- 1Department of Spine Surgery, Xiangya Hospital, Central South University, Changsha, China
- 2Key Laboratory of Organ Injury, Aging and Regenerative Medicine of Hunan Province, Xiangya Hospital, Central South University, Changsha, China
- 3Department of Obstetrics, Xiangya Hospital, Central South University, Changsha, China
- 4Department of Sports Medicine, Xiangya Hospital, Central South University, Changsha, China
Neuropathic pain (NP) is among the most intractable comorbidities of spinal cord injury. Dysregulation of non-coding RNAs has also been implicated in the development of neuropathic pain. Here, we identified a novel lncRNA, PKIA-AS1, by using lncRNA array analysis in spinal cord tissue of spinal nerve ligation (SNL) model rats, and investigated the role of PKIA-AS1 in SNL-mediated neuropathic pain. We observed that PKIA-AS1 was significantly upregulated in SNL model rats and that PKIA-AS1 knockdown attenuated neuropathic pain progression. Alternatively, overexpression of PKIA-AS1 was sufficient to induce neuropathic pain-like symptoms in uninjured rats. We also found that PKIA-AS1 mediated SNL-induced neuropathic pain by directly regulating the expression and function of CDK6, which is essential for the initiation and maintenance of neuroinflammation and neuropathic pain. Therefore, our study identifies PKIA-AS1 as a novel therapeutic target for neuroinflammation related neuropathic pain.
Introduction
Neuropathic pain (NP) is among the most intractable comorbidities of spinal cord injury (SCI). It can manifest as multiple forms of chronic pain in areas where skin sensation is lost below the injury plane (Dones and Levi, 2018). The incidence of NP in patients with SCI is increasing annually (Wayne, 2018). There are currently no curative therapies for SCI-associated NP, and clinical management is limited to symptomatic treatment. Further, treatment efficacy is often not ideal, which seriously affects patient quality of life. Indeed, many patients experience depression and anxiety, develop drug addictions, and even contemplate or attempt suicide.
The release of inflammatory cytokines from activated astrocytes and microglia is strongly implicated in the development of neuropathic pain (McKelvey et al., 2015). As inhibition of neuroinflammation by inactivation of astrocytes and microglia can ameliorate pain-related behavior (Walters, 2014). In addition, genetic deletion of cell cycle-related proteins such as cyclin-dependent kinases (CDKs) has also been shown to reverse pain-related behavior (Gwak and Hulsebosch, 2011). Nociception is an integral component of host defense, but prolonged nociceptive input can drive central neuroinflammation and induce neuroplastic changes within pain pathways (Gwak et al., 2012). It has been suggested that spinal injury can activate nociceptor-mediated host defense responses via neuroinflammatory signaling to produce chronic pain (Coraggio et al., 2018).
Non-coding RNAs (ncRNAs), including microRNAs and long non-coding RNAs (lncRNAs), contribute to diverse biological processes by regulating the expression of mRNA targets at the post-transcriptional level (Bali and Kuner, 2014; Zhou et al., 2017) by targeting several pain pathways (Tan et al., 2015; Plassais et al., 2016; Shao et al., 2018). Dysregulation of non-coding RNAs has also been implicated in the development of neuropathic pain (Liu et al., 2017; Vieira et al., 2018). For example, lncRNA BC168687 is associated with diabetic neuropathic pain (Liu et al., 2018), while lncRNA MRAK009713, NEAT1, and XIST can regulate chronic constriction injury-induced neuropathic pain (Li et al., 2017; Xia et al., 2018; Yan et al., 2018).
Here, we identified a novel lncRNA, PKIA-AS1, by using lncRNA array analysis in spinal cord tissue of spinal nerve ligation (SNL) model rats, and investigated the role of PKIA-AS1 in SNL-mediated neuropathic pain. We observed that PKIA-AS1 was significantly upregulated in SNL model rats and that PKIA-AS1 knockdown attenuated neuropathic pain progression. Alternatively, overexpression of PKIA-AS1 was sufficient to induce neuropathic pain-like symptoms in uninjured rats. We also found that PKIA-AS1 mediated SNL-induced neuropathic pain by directly regulating the expression and function of CDK6, which is essential for the initiation and maintenance of neuroinflammation and neuropathic pain. Therefore, our study identifies PKIA-AS1 as a novel therapeutic target for neuroinflammation related neuropathic pain.
Materials and Methods
Animal Model
Male Sprague-Dawley (SD) rats (∼200 g) were obtained from the Experimental Animal Central of Central South University. The animal experiments were approved by the Animal Care and Use Committee of Central South University. Spinal nerve ligation (SNL)-induced neuropathic pain model was established as previous described (Zhang et al., 2013). Briefly, following anesthetized with 1% sodium pentobarbital (40 mg/kg i.p.), the fifth lumbar (L5) spinal nerves were isolated and ligated tightly with 6–0 silk suture. For sham-operated mice, rats received identical surgical procedure without ligation. Mechanical withdrawal threshold (MWT) and thermal withdrawal latency (TWL) were used to validate the SNL model.
Animal Infection
To test the effect of lentivirus shPKIA-AS1 (Lv-shPKIA-AS1, sequence: CCGGGACGACCCTGACCATGGTCGCGATATCTCGAGATATTTGAGGTCAGGGTCGTCAATTTG, Sigma Aldrich), SNL rats were received intrathecal injections of either Lv-shPKIA-AS1 (MOI = 100) or the lentivirus with scramble sequence on the third day after surgery.
To assess the effect of PKIA-AS1 overexpression, rats were received intrathecal injections of either Lv-PKIA-AS1 [multiplicity of infection (MOI) = 100] or the lentivirus with scramble sequence on the fifth day of observation.
To assess the effect of CDK6 overexpression, rats were received intrathecal injections of either Lv-CDK6 (MOI = 100) or the lentivirus with scramble sequence on the third day after surgery. Intrathecal injection process was performed as previously described (Zhang et al., 2013). Briefly, rats were placed in a prone position after anesthesia. After creating a small opening at the intervertebral space between the L4–L6 vertebrae, a sterile PE10 intrathecal catheter was inserted through the opening into the lumbar enlargement. After catheterization, all rats were allowed to recover for two days before being used in other experiments.
Pain Threshold Assessment
Mechanical allodynia was evaluated by paw withdrawal threshold (PWT) using von Frey filaments. Briefly, rats were put into a transparent plastic box (22 cm × 12 cm × 22 cm). The box has a metal mesh floor. The calibrated von Frey filaments (IITC, Woodland Hills, CA, United States) were used to make pressure on the plantar surface of rat hind paw. The investigators recorded the size of the filaments when paw withdrawal. Paw withdrawal latency (PWL) was used to evaluated thermal hyperalgesia by the Plantar Test Instrument (Hargreave’s Method). The hind paws were tested alternately at 5-min intervals. The investigators recorded the duration between stimuli and paw withdrawal. The cut-off time was at 30 s. All tests were performed blindly.
LncRNA Microarray Analysis
On the 15th day after spinal nerve ligation (SNL) or sham operation, rats were anesthetized with pentobarbital sodium (150 mg/kg IV) and their spinal cord tissues (L5) were removed and frozen by liquid nitrogen immediately. RNeasy Mini Kit (Qiagen, GmBH, Hilden, Germany) was used to extract total RNA from spinal cord tissues according to the manufacturer’s instructions. NanoDrop 2000 spectrophotometer was used to quantify purified total RNA. To analyze lncRNA expression profiles (Arraystar Rat LncRNA Expression Array V2.0), the purified total RNA was sent to Novogene Co. Ltd. (Beijing, China). Differentially expressed lncRNAs were identified through fold change (>2) and P-value (<0.05).
RNA Extraction and qRT-PCR
TRIzol reagent (Invitrogen) was used to extract total RNA from cells or tissues. Maxima First Strand cDNA Synthesis kit (cat no. K1642; Thermo Fisher Scientific, Inc.) was used for reverse transcription according to the manufacturer’s protocol. Quantitative PCR amplification was performed with an CFX96 TouchTM Deep Well Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). Expression of lncRNAs, PKIA-AS1, IL-1β, IL-6, IL-12, TNFα, and CDK-6 was detected via UltraSYBR Mixture (cat. no. CW2602; CWBio) according to the manufacturer’s protocol. QPCR was performed at the condition: 95.0°C for 3 min, and 39 circles of 95.0°C for 10 s and 60°C for 30 s. Expression of β-actin was used as an endogenous control. Relative gene expression was calculated using 2-ΔΔCt method. The real-time PCR primer sequences are shown in Supplementary Table S1.
Western Blot
Radio immunoprecipitation assay (RIPA) lysis buffer (Boster, Wuhan, China) was used to extract protein from spinal cord tissues (L5). BCA Protein assay kit (Thermo Scientific, Waltham, MA, United States) was used to measure protein concentrations. Proteins (60 μg) were separated by 10% SDS/PAG. The membranes were blocked by 5% non-fat milk for 30 min at room temperature and then immunoblotted with the following primary antibodies: CDK6 antibody (cat no. 13331, Rabbit monoclonal, diluted at 1:1000), DNMT1 antibody (cat no. 5032, Rabbit monoclonal, diluted at 1:1000), DNMT3A antibody (cat no. 32578, Rabbit monoclonal, diluted at 1:1000), DNMT3B antibody (cat no. 57868, Rabbit monoclonal, diluted at 1:1000), GAPDH antibody (cat no. 5174, Rabbit monoclonal, diluted at 1:1000). All antibodies were purchased from Cell Signaling Technology. Membranes were then incubated with peroxidase-conjugated secondary antibody, and specific bands were detected with a Bio-Rad (Hercules, CA, United States) imaging system.
Immunofluorescence Assays
Spinal cord tissues were cut into twenty-μm thick sections for immunofluorescence assays. After permeabilized with 0.5% Triton-100, the sections were blocked with BSA for 60 min at 37°C. The sections then were incubated with primary antibody (cat no. ab68428, anti-GFAP, 1:500; cat no. ab15690, anti-Iba-1, 1:200, Abcam, Cambridge, United Kingdom) at 4°C overnight. After that, sections were washed by PBS and secondary antibody was added to incubation at 37°C for 1 h. The coverslips were stained with DAPI (1:1000, Santa Cruz Biotechnology) for 2 min at room temperature and mounted. Fluorescence microscope (Nikon ECLIPSE 80i, Nikon Corporation, Tokyo, Japan) was used to immunofluorescence acquire images. ImageJ (version 1.8) was used to quantify the fluorescence intensity on 8 sections per animal at 40× magnification.
Proteomics Analysis
Total proteins were extracted and purified from fresh spinal cord tissues (L5) of Lv-PKIA-AS1 infected mice and the control mice using a ReadyPrepTM Protein Extraction kit (Bioscience) following the procedure recommended by the manufacturer. The proteomics analysis was performed as previously described (Yang et al., 2014). Protein samples were separated by two-dimensional polyacrylamide gel electrophoresis (2-DE). Samples containing 150 μg of protein were diluted to 450 μl with rehydration solution and used for isoelectric focusing. The proteins were electrophoresed in SDS–PAGE and then stained with coomassie brilliant blue dye. Spot-detect and determine the quantity were analyzed by DeCyder software version 6.5 (GE). The statistical significance was assessed by using a one-way ANOVA analysis. Protein spots were selected as the mean ratio was greater than 1.5-fold or less than -1.5-fold.
Cell Culture and Cell Infection
PC12 cells were purchased from the American Type Culture Collection (Manassas, VA, United States). The cells were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, United States) and 1% penicillin/streptomycin in a 5% CO2 incubator at 37°C.
The cells were infected with the lentiviral transduction particles for PKIA-AS1. Briefly, when the cells confluence reached 80–90% in 6-well plates, the cells were infected with lentivirus [multiplicity of infection (MOI) = 50] in the presence of 5 mg/ml polybrene (Sigma-Aldrich, St. Louis, MO, United States) for 48 h.
Luciferase Reporter Assay
To test the promoter activity, CDK6 promoter was cloned into the pGL3-Basic vector using the Fast-FusionTM Cloning Kit (FulenGen, Guangzhou, China). Luciferase reporter constructs were co-transfected with pRL-TK (Promega) into PC12 cells, followed by infection with Lv-PKIA-AS1. Dual Luciferase Reporter Assay Kit (Promega) was used to measure the luciferase activity according to the manufacturer’s instruction.
RNA-Binding Protein Immunoprecipitation Assay
RNA-binding protein immunoprecipitation assay was conducted using Magna RIP Kit (EMD Millipore, Billerica, MA, United States) according to the manufacturer’s instruction. After the cells were lysed by RIP lysis buffer, magnetic beads conjugated to human anti-Ago2 antibody (Millipore) or control antibody (normal mouse IgG; Millipore) were added into cell lysate for incubation at 4°C overnight. CDK6 and PKIA-AS1 expression were measured by qRT-PCR.
Quantitative Methylation-Specific PCR
CDK6 promoter (-2000–+150) methylation status was measured by MSP. Genomic DNA (1 μg per sample) extracted by the Qiagen FFPE DNA Kit (Qiagen, CA, United States) was modified with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo, Orange County, CA, United States) according to the manufacturer’s instructions. Bisulfate-treated DNA was used to perform quantitative methylation-specific PCR (MSP). The qPCR steps were as following: initial denaturation at 95.0°C for 3 min; 39 cycles of 95.0°C for 10 s and 60°C for 30 s. The primers were as following: methylated-specific primer, forward, 5′-AGGCGGTTGTAGTTTTTGTAGTC-3′, reverse, 5′-ATTATTATTATTACTTTCCCACGCT-3′; unmethylated-specific primer, forward, 5′-GGAGGTGGTTGTAGTTTTTGTAGTT-3′, reverse, 5′-ATTATTATTATTACTTTCCCACACT-3′.
ELISA
Rat IL-1β, IL-6, IL-12, and TNFα ELISA kit was purchased from Cwbiotech (Beijing, China). The lumbar spinal cord segments (L5) were removed and were homogenized in a lysis buffer. ELISA was performed according to manufacturer’s protocol.
Statistical Analysis
The experiments repeated at least three times. Data are expressed as means ± SEM. Statistical analyses were processed on GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, United States). Student’s t-test and one-way ANOVA with Bonferroni test were used to assess the statistical significance of the differences between two groups and multiple groups, respectively. Two-way repeated measure ANOVA with Bonferroni post-test for multiple comparisons at each time point was used to analyze PWT and PWL. P less than 0.05 was considered statistically significant.
Results
Identification of Differentially Expressed lncRNAs in a Rat Spinal Nerve Ligation (SNL) Model of NP
To investigate the role of lncRNAs in neuropathic pain induced by spinal cord injury, we established a SNL rat model (Figures 1A,B). Total RNA was extracted from spinal cord tissues of these SNL rats and sham-operated control rats to perform lncRNA array analysis. We successfully identified 1259 downregulated lncRNAs and 2473 upregulated lncRNAs in SNL rats compared to controls (Figure 1C). Table 1 presents the detail information on the top 10 most upregulated and top 10 most downregulated lncRNAs. We then investigated the profiles of the 10 most highly upregulated lncRNAs in the spinal cord of SNL rats by real-time quantitative PCR. These substantially upregulated lncRNAs, included PKIA-AS1 (fold change >15), SNHG4 (fold change >5), STAC3 (fold change >5), and CIRBP-AS1 (fold change >5) (Figure 1D). Specific overexpression in the SNL model strongly suggests that these differentially expressed lncRNAs are involve in the pathogenesis of neuropathic pain. In this study, we focused on the contributions of PKIA-AS1 to SNL-associated NP development as it exhibited the greatest upregulation.
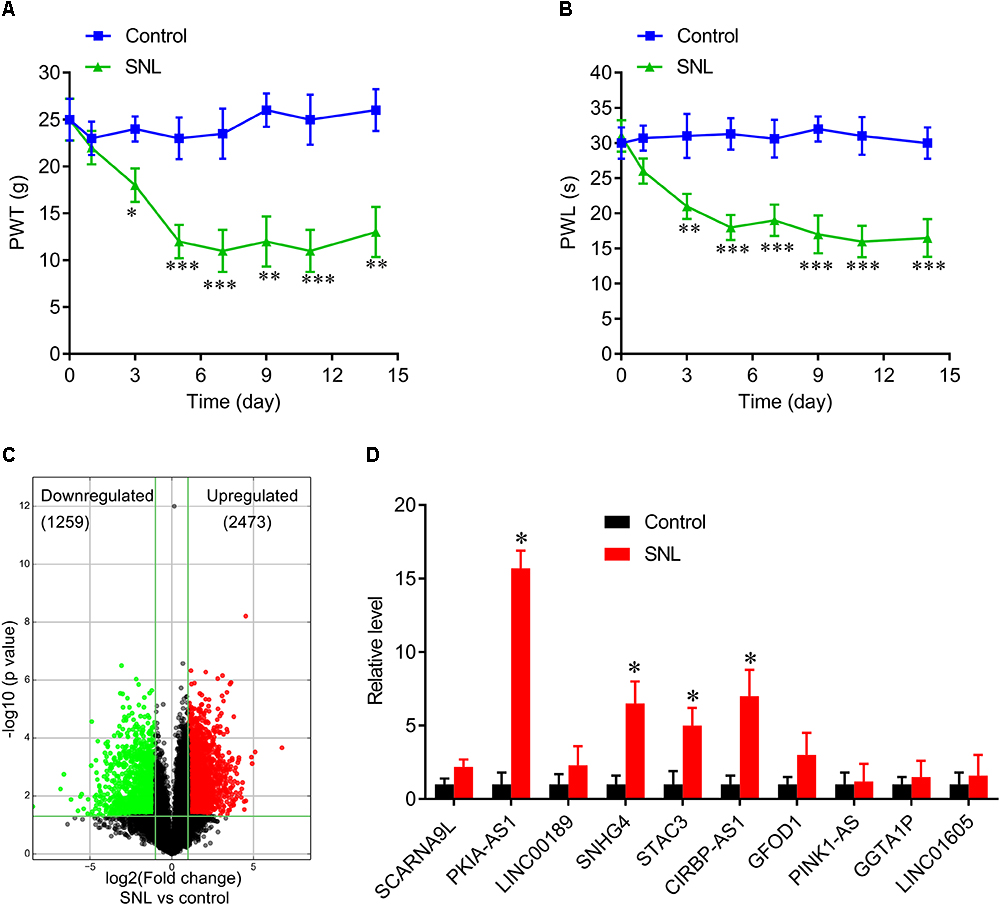
Figure 1. Differentially expressed lncRNAs in spinal cord of SNL rats. (A,B) Spinal never ligation induced mechanical hyperalgesia (A) and thermal hyperalgesia (B) in rats. N = 8 for each group, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. control group. (C) Volcano plot shown the downregulated and upregulated lncRNAs. (D) Real-time PCR data confirmed the expressions of top 10 upregulated lncRNAs in spinal cord of SNL rats and control group. ∗P < 0.05 vs. control group. SNL, spinal never ligation; PWT, paw withdrawal threshold; PWL, paw withdrawal latency; lncRNA, long non-coding RNA.
Effects of PKIA-AS1 on Spinal Cord Ligation-Induced Neuropathic Pain
Similar to NP development, expression of PKIA-AS1 increased over time following SNL (Figure 2A). Therefore, we investigated whether reducing PKIA-AS1 expression in SNL rats by shRNA knockdown can attenuate SNL induced neuropathic pain. Real-time PCR demonstrated that PKIA-AS1 expression in spinal cord of SNL rats was markedly downregulated 3 days following infection with a lentivirus vector targeting PKIA-AS1 (Lv-PKIA-AS1) compared with the SNL rats infected with control lentivirus (P < 0.05, N = 5; Figure 2B).
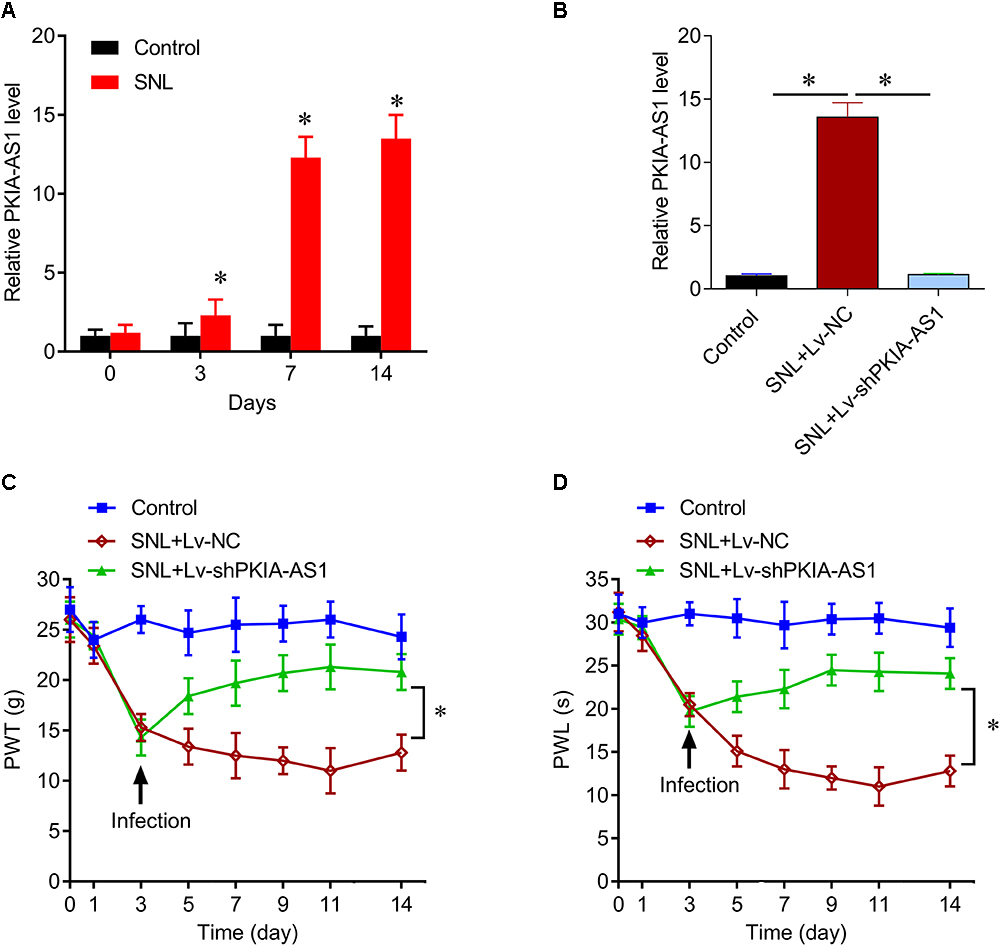
Figure 2. lncRNA PKIA-AS1 expression is increased in the SNL spinal cord and contributes to the SNL-induced neuropathic pain. (A) Real-time PCR data show the expressions of PKIA-AS1 in the SNL spinal cord on different time points. (B) Real-time PCR data show that the expressions of PKIA-AS1 in the SNL spinal cord treated with shPKIA-AS1 lentivirus were significantly lower than that of the SNL+Lv-NC group. (C,D) shRNA silencing of PKIA-AS1 alleviates mechanical hyperalgesia (C) and thermal hyperalgesia (D) in the SNL rats. N = 8 for each group, ∗P < 0.05 vs. control group. SNL, spinal never ligation; PWT, paw withdrawal threshold; PWL, paw withdrawal latency; lncRNA, long non-coding RNA.
The effects of PKIA-AS1 shRNA on both mechanical and thermal hyperalgesia were tested by measuring the paw withdrawal threshold (PWT) and latency (PWL). Animals developed progressive mechanical hyperalgesia following SNL as evidenced by reduced PWT that plateaued on the 7th day after surgery. At 3 days post-surgery, PWT was significantly lower in rats earmarked for Lv-NC and Lv-shPKIA-AS1 infection (SNL+Lv-NC and SNL+Lv-shPKIA-AS1 groups, respectively) compared to sham-operated controls. The PWT of the SNL+Lv-shPKIA-AS1 group rose significantly above that in the SNL+Lv-NC group following infection (P < 0.05, N = 8 rats per group), indicating reduced hyperalgesia. However, the PWT of the SNL+ Lv-shPKIA-AS1 group remained slightly lower than that of the control group (Figure 2C). Similarly, the PWL was significantly shorter in the SNL+Lv-NC, and SNL+ Lv-shPKIA-AS1 groups compared with the control group on the 3th day after surgery, but was significantly higher in the SNL+Lv-shPKIA-AS1 group than the SNL+Lv-NC group following infection (P < 0.05, N = 8 rats per group) (Figure 2D). These results suggest that knockdown of PKIA-AS1 attenuates SNL-induced neuropathic pain.
Next, we assessed whether PKIA-AS1 overexpression alone is sufficient to cause neuropathic pain–like symptoms in uninjured control rats. Seven days after infection with Lv-PKIA-AS1, real-time PCR showed that expression of PKIA-AS1 in the spinal cord was fivefold higher than in control rats or rats infected with the control lentivirus (P < 0.05, N = 5) (Figure 3A). The PWT in rats infected with Lv-PKIA-AS1 started to decrease by the second day post-infection and was even lower on days 4, 6, and 9 (P < 0.01, N = 8 rats per group) compared with rats infected with control lentivirus (Figure 3B), indicating progressive development of hyperalgesia. Similarly, PWL was significantly shorter in rats following Lv-PKIA-AS1 infection compared to rats infected with the control lentivirus (P < 0.05, N = 8 rats for each group) 2, 4, 6, and 9 days after infection (Figure 3C). These results suggest that overexpression of PKIA-AS1 alone is sufficient to produce hyperalgesia.
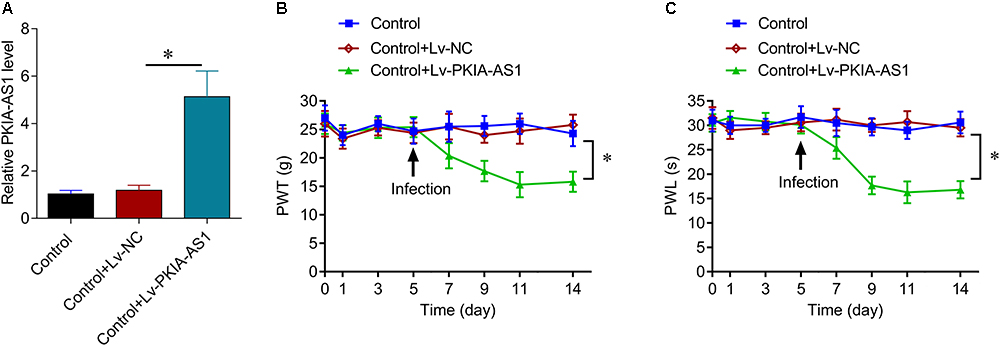
Figure 3. Overexpression of PKIA-AS1 is sufficient to cause pain behavior in rats. (A) Real-time PCR show that the expression of PKIA-AS1 in the control rats 6 days after infection of PKIA-AS1 lentivirus was significantly higher than that of the control group. ∗P < 0.05. (B,C) Overexpression of PKIA-AS1 produced mechanical hyperalgesia (B) and thermal hyperalgesia (C) in the control rats. N = 8 for each group, ∗P < 0.05. PWT, paw withdrawal threshold; PWL, paw withdrawal latency.
PKIA-AS1-Induced Neuropathic Pain Is Associated With Neuroinflammation
To investigate the effects of PKIA-AS1 on neuroinflammation associated with NP, we evaluated the activation of astrocyte and microglia as well as the expression levels of inflammatory cytokines interleukin (IL)-6, IL-1β, IL-12 and tumor necrosis factor (TNF-)-α expression in spinal cord. As expected, SNL induced astrocytic and microglial activation as evaluated by increased GFAP and Iba-1 expression levels, respectively, while PKIA-AS1 knockdown substantially reduced glial activation (Figure 4A). PKIA-AS1 knockdown also inhibited SNL-mediated upregulation of IL-6, IL-1β, and TNF-α at both mRNA and protein levels in spinal cord (Figures 4B,C). These findings suggest that knockdown of PKIA-AS1 represses neuropathic pain by inhibiting neuroinflammation.
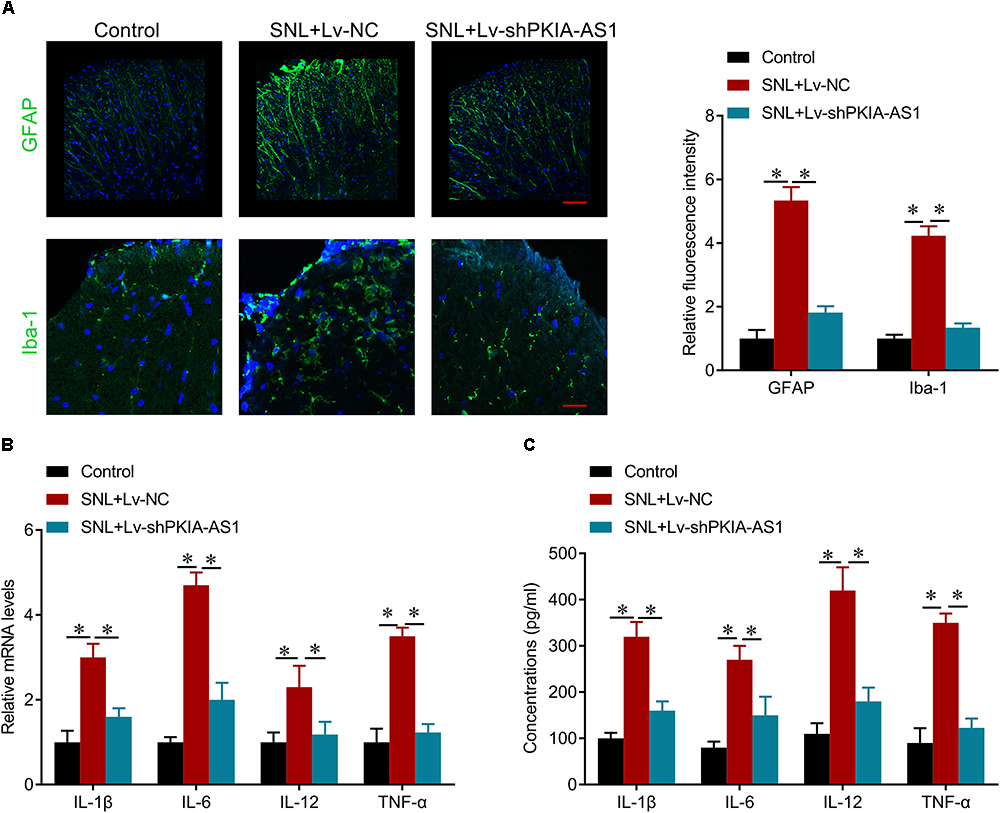
Figure 4. shRNA silencing of PKIA-AS1 repressed neuroinflammation in SNL rats. (A) Immunofluorescence assay results show that silencing of PKIA-AS1 repressed SNL-mediated astrocyte and microglia activation evaluated by GFAP and Iba-1, respectively. Scale bar: 100 μm, N = 8 for each group. (B) mRNA levels of IL-1β, IL-6, IL-12, and TNF-α in L5 spinal cord tissues in the rats 6 days after infection of PKIA-AS1 lentivirus. (C) The protein levels of IL-1β, IL-6, IL-12, and TNF-α in the L5 spinal cord of rats were tested by ELISA at postoperative day 6. N = 8 for each group; ∗P < 0.05.
PKIA-AS1 Interacts With CDK6
The potential mechanisms underlying regulation of neuropathic pain by PKIA-AS1 were then investigated. To identify possible targets of PKIA-AS1, we screened numerous differentially expressed proteins in spinal cord tissues after PKIA-AS1 overexpression (Figure 5A). The detail information on the top 10 most upregulated and downregulated proteins is listed in Table 2. One of the most upregulated proteins is CDK6, which has been strongly implicated in inflammation (Handschick et al., 2014). We then confirmed the expression of CDK6 in spinal cord tissues from rats infected with PKIA-AS1. Indeed, Lv-PKIA-AS1 infection induced CDK6 overexpression at both mRNA and protein levels (Figures 5B,C). We further demonstrated that Lv-PKIA-AS1 infection upregulated CDK6 gene promotor activity (Figure 5D). We next tested whether PKIA-AS1 interacts with CDK6 promoter by conducting RNA pulldown assays in PC12 cells infected with Lv-PKIA-AS1 or control lentivirus. Both PKIA-AS1 and CDK6 promoter were more abundant in the Ago2 pellet than in the IgG pellet (Figure 5E), demonstrating that the CDK6 promoter interacts PKIA-AS1. We further tested how PKIA-AS1 regulates CDK6 promoter activity. The methylation status of CDK6 promoter was evaluated by quantitative methylation-specific PCR (qMSP) of the PC12 cells following Lv-PKIA-AS1 infection. Overexpression of PKIA-AS1 downregulated methylation of CDK6 promoter, suggesting that enhanced CDK6 expression in the SNL spinal cord is mediated by promoter hypomethylation induced by PKIA-AS1 (Figure 5F). In addition, PKIA-AS1 overexpression significantly inhibited DNMT1 expression, but not DNMT3A and DNMT3B expression (Figure 5G), suggesting that PKIA-AS1 regulates CDK6 expression by decreasing DNMT1-catalyzed methylation of CDK6 promoter.
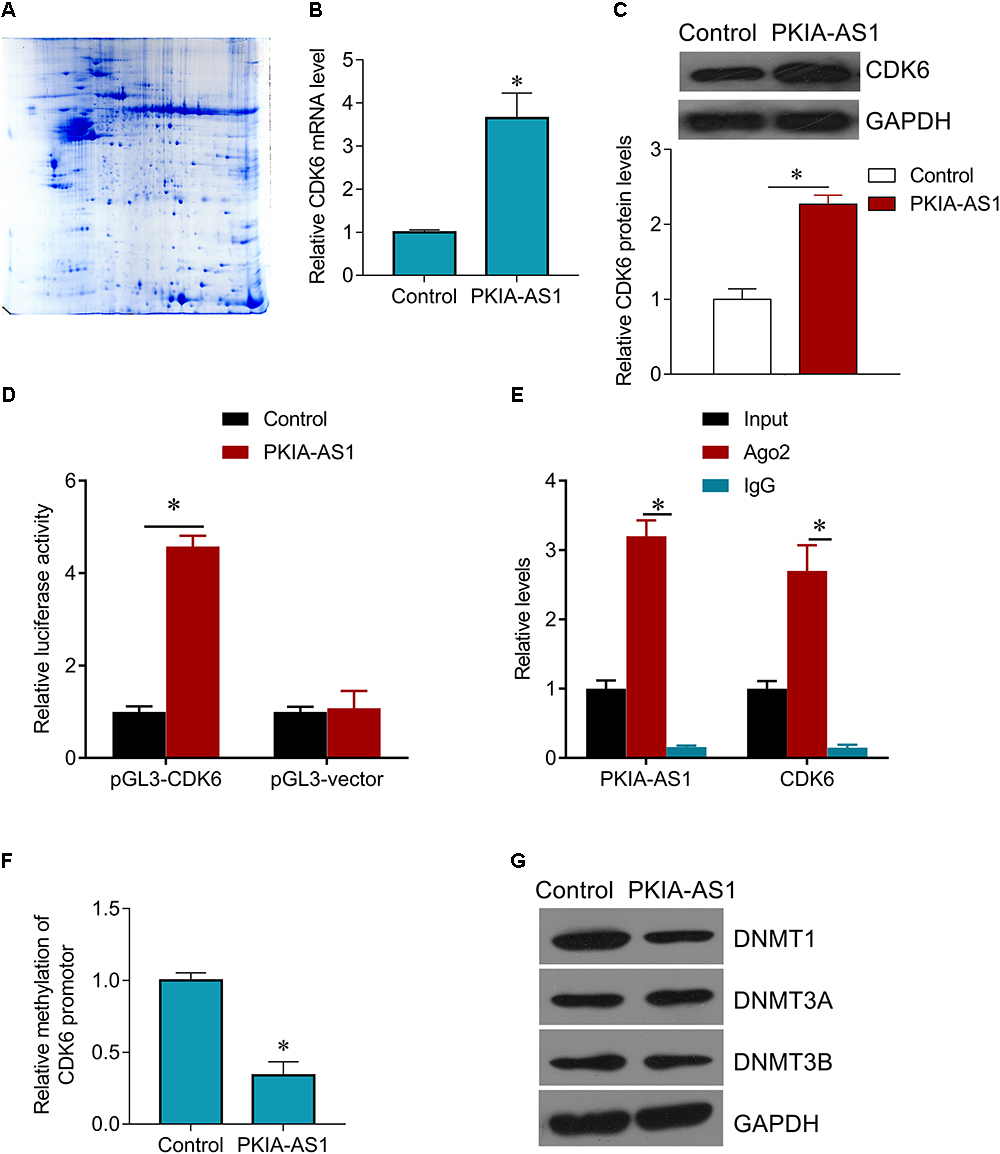
Figure 5. PKIA-AS1 epigenetically regulates CDK6 by hypomethylating its promotor. (A) The differential protein spots from 2-DE gels by differential proteomics analysis. The gel was stained with coomassie brilliant blue. 2-DE, two-dimensional polyacrylamide gel electrophoresis. (B) CDK6 mRNA expression in PC12 cells infected with Lv-PKIA-AS1. (C) CDK6 protein expression in PC12 cells infected with Lv-PKIA-AS1. (D) Dual luciferase assay shown that PKIA-AS1 induced CDK6 promotor activity. (E) The correlation between PKIA-AS1, CDK6 promotor and Ago2 was detected by RIP assay. Cellular lysates were immunoprecipitated using Ago2 antibody or IgG. PKIA-AS1 and CDK6 promotor expression was tested by qRT-PCR. (F) PC12 cells were infected with PKIA-AS1 lentivirus. qMSP was perform to analyze CDK6 promoter methylation. (G) DNMT1, DNMT3a, and DNMT3b protein levels were assessed by western blot in PC12 cells infected with Lv-PKIA-AS1; ∗P < 0.05.
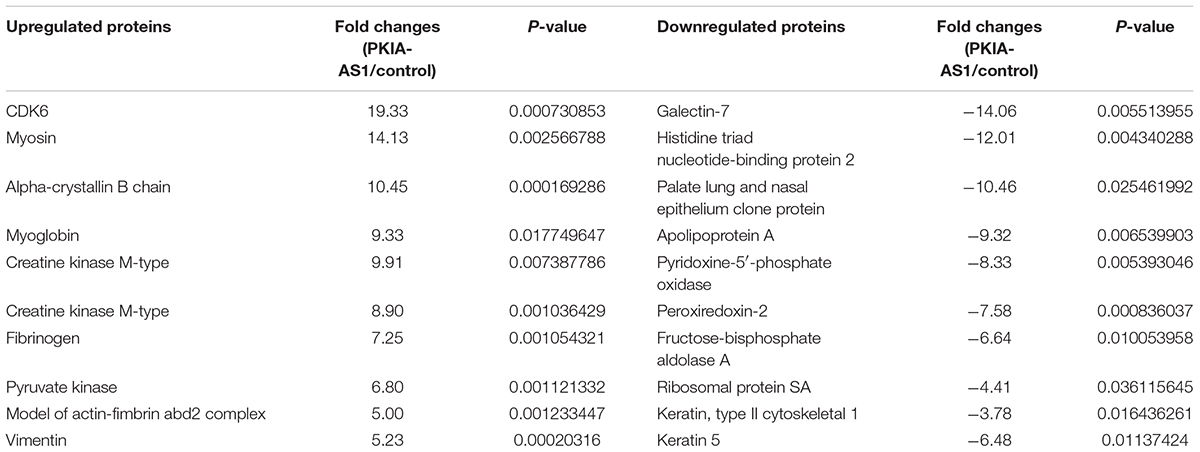
Table 2. The detail information of the top 10 most up-regulated and 10 most down-regulated proteins.
PKIA-AS1 Regulates Neuropathic Pain Progression Through CDK6
We further investigated whether PKIA-AS1 controls neuropathic pain via CDK6. Expression of CDK6 was significantly increased in SNL model rats compared with control rats, whereas CDK6 expression was inhibited by PKIA-AS1 knockdown. In addition, PKIA-AS1 knockdown-mediated inhibition of CDK6 was reversed by Lv-CDK6 infection (Figures 6A,B). We found that Lv-CDK6 infection reversed the attenuation of mechanical allodynia and thermal hyperalgesia by Lv-shPKIA-AS1 infection following SNL (Figures 6C,D). Moreover, Lv-CDK6 reversed the suppression of IL-1β, IL-6, IL-12, and TNF-α mRNA and protein expression by Lv-shPKIA-AS1 (Figures 7A,B). Collectively, these results indicate that an epigenetic regulatory pathway involving PKIA-AS1 and CDK6 mediates SNL-induced neuropathic pain.
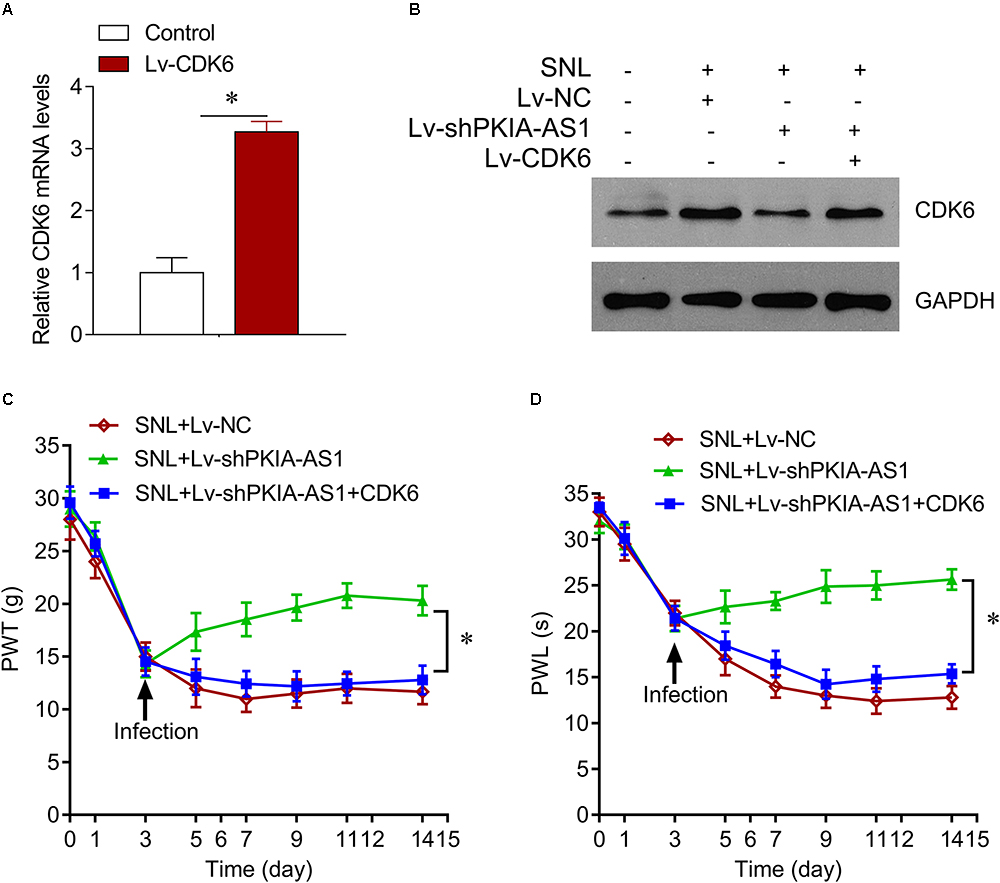
Figure 6. PKIA-AS1 modulated neuropathic pain by regulating CDK6. (A) CDK6 mRNA expression in SNL rat models infected with Lv-CDK6 6 days after lentivirus infection. (B) CDK6 protein expression in the spinal cord tissues of rats infected with indicated viruses. (C) The effect of CDK6 on mechanical allodynia was evaluated by PWT. (D) The effect of CDK6 on thermal hyperalgesia was assessed by PWL. N = 8 for each group; ∗P < 0.05. PWT, paw withdrawal threshold; PWL, paw withdrawal latency.
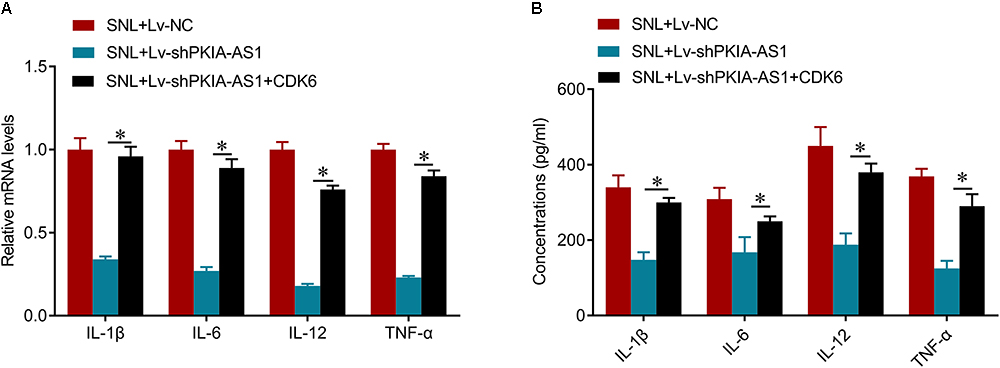
Figure 7. shRNA silencing of PKIA-AS1 inhibits neuroinflammation by regulating CDK6 in SNL rats. (A) mRNA levels of IL-1β, IL-6, IL-12, and TNF-α in L5 spinal cord tissues in the rats 6 days after lentivirus infection. (B) The protein levels of IL-1β, IL-6, IL-12, and TNF-α in the L5 spinal cord of rats were tested by ELISA 6 days after lentivirus infection. N = 8 for each group; ∗P < 0.05.
Discussion
We have identified the lncRNA PKIA-AS1 as a major regulator of SNL associated with NP. First, PKIA-AS1 was significantly upregulated in the SNL model rat spinal cord. Furthermore, both gain-of-function and loss-of-function studies confirmed that PKIA-AS1 was necessary for generating SNL-induced neuropathic pain. Our results suggest that PKIA-AS1 is a critical regulator of SNL-induced neuropathic pain, and that manipulation of PKIA-AS1 expression could potentially be used to improve spinal nerve ligation-induced neuropathic pain.
Peripheral inflammation and nerve injury can alter the expression of numerous genes, including lncRNAs, in affected pain pathways (Jiang et al., 2015; Wu et al., 2018). Moreover, numerous lncRNAs have been identified in the pain-related regions, such as dorsal root ganglion and spinal cord of mouse, rat, and human (Hu et al., 2016; Dou et al., 2017; Mo et al., 2018). The spinal cord dorsal horn is responsible for relaying and modulating pain-related signals from nociceptors to the supraspinal brain regions. The expression of the lncRNA colon cancer associated transcript-1 (CCAT1) was found to decrease in the spinal cord, dorsal root ganglion, hippocampus, and anterior cingulate cortex from day 1 to day 5 after chronic sciatic nerve injury (CCI). Over-expression of lncRNA CCAT1 alleviated CCI-induced mechanical allodynia (Dou et al., 2017). In the present study, the expression of another lncRNA, PKIA-AS1 was markedly increased in the spinal cord following SNL and was associated with heightened pain sensitivity, whereas shRNA-mediated knockdown of PKIA-AS1 alleviated thermal and mechanical hypersensitivity caused by SNL. These results suggest that upregulation of PKIA-AS1 in the spinal cord may be an important contributor to SNL-induced neuropathic pain. This possibility is further supported by our findings that PKIA-AS1 overexpression in spinal cord of uninjured rats was sufficient to induce pain-related behaviors. In the present study, the expression profiles of lncRNA were obtained from whole, unperfused spinal cord tissues after SNL so the specific cells overexpressing this lncRNA are currently unknown. Additional studies are needed to assess whether PKIA-AS1 and the other lncRNAs differentially expressed in the SNL spinal cord are altered in neurons, microglia. and (or) astrocytes.
We identified many possible targets of PKIA-AS1 regulation by proteomic assay, and confirmed that CDK6 is an interaction partner of PKIA-AS1. We further demonstrated that PKIA-AS1 interacts with CDK6 promoter and enhances promoter activity by modulating its methylation status. Our results strongly support the notion that PKIA-AS1 interacts with CDK6 promoter to enhance its expression and function. Cyclin-dependent kinases (CDKs) control diverse biological processes via the regulation of the cell cycle and gene expression (Peng et al., 2013). CDKs transcriptionally induce proinflammatory genes during the G1 phase of cell cycle. Further, cytokine-induced recruitment of CDK6 to the nuclear chromatin fraction is associated with activation of NF-κB, STAT, and AP-1 transcription factor families. CDK6 can recruit NF-κB subunit p65 to its target sites (Schmitz and Kracht, 2016). The demonstrated involvement of CDKs in proinflammatory gene expression makes these proteins promising target for treatment of chronic inflammatory diseases. CDK6 has been implicated in the proliferation, differentiation, and survival of neurons (Beukelaers et al., 2011; Mi et al., 2013; Caron et al., 2018; Hasenpusch-Theil et al., 2018). For example, preventing CDK6 over-activation can reduce 6-hydroxy-dopamine-induced neuronal death (Alquezar et al., 2015). According to previous studies, CDK6 is localized to the cytoplasm (Kohrt et al., 2009). CDK6 also regulates astrocyte proliferation and microglial activation (Zhao et al., 2013; Gu et al., 2016), suggesting that CDK6 is involved with inflammatory pain. In our study, we demonstrate for the first time that PKIA-AS1 epigenetically controls CDK6 expression by modulating promoter methylation, thereby regulating proinflammatory cytokine release, and ultimately controlling neuropathic pain.
In conclusion, PKIA-AS1 plays an important role in the pathogenesis of neuropathic pain through direct interaction with CDK6. Silencing PKIA-AS1 may be a complementary approach to reduce neuropathic pain induced by spinal nerve ligation through downregulation of CDK6 expression.
Author Contributions
J-ZH and H-BL designed the study. JZH and Z-JR performed cell biological experiments and in vivo experiments. ML, PL, L-YJ, Z-XL, C-YD and YC analyzed and interpreted the data. All authors contributed to writing the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 81672174 and 81874004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2019.00050/full#supplementary-material
References
Alquezar, C., Barrio, E., Esteras, N., de la Encarnacion, A., Bartolome, F., Molina, J. A., et al. (2015). Targeting cyclin D3/CDK6 activity for treatment of Parkinson’s disease. J. Neurochem. 133, 886–897. doi: 10.1111/jnc.13070
Bali, K. K., and Kuner, R. (2014). Noncoding RNAs: key molecules in understanding and treating pain. Trends Mol. Med. 20, 437–448. doi: 10.1016/j.molmed.2014.05.006
Beukelaers, P., Vandenbosch, R., Caron, N., Nguyen, L., Belachew, S., Moonen, G., et al. (2011). Cdk6-dependent regulation of G(1) length controls adult neurogenesis. Stem Cells 29, 713–724. doi: 10.1002/stem.616
Caron, N., Genin, E. C., Marlier, Q., Verteneuil, S., Beukelaers, P., Morel, L., et al. (2018). Proliferation of hippocampal progenitors relies on p27-dependent regulation of Cdk6 kinase activity. Cell. Mol. Life Sci. 75, 3817–3827. doi: 10.1007/s00018-018-2832-x
Coraggio, V., Guida, F., Boccella, S., Scafuro, M., Paino, S., Romano, D., et al. (2018). Neuroimmune-driven neuropathic pain establishment: a focus on gender differences. Int. J. Mol. Sci. 19:E281. doi: 10.3390/ijms19010281
Dones, I., and Levi, V. (2018). Spinal cord stimulation for neuropathic pain: current trends and future applications. Brain Sci. 8:E138. doi: 10.3390/brainsci8080138
Dou, L., Lin, H., Wang, K., Zhu, G., Zou, X., Chang, E., et al. (2017). Long non-coding RNA CCAT1 modulates neuropathic pain progression through sponging miR-155. Oncotarget 8, 89949–89957. doi: 10.18632/oncotarget.21192
Gu, J., Ni, Y., Xu, L., Xu, H., and Cai, Z. (2016). Nanog interact with CDK6 to regulates astrocyte cells proliferation following spinal cord injury. Biochem. Biophys. Res. Commun. 469, 1097–1103. doi: 10.1016/j.bbrc.2015.12.053
Gwak, Y. S., and Hulsebosch, C. E. (2011). GABA and central neuropathic pain following spinal cord injury. Neuropharmacology 60, 799–808. doi: 10.1016/j.neuropharm.2010.12.030
Gwak, Y. S., Kang, J., Unabia, G. C., and Hulsebosch, C. E. (2012). Spatial and temporal activation of spinal glial cells: role of gliopathy in central neuropathic pain following spinal cord injury in rats. Exp. Neurol. 234, 362–372. doi: 10.1016/j.expneurol.2011.10.010
Handschick, K., Beuerlein, K., Jurida, L., Bartkuhn, M., Muller, H., Soelch, J., et al. (2014). Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-kappaB-dependent gene expression. Mol. Cell 53, 193–208. doi: 10.1016/j.molcel.2013.12.002
Hasenpusch-Theil, K., West, S., Kelman, A., Kozic, Z., Horrocks, S., McMahon, A. P., et al. (2018). Gli3 controls the onset of cortical neurogenesis by regulating the radial glial cell cycle through Cdk6 expression. Development 145:dev163147. doi: 10.1242/dev.163147
Hu, J., Ni, S., Cao, Y., Zhang, T., Wu, T., Yin, X., et al. (2016). The angiogenic effect of microRNA-21 targeting TIMP3 through the regulation of MMP2 and MMP9. PLoS One 11:e0149537. doi: 10.1371/journal.pone.0149537
Jiang, B. C., Sun, W. X., He, L. N., Cao, D. L., Zhang, Z. J., and Gao, Y. J. (2015). Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol. Pain 11:43. doi: 10.1186/s12990-015-0047-9
Kohrt, D. M., Crary, J. I., Gocheva, V., Hinds, P. W., and Grossel, M. J. (2009). Distinct subcellular distribution of cyclin dependent kinase 6. Cell Cycle 8, 2837–2843. doi: 10.4161/cc.8.17.9521
Li, G., Jiang, H., Zheng, C., Zhu, G., Xu, Y., Sheng, X., et al. (2017). Long noncoding RNA MRAK009713 is a novel regulator of neuropathic pain in rats. Pain 158, 2042–2052. doi: 10.1097/j.pain.0000000000001013
Liu, C., Li, C., Deng, Z., Du, E., and Xu, C. (2018). Long non-coding RNA BC168687 is involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience 374, 214–222. doi: 10.1016/j.neuroscience.2018.01.049
Liu, Z., Liang, Y., Wang, H., Lu, Z., Chen, J., Huang, Q., et al. (2017). LncRNA expression in the spinal cord modulated by minocycline in a mouse model of spared nerve injury. J. Pain Res. 10, 2503–2514. doi: 10.2147/JPR.S147055
McKelvey, R., Berta, T., Old, E., Ji, R. R., and Fitzgerald, M. (2015). Neuropathic pain is constitutively suppressed in early life by anti-inflammatory neuroimmune regulation. J. Neurosci. 35, 457–466. doi: 10.1523/JNEUROSCI.2315-14.2015
Mi, D., Carr, C. B., Georgala, P. A., Huang, Y. T., Manuel, M. N., Jeanes, E., et al. (2013). Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb. Neuron 78, 269–284. doi: 10.1016/j.neuron.2013.02.012
Mo, C., Xu, M., Wen, C., Chang, R., Huang, C., Zou, W., et al. (2018). Normalizing JMJD6 expression in rat spinal dorsal horn alleviates hyperalgesia following chronic constriction injury. Front. Neurosci. 12:542. doi: 10.3389/fnins.2018.00542
Peng, B., Cao, J., Yi, S., Wang, C., Zheng, G., and He, Z. (2013). Inhibition of proliferation and induction of G1-phase cell-cycle arrest by dFMGEN, a novel genistein derivative, in lung carcinoma A549 cells. Drug Chem. Toxicol. 36, 196–204. doi: 10.3109/01480545.2012.710620
Plassais, J., Lagoutte, L., Correard, S., Paradis, M., Guaguere, E., Hedan, B., et al. (2016). A point mutation in a lincRNA upstream of GDNF is associated to a canine insensitivity to pain: a spontaneous model for human sensory neuropathies. PLoS Genet. 12:e1006482. doi: 10.1371/journal.pgen.1006482
Schmitz, M. L., and Kracht, M. (2016). Cyclin-dependent kinases as coregulators of inflammatory gene expression. Trends Pharmacol. Sci. 37, 101–113. doi: 10.1016/j.tips.2015.10.004
Shao, J., Wang, J., Huang, J., Liu, C., Pan, Y., Guo, Q., et al. (2018). Identification of lncRNA expression profiles and ceRNA analysis in the spinal cord of morphine-tolerant rats. Mol. Brain 11:21. doi: 10.1186/s13041-018-0365-8
Tan, Y., Yang, J., Xiang, K., Tan, Q., and Guo, Q. (2015). Suppression of microRNA-155 attenuates neuropathic pain by regulating SOCS1 signalling pathway. Neurochem. Res. 40, 550–560. doi: 10.1007/s11064-014-1500-2
Vieira, A. S., Dogini, D. B., and Lopes-Cendes, I. (2018). Role of non-coding RNAs in non-aging-related neurological disorders. Braz. J. Med. Biol. Res. 51:e7566. doi: 10.1590/1414-431X20187566
Walters, E. T. (2014). Neuroinflammatory contributions to pain after SCI: roles for central glial mechanisms and nociceptor-mediated host defense. Exp. Neurol. 258, 48–61. doi: 10.1016/j.expneurol.2014.02.001
Wayne, N. P. (2018). Severe chronic pain following spinal cord damage: a pragmatic perspective for prescribing opioids. Spinal Cord. Ser. Cases 4:65. doi: 10.1038/s41394-018-0094-3
Wu, S., Bono, J., and Tao, Y. X. (2018). Long noncoding RNA (lncRNA): a target in neuropathic pain. Expert Opin. Ther. Targets 23, 15–20. doi: 10.1080/14728222.2019.1550075
Xia, L. X., Ke, C., and Lu, J. M. (2018). NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J. Cell. Physiol. 233, 7103–7111. doi: 10.1002/jcp.26526
Yan, X. T., Lu, J. M., Wang, Y., Cheng, X. L., He, X. H., Zheng, W. Z., et al. (2018). XIST accelerates neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models. J. Cell. Physiol. 233, 6098–6106. doi: 10.1002/jcp.26453
Yang, J., Zhou, M., Zhao, R., Peng, S., Luo, Z., Li, X., et al. (2014). Identification of candidate biomarkers for the early detection of nasopharyngeal carcinoma by quantitative proteomic analysis. J. Proteomics 109, 162–175. doi: 10.1016/j.jprot.2014.06.025
Zhang, Z. J., Cao, D. L., Zhang, X., Ji, R. R., and Gao, Y. J. (2013). Chemokine contribution to neuropathic pain: respective induction of CXCL1 and CXCR2 in spinal cord astrocytes and neurons. Pain 154, 2185–2197. doi: 10.1016/j.pain.2013.07.002
Zhao, H., Wang, J., Gao, L., Wang, R., Liu, X., Gao, Z., et al. (2013). MiRNA-424 protects against permanent focal cerebral ischemia injury in mice involving suppressing microglia activation. Stroke 44, 1706–1713. doi: 10.1161/STROKEAHA.111.000504
Keywords: neuropathic pain, long non-coding RNA, spinal cord injury, neuroinflammation, CDK6
Citation: Hu J-Z, Rong Z-J, Li M, Li P, Jiang L-Y, Luo Z-X, Duan C-Y, Cao Y and Lu H-B (2019) Silencing of lncRNA PKIA-AS1 Attenuates Spinal Nerve Ligation-Induced Neuropathic Pain Through Epigenetic Downregulation of CDK6 Expression. Front. Cell. Neurosci. 13:50. doi: 10.3389/fncel.2019.00050
Received: 06 September 2018; Accepted: 01 February 2019;
Published: 20 February 2019.
Edited by:
Thomas Fath, Macquarie University, AustraliaReviewed by:
Huiyin Tu, Zhengzhou University, ChinaJustin Lees, University of New South Wales, Australia
Alexandre Charlet, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2019 Hu, Rong, Li, Li, Jiang, Luo, Duan, Cao and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Bin Lu, aG9uZ2Jpbmx1QGhvdG1haWwuY29t
 Jian-Zhong Hu1,2
Jian-Zhong Hu1,2 Zi-Jie Rong
Zi-Jie Rong Hong-Bin Lu
Hong-Bin Lu