- Beaumont Research Institute, Department of Neurology, Beaumont Health, Royal Oak, MI, United States
Misfolded proteins are pathological findings in some chronic neurodegenerative disorders including Alzheimer’s, Parkinson’s, and Huntington’s diseases. Aging is a major risk factor for these disorders, suggesting that the mechanisms responsible for clearing misfolded proteins from the brain, the ubiquitin-proteasome system and the autophagy-lysosomal pathway, may decline with age. Although autophagic mechanisms have been found to decrease with age in many experimental models, whether they do so in the brain is unclear. This review examines the literature with regard to age-associated changes in macroautophagy and chaperone-mediated autophagy (CMA) in the central nervous system (CNS). Beclin 1, LC3-II, and the LC3-II/LC3-I ratio have frequently been used to examine changes in macroautophagic activity, while lamp2a and HSPA8 (also known as hsc70) have been used to measure CMA activity. Three gene expression analyses found evidence for an age-related downregulation of macroautophagy in human brain, but no published studies were found of age-related changes in CMA in human brain, although cerebrospinal fluid concentrations of HSPA8 were reported to decrease with age. Most studies of age-related changes in brain autophagy in experimental animals have found age-related declines in macroautophagy, and macroautophagy is necessary for normal lifespan in Caenorhabditis elegans, Drosophila, and mice. However, the few studies of age-related changes in brain CMA in experimental animals have produced conflicting results. Investigations of the influence of aging on macroautophagy in experimental animals in systems other than the CNS have generally found an age-related decrease in Beclin 1, but conflicting results for LC3-II and the LC3-II/LC3-I ratio, while CMA decreases with age in most models. CONCLUSION: while indirect evidence suggests that brain autophagy may decrease with normal aging, this issue has not been investigated sufficiently, particularly in human brain. Measuring autophagic activity in the brain can be challenging because of differences in basal autophagic activity between experimental models, and the inability to include lysosomal inhibitors when measuring the LC3-II/LC3-I ratio in postmortem specimens. If autophagy does decrease in the brain with aging, then pharmacological interventions and/or lifestyle alterations to slow this decline could reduce the risk of developing age-related neurodegenerative disorders.
Introduction
More than 30% of newly synthesized proteins are misfolded because of errors in translation or post-translational processes (Schubert et al., 2000). Molecular chaperones, primarily heat shock proteins, recognize these proteins and attempt to facilitate their refolding (Ciechanover and Kwon, 2017). Two proteolytic mechanisms, the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP), are responsible for cellular clearance of misfolded proteins which are unable to be refolded. The ALP includes three autophagic processes in mammals, namely macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy (Orenstein and Cuervo, 2010), which use different mechanisms to deliver their cargo to lysosomes for enzymatic degradation. Misfolded proteins are initially subjected to removal by the UPS and selected proteins may also be removed by CMA. Misfolded proteins which avoid clearance by these mechanisms may be removed by macroautophagy (Ciechanover and Kwon, 2015). Protein oligomers, soluble aggregates, and intact organelles are too large to enter the proteasome’s narrow opening, so they can be degraded by the ALP but not by the UPS (Bence et al., 2001; Wu et al., 2015). Cross-talk is present between CMA and macroautophagy so that if one mechanism is impaired, the activity of the other may increase in an effort to maintain protein homeostasis (Massey et al., 2006a; Wu et al., 2015). Clearance of misfolded proteins is critical for cell survival because misfolding alters a protein’s three-dimensional structure, impairing its biological activities and increasing its propensity to form toxic aggregates (Hartl, 2017). Misfolded and aggregated proteins are key pathological findings in several chronic neurodegenerative diseases including Alzheimer’s disease (AD) (Selkoe, 2004), Parkinson’s disease (PD) (Tan et al., 2009), Huntington’s disease (Hatters, 2008), amyotrophic lateral sclerosis (Mulligan and Chakrabartty, 2013), and prion disorders (Kupfer et al., 2009); not surprisingly, dysregulation of the UPS and the ALP has been reported in some of these disorders (Bandyopadhyay and Cuervo, 2007; Wang et al., 2009; Alvarez-Erviti et al., 2010, 2013; Salminen et al., 2013; Murphy et al., 2015; Zheng et al., 2016; Budini et al., 2017; Guo et al., 2018). Because aging is the most consistent known risk factor for progressive neurodegenerative disorders (Jeppesen et al., 2011), the impairment of proteolytic mechanisms in these disorders may be due in part to an age effect (Lipinski et al., 2010; Xilouri and Stefanis, 2016).
The biological changes that occur in aging cells include alterations in proteostatic mechanisms, chromosome regulation, transcriptional regulation, protein translation, mitochondrial functioning, and cytoskeletal integrity (DiLoreto and Murphy, 2015). This review will focus on studies that have examined age-related changes in macroautophagy and CMA in the central nervous system (CNS) including the brain, cerebrospinal fluid (CSF), and retina. For comparative purposes, findings from studies which have investigated age-associated changes in these processes in non-CNS experimental systems will also be summarized.
Autophagy Overview
Autophagy involves degradation of cytoplasmic contents within the lysosome, followed by recycling of the resulting macromolecular constituents including amino acids, sugars, lipids, and nucleic acids (Feng et al., 2014; Wu et al., 2015; Hewitt and Korolchuk, 2017). Macroautophagy (Figure 1), the best studied of these mechanisms, involves sequestering of cytoplasmic cargo into an autophagosome followed by lysosomal degradation of the cargo (Klionsky et al., 2016). A double-membraned structure, the phagophore, initially forms around bulk cytoplasm and its constituents (“non-selective macroautophagy”) or specific cytoplasmic targets including organelles (“selective macroautophagy”). The origin of the components which comprise the phagophore membrane is unclear; it has been suggested to derive from multiple sources including plasma membrane, endoplasmic reticulum, mitochondria, and the Golgi apparatus (Mari et al., 2011). Once autophagy is initiated, double-membrane nucleation allows formation of a phagophore which elongates, sequesters the cargo, and closes to form an autophagosome. The autophagosome then fuses with a lysosome to form an autolysosome (Seglen et al., 1990; Mizushima, 2007; Rubinsztein et al., 2011). Sequestered constituents are rapidly degraded by lysosomal hydrolytic enzymes. The term “autophagic flux,” which is a measure of autophagic degradation activity (Loos et al., 2014), is used to refer to the complete process which starts with autophagosome formation and ends with the release of macromolecules into the cytosol (Zhang et al., 2013). Quantification of autophagic flux requires multiple measurements of macroautophagy markers over time to determine the rate of the process (Loos et al., 2014).
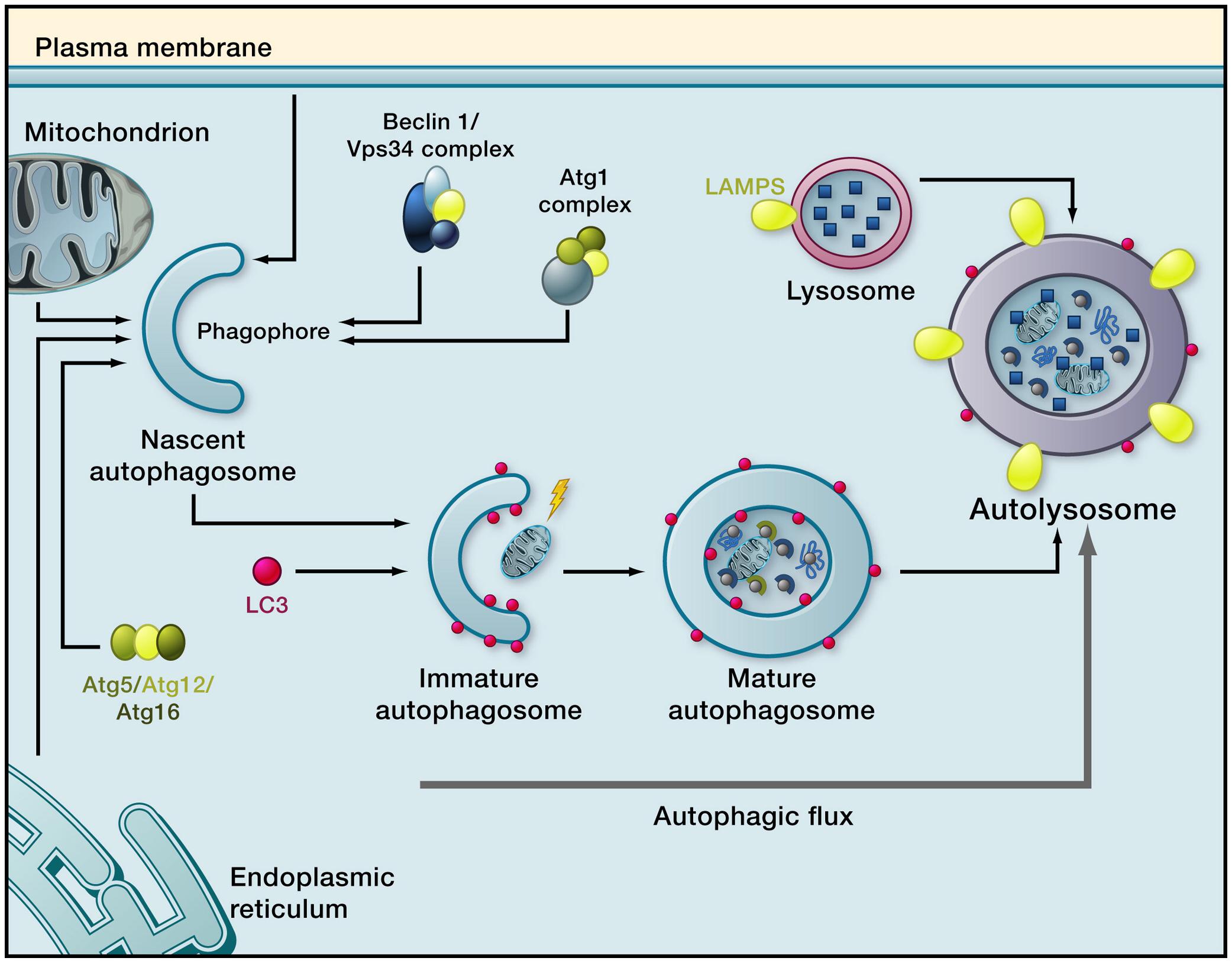
Figure 1. Macroautophagy. Induction of macroautophagy results in phagophore formation, which requires the kinase Vps34 (functioning in a complex that contains Atg6, whose mammalian homolog is Beclin 1). This is followed by phagophore membrane elongation, which is regulated by LC3-II. The phagophore closes around sequestered cargo, resulting in formation of a mature autophagosome. The autophagosome moves along microtubules to a lysosome and fuses with it. Cargo is then degraded by lysosomal enzymes and breakdown products are recycled to the cytosol. The complete process is termed autophagy flux (Reprinted from Rubinsztein et al., 2011).
Much of what is known about the induction of macroautophagy comes from studies with yeast, where 41 autophagy-related proteins, known as Atgs, have been identified to date (Noda and Mizushima, 2016; Pöggeler et al., 2018). Approximately half of these proteins are known to have homologs in higher eukaryotes (Klionsky and Codogno, 2013). The proteins necessary for autophagosome formation are referred to as “core molecular machinery” (Xie and Klionsky, 2007). The proteins involved in macroautophagy in mammals are similar but not identical to those in yeast; several reviews of the process in mammalian systems have appeared (Eskelinen, 2008; Mehrpour et al., 2010; Yang and Klionsky, 2010; Abounit et al., 2012). The core molecular machinery has been divided into functional groups (Yang and Klionsky, 2010; Noda and Mizushima, 2016): (a) the Atg1 kinase complex [in mammals, the Atg1/unc-51-like kinase (ULK) complex], (b) the Atg12-conjugation system, (c) the Atg8/microtubule-associated protein light chain 3 (LC3)-conjugation system, (d) the Vps34 phosphoinositide 3-kinase (PI3 kinase) complex, (e) the Atg9 and Atg2-Atg18 complex, and (f) vacuole-membrane-protein-1 (VMP1). ULK-1 and ULK-2 are mammalian proteins with close homology to Atg1. The Atg1/ULK complex, composed of ULK1 or ULK2, mAtg13 (the mammalian homolog of Atg13), Atg101, and FIP200 (the homolog of yeast Atg17), regulates macroautophagic activity by detecting changes in concentrations of constituents such as glucose, growth factors, nitrogen, amino acids, and reactive oxygen species (Kim et al., 2013). When nutrients are sufficient, mammalian TOR complex 1 (mTORC1) is included in this complex, but mTORC1 dissociates from it when nutrients are limiting. Macroautophagy can be induced by multiple mechanisms (Rubinsztein et al., 2011) including caloric restriction or treatment with rapamycin, both of which inhibit mTORC1. A decrease in mTOR’s kinase activity results in dephosphorylation of ULK1, ULK2, and mAtg13, which activates ULK1 and ULK2. ULK1 and ULK2 phosphorylate mAtg13 and FIP200, which initiates autophagosome formation (Hosokawa et al., 2009). Beclin 1, the mammalian homolog of Atg6, participates with other subunits in forming the PI3 kinase complex (Wu et al., 2015). The Atg12 and Atg8/LC3 conjugation systems are involved with elongation and expansion of the phagophore membrane (Yang and Klionsky, 2010). The Atg8-conjugation system conjugates Atg8 (and, in mammals, LC3) to phosphatidylethanolamine (PE). PE-conjugated LC3 is referred to as LC3-II, whereas its unconjugated counterpart is known as LC3-I. LC3-II regulates phagophore membrane elongation (Abeliovich et al., 2000) and facilitates membrane tethering and hemifusion (Nakatogawa et al., 2007). The Vps34 PI3 kinase complex is a positive regulator of macroautophagy; its activity generates phosphatidylinositol 3-phosphate, which recruits the Atg2–Atg18 complex to bind to Atg9 on autophagic membranes (Obara and Ohsumi, 2008). Atg9 also carries membranes from donor organelles between the developing autophagosome and the periphery. VMP1, a mammalian protein which does not appear to have an equivalent in yeast, induces autophagy even when nutrients are adequate by binding to Beclin 1, leading to the formation of a complex with Vps34 (Molejon et al., 2013). UV radiation-associated resistance gene (UVRAG) and Beclin 1-associated autophagy related key regulator (Barkor) stabilize the Beclin 1/Vps34 complex (Liang et al., 2008; Sun et al., 2008). Other important regulators of macroautophagy include AMP-activated protein kinase (AMPK) (Meley et al., 2006) and the PI3K/Akt/mTOR pathway (Woo et al., 2017). AMPK is activated by low ATP or glucose concentrations. Its activation can induce macroautophagy by inactivating TORC1 and by phosphorylating ULK1 (Hardie, 2011). Conversely, signaling from the PI3K/Akt/mTOR complex leads to phosphorylation, and thus activation, of mTOR (Triplett et al., 2015), which inhibits macroautophagy (Cao et al., 2014).
Markers which have been used to measure macroautophagy (reviewed by Klionsky et al., 2016) include the proteins encoded by Atg5 (Codogno and Meijer, 2006) and Atg12 (Hanada and Ohsumi, 2005), Beclin 1 (Kang et al., 2011), mTOR (Brown et al., 1994), and LC3 (Sugawara et al., 2004), including LC3-I, LC3-II, and the LC3-II/LC3-I ratio. LC3-II is initially found on inner and outer layers of the phagophore membrane. During autophagosome maturation, it is removed from the outer (cytosolic) surface due to cleavage of PE by Atg4 (Yu et al., 2012), but remains on the inner (lumenal) surface of the autophagosome. LC3-II levels therefore correlate with autophagosome numbers (Mizushima and Yoshimori, 2007; Wu et al., 2015). The LC3-II/LC3-I ratio is often examined by Western blot at a single time point to assess changes in macroautophagic activity, but this procedure does not actually measure autophagic flux because no rate of change is measured (Loos et al., 2014). Alterations in LC3-II and in the LC3-II/LC3-I ratio can be difficult to interpret, because an increase in the LC3-II level may be due to either activation of macroautophagy or a decrease in autophagosomal-lysosomal fusion or lysosomal degradation of cargo; similarly, a reduction in LC3-II may be due to inhibition of macroautophagy or to an increase in autophagic flux (Linton et al., 2015). Preventing lysosomal degradation of autophagosomal LC3-II, by including inhibitors of autophagosome-lysosome fusion or of lysosomal proteases in the experimental system, may be necessary to interpret LC3-II and LC3-II/LC3-I measurements (Klionsky et al., 2016; Orhon and Reggiori, 2017). Other markers that have been used to investigate changes in macroautophagy include the lysosomal protease cathepsin D (Tatti et al., 2012) and the autophagic adaptor proteins p62/Sequestosome-1 (known as p62/SQSTM1 or p62) (Lippai and Löw, 2014) and its functional homolog, neighbor of BRCA1 (NBR1) (Kirkin et al., 2009). The latter two proteins are macroautophagy substrates; however, similar to the situation with LC3-II, single measurements of these proteins provides no information about their rate of turnover (Loos et al., 2014).
Chaperone-mediated autophagy (Figure 2), a more selective autophagic mechanism than macroautophagy which has been identified only in mammals, does not involve vesicle formation (Cuervo, 2010). Its selectivity is due to the requirement for its substrate proteins to contain amino acid sequences biochemically similar to Lys-Phe-Glu-Arg-Gln (KFERQ), which is present in approximately 30% of cytosolic proteins (Dice et al., 1986; Chiang and Dice, 1988; Chiang et al., 1989). Proteins bearing this sequence are bound by the molecular chaperone heat shock 70 kDa protein 8 (HSPA8, also known as hsc70 and hsc73; hereafter, “HSPA8/hsc70”) in association with co-chaperones (Agarraberes and Dice, 2001). The protein-HSPA8/hsc70 complex then binds to a lysosomal surface receptor, lysosome-associated membrane glycoprotein 2a (lamp2a), which facilitates its translocation into the lysosomal lumen. Binding of the protein-HSPA8/hsc70 complex to lamp2a is rate-limiting for CMA (Cuervo and Dice, 1996) so the concentration of lamp2a on lysosomal membranes is considered to be a marker for CMA (Cuervo and Dice, 2000b; Patel and Cuervo, 2015). In CMA, substrate proteins must unfold before their translocation into the lysosomal lumen. This unfolding is facilitated by HSPA8/hsc70 and its co-chaperones (Salvador et al., 2000). Only lysosomes containing HSPA8/hsc70 on the lumenal side of the lysosomal membrane are capable of performing CMA (Patel and Cuervo, 2015). The percentage of CMA-competent lysosomes in the rat liver is 20–30% under basal conditions but can increase to 80% during starvation (Cuervo et al., 1997).
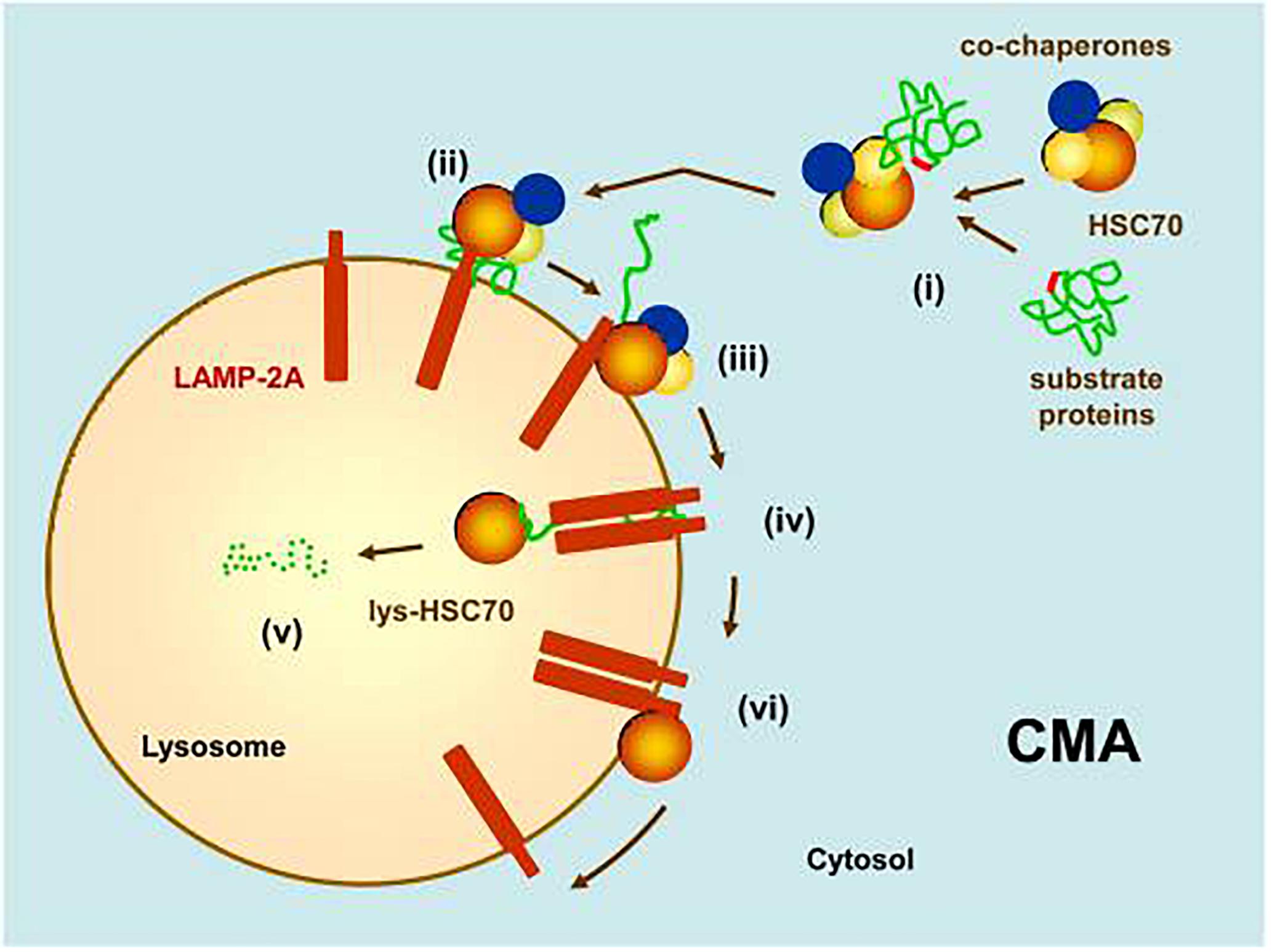
Figure 2. Chaperone-mediated autophagy (CMA). (i) Substrate proteins with an amino acid sequence biochemically similar to KFERQ are recognized by HSPA8/hsc70 in association with co-chaperones. (ii) This complex binds to the monomeric form of a lysosomal surface receptor, lamp2a, which will facilitate its translocation into the lumen of the lysosome. (iii) Binding of the substrate to lamp2a causes lamp2a to multimerize into a translocation complex. (iv) After unfolding, the substrate translocates into the lysosomal lumen. The presence of a lysosomal luminal form of HSPA8/hsc70 is required for this translocation to occur. (v) The substrate is quickly degraded by lysosomal hydrolytic enzymes. (vi) Lamp2a disassembles from its translocation complex, facilitated by HSPA8/hsc70 (Reprinted from Cuervo, 2010).
Microautophagy has been less studied than the other autophagic mechanisms. In this process the lysosomal membrane directly engulfs portions of cytoplasm and the constituents within it. This process can be non-selective (degradation of randomly sequestered cytosol) or selective (degradation of specific organelles such as mitochondria, portions of the nucleus, or peroxisomes) (Farré et al., 2009; Mijaljica et al., 2011). Little is known about the factors that regulate microautophagy in mammalian cells (Mijaljica et al., 2011).
Cross-Talk Between Autophagic Mechanisms
Both CMA and macroautophagy are maximally activated by stress conditions such as starvation and oxidative stress (Kaushik et al., 2008). Cross-talk occurs between these two autophagic mechanisms (Figure 3) (reviewed by Wu et al., 2015); if the activity of one is reduced, the activity of the other may increase in an effort to maintain protein homeostasis (Cuervo et al., 2004; Massey et al., 2006a,b, 2008; Kaushik et al., 2008; Wang et al., 2010). For example, while macroautophagy is initially upregulated due to nutrient deprivation, if this condition continues it becomes downregulated, with compensatory activation of CMA (Cuervo et al., 1995; Massey et al., 2006a). Conversely, blockage of CMA by inhibitory RNA targeting of lamp2a initially results in a decline in macroautophagy due to increased mTOR activity, but if CMA blockage continues then macroautophagy becomes activated as indicated by increased levels of Beclin 1. In a cell model of tauopathy, failure of CMA to clear mutated tau led to formation of tau aggregates, which could be cleared by macroautophagy (Wang et al., 2010). The signaling mechanisms in this cross-talk are incompletely understood; proteins suggested by Wu et al. (2015) to be involved in this process include HSPA8/hsc70 (Vakifahmetoglu-Norberg et al., 2013), p53 (Tasdemir et al., 2008), nuclear factor (erythroid-derived 2)-like 2 (Nrf2) (Gan et al., 2012), and ubiquilin (Rothenberg et al., 2010). Reduced phosphorylation of mTOR may play a role in the compensatory increase in macroautophagy when CMA is impaired (Massey et al., 2008), and when macroautophagy is inhibited, compensatory upregulation of CMA can occur through increasing lysosomal concentrations of lamp2a and HSPA8/hsc70 (Kaushik et al., 2008; Koga et al., 2011). Upregulation of CMA may not fully compensate for impairment in macroautophagy, and vice versa (Massey et al., 2006b; Wu et al., 2015). The cross-talk between the autophagic processes is not always bidirectional. Rodríguez-Muela et al. (2013) found an age-associated decrease in macroautophagy (decreased LC3-II/LC3-I ratio and reduced mRNA expression of Beclin 1 and Atg7) in the mouse retina, which was accompanied by an increase in CMA (increased lysosomal levels of lamp2a and HSPA8/hsc70). A similar result was obtained in vitro by shRNA-mediated knockdown of Atg7 expression in the 661W mouse retinal cell line. However, inhibition of CMA in these cells (by downregulating lamp2a) did not result in an increase in macroautophagy. The interactions between CMA and macroautophagy have been suggested to delay the accumulation of abnormal proteins, perhaps contributing to the association between aging and neurodegenerative disorders (Cuervo and Wong, 2014).
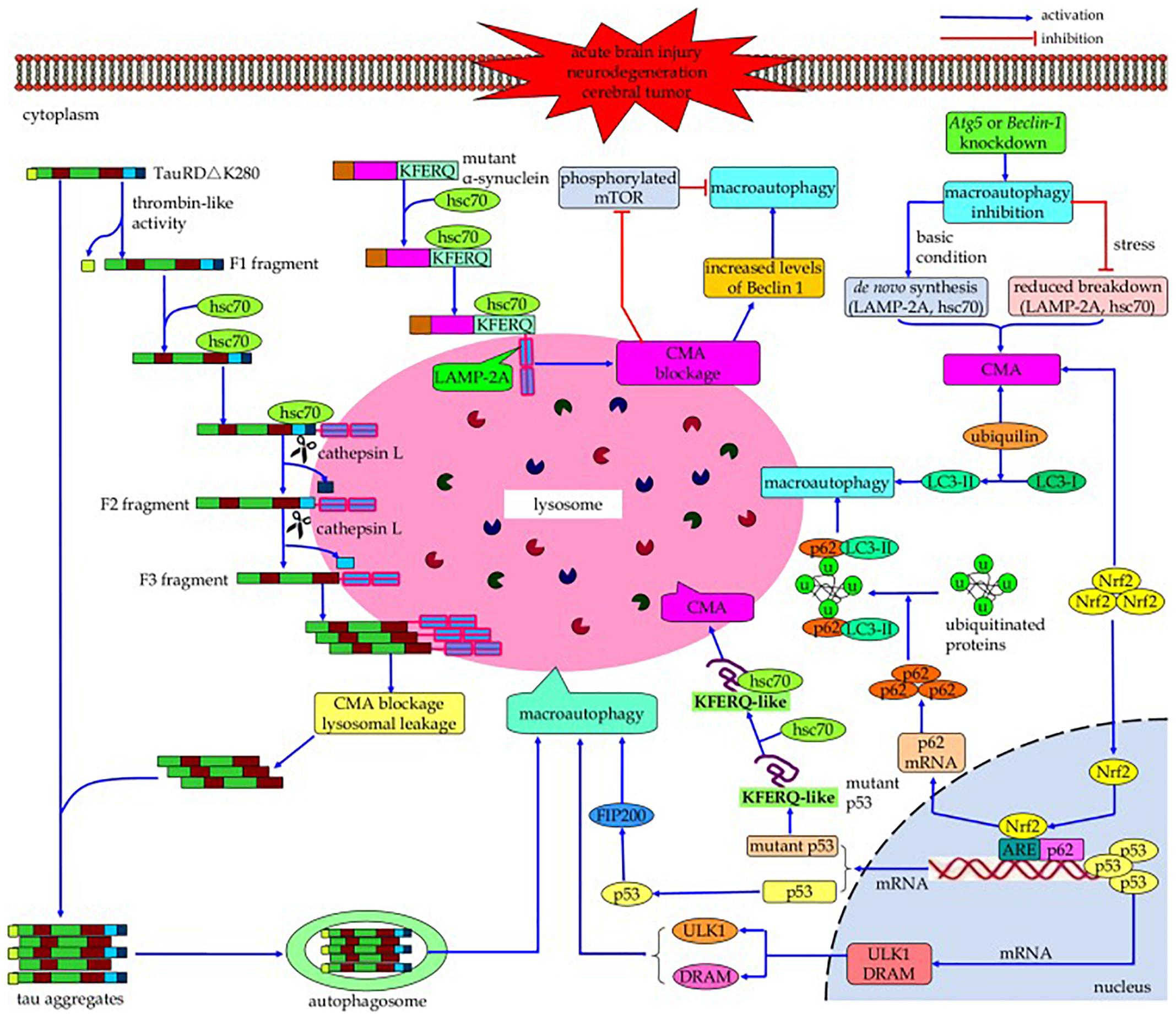
Figure 3. Cross-talk between macroautophagy and CMA. If the activity of one of these autophagic processes is reduced, the activity of the other may increase in a compensatory manner. Continued blockage of CMA by inhibitory RNA targeting of lamp2a results in activation of macroautophagy as indicated by increased levels of Beclin 1; conversely, while nutrient deprivation initially upregulates macroautophagy, continued nutrient deprivation downregulates it with compensatory activation of CMA. However, this compensation may not be bi-directional, and the upregulation of one mechanism may not fully compensate for impairment of the other one. The signaling mechanisms involved in this cross-talk, while poorly understood, may include HSPA8/hsc70, p53, Nrf2, and/or ubiquilin (Reprinted from Wu et al., 2015).
Influence of Normal Aging on Autophagy in Human Brain and CSF
Few studies have been performed on the effects of normal aging on autophagy in human brain or CSF. The findings in these studies are summarized in Table 1. Three gene expression studies with human brain have found age-associated reductions in macroautophagy markers. Shibata et al. (2006) found downregulation of Beclin 1, and Lipinski et al. (2010) found decreases in autophagy-regulating genes including Atg5 and Atg7. Guebel and Torres (2016), investigating the effects of gender and aging on gene transcription in the hippocampus, reported decreased expression of LC3, HDAC6 (a deacetylase required for autophagosomal maturation and fusion with lysosomes) (Lee J.H. et al., 2010), and PINK1 (a mitochondrial kinase whose activity is crucial to mitochondrial function) (Qu et al., 2015) in older women. In older men expression of Bcl-2, which inhibits Beclin 1 (Liang et al., 1999), was increased, suggesting a decrease in macroautophagic activity. Conversely, expression of BAG-2, which inhibits PINK1 degradation by blocking its ubiquitination, and therefore triggers PARKIN-mediated mitophagy (Qu et al., 2015), was also increased in older men, suggesting activation of mitophagy. Notably, no studies were found of the effects of normal aging on CMA activity in human brain.
Three studies have examined the effects of normal aging on autophagy-related proteins in CSF. A proteomics analysis found CSF concentrations of three proteins with the gene ontology classification of “autophagy,” namely myoglobin, MMP8, and HMW kininogen, to be positively correlated with age (Baird et al., 2012). Conversely, the CSF concentration of HSPA8/hsc70 was found to decrease with age (Loeffler et al., 2016) while the CSF level of lamp2 changed little (Loeffler et al., 2018). Lamp2 is not a marker for a specific autophagic process, because in addition to the previously discussed role of lamp2a in CMA (Cuervo and Dice, 2000a), lamp2 has two other isoforms which also participate in autophagic processes (Nishino et al., 2000; Fujiwara et al., 2015).
Influence of Normal Aging on Autophagy in the CNS in Experimental Models
Studies of the relationship between aging and brain macroautophagy, including age-related changes in macroautophagy markers and the effects of impairing macroautophagy (for example, by introducing mutations in Atg genes) on lifespan, are shown in Table 2 for experimental animal models. This table is not all-inclusive; additional references can be found in recent reviews by Hansen et al. (2018) and Nakamura and Yoshimori (2018). Macroautophagy has been shown to be required for normal lifespan in Caenorhabditis elegans and Drosophila (Meléndez et al., 2003; Hars et al., 2007; Simonsen et al., 2008; Minnerly et al., 2017) and enhancement of macroautophagic activity, for example by upregulation of AMPK or rapamycin-mediated inhibition of TOR, increases lifespan in these models (Vellai et al., 2003; Simonsen et al., 2008; Bjedov et al., 2010; Ulgherait et al., 2014). However, conflicting results were found by Hashimoto et al. (2009), who reported that knockout of some autophagy genes in daf-2 mutant C. elegans mutants increased rather than decreased their lifespan. Also in C. elegans, Saha et al. (2015) found an age-associated loss of macroautophagic function in dopamine neurons. A recent review by Nakamura and Yoshimori (2018) concluded that “basal level of autophagic activity is elevated in many longevity paradigms and the activity is required for lifespan extension.” The effects of aging on brain macroautophagy have also been investigated in mice (Hara et al., 2006; Komatsu et al., 2006; Gamerdinger et al., 2009; Kaushik et al., 2012; Triplett et al., 2015; Ott et al., 2016), rats (Yu et al., 2017), and cows (De Biase et al., 2017). Mice lacking Atg7 in the CNS developed neuronal loss and died within 28 weeks (Komatsu et al., 2006), and mice deficient for Atg5 in neural cells developed progressive motor deficits in conjunction with neuronal inclusion bodies (Hara et al., 2006). An association was reported between activation of macroautophagy and retention of spatial memory during aging; mice that were subjected to caloric restriction for 10 months, beginning at 6 weeks of age, performed better on Morris water maze testing than mice fed a high calorie diet, and the mice subjected to caloric restriction had higher hippocampal levels of Beclin 1, LC3, and cathepsin B and lower levels of mTOR and p62 than the high-calorie group (Dong et al., 2015). Kaushik et al. (2012) and Ott et al. (2016) reported age-related decreases in macroautophagy markers in mouse brain, and similar findings were reported by Yu et al. (2017) in rat brain. Brain levels of Beclin 1 were found to decline in old naked mole-rats, the longest-lived rodent, suggesting an age-associated decrease in macroautophagy in this species as well (Triplett et al., 2015). Rodríguez-Muela et al. (2013), examining age-related changes in autophagic mechanisms in mouse retina, found lower Beclin 1, and lower LC3-II after lysosomal blockage, in older mice, and De Biase et al. (2017) found lower Beclin 1 but an increase in the LC3-II/LC3-I ratio in old cow brain specimens. In conflict with these results is a study by Gamerdinger et al. (2009) which reported increased total cathepsin and cathepsin B activity in old mouse brain. In that study, aging was associated with a switch of regulatory control over degradation of polyubiquitinated proteins in mouse brain from Bcl-2-associated athanogene-1 (BAG1), a regulator of proteasomal degradation pathways (Lüders et al., 2000), to BAG3, a stimulator of macroautophagy (Carra et al., 2008), suggesting that aged cells may rely more on macroautophagy for degrading polyubiquitinated proteins.
Few studies have investigated the effects of normal aging on CMA in the CNS in experimental models. The results from these studies are summarized in Table 3. Three studies examined the influence of normal aging on the concentration of HSPA8/hsc70 in rat brain. Two of these reported an increase (Unno et al., 2000; Calabrese et al., 2004) while the third one found a decrease (Gleixner et al., 2014). In the study discussed above in which an age-related decrease in macroautophagy was found in mouse retina (Rodríguez-Muela et al., 2013), CMA was reported to increase with normal aging, possibly to compensate for the decrease in macroautophagy.
Influence of Normal Aging on Autophagy in Experimental Systems Other Than the CNS
The experimental models that have been used most frequently to examine the effects of aging on macroautophagy in non-CNS systems are human fibroblast cultures (Tashiro et al., 2014; Ott et al., 2016; Pernodet et al., 2016; Romero et al., 2016) and rat liver, in which Bergamini and colleagues have extensively investigated the effects of caloric restriction on macroautophagy and its endocrine regulation (Vittorini et al., 1999; Cavallini et al., 2001, 2007; Donati et al., 2001a,b, 2008, 2009; Del Roso et al., 2003). Studies have also been performed in mouse liver (Terman, 1995), rat heart (Wohlgemuth et al., 2007), mouse muscle (Masiero et al., 2009; Carnio et al., 2014), mouse heart (Inuzuka et al., 2009; Taneike et al., 2010; Boyle et al., 2011; Zhou et al., 2017), rat nucleus pulposis (Ye et al., 2011), and human muscle biopsies (Carnio et al., 2014). The results in these studies are summarized in Table 4. Most studies using Beclin 1 as a marker for macroautophagy have found an age-related decrease, while conflicting results have been obtained with measurements of LC3-II and LC3-II/LC3-I.
Studies on the effects of aging on CMA in experimental systems other than the CNS are summarized in Table 5. The initial studies on CMA were done by Dice and his colleagues (Dice, 1982; Okada and Dice, 1984; McElligott et al., 1985; Dice et al., 1986; Chiang and Dice, 1988; Chiang et al., 1989; Cuervo and Dice, 1996). Cuervo and Dice (2000a) reported that CMA decreased in lysosomes from aged rat livers and late-passage human fibroblasts, in association with an age-associated decrease in lamp2a on lysosomal membranes. Their finding of an age-related decrease in CMA in rat liver has been confirmed and extended in subsequent studies (Kaushik et al., 2007; Kiffin et al., 2007; Zhang and Cuervo, 2008; Bandyopadhyay et al., 2010; Rodríguez-Navarro and Cuervo, 2012; Schneider et al., 2015). Age-related changes in CMA have also been studied in mouse skeletal and cardiac muscle (Zhou et al., 2017) and rat nucleus pulposis (Ye et al., 2011), with conflicting results. An age-associated decrease in lamp2 gene expression was reported for human leukocytes (Huang et al., 2012) but as discussed above, lamp2 measurements are not specific for CMA.
Influence of Normal Aging on Lysosomal Activity
Normal aging has been associated with decreased lysosomal activity (Reeg and Grune, 2015; Carmona-Gutierrez et al., 2016). Successful autophagic removal of proteins requires an efficiently functioning lysosomal pool, so a decline in lysosomal activity could play a prominent role in age-related deficits in macroautophagy and CMA. Lysosomal hydrolytic enzymes include proteases, nucleases, lipases, sulfatases, and phosphatases, whose optimal pH range is 4.5–5 (Carmona-Gutierrez et al., 2016). In yeast, lysosomal pH is a regulator of lifespan and mitochondrial function (Hughes and Gottschling, 2012; Ruckenstuhl et al., 2014). Lysosome numbers and their size increase in senescent cells (Robbins et al., 1970; Brunk et al., 1973; Kurz et al., 2000), possibly due to accumulation of non-degradable constituents such as lipofuscin in autophagic vacuoles (Brunk and Terman, 1999). Lipofuscin accumulates during normal aging in lysosomes of neurons and other post-mitotic cells, where it may exert deleterious effects on autophagy if lysosomal enzymatic activity is expended in an effort to degrade it (Brunk and Terman, 2002). The literature contains conflicting results with regard to age-related changes in the activities of different lysosomal proteases in the CNS. Kenessey et al. (1989) and Nakamura et al. (1989) reported that cathepsin D activity increased in the aged rat brain, and Nakanishi et al. (1994) found increases in cathepsin D, E, and B activities, but decreased cathepsin L activity (although not in cathepsin L protein), in brain regions from older rats. Cathepsin D localized primarily to lysosomes in young rat cerebral cortical neurons, but diffuse cytosolic immunoreactivity was also present in older rats, possibly contributing to age-related cell death (Jung et al., 2010). (See Stoka et al., 2016 for a review of the regulation of lysosomal cathepsins in aging and neurodegeneration and their contribution to apoptotic cell death.) Conversely, Sarkis et al. (1988) reported age-related decreases in the activities of cathepsins D, Ce1, and Ce2 in C. elegans. Finally, Banay-Schwartz et al. (1992), measuring cathepsin D in 50 brain regions of adult and older individuals, found the levels of this enzyme to be significantly increased with age in 14 regions and decreased in two regions. The findings in these studies suggest that an age-related decline in lysosomal function is not necessarily accompanied by decreases in the activities of lysosomal proteases.
Discussion and Conclusion
Autophagic processes have often been stated to decrease with age (Martinez-Lopez et al., 2005; Martinez-Vicente et al., 2005; Cuervo, 2008; He et al., 2013; Cuervo and Wong, 2014; Schroeder et al., 2014). This conclusion is supported by most studies that have examined this issue in experimental systems other than the CNS (Tables 4, 5). The literature also suggests an age-related decline in macroautophagy in the CNS in experimental models, particularly C. elegans, Drosophila, and mice (Table 2). But because of the few studies on this issue in the human CNS (Table 1), no conclusion is currently possible as to whether autophagic processes in human brain decrease with normal aging, despite indirect evidence for this possibility from the studies in experimental systems. A similar conclusion was reached in a recent review by Hansen et al. (2018) of the connection between macroautophagy and longevity: “…further work is required to more rigorously test the hypothesis that autophagic activity may decline in an age-dependent fashion in the nervous system.”
The three gene expression studies which have investigated this issue in human brain found age- associated decreases in macroautophagy proteins, although the study by Guebel and Torres (2016) suggested differences between men and women for these parameters. However, confirmation that macroautophagy decreases in human brain during normal aging requires similar results with other methodologies. The lack of studies on age-associated changes in CMA in human brain specimens is a critical gap in the literature; unfortunately, the few CSF studies on this issue do not help to resolve the situation. Although the study by Baird et al. (2012) found the concentrations of three autophagy-related proteins to correlate with age, none of the proteins plays a major role in autophagy. The reports of an age-associated decrease in CSF HSPA8/hsc70 (Loeffler et al., 2016) and no change with normal aging in CSF lamp2 levels (Loeffler et al., 2018) provide no information about the effects of aging on specific autophagic mechanisms in the CNS. HSPA8/hsc70 participates in many processes in addition to CMA (Stricher et al., 2013; Liao and Tang, 2014) and measurement of total lamp2 is not specific for a particular autophagic mechanism. CSF studies are unlikely to be of value for determining the effects of aging on brain autophagy because (a) changes in the CSF concentration of a protein may not correlate with changes in its brain concentration, (b) the levels of autophagy-related proteins in CSF provide no information about their concentrations on lysosomal membranes; (c) it is unclear if the markers commonly used to assess changes in autophagic processes can be detected in CSF; and (d) measurement of the LC3-II/LC3-I ratio in the presence of inhibitors of lysosomal proteases or autophagosomal-lysosomal fusion is not possible in humans.
Some of these same issues raise concerns about the findings in the studies that have examined the effects of aging on brain autophagic processes in vertebrate models. Many of the studies on age-related changes in macroautophagy in experimental systems (Table 2) included measurements of total LC3, LC3-II, or the LC3-II/LC3-I ratio. The results in most of these studies are difficult to interpret with regard to autophagic flux because autophagic inhibitors were not included (exceptions: Kaushik et al., 2012; Rodríguez-Muela et al., 2013; Tashiro et al., 2014; Ott et al., 2016), and because they represent findings at only one time point. None of the studies on rat brain listed in Table 3 (effects of normal aging on CNS CMA in experimental systems) were performed on lysosome-enriched fractions, so the extent to which the reported levels of HSPA8/hsc70 reflected lysosomal concentrations of this protein is unknown. An inherent difficulty with comparing autophagy studies between different models is that basal autophagic activity in experimental animals can vary according to age, sex, and genetic background (Klionsky et al., 2016).
Comprehensive reviews of methods for assessing macroautophagy and CMA have recently been published (Patel and Cuervo, 2015; Klionsky et al., 2016). Appropriate procedures for investigating the effects of aging on these processes in the brain include transmission electron microscopy (TEM), western blot, immunohistochemical and immunofluorescent staining, and gene expression analyses. Combinations of these methods should be used (Klionsky et al., 2016). In addition to potential difficulties with data interpretation resulting from agonal changes and postmortem autolysis, studies on postmortem brain specimens provide only a “snapshot” of autophagic activity at a single time point, with no information about autophagic flux. TEM can detect and quantify morphological changes in autophagosomes, autolysosomes, amphisomes, and sequestered organelles (Eskelinen et al., 2011). Western blot is often used for examining changes in LC3-I and LC3-II levels (Tanida et al., 2005), but there are some caveats to the use of this procedure: (a) the concentration of LC3-I is higher than LC3-II in the brain, so LC3-II may be difficult to detect by western blot in crude homogenates (Chu et al., 2009; Yang et al., 2011); (b) LC3-I is more susceptible than LC3-II to degradation in SDS sample buffer or by exposure to freeze/thaw cycles; and (c) LC3-II levels measured by western blot should be compared to the levels of one or more loading control proteins rather than to LC3-I (Klionsky et al., 2016). Specific markers for CMA are presently limited to lamp2a and HSPA8/hsc70, but only the lysosomal membrane levels of these proteins correlate with CMA activity so their detection by Western blot or RT-PCR (Zhang et al., 2006; Jesus et al., 2013) should optimally be performed on lysosome-enriched fractions (Klionsky et al., 2016). Immunodetection of lamp2a should be done with antibodies which are specific for lamp2a (Patel and Cuervo, 2015).
Determining whether autophagic mechanisms decrease in the brain during normal aging has important clinical ramifications. Lifestyle modifications such as caloric restriction (Morselli et al., 2010; Rodríguez-Navarro and Cuervo, 2012; Rodríguez-Navarro et al., 2012) and exercise (He et al., 2012) have been shown to increase autophagic activity in experimental animals. Exercise reduces brain pathology in some mouse models of PD (Lau et al., 2011; Patki and Lau, 2011) and AD (Adlard et al., 2005; Nichol et al., 2008; Um et al., 2008), perhaps by induction of autophagy. Note, however, that a study in the P301S mouse model of tauopathy which examined the influence of long-term exercise on autophagy as well as tau pathology found reduced levels of full-length and hyperphosphorylated tau in the hippocampus and spinal cord despite no significant changes in the concentrations of LC3-II or p62/SQSTM1 (Ohia-Nwoko et al., 2014), suggesting that the neuroprotective effects of exercise in that study may have been due to factors other than induction of macroautophagy. Exercise can also exert neuroprotective effects by increasing synaptic protein levels, cell proliferation, and neurotrophic factors in the brain (Molteni et al., 2002; Naylor et al., 2005; Huang et al., 2006; Marlatt et al., 2012). In transgenic mouse models of AD, reversal of autophagic deficits reduces brain Aβ deposition and prevents learning and memory deficits (Caccamo et al., 2010; Spilman et al., 2010; Yang et al., 2011; Steele et al., 2013), and a reduction in α-synuclein aggregation and mitochondrial dysfunction has been found when autophagy is increased in experimental animal models of PD (reviewed by Moors et al., 2017). Taken together, these findings suggest that pharmacologic interventions and/or lifestyle alterations which increase brain autophagy might be useful for treatment and possibly even prevention of age-associated neurodegenerative disorders. The development of autophagy-inducing agents is an active field of research (reviewed by Levine et al., 2015; Vakifahmetoglu-Norberg et al., 2015). This approach is not without risk, because autophagy has been associated with increased tumor cell survival in some studies (reviewed by Gelino and Hansen, 2012). Further, in patients with impairments in autophagosomal-lysosomal fusion, such as AD patients with presenilin gene mutations (Lee J.Y. et al., 2010), treatment with autophagy- inducing agents might produce further accumulation of neurotoxic polyubiquitinated protein aggregates (Levine et al., 2015). Nevertheless, if autophagic mechanisms can be conclusively shown to decrease in the brain during normal aging, then interventions to prevent this decrease might reduce the risk for some age-related neurodegenerative disorders, as suggested by Cuervo and Wong (2014) and Loos et al. (2017).
Author Contributions
DL planned and wrote the manuscript.
Funding
This review was supported by a generous donation to the Beaumont Foundation by Ms. Marilyn Bishop.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abeliovich, H., Dunn, W. A. Jr., Kim, J., and Klionsky, D. J. (2000). Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025–1034. doi: 10.1083/jcb.151.5.1025
Abounit, K., Scarabelli, T. M., and McCauley, R. B. (2012). Autophagy in mammalian cells. World J. Biol. Chem. 3, 1–6. doi: 10.4331/wjbc.v3.i1.1
Adlard, P. A., Perreau, V. M., Pop, V., and Cotman, C. W. (2005). Voluntary exercise decreases amyloid load in a transgenic model of Alzheimer’s disease. J. Neurosci. 25, 4217–4221. doi: 10.1523/JNEUROSCI.0496-05.2005
Agarraberes, F. A., and Dice, J. F. (2001). A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114, 2491–2499.
Alvarez-Erviti, L., Rodriguez-Oroz, M. C., Cooper, J. M., Caballero, C., Ferrer, I., Obeso, J. A., et al. (2010). Chaperone-mediated autophagy markers in Parkinson disease brains. Arch. Neurol. 67, 1464–1472. doi: 10.1001/archneurol.2010.198
Alvarez-Erviti, L., Seow, Y., Schapira, A. H., Rodriguez-Oroz, M. C., Obeso, J. A., and Cooper, J. M. (2013). Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell Death Dis. 4:e545. doi: 10.1038/cddis.2013.73
Baird, G. S., Nelson, S. K., Keeney, T. R., Stewart, A., Williams, S., Kraemer, S., et al. (2012). Age-dependent changes in the cerebrospinal fluid proteome by slow off-rate modified aptamer array. Am. J. Pathol. 180, 446–456. doi: 10.1016/j.ajpath.2011.10.024
Banay-Schwartz, M., DeGuzman, T., Kenessey, A., Palkovits, M., and Lajtha, A. (1992). The distribution of cathepsin D activity in adult and aging human brain regions. J. Neurochem. 58, 2207–2211. doi: 10.1111/j.1471-4159.1992.tb10965.x
Bandyopadhyay, U., and Cuervo, A. M. (2007). Chaperone-mediated autophagy in aging and neurodegeneration: lessons from alpha-synuclein. Exp. Gerontol. 42, 120–128. doi: 10.1016/j.exger.2006.05.019
Bandyopadhyay, U., Sridhar, S., Kaushik, S., Kiffin, R., and Cuervo, A. M. (2010). Identification of regulators of chaperone-mediated autophagy. Mol. Cell 39, 535–547. doi: 10.1016/j.molcel.2010.08.004
Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001). Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555. doi: 10.1126/science.292.5521.1552
Bjedov, I., Toivonen, J. M., Kerr, F., Slack, C., Jacobson, J., Foley, A., et al. (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46. doi: 10.1016/j.cmet.2009.11.010
Boyle, A. J., Shih, H., Hwang, J., Ye, J., Lee, B., Zhang, Y., et al. (2011). Cardiomyopathy of aging in the mammalian heart is characterized by myocardial hypertrophy, fibrosis and a predisposition towards cardiomyocyte apoptosis and autophagy. Exp. Gerontol. 46, 549–559. doi: 10.1016/j.exger.2011.02.010
Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S., et al. (1994). A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758. doi: 10.1038/369756a0
Brunk, U. T., Ericsson, J. L. E., Ponten, J., and Westermark, B. (1973). Residual bodies and ‘aging’ in cultured human glia cells. Exp. Cell Res. 79, 1–14. doi: 10.1016/0014-4827(73)90484-9
Brunk, U. T., and Terman, A. (1999). “The mitochondrial-lysosomal axis theory of cellular aging,” in Understanding the Process of Aging: The Roles of Mitochondria, Free Radicals and Antioxidants, eds E. Cadenas and L. Packer (New York, NY: Marcel Dekker Inc), 229–249.
Brunk, U. T., and Terman, A. (2002). Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic. Biol Med. 33, 611–619. doi: 10.1016/S0891-5849(02)00959-0
Budini, M., Buratti, E., Morselli, E., and Criollo, A. (2017). Autophagy and its impact on neurodegenerative diseases: new roles for TDP-43 and C9orf72. Front. Mol. Neurosci. 10:170. doi: 10.3389/fnmol.2017.00170
Caccamo, A., Majumder, S., Richardson, A., Strong, R., and Oddo, S. (2010). Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments. J. Biol. Chem. 285, 13107–13120. doi: 10.1074/jbc.M110.100420
Calabrese, V., Scapagnini, G., Ravagna, A., Colombrita, C., Spadaro, F., Butterfield, D. A., et al. (2004). Increased expression of heat shock proteins in rat brain during aging: relationship with mitochondrial function and glutathione redox state. Mech. Ageing Dev. 125, 325–335. doi: 10.1016/j.mad.2004.01.003
Cao, B., Li, J., Zhou, X., Juan, J., Han, K., Zhang, Z., et al. (2014). Clioquinol induces pro-death autophagy in leukemia and myeloma cells by disrupting the mTOR signaling pathway. Sci. Rep. 4:5749. doi: 10.1038/srep05749
Carmona-Gutierrez, D., Hughes, A. L., Madeo, F., and Ruckenstuhl, C. (2016). The crucial impact of lysosomes in aging and longevity. Ageing Res. Rev. 32, 2–12. doi: 10.1016/j.arr.2016.04.009
Carnio, S., LoVerso, F., Baraibar, M. A., Longa, E., Khan, M. M., Maffei, M., et al. (2014). Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 8, 1509–1521. doi: 10.1016/j.celrep.2014.07.061
Carra, S., Seguin, S. J., Lambert, H., and Landry, J. (2008). HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J. Biol. Chem. 283, 1437–1444. doi: 10.1074/jbc.M706304200
Cavallini, G., Donati, A., Gori, Z., Pollera, M., and Bergamini, E. (2001). The protection of rat liver autophagic proteolysis from the age-related decline co-varies with the duration of anti-ageing food restriction. Exp. Gerontol. 36, 497–506. doi: 10.1016/S0531-5565(00)00224-2
Cavallini, G., Donati, A., Taddei, M., and Bergamini, E. (2007). Evidence for selective mitochondrial autophagy and failure in aging. Autophagy 3, 26–27. doi: 10.4161/auto.3268
Chiang, H. I., and Dice, J. F. (1988). Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J. Biol. Chem. 263, 6797–6805.
Chiang, H. L., Terlecky, S. R., Plant, C. P., and Dice, J. F. (1989). A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385. doi: 10.1126/science.2799391
Chu, C. T., Plowey, E. D., Dagda, R. K., Hickey, R. W., Cherra, S. J. III, and Clark, R. S. (2009). Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 453, 217–249. doi: 10.1016/S0076-6879(08)04011-1
Ciechanover, A., and Kwon, Y. T. (2015). Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp. Mol. Med. 47:e147. doi: 10.1038/emm.2014.117
Ciechanover, A., and Kwon, Y. T. (2017). Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 11:185. doi: 10.3389/fnins.2017.00185
Codogno, P., and Meijer, A. J. (2006). Atg5: more than an autophagy factor. Nat. Cell Biol. 8, 1045–1047. doi: 10.1038/ncb1006-1045
Cuervo, A., Dice, J., and Knecht, E. (1997). A lysosomal population responsible for the hsc73-mediated degradation of cytosolic proteins in lysosomes. J. Biol. Chem. 272, 5606–5615. doi: 10.1074/jbc.272.9.5606
Cuervo, A. M. (2008). Autophagy and aging: keeping that old broom working. Trends Genet. 24, 604–612. doi: 10.1016/j.tig.2008.10.002
Cuervo, A. M. (2010). Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol. Metab. 21, 142–150. doi: 10.1016/j.tem.2009.10.003
Cuervo, A. M., and Dice, J. F. (1996). A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503. doi: 10.1126/science.273.5274.501
Cuervo, A. M., and Dice, J. F. (2000a). Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505–31513.
Cuervo, A. M., and Dice, J. F. (2000b). Regulation of lamp2a levels in the lysosomal membrane. Traffic 1, 570–583. doi: 10.1034/j.1600-0854.2000.010707.x
Cuervo, A. M., Knecht, E., Terlecky, S. R., and Dice, J. F. (1995). Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 269, C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200
Cuervo, A. M., Stefanis, L., Fredenburg, R., Lansbury, P. T., and Sulzer, D. (2004). Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295. doi: 10.1126/science.1101738
Cuervo, A. M., and Wong, E. (2014). Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24, 92–104. doi: 10.1038/cr.2013.153
De Biase, D., Costagliola, A., Pagano, T. B., Piegari, G., Wojcik, S., Dziewiątkowski, J., et al. (2017). Amyloid precursor protein, lipofuscin accumulation and expression of autophagy markers in aged bovine brain. BMC Vet. Res. 13:102. doi: 10.1186/s12917-017-1028-1
Del Roso, A., Vittorini, S., Cavallini, G., Donati, A., Gori, Z., Masini, M., et al. (2003). Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp. Gerontol. 38, 519–527. doi: 10.1016/S0531-5565(03)00002-0
Dice, J. F. (1982). Altered degradation of proteins microinjected into senescent human fibroblasts. J. Biol. Chem. 257, 14624–14627.
Dice, J. F., Chiang, H. L., Spencer, E. P., and Backer, J. M. (1986). Regulation of catabolism of microinjected ribonuclease A. Identification of residues 7-11 as the essential pentapeptide. J. Biol. Chem. 261, 6853–6859.
DiLoreto, R., and Murphy, C. T. (2015). The cell biology of aging. Mol. Biol. Cell 26, 4524–4531. doi: 10.1091/mbc.E14-06-1084
Donati, A., Cavallini, G., and Bergamini, E. (2009). Methods for inducing and monitoring liver autophagy relative to aging and antiaging caloric restriction in rats. Methods Enzymol. 452, 441–455. doi: 10.1016/S0076-6879(08)03626-4
Donati, A., Cavallini, G., Paradiso, C., Vittorini, S., Pollera, M., Gori, Z., et al. (2001a). Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J. Gerontol. A Biol. Sci. Med. Sci. 56, B288–B293. doi: 10.1093/gerona/56.7.B288
Donati, A., Cavallini, G., Paradiso, C., Vittorini, S., Pollera, M., Gori, Z., et al. (2001b). Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J. Gerontol. A Biol. Sci. Med. Sci. 56, B375–B383.
Donati, A., Recchia, G., Cavallini, G., and Bergamini, E. (2008). Effect of aging and anti-aging caloric restriction on the endocrine regulation of rat liver autophagy. J. Gerontol. A Biol. Sci. Med. Sci. 63, 550–555. doi: 10.1093/gerona/63.6.550
Dong, W., Wang, R., Ma, L. N., Xu, B. L., Zhang, J. S., Zhao, Z. W., et al. (2015). Autophagy involving age-related cognitive behavior and hippocampus injury is modulated by different caloric intake in mice. Int. J. Clin. Exp. Med. 8, 11843–11853.
Eskelinen, E. L. (2008). New insights into the mechanisms of macroautophagy in mammalian cells. Int. Rev. Cell Mol. Biol. 266, 207–247. doi: 10.1016/S1937-6448(07)66005-5
Eskelinen, E. L., Reggiori, F., Baba, M., Kovács, A. L., and Seglen, P. O. (2011). Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 7, 935–956. doi: 10.4161/auto.7.9.15760
Farré, J. C., Krick, R., Subramani, S., and Thumm, M. (2009). Turnover of organelles by autophagy in yeast. Curr. Opin. Cell Biol. 21, 522–530. doi: 10.1016/j.ceb.2009.04.015
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. doi: 10.1038/cr.2013.168
Fujiwara, Y., Hase, K., Wada, K., and Kabuta, T. (2015). An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochem. Biophys. Res. Commun. 460, 281–286. doi: 10.1016/j.bbrc.2015.03.025
Gamerdinger, M., Hajieva, P., Kaya, A. M., Wolfrum, U., Hartl, F. U., and Behl, C. (2009). Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28, 889–901. doi: 10.1038/emboj.2009.29
Gan, L., Vargas, M. R., Johnson, D. A., and Johnson, J. A. (2012). Astrocyte-specific overexpression of Nrf2 delays motor pathology and synuclein aggregation throughout the CNS in the alpha-synuclein mutant (A53T) mouse model. J. Neurosci. 32, 17775–17787. doi: 10.1523/JNEUROSCI.3049-12.2012
Gelino, S., and Hansen, M. (2012). Autophagy - an emerging anti-aging mechanism. J. Clin. Exp. Pathol. Suppl 4:006.
Gleixner, A. M., Pulugulla, S. H., Pant, D. B., Posimo, J. M., Crum, T. S., and Leak, R. K. (2014). Impact of aging on heat shock protein expression in the substantia nigra and striatum of the female rat. Cell Tissue Res. 357, 43–54. doi: 10.1007/s00441-014-1852-6
Guebel, D. V., and Torres, N. V. (2016). Sexual dimorphism and aging in the human hyppocampus: identification, validation, and impact of differentially expressed genes by factorial microarray and network analysis. Front. Aging Neurosci. 8:229. doi: 10.3389/fnagi.2016.00229
Guo, F., Liu, X., Cai, H., and Le, W. (2018). Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain Pathol. 28, 3–13. doi: 10.1111/bpa.12545
Hanada, T., and Ohsumi, Y. (2005). Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy 1, 110–118. doi: 10.4161/auto.1.2.1858
Hansen, M., Rubinsztein, D. C., and Walker, D. W. (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593. doi: 10.1038/s41580-018-0033-y
Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., et al. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. doi: 10.1038/nature04724
Hardie, D. G. (2011). AMPK and autophagy get connected. EMBO J. 30, 634–635. doi: 10.1038/emboj.2011.12
Hars, E. S., Qi, H., Ryazanov, A. G., Jin, S., Cai, L., Hu, C., et al. (2007). Autophagy regulates ageing in C. elegans. Autophagy 3, 93–95. doi: 10.4161/auto.3636
Hartl, F. U. (2017). Protein misfolding diseases. Annu. Rev. Biochem. 86, 21–26. doi: 10.1146/annurev-biochem-061516-044518
Hashimoto, Y., Ookuma, S., and Nishida, E. (2009). Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes Cells 14, 717–726. doi: 10.1111/j.1365-2443.2009.01306.x
Hatters, D. M. (2008). Protein misfolding inside cells: the case of huntingtin and Huntington’s disease. IUBMB Life 60, 724–728. doi: 10.1002/iub.111
He, C., Sumpter, R. Jr., and Levine, B. (2012). Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 8, 1548–1551. doi: 10.4161/auto.21327
He, L. Q., Lu, J. H., and Yue, Z. Y. (2013). Autophagy in ageing and ageing-associated diseases. Acta Pharmacol. Sin. 34, 605–611. doi: 10.1038/aps.2012.188
Hewitt, G., and Korolchuk, V. I. (2017). Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends Cell Biol. 27, 340–351. doi: 10.1016/j.tcb.2016.11.011
Hosokawa, N., Hara, T., Kaizuka, T., Kishi, C., Takamura, A., Miura, Y., et al. (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991. doi: 10.1091/mbc.E08-12-1248
Huang, A. M., Jen, C. J., Chen, H. F., Yu, L., Kuo, Y. M., and Chen, H. I. (2006). Compulsive exercise acutely upregulates rat hippocampal brain-derived neurotrophic factor. J. Neural Transm. 113, 803–811. doi: 10.1007/s00702-005-0359-4
Huang, J., Xu, J., Pang, S., Bai, B., and Yan, B. (2012). Age-related decrease of the LAMP-2 gene expression in human leukocytes. Clin. Biochem. 45, 1229–1232. doi: 10.1016/j.clinbiochem.2012.06.01
Hughes, A. L., and Gottschling, D. E. (2012). An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265. doi: 10.1038/nature11654
Inuzuka, Y., Okuda, J., Kawashima, T., Kato, T., Niizuma, S., Tamaki, Y., et al. (2009). Suppression of phosphoinositide 3-kinase prevents cardiac aging in mice. Circulation 120, 1695–1703. doi: 10.1161/CIRCULATIONAHA.109.871137
Jeppesen, D. K., Bohr, V. A., and Stevnsner, T. (2011). DNA repair deficiency in neurodegeneration. Prog. Neurobiol. 94, 166–200. doi: 10.1016/j.pneurobio.2011.04.013
Jesus, T. F., Inácio, A., and Coelho, M. C. (2013). Different levels of hsp70 and hsc70 mRNA expression in Iberian fish exposed to distinct river conditions. Genet. Mol. Biol. 36, 61–69. doi: 10.1590/S1415-47572013000100009
Jung, C. H., Ro, S. H., Cao, J., Otto, N. M., and Kim, D. H. (2010). mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295. doi: 10.1016/j.febslet.2010.01.017
Kang, R., Zeh, H. J., Lotze, M. T., and Tang, D. (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18, 571–580. doi: 10.1038/cdd.2010.191
Kaushik, S., Arias, E., Kwon, H., Lopez, N. M., Athonvarangkul, D., Sahu, S., et al. (2012). Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 13, 258–265. doi: 10.1038/embor.2011.260
Kaushik, S., Kiffin, R., and Cuervo, A. M. (2007). Chaperone-mediated autophagy and aging: a novel regulatory role of lipids revealed. Autophagy 3, 387–389. doi: 10.4161/auto.4246
Kaushik, S., Massey, A. C., Mizushima, N., and Cuervo, A. M. (2008). Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell 19, 2179–2192. doi: 10.1091/mbc.e07-11-1155
Kenessey, A., Banay-Schwartz, M., DeGuzman, T., and Lajtha, A. (1989). Increase in cathepsin D activity in rat brain in aging. J. Neurosci. Res. 23, 454–456. doi: 10.1002/jnr.490230412
Kiffin, R., Kaushik, S., Zeng, M., Bandyopadhyay, U., Zhang, C., Massey, A. C., et al. (2007). Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J. Cell Sci. 120(Pt. 5), 782–791. doi: 10.1242/jcs.001073
Kim, S. G., Buel, G. R., and Blenis, J. (2013). Nutrient regulation of the mTOR complex 1 signaling pathway. Mol. Cells 35, 463–473. doi: 10.1007/s10059-013-0138-2
Kirkin, V., Lamark, T., Sou, Y. S., Bjørkøy, G., Nunn, J. L., Bruun, J. A., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516. doi: 10.1016/j.molcel.2009.01.020
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Klionsky, D. J., and Codogno, P. (2013). The mechanism and physiological function of macroautophagy. J. Innate Immun. 5, 427–433. doi: 10.1159/000351979
Koga, H., Martinez-Vicente, M., Arias, E., Kaushik, S., Sulzer, D., and Cuervo, A. M. (2011). Constitutive upregulation of chaperone-mediated autophagy in Huntington’s disease. J. Neurosci. 31, 18492–18505. doi: 10.1523/JNEUROSCI.3219-11.2011
Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J., Tanida, I., et al. (2006). Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884. doi: 10.1038/nature04723
Kupfer, L., Hinrichs, W., and Groschup, M. (2009). Prion protein misfolding. Curr. Mol. Med. 9, 826–835. doi: 10.2174/156652409789105543
Kurz, D. J., Decary, S., Hong, Y., and Erusalimsky, J. D. (2000). Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J. Cell Sci. 113, 3613–3622.
Lau, Y. S., Patki, G., Das-Panja, K., Le, W. D., and Ahmad, S. O. (2011). Neuroprotective effects and mechanisms of exercise in a chronic mouse model of Parkinson’s disease with moderate neurodegeneration. Eur. J. Neurosci. 33, 1264–1274. doi: 10.1111/j.1460-9568.2011.07626.x
Lee, J. H., Yu, W. H., Kumar, A., Lee, S., Mohan, P. S., Peterhoff, C. M., et al. (2010). Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158. doi: 10.1016/j.cell.2010.05.008
Lee, J. Y., Koga, H., Kawaguchi, Y., Tang, W., Wong, E., Gao, Y. S., et al. (2010). HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29, 969–980. doi: 10.1038/emboj.2009.405
Levine, B., Packer, M., and Codogno, P. (2015). Development of autophagy inducers in clinical medicine. J. Clin. Invest. 125, 14–24. doi: 10.1172/JCI73938
Liang, C., Lee, J. S., Inn, K. S., Gack, M. U., Li, Q., Roberts, E. A., et al. (2008). Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787. doi: 10.1038/ncb1740
Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., et al. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676. doi: 10.1038/45257
Liao, Y., and Tang, L. (2014). The critical roles of HSC70 in physiological and pathological processes. Curr. Pharm. Des. 20, 101–107. doi: 10.2174/13816128113199990585
Linton, P. J., Gurney, M., Sengstock, D., Mentzer, R. M. Jr., and Gottlieb, R. A. (2015). This old heart: Cardiac aging and autophagy. J. Mol. Cell. Cardiol. 83, 44–54. doi: 10.1016/j.yjmcc.2014.12.017
Lipinski, M. M., Zheng, B., Lu, T., Yan, Z., Py, B. F., Ng, A., et al. (2010). Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 107, 14164–14169. doi: 10.1073/pnas.1009485107
Lippai, M., and Löw, P. (2014). The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed. Res. Int. 2014:832704. doi: 10.1155/2014/832704
Loeffler, D. A., Klaver, A. C., Coffey, M. P., and Aasly, J. O. (2018). Cerebrospinal fluid concentration of key autophagy protein Lamp2 changes little during normal aging. Front. Aging Neurosci. 10:130. doi: 10.3389/fnagi.2018.00130
Loeffler, D. A., Klaver, A. C., Coffey, M. P., Aasly, J. O., and LeWitt, P. A. (2016). Age-related decrease in heat shock 70-kDa protein 8 in cerebrospinal fluid is associated with increased oxidative stress. Front. Aging Neurosci. 8:178. doi: 10.3389/fnagi.2016.00178
Loos, B., du Toit, A., and Hofmeyr, J. H. (2014). Defining and measuring autophagosome flux -concept and reality. Autophagy 10, 2087–2096. doi: 10.4161/15548627.2014.973338
Loos, B., Klionsky, D. J., and Wong, E. (2017). Augmenting brain metabolism to increase macro- and chaperone-mediated autophagy for decreasing neuronal proteotoxicity and aging. Prog. Neurobiol. 156, 90–106. doi: 10.1016/j.pneurobio.2017.05.001
Lüders, J., Demand, J., and Höhfeld, J. (2000). The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275, 4613–4617. doi: 10.1074/jbc.275.7.4613
Mari, M., Tooze, S. A., and Reggiori, F. (2011). The puzzling origin of the autophagosomal membrane. F1000 Biol. Rep. 3:25. doi: 10.3410/B3-25
Marlatt, M. W., Potter, M. C., Lucassen, P. J., and van Praag, H. (2012). Running throughout middle-age improves memory function, hippocampal neurogenesis, and BDNF levels in female C57BL/6J mice. Dev. Neurobiol. 72, 943–952. doi: 10.1002/dneu.22009
Martinez-Lopez, N., Martinez-Vicente, M., Sovak, G., and Cuervo, A. M. (2005). Protein degradation and aging. Exp. Gerontol. 40, 622–633. doi: 10.1016/j.exger.2005.07.005
Martinez-Vicente, M., Sovak, G., and Cuervo, A. M. (2005). Protein degradation and aging. Exp. Gerontol. 40, 622–633. doi: 10.1016/j.exger.2005.07.005
Masiero, E., Agatea, L., Mammucari, C., Blaauw, B., Loro, E., Komatsu, M., et al. (2009). Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515. doi: 10.1016/j.cmet.2009.10.008
Massey, A. C., Follenzi, A., Kiffin, R., Zhang, C., and Cuervo, A. M. (2008). Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy 4, 442–456. doi: 10.4161/auto.5654
Massey, A. C., Kaushik, S., and Cuervo, A. M. (2006a). Lysosomal chat maintains the balance. Autophagy 2, 325–327.
Massey, A. C., Kaushik, S., Sovak, G., Kiffin, R., and Cuervo, A. M. (2006b). Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U.S.A. 103, 5805–5810. doi: 10.1073/pnas.0507436103
McElligott, M. A., Miao, P., and Dice, J. F. (1985). Lysosomal degradation of ribonuclease A and ribonuclease S-protein microinjected into the cytosol of human fibroblasts. J. Biol. Chem. 260, 11986–11993.
Mehrpour, M., Esclatine, A., Beau, I., and Codogno, P. (2010). Overview of macroautophagy regulation in mammalian cells. Cell Res. 20, 748–762. doi: 10.1038/cr.2010.82
Meléndez, A., Tallóczy, Z., Seaman, M., Eskelinen, E. L., Hall, D. H., and Levine, B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391. doi: 10.1126/science.1087782
Meley, D., Bauvy, C., Houben-Weerts, J. H., Dubbelhuis, P. F., Helmond, M. T., Codogno, P., et al. (2006). AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 281, 34870–34879. doi: 10.1074/jbc.M605488200
Mijaljica, D., Prescott, M., and Devenish, R. J. (2011). Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7, 673–682. doi: 10.4161/auto.7.7.14733
Minnerly, J., Zhang, J., Parker, T., Kaul, T., and Jia, K. (2017). The cell non-autonomous function of ATG-18 is essential for neuroendocrine regulation of Caenorhabditis elegans lifespan. PLoS Genet. 13:e1006764. doi: 10.1371/journal.pgen.1006764
Mizushima, N. (2007). Autophagy: process and function. Genes Dev. 21, 2861–2873. doi: 10.1101/gad.1599207
Mizushima, N., and Yoshimori, T. (2007). How to interpret LC3 immunoblotting. Autophagy 3, 542–545. doi: 10.4161/auto.4600
Molejon, M. I., Ropolo, A., Re, A. L., Boggio, V., and Vaccaro, M. I. (2013). The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 3:1055. doi: 10.1038/srep01055
Molteni, R., Ying, Z., and Gómez-Pinilla, F. (2002). Differential effects of acute and chronic exercise on plasticity-related genes in the rat hippocampus revealed by microarray. Eur. J. Neurosci. 16, 1107–1116. doi: 10.1046/j.1460-9568.2002.02158.x
Moors, T. E., Hoozemans, J. J., Ingrassia, A., Beccari, T., Parnetti, L., Chartier-Harlin, M. C., et al. (2017). Therapeutic potential of autophagy-enhancing agents in Parkinson’s disease. Mol. Neurodegener. 12:11. doi: 10.1186/s13024-017-0154-3
Morselli, E., Maiuri, M. C., Markaki, M., Megalou, E., Pasparaki, A., Palikaras, K., et al. (2010). Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction f autophagy. Cell Death Dis. 1:e10. doi: 10.1038/cddis.2009.8
Mulligan, V. K., and Chakrabartty, A. (2013). Protein misfolding in the late-onset neurodegenerative diseases: common themes and the unique case of amyotrophic lateral sclerosis. Proteins 81, 1285–1303. doi: 10.1002/prot.24285
Murphy, K. E., Gysbers, A. M., Abbott, S. K., Spiro, A. S., Furuta, A., Cooper, A., et al. (2015). Lysosomal-associated membrane protein 2 isoforms are differentially affected in early Parkinson’s disease. Mov. Disord. 30, 1639–1647. doi: 10.1002/mds.26141
Nakamura, S., and Yoshimori, T. (2018). Autophagy and longevity. Mol. Cells 41, 65–72. doi: 10.14348/molcells.2018.2333
Nakamura, Y., Takeda, M., Suzuki, H., Morita, H., Tada, K., Hariguchi, S., et al. (1989). Age-dependent change in activities of lysosomal enzymes in rat brain. Mech. Ageing Dev. 50, 215–225. doi: 10.1016/0047-6374(89)90101-2
Nakanishi, H., Tominaga, K., Amano, T., Hirotsu, I., Inoue, T., and Yamamoto, K. (1994). Age-related changes in activities and localizations of cathepsins D, E, B, and L in the rat brain tissues. Exp. Neurol. 126, 119–128. doi: 10.1006/exnr.1994.1048
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Naylor, M., Bowen, K. K., Sailor, K. A., Dempsey, R. J., and Vemuganti, R. (2005). Preconditioning-induced ischemic tolerance stimulates growth factor expression and neurogenesis in adult rat hippocampus. Neurochem. Int. 47, 565–572. doi: 10.1016/j.neuint.2005.07.003
Nichol, K. E., Poon, W. W., Parachikova, A. I., Cribbs, D. H., Glabe, C. G., and Cotman, C. W. (2008). Exercise alters the immune profile in Tg2576 Alzheimer mice toward a response coincident with improved cognitive performance and decreased amyloid. J. Neuroinflammation 5:13. doi: 10.1186/1742-2094-5-13
Nishino, I., Fu, J., Tanji, K., Yamada, T., Shimojo, S., Koori, T., et al. (2000). Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906–910. doi: 10.1038/35022604
Noda, N. N., and Mizushima, N. (2016). Atg101: not just an accessory subunit in the autophagy-initiation complex. Cell Struct. Funct. 41, 13–20. doi: 10.1247/csf.15013
Obara, K., and Ohsumi, Y. (2008). Dynamics and function of PtdIns(3)P in autophagy. Autophagy 4, 952–954. doi: 10.4161/auto.6790
Ohia-Nwoko, O., Montazari, S., Lau, Y. S., and Eriksen, J. L. (2014). Long-term treadmill exercise attenuates tau pathology in P301S tau transgenic mice. Mol. Neurodegener. 9:54. doi: 10.1186/1750-1326-9-54
Okada, A. A., and Dice, J. F. (1984). Altered degradation of intracellular proteins in aging human fibroblasts. Mech. Ageing Dev. 26, 341–356. doi: 10.1016/0047-6374(84)90105-2
Orenstein, S. J., and Cuervo, A. M. (2010). Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 21, 719–726. doi: 10.1016/j.semcdb.2010.02.005
Orhon, I., and Reggiori, F. (2017). Assays to monitor autophagy progression in cell cultures. Cells 6:E20. doi: 10.3390/cells6030020
Ott, C., König, J., Höhn, A., Jung, T., and Grune, T. (2016). Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 10, 266–273. doi: 10.1016/j.redox.2016.10.015
Patel, B., and Cuervo, A. M. (2015). Methods to study chaperone-mediated autophagy. Methods 75, 133–140. doi: 10.1016/j.ymeth.2015.01.003
Patki, G., and Lau, Y. S. (2011). Impact of exercise on mitochondrial transcription factor expression and damage in the striatum of a chronic mouse model of Parkinson’s disease. Neurosci. Lett. 505, 268–272. doi: 10.1016/j.neulet.2011.10.036
Pernodet, N., Dong, K., and Pelle, E. (2016). Autophagy in human skin fibroblasts: Comparison between young and aged cells and evaluation of its cellular rhythm and response to Ultraviolet A radiation. J. Cosmet. Sci. 67, 13–20.
Pöggeler, S., Nowrousian, M., Teichert, I., Beier, A., and Kück, U. (2018). “Fruiting-body development in ascomycetes,” in The Mycota. A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research. XV. Physiology and Genetics: Selected Basic and Applied Aspects, 2nd Edn, eds T. Anke and A. Schüffler (Basel: Springer International Publishing AG), 26. doi: 10.1007/978-3-319-71740-1_1
Qu, D., Hage, A., Don-Carolis, K., Huang, E., Joselin, A., Safarpour, F., et al. (2015). BAG2 gene-mediated regulation of PINK1 protein is critical for mitochondrial translocation of PARKIN and neuronal survival. J. Biol. Chem. 290, 30441–30452. doi: 10.1074/jbc.M115.677815
Reeg, S., and Grune, T. (2015). Protein oxidation in aging: does it play a role in aging progression? Antioxid. Redox Signal. 23, 239–255. doi: 10.1089/ars.2014.6062
Robbins, E., Levine, E. M., and Eagle, H. (1970). Morphologic changes accompanying senescence of cultured human diploid cells. J. Exp. Med. 131, 1211–1222. doi: 10.1084/jem.131.6.1211
Rodríguez-Muela, N., Koga, H., García-Ledo, L., de la Villa, P., de la Rosa, E. J., Cuervo, A. M., et al. (2013). Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 12, 478–488. doi: 10.1111/acel.12072
Rodríguez-Navarro, J. A., and Cuervo, A. M. (2012). Dietary lipids and aging compromise chaperone-mediated autophagy by similar mechanisms. Autophagy 8, 1152–1154. doi: 10.4161/auto.20649
Rodríguez-Navarro, J. A., Kaushik, S., Koga, H., Dall’Armi, C., Shui, G., Wenk, M. R., et al. (2012). Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad Sci. U.S.A. 109, E705–E714. doi: 10.1073/pnas.1113036109
Romero, Y., Bueno, M., Ramirez, R., Álvarez, D., Sembrat, J. C., Goncharova, E. A., et al. (2016). mTORC1 activation decreases autophagy in aging and idiopathic pulmonary fibrosis and contributes to apoptosis resistance in IPF fibroblasts. Aging Cell 15, 1103–1112. doi: 10.1111/acel.12514
Rothenberg, C., Srinivasan, D., Mah, L., Kaushik, S., Peterhoff, C. M., Ugolino, J., et al. (2010). Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 19, 3219–3232. doi: 10.1093/hmg/ddq231
Rubinsztein, D. C., Mariño, G., and Kroemer, G. (2011). Autophagy and aging. Cell 146, 682–695. doi: 10.1016/j.cell.2011.07.030
Ruckenstuhl, C., Netzberger, C., Entfellner, I., Carmona-Gutierrez, D., Kickenweiz, T., and Stekovic, S. (2014). Autophagy extends lifespan via vacuolar acidification. Microb. Cell 1, 160–162. doi: 10.15698/mic2014.05.147
Saha, S., Ash, P. E., Gowda, V., Liu, L., Shirihai, O., and Wolozin, B. (2015). Mutations in LRRK2 potentiate age-related impairment of autophagic flux. Mol. Neurodegener. 10:26. doi: 10.1186/s13024-015-0022-y
Salminen, A., Kaarniranta, K., Kauppinen, A., Ojala, J., Haapasalo, A., Soininen, H., et al. (2013). Impaired autophagy and APP processing in Alzheimer’s disease: the potential role of Beclin 1 interactome. Prog. Neurobiol. 106-107, 33–54. doi: 10.1016/j.pneurobio.2013.06.002
Salvador, N., Aguado, C., Horst, M., and Knecht, E. (2000). Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J. Biol. Chem. 275, 27447–27456. doi: 10.1074/jbc.M001394200
Sarkis, G. J., Ashcom, J. D., Hawdon, J. M., and Jacobson, L. A. (1988). Decline in protease activities with age in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 45, 191–201. doi: 10.1016/0047-6374(88)90001-2
Schneider, J. L., Villarroya, J., Diaz-Carretero, A., Patel, B., Urbanska, A. M., Thi, M. M., et al. (2015). Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264. doi: 10.1111/acel.12310
Schroeder, S., Zimmermann, A., Carmona-Gutierrez, D., Eisenberg, T., Ruckenstuhl, C., Andryushkova, A., et al. (2014). Metabolites in aging and autophagy. Microb. Cell 1, 110–114. doi: 10.15698/mic2014.04.142
Schubert, U., Antón, L. C., Gibbs, J., Norbury, C. C., Yewdell, J. W., and Bennink, J. R. (2000). Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774. doi: 10.1038/35008096
Seglen, P. O., Gordon, P. B., and Holen, I. (1990). Non-selective autophagy. Semin. Cell Biol. 1, 441–448.
Selkoe, D. J. (2004). Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat. Cell Biol. 6, 1054–1061. doi: 10.1038/ncb1104-1054
Shibata, M., Lu, T., Furuya, T., Degterev, A., Mizushima, N., Yoshimori, T., et al. (2006). Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 281, 14474–14485. doi: 10.1074/jbc.M600364200
Simonsen, A., Cumming, R. C., Brech, A., Isakson, P., Schubert, D. R., and Finley, K. D. (2008). Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. doi: 10.4161/auto.5269
Spilman, P., Podlutskaya, N., Hart, M. J., Debnath, J., Gorostiza, O., Bredesen, D., et al. (2010). Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One 5:e9979. doi: 10.1371/journal.pone.0009979
Steele, J. W., Lachenmayer, M. L., Ju, S., Stock, A., Liken, J., Kim, S. H., et al. (2013). Latrepirdine improves cognition and arrests progression of neuropathology in an Alzheimer’s mouse model. Mol. Psychiatry 18, 889–897. doi: 10.1038/mp.2012.106
Stoka, V., Turk, V., and Turk, B. (2016). Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res. Rev. 32, 22–37. doi: 10.1016/j.arr.2016.04.010
Stricher, F., Macri, C., Ruff, M., and Muller, S. (2013). HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy 9, 1937–1954. doi: 10.4161/auto.26448
Sugawara, K., Suzuki, N. N., Fujioka, Y., Mizushima, N., Ohsumi, Y., and Inagaki, F. (2004). The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9, 611–618. doi: 10.1111/j.1356-9597.2004.00750.x
Sun, Q., Fan, W., Chen, K., Ding, X., Chen, S., and Zhong, Q. (2008). Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216. doi: 10.1073/pnas.0810452105
Tan, J. M., Wong, E. S., and Lim, K. L. (2009). Protein misfolding and aggregation in Parkinson’s disease. Antioxid. Redox Signal. 11, 2119–2134. doi: 10.1089/ARS.2009.2490
Taneike, M., Yamaguchi, O., Nakai, A., Hikoso, S., Takeda, T., Mizote, I., et al. (2010). Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 6, 600–606. doi: 10.4161/auto.6.5.11947
Tanida, I., Minematsu-Ikeguchi, N., Ueno, T., and Kominami, E. (2005). Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84–91. doi: 10.4161/auto.1.2.1697
Tasdemir, E., Chiara Maiuri, M., Morselli, E., Criollo, A., D’Amelio, M., Djavaheri-Mergny, M., et al. (2008). A dual role of p53 in the control of autophagy. Autophagy 4, 810–814. doi: 10.4161/auto.6486
Tashiro, K., Shishido, M., Fujimoto, K., Hirota, Y., Yo, K., Gomi, T., et al. (2014). Age-related disruption of autophagy in dermal fibroblasts modulates extracellular matrix components. Biochem. Biophys. Res. Commun. 443, 167–172. doi: 10.1016/j.bbrc.2013.11.066
Tatti, M., Motta, M., Di Bartolomeo, S., Scarpa, S., Cianfanelli, V., Cecconi, F., et al. (2012). Reduced cathepsins B and D cause impaired autophagic degradation that can be almost completely restored by overexpression of these two proteases in Sap C-deficient fibroblasts. Hum. Mol. Genet. 21, 5159–5173. doi: 10.1093/hmg/dds367
Terman, A. (1995). The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology 41(Suppl. 2), 319–326. doi: 10.1159/000213753
Triplett, J. C., Tramutola, A., Swomley, A., Kirk, J., Grimes, K., Lewis, K., et al. (2015). Age-related changes in the proteostasis network in the brain of the naked mole-rat: Implications promoting healthy longevity. Biochim. Biophys. Acta 1852, 2213–2224. doi: 10.1016/j.bbadis.2015.08.002
Ulgherait, M., Rana, A., Rera, M., Graniel, J., and Walker, D. W. (2014). AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 8, 1767–1780. doi: 10.1016/j.celrep.2014.08.006
Um, H. S., Kang, E. B., Leem, Y. H., Cho, I. H., Yang, C. H., Chae, K. R., et al. (2008). Exercise training acts as a therapeutic strategy for reduction of the pathogenic phenotypes for Alzheimer’s disease in an NSE/APPsw-transgenic model. Int. J. Mol. Med. 22, 529–539.
Unno, K., Asakura, H., Shibuya, Y., Kaiho, M., Okada, S., and Oku, N. (2000). Increase in basal level of Hsp70, consisting chiefly of constitutively expressed Hsp70 (Hsc70) in aged rat brain. J. Gerontol. A Biol. Sci. Med. Sci. 55, B329–B335. doi: 10.1093/gerona/55.7.b329
Vakifahmetoglu-Norberg, H., Kim, M., Xia, H. G., Iwanicki, M. P., Ofengeim, D., Coloff, J. L., et al. (2013). Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 27, 1718–1730. doi: 10.1101/gad.220897.113
Vakifahmetoglu-Norberg, H., Xia, H. G., and Yuan, J. (2015). Pharmacologic agents targeting autophagy. J. Clin. Invest. 125, 5–13. doi: 10.1172/JCI73937
Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A. L., Orosz, L., and Müller, F. (2003). Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426:620. doi: 10.1038/426620a
Vittorini, S., Paradiso, C., Donati, A., Cavallini, G., Masini, M., Gori, Z., et al. (1999). The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J. Gerontol. A Biol. Sci. Med. Sci. 54, B318–B323. doi: 10.1093/gerona/54.8.B318
Wang, Y., Martinez-Vicente, M., Krüger, U., Kaushik, S., Wong, E., Mandelkow, E. M., et al. (2009). Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18, 4153–4170. doi: 10.1093/hmg/ddp367
Wang, Y., Martinez-Vicente, M., Kruger, U., Kaushik, S., Wong, E., Mandelkow, E. M., et al. (2010). Synergy and antagonism of macroautophagy and chaperone-mediated autophagy in a cell model of pathological tau aggregation. Autophagy 6, 182–183. doi: 10.4161/auto.6.1.10815
Wohlgemuth, S. E., Julian, D., Akin, D. E., Fried, J., Toscano, K., Leeuwenburgh, C., et al. (2007). Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 10, 281–292. doi: 10.1089/rej.2006.0535
Woo, S. U., Sangai, T., Akcakanat, A., Chen, H., Wei, C., and Meric-Bernstam, F. (2017). Vertical inhibition of the PI3K/Akt/mTOR pathway is synergistic in breast cancer. Oncogenesis 6:e385. doi: 10.1038/oncsis.2017.86
Wu, H., Chen, S., Ammar, A. B., Xu, J., Wu, Q., Pan, K., et al. (2015). Crosstalk between macroautophagy and chaperone-mediated autophagy: implications for the treatment of neurological diseases. Mol. Neurobiol. 52, 1284–1296. doi: 10.1007/s12035-014-8933-0
Xie, Z., and Klionsky, D. J. (2007). Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109. doi: 10.1038/ncb1007-1102
Xilouri, M., and Stefanis, L. (2016). Chaperone mediated autophagy in aging: starve to prosper. Ageing Res. Rev. 32, 13–21. doi: 10.1016/j.arr.2016.07.001
Yang, D. S., Stavrides, P., Mohan, P. S., Kaushik, S., Kumar, A., Ohno, M., et al. (2011). Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain 134, 258–277. doi: 10.1093/brain/awq341
Yang, Z., and Klionsky, D. J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124–131. doi: 10.1016/j.ceb.2009.11.014
Ye, W., Xu, K., Huang, D., Liang, A., Peng, Y., Zhu, W., et al. (2011). Age-related increases of macroautophagy and chaperone-mediated autophagy in rat nucleus pulposus. Connect. Tissue Res. 52, 472–478. doi: 10.3109/03008207.2011.564336
Yu, Y., Feng, L., Li, J., Lan, X., A, L., Lv, X., et al. (2017). The alteration of autophagy and apoptosis in the hippocampus of rats with natural aging-dependent cognitive deficits. Behav. Brain Res. 334, 155–162. doi: 10.1016/j.bbr.2017.07.003
Yu, Z. Q., Ni, T., Hong, B., Wang, H. Y., Jiang, F. J., Zou, S., et al. (2012). Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8, 883–892. doi: 10.4161/auto.19652
Zhang, C., and Cuervo, A. M. (2008). Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14, 959–965. doi: 10.1038/nm.1851
Zhang, X. J., Chen, S., Huang, K. X., and Le, W. D. (2013). Why should autophagic flux be assessed? Acta Pharmacol. Sin. 34, 595–599. doi: 10.1038/aps.2012.184
Zhang, Z., Fan, J., Becker, K. G., Graff, R. D., Lee, G. M., and Francomano, C. A. (2006). Comparaison of gene expression profile between human chondrons and chondrocytes: a cDNA microarray study. Osteoarthr. Cartil. 14, 449–459. doi: 10.1016/j.joca.2005.11.008
Zheng, Q., Huang, T., Zhang, L., Zhou, Y., Luo, H., Xu, H., et al. (2016). Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front. Aging Neurosci. 8:303. doi: 10.3389/fnagi.2016.00303
Keywords: aging, autophagy, brain, chaperone-mediated autophagy, macroautophagy
Citation: Loeffler DA (2019) Influence of Normal Aging on Brain Autophagy: A Complex Scenario. Front. Aging Neurosci. 11:49. doi: 10.3389/fnagi.2019.00049
Received: 01 November 2018; Accepted: 19 February 2019;
Published: 11 March 2019.
Edited by:
George E. Barreto, Pontificia Universidad Javeriana, ColombiaReviewed by:
Merce Pallas, University of Barcelona, SpainAinhoa Plaza-Zabala, Achucarro Basque Center for Neuroscience, Spain
Jihye Paik, Weill Cornell Medicine, United States
Copyright © 2019 Loeffler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David A. Loeffler, ZGxvZWZmbGVyQGJlYXVtb250LmVkdQ==
 David A. Loeffler
David A. Loeffler