
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurol. , 14 June 2018
Sec. Neurodegeneration
Volume 9 - 2018 | https://doi.org/10.3389/fneur.2018.00429
This article is part of the Research Topic Biomarkers in Neurology View all 17 articles
Traumatic brain injury (TBI) is a serious problem that causes high morbidity and mortality around the world. Currently, no reliable biomarkers are used to assess the severity and predict the recovery. Many protein biomarkers were extensively studied for diagnosis and prognosis of different TBI severities such as S-100β, glial fibrillary acidic protein (GFAP), neuron-specific enolase (NSE), neurofilament light chain (NFL), cleaved tau protein (C-tau), and ubiquitin C-terminal hydrolase-L1 (UCH-L1). However, none of these candidates is currently used in the clinical practice, due to relatively low sensitivity, for the diagnosis of mild TBI (mTBI) or mild to moderate TBI (MMTBI) patients who are clinically well and do not have a detectable intracranial pathology on the scans. MicroRNAs (miRNAs or miRs) are a class of small endogenous molecular regulators, which showed to be altered in different pathologies, including TBI and for this reason, their potential role in diagnosis, prognosis and therapeutic applications, is explored. Promising miRNAs such as miR-21, miR-16 or let-7i were identified as suitable candidate biomarkers for TBI and can differentiate mild from severe TBI. Also, they might represent new potential therapeutic targets. Identification of miRNA signature in tissue or biofluids, for several pathological conditions, is now possible thanks to the introduction of new high-throughput technologies such as microarray platform, Nanostring technologies or Next Generation Sequencing. This review has the aim to describe the role of microRNA in TBI and to explore the most commonly used techniques to identify microRNA profile. Understanding the strengths and limitations of the different methods can aid in the practical use of miRNA profiling for diverse clinical applications, including the development of a point-of-care device.
Head injuries are a significant cause of disability and mortality worldwide and one of the most common reasons of emergency department visits especially among young males (1), creating a severe physical, psychological and socioeconomic burden on the patients, their families and the community (1, 2).
In particular, traumatic brain injury (TBI) is a complex pathological alteration in the neural homeostasis which is triggered by an external mechanical force resulting in a broad spectrum of temporary or permanent injuries and outcomes (3). Annual TBI incidents are estimated to be more than 10 million patients worldwide (4–6). The most frequent causes of TBI are falls, road traffic accidents, sport and recreation activities, military injuries and assault or abuse. TBI pathology can be classified as primary and secondary brain damage (7). The primary injury occurs immediately after receiving the mechanical impact which disrupts the integrity of neuronal, glial, endothelial cells and dysregulates the cerebral blood flow (CBF), whereas the secondary brain injury is due to a range of biochemical and cellular changes that causes neuronal apoptosis and death, blood-brain barrier (BBB) disruption, etc. (8–15). Controlling the development of secondary injury is the only strategy that can be beneficial to improve the outcome of the primary injury that cannot be managed medically. The heterogeneity of the disease makes an accurate assessment of the severity of trauma and the prediction of patient outcome, challenging. Clinically, head injuries are diagnosed as mild, moderate, or severe according to the Glasgow Coma Scale (GCS) score, which uses a motor, eye and verbal responses to assess the conscious level of the patient. However, this score might underestimate mild TBI (mTBI) cases (16). Computed tomography (CT) or magnetic resonance imaging (MRI) scans are also used to assess TBI according the current guidelines (17). Although these techniques show limited diagnostic ability for the detection of mild brain tissue insult with concerns for radiation risks from CT scans and the escalating costs of diagnostic imaging techniques (18, 19) in the future, imaging has the potential to complement molecular diagnostics (20).
For this reason, mTBI detection remains one of the most difficult clinical diagnoses, it accounts for 75–90% of the TBI cases in the United States (21) and 10–20% of the patients remain symptomatic and complain of post-concussive syndrome (PCS) symptoms (22). In addition, people such as military, sportive and children are at risk of repeated concussions and may develop depression (23) and neurodegenerative conditions in later life, e.g., Parkinson's disease, motor neuron disease, and chronic traumatic encephalopathy (CTE) (24, 25).
Currently, no TBI biomarkers were identified that could reliably be used in the clinical practice for diagnosis and prognosis.
Recently, the U.S. Food and Drug Administration reviewed and authorized for marketing the Banyan Brain Trauma Indicators which are ubiquitin C-terminal hydrolase-L1 (UCH-L1) and glial fibrillary acidic protein (GFAP), to evaluate mTBI in adults. These two proteins are released from the injured tissue into the blood and can be quantified within 12 h of the brain injury and can help to predict the patients with detectable intracranial lesions on the CT scan with 97.5% of accuracy. However, a biomarker able to accurately diagnose mTBI is still needed.
In the last decades, many molecules were proposed as promising TBI biomarkers, but the complicated anatomy of the brain and the disparate pathology of the TBI make it challenging to apply into the clinical practice (26).
Biofluid biomarkers would be preferable as they present various advantages such as cost-effective and minimally invasive sample collection.
Among the most extensively studied biomarkers in the serum and cerebrospinal fluid (CSF), there are S-100β and GFAP. S-100β is an extracellular protein with a short half-life of <30 min (27). However, because of its size, it does not cross an intact BBB. Besides, S-100β is not a brain-specific protein and can also be released by other organs in case of polytrauma (28–31). In 2013, the S-100β serum level was used to reduce the unnecessary CT scans in the adult mTBI patients among the Scandinavian population. However, it remains challenging to find the appropriate cut-off value of S-100β that correlates with the injury primarily because of the lower sensitivity in polytrauma patients (32, 33).
On the contrary, GFAP is a structural protein exclusively expressed in the astroglial cells and plays a pivotal role in the astrocyte's cytoskeleton as a component of the intermediate filament (IF) network (34). GFAP was found to be slightly elevated in mild TBI and when added to the clinical data, it improved the power of outcome prediction (35). Animal studies also showed GFAP to be a promising biomarker, since its cellular release is correlated to all grades of injury severities (36). The only limitation in the use of GFAP as a biomarker is related to the release into the bloodstream or CSF, which is, indeed, strictly BBB-damage dependent (35, 37).
Neuron-specific enolase (NSE), neurofilament light polypeptide (NFL), cleaved tau protein (C-tau) and UCH-L1 were also considered promising biomarkers. However, the biological significance of these biomarkers cannot be confidently declared, due to the lack of studies with adequate sample size and low sensitivity for mTBI in individuals without detectable structural brain abnormalities. A summary of papers showing the area under the curve (AUC) of representative TBI biomarkers is presented in Table 1.
Enolases are glycolytic enzymes composed by three different subunits (α, β, γ). The two most stable isoforms are γγ and αγ, which are referred to as NSE, are particularly abundant in the neuron cytoplasm, however NSE proteins can also be found in erythrocytes and platelets making the process of haemolysis a significant extracranial source when measured in trauma (46, 47). In the context of mild TBI, NSE can predict the early prognosis of patients when measured in combination with S-100β (48). However, its slow elimination from plasma, leads to difficulties in distinguishing between primary and secondary insults to the brain (49, 50).
One of the most recently identified biomarkers is NFL. It was suggested as a potential, sensitive and specific marker in detecting axonal injury in mTBI (51). One of the advantages of its clinical use is the relatively long half-life which was estimated to be ~3 weeks (52). NFL also plays a vital role in the neuro-axonal cytoskeleton (53). Therefore, increased NFL levels were found in the CSF and serum of individuals with a wide range of neurodegenerative and neuroinflammatory diseases (54, 55). Another proposed serum marker is C-tau, which is a microtubule-associated protein (MAP) primarily found in the neuronal axons and dendrites (49, 56). After an axonal injury, tau protein can be detected in the extracellular space and diffuses into the CSF after N- and C- terminals cleavage (56). In 2006, a study demonstrated that higher levels of post-traumatic CSF C-tau were associated with a poorer clinical outcome following severe TBI (sTBI) (57). However, there was no significant correlation between the levels of C-tau and the outcome following mTBI (58).
UCH-L1 was identified as highly specific to the human brain (59) and the increased levels were correlated with the TBI severity and a worse outcome. Its diagnostic value was found to be beyond the first 24 h of injury (60–63). It could also distinguish between the patients with TBI and the uninjured patients with altered GCS secondary to drugs and alcohol intoxication (50).
Recently, a new protein Cystatin D (CTS5), which inhibits lysosomal and secreted cysteine proteases, was also identified as a potential biomarker to assess the severity of TBI and its expression at very early time points, makes CTS5, an ideal biomarker for a point-of-care (PoC) device (45). To the best of our knowledge, none of the previous protein biomarkers has been successfully used in the clinical setting for diagnosis and prognosis of TBI patients.
MicroRNAs (miRNAs or miRs) are a class of molecular regulators discovered for the first time in Caenorhabditis elegans in 1993 (64). Then, dozens of miRNAs were identified in worms, flies and human suggesting that miRNAs represent a previously unknown group of molecules (65).
miRs are short (~22 nucleotides) non-coding, single-stranded RNAs that play key roles in the regulation of several biological processes such as cell proliferation and differentiation, survival, and motility via negative feedback mechanism at the post-transcriptional level by binding to the 3′-untranslated region (UTR) of the target miRs and leading to either suppression of the translation process, mRNAs degradation or both. A single miRNA can regulate multiple mRNAs and vice-versa because they do not always require a perfect complementarity for target recognition. Therefore, they can briefly interchange between the cellular programs (66).
The synthesis of miRs begins in the nucleus with transcription by the RNA polymerase II or III producing long primary miRNA transcripts (pri-miRNA) that contain functional secondary structures, termed stem-loops and carrying mature miRNA sequences. Maturation of the pri-miRNA transcripts includes several steps which are initiated by RNase III endonuclease Drosha and produces the precursor-miR (pre-miR) (67, 68). Following Drosha processing, a complex of proteins, exportin-5 (EXP5) with GTP-binding nuclear protein Ran-GTP, transports pre-miR from the nucleus into the cytoplasm where it is cleaved by Dicer and TAR RNA-binding protein (TRBP) (69, 70). This produces a double-stranded RNA molecule composed of 20–24 nucleotide miR and a complementary miR* of the same length (71). It has been found that not only the mature miR strand is biologically active, but also the miR* strand is functional and not just degraded as was previously hypothesized (72). Then, mature miRNAs bind to mRNA molecules through a process facilitated by the RNA-induced silencing complex (RISC), which consists of RNase Dicer, TRBP, PACT (protein activator of PKR) and the Argonaute proteins (68).
The resulting RISC-miRNA complex binds the complementary regions of the target mRNAs by partial or total base-pairing at the 3′ UTR. This interaction, controlled by nucleotides 2–8 at the 5′ end of the miR and known as “seed sequence” (73), reduces protein production by translation inhibition and mRNA degradation (74). However, miRNAs do not target all mRNAs because there are only binding sites in one-third of the mRNAs (75).
Currently, in the human genome, over 2,000 miRNAs were identified and numerous studies were mainly focused on the miRNA profiling in various tissues and biofluids that can aid the diagnosis of a wide range of diseases, including cancer, cardiovascular, nervous system disorders and many other disorders (76, 77, 147). Since miRs are relatively abundant and stable in the human biofluids, they are considered to be better than protein biomarkers and therefore are now being investigated as the new class of markers for numerous pathologies including but not limited to neurodegenerative diseases. However, a better understanding of the biological mechanisms of miRNAs in these diseases is required to improve their application as biomarkers (78).
With the discovery of miRNAs and its critical role as regulators in various diseases, it is now possible to investigate their role as biomarkers and emerging therapeutic targets. Based on the antisense technology, very potent oligonucleotides targeted against miRNA known as anti-miR were developed (79, 80).
TBI research associated with the changes in miRNA expression is only at the beginning to be understood. Few studies showed the miRNA profile in serum plasma and CSF after different TBI severities and at different time points (81–85).
Redell et al. found a downregulation of miR-16 and miR-92a in severe TBI patients and an upregulation of miR-765 in mild and severe TBI, within the first 24 h and by using a microarray approach (81). Bhomia and collaborators analyzed microRNA profile in serum and CSF of patients grouped in three different categories, mild-moderate TBI (MM-TBI), severe TBI and orthopedic injury patients with samples collected within 48 h from injury and compared to healthy volunteers. Eighteen and 20 miRNAs were observed in MMTBI and sTBI groups respectively and among these, 10 miRNAs were present at both TBI severities. Finally, four of these 10 miRNAs were also found in CSF (85). Di Pietro et al. screened 754 microRNAs using TaqMan Array Human MicroRNA A+B cards in mTBI+EC (extra-cranial injury), sTBI+EC, EC only groups and compared the results to healthy volunteers at different time points. Particularly interesting were the results obtained within the first hour from injury, in serum of mTBI+EC. These data reported two microRNAs, miR-425-5p and miR-502, having high diagnostic accuracy (AUC > 0.9) in differentiating mTBI from sTBI (84).
Recently, saliva was also explored as potential source of biomarkers for TBI. Salivary microRNA changes were found to be associated with prolonged concussion symptoms in pediatrics (86). Five miRs (miR-320c-1, miR-133a-5p, miR-769-5p, miR-1307-3p and let-7a-3p) were detected in the patients with prolonged post-concussive symptoms, and three of them; miR-320c-1, miR-629, and let-7b-5p were associated with memory problems, headache and fatigue that were developed 4 weeks after head injury. The same group has also matched miRNA changes in saliva and CSF, identifying six miRs (miR-182-5p, 221-3p, 26b-5p, 320c, 29c-3p, and 30e-5p) with similar changes in both biofluids (87).
A completed list of microRNA detected in different biofluids in TBI patients can be found in Table 2. Results presented, were not always consistent. However, it is not always possible to compare these studies, since sample collection timing or the different biofluid analyzed, play a relevant role to uniform the biomarker discovery.
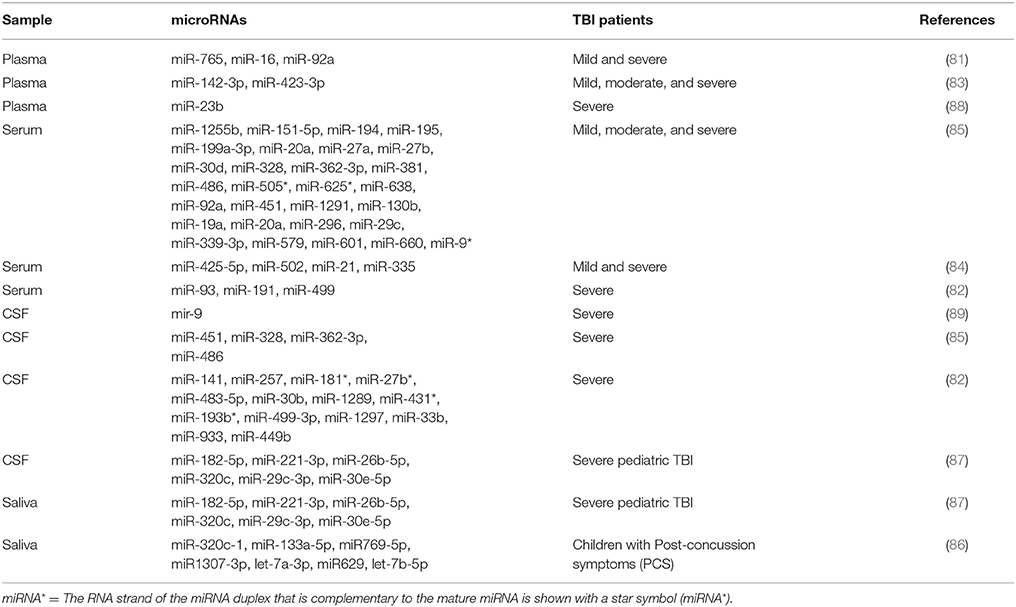
Table 2. MicroRNA differentially expressed according the severity of TBI and the different human biofluid.
Many microRNAs were also described in the brain of injured animals by using different models of TBI. Some of these studies have also investigated the potential pathobiology of the microRNAs differentially expressed in tissue.
Human miR-21 is one of the most studied miRs in TBI. It is a polycistronic miR (chromosome 17q23.2), and it overlaps with the Vacuole Membrane Protein 1 (VMP1) coding gene, also known as Transmembrane Protein 49 (TMEM49) (90).
Recent studies have demonstrated high miR-21 expression levels after TBI. Also, it has been found to improve the neurological outcome through inhibiting apoptosis and targeting angiogenesis molecules. In particular, the upregulation of miR-21 was found to reduce brain oedema derived by BBB-leakage. Hence, ago-miR-21 treatment was proposed as a potential therapy to decrease BBB damage (91) by inhibiting the loss of occludin and claudin-5 among other tight junction proteins. It also increases the levels of Angiopoietin-1 and its Tie-2 receptor, which maintain the normal BBB condition. MiR-21 was also found to improve experimental TBI mice cognition after the running wheel exercise (92, 103). The therapeutic role of miR-21 might also be due to inhibiting apoptotic cell loss by targeting the phosphatase and tensin homolog (PTEN)-Akt pathway (91). In an interesting study, extracellular vesicles (EV) were isolated from the brain of injured mice and controls, and the expression of miR-21 was found significantly increased with the injury. Concomitantly, an increase of miR-21 in neurons was observed, suggesting miR-21 secretion from neurons by EV cargo (92). Further support via the upregulation of miR-21 was also found in the serum of sTBI patients but not of mTBI, at very early time points and up to 15 days from injury. Also, no increase was found in the musculoskeletal injured patients, and for this reason, miR-21 was considered as a potential new TBI biomarker and a future therapeutic target for TBI (84).
Another exciting miR associated with TBI is miR-16, involved in the regulation of several biological processes activated after TBI; such as being involved in apoptosis by targeting BCL-2 (93) and in the cell cycle by targeting CDK6 (cyclin-dependent kinase 6), CDC27 (cell division cycle 27) and CARD10 (caspase recruitment domain 10) (94, 95, 148). Also, miR-16 was significantly increased within the first 24 h in the mild TBI patients and significantly decreased in the severe TBI patients compared to the healthy volunteers (81). MiR-107 was found to be underexpressed in cortex and hippocampus of a rat model of severe controlled cortical impact (CCI) (96). MiR-107 can regulate granulin (GRN) mRNA, suggesting a role in inflammatory process, energy metabolism and neuron regeneration (104). MiR-27a and miR-23a were downregulated in mouse cortex in a moderate model of CCI and was found to regulate pro-apoptotic Bcl-2 family members (97). Furthermore, miR-711, was upregulated in hippocampus after severe CCI, (96) and was found to reduce the neuronal cell death and lesion volume via Akt-pathway.Let-7i is another exciting biomarker with potential implications in TBI. It was upregulated in the serum and CSF of the rodent model of mild to moderate blast overpressure wave. It might be a potential regulator of many proteins and inflammatory cytokines, including S-100β and UCH-L1 (98). A detailed list of the pathobiology for miRNA differentially regulated in animal models of TBI can be found in Table 3.
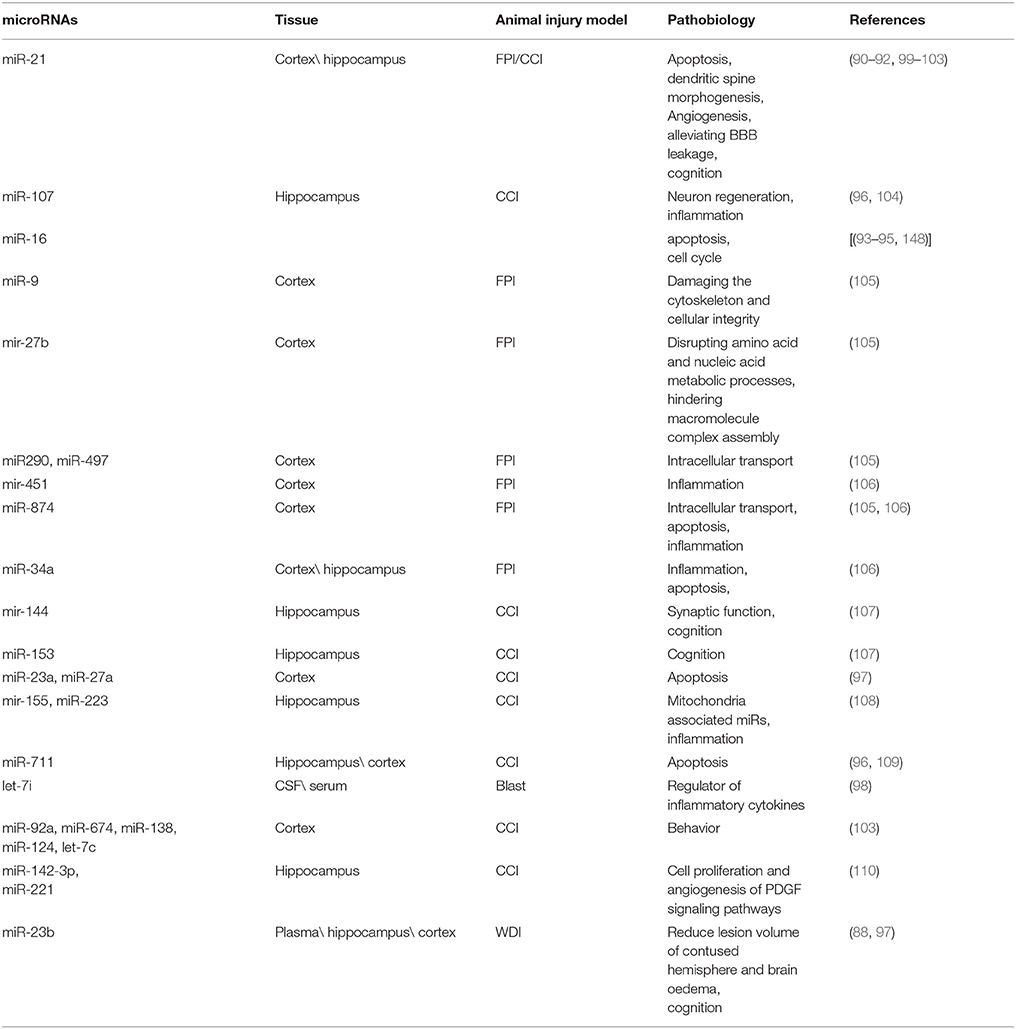
Table 3. Pathobiology of the main differentially expressed microRNAs in brain of different animal injury models.
Numerous studies investigated the global profiling of miRNAs in human diseases with the aim to identify a variety of biomarkers when compared the normal and affected tissues, which can further be correlated with the prognosis or the therapeutic response. MicroRNA can be extracted from a variety of sources, including cell lines, fresh tissues, formailin-fixed paraffin embedded (FFPE) tissues and also biofluids such as plasma, serum, urine, saliva and CSF.
Many are the techniques used to analyze microRNAs. Generally, qPCR is suitable to investigate one or two miRNAs, whereas for larger studies examining multiple miRNAs at once, platforms such as TaqMan™Array Microfluidic Cards, miScript miRNA PCR Array or nCounter® microRNA panels, are more suitable. Finally, to discover new miRNA variants, the Next Generation Sequence (NGS) solution results are more appropriate. In Figure 1 a decision making chart is represented.
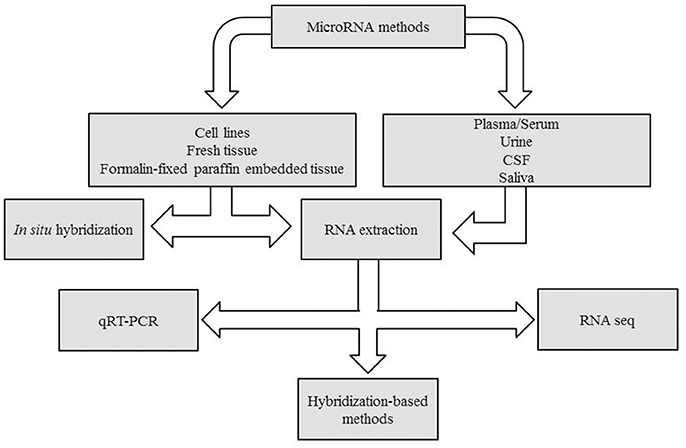
Figure 1. Decision making chart. MiRNAs can be extracted from different sample types, such as tissues and body fluids. The experimental design determines the methodology chosen for miRNA detection.
Sample processing and storage is the first step to perform miRNA profiling. This step is particularly crucial in order to obtain high-quality microRNA, especially for the determination of miRNA expression in biofluids. MiRNAs are stable in biofluids because of their molecular size and because they are protected within protein complexes or contained within EVs (microvescicles or exosomes). However, an immediate separation of cells is required to prevent lysis of cells and to avoid RNA contamination.
In addition, caution must be taken when collecting plasma or serum. Heparin-plasma for example, is a potent PCR inhibitor (111). Differentially, plasma-EDTA does not affect PCR and can overcome clotting due to platelet activation. In addition, plasma content of miRNA is higher than serum which is confirmed by slightly lower Ct value in the plasma (112).
Different kits are commercially available for miRNA extraction from different tissues or biofluids such as miRNeasy (Qiagen), mirVana TM (Ambion) or PureLink TM (Invitorgen) miRNA. The most commonly used kits are based on two main steps. The first one, is a chemical extraction with guanidine thiocynate (e.g., Trizol and QIAzol reagents); the second one, is an extraction procedure based on silica columns. New phenol-free kits were also recently developed such as ISOLATE II miRNA (Bioline) or ReliaPrep™miRNA (Promega).
Alternative strategies apply magnetic bead–based technology to purify samples, such as TaqMan™miRNA ABC Purification Kit (Thermo Fisher). All these kits differ, compared to those used for total RNA extraction, for additional steps to enrich the smallRNA fraction (113). Extraction from biofluid samples are particularly challenging, compared to extraction from tissues or cells, because of the lower RNA content, the possibility of hemolysis or platelet contamination and presence of serum proteins (such as RNases and PCR inhibitors). In addition the lack of well-established reference genes makes it difficult to analyze and interpret the data (114).
Nevertheless, several strategies can be used to maximize RNA yield. Among these, RNase-free glycogen, which acts as nucleic acid carrier can be added during the extraction (112). Similarly, other RNA carriers, such as the bacteriophage MS2 RNA, can also help to maximize RNA recovery (115). Therefore, monitoring the efficiency of the RNA extraction by addition of a known amount of synthetic miRNA spike-in is recommended (116). Alternatively, isolation of exosomes from biological fluid can help to increase the amount of retrieved RNA.
Exosomes are vesicles with diameter between 30 and 100 nm, originated from multivesiculated body (MVBs) and released into the extracellular space. The exosomes are able to carry different molecules as mRNAs, miRNAs, lipids and proteins and to transfer their contents to recipient cells, therefore influencing different physiologic and pathologic processes (117). The current techniques to separate exosomes from biological fluids include methods based on exosome size differences, as ultracentrifugation or size exclusion chromatography, or on identification of specific surface markers as immunoaffinity capture-based techniques (118). In ultracentrifugation procedure, the force used ranges from ~100,000 to 120,000 × g. After the centrifugation step, the exosome pellet is dissolved in phosphate buffered saline (PBS) and subjected to subsequent centrifugation runs with increasing force. Finally, isolated exosomes can be stored at −80°C until further analysis or in Trizol for RNA extraction. Based on their size, exosomes can also be purified by using membrane filters with 0.2 μm of diameter. This method, although widely used, can result in samples contaminated by others EVs and a large sample volume is requested. Other techniques were also developed to isolate exosomes. The presence of tetraspanins as exosomal surface markers, for example, is used for immunoaffinity reactions and different companies have already developed specific kit, based on affinity spin columns for exosome purification (Invitrogen, Qiagen).
The measurement of RNA concentration by using conventional spectrophotometers, such as nanodrop, is not possible for miRNA quantification and quality control (119). However, RNA integrity can be checked by spectrophotometry and automated capillary electrophoresis instruments such as the Bioanalyzer 2100 (Agilent) and Experion (Bio-Rad). In particular, the Bioanalyzer 2100, can also estimate miRNA concentration as the result of the ratio of 15-40nt RNA fragments and the total RNA, (120) providing that RNA integrity is very high. For this reason; it is a common practice to perform the analysis using established volume and not concentration of RNA extracted from the same volume of biofluid or tissue. However, accurate strategies to relatively quantify the sample are still necessary.
The most widely and well established approaches used to determine microRNA profile can be divided into three main categories: quantitative real time PCR (qRT-PCR), hybridisation-based methods (i.e., Microarrays, Nanostring) and high-throughput next generation sequencing. The main advantages or disadvantages in using the above techniques are reported in Table 4.
qRT-PCR is the most popular technique to accurately assess miRNAs.
The single assay is primarily used to efficiently validate the results of large screening studies or for relatively small experiments.
The technique is relatively expensive and can be divided in two main steps: the conversion of miRNA into cDNA and quantitative polymerase chain reaction.
Because of the length of miRNAs and the lack of a common sequence such as a poly(A) tail that can be used for reverse transcription, cDNA synthesis presents its own challenges.
Two main strategies are used to generate cDNA:
1) The use of a stem–loop RT primer which first hybridizes with the miRNA strand, followed by reverse transcription using MultiScribe reverse transcriptase. cDNA products are then amplified using conventional TaqMan PCR.
2) The addition of a poly(a) tail using E. coli poly(A) polymerase (assay). An oligo-dt primer is then used to pair the miRNA tailed and allows the retro-transcription of the resulting cDNA, which is further amplified using specific primers and detected by the use of a fluorescent dye such as SYBER green.
However, large experiments using qRT-PCR can become quite laborious to perform. In order to overcome this problem, reactions can also be carried out in high-throughput form.
Pre-plated PCR primers, for example, are commercially available and distributed typically across multiwall dishes, or alternatively microfluidic cards containing nanoliter-scale wells. However, performing highly parallel qRT-PCR might present some challenge due to differences in primer annealing temperatures. However, it is still possible to solve this issue by using the locked nucleic acids (LNAs) into primers and allowing the optimal hybridisation conditions for several PCR assays to be run simultaneously (114).
qRT-PCR allows both absolute and relative quantification. In the first one, a standard curve from serial dilutions of known concentrations of synthetic miRNA is generated and used to calculate the number of copies of a specific miRNA. In the second case, before setting up the microRNA expression analysis, an endogenous normalizer (reference gene) has to be chosen, among several control candidates tested. These candidates offer stable expression over the whole range of samples, and are selected based on the literature or pre-existing data.
Hsa-miR-16-5p is widely used in the literature as an endogenous miR, despite the lack of a panel of endogenous miRNA consensus (121). Hsa-miR-223 (116) hsa-let-7d-5p (122) hsa-miR-484 (123), hsa-miR-191-5p (124), and hsa-miR-423 (125) were also described as relatively invariant reference genes in plasma/serum. MiR-331 and miR-223 were identified as the most stables in traumatic brain injury patients (84). MiR-202 was also used as normalizer gene in CSF of TBI patients (85).
In addition, to identifying the appropriate endogenous controls, it is also possible to use some software as geNorm Algorithm (https://genorm.cmgg.be/) and DataAssist v.3 software (Applied Biosystems). GeNorm is used to normalize the data from a large and unbiased set of miRNAs. DataAssist is useful to quantify gene expression in samples when using the comparative CT (ΔΔCT) method (126, 127). However, it is always preferable to add a spike-in control during the RNA extraction and to normalize the microRNA using an exogenous control (e.g., cell-miR-39).
Several hybridization-based methods exist to identify microRNA abundance. In situ hybridization (ISH) is the most used method to localize DNA or RNA using labeled complementary nucleic acid probes in tissue section or fixed cells (128). However, this technique is not suitable for miRNA detection because of their length, but the introduction of LNA showed a significant improvement in the sensitivity and specificity of this technique applied to miRNAs detection (129). Microarray-based technique is another powerful high-throughput method extensively used for microRNA profiling, because of their ability to screen large number of miRs simultaneously in large variety of samples (from tissue to biofluid). MiRNA microarray is a nucleic acid hybridization technique which uses amino-modified 5' termininal complementary probes immobilized onto glass slides through covalent crosslinking between the amino-groups and the self-assembling monolayer (130). After RNA purification, miRNAs are tagged with fluorophore-labeled nucleotides at their 3′ end. LNAs can also be incorporated into capture probes to increase specificity and sensitivity (131). The main advantages of using micrarray are the low costs and the parallel measurements. Typically microarray involves a comparison between two or more groups and cannot be used to determine absolute quantification. Because of limited specificity, data obtained are typically validated by a qRT-PCR.
A new technology, the Nanostring nCounter Analysis System, was recently developed to allow the quantification of more than 800 RNA molecules in 12 samples, in a single assay. The nCounter Analysis System is a very new technology which uses digital color-coded barcode for precise multiplexed measurement of the gene expression (<1 copy per cell). This system is more sensitive than microarrays and as sensitive and accurate as qRT-PCR. The combination of color-coded barcode attached to a single target-specific probe corresponding to a gene of interest and the single molecule imaging, allows detecting and counting hundreds of unique transcripts in a single reaction. Each color-coded barcode represents a single target molecule. No amplification is required (132).
Finally, to identify the significant differentially expressed miRNAs in a large genomic data, such as the microarray data but also the microfluidic card and the RNA sequencing data, the most frequently used method is the Significance Analysis of Microarrays (SAM) computed by Multi Experiment Viewer (MEV) v4.8.1 (http://www.tm4.org).
The introduction of the next generation sequencing has become increasingly popular in biomedical research, overcoming the limitations of the microarray analysis (133). While it cannot quantify miRNA levels with the same resolution of qPCR, it still has the advantage to detect all known or unknown miRNAs present in a sample and to precisely distinguish all isoforms in the absence of background and cross-hybridization problems. IsomiRNAs, indeed are miRNA containing sequence variations, typically by shortening or lengthening of the 3′ end. Over 3,300 miRNA variants were identified and reported at the following website http://galas.systemsbiology.net/cgi-bin/isomir/find.pl. However, one or two isomers contribute to >90% of the signal detected, while the remaining variants are not abundant enough to be revealed.
The procedure consists in a small-RNA cDNA library preparation followed by “massive parallel” sequencing on a single run. First of all, miRNA fragments are extracted from total RNA. Running the sample on an agarose gel and cutting out the band corresponding to the miRNA size is the second step. Then, the selected RNA fragments are ligated to sequencing adapters and transcribed into cDNA by ~12–15 RT-PCR cycles of amplification and using a reverse transcription primer which hybridizes to the 3′ adapter.
At this point, another run on agarose gel of the obtained cDNA library is performed and the band with size corresponding to the length of adapter sequences plus the miRNA insert of ~20–30 bases (for a total length of 120 bp) is cut out and ready for sequencing. The gel size selection is particularly crucial because of the potential presence of adapter dimer side products created during the ligation step as well as higher molecular weight products generated from ligation of other RNA fragments, such as tRNA and snoRNA, containing 5′ phosphate groups.
Significant computational resources and bioinformatics expertise are required for data interpretation not only for known miRs but also for the newly discovered miRs. Initially all generated reads are aligned to the reference genome of the sequenced organisms. Short read aligner tools are available to process the reads such as maq (http://maq.sourceforge.net/maq-man.shtml), sop (http://soap.genomics.org.com) or bwa (http://bio-bwa.souceforge.net/). In addition, it is also important to filter out reads that align against other non-coding small RNA species and RNA degradation products which sequences are available on the University of Santa Cruz (UCSC) Genome Browser.
Another bioinformatics challenge is the relative quantification. Expression levels are analyzed on the base of the read counts for each sequenced sample. The number of reads of each individual molecule is normalized against the total number of reads produced in the same sample (134).
Different tools are also available to predict novel miRNAs from generated data. One of the commonly used is mirDeep (https://www.mdc-berlin.de/8551903/en/) (135).
Although the NGS is one of the most advanced techniques currently used, other challenges, beside the bioinformatic support, need to be faced. One of these is the cost required for equipment, software and consumables. In addition a high quality of purified RNA and a large amount of RNA, usually 5 μg, are required for the analysis. Validation is another important aspect to address in order to use this technology for diagnosis and prognosis of diseases.
Since miRNAs control the regulation of several genes and they are linked to many disorders, it is also possible to reliably predict potential miRNA targets which can be involved in these pathologies. The prediction of the mRNA targets is based on the partial complementary sequences between the mature miR and the mRNA candidate target. This search is generated by miRNA target prediction algorithms able to seek for putative binding sites in the 3′UTRs of the candidate mRNAs (i.e., PicTar, TargetScan, DIANA-microT, miRanda, rna22). High complementarity between the miRNA and the target binding region results in the degradation of the target, whereas the presence of mismatches represses the translational process. However, results of their applications are often not consistent and must be experimentally validated. Many lab-based techniques can be used to overcome the challenge of target validation such as the inverse correlation between the expression of miRNA and its target, the effect on protein expression /function or a direct validation by using the luciferase assay or their functional effects (proliferation, differentiation, apoptosis) on a cell culture system.
In addition, databases such as mirTarBase or miRewcords collect both predicted and experimentally confirmed miRNA targets.
Finally, functional analysis of miRNAs or miRNA high-throughput data sets can also been performed. For example, Gene Ontology (GO) analysis is commonly used to identify pathways and processes from a list of genes provided, for example from results obtained using gene expression microarray (136) or generated from a target prediction tools in the case of miRNA (137).
The use of miRNA signature as a novel diagnostic/prognostic tool is still in the descriptive phase. Numerous data have been collected so far, in various disease states; however their translation in clinical applicability requires much larger studies and universally implemented guidelines.
First of all, the lack of methodological details in published papers makes it difficult to directly compare the results, and lead to inconsistent or even contradictory results.
Standard protocols must be achieved for the different steps of miRNA analysis such as sample processing, RNA extraction and expression measurement/assessment methods as well as differences in specimen type, for example FFPE vs. fresh frozen samples, must be considered.
In addition, the research of miRNA profile in biofluids is particularly challenging as miRNAs, circulate either associated with proteins, lipoproteins or EVs, and this might require specific precautions during the extraction or analysis processes. Moreover, it is good practice to check the presence of small clots and hemolysis in plasma/serum which may contribute to the variability in miRNA expression.
Furthermore, we are not aware if miRNA expression varies at specific conditions such as: fasting or circadian rhythm, thus, standardization and annotation of these protocol details is necessary in order to minimize variability of unknown factors.
Data normalization, identification of well-characterized endogenous miRs specific to biofluid and pathology of interest, as well as characterization of baseline levels for miRs described as potential biomarkers are other crucial points in obtaining accurate results.
Certainly, a common information infrastructure for data exchange, analysis and protocols used would facilitate research in the miRNA biomarker discovery.
Besides the challenge of biomarker discovery, there lies the challenge of rapidly detecting them with clinically relevant sensitivity and specificity using a low-cost and easy point-of-care injury test.
In the case of traumatic brain injury, a PoC technology would have several applications. This is particularly true for mTBI which represents a serious problem in military, and contact sports that has led to reduction in the sport participation in younger age groups.
The development of a pitch-side or “pre-hospital,” portable TBI diagnostic devices, would implement the current guidelines in the management of mTBI (17, 138) in different ways:
1) In the initial pre-hospital assessment to determine whether patients should be transferred to a Major Trauma Centre or a local Trauma Unit.
2) In the Emergency Department (ED), to determine the need for a CT brain scan.
3) Pitch-side, to assist decision making as to removal from play and assessment of the need to take the player to the ED.
4) In sports clinics, to diagnose a concussive event and guide return to play.
5) In combat theaters, to determine the need to dispatch a rescue team.
So far, proteins were widely explored as biomarkers and immunoassays are extensively used as method of detection, although not often very sensitive and prone to false positives (139).
The PCR amplification method has played an important role in diagnostics over the last years because of its ability to detect few molecules (140) and the fact that microRNAs are particularly stable in biofluids, positions them as a new valid potential biomarkers to explore. MicroRNAs are also particularly suitable for these clinical applications as they are molecular switch regulators and for this reason their early expression anticipates the molecular mechanisms trigged by TBI.
Several companies are now working on point-of-care device that can measure microRNAs in the field. This is quite challenging, although not insurmountable, as microRNAs are present in femtomolar and picomolar concentrations and need to be extracted from the biofluid first.
Micorfluidics is another challenging problem. Transporting the methodology in a portable device, reducing the volume to few microliters over a few millimeters and mixing the rinsing solutions and all reagents are main issues.
Detection is another important point to discuss; various strategies were developed to improve the detection of miRNA (141).
Nanoparticles(NPs)-based biosensors, for example, are widely studied. The use of this biosensor has the potential to miniaturize the equipment and reduce the cost. In particular, carbon and metal-based NPs, such as gold nanoparticles (AuNPs) are excellent miRNA carriers that can be used to accelerate the signal transduction enhancing a rapid analysis and lowering the detection limit. Recently, a dual-function gold nanoparticle biolaben was used to detect miR-21 in serum (142).
Magnetic nanoparticles are also very popular. Wanunu et al. (143) developed a protocol using probe:miRNA duplex binded to p19-functionalized magnetic beads, which are first eluted and electronically detected using a nanopore (143).
Optical detection in combination with NP probes was also explored in the development of a novel highly specific and reproducible platform, the Scanometric MicroRNA (Scano-miR), to detect low concentrations of miRNAs (144).
Surface plasmon resonance (SPR) biosensors, is another example of label-free optical biosensing technologies. This method is based on optical measurement of refractive index changes given by the binding of analyte molecules present in samples to specific receptors immobilized on the SPR sensor. This method showed to be able to detect miRNA in <30 min at concertation down to 2 pM (145).
Finally, enzyme catalytic amplification-based electrochemical assay are also developed for this purpose (146).
However, hard work is still required to develop a reliable portable PoC device.
miRNA profiling and detection provide valuable information on their essential roles in normal cellular function and disease, projecting their use in the clinical practice for the diagnosis and prognosis of several pathologies. With this review, our aim was to provide insights into the miRNA expression in TBI, the main commonly used detection methods to discover new biomarkers and the state-of-the art of the PoC development.
Despite their limited use as routine biomarkers, several companies already offer miRNA-based diagnostic assays.
In addition, there are new emerging classes of non-coding RNA such as piwi-interacting RNAs, and long non-coding RNA (lncRNA) that have important role in cellular physiology.
In the future, profiling methods that have the potential to detect all the RNA classes are likely to improve the understanding of the whole transcriptome and provide new valid information for the diagnosis, prognosis and therapy of several pathologies, including TBI.
VD drafting the article and final approval of the version to be published. KY drafting the article. US drafting the article. CD critical revision of the article. AB critical revision of the article.
The University of Birmingham has intellectual property associated with miRNA listed in this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Taylor CA, Bell JM, Breiding MJ, Xu L. Traumatic brain injury-related emergency department visits, hospitalizations, and deaths - United States, 2007 and 2013. MMWR Surveill Summ. (2017) 66:1–16. doi: 10.15585/mmwr.ss6609a1
2. De Guzman E, Ament A. Neurobehavioral management of traumatic brain injury in the critical care setting: an update. Crit Care Clin. (2017) 33:423–40. doi: 10.1016/j.ccc.2017.03.011
3. Menon DK, Schwab K, Wright DW, Maas AI. Position statement: definition of traumatic brain injury. Arch Phys Med Rehabil. (2010) 91:1637–40. doi: 10.1016/j.apmr.2010.05.017
4. Hyder AA, Wunderlich CA, Puvanachandra P, Gururaj G, Kobusingye OC. The impact of traumatic brain injuries: a global perspective. NeuroRehabilitation (2007) 22:341–53.
5. Corrigan JD, Selassie AW, Orman JAL. The epidemiology of traumatic brain injury. J Head Trauma Rehabil. (2010) 25:72–80. doi: 10.1097/HTR.0b013e3181ccc8b4
6. Feigin VL, Theadom A, Barker-Collo S, Starkey NJ, McPherson K, Kahan M, et al. Incidence of traumatic brain injury in New Zealand: a population-based study. Lancet Neurol. (2013) 12:53–64. doi: 10.1016/S1474-4422(12)70262-4
7. Stoica BA, Faden AI. Cell death mechanisms and modulation in traumatic brain injury. Neurotherapeutics (2010) 7:3–12. doi: 10.1016/j.nurt.2009.10.023
8. Amorini AM, Lazzarino G, Di Pietro V, Signoretti S, Lazzarino G, Belli A, et al. Metabolic, enzymatic and gene involvement in cerebral glucose dysmetabolism after traumatic brain injury. Biochim Biophys Acta (2016) 1862:679–87. doi: 10.1016/j.bbadis.2016.01.023
9. Amorini AM, Lazzarino G, Di Pietro V, Signoretti S, Lazzarino G, Belli A, et al. Severity of experimental traumatic brain injury modulates changes in concentrations of cerebral free amino acids. J Cell Mol Med. (2017) 21:530–42. doi: 10.1111/jcmm.12998
10. Di Pietro V, Amin D, Pernagallo S, Lazzarino G, Tavazzi B, Vagnozzi R, et al. Transcriptomics of traumatic brain injury: gene expression and molecular pathways of different grades of insult in a rat organotypic hippocampal culture model. J Neurotrauma (2010) 27:349–59. doi: 10.1089/neu.2009.1095
11. Di Pietro V, Amorini AM, Tavazzi B, Hovda DA, Signoretti S, Giza CC, et al. Potentially neuroprotective gene modulation in an in vitro model of mild traumatic brain injury. Mol Cell Biochem. (2013) 375:185–98. doi: 10.1007/s11010-012-1541-2
12. Di Pietro V, Amorini AM, Tavazzi B, Vagnozzi R, Logan A, Lazzarino G, et al. The molecular mechanisms affecting N-acetylaspartate homeostasis following experimental graded traumatic brain injury. Mol Med. (2014) 20:147. doi: 10.2119/molmed.2013.00153
13. Di Pietro V, Lazzarino G, Amorini AM, Tavazzi B, D'Urso S, Longo S, et al. Neuroglobin expression and oxidant/antioxidant balance after graded traumatic brain injury in the rat. Free Radic Biol. Med. (2014) 69:258–64. doi: 10.1016/j.freeradbiomed.2014.01.032
14. Tavazzi B, Vagnozzi R, Signoretti S, Amorini AM, Finocchiaro A, Cimatti M, et al. Temporal window of metabolic brain vulnerability to concussions: oxidative and nitrosative stresses—part II. Neurosurgery (2007) 61:390–6. doi: 10.1227/01.NEU.0000255525.34956.3F
15. Vagnozzi R, Tavazzi B, Signoretti S, Amorini AM, Belli A, Cimatti M, et al. Temporal window of metabolic brain vulnerability to concussions: mitochondrial-related impairment—part I. Neurosurgery (2007) 61:379–89. doi: 10.1227/01.NEU.0000280002.41696.D8
16. Duncan CC, Summers AC, Perla EJ, Coburn KL, Mirsky AF. Evaluation of traumatic brain injury: brain potentials in diagnosis, function, and prognosis. Int J Psychophysiol. (2011) 82:24–40. doi: 10.1016/j.ijpsycho.2011.02.013
17. Vos PE, Alekseenko Y, Battistin L, Ehler E, Gerstenbrand F, Muresanu DF, et al. Mild traumatic brain injury. Eur J Neurol. (2012) 19:191–8. doi: 10.1111/j.1468-1331.2011.03581.x
18. Brenner DJ, Hall EJ. Computed tomography—an increasing source of radiation exposure. N Engl J Med. (2007) 357:2277–84. doi: 10.1056/NEJMra072149
19. Choi SJ, Kim EY, Kim HS, Choi H-Y, Cho J, Yang HJ, et al. Cumulative effective dose associated with computed tomography examinations in adolescent trauma patients. Pediatr Emerg Care (2014) 30:479–82. doi: 10.1097/PEC.0000000000000165
20. Gatto R, Chauhan M, Chauhan N. Anti-edema effects of rhEpo in experimental traumatic brain injury. Restor Neurol Neurosci. (2015) 33:927–41. doi: 10.3233/RNN-150577
21. Marin JR, Weaver MD, Yearly DM, Mannix RC. Trends in visits for traumatic brain injury to emergency departments in the United States. JAMA (2014) 311:1917–19. doi: 10.1001/jama.2014.3979
22. Hadanny A, Efrati S. Treatment of persistent post-concussion syndrome due to mild traumatic brain injury: current status and future directions. Expert Rev Neurother. (2016) 16:875–87. doi: 10.1080/14737175.2016.1205487
23. Walker WC, Franke LM, Sima AP, Cifu DX. Symptom trajectories after military blast exposure and the influence of mild traumatic brain injury. J Head Trauma Rehabil. (2017) 32:E16–26. doi: 10.1097/HTR.0000000000000251
24. Gaetz M. The multi-factorial origins of Chronic Traumatic Encephalopathy (CTE) symptomology in post-career athletes: the athlete post-career adjustment (AP-CA) model. Med Hypotheses (2017) 102:130–43. doi: 10.1016/j.mehy.2017.03.023
25. Omalu BI, Fitzsimmons RP, Hammers J, Bailes J. Chronic traumatic encephalopathy in a professional American wrestler. J Forensic Nurs. (2010) 6:130–6. doi: 10.1111/j.1939-3938.2010.01078.x
26. Azar S, Hasan A, Younes R, Najdi F, Baki L, Ghazale H, et al. Biofluid proteomics and biomarkers in traumatic brain injury. Methods Mol Biol. (2017) 1598:45–63. doi: 10.1007/978-1-4939-6952-4_3
27. Jönsson H, Johnsson P, Höglund P, Alling C, Blomquist S. Elimination of S100B and renal function after cardiac surgery. J Cardiothorac Vasc Anesth. (2000) 14:698–701. doi: 10.1053/jcan.2000.18444
28. Sen J, Belli A. S100B in neuropathologic states: the CRP of the brain? J Neurosci Res. (2007) 85:1373–80. doi: 10.1002/jnr.21211
29. Goyal A, Failla MD, Niyonkuru C, Amin K, Fabio A, Berger RP, et al. S100b as a prognostic biomarker in outcome prediction for patients with severe traumatic brain injury. J Neurotrauma (2013) 30:946–57. doi: 10.1089/neu.2012.2579
30. Anderson RE, Hansson LO, Nilsson O, Dijlai-Merzoug R, Settergren G. High serum S100B levels for trauma patients without head injuries. Neurosurgery (2001) 48:1255–60.
31. Pelinka LE, Toegel E, Mauritz W, Redl H. Serum S 100 B: a marker of brain damage in traumatic brain injury with and without multiple trauma. Shock (2003) 19:195–200. doi: 10.1097/00024382-200303000-00001
32. Undén J, Ingebrigtsen T, Romner B. Scandinavian guidelines for initial management of minimal, mild and moderate head injuries in adults: an evidence and consensus-based update. BMC Med. (2013) 11:50. doi: 10.1186/1741-7015-11-50
33. Undén L, Calcagnile O, Undén J, Reinstrup P, Bazarian J. Validation of the Scandinavian guidelines for initial management of minimal, mild and moderate traumatic brain injury in adults. BMC Med. (2015) 13:292. doi: 10.1186/s12916-015-0533-y
34. Middeldorp J, Hol E. GFAP in health and disease. Prog Neurobiol. (2011) 93:421–43. doi: 10.1016/j.pneurobio.2011.01.005
35. Czeiter E, Mondello S, Kovacs N, Sandor J, Gabrielli A, Schmid K, et al. Brain injury biomarkers may improve the predictive power of the IMPACT outcome calculator. J Neurotrauma (2012) 29:1770–8. doi: 10.1089/neu.2011.2127
36. Di Pietro V, Amorini AM, Lazzarino G, Yakoub KM, D'Urso S, Lazzarino G. S100B and glial fibrillary acidic protein as indexes to monitor damage severity in an in vitro model of traumatic brain injury. Neurochem Res. (2015) 40:991–9. doi: 10.1007/s11064-015-1554-9
37. Yang Z, Wang KK. Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. (2015) 38:364–74. doi: 10.1016/j.tins.2015.04.003
38. Borg K, Bonomo J, Jauch EC, Kupchak P, Stanton EB, Sawadsky B. Serum level of biochemical markers of traumatic brain injury. ISRN Emer Med. (2012) 2012:417313. doi: 10.5402/2012/417313
39. Shahim P, Tegner Y, Wilson D, Randall J, Skillback T, Pazooki D, et al. Blood biomarkers for brain injury in concussed professional ice hockey players. JAMA Neurol. (2014) 71:684–92. doi: 10.1001/jamaneurol.2014.367
40. Wolf H, Frantal S, Pajenda GS, Salameh O, Widhalm H, Hajdu S, et al. Predictive value of neuromarkers supported by a set of clinical criteria in patients with mild traumatic brain injury: S100B protein and neuron-specific enolase on trial: clinical article. J Neurosurg. (2013) 118:1298–303. doi: 10.3171/2013.1.JNS121181
41. Siman R, Shahim P, Tegner Y, Blennow K, Zetterberg H, Smith DH, et al. Serum SNTF increases in concussed professional ice hockey players and relates to the severity of postconcussion symptoms. J Neurotrauma (2015) 32:1294–300. doi: 10.1089/neu.2014.3698
42. Papa L, Silvestri S, Brophy GM, Giordano P, Falk JL, Braga CF, et al. GFAP out-performs S100β in detecting traumatic intracranial lesions on computed tomography in trauma patients with mild traumatic brain injury and those with extracranial lesions. J Neurotrauma (2014) 31:1815–22. doi: 10.1089/neu.2013.3245
43. Papa L, Lewis LM, Silvestri S, Falk JL, Giordano P, Brophy GM, et al. Serum levels of ubiquitin C-terminal hydrolase distinguish mild traumatic brain injury from trauma controls and are elevated in mild and moderate traumatic brain injury patients with intracranial lesions and neurosurgical intervention. J Trauma Acute Care Surg. (2012) 72:1335–44. doi: 10.1097/TA.0b013e3182491e3d
44. Mondello S, Buki A, Barzo P, Randall J, Provuncher G, Hanlon D, et al. CSF and plasma amyloid-β temporal profiles and relationships with neurological status and mortality after severe traumatic brain injury. Sci Rep. (2014) 4:6446. doi: 10.1038/srep06446
45. Hill LJ, Di Pietro V, Hazeldine J, Davies D, Tomman E, Logan A, et al. Cystatin D (CST5): an ultra-early inflammatory biomarker of traumatic brain injury. Sci Rep. (2017) 7:5002. doi: 10.1038/s41598-017-04722-5
46. Svetlov SI, Larner SF, Kirk DR, Atkinson J, Hayes RL, Wang KK. Biomarkers of blast-induced neurotrauma: profiling molecular and cellular mechanisms of blast brain injury. J Neurotrauma (2009) 26:913–21. doi: 10.1089/neu.2008.0609
47. Johnsson P. Markers of cerebral ischemia after cardiac surgery. J Cardiothorac Vasc Anesth. (1996) 10:120–6. doi: 10.1016/S1053-0770(96)80187-X
48. Topolovec-Vranic J, Pollmann-Mudryj M-A, Ouchterlony D, Klein D, Spence J, Romaschin A, et al. The value of serum biomarkers in prediction models of outcome after mild traumatic brain injury. J Trauma Acute Care Surg. (2011) 71:S478–86. doi: 10.1097/TA.0b013e318232fa70
49. Toman E, Harrisson S, Belli T. Biomarkers in traumatic brain injury: a review. Army Med Corps (2016) 162:103–8. doi: 10.1136/jramc-2015-000517
50. Mrozek S, Dumurgier J, Citerio G, Mebazaa A, Geeraerts T. Biomarkers and acute brain injuries: interest and limits. Crit Care (2014) 18:220. doi: 10.1186/cc13841
51. Oliver JM, Jones MT, Kirk KM, Gable DA, Repshas JT, Johnson TA, et al. Serum neurofilament light in American football athletes over the course of a season. J Neurotrauma (2016) 33:1784–9. doi: 10.1089/neu.2015.4295
52. Millecamps S, Gowing G, Corti O, Mallet J, Julien JP. Conditional NF-L transgene expression in mice for in vivo analysis of turnover and transport rate of neurofilaments. J Neuroscience (2007) 27:4947–56. doi: 10.1523/JNEUROSCI.5299-06.2007
53. Perrot R, Berges R, Bocquet A, Eyer J. Review of the multiple aspects of neurofilament functions, and their possible contribution to neurodegeneration. Mol Neurobiol. (2008) 38:27–65. doi: 10.1007/s12035-008-8033-0
54. Gaiottino J, Norgren N, Dobson R, Topping J, Nissim A, Malaspina A, et al. Increased neurofilament light chain blood levels in neurodegenerative neurological diseases. PLoS ONE (2013) 8:e75091. doi: 10.1371/journal.pone.0075091
55. Tortelli R, Ruggieri M, Cortese R, D'errico E, Capozzo R, Leo A, et al. Elevated cerebrospinal fluid neurofilament light levels in patients with amyotrophic lateral sclerosis: a possible marker of disease severity and progression. Eur J Neurol. (2012) 19:1561–7. doi: 10.1111/j.1468-1331.2012.03777.x
56. Pandey S, Singh K, Sharma V, Pandey D, Jha RP, Rai SK, et al. A prospective pilot study on serum cleaved tau protein as a neurological marker in severe traumatic brain injury. Br J Neurosurg. (2017) 31:356–3. doi: 10.1080/02688697.2017.1297378
57. Öst M, Nylen K, Csajbok L, Öhrfelt AO, Tullberg M, Wikkelsö C, et al. Initial CSF total tau correlates with 1-year outcome in patients with traumatic brain injury. Neurology (2006) 67:1600–4. doi: 10.1212/01.wnl.0000242732.06714.0f
58. Bazarian JJ, Zemlan FP, Mookerjee S, Stigbrand T. Serum S-100B and cleaved-tau are poor predictors of long-term outcome after mild traumatic brain injury. Brain Inj. (2006) 20:759–65. doi: 10.1080/02699050500488207
59. Wang KK, Yang Z, Sarkis G, Torres I, Raghavan V. Ubiquitin C-terminal hydrolase-L1 (UCH-L1) as a therapeutic and diagnostic target in neurodegeneration, neurotrauma and neuro-injuries. Expert Opin Ther Targets (2017) 21:627–38. doi: 10.1080/14728222.2017.1321635
60. Papa L, Brophy GM, Welch RD, Lewis LM, Braga CF, Tan CN, et al. Time course and diagnostic accuracy of glial and neuronal blood biomarkers GFAP and UCH-L1 in a large cohort of trauma patients with and without mild traumatic brain injury. JAMA Neurol. (2016) 73:551–60. doi: 10.1001/jamaneurol.2016.0039
61. Posti JP, Takala RS, Runtti H, Newcombe VF, Outtrim J, Katila AJ, et al. The levels of glial fibrillary acidic protein and ubiquitin C-terminal hydrolase-L1 during the first week after a traumatic brain injury: correlations with clinical and imaging findings. Neurosurgery (2016) 79:456–64. doi: 10.1227/NEU.0000000000001226
62. Takala RS, Posti JP, Runtti H, Newcombe VF, Outtrim J, Katila AJ, et al. Glial fibrillary acidic protein and ubiquitin C-terminal hydrolase-L1 as outcome predictors in traumatic brain injury. World Neurosurg. (2016) 87:8–20. doi: 10.1016/j.wneu.2015.10.066
63. Meier T, Nelson LD, Huber DL, Bazarian J, Hayes RL, McCrea M. Prospective assessment of acute blood markers of brain injury in sport-related concussion. J Neurotrauma (2017) 34:3134–42. doi: 10.1089/neu.2017.5046
64. Lee RC, Feinbaum RL, Ambros V. The, C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y
65. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science (2001) 294:853–8. doi: 10.1126/science.1064921
66. Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia (2010) 14:236–40.
67. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. (2002) 21:4663–70. doi: 10.1093/emboj/cdf476
68. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. (2009) 11:228–34. doi: 10.1038/ncb0309-228
69. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. (2003) 17:3011–6. doi: 10.1101/gad.1158803
70. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature (2005) 436:740–4. doi: 10.1038/nature03868
71. Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell (2003) 115:199–208. doi: 10.1016/S0092-8674(03)00759-1
72. Long JM, Lahiri DK. Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders. Exp Neurol. (2012) 235:402–18. doi: 10.1016/j.expneurol.2011.12.043
73. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
74. Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. (2012) 19:586–93. doi: 10.1038/nsmb.2296
75. Esquela-Kerscher A, Slack FJ. Oncomirs–microRNAs with a role in cancer. Nat Rev Cancer (2006) 6:259–69. doi: 10.1038/nrc1840
76. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
77. Vijayan M, Reddy PH. Peripheral biomarkers of stroke: focus on circulatory microRNAs. Biochim Biophys Acta (2016) 1862:1984–93. doi: 10.1016/j.bbadis.2016.08.003
78. Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. (2016) 231:25–30. doi: 10.1002/jcp.25056
79. Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. (2010) 70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010
80. Czech MP. MicroRNAs as therapeutic targets. N Engl J Med. (2006) 354:1194–5. doi: 10.1056/NEJMcibr060065
81. Redell JB, Moore AN, Ward NH III, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microRNA levels. J Neurotrauma (2010) 27:2147–56. doi: 10.1089/neu.2010.1481
82. Yang T, Song J, Bu X, Wang C, Wu J, Cai J, et al. Elevated serum miR-93, miR-191, and miR-499 are noninvasive biomarkers for the presence and progression of traumatic brain injury. J Neurochem. (2016) 137:122–9. doi: 10.1111/jnc.13534
83. Mitra B, Rau TF, Surendran N, Brennan JH, Thaveenthiran P, Sorich E, et al. Plasma micro-RNA biomarkers for diagnosis and prognosis after traumatic brain injury: a pilot study. J Clin Neurosci. (2017) 38:37–42. doi: 10.1016/j.jocn.2016.12.009
84. Di Pietro V, Ragusa M, Davies D, Su Z, Hazeldine J, Lazzarino G, et al. MicroRNAs as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. J Neurotrauma (2017) 34:1948–56. doi: 10.1089/neu.2016.4857
85. Bhomia M, Balakathiresan NS, Wang KK, Papa L, Maheshwari RK. A panel of serum MiRNA biomarkers for the diagnosis of severe to mild traumatic brain injury in humans. Sci Rep. (2016) 6:28148. doi: 10.1038/srep28148
86. Johnson JJ, Loeffert AC, Stokes J, Olympia RP, Bramley H, Hicks SD. Association of salivary MicroRNA changes with prolonged concussion symptoms. JAMA Pediatr. (2018) 172:65–73. doi: 10.1001/jamapediatrics.2017.3884
87. Hicks SD, Johnson J, Carney MC, Bramley H, Olympia RP, Loeffert AC, et al. Overlapping MicroRNA expression in saliva and cerebrospinal fluid accurately identifies pediatric traumatic brain injury. J Neurotrauma (2018) 35:64–72. doi: 10.1089/neu.2017.5111
88. Sun L, Liu A, Zhang J, Ji W, Li Y, Yang X, et al. miR-23b improves cognitive impairments in traumatic brain injury by targeting ATG12-mediated neuronal autophagy. Behav Brain Res. (2016) 340:126–36. doi: 10.1016/j.bbr.2016.09.020
89. Patz S, Trattnig C, Grunbacher G, Ebner B, Gülly C, Novak A, et al. More than cell dust: microparticles isolated from cerebrospinal fluid of brain injured patients are messengers carrying mRNAs, miRNAs, and proteins. J Neurotrauma (2013) 30:1232–42. doi: 10.1089/neu.2012.2596
90. Liu RH, Ning B, Ma XE, Gong WM, Jia TH. Regulatory roles of microRNA-21 in fibrosis through interaction with diverse pathways. Mol Med Rep. (2016) 13:2359–66. doi: 10.3892/mmr.2016.4834
91. Ge X, Han Z, Chen F, Wang H, Zhang B, Jiang R, et al. MiR-21 alleviates secondary blood–brain barrier damage after traumatic brain injury in rats. Brain Res. (2015) 1603:150–7. doi: 10.1016/j.brainres.2015.01.009
92. Harrison EB, Hochfelder CG, Lamberty BG, Meays BM, Morsey BM, Kelso ML, et al. Traumatic brain injury increases levels of miR-21 in extracellular vesicles: implications for neuroinflammation. FEBS Open Bio (2016) 6:835–46. doi: 10.1002/2211-5463.12092
93. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
94. Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. (2007) 27:2240–52. doi: 10.1128/MCB.02005-06
95. Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. (2008) 105:5166–71. doi: 10.1073/pnas.0800121105
96. Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. (2009) 87:1435–48. doi: 10.1002/jnr.21945
97. Sabirzhanov B, Zhao Z, Stoica BA, Loane DL, Wu J, Borroto C, et al. Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci. (2014) 34:10055–71. doi: 10.1523/JNEUROSCI.1260-14.2014
98. Balakathiresan N, Bhomia M, Chandran R, Chavko M, McCarron RM, Maheshwari RK. MicroRNA let-7i is a promising serum biomarker for blast-induced traumatic brain injury. J Neurotrauma (2012) 29:1379–87. doi: 10.1089/neu.2011.2146
99. Hu T, Zhou FJ, Chang YF, Li YS, Liu GC, Hong Y, et al. MiR21 is associated with the cognitive improvement following voluntary running wheel exercise in TBI mice. J Mol Neurosci. (2015) 57:114–22. doi: 10.1007/s12031-015-0584-8
100. Redell JB, Zhao J, Dash PK. Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J Neurosci Res. (2011) 89:212–21. doi: 10.1002/jnr.22539
101. Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, et al. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. (2014) 4:6718. doi: 10.1038/srep06718
102. Sandhir R, Gregory E, Berman NEJ. Differential response of miRNA-21 and its targets after traumatic brain injury in aging mice. Neurochem Int. (2014) 78:117–21. doi: 10.1016/j.neuint.2014.09.009
103. Miao W, Bao T, Han J, Yin M, Yan Y, Wang W, et al. Voluntary exercise prior to traumatic brain injury alters miRNA expression in the injured mouse cerebral cortex. Braz J Med Biol Res. (2015) 48:433–9. doi: 10.1590/1414-431X20144012
104. Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, et al. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. (2010) 177:334–45. doi: 10.2353/ajpath.2010.091202
105. Truettner JS, Alonso OF, Bramlett HM, Dietrich WD. Therapeutic hypothermia alters microRNA responses to traumatic brain injury in rats. J Cereb Blood Flow Metab. (2011) 31:1897–907. doi: 10.1038/jcbfm.2011.33
106. Truettner JS, Motti D, Dietrich WD. MicroRNA overexpression increases cortical neuronal vulnerability to injury. Brain Res. (2013) 1533:122–30. doi: 10.1016/j.brainres.2013.08.011
107. Liu L, Sun T, Liu Z, Chen X, Zhao L, Qu G, et al. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS ONE (2014) 8:e103948. doi: 10.1371/journal.pone.0103948
108. Wang WX, Visavadiya NP, Pandya JD, Nelson PT, Sullivan GP, Springer JE. Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury. Exp Neurol. (2015) 265:84–93. doi: 10.1016/j.expneurol.2014.12.018
109. Sabirzhanov B, Stoica BA, Zhao Z, Loane DJ, Wu J, Dorsey SG, et al. miR-711 upregulation induces neuronal cell death after traumatic brain injury. Cell Death Differ. (2016) 23:654–68. doi: 10.1038/cdd.2015.132
110. Sun TY, Chen XR, Liu ZL, Zhao LL, Jiang YX, Qu GQ, et al. Expression profiling of microRNAs in hippocampus of rats following traumatic brain injury. J Huazhong Univ Sci Technolog Med Sci. (2014) 34:548–53. doi: 10.1007/s11596-014-1313-1
111. Yokota M, Tatsumi N, Nathalang O, Yamada T, Tsuda I. Effects of heparin on polymerase chain reaction for blood white cells. J Clin Lab Anal. (1999) 13:133–40. doi: 10.1002/(SICI)1098-2825(1999)13:3<133::AID-JCLA8>3.0.CO;2-0
112. Roberts TC, Coenen-Stass AM, Betts CA, Wood MJ. Detection and quantification of extracellular microRNAs in murine biofluids. Biol Proced Online (2014) 16:5. doi: 10.1186/1480-9222-16-5
113. Accerbi M, Schmidt SA, De Paoli E, Park S, Jeong D-H, Green PJ. Methods for isolation of total RNA to recover miRNAs and other small RNAs from diverse species. Methods Mol Biol. (2010) 592:31–50. doi: 10.1007/978-1-60327-005-2_3
114. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. (2012) 13:358–69. doi: 10.1038/nrg3198
115. Blondal T, Nielsen SJ, Baker A, Andreasen D, Mouritzen P, Teilum MW, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods (2013) 59:S1–6. doi: 10.1016/j.ymeth.2012.09.015
116. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods (2010) 50:298–301. doi: 10.1016/j.ymeth.2010.01.032
117. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
118. Li P, Kaslan M, Lee SH, Yao Y, Gao Z. Progress in exosome isolation techniques. Theranostics (2017) 7:789–804. doi: 10.7150/thno.18133
119. Jones LJ, Yue ST, Cheung CY, Singer VL. RNA quantitation by fluorescence-based solution assay: RiboGreen reagent characterization. Anal Biochem. (1998) 265:368–74. doi: 10.1006/abio.1998.2914
120. Becker C, Hammerle-Fickinger A, Riedmaier I, Pfaffl M. mRNA and microRNA quality control for RT-qPCR analysis. Methods (2010) 50:237–43. doi: 10.1016/j.ymeth.2010.01.010
121. Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. (2014) 18:371–90. doi: 10.1111/jcmm.12236
122. Chen X, Liang H, Guan D, Wang C, Hu X, Cui L, et al. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS ONE (2013) 8:e79652. doi: 10.1371/journal.pone.0079652
123. Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis (2012) 33:828–34. doi: 10.1093/carcin/bgs030
124. Zheng G, Wang H, Zhang X, Yang Y, Wang L, Du L, et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS ONE (2013) 8:e83025. doi: 10.1371/journal.pone.0083025
125. Gharbi S, Shamsara M, Khateri S, Soroush MR, Ghorbanmehr N, Tavallaei M, et al. Identification of reliable reference genes for quantification of microRNAs in serum samples of sulfur mustard-exposed veterans. Cell J. (2015) 17:494–501.
126. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
127. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
128. McDougall J, Dunn A, Jones K. In situ hybridization of adenovirus RNA and DNA. Nature (1972) 236:346–8. doi: 10.1038/236346a0
129. Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods (2006) 3:27–9. doi: 10.1038/nmeth843
130. Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. (2009) 394:1117–24. doi: 10.1007/s00216-008-2570-2
131. Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA (2006) 12:913–20. doi: 10.1261/rna.2332406
132. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. (2008) 26:317–25. doi: 10.1038/nbt1385
133. Metzker ML. Sequencing technologies–the next generation. Nat Rev Genet. (2010) 11:31–46. doi: 10.1038/nrg2626
134. Motameny S, Wolters S, Nürnberg P, Schumacher B. Next generation sequencing of miRNAs–strategies, resources and methods. Genes (2010) 1:70–84. doi: 10.3390/genes1010070
135. Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. (2008) 26:407–15. doi: 10.1038/nbt1394
136. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. (2008) 37:1–13. doi: 10.1093/nar/gkn923
137. Liu G, Ding M, Chen J, Huang J, Wang H, Jing Q, et al. Computational analysis of microRNA function in heart development. Acta Biochim Biophys Sin. (2010) 42:662–70. doi: 10.1093/abbs/gmq072
138. McCulloch KL, Goldman LS, Lowe L, Radomski MV, Reynolds J, Shapiro CR, et al. Development of clinical recommendations for progressive return to activity after military mild traumatic brain injury: Guidance for rehabilitation providers. J Head Trauma Rehabil. (2015) 30:56–67. doi: 10.1097/HTR.0000000000000104
139. Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ, et al. Advances in paper-based point-of-care diagnostics. Biosens Bioelectron. (2014) 54:585–97. doi: 10.1016/j.bios.2013.10.075
140. Maurer JJ. Rapid detection and limitations of molecular techniques. Annu Rev Food Sci Technol. (2011) 2:259–79. doi: 10.1146/annurev.food.080708.100730
141. Haifeng D, Jianping L, Lin D, Yongqiang W, Huangxian J, Xueji Z. MicroRNA: function, detection, and bioanalysis. Chem Rev. (2013) 113:6207–33. doi: 10.1021/cr300362f
142. Fredj Z, Azzouzi S, Turner A, Ali M, Cheung W. Neutravidin biosensor for direct capture of dual-functional biotin-moleculat beacon AuNP probe for sensitive volatametric detection of microRNA. Sensors Actuators B Chem. (2017) 248:77–84. doi: 10.1016/j.snb.2017.03.160
143. Wanunu M, Dadosh T, Vishva R, Jingmin J, McReynolds L, Drndić M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. (2010) 5:807–14. doi: 10.1038/nnano.2010.202
144. Alhasan AH, Kim DY, Daniel WL, Watson E, Meeks JJ, Thaxton CS, et al. Scanometric MicroRNA array profiling of prostate cancer markers using spherical nucleic acid–gold nanoparticle conjugates. Anal Chem. (2012) 84:4153–4160. doi: 10.1021/ac3004055
145. Xiaojuan D, Yurong Y, Shengqiang L, Ye Z, Wei C, Quan C, et al. Surface plasmon resonance biosensor for highly sensitive detection of microRNA based on DNA super-sandwich assemblies and streptavidin signal amplification. Anal Chim Acta (2015) 874:59–65. doi: 10.1016/j.aca.2015.03.021
146. Hu T, Zhang L, Wen W, Zhang X, Wang S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosens Bioelectron. (2016) 77:451–6. doi: 10.1016/j.bios.2015.09.068
147. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. (2008) 18:997. doi: 10.1038/cr.2008.282
Keywords: traumatic brain injury, biomarkers, microRNA, diagnosis, prognosis, therapy, high-throughput technology, point-of-care
Citation: Di Pietro V, Yakoub KM, Scarpa U, Di Pietro C and Belli A (2018) MicroRNA Signature of Traumatic Brain Injury: From the Biomarker Discovery to the Point-of-Care. Front. Neurol. 9:429. doi: 10.3389/fneur.2018.00429
Received: 07 March 2018; Accepted: 22 May 2018;
Published: 14 June 2018.
Edited by:
Stefania Mondello, Università degli Studi di Messina, ItalyReviewed by:
Rodolfo Gabriel Gatto, University of Illinois at Chicago, United StatesCopyright © 2018 Di Pietro, Yakoub, Scarpa, Di Pietro and Belli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valentina Di Pietro, di5kaXBpZXRyb0BiaGFtLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.