- 1Department of Laboratory Medicine and Pathology, University of Albert Hospital, Edmonton, AB, Canada
- 2Department of Pediatrics, Stollery Children's Hospital, University of Alberta Hospital, Edmonton, AB, Canada
The recent pandemic of coronavirus infectious illness 2019 (COVID19) triggered by SARS-CoV-2 has rapidly spread around the globe, generating in severe events an acute, highly lethal pneumonia and death. In the past two hitherto similar CoVs, the severe acute respiratory syndrome CoV (SARS-CoV-1) and Middle East respiratory syndrome CoV (MERS-CoV) also gained universal attention as they produced clinical symptoms similar to those of SARS-CoV-2 utilizing angiotensin-converting enzyme 2 (ACE2) receptor and dipeptidyl peptidase 4 (DPP4) to go into the cells. COVID-19 may also present with overtly neurological symptoms. The proper understanding of the expression and dissemination of ACE2 in central and peripheral nerve systems is crucial to understand better the neurological morbidity caused by COVID-19. Using the STRING bioinformatic tool and references through text mining tools associated to Coronaviruses, we identified SAMHD1 as the probable link to neurological symptoms. Paralleled to the response to influenza A virus and, specifically, respiratory syncytial virus, SARS-CoV-2 evokes a response that needs robust induction of a subclass of cytokines, including the Type I and, obviously, Type III interferons as well as a few chemokines. We correlate ACE2 to the pathogenesis and neurologic complications of COVID-19 and found that SAMHD1 links to NF-κB pathway. No correlation was found with other molecules associated with Coronavirus infection, including ADAR, BST2, IRF3, IFITM3, ISG15, MX1, MX2, RNASEL, RSAD2, and VPRBP. We suggest that SAMHD1 is the molecule that may be behind the mechanisms of the neurological complications associated with COVID-19.
Introduction
Coronaviruses (CoVs), which belong to Coronaviridae family, are enveloped RNA viruses that have been associated with respiratory and extra-respiratory (e.g., enteric and neurological) diseases in various animal species (1). Human coronaviruses (HCoVs) are commonly known to be accountable for both lower and upper infections of the respiratory tract (2). In the last two decades, SARS-CoV-1 and MERS-CoV caused severe acute respiratory syndrome coronavirus and the Middle East respiratory syndrome, which triggered a large-scale public health response (3). In December 2019, another novel coronavirus SARS-CoV-2-, labeled astempestively as COVID 19, emerged and has been the cause of a worldwide pandemic (4). SARS-CoV-2 is found to have 75–80% remarkably genomic similarity to SARS-CoV-1 and 50% genomic similarity to the Middle East Respiratory Syndrome coronavirus (MERS-CoV) (5). All three of these viruses belong to the family of β-coronaviruses, with bats being the possible common reservoir (6). These β-coronaviruses typically produce respiratory and gastrointestinal symptoms in human and animal hosts, respectively (5–7). However, there is accumulating evidence that in addition to these two systems, coronaviruses may conquer the central nervous system (CNS) as well causing diseases affecting the neurological system. In fact, patients of COVID-19 have been reported to exhibit some symptoms of Guillain Barre syndrome (GBS) as well as other neurological diseases (8–10). Pondering the high similarity between SARS-CoV-1 and SARS-CoV-2, some preliminary studies are suggesting that the potential CNS invasion may partially be responsible for the mortality (other than morbidity) associated with COVID-19 infection and have some late neurologic sequelae (8, 11).
COVID-19 Virus Obtain Entry In The Interior of The Host Cells Via ACE2 Receptor
These enveloped viruses harbor a positive-strand RNA genome with dimensions of up to 31 kb, which is representative of the largest known genome including all RNA viruses (9). This genome consists of specific genes coding numerous structural and non-structural proteins. Along With these proteins, the Spike or S protein possesses a unique biological significance (9). It has been associated with properties such as tropism and its modulation (9, 10). It is important to remember that the S units of coronaviruses are crucial in aiming for a cellular receptor that mediates infection in the target cells (10). Similar to SARS, angiotensin-converting enzyme 2 (ACE2) has been recognized as the functional receptor for SARS-CoV-2 as well (7, 12). The intracellular penetration of SARS-CoV-1 and MERS-CoV utilize the ACE2 receptor and DPP4, respectively. With the mRNA programming several other proteins, SARS-CoV-2, uses S1 to enable the attachment of the virion to the cellular membrane by networking with the host ACE2 receptor (13). It has also been elucidated that spike proteins belonging to all three CoV are not identical, even though they are considered similar. The ACE2 binding affinity identified in the 2019-nCoV spike protein ectodomain is roughly 10–20-fold higher as compared to that of the SARS-CoV spike protein, thus justifying the higher required affinity of the COVID-19 spike protein to the human counterpart (ACE2) receptor (13). ACE2, which is produced in multiple human organs, including human upper airway epithelia, lung, kidney, and gut parenchyma as well as vascular endothelia, is also present in the brain (13–16).
The brain expression of ACE2 is indicative of the fact that SARS-CoV-2 may obviously cause some symptoms of neurological type via direct or indirect mechanisms (13). Investigations have reported the occurrance of various neurological symptoms. In the following section, we discuss the distribution of ACE2 receptors in the brain and attempt to address potential short term and long-term implications of brain involvement of this infection using STRING as a bioinformatic tool (17).
Clinical Characteristics
COVID-19 patients have been delineated to exhibit symptoms such as fever, myalgia, diarrhea, and cough (18). Typical clinical presentation of SARS-CoV-2 consists of fever, dry cough, and progressive respiratory distress at the onset of illness, which may lead to lethal pneumonia. It has been estimated that more than half of patients exhibiting dyspnea end up needing intensive care (8, 18, 19). Respiratory failure, infections, and cardiovascular events are the leading cause of mortality (8, 11, 20). Importantly, recent studies suggest that, in COVID-19 patients, in some cases, neurological symptoms occur as well (14, 19, 21). Furthermore, when present, neurological manifestation of COVID-19 infection has been categorized as CNS and peripheral nervous system (PNS) symptoms. CNS symptoms consist of headache, dizziness, impaired consciousness, and even, acute cerebrovascular disease (14, 22–26). PNS symptoms may consist of hyposmia, hypoplasia, neuralgia, and hypogeusia. Moreover, very recently, a patient with viral encephalitis has been reported (26). Overall, these findings support the notion that SARS-CoV-2 does possess neuroinvasive properties. It is crucial to gain knowledge of the molecule, which is central in its neurotropism.
Neurotropism is One Common Feature of CoVs
Several studies have elaborated on the neuroinvasive nature of CoVs (27). Animal studies have identified the brain as a primary focus organ for infection in rodents (mice) that are transgenic for the SARS-CoV receptor (ACE2) (28, 29). Additionally, further studies of this model elucidated that that the virus may enter the brain via the unique path of the olfactory bulb, causing infection with a rapid, spread (transneuronal) to connected cerebral areas. Death of the animal was assumed to be from non-functionality and/or destruction of infected neurons, notably those located in cardiorespiratory centers located in the medulla oblongata. Studies demonstrated that neurons are a highly vulnerable target for SARS-CoV (28). Neurological damage has been confirmed in the infection of coronavirus, such as in SARS-CoV and MERS-CoV, as well as, currently, SARS-CoV-2 (30). Studies on MERS-CoV demonstrated that low inoculum dosages of MERS-CoV viral particles were discovered only in the brain of mice, while none in the lung, indicative of the fact that mortality may have been associated with infection in the CNS (31–33). In particular, the brainstem has been shown to be severely infected by SARS-CoV (34, 35) or MERS-CoV (20). Additionally, the autopsy findings in patients who suffered from SARS-CoV infections have demonstrated robust evidence of the existence of SARS-CoV by SARS genome sequences being restricted to the cytoplasm of various neurons in the hypothalamus and brain cortex. Also, edema (cytologic hydropic degeneration) and scattered red disintegration of the neurons was reported in autopsies as well as suggestive of the fact that these viruses are capable of invading the nervous system (34).
Neuroinvasion of SARS-CoV-2 and SAMHD1
The idea has been put forth concerning how specific viruses enter into the nervous system. Numerous studies suggest that at first, certain CoVs colonize peripheral nerve terminals, and subsequently enter CNS via a synapse-connected path (35–38). Some other CoVs have been reported to enter the brain through trans-synaptic transfer (8). As far as SARS-CoV and MERS-CoV are concerned, experimental studies of animal models showed that SARS-CoV and MERS-CoV, when given through the nose, possibly invade the cerebrum via the olfactory nerves, spreading to some regions of the brain including thalamus and brainstem (8, 20, 34). Notably, it has been suggested that ACE2 olfactory (epithelial) support cells and stem cells produce ACE2 genes, as do cells located in the epithelium of the nasal respiratory tract in patients afflicted with COVID-19, which may show some parallels with attention-deficit/hyperactivity disorder (39–41). So far, in the light of the evidence provided enough, it is conceivable that SARS-CoV-2 is neuroinvasive, and it uses ACE2 as receptors to penetrate the cell (13). We target the ACE2 distribution in the brain, based on literature and discuss various associated pathways, which can potentially cause mortality in COVID-19 infected patients.
Evaluating protein -protein interactions can be tremendously essential in understanding the underlying biological mechanisms of any disease (42). Protein-protein interactions possess an integral role in several biological functions which include metabolic and signaling pathways' regulation, replication of DNA, and immunological recognition, among others (43). In addition to providing a reasonable analysis of interactions, STRING also offers a full set of images of the interaction network. It utilizes domains, pathways, and Gene Ontology annotations in order to support the functional enrichment of network proteins (44, 45). It is a bioinformatic tool and Text-Mining tools belonging to the Swiss Institute of Bioinformatics (SIB), European Molecular Biology Laboratory (EMBL), and Novo Nordisk Foundation Center Protein Research (CPR). Therefore, we used the bioinformatic tool STRING and references through text mining tools that has been correlated with Coronaviruses. Using STRING, we discovered SAMHD1, which is the acronyme for “sterile-α-motif and HD domain-containing protein 1” as the probable link to neurological symptoms. Equated to the response to influenza A virus and, specifically, respiratory syncytial virus, SARS-CoV-2 provokes a response that lacks induction of a subgroup of cytokines, including types I and III interferons as well as a few chemokines. In the setting of pathogenesis and neurologic complications of COVID-19, we could not find that SAMHD1 had links to ADAR, BST2, IRF3, IFITM3, ISG15, MX1, MX2, RNASEL, RSAD2, and VPRBP. SAMHD1 plays a role in the control of the innate immune response. The coded protein is upregulated in reaction to viral infection and may be involved in the intercession of tumor necrosis factor (TNF)-α pro-inflammatory responses.
Potential Host Responses To COVID-19
So far, we have established the potential invasion of the brain by COVID 19. At this point, It is also essential to consider the host response to this virus from the therapeutic perspective (2). It is commonly known that one core function of the innate immune system is to find virologic infections and induce antiviral effectors to prevent the spread of disease and activates antigen-specific adaptive response (46). As indicated above, COVID 19 lung tissue shows that SARS-CoV-2 evokes a distinctive response of transcriptional type, which lacks Types I and III interferons (IFN-I and IFN-III) expression (47). Along with this, there is an induction of well-categorized direct effectors of the innate immunologic response, including SAMHD1, MX1, IFITM3, and TRIM25, as well as the induction of viral sensors of RNA type such as RIG-I and the OAS1-3 genes (47). It is to note that previous studies on CoVs have also indicated that CoVs prevent IFN induction at transcription level as well as subsequent antiviral gene activation (48). Typically speaking, the innate response consists of signaling cascades that are activated upon the detection of the foreign pathogens in the host (49). Generally, during a viral infection, cytoplasmic sensors of protein type, such as RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated protein 5), bind to viral single-stranded RNA and/or, remarkably, double-stranded RNA (dsRNA) and signal through a protein called MAVS (mitochondrial antiviral signaling protein) (42, 43). Initiation of MAVS results in the subsequent activation of TBK1/IKKε, which are TANK-binding kinase 1 (TBK1) as well as the homolog IκB kinase (IKK) epsilon (IKKε, originally IKKi), two kinases which phosphorylate IRF3 (Interferon Regulatory Factor 3) to induce its dimerization, and import into the nucleus (43). TBK1/IKKε have been studied profusely in relation to their functions in inducing the type I interferon response. Nuclear IRF3, along with several other proteins, leads to transcription and induction of type I IFN to warn neighboring cells of the infection (44). As soon as IRF3 induces transcription of IFN-I, this potent cytokine is emitted from the cells. The NF-κB pathway is also remarkably activated via these mediators. Activation of classical NF-κB is essential for a successful immune response as well as for the survival and proliferation of cells (45). Interestingly, exogenous SAMHD1 expression in cells and, alternatively, SAMHD1 reconstitution in knockout cells have been shown to quell NF-κB activation and IFN-I induction, as specifically reported (50).
SAMHD1 is a vertebrate protein. It is engaged in the control of cellular deoxyribonucleoside triphosphates (dNTP) pool by catalyzing the hydrolysis of dNTP into 20-deoxynucleoside and triphosphate products (51). SAMDH1 has been reported to be a key mechanism of cell proliferation and is an essential player in dNTP homeostasis. SAMHD1 is a dNTP hydrolase which reduces the concentration of intracellular dNTP pools (52, 53). It has been suggested that the inhibitory effect of SAMHD1 on HIV-1 replication is due to its dNTPase action, or it has also been suggested that SAMHD1, or an associated protein, may limit HIV-1 replication through its RNase activity (54). In Figure 1 is shown an interactive panel of a number of proteins linked to SAMHD1 using version 11 of STRING. In mammalian cells, the rates of dNTPs are thought to be controlled by coaction of synthesis and degradation. In this process, synthesis of dNTPs involves two corridors, (1) de novo synthesis of dNDPs by ribonucleotide reductase (RNR) in the cytosol followed by (2) phosphorylation to dNTPs by a nucleoside-diphosphate kinase (55). Rescue of deoxyribonucleosides occurs by two parallel sets of deoxy-nucleoside and -nucleotide kinases in both the mitochondria and cytosol (55).
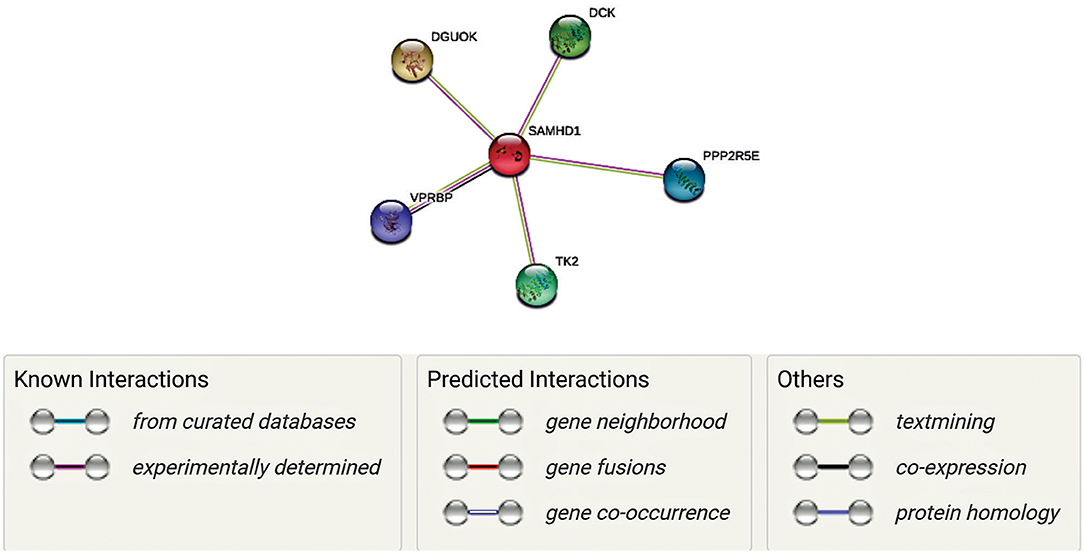
Figure 1. Network representation of proteins linked to SAMHD1 using version 11 of STRING. Each gene has a unique function as well as interacts with SAMDH1 in maintaining the concentrations of deoxyribonucleoside triphosphates (dNTPs) by regulating synthesis and degradation in DNA synthesis. Mutations of some of these genes have been correlated with AGs (Aicardi-Goutières syndrome (AGS) is a rare disease, characterized by genetically determined early-onset progressive encephalopathy). Thus, we hypothesize that these interactions are essential in the context of neurological symptoms associated with COVID 19.
Along with SAMDH1, DCK (deoxycytidine kinase), DGUOK (deoxyguanosine kinase), PPP2R5E (Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform), VPRBP (Vpr (HIV-1) binding protein), and TK2 (thymidine kinase 2) may play unique roles in this process. In fact, VPRBP has a role in cell growth, as it regulates the G1 phase of cell cycle and is crucial for the achievement of DNA replication in the S phase (56, 57). PPP2R5E has been demonstrated to have multiple signaling pathways and is vital during early development (58). While TK2, DGOUK, and DCK are involved in the deoxynucleoside salvage pathway (59–61), it is to note that VprBP has been linked with SAMDH1 in the context of its antiviral activity against HIV. Mutations of TK2 and DGOUK have been linked with SAMDH1 in the context of AGS elated neurological symptoms (56, 59). Interestingly, regulation of their function and the DGOUK mutations have also been known to cause a specific type of mitochondrial encephalopathy (61). Thus, it is conceivable that a SAMDH1 and its interactions with these proteins, as well as the potential impairment of these interactions, are essential in the antiviral response as well as neurological symptoms associated with might be involved in COVID 19.
Current evidence suggests SAMHD1 is an effective antiviral restriction factor suitably targeting numerous other medically relevant viruses (62). However, it is to note that the study on CHIKV and ZIKV reported that SAMHD1 might be proviral as it promotes their replication. This study found that Vpx/VLP-mediated SAMHD1 deterioration leads to a conspicuous decrease in the replication and, notably, virion production by both viruses. While the same treatment reduced the replication of CHIKV (Chikungunya virus, an alphavirus, replicon, it was also observed that the over-expression of SAMHD1 facilitated CHIKV and ZIKV (Zika virus) replication (63). It is worth mentioning that unlike HIV-1 and HBV, in CHIKV and ZIKV, the reverse transcription step is not needed for their replication, hence could be the reason for such contrasting results. Also, unlike HIV, in CHIKV and ZIKV the presence of dNTP (deoxynucleoside triphosphate) is not required. Furthermore, it has also been demonstrated that SAMHD1 interacts with the IKKε and IFN regulatory factor 7 (IRF7), thus causes the suppression of the IFN-I induction pathway by reducing IKKε-mediated IRF7 phosphorylation. Also, SAMHD1 inhibits NF-κB activation by interacting with NF-κB1/2, thereby reducing phosphorylation of the NF-κB inhibitory protein IκB (50, 63).
Mechanistically, SAMHD1 promotes an inhibiton of the NF-κB activation by interacting with NF-κB1/2 and decreasing phosphorylation of the NF-κB inhibitory protein IκBα. Also, SAMHD1 interacts with IKKε and IRF7, leading to the suppression of the IFN-I induction pathway by actively reducing IKKε -mediated IRF7 phosphorylation. The intimate interactions of endogenous SAMHD1 with NF-κB and IFN-I pathway proteins have been specifically validated in both human monocytic cells and primary macrophages (63).
Notably CHIKV and ZIKV both show neurological symptoms as well (63). It is important to recall that numerous patients with Zika fever have also been reported to have symptoms of Guillain-Barre Syndrome (GBS) (64). Hence it can be assumed that SAMDH1 might be a commonality and a causative factor among these viral infections. Along with them, SAMDH1 has been associated with several other disorders that show signs and symptoms of neurological involvement (65–68). These include Aicardi-Goutieres syndrome (AGS), which is an autoimmune condition. Aicardi-Goutieres syndrome (AGS), which is caused by irregular type I IFN responses, results in physical and intellectual disabilities. AGS patients end up with mild to severe mental or physical impairments. Additionally, SAMHD1 mutation has been reported to play essential roles in immunoregulation and cerebral vascular homeostasis. It has been thought that cerebrovascular events (e.g., stroke) associated with specific mutations in SAMHD1 extend the phenotypic spectrum of AGS (69). Various inflammatory vasculopathies of the brain that may lead to cerebrovascular stenoses and early stroke have been correlated with SAMDH1 as well (70). Moreover, SAMHD1 expression has been associated with neurological symptoms of HIV as well (71). Also, mutations of SAMDH1 have been correlated with the pathogenesis of neurogenerative disorder as well (64). Overall, these findings suggest that SAMDH1 is involved in various diseases related to the brain.
Like other innate immunity proteins, SAMDH1 shows a crucial role in shaping innate and adaptive immunological responses to protect animals against viruses by disrupting the virus' life cycle (52). In response to that, viruses generate proteins that bind these host factors and alter their activity. This setting leads to an evolutionary conflictual event as immunity and virus proteins exquisitely adapt to prevent and restore binding, respectively (52, 54). One such adaptation has been reported to occur in a region of SAMHD1 that controls its activity, which influences its enzymatic properties suggesting that evolutionary conflictual eventhas engaged modulation of SAMHD1 regulation and function and has swayed both SAMHD1's dNTPase and antiviral actions (52, 54, 63).
There is evidence of SAMDH1 being both antiviral and proviral (50, 63). Thus, in the perspective of COVID 19 infection, it is profoundly essential to improve the knowledge of how SAMHD1 executes its activity in CNS as well as on the level of the whole organism. SAMHD1 plays a crucial role in shaping innate and adaptive immune responses. Since SAMHD1 has been demonstrated to efficiently inhibit a wide variety of viruses (52), many of these viruses, in turn, have developed various mechanisms to conquer SAMHD1 imposed block to viral replication (52, 63). Moreover, the NFκB signaling pathway has been shown to play a role in previous CoVs (72, 73). The fact that SAMDH1 has been shown to have proviral activities by suppressing the NFκB signaling pathway is significant. It is imperative to understand the full mechanisms in the pathogenesis of COVID 19, which may help develop new drugs and modulate SAMHD1 activity.
ACE2 Localization In The Human Brain
Evidence of Expression and Distribution of ACE2 in Brain
ACE2 that has been established as the functional receptor for SARS-CoV-2 is a transmembrane monocarboxypeptidase (8, 13, 74). It is significantly important to note that in addition to other human organs, kidneys, lungs, heart, and testes, it has been reported to be expressed in the brain in several databases as well as in the scientific literature (75). Initially, ACE2 was sequenced and cloned from the human failing heart and human lymphoma cDNA libraries (76, 77). The distribution of ACE2 in the brain was disputed in the beginning since original reports were unable to detect carboxypeptidase in this tissue (78). Later, studies of animal models exhibited the widespread presence of ACE2 mRNA and protein in the mouse brain (75). Animal model studies suggested that the ACE2 presence in nuclei has been identified in the central management of cardiovascular function including subfornical organ, paraventricular nucleus (PVN), the nucleus of the tractus solitarius (NTS), and rostral ventrolateral medulla (RVLM) (79, 80). The presence of ACE2 was also reported in non-cardiovascular areas, which included the motor cortex and raphe. Another study reported the occurrence of ACE2 mRNA and protein in the mouse brainstem (81). In addition, recent studies have also described the presence of ACE2 mRNA in the rat medulla oblongata and mouse brain (82, 83).
ACE2, a Member of Brain RAS May Have Cardioprotective, and Cerebro-Protective Effects
It is well-established the fact that ACE2 is an elegant and integral member of the renin-angiotensin system (RAS) (84). In the body, RAS functions to regulate blood pressure, kidney function, and salt and water homeostasis (84). Importantly, there is a RAS in the brain, as well (85). Brain RAS is similar to the other tissue RAS (86). ACE2 has been reported to be expressed specifically in the areas controlling central blood arterial pressure (78). Several studies have revealed that brain ACE2 has been involved in the progression of neurogenic hypertension, the molecular mechanisms, by which this protective effect occurs, seem to include the regulation of angiotensin (AT) receptors expression. ACE2 is capable of adjusting the AT2/AT1 and Mas/AT1 ratios so that it opposing the promotion of hypertension. In addition, animal model demonstrated that nitric oxide (NO) signaling pathways may also be influenced by ACE2 over-expression in the CNS as demonstrated by an overall reduction of hypertension of neurogenic type in syn-hACE2 mice (78, 79, 84). Therefore, it has been suggested that since ACE2 possesses a monitoring role in the central regulation of cardiovascular function and blood pressure (86), ACE2 in the brain is a potential therapeutic target for hypertension and other cardiovascular diseases that result from an overactive RAS (80). This fact is significantly crucial in the context of COVID-19 because, apart from pneumonia, COVID-19 leads to acute myocardial injury and, progressively, chronic damage to the cardiovascular system (87). It is conceivable that cardiovascular protection is a vital part of treatment for COVID-19 (87, 88). Targeting ACE2 on the central regulation of cardiovascular function may at least partially be helpful in the treatment of COVID-19.
Also, ACE2 has a peripheral other than cardio-cerebral vascular protection effect (86, 89). The significance of peripheral ACE2 in regulating blood pressure is well-established, but fewer studies emphasize that ACE2 plays a key role in regulating blood pressure (15, 16, 90). Uncharacteristically elevated blood pressure increases the risk of cerebral hemorrhage (26). It is to note that death associated with cerebrovascular disease has been reported in patients suffering from COVID-19 (14). Activation of ACE2/Ang (1–7)/MASR axis has also been shown to improve neurological deficits as it has anti-inflammatory and antioxidative effects in the ischemic insult. Thus, ACE2/Ang (1–7)/MASR axis is considered to play a protective role in the treatment of ischemic stroke, as well as other cerebrovascular events (86). Currently, the neuroinvasive propensity of COVID19 is well established (8, 13, 28, 91, 92). Also, there is evidence from animal studies that ACE2 receptors are present in areas such as medulla and brainstem (82, 83). Therefore, as suggested previously, the pathogenesis SARS-CoV-2-induced respiratory failure may also partially stem from the brain.
Long Term Complication of COVID 19 Invasion in the Brain
Although they are known as respiratory pathogens, the neuroinvasive and neurotropic properties of CoVs have been demonstrated in mice (93–95). Also, studies reveal that a human CoV (OC43) is capable of infecting and then persisting in human neuronal and glial cells, subsequently activating glial cells to deliver pro-inflammatory mediators and causing neurodegeneration (93–97). COVID-19 appears to be neuroinvasive using the ACE2 functional receptor (20–23). As mentioned above, ACE2 is an important component of RAS, and pathways associated with it have been correlated with Alzheimer disease as well as other neurodegenerative diseases (74, 81, 91). Dysregulated brain RAS has been associated with neurodegeneration due to the strong evidence for initiating a cascade of events leading to increase in oxidative stress, apoptosis, and neuroinflammation causing progressively neurodegeneration. Degenerative diseases of the brain that have been linked to altered RAS include Parkinson disease, Alzheimer disease, Huntington disease, multiple sclerosis, amyotrophic lateral sclerosis, traumatic brain injury, other than stroke as mentioned above (86, 98, 99). Moreover, the dysregulation of the antiviral response is another factor that has been associated with neurodegenerative disease (100, 101). It has been shown that products of certain genes may have potent antiviral activities, and may have deleterious effects when their expression is not appropriately regulated. There is evidence of mutations that have been linked to neurological conditions occurring in genes related to the antiviral response. Among such genes, SAMDH1 is probably the one. Notably, SAMDH1 has been related to Aicardi-Goutieres syndrome—a congenital disease that includes neurodegeneration as one of its main symptoms (62). As mentioned above, SAMDH1 appears to control the NF-κB pathway. Importantly, NF-κB has been engaged in the pathogenesis of a range of neurodegenerative disorders, including Alzheimer disease, Parkinson disease, Huntington disease, diabetic neuropathy, AIDS-dementia, and amyotrophic lateral sclerosis (102–104). Taken together, based on its similarity with other CoVs, and the fact that COVID 29 is neuroinvasive and its functional receptor is ACE2, and also because SAMDH1 is upregulated in the pathogenesis of COVID 19, it can be hypothesized that the SARS-CoV-2 entry into the brain will result in long-term neurological sequelae and may ultimately lead to neurodegenerative changes.
Although text mining and databases are useful to suggest mechanisms, there is one limitation of our study is that it relies on computational methods. Despite a sophisticated analysis was carried out, this study may not obviously predict the postulated interactions with 100% accuracy (42–45). Since the pandemic has been declared in March 2020, almost 19 million global cases and over 700,000 global deaths have been recorded on August 6, 2020. In these months several laboratories have also collected plasma as well as other body fluids that will be key in testing several hypotheses and confirm the presumed role of SAMDH1 in triggering the neurological symptoms of COVID 19 infection.
Conclusions
There is accumulating evidence that COVID-19 involves the brain. Not only studies have reported mild neurological symptoms, but encephalitis and cerebrovascular disease have been reported. Most struggle in the human war against this new virus revolves around the fact that COVID 19 is a respiratory pathogen. Mortality associated with this virus is because of respiratory failure, cardiovascular damage, and cerebrovascular accidents. In this study, we have explored the role of SAMDH1 as the potential link between SARS-CoV-2 infection and neurological illness. We discussed the distribution of ACE2 in CNS and its vital role regulation of cardiovascular function and, specifically, blood pressure in the brain (centrally) and at the periphery. We assume that detailed neurological and neuropathological investigations are ongoing. Establishing a therapeutic strategy that targets the brain ACE2 may improve the outcome of COVID-19 patients. Interestingly, the same SAMDH1 has been associated with various neurological diseases, including Aicardi-Goutieres syndrome (AGS) as well as GBS, which has been associated with Zika Virus infection. We strongly suggest that SAMDH1-targeting COVID-19 may be at the basis of both early neurological symptoms and late neurodegeneration and our neuropathological team is open to any cooperation.
Author Contributions
AK conceptualized the study, collected data, and drafted the initial manuscript. CS designed the study and revised the manuscript. All authors meet the ICMJE requirements for authorship, approved the final manuscript as submitted, and agree to be accountable for all aspects of the work.
Funding
This research has been funded by the generosity of the Stollery Children's Hospital Foundation and supporters of the Lois Hole Hospital for Women through the Women and Children's Health Research Institute (WCHRI, Grant ID #: 2096), Hubei Province Natural Science Funding for Hubei University of Technology (100-Talent Grant for Recruitment Program of Foreign Experts Total Funding: Digital PCR and NGS-based diagnosis for infection and oncology, 2017-2022).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Glass WG, Subbarao K, Murphy B, Murphy PM. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol. (2004) 173:4030–9. doi: 10.4049/jimmunol.173.6.4030
2. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
3. Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. (2015) 23:468–78. doi: 10.1016/j.tim.2015.06.003
4. Novel Coronavirus Pneumonia Emergency Response Epidemiology T. [The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China]. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:145–51. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003
5. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. (2020) 38:1–9. doi: 10.12932/AP-200220-0772
6. Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. (2019) 11:59. doi: 10.3390/v11010059
7. Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol. (2020) 92:491–4. doi: 10.1002/jmv.25709
8. Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. (2020) 92:552–5. doi: 10.1002/jmv.25728
9. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
10. Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J Virol. (2003) 77:2530–8. doi: 10.1128/JVI.77.4.2530-2538.2003
11. Li Y, Bai WZ, Hashikawa T. Response to Commentary on: “The neuroinvasive potential of SARS-CoV-2 may play a role in the respiratory failure of COVID-19 patients.” J Med Virol. (2020) 92:707–9. doi: 10.1002/jmv.25824
12. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. (2003) 426:450–4. doi: 10.1038/nature02145
13. Baig AM, Khaleeq A, Ali U, Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. (2020) 11:995–8. doi: 10.1021/acschemneuro.0c00122
14. Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. (2020) 77:683–69. doi: 10.1001/jamaneurol.2020.1127
15. Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, et al. Angiotensin converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ Res. (2020) 126:1456–74. doi: 10.1161/CIRCRESAHA.120.317015
16. Oudit GY, Kassiri Z, Jiang C, Liu PP, Poutanen SM, Penninger JM, et al. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest. (2009) 39:618–25. doi: 10.1111/j.1365-2362.2009.02153.x
17. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2019) 47:D607–D13. doi: 10.1093/nar/gky1131
18. Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, et al. COVID-19 outbreak: an overview. Chemotherapy. (2019) 64:215–23. doi: 10.1159/000507423
19. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061–9. doi: 10.1001/jama.2020.1585
20. Li K, Wohlford-Lenane C, Perlman S, Zhao J, Jewell AK, Reznikov LR, et al. Middle east respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis. (2016) 213:712–22. doi: 10.1093/infdis/jiv499
21. Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D, et al. Clinical characteristics of imported cases of COVID-19 in jiangsu province: a multicenter descriptive study. Clin Infect Dis. (2020) 71:706–12. doi: 10.1093/cid/ciaa199
22. Burke JF, Chan AK, Mummaneni V, Chou D, Lobo EP, Berger MS, et al. Letter: the coronavirus disease 2019 global pandemic: a neurosurgical treatment algorithm. Neurosurgery. (2020) 87:E50–E56. doi: 10.1093/neuros/nyaa116
23. Du Y, Tu L, Zhu P, Mu M, Wang R, Yang P, et al. Clinical features of 85 fatal cases of COVID-19 from Wuhan. A retrospective observational study. Am J Resp Crit Care Med. (2020) 201:1372–9. doi: 10.1164/rccm.202003-0543OC
24. Federico A. Brain Awareness Week, CoVID-19 infection and Neurological Sciences. Neurol Sci. (2020) 41:747–48. doi: 10.1007/s10072-020-04338-0
25. Papa SM, Brundin P, Fung VSC, Kang UJ, Burn DJ, Colosimo C, et al. Impact of the COVID-19 pandemic on Parkinson's disease and movement disorders. Mov Disord. (2020) 35:711–5. doi: 10.1002/mds.28067
26. Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, et al. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun. (2020) 87:18–22. doi: 10.1016/j.bbi.2020.03.031
27. Talbot PJ, Ekande S, Cashman NR, Mounir S, Stewart JN. Neurotropism of human coronavirus 229E. Adv Exp Med Biol. (1993) 342:339–46. doi: 10.1007/978-1-4615-2996-5_52
28. Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol. (2008) 82:7264–75. doi: 10.1128/JVI.00737-08
29. McCray PB Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. (2007) 81:813–21. doi: 10.1128/JVI.02012-06
30. Zhai P, Ding Y, Wu X, Long J, Zhong Y, Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents 55:105955. doi: 10.1016/j.ijantimicag.2020.105955
31. Arabi YM, Al-Omari A, Mandourah Y, Al-Hameed F, Sindi AA, Alraddadi B, et al. Critically ill patients with the middle east respiratory syndrome: a multicenter retrospective cohort study. Crit Care Med. (2017) 45:1683–95. doi: 10.1097/CCM.0000000000002621
32. Arabi YM, Fowler R, Hayden FG. Critical care management of adults with community-acquired severe respiratory viral infection. Intensive Care Med. (2020) 46:315–28. doi: 10.1007/s00134-020-05943-5
33. Arabi YM, Balkhy HH, Hayden FG, Bouchama A, Luke T, Baillie JK, et al. Middle east respiratory syndrome. N Engl J Med. (2017) 376:584–94. doi: 10.1056/NEJMsr1408795
34. Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. (2005) 202:415–24. doi: 10.1084/jem.20050828
35. Li YC, Bai WZ, Hirano N, Hayashida T, Hashikawa T. Coronavirus infection of rat dorsal root ganglia: ultrastructural characterization of viral replication, transfer, and the early response of satellite cells. Virus Res. (2012) 163:628–35. doi: 10.1016/j.virusres.2011.12.021
36. Andries K, Pensaert MB. Immunofluorescence studies on the pathogenesis of hemagglutinating encephalomyelitis virus infection in pigs after oronasal inoculation. Am J Vet Res. (1980) 41:1372–8.
37. Li YC, Bai WZ, Hirano N, Hayashida T, Taniguchi T, Sugita Y, et al. Neurotropic virus tracing suggests a membranous-coating-mediated mechanism for transsynaptic communication. J Comp Neurol. (2013) 521:203–12. doi: 10.1002/cne.23171
38. Matsuda K, Park CH, Sunden Y, Kimura T, Ochiai K, Kida H, et al. The vagus nerve is one route of transneural invasion for intranasally inoculated influenza a virus in mice. Vet Pathol. (2004) 41:101–7. doi: 10.1354/vp.41-2-101
39. Brann D, Tsukahara T, Weinreb C, Lipovsek M, Van den Berge K, Gong B, et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. bioRxiv. (2020) 6:eabc5801. doi: 10.1126/sciadv.abc5801
40. Brann JH, Firestein SJ. A lifetime of neurogenesis in the olfactory system. Front Neurosci. (2014) 8:182. doi: 10.3389/fnins.2014.00182
41. Karsz FR, Vance A, Anderson VA, Brann PG, Wood SJ, Pantelis C, et al. Olfactory impairments in child attention-deficit/hyperactivity disorder. J Clin Psychiatry. (2008) 69:1462–8. doi: 10.4088/JCP.v69n0914
42. Lopez-Rodriguez R, Hernandez-Bartolome A, Borque MJ, Rodriguez-Munoz Y, Martin-Vilchez S, Garcia-Buey L, et al. Interferon-related genetic markers of necroinflammatory activity in chronic hepatitis C. PLoS ONE. (2017) 12:e0180927. doi: 10.1371/journal.pone.0180927
43. Yang S, Zhan Y, Zhou Y, Jiang Y, Zheng X, Yu L, et al. Interferon regulatory factor 3 is a key regulation factor for inducing the expression of SAMHD1 in antiviral innate immunity. Sci Rep. (2016) 6:29665. doi: 10.1038/srep29665
44. Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, et al. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type i interferon production by interfering with TRIM25-mediated RIG-I Ubiquitination. J Virol. (2017) 91:JVI.02143-16. doi: 10.1128/JVI.02143-16
45. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. (2014) 5:461. doi: 10.3389/fimmu.2014.00461
46. El Asmi F, Brantis-de-Carvalho CE, Blondel D, Chelbi-Alix MK. Rhabdoviruses, Antiviral Defense, and SUMO Pathway. Viruses. (2018) 10:686. doi: 10.3390/v10120686
47. Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Møller R, Panis M, Sachs D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. (2020) 181:1036–45. doi: 10.1016/j.cell.2020.04.026
48. Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. (2007) 361:18–26. doi: 10.1016/j.virol.2007.01.020
49. Schoggins JW. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol. (2014) 6:40–6. doi: 10.1016/j.coviro.2014.03.006
50. Chen S, Bonifati S, Qin Z, St Gelais C, Kodigepalli KM, Barrett BS, et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kappaB and interferon pathways. Proc Natl Acad Sci USA. (2018) 115:E3798–E807. doi: 10.1073/pnas.1801213115
51. Monit C, Morris ER, Ruis C, Szafran B, Thiltgen G, Tsai MC, et al. Positive selection in dNTPase SAMHD1 throughout mammalian evolution. Proc Natl Acad Sci USA. (2019) 116:18647–54. doi: 10.1073/pnas.1908755116
52. Roux A, Leroy H, De Muylder B, Bracq L, Oussous S, Dusanter-Fourt I, et al. FOXO1 transcription factor plays a key role in T cell-HIV-1 interaction. PLoS Pathog. (2019) 15:e1007669. doi: 10.1371/journal.ppat.1007669
53. Sergi C, Shen F, Liu SM. Insulin/IGF-1R, SIRT1, and FOXOs pathways-an intriguing interaction platform for bone and osteosarcoma. Front Endocrinol. (Lausanne). (2019) 10:93. doi: 10.3389/fendo.2019.00093
54. Buffone C, Kutzner J, Opp S, Martinez-Lopez A, Selyutina A, Coggings SA, et al. The ability of SAMHD1 to block HIV-1 but not SIV requires expression of MxB. Virology. (2019) 531:260–8. doi: 10.1016/j.virol.2019.03.018
55. Franzolin E, Salata C, Bianchi V, Rampazzo C. The deoxynucleoside triphosphate triphosphohydrolase activity of SAMHD1 protein contributes to the mitochondrial DNA depletion associated with genetic deficiency of deoxyguanosine kinase. J Biol Chem. (2015) 290:25986–96. doi: 10.1074/jbc.M115.675082
56. Transy C, Margottin-Goguet F. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. (2009) 8:2489–90. doi: 10.4161/cc.8.16.9129
57. Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. (2007) 6:182–8. doi: 10.4161/cc.6.2.3732
58. Liu X, Liu Q, Fan Y, Wang S, Liu X, Zhu L, et al. Downregulation of PPP2R5E expression by miR-23a suppresses apoptosis to facilitate the growth of gastric cancer cells. FEBS Lett. (2014) 588:3160–9. doi: 10.1016/j.febslet.2014.05.068
59. Leshinsky-Silver E, Malinger G, Ben-Sira L, Kidron D, Cohen S, Inbar S, et al. A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. Eur J Hum Genet. (2011) 19:287–92. doi: 10.1038/ejhg.2010.213
60. Hatzis P, Al-Madhoon AS, Jullig M, Petrakis TG, Eriksson S, Talianidis I. The intracellular localization of deoxycytidine kinase. J Biol Chem. (1998) 273:30239–43. doi: 10.1074/jbc.273.46.30239
61. Ronchi D, Garone C, Bordoni A, Gutierrez Rios P, Calvo SE, Ripolone M, et al. Next-generation sequencing reveals DGUOK mutations in adult patients with mitochondrial DNA multiple deletions. Brain. (2012) 135:3404–15. doi: 10.1093/brain/aws258
62. Majer C, Schussler JM, Konig R. Intertwined: SAMHD1 cellular functions, restriction, and viral evasion strategies. Med Microbiol Immunol. (2019) 208:513–29. doi: 10.1007/s00430-019-00593-x
63. Wichit S, Hamel R, Zanzoni A, Diop F, Cribier A, Talignani L, et al. SAMHD1 enhances chikungunya and zika virus replication in human skin fibroblasts. Int J Mol Sci. (2019) 20:1695. doi: 10.3390/ijms20071695
64. Richards RI, Robertson SA, Kastner DL. Neurodegenerative diseases have genetic hallmarks of autoinflammatory disease. Hum Mol Genet. (2018) 27:R108–R18. doi: 10.1093/hmg/ddy139
65. Crow YJ. Aicardi-Goutieres syndrome. Handb Clin Neurol. (2013) 113:1629–35. doi: 10.1016/B978-0-444-59565-2.00031-9
66. Coggins SA, Mahboubi B, Schinazi RF, Kim B. SAMHD1 Functions and Human Diseases. Viruses. (2020) 12:382. doi: 10.3390/v12040382
67. Gilani A, Adang LA, Vanderver A, Collins A, Kleinschmidt-DeMasters BK. Neuropathological findings in a case of IFIH1-related aicardi-goutieres syndrome. Pediatr Dev Pathol. (2019) 22:566–70. doi: 10.1177/1093526619837797
68. Samanta D, Ramakrishnaiah R, Crary SE, Sukumaran S, Burrow TA. Multiple autoimmune disorders in aicardi-goutieres syndrome. Pediatr Neurol. (2019) 96:37–9. doi: 10.1016/j.pediatrneurol.2019.01.017
69. Thiele H, du Moulin M, Barczyk K, George C, Schwindt W, Nurnberg G, et al. Cerebral arterial stenoses and stroke: novel features of Aicardi-Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Hum Mutat. (2010) 31:E1836–50. doi: 10.1002/humu.21357
70. Xin B, Jones S, Puffenberger EG, Hinze C, Bright A, Tan H, et al. Homozygous mutation in SAMHD1 gene causes cerebral vasculopathy and early onset stroke. Proc Natl Acad Sci USA. (2011) 108:5372–7. doi: 10.1073/pnas.1014265108
71. Buchanan EL, Espinoza DA, McAlexander MA, Myers SL, Moyer A, Witwer KW. SAMHD1 transcript upregulation during SIV infection of the central nervous system does not associate with reduced viral load. Sci Rep. (2016) 6:22629. doi: 10.1038/srep22629
72. DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeno JM, Fernandez-Delgado R, Fett C, et al. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. (2014) 88:913–24. doi: 10.1128/JVI.02576-13
73. Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol. (2009) 83:6689–705. doi: 10.1128/JVI.02220-08
74. Gaddam RR, Chambers S, Bhatia M. ACE and ACE2 in inflammation: a tale of two enzymes. Inflamm Allergy Drug Targets. (2014) 13:224–34. doi: 10.2174/1871528113666140713164506
75. Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. (2002) 532:107–10. doi: 10.1016/S0014-5793(02)03640-2
76. Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. (2000) 87:E1–9. doi: 10.1161/01.RES.87.5.e1
77. Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. (2000) 275:33238–43. doi: 10.1074/jbc.M002615200
78. Doobay MF, Talman LS, Obr TD, Tian X, Davisson RL, Lazartigues E. Differential expression of neuronal ACE2 in transgenic mice with overexpression of the brain renin-angiotensin system. Am J Physiol Regul Integr Comp Physiol. (2007) 292:R373–81. doi: 10.1152/ajpregu.00292.2006
79. Xia H, Lazartigues E. Angiotensin-converting enzyme 2 in the brain: properties and future directions. J Neurochem. (2008) 107:1482–94. doi: 10.1111/j.1471-4159.2008.05723.x
80. Xia H, Lazartigues E. Angiotensin-converting enzyme 2: central regulator for cardiovascular function. Curr Hypertens Rep. (2010) 12:170–5. doi: 10.1007/s11906-010-0105-7
81. Sakima A, Averill DB, Gallagher PE, Kasper SO, Tommasi EN, Ferrario CM, et al. Impaired heart rate baroreflex in older rats: role of endogenous angiotensin-(1-7) at the nucleus tractus solitarii. Hypertension. (2005) 46:333–40. doi: 10.1161/01.HYP.0000178157.70142.33
82. Elased KM, Cunha TS, Gurley SB, Coffman TM, Morris M. New mass spectrometric assay for angiotensin-converting enzyme 2 activity. Hypertension. (2006) 47:1010–7. doi: 10.1161/01.HYP.0000215588.38536.30
83. Lin Z, Chen Y, Zhang W, Chen AF, Lin S, Morris M. RNA interference shows interactions between mouse brainstem angiotensin AT1 receptors and angiotensin-converting enzyme 2. Exp Physiol. (2008) 93:676–84. doi: 10.1113/expphysiol.2007.041657
84. Bitker L, Burrell LM. Classic and nonclassic renin-angiotensin systems in the critically Ill. Crit Care Clin. (2019) 35:213–27. doi: 10.1016/j.ccc.2018.11.002
85. Unger T, Badoer E, Ganten D, Lang RE, Rettig R. Brain angiotensin: pathways and pharmacology. Circulation. (1988) 77:I40–54. PubMed PMID: 2836110.
86. Abiodun OA, Ola MS. Role of brain renin angiotensin system in neurodegeneration: an update. Saudi J Biol Sci. (2020) 27:905–12. doi: 10.1016/j.sjbs.2020.01.026
87. Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. (2020) 17:259–26. doi: 10.1038/s41569-020-0360-5
88. Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. (2020) 5:802–10. doi: 10.1001/jamacardio.2020.0950
89. Feng Y, Yue X, Xia H, Bindom SM, Hickman PJ, Filipeanu CM, et al. Angiotensin-converting enzyme 2 overexpression in the subfornical organ prevents the angiotensin II-mediated pressor and drinking responses and is associated with angiotensin II type 1 receptor downregulation. Circ Res. (2008) 102:729–36. doi: 10.1161/CIRCRESAHA.107.169110
90. Xia H, Feng Y, Obr TD, Hickman PJ, Lazartigues E. Angiotensin II type 1 receptor-mediated reduction of angiotensin-converting enzyme 2 activity in the brain impairs baroreflex function in hypertensive mice. Hypertension. (2009) 53:210–6. doi: 10.1161/HYPERTENSIONAHA.108.123844
91. Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. (2020) 382:1708–20.
92. Moriguchi T, Harii N, Goto J, Harada D, Sugawara H, Takamino J, et al. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int J Infect Dis. (2020) 94:55–8. doi: 10.1016/j.ijid.2020.03.062
93. Dube M, Le Coupanec A, Wong AHM, Rini JM, Desforges M, Talbot PJ. Axonal transport enables neuron-to-neuron propagation of human coronavirus OC43. J Virol. (2018) 92: e00404–18. doi: 10.1128/JVI.00404-18
94. Bleau C, Filliol A, Samson M, Lamontagne L. Brain invasion by mouse hepatitis virus depends on impairment of tight junctions and beta interferon production in brain microvascular endothelial cells. J Virol. (2015) 89:9896–908. doi: 10.1128/JVI.01501-15
95. St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol. (2004) 78:8824–34. doi: 10.1128/JVI.78.16.8824-8834.2004
96. Brison E, Jacomy H, Desforges M, Talbot PJ. Novel treatment with neuroprotective and antiviral properties against a neuroinvasive human respiratory virus. J Virol. (2014) 88:1548–63. doi: 10.1128/JVI.02972-13
97. Arbour N, Day R, Newcombe J, Talbot PJ. Neuroinvasion by human respiratory coronaviruses. J Virol. (2000) 74:8913–21. doi: 10.1128/JVI.74.19.8913-8921.2000
98. Rodriguez-Perez AI, Sucunza D, Pedrosa MA, Garrido-Gil P, Kulisevsky J, Lanciego JL, et al. Angiotensin type 1 receptor antagonists protect against alpha-synuclein-induced neuroinflammation and dopaminergic neuron death. Neurotherapeutics. (2018) 15:1063–81. doi: 10.1007/s13311-018-0646-z
99. Savaskan E. The role of the brain renin-angiotensin system in neurodegenerative disorders. Curr Alzheimer Res. (2005) 2:29–35. doi: 10.2174/1567205052772740
100. Nazmi A, Field RH, Griffin EW, Haugh O, Hennessy E, Cox D, et al. Chronic neurodegeneration induces type I interferon synthesis via STING, shaping microglial phenotype and accelerating disease progression. Glia. (2019) 67:1254–76. doi: 10.1002/glia.23592
101. Talwar P, Gupta R, Kushwaha S, Agarwal R, Saso L, Kukreti S, et al. Viral induced oxidative and inflammatory response in alzheimer's disease pathogenesis with identification of potential drug candidates: a systematic review using systems biology approach. Curr Neuropharmacol. (2019) 17:352–65. doi: 10.2174/1570159X16666180419124508
102. Zhang Y, Hu W. NFkappaB signaling regulates embryonic and adult neurogenesis. Front Biol. (Beijing). (2012) 7:10.1007/s11515-012-1233-z. doi: 10.1007/s11515-012-1233-z
103. Okun E, Griffioen KJ, Lathia JD, Tang SC, Mattson MP, Arumugam TV. Toll-like receptors in neurodegeneration. Brain Res Rev. (2009) 59:278–92. doi: 10.1016/j.brainresrev.2008.09.001
Keywords: COVID-19, neuroinvasion, ACE2, SAR-CoV2, string, bioinformatics, neurodegeneration, prognosis
Citation: Khan A and Sergi C (2020) SAMHD1 as the Potential Link Between SARS-CoV-2 Infection and Neurological Complications. Front. Neurol. 11:562913. doi: 10.3389/fneur.2020.562913
Received: 16 May 2020; Accepted: 24 August 2020;
Published: 25 September 2020.
Edited by:
Shoji Tsuji, The University of Tokyo Hospital, JapanReviewed by:
Mario Reynaldo Cornejo-Olivas, National Institute of Neurological Sciences (INCN), PeruMiguel A. Barboza, University of Costa Rica, Costa Rica
Copyright © 2020 Khan and Sergi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Consolato Sergi, c2VyZ2kmI3gwMDA0MDt1YWxiZXJ0YS5jYQ==
 Aiza Khan
Aiza Khan Consolato Sergi
Consolato Sergi