- Laboratory of Cellular and Molecular Neurobiology—Stem Cells, Department of Neurobiology, Hellenic Pasteur Institute, Athens, Greece
Identification of the unique features of human brain development and function can be critical towards the elucidation of intricate processes such as higher cognitive functions and human-specific pathologies like neuropsychiatric and behavioral disorders. The developing primate and human central nervous system (CNS) are distinguished by expanded progenitor zones and a protracted time course of neurogenesis, leading to the expansion in brain size, prominent gyral anatomy, distinctive synaptic properties, and complex neural circuits. Comparative genomic studies have revealed that adaptations of brain capacities may be partly explained by human-specific genetic changes that impact the function of proteins associated with neocortical expansion, synaptic function, and language development. However, the formation of complex gene networks may be most relevant for brain evolution. Indeed, recent studies identified distinct human-specific gene expression patterns across developmental time occurring in brain regions linked to cognition. Interestingly, such modules show species-specific divergence and are enriched in genes associated with neuronal development and synapse formation whilst also being implicated in neuropsychiatric diseases. microRNAs represent a powerful component of gene-regulatory networks by promoting spatiotemporal post-transcriptional control of gene expression in the human and primate brain. It has also been suggested that the divergence in miRNA expression plays an important role in shaping gene expression divergence among species. Primate-specific and human-specific miRNAs are principally involved in progenitor proliferation and neurogenic processes but also associate with human cognition, and neurological disorders. Human embryonic or induced pluripotent stem cells and brain organoids, permitting experimental access to neural cells and differentiation stages that are otherwise difficult or impossible to reach in humans, are an essential means for studying species-specific brain miRNAs. Single-cell sequencing approaches can further decode refined miRNA-mRNA interactions during developmental transitions. Elucidating species-specific miRNA regulation will shed new light into the mechanisms that control spatiotemporal events during human brain development and disease, an important step towards fostering novel, holistic and effective therapeutic approaches for neural disorders. In this review, we discuss species-specific regulation of miRNA function, its contribution to the evolving features of the human brain and in neurological disease, with respect also to future therapeutic approaches.
Introduction
Human brain development presents unique features and underlies the intricately coordinated spatiotemporal expression of thousands of genes. This elaborate mechanism entails the timely acquisition of diverse cellular identities further orchestrating regional specialization and inter-connectivity in the brain. microRNAs (miRNAs) are powerful post-transcriptional regulators, increasingly recognized as important components of fundamental neurodevelopmental processes and related disorders (Adlakha and Saini, 2014). miRNAs act to repress the translation or degrade their mRNA targets to modulate and fine-tune gene expression levels. One of the early roles ascribed to miRNAs was their contribution to developmental transitions by suppressing transcripts associated with the previous stage. An alternative, but complementary hypothesis suggests that miRNAs act reduce the variance in the expression level of their target genes, conferring increased robustness to signaling decisions during development (Hornstein and Shomron, 2006; Ebert and Sharp, 2012). Importantly, it is appreciated that miRNA-driven regulation contributed critically to gene expression changes on the human evolutionary lineage affecting genes involved in progenitor proliferation and neuronal generation and function (Nowakowski et al., 2013; Arcila et al., 2014). miRNA-mediated regulation in the developing brain presents primate distinct aspects, including over 100 primate-specific and 14 human-specific miRNAs that have been identified (Berezikov, 2011; Hu et al., 2011).
In this review we present current data on species-specific miRNA regulation and discuss miRNAs as hubs of critical brain transcriptional processes during human neural development. Finally, we stress the requirement to delineate the functional significance of miRNA-driven transcriptome changes at the single-cell level, as an important step towards resolving the complex regulatory network operating during human neurogenesis. Towards this direction, brain organoids and application of single-cell sequencing methodologies constitute invaluable tools to address causality between the emergence of novel miRNAs and rewiring of transcriptional programs during the evolution of brain complexity.
Distinct Features of Human Brain Development
Although brain development follows the same principles across mammals, the primate and human central nervous system (CNS) is distinguished by highly derived features. These include expanded progenitor zones (Smart et al., 2002) accompanied by enhanced and tightly controlled proliferative and/or neurogenic potential of progenitor cells (Otani et al., 2016; Sousa et al., 2017a,b), further associated with molecular changes and an increased diversity of neural cell types (Bystron et al., 2006; Lui et al., 2011; Gulden and Šestan, 2014; Taverna et al., 2014; Bae et al., 2015; Dehay et al., 2015). Human brain development is characterized by a relatively protracted time course of neurogenesis, followed by an extraordinary numeric expansion of the neuronal cell population (Rockel et al., 1980; Hutsler et al., 2005; Marín-Padilla, 2014; Otani et al., 2016) and the emergence of sophisticated neural circuits of connectivity reflected in prominent changes in gyral anatomy (Rogers et al., 2010; Hofman, 2012; Figure 1).
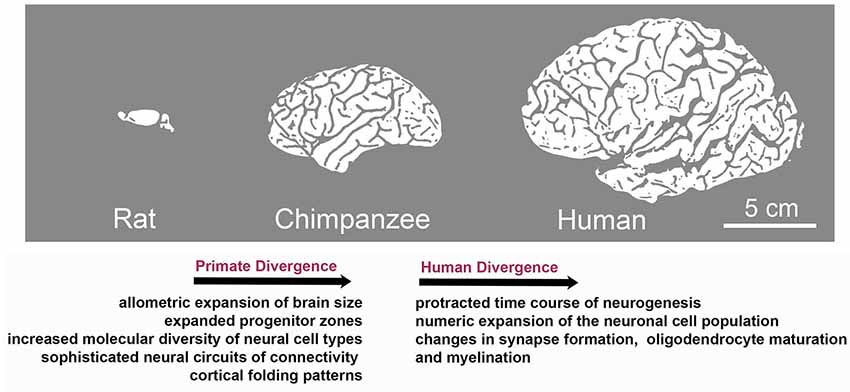
Figure 1. Species-specific evolution of traits associated with brain development during the divergence to primate and human lineages. Although brain development follows the same basic principles across mammals, evolution has resulted in the appearance of species-specific features. Characteristics of primate divergence include allometric increase in brain size, expansion of progenitor zones along with increased diversity of neural cell types and sophistication of neural circuits reflected in enhanced gyral anatomy. Human brain development is further distinguished by a relatively protracted period of neurogenesis, followed by an extraordinary numeric expansion of the neuronal cell population and by the heterochronic or heterotopic expression of genes associated with synapse formation and myelination in brain regions including the prefrontal cortex, which is central to human cognition and behavior.
Brain evolution produced changes in morphology, abundance, and function of cell types. For example, differences between rodent and human neuronal features include distinctive membrane (Wang et al., 2015; Eyal et al., 2016) and synaptic properties Molnár et al., 2008, 2016; Testa-Silva et al., 2010; Verhoog et al., 2013; Szegedi et al., 2016). Deviations between humans and non-human primates (NHPs) have also been reported regarding the morphology and number of glial cells in the brain (Oberheim et al., 2012; Bianchi et al., 2013; Herculano-Houzel et al., 2016). In addition, excitatory projection neurons in humans show more elaborate dendritic arborization, and contain a greater number and density of spines compared to non-human primates (NHPs; Duan et al., 2003; Elston et al., 2011). Along this line, a subgroup of modified pyramidal neurons, known as spindle or von Economo neurons, mainly found in the fronto-insular and anterior cingulate cortex, are larger and more numerous in humans than in other apes (Allman et al., 2010). Although their function remains elusive, they have been implicated in brain disorders with social-emotional deficits, such as autism spectrum disorders (ASDs), schizophrenia (SCZ), frontotemporal dementia and Alzheimer’s disease (AD; Yang et al., 2019). Evidence also exists for species-specific differences in serotonergic transmission, dopamine innervation and the regional localization and abundance of certain subclasses of inhibitory neurons and their axonal projections in the neocortex of humans and NHPs (Sherwood et al., 2004; Raghanti et al., 2008, 2016). Similarly, a specialized GABAergic neuron subtype has recently been identified with a molecular and anatomical signature specific to humans (Boldog et al., 2018).
Gene Networks Associated With Primate- and Human-Specific Brain Adaptations
Comparative genomic studies revealed that adapted brain specializations and capacities may be partly explained by human-specific gene conversions that impact the function of proteins associated with neocortical expansion, synaptic function, and language development. Characteristic examples include genes that have undergone human-specific duplication like the cortical development gene Slit-Robo Rho GTPase activating protein 2 (SRGAP2) which induces branching of neurons and neurite outgrowth (Dennis et al., 2012). Another case is ARHGAP11 which is expressed in basal progenitors and promotes neocortical expansion by increasing neuron numbers and brain folding (Florio et al., 2015). FOXP2 is also a gene that has undergone surprisingly rapid evolution in the primate lineage leading to humans and encodes a homeodomain protein essential for normal human speech (Lai et al., 2001). Similarly, ASPM (Gai et al., 2016), Microcephalin (Evans et al., 2004) and AHI1 genes (Ferland et al., 2004) that have undergone “positive selection,” encode for human proteins that associate with normal cerebral cortical size and axon guidance.
Importantly, gene co-expression analyses have revealed that transcriptional regulation and complexity in the neocortex have dramatically increased on the human lineage (Konopka et al., 2012; Silbereis et al., 2016). The allometric expansion of the brain is primarily accompanied by changes in patterns of gene networks and neuronal activity leading to the structural reorganization of the connectome and possibly harboring new behavioral and cognitive phenotypes (Buckner and Krienen, 2013). In this regard, recent transcriptome studies have provided crucial information on divergent patterns of molecular expression that are most relevant in evolutionary terms and can be used to uncover human specializations of brain structure and function.
Specifically, fetal gene co-expression modules have been identified that showed substantial regional differences in the developing human neocortex (Johnson et al., 2009; Miller et al., 2014; Pletikos et al., 2014). Enriched genes were shown to be critical for neuronal processes, such as differentiation, maturation, axonal projection, and synapse formation. These molecular networks also displayed divergence between humans and rhesus macaques (Figure 1; Pletikos et al., 2014), highlighting evolving biological processes involved in the patterning and differentiation of neural circuits in discrete areas (Miller et al., 2014; Hoerder-Suabedissen and Molnár, 2015).
Further work revealed human-distinct temporal progression of neurodevelopmental processes predominantly during the early and mid-fetal period. The identified gene modules were linked to synapse formation, neuronal differentiation, oligodendrocyte maturation, and myelination, and exhibited heterochronic or heterotopic expression in brain regions including the prefrontal cortex, which is central to human cognition and behavior (Figure 1). Interestingly, divergent spatiotemporal expression patterns included also genes associated with ASDs and schizophrenia (Konopka et al., 2012; Zhu et al., 2018). These studies highlight gene expression regulation in space and time as an evolution-modified feature that impacts on neurodevelopmental processes as well as in brain complexity and disease. Diverse expression patterns responsible for interspecies differences can be attributed to changes in DNA methylation, histone modifications (Maze et al., 2014), alternative splicing events, promoter-driven transcription regulation (Davuluri et al., 2008; Nilsen and Graveley, 2010) and non-coding RNAs.
microRNAs Are Powerful Post-Transcriptional Regulators of Gene Expression in the Brain
microRNAs (miRNAs) are a class of short non-coding RNAs of ~22 nucleotides in length that constitute an important component of the regulatory circuitry determining expression patterns. Mature miRNAs mediate post-transcriptional regulation of gene expression through direct degradation of their target mRNA and/or suppression of translation. miRNAs bind to their mRNA targets by partial complementarity between the mRNA’s 3′UTR and a 6–8 nucleotides long “seed” sequence at the 5′ end of the microRNA. Therefore, a single microRNA can target multiple mRNAs simultaneously, while a single mRNA may be regulated by different microRNAs (Klein et al., 2005; Kosik, 2006; Saliminejad et al., 2019).
miRNA biogenesis begins with the transcription of double-stranded primary miRNA (pri-miRNA) short hairpin structures by RNA polymerase II. The pri-miRNA is then cleaved by the RNase-III enzyme, Drosha, producing ~70-bp pre-miRNAs that are exported from the nucleus into the cytoplasm by Exportin 5 (Exp5), a Ran-GTP dependent Nucleo/cytoplasmic cargo transporter. The Dicer enzyme cleaves pre-miRNA sequences into 21–23 nt mature miRNA double-stranded duplexes which are loaded into a pre-RISC (pre-miRNA-induced silencing complex) containing Argonaute (Ago) and other proteins. In the mature miRISC complex the “passenger” strand (complementary strand) is removed leaving just the “guide” strand (mature miRNA strand) which will bind to the mRNA target and instigate inhibition of its expression, reviewed in Winter et al. (2009) and Davis et al. (2015).
The concentration of miRNAs within cells is regulated at different levels. It has been shown that Ago proteins are not only critical for miRNA biogenesis and function, but they also regulate the abundance of mature miRNAs by increasing their stability (Grishok et al., 2001; Diederichs and Haber, 2007; Winter and Diederichs, 2011). Association with mRNA targets also enhances miRNA stability, a phenomenon known as target-mediated miRNA protection, while the introduction of additional target sites can also promote miRNA accumulation (Chatterjee et al., 2011). On the other hand, miRNAs are subject to degradation. It has been shown that miRNA-mRNA interactions not only stabilize but can also destabilize the miRNA and promote its degradation through a process known as target RNA directed miRNA degradation (Ameres et al., 2010; Fuchs Wightman et al., 2018). The degree of sequence complementarity infers the outcome of the miRNA-mRNA interaction, with higher complementarity favoring miRNA degradation and lower complementarity favoring miRNA stabilization. As demonstrated, target mRNAs promote posttranscriptional modifications to the 3′ end of the miRNA involving either the addition of non-templated nucleotides, a process known as “3′-end tailing” or elimination of nucleotides via 3′-to-5′ trimming, both of which control the rate of miRNA decay (Baccarini et al., 2011; Marcinowski et al., 2012).
The ability of miRNAs to fine-tune gene expression levels (Schratt, 2009) constitutes a critical property for controlling spatiotemporal events during brain development. Of the 2,500 mature miRNAs that have been identified in humans (Friedländer et al., 2014), an estimated 70% is expressed in the nervous system (Adlakha and Saini, 2014). miRNAs have emerged as important post-transcriptional regulators of gene expression involved in neurogenesis and neural function in mammalian species (Davis et al., 2015; Nowakowski et al., 2018). A relatively small number of brain miRNAs are well characterized, including miR-92 which targets EOMES (TBR2), a T-box transcription factor that is preferentially expressed in cortical intermediate progenitors and regulates cortical neuron production and expansion, thereby affecting the thickness of the cerebral cortex (Nowakowski et al., 2013). miR-124 and miR-9 have also been shown to affect neural lineage differentiation by downregulating multiple mRNAs (Krichevsky et al., 2006). MiR-9 is a highly brain enriched miRNA that is involved in a negative feedback loop with TLX, a nuclear receptor (Zhao et al., 2009) which controls stem cell proliferation in developing and adult brain (Shi et al., 2004; Liu et al., 2008; Zhang et al., 2008). MiR-9 together with miR-124, one of the most abundant miRNAs in the brain, target REST which opposes neuronal differentiation (Conaco et al., 2006; Visvanathan et al., 2007), while REST itself acts as an inhibitor of miR-124 expression. Moreover, both miRNAs act synergistically to repress BAF53a, a subunit of the neural-progenitor-specific BAF (npBAF) chromatin-remodeling complex, operating during the post-mitotic phase of neuronal development (Yoo et al., 2009). Finally, miR-124 downregulates the RNA-binding protein Ptbp1, a repressor of neuron-specific splicing (Makeyev et al., 2007).
Additionally, miRNAs have been shown to be essential for neural subtype specification. For example, miR-7a promotes oligodendrocyte generation by targeting Pax6 and NeuroD4 (Zhao et al., 2012), while miR-218 is required to establish motor neuron fate (Thiebes et al., 2015). miRNAs have also important roles in synapse formation and plasticity. These include miR-125 which targets the post-synaptic protein PSD-95 in cortical neurons (Muddashetty et al., 2011), the neuron-specific miR-129 which represses Kv1.1, a voltage-gated potassium channel that regulates excitability (Sosanya et al., 2013), and miR-219 which downregulates CamKII, a the major mediator of Long Term Potentiation (LTP) and N-methyl-D-aspartate receptor (NMDA) signaling (Kocerha et al., 2009). Other miRNAs modulate synaptic function upon activation. miR-485 expression is increased following neuronal stimulation to regulate the pre-synaptic protein SV2A and inhibit neurotransmitter release (Cohen et al., 2011). miR-132, on the other hand, accumulates in response to activity in forebrain neurons to regulate dendritic growth, activity-induced spine growth and spine morphology (Magill et al., 2010; Nudelman et al., 2010).
The Evolving Role of Mirnas in the Primate and Human Brain
The Evolution of miRNAs
Evolution of miRNAs is an ongoing process and experimental evidence suggests that along with highly conserved miRNAs, a number of new brain miRNAs have emerged. Many of these are not conserved beyond primates, indicating their recent origin (Berezikov et al., 2006). Following evolutionary adaptations, more than 100 primate-specific miRNAs (that is, miRNAs present only in humans and non-human primates) and 14 human-specific miRNAs (Table 1) have been identified in the developing brain (Berezikov, 2011). Novel miRNAs arise either by the appearance of transcribed hairpin structures or by mutations in the miRNA seed region (Lu et al., 2008). The genomic sources for the acquisition of novel miRNAs are reviewed in detail in “Evolution of microRNA diversity and regulation in animals” (Berezikov, 2011). Briefly, novel miRNAs can emerge by duplication of existing miRNA genes. Alternatively, introns are a frequent source of unstructured transcripts that can gradually evolve into novel intronic miRNAs. De novo emergence of miRNAs can also occur, where an evolved transcriptional unit provides a source of the initially unstructured transcript that transitions through the miRNA-like hairpin stage and evolves into a novel miRNA gene. In addition, transposable elements or structured transcripts, such as tRNA and small nucleolar RNA (snoRNA), can provide novel transcriptional units for the evolution of miRNA-like hairpins into novel miRNA genes. Finally, antisense transcription of existing miRNA loci can lead to the formation of miRNA hairpins with novel mature miRNA sequences.

Table 1. List of human-specific miRNAs identified so far according to the study of Hu et al. (2012).
miRNAs as Key Regulators in Shaping Patterns of Gene Expression in the Developing and Adult Primate and Human Brain
A series of observations support the notion that miRNAs are instrumental contributors to the alterations that may account for the accelerated evolution of the human brain. First, elevated production of mature miRNAs has been observed in the human brain compared to other species, which is attributed to the higher processing efficiency of miRNA precursors in humans (Chakraborty et al., 2018).
It has been proposed that the evolution of miRNA-mediated regulatory networks has contributed to organismal complexity (Berezikov, 2011). This can mechanistically be explained by the ability of miRNAs to advance network functionalities by delivering the extra precision required to constrain the intrinsically noisy gene expression process (Raj and van Oudenaarden, 2008; Herranz and Cohen, 2010). In support, the work of Arcila et al. (2014) on primate-specific miRNAs suggests that integration of novel miRNAs into ancient gene circuitry exerted additional regulation over-proliferation of neural progenitors in cortical germinal areas (Figure 2), a region that demonstrates significant expansion across brain evolution (Arcila et al., 2014).
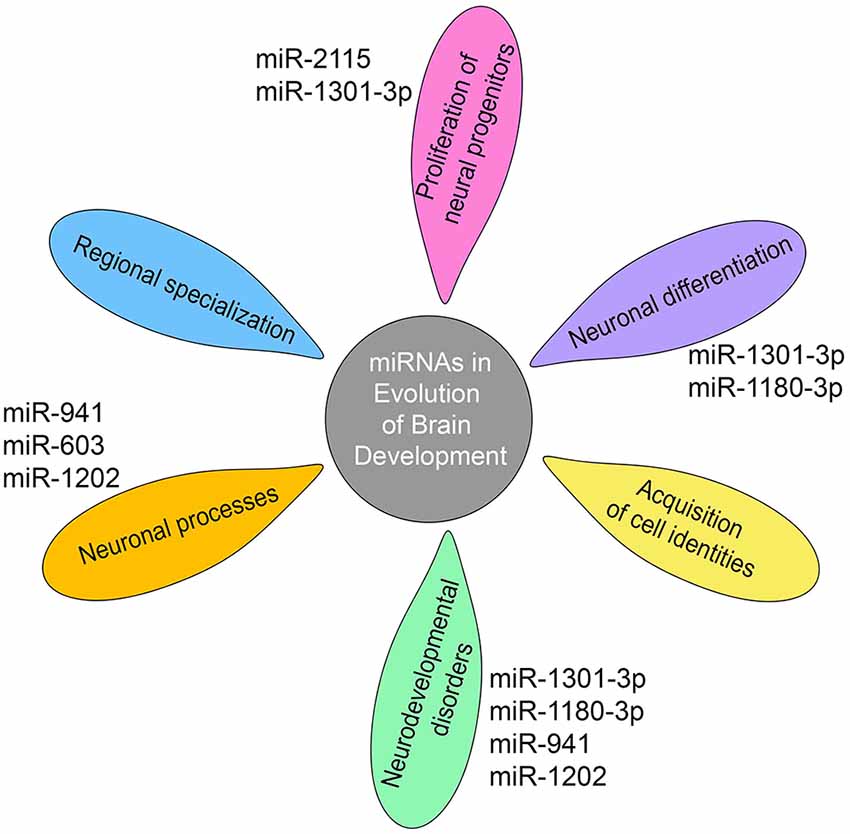
Figure 2. miRNAs shape gene networks during the evolution of human and non-human primate brain development. Transcriptomic studies show that miRNA-mediated regulation during primate brain evolution contributed critically in shaping gene networks associated with progenitor proliferation, neuronal differentiation, and acquisition of cell identities, an extension of neuronal processes, regional specialization and neurodevelopmental disorders as depicted on the petals. A limited number of primate- or human-specific miRNAs are indicated next to the petals, for which separate studies exist so far to demonstrate their individual involvement in the respective biological processes.
Furthermore, evolving miRNA-mediated regulation shows critical implications in shaping gene networks and even determining anatomical regions during brain development. Work on primates has revealed that miRNA profiles can resolve discrete areas within the developing cortex while dominant differences were observed between the germinal zone and the differentiated cells in the cortical plate (Arcila et al., 2014). Consistently, studies on the transition from infant to adolescent human brain show differential expression of miRNAs within and between brain regions, with the prefrontal cortex, the region mostly connected with human cognition, exhibiting the greatest number of differentially expressed miRNAs (Ziats and Rennert, 2014). Moreover, miRNA expression displays increased deviations between brain regions over time, indicating the implication of miRNAs in the regional specialization as the brain matures (Ziats and Rennert, 2014). Importantly, common neurodevelopmental disorders associated with genes targeted by these miRNAs (Figure 2; Ziats and Rennert, 2014).
Recent elegant work highlights the impact of miRNAs in regulating regionally divergent transcriptional states in the developing human cortex. To characterize the landscape of miRNA–mRNA interactions during human brain development, Nowakovski et al developed a new single-cell approach for combined mRNA and miRNA profiling in the same cell across human fetal tissue samples corresponding to peak neurogenesis [Gestational week (GW) 15 and 16.5] and early gliogenesis (GW 19–20.5). Their study revealed that major regulatory molecules like transcription factors, chromatin modifiers, and signaling components are enriched among miRNA targets. Reconstruction of gene-regulatory networks uncovered that miRNA-mRNA interactions often correspond to the acquisition of cell-type identities and undergo dynamic transitions even among closely related cell types during neuronal differentiation and maturation (Figure 2). Further strengthening previous studies, the authors demonstrate that different pathways driven by brain-specific miRNAs are related to developmental stage and cortical area specificity (Nowakowski et al., 2018).
Transcriptomic studies founded the notion that miRNA divergence correlates with the divergence of gene expression patterns in the prefrontal cortex and cerebellum among humans, chimpanzees, and macaques. Accordingly, it has been reported that a significant inverse relationship exists between human and chimpanzee miRNA expression divergence and expression divergence of their predicted target genes at both mRNA and protein levels (Hu et al., 2011). Inline, Svante Pääbo and colleagues placed miRNAs among the key regulators that remodeled cortical development (Somel et al., 2011). Using a computational method of analysis, they show that trans-acting regulators and particularly miRNAs drive the pronounced gene expression changes observed in the human prefrontal cortex. As deduced, the developmental profiles of miRNAs, as well as their target genes, show the fastest rates of human-specific evolutionary change.
Human- and Primate-Specific Mirnas in Neural Development, Physiology and Pathology
Expression, even if at low levels, has been detected in the prefrontal cortex and cerebellum for the majority of the 14 human-specific miRNAs identified so far (Hu et al., 2012). Enrichment analysis of the predicted target genes of human-specific miRNAs attests that these relate to neuronal components and processes, including cytoskeletal elements, metal ion binding, and postsynaptic, dendritic, and somatic functions (Barbash et al., 2014). Separate studies on individual primate- and human-specific miRNAs are limited and confined in human tissue examination complemented by functional analysis using cell lines or mouse models in one case (Table 2). Nevertheless, data so far show that primate-specific miRNAs are principally involved in the regulation of cell cycle dynamics operating during progenitor proliferation and neuronal differentiation while they further associate with neurodevelopmental disorders (Figure 2).
A great ape specific miRNA, miR-2115, is enriched in radial glia and becomes prominently upregulated at GW19–20 in the human germinal zones. It controls cell-cycle dynamics during human cortical development by fine-tuning the expression of ORC4, a known regulator of DNA replication (Nowakowski et al., 2018). miR-1301-3p and miR-1180-3p are primate-specific miRNAs that have been identified in the germinal zones of the visual cortex of the macaque developing brain. miR-1301-3p regulates the mRNA for histone-lysine N-methyltransferases mll1 and mll2 (MixedLineage, Leukemia) that function by resolving silenced bivalent loci in neural precursors for induction of neurogenesis. Another target of miR-1301-3p is the transcription factor TCF4 which patterns progenitor cells in the developing CNS and has been linked to schizophrenia and intellectual disability (Arcila et al., 2014). miR-1180-3p targets kansl1 and dlx1. Kansl1 is a chromatin regulator that when haploinsufficient, causes intellectual disability, hypotonia, and distinctive facial features associated with the 17q21.31microdeletion syndrome, while dlx1 is a homeodomain transcription factor that controls GABAergic neurogenesis and has been associated with autism (Arcila et al., 2014).
In addition, a number of species-specific miRNAs associate with human cognition and neurodegenerative diseases (Table 2). miR-941 is the only human-specific miRNA expressed highly in the prefrontal cortex and cerebellum and has been proposed to be associated with longevity and neurotransmitter signaling. Individuals containing a microdeletion in the chromosomal region containing pre-miR-941 display developmental delay and disruption of cognitive functions including language and speech (Figure 2; Hu et al., 2012). miR-1202, a miRNA specific to primates, and enriched in the human brain is associated with the pathophysiology of depression. It is differentially expressed in depressed individuals and has been shown to target the Metabotropic Glutamate Receptor 4 (GRM4; Lindsley and Hopkins, 2012), a synaptic molecule modulating neurotransmission that also constitutes an attractive therapeutic target for Parkinson’s disease and schizophrenia (Figure 2; Lopez et al., 2014).
Another primate-specific miRNA, miR-603, is a novel intronic miRNA of the gene KIAA1217, which is highly expressed in human brain. miR-603 directly downregulates the key neuronal apoptotic component E2F1 and can prevent cells from undergoing apoptosis. In addition, miR-603 targets LRPAP1 involved in Aβ amyloid peptide clearance and the pathogenesis of AD. Finally, the rs11014002 SNP in precursor pre-miR-603 increases the expression of mature miR-603, which may account for its association with reduced risk for AD (Zhang et al., 2016; Figure 2).
Human-Specific Regulation of Mirna Function in the Brain
Evolutionary changes and consequently species distinct features also exist for the binding partners of miRNAs. The evolution of miRNA regulation is intimately intertwined with the evolution of their targets, as newly emerging miRNAs integrate into preexisting gene expression circuitries (Arcila et al., 2014). This is particularly pronounced in the brain where neuronal transcripts not only have longer 3′-untranslated regions (UTRs) which are the main target region for miRNAs (Meunier et al., 2013), but also display an increased density of potential binding sites, enhancing their selective advantage for acquiring miRNA-mediated regulation (Cacchiarelli et al., 2008; Barbash et al., 2014). These findings in conjunction with the appearance of increased new variants around target genes of human-specific miRNAs are consistent with the theory that the speciation of hominids was accompanied by an enhancement in the capacity of newly evolved miRNAs to modulate gene expression (Barbash et al., 2014).
Interestingly Hu et al identified five miRNAs, namely miR-184, miR-487a, miR-383, miR-34c-5p, and miR-299-3p, with high sequence conservation among species, which nevertheless show significantly different levels of expression in humans, while two of these (miR-299-3p and miR-184) show preferential expression in cortical neurons (Hu et al., 2012). Functional analysis combining the targets of all five miRNAs revealed enrichment in genes associated with neural processes and specifically with cell proliferation and differentiation, synaptic transmission and neuronal function. Moreover, miR-299-3p targets associate with axon guidance, while miR-184 targets are related to long-term potentiation, which is directly linked to learning and memory formation (Hu et al., 2011). Human-specific miRNA-mediated regulation of certain genes with a critical role in brain function has also been reported. For instance, miR-483–5p, binds in a sequence-exclusive manner to the human epigenetic regulator methyl CpG binding protein 2 (MeCP2) that controls proper neurological function, to modulate its levels in the fetal cortex (Han et al., 2013).
Human Experimental Models for Investigation of Mirna-Specific Features of Brain Development and Associated Disorders
Access to human tissue in combination with high-throughput sequencing techniques provided important insight into the significance of the miRNA-driven transcriptome changes across brain development and evolution. However, it is necessary to validate predicted targets and clarify cell type association, stage-specificity and mode of action for individual miRNAs. As experimental animals cannot fully simulate human brain development and disease, human embryonic stem cells (hESCs) or induced pluripotent stem cells (iPSCs) represent valuable means for advancing human studies (Chambers et al., 2009; Shi et al., 2012). Directed cortical differentiation of human, chimpanzee, and macaque iPSCs (Otani et al., 2016) clearly depicted the divergence in the timing of key developmental events and highlighted the extended proliferation of chimpanzee and human progenitors as compared to macaque, emphasizing the validity of such systems in reproducing species-specific features.
Further studies on hESCs accompanied by miRNA profiling during differentiation of various neuronal subtypes underlined the effect of known miRNAs, such as let-7, miR-124, miR-7, miR-125 and miR-9 in progenitor proliferation, cell fate specification and neuronal commitment and maturation (Delaloy et al., 2010; Boissart et al., 2012; Liu et al., 2012; Cimadamore et al., 2013; Tu et al., 2018). miRNA research on hESC models has also uncovered new targets and roles for these miRNAs. For example, miR-9 which has been described in mouse to promote neuronal lineage differentiation by targeting REST, was shown to also target Stathmin, a protein involved in microtubule stability, and to coordinate proliferation and migration during the early stage of maturation of neural progenitors. Similarly, miR-125 with reported neuronal function, was shown to act in earlier stages to promote exit from pluripotency and potentiate neural specification by targeting SMAD4. Moreover, research on patient induced pluripotent stem cells and their neuronal derivatives uncovered the role of certain miRNAs in autism and schizophrenia (Halevy et al., 2015; Murai et al., 2016; Mellios et al., 2018).
Notably, given the complexity of the human brain, 3D tissue culture models that embody cellular diversity and spatial organization that mimics brain architecture, are more relevant to understand the critical input of miRNAs in shaping intricate brain gene networks. Brain organoids that grow as 3D aggregates from pluripotent stem cells comprise an exciting new tool for modeling human brain physiology and pathology, for uncovering human-specific traits and for drug discovery and testing. Despite current limitations, particularly batch heterogeneity that impedes consistency, organoids resemble human brain not only at the cellular level, but also in terms of general tissue structure, developmental trajectories and neuronal functionalities (Lancaster et al., 2013; Paşca et al., 2015; Qian et al., 2016; Giandomenico et al., 2019). Cortical organoids in particular that have been more extensively studied, recapitulate the organization of neural progenitor zones to a considerable degree, reflecting developmental events during embryonic stages in vivo (Lancaster et al., 2013; Qian et al., 2016). Such systems can be valuable for functional gain- or loss-of-function studies to elucidate the contribution of ancient or novel miRNAs in primate and/or human-specific processes. Nevertheless, studying neuronal network formation among different brain regions, especially distant ones, remains a challenge. Recent advances in the generation of fused brain organoids from co-culture of individual ones with distinct regional identities may provide a closer-to-the-in-vivo-situation model to investigate the basis of human neural circuitry formation in health and disease (Birey et al., 2017; Xiang et al., 2017, 2019). Alternatively, it is possible to exploit the inherent intrinsic heterogeneity observed within single organoids, as for example in brain organoids containing retinal regions that can respond to light stimuli (Quadrato et al., 2017). However in all cases absence of vascularization restricts oxygen and nutrient supplies limiting organoid survival, consequently affecting the time-scale of such studies. Therefore a combination of different approaches and technologies, including 2D cultures derived from pluripotent stem cells, brain organoids, single-cell transcriptomics, potentially along with miRNA target degradation kinetics analysis to elucidate the miRNA-driven evolutionary variants of the neurodevelopmental program should prove more rewarding.
In relevance, single-cell RNA-sequencing technology has greatly facilitated research efforts in resolving the diversity and developmental trajectories of human brain cell types providing transformative insights into the developmental lineages and functional states of individual cells (Tasic et al., 2016, 2018; Nowakowski et al., 2017; Paul et al., 2017; Hrvatin et al., 2018; Lake et al., 2018; Zhong et al., 2018). Importantly, a number of studies have illustrated the potential to integrate spatial information at single-cell resolution (Ke et al., 2013; Lubeck et al., 2014; Chen et al., 2015; Satija et al., 2015; Salmén et al., 2018). However, when designing such experiments, consideration of the tradeoff between a number of cells sequenced and the read depth per cell is a factor that has to be seriously considered, not the least because of budgetary constraints (Menon, 2018). Additionally, such developments still necessitate the formulation of new computational modalities to integrate the rapidly expanding data sets generated from diverse sources so that biological sense is made (Butler et al., 2018; Stuart and Satija, 2019). Improvements in algorithm design and data analysis are essential to allow unsupervised methods for tracking gene expression and correctly reconstructing trajectories that map cell lineages and developmental transition of cells.
Towards miRNA Based Therapeutics
The more we understand miRNA biology and its contribution to the intricate regulatory networks of gene expression during development, the better will we be able to uncover related disturbances underlying the origin of various neuropsychiatric and behavioral disorders. Indeed, compelling evidence regarding the involvement of miRNAs in human disease has instigated attention into their potential use as therapeutics (van Rooij et al., 2012). miRNA-based strategies include miRNA mimics and inhibitors (antagomiRs) to respectively increase and decrease their expression or, conversely, their target genes. The field currently progresses rapidly as in 2018 the FDA approved the first therapy based on miRNA administration for the treatment of rare progressive polyneuropathy caused by hereditary transthyretin-mediated amyloidosis (Adams et al., 2018; Wood, 2018). Understanding the functions of miRNAs in specific cell types and during different stages throughout life under normal and pathological conditions could improve the specificity and efficacy of miRNA therapeutic strategies. Moreover, delivery to the brain is challenging and the pleiotropic nature of miRNA functions undeniably makes off-target biological effects an important limitation (Junn and Mouradian, 2012). Novel approaches are in the pipeline to design new chemical formulations in order to increase miRNA stability and improve their specificity and permeability, while delivery methods are also being developed to target the brain and decrease unwanted side effects (van Rooij et al., 2012; Søkilde et al., 2015; Rupaimoole and Slack, 2017). In this regard, decoding species-specific regulation of miRNA function will prove an important step for fostering novel, holistic and effective therapeutic approaches for human disorders.
Conclusions and Future Perspectives
Accumulating evidence highlights the emergent role of miRNAs as critical regulators that influence the overall transcriptional landscape in the human brain. In addition, primate- and human-specific regulation by miRNAs during neurodevelopment argues in favor of their involvement in the establishment of species-specific features during brain evolution. A comprehensive understanding of the spatiotemporal miRNA mediated regulation will help resolve the intricate regulatory network operating during human neurogenesis and will grant important cues for the causality of neurodevelopmental disorders. Down this line, 2D- and 3D-human pluripotent-derived experimental setups constitute a useful system for modeling species-specific features of development and for capturing miRNA mediated rewiring of the transcriptional program. In conjunction, single-cell sequencing approaches can decode the refined miRNA-mRNA interactions during cell developmental transitions. Taken together, miRNAs constitute an indispensable component of the extended gene regulatory network and evaluating their functional significance in the evolving brain can determine key features of human neuronal development and disease.
Author Contributions
KP and RM planned and wrote the manuscript.
Funding
We acknowledge funding from Stavros Niarchos Foundation to the Hellenic Pasteur Institute as part of the Foundation’s initiative to support the Greek Research Center Ecosystem and the Hellenic Foundation for Research and Innovation 899-PARKINSynapse grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the Matsas Lab for stimulating discussions and comments.
References
Adams, D., Gonzalez-Duarte, A., O’Riordan, W. D., Yang, C. C., Ueda, M., Kristen, A. V., et al. (2018). Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21. doi: 10.1056/NEJMoa1716153
Adlakha, Y. K., and Saini, N. (2014). Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol. Cancer 13:33. doi: 10.1186/1476-4598-13-33
Allman, J. M., Tetreault, N. A., Hakeem, A. Y., Manaye, K. F., Semendeferi, K., Erwin, J. M., et al. (2010). The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct. Funct. 214, 495–517. doi: 10.1007/s00429-010-0254-0
Ameres, S. L., Horwich, M. D., Hung, J. H., Xu, J., Ghildiyal, M., Weng, Z., et al. (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539. doi: 10.1126/science.1187058
Arcila, M. L., Betizeau, M., Cambronne, X. A., Guzman, E., Doerflinger, N., Bouhallier, F., et al. (2014). Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns. Neuron 81, 1255–1262. doi: 10.1016/j.neuron.2014.01.017
Baccarini, A., Chauhan, H., Gardner, T. J., Jayaprakash, A. D., Sachidanandam, R., and Brown, B. D. (2011). Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol. 21, 369–376. doi: 10.1016/j.cub.2011.01.067
Bae, B. I., Jayaraman, D., and Walsh, C. A. (2015). Genetic changes shaping the human brain. Dev. Cell 32, 423–434. doi: 10.1016/j.devcel.2015.01.035
Barbash, S., Shifman, S., and Soreq, H. (2014). Global coevolution of human microRNAs and their target genes. Mol. Biol. Evol. 31, 1237–1247. doi: 10.1093/molbev/msu090
Berezikov, E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. doi: 10.1038/nrg3079
Berezikov, E., Thuemmler, F., van Laake, L. W., Kondova, I., Bontrop, R., Cuppen, E., et al. (2006). Diversity of microRNAs in human and chimpanzee brain. Nature genetics 38, 1375–1377. doi: 10.1038/ng1914
Bianchi, S., Stimpson, C. D., Bauernfeind, A. L., Schapiro, S. J., Baze, W. B., McArthur, M. J., et al. (2013). Dendritic morphology of pyramidal neurons in the chimpanzee neocortex: regional specializations and comparison to humans. Cereb. Cortex 23, 2429–2436. doi: 10.1093/cercor/bhs239
Birey, F., Andersen, J., Makinson, C. D., Islam, S., Wei, W., Huber, N., et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. doi: 10.1038/nature22330
Boissart, C., Nissan, X., Giraud-Triboult, K., Peschanski, M., and Benchoua, A. (2012). miR-125 potentiates early neural specification of human embryonic stem cells. Development 139, 1247–1257. doi: 10.1242/dev.073627
Boldog, E., Bakken, T. E., Hodge, R. D., Novotny, M., Aevermann, B. D., Baka, J., et al. (2018). Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 21, 1185–1195. doi: 10.1038/s41593-018-0205-2
Buckner, R. L., and Krienen, F. M. (2013). The evolution of distributed association networks in the human brain. Trends Cogn. Sci. 17, 648–665. doi: 10.1016/j.tics.2013.09.017
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. doi: 10.1038/nbt.4096
Bystron, I., Rakic, P., Molnar, Z., and Blakemore, C. (2006). The first neurons of the human cerebral cortex. Nat. Neurosci. 9, 880–886. doi: 10.1038/nn1726
Cacchiarelli, D., Santoni, D., and Bozzoni, I. (2008). MicroRNAs as prime players in a combinatorial view of evolution. RNA Biol. 5, 120–122. doi: 10.4161/rna.5.3.6569
Chakraborty, S., Islam, M. R., Ali, M. M., and Nabi, A. (2018). Evolutionary divergence of brain-specific precursor miRNAs drives efficient processing and production of mature miRNAs in human. Neuroscience 392, 141–159. doi: 10.1016/j.neuroscience.2018.09.010
Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M., and Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280. doi: 10.1038/nbt.1529
Chatterjee, S., Fasler, M., Büssing, I., and Grosshans, H. (2011). Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell 20, 388–396. doi: 10.1016/j.devcel.2011.02.008
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S., and Zhuang, X. (2015). RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348:aaa6090. doi: 10.1126/science.aaa6090
Cimadamore, F., Amador-Arjona, A., Chen, C., Huang, C. T., and Terskikh, A. V. (2013). SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. U S A 110, E3017–E3026. doi: 10.1073/pnas.1220176110
Cohen, J. E., Lee, P. R., Chen, S., Li, W., and Fields, R. D. (2011). MicroRNA regulation of homeostatic synaptic plasticity. Proc. Natl. Acad. Sci. U S A 108, 11650–11655. doi: 10.1073/pnas.1017576108
Conaco, C., Otto, S., Han, J. J., and Mandel, G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U S A 103, 2422–2427. doi: 10.1073/pnas.0511041103
Davis, G. M., Haas, M. A., and Pocock, R. (2015). MicroRNAs: not “fine-tuners” but key regulators of neuronal development and function. Front. Neurol. 6:245. doi: 10.3389/fneur.2015.00245
Davuluri, R. V., Suzuki, Y., Sugano, S., Plass, C., and Huang, T. H. (2008). The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 24, 167–177. doi: 10.1016/j.tig.2008.01.008
Dehay, C., Kennedy, H., and Kosik, K. S. (2015). The outer subventricular zone and primate-specific cortical complexification. Neuron 85, 683–694. doi: 10.1016/j.neuron.2014.12.060
Delaloy, C., Liu, L., Lee, J. A., Su, H., Shen, F., Yang, G. Y., et al. (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335. doi: 10.1016/j.stem.2010.02.015
Dennis, M. Y., Nuttle, X., Sudmant, P. H., Antonacci, F., Graves, T. A., Nefedov, M., et al. (2012). Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922. doi: 10.1016/j.cell.2012.03.033
Diederichs, S., and Haber, D. A. (2007). Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131, 1097–1108. doi: 10.1016/j.cell.2007.10.032
Duan, H., Wearne, S. L., Rocher, A. B., Macedo, A., Morrison, J. H., and Hof, P. R. (2003). Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb. Cortex 13, 950–961. doi: 10.1093/cercor/13.9.950
Ebert, M. S., and Sharp, P. A. (2012). Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524. doi: 10.1016/j.cell.2012.04.005
Elston, G. N., Benavides-Piccione, R., Elston, A., Manger, P. R., and Defelipe, J. (2011). Pyramidal cells in prefrontal cortex of primates: marked differences in neuronal structure among species. Front. Neuroanat. 5:2. doi: 10.3389/fnana.2011.00002
Evans, P. D., Anderson, J. R., Vallender, E. J., Choi, S. S., and Lahn, B. T. (2004). Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 13, 1139–1145. doi: 10.1093/hmg/ddh126
Eyal, G., Verhoog, M. B., Testa-Silva, G., Deitcher, Y., Lodder, J. C., Benavides-Piccione, R., et al. (2016). Unique membrane properties and enhanced signal processing in human neocortical neurons. Elife 5:e16553. doi: 10.7554/eLife.16553
Ferland, R. J., Eyaid, W., Collura, R. V., Tully, L. D., Hill, R. S., Al-Nouri, D., et al. (2004). Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 36, 1008–1013. doi: 10.1038/ng1419
Florio, M., Albert, M., Taverna, E., Namba, T., Brandl, H., Lewitus, E., et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470. doi: 10.1126/science.aaa1975
Friedländer, M. R., Lizano, E., Houben, A. J., Bezdan, D., Banez-Coronel, M., Kudla, G., et al. (2014). Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 15:R57. doi: 10.1186/gb-2014-15-4-r57
Fuchs Wightman, F., Giono, L. E., Fededa, J. P., and de la Mata, M. (2018). Target RNAs strike back on MicroRNAs. Front. Genet. 9:435. doi: 10.3389/fgene.2018.00435
Gai, M., Bianchi, F. T., Vagnoni, C., Vernì, F., Bonaccorsi, S., Pasquero, S., et al. (2016). ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep. 17, 1396–1409. doi: 10.15252/embr.201541823
Giandomenico, S. L., Mierau, S. B., Gibbons, G. M., Wenger, L. M. D., Masullo, L., Sit, T., et al. (2019). Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 22, 669–679. doi: 10.1038/s41593-019-0350-2
Grishok, A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., et al. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. doi: 10.1016/s0092-8674(01)00431-7
Gulden, F. O., and Šestan, N. (2014). Neurobiology: building a bigger brain. Nature 515, 206–207. doi: 10.1038/515206a
Halevy, T., Czech, C., and Benvenisty, N. (2015). Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Rep. 4, 37–46. doi: 10.1016/j.stemcr.2014.10.015
Han, K., Gennarino, V. A., Lee, Y., Pang, K., Hashimoto-Torii, K., Choufani, S., et al. (2013). Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483–5p. Genes Dev. 27, 485–490. doi: 10.1101/gad.207456.112
Herculano-Houzel, S., Kaas, J. H., and de Oliveira-Souza, R. (2016). Corticalization of motor control in humans is a consequence of brain scaling in primate evolution. J. Comp. Neurol. 524, 448–455. doi: 10.1002/cne.23792
Herranz, H., and Cohen, S. M. (2010). MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24, 1339–1344. doi: 10.1101/gad.1937010
Hoerder-Suabedissen, A., and Molnár, Z. (2015). Development, evolution and pathology of neocortical subplate neurons. Nat. Rev. Neurosci. 16, 133–146. doi: 10.1038/nrn3915
Hofman, M. A. (2012). Design principles of the human brain: an evolutionary perspective. Prog. Brain Res. 195, 373–390. doi: 10.1016/B978-0-444-53860-4.00018-0
Hornstein, E., and Shomron, N. (2006). Canalization of development by microRNAs. Nat. Genet. 38, S20–S24. doi: 10.1038/ng1803
Hrvatin, S., Hochbaum, D. R., Nagy, M. A., Cicconet, M., Robertson, K., Cheadle, L., et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129. doi: 10.1038/s41593-017-0029-5
Hu, H. Y., Guo, S., Xi, J., Yan, Z., Fu, N., Zhang, X., et al. (2011). MicroRNA expression and regulation in human, chimpanzee and macaque brains. PLoS Genet. 7:e1002327. doi: 10.1371/journal.pgen.1002327
Hu, H. Y., He, L., Fominykh, K., Yan, Z., Guo, S., Zhang, X., et al. (2012). Evolution of the human-specific microRNA miR-941. Nat. Commun. 3:1145. doi: 10.1038/ncomms2146
Hutsler, J. J., Lee, D. G., and Porter, K. K. (2005). Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res. 1052, 71–81. doi: 10.1016/j.brainres.2005.06.015
Johnson, M. B., Kawasawa, Y. I., Mason, C. E., Krsnik, Z., Coppola, G., Bogdanovic, D., et al. (2009). Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509. doi: 10.1016/j.neuron.2009.03.027
Junn, E., and Mouradian, M. M. (2012). MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol. Ther. 133, 142–150. doi: 10.1016/j.pharmthera.2011.10.002
Ke, R., Mignardi, M., Pacureanu, A., Svedlund, J., Botling, J., Wahlby, C., et al. (2013). in situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860. doi: 10.1038/nmeth.2563
Klein, M. E., Impey, S., and Goodman, R. H. (2005). Role reversal: the regulation of neuronal gene expression by microRNAs. Curr. Opin. Neurobiol. 15, 507–513. doi: 10.1016/j.conb.2005.08.011
Kocerha, J., Faghihi, M. A., Lopez-Toledano, M. A., Huang, J., Ramsey, A. J., Caron, M. G., et al. (2009). MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. U S A 106, 3507–3512. doi: 10.1073/pnas.0805854106
Konopka, G., Friedrich, T., Davis-Turak, J., Winden, K., Oldham, M. C., Gao, F., et al. (2012). Human-specific transcriptional networks in the brain. Neuron 75, 601–617. doi: 10.1016/j.neuron.2012.05.034
Kosik, K. S. (2006). The neuronal microRNA system. Nat. Rev. Neurosci. 7, 911–920. doi: 10.1038/nrn2037
Krichevsky, A. M., Sonntag, K. C., Isacson, O., and Kosik, K. S. (2006). Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24, 857–864. doi: 10.1634/stemcells.2005-0441
Lai, C. S., Fisher, S. E., Hurst, J. A., Vargha-Khadem, F., and Monaco, A. P. (2001). A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523. doi: 10.1038/35097076
Lake, B. B., Chen, S., Sos, B. C., Fan, J., Kaeser, G. E., Yung, Y. C., et al. (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80. doi: 10.1038/nbt.4038
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Lindsley, C. W., and Hopkins, C. R. (2012). Metabotropic glutamate receptor 4 (mGlu4)-positive allosteric modulators for the treatment of Parkinson’s disease: historical perspective and review of the patent literature. Expert Opin. Ther. Pat. 22, 461–481. doi: 10.1517/13543776.2012.679437
Liu, H. K., Belz, T., Bock, D., Takacs, A., Wu, H., Lichter, P., et al. (2008). The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev. 22, 2473–2478. doi: 10.1101/gad.479308
Liu, J., Githinji, J., McLaughlin, B., Wilczek, K., and Nolta, J. (2012). Role of miRNAs in neuronal differentiation from human embryonic stem cell-derived neural stem cells. Stem Cell Rev. Rep. 8, 1129–1137. doi: 10.1007/s12015-012-9411-6
Lopez, J. P., Lim, R., Cruceanu, C., Crapper, L., Fasano, C., Labonte, B., et al. (2014). miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 20, 764–768. doi: 10.1038/nm.3582
Lu, J., Shen, Y., Wu, Q., Kumar, S., He, B., Shi, S., et al. (2008). The birth and death of microRNA genes in Drosophila. Nat. Genet. 40, 351–355. doi: 10.1038/ng.73
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M., and Cai, L. (2014). Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361. doi: 10.1038/nmeth.2892
Lui, J. H., Hansen, D. V., and Kriegstein, A. R. (2011). Development and evolution of the human neocortex. Cell 146, 18–36. doi: 10.1016/j.cell.2011.06.030
Magill, S. T., Cambronne, X. A., Luikart, B. W., Lioy, D. T., Leighton, B. H., Westbrook, G. L., et al. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U S A 107, 20382–20387. doi: 10.1073/pnas.1015691107
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448. doi: 10.1016/j.molcel.2007.07.015
Marcinowski, L., Tanguy, M., Krmpotic, A., Radle, B., Lisnic, V. J., Tuddenham, L., et al. (2012). Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8:e1002510. doi: 10.1371/journal.ppat.1002510
Marín-Padilla, M. (2014). The mammalian neocortex new pyramidal neuron: a new conception. Front. Neuroanat. 7:51. doi: 10.3389/fnana.2013.00051
Maze, I., Chaudhury, D., Dietz, D. M., Von Schimmelmann, M., Kennedy, P. J., Lobo, M. K., et al. (2014). G9a influences neuronal subtype specification in striatum. Nat. Neurosci. 17, 533–539. doi: 10.1038/nn.3670
Mellios, N., Feldman, D. A., Sheridan, S. D., Ip, J. P. K., Kwok, S., Amoah, S. K., et al. (2018). MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatry 23, 1051–1065. doi: 10.1038/mp.2017.86
Menon, V. (2018). Clustering single cells: a review of approaches on high-and low-depth single-cell RNA-seq data. Brief. Funct. Genomics 17, 240–245. doi: 10.1093/bfgp/elx044
Meunier, J., Lemoine, F., Soumillon, M., Liechti, A., Weier, M., Guschanski, K., et al. (2013). Birth and expression evolution of mammalian microRNA genes. Genome Res. 23, 34–45. doi: 10.1101/gr.140269.112
Miller, J. A., Ding, S. L., Sunkin, S. M., Smith, K. A., Ng, L., Szafer, A., et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. doi: 10.1038/nature13185
Molnár, G., Oláh, S., Komlósi, G., Füle, M., Szabadics, J., Varga, C., et al. (2008). Complex events initiated by individual spikes in the human cerebral cortex. PLoS Biol. 6:e222. doi: 10.1371/journal.pbio.0060222
Molnár, G., Rózsa, M., Baka, J., Holderith, N., Barzó, P., Nusser, Z., et al. (2016). Human pyramidal to interneuron synapses are mediated by multi-vesicular release and multiple docked vesicles. Elife 5:e18167. doi: 10.7554/eLife.18167
Muddashetty, R. S., Nalavadi, V. C., Gross, C., Yao, X., Xing, L., Laur, O., et al. (2011). Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation and mGluR signaling. Mol. Cell 42, 673–688. doi: 10.1016/j.molcel.2011.05.006
Murai, K., Sun, G., Ye, P., Tian, E., Yang, S., Cui, Q., et al. (2016). The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat. Commun. 7:10965. doi: 10.1038/ncomms10965
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. doi: 10.1038/nature08909
Nowakowski, T. J., Bhaduri, A., Pollen, A. A., Alvarado, B., Mostajo-Radji, M. A., Di Lullo, E., et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. doi: 10.1126/science.aap8809
Nowakowski, T. J., Fotaki, V., Pollock, A., Sun, T., Pratt, T., and Price, D. J. (2013). MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc. Natl. Acad. Sci. U S A 110, 7056–7061. doi: 10.1073/pnas.1219385110
Nowakowski, T. J., Rani, N., Golkaram, M., Zhou, H. R., Alvarado, B., Huch, K., et al. (2018). Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat. Neurosci. 21, 1784–1792. doi: 10.1038/s41593-018-0265-3
Nudelman, A. S., DiRocco, D. P., Lambert, T. J., Garelick, M. G., Le, J., Nathanson, N. M., et al. (2010). Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20, 492–498. doi: 10.1002/hipo.20646
Oberheim, N. A., Goldman, S. A., and Nedergaard, M. (2012). Heterogeneity of astrocytic form and function. Methods Mol. Biol. 814, 23–45. doi: 10.1007/978-1-61779-452-0_3
Otani, T., Marchetto, M. C., Gage, F. H., Simons, B. D., and Livesey, F. J. (2016). 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell 18, 467–480. doi: 10.1016/j.stem.2016.03.003
Paşca, A. M., Sloan, S. A., Clarke, L. E., Tian, Y., Makinson, C. D., Huber, N., et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678. doi: 10.1038/nmeth.3415
Paul, A., Crow, M., Raudales, R., He, M., Gillis, J., and Huang, Z. J. (2017). Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171, 522.e20–539.e20. doi: 10.1016/j.cell.2017.08.032
Pletikos, M., Sousa, A. M., Sedmak, G., Meyer, K. A., Zhu, Y., Cheng, F., et al. (2014). Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81, 321–332. doi: 10.1016/j.neuron.2013.11.018
Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., et al. (2016). Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254. doi: 10.1016/j.cell.2016.04.032
Quadrato, G., Nguyen, T., Macosko, E. Z., Sherwood, J. L., Min Yang, S., Berger, D. R., et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53. doi: 10.1038/nature22047
Raghanti, M. A., Edler, M. K., Stephenson, A. R., Wilson, L. J., Hopkins, W. D., Ely, J. J., et al. (2016). Human-specific increase of dopaminergic innervation in a striatal region associated with speech and language: a comparative analysis of the primate basal ganglia. J. Comp. Neurol. 524, 2117–2129. doi: 10.1002/cne.23937
Raghanti, M. A., Stimpson, C. D., Marcinkiewicz, J. L., Erwin, J. M., Hof, P. R., and Sherwood, C. C. (2008). Differences in cortical serotonergic innervation among humans, chimpanzees and macaque monkeys: a comparative study. Cereb. Cortex 18, 584–597. doi: 10.1093/cercor/bhm089
Raj, A., and van Oudenaarden, A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. doi: 10.1016/j.cell.2008.09.050
Rockel, A. J., Hiorns, R. W., and Powell, T. P. (1980). The basic uniformity in structure of the neocortex. Brain 103, 221–244. doi: 10.1093/brain/103.2.221
Rogers, J., Kochunov, P., Zilles, K., Shelledy, W., Lancaster, J., Thompson, P., et al. (2010). On the genetic architecture of cortical folding and brain volume in primates. Neuroimage 53, 1103–1108. doi: 10.1016/j.neuroimage.2010.02.020
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222. doi: 10.1038/nrd.2016.246
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: biology, functions, therapeutics and analysis methods. J. Cell. Physiol. 234, 5451–5465. doi: 10.1002/jcp.27486
Salmén, F., Ståhl, P. L., Mollbrink, A., Navarro, J. F., Vickovic, S., Frisen, J., et al. (2018). Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nat. Protoc. 13, 2501–2534. doi: 10.1038/s41596-018-0045-2
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F., and Regev, A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. doi: 10.1038/nbt.3192
Schratt, G. (2009). Fine-tuning neural gene expression with microRNAs. Curr. Opin. Neurobiol. 19, 213–219. doi: 10.1016/j.conb.2009.05.015
Sherwood, C. C., Holloway, R. L., Erwin, J. M., and Hof, P. R. (2004). Cortical orofacial motor representation in Old World monkeys, great apes and humans: II. Stereologic analysis of chemoarchitecture. Brain Behav. Evol. 63, 82–106. doi: 10.1159/000075673
Shi, Y., Chichung Lie, D., Taupin, P., Nakashima, K., Ray, J., Yu, R. T., et al. (2004). Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427, 78–83. doi: 10.1038/nature02211
Shi, Y., Kirwan, P., Smith, J., Robinson, H. P., and Livesey, F. J. (2012). Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 15, 477–486. doi: 10.1038/nn.3041
Silbereis, J. C., Pochareddy, S., Zhu, Y., Li, M., and Sestan, N. (2016). The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268. doi: 10.1016/j.neuron.2015.12.008
Smart, I. H., Dehay, C., Giroud, P., Berland, M., and Kennedy, H. (2002). Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 12, 37–53. doi: 10.1093/cercor/12.1.37
Søkilde, R., Newie, I., Persson, H., Borg, Å., and Rovira, C. (2015). Passenger strand loading in overexpression experiments using microRNA mimics. RNA Biol. 12, 787–791. doi: 10.1080/15476286.2015.1020270
Somel, M., Liu, X., Tang, L., Yan, Z., Hu, H., Guo, S., et al. (2011). MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 9:e1001214. doi: 10.1371/journal.pbio.1001214
Sosanya, N. M., Huang, P. P., Cacheaux, L. P., Chen, C. J., Nguyen, K., Perrone-Bizzozero, N. I., et al. (2013). Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J. Cell Biol. 202, 53–69. doi: 10.1083/jcb.201212089
Sousa, A. M. M., Meyer, K. A., Santpere, G., Gulden, F. O., and Sestan, N. (2017a). Evolution of the human nervous system function, structure and development. Cell 170, 226–247. doi: 10.1016/j.cell.2017.06.036
Sousa, A. M. M., Zhu, Y., Raghanti, M. A., Kitchen, R. R., Onorati, M., Tebbenkamp, A. T. N., et al. (2017b). Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032. doi: 10.1126/science.aan3456
Stuart, T., and Satija, R. (2019). Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272. doi: 10.1038/s41576-019-0093-7
Szegedi, V., Paizs, M., Csakvari, E., Molnar, G., Barzo, P., Tamas, G., et al. (2016). Plasticity in single axon glutamatergic connection to GABAergic interneurons regulates complex events in the human neocortex. PLoS Biol. 14:e2000237. doi: 10.1371/journal.pbio.2000237
Tasic, B., Menon, V., Nguyen, T. N., Kim, T. K., Jarsky, T., Yao, Z., et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346. doi: 10.1038/nn.4216
Tasic, B., Yao, Z., Graybuck, L. T., Smith, K. A., Nguyen, T. N., Bertagnolli, D., et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. doi: 10.1038/s41586-018-0654-5
Taverna, E., Götz, M., and Huttner, W. B. (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502. doi: 10.1146/annurev-cellbio-101011-155801
Testa-Silva, G., Verhoog, M. B., Goriounova, N. A., Loebel, A., Hjorth, J., Baayen, J. C., et al. (2010). Human synapses show a wide temporal window for spike-timing-dependent plasticity. Front. Synaptic Neurosci. 2:12. doi: 10.3389/fnsyn.2010.00012
Thiebes, K. P., Nam, H., Cambronne, X. A., Shen, R., Glasgow, S. M., Cho, H. H., et al. (2015). miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat. Commun. 6:7718. doi: 10.1038/ncomms8718
Tu, J., Cao, D., Li, L., Cheung, H. H., and Chan, W. Y. (2018). MicroRNA profiling during directed differentiation of cortical interneurons from human-induced pluripotent stem cells. FEBS Open Bio 8, 502–512. doi: 10.1002/2211-5463.12377
van Rooij, E., Purcell, A. L., and Levin, A. A. (2012). Developing microRNA therapeutics. Circ. Res. 110, 496–507. doi: 10.1161/CIRCRESAHA.111.247916
Verhoog, M. B., Goriounova, N. A., Obermayer, J., Stroeder, J., Hjorth, J. J., Testa-Silva, G., et al. (2013). Mechanisms underlying the rules for associative plasticity at adult human neocortical synapses. J. Neurosci. 33, 17197–17208. doi: 10.1523/jneurosci.3158-13.2013
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749. doi: 10.1101/gad.1519107
Wang, B., Yin, L., Zou, X., Ye, M., Liu, Y., He, T., et al. (2015). A subtype of inhibitory interneuron with intrinsic persistent activity in human and monkey neocortex. Cell Rep. 10, 1450–1458. doi: 10.1016/j.celrep.2015.02.018
Winter, J., and Diederichs, S. (2011). Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 8, 1149–1157. doi: 10.4161/rna.8.6.17665
Winter, J., Jung, S., Keller, S., Gregory, R. I., and Diederichs, S. (2009). Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234. doi: 10.1038/ncb0309-228
Wood, H. (2018). FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 14:570. doi: 10.1038/s41582-018-0065-0
Xiang, Y., Tanaka, Y., Cakir, B., Patterson, B., Kim, K. Y., Sun, P., et al. (2019). hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 24, 487.e7–497.e7. doi: 10.1016/j.stem.2018.12.015
Xiang, Y., Tanaka, Y., Patterson, B., Kang, Y. J., Govindaiah, G., Roselaar, N., et al. (2017). Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383.e7–398.e7. doi: 10.1016/j.stem.2017.07.007
Yang, L., Yang, Y., Yuan, J., Sun, Y., Dai, J., and Su, B. (2019). Transcriptomic landscape of von economo neurons in human anterior cingulate cortex revealed by microdissected-cell RNA sequencing. Cereb. Cortex 29, 838–851. doi: 10.1093/cercor/bhy286
Yoo, A. S., Staahl, B. T., Chen, L., and Crabtree, G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646. doi: 10.1038/nature08139
Zhang, C., Lu, J., Liu, B., Cui, Q., and Wang, Y. (2016). Primate-specific miR-603 is implicated in the risk and pathogenesis of Alzheimer’s disease. Aging 8, 272–290. doi: 10.18632/aging.100887
Zhang, C. L., Zou, Y., He, W., Gage, F. H., and Evans, R. M. (2008). A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451, 1004–1007. doi: 10.1038/nature06562
Zhao, C., Sun, G., Li, S., and Shi, Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371. doi: 10.1038/nsmb.1576
Zhao, X., Wu, J., Zheng, M., Gao, F., and Ju, G. (2012). Specification and maintenance of oligodendrocyte precursor cells from neural progenitor cells: involvement of microRNA-7a. Mol. Biol. Cell 23, 2867–2878. doi: 10.1091/mbc.e12-04-0270
Zhong, S., Zhang, S., Fan, X., Wu, Q., Yan, L., Dong, J., et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. doi: 10.1038/nature25980
Zhu, Y., Sousa, A. M. M., Gao, T., Skarica, M., Li, M., Santpere, G., et al. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362:eaat8077. doi: 10.1126/science.aat8077
Keywords: RNA sequencing (RNAseq), primate, neurogenesis, evolution, gene networks, brain, miRNAs, human
Citation: Prodromidou K and Matsas R (2019) Species-Specific miRNAs in Human Brain Development and Disease. Front. Cell. Neurosci. 13:559. doi: 10.3389/fncel.2019.00559
Received: 28 September 2019; Accepted: 04 December 2019;
Published: 18 December 2019.
Edited by:
Daniella Rylander Ottosson, Lund University, SwedenReviewed by:
Hermona Soreq, Hebrew University of Jerusalem, IsraelDavide Lecca, University of Milan, Italy
Copyright © 2019 Prodromidou and Matsas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kanella Prodromidou, a3Byb2Ryb21pZG91QHBhc3RldXIuZ3I=
 Kanella Prodromidou
Kanella Prodromidou Rebecca Matsas
Rebecca Matsas