
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 30 November 2018
Sec. Cellular Neurophysiology
Volume 12 - 2018 | https://doi.org/10.3389/fncel.2018.00460
This article is part of the Research Topic Paradigm shifts and innovations in Neuroscience View all 89 articles
A correction has been applied to this article in:
Corrigendum: Potentials of Cellular Reprogramming as a Novel Strategy for Neuroregeneration
 Lyujie Fang1,2,3†
Lyujie Fang1,2,3† Layal El Wazan1,2†
Layal El Wazan1,2† Christine Tan1,2
Christine Tan1,2 Tu Nguyen1,2
Tu Nguyen1,2 Sandy S. C. Hung1
Sandy S. C. Hung1 Alex W. Hewitt1,4
Alex W. Hewitt1,4 Raymond C. B. Wong1,2,5*
Raymond C. B. Wong1,2,5*Cellular reprogramming technology holds great potential for tissue repair and regeneration to replace cells that are lost due to diseases or injuries. In addition to the landmark discovery of induced pluripotent stem cells, advances in cellular reprogramming allow the direct lineage conversion of one somatic cell type to another using defined transcription factors. This direct reprogramming technology represents a rapid way to generate target cells in the laboratory, which can be used for transplantation and studies of biology and diseases. More importantly, recent work has demonstrated the exciting application of direct reprogramming to stimulate regeneration in vivo, providing an alternative approach to transplantation of donor cells. Here, we provide an overview of the underlying concept of using cellular reprogramming to convert cell fates and discuss the current advances in cellular reprogramming both in vitro and in vivo, with particular focuses on the neural and retinal systems. We also discuss the potential of in vivo reprogramming in regenerative medicine, the challenges and potential solutions to translate this technology to the clinic.
During development, cellular identity, and differentiation potential are largely determined by the lineage history of the specific cell. Generally, the cell identity or the differentiated state of an adult cell is remarkably stable, and only stem cells with multipotent/pluripotent potential possess the ability to turn into another cell type(s). By transferring the somatic cell nucleus into an enucleated oocyte in Xenopus, the Nobel Prize-winning work of Gurdon (1962) first demonstrated that mature cells can be reprogrammed back to an embryonic state and became pluripotent. Similar nuclear transfer was subsequently demonstrated in mammals, including Dolly the cloned sheep. These pioneering studies provided evidence that reprogramming factors in the oocyte cytoplasm can overwrite the cellular identity encoded in the nucleus of a fully differentiated cell (Campbell et al., 1996). However, the precise factors that enables cell fate conversion in mature cells remain largely elusive. The seminal work of Shinya Yamanaka’s group identified the precise signals required for cellular reprogramming, and showed that a cocktail of four transcription factors (Oct4, Sox2, c-Myc, and Klf4) are sufficient to reprogram skin fibroblasts into induced pluripotent stem (iPS) cells that resemble a primitive embryonic state (Takahashi and Yamanaka, 2006). Since then, there has been intensive research on the identification of the precise transcription factors to alter and reprogram cell fates.
Beyond iPS cell reprogramming, direct conversion of one somatic cell type to an unrelated cell type can occur without passing through an intermediate multipotent state. This direct reprogramming approach, also known as ‘transdifferentiation,’ was first demonstrated with the conversion of fibroblasts into myoblasts by overexpression of a single transcription factor MyoD. This highlighted the potential power of controlling cell fates using genetic factors (Davis et al., 1987). Another early example of direct reprogramming was conducted by Kulessa et al. (1995) who successfully reprogrammed myoblasts into eosinophils and thromboblasts by forceful overexpressing of GATA1.
In recent years, direct reprogramming has attracted enormous interest, yielding promising results in the conversion across multiple lineages to generate neurons, oligodendrocytes, cardiomyocytes, muscle cells, blood progenitors, and hepatocytes (Ladewig et al., 2013). Notably, this direct reprogramming process is potentially much faster than generating iPS cells and subsequent differentiation into the target cell types, which could take months. Thus, it represents a faster and more cost-effective method to generate cells in vitro. The concept of taking patient’s cells and turning them into a different cell type is very attractive for tissue engineering and regenerative medicine, as it eliminates the risk of immune-rejection of the reprogrammed cells following transplantation. This direct reprogramming approach also has the potential to generate disease models using the patient’s cells, allowing the study of disease mechanism as well as providing an in vitro platform for drug discovery, toxicology study and testing of gene therapy (Figure 1A). Moreover, the application of direct reprogramming in vivo would allow us to reprogram endogenous cells within the body to become new cell types, providing a novel strategy to regenerate cells that are lost in diseases or injuries (Figure 1B).
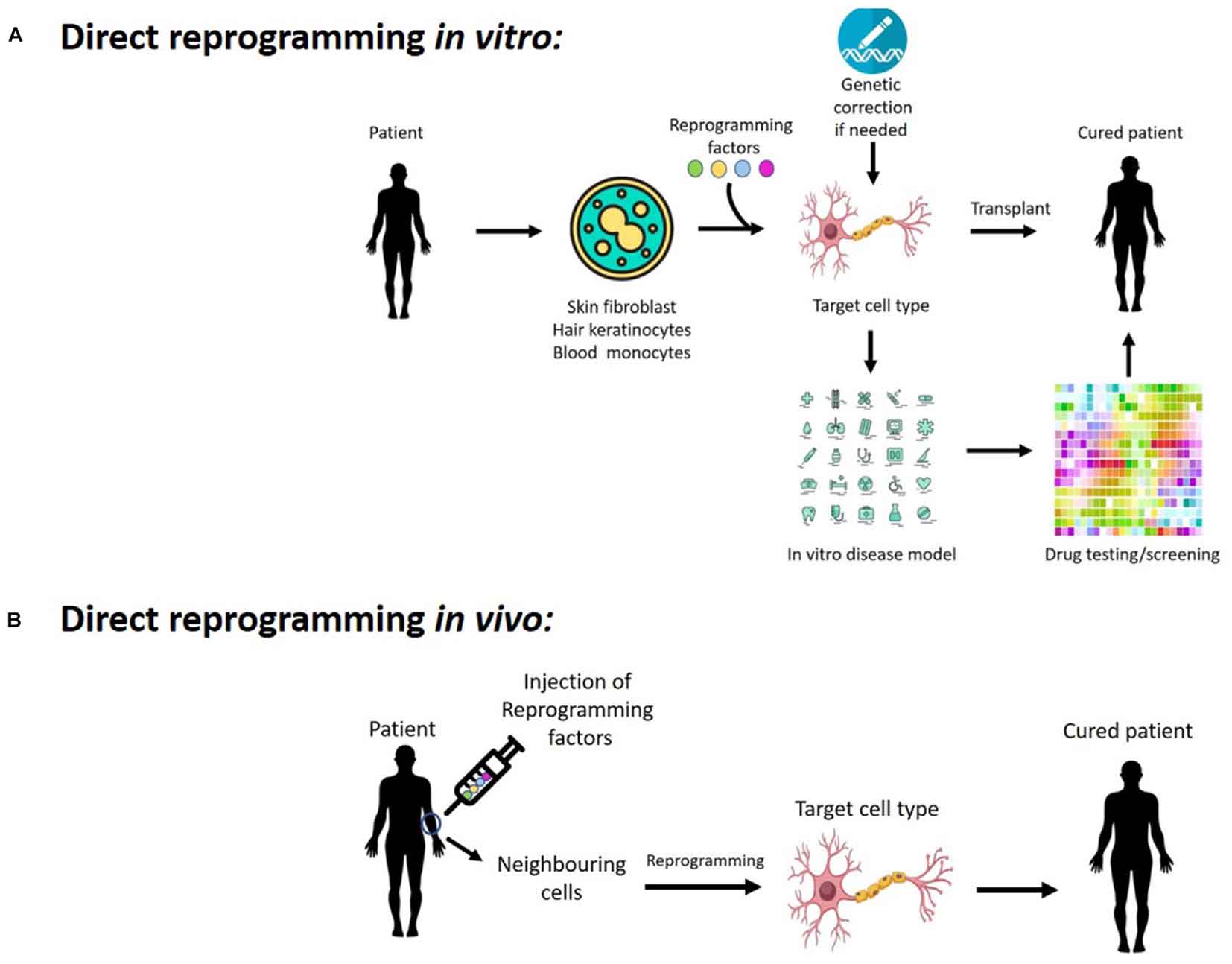
FIGURE 1. Potentials of cellular reprogramming (A) in vitro and (B) in vivo for regenerative medicine, disease modeling, as well as drug discovery and testing gene therapy.
In this review we will discuss both the in vitro and in vivo application of direct reprogramming, its clinical implementation in regenerative medicine, the future directions and the current challenges to translate this technology into clinical practice.
The underlying concept of cellular reprogramming is that the transcriptome plays an important role in defining cellular identity, hence alteration of the transcriptome to a profile specific to the target cell type would allow us to control and convert cell fate. Many reprogramming studies utilized a ‘defined transcription factors’ approach, where a combination of transcription factors, usually master regulators during development, were used to alter the transcriptome and effectively reprogram cell fates. Here we will discuss recent reprogramming studies in conversion of cell lineages, with a focus on transcription factor-based reprogramming studies. However, it should be noted that other reprogramming approaches have also been described, such as the use of microRNA and small molecules for cellular reprogramming which were reviewed in details recently by others (Janowska et al., 2018; Li et al., 2018; Lu and Yoo, 2018).
In a landmark study, Vierbuchen et al. (2010) showed that Ascl1, a proneural master regulator in neuron specification, can be used in combination with two other factors (Brn2, Mytl1), to reprogram mouse fibroblasts into induced Neurons (iN). These iN expressed neuronal markers and have the ability to form functional synapses (Vierbuchen et al., 2010). Subsequent studies demonstrated the generation of iN from human embryonic fibroblast, using ASCL1, BRN2, MYTL1 with the addition of NEUROD1 (Pang et al., 2011), which is a helix-loop-helix transcription factor that plays an important role in neuronal development. Notably, the derived human iN are capable of synapse formation and possess a functional electrophysiological profile indicative of immature neurons. Further study demonstrated that Ascl1 solely is sufficient to reprogram fibroblasts to excitatory iN (Chanda et al., 2014). These results established Ascl1 as a powerful reprogramming factor for iN generation. Although Brn2 and Mytl1 are not required for iN reprogramming, the two factors play a role in enhancing early maturation of iN reprogramming.
It should be noted that the derived iN are often a mixed population of neuron subtypes, often a mix of GABAergic or glutamatergic neurons. Other transcription factors have been identified for direct reprogramming into neuronal subtypes. For instance, both human and mouse fibroblasts can be reprogrammed into dopaminergic neurons with functional electrophysiology using Ascl1, Nurr1, and Lmx1a (Caiazzo et al., 2011). Importantly, this reprogramming process has been used to generate dopaminergic neurons from a Parkinson’s disease patient, which demonstrated the potential to use direct reprogramming for disease modeling (Figure 1A). Other studies have identified additional reprogramming factors for dopaminergic neuron reprogramming, including Foxa2, Brn2 (Pfisterer et al., 2011) and Pitx3 (Kim et al., 2011). Moreover, fibroblasts can be reprogrammed into dopaminergic neurons using a cocktail of five transcription factors, ASCL1, NGN2, SOX2, NURR1 and PITX3, with a reported 40% efficiency in generation of dopaminergic neuron-like cells positive for dopa decarboxylase expression. The reprogrammed neurons exhibited an electrophysiological profile that is dependent of the D2 autoreceptor, thus providing evidence that the reprogrammed neurons are functionally similar to dopaminergic neurons (Liu et al., 2012). In another example, motor neurons can be generated using a similar direct reprogramming strategy. Son et al. (2011) demonstrated that a complex mix of Ascl1, Brn2, Mytl1, Ngn2, Lhx3, Isl1, and Hb9 or Neurod1 allow the reprogramming of fibroblasts into motor neurons. Additional work showed the role of Sox11 in facilitating this reprogramming process (Liu et al., 2013). Similarly, the feasibility of generating sensory neurons has also been demonstrated using direct reprogramming. Brn3a, Ngn1, or Ngn2 allowed the reprogramming of fibroblasts into peripheral sensory neurons that include a subset that respond similarly to itch- and pain-sensing neurons (Blanchard et al., 2015). On the other hand, Ascl1, Brn2, Mytl1, Ngn1, Isl2, and Klf7 have been utilized for direct reprogramming to generate noxious stimulus-detecting neurons with a characteristic inflammatory peripheral sensitization response (Wainger et al., 2015).
Also, emerging evidences have supported feasibility of using miRNAs for cellular reprogramming. Similar to transcription factors, miRNAs are also able to target large networks of genes and epigenetic regulators of the chromatin (Rajman and Schratt, 2017). In particular, overexpression of the neural-enriched miRNA, miR-9/9∗ and miR-124, allowed the direct reprogramming of human fibroblasts into functional neurons (Yoo et al., 2011). This reprogramming process can be further enhanced by the addition of NEUROD2, ASCL1 and MYT1L, which supported an important role of the miR-9/9∗-124 signaling in neuronal reprogramming. Furthermore, miRNA-mediated reprogramming to generate specific neuronal subtypes is also possible. A follow-up study by the same group showed that this miRNA-mediated reprogramming process can be directed to generate striatal medium spiny neurons, by addition of four other transcription factors: BCL11B, DLX1, DLX2, and MYT1L (Victor et al., 2014). On the other hand, co-expression of miR-9/9∗-124 with two other transcription factors, ISL1 and LHX3, promote reprogramming of human fibroblasts to motor neurons (Abernathy et al., 2017). Similarly, miR124 can also be used with other transcription factors (ASCL1, NURR1, LMX1A) for neuronal reprogramming to generate human dopaminergic neurons (Jiang et al., 2015). Recent studies have provided insights into the mechanism by which miR-9/9∗-124 promote neuronal reprogramming. There are evidences to support that miR09/9∗-124 promote neuronal reprogramming by reducing activity of the transcriptional repressor REST, which in turn reconfigure chromatin accessibility to allow expression of neuronal genes (Abernathy et al., 2017; Drouin-Ouellet et al., 2017; Lee et al., 2018). Collectively, these studies support an important role of miRNA in the conversion of cell fate to neurons.
Furthermore, small molecules have been identified to complement transcription factor-based direct reprogramming. This includes Forskolin, an activator of adenylate cyclase resulting in an increase in c-AMP, as well as Dorsomorphin, an inhibitor of BMP signaling. The combination of these two small molecules has been demonstrated to enhance neuronal reprogramming using NGN2 or SOX11 (Liu et al., 2013). In addition, inhibition of SMAD signaling (SB431542 and noggin) and GSK3B signaling (CHIR99021) have also been shown to promote neuronal reprogramming (Ladewig et al., 2012). An important milestone was achieved in 2015, when HongKui Deng’s group developed an all chemical approach to convert mouse fibroblasts into induced neurons that doesn’t require genetic manipulation (Li et al., 2015). The authors demonstrated that the use of five small molecules (ISX9, SB431542, Forskolin, CHIR99021, I-BET151) was sufficient to rewrite the fibroblast-specific transcriptome, activate neuronal-related genes and effectively convert fibroblast into neurons with up to 90% efficiency in 16 days. Subsequent work by Hu et al. (2015) extended this all-chemical approach for neuronal reprogramming of human fibroblasts, using a cocktail of seven small molecules: valproic acid, CHIR99021, REPSOX, Forskolin, Sp600125, GO6983, and Y27632. This reprogramming method allowed derivation of human iN containing a mix of GABAergic, cholinergic and dopaminergic neurons, and provided an alternative approach for neuronal reprogramming without the aid of transcription factors.
As direct reprogramming allows the conversion of cells from distinct lineages, it is postulated that this reprogramming process would potentially be more efficient to convert cell fates across lineages that are closely related and more similar in nature, compared to lineages that are distant and markedly different. For instance, iPS cell generation using keratinocytes exhibits a much higher efficiency compared to using fibroblasts (Aasen et al., 2008; Piao et al., 2014). One reason for the increased efficiency can be attributed to the nature of fibroblasts, which go through a mesenchymal-to-epithelial transition in the process of iPS cell reprogramming (Li et al., 2010; Takahashi et al., 2014), whereas keratinocytes of ectodermal origin may not require that transition. Another cellular reprogramming example across related lineages is the conversion of glia to neurons. Early work by Heins et al. (2002) showed that overexpression of Pax6, a master regulator of neural specification, can induce neurogenesis in glial cells. Further work by Heinrich and colleagues identified Ngn2 as a reprogramming factor to convert astroglia into glutamatergic neurons, while expression of Dlx2 induced GABAergic neurons with mature dendrites. The reprogrammed cells expressed pan-neuronal markers (βIII tubulin, MAP2) while the glial marker GFAP was downregulated. Importantly, the reprogrammed neurons were capable of establishing synapses and possessed functional electrophysiology (Heinrich et al., 2010). Similarly, others have highlighted the feasibility of using small molecules to reprogram human fetal astrocytes into neurons (Zhang et al., 2015; Gao et al., 2017).
The prospect of using direct reprogramming to convert endogenous cells in vivo into the target cells that are lost in disease or injury provides an exciting approach for regenerative medicine (Figure 1B). This is an emerging area of research that is attracting enormous interest for its therapeutic potential (Srivastava and DeWitt, 2016; Wong and Nguyen, 2017). Notably, this approach bypasses many of the major obstacles posed by transplantation-based regenerative strategies, such as issues with immune rejection and the high cost involved in production of clinical grade cells for transplantation. Instead, in vivo reprogramming can be promoted by viral delivery of reprogramming factors, such as an injection of adeno-associated viruses (AAVs) to the patient. This strategy is substantially cheaper to produce, easier to store and often an easier clinical procedure compared to cell transplantation.
Early pioneering work has demonstrated the feasibility of in vivo reprogramming to convert cell fates in the pancreas and cardiac systems. A pioneering study by Zhou et al. (2008) showed the reprogramming of pancreatic exocrine cells into β-cells in diabetic adult mice by using a combination of three transcription factors (Ngn3, Pdx1, and Mafa). Remarkably, the induced β-cells improve insulin production in the diabetic mice, demonstrating a great potential for using direct reprogramming as a regenerative strategy to treat diabetes (Zhou et al., 2008). A subsequent study has identified the use of Ngn3 and Mafa for in vivo reprogramming to convert pancreatic acinar cells into δ- or α-like endocrine cells (Li et al., 2014) and advanced the discovery of transcription factors that allow direct reprogramming to generate a panel of pancreatic cell types in vivo.
In the cardiac system, Qian et al. (2012) demonstrated that cardiac fibroblasts can be reprogrammed into cardiomyocytes in vivo by using transcription factors Gata4, Mef2c, and Tbx5. The induced cardiomyocytes possess similar characteristics of binucleation, sarcomere assembly, electrical coupling, and gene expression profile typical of cardiomyocytes. Using a mouse myocardial infarction model, the authors injected a retrovirus carrying GMT into the infarct site and found that the infarct area was diminished. Notably, about 35% of the cardiomyocytes in the marginal area of the infarct were reprogrammed from cardiac fibroblasts. The induced cardiomyocytes can express the functional characteristics of adult ventricular muscles; about half of them have an organized sarcomere structure that improves cardiac function 2–3 months after myocardial infarction. Concurrently, Song et al. (2012) showed the use of a similar cocktail (Gata4, Mef2c, Hand2, Tbx5) to successfully reprogram non-cardiomyocytes into induced cardiomyocytes in mice. Similarly, in vivo reprogramming of cardiac fibroblasts to induced cardiomyocytes can also be achieved with miRNAs (Jayawardena et al., 2012). The induced cardiomyocytes can attenuate cardiac dysfunction after myocardial infarction. Collectively, these studies demonstrated the use of in vivo reprogramming for cardiomyocyte regeneration which would have important therapeutic implications for myocardial infarction.
Another interesting research direction is the use of in vivo reprogramming technologies to rejuvenate aging cells. It is widely established that the use of the Yamanaka factors (Oct4, Sox2, Klf4, and c-Myc) can reprogram somatic cells into iPS cells – a reprogramming process that reverses many aspects of aging signatures in the target cells, including reorganization of mitochondria and reconstituted telomerase activity (Rohani et al., 2014). Overexpression of the Yamanaka factors in vivo allowed generation of iPS cells in mice, however this resulted in teratoma formation in multiple organs (Abad et al., 2013). Ocampo et al. (2016) overcame this issue of tumorigenicity using an inducible system to repeatedly induce short-term expression of the Yamanaka factors in mice. Interestingly, this strategy prevented teratoma formation in the mice. Using a premature aging mouse model for Hutchinson-Gilford progeria syndrome, the author showed that this partial reprogramming strategy reversed the cellular and physiological hallmarks of aging, such as DNA damage and epigenetic dysregulation, and strikingly extended the lifespan of these mice by ∼30%. Furthermore, this reprogramming approach also allowed wild type aged mice to increase resistance to metabolic disease and muscle injury. These results supported the use of short-term induction of Yamanaka factors for partial reprogramming to rejuvenate aged cells, and provided exciting possibilities of using regenerative medicine for anti-aging.
In vivo reprogramming in the nervous system has advanced rapidly since 2013. Transcription factor-based reprogramming to induce neurogenesis in the brain in vivo has tremendous potential as a novel approach for neural repair in an injury or disease setting. For instance, Grande et al. (2013) showed that Neurog2 can be used for in vivo reprogramming to generate new neurons in the neocortex and striatum following a stab wound or ischemia. The addition of FGF2 and EGF was found to enhance this process and generate more neurons in vivo. Interestingly, the outcome of this neuronal reprogramming in vivo was highly dependent on location; in vivo reprogramming in the striatum only induced a small number of neurons, whereas in vivo reprogramming in the neocortex induced a large number of immature neurons, but only a small number of neurons matured to a later stage. However, the precise cell types that were targeted for reprogramming remain unclear in this study, as the retroviruses used in this study did not deliver reprogramming factors to a specific cell type in the brain. Future studies with the use of cell type-specific promoter to induce reprogramming will help address this issue.
In the nervous system, many studies have targeted the glial cells for in vivo reprogramming into neurons. An early study by Niu et al. (2013) demonstrated in vivo neuronal reprogramming with lentiviral delivery of a single transcription factor – Sox2. Sox2 alone was sufficient to reprogram striatal astrocytes into neuroblasts within the adult mouse brain. Subsequent addition of BDNF and noggin can direct the induced neuroblasts to develop into electrophysiologically mature neurons (Niu et al., 2013). Concurrently, Torper et al. (2013) reported a glial-neuron reprogramming approach that did not require treatment with neurotrophic factors. The authors demonstrated that lentiviral delivery of Ascl1, Brn2, and Myt1l into the striatal astrocytes of the mouse brain converted them into neurons, with a reported efficiency of up to ∼6% (Torper et al., 2013). The induced neurons exhibit neuronal morphology and express NeuN, but are otherwise not well-characterized. A follow up study by the same group reported an improved method for neuronal reprogramming (Torper et al., 2015). This improved method utilized AAV delivery of Ascl1, Lmx1a, and Nurr1 into striatal NG2 glia, which resulted in induced neurons with functional electrophysiological properties. Using modified rabies virus to trace cell-cell connectivity, the authors demonstrated that the induced neurons successfully integrated into the existing host circuitry, with an average of innervation by 3–4 host neurons. This confirmed synaptogenesis between existing host afferent inputs and the newly formed in vivo reprogrammed neurons (Torper et al., 2015). Subsequent study by Pereira et al. (2017) showed that the NG2 glia-derived induced neurons reach functional maturation after 12 weeks following in vivo reprogramming, with properties similar to fast-spiking, parvalbumin-containing interneurons.
Whilst the in vivo reprogramming studies discussed above targeted quiescent glial cells, other studies targeted the reactive glial cells, which are apparent in disease or injury settings. Using a stab wound injury model to elicit activation of glial cells, Heinrich et al. (2014) showed that sole addition of Sox2 can induce conversion of reactive glia into induced neurons in the adult mouse cerebral cortex. This demonstrated the potential for using in vivo reprogramming for cortical repair of acute injury. Although the induced neurons possess functional electrophysiological properties, the authors noted that many of the induced neurons lack neuronal subtype-specific traits such as neurotransmitter release, which may suggest that the reprogrammed cells are in an immature state. Similarly, Guo et al. (2014) provided experimental evidence that overexpression of Neurod1 can reprogram reactive glial cells to neurons in the cortex of a stab-injured or Alzheimer’s disease model. This Neurod1-mediated reprogramming is highly efficient, with a reported efficiency of > 90%. Interestingly, the authors showed a differential effect of Neurod1 in the reprogramming of different glial cells. While astrocytes were mostly reprogrammed into glutamatergic neurons, NG2 glial cells were reprogrammed into both glutamatergic and GABAergic neurons. Collectively, these studies demonstrate a potential therapeutic approach to regenerate neurons and restore lost neuronal function in an injured or diseased brain. Moreover, in vivo reprogramming strategies targeting reactive glial cells for conversion into neurons may also be used to reduce reactive gliosis and reduce glial scarring following injury in the nervous system (Li and Chen, 2016).
Similar advances have been made for in vivo reprogramming in the retina. The major glial cells in the retina, Müller glia (MG), have been shown in a number of species to have the ability to convert into new neurons. In teleost fish and postnatal chickens, upon retinal stresses, MG can dedifferentiate to multipotent progenitors and give rise to all retinal neural cell types to promote retinal regeneration (Fischer and Reh, 2001; Goldman, 2014). In rodents, acute retinal stresses can stimulate MG to proliferate and produce a small number of neurons in vivo (Jadhav et al., 2009). These studies support the notion that, given the appropriate stimuli, the MG possess the plasticity to become retinal neurons, and as such are an ideal target for testing the feasibility of in vivo reprogramming to regenerate retinal neurons.
Pioneering studies from the Reh laboratory have paved ways for using transcription factor-based reprogramming of MG into retinal neurons in vivo. Their work showed that following retinal injury, overexpression of Ascl1 in transgenic mice can reprogram MG into retinal neurons, including photoreceptors, bipolar and amacrine cells (Ueki et al., 2015). However, this reprogramming is only observed in young mice (day 7) and the neurogenic capacity of MG is lost by day 16. Subsequently, an important improvement was reported by the same laboratory. The addition of a histone deacetylase inhibitor Trichostatin A, combined with Ascl1 overexpression following retinal injury, allowed in vivo reprogramming of MG to neurons in adult mice up to 8 months old (Jorstad et al., 2017). Using an elegant Cre-loxP system, the authors expressed Ascl1 specifically in MG and traced their conversion to retinal neurons in vivo. The authors also provided evidence that addition of Trichostatin A enabled epigenetic changes that allow expression of other retinal neuronal genes, such as Otx2, that presumably facilitate this in vivo reprogramming process. Notably, the reprogrammed neurons are mostly bipolar cells and are able to form synaptic connections with the existing retinal circuitry and respond to visual stimuli. These results provide a major step toward using in vivo reprogramming to regenerate the retina.
A more recent study by Yao et al. (2018) built on this in vivo reprogramming strategy to direct MG into rod photoreceptors. Using a two-step reprogramming method, the authors used AAV to first overexpress beta catenin to stimulate MG proliferation, then overexpress a cocktail of Otx2, Crx, and Nrl for differentiation into rod photoreceptors. The MG-derived rod photoreceptors formed functional synapses and are able to functionally integrate into the visual pathway. Although the reprogramming efficiency is difficult to assess in vivo, remarkably the authors showed that this reprogramming process regenerated sufficient rod photoreceptors to restore visual functions in a photoreceptor degeneration mouse model, which included restoration of responses to light and transmission of visual signals to the primary visual cortex. Notably, the demonstration of using AAV for MG reprogramming, as well as functional recovery in a disease model provided a major step toward clinical development of cell therapies to replace retinal neurons, which have important implications in treatment of retinal degenerative diseases.
It is worth noting that the therapeutic potential of in vivo reprogramming is not limited to generating cells that were lost in injury or disease. In vivo reprogramming can also be used to intervene disease progression. For instance, Montana et al. (2013) demonstrated that transgenic knockout of Nrl, an essential transcription factor for the formation of rod photoreceptors during development, can partially reprogram adult mouse rods into cones in vivo. Interestingly, by preventing secondary cone loss, this reprogramming strategy rescued cone function and achieved an improvement in the vision of a mouse model of retinitis pigmentosa. Since then, two follow-up studies have confirmed this finding and improved the reprogramming method, by using AAV-mediated CRISPR/Cas9 to knockout Nrl and preserve cone functions in multiple mouse models of photoreceptor degeneration (Yu et al., 2017; Zhu et al., 2017). Based on this concept of converting rod into cone photoreceptors, another study has highlighted the potential of developing small molecules to inhibit the rod photoreceptor gene Nr2e3 to treat retinitis pigmentosa (Nakamura et al., 2016). Collectively, this cone-to-rod reprogramming approach represents an exciting direction with promising results that warrant further investigation.
In degenerative diseases or injuries that affect the central nervous system, often there is no cure or effective treatment once the neural cells are lost. Regenerative medicine offers a promising approach to replace or regenerate the lost cells to restore normal functions in the patients. Several strategies have been proposed for neuroregeneration (Table 1), including cell replacement therapies using cells from donors, cells derived from pluripotent stem cells, or cells derived from direct reprogramming in vitro. Compared to pluripotent stem cell differentiation, direct reprogramming in vitro offers a potentially faster and more cost-effective approach to generate patient-specific cells for transplantation. Also, the recent development of in vivo reprogramming represents a novel approach for regenerative medicine, by stimulating endogenous cells to replace the cells lost in diseases/injuries. Notably, transcription factor-based reprogramming in vivo would follow the clinical development pathway of gene therapy which is highly attractive commercially (de Wilde et al., 2016), as opposed to cell replacement therapy which incurs high cost associated with production of clinical grade cells. It is clear that there are great opportunities of developing novel regenerative strategies using in vivo reprogramming. However, there are also significant hurdles that need to be overcome before this technology can translate into the clinics. Here we will discuss some of these hurdles for in vivo reprogramming and potential solutions for these issues.
In order to achieve functional rescue in disease or injury, often a sizeable number of cells must be regenerated. Understandably, this may lead to the depletion of endogenous cells that were reprogrammed. This is an important consideration for using in vivo reprogramming for regeneration. To what extent, if any, will normal function be compromised with the repurposing of these cells? Will the remaining population be sufficient to carry out their functional roles? Careful selection of the target cell for reprogramming is therefore necessary. For instance, targeting cell types with a large number of cells, or a cell type whose pool is continually replenished under homeostasis, would be advantageous in the application of in vivo reprogramming.
In degenerative diseases that are caused by genetic mutation(s), it is important to consider that while in vivo reprogramming may regenerate cells that were lost, the reprogrammed cells will still harbor the disease-causing mutation and can be prone to degeneration in the future. In this regard, it can be argued that disease-mediated degeneration may take years to manifest, and as such in vivo reprogramming can be used as a temporal treatment. Alternatively, a combination of gene therapy and in vivo reprogramming, as demonstrated by Yao et al. (2018) in retinal glial cells, will allow for both the correction of genetic mutation(s) and cell replacement in hereditary degenerative diseases.
A major limitation is the identification of reprogramming factors that promote conversion to a specific target cell type. The conventional strategy is to screen through a list of transcription factors that are important for the development of the target cells to identify the factors that allow cellular reprogramming. However, this is a time-consuming and laborious process. To address this issue, several groups have reported on the development of computational algorithm to analyze gene regulatory networks to predict the transcription factors that promote cellular reprogramming. One example is Mogrify, which predicts and ranks transcription factors based on their transcriptional regulatory influence on the target cell type by enrichment and network topology analysis (Rackham et al., 2016). Another recent algorithm incorporates cell cycle features to improve the prediction of reprogramming factors and ideal timing for the introduction of these factors during the direct reprogramming process (Ronquist et al., 2017).
Another consideration for the clinical translation of in vivo reprogramming is the gene delivery system. Ideally, delivery of reprogramming factors should be safe, effective, and specific to the target cells. A marker-specific promoter can be incorporated into the vector design to achieve cell specific expression of reprogramming factors and minimize off-target effects of in vivo reprogramming. There are numerous methods commonly used to deliver the genes, including lentivirus/retrovirus, adenovirus, AAV, and liposomes. Lentivirus and retrovirus allow delivery of larger insert, but given their nature to integrate into the host genome they are less desirable to be used for gene therapy. Of the integration-free viral delivery systems, AAV is a very attractive gene delivery system for gene therapy, with mounting evidence supporting its safety and effectiveness in preclinical and clinical settings (Naso et al., 2017). Moreover, AAV with different serotypes show varying transduction efficiency that are cell type dependent (Balaji et al., 2013), a characteristic which can be used to promote specific gene expression in a target cell type. However, AAV has a ∼5 kb limit on the insert size (Björklund et al., 2000), thus often this is not suitable for co-expression of more than one transgene. Further development and modification of AAV will be an exciting direction to improve the specificity of transduction in target cells.
Alternatively, another potential solution is to use small molecules to replace transcription factors for in vivo reprogramming (Li et al., 2018). Although in vivo reprogramming in the neural systems with an all-chemical approach is still in its infancy, this is an attractive reprogramming strategy for clinical translation, as small molecules can be cost-effective, non-immunogenic, produced in high quantity and efficiently delivered across the membrane into cells. Future studies in this direction would be very exciting with high potentials for clinical translation.
It is imperative to distinguish newly reprogrammed cells from the resident cells within the tissue. To address this, the gene delivery vector can be designed to incorporate a fluorescent tag, such as the Cre-loxP system, for lineage tracing of the cell conversion in vivo. In many cases, the reprogramming efficiency in vivo may be low, thus the analysis techniques utilized have to be sufficiently sensitive to address this. The emergence of single cell analysis provides an exciting technology to understand the process of in vivo reprogramming. For instance, the recent advances in microfluidic technology have greatly enhanced the throughput of single cell transcriptome analysis (Stubbington et al., 2017). Single cell transcriptome analysis is highly suitable to analyze the process of cellular reprogramming which is often inefficient. We expect to see more utilization of single cell RNA sequencing in the coming years.
Rapid advances in the field of cellular reprogramming have created a new paradigm for regenerative medicine. Direct reprogramming provides a feasible method to generate cells in vitro for biological studies, tissue engineering or transplantation. Moreover, the application of direct reprogramming in vivo provides an exciting regenerative strategy to reprogram and repurpose endogenous cells within damaged tissues, which overcomes limitations associated with transplantation of donor cells. The studies outlined here demonstrate advances in their respective fields and collectively demonstrate an exciting progress of applying cellular reprogramming for regenerative medicine. Future studies to test the therapeutic potential of in vivo reprogramming in animal disease models or injury models would be of high interest. We expect that the development of better computational methods would improve prediction of transcription factors to convert cell fates and advance the field of cellular reprogramming. Other emerging new technologies, such as single cell transcriptome analysis and new CRISPR/Cas systems to modulate gene expression, would provide new tools to better understand the cellular reprogramming process, improve the specificity and efficiency of cellular reprogramming and facilitate the clinical application of this technology for regenerative medicine.
LF, LEW, and RW provided conceptual framework of this manuscript. LF, LEW, CT, TN, SH, and AH contributed to writing of manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Some of the graphics used in Figure 1 are designed by Freepik and used under the Creative Commons BY 3.0 license. RW is supported by funding from the Ophthalmic Research Institute of Australia, The University of Melbourne De Brettville Trust, Centre for Eye Research Foundation and the Kel and Rosie Day Foundation. The Centre for Eye Research Australia receives operational infrastructure support from the Victorian Government.
Aasen, T., Raya, A., Barrero, M. J., Garreta, E., Consiglio, A., Gonzalez, F., et al. (2008). Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284. doi: 10.1038/nbt.1503
Abad, M., Mosteiro, L., Pantoja, C., Cañamero, M., Rayon, T., Ors, I., et al. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345. doi: 10.1038/nature12586
Abernathy, D. G., Kim, W. K., McCoy, M. J., Lake, A. M., Ouwenga, R., Lee, S. W., et al. (2017). MicroRNAs Induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 21, 332–348. doi: 10.1016/j.stem.2017.08.002
Balaji, S., King, A., Dhamija, Y., Le, L. D., Shaaban, A. F., Crombleholme, T. M., et al. (2013). Pseudotyped adeno-associated viral vectors for gene transfer in dermal fibroblasts: implications for wound-healing applications. J. Surg. Res. 184, 691–698. doi: 10.1016/j.jss.2013.03.051
Björklund, A., Kirik, D., Rosenblad, C., Georgievska, B., Lundberg, C., and Mandel, R. J. (2000). Towards a neuroprotective gene therapy for parkinson’s disease: use of adenovirus, AAV and lentivirus vectors for gene transfer of GDNF to the nigrostriatal system in the rat parkinson model. Brain Res. 886, 82–98. doi: 10.1016/S0006-8993(00)02915-2
Blanchard, J. W., Eade, K. T., Szűcs, A., Lo Sardo, V., Tsunemoto, R. K., Williams, D., et al. (2015). Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 18, 25–35. doi: 10.1038/nn.3887
Caiazzo, M., Dell’Anno, M. T., Dvoretskova, E., Lazarevic, D., Taverna, S., Leo, D., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227. doi: 10.1038/nature10284
Campbell, K. H., McWhir, J., Ritchie, W. A., and Wilmut, I. (1996). Sheep cloned by nuclear transfer from a cultured cell line. Nature 380, 64–66. doi: 10.1038/380064a0
Chanda, S., Ang, C. E., Davila, J., Pak, C., Mall, M., Lee, Q. Y., et al. (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3, 282–296. doi: 10.1016/j.stemcr.2014.05.020
Davis, R. L., Weintraub, H., and Lassar, A. B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. doi: 10.1016/0092-8674(87)90585-X
de Wilde, S., Guchelaar, H.-J., Zandvliet, M. L., and Meij, P. (2016). Clinical development of gene- and cell-based therapies: overview of the european landscape. Mol. Ther. Methods Clin. Dev. 3:16073. doi: 10.1038/mtm.2016.73
Drouin-Ouellet, J., Lau, S., Brattås, P. L., Rylander Ottosson, D., Pircs, K., Grassi, D. A., et al. (2017). REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol. Med. 9, 1117–1131. doi: 10.15252/emmm.201607471
Fischer, A. J., and Reh, T. A. (2001). Müller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat. Neurosci. 4, 247–252. doi: 10.1038/85090
Gao, L., Guan, W., Wang, M., Wang, H., Yu, J., Liu, Q., et al. (2017). Direct generation of human neuronal cells from adult astrocytes by small molecules. Stem Cell Rep. 8, 538–547. doi: 10.1016/j.stemcr.2017.01.014
Goldman, D. (2014). Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 15, 431–442. doi: 10.1038/nrn3723
Grande, A., Sumiyoshi, K., López-Juárez, A., Howard, J., Sakthivel, B., Aronow, B., et al. (2013). Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat. Commun. 4:2373. doi: 10.1038/ncomms3373
Guo, Z., Zhang, L., Wu, Z., Chen, Y., Wang, F., and Chen, G. (2014). in vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an alzheimer’s disease model. Cell Stem Cell 14, 188–202. doi: 10.1016/j.stem.2013.12.001
Gurdon, J. B. (1962). The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 10, 622–640.
Heinrich, C., Bergami, M., Gascón, S., Lepier, A., Viganò, F., Dimou, L., et al. (2014). Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Rep. 3, 1000–1014. doi: 10.1016/j.stemcr.2014.10.007
Heinrich, C., Blum, R., Gascón, S., Masserdotti, G., Tripathi, P., Sánchez, R., et al. (2010). Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 8:e1000373. doi: 10.1371/journal.pbio.1000373
Heins, N., Malatesta, P., Cecconi, F., Nakafuku, M., Tucker, K. L., Hack, M. A., et al. (2002). Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 5, 308–315. doi: 10.1038/nn828
Hu, W., Qiu, B., Guan, W., Wang, Q., Wang, M., Li, W., et al. (2015). Direct conversion of normal and alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17, 204–212. doi: 10.1016/j.stem.2015.07.006
Jadhav, A. P., Roesch, K., and Cepko, C. L. (2009). Development and neurogenic potential of müller glial cells in the vertebrate retina. Prog. Retin. Eye Res. 28, 249–262. doi: 10.1016/j.preteyeres.2009.05.002
Janowska, J., Gargas, J., Ziemka-Nalecz, M., Teresa, Z., Buzanska, L., and Sypecka, J. (2018). Directed glial differentiation and transdifferentiation for neural tissue regeneration. Exp. Neurol. doi: 10.1016/j.expneurol.2018.08.010 [Epub ahead of print].
Jayawardena, T. M., Egemnazarov, B., Finch, E. A., Zhang, L., Payne, J. A., Pandya, K., et al. (2012). MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110, 1465–1473. doi: 10.1161/CIRCRESAHA.112.269035
Jiang, H., Xu, Z., Zhong, P., Ren, Y., Liang, G., Schilling, H. A., et al. (2015). Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat. Commun. 6:10100. doi: 10.1038/ncomms10100
Jorstad, N. L., Wilken, M. S., Grimes, W. N., Wohl, S. G., VandenBosch, L. S., Yoshimatsu, T., et al. (2017). Stimulation of functional neuronal regeneration from müller glia in adult mice. Nature 548, 103–107. doi: 10.1038/nature23283
Kim, J., Su, S. C., Wang, H., Cheng, A. W., Cassady, J. P., Lodato, M. A., et al. (2011). Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell 9, 413–419. doi: 10.1016/j.stem.2011.09.011
Kulessa, H., Frampton, J., and Graf, T. (1995). GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9, 1250–1262. doi: 10.1101/gad.9.10.1250
Ladewig, J., Koch, P., and Brüstle, O. (2013). Leveling waddington: the emergence of direct programming and the loss of cell fate hierarchies. Nat. Rev. Mol. Cell Biol. 14, 225–236. doi: 10.1038/nrm3543
Ladewig, J., Mertens, J., Kesavan, J., Doerr, J., Poppe, D., Glaue, F., et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9, 575–578. doi: 10.1038/nmeth.1972
Lee, S. W., Oh, Y. M., Lu, Y.-L., Kim, W. K., and Yoo, A. S. (2018). MicroRNAs overcome cell fate barrier by reducing EZH2-controlled REST stability during neuronal conversion of human adult fibroblasts. Dev. Cell 46, 73–84. doi: 10.1016/j.devcel.2018.06.007
Li, H., and Chen, G. (2016). in vivo reprogramming for CNS repair: regenerating neurons from endogenous glial cells. Neuron 91, 728–738. doi: 10.1016/j.neuron.2016.08.004
Li, R., Liang, J., Ni, S., Zhou, T., Qing, X., Li, H., et al. (2010). A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63. doi: 10.1016/j.stem.2010.04.014
Li, W., Nakanishi, M., Zumsteg, A., Shear, M., Wright, C., Melton, D. A., et al. (2014). In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. Elife 3:e01846. doi: 10.7554/eLife.01846
Li, X., Xu, J., and Deng, H. (2018). Small molecule-induced cellular fate reprogramming: promising road leading to rome. Curr. Opin. Genet. Dev. 52, 29–35. doi: 10.1016/j.gde.2018.05.004
Li, X., Zuo, X., Jing, J., Ma, Y., Wang, J., Liu, D., et al. (2015). Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203. doi: 10.1016/j.stem.2015.06.003
Liu, M.-L., Zang, T., Zou, Y., Chang, J. C., Gibson, J. R., Huber, K. M., et al. (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4:2183. doi: 10.1038/ncomms3183
Liu, X., Li, F., Stubblefield, E. A., Blanchard, B., Richards, T. L., Larson, G. A., et al. (2012). Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 22, 321–332. doi: 10.1038/cr.2011.181
Lu, Y.-L., and Yoo, A. S. (2018). Mechanistic insights into microRNA-induced neuronal reprogramming of human adult fibroblasts. Front. Neurosci. 12:522. doi: 10.3389/fnins.2018.00522
Montana, C. L., Kolesnikov, A. V., Shen, S. Q., Myers, C. A., Kefalov, V. J., and Corbo, J. C. (2013). Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proc. Natl. Acad. Sci. U. S. A. 110, 1732–1737. doi: 10.1073/pnas.1214387110
Nakamura, P. A., Tang, S., Shimchuk, A. A., Ding, S., and Reh, T. A. (2016). Potential of small molecule–mediated reprogramming of rod photoreceptors to treat retinitis pigmentosa. Invest. Opthalmol. Vis. Sci. 57:6407. doi: 10.1167/iovs.16-20177
Naso, M. F., Tomkowicz, B., Perry, W. L. III, and Strohl, W. R. (2017). Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31, 317–334. doi: 10.1007/s40259-017-0234-5
Niu, W., Zang, T., Zou, Y., Fang, S., Smith, D. K., Bachoo, R., et al. (2013). In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175. doi: 10.1038/ncb2843
Ocampo, A., Reddy, P., Martinez-Redondo, P., Platero-Luengo, A., Hatanaka, F., Hishida, T., et al. (2016). In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167, 1719–1733. doi: 10.1016/j.cell.2016.11.052
Pang, Z. P., Yang, N., Vierbuchen, T., Ostermeier, A., Fuentes, D. R., Yang, T. Q., et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223. doi: 10.1038/nature10202
Pereira, M., Birtele, M., Shrigley, S., Benitez, J. A., Hedlund, E., Parmar, M., et al. (2017). direct reprogramming of resident ng2 glia into neurons with properties of fast-spiking parvalbumin-containing interneurons. Stem Cell Rep. 9, 742–751. doi: 10.1016/j.stemcr.2017.07.023
Pfisterer, U., Kirkeby, A., Torper, O., Wood, J., Nelander, J., Dufour, A., et al. (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U. S. A. 108, 10343–10348. doi: 10.1073/pnas.1105135108
Piao, Y., Hung, S. S.-C., Lim, S. Y., Wong, R. C.-B., and Ko, M. S. H. (2014). Efficient generation of integration-free human induced pluripotent stem cells from keratinocytes by simple transfection of episomal vectors. Stem Cells Transl. Med. 3, 787–791. doi: 10.5966/sctm.2013-0036
Qian, L., Huang, Y., Spencer, C. I., Foley, A., Vedantham, V., Liu, L., et al. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598. doi: 10.1038/nature11044
Rackham, O. J., Firas, J., Fang, H., Oates, M. E., Holmes, M. L., Knaupp, A. S., et al. (2016). A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 48, 331–335. doi: 10.1038/ng.3487
Rajman, M., and Schratt, G. (2017). MicroRNAs in neural development: from master regulators to fine-tuners. Development 144, 2310–2322. doi: 10.1242/dev.144337
Rohani, L., Johnson, A. A., Arnold, A., and Stolzing, A. (2014). The aging signature: a hallmark of induced pluripotent stem cells? Aging Cell 13, 2–7. doi: 10.1111/acel.12182
Ronquist, S., Patterson, G., Muir, L. A., Lindsly, S., Chen, H., Brown, M., et al. (2017). Algorithm for cellular reprogramming. Proc. Natl. Acad. Sci. U.S.A. 114, 11832–11837. doi: 10.1073/pnas.1712350114
Son, E. Y., Ichida, J. K., Wainger, B. J., Toma, J. S., Rafuse, V. F., Woolf, C. J., et al. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218. doi: 10.1016/j.stem.2011.07.014
Song, K., Nam, Y.-J., Luo, X., Qi, X., Tan, W., Huang, G. N., et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604. doi: 10.1038/nature11139
Srivastava, D., and DeWitt, N. (2016). In vivo cellular reprogramming: the next generation. Cell 166, 1386–1396. doi: 10.1016/j.cell.2016.08.055
Stubbington, M. J. T., Rozenblatt-Rosen, O., Regev, A., and Teichmann, S. A. (2017). Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63. doi: 10.1126/science.aan6828
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Sasaki, A., Yamamoto, M., et al. (2014). Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 5:3678. doi: 10.1038/ncomms4678
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Torper, O., Ottosson, D. R., Pereira, M., Lau, S., Cardoso, T., Grealish, S., et al. (2015). In vivo reprogramming of striatal NG2 glia into functional neurons that integrate into local host circuitry. Cell Rep. 12, 474–481. doi: 10.1016/j.celrep.2015.06.040
Torper, O., Pfisterer, U., Wolf, D. A., Pereira, M., Lau, S., Jakobsson, J., et al. (2013). Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 7038–7043. doi: 10.1073/pnas.1303829110
Ueki, Y., Wilken, M. S., Cox, K. E., Chipman, L., Jorstad, N., Sternhagen, K., et al. (2015). Transgenic expression of the proneural transcription factor ascl1 in müller glia stimulates retinal regeneration in young mice. Proc. Natl. Acad. Sci. U.S.A. 112, 13717–13722. doi: 10.1073/pnas.1510595112
Victor, M. B., Richner, M., Hermanstyne, T. O., Ransdell, J. L., Sobieski, C., Deng, P.-Y., et al. (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84, 311–323. doi: 10.1016/j.neuron.2014.10.016
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Südhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. doi: 10.1038/nature08797
Wainger, B. J., Buttermore, E. D., Oliveira, J. T., Mellin, C., Lee, S., Saber, W. A., et al. (2015). Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 18, 17–24. doi: 10.1038/nn.3886
Wong, R.-B., and Nguyen, T. (2017). Neuroregeneration using in vivo cellular reprogramming. Neural Regen. Res. 12:1073. doi: 10.4103/1673-5374.211182
Yao, K., Qiu, S., Wang, Y. V., Park, S. J. H., Mohns, E. J., Mehta, B., et al. (2018). Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 560, 484–488. doi: 10.1038/s41586-018-0425-3
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. doi: 10.1038/nature10323
Yu, W., Mookherjee, S., Chaitankar, V., Hiriyanna, S., Kim, J.-W., Brooks, M., et al. (2017). Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun. 8:14716. doi: 10.1038/ncomms14716
Zhang, L., Yin, J.-C., Yeh, H., Ma, N.-X., Lee, G., Chen, X. A., et al. (2015). Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 17, 735–747. doi: 10.1016/j.stem.2015.09.012
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J., and Melton, D. A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455, 627–632. doi: 10.1038/nature07314
Keywords: cell reprogramming, retina, neuroregeneration, direct reprogramming, in vivo reprogramming, regenerative medicine, gene therapeutics
Citation: Fang L, El Wazan L, Tan C, Nguyen T, Hung SSC, Hewitt AW and Wong RCB (2018) Potentials of Cellular Reprogramming as a Novel Strategy for Neuroregeneration. Front. Cell. Neurosci. 12:460. doi: 10.3389/fncel.2018.00460
Received: 22 September 2018; Accepted: 12 November 2018;
Published: 30 November 2018.
Edited by:
Ioan Opris, University of Miami, United StatesReviewed by:
Xiang Li, University of Wisconsin-Madison, United StatesCopyright © 2018 Fang, El Wazan, Tan, Nguyen, Hung, Hewitt and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raymond C. B. Wong, d29uZ2NiQHVuaW1lbGIuZWR1LmF1
†Equal first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.