- Department of Entomology, The Hebrew University of Jerusalem, Rehovot, Israel
Mosquitoes exhibit highly diverse and fast evolving odorant receptors (ORs). The indole-sensitive OR gene clade, comprised of Or2 and Or10 is a notable exception on account of its conservation in both mosquito subfamilies. This group of paralogous genes exhibits a complex developmental expression pattern in Aedes aegypti: AaegOr2 is expressed in both adults and larvae, AaegOr10 is adult-specific and a third member named AaegOr9 is larva-specific. OR2 and OR10 have been deorphanized and are selectively activated by indole and skatole, respectively. Using the two-electrode voltage clamp of Xenopus oocytes expressing Ae. aegypti ORs, we show that AaegOR9 is supersensitive and narrowly tuned to skatole. Our findings suggest that Ae. aegypti has evolved two distinct molecular strategies to detect skatole in aquatic and terrestrial environments, highlighting the central ecological roles of indolic compounds in the evolutionary and life histories of these insects.
Introduction
Aromatic and heterocyclic compounds play an important role in the ecology of adult mosquitoes as indicated by the odor space of the Anopheles gambiae odorant receptor (OR) repertoire (Carey et al., 2010; Wang et al., 2010). Specifically, indole (IUPAC name, 1H-indole) and skatole (IUPAC name, 3-methylindole) are respectively detected by the narrowly tuned Or2 and Or10 paralogous genes found in Culex quinquefasciatus (Hughes et al., 2010; Pelletier et al., 2010), Aedes aegypti and An. gambiae (Carey et al., 2010; Wang et al., 2010) reflecting their ancestral origin (Bohbot et al., 2011). Due to the sensitive and selective nature of the OR2-indole and OR10-skatole interactions, they have been referred to as the “indolergic” receptors (Bohbot and Pitts, 2015).
Indole and skatole are released by a wide variety of organisms but are mainly synthesized by bacteria (Elgaali et al., 2002; Schulz and Dickschat, 2007; Lindh et al., 2008; Hubbard et al., 2015), fungi (Chen et al., 2014; Tomberlin et al., 2017) and plants (Turlings et al., 1991; Frey et al., 2000; Ober, 2005). In adult mosquitoes, both compounds have been proposed to mediate oviposition site (Blackwell and Johnson, 2000) and host-locating behaviors (Cork, 1996). However, their exact ecological role(s) remain complex since indoles are major constituents of floral (Knudsen et al., 2006) and animal scents (Meijerink et al., 2001; Lee et al., 2015). Interestingly, indolic compounds play additional ecological roles in mosquito larvae (Xia et al., 2008; Scialò et al., 2012).
Or2 is expressed in the adult and larval stages of Ae. aegypti (Bohbot et al., 2007) and An. gambiae (Hill et al., 2002; Xia et al., 2008). Or10 expression is more complex: in An. gambiae, Or10 is expressed both in larvae and adults. In Ae. Aegypti, Or10 is only expressed in adults, while a third paralog named Or9, is expressed in the larval antenna (Bohbot et al., 2007). Based on pharmacological studies, we have suggested that receptor sensitivity towards odorants in the nanomolar concentration range is a predictor of OR-semiochemical relationships (Bohbot and Pitts, 2015). The activation of AaegOR9 by indole in the low micromolar concentration range (Bohbot et al., 2011) indicated that a more potent indolic cognate ligand selectively activates this receptor.
Using a reverse chemical ecology approach, we set out to identify a potential cognate ligand for this larval-expressed Or9 gene (Supplementary Table 1). First, we used a panel of 31 indole derivatives from plants and microbes to identify a potent activator of AaegOR9, then we showed that AaegOR9 is narrowly tuned to skatole in the low nanomolar concentration range. Our findings suggest that Culicinae have developed a supersensitive skatole receptor that operates in water where this compound exhibits low solubility. The occurrence of two skatole receptors, each assigned to a different developmental stage indicates the central role of this odorant in the Ae. aegypti life cycle. The deorphanization of AaegOR9: (i) provides a molecular target for future larval behavioral disruption studies; (ii) improves our understanding of insect OR coding; and (iii) raises questions on the possible ecological roles of mosquito indolergic receptors.
Materials and Methods
Chemical Reagents
The chemicals (Supplementary Table 1) used for the deorphanization of AaegOR9 were obtained from Sigma-Aldrich (Milwaukee, WI, USA), ChemCruz (Dallas, TX, USA), Glentham Life Sciences (Corsham, UK), FluoroChem (Hadfield, UK), SL Moran (Jerusalem, Israel), Holland Moran (Yehun, Israel), Alfa Aesar (Ward Hill, MA, USA) and from the generous contribution of the Dr. Kolodkin-Gal Lab (Weizmann Institute of Science, Israel).
Two-Electrode Voltage Clamp of Xenopus Oocytes Expressing ORs
The methodologies and protocols have been described in details elsewhere (Bohbot and Dickens, 2009). AaegOr9 and Aaeg-ORco cRNAs (Bohbot et al., 2011) were synthesized from linearized pSP64DV expression vectors using the mMESSAGE mMACHINE® SP6 kit (Life Technologies). Stage V-VII oocytes were harvested from Xenopus laevis females, mechanically separated, treated with collagenase (8 mg/mL, 30 min, 18°C) and rinsed in washing solution (96 mM NaCl, 2 mM KCl, 5 mM MgCl2 and 5 mM HEPES, pH 7.6). Oocytes were microinjected with 27.6 ng AaegOr9 and AaegORco cRNAs, incubated at 18°C for 3–4 days in ND96 solution (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, 0.8 mM CaCl2 and 5 mM HEPES, pH 7.6), supplemented with 5% dialyzed horse serum, 50 μg/mL tetracycline, 100 μg/mL streptomycin and 550 μg/mL sodium pyruvate. Whole-cell currents were recorded using the two-microelectrode voltage-clamp technique. During recording sessions, the holding potential was maintained at −80 mV using an OC-725C oocyte clamp (Warner Instruments, LLC, Hamden, CT, USA). Oocytes placed in a RC-3Z oocyte recording chamber (Warner Instruments, LLC, Hamden, CT, USA) were exposed to odorants for 8 s. Current was allowed to return to baseline between odorant applications. Data acquisition and concentration-response analyses were carried out with a Digidata 1550A and the pCLAMP10 software (Molecular Devices, Sunnyvale, CA, USA), and analyzed using GraphPad Prism 7 (GraphPad Software Inc., La Jolla, CA, USA). Stock concentration of odorants (10−2 M) were dissolved in ringer solution containing 2% dimethyl sulfoxide (DMSO) in order to solubilize the hydrophobic indolic compounds.
Pharmacological Characterization
The response profile was established using multiple sessions, each including six compounds at a time and indole as an internal reference. The order in which these compounds were administered was reversed within a session to mitigate against any potential sequence effects between compounds (none were observed). All the response values were normalized to the indole reference in each recording session (Supplementary Figure 1).
For the establishment of the concentration-response curves, oocytes were exposed to increasing concentrations of indole, skatole and indole-3-carboxaldehyde (I3C; Supplementary Figure 2). Quantitative characterization of OR sensitivity was estimated using the averaged effective concentration at 50% of the maximal response (EC50) over the sample population. The data to establish the concentration response curve and EC50 of AaegOR10-skatole was extracted from a previous study (Bohbot and Dickens, 2012).
Phylogeny OR Intron-Exon Structure and Phylogeny
All the sequences used in our phylogenic analysis were obtained from the VectorBase and NCBI databases using AaegOr2/9/10 as query (for accession numbers, see Supplementary Table 3). DNA sequences for Toxorhynchites Or2 and Or10 can be accessed here: http://dx.doi.org/10.6084/m9.figshare.1092617. MAFFT version 7 (Nakamura et al., 2018) was used for multiple amino-acid sequence alignment. The phylogenic software IQ-TREE (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017; Hoang et al., 2018) and the FigTree editor1 were used for building the mosquito indolergic receptor phylogenic tree based on the maximum likelihood method (Model: JC, UFbootstrap: 5,000). Using MAFFT (default parameters) and the Vectorbase database, we located the intron positions on the indolergic receptor genes.
Results
AaegOR9 Is Narrowly Tuned to Skatole
Based on its larval expression (Bohbot et al., 2007) and functional characterization (Bohbot et al., 2011), we initially surmised that AaegOR9 would be narrowly tuned to a water-soluble indolic ligand. To test this hypothesis, we screened AaegOR9 with a panel of 31 indolic derivatives (Supplementary Table 1) exhibiting some degree of water solubility using the two electrodes voltage clamp of Xenopus laevis oocytes. We also included indole and skatole as references (Bohbot et al., 2011). The screen was carried out using a low odorant concentration (500 nM) in order to mitigate the caveats associated with high ligand concentrations, including receptor adaptation, antagonist effects and technical artifacts such as broad molecular receptive ranges (Bohbot and Pitts, 2015).
At this concentration, skatole was decisively the most potent ligand, eliciting responses five times higher than indole (Figure 1) confirming the hypothesis that the potential cognate ligand of AaegOR9 is an indole derivative. However, this result contradicts our water-soluble ligand hypothesis. Indeed, skatole is about seven times less water soluble (0.5 mg/mL, ChemIDplus) than indole (3.56 mg/mL, ChemIDplus; Figure 1) and is considered rather insoluble in water as a result.
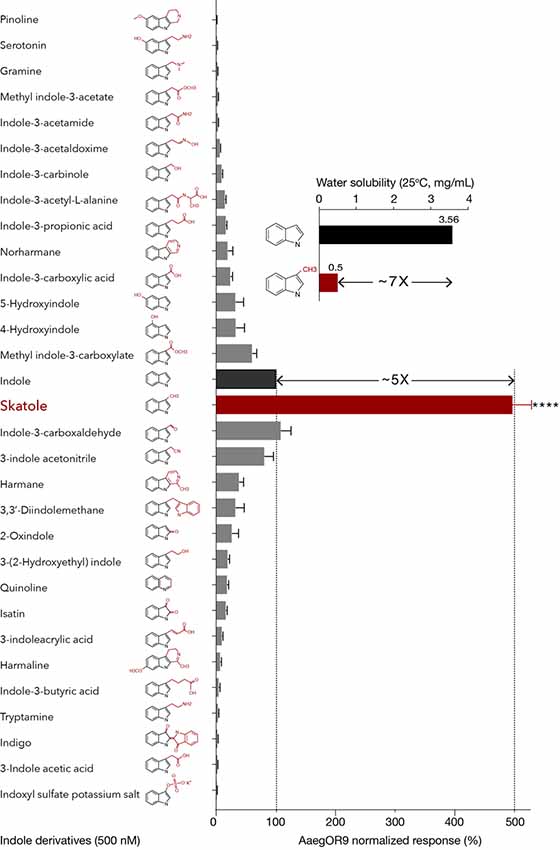
Figure 1. Aedes aegypti OR9 (AaegOR9) is narrowly tuned to skatole. Indole-tuning curve of AaegOR9 to an odor panel comprised of 31 indole derivatives (kurtosis value: 17.75). Skatole is five times more potent than indole (t-test: ****p < 0.0001; n = 10; 100% and 500% response thresholds shown as dotted lines), which is inversely correlated with their respective water solubility (inset). Error bars of average responses indicate standard errors.
I3C, indole, 3-indole acetonitrile and methyl indole-3-carboxylate were the next most potent ligands suggesting that indole derivatives with a short side chain on position C3 can fit into the binding pocket of AaegOR9. However, comparable small indolic compounds such as indole-3-carbinole or gramine were among the least potent ligands (Figure 1). Overall, the AaegOR9 response profile was narrow (kurtosis value of 17.75), especially considering that our odorant panel was restricted to indole derivatives.
AaegOR9 Is Supersensitive to Skatole
To characterize the pharmacological sensitivity of AaegOR9, we measured the amplitudes of the current responses of this receptor when exposed to increasing concentrations (100 pM to 10 μM) of skatole, indole and I3C (Figure 2A). Compound sensitivities were determined using the extrapolated EC50 values. This analysis confirmed that AaegOR9 is 176 times more sensitive to skatole than to indole. AaegOR9 is a significantly more sensitive skatole receptor (EC50 ≃ 5 nM) than AaegOR10 (EC50 = 100 nM; Figure 2B). Although I3C and indole elicited comparable responses in our tuning curve experiment (Figure 1), their EC50 values were significantly different (Figure 2A), underscoring the caveat of inferring receptor sensitivity based on tuning curves.
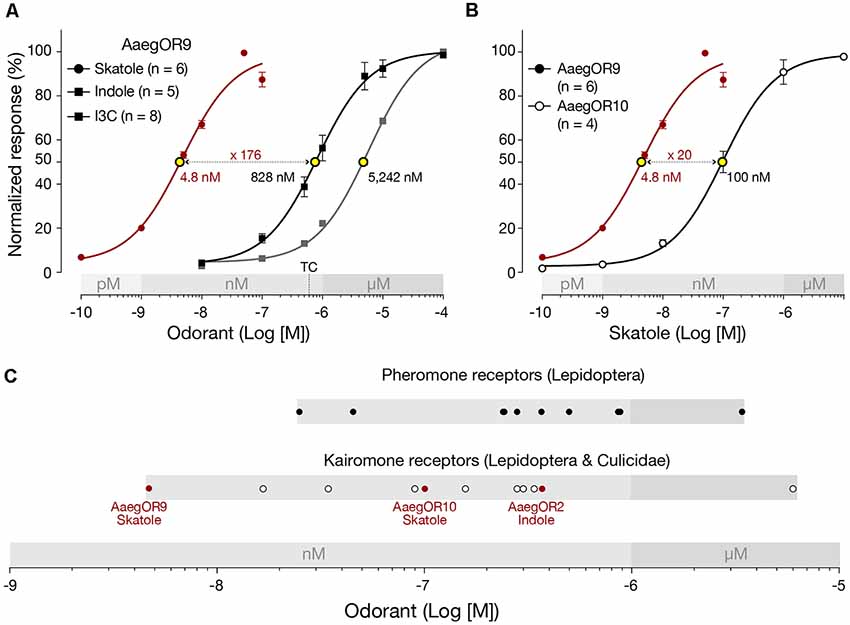
Figure 2. Aedes aegypti OR9 (AaegOR9) is a supersensitive skatole receptor. (A) Based on their respective EC50 values (yellow dots), AaegOR9 is significantly (one-way ANOVA followed by Tukey’s post test; p < 0.0001) more sensitive to skatole than to indole or to indole-3-carboxaldehyde (I3C). The concentration (500 nM) to which the tuning curve is based on is indicated by “TC.” (B) AaegOR9 is a more sensitive skatole receptor than AaegOR10 (t-test; p < 0.01). (C) Sensitivity ranking (according to EC50 values of cognate receptor-semiochemical interactions) of pheromone and kairomone receptors (Supplementary Table 2).
Plotting the EC50 values of AaegOR9 against previously characterized OR-cognate odorant pairs (Supplementary Table 2) using our pharmacological platform (Bohbot and Pitts, 2015) reveals that the AaegOR9-skatole pair is the most sensitive indolergic OR (Figure 2C) and the most sensitive OR-ligand pair identified so far, outperforming the most sensitive pheromone receptor.
OR9 Is a Culicinae-Specific Gene Expansion
The conserved Or2, Or9, Or10 genes were initially identified from the An. gambiae and Ae. aegypti genomes (Hill et al., 2002; Bohbot et al., 2007, 2011). These genes encode proteins with high amino-acid sequence identity considering the sequence divergence characteristic of this family (Figure 3A). We have extended our previous analysis (Bohbot et al., 2011) by including additional Anopheles species, Ae. albopictus and Toxorhynchites amboinensis. We confirm that the indolergic receptor clade is divided into the OR2 and OR10 subgroups indicating their ancestral origin (Figure 3A). Or9 emerged in the Culicinae lineage 52–54 mya (Arensburger et al., 2010), which includes Ae. albopictus and Cu. quinquefasciatus. There is no information regarding Or expression in To. amboinensis. We failed to identify any signature of other paralogs in An. gambiae and Ae. aegypti consistent with the hypothesis that the Or9-Or10 split occurred in Culicinae.
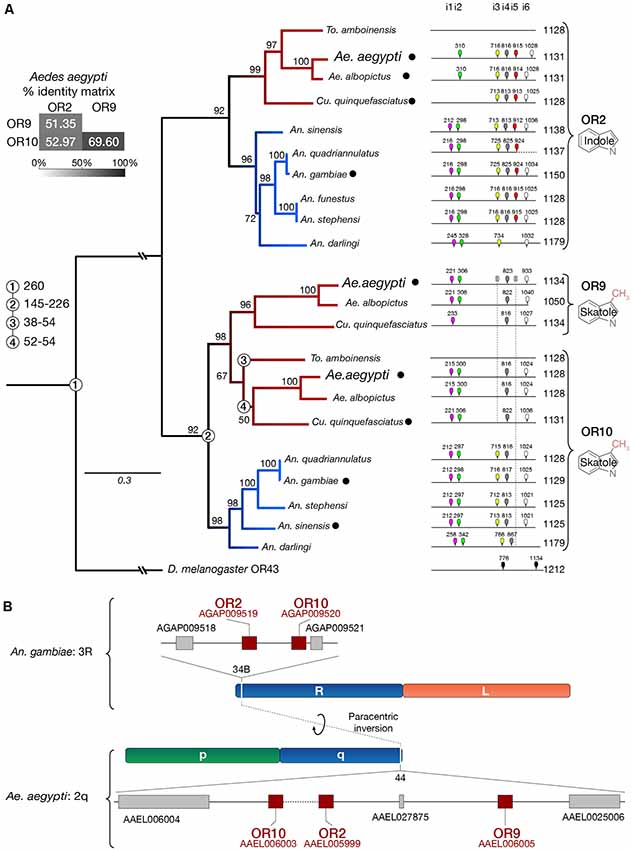
Figure 3. Or9 is a Culicinae-specific gene expansion. (A) DNA sequence identity, substitution rates, intron locations and odorant ligands (deorphanized receptors are labeled with a black dot, see Supplementary Table 2) suggest that Or9 is a Culicinae-specific gene expansion while Or2 and Or10 are present in both Culicinae (red branches) and Anophelinae (blue branches). Intron locations are color-coded and numbered from 1 to 6 (i1–i6). Missing introns are indicated by a crossed intron with a dotted lines underneath. Bootstrap values (%) are based on 5,000 replicates. Numbered circles on branch points indicate lineage splits in million years (MY). (B) Indolergic receptors are located on the q arm of chromosome 2 and on the R arm of chromosome 3 in Ae. aegypti and An. gambiae, respectively. Transcript numbers are shown for An. gambiae (AGAP#) and Ae. aegypti (AAEL#).
The greater similarity between AaegOR9 and AaegOR10 not only includes sequence conservation and amino-acid substitution rates but is also reflected by the patterns of conserved introns (Figure 3A). Both Culicinae Or10 and Or9 genes are missing introns 3 and 5. The absence of intron 5 is also a conserved feature within the Anophelinae Or10 clade. Or2, Or9, Or10 are clustered together in region 44 of the q arm of chromosome 2 in Ae. aegypti (Figure 3B). This region corresponds to region 34B of the orthologous chromosome 3R in An. gambiae, reflecting the extensive paracentric inversion events that occurred within chromosome segments between these two species (Severson et al., 2004; Arensburger et al., 2010).
Discussion
We had originally reported AaegOR9 as a broadly tuned receptor sensitive to indole in the low micromolar range suggesting that its cognate ligand, likely an indole derivative, remained to be identified (Bohbot et al., 2011). Here, we have shown that skatole selectively and reversibly activates AaegOR9 in the high picomolar (below 1 nM) to low nanomolar (between 1 and 10 nM) concentration range consistent with the idea that it is the cognate odorant ligand for this receptor (Bohbot and Pitts, 2015).
Based on our analyzes and on the principle of parsimony, we propose that Or2, Or9 and Or10 derive from two gene duplication events, one preceding the Anophelinae-Culicinae split that occurred 145–226 mya (Krzywinski et al., 2006; Reidenbach et al., 2009) and a second one that occurred in the Culicinae lineage (Figure 4A). The most common recent ancestors of Or2 and Or10 were likely indole and skatole receptors since this function is still conserved in the two mosquito subfamilies (Bohbot et al., 2007). We hypothesize, that the original ancestral receptor was sensitive to an indolic compound, perhaps indole or skatole. Although it is unlikely, due to the nanomolar sensitivity of OR10 for skatole, it is conceivable that the cognate ligand for OR10 remains to be identified. It is remarkable that during their evolution, mosquitoes have retained the function of discriminating between close structural chemical analogs, differing by a methyl group on position C3.
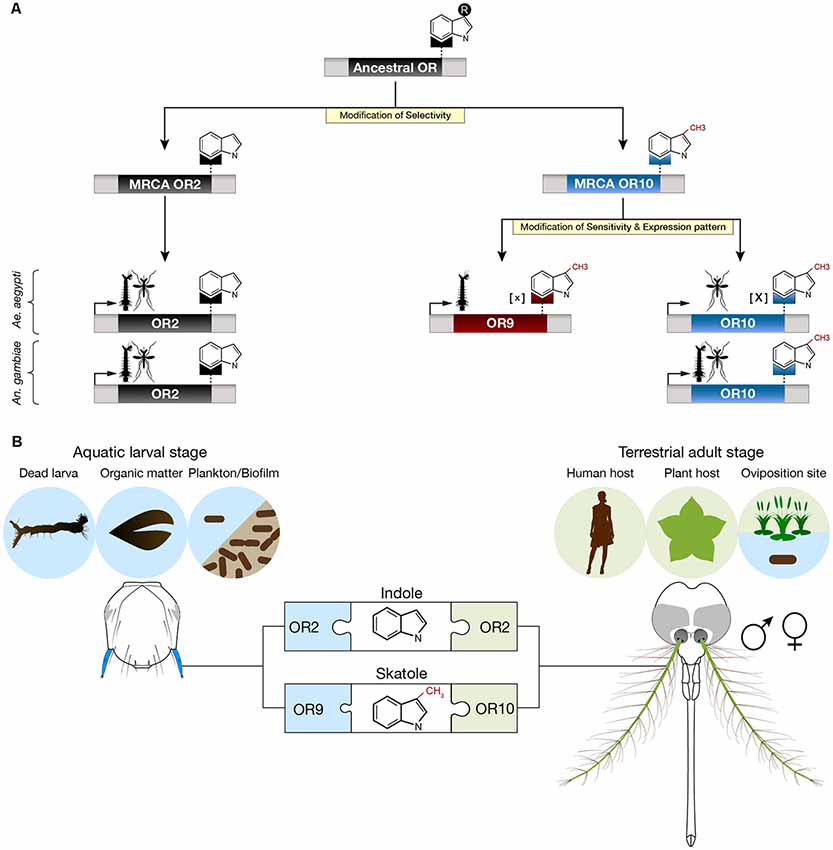
Figure 4. Proposed evolution and ecological roles of Aedes aegypti indolergic receptors. (A) The Or2, 9 and 10 gene clade derives from two successive gene duplication events followed by neofunctionalization (yellow highlights) consisting of modifications of ligand selectivity, sensitivity and developmental expression patterns. The most parsimonious hypothesis is that the most recent common ancestors (MRCAs) of Or2 and Or10 derived from a gene duplication event prior to the Anophelinae-Culicinae split. This event lead to neofunctionalization by means of selective detection of indole derivatives. The second duplication event arose in the Culicinae subfamily (e.g., Ae. aegypti) leading to the emergence of a low [X] and high [x] sensitivity skatole receptor, each expressed in a distinct developmental stage. (B) Putative ecological roles of indole and skatole in adults and larvae. Adults may detect both indole compounds to identify suitable hosts and oviposition sites while larvae may use these compounds to locate food sources, including dead larvae, decomposing organic matter and microbes.
While both AaegOR10 and AaegOR9 act as skatole receptors, they have diverged in function: larval AaegOR9 has a significantly higher sensitivity for this compound than adult AaegOR10. In addition, the expression pattern of AaegOr10 and AaegOr9 also diverged, allocating the role of skatole detection to the former in adults and to the latter in larvae (Figure 4A). It is interesting that other insects, including Drosophila melanogaster larvae and adults detect the same cues using different ORs (Dweck et al., 2015). In our case, the role for this increased sensitivity and distinct expression patterns remain unclear.
Skatole seems to be playing a unique role in Ae. aegypti. The increased sensitivity of OR9 to skatole may correspond to an evolutionary adaptation to the reduced water solubility of this compound, conferring a fitness advantage at the larval stage. Being outperformed, larval Or10 expression may have been relegated to the adult stage where skatole can occur at much higher concentrations due to its high volatility. It is interesting that both larva and adult An. gambiae detect skatole using the same receptor. Considering the differential sensitivity and developmental expression pattern of AaegOr9 and AaegOr10, we do not think this is a case of gene redundancy.
During the terrestrial stage, adult Ae. aegypti use olfaction to locate nectar sources, suitable oviposition sites and animal hosts using about 80 Or genes. The aquatic larva has a more limited range of behaviors (mostly feeding and escape behaviors), occupy one type of habitat and express 23 Or genes of which 15 are larval specific. Based on our study, it will be interesting to explore whether other larval-specific receptors have evolved enhanced sensitivity to poorly soluble volatile organic compounds.
Larvae mainly graze on biofilm (fungi, bacteria, algae), dead larvae (Kinney et al., 2014) and other decomposing organic matter including plant materials, which are sources of indolic compounds (Figure 4B). The exact roles of indole in adult mosquitoes, while traditionally ascribed to oviposition (Blackwell and Johnson, 2000), is much more complex than previously thought as male adult mosquitoes, as well as larvae, have relied on these compounds in the last 200 millions of years of evolution. These findings underscore the conserved and different roles indoles assume in the life histories of mosquitoes. It now remains to untangle the physiological, behavioral and ecological roles of these interesting compounds in the life of these insects.
Data Availability
The datasets generated for this study can be found in Figshare, http://dx.doi.org/10.6084/m9.figshare.1092617.
Ethics Statement
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed (NIH approval number: OPRR-A01-5011).
Author Contributions
JB designed and wrote the manuscript. EY provided technical support. DR carried out the experiment, analyzed the data and participated in the writing of the manuscript.
Funding
This work was supported by the Israel Science Foundation (grant no. 1990/16).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ronald J. Pitts for his critical reading of the manuscript, Dr. Osnat Malka for her insights and Dr. Diego Santos-Garcia for his expertise in computational biology.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2018.00533/full#supplementary-material
FIGURE S1 | Examples of current traces of AaegOR9-ORco in response to 500 nM of 31 indole derivatives.
FIGURE S2 | Examples of current traces of AaegOR9-ORco in response to increasing concentrations (Log units) of indole, skatole and indole-3-carboxaldehyde.
TABLE S1 | Indole derivatives ranked according to their potency (see Figure 1).
TABLE S2 | List of cognate pheromone and kairomone receptors deorphanized in the Xenopus laevis and other functional bioassays.
TABLE S3 | Accession numbers of indolergic receptors used in Figure 3 (Ae., Aedes; Cu., Culex; An., Anopheles; To., Toxorhynchites; D., Drosophila).
References
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Arensburger, P., Megy, K., Waterhouse, R. M., Abrudan, J., Amedeo, P., Antelo, B., et al. (2010). Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330, 86–88. doi: 10.1126/science.1191864
Blackwell, A., and Johnson, S. (2000). Electrophysiological investigation of larval water and potential oviposition chemo-attractants for Anopheles gambiae s.s. Ann. Trop. Med. Parasitol. 94, 389–398. doi: 10.1080/00034983.2000.11813554
Bohbot, J. D., and Dickens, J. C. (2009). Characterization of an enantioselective odorant receptor in the yellow fever mosquito Aedes aegypti. PLoS One 4:e7032. doi: 10.1371/journal.pone.0007032
Bohbot, J. D., and Dickens, J. C. (2012). Odorant receptor modulation: ternary paradigm for mode of action of insect repellents. Neuropharmacology 62, 2086–2095. doi: 10.1016/j.neuropharm.2012.01.004
Bohbot, J. D., Jones, P. L., Wang, G., Pitts, R. J., Pask, G. M., and Zwiebel, L. J. (2011). Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem. Senses 36, 149–160. doi: 10.1093/chemse/bjq105
Bohbot, J. D., and Pitts, R. J. (2015). The narrowing olfactory landscape of insect odorant receptors. Front. Ecol. Evol. 3:39. doi: 10.3389/fevo.2015.00039
Bohbot, J., Pitts, R. J., Kwon, H. W., Rutzler, M., Robertson, H. M., and Zwiebel, L. J. (2007). Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol. Biol. 16, 525–537. doi: 10.1111/j.1365-2583.2007.00748.x
Carey, A. F., Wang, G., Su, C.-Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1038/nature08834
Chen, G., Zhang, R.-R., Liu, Y., and Sun, W.-B. (2014). Spore dispersal of fetid by feces of mycophagous insects. J. Chem. Ecol. 40, 893–899. doi: 10.1007/s10886-014-0481-6
Chen, X.-G., Jiang, X., Gu, J., Xu, M., Wu, Y., Deng, Y., et al. (2015). Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. U S A 112, E5907–E5915. doi: 10.1073/pnas.1516410112
Cork, A. (1996). “Olfactory basis of host location by mosquitoes and other haematophagous Diptera,” in Olfaction in Mosquito Host Interactions (Ciba Foundation Symposium 200), eds G. R. Bock and G. Cardew (Chichester: Weily), 71–88.
Dweck, H. K. M., Ebrahim, S. A. M., Farhan, A., Hansson, B. S., and Stensmyr, M. C. (2015). Olfactory proxy detection of dietary antioxidants in Drosophila. Curr. Biol. 25, 455–466. doi: 10.1016/j.cub.2014.11.062
Elgaali, H., Hamilton-Kemp, T. R., Newman, M. C., Collins, R. W., Yu, K., and Archbold, D. D. (2002). Comparison of long-chain alcohols and other volatile compounds emitted from food-borne and related Gram positive and Gram negative bacteria. J. Basic Microbiol. 42, 373–380. doi: 10.1002/1521-4028(200212)42:6<373::aid-jobm373>3.0.co;2-4
Frey, M., Stettner, C., Pare, P. W., Schmelz, E. A., Tumlinson, J. H., and Gierl, A. (2000). An herbivore elicitor activates the gene for indole emission in maize. Proc. Natl. Acad. Sci. U S A 97, 14801–14806. doi: 10.1073/pnas.260499897
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., et al. (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178. doi: 10.1126/science.1076196
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., et al. (2002). The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129–149. doi: 10.1126/science.1076181
Hubbard, T. D., Murray, I. A., and Perdew, G. H. (2015). Indole and tryptophan metabolism: endogenous and dietary routes to ah receptor activation. Drug Metab. Dispos. 43, 1522–1535. doi: 10.1124/dmd.115.064246
Hughes, D. T., Pelletier, J., Luetje, C. W., and Leal, W. S. (2010). Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J. Chem. Ecol. 36, 797–800. doi: 10.1007/s10886-010-9828-9
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kinney, M. P., Panting, N. D., and Clark, T. M. (2014). Modulation of appetite and feeding behavior of the larval mosquito Aedes aegypti (L.) by the serotonin-selective reuptake inhibitor paroxetine: shifts between distinct feeding modes and the influence of feeding status. J. Exp. Biol. 217, 935–943. doi: 10.1242/jeb.094904
Knudsen, J. T., Eriksson, R., Gershenzon, J., and Ståhl, B. (2006). Diversity and distribution of floral scent. Bot. Rev. 72, 1–120. doi: 10.1663/0006-8101(2006)72[1:dadofs]2.0.co;2
Krzywinski, J., Grushko, O. G., and Besansky, N. J. (2006). Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol. Phylogenet. Evol. 39, 417–423. doi: 10.1016/j.ympev.2006.01.006
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E. Jr., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U S A 109, 14081–14086. doi: 10.1073/pnas.1204661109
Lee, J.-H., Wood, T. K., and Lee, J. (2015). Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718. doi: 10.1016/j.tim.2015.08.001
Lindh, J. M., Kännaste, A., Knols, B. G. J., Faye, I., and Borg-Karlson, A.-K. (2008). Oviposition responses of Anopheles gambiae s.s. (Diptera: Culicidae) and identification of volatiles from bacteria-containing solutions. J. Med. Entomol. 45, 1039–1049. doi: 10.1603/0022-2585(2008)45[1039:oroags]2.0.co;2
Liu, H., Liu, L., Cheng, P., Huang, X., and Gong, M. (2018). An odorant receptor from Anopheles sinensis in China is sensitive to oviposition attractants. Malar. J. 17:348. doi: 10.1186/s12936-018-2501-4
Liu, C., Liu, Y., Guo, M., Cao, D., Dong, S., and Wang, G. (2014). Narrow tuning of an odorant receptor to plant volatiles in Spodoptera exigua (Hübner). Insect Mol. Biol. 23, 487–496. doi: 10.1111/imb.12096
Liu, Y., Liu, C., Lin, K., and Wang, G. (2013). Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS One 8:e62094. doi: 10.1371/journal.pone.0062094
Marinotti, O., Cerqueira, G. C., de Almeida, L. G. P., Ferro, M. I. T., Loreto, E. L. D. S., Zaha, A., et al. (2013). The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res. 41, 7387–7400. doi: 10.1093/nar/gkt484
Meijerink, J., Braks, M. A., and Van Loon, J. J. (2001). Olfactory receptors on the antennae of the malaria mosquito Anopheles gambiae are sensitive to ammonia and other sweat-borne components. J. Insect Physiol. 47, 455–464. doi: 10.1016/s0022-1910(00)00136-0
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Nakamura, T., Yamada, K. D., Tomii, K., and Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Mol. Biol. Evol. 34, 2490–2492. doi: 10.1093/bioinformatics/bty121
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ober, D. (2005). Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 10, 444–449. doi: 10.1016/j.tplants.2005.07.007
Pelletier, J., Hughes, D. T., Luetje, C. W., and Leal, W. S. (2010). An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS One 5:e10090. doi: 10.1371/journal.pone.0010090
Reidenbach, K. R., Cook, S., Bertone, M. A., Harbach, R. E., Wiegmann, B. M., and Besansky, N. J. (2009). Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol. Biol. 9:298. doi: 10.1186/1471-2148-9-298
Schulz, S., and Dickschat, J. S. (2007). Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24, 814–842. doi: 10.1039/b507392h
Scialò, F., Hansson, B. S., Giordano, E., Polito, C. L., and Digilio, F. A. (2012). Molecular and functional characterization of the odorant receptor2 (or2) in the tiger mosquito Aedes albopictus. PLoS One 7:e36538. doi: 10.1371/journal.pone.0036538
Severson, D. W., Debruyn, B., Lovin, D. D., Brown, S. E., Knudson, D. L., and Morlais, I. (2004). Comparative genome analysis of the yellow fever mosquito Aedes aegypti with Drosophila melanogaster and the malaria vector mosquito Anopheles gambiae. J. Hered. 95, 103–113. doi: 10.1093/jhered/esh023
Sun, M., Liu, Y., Walker, W. B., Liu, C., Lin, K., Gu, S., et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS One 8:e62098. doi: 10.1371/journal.pone.0062098
Tanaka, K., Uda, Y., Ono, Y., Nakagawa, T., Suwa, M., Yamaoka, R., et al. (2009). Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890. doi: 10.1016/j.cub.2009.04.035
Tomberlin, J. K., Crippen, T. L., Wu, G., Griffin, A. S., Wood, T. K., and Kilner, R. M. (2017). Indole: an evolutionarily conserved influencer of behavior across kingdoms. Bioessays 39:1600203. doi: 10.1002/bies.201600203
Turlings, T. C., Tumlinson, J. H., Heath, R. R., Proveaux, A. T., and Doolittle, R. E. (1991). Isolation and identification of allelochemicals that attract the larval parasitoid, Cotesia marginiventris (Cresson), to the microhabitat of one of its hosts. J. Chem. Ecol. 17, 2235–2251. doi: 10.1007/bf00988004
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U S A 107, 4418–4423. doi: 10.1073/pnas.091339
Wang, G., Vásquez, G. M., Schal, C., Zwiebel, L. J., and Gould, F. (2011). Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol. 20, 125–133. doi: 10.1111/j.1365-2583.2010.01045.x
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E. Jr., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One 5:e8685. doi: 10.1371/journal.pone.0008685
Wanner, K. W., Nichols, A. S., Walden, K. K., Brockmann, A., Luetje, C. W., and Robertson, H. M. (2007). A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc. Natl. Acad. Sci. U S A 104, 14383–14388. doi: 10.1073/pnas.0705459104
Xia, Y., Wang, G., Buscariollo, D., Pitts, R. J., Wenger, H., and Zwiebel, L. J. (2008). The molecular and cellular basis of olfactory-driven behavior in Anopheles gambiae larvae. Proc. Natl. Acad. Sci. U S A 105, 6433–6438. doi: 10.1073/pnas.0801007105
Xu, P., Garczynski, S. F., Atungulu, E., Syed, Z., Choo, Y. M., Vidal, D. M., et al. (2012). Moth sex pheromone receptors and deceitful parapheromones. PLoS One 7:e41653. doi: 10.1371/journal.pone.0041653
Keywords: odorant receptor, indole, skatole, mosquito, Aedes aegypti
Citation: Ruel DM, Yakir E and Bohbot JD (2019) Supersensitive Odorant Receptor Underscores Pleiotropic Roles of Indoles in Mosquito Ecology. Front. Cell. Neurosci. 12:533. doi: 10.3389/fncel.2018.00533
Received: 05 December 2018; Accepted: 31 December 2018;
Published: 24 January 2019.
Edited by:
Dieter Wicher, Max Planck Institute for Chemical Ecology, GermanyReviewed by:
Merid Negash Getahun, International Centre of Insect Physiology and Ecology (ICIPE), KenyaRicardo C. Araneda, University of Maryland, College Park, United States
Copyright © 2019 Ruel, Yakir and Bohbot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan D. Bohbot, am9uYXRoYW4uYm9oYm90QG1haWwuaHVqaS5hYy5pbA==
 David M. Ruel
David M. Ruel Esther Yakir
Esther Yakir Jonathan D. Bohbot
Jonathan D. Bohbot