- Department of Anesthesiology, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, China
Long noncoding RNAs (lncRNAs) refer to a group of noncoding RNAs (ncRNAs) that has a transcript of more than 200 nucleotides in length in eukaryotic cells. The lncRNAs regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels by multiple action modes. In this review, we describe the diverse roles reported for lncRNAs, and discuss how they could mechanistically be involved in the development of central nervous system (CNS) and neurodegenerative diseases. Further studies on the function of lncRNAs and their mechanism will help deepen our understanding of the development, function, and diseases of the CNS, and provide new ideas for the design and development of some therapeutic drugs.
Introduction
Over the past decade, the extensive applications of second-generation sequencing technology have led to the discovery of tens of thousands of RNA transcripts that have similar properties to mRNAs, but are not translated into proteins. Long noncoding RNA (lncRNA) is a kind of noncoding RNA (ncRNA), with a length of longer than 200 nucleotides, that lacks a significant open reading frame (ORF) encoding a protein (Sun and Kraus, 2015). The central nervous system (CNS) is the most highly evolved and sophisticated biological system. The development of the CNS is a complex arrangement of stem cells, growth/differentiation factors, transcription factors, and epigenetic control. It consists of a large number of neuronal and glial cell subtypes distributed in rigorous and precise regions, forming a dynamic neural network that responds to internal signals and external stimuli and then is responsible for mediating the complex functional repertoire of the CNS including performing higher level functions, for example, cognition and behavior (Graff and Mansuy, 2008). As one of the most abundant ncRNA classes, lncRNAs are derived from different locations in the genome for transcription and are highly expressed in the brain (Mercer et al., 2008; Qureshi et al., 2010). The role of lncRNAs has been validated in brain development, neuronal function, maintenance, and differentiation.
Neurodegenerative diseases are associated with multiple clinical manifestations, brain pathologies, and health consequences (Quan et al., 2017). They include relatively well-known conditions like Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD). In particular, AD and PD are a group of typically late-onset, progressive disorders that lead to cognitive and/or movement disorders (Peden and Ironside, 2012). Although drug therapy and/or surgery can delay the progression of these diseases, most neurodegenerative diseases remain untreatable. In addition, neurodegenerative diseases represent an increasing financial burden on health care systems, which attempt to respond to an aging population. Therefore, there is an urgent need to develop methods for preventing or curing neurodegenerative diseases. Some efforts carried out by the scientific community revealed important insights into the molecular bases of these disease, but the specific mechanism remains unknown. Increasing evidence has suggested that lncRNAs are involved in the pathogenesis of neurodegenerative diseases (Johnson, 2012; Briggs et al., 2015; Riva et al., 2016). This review summarizes data on lncRNA expression in the central nervous system (CNS) and neurodegenerative diseases and focuses on the role of some specific lncRNAs, which may provide new insights into our understanding of the etiology and pathophysiology of the neurodegenerative diseases.
Biology of lncRNAs
Definition of lncRNAs
Genomewide analysis of the eukaryotic transcriptome revealed that up to 90% of the human genome IS transcribed. However, the GENCODE-annotated exon of the protein-encoding gene covers only 2.94% of the genome, while the rest are ncRNAs (Djebali et al., 2012). Noncoding transcripts are further divided into house-keeping ncRNAs and regulatory ncRNAs. House-keeping ncRNAs include ribosomes, metastasis, small nuclei, and small nucleolar RNA. Regulatory ncRNAs are generally divided into two classes based on nucleotide length. Those <200 nucleotides are commonly referred to as short/small ncRNAs, including microRNAs (miRNAs), small interfering RNAs, and Piwi-related RNAs, and those >200 nucleotides are known as lncRNAs (Nagano and Fraser, 2011).
The lncRNA transcripts are partially similar to messenger RNAs (mRNAs) as they are frequently transcribed by RNA polymerase II, contain classical splice sites (GU/AG), have an mRNA-like structure that contains intron and exon structures, exhibit alternative splicing, no open reading frame (ORF) in the sequence, and are associated with the same types of histone modifications as protein-coding genes (PCGs)(Ponting et al., 2009). They also have a specific secondary structure that provides multiple sites for protein binding or the specific binding between DNA and RNA by the principle of base pair complementarity. The main sources of lncRNAs are from PCG-related regions (Magistri et al., 2012), gene regulatory regions (Hung et al., 2011; Mercer et al., 2011), and specific chromosomal regions (Azzalin et al., 2007). The lncRNA was originally thought to be the “noise” of genomic transcription and as not having biological functions. However, recent studies have shown that lncRNAs can regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels, and participate in X-chromosome silencing, genome imprinting, and chromatin modification, transcriptional activation, and many other important biological processes (Singh and Prasanth, 2013; Goff and Rinn, 2015; Kazemzadeh et al., 2015). Until now, only very few lncRNAs have been validated by experiment, while most of the lncRNAs have been annotated via bioinformatics and still need further experimental verification.
Expression and Sequence Conservation of lncRNAs
In 2002, Japanese scientists revealed that in large-scale sequencing of a mouse full-length complementary DNA (cDNA) library, a large number of ncRNA transcripts were identified (Okazaki et al., 2002). However, due to lack of functional annotation, these RNA transcripts did not attract the attention of researchers during subsequent periods. Not until 2007 did the situation change. Rinn et al. in Stanford University reported a 2.2-Kb functional lncRNA gene (Hox transcript antisense intergenic RNA, HOTAIR) (Rinn et al., 2007). It was found that HOTAIR could interact with the protein complex polycomb, which can modify chromatin, inhibit the transcription of the Hox gene, and regulate the growth and development of organisms. In 2008, Mercer et al. used in situ hybridization to identify the expression of a large amount of lncRNAs in mouse brain (Mercer et al., 2008). The expression levels of these lncRNAs are associated with specific neuroanatomical locations, cell types, and subcellular locations. For example, Evf2 is mainly expressed in the ventral forebrain. The tissue-specific expression of lncRNAs also includes Hox transcript antisense intergenic RNA myeloid1 (HOTAIRM1), which is specifically expressed in the bone marrow (Zhang et al., 2009) and Msx1 antisense RNA (Msx1-AS RNA), which is expressed only in differentiated teeth and bone cells (Coudert et al., 2005). The expression level of Msx1-AS RNA is negatively correlated with the content of Msx1 protein (Babajko et al., 2009).
The lncRNAs can appear in different subcellular structures, and the proportion of those lncRNAs located in the nucleus is the largest. For example, lncRNA MEN ε/β is mainly located in the nucleus, and is an important component of nuclear substructure paraspeckles (Sasaki et al., 2009; Sunwoo et al., 2009). Furthermore, metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) and nuclear-enriched abundant transcript 1 (Neat1) are localized mainly in the nuclear speckle of the nucleus, and are associated with the cleavage of RNA precursors (Hutchinson et al., 2007; Tripathi et al., 2010). In addition, Cesana et al. reported that linc-MD1 is mainly expressed in the cytoplasm of differentiated muscle cells, and regulates the differentiation of skeletal muscles as a competing endogenous RNA (ceRNA) (Cesana et al., 2011). In 2011, Rackham et al. first identified three lncRNAs (lncND5, lncND6, and lncCytb) encoded by mitochondrial genome DNA in the analysis of high-throughput sequencing data (Rackham et al., 2011). This shows that lncRNAs may exist in many subcellular structures. Hence, special subcellular localization plays an important role in the biological function of lncRNAs.
The sequence conservation of lncRNAs is low. The sequence similarity is close to the intron region of the PCG, lower than 70% in humans and mice, and slightly lower than that in the 5′UTR or 3′UTR of genes (Pang et al., 2006). It was revealed in some studies that low sequence conservation did not affect the functional conservation of lncRNAs. Illustrated by the examples of Xist and HOTAIR expressed in mammals, although they are not highly conservative in sequence, the roles in X chromosome dosage compensation and epigenetic silencing are the same (Braidotti et al., 2004; Pauler et al., 2005). In addition, some lncRNAs with high sequence homology have also been found such as MALAT1 and Neat1 (Nakagawa et al., 2012; Zhang et al., 2012).
The lncRNAs in CNS Development
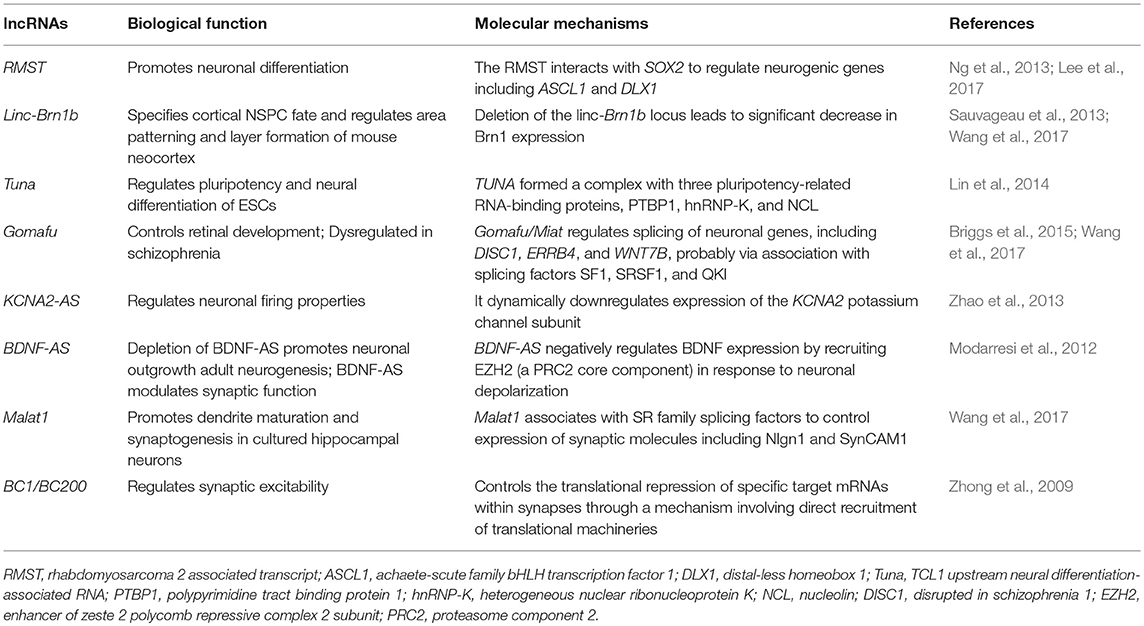
The CNS development is a complex and stereotyped process that requires the precise spatiotemporal regulation of pluripotent stem cell proliferation and differentiation. Neurons are able to change their set of synaptic connections and the relative strength of each of these connections over time in response to sensory experience and other environmental cues. The dynamic expression of lncRNAs plays an important role in controlling these processes (Table 1).
Molecular Mechanisms of lncRNAs in Cell Proliferation and Differentiation
A study has revealed that approximately 40% of lncRNAs are distributed in the CNS (equivalent to 4,000–20,000 lncRNA genes) (Briggs et al., 2015). It may be that brain complexity requires more regulatory RNA to maintain normal brain development and function. The lncRNAs are involved in the regulation of the proliferation and development of the nervous system, and enable the nervous system to proliferate and differentiate according to normal time and spatial orders (Amaral et al., 2013).
The lncRNAs are involved in the differentiation of embryonic cells into neural cells during the embryonic period (Klattenhoff et al., 2013). A genetic analysis of the embryo revealed that lncRNAs are closely associated with the coding genes involved in neuronal differentiation and cell morphological maintenance, such as brain-derived neurotrophic factor (BDNF), developing brain homeobox 1 (DBX1), neuron–glia-related cell-adhesion molecule (Nrcam), etc. (Lv et al., 2013). Studies on neural stem cells revealed that lncRNAs are involved in regulating the differentiation of stem cells into neural cells (Ng et al., 2012; Antoniou et al., 2014; Ramos et al., 2016). For example, Ng et al. revealed that lncRNA-ES1, lncRNA-ES2, and lncRNA-ES are associated with the maintenance and differentiation of neural stem cells. Guttman et al. discovered at least 1,000 conserved intergenic lncRNAs genes by analyzing the chromatin of mouse neurons (including neural precursor cells) in 2009; they also primarily hypothesized and verified the role of lncRNAs in the maintenance of embryonic stem cells (Guttman et al., 2009). Subsequently, functional genomic analysis revealed that these intergenic lncRNA genes were not only involved the differentiation of mouse ventral forebrain-derived neural stem cells, but also in brain aging, mouse hippocampal development, differentiation of gamma aminobutyric acid (GABA)-ergic neurons, oligodendrocyte myelination, and the calcineurin-dependent signaling pathway, by regulating the expression of some important genes (Mercer et al., 2010).
A recent study has revealed that lncRNAs are associated with PCGs in neural development and play an important role in maintaining the intrinsic morphology and characteristics of neurons (Roberts et al., 2014). For example, the sex-determining region Y-box 2 (Sox2) is an important regulator of neural stem cell differentiation and nerve growth. Sox2OT encodes a sense-orientation transcript that overlaps with the Sox2. The genomic proximity of Sox2OT and Sox2 suggested a possible regulatory role for Sox2OT in the neural stem cell differentiation and regeneration of neural cells.
The lncRNAs and Synaptic Plasticity
Synaptic plasticity is the basis of learning and memory and plays a key role in maintaining the stability of the nerve pathway. Some studies found that many lncRNAs might be involved in the regulation of synaptic plasticity (Leal et al., 2014; Panja and Bramham, 2014; Maag et al., 2015). Angelman syndrome (AS) is caused by the deletion of or an inactivating mutation in the maternal E3 ubiquitin ligase (UBE3A) gene and is characterized by intellectual disability, severe developmental delays, and speech impairment (Mabb et al., 2011). Furthermore, due to the expression of lncRNA UBE3A-ATS, the paternal allele of UBE3A undergoes silencing. A study found that hippocampal long-term potentiation (LTP) is defective in the AS-mutant mouse model (Jiang et al., 1998). Similar plasticity deficits may be the cause of learning disabilities observed in AS patients. This important role of UBE3A-ATS in neuronal function highlights the possible role of lncRNA in memory formation, while further studies on the role of other lncRNAs in these processes are necessary. The normal development of gamma-aminobutyric acid-ergic (GABAergic) inhibitory interneurons in the hippocampus is the key to learning in embryonic and adult brains. The Evf-2 lncRNA, which transcribed from the Dlx-5/6 ultraconserved region, is essential for GABAergic neuron development. The Evf-2 acts through the Dlx-2 transcriptional coactivator to increase the transcriptional activity of Dlx-5/6 and glutamate decarboxylase 1 (Gad1 required for the conversion of glutamate to GABA) (Colasante et al., 2008), and then regulates the gene expression of GABAergic interneurons in the developing mouse brain. The Evf-2 silencing leads to abnormal synaptic activity in mice through abnormal formation of GABAergic circuits in the hippocampus and dentate gyrus (Bond et al., 2009).
In neurons, local protein synthesis in synaptodendritic microdomains has been implicated in the growth and plasticity of synapses. The lncRNA brain cytoplasmic 200 (BC200) is selectively located in the dendrites of postsynaptic neurons, and regulates local protein synthesis by blocking transcription initiation, thereby, controlling signal transduction (Kondrashov et al., 2005). In addition, BC1 is an lncRNA located in the dendrites of neurons, which is a specific repressor of translation. Experimental use of internal ribosome entry mechanisms and sucrose density gradient centrifugation showed that BC1-mediated repression targets translation at the level of initiation. Specifically, BC1 RNA inhibited formation of the 48S preinitiation complex, i.e., recruitment of the small ribosomal subunit to the mRNA (Wang et al., 2002). After that, the authors demonstrated that the lack of BC1 could induce nerve overexcitation (Wang et al., 2005).
Neurons can change their connections when they face changes in the environment. Alterations in components of ion channels or signal proteins may affect neuronal excitability or neuronal function. For example, the magnitude of the action potential can be affected by changing the subunit stoichiometry of potassium channels. Potassium voltage-gated channel subfamily A member 2 (KCNA2) is a major potassium channel subunit, and its expression is regulated by overlapping antisense RNA when facing peripheral nerve injury or neuralgia. In a healthy rat model, KCNA2 antisense (KCNA2-AS) is expressed in few dorsal root ganglia (DRG) neurons (<20% of DRG), and KCNA2 is expressed in most DRG neurons. However, in the face of peripheral nerve injury, a large amount of KCNA2-AS is induced by transcription factor myeloid zinc finger 1 in DRG neurons. In vivo and in vitro experiments have revealed that the overexpression of KCNA2-AS could decrease KCNA2 mRNA and protein content. The mechanism may be that it binds with competing DNA- or RNA-binding factors, thereby, regulating the expression of KCNA2 (Zhao et al., 2013). This regulation alters the function of DRG neurons. Therefore, KCNA2-AS can respond to peripheral nerve damage by altering synaptic plasticity. The lncRNA NEAT1 provides a scaffolding function in the nucleus that releases regulatory proteins after neuronal activation to finetune excitatory responses and correlate with pathological seizure states. In addition, downregulation of lncRNA NEAT1 results in changes in the expression of multiple gene transcripts involved in ion channel function following neuronal activation (Barry et al., 2017).
The BDNF is a class of secreted growth factors that are essential for neuronal growth, synaptic plasticity, and participation in learning and memory processes (Leal et al., 2014; Ninan, 2014; Zagrebelsky and Korte, 2014). The BDNF is an important growth factor, not only being regulated by miRNA, but also regulated by lncRNA. Dissection of the human BDNF locus revealed that antisense transcription of the BDNF gene from anti-BDNF (BDNF-AS, also annotated as BDNF-OS) in the brain takes place to form dsRNA duplexes with BDNF mRNA. Inhibition of BDNF-AS by antagoNAT in vivo or siRNA in vitro both resulted in increased BDNF mRNA and protein levels, which promoted neurite outgrowth and maturation, suggesting that anti-BDNF plays an important role in BDNF function (Lipovich et al., 2012; Modarresi et al., 2012). Further studies revealed that BDNF-AS inhibits BDNF transcription by recruiting the zeste homolog 2 (EZH2) and polycomb suppression complex 2 (PRC2) enhancers to the BDNF promoter region (Pruunsild et al., 2007).
The lncRNA Gomafu is widely expressed in the brain, (Mercer et al., 2008) and has recently been shown to modulate alternative splicing of the schizophrenia-associated genes DISC1 and ERBB4 (Barry et al., 2014). Since deletion of Erbb4 in mice enhances LTP in the hippocampus, it is important to determine whether Gomafu-regulated splice isoforms affect normal synaptic plasticity (Pitcher et al., 2008; Shamir et al., 2012). In addition, lncRNA MALAT1 mediates spinal cord maturation and synapse formation by recruiting splicing factors. In cultured hippocampal neurons, knock-down of Malat1 reduces synaptic density, whereas overexpression leads to autonomous increase in cells at synaptic density (Bernard et al., 2010).
These findings suggest that lncRNAs can regulate the synaptic plasticity, and thus, the fidelity of cognitive and memory processes by dynamically monitoring and integrating multiple transcriptional and post-transcriptional events.
The Role of lncRNAs in Neurodegenerative Diseases
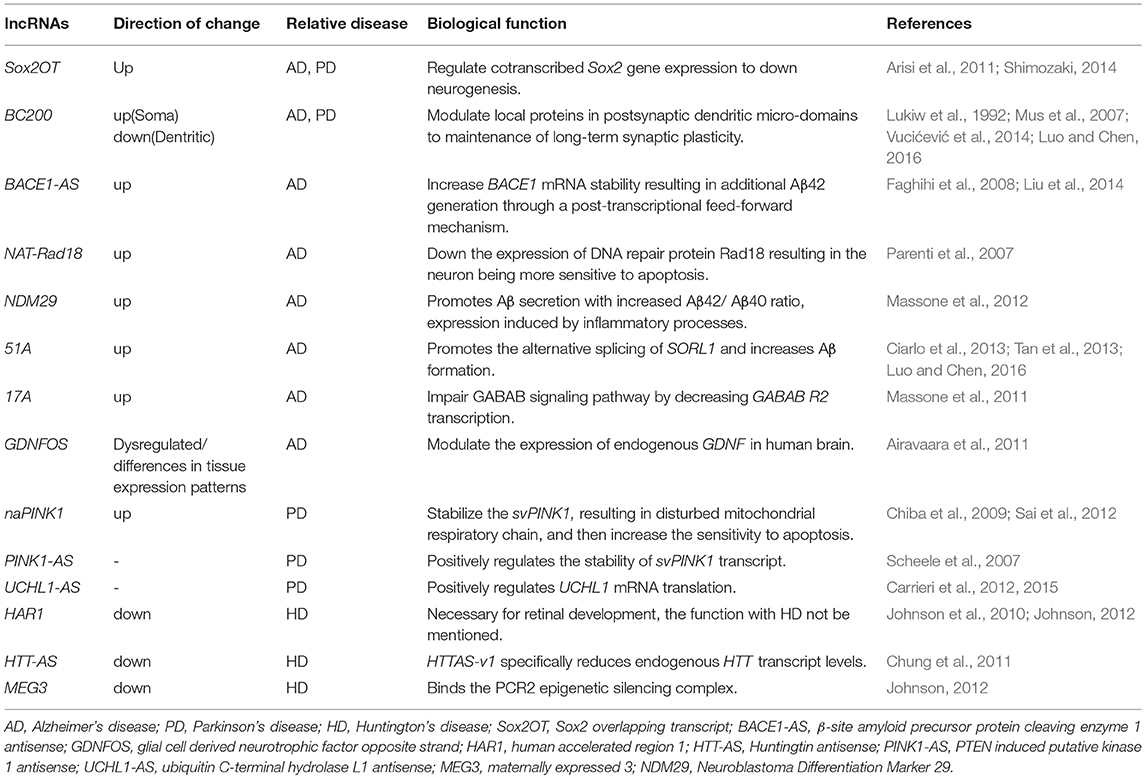
The lncRNAs have broad-spectrum functions in the normal brain development and function maintenance. It is not surprising that dysregulation of lncRNAs might play a pivotal role in neurodegenerative diseases. This view has been reinforced by the identification of a growing number of lncRNAs that directly regulate the expression of genes associated with neurodegenerative disorders, including AD, PD, HD, respectively.
The Correlation of lncRNAs and Alzheimer's Disease
The AD is a neurodegenerative disease with cognitive decline as the main clinical manifestation. Its pathological mechanism has not been completely understood to date. The main pathological characteristic of AD is the progressive disease of neurons accompanied by the loss of neurons, senile plaques (SPs), and neurofibrillary tangles (Dewachter et al., 2000; Ghosal et al., 2009). Senile plaques mainly consist of β-amyloid (Aβ). Amyloid precursor protein was abnormally cleaved by β-amyloid precursor protein cleaving enzyme 1 (BACE1) to produce Aβ. The excessive accumulation of Aβ produces neurotoxicity. The BACE1-AS is an lncRNA transcribed from the antisense strand of the BACE1 gene, which is highly expressed in the brain of patients with AD. A study revealed that the expression of BACE1-AS was significantly increased under extracellular stimulation, such as Aβ42 (Faghihi et al., 2008). However, BACE1-AS does not inhibit the transcription of mRNA by forming dimers through binding with the coding genes as general natural antisense transcripts (NATs). In contrast, BACE1-AS covers the binding site of miR-485-5p on BACE1 mRNA, thereby, silencing the inhibition of miR-485-5p on BACE1 mRNA and increasing the stability of BACE1 mRNA (Faghihi et al., 2010). This leads to the production of more Aβ42 and an increase in the formation of SPs in the brain in AD patients, aggravating the development of the disease. Feng et al. found that the level of the BACE1 is increased in the plasma of AD patients and that it has a high specificity (88%) for AD, indicating BACE1 may be a potential candidate biomarker to predict AD (Feng et al., 2018). Recently, Yang et al., investigated the hippocampal expression patterns of dysregulated lncRNAs in a rat model of AD through microarray (Yang et al., 2017). The authors identified a total of 315 lncRNAs and 311 mRNAs significantly dysregulated in the AD model, such as BC158567, MRAK050857, Mrak033976, etc. These differentially expressed genes are involved in synaptic transmission regulation, cholinergic regulation, and CNS neuron differentiation, all of which are important in learning and memory, as well as the development of AD. Furthermore, the dysregulated lncRNAs in the AD group are involved in the neuroactive ligand–receptor interaction, the renin–angiotensin system, axon guidance, and the PI3K–Akt, MAPK, and mTOR signaling pathways. Among these, the PI3K–Akt, MAPK, and mTOR signaling pathways play important roles in long-term learning and memory.
The BC200 RNA is selectively located in neuronal synapses, which regulates the synthesis of proteins surrounding the postsynaptic membrane. Mus et al. revealed that the expression of BC200 RNA was significantly downregulated in the prefrontal cortical area in normal elderly subjects (49–86 years old, by autopsy), and was significantly increased in the brain of AD patients. Further studies have revealed that the distribution of BC200 RNA in the brain of AD patients has changed. The BC200 RNA in the brain of AD patients is located in the perinuclear area in the cluster, instead of the synaptic terminals. Hence, it loses its regulatory function on proteins surrounding the postsynaptic membrane. The overexpression and error spatial localization of BC200 RNA may excessively inhibit the synthesis of cytoplasmic proteins, thereby, aggravating the pathological changes of AD (Mus et al., 2007).
The 17A is also an lncRNA that has been recently discovered. It is located in intron 3 of the G protein-coupled receptor-51 (GRP51) gene, regulates the production of GRP51 variable transcript, and inhibits the canonical transcription of GAGA(B2) receptors, thereby, significantly affecting the GABA-B signaling pathway. The inflammatory reaction in the brain in AD patients can activate the expression of 17A. This increases the secretion of Aβ and the Aβ42/Aβ40 ratio, aggravating the progress of the disease (Massone et al., 2011). Another lncRNA 51A overlapping with SORL1 (antisense) was also shown to affect Aβ formation and upregulated in AD (Ciarlo et al., 2013).
In addition, neurotrophic factors (NTFs) play an important role in maintaining nervous system function. The expression level of BDNF is significantly changed in neurodegenerative diseases, psychosis, and neurodevelopmental disorders (Lu et al., 2013, 2014; Song et al., 2015). The inhibition of BDNF-AS expression can increase the expression of BDNF in the brain, which has a wide prospect for the treatment of neurodegenerative diseases. Glial cell-derived neurotrophic factor opposite strand (GDNFOS) is transcribed from the antisense strand of the GDNF gene. A study revealed that there was a difference in the expression of GDNFOS subunit in AD brains compared with normal brains, but the underlying mechanisms remain unknown at present (Airavaara et al., 2011). Furthermore, another study found that Sox2OT and 1810014B01Rik could serve as biomarkers in the early and late stages of neurodegenerative diseases (Arisi et al., 2011). Tremendous efforts have been put into their translational applications by identifying specific lncRNAs that are changed in Alzheimer's disease in order to provide biomarkers and better illustrate molecular pathways.
The Correlation Between lncRNAs and Parkinson's Disease
Parkinson's disease (PD) is a neurodegenerative disease that commonly occurs in the elderly. Its pathological characteristic is the degeneration of dopaminergic neurons in the substantia nigra-striatum system, which decreases dopamine secretion, resulting in a series of extrapyramidal responses. The maintenance of mitochondrial homeostasis plays an important role in the progression of PD (Moreira et al., 2010; Jin et al., 2014; Luo et al., 2015). Gene studies revealed that PD family-related genes such as α-synuclein, parkin, PTEN-induced putative kinase 1 (PINK1), DJ-1, and leucine-rich repeat kinase 2 (LRRK2) were closely related to mitochondrial function (Puspita et al., 2017). Endogenous PINK1 is localized in the mitochondrial membrane and plays an important role in energy metabolism in neurons and muscle cells. In addition, PINK1 can also inhibit the release of cytochrome C from mitochondria and decrease the occurrence of apoptosis. The inhibition or overexpression of PINK1 can lead to abnormal mitochondrial morphology and affect the release of dopamine, resulting in behavior defects (Petit et al., 2005). Study found that lncRNA NEAT1 was significantly upregulated in the midbrain of PD mice, and that lncRNA NEAT1 promoted MPTP-induced autophagy in PD by stabilizing PINK1 protein (Yan et al., 2018). Noncoding antisense PTEN-induced putative kinase 1 (naPINK1) is an lncRNA transcribed from the antisense strand of the PINK1 gene, which can stabilize the expression of the PINK1 variable transcript svPINK1. The silencing of naPINK1 leads to the decrease in svPINK1 in neurons. This suggests that the PD process can be improved by regulating the PINK1 locus (Scheele et al., 2007).
Ubiquitin carboxy-terminal hydrolase L1 (Uchl1) is a neuron-restricted protein acting as a de-ubiquitinating enzyme or a monoubiquitin stabilizer. The UCHL1 gene mutations have been discovered to be related to familial PD and the oxidative inactivation of Uchl1 protein has been reported in PD and AD brains (Choi et al., 2004). The Uchl1-AS is a nuclear-enriched lncRNA that is transcribed antisense to the mouse Uchl1. The Uchl1-AS increases the protein synthesis of UCHL1 at the post-transcriptional level and then regulates the progression of PD (Carrieri et al., 2015). Furthermore, the activity of Uchl1-AS is controlled by the signaling pathway. The Uchl1 mRNA is mainly localized in the cytoplasm, while Uchl1-AS is abundant in the nucleus of dopaminergic neurons. Interestingly, the mTOR inhibitor-Rapamycin treatment resulted in the induction of Uchl1 protein by association of shuttling Uchl1-AS from the nucleus to the cytoplasm, suggesting that the interaction between Uchl1–ncRNA–mTOR may be critical for PD development (Carrieri et al., 2012; Vucićević et al., 2014).
Correlation Between lncRNAs and Huntington's Disease
Huntington's disease (HD) is a rare autosomal dominant inherited neurodegenerative disease. Its pathological change is the loss of neurons in the striatum and cortex in brain. There is a CAG repetitive sequence in exon 1 of its pathogenic gene huntingtin (HTT), which encodes polyglutamine. The excessive repeat amplification of CAG in the gene-coding region induces the prolongation of the polyglutamine chain in protein, thereby inducing lesions (Aziz et al., 2011; De Souza and Leavitt, 2015). The HTT can regulate the nuclear translocation of transcription inhibiting factor RE-1 silencing transcription factor (REST), which is also known as neuron-restrictive silencer factor (NRSF)(Shimojo, 2008). Mutation in HTT leads to the abnormal nuclear/interstitial translocation of REST/NRSF, thereby, inducing the abnormal expression of the downstream target gene of REST/NRSF, including PCGs and ncRNA. Through studies on the expression profile of brain tissue in HD patients, Johnson et al. revealed that the expression of HAR1 lncRNA in striatum decreased significantly, and the main cause was that REST inhibited HAR1 transcription by locating in the HAR1 locus via the specific DNA regulatory element (Johnson et al., 2010). Chung et al. discovered an lncRNA-HTTAS transcribed from the antisense strand of HTT. This has two types of transcripts: HTTAS-v1 (exons 1 and 3) and HTTAS-v2 (exons 2 and 3), where exon 1 contains repeated gene loci. Cell level verification results revealed that the overexpression of HTTAS-v1 could significantly decrease the transcription level of HTT, while HTT transcription level significantly increased after HTTAS-v1 was disrupted by siRNA. Furthermore, HTTAS-v1 expression was found to be downregulated in frontal cortex of HD patients, and this strongly suggests that the change in HTTAS may play a certain role in the progress of HD (Chung et al., 2011).
The lncRNA TUG1 has been shown to be a direct downstream target of p53, which is known to be upregulated in HD itself. Therefore, TUG1 appears to be a pro-survival factor in neurons. The upregulation we observed in HD - probably through p53 activation–actually acts against mutHTT cytotoxicity (Khalil et al., 2009). The NEAT1 is a nuclear-enrich lncRNA that is essential for the formation and maintenance of paraspeckles, which are subnuclear bodies found in mammalian cells (Clemson et al., 2009). Sunwoo et al. found that the levels of NEAT1 were increased in R6/2 mice and HD patients. In order to determine the biological effects of NEAT1 on neuronal survival, the authors transfected neuro2A cells with the NEAT1 short isotype vector and subjected them to H2O2-induced damage. The NEAT1 transfected cells showed enhanced viability under oxidative stress, confirming that upregulation of NEAT1 contributes to neuroprotective mechanisms against neuronal damage rather than pathology of neurodegenerative diseases (Sunwoo et al., 2017). It has been reported that MEG3 is a REST target and is dynamically expressed during neurodevelopment and associated with PRC2 chromatin regulators. This supporting MEG3 might be participating in chromatin regulation, noncoding transcription, and neurodevelopment in HD (Johnson, 2012). Although functional studies on DGCR5 have not been performed, the fact that this neuro-specific disease-related transcript is directly targeted by REST suggests that it has important functions in the human nervous system (Sutherland et al., 1996; Johnson, 2012).
Summary of lncRNAs in Neurodegenerative Diseases
An increasing number of studies report on lncRNAs as being implicated in neurodegenerative diseases, including AD, PD, and HD (Table 2).
The roles of BACE1-AS lncRNA have been widely defined in AD. The BACE1-AS levels are upregulated in AD brains, where BACE1-AS acts by stabilizing BACE1 mRNA, thereby, increasing BACE1 protein content and Aβ42 formation. While lncRNA BC200, 17A, BC158567, MRAK050857, Mrak033976, BDNF-AS, Sox2OT, and 1810014B01Rik are also involved in AD. In PD, the lncRNA UCHL1-AS1 acts by directly promoting translation of UCHL1 protein leading to perturbation of the ubiquitin–proteasome system. Different lncRNAs, such as naPINK1, NEAT1 PINK1-AS, BC200, and Sox2OT, were found to be dysregulated in their expression also in PD. Several studies reported altered expression levels of known lncRNAs HTT-AS, TUG1, NEAT1, MEG3, and DGCR5 in the brains of HD patients. The emerging role of lncRNAs in neurodegenerative diseases suggests that their dysregulation may trigger neuronal death through an unexplored RNA-based regulatory mechanism that needs to be further investigated.
Challenges and Perspectives
With the continuous deepening of gene regulation research and the emergence of more biological methods, lncRNA function and mechanism will be further elucidated.
This relatively poorly characterized class of RNAs, with little or no coding capacity, has been implicated in the growth and development of the nervous system, the differentiation of neurons, synaptic plasticity, and the occurrence and progress of many diseases. Although many studies have been performed in clinical patients using various disease models, the exact role and impact of lncRNAs in disease pathogenesis still remain obscure. At present, there are still challenges for our understanding of lncRNAs.
Our lack of understanding of the molecular mechanisms of lncRNA action makes it particularly difficult for us to recognize the biological function of lncRNA. As we move forward from the annotation of lncRNAs to more emphasis on molecular function and biology, we still do not fully understand the biological significance of lncRNAs as a group. We need better tools to track lncRNA localization across the genome, monitor lncRNA interactions with proteins and nucleic acids, and determine the structure and elucidate the key structure–function relationships of lncRNAs, especially how they interact with proteins. Furthermore, unlike PCGs with systemic functional annotation systems, the lack of an annotation system for lncRNA function makes it difficult to evaluate computational algorithms for functional prediction. Finally, is it useful to find lncRNAs in neurodegenerative diseases? Two major long-term challenges in disease research are (a) the development of noninvasive diagnostic methods for monitoring the progression of neurodegenerative diseases, and (b) the development of treatments to cure and reverse disease processes.
In fact, more and more attention has been paid to lncRNAs as potential targets for disease biomarkers or therapeutic strategies. Indeed, several commercial entities targeting lncRNAs (such as OPKO-CURNA and RaNA therapeutic agents) have been developed to design and develop oligonucleotide therapeutics for the treatment of neurodegenerative diseases (Qureshi and Mehler, 2013). Hereby, further investigation into the role of lncRNAs will provide a better understanding of how the brain functions and how diseases develop, and lead to greater insights into further therapeutic development for neurodegenerative diseases based on manipulations of lncRNA functions.
Author Contributions
C-WW substantial contributions to the conception and design of the work, the acquisition, analysis, interpretation of data for the work. C-WW drafting the work and revising it critically for important intellectual content. A-SW final approval of the version to be published. TL and S-SZ agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81371199, 81771139).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Airavaara, M., Pletnikova, O., Doyle, M. E., Zhang, Y. E., Troncoso, J. C., and Liu, Q. R. (2011). Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J. Biol. Chem. 286, 45093–45102. doi: 10.1074/jbc.M111.310250
Amaral, P. P., Dinger, M. E., and Mattick, J. S. (2013). Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief. Funct. Genomics 12, 254–278. doi: 10.1093/bfgp/elt016
Antoniou, D., Stergiopoulos, A., and Politis, P. K. (2014). Recent advances in the involvement of long non-coding RNAs in neural stem cell biology and brain pathophysiology. Front. Physiol. 5:155. doi: 10.3389/fphys.2014.00155
Arisi, I., D'Onofrio, M., Brandi, R., Felsani, A., Capsoni, S., Drovandi, G., et al. (2011). Gene expression biomarkers in the brain of a mouse model for Alzheimer's disease: mining of microarray data by logic classification and feature selection. Biochim. Biophys. Acta 24, 721–738. doi: 10.3233/JAD-2011-101881
Aziz, N. A., van Belzen, M. J., Coops, I. D., Belfroid, R. D., and Roos, R. A. (2011). Parent-of-origin differences of mutant HTT CAG repeat instability in Huntington's disease. Eur. J. Med. Genet. 54, e413–e418. doi: 10.1016/j.ejmg.2011.04.002
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., and Lingner, J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. doi: 10.1126/science.1147182
Babajko, S., Petit, S., Fernandes, I., Meary, F., LeBihan, J., Pibouin, L., et al. (2009). Msx1 expression regulation by its own antisense RNA: consequence on tooth development and bone regeneration. Cells Tissues Organs 189, 115–121. doi: 10.1159/000151748
Barry, G., Briggs, J. A., Hwang, D. W., Nayler, S. P., Fortuna, P. R., Jonkhout, N., et al. (2017). The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 7:40127. doi: 10.1038/srep40127
Barry, G., Briggs, J. A., Vanichkina, D. P., Poth, E. M., Beveridge, N. J., Ratnu, V. S., et al. (2014). The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494. doi: 10.1038/mp.2013.45
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Bond, A. M., Vangompel, M. J., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. doi: 10.1038/nn.2371
Braidotti, G., Baubec, T., Pauler, F., Seidl, C., Smrzka, O., Stricker, S., et al. (2004). The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb. Symp. Quant. Biol. 69, 55–66. doi: 10.1101/sqb.2004.69.55
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L., and Barry, G. (2015). Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 88, 861–877. doi: 10.1016/j.neuron.2015.09.045
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. doi: 10.1038/nature11508
Carrieri, C., Forrest, A. R., Santoro, C., Persichetti, F., Carninci, P., Zucchelli, S., et al. (2015). Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells' differentiation in vitro and in neurochemical models of Parkinson's disease. Front. Cell. Neurosci. 9:114. doi: 10.3389/fncel.2015.00114
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chiba, M., Kiyosawa, H., Hiraiwa, N., Ohkohchi, N., and Yasue, H. (2009). Existence of Pink1 antisense RNAs in mouse and their localization. Cytogenet. Genome Res. 126, 259–270. doi: 10.1159/000251963
Choi, J., Levey, A. I., Weintraub, S. T., Rees, H. D., Gearing, M., Chin, L. S., et al. (2004). Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J. Biol. Chem. 279, 13256–13264. doi: 10.1074/jbc.M314124200
Chung, D. W., Rudnicki, D. D., Yu, L., and Margolis, R. L. (2011). A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477. doi: 10.1093/hmg/ddr263
Ciarlo, E., Massone, S., Penna, I., Nizzari, M., Gigoni, A., Dieci, G., et al. (2013). An intronic ncRNA-dependent regulation of SORL1 expression affecting Abeta formation is upregulated in post-mortem Alzheimer's disease brain samples. Dis. Model. Mech. 6, 424–433. doi: 10.1242/dmm.009761
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. doi: 10.1016/j.molcel.2009.01.026
Colasante, G., Collombat, P., Raimondi, V., Bonanomi, D., Ferrai, C., Maira, M., et al. (2008). Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J. Neurosci. 28, 10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008
Coudert, A. E., Pibouin, L., Vi-Fane, B., Thomas, B. L., Macdougall, M., Choudhury, A., et al. (2005). Expression and regulation of the Msx1 natural antisense transcript during development. Nucleic Acids Res. 33, 5208–5218. doi: 10.1093/nar/gki831
De Souza, R. A., and Leavitt, B. R. (2015). Neurobiology of Huntington's disease. Curr. Top. Behav. Neurosci. 22:81–100. doi: 10.1007/7854_2014_353
Dewachter, I., van Dorpe, J., Spittaels, K., Tesseur, I., Van Den Haute, C., Moechars, D., et al. (2000). Modeling Alzheimer's disease in transgenic mice: effect of age and of presenilin1 on amyloid biochemistry and pathology in APP/London mice. Exp. Gerontol. 35, 831–841. doi: 10.1016/S0531-5565(00)00149-2
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van der Brug, M. P., Nalls, M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11:R56. doi: 10.1186/gb-2010-11-5-r56
Feng, L., Liao, Y. T., He, J. C., Xie, C. L., Chen, S. Y., Fan, H. H., et al. (2018). Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol. 18:4. doi: 10.1186/s12883-017-1008-x
Ghosal, K., Vogt, D. L., Liang, M., Shen, Y., Lamb, B. T., and Pimplikar, S. W. (2009). Alzheimer's disease-like pathological features in transgenic mice expressing the APP intracellular domain. Proc. Natl. Acad. Sci. U.S.A. 106, 18367–18372. doi: 10.1073/pnas.0907652106
Goff, L. A., and Rinn, J. L. (2015). Linking RNA biology to lncRNAs. Genome Res. 25, 1456–1465. doi: 10.1101/gr.191122.115
Graff, J., and Mansuy, I. M. (2008). Epigenetic codes in cognition and behaviour. Behav. Brain Res. 192, 70–87. doi: 10.1016/j.bbr.2008.01.021
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Hung, T., Wang, Y., Lin, M. F., Koegel, A. K., Kotake, Y., Grant, G. D., et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629. doi: 10.1038/ng.848
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B., and Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. doi: 10.1186/1471-2164-8-39
Jiang, Y. H., Armstrong, D., Albrecht, U., Atkins, C. M., Noebels, J. L., Eichele, G., et al. (1998). Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21, 799–811. doi: 10.1016/S0896-6273(00)80596-6
Jin, H., Kanthasamy, A., Ghosh, A., Anantharam, V., Kalyanaraman, B., and Kanthasamy, A. G. (2014). Mitochondria-targeted antioxidants for treatment of Parkinson's disease: preclinical and clinical outcomes. Biochim. Biophys. Acta 1842, 1282–1294. doi: 10.1016/j.bbadis.2013.09.007
Johnson, R. (2012). Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 46, 245–254. doi: 10.1016/j.nbd.2011.12.006
Johnson, R., Richter, N., Jauch, R., Gaughwin, P. M., Zuccato, C., Cattaneo, E., et al. (2010). Human accelerated region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol. Genomics 41, 269–274. doi: 10.1152/physiolgenomics.00019.2010
Kazemzadeh, M., Safaralizadeh, R., and Orang, A. V. (2015). LncRNAs: emerging players in gene regulation and disease pathogenesis. J. Genet. 94, 771–784. doi: 10.1007/s12041-015-0561-6
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. doi: 10.1016/j.cell.2013.01.003
Kondrashov, A. V., Kiefmann, M., Ebnet, K., Khanam, T., Muddashetty, R. S., and Brosius, J. (2005). Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353, 88–103. doi: 10.1016/j.jmb.2005.07.049
Leal, G., Comprido, D., and Duarte, C. B. (2014). BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology 76(Pt C), 639–656. doi: 10.1016/j.neuropharm.2013.04.005
Lee, S., Seo, H. H., Lee, C. Y., Lee, J., Shin, S., Kim, S. W., et al. (2017). Human long noncoding RNA regulation of stem cell potency and differentiation. Stem Cells Int. 2017:6374504. doi: 10.1155/2017/6374504
Lin, N., Chang, K. Y., Li, Z., Gates, K., Rana, Z. A., Dang, J., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019. doi: 10.1016/j.molcel.2014.01.021
Lipovich, L., Dachet, F., Cai, J., Bagla, S., Balan, K., Jia, H., et al. (2012). Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 192, 1133–1148. doi: 10.1534/genetics.112.145128
Liu, T., Huang, Y., Chen, J., Chi, H., Yu, Z., Wang, J., et al. (2014). Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1AS expression. Mol. Med. Rep. 10, 1275–1281. doi: 10.3892/mmr.2014.2351
Lu, B., Nagappan, G., Guan, X., Nathan, P. J., and Wren, P. (2013). BDNF-based synaptic repair as a disease-modifying strategy for neurodegenerative diseases. Nat. Rev. Neurosci. 14, 401–416. doi: 10.1038/nrn3505
Lu, B., Nagappan, G., and Lu, Y. (2014). BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb. Exp. Pharmacol. 220:223–250. doi: 10.1007/978-3-642-45106-5_9
Lukiw, W. J., Handley, P., Wong, L., and Crapper McLachlan, D. R. (1992). BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem. Res. 17, 591–597. doi: 10.1007/BF00968788
Luo, Q., and Chen, Y. (2016). Long noncoding RNAs and Alzheimer's disease. Clin. Interv. Aging 11:867–872. doi: 10.2147/CIA.S107037
Luo, Y., Hoffer, A., Hoffer, B., and Qi, X. (2015). Mitochondria: a therapeutic target for Parkinson's disease?. Int. J. Mol. Sci. 16, 20704–20730. doi: 10.3390/ijms160920704
Lv, J., Liu, H., Huang, Z., Su, J., He, H., Xiu, Y., et al. (2013). Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res. 41, 10044–10061. doi: 10.1093/nar/gkt818
Maag, J. L., Panja, D., Sporild, I., Patil, S., Kaczorowski, D. C., Bramham, C. R., et al. (2015). Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front. Neurosci. 9:351. doi: 10.3389/fnins.2015.00351
Mabb, A. M., Judson, M. C., Zylka, M. J., and Philpot, B. D. (2011). Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 34, 293–303. doi: 10.1016/j.tins.2011.04.001
Magistri, M., Faghihi, M. A., St Laurent, G. III., and Wahlestedt, C. (2012). Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 28, 389–396. doi: 10.1016/j.tig.2012.03.013
Massone, S., Ciarlo, E., Vella, S., Nizzari, M., Florio, T., Russo, C., et al. (2012). NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid beta secretion. Biochim. Biophys. Acta 1823, 1170–1177. doi: 10.1016/j.bbamcr.2012.05.001
Massone, S., Vassallo, I., Fiorino, G., Castelnuovo, M., Barbieri, F., Borghi, R., et al. (2011). 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317. doi: 10.1016/j.nbd.2010.09.019
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721. doi: 10.1073/pnas.0706729105
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G., Mattick, J. S., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Mercer, T. R., Wilhelm, D., Dinger, M. E., Solda, G., Korbie, D. J., Glazov, E. A., et al. (2011). Expression of distinct RNAs from 3' untranslated regions. Nucleic Acids Res. 39, 2393–2403. doi: 10.1093/nar/gkq1158
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. doi: 10.1038/nbt.2158
Moreira, P. I., Zhu, X., Wang, X., Lee, H. G., Nunomura, A., Petersen, R. B., et al. (2010). Mitochondria: a therapeutic target in neurodegeneration. Biochim. Biophys. Acta 1802, 212–220. doi: 10.1016/j.bbadis.2009.10.007
Mus, E., Hof, P. R., and Tiedge, H. (2007). Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 104, 10679–10684. doi: 10.1073/pnas.0701532104
Nagano, T., and Fraser, P. (2011). No-nonsense functions for long noncoding RNAs. Cell 145, 178–181. doi: 10.1016/j.cell.2011.03.014
Nakagawa, S., Ip, J. Y., Shioi, G., Tripathi, V., Zong, X., Hirose, T., et al. (2012). Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–1499. doi: 10.1261/rna.033217.112
Ng, S. Y., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi: 10.1016/j.molcel.2013.07.017
Ng, S. Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Ninan, I. (2014). Synaptic regulation of affective behaviors; role of BDNF. Neuropharmacology 76(Pt C), 684–695. doi: 10.1016/j.neuropharm.2013.04.011
Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., et al. (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573. doi: 10.1038/nature01266
Pang, K. C., Frith, M. C., and Mattick, J. S. (2006). Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22, 1–5. doi: 10.1016/j.tig.2005.10.003
Panja, D., and Bramham, C. R. (2014). BDNF mechanisms in late LTP formation: a synthesis and breakdown. Neuropharmacology 76(Pt C), 664–676. doi: 10.1016/j.neuropharm.2013.06.024
Parenti, R., Paratore, S., Torrisi, A., and Cavallaro, S. (2007). A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur. J. Neurosci. 26, 2444–2457. doi: 10.1111/j.1460-9568.2007.05864.x
Pauler, F. M., Stricker, S. H., Warczok, K. E., and Barlow, D. P. (2005). Long-range DNase I hypersensitivity mapping reveals the imprinted Igf2r and Air promoters share cis-regulatory elements. Genome Res. 15, 1379–1387. doi: 10.1101/gr.3783805
Peden, A. H., and Ironside, J. W. (2012). Molecular pathology in neurodegenerative diseases. Curr. Drug Targets 13, 1548–1559. doi: 10.2174/138945012803530134
Petit, A., Kawarai, T., Paitel, E., Sanjo, N., Maj, M., Scheid, M., et al. (2005). Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J. Biol. Chem. 280, 34025–34032. doi: 10.1074/jbc.M505143200
Pitcher, G. M., Beggs, S., Woo, R. S., Mei, L., and Salter, M. W. (2008). ErbB4 is a suppressor of long-term potentiation in the adult hippocampus. Neuroreport 19, 139–143. doi: 10.1097/WNR.0b013e3282f3da10
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Pruunsild, P., Kazantseva, A., Aid, T., Palm, K., and Timmusk, T. (2007). Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90, 397–406. doi: 10.1016/j.ygeno.2007.05.004
Puspita, L., Chung, S. Y., and Shim, J. W. (2017). Oxidative stress and cellular pathologies in Parkinson's disease. Mol. Brain 10:53. doi: 10.1186/s13041-017-0340-9
Quan, Z., Zheng, D., and Qing, H. (2017). Regulatory roles of long non-coding RNAs in the central nervous system and associated neurodegenerative diseases. Front. Cell. Neurosci. 11:175. doi: 10.3389/fncel.2017.00175
Qureshi, I. A., Mattick, J. S., and Mehler, M. F. (2010). Long non-coding RNAs in nervous system function and disease. Brain Res. 1338c, 20–35. doi: 10.1016/j.brainres.2010.03.110
Qureshi, I. A., and Mehler, M. F. (2013). Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 10, 632–646. doi: 10.1007/s13311-013-0199-0
Rackham, O., Shearwood, A. M., Mercer, T. R., Davies, S. M., Mattick, J. S., and Filipovska, A. (2011). Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093. doi: 10.1261/rna.029405.111
Ramos, A. D., Attenello, F. J., and Lim, D. A. (2016). Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 625, 70–79. doi: 10.1016/j.neulet.2015.12.025
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Riva, P., Ratti, A., and Venturin, M. (2016). The Long Non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr. Alzheimer Res. 13, 1219–1231. doi: 10.2174/1567205013666160622112234
Roberts, T. C., Morris, K. V., and Wood, M. J. (2014). The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130507. doi: 10.1098/rstb.2013.0507
Sai, Y., Zou, Z., Peng, K., and Dong, Z. (2012). The Parkinson's disease-related genes act in mitochondrial homeostasis. Neurosci. Biobehav. Rev. 36, 2034–2043. doi: 10.1016/j.neubiorev.2012.06.007
Sasaki, Y. T., Ideue, T., Sano, M., Mituyama, T., and Hirose, T. (2009). MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U.S.A. 106, 2525–2530. doi: 10.1073/pnas.0807899106
Sauvageau, M., Goff, L. A., Lodato, S., Bonev, B., Groff, A. F., Gerhardinger, C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2:e01749. doi: 10.7554/eLife.01749
Scheele, C., Petrovic, N., Faghihi, M. A., Lassmann, T., Fredriksson, K., Rooyackers, O., et al. (2007). The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 8:74. doi: 10.1186/1471-2164-8-74
Shamir, A., Kwon, O. B., Karavanova, I., Vullhorst, D., Leiva-Salcedo, E., Janssen, M. J., et al. (2012). The importance of the NRG-1/ErbB4 pathway for synaptic plasticity and behaviors associated with psychiatric disorders. J. Neurosci. 32, 2988–2997. doi: 10.1523/JNEUROSCI.1899-11.2012
Shimojo, M. (2008). Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued. J. Biol. Chem. 283, 34880–34886. doi: 10.1074/jbc.M804183200
Shimozaki, K. (2014). Sox2 transcription network acts as a molecular switch to regulate properties of neural stem cells. World J. Stem Cells 6, 485–490. doi: 10.4252/wjsc.v6.i4.485
Singh, D. K., and Prasanth, K. V. (2013). Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res. 21, 695–711. doi: 10.1007/s10577-013-9391-7
Song, J. H., Yu, J. T., and Tan, L. (2015). Brain-derived neurotrophic factor in Alzheimer's disease: risk, mechanisms, and therapy. Mol. Neurobiol. 52, 1477–1493. doi: 10.1007/s12035-014-8958-4
Sun, M., and Kraus, W. L. (2015). From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 36, 25–64. doi: 10.1210/er.2014-1034
Sunwoo, H., Dinger, M. E., Wilusz, J. E., Amaral, P. P., Mattick, J. S., and Spector, D. L. (2009). MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359. doi: 10.1101/gr.087775.108
Sunwoo, J. S., Lee, S. T., Im, W., Lee, M., Byun, J. I., Jung, K. H., et al. (2017). Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Mol. Neurobiol. 54, 1577–1586. doi: 10.1007/s12035-016-9928-9
Sutherland, H. F., Wadey, R., McKie, J. M., Taylor, C., Atif, U., Johnstone, K. A., et al. (1996). Identification of a novel transcript disrupted by a balanced translocation associated with DiGeorge syndrome. Am. J. Hum. Genet. 59, 23–31.
Tan, L., Yu, J. T., Hu, N., and Tan, L. (2013). Non-coding RNAs in Alzheimer's disease. Mol. Neurobiol. 47, 382–393. doi: 10.1007/s12035-012-8359-5
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Vucićević, D., Schrewe, H., and Orom, U. A. (2014). Molecular mechanisms of long ncRNAs in neurological disorders. Front. Genet. 5:48. doi: 10.3389/fgene.2014.00048
Wang, A., Wang, J., Liu, Y., and Zhou, Y. (2017). Mechanisms of long non-coding RNAs in the assembly and plasticity of neural circuitry. Front. Neural Circuits 11:76. doi: 10.3389/fncir.2017.00076
Wang, H., Iacoangeli, A., Lin, D., Williams, K., Denman, R. B., Hellen, C. U., et al. (2005). Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 171, 811–821. doi: 10.1083/jcb.200506006
Wang, H., Iacoangeli, A., Popp, S., Muslimov, I. A., Imataka, H., Sonenberg, N., et al. (2002). Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 22, 10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002
Yan, W., Chen, Z. Y., Chen, J. Q., and Chen, H. M. (2018). LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson's disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 496, 1019–1024. doi: 10.1016/j.bbrc.2017.12.149
Yang, B., Xia, Z. A., Zhong, B., Xiong, X., Sheng, C., Wang, Y., et al. (2017). Distinct hippocampal expression profiles of long non-coding RNAs in an Alzheimer's disease model. Mol. Neurobiol. 54, 4833–4846. doi: 10.1007/s12035-016-0038-5
Zagrebelsky, M., and Korte, M. (2014). Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology 76(Pt C), 628–638. doi: 10.1016/j.neuropharm.2013.05.029
Zhang, B., Arun, G., Mao, Y. S., Lazar, Z., Hung, G., Bhattacharjee, G., et al. (2012). The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123. doi: 10.1016/j.celrep.2012.06.003
Zhang, X., Lian, Z., Padden, C., Gerstein, M. B., Rozowsky, J., Snyder, M., et al. (2009). A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113, 2526–2534. doi: 10.1182/blood-2008-06-162164
Zhao, X., Tang, Z., Zhang, H., Atianjoh, F. E., Zhao, J. Y., Liang, L., et al. (2013). A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16, 1024–1031. doi: 10.1038/nn.3438
Keywords: long noncoding RNAs, central nervous system, neurodegenerative diseases, regulatory mechanism, gene expression
Citation: Wei C-W, Luo T, Zou S-S and Wu A-S (2018) The Role of Long Noncoding RNAs in Central Nervous System and Neurodegenerative Diseases. Front. Behav. Neurosci. 12:175. doi: 10.3389/fnbeh.2018.00175
Received: 09 March 2018; Accepted: 27 July 2018;
Published: 28 September 2018.
Edited by:
Ioan Opris, University of Miami, United StatesReviewed by:
Neha Nagpal, Boston Children's Hospital, United StatesTimothy J. Jarome, Virginia Tech, United States
Copyright © 2018 Wei, Luo, Zou and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: An-Shi Wu, d3VhbnNoaTg4MDkyM0AxNjMuY29t
 Chang-Wei Wei
Chang-Wei Wei An-Shi Wu
An-Shi Wu