- Gamaleya Research Centre for Epidemiology and Microbiology, Moscow, Russia
Legionella is a gram-negative microorganism and an infectious agent of pneumonia in humans. It is an intracellular pathogen and multiplies in different eukaryotic cells like amoebae, ciliated protozoa, macrophages, monocytes, and lung epithelial cells. Proliferation of L. pneumophila in eukaryotic cells depends on its type 4 secretion system, which delivers an arsenal of bacterial effector proteins to cytoplasm of its host. Once within the cytoplasm, effectors modify a broad range of host activities, including mRNA translation. Translation is inhibited by Legionella through the action of several effector proteins including Lgt1, Lgt2, Lgt3, SidI, LegK4, SidL, and RavX. Lgt1-3 and SidI target elongation factors: Lgt1-3 mono-glucosylate elongation factor eEF1A, while SidI binds eEF1A, and eEF1Bγ. Effector LegK4 inhibits protein synthesis by phosphorylating Hsp70 proteins, while SidL and RavX have no defined targets in protein synthesis machinery thus far. In addition to direct inhibition of protein synthesis, SidI also affects the stress response, whereas Lgt1-3 – unfolded protein response and cell-cycle progression of host cells. Whether manipulation of these processes is linked to canonical or non-canonical function(s) of targeted elongation factors remains unknown.
Introduction
Protein synthesis is vitally important to eukaryotic cells. Therefore, it is not surprising that mRNA translation has been targeted by pathogens throughout evolution. The first bacterial molecule capable of inhibiting protein synthesis, identified in the late 1950s and early 1960s, was the “diphtheriae toxin” of Corynebacterium diphtheriae (Strauss and Hendee, 1959; Kato and Pappenheimer, 1960). Some years later, another toxin with similar activity was found in cultures of Pseudomonas aeruginosa and was named “exotoxin A” (Liu, 1966). Soon thereafter, the molecular mechanism of translational inhibition was revealed for both identified toxins, which involved the mono-adenosine 5′-diphosphate (ADP) ribosylation of eukaryotic translational elongation factor 2 (eEF2; Honjo et al., 1968; Gill et al., 1969; Iglewski and Kabat, 1975). The site of modification was determined to be a diphthamide residue, which involved post-translationally modified histidine-699 of eEF2 (Van Ness et al., 1980).
In subsequent studies, multiple research groups were able to show that proliferation of certain medically important microorganisms like Shigella, Salmonella, Chlamydia or Legionella within host cells resulted in the inhibition of eukaryotic protein synthesis (Hale and Formal, 1980, 1981; McCusker et al., 1991; Ohmer et al., 2019). In Shigella and Salmonella, observed cytotoxic effects were attributed to the production of cytotoxins, Shiga or Shiga-like toxins by the pathogens (Brown et al., 1980; Koo et al., 1984). The latter toxins specifically cleaved 28S rRNA to prevent the elongation of peptide chains on eukaryotic ribosomes (Endo et al., 1988). However, observed translational inhibition in target cells by Legionella was not explained until our knowledge of bacterial secretion systems improved (Segal et al., 1998; Vogel et al., 1998). It turned out that per se non-toxic protein effectors, been delivered directly into eukaryotic cytoplasm by specialized secretion machinery, became powerful virulence factors with diverse intracellular activities. These advancements revolutionized the field of bacterium-host interactions and created a platform for the identification of novel toxic proteins and the elucidation of sophisticated virulence mechanisms.
Intracellular Biology of L. pneumophila
Legionella pneumophila is a gram-negative bacterium and an infectious agent of legionellosis, a most known form of which (Legionnaires’ Disease) is characterized by severe pneumonia in humans (McDade et al., 1977). In natural environment the pathogen multiplies in a free-living unicellular organism like amoebae and ciliated protozoa (Rowbotham, 1983). During the infection process in humans the microorganisms predominantly invade macrophages, monocytes and lung epithelial cells (Richards et al., 2013). After uptake by host cells, legionellae multiply within a specialized phagosome-derived replicative vacuole, which avoids fusion with the lysosome and subsequent degradation (Isberg et al., 2009). Formation of a replicative vacuole by L. pneumophila is dependent upon the bacterial type 4 secretion system (T4SS), which translocates a plethora of bacterial effector proteins to the eukaryotic target cell. The highly specialized activities of this arsenal of Legionella factors are prerequisites for the successful proliferation of the infectious agent within its host (Escoll et al., 2016).
The range of eukaryotic organelles and host processes targeted by the Legionella effectors is amazingly broad (Escoll et al., 2016). The largest group of Legionella effectors manipulates eukaryotic small GTPases, which are involved in vesicular trafficking and membrane maturation in host cells [reviewed in Goody and Itzen (2013), Sherwood and Roy (2013), Isaac and Isberg (2014), Hilbi et al. (2017), Spano and Galan (2018)]. However, in addition to modifying endocytic machinery, a vast number of other cellular processes are affected throughout Legionella replication within host cells. These include apoptosis (Misch, 2016), autophagy (Sherwood and Roy, 2016; Siqueira et al., 2018), DNA transcription (Li et al., 2013; Rolando et al., 2013; Lee and Machner, 2018; Schuelein et al., 2018; Von Dwingelo et al., 2019), cytoskeleton functioning (Guo et al., 2014; Michard et al., 2015; Roy et al., 2017; He et al., 2019), mitochondrial dynamics (Arasaki et al., 2017), and phospholipid biosynthesis (Viner et al., 2012). Importantly, several L. pneumophila effectors have been shown to inhibit eukaryotic protein synthesis by targeting mRNA translation either by directly attacking translational factors or by phosphorylation of ribosome-associated chaperones (Belyi et al., 2006; Shen et al., 2009; Moss et al., 2019).
L. pneumophila Effectors That Target Protein Synthesis in Eukaryotic Cell
The first identified L. pneumophila effector that has been shown to inhibit protein synthesis was glucosyltransferase Lgt1 [Legionella glucosyltransferase 1, (GeneBank identification code is Lpg1236)] (Belyi et al., 2003). Later, in silico analysis of available genomic sequences from L. pneumophila strains revealed that several open reading frames exhibited significant sequence similarity to Lgt1. Based on their amino acid sequences, gene products were grouped into three subfamilies [Lgt1, Lgt2 (Lpg2862), and Lgt3 (Lpg1488)]. Some strains of L. pneumophila (e.g., Philadelphia-1) contained lgt1, lgt2, and lgt3, whereas others (e.g., Paris, Corby, Lens) possessed only lgt1 and lgt3 (Belyi et al., 2008). Interestingly, the prevalence of Lgt2 was higher within clinical strains of the bacteria than in environmental L. pneumophila isolates (Sadretdinova et al., 2012). Therefore, one can speculate that the addition of Lgt2 to the repertoire of glucosyltransferases in Legionella increases the virulence of the pathogen and/or broadens its potential host range.
Production of Lgt1 and Lgt2 by microbial cells was strongly increased during the stationary phase of bacterial growth in vitro, while Lgt3 was detectable in throughout the pre-logarithmic growth phase. Similar results were obtained in vivo, when the protozoan Acanthamoeba castellanii was used as a host of L. pneumophila. In this study, levels of mRNA encoding Lgt1 were maximal at late timepoints of bacteria-amoeba co-infection, while lgt3 was expressed mainly at the initial stage of the interaction between Legionella and A. castellanii (Belyi et al., 2008). These experiments suggested that glucosyltransferase activity was differentially regulated in L. pneumophila and indicated that each enzyme had specific role in promoting bacterial virulence. Speculatively, Lgt3 could be important for the initiation of the infection cycle, while Lgt1-2 might be necessary for Legionella egress from the host cell.
The crystal structure of Lgt1 is available (Hurtado-Guerrero et al., 2010; Lü et al., 2010). The effector is classified as a GT-A type glucosyltransferase family protein in the carbohydrate-active enzymes database http://www.cazy.org/GT88.html (Lombard et al., 2014). Several conserved amino acid residues of the catalytic core have been identified, including two aspartic amino acid residues (D-246 and D-248), which form a DXD motif that is typical of glycosyltransferases. The motif is crucial for divalent cation binding and coordination of the co-substrate (Busch et al., 1998; Wiggins and Munro, 1998). Lgt1 also possesses a flexible loop at its COOH-terminus, which is important for the proper arrangement of the donor substrate binding site, the accommodation of uridine diphospho (UDP)-glucose in the catalytic center and the release of reaction products after catalysis. Lgt1 uses UDP-glucose as a donor for the glucosylation reaction. The first identified target of Lgt1-3 was reported to be eukaryotic elongation factor eEF1A.
When mRNA is translated, eEF1A cycles between forming a translationally active ternary complex (eEF1A•GTP•aminoacyl-tRNA) and its inactive, GDP-bound conformation. The NH2-terminal G-domain of eEF1a, which is necessary for GTP binding and hydrolysis, plays a major role in the transition between inactive and active forms. Lgt1-3 modify an amino acid residue serine-53 located near the Switch-1 region of the G domain of the elongation factor that lies within a protruding loop that connects helices A∗ and A’ (Pedersen et al., 2001). Our studies have shown that eEF1A within a ternary complex, or truncated versions of the elongation factor were glucosylated several orders of magnitude higher than the full-length, apo-form of eEF1A (Belyi et al., 2009; Tzivelekidis et al., 2011). These data indicate that glucosylation efficiency appears to depend on the specific conformation of the factor. Indeed, the Switch-1 region undergoes conformational change as it alternates between GDP- and GTP-bound states (Figure 1). This change in conformation may influence modification rates and confer “super-specificity” to Lgt1-3-induced glucosylation, since the Legionella effectors are then able to discriminate and glucosylate not every, but preferentially translationally competent form of eEF1A within host cells.
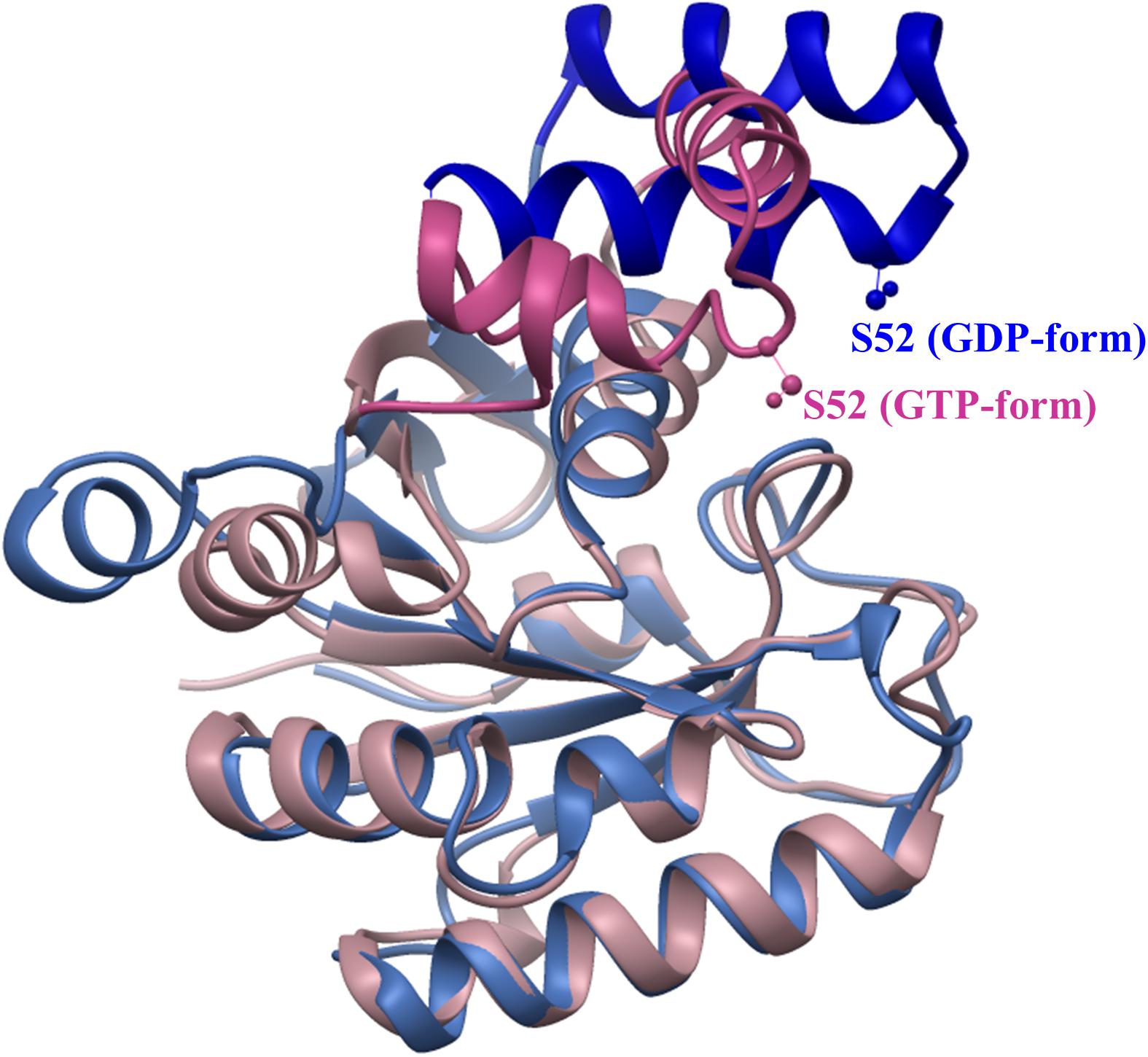
Figure 1. Conformational change of archaeal elongation factor 1A in GDP- and GTP-bound states. Structures of G-domains of aEF1A from Sulfolobus solfataricus in complex with GDP (1SKQ) and Aeropyrum pernix in complex with GTP (3AGJ) were aligned using USCF Chimera (Pettersen et al., 2004). Serine-52 (analog of serine-53 in eEF1A, which is glucosylated by Lgt1-3) is shown in “ball and stick” representation. Regions of aEF1As subjected to major change are drawn in dark blue (GDP-bound) and orchid (GTP-bound) colors, while regions of similar structure are colored in sky-blue (GDP-bound) and pink (GTP-bound).
Another substrate of Lgt1-3 is Hbs1 [Hsp70 subfamily B suppressor 1 (Nelson et al., 1992)]. Hbs1 is a conserved protein that can be found in diverse eukaryotic organisms ranging from yeast to humans and is implicated in the recycling of ribosomes stalled on an aberrant mRNA (Hilal et al., 2016). Yeast Hbs1 was modified on serine-213, located within a region structurally similar to the substrate sequence in eEF1A (Belyi et al., 2009).
Eukaryotic substrates modified by Lgt1-3 (eEF1A and Hbs1) include crucial components of the translational machinery. Not surprisingly, addition of this effector protein to in vitro reticulocyte translational extracts resulted in the dose-dependent inhibition of protein synthesis. Furthermore, delivery of the protein into mammalian cells, or expression of the corresponding genes in Saccharomyces cerevisiae, resulted in eEF1A glucosylation, the inhibition of protein synthesis, and cell death. Yeast strain containing eEF1A S53A but not Hbs1 S213A was insensitive to glucosylation and did not die in the presence of intracellular Lgt1 (Belyi et al., 2012). The latter experiment indicates that elongation factor eEF1A is the major target of the Legionella Lgt1 enzyme that causes toxicity in yeast.
SidI (Lpg2504) is another Legionella effector that has been shown to inhibit protein synthesis and kill eukaryotic cells (Shen et al., 2009; Joseph et al., 2020). This protein was identified during a screen of L. pneumophila genes capable of producing toxic effects in S. cerevisiae. Subsequently, the authors were able to demonstrate that SidI directly binds to both eEF1A and eEF1Bγ. The binding efficiency of wild type and point-mutated SidI correlated neither with its toxicity to eukaryotic cells nor with its inhibitory effect on mRNA translation in vitro (Shen et al., 2009). Moreover, the COOH-terminal fragment of the effector, shown to be able to bind eEF1A, only modestly inhibited in vitro translation compared to the full-sized molecule (Joseph et al., 2020). These data suggested that in addition to binding, the effector might possess another type of biochemical activity (e.g., enzymatic) that is required for its observed biological effects.
Indeed, using structure-predicting software, researchers have been able to demonstrate that SidI and GT-B fold glycosyltransferases are structurally similar. In accordance with this observation, the SidI protein has been shown to have the capacity to cleave GDP-mannose and, to a much lesser extent, UDP-glucose (Joseph et al., 2020). However, whether SidI can glycosylate any targets is not clear, since treatment of eEF1A with the effector did not result in any type of posttranslational modification of the protein (Shen et al., 2009). Interestingly, protein synthesis inhibition, accomplished by SidI is potently regulated by a metaeffector, L. pneumophila protein termed MesI (Lpg2505), which can bind to and significantly suppress enzymatic activity of the effector (Joseph et al., 2020).
LegK4 (lpl0262) is a protein kinase effector produced by Legionella that is capable of inhibiting protein synthesis in eukaryotic cells (Barry et al., 2013; Flayhan et al., 2015). LegK4 has been experimentally shown to be capable of phosphorylating conserved threonine residue of members of Hsp70 chaperone family both in vitro and in vivo. This residue (T-492 in Ssa1 of S. cerevisiae) is located within substrate binding domain and its modification by the protein kinase results in reduced chaperone ATPase activity and the concomitant inhibition of the refolding ability. Phosphorylation and subsequent inactivation of Hsp70 enhances its association with ribosomes and reduces global levels of translation. These data suggest a mechanism in which Hsp70 molecules, after being phosphorylated by LegK4, fail to fold nascent polypeptides correctly. Therefore, they remain associated with polysomes longer than usual, and block protein synthesis (Moss et al., 2019).
The least-studied effectors capable of inhibiting eukaryotic translation in vivo are SidL (Lpg0437; Fontana et al., 2011) and RavX (Lpg1489; Barry et al., 2013). Both proteins were shown to have the capacity to kill eukaryotic cells. However, the targets within eukaryotic cells and mechanisms of the translational inhibition of these effectors have not been elucidated. Surprisingly, the toxicity of SidL to yeast was completely alleviated by overexpression of profilin, one of the major cytoskeleton-related proteins. In other experiments direct interaction of the effector with actin and subsequent inhibition of actin polymerization were demonstrated (Guo et al., 2014). Bearing in mind the fact that actin cytoskeleton and components of protein synthesis machinery are functionally linked (Gross and Kinzy, 2005; Pittman et al., 2009), it would be interesting to explore the mechanism of inhibition of mRNA translation by SidL in relation to its effect upon eukaryotic cytoskeleton.
Cell Killing by Legionella Effectors and Beyond
The significance of effector-induced protein synthesis inhibition for the pathogenesis of Legionella infection remains unclear. It has been speculated that the action of bacterial inhibitors of eukaryotic mRNA translation strongly decreases cellular metabolism, and correspondingly, antibacterial activity. This makes weakened host cells more susceptible to invading bacteria (Figure 2). Another possible explanation states that global decreases in the translation of eukaryotic transcripts may provide a large pool of unused amino acids and other nutrients to Legionella. Finally, at late stages of the intracellular life cycle, Legionella escape from its host requires the killing of the eukaryotic cell. Translation-targeting effectors may facilitate such a killing of the host. The above possibilities can be attributed to as “direct” roles of the effectors (Belyi et al., 2013).
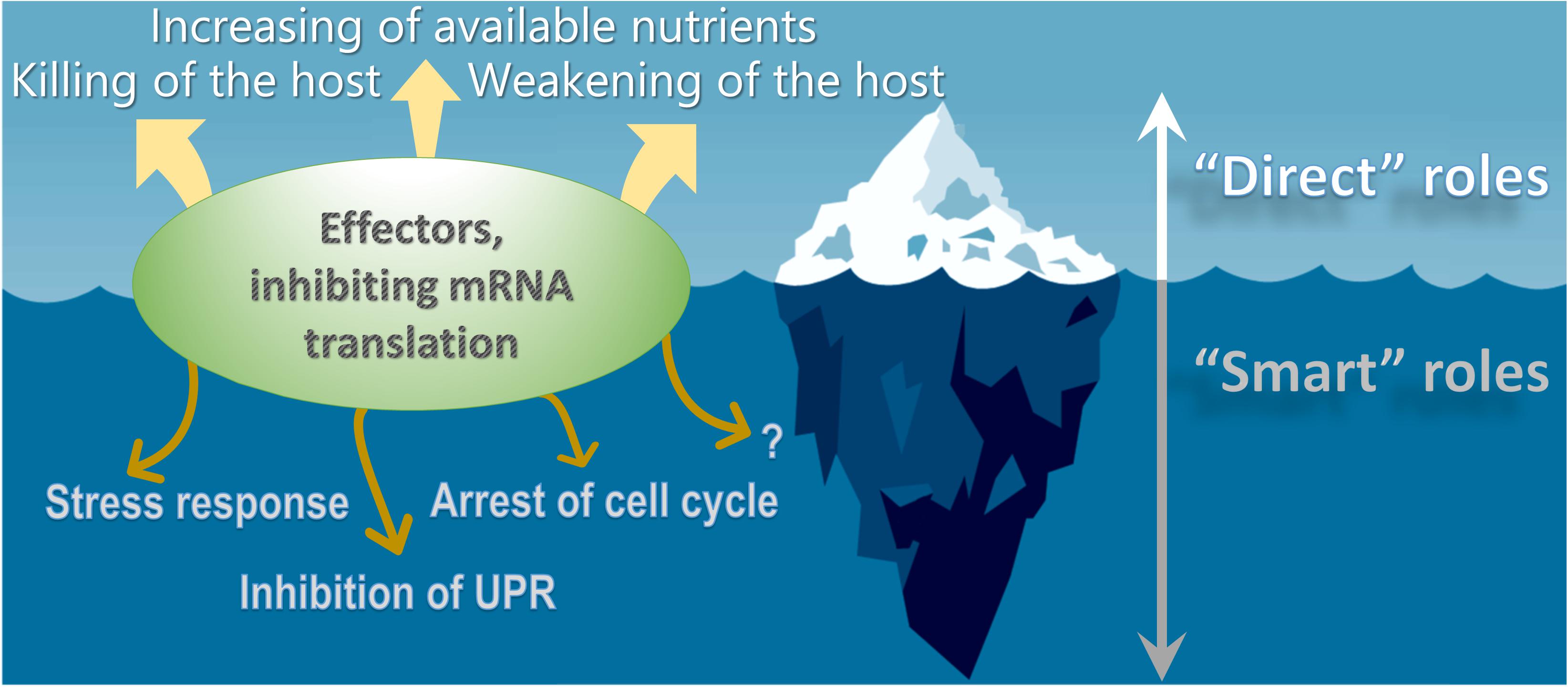
Figure 2. Different roles of Legionella effectors inhibiting eukaryotic mRNA translation. The bacterial inhibitors of eukaryotic protein synthesis may facilitate decrease in the translation of eukaryotic transcripts thus providing a large pool of unused nutrients to Legionella, weakening of antibacterial activity and ultimately – killing of the host. The above examples represent “direct” roles of the effectors. However, the latter proteins might also play “smart” roles in the virulence of Legionella that are aimed at delicately manipulating host cell functioning for the benefit of the pathogen, including regulation of stress response, suppressing of unfolded protein response, arrest of cell cycle progression and possibly some other processes, which are unknown to date.
Several lines of evidence suggest, however, that the biological importance of effectors should not be oversimplified. Accumulated data show that virulence factors might have “smart” roles that are aimed at delicately manipulating of host cell functioning for the benefit of the pathogen (Figure 2). In this case, observed cytotoxicity may be a side effect of some other pro-bacterial consequences of translational arrest (Ensminger and Isberg, 2009). One example of a smart role of Legionella effectors was reported in the investigation of the regulation of the stress response by SidI.
Stress shock response is a fundamental mechanism necessary for eukaryotic cell survival within a variety of harmful environments. The stress response in mammalian cells is controlled by a multi-component complex that consists of heat shock transcription factor 1 (Hsf1), eEF1A and non-coding RNA molecules (heat shock RNA 1, Hsr1; Shamovsky et al., 2006). Hsf1 is able to bind specific promoters (heat shock elements, Hse) and thus, induces the production of a panel of heat shock proteins necessary for rescuing eukaryotic cells that are experiencing unfavorable conditions (Sarge et al., 1991). Infection of macrophage-like cells with virulent L. pneumophila or transfection of eukaryotic cells with SidI-coding plasmids resulted in an elevated eukaryotic stress response, which researchers detected by observing elevated levels of the Hfs1/eEF1A complex, increased binding of Hsf1 to Hse and the stimulation of hsp70 expression (Shen et al., 2009). These results indicate that Hsf1 is activated during L. pneumophila infection and SidI, which was initially shown to suppress protein synthesis, mediates its activation.
Another process that is influenced by Legionella infection is the unfolded protein response (UPR). The UPR is initiated by the eukaryotic cell to cope with the accumulation of misfolded proteins within the lumen of the endoplasmic reticulum (ER; Hetz and Papa, 2018). A branch of the UPR is also linked to the innate immune response (Hasnain et al., 2012). The consequences of activating the UPR include the inhibition of global protein synthesis, upregulation of the production of ER stress proteins (e.g., luminal chaperone BiP), and the initiation of apoptotic (e.g., through induction of CHOP) and proinflammatory (e.g., activation of NF-kB) programs.
Legionella pneumophila was shown to inhibit the UPR via multiple mechanisms (Treacy-Abarca and Mukherjee, 2015). To elucidate means by which UPR was manipulated by L. pneumophila, experiments investigating IRE1, a known sensor of the UPR were conducted. Under conditions of ER stress, activated IRE1 removes an intron from XBP1 mRNA, an UPR intermediate. Thus, a spliced variant is formed that acts as a transcription factor and enhances transcription of the ER chaperone gene. The intracellularly-multiplying, wild-type L. pneumophila strain efficiently blocked effects of the production of the splice variant. The genetic inactivation of T4SS of Legionella or the five translational effectors (i.e., Δ5: Lgt1-3, SidI, and SidL) restored splicing activity in host cells, which was able to be blocked again by complementing the Δ5 mutant with plasmids encoding Lgt2 or Lgt3 (Hempstead and Isberg, 2015; Treacy-Abarca and Mukherjee, 2015).
Legionella multiplication is dependent upon the life cycle of its host. In particular, it has been shown that host cells in G1 and G2/M phases facilitate bacterial replication, while eukaryotic cells in S phase provide an environment that hinders L. pneumophila replication (de Jesús-Díaz et al., 2017). The wild type, but not the Δ5 strain, was able to arrest host cell-cycle progression in macrophages and amoebae. Strikingly, failure of the Δ5 strain to control cell-cycle progression was able to be reversed by transfecting the host with Lgt3-encoding plasmid. Moreover, cell cycle arrest was initiated by the ectopic expression of a single glucosyltransferase protein, either Lgt1 or Lgt3 (Sol et al., 2019).
Discussion
Modification of the activities of key translation factors appears to be an important virulence mechanism that aids proliferation of Legionella within its eukaryotic host. Given that translation factors have both canonical and non-canonical functions, their regulation allows the pathogen to directly or indirectly influence an array of processes within the host cell. This should be kept in mind while studying intracellular biology of L. pneumophila and pathogenesis of the infectious disease. Although this type of knowledge mainly is in the realm of basic science, it will certainly facilitate the development of new therapeutics, vaccines and diagnostic methods.
The high degree of specificity and activity of bacterial virulence factors make them powerful probes that facilitate the dissection of the functions of their eukaryotic targets. There are numerous examples of how the use of virulence factors have enhanced our knowledge of host biology. In particular, the study of Rho protein biochemistry, G-protein-guided signal transduction and retrograde transport have been elucidated by the study of glucosylating Clostridium difficile toxins, Pasteurella multocida toxin and Shiga toxin, respectively. Therefore, the addition of Legionella effectors to this experimental toolbox may facilitate the discovery of novel functions of different eukaryotic proteins and may shed light on their pathways. Elongation factors are emerging as types of eukaryotic proteins whose functions have become more obvious as a result of the study of proteins produced by L. pneumophila.
Funding
The work was supported partially by FRIAS (Freiburg Institute for Advanced Studies).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Klaus Aktories (Institute of Experimental and Clinical Pharmacology and Toxicology, University of Freiburg) and Sabine Rospert (Institute of Biochemistry and Molecular Biology, University of Freiburg) for everlasting interest in and support of my studies.
References
Arasaki, K., Mikami, Y., Shames, S. R., Inoue, H., Wakana, Y., and Tagaya, M. (2017). Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat. Commun. 8:15406. doi: 10.1038/ncomms15406
Barry, K. C., Fontana, M. F., Portman, J. L., Dugan, A. S., and Vance, R. E. (2013). IL-1alpha signaling initiates the inflammatory response to virulent Legionella pneumophila in vivo. J. Immunol. 190, 6329–6339. doi: 10.4049/jimmunol.1300100
Belyi, I., Popoff, M. R., and Cianciotto, N. P. (2003). Purification and characterization of a UDP-glucosyltransferase produced by Legionella pneumophila. Infect. Immun. 71, 181–186.
Belyi, Y., Jank, T., and Aktories, K. (2013). Cytotoxic glucosyltransferases of Legionella pneumophila. Curr. Top. Microbiol. Immunol 376, 211–226. doi: 10.1007/82_2013_338
Belyi, Y., Niggeweg, R., Opitz, B., Vogelsgesang, M., Hippenstiel, S., Wilm, M., et al. (2006). Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc. Natl. Acad. Sci. U.S.A. 103, 16953–16958. doi: 10.1073/pnas.0601562103
Belyi, Y., Stahl, M., Sovkova, I., Kaden, P., Luy, B., and Aktories, K. (2009). Region of elongation factor 1A1 involved in substrate recognition by Legionella pneumophila glucosyltransferase Lgt1: identification of Lgt1 as a retaining glucosyltransferase. J. Biol. Chem. 284, 20167–20174. doi: 10.1074/jbc.M109.008441
Belyi, Y., Tabakova, I., Stahl, M., and Aktories, K. (2008). Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J. Bacteriol. 190, 3026–3035. doi: 10.1128/JB.01798-07
Belyi, Y., Tartakovskaya, D., Tais, A., Fitzke, E., Tzivelekidis, T., Jank, T., et al. (2012). Elongation factor 1A is the target of growth inhibition in yeast caused by Legionella pneumophila glucosyltransferase Lgt1. J. Biol. Chem. 287, 26029–26037. doi: 10.1074/jbc.M112.372672
Brown, J. E., Rothman, S. W., and Doctor, B. P. (1980). Inhibition of protein synthesis in intact HeLa cells by Shigella dysenteriae 1 toxin. Infect. Immun. 29, 98–107.
Busch, C., Hofmann, F., Selzer, J., Munro, S., Jeckel, D., and Aktories, K. (1998). A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem. 273, 19566–19572.
de Jesús-Díaz, D. A., Murphy, C., Sol, A., Dorer, M., and Isberg, R. R. (2017). Host Cell S Phase Restricts Legionella pneumophila Intracellular Replication by Destabilizing the Membrane-Bound Replication Compartment. mBio 8, e2345–e2316. doi: 10.1128/mBio.02345-16
Endo, Y., Tsurugi, K., Yutsudo, T., Takeda, Y., Ogasawara, T., and Igarashi, K. (1988). Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171, 45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x
Ensminger, A. W., and Isberg, R. R. (2009). Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12, 67–73. doi: 10.1016/j.mib.2008.12.004
Escoll, P., Mondino, S., Rolando, M., and Buchrieser, C. (2016). Targeting of host organelles by pathogenic bacteria: a sophisticated subversion strategy. Nat. Rev. Microbiol. 14, 5–19. doi: 10.1038/nrmicro.2015.1
Flayhan, A., Berge, C., Bailo, N., Doublet, P., Bayliss, R., and Terradot, L. (2015). The structure of Legionella pneumophila LegK4 type four secretion system (T4SS) effector reveals a novel dimeric eukaryotic-like kinase. Sci Rep 5:14602. doi: 10.1038/srep14602
Fontana, M. F., Banga, S., Barry, K. C., Shen, X., Tan, Y., Luo, Z.-Q., et al. (2011). Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 7:e1001289–e1001289. doi: 10.1371/journal.ppat.1001289
Gill, D. M., Pappenheimer, A. M. Jr., Brown, R., and Kurnick, J. T. (1969). Studies on the mode of action of diphtheria toxin. VII. Toxin-stimulated hydrolysis of nicotinamide adenine dinucleotide in mammalian cell extracts. J. Exp. Med. 129, 1–21. doi: 10.1084/jem.129.1.1
Goody, R. S., and Itzen, A. (2013). Modulation of small GTPases by Legionella. Curr. Top. Microbiol. Immunol 376, 117–133. doi: 10.1007/82_2013_340
Gross, S. R., and Kinzy, T. G. (2005). Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12, 772–778. doi: 10.1038/nsmb979
Guo, Z., Stephenson, R., Qiu, J., Zheng, S., and Luo, Z. Q. (2014). A Legionella effector modulates host cytoskeletal structure by inhibiting actin polymerization. Microbes Infect. 16, 225–236. doi: 10.1016/j.micinf.2013.11.007
Hale, T. L., and Formal, S. B. (1980). Cytotoxicity of Shigella dysenteriae 1 for cultured mammalian cells. Am. J. Clin. Nutr. 33(Suppl. 11), 2485–2490. doi: 10.1093/ajcn/33.11.2485
Hale, T. L., and Formal, S. B. (1981). Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect. Immun. 32, 137–144.
Hasnain, S. Z., Lourie, R., Das, I., Chen, A. C. H., and McGuckin, M. A. (2012). The interplay between endoplasmic reticulum stress and inflammation. Immunol. Cell Biol. 90, 260–270. doi: 10.1038/icb.2011.112
He, L., Lin, Y., Ge, Z. H., He, S. Y., Zhao, B. B., Shen, D., et al. (2019). The Legionella pneumophila effector WipA disrupts host F-actin polymerisation by hijacking phosphotyrosine signalling. Cell. Microbiol. 21:e13014. doi: 10.1111/cmi.13014
Hempstead, A. D., and Isberg, R. R. (2015). Inhibition of host cell translation elongation by Legionella pneumophila blocks the host cell unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 112, E6790–E6797. doi: 10.1073/pnas.1508716112
Hetz, C., and Papa, F. R. (2018). The unfolded protein response and cell fate control. Mol. Cell. 69, 169–181. doi: 10.1016/j.molcel.2017.06.017
Hilal, T., Yamamoto, H., Loerke, J., Burger, J., Mielke, T., and Spahn, C. M. (2016). Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution. Nat. Commun. 7:13521. doi: 10.1038/ncomms13521
Hilbi, H., Nagai, H., Kubori, T., and Roy, C. R. (2017). “Subversion of host membrane dynamics by the Legionella Dot/Icm type IV secretion system,” in Type IV Secretion in Gram-Negative and Gram-Positive Bacteria, eds S. Backert and E. Grohmann (Cham: Springer International Publishing), 221–242.
Honjo, T., Nishizuka, Y., and Hayaishi, O. (1968). Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 243, 3553–3555.
Hurtado-Guerrero, R., Zusman, T., Pathak, S., Ibrahim, A. F. M., Shepherd, S., Prescott, A., et al. (2010). Molecular mechanism of elongation factor 1A inhibition by a Legionella pneumophila glycosyltransferase. Biochem. J. 426, 281–292. doi: 10.1042/BJ20091351
Iglewski, B. H., and Kabat, D. (1975). NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc. Natl. Acad. Sci. U.S.A. 72, 2284–2288. doi: 10.1073/pnas.72.6.2284
Isaac, D. T., and Isberg, R. (2014). Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Fut. Microbiol. 9, 343–359. doi: 10.2217/fmb.13.162
Isberg, R. R., O’Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24.
Joseph, A. M., Pohl, A. E., Ball, T. J., Abram, T. G., Johnson, D. K., Geisbrecht, B. V., et al. (2020). The Legionella pneumophila metaeffector Lpg2505 (MesI) regulates SidI-mediated translation inhibition and novel glycosyl hydrolase activity. Infect Immun. doi: 10.1128/IAI.00853-19 [Epub ahead of print].
Kato, I., and Pappenheimer, A. M. Jr. (1960). An early effect of diphtheria toxin on the metabolism of mammalian cells growing in culture. J. Exp. Med. 112, 329–349. doi: 10.1084/jem.112.2.329
Koo, F. C., Peterson, J. W., Houston, C. W., and Molina, N. C. (1984). Pathogenesis of experimental salmonellosis: inhibition of protein synthesis by cytotoxin. Infect. Immun. 43, 93–100.
Lee, P.-C., and Machner, M. P. (2018). The Legionella effector kinase LegK7 hijacks the host Hippo pathway to promote infection. Cell Host & Microbe 24, 429–438. doi: 10.1016/j.chom.2018.08.004 .e426.
Li, T., Lu, Q., Wang, G., Xu, H., Huang, H., Cai, T., et al. (2013). SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO Rep. 14, 733–740. doi: 10.1038/embor.2013.86
Liu, P. V. (1966). The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. 3. Identity of the lethal toxins produced in vitro and in vivo. J. Infect. Dis. 116, 481–489. doi: 10.1093/infdis/116.4.481
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Lü, W., Du, J., Stahl, M., Tzivelekidis, T., Belyi, Y., Gerhardt, S., et al. (2010). Structural basis of the action of glucosyltransferase Lgt1 from Legionella pneumophila. J. Mol. Biol. 396, 321–331. doi: 10.1016/j.jmb.2009.11.044
McCusker, K. T., Braaten, B. A., Cho, M. W., and Low, D. A. (1991). Legionella pneumophila inhibits protein synthesis in Chinese hamster ovary cells. Infect. Immun. 59, 240–246.
McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A., and Dowdle, W. R. (1977). Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203. doi: 10.1056/NEJM197712012972202
Michard, C., Sperandio, D., Bailo, N., Pizarro-Cerda, J., LeClaire, L., Chadeau-Argaud, E., et al. (2015). The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. MBio 6:e354-15.
Misch, E. A. (2016). Legionella: virulence factors and host response. Curr. Opin. Infect. Dis. 29, 280–286. doi: 10.1097/QCO.0000000000000268
Moss, S. M., Taylor, I. R., Ruggero, D., Gestwicki, J. E., Shokat, K. M., and Mukherjee, S. (2019). A Legionella pneumophila kinase phosphorylates the Hsp70 chaperone family to inhibit eukaryotic protein synthesis. Cell Host Microbe 25, 454–462.e6. doi: 10.1016/j.chom.2019.01.006
Nelson, R. J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M., and Craig, E. A. (1992). The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71, 97–105.
Ohmer, M., Tzivelekidis, T., Niedenführ, N., Volceanov-Hahn, L., Barth, S., Vier, J., et al. (2019). Infection of HeLa cells with Chlamydia trachomatis inhibits protein synthesis and causes multiple changes to host cell pathways. Cell Microbiol. 21:e12993. doi: 10.1111/cmi.12993
Pedersen, L., Andersen, G. R., Knudsen, C. R., Kinzy, T. G., and Nyborg, J. (2001). Crystallization of the yeast elongation factor complex eEF1A-eEF1B alpha. Acta Crystallogr. D. Biol. Crystallogr. 57(Pt 1), 159–161.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Pittman, Y. R., Kandl, K., Lewis, M., Valente, L., and Kinzy, T. G. (2009). Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha. J. Biol. Chem. 284, 4739–4747. doi: 10.1074/jbc.M807945200
Richards, A. M., Von Dwingelo, J. E., Price, C. T., and Abu Kwaik, Y. (2013). Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 4, 307–314. doi: 10.4161/viru.24290
Rolando, M., Sanulli, S., Rusniok, C., Gomez-Valero, L., Bertholet, C., Sahr, T., et al. (2013). Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13, 395–405. doi: 10.1016/j.chom.2013.03.004
Rowbotham, T. J. (1983). Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36, 978–986. doi: 10.1136/jcp.36.9.978
Roy, C. R., Liu, Y., Zhu, W., Tan, Y., Nakayasu, E. S., Staiger, C. J., et al. (2017). A Legionella effector disrupts host cytoskeletal structure by cleaving actin. PLoS Pathog. 13:e1006186. doi: 10.1371/journal.ppat.1006186
Sadretdinova, O. V., Liuk, K., Karpova, T. I., Belyi, Iu, F., and Tartakovskii, I. S. (2012). [Prevalence of glucosyl transferase Lgt among Legionella pneumophila strains isolated from various sources]. Z. Mikrobiol. Epidemiol. Immunobiol. 3, 8–12.
Sarge, K. D., Zimarino, V., Holm, K., Wu, C., and Morimoto, R. I. (1991). Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 5, 1902–1911.
Schuelein, R., Spencer, H., Dagley, L. F., Li, P. F., Luo, L., Stow, J. L., et al. (2018). Targeting of RNA Polymerase II by a nuclear Legionella pneumophila Dot/Icm effector SnpL. Cell Microbiol. 20:e12852. doi: 10.1111/cmi.12852
Segal, G., Purcell, M., and Shuman, H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674. doi: 10.1073/pnas.95.4.1669
Shamovsky, I., Ivannikov, M., Kandel, E. S., Gershon, D., and Nudler, E. (2006). RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–560. doi: 10.1038/nature04518
Shen, X., Banga, S., Liu, Y., Xu, L., Gao, P., Shamovsky, I., et al. (2009). Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 11, 911–926. doi: 10.1111/j.1462-5822.2009.01301.x
Sherwood, R. K., and Roy, C. R. (2013). A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14, 256–268. doi: 10.1016/j.chom.2013.08.010
Sherwood, R. K., and Roy, C. R. (2016). Autophagy evasion and endoplasmic reticulum subversion: the Yin and Yang of Legionella intracellular infection. Annu. Rev. Microbiol. 70, 413–433. doi: 10.1146/annurev-micro-102215-095557
Siqueira, M. D. S., Ribeiro, R. M., and Travassos, L. H. (2018). Autophagy and its interaction with intracellular bacterial pathogens. Front. Immunol. 9:935. doi: 10.3389/fimmu.2018.00935
Sol, A., Lipo, E., de Jesus-Diaz, D. A., Murphy, C., Devereux, M., and Isberg, R. R. (2019). Legionella pneumophila translocated translation inhibitors are required for bacterial-induced host cell cycle arrest. Proc. Natl. Acad. Sci. U.S.A. 116, 3221–3228. doi: 10.1073/pnas.1820093116
Spano, S., and Galan, J. E. (2018). Taking control: hijacking of Rab GTPases by intracellular bacterial pathogens. Small GTPases 9, 182–191. doi: 10.1080/21541248.2017.1336192
Strauss, N., and Hendee, E. D. (1959). The effect of diphtheria toxin on the metabolism of HeLa cells. J. Exp. Med. 109, 145–163. doi: 10.1084/jem.109.2.145
Treacy-Abarca, S., and Mukherjee, S. (2015). Legionella suppresses the host unfolded protein response via multiple mechanisms. Nat. Communi. 6:7887–7887. doi: 10.1038/ncomms8887
Tzivelekidis, T., Jank, T., Pohl, C., Schlosser, A., Rospert, S., Knudsen, C. R., et al. (2011). Aminoacyl-tRNA-charged eukaryotic elongation factor 1A is the bona fide substrate for Legionella pneumophila effector glucosyltransferases. PLoS ONE 6:e29525. doi: 10.1371/journal.pone.0029525
Van Ness, B. G., Howard, J. B., and Bodley, J. W. (1980). ADP-ribosylation of elongation factor 2 by diphtheria toxin. NMR spectra and proposed structures of ribosyl-diphthamide and its hydrolysis products. J. Biol. Chem. 255, 10710–10716.
Viner, R., Chetrit, D., Ehrlich, M., and Segal, G. (2012). Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog. 8:e1002988. doi: 10.1371/journal.ppat.1002988
Vogel, J. P., Andrews, H. L., Wong, S. K., and Isberg, R. R. (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science (New York, N.Y.) 279, 873–876. doi: 10.1126/science.279.5352.873
Von Dwingelo, J., Chung, I. Y. W., Price, C. T., Li, L., Jones, S., Cygler, M., et al. (2019). Interaction of the ankyrin H core effector of Legionella with the host LARP7 component of the 7SK snRNP complex1. MBio 10:e1942-19. doi: 10.1128/mBio.01942-19
Keywords: Legionella, protein synthesis, inhibition, elongation factor eEF1A, glycosylation
Citation: Belyi Y (2020) Targeting Eukaryotic mRNA Translation by Legionella pneumophila. Front. Mol. Biosci. 7:80. doi: 10.3389/fmolb.2020.00080
Received: 14 February 2020; Accepted: 07 April 2020;
Published: 29 April 2020.
Edited by:
Burcu Biterge Süt, Ni ð de Ömer Halisdemir University, TurkeyReviewed by:
Stephanie Rochelle Shames, Kansas State University, United StatesAyten Ozturk, Ni ð de Ömer Halisdemir University, Turkey
Copyright © 2020 Belyi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yury Belyi, YmVseWlAZ2FtYWxleWEub3Jn; eV9iZWx5aUB5YWhvby5jb20=
 Yury Belyi
Yury Belyi