
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 02 September 2020
Sec. RNA Networks and Biology
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.555372
This article is part of the Research Topic Role of Ribonucleoprotein Complexes in Neurodevelopment and in the Physiopathology of Neurological Diseases View all 9 articles
RNA modifications termed epitranscriptomics represent an additional layer of gene regulation similar to epigenetic mechanisms operating on DNA. The dynamic nature and the increasing number of RNA modifications offer new opportunities for a rapid fine-tuning of gene expression in response to specific environmental cues. In cooperation with a diverse and versatile set of effector proteins that “recognize” them, these RNA modifications have the ability to mediate and control diverse fundamental cellular functions, such as pre-mRNA splicing, nuclear export, stability, and translation. N6-methyladenosine (m6A) is the most abundant of these RNA modifications, particularly in the nervous system, where recent studies have highlighted it as an important post-transcriptional regulator of physiological functions from development to synaptic plasticity, learning and memory. Here we review recent findings surrounding the role of m6A modification in regulating physiological responses of the mammalian nervous system and we discuss its emerging role in pathological conditions such as neuropsychiatric and neurodegenerative disorders.
Epitranscriptomics or “RNA epigenetics” refers to post-transcriptional RNA modifications that mediate several cellular processes necessary for maintaining RNA homeostasis. To date approximately 170 distinct modifications have been identified in coding and non-coding RNA (Boccaletto et al., 2018), with the most prominent and best characterized being N6-methyladenosine (m6A) methylation. m6A, the deposition of a methyl group on the adenosine base at the nitrogen-6 position (Bokar et al., 1997), was first discovered in 1974 (Desrosiers et al., 1974; Perry et al., 1975). However, it has only recently garnered increased attention due to advances in transcriptome-wide mapping methods that provide a greater resolution of its distribution on m6A-modified transcripts (Dominissini et al., 2012; Meyer et al., 2012; Linder et al., 2015) and the identification of enzymes that could reverse this modification (Jia et al., 2011; Zheng et al., 2013).
m6A is the most abundant internal RNA modification in mammalian mRNA with approximately 25% of mammalian mRNAs being modified, with an average of 1–3 modifications per transcript. It is preferentially installed near stop codons and 5′ and 3′ UTRs, as well as in introns and long internal exons but to a lesser extent, with a high degree of conservation between human and mouse (Dominissini et al., 2012; Meyer et al., 2012). m6A is installed by a methyltransferase complex termed the writer complex and removed by demethylases termed erasers. Effector proteins termed readers recognize m6A and mediate multifaceted functions of mRNA metabolism including splicing, nuclear export, stability, translation, microRNA processing, and subcellular targeting of m6A-modified mRNAs (Meyer and Jaffrey, 2017; Shi et al., 2019; Zaccara et al., 2019). To date a plethora of studies have implicated m6A in diverse biological functions including stem cell proliferation and differentiation (Heck and Wilusz, 2019), circadian rhythm (Hastings, 2013), spermatogenesis (Lin and Tong, 2019), hematopoiesis (Weng et al., 2019), immunoregulation (Zhang et al., 2019), metabolism (Zhong et al., 2020), and cancer (Huang et al., 2020).
Proper development of the nervous system is a highly coordinated, multi-step process that requires tight regulation to ensure proper function. As a consequence, deficits in neurodevelopment have been implicated in several brain disorders (Kiser et al., 2015; Meredith, 2015; Cardoso et al., 2019). Interestingly, m6A is highly enriched in the mammalian brain and is developmentally regulated with its expression peaking during adulthood (Meyer et al., 2012; Nainar et al., 2016; Chang et al., 2017). Several studies have highlighted the functional significance of m6A regulation in the physiology of the nervous system both during early development and in adulthood (Widagdo and Anggono, 2018; Li et al., 2019; Livneh et al., 2020). However, the role of m6A in maladapted responses of the nervous system is still poorly understood and has been the focus of recent studies that implicate m6A in neurodevelopmental, neuropsychiatric, and neurodegenerative disorders (Engel and Chen, 2018; Shafik et al., 2020). In this review, we will summarize the current body of knowledge surrounding the m6A effectors, the regulatory mechanisms that they govern, as well as their biological consequences in both physiology and pathology of the mammalian nervous system.
m6A is installed co-transcriptionally in nascent transcripts by the writer complex. The core elements of this complex consist of methyltransferase-like 3 (METTL3), the catalytic component, and methyltransferase-like 14 (METTL14), which activates METTL3’s enzymatic activity and facilitates RNA substrate binding (Figure 1) (Schwartz et al., 2014; Wang X. et al., 2014; Liu K. et al., 2016; Sledz and Jinek, 2016; Wang et al., 2016). Additional regulatory subunits contribute to the activity and specificity of the writer complex. Wilms tumor 1-associating protein (WTAP) is essential for optimal substrate recruitment and METTL3/14 localization to the nuclear speckles (Zhong et al., 2008; Ping et al., 2014; Schwartz et al., 2014). KIAA1429 or vir-like m6A methyltransferase associated (VIRMA) mediates preferential m6A-deposition in the 3’ UTR and near stop codons (Yue et al., 2018). Cbl-proto-oncogene-like protein 1 (CBLL1, also known as HAKAI) is required for m6A deposition on mRNA-targets (Ruzicka et al., 2017). Zinc finger CCCH-type containing 13 (ZC3H13) is required for nuclear localization of the writer complex (Wen et al., 2018). RNA binding motif protein 15/15B (RBM15/15B) directs m6A-deposition in specific sites of mRNA substrates (Patil et al., 2016).
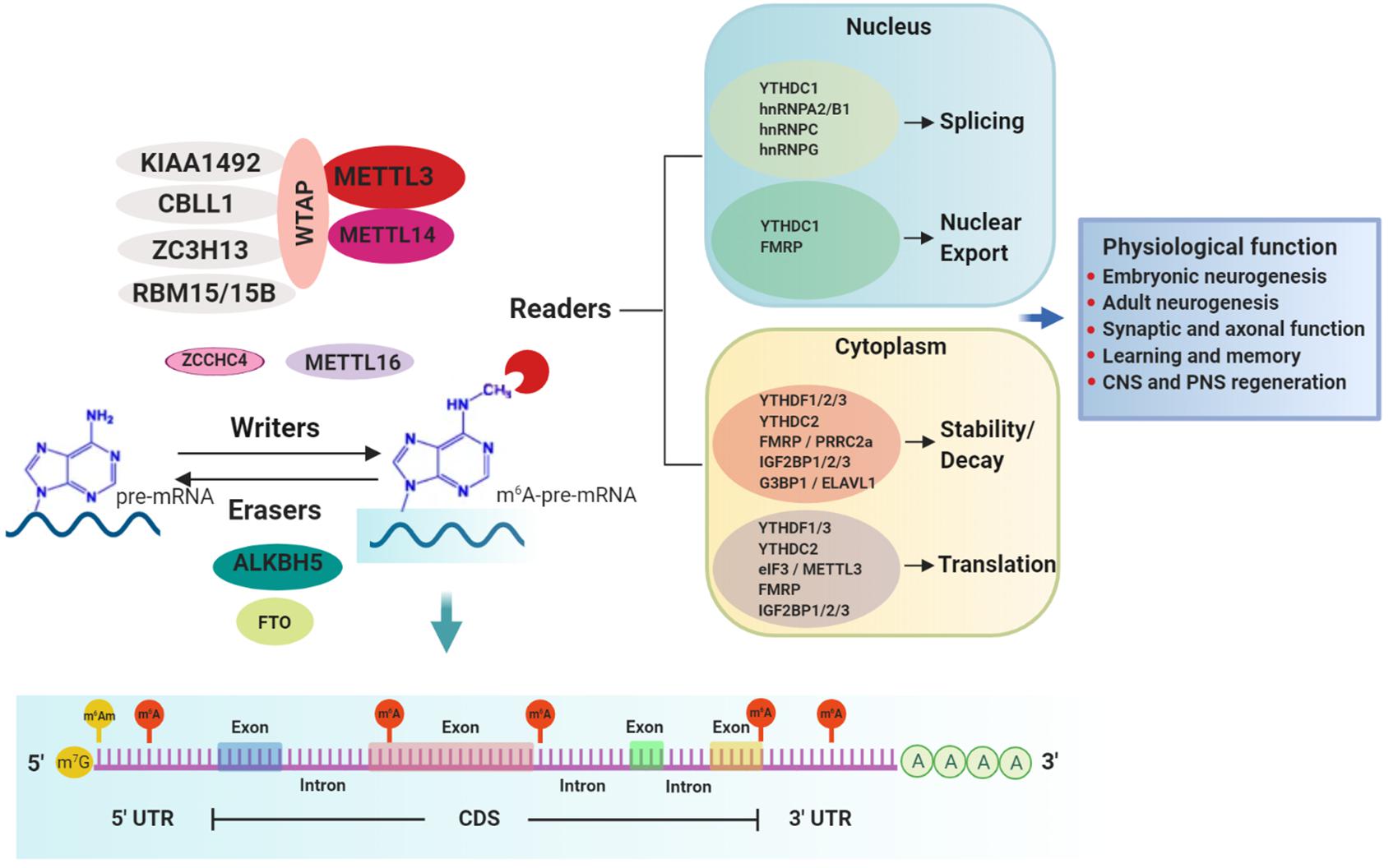
Figure 1. m6A-mediated mRNA regulation and physiological responses in the nervous system. m6A methylation in mRNA substrates is performed by writers. The best-characterized writer is a multi-subunit complex consisting of the core enzymes METTL3 (catalytic enzyme) and METTL14 and other regulatory proteins such as WTAP, KIAA1492, ZC3H13, RBM15/15B, and CBLL1. Other less characterized writer proteins are METTL16 and the recently discovered ZCCHC4. Two erasers, FTO and ALKBH5, mediate the reverse process of demethylation. m6A is preferentially installed near stop codons and 5′ and 3′ UTRs, as well as in introns (near 5′ and 3′ splice sites) and long internal exons but to a lesser extent. Conversely, m6Am, a modification regulated also by FTO, occurs only adjacent to the m7G (N7-methylguanosine) cap. m6A-modified mRNAs are recognized by a diverse group of RNA binding proteins called readers which regulate several aspects of the mRNA metabolism including splicing, nuclear export, decay, stability and translation, through which different physiological processes in the nervous system are accomplished.
While METTL3/14 writer complex typically installs m6A in a sequence motif of DRACH (D = A, G or U; R = A or G; H = A,C or U) (Liu et al., 2014), another m6A writer, methyltransferase-like 16 (METTL16), places m6A in a sequence context of UAC(m6A)GAGAA on top of a hairpin RNA structure in selected mRNA targets, including MAT2A (Pendleton et al., 2017; Doxtader et al., 2018; Mendel et al., 2018). More recently, a new m6A writer, ZCCHC4, was identified to methylate primarily the 28S rRNA and also to interact with a subset of mRNAs (Ma et al., 2019). The proteins comprising the m6A-writer complex are mostly nuclear (Jia et al., 2011; Ping et al., 2014). However, several studies have reported these proteins can also be located in the cytoplasm where they mediate m6A deposition in mature mRNAs as well as being involved in methylation-independent functions (Alarcon et al., 2015b; Chen et al., 2015; Lin et al., 2016).
m6A can be reversed by erasers, of which only two have been identified to date, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) (Figure 1). FTO, the first RNA demethylase identified, was reported to remove the methyl group of m6A in mammalian mRNA (Jia et al., 2011; Fu et al., 2013). However, further characterization of substrate spectrum of FTO showed that its demethylase activity is not only selective toward internal m6A in mRNA but also toward m1A in specific tRNAs, cap-m6Am in mRNA as well as m6Am in some snRNAs (Wei et al., 2018). Therefore, it is important to consider that FTO’s contribution to physiological and disease phenotypes that are discussed below should not be interpreted solely in conjunction with its function as m6A eraser. Conversely, ALKBH5 preferentially demethylates m6A in a consensus DRACH motif-dependent manner in specific transcripts and has no other known substrates (Zheng et al., 2013; Xu et al., 2014; Zhang et al., 2016, 2017; Tang et al., 2018). The expression patterns of ALKBH5 and FTO are distinct among tissues. ALKBH5 is highly expressed in testis and is essential for spermatogenesis (Zheng et al., 2013), whereas FTO, despite being ubiquitously expressed, is enriched in the brain, specifically in neurons (Gerken et al., 2007; McTaggart et al., 2011; Aas et al., 2017), implicating a key regulatory role in the nervous system.
m6A-dependent post-transcriptional gene regulation is mediated by RNA binding proteins (RBPs), termed readers, that recognize and decode the m6A epimark (Figure 1 and Table 1). Characterization of m6A readers has revealed the presence of different classes of RBPs, divergent binding modes and multifaceted downstream biological functions (Dominissini et al., 2012; Edupuganti et al., 2017). Direct RNA binding relies on a dedicated domain within the reader protein that recognizes the modification within a sequence motif. For example, proteins containing the YT521-B homology (YTH) domain, including cytoplasmic YTH domain family 1–3 (YTHDF1–3) and nuclear YTH domain containing 1–2 (YTHDC1–2) in mammals directly bind methylated transcripts to exert their effects on gene expression (Zhang et al., 2010). A different group of readers utilizes common RNA binding domains to bind m6A-containing RNAs, conferring greater access to RBPs through methylation-induced remodeling of local RNA structures (Liu et al., 2015). Several proteins fall into this category, including the heterogeneous nuclear ribonucleoproteins (hnRNPs) (Alarcon et al., 2015a; Liu et al., 2015; Liu N. et al., 2017; Wu et al., 2018), the fragile X mental retardation protein (FMRP) (Edupuganti et al., 2017; Zhang Z. et al., 2018), the insulin-like growth factor 2 mRNA-binding proteins 1–3 (IGF2BP1-3) (Huang et al., 2020) and the proline rich coiled-coil 2A (PRRC2A) protein (Wu et al., 2019). Not only does m6A attract RBPs, but recent studies have also identified RBPs that are instead repelled by the presence of m6A, such as Ras GTPase-activating protein-binding protein 1 (G3BP1) and G3BP2 proteins (Arguello et al., 2017; Edupuganti et al., 2017), adding another layer of complexity to m6A-dependent gene expression regulation.
The fact that m6A is present within exons (Dominissini et al., 2012) and further associated with pre-mRNA splicing factors (SRSFs) (Zhao et al., 2014; Xiao et al., 2016) strengthened the view that it may regulate splicing. YTHDC1 was shown to recruit SRSF3 to promote exon inclusion in an m6A-dependent manner, while blocking SRSF10, which favors exon skipping (Xiao et al., 2016). Several hnRNPs, which are known to control different aspects of mRNA metabolism, such as hnRNPC, hnRNPG, and hnRNPA2-B1 have also been described to interact with m6A-modified mRNAs through multiple proteomic screenings (Edupuganti et al., 2017; Liu N. et al., 2017). A study by Liu et al. (2015) described the presence of an “m6A-switch” mechanism where m6A alters the local structure in mRNA and long non-coding RNA to facilitate the binding of hnRNPC and promote alternative splicing of target mRNAs. hnRNPA2-B1 was also shown to bind the m6A-consensus motif in vitro and in vivo and was suggested to mediate, in part, the effects of m6A/METTL3 alternative splicing as well as microRNA processing (Alarcon et al., 2015a).
Apart from its m6A-mediated splicing role, YTHDC1 can interact with the SRSF3 to facilitate the nuclear RNA export factor 1 (NXF1)-mediated nuclear export of m6A-modified mRNAs (Roundtree et al., 2017). Toward the same direction, FMRP, an m6A reader with a yet undetermined binding mode, can also facilitate the nuclear export of m6A-modified mRNAs through the nuclear export protein CRM1 during neural differentiation (Edens et al., 2019).
A possible link between m6A and mRNA stability was first suggested in the late 1970’s (Sommer et al., 1978), a finding that was further confirmed by later studies in which proteins of the writer complex (METTL3 and WTAP) were disrupted in both human and mouse cells (Batista et al., 2014; Liu et al., 2014; Schwartz et al., 2014). Thus far, the best characterized decay-inducing reader is YTHDF2. YTHDF2 can directly mediate mRNA decay either by binding to m6A-containing mRNAs and targeting them to P-bodies (Wang X. et al., 2014; Ries et al., 2019), or by recruiting the CCR4-NOT deadenylase complex to promote m6A-mRNA degradation independently from P-bodies (Du et al., 2016). YTHDC2, which can be both nuclear and cytoplasmic, has a dual role during early spermatogenesis, where it both enhances the translation efficiency of its targets while decreasing their mRNA abundance (Hsu et al., 2017). An additional route for YTHDF2-dependent mRNA decay has been recently reported, where m6A-containing RNAs are subjected to endoribonucleolytic cleavage via HRSP12 (adaptor protein) and RNase P/MRP (endoribonucleases) (Park et al., 2019). YTHDF2 was also suggested to act synergistically with YTHDF3, which might control the mRNA binding specificity, to promote m6A-mediated mRNA decay (Shi et al., 2017). However, the view that YTHDF proteins bind to specific m6A sites on selected mRNAs has been challenged by a new study proposing a different model to explain how YTHDF proteins influence the fate of m6A-modified mRNAs (Zaccara and Jaffrey, 2020). According to the authors, all three YTHDF proteins bind to all m6A site in a similar manner and they work in concert to promote mRNA degradation (Zaccara and Jaffrey, 2020).
While YTHDF proteins have been proposed to promote mRNA degradation, several indirect readers have been reported to enhance mRNA stability, such as FMRP, which likely competes with YTHDF2 for binding to m6A-containing mRNAs and the stress granule component G3BP1, which is repelled by m6A (Edupuganti et al., 2017; Zhang F. et al., 2018). IGF2BP1-3 were also demonstrated to stabilize m6A-containing mRNAs and enhance translation (Huang et al., 2018). In addition, a recent report added PRRC2A to the list of m6A readers (Wu et al., 2019), showing that recombinant PRRC2A prefers to bind a methylated probe and further stabilizing a critical m6A modified transcript required for myelination. Lastly, the stabilizing/destabilizing effect of m6A may also be promoted by human antigen R (HuR; AKA ELAV-like protein 1, ELAVL1), depending on the position of m6A and HuR-binding site in target mRNAs (Wang Y. et al., 2014).
Methylation at either the 5′ UTR or 3′ UTR near the stop codon of mRNAs has been reported to promote translation in mammalian cell systems. Two readers of the cytoplasmic YTH-domain family, YTHDF1 and YTHDF3, were reported to mediate cap-dependent translation, presumably via 3′ UTR methylation of target transcripts in mammalian cells, by recruiting translation initiation factors (Wang et al., 2015; Li et al., 2017; Shi et al., 2018). Although both YTHDF1 and YTHDF3 can functionally associate with ribosomes, only YTHDF1 can directly interact with the translation initiation factor complex 3 (eIF3) (Li et al., 2017; Shi et al., 2018). However, a recent study challenged the prevailing model that YTHDF1 and YTHDF3 function as mRNA translation enhancers by proposing that all three YTHDF proteins act redundantly to promote m6A-mediated mRNA degradation (Zaccara and Jaffrey, 2020).
Irrespective of the controversial role of YTHDF1 and YTHDF3 on mRNA translation, m6A at the 5′ UTR of mRNAs can directly interact with the eIF3 and promote cap-independent translation, especially under stress-response conditions where the cap-dependent translation is repressed (Meyer et al., 2015; Zhou et al., 2015). Interestingly, METTL3 itself has also been suggested to act in the cytoplasm as an m6A reader and promote translation cooperatively with eIF3 in human cancer cells (Lin et al., 2016). Translational control by m6A has also been suggested via its interaction with the FMRP protein in HeLa cells (Arguello et al., 2017; Edupuganti et al., 2017). Further supporting m6A’s role in regulating translation, a recent study introduced a mechanism where m6A methylation in coding sequences promotes translational elongation by reducing ribosomal pausing via recruitment of the YTHDC2 reader (Mao et al., 2019).
RNA modifications, and m6A in particular, have been shown to influence the spatial distribution of their target mRNAs to exert their subcellular-specific functions. There is currently limited knowledge regarding how this process is being mediated through m6A and which proteins are involved in the guidance of the m6A-modified mRNAs in their final subcellular destination. The first evidence of m6A being involved in the spatial regulation of its mRNA targets came from a study by Zheng et al. (2013) where the demethylation activity of ALKBH5 significantly altered mRNA nuclear export. In this study ALKBH5-deficient cells showed increased cytoplasmic mRNA levels due to accelerated export in contrast to control cells where mRNAs were mainly accumulated in the nucleus, suggesting that m6A might be required for mRNA export.
Studies have linked cytoplasmic m6A with specific structures such as P-bodies and stress granules (SGs) (Gao et al., 2019; Ries et al., 2019; Fu and Zhuang, 2020) which constitute assemblies of mRNAs and RBPs involved in mRNA turnover or translation (Decker and Parker, 2012). m6A-modified mRNAs can interact with P-bodies via the YTHDF2 reader to promote their decay (Wang X. et al., 2014; Ries et al., 2019). Notably, Ries et al. (2019) demonstrated that YTHDF proteins can undergo liquid–liquid phase separation in vitro and in cells, and that this process is markedly enhanced by the presence of poly-methylated m6A-mRNAs rather than single m6A-mRNAs. These m6A-mRNAs/YTHDF complexes can then partition into different endogenous phase-separated compartments, such as P-bodies, SGs or neuronal RNA granules (Ries et al., 2019). Along the same line, another study showed that poly-methylated m6A-mRNA rather than single m6A motifs can dramatically enhance the phase separation of YTHDF proteins (Gao et al., 2019). On the contrary, G3BP1 and G3BP2, two proteins that contribute to the formation of stress granules that have been suggested to act as m6A-readers, have been shown to be repelled by the presence of m6A modification in mRNA substrates (Arguello et al., 2017; Edupuganti et al., 2017). These studies underscore m6A’s complex role in regulating the fate of cytosolic mRNAs depending on the state of a cell (normal versus stress-induced conditions), the number of m6A-modified sites (poly-methylated versus mono-methylated), and the nature of the RBPs that interact with these modified mRNAs.
Further insights on m6A’s role in spatial regulation in neurons derived from studies showing that synapses are preferentially enriched both in m6A-modified transcripts and m6A regulatory proteins such as FTO, YTHDF1-3 family, and FMRP. Importantly, the presence of these m6A-transcripts as well as the m6A-regulatory proteins in the synapses was essential for their maintenance and function (Chang et al., 2017; Merkurjev et al., 2018), indicating that the m6A-effector proteins might regulate not only the fate of m6A-modified targets but also their subcellular distribution.
The widespread distribution of m6A modification throughout the mammalian brain in conjunction with functional studies targeting the m6A effectors in neural cells highlight m6A as an important post-transcriptional regulator of the nervous system (Meyer et al., 2012; Hess et al., 2013; Schwartz et al., 2014; Nainar et al., 2016; Widagdo et al., 2016; Aas et al., 2017; Chang et al., 2017). To date, in the mammalian nervous system, m6A regulation has been demonstrated in several physiological functions, including neurodevelopment, learning and memory, synaptic function, as well as injury (Figure 1).
Accumulating evidence emphasizes m6A’s central role in shaping the transcriptome of both neuronal progenitor cells and adult stem cells during neurogenesis in the mammalian brain. In rodents, deletions of writers (Mettl3, Mettl14), erasers (Fto) and readers (Ythdf2, Fmr1) have profound developmental effects (Fischer et al., 2009; Gao et al., 2010; Yoon et al., 2017; Li et al., 2018). Deletion of Mettl3 in mouse and human embryonic stem cells leads to a significant decrease in m6A levels and severely impairs stem cells transition from self-renewal to differentiation (Batista et al., 2014; Wang Y. et al., 2014; Chen et al., 2015; Geula et al., 2015; Chen J. et al., 2019). Notably, germline deletion of Mettl3 in mice causes early embryonic lethality (Geula et al., 2015). Conditional knockout of Mettl14 in the developing mouse nervous system prolongs the cell cycle of radial glia cells and extends cortical neurogenesis into postnatal stages (Yoon et al., 2017). Remarkably, m6A-tagged transcripts derived from early embryonic mice forebrains were enriched in genes related to cell cycle and neurogenesis, whereas inhibition of m6A attenuated the decay of these transcripts (Yoon et al., 2017). Deletion of Ythdf2 in mice caused lethality at late embryonic developmental stages, with embryos characterized by delayed cortical neurogenesis (Li et al., 2018). This phenotype is attributed to hindered degradation of m6A-tagged genes associated with neural development and differentiation in Ythdf2 knockout mice, reminiscent of the Mettl14 knockout defects in cortical neurogenesis (Yoon et al., 2017). Similarly, ablation of FMRP leads to neural differentiation defects due to m6A-dependent mRNA nuclear export impairment in Fmr1 knockout mice (Edens et al., 2019).
Aside from its role in controlling embryonic neurogenesis in the mouse brain, m6A was also proposed to affect adult neurogenesis in Fto knock-out mice (Li et al., 2018). In particular, loss of FTO was accompanied by a reduced proliferation and differentiation of adult neural stem cells (aNSCs) in the subventricular zone of the dentate gyrus in the hippocampus and this reduction was concomitant with impairments in learning and memory (Li et al., 2018). A more recent study provided further support on the role of m6A in adult neurogenesis by demonstrating that depletion of Mettl3 significantly reduced m6A levels in aNSCs, inhibited their proliferation, and affected the morphological maturation of newborn neurons in the adult brain (Chen X. et al., 2019). Importantly, using methylated RNA immunoprecipitation sequencing (Me-RIP-seq) this study uncovered the presence of a crosstalk between m6A and histone modifications in aNSCs. Specifically, Me-RIP-seq revealed that m6A was present on the transcripts of the histone methyltransferase Ezh2 (Chen J. et al., 2019). Reduction of m6A following Mettl3 depletion decreased both EZH2 protein expression and the levels of its target H3K27me3, leading to defects in neuronal development, which could be rescued by Ezh2 overexpression (Chen J. et al., 2019). In support of this crosstalk between m6A and histone modifications, a study by Wang Y. et al. (2018) reported that m6A can regulate histone modifications directly by destabilizing transcripts encoding histone modifiers, such as the H3K27 acetyltransferase CBP/p300. Therefore, these studies proposed a novel mechanism for gene regulation where m6A and epigenetic mechanisms work in unison to safeguard normal embryonic and adult mouse brain neurogenesis.
Several studies in neurons have reported that synapses are enriched not only of m6A-modified mRNAs but also of m6A-effectors (Merkurjev et al., 2018; Yu et al., 2018; Zhuang et al., 2019). Transcriptome-wide m6A profiling in the mouse cerebellum and cerebral cortex revealed that m6A signatures/peaks were enriched in functions pertaining to transcriptional regulation, axon guidance, synapse assembly and organization (Chang et al., 2017). Notably, m6A-sequencing of synaptosomal RNA from healthy adult mouse forebrains revealed that the synaptic m6A epitranscriptome is functionally enriched in genes involved in the synthesis and modulation of the “tripartite” synapses, a term referring to presynaptic terminals, postsynaptic terminals, and glia cells (Perea et al., 2009). In the same study, m6A-regulatory proteins such as YTHDF1-3, and FTO were localized in extrasomatic regions and in dendrites in adult mouse brain slices and in hippocampal neuronal cultures, respectively. Selective knockdown of readers Ythdf1 and Ythdf3 in hippocampal neurons resulted in synaptic dysfunction including immature spine morphology and dampened excitatory synaptic transmission (Merkurjev et al., 2018), suggesting a functional role for m6A readers in these synapses.
FTO was shown to be enriched in the axons and most importantly to be locally translated (Yu et al., 2018). Axon-specific inhibition of FTO led to generally increased m6A levels and, specifically, m6A levels within axonal growth-associated protein-43 (GAP-43) mRNA, whose expression is required for axon elongation (Skene and Willard, 1981). This increase in m6A-GAP-43 mRNA suppressed translation of axonal GAP-43 mRNA, which repressed axon elongation in mouse dorsal root ganglia neurons (Yu et al., 2018). In addition to its synaptic function, YTHDF1 was recently reported to regulate axonal guidance in spinal cord through translational control of the axon guidance receptor Robo3.1 (Zhuang et al., 2019). Likewise, other RBPs, such as FMRP (Edupuganti et al., 2017), can act as sequence-context-dependent m6A readers and form RNA transport granules, which regulate dendritic localization of RNAs as well as inhibit local synaptic transcript translation (Dictenberg et al., 2008; Kao et al., 2010), highlighting m6A’s diverse role in promoting regulation of synaptic function and growth.
Several studies focusing on m6A’s role in the nervous system, following perturbations of its machinery, have underlined its involvement in the process of learning and memory. Evidence derived from early genome-wide studies in humans indicates the importance of FTO in memory processing (Ho et al., 2010; Benedict et al., 2011). In line with this observation, Fto downregulation in specific brain regions can lead to changes in the learning capacity of mice (Widagdo et al., 2016; Walters et al., 2017). In particular, m6A levels were shown to be dynamically regulated in the mouse medial prefrontal cortex following post-behavioral training, in fear-conditioned mice, with Fto knockdown increasing m6A levels and resulting in enhanced consolidation of cued fear memory (Widagdo et al., 2016). Similarly, downregulation of Fto in the dorsal hippocampus enhanced contextual fear memory (Walters et al., 2017). Studies on METTL3 have highlighted its direct role in regulating the efficacy of hippocampus-dependent memory consolidation, likely by promoting the translation of neuronal early-responsive genes (Zhang Z. et al., 2018). Interestingly, conditional deletion of Mettl3 or Fto in adult excitatory neurons in mice can increase fear memory and induce transcriptomic alterations in several genes involved in neuronal circuit function, suggesting m6A contributes to neuronal circuit regulation (Engel et al., 2018). Deletion of another core writer subunit, Mettl14, in the striatum lead to increased neuronal excitability and severely impairs striatal-mediated behaviors (Koranda et al., 2018), highlighting m6A’s important role in normal striatal function in adult mice.
m6A readers have also been linked to learning and memory related processes (Shi et al., 2019; Wu et al., 2019). YTHDF1-dependent m6A protein synthesis of target transcripts, in response to neuronal stimuli, facilitates learning and memory in the adult mouse hippocampus. Mice with genetic deletion of Ythdf1 exhibit learning and memory defects as well as impaired hippocampal synaptic transmission and long-term potentiation (Shi et al., 2019). Lastly, neural or oligodendroglia-specific knockout of Prrc2a, a novel m6A reader that controls the specification and myelination of oligodendrocytes, was also reported to be associated with locomotive and cognitive defects in a mouse model (Wu et al., 2019).
Injuries in the adult peripheral nervous system can induce regeneration processes, which are accompanied by de novo gene transcription and protein translation of regeneration-associated genes (RAGs). Using a sciatic nerve injury model in adult mice, Weng et al. (2018) reported upregulation of m6A-tagged transcripts, encoding many RAGs, to occur up to 3 days post-injury in the dorsal root ganglion neurons (DRGs). Deletion of either Mettl14 or Ythdf1 attenuated injury-induced protein translation in DRGs, which consequently hindered the regeneration process in the mouse peripheral nervous system (Weng et al., 2018). Similarly, in the mouse adult central nervous system, regeneration of retinal ganglion cells post-injury was also shown to implicate m6A-dependent function (Weng et al., 2018). Recently, m6A profiling in the rat cortex following traumatic brain injury (TBI) showed that Mettl14 and Fto were significantly reduced and functional FTO was necessary to repair the neurological damage caused by TBI (Yu et al., 2020). These early studies suggest that m6A might exert a broader role in response to pathological stimuli in the adult mammalian nervous system.
Considering the widespread effects of m6A modification on gene expression, it is not surprising that aberrant methylation could underlie the disruption of RNA processing events that are increasingly implicated in neurological diseases (Nussbacher et al., 2019). Initial indication that m6A modification may be involved in the etiology of neurological diseases comes from genome-wide association studies in which several variants in genes encoding for components of the m6A machinery were associated with the risk of developing neurological disorders, such as intellectual disability, depression, schizophrenia, and Alzheimer’s disease (Engel and Chen, 2018). In this section, we will review the link between altered m6A deposition on mRNA and susceptibility to neurological diseases, broadly classified as neurodevelopmental, mood, and neurodegenerative disorders.
Neurodevelopmental disorders (NDDs) are a group of disabilities which typically manifest early in development. They are characterized by brain dysfunction which can lead to neuropsychiatric problems or impairments in cognition, language, behavior, memory, motor skills or other neurological functions (Reiss, 2009). While NDD symptoms and behaviors often evolve as a child grows older, some disabilities such as attention deficit hyperactivity disorder (ADHD) and autism spectrum disorder can be lifelong conditions (Thapar et al., 2017). Treatment of these disorders can be complex and it often involves a combination of behavioral therapy and medications.
Initial indication that altered RNA methylation could be linked to the etiology of NDDs stems from the observation that a loss-of-function mutation in the FTO gene was linked with structural and functional brain abnormalities, as well as severe motor delay (Boissel et al., 2009). ADHD is the most common NDD observed in childhood with symptoms that include inattention, hyperactivity, and impulsivity (Doernberg and Hollander, 2016). Interestingly, a duplication of the FTO gene results in intellectual disability and ADHD (van den Berg et al., 2010). Velders et al. (2012) provided the first indication that the FTO allele at rs9939609 is linked with decreased risk for symptoms of ADHD and fewer challenges with emotional control in young children. Choudhry et al. (2013) corroborated these observations implicating FTO in the modulation of ADHD phenotype with a more obvious effect in those children who were not exposed to smoke during gestation.
Notably, m6A is enriched at synapses (Shafik et al., 2020) and aberrant translation at synapses has long been associated with autism and other intellectual disorders such as the Fragile X syndrome (FXR). FXR is the most common form of inherited intellectual disability caused by the expansion of trinucleotide repeats in the 5′ UTR of the FMR1 gene, resulting in reduced levels of the encoded protein FMRP (Santoro et al., 2012). Suggestive of a role for m6A in these forms of intellectual disabilities, mutations in the YTHDC2 gene have been implicated as a risk factor for autism spectrum disorder (Liu X. et al., 2016). In addition, in line with a likely association between m6A regulation and intellectual disability, a study found that m6A is present in many mRNAs that are known synaptic targets of FMRP (Chang et al., 2017). Moreover, FMRP has been shown to stimulate nuclear export during neural differentiation by binding to m6A-modified sites on several mRNA (Edens et al., 2019; Hsu et al., 2019).
Interestingly, Yoon et al. (2017) also reported that several transcripts associated with schizophrenia−a mental disorder with roots in early brain development and characterized by disconnection with the reality (Stepnicki et al., 2018)−are m6A-modified in human, but not in mouse cultures of neuronal progenitor cells, suggesting a specific role for m6A in this disease (Yoon et al., 2017). In line with this observation, polymorphisms in the ZC3H13 gene have also been linked with schizophrenia (Oldmeadow et al., 2014).
Mood disorders are a group of emotional disturbances characterized by prolonged periods of excessive happiness, excessive sadness, or both. Mood disorders can cause behavioral changes that can impact pragmatic and executive functioning (Sekhon and Gupta, 2020). Two of the most common mood disorders are major depression disorder (MDD) and bipolar disorder. Psychotherapy, antidepressants, and support can help treat mood disorders. In particular, there is evidence of an impairment of glucocorticoid receptor-mediated negative feedback on the hypothalamic-pituitary-adrenal (HPA) axis resulting in a glucocorticoid resistance in MDD (Holsboer, 2000). Importantly, successful treatment with antidepressant can lead to resolution of glucocorticoid resistance (Heuser et al., 1996).
A survey of Chinese Han people has shown an association between specific ALKBH5 polymorphisms and MDD, raising the possibility that ALKBH5 is involved in conferring a higher risk of this mood disorder (Du et al., 2016). In addition, altered levels of m6A were observed in depressed patients after glucocorticoid stimulation (Engel et al., 2018; Spychala and Ruther, 2019). Specifically, Spychala and Ruther (2019) reported that Fto loss-of-function results in increased stress parameters such as corticosterone in the blood, suggesting hypersensitivity of the HPA axis. A processing defect of brain-derived neurotrophic factor (BDNF)−a neurotrophic factor involved in the pathophysiology of MDD (Lee and Kim, 2010)−was found in the hippocampus of these Fto knockout mice, which presented with deficits in working memory and increased anxiety (Spychala and Ruther, 2019). Engel et al. (2018) further elucidated the role of m6A in the context of stress response in adult mice. By examining mice with Mettl3 and Fto depletion in excitatory neurons they found alterations on m6A profiles with concomitant changes in transcriptome regulation, behavior, and electrophysiological properties (Engel et al., 2018). Importantly, they also reported that m6A regulation in blood could be used as a peripheral proxy of the brain’s m6A responses that are impaired in MDD patients (Engel et al., 2018).
Neurodegenerative disorders are a heterogeneous group of illnesses that are characterized by the progressive demise of neurons in different regions of the nervous system. As neurons deteriorate, neurological signs and symptoms characteristic of each of these disorders arise and progressively worsen. In some cases, patients may lose the ability to walk independently, think clearly, or perform their daily functions. Ultimately, many of these diseases are fatal (Dugger and Dickson, 2017). Neurodegenerative disorders affect millions of people worldwide, with Alzheimer’s disease (AD) and Parkinson’s disease (PD) being the most common. Although treatments may help relieve some of the physical or neurological symptoms associated with neurodegenerative disorders, currently there is no cure.
In older adults, AD is the most common form of dementia that is caused by damage in several areas of the brain, which include the hippocampus and the frontal lobe. As a result, patients have difficulties to form new memories and eventually have problems with intellect, judgment, and behavior (Weller and Budson, 2018). Several lines of evidence link genetic variations in the FTO gene with a greater risk of Alzheimer’s disease (AD) and related dementias. First, in a large magnetic resonance imaging (MRI) study of healthy elderly subjects, Ho et al. (2010) reported substantial brain tissue deficits in carriers of the FTO rs9939609 risk allele. Subjects carrying one copy or more of the FTO rs9939609 allele presented with brain atrophy in the frontal lobe, an area important for language and experience-dependent functions of the brain (Ho et al., 2010). Second, studying a cohort of elderly subjects who had no clinically overt cognitive dysfunction, Benedict et al. (2011) found that only obese and overweight but not normal weight FTO risk allele carriers showed reduced language performance relative to non-carriers. However, general cognitive performance remained unaffected in the obese and overweight subjects, suggesting that the FTO gene influences mainly frontal lobe-dependent brain functions (Benedict et al., 2011). Third, in a longitudinal cohort study, Keller et al. (2011) found an association between the FTO risk allele and the risk of developing AD and dementia. These findings were confirmed and extended in another study that reported a link between certain polymorphisms in intron 1, exon 2, or intron 2 of the FTO gene and AD in Caucasians of European ancestry as well as in Caribbean Hispanics (Reitz et al., 2012). In addition, in two independent datasets they found significantly lower levels of FTO in AD cases compared to controls and a positive association between the polymorphism residing in intron 2 and FTO expression levels (Reitz et al., 2012). Finally, a recent study reported that the expression of Mettl3 increased in AD mice relative to control mice, while the expression of Fto decreased, which was in line with the increased methylation observed in the AD group (Han et al., 2020). Genes related to synaptic function were the most abundant group of transcripts among those with changes in m6A RNA methylation levels, providing further support for a role of altered m6A regulation in the etiology of AD and related dementias (Han et al., 2020).
Parkinson’s Disease is a progressive disorder that affects predominately dopamine-producing neurons in the area of the brain which controls movement, called the substantia nigra. Normally, dopamine functions in a delicate balance with other neurotransmitters to help coordinate the neural circuit in the basal ganglia that controls movement. Without enough dopamine, this balance is altered, resulting over the years in slowness of movement, lack of spontaneous motility, resting tremor and rigidity (Przedborski, 2017). Because glutamate receptors (NMDARs) modulate synaptic transmission throughout the basal ganglia, and pharmacological manipulation of these receptors can correct aberrant neurotransmission such as that observed in the parkinsonian brain, NMDAR have been proposed as therapeutic targets for treating PD (Johnson et al., 2009). Evidence is also emerging in support of an involvement of FTO and m6A aberrant regulation with the occurrence of PD. For instance, electrophysiological analysis of both conventional and conditional Fto deficient mice showed an impaired dopamine type 2 (DR2) and 3 (DR3) receptor response characterized by a reduced cocaine-mediated suppression of firing rate (Hess et al., 2013). This view is further substantiated by the finding that selective deletion of Fto in dopaminergic neurons resulted in an increased sensitivity toward cocaine-induced locomotor activity (Hess et al., 2013). While these findings involve FTO and m6A in the control of D2R- and D3R- dependent signaling, whether humans carrying polymorphisms in the FTO gene have aberrant D2R- and D3R-dependent behaviors is still unknown. In a recent study using PC12 cells and rats treated with 6-OHDA to model the disease, Chen X. et al. (2019) more closely investigated the role of m6A modification in the pathogenesis of PD. They found that m6A modification is downregulated in PC12 cells and in the substantia nigra of rats after treatment with 6-OHDA. They also reported an increase in the expression level of glutamate receptor subunit 1 (Nmdar1), which could be a consequence of reduced m6A modification. Consistent with this hypothesis, they reported that Fto deficiency in PC12 cells caused a reduction in Nmdar1 levels and prevented cell death. Altogether, these observations support a critical role for m6A modification in the pathogenesis of PD (Chen X. et al., 2019).
Amyotrophic Lateral Sclerosis (ALS) is a fatal neurodegenerative disorder which combines clinical evidence of both upper and lower motor neuron degeneration with patients resulting in dysfunction of the somatic muscles (Rowland et al., 2010; Saberi et al., 2015; Grad et al., 2017). About 90% of all cases of ALS are sporadic (sALS), while the remaining ∼10% is familial (fALS). Familial cases are inherited through an autosomal dominant pattern due to mutations in several genes (Alsultan et al., 2016; Grad et al., 2017). Usually, ALS is a disease of mid and late life with only 10% beginning before the age of 40 (Rowland et al., 2010). However, the onset is highly variable among patient and often earlier in fALS compared to sALS cases, despite the fact that the two forms of the disease are phenotypically indistinguishable (Swinnen and Robberecht, 2014; Grad et al., 2017). The rate of progression is also highly variable (Swinnen and Robberecht, 2014) even though the mean survival from the time of diagnosis is approximately 4 years with less than 20% surviving more than 5 years (Rowland et al., 2010). Evidence of m6A involvement in the pathogenesis of ALS is also emerging. In addition to its wide expression in brain, FTO has been shown to be highly expressed in lower motor neurons (Mitropoulos et al., 2017). Whole-genome sequencing analysis of Greek patients with sporadic ALS revealed a positive association between FTO gene variants and the disease (Mitropoulos et al., 2017). Moreover, a targeted gene sequencing of RBPs in ALS patients identified rare deleterious polymorphisms at a significantly higher rate than control in the RBM15 gene and in its paralog RMB15B (Cooper-Knock et al., 2017). In addition to being part of the m6A methyltransferase complex, Nito, the Drosophila homolog of human RBM15, was reported to regulate axon outgrowth, branching and to control synaptogenesis via the activity of the bursicon-expressing neurons (Gu et al., 2017). One further observation that encourages investigation on the potential involvement of m6A in motor neuron degeneration is that mutations in the gene coding for the m6A reader hnRNPA2-B1 can cause ALS (Kim et al., 2013).
Spinocerebellar ataxias (SCAs) are a group of hereditary disorders that are characterized by loss of balance and coordination accompanied by slurred speech. SCAs are due to degenerative changes in Purkinje neurons in the cerebellum, with resulting cerebellar atrophy. In some cases, other parts of the nervous system, such as the spinal cord, basal ganglia and pontine nuclei, can be affected in the disease (Klockgether et al., 2019). A link between m6A and SCA is suggested by studies reporting an essential role for m6A in the regulation of postnatal cerebellar development (Ma et al., 2018; Wang C. X. et al., 2018). In particular, one study reported that selective Mettl3 deletion in the nervous system leads to a severe reduction of m6A together with prominent developmental defects in the cerebellum of these mice that present typical features of cerebellar ataxia (Wang C. X. et al., 2018). Wang C. X. et al. (2018) further reported that Mettl3 and m6A participate to cerebellar development by modulating RNA stability of development- and apoptosis-associated genes and by controlling the alternative splicing of synapse-associated transcripts. Ma et al. (2018) substantiated these observations showing that aberrant expression of METTL3 and ALKBH5 resulted in imbalanced m6A, which in turn led to aberrant cerebellar development (Ma et al., 2018). Interestingly, in line with a potential involvement of m6A in the pathogenesis of SCA, reduced level of Ythdc1 was found to enhance neurodegeneration in a Drosophila model of SCA-1 (Fernandez-Funez et al., 2000).
The nervous system requires multiple levels of regulation to establish proper circuit assembly and function. RNA modifications (epitranscriptomics) are a new addition to the complex world of post-transcriptional gene regulation that can control the fate and function of both coding and non-coding RNAs. RNA modifications are preferentially enriched in neurons and are increasingly becoming critical determinants of neuronal function. The fast turnover and pervasiveness of RNA molecules, combined with the dynamic nature of various RNA modifications, allow epitranscriptomics to become a primary regulatory mechanism of cellular response to environmental cues. Based on indications that have linked aberrant m6A methylation with brain disorders, this specific RNA modification is emerging as an important regulator of neural cell fate decisions and plasticity. Despite these discoveries, our mechanistic understanding of m6A signaling in neuronal physiology and its contribution to pathology are still largely unexplored.
Due to the abundance of m6A-modified mRNAs in neurons, one of the major challenges is to identify the RNAs that are targeted by m6A in individual cells, characterize the effect of the modification on gene expression, and then link the m6A-dependent effect with normal and pathological behavior in vivo. The availability of conditional knockout mice for most of the m6A writers, erasers and readers is facilitating our understanding of the consequences of manipulation of m6A at the cellular and behavioral levels. However, mutation of a single m6A site, rather than manipulation of common m6A enzymes, is required to functionally uncover the consequences of site-specific methylation. Unfortunately, the lack of tools for a specific spatiotemporal manipulation of single m6A sites in vivo still limits the causal investigation of the functional outcomes of covalent RNA modulation. Several clustered regularly interspaced short palindromic repeats (CRISPR)-based approaches that in addition to targeting mRNA directly are also carrying RNA-modifying enzymes to specific targets may soon become a valuable toolkit toward this goal (Abudayyeh et al., 2016; East-Seletsky et al., 2016; Liu L. et al., 2017).
Along the same line, reliable quantification of m6A dynamics in different brain cells will be crucial to identify the downstream consequences of specific methylation events. Currently, methods to precisely discriminate “regulated” from “constitutive” m6A sites require large quantities of material or are limited to global or low-throughput gene- and site-specific determination. Recent development of new methods to enable analysis of m6A stoichiometry, such as RNA digestion via m6A sensitive RNase (MAZTER-seq) (Garcia-Campos et al., 2019) and Nanopore (Garalde et al., 2018), may overcome these limitations and will allow to determine to which extent the RNA methylation can be dynamically regulated in order to control gene expression. For this purpose, it will also be crucial to integrate findings of mRNA methylation profiles with analysis of RNA levels, alternative polyadenylation, alternative splicing, localization, and translation efficiency. By linking m6A-mediated effects on gene expression to specific neurological defects, these investigations have the potential not only to advance our mechanistic understanding of brain disorders but also to discover and develop new therapeutics based on rational manipulation of specific epitranscriptomic processes.
GD and FL researched the data for the review and wrote, reviewed, and edited the manuscript before submission. Both authors contributed to the article and approved the submitted version.
Work in the Lotti laboratory was supported by grants from Cure SMA, Thompson Family Foundation Initiative (TFFI), and NIH grant R21NS101575.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the members of the Lotti laboratory for comments and suggestions and Benjamin Hoover for critical reading of the manuscript.
Aas, A., Isakson, P., Bindesboll, C., Alemu, E. A., Klungland, A., and Simonsen, A. (2017). Nucleocytoplasmic shuttling of FTO does not affect starvation-induced autophagy. PLoS One 12:e0168182. doi: 10.1371/journal.pone.0168182
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573
Alarcon, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015a). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. doi: 10.1038/nature14281
Alsultan, A. A., Waller, R., Heath, P. R., and Kirby, J. (2016). The genetics of amyotrophic lateral sclerosis: current insights. Degener. Neurol. Neuromuscul. Dis. 6, 49–64. doi: 10.2147/dnnd.s84956
Arguello, A. E., DeLiberto, A. N., and Kleiner, R. E. (2017). RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 139, 17249–17252. doi: 10.1021/jacs.7b09213
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi: 10.1016/j.stem.2014.09.019
Benedict, C., Jacobsson, J. A., Ronnemaa, E., Sallman-Almen, M., Brooks, S., Schultes, B., et al. (2011). The fat mass and obesity gene is linked to reduced verbal fluency in overweight and obese elderly men. Neurobiol. Aging 32, e1151–e1155. doi: 10.1016/j.neurobiolaging.2011.02.006
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Boissel, S., Reish, O., Proulx, K., Kawagoe-Takaki, H., Sedgwick, B., Yeo, G. S., et al. (2009). Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 85, 106–111. doi: 10.1016/j.ajhg.2009.06.002
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247.
Cardoso, A. R., Lopes-Marques, M., Silva, R. M., Serrano, C., Amorim, A., Prata, M. J., et al. (2019). Essential genetic findings in neurodevelopmental disorders. Hum. Genomics 13:31. doi: 10.1186/s40246-019-0216-4
Chang, M., Lv, H., Zhang, W., Ma, C., He, X., Zhao, S., et al. (2017). Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7:170166. doi: 10.1098/rsob.170166
Chen, J., Zhang, Y. C., Huang, C., Shen, H., Sun, B., Cheng, X., et al. (2019). m(6)A regulates neurogenesis and neuronal development by modulating histone Methyltransferase Ezh2. Genomics Proteomics Bioinform. 17, 154–168. doi: 10.1016/j.gpb.2018.12.007
Chen, T., Hao, Y. J., Zhang, Y., Li, M. M., Wang, M., Han, W., et al. (2015). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301. doi: 10.1016/j.stem.2015.01.016
Chen, X., Yu, C., Guo, M., Zheng, X., Ali, S., Huang, H., et al. (2019). Down-Regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10, 2355–2363. doi: 10.1021/acschemneuro.8b00657
Choudhry, Z., Sengupta, S. M., Grizenko, N., Thakur, G. A., Fortier, M. E., Schmitz, N., et al. (2013). Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring) 21, E738–E744. doi: 10.1002/oby.20444
Cooper-Knock, J., Robins, H., Niedermoser, I., Wyles, M., Heath, P. R., Higginbottom, A., et al. (2017). Targeted genetic screen in amyotrophic lateral sclerosis reveals novel genetic variants with synergistic effect on clinical phenotype. Front. Mol. Neurosci. 10:370. doi: 10.3389/fnmol.2017.00370
Decker, C. J., and Parker, R. (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4:a012286. doi: 10.1101/cshperspect.a012286
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Dictenberg, J. B., Swanger, S. A., Antar, L. N., Singer, R. H., and Bassell, G. J. (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939. doi: 10.1016/j.devcel.2008.04.003
Doernberg, E., and Hollander, E. (2016). Neurodevelopmental disorders (ASD and ADHD): DSM-5, ICD-10, and ICD-11. CNS Spectr. 21, 295–299. doi: 10.1017/s1092852916000262
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Doxtader, K. A., Wang, P., Scarborough, A. M., Seo, D., Conrad, N. K., and Nam, Y. (2018). Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell 71:e1004. doi: 10.1016/j.molcel.2018.07.025
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626. doi: 10.1038/ncomms12626
Dugger, B. N., and Dickson, D. W. (2017). Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 9:a028035. doi: 10.1101/cshperspect.a028035
East-Seletsky, A., O’Connell, M. R., Knight, S. C., Burstein, D., Cate, J. H., Tjian, R., et al. (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. doi: 10.1038/nature19802
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep. 28:e845. doi: 10.1016/j.celrep.2019.06.072
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi: 10.1038/nsmb.3462
Engel, M., and Chen, A. (2018). The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 17:e12428. doi: 10.1111/gbb.12428
Engel, M., Eggert, C., Kaplick, P. M., Eder, M., Roh, S., Tietze, L., et al. (2018). The Role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99:e389. doi: 10.1016/j.neuron.2018.07.009
Fernandez-Funez, P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., et al. (2000). Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408, 101–106. doi: 10.1038/35040584
Fischer, J., Koch, L., Emmerling, C., Vierkotten, J., Peters, T., Bruning, J. C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458, 894–898. doi: 10.1038/nature07848
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4:1798. doi: 10.1038/ncomms2822
Fu, Y., and Zhuang, X. (2020). m(6)A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16, 955–963. doi: 10.1038/s41589-020-0524-y
Gao, X., Shin, Y. H., Li, M., Wang, F., Tong, Q., and Zhang, P. (2010). The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One 5:e14005. doi: 10.1371/journal.pone.0014005
Gao, Y., Pei, G., Li, D., Li, R., Shao, Y., Zhang, Q. C., et al. (2019). Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 29, 767–769. doi: 10.1038/s41422-019-0210-3
Garalde, D. R., Snell, E. A., Jachimowicz, D., Sipos, B., Lloyd, J. H., Bruce, M., et al. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206. doi: 10.1038/nmeth.4577
Garcia-Campos, M. A., Edelheit, S., Toth, U., Safra, M., Shachar, R., Viukov, S., et al. (2019). Deciphering the “m(6)A Code” via antibody-independent quantitative profiling. Cell 178:e716. doi: 10.1016/j.cell.2019.06.013
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. doi: 10.1126/science.1151710
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006. doi: 10.1126/science.1261417
Grad, L. I., Rouleau, G. A., Ravits, J., and Cashman, N. R. (2017). Clinical spectrum of amyotrophic lateral sclerosis (ALS). Cold Spring Harb. Perspect. Med. 7:a024117.
Gu, T., Zhao, T., Kohli, U., and Hewes, R. S. (2017). The large and small SPEN family proteins stimulate axon outgrowth during neurosecretory cell remodeling in Drosophila. Dev. Biol. 431, 226–238. doi: 10.1016/j.ydbio.2017.09.013
Han, M., Liu, Z., Xu, Y., Liu, X., Wang, D., Li, F., et al. (2020). Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 14:98. doi: 10.3389/fnins.2020.00098
Hastings, M. H. (2013). m(6)A mRNA methylation: a new circadian pacesetter. Cell 155, 740–741. doi: 10.1016/j.cell.2013.10.028
Heck, A. M., and Wilusz, C. J. (2019). Small changes, big implications: the impact of m(6)A RNA methylation on gene expression in pluripotency and development. Biochim. Biophys. Acta Gene Regul. Mech. 1862:194402. doi: 10.1016/j.bbagrm.2019.07.003
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A., Koch, L., Bronneke, H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048. doi: 10.1038/nn.3449
Heuser, I. J., Schweiger, U., Gotthardt, U., Schmider, J., Lammers, C. H., Dettling, M., et al. (1996). Pituitary-adrenal-system regulation and psychopathology during amitriptyline treatment in elderly depressed patients and normal comparison subjects. Am. J. Psychiatry 153, 93–99. doi: 10.1176/ajp.153.1.93
Ho, A. J., Stein, J. L., Hua, X., Lee, S., Hibar, D. P., Leow, A. D., et al. (2010). A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc. Natl. Acad. Sci. U.S.A. 107, 8404–8409.
Holsboer, F. (2000). The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology 23, 477–501. doi: 10.1016/s0893-133x(00)00159-7
Hsu, P. J., Shi, H., Zhu, A. C., Lu, Z., Miller, N., Edens, B. M., et al. (2019). The RNA-binding protein FMRP facilitates the nuclear export of N (6)-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895. doi: 10.1074/jbc.ac119.010078
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Huang, H., Weng, H., and Chen, J. (2020). m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell 37, 270–288. doi: 10.1016/j.ccell.2020.02.004
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Johnson, K. A., Conn, P. J., and Niswender, C. M. (2009). Glutamate receptors as therapeutic targets for Parkinson’s disease. CNS Neurol. Disord. Drug Targets 8, 475–491. doi: 10.2174/187152709789824606
Kao, D. I., Aldridge, G. M., Weiler, I. J., and Greenough, W. T. (2010). Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc. Natl. Acad. Sci. U.S.A. 107, 15601–15606. doi: 10.1073/pnas.1010564107
Keller, L., Xu, W., Wang, H. X., Winblad, B., Fratiglioni, L., and Graff, C. (2011). The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study. J. Alzheimers Dis. 23, 461–469. doi: 10.3233/jad-2010-101068
Kim, H. J., Kim, N. C., Wang, Y. D., Scarborough, E. A., Moore, J., Diaz, Z., et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. doi: 10.1038/nature11922
Kiser, D. P., Rivero, O., and Lesch, K. P. (2015). Annual research review: the (epi)genetics of neurodevelopmental disorders in the era of whole-genome sequencing–unveiling the dark matter. J. Child Psychol. Psychiatry 56, 278–295. doi: 10.1111/jcpp.12392
Klockgether, T., Mariotti, C., and Paulson, H. L. (2019). Spinocerebellar ataxia. Nat. Rev. Dis. Primers 5:24.
Koranda, J. L., Dore, L., Shi, H., Patel, M. J., Vaasjo, L. O., Rao, M. N., et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99:e285.
Lee, B. H., and Kim, Y. K. (2010). The roles of BDNF in the pathophysiology of major depression and in antidepressant treatment. Psychiatry Investig. 7, 231–235. doi: 10.4306/pi.2010.7.4.231
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. doi: 10.1038/cr.2017.10
Li, J., Yang, X., Qi, Z., Sang, Y., Liu, Y., Xu, B., et al. (2019). The role of mRNA m(6)A methylation in the nervous system. Cell Biosci. 9:66.
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19:69.
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. doi: 10.1016/j.molcel.2016.03.021
Lin, Z., and Tong, M. H. (2019). m(6)A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 403–411. doi: 10.1016/j.bbagrm.2018.10.016
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi: 10.1038/nmeth.3453
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, K., Ding, Y., Ye, W., Liu, Y., Yang, J., Liu, J., et al. (2016). Structural and functional characterization of the proteins responsible for N(6)-methyladenosine modification and recognition. Curr. Protein Pept. Sci. 17, 306–318. doi: 10.2174/1389203716666150901113553
Liu, L., Li, X., Ma, J., Li, Z., You, L., Wang, J., et al. (2017). The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170:e710.
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063. doi: 10.1093/nar/gkx141
Liu, X., Shimada, T., Otowa, T., Wu, Y. Y., Kawamura, Y., Tochigi, M., et al. (2016). Genome-wide association study of autism spectrum disorder in the East Asian populations. Autism Res. 9, 340–349. doi: 10.1002/aur.1536
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N., Rechavi, G., and Dominissini, D. (2020). The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21, 36–51. doi: 10.1038/s41583-019-0244-z
Ma, C., Chang, M., Lv, H., Zhang, Z. W., Zhang, W., He, X., et al. (2018). RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19:68.
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94. doi: 10.1038/s41589-018-0184-3
Mao, Y., Dong, L., Liu, X. M., Guo, J., Ma, H., Shen, B., et al. (2019). m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10:5332.
McTaggart, J. S., Lee, S., Iberl, M., Church, C., Cox, R. D., and Ashcroft, F. M. (2011). FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PLoS One 6:e27968. doi: 10.1371/journal.pone.0027968
Mendel, M., Chen, K. M., Homolka, D., Gos, P., Pandey, R. R., McCarthy, A. A., et al. (2018). Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell 71:e1011.
Meredith, R. M. (2015). Sensitive and critical periods during neurotypical and aberrant neurodevelopment: a framework for neurodevelopmental disorders. Neurosci. Biobehav. Rev. 50, 180–188. doi: 10.1016/j.neubiorev.2014.12.001
Merkurjev, D., Hong, W. T., Iida, K., Oomoto, I., Goldie, B. J., Yamaguti, H., et al. (2018). Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014. doi: 10.1038/s41593-018-0173-6
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi: 10.1146/annurev-cellbio-100616-060758
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5’ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’, UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Mitropoulos, K., Merkouri Papadima, E., Xiromerisiou, G., Balasopoulou, A., Charalampidou, K., Galani, V., et al. (2017). Genomic variants in the FTO gene are associated with sporadic amyotrophic lateral sclerosis in Greek patients. Hum. Genomics 11:30.
Nainar, S., Marshall, P. R., Tyler, C. R., Spitale, R. C., and Bredy, T. W. (2016). Evolving insights into RNA modifications and their functional diversity in the brain. Nat. Neurosci. 19, 1292–1298. doi: 10.1038/nn.4378
Nussbacher, J. K., Tabet, R., Yeo, G. W., and Lagier-Tourenne, C. (2019). Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320. doi: 10.1016/j.neuron.2019.03.014
Oldmeadow, C., Mossman, D., Evans, T. J., Holliday, E. G., Tooney, P. A., Cairns, M. J., et al. (2014). Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J. Psychiatr. Res. 52, 44–49. doi: 10.1016/j.jpsychires.2014.01.011
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell 74:e498.
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A Methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169:e814.
Perea, G., Navarrete, M., and Araque, A. (2009). Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 32, 421–431. doi: 10.1016/j.tins.2009.05.001
Perry, R. P., Kelley, D. E., Friderici, K., and Rottman, F. (1975). The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5’ terminus. Cell 4, 387–394. doi: 10.1016/0092-8674(75)90159-2
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Przedborski, S. (2017). The two-century journey of Parkinson disease research. Nat. Rev. Neurosci. 18, 251–259. doi: 10.1038/nrn.2017.25
Reiss, A. L. (2009). Childhood developmental disorders: an academic and clinical convergence point for psychiatry, neurology, psychology and pediatrics. J. Child Psychol. Psychiatry 50, 87–98. doi: 10.1111/j.1469-7610.2008.02046.x
Reitz, C., Tosto, G., Mayeux, R., Luchsinger, J. A., and Group, N.-L. N. F. S. Alzheimer’s Disease Neuroimaging, Initiative (2012). Genetic variants in the fat and obesity associated (FTO) gene and risk of Alzheimer’s disease. PLoS One 7:e50354. doi: 10.1371/journal.pone.0050354
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m(6)A enhances the phase separation potential of mRNA. Nature 571, 424–428. doi: 10.1038/s41586-019-1374-1
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6:e31311.
Rowland, L. P., Mitsumoto, H., and Przedborski, S. (2010). “Amyotrophic lateral sclerosis, progressive musclar atrophy and primary lateral sclerosis,” in Merritt’s Neurology, eds L. P. Rowland and T. A. Pedley (Philadelphia, PA: Lippincott, Williams & Wilkins), 802–808.
Ruzicka, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Saberi, S., Stauffer, J. E., Schulte, D. J., and Ravits, J. (2015). Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol. Clin. 33, 855–876. doi: 10.1016/j.ncl.2015.07.012
Santoro, M. R., Bray, S. M., and Warren, S. T. (2012). Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 7, 219–245. doi: 10.1146/annurev-pathol-011811-132457
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296. doi: 10.1016/j.celrep.2014.05.048
Sekhon, S., and Gupta, V. (2020). “Mood disorder,” in StatPearls, ed. StatPearls Publishing (Treasure Island, FL: StatPearls Publishing).
Shafik, A. M., Allen, E. G., and Jin, P. (2020). Dynamic N6-methyladenosine RNA methylation in brain and diseases. Epigenomics 12, 371–380. doi: 10.2217/epi-2019-0260
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Shi, H., Zhang, X., Weng, Y. L., Lu, Z., Liu, Y., Lu, Z., et al. (2018). m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253. doi: 10.1038/s41586-018-0666-1
Skene, J. H., and Willard, M. (1981). Axonally transported proteins associated with axon growth in rabbit central and peripheral nervous systems. J. Cell Biol. 89, 96–103. doi: 10.1083/jcb.89.1.96
Sledz, P., and Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5:e18434.
Sommer, S., Lavi, U., and Darnell, J. E. Jr. (1978). The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124, 487–499. doi: 10.1016/0022-2836(78)90183-3
Spychala, A., and Ruther, U. (2019). FTO affects hippocampal function by regulation of BDNF processing. PLoS One 14:e0211937. doi: 10.1371/journal.pone.0211937
Stepnicki, P., Kondej, M., and Kaczor, A. A. (2018). Current concepts and treatments of schizophrenia. Molecules 23:2087. doi: 10.3390/molecules23082087
Swinnen, B., and Robberecht, W. (2014). The phenotypic variability of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 10, 661–670. doi: 10.1038/nrneurol.2014.184
Tang, C., Klukovich, R., Peng, H., Wang, Z., Yu, T., Zhang, Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 115, E325–E333.
Thapar, A., Cooper, M., and Rutter, M. (2017). Neurodevelopmental disorders. Lancet Psychiatry 4, 339–346.
van den Berg, L., de Waal, H. D., Han, J. C., Ylstra, B., Eijk, P., Nesterova, M., et al. (2010). Investigation of a patient with a partial trisomy 16q including the fat mass and obesity associated gene (FTO): fine mapping and FTO gene expression study. Am. J. Med. Genet. A 152A, 630–637. doi: 10.1002/ajmg.a.33229
Velders, F. P., De Wit, J. E., Jansen, P. W., Jaddoe, V. W., Hofman, A., Verhulst, F. C., et al. (2012). FTO at rs9939609, food responsiveness, emotional control and symptoms of ADHD in preschool children. PLoS One 7:e49131. doi: 10.1371/journal.pone.0049131
Walters, B. J., Mercaldo, V., Gillon, C. J., Yip, M., Neve, R. L., Boyce, F. M., et al. (2017). The role of The RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42, 1502–1510. doi: 10.1038/npp.2017.31
Wang, C. X., Cui, G. S., Liu, X., Xu, K., Wang, M., Zhang, X. X., et al. (2018). METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 16:e2004880. doi: 10.1371/journal.pbio.2004880
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198. doi: 10.1038/ncb2902
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206. doi: 10.1038/s41593-017-0057-1
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71:e975.
Weller, J., and Budson, A. (2018). Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Res 7:F1000 Faculty Rev-1161.
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69:e1026.
Weng, H., Huang, H., and Chen, J. (2019). RNA N (6)-methyladenosine modification in normal and malignant hematopoiesis. Adv. Exp. Med. Biol. 1143, 75–93. doi: 10.1007/978-981-13-7342-8_4
Weng, Y. L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97:e316.
Widagdo, J., and Anggono, V. (2018). The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J. Neurochem. 147, 137–152. doi: 10.1111/jnc.14481
Widagdo, J., Zhao, Q. Y., Kempen, M. J., Tan, M. C., Ratnu, V. S., Wei, W., et al. (2016). Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777. doi: 10.1523/jneurosci.4053-15.2016
Wu, B., Su, S., Patil, D. P., Liu, H., Gan, J., Jaffrey, S. R., et al. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9:420.
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41. doi: 10.1038/s41422-018-0113-8
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Xu, C., Liu, K., Tempel, W., Demetriades, M., Aik, W., Schofield, C. J., et al. (2014). Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J. Biol. Chem. 289, 17299–17311. doi: 10.1074/jbc.m114.550350
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171:e817.
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423. doi: 10.1093/nar/gkx1182
Yu, J., Zhang, Y., Ma, H., Zeng, R., Liu, R., Wang, P., et al. (2020). Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain 13:11.
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10.
Zaccara, S., and Jaffrey, S. R. (2020). A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell 181:e1518.
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Zhang, C., Fu, J., and Zhou, Y. (2019). A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 10:922. doi: 10.3389/fimmu.2019.00922
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 113, E2047–E2056.
Zhang, F., Kang, Y., Wang, M., Li, Y., Xu, T., Yang, W., et al. (2018). Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950.
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31:e596.
Zhang, Z., Theler, D., Kaminska, K. H., Hiller, M., de la Grange, P., Pudimat, R., et al. (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285, 14701–14710.
Zhang, Z., Wang, M., Xie, D., Huang, Z., Zhang, L., Yang, Y., et al. (2018). METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061. doi: 10.1038/s41422-018-0092-9
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419. doi: 10.1038/cr.2014.151
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zhong, H., Tang, H. F., and Kai, Y. (2020). N6-methyladenine RNA modification (m6A): an emerging regulator of metabolic diseases. Curr. Drug Targets. doi: 10.2174/1389450121666200210125247 [Epub ahead of print].
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. doi: 10.1105/tpc.108.058883
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. doi: 10.1038/nature15377
Keywords: RNA modifications, N6-methyladenosine, m6A, RNA metabolism, neurodevelopment, mood disorders, neurodevelopmental disorders, neurodegenerative disorders
Citation: Dermentzaki G and Lotti F (2020) New Insights on the Role of N6-Methyladenosine RNA Methylation in the Physiology and Pathology of the Nervous System. Front. Mol. Biosci. 7:555372. doi: 10.3389/fmolb.2020.555372
Received: 24 April 2020; Accepted: 12 August 2020;
Published: 02 September 2020.
Edited by:
Florence Rage, Délégation Languedoc Roussillon (CNRS), FranceReviewed by:
Sunny Sharma, Rutgers, The State University of New Jersey, United StatesCopyright © 2020 Dermentzaki and Lotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Lotti, fl2219@columbia.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.