- 1Department of Radiology and BRIC, University of North Carolina, Chapel Hill, NC, United States
- 2School of Automation, Northwestern Polytechnical University, Xi'an, China
- 3Department of Brain and Cognitive Engineering, Korea University, Seoul, South Korea
Groupwise image registration tackles biases that can potentially arise from inappropriate template selection. It typically involves simultaneous registration of a cohort of images to a common space that is not specified a priori. Existing groupwise registration methods are computationally complex and are only effective for image populations without large anatomical variations. In this paper, we propose a deep learning framework to rapidly estimate large deformations between images to significantly reduce structural variability. Specifically, we employ a multi-level graph coarsening method to agglomerate similar images into clusters, each represented by an exemplar image. We then use a deep learning framework to predict the initial deformations between images. Warping with the estimated deformations brings the images closer in the image manifold and their alignment can be further refined using conventional groupwise registration algorithms. We evaluated the effectiveness of our method in groupwise registration of MR brain images and compared it against state-of-the-art groupwise registration methods. Experimental results indicate that deformation initialization enables groupwise registration to converge significantly faster with competitive accuracy, therefore facilitates large-scale imaging studies.
1. Introduction
Deformable image registration plays a crucial role in applications, such as dose planning in radiation therapy (Castadot et al., 2008; Velec et al., 2011; Gu et al., 2013; Fortin et al., 2014; Cunliffe et al., 2015; König et al., 2016; Samavati et al., 2016; Brock et al., 2017; Flower et al., 2017; Oh and Kim, 2017), motion and deformation modeling of organs (Yang et al., 2008; Cammin and Taguchi, 2010; Schmidt-Richberg et al., 2012; Risser et al., 2013; Li et al., 2016; Meschini et al., 2016), and automatic delineation of the anatomical structures (Gorthi et al., 2011; Arabi and Zaidi, 2017; Wang et al., 2017). In addition, medical practitioners rely on deformable registration for morphometric analysis of anatomical structures (Shi et al., 2009; Matsuda, 2013; Agnello et al., 2016; Joshi et al., 2016), intra-subject structural changes in longitudinal studies (Wu et al., 2012; Csapo et al., 2013; Lee et al., 2017) and analysis of inter-subject anatomical variability (Chen et al., 2017). To date, numerous techniques have been developed for pairwise registration of a moving image and a reference image (Vercauteren et al., 2009; Suh et al., 2011; Hu et al., 2012; Csapo et al., 2013; Razlighi and Kehtarnavaz, 2014; Onofrey et al., 2015; Heinrich et al., 2016; Aganj et al., 2017; Sun et al., 2017; Yang et al., 2017). However, these pairwise registration methods require selecting a particular image as the reference, to which subsequent analyses are biased (Toga and Thompson, 2001).
Groupwise registration methods do not require a pre-specified reference image, but instead automatically determine the hidden common space in an unbiased manner. Groupwise registration techniques typically simultaneously align a cohort of images to a common space (Sabuncu et al., 2009; Spiclin et al., 2012; Wachinger and Navab, 2013). For example, in Joshi et al. (2004) an initial group center is defined by the average of all affine-registered images. The group center is iteratively updated with the average of images registered to it. While this method mitigates bias, it leads to registration inaccuracy as the initial group center is fuzzy. This limitation was addressed in Wu et al. (2011) by constructing a “Sharp-Mean” group center by weighted averaging of the registered images. ABSORB (atlas-building by self organized registration and bundling) (Jia et al., 2010) is a groupwise registration algorithm that warps the images based on their neighboring images. However, ABSORB does not consider the whole image distribution and takes into account only the immediate neighbors of an image. HUGS (hierarchical unbiased graph shrinkage) (Ying et al., 2014) models the image distribution using a graph and formulates groupwise registration as a dynamic graph shrinkage problem where images, represented as nodes, are warped along graph edges. Yet another groupwise registration strategy is by constructing a minimal spanning tree with a root node that gives a minimum overall edge length to all other nodes. The image deformation is estimated by composing all the transformations along the path from a leaf node to the root node (Hamm et al., 2009).
The aforementioned methods assume a single common space and are not designed to deal with heterogeneous populations with large anatomical variations. An inhomogeneous population with large deformations is better represented using multiple group centers and directly warping the images to a single group center is ineffective and inaccurate (Sabuncu et al., 2008; Liao et al., 2012). As a remedy, the population is typically divided into multiple homogeneous subgroups with an atlas constructed for each subgroup for registration (Sabuncu et al., 2009; Ribbens et al., 2010). For example, Wang et al. (2010) cluster the population into subgroups and perform groupwise registration within each subgroup. The center images of the subgroups are then registered using a pyramidal hierarchy. While effective, these methods are computationally expensive and not scalable to large datasets.
In this paper, we present a novel deformation initialization framework to reduce anatomical variations prior to groupwise registration. This removes large structural variations in an inhomogeneous image population so that conventional groupwise registration algorithms can be applied more effectively and accurately. Our initialization framework is formulated as a two-step process: (i) graph coarsening and (ii) deep learning deformation prediction. In the first step, the images are represented using a graph and are clustered via iterative graph coarsening. In the second step, deep learning is employed to estimate the deformations between images in the population according to the hierarchical structure resulting from graph coarsening.
2. Methods
Given a diverse dataset of MR brain images with large inter-subject variability, our objectives are to (i) reduce the anatomical variability in a dataset such that the images can be simultaneously registered to a single latent common space and (ii) speed up groupwise registration so that it is scalable to large-scale datasets. To achieve these objectives, we will employ deep learning for predicting large deformations to reduce structural variations so that the images are close enough to be registered efficiently and accurately to a common space.
2.1. Deformation Initialization
2.1.1. Multi-Level Graph Coarsening Based Image Clustering
We propose to use multi-level graph coarsening for image clustering. Graph coarsening is used in multi-level graph partitioning to construct smaller graphs by hierarchically combining neighboring vertices (Xiao et al., 2013; Safro et al., 2015). That is, for an h-level coarsening, we have |G0| < |G1| < … < |Gh|, where |Gh| denotes the size of the graph at level h. Graph coarsening can generally be accomplished by either (i) contraction or (ii) algebraic multigrid (AMG) (Safro et al., 2015) scheme. In the current work, we use AMG graph coarsening (Ruge and Stüben, 1987; Rakai et al., 2012) to split the dataset into image clusters. Let G0 = (V0, E0) be the original graph consisting of a vertex set , representing images , and an edge set , representing the similarity between the images. Each edge eij is defined for images Ii and Ij and is calculated via normalized cross correlation (NCC) as
where x0 is a voxel location in the brain region and and are mean intensity values of images Ii and Ij, respectively. If registration needs to be performed across modalities, information theoretic measures, such as mutual information, can be used. The fine graph G0 is progressively coarsened with the coarsened graph at level l is denoted as . The coarsening algorithm is detailed below:
Step 1: The connection of vertex j with respect to vertex i is considered strong if for given ρ ∈ (0, 1]:
where Nl is the total number of vertices at level l. Note that this criterion is not symmetric with respect to i and j. With ρ = 1, the connection of the vertex with maximum similarity with vertex i is considered strong. This results in a large number of clusters, each with few images. On the other hand, if the value of ρ is too small, then we get too few clusters. We set ρ = 0.95 so that connections with above 95% of the maximum similarity value are considered strong.
Step 2: The desirability ψi of a vertex to be selected as a coarse vertex is computed as the total number of strong connections with respect to the vertex. The vertex with the largest desirability is designated as a coarse vertex and all vertices that are strongly connected with respect to this coarse vertex are designated as fine vertices.
Step 3: The desirability values of vertices strongly connected with respect to fine vertices are increased by 1. The desirability values of vertices strongly connected with respect to coarse vertices are decreased by 1.
Step 4: Steps 2 and 3 are repeated until all the vertices are designated as either coarse or fine.
Step 5: Steps 1 − 4 are repeated with the coarse vertices with l ← l + 1 until we get stable graphs (i.e., when the size of the two consecutive graphs is same |Gl| = |Gl − 1|).
The coarsening process is illustrated in Figure 1. Eventually, images are hierarchically grouped into clusters, where at each level the cluster exemplars are represented by coarse vertices and cluster members are represented by fine vertices. Cluster exemplars at the highest level will be used for deformation initialization.
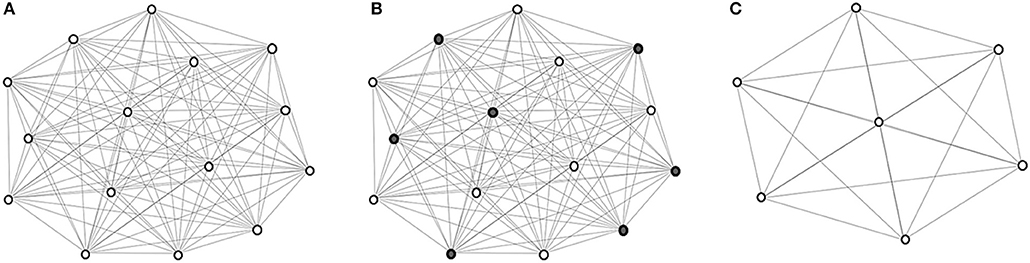
Figure 1. Graph coarsening. (A) The initial graph G0 with vertex set V0 representing the images in and edge set E0 representing the edges between image pairs, computed using (1). (B) Coarse vertices (shaded). (C) Coarse vertices at a subsequent level.
2.1.2. Deep Learning Based Registration
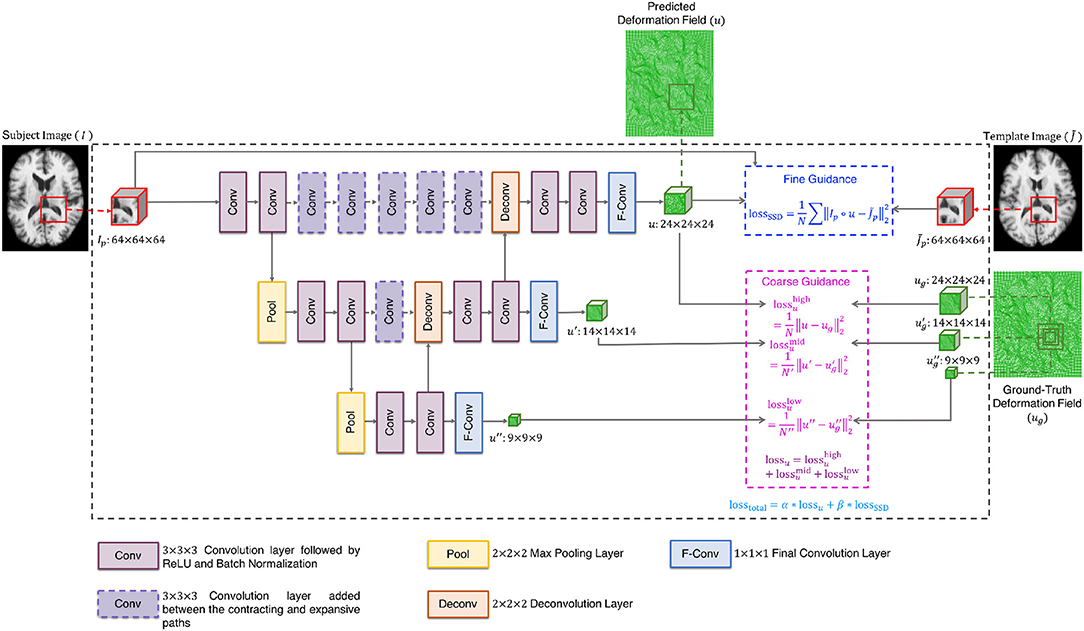
The cluster exemplars obtained at the highest level (h) of graph coarsening, , will be used to deform all the images in the dataset. Registration is performed using a convolutional neural network (CNN) (Fan et al., 2018) with these exemplars regarded as fixed templates. The CNN is based on U-Net (Ronneberger et al., 2015), with additional convolutional layers at same levels of contracting and expansive paths to learn high-level features that are helpful in predicting the deformation fields (Figure 2). More specifically, the network consists of (i) 3 × 3 × 3 convolutional layers followed by ReLU and batch normalization, (ii) 2 × 2 × 2 max pooling layers, (iii) 2 × 2 × 2 deconvolutional layers, (iv) 1 × 1 × 1 final convolutional layers, and (v) 3 × 3 × 3 convolutional layers added between the contracting and expansive paths. In addition, a loss function is added in each layer to ensure that the parameters of the frontal convolutional layers are updated. This strategy helps to avoid over-fitting caused by the more frequent parameter update of the later convolutional layers. The registration network takes the overlapping 64 × 64 × 64 patches as input, and outputs 24 × 24 × 24 deformation field patches. In order to obtain a deformation field that is equal in size to the input image, we extract the predicted deformation field patches with a step size of 24 without overlap. A CNN is associated with each template. To train the CNNs, we first select the template which is most similar to all other templates, based on the following criterion:
The CNN is trained using dual-guidance: (i) coarse guidance from deformation fields estimated using an existing registration method and (ii) fine guidance using image dissimilarity between and the warped subject images. The latter ensures that the training does not completely depend on the guidance from ground-truth deformation fields estimated from the existing registration method. We used diffeomorphic Demons (Vercauteren et al., 2009) to estimate the ground-truth deformation fields. Our learning model is therefore semi-supervised with loss function consisting of two components: (i) the Euclidean distance between the predicted and the ground-truth deformation fields (lossu) and (ii) the sum of squared intensity difference between and the subject image warped using the predicted deformation field (lossSSD). As shown in Figure 2, the deformation field is predicted at three different resolution levels, therefore lossu is comprised of the loss functions computed at each level i.e., . The two components of the total loss function were dynamically balanced during the training stage (losstotal = α * lossu + β *lossSSD). Initially the first component is given a higher weight α to converge quickly and then the prediction is refined by giving more weight β to the second component of the loss function. At each epoch, the sum of the weights of the two components was equal to 1. We trained the network for 10 epochs, which we found enough for convergence.
We used a 75:25 train-test split of the dataset (excluding the templates) and trained the network using the ADAM optimizer with a learning rate of 1 × 10−2. Once the network was trained with respect to , the networks for the other templates were trained using transfer learning by initializing the weights with those of the network trained with respect to and updating the weights using the ADAM optimizer with an overall learning rate of 1 × 10−7. We kept a small learning rate as all the CNNs have a common task domain. Transfer learning allows the training of the CNNs to be expedited.
Once the networks have been trained, each of them is used to register the images to the templates, producing a set of deformation fields . Our goal is to warp each image to a hidden common space using the average deformation computed with respect to the templates. To achieve this, we first invert the deformation fields as . The deformation field for an image Ii is computed as
and is used to warp the image Ii via (see Figure 3). This process is repeated for all the images, producing a set of warped .
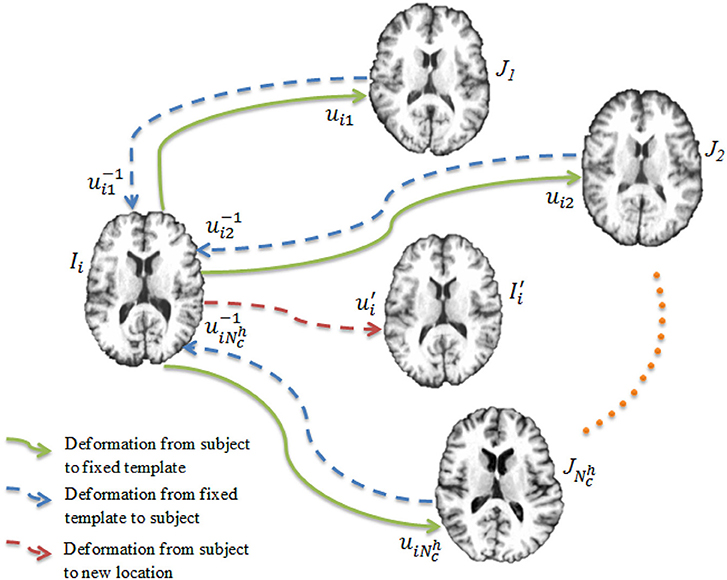
Figure 3. Deep learning registration. Image Ii is registered to templates using CNNs. The image is warped from Ii using the average deformation field computed using Equation (4).
The alignment of the images in can be improved using groupwise registration algorithms. Since the differences between the images are smaller, they can be brought to a common space more efficiently in a smaller amount of time.
3. Results and Discussion
3.1. Evaluation of Registration Performance
The efficacy of our method was evaluated both qualitatively and quantitatively, in comparison with SharpMean (Wu et al., 2011), ABSORB (Jia et al., 2010), and GroupMean (Joshi et al., 2004). With initialization, the methods are denoted as iSharpMean, iABSORB, and iGroupMean.
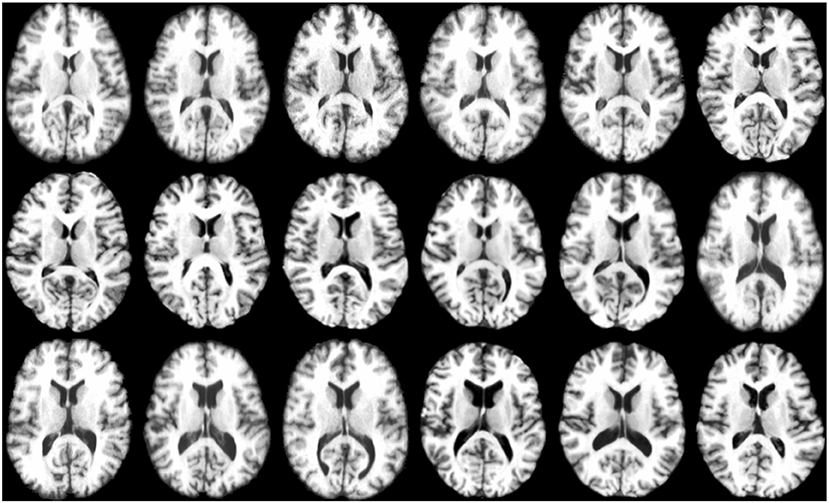
The experiments were conducted by combining all the T1 weighted MR images from LONI LPBA40 (Shattuck et al., 2008) and IXI1 datasets. As LONI LPBA40 has 40 images and IXI has 30 images, in our dataset we have a total of 70 images that we registered jointly. All the images have 184 slices of 220 × 220 pixels with isotropic voxel size of 1mm3. The age range for LPBA40 is 29.20 ± 6.30 years and IXI is 20–54 years. The union of two datasets ensures that the images exhibit large inter-subject variability characterized by the presence of different age groups (young adults and elderly). Figure 4 shows some typical images from this dataset, indicating significant inter-subject differences. All the images were histogram matched and affine registered using ANTs (Avants et al., 2008). The image which is most similar to the rest of the images in the dataset is used as template for affine registration. Also, for training the deep learning based registration network, we had a total of 69 images (excluding the template), which were divided using a 75:25 train-test split. This means that 52 images were used for training and the remaining 17 images were used for testing.
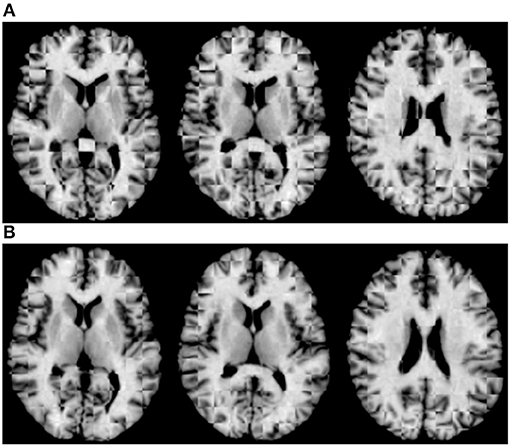
For qualitative assessment, we used checkerboard images to simultaneously display two images so that structural boundaries can be compared. In the ideal situation where two images are perfectly aligned, the checkerboard image will be seamless. Figure 5A shows the axial-view checkerboard image of two randomly selected images before initialization, showing apparent misalignments especially in the lateral ventricular region. In contrast, Figure 5B shows that deformation initialization reduces variability across images.
Figure 6 shows the 3D surface renderings of the group mean images given by different methods. It can be seen that groupwise registration with initialization improves sharpness of the group mean images of SharpMean, ABSORB, and GroupMean.
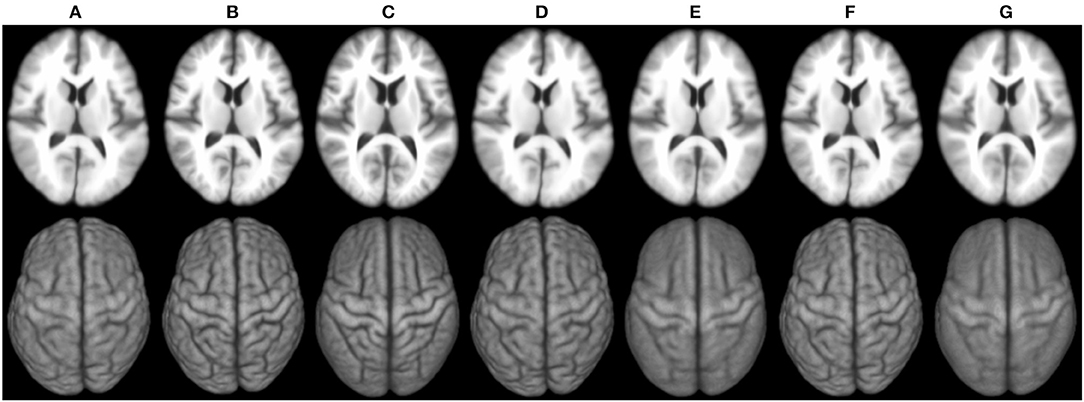
Figure 6. 3D surface renderings (bottom row) of the group mean images (top row) generated by (A) deformation initialization, (B) iSharpMean, (C) SharpMean, (D) iABSORB, (E) ABSORB, (F) iGroupMean, and (G) GroupMean.
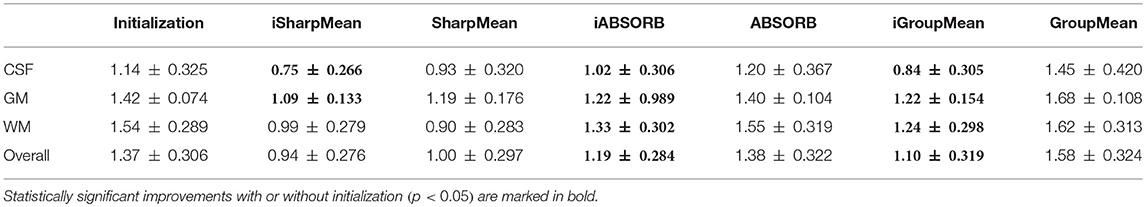
For quantitative evaluation, we computed the NCCs between all the warped images with respect to the group mean image generated by each method. The results, shown in Table 1, indicate that deformation initialization moves the images closer and groupwise registration with initialization yields higher mean NCC values (statistically significant with paired t-tests, p < 0.05), than without initialization.
Evaluation was also performed based on Dice ratio of hippocampus and brain tissue segmentation, i.e., cerebrospinal fluid (CSF), gray matter (GM), and white matter (WM). The Dice ratio (D) is given by
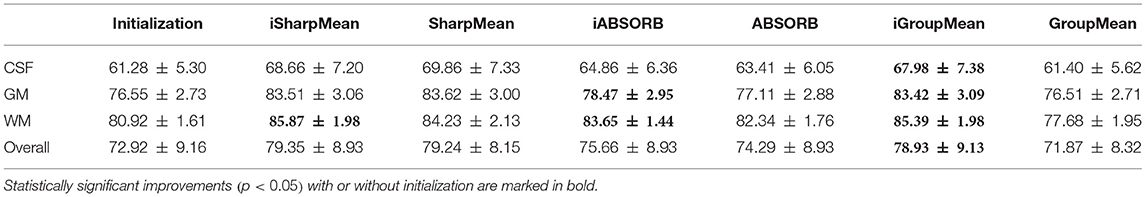
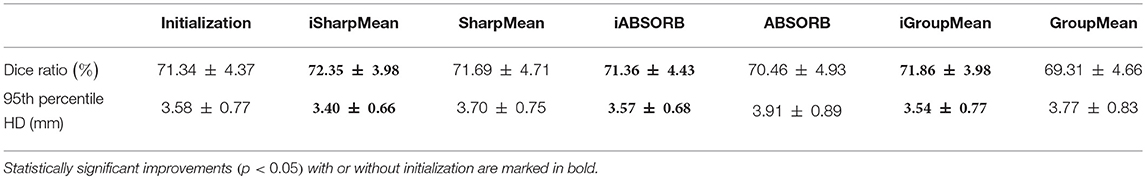
where V1 is the volume of a segmented tissue or hippocampus in the subject image domain and V2 is the volume of a segmented tissue or hippocampus in the reference image domain. The reference image for hippocampus and brain tissues is obtained in the common space, respectively by majority voting based on the hippocampus and tissue segmentation of all the warped images. The results for different brain tissues are summarized in Table 2. The overall Dice ratio achieved by deformation initialization is 72.92(±9.16)%. iSharpMean and SharpMean registration methods achieve comparable results with overall values of 79.35(±8.93) and 79.24(±8.15)%, respectively. The differences are not statistically significant (p > 0.05). The results for iABSORB and ABSORB are comparable, i.e., 75.66(±8.93) and 74.29(±8.93)%, respectively, and the differences are not statistically significant. iGroupMean [78.93(±9.13)%] yields higher Dice ratios with statistical significance (p < 0.05) than GroupMean [71.87(±8.32)%]. The results are summarized using box plots in Figure 7A for CSF, GM, and WM. The Dice ratios for hippocampus are summarized in Table 4. The groupwise registration methods with initialization show improved Dice ratio (statistically significant with p < 0.05) as compared to no initialization.
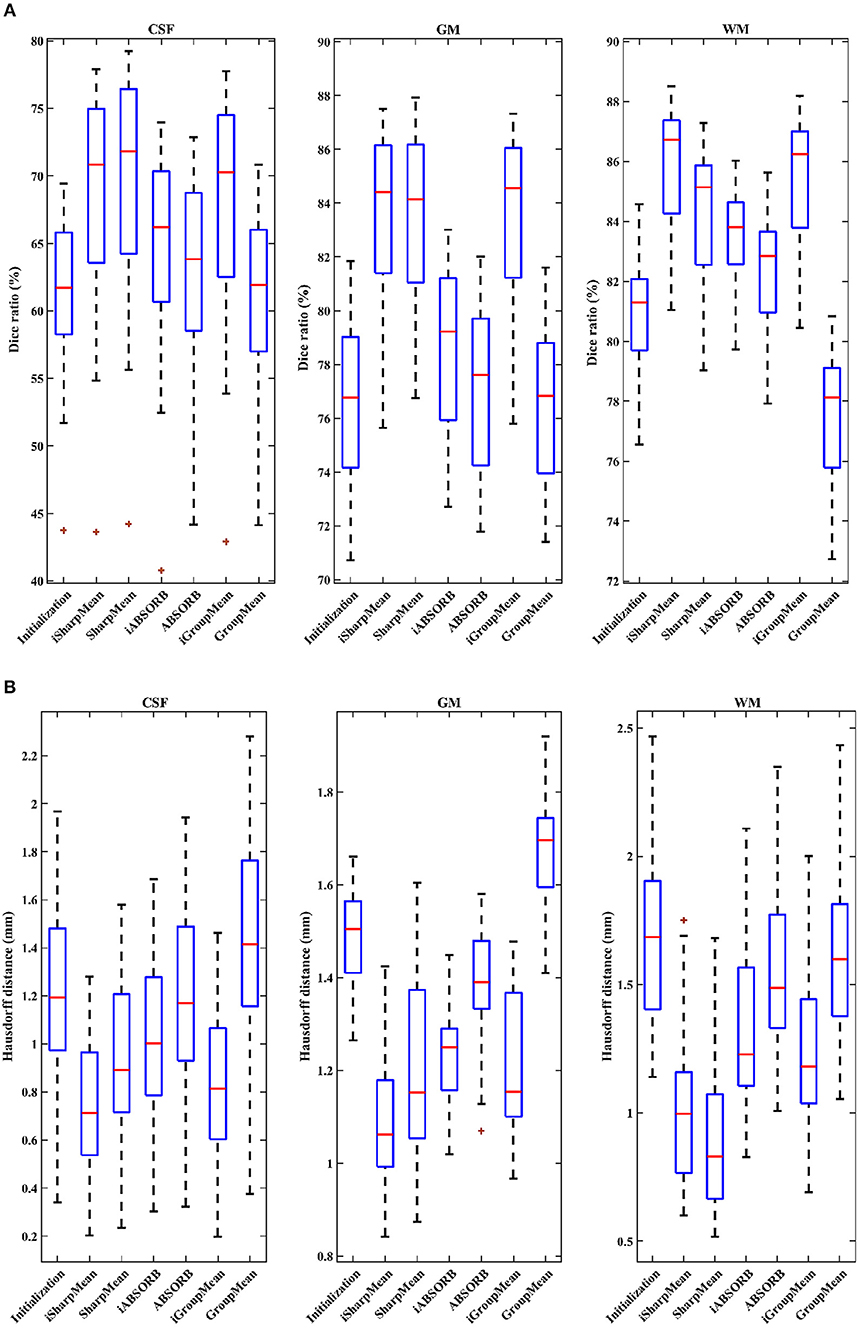
Figure 7. (A) Dice ratios (%) and (B) 95th percentile of Hausdorff distances for different tissue types (CSF, GM, WM).
We computed the 95th percentile of Hausdorff distance for performance evaluation. The Hausdorff distance (HD) is given by
where R and S are the 3D point sets of the boundaries of the tissue segmentations or hippocampus of the reference image and subject image, respectively. d(r, s) is the Euclidean distance between two finite point sets. We reported the 95th percentile of HD since it is less sensitive to outliers. Table 3 summarizes the results for different brain tissue types. The overall value yielded by deformation initialization is 1.37(±0.306)mm thus confirming its usefulness in the reduction of anatomical variability. In addition, deformation initialization improves groupwise registration. Box plots are shown in Figure 7B for evaluation. Table 4 summarizes the 95th percentile of HD for hippocampus. We can see that the initialized groupwise registration significantly decreased the Hausdorff distance as compared to without initialization.
Table 5 summarizes the computational times of all the methods along with the number of iterations needed for convergence. Groupwise registration methods with initialization are faster and converge very quickly, compared with no initialization. More specifically, SharpMean took around 18 h, whereas iSharpMean converged within 3 h with comparable accuracy. iABSORB achieved results comparable to ABSORB and requires 21 h less. iGroupMean took just half an hour to converge, compared with 13.5 h taken by GroupMean. These results indicate that initialization improves registration accuracy by reducing anatomical variability and is hence important for detection of subtle changes associated with aging and disorders.
3.2. Significance of Graph Coarsening
To investigate the impact of graph coarsening on template selection, we performed two experiments.
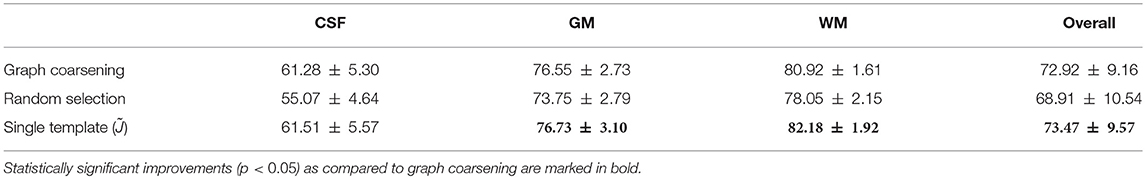
In the first experiment, instead of utilizing graph coarsening, we used randomly selected templates for deformation initialization. The number of selected templates was kept consistent with that given by graph coarsening. It can be observed from Table 6 that the accuracy decreases in comparison with initialization using graph coarsening. This demonstrates the importance of taking into consideration the image distribution in template selection.
In the second experiment, we evaluated the effects of the number of templates. Using a single template (i.e., ), although giving good alignment (Table 6), will affect subsequent population analysis [e.g., voxel-based morphometry (VBM)] with bias toward the selected template and neglecting inter-subject variation. Moreover, if the selected template image is an outlier, population analysis can be severely affected. Graph coarsening takes into account inter-subject heterogeneity and determines multiple images that are representative of image sub-populations. The higher Dice ratios given by single template case is partially due to the greater image sharpness when no averaging is performed.
3.3. Generalizability
To assess generalizability, we conducted two experiments. In the first experiment, we trained the registration network with the LONI LPBA40 dataset and tested it with the IXI dataset. In the second experiment, we trained the registration network with both LONI LPBA40 and IXI datasets and tested it on the IXI dataset. Figure 8 shows the Dice ratios for 78 ROIs (see Table 7) of the IXI dataset. The overall Dice ratio achieved in first and second experiment is 73.07(±9.91) and 74.29(±9.84)%, respectively (p > 0.05), indicating generalizability of our method to the unseen image datasets.
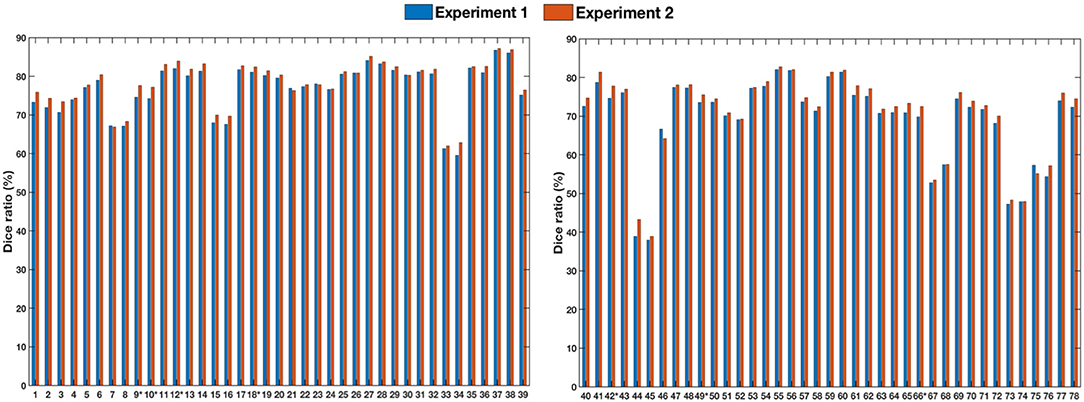
Figure 8. Dice ratios for different ROIs of IXI dataset. “*” indicates statistically significant improvements (p < 0.05).
4. Conclusion
In this paper, we presented an effective and efficient deformation initialization method for groupwise registration of images with large anatomical differences. Deformation initialization decreases structural discrepancies and brings the images closer to the common space. The results validated that deformation initialization improves alignment accuracy and significantly reduces computation times.
Author Contributions
SA implemented the code, performed experiments, and prepared the manuscript draft. JF contributed in code implementation. PD and XC helped in designing the experiments. JF, PD, and XC participated in idea discussion. P-TY contributed in algorithm development and critical revision of the manuscript. DS designed the project, led the team, and revised the manuscript.
Funding
This work was supported in part by NIH grants (EB006733, EB008374, MH100217, AG053867).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1. ^ Available online at http://brain-development.org/ixi-dataset/
References
Aganj, I., Iglesias, J. E., Reuter, M., Sabuncu, M. R., and Fischl, B. (2017). Mid-space-independent deformable image registration. Neuroimage 152, 158–170. doi: 10.1016/j.neuroimage.2017.02.055
Agnello, L., Comelli, A., Ardizzone, E., and Vitabile, S. (2016). Unsupervised tissue classification of brain MR images for voxel-based morphometry analysis. Int. J. Imag. Syst. Tech. 26, 136–150. doi: 10.1002/ima.22168
Arabi, H., and Zaidi, H. (2017). Comparison of atlas-based techniques for whole-body bone segmentation. Med. Image Anal. 36, 98–112. doi: 10.1016/j.media.2016.11.003
Avants, B. B., Epstein, C. L., Grossman, M., and Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41. doi: 10.1016/j.media.2007.06.004
Brock, K. K., Mutic, S., McNutt, T. R., Li, H., and Kessler, M. L. (2017). Use of image registration and fusion algorithms and techniques in radiotherapy: report of the AAPM radiation therapy committee task group no. 132. Med. Phys. 44, e43–e76. doi: 10.1002/mp.12256
Cammin, J., and Taguchi, K. (2010). “Image-based motion estimation for cardiac CT via image registration,” in Proc. SPIE, Vol. 7623, eds, B. M. Dawant, and D. R. Haynor (San Diego, CA: SPIE), 76232G–10.
Castadot, P., Lee, J. A., Parraga, A., Geets, X., Macq, B., and Grégoire, V. (2008). Comparison of 12 deformable registration strategies in adaptive radiation therapy for the treatment of head and neck tumors. Radiother. Oncol. 89, 1–12. doi: 10.1016/j.radonc.2008.04.010
Chen, H., Zhao, Y., Li, Y., Lv, J., and Liu, T. (2017). “Inter-subject fMRI registration based on functional networks,” in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) (Melbourne, QLD), 863–867.
Csapo, I., Davis, B., Shi, Y., Sanchez, M., Styner, M., and Niethammer, M. (2013). Longitudinal image registration with temporally-dependent image similarity measure. IEEE Trans. Med. Imaging 32, 1939–1951. doi: 10.1109/TMI.2013.2269814
Cunliffe, A. R., Contee, C., Armato, S. G., White, B., Justusson, J., Malik, R., et al. (2015). Effect of deformable registration on the dose calculated in radiation therapy planning CT scans of lung cancer patients. Med. Phys. 42, 391–399. doi: 10.1118/1.4903267
Fan, J., Cao, X., Yap, P.-T., and Shen, D. (2018). BIRNet: brain image registration using dual-supervised fully convolutional networks. arXiv:1802.04692.
Flower, E., Do, V., Sykes, J., Dempsey, C., Holloway, L., Summerhayes, K., et al. (2017). Deformable image registration for cervical cancer brachytherapy dose accumulation: organ at risk dose-volume histogram parameter reproducibility and anatomic position stability. Brachytherapy 16, 387–392. doi: 10.1016/j.brachy.2016.12.006
Fortin, D., Basran, P. S., Berrang, T., Peterson, D., and Wai, E. S. (2014). Deformable versus rigid registration of PET/CT images for radiation treatment planning of head and neck and lung cancer patients: a retrospective dosimetric comparison. Radiat. Oncol. 9:50. doi: 10.1186/1748-717X-9-50
Gorthi, S., Duay, V., Bresson, X., Cuadra, M. B., Castro, F. J., Pollo, C., et al. (2011). Active deformation fields: dense deformation field estimation for atlas-based segmentation using the active contour framework. Med. Image Anal. 15, 787–800. doi: 10.1016/j.media.2011.05.008
Gu, X., Dong, B., Wang, J., Yordy, J., Mell, L., Jia, X., et al. (2013). A contour-guided deformable image registration algorithm for adaptive radiotherapy. Phys. Med. Biol. 58, 1889–1901. doi: 10.1088/0031-9155/58/6/1889
Hamm, J., Davatzikos, C., and Verma, R. (2009). “Efficient large deformation registration via geodesics on a learned manifold of images,” in Medical Image Computing and Computer-Assisted Intervention MICCAI 2009, eds G.-Z. Yang, D. Hawkes, D. Rueckert, A. Noble, and C. Taylor (London; Berlin; Heidelberg: Springer), 680–687.
Heinrich, M. P., Simpson, I. J., Papież, B. W., Brady, S. M., and Schnabel, J. A. (2016). Deformable image registration by combining uncertainty estimates from supervoxel belief propagation. Med. Image Anal. 27, 57–71. doi: 10.1016/j.media.2015.09.005
Hu, Y., Ahmed, H. U., Taylor, Z., Allen, C., Emberton, M., Hawkes, D., et al. (2012). MR to ultrasound registration for image-guided prostate interventions. Med. Image Anal. 16, 687–703. doi: 10.1016/j.media.2010.11.003
Jia, H., Wu, G., Wang, Q., and Shen, D. (2010). ABSORB: atlas building by self-organized registration and bundling. Neuroimage 51, 1057–1070. doi: 10.1016/j.neuroimage.2010.03.010
Joshi, A. A., Leahy, R. M., Badawi, R. D., and Chaudhari, A. J. (2016). Registration-based bone morphometry for shape analysis of the bones of the human wrist. IEEE Trans. Med. Imaging 35, 416–426. doi: 10.1109/TMI.2015.2476817
Joshi, S., Davis, B., Jomier, M., and Gerig, G. (2004). Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage 23, S151–S160. doi: 10.1016/j.neuroimage.2004.07.068
König, L., Derksen, A., Papenberg, N., and Haas, B. (2016). Deformable image registration for adaptive radiotherapy with guaranteed local rigidity. Radiat. Oncol. 11:122. doi: 10.1186/s13014-016-0697-4
Lee, C.-Y., Wang, H.-J., Lai, J.-H., Chang, Y.-C., and Huang, C.-S. (2017). Automatic marker-free longitudinal infrared image registration by shape context based matching and competitive winner-guided optimal corresponding. Sci. Rep. 7:39834. doi: 10.1038/srep39834
Li, M., Miller, K., Joldes, G. R., Kikinis, R., and Wittek, A. (2016). Biomechanical model for computing deformations for whole-body image registration: a meshless approach. Int. J. Numer. Method Biomed. Eng. 32, e02771–e02788. doi: 10.1002/cnm.2771
Liao, S., Wu, G., and Shen, D. (2012). A statistical framework for inter-group image registration. Neuroinformatics 10, 367–378. doi: 10.1007/s12021-012-9156-z
Matsuda, H. (2013). Voxel-based morphometry of brain MRI in normal aging and Alzheimer's disease. Aging Dis. 4, 29–37. Available online at: http://www.aginganddisease.org/EN/Y2013/V4/I1/29
Meschini, G., Seregni, M., Pella, A., Baroni, G., and Riboldi, M. (2016). SU-F-J-80: deformable image registration for residual organ motion estimation in respiratory gated treatments with scanned carbon ion beams. Med. Phys. 43(6Part10), 3424–3425. doi: 10.1118/1.4955988
Oh, S., and Kim, S. (2017). Deformable image registration in radiation therapy. Radiat. Oncol. J. 35, 101–111. doi: 10.3857/roj.2017.00325
Onofrey, J. A., Papademetris, X., and Staib, L. H. (2015). Low-dimensional non-rigid image registration using statistical deformation models from semi-supervised training data. IEEE Trans. Med. Imaging 34, 1522–1532. doi: 10.1109/TMI.2015.2404572
Rakai, L., Behjat, L., Martin, S., and Aguado, J. (2012). An algebraic multigrid-based algorithm for circuit clustering. Appl. Math. Comput. 218, 5202–5216. doi: 10.1016/j.amc.2011.10.084
Razlighi, Q. R., and Kehtarnavaz, N. (2014). Spatial mutual information as similarity measure for 3-D brain image registration. IEEE J. Transl. Eng. Health Med. 2, 27–34. doi: 10.1109/JTEHM.2014.2299280
Ribbens, A., Hermans, J., Maes, F., Vandermeulen, D., and Suetens, P. (2010). “SPARC: inified framework for automatic segmentation, probabilistic atlas construction, registration and clustering of brain MR images,” in 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (Rotterdam), 856–859.
Risser, L., Vialard, F.-X., Baluwala, H. Y., and Schnabel, J. A. (2013). Piecewise-diffeomorphic image registration: application to the motion estimation between 3D CT lung images with sliding conditions. Med. Image Anal. 17, 182–193. doi: 10.1016/j.media.2012.10.001
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-Net: convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, eds N. Navab, J. Hornegger, W. M. Wells, and A. F. Frangi (Munich; Cham: Springer), 234–241.
Ruge, J. W., and Stüben, K. (1987). “Chapter 4. Algebraic multigrid,” in Multigrid Methods, ed S. F. McCormick (Philadelphia, PA: SIAM), 73–130.
Sabuncu, M. R., Balci, S. K., and Golland, P. (2008). “Discovering modes of an image population through mixture modeling,” in Medical Image Computing and Computer-Assisted Intervention MICCAI 2008, eds D. Metaxas, L. Axel, G. Fichtinger, and G. Székely (New York, NY; Berlin; Heidelberg: Springer), 381–389.
Sabuncu, M. R., Balci, S. K., Shenton, M. E., and Golland, P. (2009). Image-driven population analysis through mixture modeling. IEEE Trans. Med. Imaging 28, 1473–1487. doi: 10.1109/TMI.2009.2017942
Safro, I., Sanders, P., and Schulz, C. (2015). Advanced coarsening schemes for graph partitioning. J. Exp. Algorithmics 19, 369–380. doi: 10.1007/978-3-642-30850-5_32
Samavati, N., Velec, M., and Brock, K. K. (2016). Effect of deformable registration uncertainty on lung SBRT dose accumulation. Med. Phys. 43, 233–240. doi: 10.1118/1.4938412
Schmidt-Richberg, A., Werner, R., Handels, H., and Ehrhardt, J. (2012). Estimation of slipping organ motion by registration with direction-dependent regularization. Med. Image Anal. 16, 150–159. doi: 10.1016/j.media.2011.06.007
Shattuck, D. W., Mirza, M., Adisetiyo, V., Hojatkashani, C., Salamon, G., and Narr, K. L., et al. (2008). Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39, 1064–1080. doi: 10.1016/j.neuroimage.2007.09.031
Shi, L., Wang, D., Chu, W. C., Burwell, R. G., Freeman, B. J., and Heng, P. A., et al. (2009). Volume-based morphometry of brain MR images in adolescent idiopathic scoliosis and healthy control subjects. Am. J. Neuroradiol. 30, 1302–1307. doi: 10.3174/ajnr.A1577
Spiclin, Z., Likar, B., and Pernus, F. (2012). Groupwise registration of multimodal images by an efficient joint entropy minimization scheme. IEEE Trans. Image Process. 21, 2546–2558. doi: 10.1109/TIP.2012.2186145
Suh, J. W., Scheinost, D., Dione, D. P., Dobrucki, L. W., Sinusas, A. J., and Papademetris, X. (2011). A non-rigid registration method for serial lower extremity hybrid SPECT/CT imaging. Med. Image Anal. 15, 96–111. doi: 10.1016/j.media.2010.08.002
Sun, W., Poot, D. H. J., Smal, I., Yang, X., Niessen, W. J., and Klein, S. (2017). Stochastic optimization with randomized smoothing for image registration. Med. Image Anal. 35, 146–158. doi: 10.1016/j.media.2016.07.003
Toga, A. W., and Thompson, P. M. (2001). The role of image registration in brain mapping. Image Vis. Comput. 19, 3–24. doi: 10.1016/S0262-8856(00)00055-X
Velec, M., Moseley, J. L., Eccles, C. L., Craig, T., Sharpe, M., and Dawson, L. A., et al. (2011). Effect of breathing motion on radiotherapy dose accumulation in the abdomen using deformable registration. Int. J. Radiat. Oncol. Biol. Phys. 80, 265–272. doi: 10.1016/j.ijrobp.2010.05.023
Vercauteren, T., Pennec, X., Perchant, A., and Ayache, N. (2009). Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 45(1 Suppl. 1), S61–S72. doi: 10.1016/j.neuroimage.2008.10.040
Wachinger, C., and Navab, N. (2013). Simultaneous registration of multiple images: similarity metrics and efficient optimization. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1221–1233. doi: 10.1109/TPAMI.2012.196
Wang, Q., Chen, L., Yap, P.-T., Wu, G., and Shen, D. (2010). Groupwise registration based on hierarchical image clustering and atlas synthesis. Hum. Brain Mapp. 31, 1128–1140. doi: 10.1002/hbm.20923
Wang, Y., Seguro, F., Kao, E., Zhang, Y., Faraji, F., Zhu, C., et al. (2017). Segmentation of lumen and outer wall of abdominal aortic aneurysms from 3D black-blood MRI with a registration based geodesic active contour model. Med. Image Anal. 40, 1–10. doi: 10.1016/j.media.2017.05.005
Wu, G., Jia, H., Wang, Q., and Shen, D. (2011). Sharpmean: groupwise registration guided by sharp mean image and tree-based registration. Neuroimage 56, 1968–1981. doi: 10.1016/j.neuroimage.2011.03.050
Wu, G., Wang, Q., and Shen, D. (2012). Registration of longitudinal brain image sequences with implicit template and spatial-temporal heuristics. Neuroimage 59, 404–421. doi: 10.1016/j.neuroimage.2011.07.026
Xiao, B., Cheng, J., and Hancock, E. (2013). Graph-based Methods in Computer Vision: Developments and Applications. Information Science Reference (Hershey, PA: IGI Global).
Yang, D., Lu, W., Low, D. A., Deasy, J. O., Hope, A. J., and El Naqa, I. (2008). 4D-CT motion estimation using deformable image registration and 5D respiratory motion modeling. Med. Phys. 35, 4577–4590. doi: 10.1118/1.2977828
Yang, X., Kwitt, R., Styner, M., and Niethammer, M. (2017). Quicksilver: fast predictive image registration–a deep learning approach. Neuroimage 158, 378–396. doi: 10.1016/j.neuroimage.2017.07.008
Keywords: groupwise registration, graph coarsening, deep learning, convolutional neural network, MRI, brain templates
Citation: Ahmad S, Fan J, Dong P, Cao X, Yap P-T and Shen D (2019) Deep Learning Deformation Initialization for Rapid Groupwise Registration of Inhomogeneous Image Populations. Front. Neuroinform. 13:34. doi: 10.3389/fninf.2019.00034
Received: 15 October 2018; Accepted: 23 April 2019;
Published: 14 May 2019.
Edited by:
Pierre Bellec, Université de Montréal, CanadaReviewed by:
Carlton Chu, Google, United KingdomStavros I. Dimitriadis, Cardiff University School of Medicine, United Kingdom
Copyright © 2019 Ahmad, Fan, Dong, Cao, Yap and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pew-Thian Yap, cHR5YXBAbWVkLnVuYy5lZHU=
Dinggang Shen, ZGdzaGVuQG1lZC51bmMuZWR1
 Sahar Ahmad
Sahar Ahmad Jingfan Fan
Jingfan Fan Pei Dong1
Pei Dong1 Xiaohuan Cao
Xiaohuan Cao Pew-Thian Yap
Pew-Thian Yap Dinggang Shen
Dinggang Shen