLRFamide in a recent study (McConn et al., 2014). GnIH was named “gonadotropin-inhibitory hormone” because it decreased gonadotropin release from cultured quail anterior pituitary gland and located in the hypothalamo-hypophysial system (Tsutsui et al., 2000; for reviews see, Tsutsui et al., 2015, 2017; Ubuka et al., 2016).
Structure of GNIH Peptides and Their Positions in Their Precursor Proteins
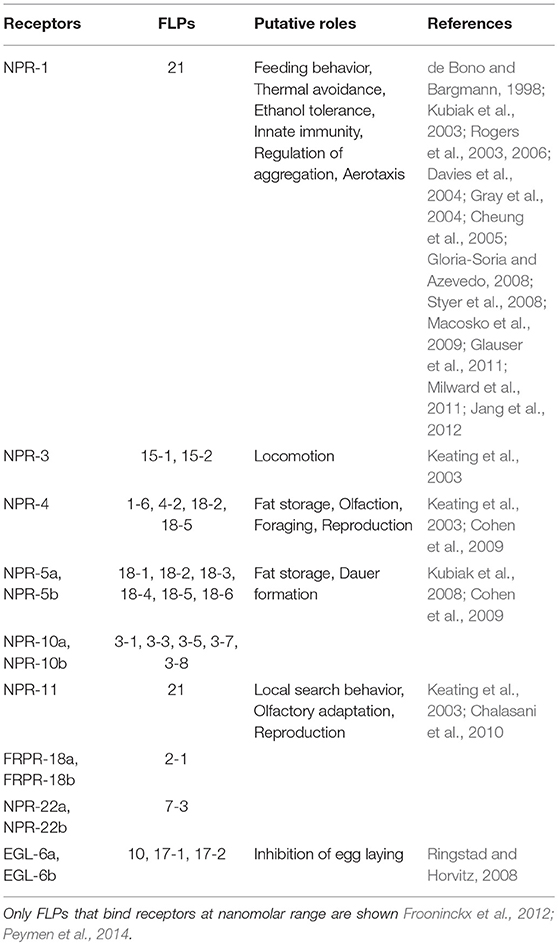
In the following year of the discovery of quail GnIH, the precursor cDNA of quail GnIH was cloned and sequenced (Satake et al., 2001). GnIH precursor is composed of 173 amino acid residues encompassing GnIH as well as two GnIH-related peptides (GnIH-RP-1 and GnIH-RP-2; Figure 1, Supplementary Figure 1, Table 1). GnIH and GnIH-related peptide sequences are flanked by an amidation signal glycine at the C-terminal as well as basic amino acids (arginine or lysine) as endoproteolytic sites on each end (Supplementary Figure 1). The translated and processed GnIH and GnIH-RPs all possess a C-terminal LPXRFamide (X = L or Q) sequence (Figure 1, Supplementary Figure 1, Table 1). Mass spectrometry has also identified the mature peptide structure of GnIH-RP-2 in addition to GnIH (Satake et al., 2001). Within the class of birds, mature GnIH peptides were also isolated in European starlings (Ubuka et al., 2008), zebra finch (Tobari et al., 2010), and chicken (McConn et al., 2014; Ubuka et al., 2016). A cDNA encoding LPXRFamide peptides in the brain of red-eared slider was recently cloned and immunoaffinity purification and mass spectrometry identified three mature LPXRFamide peptides that were encoded in the precursor protein (Ukena et al., 2016).
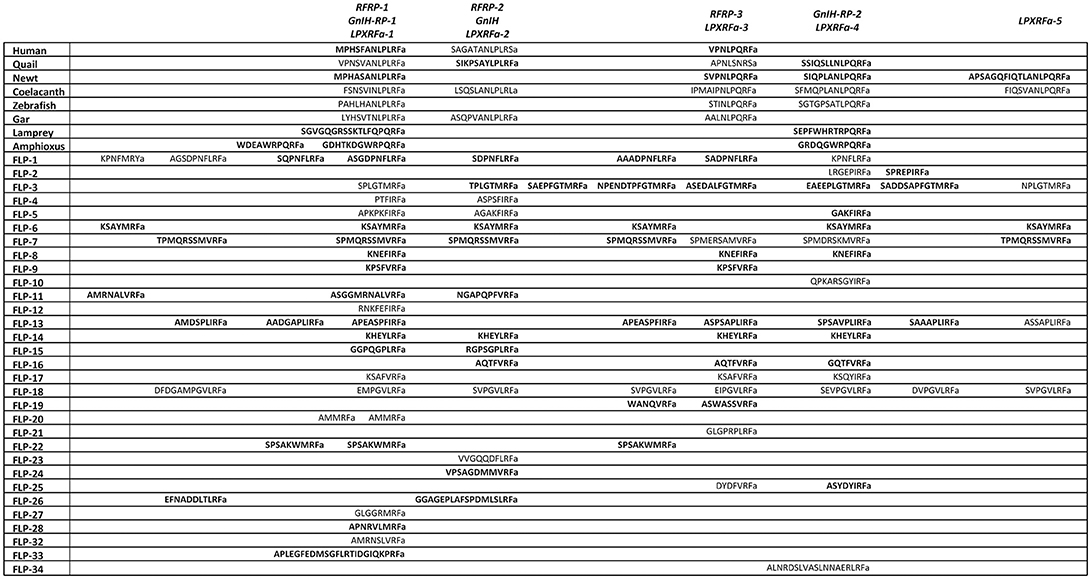
Figure 1. A schematic representation of the multiple sequence alignment of human, quail, newt, coelacanth, zebrafish, gar, lamprey, amphioxus GnIH and C. elegans FMRFamide-like peptide (FLP) precursors highlighting the sequences of identified and predicted biologically active peptides. Human, quail, newt, coelacanth, zebrafish, gar, lamprey, amphioxus GnIH and C. elegans FLP precursor polypeptides were aligned by EMBL-EBI Clustal Omega Multiple Sequence Alignment software. The positions of the identified or predicted endogenous peptide sequences in their precursors are shown in bold or normal font, respectively. The full alignment of the precursors are shown in Supplementary Figure 1. Accession numbers are human (Homo sapiens) GnIH precursor (NP_071433.3), Japanese quail (Coturnix japonica) GnIH precursor (XP_015709159.1), Japanese fire belly newt (Cynops pyrrhogaster) GnIH precursor (BAJ78290.1), West Indian Ocean coelacanth (Latimeria chalumnae) GnIH precursor (XP_005993154.1), zebrafish (Danio rerio) GnIH precursor (NP_001076418.1), spotted gar (Lepisosteus oculatus) GnIH precursor (XP_015213317.1), sea lamprey (Petromyzon marinus) GnIH precursor (BAL52329.1), Japanese amphioxus (Branchiostoma japonicum) GnIH precursor (BAO77760.1), C. elegans (Caenorhabditis elegans) FLP-1 precursor (AAC46464.1), FLP-2 precursor (NP_001024945.1), FLP-3 precursor (AAC08940.1), FLP-4 precursor (AAC08941.1), FLP-5 precursor (AAC08942.1), FLP-6 precursor (AAC08943.1), FLP-7 precursor (AAC08944.1), FLP-8 precursor (AAC08945.1), FLP-9 precursor (AAC08946.1), FLP-10 precursor (AAC08947.1), FLP-11 precursor (NP_001024752.1), FLP-12 precursor (AAC08950.1), FLP-13 precursor (AAC08951.1), FLP-14 precursor (NP_499682.2), FLP-15 precursor (NP_499820.1), FLP-16 precursor (NP_001022091.1), FLP-17 precursor (NP_503051.1), FLP-18 precursor (NP_508514.2), FLP-19 precursor (NP_509776.1), FLP-20 precursor (NP_509574.2), FLP-21 precursor (NP_505011.2), FLP-22 precursor (NP_492344.2), FLP-23 precursor (AAY18633.1), FLP-24 precursor (AAW78866.1), FLP-25 precursor (NP_001022665.1), FLP-26 precursor (NP_741827.1), FLP-27 precursor (NP_495111.1), FLP-28 precursor (NP_001024947.1), FLP-32 precursor (NP_510551.2), FLP-33 precursor (NP_871818.1), FLP-34 precursor isoform 1 (FLP-34; NP_001300170.1), FLP-34 precursor isoform 2 (FLP-34'; NP_503365.1).
In mammals, cDNAs encoding C-terminal LPXRFamide peptides were found in the gene database (Hinuma et al., 2000). Mammalian LPXRFamide peptides and related peptides were named RFamide related peptides (RFRPs) from their C-terminal structure (Hinuma et al., 2000). Human LPXRFamide precursor cDNA encodes three RFRPs (RFRP-1, RFRP-2, and RFRP-3). However, although RFRP-1 and RFRP-3 possess a C-terminal LPXRFamide (X = L or Q) motif, the predicted C-terminal sequence of RFRP-2 is RSamide (Hinuma et al., 2000; Ubuka et al., 2009c; Figure 1, Supplementary Figure 1, Table 1). The LPLRF, LPLRS, and LPQRF sequences are followed by glycine as an amidation signal, followed by arginine or lysine at the C-terminus in the human LPXRFamide peptide precursor as in birds (Supplementary Figure 1). When human GnIH (RFRP) precursor protein was aligned with GnIH precursor protein of quail, human RFRP-1, and human RFRP-2 align with quail GnIH-RP-1 and quail GnIH, respectively (Figure 1, Supplementary Figure 1). Human RFRP-3 aligns with an LPXRFa-like peptide that has a C-terminal LSNRSamide sequence but not with quail GnIH-RP-2 (Figure 1, Supplementary Figure 1). In mammals, bovine RFRP-1 (Fukusumi et al., 2001) and -3 (Yoshida et al., 2003), rat RFRP-3 (Ukena et al., 2002), Siberian hamster RFRP-1 and -3 (Ubuka et al., 2012a), macaque RFRP-3 (Ubuka et al., 2009b), and human RFRP-1 and -3 (Ubuka et al., 2009c) are identified as mature peptides by biochemical experiments (Ubuka et al., 2016).
In amphibians, bullfrog LPXRFamide peptide was identified and named as frog growth hormone-releasing hormone (fGRP) as the peptide stimulated growth hormone release (Koda et al., 2002). The precursor cDNA encoded four LPXRFamide peptides that were named as fGRP, fGRP-RP-1, fGRP-RP-2, and fGRP-RP-3 (Sawada et al., 2002a) and all peptides were identified as mature peptides (Ukena et al., 2003a). In the same year fGRP was independently identified in European green frog and named as Rana RFamide (R-RFa) Chartrel et al., 2002). LPXRFamide peptide precursor cDNA was also cloned from Japanese red-bellied newt (Chowdhury et al., 2011). The deduced precursor protein encompasses four LPXRFamide peptides that were named as nLPXRFa-1,-2,-3,-4, which were all identified as endogenous mature peptides (Chowdhury et al., 2011; Figure 1, Supplementary Figure 1, Table 1). Newt LPXRFa-2, LPXRFa-1, and LPXRFa-3 align with human RFRP-1/quail GnIH-RP-1, human RFRP-3, and quail GnIH-RP-2, respectively (Figure 1, Supplementary Figure 1, Table 1). Coelacanth is a lobe-finned fish that is evolutionarily related to tetrapods. Coelacanth LPXRFamide precursor encompasses four LPXRFamide peptides and one LPXRFamide-like peptide (Figure 1, Supplementary Figure 1, Table 1). Coelacanth LPXRFa-1, LPXRFa-2, LPXRFa-3, LPXRFa-4 align to human RFRP-1/quail GnIH-RP-1, human RFRP-2/quail GnIH, human RFRP-3, and quail GnIH-RP-2, respectively (Figure 1, Supplementary Figure 1, Table 1). Coelacanth LPXRFa-5 aligns with newt nLPXRFa-4 (Figure 1, Supplementary Figure 1, Table 1).
In teleost fishes, a cDNA encoding three LPXRFamide peptides [goldfish (gf) LPXRFa-1,-2, and -3] was first cloned from goldfish and gf LPXRFa-3 was identified as a mature peptide (Sawada et al., 2002b). The zebrafish LPXRFamide precursor gene also encodes three LPXRFamide peptides (Figure 1, Supplementary Figure 1, Table 1). Zebrafish LPXRFa-1, LPXRFa-2, and LPXRFa-3 align with human RFRP-1/quail GnIH-RP-1, human RFRP-3, and quail GnIH-RP-2, respectively (Figure 1, Supplementary Figure 1, Table 1). Spotted gar a ray-finned fish that diverged from teleost fishes before teleost specific genome duplication is regarded as an ideal model to study the evolution of fish (Braasch et al., 2016; Muñoz-Cueto et al., 2017). Three LPXRFamide peptides are found in the gar LPXRFa precursor polypeptide. Gar LPXRFa-1, LPXRFa-2, and LPXRFa-3 align to human RFRP-1/quail GnIH-RP-1, human RFRP-2/quail GnIH, and human RFRP-3, respectively (Figure 1, Supplementary Figure 1, Table 1). Lamprey is a jawless fish (agnathan) that is regarded as one of the most basal vertebrates. The lamprey precursor protein possesses C-terminal QPQRFamide and RPQRFamide peptides, but not an LPXRFamide (X = L or Q) peptide. Lamprey LPXRFa-1a and LPXRFa-2 align with human RFRP-1/quail GnIH-RP-1 and quail GnIH-RP-2, respectively (Osugi et al., 2012; Figure 1, Supplementary Figure 1, Table 1). LPXRFamide peptide precursor gene was further searched in amphioxus, one of the most basal chordates (protochordates; Osugi et al., 2014). The C-termini of the amphioxus LPXRFamide-like peptides were RPQRFamide, and again LPXRFamide peptide (X = L or Q) was not found. Three of the RPQRFamide peptides were identified as mature peptides (Osugi et al., 2014; Table 1). Amphioxus PQRFa-2 and PQRFa-3 align with human RFRP-1/quail GnIH-RP-1 and quail GnIH-RP-2, respectively (Figure 1, Supplementary Figure 1, Table 1). These results suggest that the C-terminal LPXRFamide (X = L or Q) motif was evolved in gnathostomes (jawed vertebrates; Table 1).
Neuropeptide FF (NPFF, FLFQPQRFamide) was originally isolated from the bovine brain using the antibody against FMRFamide (Yang et al., 1985). The antibody also isolated a related peptide (AGEGLSSPFWSLAAPQRFamide) from the bovine brain, which was named neuropeptide AF (NPAF; Yang et al., 1985). Both NPFF and NPAF were shown to modulate pain sensitivity in rats (Yang et al., 1985). The NPFF precursor gene was identified in the human gene (Perry et al., 1997). There are two isoforms in the human NPFF precursor protein but both proteins encode the same two C-terminal PQRFamide peptides (Figure 2). The peptides having the sequences of SQAFLFQPQRFamide (named NPSF in Perry et al., 1997) and AGEGLNSQFWSLAAPQRFamide (named NPAF in Perry et al., 1997 and Burlet-Schiltz et al., 2002) were predicted (Perry et al., 1997; Figure 2). However, only NPAF and the partial NPAF which has a sequence of SLAAPQRFamide (named NPSF in Burlet-Schiltz et al., 2002) were isolated in the human cerebrospinal fluid and their structure was determined by mass spectrometry (Burlet-Schiltz et al., 2002; Figure 2). SQAFLFQPQRFamide (named NPSF in Perry et al., 1997) or its partial FLFQPQRFamide (the same sequence as bovine NPFF) was not isolated in the human cerebrospinal fluid (Burlet-Schiltz et al., 2002; Figure 2). When human NPFF precursor isoforms were aligned with human GnIH precursor, NPSF (Perry et al., 1997) that includes bovine NPFF sequence aligns with human RFRP-1 (Ubuka et al., 2009c; Figure 2). NPAF (Perry et al., 1997; Burlet-Schiltz et al., 2002) aligns with human RFRP-3 (Ubuka et al., 2009c; Figure 2).

Figure 2. A multiple sequence alignment of human GnIH and NPFF precursors. Human GnIH and NPFF precursor polypeptides were aligned by CLUSTAL W Multiple Sequence Alignment. Multiple alignment parameters were as follows: Gap open penalty 10, Gap extension penalty 0.05, Hydrophilic residues GPSNDQERK, Weight matrix GONNET. Identified or predicted endogenous peptide sequences are underlined. Biochemically identified endogenous peptide sequences are shown in bold. Glycine (G) is an amidation signal. Lysine (K) and/or arginine (R) as endoproteolytic basic amino acids are Italicized. Accession numbers are human (Homo sapiens) GnIH precursor [Human_GnIH (RFRP), NP_071433.3], human NPFF precursor isoform 1 (Human_NPFF_isoform_1, NP_003708.1), human NPFF precursor isoform 2 (Human_NPFF_isoform_2, NP_001307225.1). Asterisk (*) indicates positions which have a single, fully conserved residue. Colon (:) indicates conservation between groups of strongly similar properties – scoring >0.5 in the Gonnet PAM 250 matrix. Period (.) indicates conservation between groups of weakly similar properties—scoring 60.5 in the Gonnet PAM 250 matrix.
GNIH Receptor and Cell Signaling
Hinuma et al. (2000) identified a specific receptor for GnIH (RFRP) and named it OT7T022, which is identical to GPR147. Bonini et al. (2000) reported two G-protein coupled receptor (GPCR) for NPFF, and named them as NPFF1 which is identical to GPR147 and NPFF2 which is identical to GPR74. GPR147 and GPR74 are paralogous (Fredriksson et al., 2003). The binding affinities and their signal transduction pathways were tested for GPR147 and GPR74, using GnIHs (RFRPs) and NPFF. GnIHs (RFRPs) had a higher affinity for GPR147, whereas NPFF had a potent agonistic activity for GPR74 (Bonini et al., 2000; Liu et al., 2001; Yoshida et al., 2003; Ikemoto and Park, 2005; Yin et al., 2005), suggesting that GPR147 (NPFF1, OT7T022) is the primary receptor for GnIH.
GnIH (RFRPs) suppresses cAMP production in cells transfected with chicken and rat GPR147 suggesting that GPR147 couples to Gαi protein which inhibits adenylate cyclase (AC) (Hinuma et al., 2000; Shimizu and Bédécarrats, 2010). The GnIH cell signaling pathway has been precisely investigated in LβT2 cells, a mouse gonadotrope cell line (Son et al., 2012). Mouse GnIHs (RFRPs) suppressed gonadotropin-releasing hormone (GnRH)-induced cAMP signaling, extracellular signal-regulated kinase (ERK) phosphorylation as well as gonadotropin subunit gene transcription by inhibiting the protein kinase A (PKA) pathway (Son et al., 2012; Ubuka et al., 2013). Because GnIH neurons extend their axons to GnRH neurons and GnRH neurons express GPR147 in birds and mammals (Ubuka et al., 2008, 2012a), GnIH cell signaling pathway was further investigated in GT1-7, a mouse GnRH neuronal cell line (Son et al., 2016). It was found that GnIH suppresses the effect of vasoactive intestinal polypeptide on AC activity, p38 and ERK phosphorylation, and c-Fos mRNA expression in GT1-7 cells (Son et al., 2016). These results suggest that GnIH specifically inhibits the AC/cAMP/PKA pathway in gonadotropes and GnRH neurons at least in birds and mammals (Son et al., 2012, 2016).
GnIH (RFRP-3) rapidly and repeatedly inhibits the firing of GnRH neurons as well, which was shown in the adult mice (Ducret et al., 2009). It was further shown that GnIH (RFRP-3) produces a non-desensitizing hyperpolarization in vesicular glutamate transporter 2 (vGluT2)-GnRH neurons by a direct postsynaptic Ba2+-sensitive K+ current mechanism (Wu et al., 2009).
Regulation of Physiology and Behavior by GNIH in Chordates
Location of GnIH Neurons and GnIH Receptors in the Brain
GnIH neuronal cell bodies are clustered in the paraventricular nucleus (PVN) of the hypothalamus in quail (Ubuka et al., 2003; Ukena et al., 2003b), house and song sparrows (Bentley et al., 2003), white-crowned sparrows (Osugi et al., 2004; Ubuka et al., 2012b), European starlings (Ubuka et al., 2008), and zebra finch (Tobari et al., 2010). GnIH-immunoreactive (ir) neuronal fibers are distributed in the diencephalic and mesencephalic areas in the brain in quail (Ukena et al., 2003b), European starlings (Ubuka et al., 2008), zebra finch (Tobari et al., 2010), and white-crowned sparrows (Ubuka et al., 2012b). GnIH neuronal axons terminate on GnRH-1 neurons in the preoptic area (POA), which release GnRH-1 at the median eminence and stimulate gonadotropin secretion from the anterior pituitary gland (King and Millar, 1982; Miyamoto et al., 1982; Sharp et al., 1990; Ubuka and Bentley, 2009, 2010; Ubuka et al., 2009a). GnIH neuronal axons also terminate on GnRH-2 neurons in the midbrain of European starlings and house sparrows (Bentley et al., 2003; Ubuka et al., 2008; Ubuka and Bentley, 2010), which stimulates reproductive behavior (Miyamoto et al., 1984; Maney et al., 1997; Temple et al., 2003; Barnett et al., 2006). It was clearly shown that GPR147 mRNA is expressed in GnRH-1 and GnRH-2 neuronal cell bodies in European starling (Ubuka et al., 2008).
GnIH neuronal cell bodies are clustered in the medial hypothalamic area, but in different brain regions or nuclei in mammals. In hamsters and mice, a cluster of GnIH neuronal cell bodies exists in the dorsomedial hypothalamic area (DMH) (Kriegsfeld et al., 2006; Ubuka et al., 2012a), whereas in rats it is in the periventricular nucleus (PerVN), and the portion between the dorsomedial (DMN) and the ventromedial (VMN) nuclei of the hypothalamus (Hinuma et al., 2000; Legagneux et al., 2009). In the macaque brain, a cluster of GnIH neuronal cell bodies principally exists in the intermediate periventricular nucleus (IPe) of the hypothalamus (Ubuka et al., 2009b), whereas in sheep it exists in the DMN and PVN (Clarke et al., 2008). GnIH-ir neuronal fibers are also widely located in the diencephalic, mesencephalic and limbic brain regions in mammals (Yano et al., 2003; Johnson et al., 2007; Ubuka et al., 2009b, 2012a). Studies in macaque, sheep and mice showed that GnIH-ir fibers are in close proximity to GnRH-1, dopamine, proopiomelanocortin (POMC), GnRH-2, neuropeptide Y (NPY), orexin, melanin-concentrating hormone (MCH), corticotrophin-releasing hormone (CRH), oxytocin, and kisspeptin neurons (Qi et al., 2009; Ubuka et al., 2009b; Poling et al., 2013). Five to ten percentage of kisspeptin neurons in the anteroventral periventricular (AVPV) region express GPR147 or GPR74, whereas approximately 25% express GPR147 or GPR74 in kisspeptin neurons in the arcuate nucleus in mice (Poling et al., 2013).
GnIH neuronal cell bodies are located in the nucleus accumbens, PVN, and upper medulla, and fibers contact the lateral processes of serotonin-ir neurons in the paraventricular organ (PVO) in the Japanese grass lizard (Kawano et al., 2006). On the other hand, GnIH neuronal cell bodies are restricted to the periventricular hypothalamic nucleus, and the fibers are densely distributed in the median eminence of red-eared slider turtle (Ukena et al., 2016).
GnIH neuronal cell bodies constitute two subpopulations in the telencephalon and diencephalon and the highest number of cell bodies are located in the POA and suprachiasmatic areas of the anuran amphibian Rana esculenta (Pinelli et al., 2015). GnIH neuronal cell bodies also exist in the medial septum, anterior commissure, dorsal hypothalamus, PerVN of the hypothalamus, and posterior tuberculum. GnIH neuronal fibers are only occasionally present in the median eminence. GnIH neuronal fibers exist in close proximity to GnRH cell bodies (Pinelli et al., 2015). GnIH (LPXRFa) neuronal cell bodies are located in the nucleus posterioris periventricularis and the nervus terminalis, and the fibers extend to nucleus lateralis tuberis pars posterioris and pituitary in goldfish and sockeye salmon (Sawada et al., 2002b; Amano et al., 2006). Sea bass GnIH (sbLPXRFa) neuronal cell bodies exist in the olfactory bulbs-terminal nerve, ventral telencephalon, caudal POA, dorsal mesencephalic tegmentum and rostral rhombencephalon, and fibers are widely distributed including the pituitary. GnIH (sbLPXRFa) neuronal fibers exist close to luteinizing hormone (LH), follicle-stimulating hormone (FSH), and growth hormone (GH) cells (Paullada-Salmerón et al., 2016a).
Regulation of Reproductive Activity
Abundant GnIH neuronal fibers exist at the median eminence in quail (Tsutsui et al., 2000; Ubuka et al., 2003; Ukena et al., 2003b) as well as in other birds (Bentley et al., 2003; Osugi et al., 2004), suggesting direct action of GnIH on gonadotropin secretion from the pituitary in birds (Tsutsui et al., 2000; Ciccone et al., 2004; Osugi et al., 2004; Ubuka et al., 2006). GPR147-positive cells are co-localized with LHβ or FSHβ mRNA containing cells the chicken pituitary (Maddineni S. et al., 2008). Abundant GnIH neuronal fibers also exist in the median eminence of sheep (Clarke et al., 2008), macaque (Ubuka et al., 2009b), and humans (Ubuka et al., 2009c), and GPR147 mRNA is expressed in the gonadotropes in the human pituitary (Ubuka et al., 2009c), suggesting that GnIH also directly inhibits gonadotropin secretion in mammals (Clarke et al., 2008; Murakami et al., 2008; Kadokawa et al., 2009; Sari et al., 2009; Pineda et al., 2010a; Smith et al., 2012). However, GnIH may not directly inhibit gonadotropin secretion in some birds and rodents, because there are only few or no GnIH neuronal fibers in the median eminence in Rufous-winged sparrows (Small et al., 2008), hamsters (Kriegsfeld et al., 2006; Ubuka et al., 2012a), and rats (Rizwan et al., 2009).
GnIH neuronal axon terminals are found in close contact with GnRH neurons in birds (Bentley et al., 2003; Ubuka et al., 2008; Tobari et al., 2010), rodents (Kriegsfeld et al., 2006; Ubuka et al., 2012a), monkeys (Ubuka et al., 2009b), and humans (Ubuka et al., 2009c), and GnIH receptor mRNA or protein is expressed in GnRH neurons in birds (Ubuka et al., 2008) and mammals (Ubuka et al., 2012a). Therefore, in addition to directly regulating the pituitary, GnIH may inhibit gonadotropins secretion by decreasing the activity of GnRH neurons (Kriegsfeld et al., 2006; Johnson et al., 2007; Anderson et al., 2009; Ducret et al., 2009; Wu et al., 2009; Pineda et al., 2010a,b; Ubuka et al., 2012a; Fraley et al., 2013; Glanowska et al., 2014; Gojska et al., 2014; Xiang et al., 2015). GnIH may also act on kisspeptin neurons to modulate reproduction in mammals (Rizwan et al., 2012).
By controlling gonadotropin secretion, GnIH regulates reproductive development and maintenance in birds (Ubuka et al., 2003, 2006; Shimizu and Bédécarrats, 2006; Maddineni S. et al., 2008; Joseph et al., 2009) and mammals (Yano et al., 2004; Quennell et al., 2010; Sethi et al., 2010; Iwasa et al., 2012; Losa-Ward et al., 2012; Poling et al., 2012; Jørgensen et al., 2014; León et al., 2014; Soga et al., 2014; Zhao et al., 2014; Semaan and Kauffman, 2015; Xiang et al., 2015). GnIH regulates estrous or menstrual cycle in female mammals (Kriegsfeld et al., 2006; Gibson et al., 2008; Smith et al., 2010; Molnár et al., 2011; Clarke et al., 2012; Li et al., 2012; Salehi et al., 2013; Jørgensen et al., 2014; Russo et al., 2015). GnIH may also regulate seasonal reproduction in birds (Bentley et al., 2003; Ubuka et al., 2005; Small et al., 2008; Chowdhury et al., 2010; Surbhi et al., 2015) and mammals (Dardente et al., 2008; Revel et al., 2008; Smith et al., 2008; Gingerich et al., 2009; Mason et al., 2010; Ubuka et al., 2012a; Harbid et al., 2013; Henson et al., 2013; Janati et al., 2013; Klosen et al., 2013; Ikeno et al., 2014; Jafarzadeh Shirazi et al., 2014; Piekarski et al., 2014; Sáenz de Miera et al., 2014).
GnIH neuronal cell bodies exist in the posterior periventricular nucleus (NPPv) of teleost fishes (Ogawa and Parhar, 2014; Biswas et al., 2015; Di Yorio et al., 2016; Ogawa et al., 2016; Paullada-Salmerón et al., 2016a). They also exist in the ventral zone of the periventricular hypothalamus of zebrafish (Spicer et al., 2017), the periventricular preoptic nucleus (NPP) as well as magnocellular preoptic nucleus (NPOm) of Indian major carp (Biswas et al., 2015). In goldfish, Indian major carp, cichlid and sea bass, GnIH cells exist in the terminal nerve/olfactory bulbs (Biswas et al., 2015; Di Yorio et al., 2016; Paullada-Salmerón et al., 2016a). GnIH cells also exist in the dorsal mesencephalic tegmentum and rhombencephalon of sea bass as well as Indian major carp (Biswas et al., 2015; Paullada-Salmerón et al., 2016a). GnIH neuronal fibers innervate broad brain area in fishes, such as olfactory bulb, ventral and dorsal telencephalon, preoptic area, hypothalamus, and midbrain area (Ogawa and Parhar, 2014; Biswas et al., 2015; Di Yorio et al., 2016; Ogawa et al., 2016; Paullada-Salmerón et al., 2016a). GnIH neuronal fibers also innervate pituitary of teleost fishes (Ogawa and Parhar, 2014; Biswas et al., 2015; Ogawa et al., 2016; Paullada-Salmerón et al., 2016a). It was shown that GnIH fibers interact with GnRH-3 cell bodies in the preoptic area of zebrafish (Spicer et al., 2017).
Inhibitory effect of GnIH on the HPG axis was shown in vivo in European sea bass (Paullada-Salmerón et al., 2016b,c), goldfish (Zhang et al., 2010; Moussavi et al., 2012, 2013; Qi et al., 2013), cinnamon clownfish (Choi et al., 2016), orange-spotted grouper (Wang et al., 2015) and flatfish (Aliaga-Guerrero et al., 2017). However, stimulatory effect of GnIH on the HPG axis was also observed in vivo (Moussavi et al., 2012, 2013; Osugi et al., 2012; Wang et al., 2015; Paullada-Salmerón et al., 2016b). Both inhibitory and stimulatory effects of GnIH administration were observed on gonadotropin expression and release from cultured fish pituitaries (Amano et al., 2006; Shahjahan et al., 2011; Moussavi et al., 2012; Qi et al., 2013; Biran et al., 2014; Di Yorio et al., 2016; Spicer et al., 2017). On the other hand, forskolin-induced CRE-luciferase activity was suppressed by GnIH administration in COS-7 cells transfected with GnIH receptors in orange spotted grouper (Wang et al., 2015) and amphioxus (Osugi et al., 2014).
Regulation of GH Secretion
GnIH (RFRP-3) increases GH-releasing hormone mRNA expression in the hypothalamus and GH release in rats (Johnson et al., 2007; Johnson and Fraley, 2008). In bullfrog, LPXRFamide peptide (fGRP) also stimulates GH release (Koda et al., 2002). On the other hand, GnIH (LPXRFa peptide) has both stimulatory and inhibitory effects on GH release and/or expression in teleost fishes (Amano et al., 2006; Moussavi et al., 2014). Goldfish GnIH (gfLPXRFa) peptides stimulate GH release from cultured sockeye salmon pituitary cells (Amano et al., 2006). On the other hand, injection of gfLPXRFa to goldfish reduces basal serum GH levels but increases pituitary GH mRNA levels (Moussavi et al., 2014). Injection of gfLPXRFa reduces serum GH and pituitary GH mRNA levels stimulated by goldfish GnRH (sGnRH and cGnRH-II) (Moussavi et al., 2014). However, administration of gfLPXRFa to goldfish pituitary cells for 24-h generally increases basal GH release and attenuates sGnRH-induced changes in GH mRNA depending on the reproductive stage (Moussavi et al., 2014). These results indicate that the effect of GnIH (LPXRFa peptide) on GH release and/or expression depends on reproductive condition in teleost fishes.
Regulation of Cardiac Contraction
Although expression levels of GnIH and GPR147 mRNAs are under detectable levels in the heart of rats, it was reported that human RFRP-1 and rat RFRP-1 rapidly and reversibly decrease the shortening and relaxation of cardiac myocytes in rats and rabbits (Nichols et al., 2010, 2012). Human RFRP-1 decreases the heart rate, stroke volume, ejection fraction, as well as cardiac output in mice (Nichols et al., 2010). It was suggested that human RFRP-1 impairs myocyte shortening by enhancing myofilament protein phosphorylation by protein kinase C (Nichols et al., 2012).
Regulation of Stress Responses
Acute and chronic immobilization stress both up-regulates GnIH expression in the DMH of rats associated with inhibition of downstream HPG activity (Kirby et al., 2009). Endotoxin administration increases GnIH and GPR147 mRNA expression in rats (Iwasa et al., 2014). The effect of short-term fasting and high-fat diet on gonadotropin suppression was less effective in GPR147-deficient male mice, suggesting the involvement of GnIH-GPR147 pathway in the suppression of gonadotropin secretion by metabolic stress (León et al., 2014). Capture-handling stress and high ambient temperature increase GnIH expression in house sparrows (Calisi et al., 2008) and chicks (Chowdhury et al., 2012), respectively. Female presence also increases GnIH mRNA expression and GnIH release by rapid increase in norepinephrine in the PVN in male quail (Tobari et al., 2014). It was demonstrated that corticosterone increases GnIH mRNA expression via glucocorticoid receptor expressed in GnIH neurons in birds and mammals (Ahmed et al., 2014; Gojska and Belsham, 2014; Son et al., 2014). Neonatal dexamethasone exposure increases GnIH cell numbers in the DMH and GPR147 mRNA in the POA in female mice with delayed vaginal opening, irregular estrous cycles and lower GnRH expression in the POA (Soga et al., 2012). GnIH (RFRP) administration induces anxiety-related behavior in rats (Kaewwongse et al., 2011), suggesting that GnIH also mediates behavioral stress responses (Ubuka et al., 2018).
Regulation of Reproductive and Aggressive Behaviors
GnIH (RFRP-3) suppresses sexual behavior of male rats (Johnson et al., 2007) and proceptive sexual behavior and motivation of female hamsters (Piekarski et al., 2013). GnIH also inhibits copulation solicitation of female white-crowned sparrows exposed to male song (Bentley et al., 2006). RNA interference (RNAi) of the GnIH gene (GnIH RNAi) reduces resting time, spontaneous production of complex vocalizations, and enhances spontaneous brief agonistic vocalizations and song production of short duration in male birds when they were exposed to playbacks of novel male songs in white-crowned sparrows, suggesting that GnIH suppresses locomotor activity and aggressiveness (Ubuka et al., 2012b). GnIH directly activates P450arom and increases neuroestrogen synthesis in the brain and suppresses socio-sexual behavior of male birds (Ubuka et al., 2014).
Regulation of Feeding Behavior
GnIH (RFRP-3) increases food intake in male rats (Johnson et al., 2007) and sheep (Clarke et al., 2012). GnIH mRNA levels are decreased in adult obese mice of both sexes (Poling et al., 2014). GnIH also stimulates food intake in chicks (Tachibana et al., 2005, 2008; McConn et al., 2014) and adult Pekin drakes (Fraley et al., 2013; see, Tsutsui and Ubuka, 2016) for a review). Studies in chicks suggested that the orexigenic effects of GnIH involves NPY, MCH, POMC neurons and opioid mu-receptor in the brain (Tachibana et al., 2008; McConn et al., 2014). Histological and physiological studies showed that NPY and POMC neurons are also regulated by GnIH in mammals (Qi et al., 2009; Ubuka et al., 2009b; Fu and van den Pol, 2010; Jacobi et al., 2013). The stimulatory effect of stress on GnIH neurons as well as the effect of GnIH on gonadotropin secretion, GH synthesis and release, and feeding activities suggest that GnIH neurones coordinate reproduction, growth and feeding activities in response to stress.
Gonadal GnIH
GnIH and GPR147 exist in the testis of Syrian hamster (Zhao et al., 2010) and the ovary of mice (Singh et al., 2011) and humans (Oishi et al., 2012). GnIH (RFRP-3) suppresses spermatogenesis, follicular development and/or steroidogenesis in mice (Singh et al., 2011; Anjum et al., 2014), pigs (Zheng et al., 2015), and humans (Oishi et al., 2012).
GnIH and GPR147 also exist in the gonads and accessory reproductive organs in passeriform and galliform birds (Bentley et al., 2008; Maddineni S. R. et al., 2008; McGuire and Bentley, 2010; McGuire et al., 2011). GnIH suppresses testosterone secretion from the testis in house sparrow (McGuire and Bentley, 2010) and European starling (McGuire et al., 2011). Corticosterone up-regulates GnIH expression in the testis of European starlings, while metabolic stress up-regulates GnIH expression in the ovaries of European starlings (McGuire et al., 2013). Restrain stress also increases GnIH mRNA expression in the testes in zebra finches (Ernst et al., 2016).
Evolutionary History of GnIH and Its Receptor
We previously searched for receptors that are structurally similar to GPR147 in the genome of mammals, birds, reptiles, amphibians, fishes, hemichordates, echinoderms, mollusks, insects, and cnidarians, to infer the evolutionary history of the GnIH system (Ubuka and Tsutsui, 2014). Neighbor joining (NJ) and maximum likelihood (ML) analyses of the amino acid sequences of the receptors grouped the receptors of vertebrates into GPR147 and GPR74. The receptors of insects were grouped into the receptor for SIFamide peptides that have a C-terminal YRKPPFNGSIFamide motif (Ubuka and Tsutsui, 2014). Human, quail, and zebrafish GPR147 was most structurally similar to SIFamide receptor within the C-terminal Famide peptide (SIFamide, FMRFamide, neuropeptide F, short neuropeptide F, drosulfakinin, myosuppressin) receptor families of fruit fly (Ubuka and Tsutsui, 2014). On the other hand, the amino acid sequences and the peptide coding regions of GnIH precursors were most similar to the FMRFamide precursor of fruit fly (Ubuka and Tsutsui, 2014). Chromosome synteny analysis of human, quail and zebrafish GnIH precursor genes and fruit fly Famide peptides precursor genes further identified a conserved synteny in vertebrate GnIH and fruit fly FMRFamide peptide precursor genes (Ubuka and Tsutsui, 2014). These results suggest that GnIH and its receptor pair have evolved from ancestral FMRFamide peptide and its receptor pair during the diversification and evolution of deuterostomian and protostomian species (Ubuka and Tsutsui, 2014).
Similarity-based clustering of various neuropeptide precursors in metazoan species forms one central cluster (Jékely, 2013). The core of the central cluster contains FMRFamide precursor of mollusk and nematode and GnIH precursor of vertebrates is evolutionarily directly related to FMRFamide precursor of mollusk and nematode (Jékely, 2013). FMRFamide-like peptides (FLPs) are the largest family of neuropeptides identified since the FMRFamide peptide was found in the venus clam Macrocallista nimbosa (Price and Greenberg, 1977; McVeigh et al., 2006). In order to further infer the evolutionary history of the GnIH-GnIH receptor system we investigate the structural similarities between GnIH and its receptor and well-studied nematode Caenorhabditis elegans (C. elegans) FLPs and their receptors and compare the functions of FLPs of nematode with GnIH of chordates.
C. elegans FLP System
The sequencing of C. elegans genome (The C. elegans Sequencing Consortium, 1998) revealed 119 neuropeptide precursor genes which is subdivided into three major families, 31 flp gene family, 40 insulin-like peptide (ins) gene family, and 48 other neuropeptide-like protein (nlp) genes (Nelson et al., 1998; Li et al., 1999; Kim and Li, 2004; Frooninckx et al., 2012). Each FLP precursor gene of C. elegans encodes one to eight FLPs (Figure 1, Supplementary Figure 1, Table 2). Flp-29 and flp-30 previously reported as parasite specific genes (McVeigh et al., 2006) are orthologous to flp-28 and flp-2, respectively (McCoy et al., 2014). Flp-31 is specific to plant parasitic nematodes (McCoy et al., 2014). Therefore, C. elegans do not have flp-29, 30, 31 (Table 2). The mature sequences of 41 FLPs were biochemically identified within total 71 FLPs encoded in the 31 flp genes (McCoy et al., 2014; Peymen et al., 2014; Table 2). FMRFamide peptide is not encoded in C. elegans flp genes unlike that of mollusks. The relatedness of FLPs is still unclear because of the large diversity of the FLP sequences (Peymen et al., 2014). It was suggested that flp-27 encodes C-terminal RXRFamide motif that is characteristic of NPF family of invertebrates (Clynen et al., 2009). FLPs are expressed in the majority of 302 neurons including sensory neurons, interneurons, and motor neurons in adult hermaphrodites and they are involved in neuroendocrine activity, locomotion, reproduction, and feeding (Peymen et al., 2014; Table 3). Predicted C. elegans neuropeptide receptors belong to rhodopsin and secretin GPCR families and 9 receptors that belong to rhodopsin family GPCR are activated by FLPs at EC50 of 1–100 nM (Frooninckx et al., 2012; Peymen et al., 2014; Table 3).
Evolutionary History of Chordate GNIH, NPFF and C. elegans FLP Precursors and Peptides
A multiple sequence alignment of the protein sequences of human, Japanese quail, fire belly newt, coelacanth, zebrafish, spotted gar, lamprey, amphioxus GnIH (RFRP, LPXRFa) peptide, and C. elegans FLP precursors was performed by EMBL-EBI Clustal Omega Multiple Sequence Alignment software (Figure 1, Supplementary Figure 1). GnIH (RFRP, LPXRFa) peptides of human, Japanese quail, fire belly newt, coelacanth, zebrafish, spotted gar aligned into five groups named from the N-terminal of their precursors, which are RFRP-1/GnIH-RP-1/LPXRFa-1, RFRP-2/GnIH/LPXRFa-2, RFRP-3/LPXRFa-3, GnIH-RP-2/LPXRFa-4, and LPXRFa-5 peptide groups (Figure 1, Supplementary Figure 1). The RFRP-1/GnIH-RP-1/LPXRFa-1 group consists of human RFRP-1, quail GnIH-RP-1, newt nLPXRFa-2, coelacanth LPXRFa-1, zebrafish LPXRFa-1, and gar LPXRFa-1, which all peptides have LPLRFamide sequence at their C-termini (Figure 1, Supplementary Figure 1, Table 1). The RFRP-2/GnIH/LPXRFa-2 group consists of human RFRP-2, quail GnIH, coelacanth LPXRFa-2, and gar LPXRFa-2, which only quail GnIH and gar LPXRFa-2 have LPLRFamide sequence at their C-termini (Figure 1, Supplementary Figure 1, Table 1). Human RFRP-2 and coelacanth LPXRFa-2 have C-terminal LPLRSamide and LPLRLamide sequences, respectively (Figure 1, Supplementary Figure 1, Table 1). The RFRP-3/LPXRFa-3 group consists of human RFRP-3, newt LPXRFa-1, coelacanth LPXRFa-3, zebrafish LPXRFa-2, and gar LPXRFa-3, which all peptides have LPQRFamide sequence at their C-termini (Figure 1, Supplementary Figure 1, Table 1). A peptide that has a sequence of APNLSNRSamide can be predicted to be produced from quail GnIH precursor, which aligns to this peptide group (Figure 1, Supplementary Figure 1, Table 1). The GnIH-RP-2/LPXRFa-4 group consists of quail GnIH-RP-2, newt LPXRFa-3, coelacanth LPXRFa-4, zebrafish LPXRFa-3, which all peptides also have LPQRFamide sequence at their C-termini (Figure 1, Supplementary Figure 1, Table 1). The LPXRFa-5 group only consists of newt LPXRFa-4 and coelacanth LPXRFa-5, which both peptides have LPQRFamide sequence at their C-termini (Figure 1, Supplementary Figure 1, Table 1). Lamprey LPXRFa-1a and amphioxus PQRFa-2 align to the RFRP-1/GnIH-RP-1/LPXRFa-1 group peptides, but these peptides have C-terminal PQRFamide sequence instead of LPLRFamide sequence (Figure 1, Supplementary Figure 1, Table 1). Lamprey lLPXRFa-2 and amphioxus PQRFa-3 align to the GnIH-RP-2/LPXRFa-4 group peptides which have C-terminal LPQRFamide sequence (Figure 1, Supplementary Figure 1, Table 1).
Twenty one, fourteen, thirteen, sixteen, and five C. elegans FLPs align to RFRP-1/GnIH-RP-1/LPXRFa-1, RFRP-2/GnIH/LPXRFa-2, RFRP-3/LPXRFa-3, GnIH-RP-2/LPXRFa-4, and LPXRFa-5 peptide groups, respectively (Figure 1, Supplementary Figure 1, Table 2). However, the C-terminal sequences of the aligned FLPs were not identical besides the RFamide sequence. There were twenty-four FLPs that did not align to either groups (Figure 1, Supplementary Figure 1, Table 2).
We next performed a phylogenetic analysis of human, quail, newt, coelacanth, zebrafish, gar, lamprey, amphioxus GnIH, human, quail, turtle, zebrafish, gar, lamprey, amphioxus NPFF, fruit fly FMRFamide, and C. elegans FLP precursors (Figure 3, Supplementary Figure 2). Molecular phylogenetic analysis was performed by ML method using MEGA7 (Kumar et al., 2016). The subclades of human, quail, newt, coelacanth, zebrafish, gar, lamprey GnIH (Osugi et al., 2012, LPXRFamide), and human, quail, turtle, zebrafish, gar, lamprey NPFF (Osugi et al., 2006, PQRFamide) precursors formed a larger clade with FLP-1, 3, 5, 8, 12, 14, 17, 18, 19, 21, 24, 27, 28, 34 (Figure 3). Amphioxus RFamide peptide precursors and fruit fly FMRFamide precursors formed a different clade with FLP-2, 6, 7, 11, 15, 16, 22, 23, 26, 32, 33 (Figure 3). These results suggest that GnIH (LPXRFamide peptide) and NPFF (PQRFamide peptide) precursor genes of vertebrates are diverged from their common ancestral gene by gene duplication (Osugi et al., 2006, 2012, 2014).
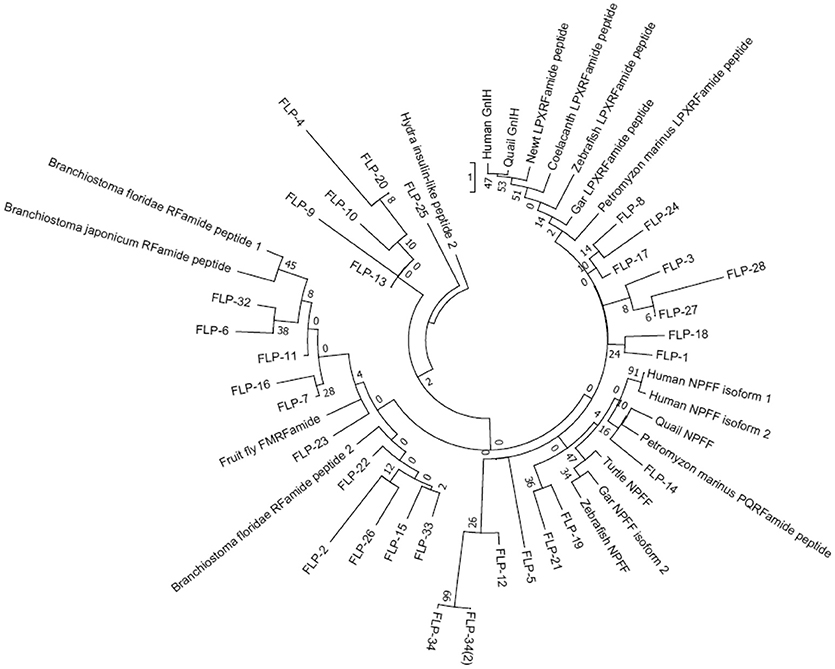
Figure 3. A phylogenetic analysis of human, quail, newt, coelacanth, zebrafish, gar, lamprey, amphioxus GnIH, human, quail, turtle, zebrafish, gar, lamprey, amphioxus NPFF, fruit fly FMRFamide and C. elegans FMRFamide-like peptide (FLP) precursors. Human, quail, newt, coelacanth, zebrafish, gar, lamprey, amphioxus GnIH, human, quail, turtle, zebrafish, gar, lamprey, amphioxus NPFF, fruit fly FMRFamide, and C. elegans FLP precursor polypeptides were aligned by CLUSTALW Multiple Sequence Alignment. Multiple alignment parameters were as follows: Gap open penalty 10, Gap extension penalty 0.2, Protein weight matrix GONNET with residue-specific and hydrophilic penalties. Molecular phylogenetic analysis was performed by Maximum Likelihood method using MEGA7 (Kumar et al., 2016). The Maximum Likelihood method was based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood is shown. Initial tree for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated by using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn with branch lengths measured in the number of substitutions per site. The analysis involved 51 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 32 positions in the final dataset. The phylogeny was tested by 50 Bootstrap replications. Accession numbers are human (Homo sapiens) GnIH precursor (Human GnIH; NP_071433.3), Japanese quail (Coturnix japonica) GnIH precursor (Quail GnIH; XP_015709159.1), Japanese fire belly newt (Cynops pyrrhogaster) GnIH precursor (Newt LPXRFamide peptide; BAJ78290.1), West Indian Ocean coelacanth (Latimeria chalumnae) GnIH precursor (Coelacanth LPXRFamide peptide; XP_005993154.1), zebrafish (Danio rerio) GnIH precursor (Zebrafish LPXRFamide peptide, NP_001076418.1), spotted gar (Lepisosteus oculatus) GnIH precursor (Gar LPXRFamide peptide; XP_015213317.1), sea lamprey (Petromyzon marinus) GnIH precursor (Petromyzon marinus LPXRFamide peptide; BAL52329.1), Japanese amphioxus (Branchiostoma japonicum) GnIH precursor (Branchiostoma japonicum RFamide peptide; BAO77760.1), human NPFF precursor isoform 1 (Human NPFF isoform 1; NP_003708.1), human NPFF precursor isoform 2 (Human NPFF isoform 2; NP_001307225.1), Japanese quail NPFF precursor (Quail NPFF; XP_015705838.1), Western painted turtle (Chrysemys picta bellii) NPFF precursor (Turtle NPFF; XP_005307776.1), zebrafish NPFF precursor (Zebrafish NPFF; BAF34891.1), spotted gar NPFF precursor isoform X2 (Gar NPFF isoform 2; XP_015199730.1), sea lamprey NPFF precursor (Petromyzon marinus PQRFamide peptide; BAE79779.1), Florida lancelet (Branchiostoma floridae) RFamide precursor 1 (Branchiostoma floridae RFamide peptide 1; XP_002599251.1), Florida lancelet RFamide precursor 2 (Branchiostoma floridae RFamide peptide 2; XP_002609543.1), Fruit fly (Drosophila melanogaster) FMRFamide precursor (Fruit fly FMRFamide; NP_523669.2), C. elegans (Caenorhabditis elegans) FLP-1 precursor (AAC46464.1), FLP-2 precursor (NP_001024945.1), FLP-3 precursor (AAC08940.1), FLP-4 precursor (AAC08941.1), FLP-5 precursor (AAC08942.1), FLP-6 precursor (AAC08943.1), FLP-7 precursor (AAC08944.1), FLP-8 precursor (AAC08945.1), FLP-9 precursor (AAC08946.1), FLP-10 precursor (AAC08947.1), FLP-11 precursor (NP_001024752.1), FLP-12 precursor (AAC08950.1), FLP-13 precursor (AAC08951.1), FLP-14 precursor (NP_499682.2), FLP-15 precursor (NP_499820.1), FLP-16 precursor (NP_001022091.1), FLP-17 precursor (NP_503051.1), FLP-18 precursor (NP_508514.2), FLP-19 precursor (NP_509776.1), FLP-20 precursor (NP_509574.2), FLP-21 precursor (NP_505011.2), FLP-22 precursor (NP_492344.2), FLP-23 precursor (AAY18633.1), FLP-24 precursor (AAW78866.1), FLP-25 precursor (NP_001022665.1), FLP-26 precursor (NP_741827.1), FLP-27 precursor (NP_495111.1), FLP-28 precursor (NP_001024947.1), FLP-32 precursor (NP_510551.2), FLP-33 precursor (NP_871818.1), FLP-34 precursor isoform 1 (FLP-34; NP_001300170.1), FLP-34 precursor isoform 2 (FLP-34'; NP_503365.1). Hydra (Hydra vulgaris) insulin-like peptide 2 precursor sequence (Hydra insulin-like peptide 2; ADA67986.1) served as outgroup (root) of the tree.
We then investigated the structural similarity of the predicted or biochemically identified human, Japanese quail, fire belly newt, coelacanth, zebrafish, spotted gar, lamprey, and amphioxus GnIH (RFRP, LPXRFamide) peptides and C. elegans FLPs by using NJ method (Saitou and Nei, 1987). This evolutionary analyses were also conducted using an online software MEGA7 (Kumar et al., 2016). Interestingly, 97 peptides analyzed were grouped into five groups that have characteristic C-terminals, MRFa, LRFa, VRFa, PQRFa, and IRFa (Figure 4, Table 4). Gar LPXRFa-1 and 2, zebrafish LPXRFa-1, coelacanth LPXRFa-1 and 2, newt nLPXRFa-2, quail GnIH and GnIH-RP-1, human RFRP-1 and RFRP-2 were grouped in the LRFa group with FLP-1-1, 2, 3, 4, 5, 6, 7, FLP-14, FLP-15-1, 2, FLP-18-1, 2, 3, 4, 5, 6, FLP-21, FLP-23, FLP-26-1, 2, FLP-33, and FLP-34 (Figure 4, Table 4). Amphioxus PQRFa-1, 2, 3, lamprey lLPXRFa-1a, 2, gar LPXRFa-3, zebrafish LPXRFa-2, 3, coelacanth LPXRFa-3, 4, 5, newt nLPXRFa-1, 3, 4, quail GnIH-RP-2 and human RFRP-3 formed a single group evolutionarily related to FLPs in the VRFa and IRFa groups (Figure 4, Table 4). FLPs produced from the same precursor were generally grouped in the same peptide groups because of the same or similar C-terminal amino acid sequences (Table 4). The exceptions are FLP-1-8 (MRFa group) and other FLP-1 peptides (LRFa group) encoded in flp-1, FLP-17-1 (VRFa group) and FLP-17-2 (IRFa group) encoded in flp-17, and FLP-25-1 (VRFa group) and FLP-25-2 (IRFa group) encoded in flp-25 (Tables 2, 4).
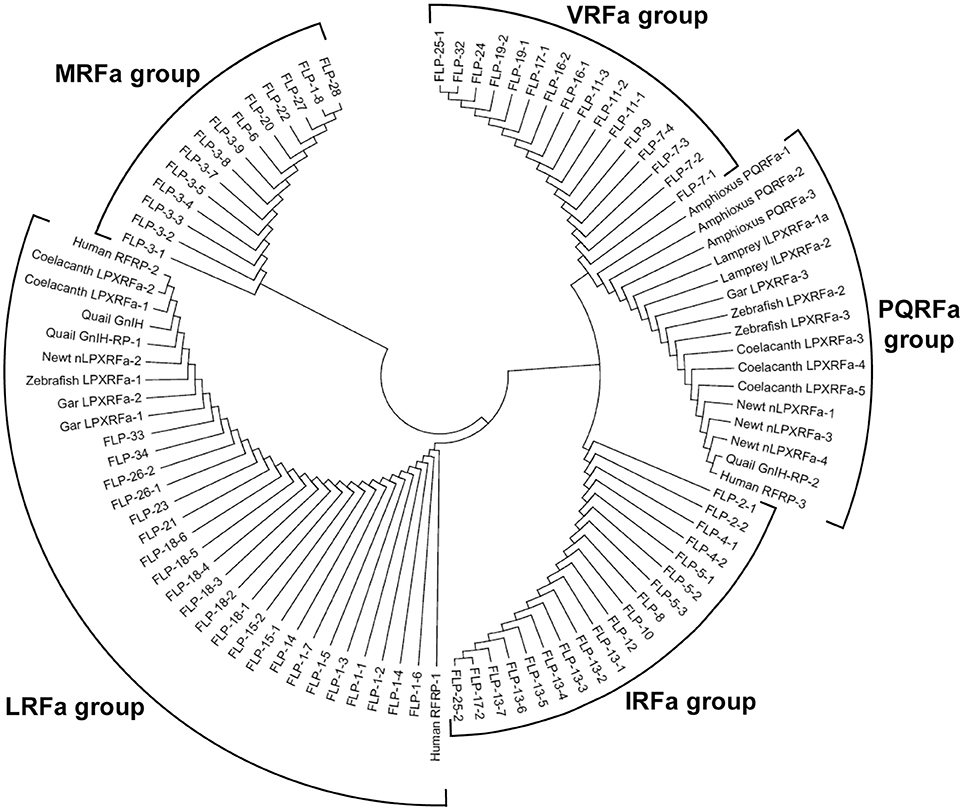
Figure 4. A phylogenetic analysis of human, quail, newt, coelacanth, zebrafish, gar, lamprey, and amphioxus GnIH (LPXRFa) peptides and C. elegans FMRFamide-like peptides (FLPs). Predicted or biochemically identified endogenous peptides shown in Table 3 were analyzed. The accession numbers of their precursor proteins are shown in the legend of Figure 1. The phylogenetic tree was constructed by Neighbor Joining method (Saitou and Nei, 1987) with the proportion of different sites statistical substitution model using MEGA 7 (Kumar et al., 2016).
Evolutionary History of Human GNIH and NPFF Receptors and C. elegans FLP Receptors
A recent phylogenomic study of GnRH receptors identified that GnIH/NPFF receptors have representation in both bilaterian and non-bilaterian lineages (Plachetzki et al., 2016) suggesting that GnIH/NPFF receptors evolved before the divergence of bilaterian and non-bilaterian species. In this study, we investigated the evolutionary history of human GnIH receptors (GPR147, GPR74) and C. elegans FLP receptors by ML method using MEGA7 (Kumar et al., 2016; Figure 5, Supplementary Figure 3). GPR147 and GPR74 were suggested to have strong evolutionary relationships with neuropeptide receptor (NPR)-22a and 22b, followed by NPR-11, NPR-1, NPR-3, NPR-5a and 5b, NPR-4a, 4b, 4c, 4d, NPR-10a and 10b (Figure 5). EGL-6a and 6b, FRPR-18a, 18b, 18c formed a different clade (Figure 5).
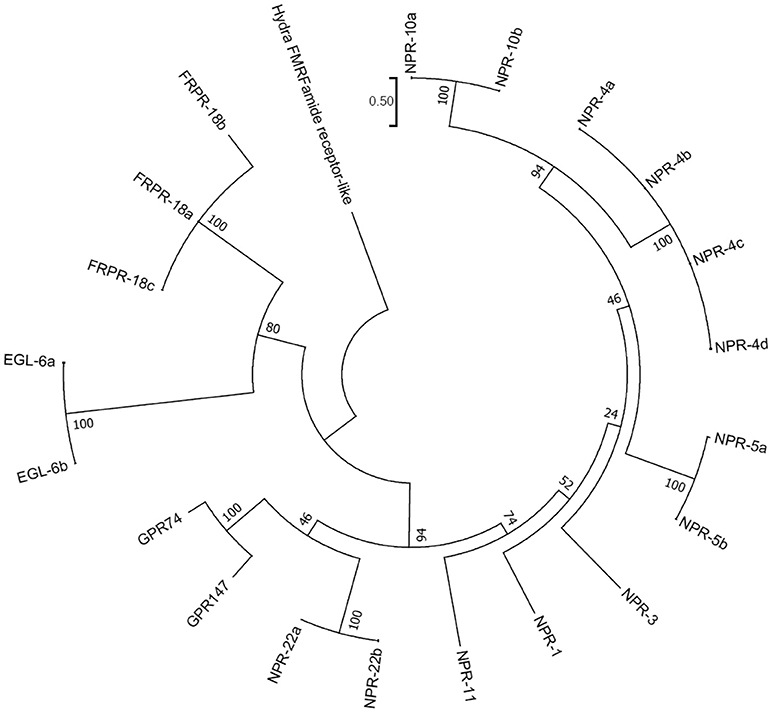
Figure 5. A phylogenetic analysis of human GnIH receptors (GPR147, GPR74) and C. elegans FMRFamide-like peptide (FLP) receptors. Human GnIH receptors (GPR147, GPR74) and C. elegans FMRFamide-like peptide (FLP) receptor amino acid sequences were aligned by CLUSTALW Multiple Sequence Alignment. Multiple alignment parameters were Gap open penalty 10, Gap extension penalty 0.2, Protein weight matrix GONNET with residue-specific and hydrophilic penalties. Molecular phylogenetic analysis was performed by Maximum Likelihood method using MEGA7 (Kumar et al., 2016). The Maximum Likelihood method was based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood is shown. Initial tree for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated by using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn with branch lengths measured in the number of substitutions per site. The analysis involved 21 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 282 positions in the final dataset. The phylogeny was tested by 50 Bootstrap replications. The accession numbers of human GnIH receptors are GPR147 (NPFF1; NP_071429.1), GPR74 (NPFF2; AAG41398.1). WormBase IDs of C. elegans FLP receptors are NPR-1 (WP:CE06941), NPR-3 (WP:CE08056), NPR-4 isoform a (WP:CE37317), NPR-4 isoform b (WP:CE50076), NPR-4 isoform c (WP:CE50063), NPR-4 isoform d (WP:CE50035), NPR-5a (WP:CE33345), NPR-5b (WP:CE36962), NPR-10a (WP:CE19767), NPR-10b (WP:CE36989), NPR-11 (WP:CE47199), FRPR-18 isoform a (WP:CE28679), FRPR-18 isoform b (WP:CE29349), FRPR-18 isoform c (WP:CE52203), NPR-22 isoform a (WP:CE31260), NPR-22 isoform b (WP:CE38456), EGL-6a (WP:CE04219), EGL-6b (WP:CE43400). Predicted hydra (Hydra vulgaris) FMRFamide receptor-like protein sequence (Hydra FMRFamide receptor-like; XP_012564736.1) served as outgroup (root) of the tree.
Roles of C. elegans FLPS and Their Receptors
NPR-1 is the first GPCR that was deorphanized in C. elegans (Kubiak et al., 2003; Rogers et al., 2003). It was shown that NPR-1 is involved in various functions, such as feeding behavior, thermal avoidance, ethanol tolerance, and innate immunity (de Bono and Bargmann, 1998; Davies et al., 2004; Gray et al., 2004; Cheung et al., 2005; Rogers et al., 2006; Gloria-Soria and Azevedo, 2008; Styer et al., 2008; Glauser et al., 2011; Milward et al., 2011; Jang et al., 2012; Table 3). NPR-1 signaling occurs through Gαi (Kubiak et al., 2003; Rogers et al., 2003). NPR-1 was suggested to suppress aggregation by inhibiting RMG neuron that is a hub of gap junction network that connects sensory neurons (Macosko et al., 2009). The RMG neuron connects five sensory neurons that are known to trigger aggregation. NPR-1 inhibits this gap junction driven activation of RMG neuron. Deletion of NPR-1 increases the threshold for heat avoidance involving RMG neuron (Glauser et al., 2011; Jang et al., 2012). NPR-1 also suppresses aerotaxis behavior in the presence of food by inhibiting ERX, AQR, PQR, and SDQ neurons (Cheung et al., 2005; Chang et al., 2006). NPR-1 binds FLP-21 with high affinity (Table 3).
It was shown that FLPs encoded by flp-18 are potent ligands of NPR-5a and NPR-5b, which are the splice variants of npr-5 (Kubiak et al., 2008; Cohen et al., 2009; Table 3). NPR-4 is also activated by FLP-18 peptides (Cohen et al., 2009; Table 3). Flp-18 loss of function or npr-4 and npr-5 deletion mutants display dauer formation, foraging defects, accumulation of excess intestinal fats and reduce aerobic metabolism (Cohen et al., 2009). It is hypothesized that detection of nutrition by sensory neurons (AWC, AFD, ASE) induces FLP-18 peptides release from AIY interneurons. FLP-18 peptides induces fat accumulation by acting on NPR-4 in intestine and NPR-5 in ciliated sensory neurons. NPR-4 in RIV and AVA neurons modulates responses to odor and foraging behavior. FLP-18 peptides also regulate dauer formation by acting on NPR-5 in ASJ neurons (Cohen et al., 2009).
Two GPCR isoforms EGL-6a and EGL-6b inhibit egg-laying (Ringstad and Horvitz, 2008). FLP-10, FLP-17-1, and FLP-17-2 activate EGL-6 and inhibit egg-laying via Gαi (Ringstad and Horvitz, 2008). Unfavorable conditions stimulate the release of FLP-17-1 and FLP-17-2 from BAG neurons, which inhibit HSN motor neurons via EGL-6. HSN motor neurons stimulate vulval muscles that are involved in egg-laying (Trent et al., 1983; White et al., 1986). FLP-10 release from vulva and spermatheca also inhibits egg-laying via EGL-6 (Kim and Li, 2004; Ringstad and Horvitz, 2008).
NPR-3 RNAi significantly impairs locomotion and some animals will be paralyzed. Green fluorescent protein reporter construct for NPR-3 indicated that NPR-3 is expressed in all excitatory and inhibitory motor neurons that have their cell bodies in the ventral nerve cord (Keating et al., 2003). NPR-3 binds FLP-15-1, 2 with high affinities (Table 3). RNAi for NPR-4 and NPR-11 significantly decreases the number of egg laid (Keating et al., 2003). NPR-4 binds FLP-1-6, FLP-4-2, FLP-18-2, 5 with high affinities (Table 3). On the other hand, NPR-11 binds FLP-21 with a high affinity (Table 3).
Conserved Function of GNIH, FLPS, and Their Receptors
As summarized earlier GnIH regulates reproductive activity in chordates by acting on GnRH neurons in the brain or gonadotropes in the pituitary or within the gonads (Hinuma et al., 2000; Tsutsui et al., 2000; Fukusumi et al., 2001; Ukena et al., 2002, 2003b; Bentley et al., 2003, 2008; Ubuka et al., 2003, 2005, 2006, 2008, 2009b,c, 2012a, 2013; Yano et al., 2003, 2004; Yoshida et al., 2003; Ciccone et al., 2004; Osugi et al., 2004, 2012; Amano et al., 2006; Kriegsfeld et al., 2006; Shimizu and Bédécarrats, 2006, 2010; Johnson et al., 2007; Clarke et al., 2008, 2012; Dardente et al., 2008; Gibson et al., 2008; Johnson and Fraley, 2008; Maddineni S. et al., 2008; Maddineni S. R. et al., 2008; Murakami et al., 2008; Revel et al., 2008; Small et al., 2008; Smith et al., 2008, 2010, 2012; Anderson et al., 2009; Ducret et al., 2009; Gingerich et al., 2009; Joseph et al., 2009; Kadokawa et al., 2009; Legagneux et al., 2009; Qi et al., 2009, 2013; Rizwan et al., 2009, 2012; Sari et al., 2009; Wu et al., 2009; Chowdhury et al., 2010; Mason et al., 2010; McGuire and Bentley, 2010; Pineda et al., 2010a,b; Quennell et al., 2010; Sethi et al., 2010; Tobari et al., 2010; Zhang et al., 2010; Zhao et al., 2010, 2014; McGuire et al., 2011; Molnár et al., 2011; Shahjahan et al., 2011; Singh et al., 2011; Iwasa et al., 2012; Li et al., 2012; Losa-Ward et al., 2012; Moussavi et al., 2012, 2013, 2014; Oishi et al., 2012; Poling et al., 2012, 2013; Son et al., 2012, 2016; Fraley et al., 2013; Harbid et al., 2013; Henson et al., 2013; Janati et al., 2013; Klosen et al., 2013; Salehi et al., 2013; Anjum et al., 2014; Biran et al., 2014; Glanowska et al., 2014; Gojska et al., 2014; Ikeno et al., 2014; Jafarzadeh Shirazi et al., 2014; Jørgensen et al., 2014; León et al., 2014; Ogawa and Parhar, 2014; Piekarski et al., 2014; Sáenz de Miera et al., 2014; Soga et al., 2014; Biswas et al., 2015; Russo et al., 2015; Semaan and Kauffman, 2015; Surbhi et al., 2015; Wang et al., 2015; Xiang et al., 2015; Zheng et al., 2015; Choi et al., 2016; Di Yorio et al., 2016; Ogawa et al., 2016; Paullada-Salmerón et al., 2016a,b,c; Aliaga-Guerrero et al., 2017; Muñoz-Cueto et al., 2017; Spicer et al., 2017). It is interesting that many of C. elegans FLP receptors such as NPR-4, NPR-11, EGL-6 and their ligands are involved in the regulation of reproductive activities (Table 3). It is especially interesting that EGL-6a and EGL-6b activated by FLP-10, FLP-17-1, and FLP-17-2 inhibit egg-laying via Gαi (Ringstad and Horvitz, 2008), which is analogous to inhibition of reproductive activities by GnIH via GPR147 in vertebrates. Expression of GnIH or GnIH neuronal activity is stimulated by stress and hence it is thought that GnIH mediates the inhibitory effects of stress on reproduction (Calisi et al., 2008; Kirby et al., 2009; Chowdhury et al., 2012; Soga et al., 2012; McGuire et al., 2013; Ahmed et al., 2014; Gojska and Belsham, 2014; Iwasa et al., 2014; Son et al., 2014; Tobari et al., 2014; Ernst et al., 2016). It was also shown in C. elegans that unfavorable conditions stimulate the release of FLP-17-1 and FLP-17-2, which inhibit egg-laying via EGL-6 (Trent et al., 1983; White et al., 1986; Kim and Li, 2004; Ringstad and Horvitz, 2008). Thus, mediation of the inhibitory effect of stress on reproductive activities may be a conserved property of GnIH and FLP systems.
GnIH is further hypothesized to be a general mediator of behavioral stress responses as GnIH suppresses locomotor activity, aggression, and reproductive behavior via GPR147 (Liu et al., 2001; Bentley et al., 2006; Kaewwongse et al., 2011; Ubuka et al., 2012b, 2014, 2018; Piekarski et al., 2013). NPR-1 activated by FLP-21 is involved in thermal avoidance behavior, ethanol tolerance and suppresses aggregation through Gαi (de Bono and Bargmann, 1998; Kubiak et al., 2003, 2008; Rogers et al., 2003, 2006; Davies et al., 2004; Gray et al., 2004; Cheung et al., 2005; Chang et al., 2006; Gloria-Soria and Azevedo, 2008; Styer et al., 2008; Macosko et al., 2009; Glauser et al., 2011; Milward et al., 2011; Jang et al., 2012; Table 3), which is analogous to the action of GnIH mediating the effect of stress on behavior (Ubuka et al., 2018). NPR-3 activated by FLP-15-1, FLP-15-2 also regulates locomotion (Keating et al., 2003; Table 3).
GnIH stimulates feeding behavior in rats (Johnson et al., 2007), sheep (Clarke et al., 2012), chicks (Tachibana et al., 2005, 2008; McConn et al., 2014), and Pekin drakes (Fraley et al., 2013) and GnIH mRNA expression is reduced in adult obese mice (Poling et al., 2014; see Tsutsui and Ubuka, 2016 for a review). Loss of function of FLP-18 or its receptors NPR-4, NPR-5 induces foraging defects, accumulation of excess intestinal fats and reduction in aerobic metabolism (Kubiak et al., 2008; Cohen et al., 2009; Table 3). Regulation of feeding behavior and metabolism may also be a conserved property of GnIH and FLPs, although the precise mechanism is not understood.
Conclusion
In order to infer the evolutionary history of the GnIH-GnIH receptor system, we compared the structures and functions of GnIH and its receptor of chordates with C. elegans FLPs and their receptors. One or two C-terminal LPLRFamide peptides and one to three C-terminal LPQRFamide peptides were encoded in the LPXRFamide (X = L or Q) precursor polypeptide genes of jawed vertebrates (human, quail, newt, coelacanth, zebrafish, gar). Orthologous LPXRFamide precursor polypeptide genes of lamprey and amphioxus encoded only two or three C-terminal PQRFamide peptides. Each FLP precursor gene encodes one to eight FLPs that have generally the same C-terminal sequences especially the last three amino acids. A multiple sequence alignment and phylogenetic analyses of GnIH, NPFF and FLP precursors (Figures 3, 6, Supplementary Figure 2) have shown that GnIH and NPFF precursors belong to different clades and there are FLPs that have structural similarities to either precursor. FLP-1, 3, 8, 17, 18, 24, 27, and 28 precursors form a clade with GnIH precursors, while FLP-14, 19, and 21 precursors form a different clade with NPFF precursors (Figures 3, 6). Although the peptide coding regions of FLP precursors in the same clade align well with those of GnIH or NPFF precursors, the sequence similarities of the peptides within the aligned precursors were weak (Figure 6). On the other hand, alignment of GnIH (LPXRFa) peptides of chordates and FLPs of C. elegans grouped the peptides into five groups according to the last C-terminal amino acid sequences, which were MRFa, LRFa, VRFa, IRFa, and PQRFa. C-terminal LPLRFamide peptides of jawed vertebrates were all in the LRFa group with other FLPs. On the other hand, C-terminal LPQRFamide peptides of jawed vertebrates and C-terminal PQRFamide peptides of lamprey and amphioxus were grouped in the PQRFa group excluding FLPs. C-terminal LPLRFamide peptides may be the original form of LPXRFamide (X = L or Q) peptides as many FLPs have the C-terminal LRFa sequence.
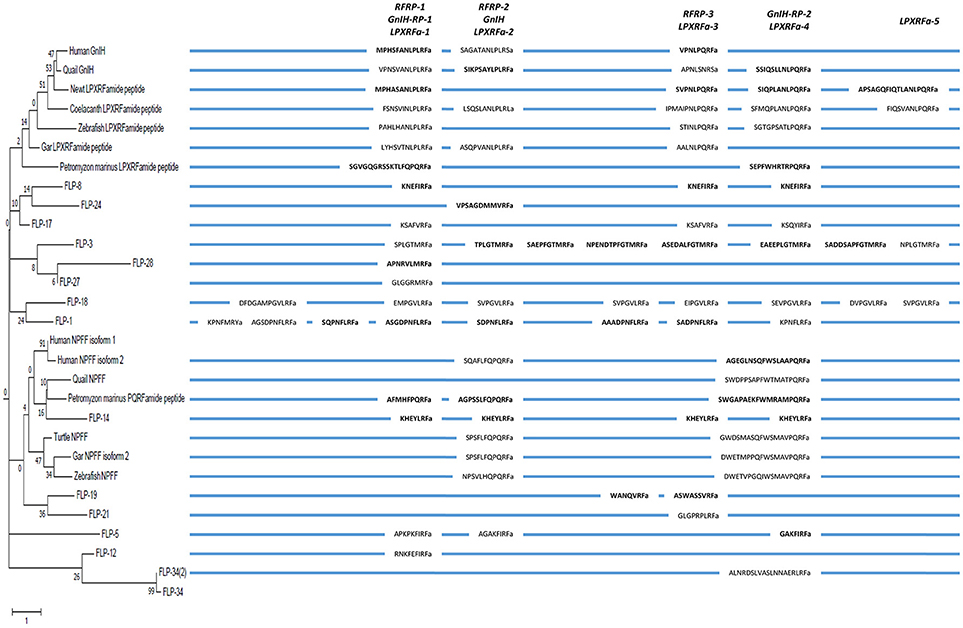
Figure 6. A schematic representation of the multiple sequence alignment of human, quail, newt, coelacanth, zebrafish, gar, lamprey GnIH, human, quail, turtle, zebrafish, gar, lamprey NPFF, and C. elegans FMRFamide-like peptide (FLP) precursors highlighting the sequences of identified and predicted biologically active peptides. Each precursor is shown next to the branch of the partial phylogenetic tree of human, quail, newt, coelacanth, zebrafish, gar, lamprey, amphioxus GnIH, human, quail, turtle, zebrafish, gar, lamprey, amphioxus NPFF, fruit fly FMRFamide and C. elegans FLP precursors (Figure 3). Note that the alignment of the peptides is based on multiple sequence alignment of all precursors (Supplementary Figure 2). Therefore, human NPSF (NPFF) is aligned to human RFRP-2 instead of human RFRP-1 (Figure 2).
Phylogenetic analysis of GnIH receptors and FLP receptors suggested that GPR147 and GPR74 have a strong evolutionary relationship with NPR-22, followed by NPR-11, NPR-1, NPR-5, NPR-4, and NPR-10. It is interesting that these receptors regulate reproduction, locomotion and feeding as GnIH and GPR147. It is also important that NPR-11 and NPR-3 bind FLP-21 and FLP-15-1,-2, respectively, peptides which all have a C-terminal PLRFamide sequence. GnIH and some FLPs mediate the effect of stress on reproduction and behavior, which may also be a conserved property of these peptide systems. Future studies are needed to investigate how neuropeptide precursor genes are mutated to evolve new neuropeptides and their inheritance.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2018.00747/full#supplementary-material
References
Ahmed, A. A., Ma, W., Ni, Y., Wang, S., and Zhao, R. (2014). Corticosterone in ovo modifies aggressive behaviors and reproductive performances through alterations of the hypothalamic-pituitary-gonadal axis in the chicken. Anim. Reprod. Sci. 146, 193–201. doi: 10.1016/j.anireprosci.2014.02.013
Aliaga-Guerrero, M., Paullada-Salmerón, J. A., Piquer, V., Mañanós, E. L., and Muñoz-Cueto, J. A. (2017). Gonadotropin-inhibitory hormone in the flatfish, Solea senegalensis: molecular cloning, brain localization and physiological effects. J. Comp. Neurol. 526, 349–370. doi: 10.1002/cne.24339
Amano, M., Moriyama, S., Iigo, M., Kitamura, S., Amiya, N., Yamamori, K., et al. (2006). Novel fish hypothalamic neuropeptides stimulate the release of gonadotrophins and growth hormone from the pituitary of sockeye salmon. J. Endocrinol. 188, 417–423. doi: 10.1677/joe.1.06494
Anderson, G. M., Relf, H. L., Rizwan, M. Z., and Evans, J. J. (2009). Central and peripheral effects of RFamide-related peptide-3 on luteinizing hormone and prolactin secretion in rats. Endocrinology 150, 1834–1840. doi: 10.1210/en.2008-1359
Anjum, S., Krishna, A., and Tsutsui, K. (2014). Inhibitory roles of the mammalian GnIH ortholog RFRP3 in testicular activities in adult mice. J. Endocrinol. 223, 79–91. doi: 10.1530/JOE-14-0333
Barnett, D. K., Bunnell, T. M., Millar, R. P., and Abbott, D. H. (2006). Gonadotropin-releasing hormone II stimulates female sexual behavior in marmoset monkeys. Endocrinology 147, 615–623. doi: 10.1210/en.2005-0662
Bentley, G. E., Jensen, J. P., Kaur, G. J., Wacker, D. W., Tsutsui, K., and Wingfield, J. C. (2006). Rapid inhibition of female sexual behavior by gonadotropin-inhibitory hormone (GnIH). Horm. Behav. 49, 550–555. doi: 10.1016/j.yhbeh.2005.12.005
Bentley, G. E., Perfito, N., Ukena, K., Tsutsui, K., and Wingfield, J. C. (2003). Gonadotropin-inhibitory peptide in song sparrows (Melospiza melodia) in different reproductive conditions, and in house sparrows (Passer domesticus) relative to chicken-gonadotropin-releasing hormone. J. Neuroendocrinol. 15, 794–802. doi: 10.1046/j.1365-2826.2003.01062.x
Bentley, G. E., Ubuka, T., McGuire, N. L., Chowdhury, V. S., Morita, Y., Yano, T., et al. (2008). Gonadotropin-inhibitory hormone and its receptor in the avian reproductive system. Gen. Comp. Endocrinol. 156, 34–43. doi: 10.1016/j.ygcen.2007.10.003
Biran, J., Golan, M., Mizrahi, N., Ogawa, S., Parhar, I. S., and Levavi-Sivan, B. (2014). LPXRFa, the piscine ortholog of GnIH, and LPXRF receptor positively regulate gonadotropin secretion in Tilapia (Oreochromis niloticus). Endocrinology 155, 4391–4401. doi: 10.1210/en.2013-2047
Biswas, S., Jadhao, A. G., Pinelli, C., Palande, N. V., and Tsutsui, K. (2015). GnIH and GnRH expressions in the central nervous system and pituitary of Indian major carp, Labeo rohita during ontogeny: an immunocytochemical study. Gen. Comp. Endocrinol. 220, 88–92. doi: 10.1016/j.ygcen.2014.06.005
Bonini, J. A., Jones, K. A., Adham, N., Forray, C., Artymyshyn, R., Durkin, M. M., et al. (2000). Identification and characterization of two G protein-coupled receptors for neuropeptide FF. J. Biol. Chem. 275, 39324–39331. doi: 10.1074/jbc.M004385200
Braasch, I., Gehrke, A. R., Smith, J. J., Kawasaki, K., Manousaki, T., Pasquier, J., et al. (2016). The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427–437. doi: 10.1038/ng.3526
Burlet-Schiltz, O., Mazarguil, H., Sol, J. C., Chaynes, P., Monsarrat, B., Zajac, J. M., et al. (2002). Identification of neuropeptide FF-related peptides in human cerebrospinal fluid by mass spectrometry. FEBS. Lett. 532, 313–318.
Calisi, R. M., Rizzo, N. O., and Bentley, G. E. (2008). Seasonal differences in hypothalamic EGR-1 and GnIH expression following capture-handling stress in house sparrows (Passer domesticus). Gen. Comp. Endocrinol. 157, 283–287. doi: 10.1016/j.ygcen.2008.05.010
Chalasani, S. H., Kato, S., Albrecht, D. R., Nakagawa, T., Abbott, L. F., and Bargmann, C. I. (2010). Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat. Neurosci. 13, 615–621. doi: 10.1038/nn.2526
Chang, A. J., Chronis, N., Karow, D. S., Marletta, M. A., and Bargmann, C. I. (2006). A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 4:e274. doi: 10.1371/journal.pbio.0040274
Chartrel, N., Dujardin, C., Leprince, J., Desrues, L., Tonon, M. C., Cellier, E., et al. (2002). Isolation, characterization, and distribution of a novel neuropeptide, Rana RFamide (R-RFa), in the brain of the European green frog Rana esculenta. J. Comp. Neurol. 448, 111–127. doi: 10.1002/cne.10253
Cheung, B. H., Cohen, M., Rogers, C., Albayram, O., and de Bono, M. (2005). Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr. Biol. 15, 905–917. doi: 10.1016/j.cub.2005.04.017
Choi, Y. J., Kim, N. N., Habibi, H. R., and Choi, C. Y. (2016). Effects of gonadotropin inhibitory hormone or gonadotropin-releasing hormone on reproduction-related genes in the protandrous cinnamon clownfish, Amphiprion melanopus. Gen. Comp. Endocrinol. 235, 89–99. doi: 10.1016/j.ygcen.2016.06.010
Chowdhury, V. S., Tomonaga, S., Nishimura, S., Tabata, S., Cockrem, J. F., Tsutsui, K., et al. (2012). Hypothalamic gonadotropin-inhibitory hormone precursor mRNA is increased during depressed food intake in heat-exposed chicks. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 162, 227–233. doi: 10.1016/j.cbpa.2012.03.009
Chowdhury, V. S., Ubuka, T., Osugi, T., Shimura, T., and Tsutsui, K. (2011). Identification, localization and expression of LPXRFamide peptides, and melatonin-dependent induction of their precursor mRNA in the newt brain. J. Endocrinol. 209, 211–220. doi: 10.1530/JOE-10-0494
Chowdhury, V. S., Yamamoto, K., Ubuka, T., Bentley, G. E., Hattori, A., and Tsutsui, K. (2010). Melatonin stimulates the release of gonadotropin-inhibitory hormone by the avian hypothalamus. Endocrinology 151, 271–280. doi: 10.1210/en.2009-0908
Ciccone, N. A., Dunn, I. C., Boswell, T., Tsutsui, K., Ubuka, T., Ukena, K., et al. (2004). Gonadotrophin inhibitory hormone depresses gonadotrophin alpha and follicle-stimulating hormone beta subunit expression in the pituitary of the domestic chicken. J. Neuroendocrinol. 16, 999–1006. doi: 10.1111/j.1365-2826.2005.01260.x
Clarke, I. J., Sari, I. P., Qi, Y., Smith, J. T., Parkington, H. C., Ubuka, T., et al. (2008). Potent action of RFamide-related peptide-3 on pituitary gonadotropes indicative of a hypophysiotropic role in the negative regulation of gonadotropin secretion. Endocrinology 149, 5811–5821. doi: 10.1210/en.2008-0575
Clarke, I. J., Smith, J. T., Henry, B. A., Oldfield, B. J., Stefanidis, A., Millar, R. P., et al. (2012). Gonadotropin-inhibitory hormone is a hypothalamic peptide that provides a molecular switch between reproduction and feeding. Neuroendocrinology 95, 305–316. doi: 10.1159/000332822
Clynen, E., Husson, S. J., and Schoofs, L. (2009). Identification of new members of the (short) neuropeptide F family in Locusts and Caenorhabditis elegans. Ann. NY Acad. Sci. 1163, 60–74. doi: 10.1111/j.1749-6632.2008.03624.x
Cohen, M., Reale, V., Olofsson, B., Knights, A., Evans, P., and de Bono, M. (2009). Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 9, 375–385. doi: 10.1016/j.cmet.2009.02.003
Dardente, H., Birnie, M., Lincoln, G. A., and Hazlerigg, D. G. (2008). RFamide-related peptide and its cognate receptor in the sheep: cDNA cloning, mRNA distribution in the hypothalamus and the effect of photoperiod. J. Neuroendocrinol. 20, 1252–1259. doi: 10.1111/j.1365-2826.2008.01784.x
Davies, A. G., Bettinger, J. C., Thiele, T. R., Judy, M. E., and McIntire, S. L. (2004). Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron 42, 731–743. doi: 10.1016/j.neuron.2004.05.004
de Bono, M., and Bargmann, C. I. (1998). Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94, 679–689.
Di Yorio, M. P., Pérez Sirkin, D. I., Delgadin, T. H., Shimizu, A., Tsutsui, K., Somoza, G. M., et al. (2016). Gonadotropin-inhibitory hormone in the cichlid fish Cichlasoma dimerus: structure, brain distribution and differential effects on the secretion of gonadotropins and growth hormone. J. Neuroendocrinol. 28:5. doi: 10.1111/jne.12377
Dockray, G. J., Reeve, J. R. Jr, Shively, J., Gayton, R. J., and Barnard, C. S. (1983). A novel active pentapeptide from chicken brain identified by antibodies to FMRFamide. Nature 305, 328–330.
Ducret, E., Anderson, G. M., and Herbison, A. E. (2009). RFamide-related peptide-3, a mammalian gonadotropin-inhibitory hormone ortholog, regulates gonadotropin-releasing hormone neuron firing in the mouse. Endocrinology 150, 2799–2804. doi: 10.1210/en.2008-1623
Ernst, D. K., Lynn, S. E., and Bentley, G. E. (2016). Differential response of GnIH in the brain and gonads following acute stress in a songbird. Gen. Comp. Endocrinol. 227, 51–57. doi: 10.1016/j.ygcen.2015.05.016
Fraley, G. S., Coombs, E., Gerometta, E., Colton, S., Sharp, P. J., Li, Q., et al. (2013). Distribution and sequence of gonadotropin-inhibitory hormone and its potential role as a molecular link between feeding and reproductive systems in the Pekin duck (Anas platyrhynchos domestica). Gen. Comp. Endocrinol. 184, 103–110. doi: 10.1016/j.ygcen.2012.11.026
Fredriksson, R., Lagerström, M. C., Lundin, L. G., and Schiöth, H. B. (2003). The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272. doi: 10.1124/mol.63.6.1256
Frooninckx, L., Van Rompay, L., Temmerman, L., Van Sinay, E., Beets, I., Janssen, T., et al. (2012). Neuropeptide GPCRs in C. elegans. Front. Endocrinol. 3:167. doi: 10.3389/fendo.2012.00167
Fu, L. Y., and van den Pol, A. N. (2010). Kisspeptin directly excites anorexigenic proopiomelanocortin neurons but inhibits orexigenic neuropeptide Y cells by an indirect synaptic mechanism. J. Neurosci. 30, 10205–19. doi: 10.1523/JNEUROSCI.2098-10.2010
Fukusumi, S., Habata, Y., Yoshida, H., Iijima, N., Kawamata, Y., Hosoya, M., et al. (2001). Characteristics and distribution of endogenous RFamide-related peptide-1. Biochim. Biophys. Acta 1540, 221–232. doi: 10.1016/S0167-4889(01)00135-5
Gibson, E. M., Humber, S. A., Jain, S., Williams, W. P. III., Zhao, S., Bentley, G. E., et al. (2008). Alterations in RFamide-related peptide expression are coordinated with the preovulatory luteinizing hormone surge. Endocrinology 149, 4958–4969. doi: 10.1210/en.2008-0316
Gingerich, S., Wang, X., Lee, P. K., Dhillon, S. S., Chalmers, J. A., Koletar, M. M., et al. (2009). The generation of an array of clonal, immortalized cell models from the rat hypothalamus: analysis of melatonin effects on kisspeptin and gonadotropin-inhibitory hormone neurons. Neuroscience 162, 1134–1140. doi: 10.1016/j.neuroscience.2009.05.026
Glanowska, K. M., Burger, L. L., and Moenter, S. M. (2014). Development of gonadotropin-releasing hormone secretion and pituitary response. J. Neurosci. 34, 15060–15069. doi: 10.1523/JNEUROSCI.2200-14.2014
Glauser, D. A., Chen, W. C., Agin, R., Macinnis, B. L., Hellman, A. B., Garrity, P. A., et al. (2011). Heat avoidance is regulated by transient receptor potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genetics 188, 91–103. doi: 10.1534/genetics.111.127100
Gloria-Soria, A., and Azevedo, R. B. (2008). npr-1 Regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr. Biol. 18, 1694–1699. doi: 10.1016/j.cub.2008.09.043
Gojska, N. M., and Belsham, D. D. (2014). Glucocorticoid receptor-mediated regulation of Rfrp (GnIH) and Gpr147 (GnIH-R) synthesis in immortalized hypothalamic neurons. Mol. Cell. Endocrinol. 384, 23–31. doi: 10.1016/j.mce.2013.12.015
Gojska, N. M., Friedman, Z., and Belsham, D. D. (2014). Direct regulation of gonadotrophin-releasing hormone (GnRH) transcription by RF-amide-related peptide-3 and kisspeptin in a novel GnRH-secreting cell line, mHypoA-GnRH/GFP. J. Neuroendocrinol. 26, 888–897. doi: 10.1111/jne.12225
Gray, J. M., Karow, D. S., Lu, H., Chang, A. J., Chang, J. S., Ellis, R. E., et al. (2004). Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430, 317–322. doi: 10.1038/nature02714
Harbid, A. A., McLeod, B. J., Caraty, A., and Anderson, G. M. (2013). Seasonal changes in RFamide-related peptide-3 neurons in the hypothalamus of a seasonally breeding marsupial species, the brushtail possum (Trichosurus vulpecula). J. Comp. Neurol. 521, 3030–3041. doi: 10.1002/cne.23328
Henson, J. R., Carter, S. N., and Freeman, D. A. (2013). Exogenous T3 elicits long day-like alterations in testis size and the RFamides Kisspeptin and gonadotropin-inhibitory hormone in short-day Siberian hamsters. J. Biol. Rhythm. 28, 193–200. doi: 10.1177/0748730413487974
Hinuma, S., Shintani, Y., Fukusumi, S., Iijima, N., Matsumoto, Y., Hosoya, M., et al. (2000). New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nat. Cell Biol. 2, 703–708. doi: 10.1038/35036326
Ikemoto, T., and Park, M. K. (2005). Chicken RFamide-related peptide (GnIH) and two distinct receptor subtypes: identification, molecular characterization, and evolutionary considerations. J. Reprod. Dev. 51, 359–377. doi: 10.1262/jrd.16087
Ikeno, T., Weil, Z. M., and Nelson, R. J. (2014). Dim light at night disrupts the short-day response in Siberian hamsters. Gen. Comp. Endocrinol. 197, 56–64. doi: 10.1016/j.ygcen.2013.12.005
Iwasa, T., Matsuzaki, T., Murakami, M., Kinouchi, R., Osugi, T., Gereltsetseg, G., et al. (2012). Developmental changes in the mammalian gonadotropin-inhibitory hormone (GnIH) ortholog RFamide-related peptide (RFRP) and its cognate receptor GPR147 in the rat hypothalamus. Int. J. Dev. Neurosci. 30, 31–37. doi: 10.1016/j.ijdevneu.2011.10.003
Iwasa, T., Matsuzaki, T., Tungalagsuvd, A., Munkhzaya, M., Kawami, T., Niki, H., et al. (2014). Hypothalamic Kiss1 and RFRP gene expressions are changed by a high dose of lipopolysaccharide in female rats. Horm. Behav. 66, 309–316. doi: 10.1016/j.yhbeh.2014.06.007
Jacobi, J. S., Coleman, H. A., Enriori, P. J., Parkington, H. C., Li, Q., Pereira, A., et al. (2013). Paradoxical effect of gonadotrophin-inhibiting hormone to negatively regulate neuropeptide Y neurones in mouse arcuate nucleus. J. Neuroendocrinol. 25, 1308–1317. doi: 10.1111/jne.12114
Jafarzadeh Shirazi, M. R., Zamiri, M. J., Salehi, M. S., Moradi, S., Tamadon, A., Namavar, M. R., et al. (2014). Differential expression of RFamide-related peptide, a mammalian gonadotrophin-inhibitory hormone orthologue, and kisspeptin in the hypothalamus of abadeh ecotype does during breeding and anoestrous seasons. J. Neuroendocrinol. 26, 186–194. doi: 10.1111/jne.12137
Janati, A., Talbi, R., Klosen, P., Mikkelsen, J. D., Magoul, R., Simonneaux, V., et al. (2013). Distribution and seasonal variation in hypothalamic RF-amide peptides in a semi-desert rodent, the jerboa. J. Neuroendocrinol. 25, 402–411. doi: 10.1111/jne.12015
Jang, H., Kim, K., Neal, S. J., Macosko, E., Kim, D., Butcher, R. A., et al. (2012). Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75, 585–592. doi: 10.1016/j.neuron.2012.06.034
Jékely, G. (2013). Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 8702–8707. doi: 10.1073/pnas.1221833110
Johnson, M. A., and Fraley, G. S. (2008). Rat RFRP-3 alters hypothalamic GHRH expression and growth hormone secretion but does not affect KiSS-1 gene expression or the onset of puberty in male rats. Neuroendocrinology 88, 305–315. doi: 10.1159/000145718
Johnson, M. A., Tsutsui, K., and Fraley, G. S. (2007). Rat RFamide-related peptide-3 stimulates GH secretion, inhibits LH secretion, and has variable effects on sex behavior in the adult male rat. Horm. Behav. 51, 171–180. doi: 10.1016/j.yhbeh.2006.09.009
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282.
Jørgensen, S. R., Andersen, M. D., Overgaard, A., and Mikkelsen, J. D. (2014). Changes in RFamide-related peptide-1 (RFRP-1)-immunoreactivity during postnatal development and the estrous cycle. Endocrinology 155, 4402–4410. doi: 10.1210/en.2014-1274
Joseph, N. T., Morgan, K., Sellar, R., McBride, D., Millar, R. P., and Dunn, I. C. (2009). The chicken type III GnRH receptor homologue is predominantly expressed in the pituitary, and exhibits similar ligand selectivity to the type I receptor. J. Endocrinol. 202, 179–190. doi: 10.1677/JOE-08-0544
Kadokawa, H., Shibata, M., Tanaka, Y., Kojima, T., Matsumoto, K., Oshima, K., et al. (2009). Bovine C-terminal octapeptide of RFamide-related peptide-3 suppresses luteinizing hormone (LH) secretion from the pituitary as well as pulsatile LH secretion in bovines. Domest. Anim. Endocrinol. 36, 219–224. doi: 10.1016/j.domaniend.2009.02.001
Kaewwongse, M., Takayanagi, Y., and Onaka, T. (2011). Effects of RFamide-related peptide (RFRP)-1 and RFRP-3 on oxytocin release and anxiety-related behaviour in rats. J. Neuroendocrinol. 23, 20–27. doi: 10.1111/j.1365-2826.2010.02077.x
Kawano, E., Takahata, Y., Oishi, T., Ukena, K., Tsutsui, K., and Tamotsu, S. (2006). Neural interaction of gonadotropin-regulating hormone immunoreactive neurons and the suprachiasmatic nucleus with the paraventricular organ in the Japanese grass lizard (Takydromus tachydromoides). Zool. Sci. 23, 277–287. doi: 10.2108/zsj.23.277
Keating, C. D., Kriek, N., Daniels, M., Ashcroft, N. R., Hopper, N. A., Siney, E. J., et al. (2003). Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr. Biol. 13, 1715–1720. doi: 10.1016/j.cub.2003.09.003
Kim, K., and Li, C. (2004). Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 475, 540–550. doi: 10.1002/cne.20189
King, J. A., and Millar, R. P. (1982). Structure of chicken hypothalamic luteinizing hormone-releasing hormone. I. Structural determination on partially purified material. J. Biol. Chem. 257, 10722–10728.
Kirby, E. D., Geraghty, A. C., Ubuka, T., Bentley, G. E., and Kaufer, D. (2009). Stress increases putative gonadotropin inhibitory hormone and decreases luteinizing hormone in male rats. Proc. Natl. Acad. Sci. U.S.A. 106, 11324–11329. doi: 10.1073/pnas.0901176106
Klosen, P., Sébert, M. E., Rasri, K., Laran-Chich, M. P., and Simonneaux, V. (2013). TSH restores a summer phenotype in photoinhibited mammals via the RF-amides RFRP3 and kisspeptin. FASEB J. 27, 2677–2686. doi: 10.1096/fj.13-229559
Koda, A., Ukena, K., Teranishi, H., Ohta, S., Yamamoto, K., Kikuyama, S., et al. (2002). A novel amphibian hypothalamic neuropeptide: isolation, localization, and biological activity. Endocrinology 143, 411–419. doi: 10.1210/endo.143.2.8630
Krajniak, K. G. (2013). Invertebrate FMRFamide related peptides. Protein Pept. Lett. 20, 647–670. doi: 10.2174/0929866511320060005
Kriegsfeld, L. J., Mei, D. F., Bentley, G. E., Ubuka, T., Mason, A. O., Inoue, K., et al. (2006). Identification and characterization of a gonadotropin-inhibitory system in the brains of mammals. Proc. Natl. Acad. Sci. U.S.A. 103, 2410–2415. doi: 10.1073/pnas.0511003103
Kubiak, T. M., Larsen, M. J., Bowman, J. W., Geary, T. G., and Lowery, D. E. (2008). FMRFamide-like peptides encoded on the flp-18 precursor gene activate two isoforms of the orphan Caenorhabditis elegans G-protein-coupled receptor Y58G8A.4 heterologously expressed in mammalian cells. Biopolymers 90, 339–348. doi: 10.1002/bip.20850
Kubiak, T. M., Larsen, M. J., Nulf, S. C., Zantello, M. R., Burton, K. J., Bowman, J. W., et al. (2003). Differential activation of “social” and “solitary” variants of the Caenorhabditis elegans G protein-coupled receptor NPR-1 by its cognate ligand AF9. J. Biol. Chem. 278, 33724–33729. doi: 10.1074/jbc.M304861200
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Legagneux, K., Bernard-Franchi, G., Poncet, F., La Roche, A., Colard, C., Fellmann, D., et al. (2009). Distribution and genesis of the RFRP-producing neurons in the rat brain: comparison with melanin-concentrating hormone- and hypocretin-containing neurons. Neuropeptides 43, 13–19. doi: 10.1016/j.npep.2008.11.001
León, S., García-Galiano, D., Ruiz-Pino, F., Barroso, A., Manfredi-Lozano, M., Romero-Ruiz, A., et al. (2014). Physiological roles of gonadotropin-inhibitory hormone signaling in the control of mammalian reproductive axis: studies in the NPFF1 receptor null mouse. Endocrinology 155, 2953–2965. doi: 10.1210/en.2014-1030
Li, C., Kim, K., and Nelson, L. S. (1999). FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res. 848, 26–34. doi: 10.1016/S0006-8993(99)01972-1
Li, X., Su, J., Lei, Z., Zhao, Y., Jin, M., Fang, R., et al. (2012). Gonadotropin-inhibitory hormone (GnIH) and its receptor in the female pig: cDNA cloning, expression in tissues and expression pattern in the reproductive axis during the estrous cycle. Peptides 36, 176–185. doi: 10.1016/j.peptides.2012.05.008
Liu, Q., Guan, X. M., Martin, W. J., McDonald, T. P., Clements, M. K., Jiang, Q., et al. (2001). Identification and characterization of novel mammalian neuropeptide FF-like peptides that attenuate morphine-induced antinociception. J. Biol. Chem. 276, 36961–36969. doi: 10.1074/jbc.M105308200
Losa-Ward, S. M., Todd, K. L., McCaffrey, K. A., Tsutsui, K., and Patisaul, H. B. (2012). Disrupted organization of RFamide pathways in the hypothalamus is associated with advanced puberty in female rats neonatally exposed to bisphenol A. Biol. Reprod. 87:28. doi: 10.1095/biolreprod.112.100826
Macosko, E. Z., Pokala, N., Feinberg, E. H., Chalasani, S. H., Butcher, R. A., Clardy, J., et al. (2009). A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458, 1171–1175. doi: 10.1038/nature07886
Maddineni, S., Ocón-Grove, O. M., Krzysik-Walker, S. M., Hendricks, G. L. III., Proudman, J. A., and Ramachandran, R. (2008). Gonadotrophin-inhibitory hormone receptor expression in the chicken pituitary gland: potential influence of sexual maturation and ovarian steroids. J. Neuroendocrinol. 20, 1078–1088. doi: 10.1111/j.1365-2826.2008.01765.x
Maddineni, S. R., Ocón-Grove, O. M., Krzysik-Walker, S. M., Hendricks, G. L. III., and Ramachandran, R. (2008). Gonadotropin-inhibitory hormone (GnIH) receptor gene is expressed in the chicken ovary: potential role of GnIH in follicular maturation. Reproduction 135, 267–274. doi: 10.1530/REP-07-0369
Maney, D. L., Richardson, R. D., and Wingfield, J. C. (1997). Central administration of chicken gonadotropin-releasing hormone-II enhances courtship behavior in a female sparrow. Horm. Behav. 32, 11–18. doi: 10.1006/hbeh.1997.1399
Mason, A. O., Duffy, S., Zhao, S., Ubuka, T., Bentley, G. E., Tsutsui, K., et al. (2010). Photoperiod and reproductive condition are associated with changes in RFamide-related peptide (RFRP) expression in Syrian hamsters (Mesocricetus auratus). J. Biol. Rhythms 25, 176–185. doi: 10.1177/0748730410368821
McConn, B., Wang, G., Yi, J., Gilbert, E. R., Osugi, T., Ubuka, T., et al. (2014). Gonadotropin-inhibitory hormone-stimulation of food intake is mediated by hypothalamic effects in chicks. Neuropeptides 48, 327–334. doi: 10.1016/j.npep.2014.09.001
McCoy, C. J., Atkinson, L. E., Zamanian, M., McVeigh, P., Day, T. A., Kimber, M. J., et al. (2014). New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteom. 3, 262–272. doi: 10.1016/j.euprot.2014.04.002
McGuire, N. L., and Bentley, G. E. (2010). A functional neuropeptide system in vertebrate gonads: Gonadotropin-inhibitory hormone and its receptor in testes of field-caught house sparrow (Passer domesticus). Gen. Comp. Endocrinol. 166, 565–572. doi: 10.1016/j.ygcen.2010.01.010
McGuire, N. L., Kangas, K., and Bentley, G. E. (2011). Effects of melatonin on peripheral reproductive function: regulation of testicular GnIH and testosterone. Endocrinology 152, 3461–3470. doi: 10.1210/en.2011-1053
McGuire, N. L., Koh, A., and Bentley, G. E. (2013). The direct response of the gonads to cues of stress in a temperate songbird species is season-dependent. PeerJ 1:e139. doi: 10.7717/peerj.139
McVeigh, P., Geary, T. G., Marks, N. J., and Maule, A. G. (2006). The FLP-side of nematodes. Trends Parasitol. 22, 385–396. doi: 10.1016/j.pt.2006.06.010
Milward, K., Busch, K. E., Murphy, R. J., de Bono, M., and Olofsson, B. (2011). Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 108, 20672–20677. doi: 10.1073/pnas.1106134109
Miyamoto, K., Hasegawa, Y., Minegishi, T., Nomura, M., Takahashi, Y., Igarashi, M., et al. (1982). Isolation and characterization of chicken hypothalamic luteinizing hormone-releasing hormone. Biochem. Biophys. Res. Commun. 107, 820–827.
Miyamoto, K., Hasegawa, Y., Nomura, M., Igarashi, M., Kangawa, K., and Matsuo, H. (1984). Identification of the second gonadotropin-releasing hormone in chicken hypothalamus: evidence that gonadotropin secretion is probably controlled by two distinct gonadotropin-releasing hormones in avian species. Proc. Natl. Acad. Sci. U.S.A. 81, 3874–3878.
Molnár, C. S., Kalló, I., Liposits, Z., and Hrabovszky, E. (2011). Estradiol down-regulates RF-amide-related peptide (RFRP) expression in the mouse hypothalamus. Endocrinology 152, 1684–1690. doi: 10.1210/en.2010-1418
Moussavi, M., Wlasichuk, M., Chang, J. P., and Habibi, H. R. (2012). Seasonal effect of GnIH on gonadotrope functions in the pituitary of goldfish. Mol. Cell. Endocrinol. 350, 53–60. doi: 10.1016/j.mce.2011.11.020
Moussavi, M., Wlasichuk, M., Chang, J. P., and Habibi, H. R. (2013). Seasonal effect of gonadotrophin inhibitory hormone on gonadotrophin-releasing hormone-induced gonadotroph functions in the goldfish pituitary. J. Neuroendocrinol. 25, 506–516. doi: 10.1111/jne.12024
Moussavi, M., Wlasichuk, M., Chang, J. P., and Habibi, H. R. (2014). Seasonal effects of GnIH on basal and GnRH-induced goldfish somatotrope functions. J. Endocrinol. 223, 191–202. doi: 10.1530/JOE-14-0441
Muñoz-Cueto, J. A., Paullada-Salmerón, J. A., Aliaga-Guerrero, M., Cowan, M. E., Parhar, I. S., and Ubuka, T. (2017). A journey through the gonadotropin-inhibitory hormone system of fish. Front. Endocrinol. 8:285. doi: 10.3389/fendo.2017.00285
Murakami, M., Matsuzaki, T., Iwasa, T., Yasui, T., Irahara, M., Osugi, T., et al. (2008). Hypophysiotropic role of RFamide-related peptide-3 in the inhibition of LH secretion in female rats. J. Endocrinol. 199, 105–112. doi: 10.1677/JOE-08-0197
Nelson, L. S., Kim, K., Memmott, J. E., and Li, C. (1998). FMRFamide-related gene family in the nematode, Caenorhabditis elegans. Mol. Brain Res. 58, 103–111. doi: 10.1016/S0169-328X(98)00106-5
Nichols, R., Bass, C., Demers, L., Larsen, B., Li, E., Blewett, N., et al. (2012). Structure-activity studies of RFamide-related peptide-1 identify a functional receptor antagonist and novel cardiac myocyte signaling pathway involved in contractile performance. J. Med. Chem. 55, 7736–7745. doi: 10.1021/jm300760m
Nichols, R., Demers, L. A., Larsen, B. M., Robinson, D., Converso, K., Russell, M. W., et al. (2010). Human RFamide-related peptide-1 diminishes cellular and integrated cardiac contractile performance. Peptides 31, 2067–2074. doi: 10.1016/j.peptides.2010.07.012
Ogawa, S., and Parhar, I. S. (2014). Structural and functional divergence of gonadotropin-inhibitory hormone from jawless fish to mammals. Front. Endocrinol. 5:177. doi: 10.3389/fendo.2014.00177
Ogawa, S., Sivalingam, M., Biran, J., Golan, M., Anthonysamy, R. S., Levavi-Sivan, B., et al. (2016). Distribution of LPXRFa, a gonadotropin-inhibitory hormone ortholog peptide, and LPXRFa receptor in the brain and pituitary of the tilapia. J. Comp. Neurol. 524, 2753–2775. doi: 10.1002/cne.23990
Oishi, H., Klausen, C., Bentley, G. E., Osugi, T., Tsutsui, K., Gilks, C. B., et al. (2012). The human gonadotropin-inhibitory hormone ortholog RFamide-related peptide-3 suppresses gonadotropin-induced progesterone production in human granulosa cells. Endocrinology 153, 3435–3445. doi: 10.1210/en.2012-1066
Osugi, T., Daukss, D., Gazda, K., Ubuka, T., Kosugi, T., Nozaki, M., et al. (2012). Evolutionary origin of the structure and function of gonadotropin-inhibitory hormone: insights from lampreys. Endocrinology 153, 2362–2374. doi: 10.1210/en.2011-2046
Osugi, T., Okamura, T., Son, Y. L., Ohkubo, M., Ubuka, T., Henmi, Y., et al. (2014). Evolutionary origin of GnIH and NPFF in chordates: insights from novel amphioxus RFamide peptides. PLoS ONE. 9:e100962. doi: 10.1371/journal.pone.0100962
Osugi, T., Ukena, K., Bentley, G. E., O'Brien, S., Moore, I. T., Wingfield, J. C., et al. (2004). Gonadotropin-inhibitory hormone in Gambel's white-crowned sparrow (Zonotrichia leucophrys gambelii): cDNA identification, transcript localization and functional effects in laboratory and field experiments. J. Endocrinol. 182, 33–42. doi: 10.1677/joe.0.1820033
Osugi, T., Ukena, K., Sower, S. A., Kawauchi, H., and Tsutsui, K. (2006). Evolutionary origin and divergence of PQRFamide peptides and LPXRFamide peptides in the RFamide peptide family. Insights from novel lamprey RFamide peptides. FEBS J. 273, 1731–1743. doi: 10.1111/j.1742-4658.2006.05187.x
Paullada-Salmerón, J. A., Cowan, M., Aliaga-Guerrero, M., Gómez, A., Zanuy, S., Mañanos, E., et al. (2016a). LPXRFa peptide system in the European sea bass: A molecular and immunohistochemical approach. J. Comp. Neurol. 524, 176–198. doi: 10.1002/cne.23833
Paullada-Salmerón, J. A., Cowan, M., Aliaga-Guerrero, M., López-Olmeda, J. F., Mañanós, E. L., Zanuy, S., et al. (2016b). Testicular steroidogenesis and socomotor activity are regulated by gonadotropin-inhibitory hormone in male European sea bass. PLoS ONE 11:e0165494. doi: 10.1371/journal.pone.0165494
Paullada-Salmerón, J. A., Cowan, M., Aliaga-Guerrero, M., Morano, F., Zanuy, S., and Muñoz-Cueto, J. A. (2016c). Gonadotropin-inhibitory hormone down-regulates the brain-pituitary reproductive axis of male European sea bass (Dicentrarchus labrax). Biol. Reprod. 94:121. doi: 10.1095/biolreprod.116.139022
Perry, S. J., Yi-Kung Huang, E., Cronk, D., Bagust, J., Sharma, R., Walker, R. J., et al. (1997). A human gene encoding morphine modulating peptides related to NPFF and FMRFamide. FEBS. Lett. 409, 426–430.
Peymen, K., Watteyne, J., Frooninckx, L., Schoofs, L., and Beets, I. (2014). The FMRFamide-like peptide family in Nematodes. Front. Endocrinol. 5:90. doi: 10.3389/fendo.2014.00090
Piekarski, D. J., Jarjisian, S. G., Perez, L., Ahmad, H., Dhawan, N., Zucker, I., et al. (2014). Effects of pinealectomy and short day lengths on reproduction and neuronal RFRP-3, kisspeptin, and GnRH in Female Turkish Hamsters. J. Biol. Rhythm. 29, 181–191. doi: 10.1177/0748730414532423
Piekarski, D. J., Zhao, S., Jennings, K. J., Iwasa, T., Legan, S. J., Mikkelsen, J. D., et al. (2013). Gonadotropin-inhibitory hormone reduces sexual motivation but not lordosis behavior in female Syrian hamsters (Mesocricetus auratus). Horm. Behav. 64, 501–510. doi: 10.1016/j.yhbeh.2013.06.006
Pineda, R., Garcia-Galiano, D., Sanchez-Garrido, M. A., Romero, M., Ruiz-Pino, F., Aguilar, E., et al. (2010a). Characterization of the inhibitory roles of RFRP3, the mammalian ortholog of GnIH, in the control of gonadotropin secretion in the rat: in vivo and in vitro studies. Am. J. Physiol. Endocrinol. Metab. 299, E39–E46. doi: 10.1152/ajpendo.00108.2010
Pineda, R., Garcia-Galiano, D., Sanchez-Garrido, M. A., Romero, M., Ruiz-Pino, F., Aguilar, E., et al. (2010b). Characterization of the potent gonadotropin-releasing activity of RF9, a selective antagonist of RF-amide-related peptides and neuropeptide FF receptors: physiological and pharmacological implications. Endocrinology 151, 1902–1913. doi: 10.1210/en.2009-1259
Pinelli, C., Jadhao, A. G., Biswas, S. P., Tsutsui, K., and D'Aniello, B. (2015). Neuroanatomical organization of the brain gonadotropin-inhibitory hormone and gonadotropin-releasing hormone systems in the frog Pelophylax esculentus. Brain. Behav. Evol. 85, 15–28. doi: 10.1159/000368594
Plachetzki, D. C., Tsai, P. S., Kavanaugh, S. I., and Sower, S. A. (2016). Ancient origins of metazoan gonadotropin-releasing hormone and their receptors revealed by phylogenomic analyses. Gen. Comp. Endocrinol. 234, 10–19. doi: 10.1016/j.ygcen.2016.06.007
Poling, M. C., Kim, J., Dhamija, S., and Kauffman, A. S. (2012). Development, sex steroid regulation, and phenotypic characterization of RFamide-related peptide (Rfrp) gene expression and RFamide receptors in the mouse hypothalamus. Endocrinology 153, 1827–1840. doi: 10.1210/en.2011-2049
Poling, M. C., Quennell, J. H., Anderson, G. M., and Kauffman, A. S. (2013). Kisspeptin neurones do not directly signal to RFRP-3 neurones but RFRP-3 may directly modulate a subset of hypothalamic kisspeptin cells in mice. J. Neuroendocrinol. 25, 876–886. doi: 10.1111/jne.12084
Poling, M. C., Shieh, M. P., Munaganuru, N., Luo, E., and Kauffman, A. S. (2014). Examination of the influence of leptin and acute metabolic challenge on RFRP-3 neurons of mice in development and adulthood. Neuroendocrinology 100, 317–333. doi: 10.1159/000369276
Price, D. A., and Greenberg, M. J. (1977). Structure of a molluscan cardioexcitatory neuropeptide. Science 197, 670–671.
Qi, X., Zhou, W., Li, S., Lu, D., Yi, S., Xie, R., et al. (2013). Evidences for the regulation of GnRH and GTH expression by GnIH in the goldfish, Carassius auratus. Mol. Cell. Endocrinol. 366, 9–20. doi: 10.1016/j.mce.2012.11.001
Qi, Y., Oldfield, B. J., and Clarke, I. J. (2009). Projections of RFamide-related peptide-3 neurones in the ovine hypothalamus, with special reference to regions regulating energy balance and reproduction. J. Neuroendocrinol. 21, 690–697. doi: 10.1111/j.1365-2826.2009.01886.x
Quennell, J. H., Rizwan, M. Z., Relf, H. L., and Anderson, G. M. (2010). Developmental and steroidogenic effects on the gene expression of RFamide related peptides and their receptor in the rat brain and pituitary gland. J. Neuroendocrinol. 22, 309–316. doi: 10.1111/j.1365-2826.2010.01963.x
Revel, F. G., Saboureau, M., Pévet, P., Simonneaux, V., and Mikkelsen, J. D. (2008). RFamide-related peptide gene is a melatonin-driven photoperiodic gene. Endocrinology 149, 902–912. doi: 10.1210/en.2007-0848
Ringstad, N., and Horvitz, H. R. (2008). FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat. Neurosci. 11, 1168–1176. doi: 10.1038/nn.2186
Rizwan, M. Z., Poling, M. C., Corr, M., Cornes, P. A., Augustine, R. A., Quennell, J. H., et al. (2012). RFamide-related peptide-3 receptor gene expression in GnRH and kisspeptin neurons and GnRH-dependent mechanism of action. Endocrinology 153, 3770–3779. doi: 10.1210/en.2012-1133
Rizwan, M. Z., Porteous, R., Herbison, A. E., and Anderson, G. M. (2009). Cells expressing RFamide-related peptide-1/3, the mammalian gonadotropin-inhibitory hormone orthologs, are not hypophysiotropic neuroendocrine neurons in the rat. Endocrinology 150, 1413–1420. doi: 10.1210/en.2008-1287
Rogers, C., Persson, A., Cheung, B., and de Bono, M. (2006). Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr. Biol. 16, 649–659. doi: 10.1016/j.cub.2006.03.023
Rogers, C., Reale, V., Kim, K., Chatwin, H., Li, C., Evans, P., et al. (2003). Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat. Neurosci. 6, 1178–1185. doi: 10.1038/nn1140
Russo, K. A., La, J. L., Stephens, S. B., Poling, M. C., Padgaonkar, N. A., Jennings, K. J., et al. (2015). Circadian control of the female reproductive axis through gated responsiveness of the RFRP-3 system to VIP signaling. Endocrinology 156, 2608–2618. doi: 10.1210/en.2014-1762
Sáenz de Miera, C., Monecke, S., Bartzen-Sprauer, J., Laran-Chich, M. P., Pévet, P., Hazlerigg, D. G., et al. (2014). A circannual clock drives expression of genes central for seasonal reproduction. Curr. Biol. 24, 1500–1506. doi: 10.1016/j.cub.2014.05.024
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Salehi, M. S., Jafarzadeh Shirazi, M. R., Zamiri, M. J., Pazhoohi, F., Namavar, M. R., Niazi, A., et al. (2013). Hypothalamic expression of KiSS1 and RFamide-related peptide-3 mRNAs during the estrous cycle of rats. Int. J. Fertil. Steril. 6, 304–309.
Sari, I. P., Rao, A., Smith, J. T., Tilbrook, A. J., and Clarke, I. J. (2009). Effect of RF-amide-related peptide-3 on luteinizing hormone and follicle-stimulating hormone synthesis and secretion in ovine pituitary gonadotropes. Endocrinology 150, 5549–5556. doi: 10.1210/en.2009-0775
Satake, H., Hisada, M., Kawada, T., Minakata, H., Ukena, K., and Tsutsui, K. (2001). Characterization of a cDNA encoding a novel avian hypothalamic neuropeptide exerting an inhibitory effect on gonadotropin release. Biochem. J. 354, 379–385. doi: 10.1042/bj3540379
Sawada, K., Ukena, K., Kikuyama, S., and Tsutsui, K. (2002a). Identification of a cDNA encoding a novel amphibian growth hormone-releasing peptide and localization of its transcript. J. Endocrinol. 174, 395–402. doi: 10.1677/joe.0.1740395
Sawada, K., Ukena, K., Satake, H., Iwakoshi, E., Minakata, H., and Tsutsui, K. (2002b). Novel fish hypothalamic neuropeptide. Eur. J. Biochem. 269, 6000–6008. doi: 10.1046/j.1432-1033.2002.03351.x
Semaan, S. J., and Kauffman, A. S. (2015). Daily successive changes in reproductive gene expression and neuronal activation in the brains of pubertal female mice. Mol. Cell Endocrinol. 401, 84–97. doi: 10.1016/j.mce.2014.11.025
Sethi, S., Tsutsui, K., and Chaturvedi, C. M. (2010). Temporal phase relation of circadian neural oscillations alters RFamide-related peptide-3 and testicular function in the mouse. Neuroendocrinology 91, 189–199. doi: 10.1159/000265760
Shahjahan, M., Ikegami, T., Osugi, T., Ukena, K., Doi, H., Hattori, A., et al. (2011). Synchronised expressions of LPXRFamide peptide and its receptor genes: seasonal, diurnal and circadian changes during spawning period in grass puffer. J. Neuroendocrinol. 23, 39–51. doi: 10.1111/j.1365-2826.2010.02081.x
Sharp, P. J., Talbot, R. T., Main, G. M., Dunn, I. C., Fraser, H. M., and Huskisson, N. S. (1990). Physiological roles of chicken LHRH-I and -II in the control of gonadotrophin release in the domestic chicken. J. Endocrinol. 124, 291–299.
Shimizu, M., and Bédécarrats, G. Y. (2006). Identification of a novel pituitary-specific chicken gonadotropin-releasing hormone receptor and its splice variants. Biol. Reprod. 75, 800–808. doi: 10.1095/biolreprod.105.050252
Shimizu, M., and Bédécarrats, G. Y. (2010). Activation of the chicken gonadotropin-inhibitory hormone receptor reduces gonadotropin releasing hormone receptor signaling. Gen. Comp. Endocrinol. 167, 331–337. doi: 10.1016/j.ygcen.2010.03.029
Singh, P., Krishna, A., Sridaran, R., and Tsutsui, K. (2011). Immunohistochemical localization of GnRH and RFamide-related peptide-3 in the ovaries of mice during the estrous cycle. J. Mol. Histol. 42, 371–381. doi: 10.1007/s10735-011-9340-8
Small, T. W., Sharp, P. J., Bentley, G. E., Millar, R. P., Tsutsui, K., Mura, E., et al. (2008). Photoperiod-independent hypothalamic regulation of luteinizing hormone secretion in a free-living Sonoran desert bird, the Rufous-winged Sparrow (Aimophila carpalis). Brain Behav. Evol. 71, 127–142. doi: 10.1159/000111459
Smith, J. T., Coolen, L. M., Kriegsfeld, L. J., Sari, I. P., Jaafarzadehshirazi, M. R., Maltby, M., et al. (2008). Variation in kisspeptin and RFamide-related peptide (RFRP) expression and terminal connections to gonadotropin-releasing hormone neurons in the brain: a novel medium for seasonal breeding in the sheep. Endocrinology 149, 5770–5782. doi: 10.1210/en.2008-0581
Smith, J. T., Shahab, M., Pereira, A., Pau, K. Y., and Clarke, I. J. (2010). Hypothalamic expression of KISS1 and gonadotropin inhibitory hormone genes during the menstrual cycle of a non-human primate. Biol. Reprod. 83, 568–577. doi: 10.1095/biolreprod.110.085407
Smith, J. T., Young, I. R., Veldhuis, J. D., and Clarke, I. J. (2012). Gonadotropin-inhibitory hormone (GnIH) secretion into the ovine hypophyseal portal system. Endocrinology 153, 3368–3375. doi: 10.1210/en.2012-1088
Soga, T., Dalpatadu, S. L., Wong, D. W., and Parhar, I. S. (2012). Neonatal dexamethasone exposure down-regulates GnRH expression through the GnIH pathway in female mice. Neuroscience 218, 56–64. doi: 10.1016/j.neuroscience.2012.05.023
Soga, T., Kitahashi, T., Clarke, I. J., and Parhar, I. S. (2014). Gonadotropin-inhibitory hormone promoter-driven enhanced green fluorescent protein expression decreases during aging in female rats. Endocrinology 155, 1944–1955. doi: 10.1210/en.2013-1786
Son, Y. L., Ubuka, T., Millar, R. P., Kanasaki, H., and Tsutsui, K. (2012). Gonadotropin-inhibitory hormone inhibits GnRH-induced gonadotropin subunit gene transcriptions by inhibiting AC/cAMP/PKA-dependent ERK pathway in LβT2 cells. Endocrinology 153, 2332–2343. doi: 10.1210/en.2011-1904
Son, Y. L., Ubuka, T., Narihiro, M., Fukuda, Y., Hasunuma, I., Yamamoto, K., et al. (2014). Molecular basis for the activation of gonadotropin-inhibitory hormone gene transcription by corticosterone. Endocrinology 155, 1817–1826. doi: 10.1210/en.2013-2076
Son, Y. L., Ubuka, T., Soga, T., Yamamoto, K., Bentley, G. E., and Tsutsui, K. (2016). Inhibitory action of gonadotropin-inhibitory hormone on the signaling pathways induced by kisspeptin and vasoactive intestinal polypeptide in GnRH neuronal cell line, GT1-7. FASEB J. 30, 2198–2210. doi: 10.1096/fj.201500055
Spicer, O. S., Zmora, N., Wong, T. T., Golan, M., Levavi-Sivan, B., Gothilf, Y., et al. (2017). The gonadotropin-inhibitory hormone (Lpxrfa) system's regulation of reproduction in the brain-pituitary axis of the zebrafish (Danio rerio). Biol. Reprod. 96, 1031–1042. doi: 10.1093/biolre/iox032
Styer, K. L., Singh, V., Macosko, E., Steele, S. E., Bargmann, C. I., and Aballay, A. (2008). Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322, 460–464. doi: 10.1126/science.1163673
Surbhi Kumari, Y., Rani, S., Tsutsui, K., and Kumar, V. (2015). Duration of melatonin regulates seasonal plasticity in subtropical Indian weaver bird, Ploceus philippinus. Gen. Comp. Endocrinol. 220, 46–54. doi: 10.1016/j.ygcen.2014.06.004
Tachibana, T., Masuda, N., Tsutsui, K., Ukena, K., and Ueda, H. (2008). The orexigenic effect of GnIH is mediated by central opioid receptors in chicks. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 150, 21–25. doi: 10.1016/j.cbpa.2008.02.018
Tachibana, T., Sato, M., Takahashi, H., Ukena, K., Tsutsui, K., and Furuse, M. (2005). Gonadotropin-inhibiting hormone stimulates feeding behavior in chicks. Brain Res. 1050, 94–100. doi: 10.1016/j.brainres.2005.05.035
Temple, J. L., Millar, R. P., and Rissman, E. F. (2003). An evolutionarily conserved form of gonadotropin-releasing hormone coordinates energy and reproductive behavior. Endocrinology 144, 13–19. doi: 10.1210/en.2002-220883
The C. elegans Sequencing Consortium (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–8. doi: 10.1126/science.282.5396.2012
Tobari, Y., Iijima, N., Tsunekawa, K., Osugi, T., Okanoya, K., Tsutsui, K., et al. (2010). Identification of gonadotropin-inhibitory hormone in the zebra finch (Taeniopygia guttata): peptide isolation, cDNA cloning and brain distribution. Peptides 31, 816–826. doi: 10.1016/j.peptides.2010.01.015
Tobari, Y., Son, Y. L., Ubuka, T., Hasegawa, Y., and Tsutsui, K. (2014). A new pathway mediating social effects on the endocrine system: female presence acting via norepinephrine release stimulates gonadotropin-inhibitory hormone in the paraventricular nucleus and suppresses luteinizing hormone in quail. J. Neurosci. 34, 9803–9811. doi: 10.1523/JNEUROSCI.3706-13.2014
Trent, C., Tsuing, N., and Horvitz, H. R. (1983). Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104, 619–647.
Tsutsui, K., Osugi, T., Son, Y. L., and Ubuka, T. (2017). Review: Structure, function and evolution of GnIH. Gen. Comp. Endocrinol. 264, 48–57. doi: 10.1016/j.ygcen.2017.07.024
Tsutsui, K., Saigoh, E., Ukena, K., Teranishi, H., Fujisawa, Y., Kikuchi, M., et al. (2000). A novel avian hypothalamic peptide inhibiting gonadotropin release. Biochem. Biophys. Res. Commun. 275, 661–667. doi: 10.1006/bbrc.2000.3350
Tsutsui, K., and Ubuka, T. (2016). GnIH control of feeding and reproductive behaviors. Front. Endocrinol. 7:170. doi: 10.3389/fendo.2016.00170
Tsutsui, K., Ubuka, T., Son, Y. L., Bentley, G. E., and Kriegsfeld, L. J. (2015). Contribution of GnIH research to the progress of reproductive neuroendocrinology. Front. Endocrinol. 6:179. doi: 10.3389/fendo.2015.00179
Ubuka, T., and Bentley, G. E. (2009). Identification, localization, and regulation of passerine GnRH-I messenger RNA. J. Endocrinol. 201, 81–87. doi: 10.1677/JOE-08-0508
Ubuka, T., and Bentley, G. E. (2010). “Neuroendocrine control of reproduction in birds,” in Hormones and Reproduction of Vertebrates, Vol. 4, eds D. O. Norris, and K. H. Lopez (London: Birds, Academic Press), 1–25.
Ubuka, T., Bentley, G. E., Ukena, K., Wingfield, J. C., and Tsutsui, K. (2005). Melatonin induces the expression of gonadotropin-inhibitory hormone in the avian brain. Proc. Natl. Acad. Sci. U.S.A. 102, 3052–3057. doi: 10.1073/pnas.0403840102
Ubuka, T., Cadigan, P. A., Wang, A., Liu, J., and Bentley, G. E. (2009a). Identification of European starling GnRH-I precursor mRNA and its seasonal regulation. Gen. Comp. Endocrinol. 162, 301–306. doi: 10.1016/j.ygcen.2009.04.001
Ubuka, T., Haraguchi, S., Tobari, Y., Narihiro, M., Ishikawa, K., Hayashi, T., et al. (2014). Hypothalamic inhibition of socio-sexual behaviour by increasing neuroestrogen synthesis. Nat. Commun. 5:3061. doi: 10.1038/ncomms4061
Ubuka, T., Inoue, K., Fukuda, Y., Mizuno, T., Ukena, K., Kriegsfeld, L. J., et al. (2012a). Identification, expression, and physiological functions of Siberian hamster gonadotropin-inhibitory hormone. Endocrinology 153, 373–385. doi: 10.1210/en.2011-1110
Ubuka, T., Kim, S., Huang, Y. C., Reid, J., Jiang, J., Osugi, T., et al. (2008). Gonadotropin-inhibitory hormone neurons interact directly with gonadotropin-releasing hormone-I and -II neurons in European starling brain. Endocrinology 149, 268–278. doi: 10.1210/en.2007-0983
Ubuka, T., Lai, H., Kitani, M., Suzuuchi, A., Pham, V., Cadigan, P. A., et al. (2009b). Gonadotropin-inhibitory hormone identification, cDNA cloning, and distribution in rhesus macaque brain. J. Comp. Neurol. 517, 841–855. doi: 10.1002/cne.22191
Ubuka, T., Morgan, K., Pawson, A. J., Osugi, T., Chowdhury, V. S., Minakata, H., et al. (2009c). Identification of human GnIH homologs, RFRP-1 and RFRP-3, and the cognate receptor, GPR147 in the human hypothalamic pituitary axis. PLoS ONE. 4:e8400. doi: 10.1371/journal.pone.0008400
Ubuka, T., Mukai, M., Wolfe, J., Beverly, R., Clegg, S., Wang, A., et al. (2012b). RNA interference of gonadotropin-inhibitory hormone gene induces arousal in songbirds. PLoS ONE. 7:e30202. doi: 10.1371/journal.pone.0030202
Ubuka, T., Parhar, I. S., and Tsutsui, K. (2018). Gonadotropin-inhibitory hormone mediates behavioral stress responses. Gen. Comp. Endocrinol. 265, 202–206. doi: 10.1016/j.ygcen.2018.03.004
Ubuka, T., Son, Y. L., Bentley, G. E., Millar, R. P., and Tsutsui, K. (2013). Gonadotropin-inhibitory hormone (GnIH), GnIH receptor and cell signaling. Gen. Comp. Endocrinol. 190, 10–17. doi: 10.1016/j.ygcen.2013.02.030
Ubuka, T., Son, Y. L., and Tsutsui, K. (2016). Molecular, cellular, morphological, physiological and behavioral aspects of gonadotropin-inhibitory hormone. Gen. Comp. Endocrinol. 227, 27–50. doi: 10.1016/j.ygcen.2015.09.009
Ubuka, T., and Tsutsui, K. (2014). Evolution of gonadotropin-inhibitory hormone receptor and its ligand. Gen. Comp. Endocrinol. 209, 48–61. doi: 10.1016/j.ygcen.2014.09.002
Ubuka, T., Ueno, M., Ukena, K., and Tsutsui, K. (2003). Developmental changes in gonadotropin-inhibitory hormone in the Japanese quail (Coturnix japonica) hypothalamo-hypophysial system. J. Endocrinol. 178, 311–318. doi: 10.1677/joe.0.1780311
Ubuka, T., Ukena, K., Sharp, P. J., Bentley, G. E., and Tsutsui, K. (2006). Gonadotropin-inhibitory hormone inhibits gonadal development and maintenance by decreasing gonadotropin synthesis and release in male quail. Endocrinology 147, 1187–1194. doi: 10.1210/en.2005-1178
Ukena, K., Iwakoshi, E., Minakata, H., and Tsutsui, K. (2002). A novel rat hypothalamic RFamide-related peptide identified by immunoaffinity chromatography and mass spectrometry. FEBS Lett. 512, 255–258. doi: 10.1016/S0014-5793(02)02275-5
Ukena, K., Iwakoshi-Ukena, E., Osugi, T., and Tsutsui, K. (2016). Identification and localization of gonadotropin-inhibitory hormone (GnIH) orthologs in the hypothalamus of the red-eared slider turtle, Trachemys scripta elegans. Gen. Comp. Endocrinol. 227, 69–76. doi: 10.1016/j.ygcen.2015.06.009
Ukena, K., Koda, A., Yamamoto, K., Kobayashi, T., Iwakoshi-Ukena, E., Minakata, H., et al. (2003a). Novel neuropeptides related to frog growth hormone-releasing peptide: isolation, sequence, and functional analysis. Endocrinology 144, 3879–3884. doi: 10.1210/en.2003-0359
Ukena, K., Ubuka, T., and Tsutsui, K. (2003b). Distribution of a novel avian gonadotropin-inhibitory hormone in the quail brain. Cell Tissue Res. 312, 73–79. doi: 10.1007/s00441-003-0700-x
Wang, Q., Qi, X., Guo, Y., Li, S., Zhang, Y., Liu, X., et al. (2015). Molecular identification of GnIH/GnIHR signal and its reproductive function in protogynous hermaphroditic orange-spotted grouper (Epinephelus coioides). Gen. Comp. Endocrinol. 216, 9–23. doi: 10.1016/j.ygcen.2015.04.016
White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340.
Wu, M., Dumalska, I., Morozova, E., van den Pol, A. N., and Alreja, M. (2009). Gonadotropin inhibitory hormone inhibits basal forebrain vGluT2-gonadotropin-releasing hormone neurons via a direct postsynaptic mechanism. J. Physiol. 587, 1401–1411. doi: 10.1113/jphysiol.2008.166447
Xiang, W., Zhang, B., Lv, F., Ma, Y., Chen, H., Chen, L., et al. (2015). The inhibitory effects of RFamide-related peptide 3 on luteinizing hormone release involves an estradiol-dependent manner in prepubertal but not in adult female mice. Biol. Reprod. 93:30. doi: 10.1095/biolreprod.115.128777
Yang, H. Y., Fratta, W., Majane, E. A., and Costa, E. (1985). Isolation, sequencing, synthesis, and pharmacological characterization of two brain neuropeptides that modulate the action of morphine. Proc. Natl. Acad. Sci. U.S.A. 82, 7757–7761.
Yano, T., Iijima, N., Hinuma, S., Tanaka, M., and Ibata, Y. (2004). Developmental expression of RFamide-related peptides in the rat central nervous system. Brain Res. Dev. Brain Res. 152, 109–120. doi: 10.1016/j.devbrainres.2004.06.008
Yano, T., Iijima, N., Kakihara, K., Hinuma, S., Tanaka, M., and Ibata, Y. (2003). Localization and neuronal response of RFamide related peptides in the rat central nervous system. Brain Res. 982, 156–167. doi: 10.1016/S0006-8993(03)02877-4
Yin, H., Ukena, K., Ubuka, T., and Tsutsui, K. (2005). A novel G protein-coupled receptor for gonadotropin-inhibitory hormone in the Japanese quail (Coturnix japonica): identification, expression and binding activity. J. Endocrinol. 184, 257–266. doi: 10.1677/joe.1.05926
Yoshida, H., Habata, Y., Hosoya, M., Kawamata, Y., Kitada, C., and Hinuma, S. (2003). Molecular properties of endogenous RFamide-related peptide-3 and its interaction with receptors. Biochim. Biophys. Acta 1593, 151–157. doi: 10.1016/S0167-4889(02)00389-0
Zhang, Y., Li, S., Liu, Y., Lu, D., Chen, H., Huang, X., et al. (2010). Structural diversity of the GnIH/GnIH receptor system in teleost: its involvement in early development and the negative control of LH release. Peptides 31, 1034–1043. doi: 10.1016/j.peptides.2010.03.003
Zhao, L., Zhong, M., Xue, H. L., Ding, J. S., Wang, S., Xu, J. H., et al. (2014). Effect of RFRP-3 on reproduction is sex- and developmental status-dependent in the striped hamster (Cricetulus barabensis). Gene 547, 273–279. doi: 10.1016/j.gene.2014.06.054
Zhao, S., Zhu, E., Yang, C., Bentley, G. E., Tsutsui, K., and Kriegsfeld, L. J. (2010). RFamide-related peptide and messenger ribonucleic acid expression in mammalian testis: association with the spermatogenic cycle. Endocrinology 151, 617–627. doi: 10.1210/en.2009-0978
Keywords: gonadotropin-inhibitory hormone (GnIH), neuropeptide FF (NPFF), FMRFamide-like peptide (FLP), GPR147, GPR74, chordates, nematodes, C. elegans
Citation: Ubuka T and Tsutsui K (2018) Comparative and Evolutionary Aspects of Gonadotropin-Inhibitory Hormone and FMRFamide-Like Peptide Systems. Front. Neurosci. 12:747. doi: 10.3389/fnins.2018.00747
Received: 23 May 2018; Accepted: 28 September 2018;
Published: 18 October 2018.
Edited by:
José A. Muñoz-Cueto, University of Cádiz, SpainReviewed by:
Hervé Tostivint, Muséum National d'Histoire naturelle, FranceGregoy Y. Bedecarrats, University of Guelph, Canada
Stacia A. Sower, University of New Hampshire, United States
Copyright © 2018 Ubuka and Tsutsui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takayoshi Ubuka, dGFrYXlvc2hpLnVidWthQGdtYWlsLmNvbQ==
Kazuyoshi Tsutsui, ay10c3V0c3VpQHdhc2VkYS5qcA==
†Present Address: Takayoshi Ubuka, Brain Research Institute Monash Sunway, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Bandar Sunway, Malaysia
 Takayoshi Ubuka
Takayoshi Ubuka Kazuyoshi Tsutsui
Kazuyoshi Tsutsui