- Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany
Brain organoids have become increasingly used systems allowing 3D-modeling of human brain development, evolution, and disease. To be able to make full use of these modeling systems, researchers have developed a growing toolkit of genetic modification techniques. These techniques can be applied to mature brain organoids or to the preceding embryoid bodies (EBs) and founding cells. This review will describe techniques used for transient and stable genetic modification of brain organoids and discuss their current use and respective advantages and disadvantages. Transient approaches include adeno-associated virus (AAV) and electroporation-based techniques, whereas stable genetic modification approaches make use of lentivirus (including viral stamping), transposon and CRISPR/Cas9 systems. Finally, an outlook as to likely future developments and applications regarding genetic modifications of brain organoids will be presented.
Introduction
The development of brain organoids (Kadoshima et al., 2013; Lancaster et al., 2013) has opened up new ways to study brain development and evolution as well as neurodevelopmental disorders. Brain organoids are multicellular 3D structures that mimic certain aspects of the cytoarchitecture and cell-type composition of certain brain regions over a particular developmental time window (Heide et al., 2018). These structures are generated by differentiation of induced pluripotent stem cells (iPSCs) or embryonic stem cells (ESCs) into embryoid bodies followed by, or combined, with neural induction (Kadoshima et al., 2013; Lancaster et al., 2013). In principle, two different classes of brain organoid protocols can be distinguished, namely: (i) the self-patterning protocols which produce whole-brain organoids; and (ii) the pre-patterning protocols which produce brain region-specific organoids (Heide et al., 2018). However, brain organoids are far from being ideal models of the brain, and notably cortical development (for a review, see Heide et al., 2018). The main issues concern reproducibility and the modeling of later stages of brain development, as mainly early stages of brain development, are modeled correctly. Very recently, approaches have been undertaken to improve the reproducibility through the optimization of protocols (Velasco et al., 2019). Moreover, modeling of later stages has been addressed by introducing organoid slice cultures grown at the air-liquid interface. This has resulted in increased neuronal survival and improved morphology as well as the generation of axonal tracts (Giandomenico et al., 2019). In future, brain organoid protocols are likely to result in even better 3D models of neural development, evolution and disease. An important step in this direction has been the very recent development of human brain organoids with a vascular-like system (Cakir et al., 2019).
However, even the best 3D model of brain development loses much of its usefulness if one cannot modify it. The ability to genetically modify brain organoids is essential for their utility as models of brain development (Birey et al., 2017), evolution (Mora-Bermúdez et al., 2016) and disease (Bian et al., 2018) as well as their evolving use as drug-screening platforms (Zhou et al., 2017). Genetic modification is a powerful tool that allows for the introduction of alterations ranging from small changes (e.g., point mutations) to the removal or integration of entire genes. This enables researchers to investigate individual genes as well as entire gene cassettes to elicit gene expression patterns, functions, and interactions. Moreover, drugs can be tested in a larger variety of disease states and in different genetic environments.
Rather than focusing on the discussion of the advantages, disadvantages and potential applications of the different brain organoid protocols, which have already been addressed in several excellent reviews (e.g., Kelava and Lancaster, 2016; Quadrato et al., 2016; Di Lullo and Kriegstein, 2017; Heide et al., 2018), the present review will focus on the genetic modification of brain organoids. An overview of the different types and methods of genetic modification in brain organoids will be given, and their advantages and disadvantages, as well as examples of their application, will be discussed.
Types of Genetic Modification of Brain Organoids
Any method of genetic modification needs to address three key issues: (i) the nature of the genetic modification; (ii) the stage, within brain organoid development, at the time of genetic modification; and (iii) the target cells of the genetic modification.
The first issue, namely the nature of the genetic modification, concerns stable vs. transient genetic modification. In a stable modification, the genetic alteration is introduced into the genome of a cell and is thus passed on to future cell generations. In contrast, in transient genetic modifications, genetic cargo (e.g., genes, short interfering RNAs, etc.) is administered to a cell without genomic insertion and the possibility of further replication. The delivered genetic cargo is then progressively degraded and diluted with each cell division. For proliferating cells, in particular, this means that the level of the administered genetic material continuously declines.
The second issue concerns the timepoint of the genetic modification relative to the stage of brain organoid development, which can range from the starting cell line over the embryoid body (EB) stage to mature brain organoids (Figure 1). The choice of this timepoint depends mainly on the purpose of the experiment and the proportion of cells to be affected by the genetic modification (Figure 2). Stable modifications are mostly performed at an early stage of brain organoid development, such as in the starting cell line, whereas transient modifications are typically performed at later stages, given the decline of genetic cargo levels due to cell proliferation (Figure 1).
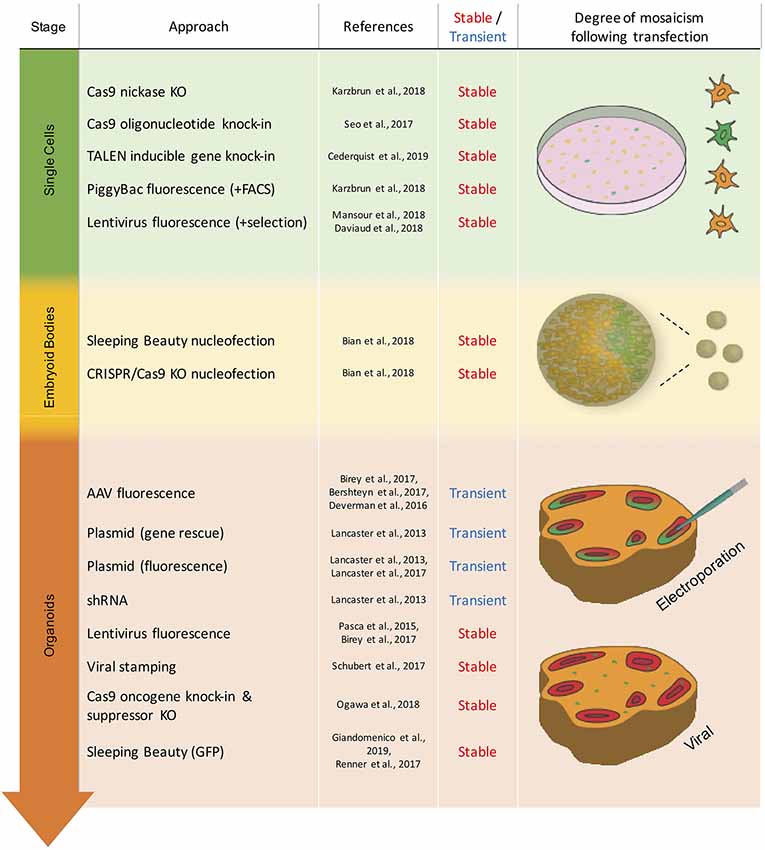
Figure 1. Overview of the transient and stable approaches for genetic modification in brain organoids. Techniques are listed by the stage of organoid development in which they have been used. Schematic representation of mosaicism and spatial distribution of genetically modified cells (green) following respective modification approaches.

Figure 2. Schematic representation of sections of brain organoids as seen subsequent to various approaches of genetic modification. Green denotes genetically modified cells. (A) Brain organoid derived from genetically modified induced pluripotent stem cells (iPSCs)/embryonic stem cells (ESCs) that did not undergo subsequent positive selection. (B) Brain organoid derived from genetically modified iPSCs/ESCs that underwent subsequent positive selection using FACS or antibiotic resistance. The vast majority of cells within the brain organoid will contain the genetic modification. (C) Brain organoids derived from an embryoid body (EB) that underwent regional genetic modification through the use of an electroporation-based approach. Note that a ventricle-like structure will only show genetic modification if transfection efficiency and cell survival is sufficiently high in the targeted region of the EB. (D) Brain organoid following genetic modifications being targeted to single cells using viral stamping. Cells both at and away from, the brain organoid surface can be targeted. (E) Brain organoid following the electroporation of genetic cargo into ventricle-like structures. (F) Brain organoid following virus-based (e.g., lentivirus or adeno-associated virus, AAV) genetic modification approaches. The proportion of genetically modified cells can vary greatly depending on incubation times of brain organoids in medium containing viral vectors.
Due to the large cellular heterogeneity of brain organoids, the third issue concerns the cell type(s) that is/are targeted within the organoid by the genetic modifications. Approaches can either target cells indiscriminately, regardless of their location and cell type or, contrariwise, target a subset of cells. This can be achieved either through direct visual identification of cells or alternatively by biologically restricting the genetic modification to specific cell types. Visual identification of target cells can be used for single-cell microinjections (Chow et al., 2016; Shull et al., 2019) or approaches such as viral stamping (to be discussed below; Schubert et al., 2018). Biological restriction, on the other hand, makes use of cell type-specific promotors to activate or suppress gene expression (Pasca et al., 2019; Birey et al., 2017). In the following, we will first discuss the various types of transient genetic modifications of brain organoids, and then the various types of stable genetic modifications.
Transient Genetic Modifications of Brain Organoids
In transient genetic modifications of brain organoids, gene vectors are used that allow protein expression or the production of short interfering RNAs (siRNAs; Lancaster et al., 2013) for a limited amount of time. Transient expression vectors have therefore been almost exclusively administered into late-stage brain organoids rather than into the cells or EBs that precede them (Figure 1). For this type of genetic modification, one of the most important aspects to consider is the mode of plasmid delivery. The two most widely used methods in the case of brain organoids are viral delivery via adeno-associated viruses (AAVs) and non-viral delivery via electroporation.
Viral Delivery—Adeno-Associated Viruses (AAVs)
AAVs are known to generally produce transient, rather than stable, expression of genetic cargo (e.g., genes or siRNAs). This is in contrast to the frequently used lentiviruses (see below), which typically produce stable transfections. However, it is important to note that AAVs can integrate genes into the genome of targeted cells at very low rates, particularly in dividing cells and at the AAV-safe loci (AAVS1; Deyle and Russell, 2009). AAVs have been successfully used in brain organoids to achieve fluorescence specifically in neurons (Deverman et al., 2016; Bershteyn et al., 2017; Birey et al., 2017). These experiments avoided targeting proliferative cell populations and primarily focused on neuronal migration within brain organoids. However, in organoid slice cultures, proliferative cell populations have been targeted using adenoviruses, which allowed for detectable fluorescence in these cells for 2–5 days (Bershteyn et al., 2017).
In most cases, AAVs are simply added to the cell culture medium, which results in scattered mosaic expression throughout the entire organoid or slice (Figure 2F). However, this mode of administration precludes a more focused targeting of individual regions within the brain organoid. Further drawbacks of AAVs include: (i) the need of special facility-requiring safety precautions; (ii) a greater amount of work in comparison to non-viral delivery due to the production and titration of virus particles; and (iii) a limit of 5 kilobases of cargo DNA (Grieger and Samulski, 2005).
Non-viral Delivery—Electroporation
An important alternative to genetic cargo delivery via AAVs is electroporation. Analogous to in utero electroporations of embryonic mouse (Saito and Nakatsuji, 2001; LoTurco et al., 2009) and ferret (Kawasaki et al., 2012) neocortex, in which plasmid DNA is injected into the brain ventricles prior to the electroporation, ventricle-like structures in brain organoids can be identified and injected with plasmid DNA and subsequently electroporated (Lancaster et al., 2013; Li et al., 2017). Here, electrical pulses lead to pore formation in the cell’s plasma membrane to allow for plasmid uptake (Kar et al., 2018). However, particularly in older brain organoids (>2 months old), targeting of ventricles by injection becomes increasingly difficult due to a reduction in tissue translucency. This can partly be overcome by confining the manipulation to the outer regions of a brain organoid where ventricles are located, which can be blindly targeted by injection. However, this approach frequently results in increased mechanical damage of the brain organoid and decreased transfection efficiency (our own observations).
When the ventricles of brain organoids can be specifically targeted, electroporation has a much higher transfection efficiency than genetic cargo delivery via AAVs. Analogous to in utero electroporation, efficiencies of up to almost half of the cells lining the ventricular surface of the targeted region can be achieved (dal Maschio et al., 2012). Such high efficiencies allow for regionally confining the area of cell transfection within a given ventricular structure of the brain organoid (Figure 2E), depending on electrode placement. Thereby a comparison between transfected and untransfected areas within the same ventricular structure in the brain organoid can be made. A further benefit of electroporation over viral delivery is that cells are transfected at the same timepoint, whereas upon viral delivery transfection occurs over a temporal range. Yet, these benefits come at the risk of cellular toxicity due to the electrical pulses that are applied. However, if the voltages for electroporation are kept low (≤80 V), cell viability usually remains high (Lancaster et al., 2013). Importantly, reflecting the decline of genetic cargo levels over time prior to EB embedding, transient genetic modification using electroporation has been found to be limited to mature brain organoids (Lancaster et al., 2013; Li et al., 2017; Figure 1).
Stable Genetic Modifications of Brain Organoids
Whilst transient genetic modification of brain organoids is suitable for many experimental approaches, it is insufficient for setups in which proliferative cells require modifications for longer periods of time. Not only can stable genetic modification approaches resolve this issue, but if applied to the brain organoid founder cells they can also greatly reduce mosaic expression patterns (Figures 2A,B). Stable genetic modification methods also heavily rely on viral delivery or electroporation (although the use of lipofection is also conceivable) in order to introduce genetic cargo into the cells. As above, genetic cargo is any nucleic acid from an exogenous source that is delivered into the cells to bring about a change in gene expression. However, unlike transient genetic modification, stable genetic modification is typically achieved by delivering a piece of machinery into the cells that subsequently will mediate the introduction of genetic modifications (from point mutations to integration of transgenes) into the host cell’s genome. A variety of tools that involve such a machinery have been developed to achieve genomic integration into the cells forming brain organoids, namely viruses with reverse transcriptase activity (lentiviruses), transposon systems, and CRISPR/Cas9.
Viral Delivery—Lentiviruses
Retrovirus-based, in particular, lentivirus-based, transduction is an efficient, fast and easy-to-use method to achieve stable genetic modification of cells. Replication-deficient lentiviral vectors are one of the most widely used tools for this purpose (Janssens et al., 2019). These commonly are human immunodeficiency viruses pseudotyped with a vesicular stomatitis virus envelope, which allows infection of most cell types (Vigna and Naldini, 2000; Merten et al., 2016). In this system, the genetic cargo (referred to as transfer plasmid) and the viral components (structural proteins and the enzymes needed for the integration of the genetic cargo, collectively referred to as packaging plasmids) are contained on different plasmids. To generate functional viral particles, these plasmids are introduced into a packaging cell line, typical HEK293, and the thus generated viral particles are then collected from the cell culture supernatant. These viral particles contain only the genetic cargo but no genes for the packaging machinery, which implies that after infection of the target cells, the genetic cargo can be integrated into the host genome, but new viral particles cannot be produced by the infected cell (Vigna and Naldini, 2000).
Lentiviral vectors have been administered to brain organoids to induce fluorescence in target cells through the use of cell-type-specific promotors (Pasca et al., 2019; Birey et al., 2017). Due to relatively low transfection efficiencies, significant mosaicism results from the use of these vectors when they are simply added to the cell culture medium of brain organoids (Figure 2A). To overcome this, brain organoid founder cells can be virally targeted, with subsequent antibiotic selection or fluorescence-activated cell sorting in order to obtain a more homogeneous cell pool for brain organoid generation (Daviaud et al., 2018; Mansour et al., 2018; Janssens et al., 2019; Figure 2B).
Moreover, researchers will in the future not only be limited to the aforementioned approaches using lentiviral vectors, which infect without cell-type specificity, on bulk cell populations or tissue. Rather, selected single cells can be targeted using such viral vectors (Schubert et al., 2018; Figure 2D). This procedure makes use of a technique termed viral stamping, in which viral vectors are brought into direct physical contact with single target cells (Schubert et al., 2018). This approach is not limited to targeting cells at the surface of brain organoids, but can also target those that lie within them. In the latter case, using a so-called shielded approach, the viral particles, which are linked to magnetic nanoparticles, are only exposed to cell surfaces upon an electromagnetic pulse. Using viral stamping, individual cells within brain organoids have been successfully targeted at a depth of up to 150 μm from the organoid’s surface, with an efficiency of 10%–25% of the targeted cells (Schubert et al., 2018).
Regardless of approach, however, lentiviral vectors have several notable drawbacks. Although lentiviral vectors have a greater size capacity for genetic cargo than AAVs, only 10–12 kb of genetic cargo have been efficiently transduced, thereby placing limits on more complex gene constructs (Kumar et al., 2001; Counsell et al., 2017). A further disadvantage is that random genomic insertion of genes can lead to unwanted and unpredictable effects, ranging from cell apoptosis to uncontrolled proliferation through activation of proto-oncogenes.
Non-viral Delivery
In contrast to viral delivery through lentiviruses, which introduces into the host cell both the genetic cargo and the machinery for its integration into the genome, non-viral delivery mediated by electroporation or lipofection may introduce only genetic cargo into the host cell. If so, non-viral delivery needs to include, or be combined with, introducing machinery into the host cell that mediates the integration of the genetic cargo into the genome. The two most often used such machinery are transposon-like systems and nuclease-based tools (nowadays mainly CRISPR/Cas9).
Transposon-Like Systems
Transposon-like systems used to obtain stable genetic modification are based on mobile DNA elements. These elements can move (“transpose”) their position within the genome using a “cut and paste” mechanism (McClintock, 1950; Grabundzija et al., 2010). This is made possible by the presence of a transposase that is able to recognize inverted terminal repeats (ITRs) flanking the transposon, which is then excised and inserted at a different site. In current transposon-like systems, the DNA cargo of interest is tagged with flanking ITRs, and the appropriate transposase is provided in trans, leading to the integration of the DNA cargo into the target genome (Ni et al., 2008). Two of the most notable transposon-like systems that have been developed are Sleeping Beauty (Ivics et al., 1997) and PiggyBac (Ding et al., 2005; Wilson et al., 2007).
With regard to brain organoids, these transposon-like systems have been used not only to achieve GFP reporter expression (Lancaster et al., 2017; Renner et al., 2017; Karzbrun et al., 2018; Giandomenico et al., 2019), but also to introduce oncogenes (Bian et al., 2018). With generally higher genomic insertion efficiencies than retroviral vectors (Ding et al., 2005; Chen et al., 2015), transposon-like systems can be delivered, typically via electroporation, not only into brain organoid founder cells, often followed by selection of expressing cells, but also into EBs (Bian et al., 2018) and even mature organoids. The aim of this approach is to achieve expression in targeted cell populations and their progeny within the brain organoid (Figure 2C).
Despite several advantages, including the potentially large size of the transposable elements of up to 14 kb, some notable drawbacks of transposon-like systems exist (Ding et al., 2005). Thus, uncertainty as to the level of gene expression when using these systems can be a major disadvantage. Specifically, transgenes may have integrated a variable number of times into the genome of a given cell type, leading to significant differences in expression from cell to cell. Furthermore, the genomic integration of the cargo DNA can interfere with the expression of endogenous genes, leading to unwanted side effects.
CRISPR/Cas9
An alternative to achieve stable genetic modification is to make use of nuclease-based tools. A variety of nuclease-based tools have been developed to induce targeted mutations and gene insertions, ranging from Transcription Activator-Like Effector Nucleases (TALENs; Boch et al., 2009) and Zinc Finger Nucleases (ZFNs; Kim et al., 1996) to the now very popular Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 (CRISPR/Cas9; Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013) system. Although there is still some use of TALENs (Cederquist et al., 2019) and ZFNs in the context of stable genetic modifications of brain organoids, these nuclease-based tools have now largely been replaced by the CRISPR/Cas9 system due to its relative ease of use and high efficiency. This system uses a guide RNA to target the nuclease Cas9 to a specific genomic locus to then cause a double-strand break, thereby activating endogenous DNA repair processes. These can be non-homologous end joining (NHEJ) or homologous-directed repair (HDR). NHEJ has a high propensity for generating small deletions or insertions during repair, thus enabling the generation of knockouts or knockdowns of targeted genes. HDR, on the other hand, is useful for introducing changes (ranging from point mutations to transgene insertions) in the presence of a DNA template to instruct the repair DNA synthesis (Doudna and Charpentier, 2014; Harrison et al., 2014; Hsu et al., 2014). It is, however, important to note that NHEJ has also been shown to play an important role in the insertion of transgenes particularly in postmitotic cells which show significantly reduced HDR (Suzuki et al., 2016; Suzuki and Belmonte, 2018). CRISPR/Cas9-mediated genetic modifications are usually conducted on brain organoid founder cells, that is, ESCs and iPSCs (Bershteyn et al., 2017; Iefremova et al., 2017; Li et al., 2017; Matsui et al., 2017; Fiddes et al., 2018; Karzbrun et al., 2018). Cells containing the desired genomic change are then selected and can be used to grow brain organoids. This approach has the advantage that all cells in the organoid will contain the previously introduced genetic modification (Figure 2B).
However, the use of CRISPR/Cas9-mediated genetic modifications is not limited to brain organoid founder cells. Plasmid vectors containing Cas9 along with one or more guide RNAs, or in vitro formed complexes of recombinant Cas9 protein and guide RNAs (Kalebic et al., 2016), can be electroporated directly into EBs or early organoids to result in loss of function mutations (Bian et al., 2018). This has been explored in organoid models of central nervous system (CNS) tumors in which tumor suppressor genes were targeted to provoke neoplasia (Bian et al., 2018). A further example is the combined use of a CRISPR/Cas9-directed tumor suppressor (TP53) knock-out combined with a CRISPR/Cas9-directed oncogene (KRAS) knock-in in 4 months-old human brain organoids (Ogawa et al., 2018). By generating cells with a glioblastoma-like proliferative potential, only a few cells needed to successfully undergo stable genetic modification.
In certain cases, especially for knock-outs of genes that exert essential functions at certain stages of brain organoid development, it would be beneficial to have temporal control of CRISPR/Cas9 activity. To this end, one may consider using CRISPR/Cas9 as an inducible system. For example, Tet- or Lac-based systems could be combined with Cas9 and guide RNA (Sun et al., 2019). Once this system is introduced into the ESC or iPSC genome, it would allow for a time-specific knockout of the gene of interest. Transcription of Cas9 would be induced by the addition of doxycycline (Tet) or isopropyl ß-D-1-thiogalactopyranoside (Lac) to the culture medium.
Of note, Cas9 activity in cells over long periods of time increases the risk of off-target effects, thereby having unforeseen effects on cell survival and phenotype. To reduce such effects, paired Cas9 nickases can be used (Ran et al., 2013). Cas9 nickases can only produce single-strand cuts rather than the double-strand cuts of wild-type Cas9. This means that two nickases need to target sequences in very close proximity to one another to result in a successful genetic modification event. This significantly increases target specificity, albeit at the cost of genetic modification efficiency.
One prime application of the CRISPR/Cas9 system in brain organoids is its use in modeling neurodevelopmental and neurodegenerative diseases. Here, either patient mutations can be introduced into control iPS cell lines, or patient iPS cells can be “repaired” to generate isogenic controls (Iefremova et al., 2017; Matsui et al., 2017; Fiddes et al., 2018). This was for example successfully applied in the modeling of retinoblastoma (Matsui et al., 2017) and Miller-Dieker Syndrome (Bershteyn et al., 2017; Iefremova et al., 2017).
Outlook and Future Applications
In summary, different modes and methods of genetic modifications have been successfully applied at various time points of brain organoid development. The spectrum of potential applications has so far ranged from the simple expression of fluorescent marker proteins (Pasca et al., 2019; Renner et al., 2017) to the study of gene function (Lancaster et al., 2013) to the modeling of disease conditions (Bershteyn et al., 2017; Iefremova et al., 2017; Matsui et al., 2017). Yet, significant untapped potential still remains.
In the case of transient genetic modification, the electroporation of mature organoids—although already successfully applied in the first report of cerebral organoids (Lancaster et al., 2013)—is not frequently employed. Many labs successfully utilize in utero electroporation of mice (Saito and Nakatsuji, 2001; LoTurco et al., 2009) and less frequently rat (Szczurkowska et al., 2016) and ferret (Kawasaki et al., 2013) developing neocortex to study gene function. Brain organoids could provide a potential replacement here, especially if their capacity to model 3D neural tissue is further improved. Moreover, the use of human or chimpanzee brain organoids (Mora-Bermúdez et al., 2016; Otani et al., 2016; Heide et al., 2018; Kanton et al., 2019; Pollen et al., 2019) for electroporations represents a huge opportunity as these organoids provide an environment that is comparable to the early stages of fetal human neocortex development in terms of gene expression and cell-type composition (Camp et al., 2015; Velasco et al., 2019). This could prove to be particularly beneficial for the study of human-specific genes that are expressed during cortical development either in progenitors or newborn neurons (Florio et al., 2018; Suzuki et al., 2018). Electroporation of chimpanzee brain organoids would be a relatively quick and powerful test to examine the function of these genes and would be one of the very few possible ways to study them in our closest living relative of the Hominidae family.
In the case of stable genetic modification, CRISPR/Cas9 is, and most likely will be, the method of choice to introduce such modifications in brain organoids. It is easy to use, does not require special safety precautions and is very efficient in comparison to other targeted genetic modification techniques. At present, a major application of CRISPR/Cas9 in brain organoids is the generation of disease models and is primarily focused on monogenic neurodevelopmental diseases as well as oncogenic mutations. A major future challenge will be to study more complex disease states that are oligo- or polygenic in nature.
Nonetheless, viral and transposon systems will still play a significant role in future brain organoid studies. This is due to the versatility of these tools and their greater genetic modification efficiencies in comparison to CRISPR/Cas9, particularly in EBs and mature brain organoids. Notably, the use of viruses in combination with viral stamping may prove useful in tracing axonal connections within single brain organoids as well as organoid assembloids (Pasca, 2019; Schwarz and Remy, 2019). Furthermore, advanced techniques to control the behavior of individual neurons, such as optogenetics, should also open up new avenues for brain organoid studies (Frank et al., 2019). In this context, the demonstration of oscillating electrical waves within cortical organoids (Trujillo et al., 2019) is of major relevance.
A major focus of future applications of stable genetic modifications of brain organoids will likely be cell lineage tracing. A multitude of methods have been devised in various systems that employ CRISPR/Cas9, viral vectors or transposons in order to track the developmental origin and fate of individual cells (Figueres-Oñate et al., 2016; Kebschull and Zador, 2018; McKenna and Gagnon, 2019). These methods make use of a differential expression of fluorophore combinations or other cell barcoding techniques. As to brain organoids, these approaches of cell lineage tracing could constitute a vital experimental avenue to confirm and complement in silico lineage analyses derived from single-cell sequencing (Camp and Treutlein, 2017; Camp et al., 2018). A significant advance in this context would be the transcriptomic analysis of defined cell subpopulations with an identified location within brain organoids, labeled via lineage-specific expression of fluorescent proteins.
Finally, combinations of stable and transient genetic modifications will likely hold great promise for studying various aspects of brain organoid development and performance in the future. For example, such combination may prove advantageous for rescue experiments of a given phenotype. On a general note, combinations of stable and transient genetic modifications will allow for the modeling and dissection of increasingly complex disease states as well as developmental and evolutionary processes in brain organoids.
Author Contributions
All authors wrote and edited the manuscript.
Funding
JF was supported by the Else-Kröner-Fresenius Foundation. Work in the laboratory of WH was supported by grants from the DFG (SFB 655, A2), the ERC (250197) and ERA-NET NEURON (MicroKin).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to all researchers whose work could not be cited due to space limitation.
References
Bershteyn, M., Nowakowski, T. J., Pollen, A. A., Di Lullo, E., Nene, A., Wynshaw-Boris, A., et al. (2017). Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20, 435.e4–449.e4. doi: 10.1016/j.stem.2016.12.007
Bian, S., Repic, M., Guo, Z., Kavirayani, A., Burkard, T., Bagley, J. A., et al. (2018). Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 15, 631–639. doi: 10.1038/s41592-018-0070-7
Birey, F., Andersen, J., Makinson, C. D., Islam, S., Wei, W., Huber, N., et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. doi: 10.1038/nature22330
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Cakir, B., Xiang, Y., Tanaka, Y., Kural, M. H., Parent, M., Kang, Y. J., et al. (2019). Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 16, 1169–1175. doi: 10.1038/s41592-019-0586-5
Camp, J. G., and Treutlein, B. (2017). Human organomics: a fresh approach to understanding human development using single-cell transcriptomics. Development 144, 1584–1587. doi: 10.1242/dev.150458
Camp, J. G., Badsha, F., Florio, M., Kanton, S., Gerber, T., Wilsch-Bräuninger, M., et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A 112, 15672–15677. doi: 10.1073/pnas.1520760112
Camp, J. G., Wollny, D., and Treutlein, B. (2018). Single-cell genomics to guide human stem cell and tissue engineering. Nat. Methods 15, 661–667. doi: 10.1038/s41592-018-0113-0
Cederquist, G. Y., Asciolla, J. J., Tchieu, J., Walsh, R. M., Cornacchia, D., Resh, M. D., et al. (2019). Specification of positional identity in forebrain organoids. Nat. Biotechnol. 37, 436–444. doi: 10.1038/s41587-019-0085-3
Chen, X., Cui, J., Yan, Z., Zhang, H., Chen, X., Wang, N., et al. (2015). Sustained high level transgene expression in mammalian cells mediated by the optimized piggyBac transposon system. Genes Dis. 2, 96–105. doi: 10.1016/j.gendis.2014.12.001
Chow, Y. T., Chen, S., Wang, R., Liu, C., Kong, C. W., Li, R. A., et al. (2016). Single cell transfection through precise microinjection with quantitatively controlled injection volumes. Sci. Rep. 6:24127. doi: 10.1038/srep24127
Cong, L., Ran, F. A., Cox, D., Lin, S. L., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Counsell, J. R., Asgarian, Z., Meng, J., Ferrer, V., Vink, C. A., Howe, S. J., et al. (2017). Lentiviral vectors can be used for full-length dystrophin gene therapy. Sci. Rep. 7:79. doi: 10.1038/s41598-017-00152-5
dal Maschio, M., Ghezzi, D., Bony, G., Alabastri, A., Deidda, G., Brondi, M., et al. (2012). High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat. Commun. 3:960. doi: 10.1038/ncomms1961
Daviaud, N., Friedel, R. H., and Zou, H. (2018). Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro 5:ENEURO.0219-18.2018. doi: 10.1523/ENEURO.0219-18.2018
Deverman, B. E., Pravdo, P. L., Simpson, B. P., Kumar, S. R., Chan, K. Y., Banerjee, A., et al. (2016). Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209. doi: 10.1038/nbt.3440
Deyle, D. R., and Russell, D. W. (2009). Adeno-associated virus vector integration. Curr. Opin. Mol. Ther. 11, 442–447.
Di Lullo, E., and Kriegstein, A. R. (2017). The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 18, 573–584. doi: 10.1038/nrn.2017.107
Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y., and Xu, T. (2005). Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483. doi: 10.1016/j.cell.2005.07.013
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Fiddes, I. T., Lodewijk, G. A., Mooring, M., Bosworth, C. M., Ewing, A. D., Mantalas, G. L., et al. (2018). Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356.e22–1369.e22. doi: 10.1016/j.cell.2018.03.051
Figueres-Oñate, M., García-Marqués, J., and López-Mascaraque, L. (2016). UbC-StarTrack, a clonal method to target the entire progeny of individual progenitors. Sci. Rep. 6:33896. doi: 10.1038/srep33896
Florio, M., Heide, M., Pinson, A., Brandl, H., Albert, M., Winkler, S., et al. (2018). Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife 7:e32332. doi: 10.7554/eLife.32332
Frank, J. A., Antonini, M. J., and Anikeeva, P. (2019). Next-generation interfaces for studying neural function. Nat. Biotechnol. 37, 1013–1023. doi: 10.1038/s41587-019-0198-8
Giandomenico, S. L., Mierau, S. B., Gibbons, G. M., Wenger, L. M. D., Masullo, L., Sit, T., et al. (2019). Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 22, 669–679. doi: 10.1038/s41593-019-0350-2
Grabundzija, I., Irgang, M., Mates, L., Belay, E., Matrai, J., Gogol-Doring, A., et al. (2010). Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 18, 1200–1209. doi: 10.1038/mt.2010.47
Grieger, J. C., and Samulski, R. J. (2005). Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 79, 9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005
Harrison, M. M., Jenkins, B. V., O’Connor-Giles, K. M., and Wildonger, J. (2014). A CRISPR view of development. Genes Dev. 28, 1859–1872. doi: 10.1101/gad.248252.114
Heide, M., Huttner, W. B., and Mora-Bermudez, F. (2018). Brain organoids as models to study human neocortex development and evolution. Curr. Opin. Cell Biol. 55, 8–16. doi: 10.1016/j.ceb.2018.06.006
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Iefremova, V., Manikakis, G., Krefft, O., Jabali, A., Weynans, K., Wilkens, R., et al. (2017). An organoid-based model of cortical development identifies non-cell-autonomous defects in wnt signaling contributing to miller-dieker syndrome. Cell Rep. 19, 50–59. doi: 10.1016/j.celrep.2017.03.047
Ivics, Z., Hackett, P. B., Plasterk, R. H., and Izsvak, Z. (1997). Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish and its transposition in human cells. Cell 91, 501–510. doi: 10.1016/s0092-8674(00)80436-5
Janssens, S., Schotsaert, M., Manganaro, L., Dejosez, M., Simon, V., Garcia-Sastre, A., et al. (2019). FACS-mediated isolation of neuronal cell populations from virus-infected human embryonic stem cell-derived cerebral organoid cultures. Curr. Protoc. Stem Cell Biol. 48:e65. doi: 10.1002/cpsc.65
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kadoshima, T., Sakaguchi, H., Nakano, T., Soen, M., Ando, S., Eiraku, M., et al. (2013). Self-organization of axial polarity, inside-out layer pattern and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. U S A 110, 20284–20289. doi: 10.1073/pnas.1315710110
Kalebic, N., Taverna, E., Tavano, S., Wong, F. K., Suchold, D., Winkler, S., et al. (2016). CRISPR/Cas9-induced disruption of gene expression in mouse embryonic brain and single neural stem cells in vivo. EMBO Rep. 17, 338–348. doi: 10.15252/embr.201541715
Kanton, S., Boyle, M. J., He, Z. S., Santel, M., Weigert, A., Sanchis-Calleja, F., et al. (2019). Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422. doi: 10.1038/s41586-019-1654-9
Kar, S., Loganathan, M., Dey, K., Shinde, P., Chang, H.-Y., Nagai, M., et al. (2018). Single-cell electroporation: current trends, applications and future prospects. J. Micromech. Microeng. 28:123002. doi: 10.1088/1361-6439/aae5ae
Karzbrun, E., Kshirsagar, A., Cohen, S. R., Hanna, J. H., and Reiner, O. (2018). Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 14, 515–522. doi: 10.1038/s41567-018-0046-7
Kawasaki, H., Iwai, L., and Tanno, K. (2012). Rapid and efficient genetic manipulation of gyrencephalic carnivores using in utero electroporation. Mol. Brain 5:24. doi: 10.1186/1756-6606-5-24
Kawasaki, H., Toda, T., and Tanno, K. (2013). In vivo genetic manipulation of cortical progenitors in gyrencephalic carnivores using in utero electroporation. Biol. Open 2, 95–100. doi: 10.1242/bio.20123160
Kebschull, J. M., and Zador, A. M. (2018). Cellular barcoding: lineage tracing, screening and beyond. Nat. Methods 15, 871–879. doi: 10.1038/s41592-018-0185-x
Kelava, I., and Lancaster, M. A. (2016). Stem cell models of human brain development. Cell Stem Cell 18, 736–748. doi: 10.1016/j.stem.2016.05.022
Kim, Y. G., Cha, J., and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U S A 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Kumar, M., Keller, B., Makalou, N., and Sutton, R. E. (2001). Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 12, 1893–1905. doi: 10.1089/104303401753153947
Lancaster, M. A., Corsini, N. S., Wolfinger, S., Gustafson, E. H., Phillips, A. W., Burkard, T. R., et al. (2017). Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666. doi: 10.1038/nbt.3906
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Li, Y., Muffat, J., Omer, A., Bosch, I., Lancaster, M. A., Sur, M., et al. (2017). Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 20, 385.e3–396.e3. doi: 10.1016/j.stem.2016.11.017
LoTurco, J., Manent, J. B., and Sidiqi, F. (2009). New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb. Cortex 19, i120–i125. doi: 10.1093/cercor/bhp033
Mali, P., Yang, L. H., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Mansour, A. A., Gonçalves, J. T., Bloyd, C. W., Li, H., Fernandes, S., Quang, D., et al. (2018). An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441. doi: 10.1038/nbt.4127
Matsui, T., Nieto-Estevez, V., Kyrychenko, S., Schneider, J. W., and Hsieh, J. (2017). Retinoblastoma protein controls growth, survival and neuronal migration in human cerebral organoids. Development 144, 1025–1034. doi: 10.1242/dev.143636
McClintock, B. (1950). The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U S A 36, 344–355. doi: 10.1073/pnas.36.6.344
McKenna, A., and Gagnon, J. A. (2019). Recording development with single cell dynamic lineage tracing. Development 146:dev169730. doi: 10.1242/dev.169730
Merten, O. W., Hebben, M., and Bovolenta, C. (2016). Production of lentiviral vectors. Mol. Ther. Methods Clin. Dev. 3:16017. doi: 10.1038/mtm.2016.17
Mora-Bermúdez, F., Badsha, F., Kanton, S., Camp, J. G., Vernot, B., Kohler, K., et al. (2016). Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife 5:e18683. doi: 10.7554/eLife.18683
Ni, J., Clark, K. J., Fahrenkrug, S. C., and Ekker, S. C. (2008). Transposon tools hopping in vertebrates. Brief. Funct. Genomic. Proteomic. 7, 444–453. doi: 10.1093/bfgp/eln049
Ogawa, J., Pao, G. M., Shokhirev, M. N., and Verma, I. M. (2018). Glioblastoma model using human cerebral organoids. Cell Rep. 23, 1220–1229. doi: 10.1016/j.celrep.2018.03.105
Otani, T., Marchetto, M. C., Gage, F. H., Simons, B. D., and Livesey, F. J. (2016). 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell 18, 467–480. doi: 10.1016/j.stem.2016.03.003
Pasca, A. M., Sloan, S. A., Clarke, L. E., Tian, Y., Makinson, C. D., Huber, N., et al. (2019). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678. doi: 10.1038/nmeth.3415
Pasca, S. P. (2019). Assembling human brain organoids. Science 363, 126–127. doi: 10.1126/science.aau5729
Pollen, A. A., Bhaduri, A., Andrews, M. G., Nowakowski, T. J., Meyerson, O. S., Mostajo-Radji, M. A., et al. (2019). Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743.e717–756.e717. doi: 10.1016/j.cell.2019.01.017
Quadrato, G., Brown, J., and Arlotta, P. (2016). The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat. Med. 22, 1220–1228. doi: 10.1038/nm.4214
Ran, F. A., Hsu, P. D., Lin, C. Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. doi: 10.1016/j.cell.2013.08.021
Renner, M., Lancaster, M. A., Bian, S., Choi, H., Ku, T., Peer, A., et al. (2017). Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 36, 1316–1329. doi: 10.15252/embj.201694700
Saito, T., and Nakatsuji, N. (2001). Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240, 237–246. doi: 10.1006/dbio.2001.0439
Schubert, R., Trenholm, S., Balint, K., Kosche, G., Cowan, C. S., Mohr, M. A., et al. (2018). Virus stamping for targeted single-cell infection in vitro and in vivo. Nat. Biotechnol. 36, 81–88. doi: 10.1038/nbt.4034
Schwarz, M. K., and Remy, S. (2019). Rabies virus-mediated connectivity tracing from single neurons. J. Neurosci. Methods 325:108365. doi: 10.1016/j.jneumeth.2019.108365
Shull, G., Haffner, C., Huttner, W. B., Kodandaramaiah, S. B., and Taverna, E. (2019). Robotic platform for microinjection into single cells in brain tissue. EMBO Rep. 20:e47880. doi: 10.15252/embr.201947880
Sun, N., Petiwala, S., Wang, R., Lu, C., Hu, M., Ghosh, S., et al. (2019). Development of drug-inducible CRISPR-Cas9 systems for large-scale functional screening. BMC Genomics 20:225. doi: 10.1186/s12864-019-5601-9
Suzuki, I. K., Gacquer, D., Van Heurck, R., Kumar, D., Wojno, M., Bilheu, A., et al. (2018). Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell 173, 1370.e1316–1384.e1316. doi: 10.1016/j.cell.2018.03.067
Suzuki, K., and Belmonte, J. C. I. (2018). In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 63, 157–164. doi: 10.1038/s10038-017-0352-4
Suzuki, K., Tsunekawa, Y., Hernandez-Benitez, R., Wu, J., Zhu, J., Kim, E. J., et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. doi: 10.1038/nature20565
Szczurkowska, J., Cwetsch, A. W., dal Maschio, M., Ghezzi, D., Ratto, G. M., and Cancedda, L. (2016). Targeted in vivo genetic manipulation of the mouse or rat brain by in utero electroporation with a triple-electrode probe. Nat. Protoc. 11, 399–412. doi: 10.1038/nprot.2016.014
Trujillo, C. A., Gao, R., Negraes, P. D., Gu, J., Buchanan, J., Preissl, S., et al. (2019). Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25, 558.e557–569.e557. doi: 10.1016/j.stem.2019.08.002
Velasco, S., Kedaigle, A. J., Simmons, S. K., Nash, A., Rocha, M., Quadrato, G., et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. doi: 10.1038/s41586-019-1289-x
Vigna, E., and Naldini, L. (2000). Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2, 308–316. doi: 10.1002/1521-2254(200009/10)2:5<308::AID-JGM131>;3.0.CO;2-3
Wilson, M. H., Coates, C. J., and George, A. L. Jr. (2007). PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15, 139–145. doi: 10.1038/sj.mt.6300028
Zhou, T., Tan, L., Cederquist, G. Y., Fan, Y., Hartley, B. J., Mukherjee, S., et al. (2017). High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274.e275–283.e275. doi: 10.1016/j.stem.2017.06.017
Keywords: brain organoids, genetic modification, adeno-associated virus, electroporation, lentivirus, transposon, CRISPR/Cas9
Citation: Fischer J, Heide M and Huttner WB (2019) Genetic Modification of Brain Organoids. Front. Cell. Neurosci. 13:558. doi: 10.3389/fncel.2019.00558
Received: 14 October 2019; Accepted: 04 December 2019;
Published: 17 December 2019.
Edited by:
Alysson Renato Muotri, University of California, San Diego, United StatesReviewed by:
Marina Guizzetti, Oregon Health & Science University, United StatesHarold MacGillavry, Utrecht University, Netherlands
Copyright © 2019 Fischer, Heide and Huttner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Heide, heide@mpi-cbg.de; Wieland B. Huttner, huttner@mpi-cbg.de
† These authors share first authorship
 Jan Fischer
Jan Fischer Michael Heide
Michael Heide Wieland B. Huttner
Wieland B. Huttner