- 1Faculty of Dentistry, Institute of Oral Biology, University of Oslo, Oslo, Norway
- 2Department of Microbiology, The Forsyth Institute, Cambridge, MA, United States
- 3Department of Biological Sciences, College of Liberal Arts and Sciences, University of Illinois at Chicago, Chicago, IL, United States
Natural transformation is regarded as an important mechanism in bacteria that allows for adaptation to different environmental stressors by ensuring genome plasticity. Since the discovery of this phenomenon in Streptococcus pneumoniae, remarkable progress has been made in the understanding of the molecular mechanisms and pathways coordinating this process. Recently, the advent of high-throughput sequencing allows the posing of questions that address the system at a larger scale but also allow for the creation of high-resolution maps of transcription. Thus, while much is already known about genetic competence in streptococci, recent studies continue to reveal intricate novel regulation pathways and components. In this perspective article, we highlight the use of transcriptional profiling and mapping as a valuable resource in the identification and characterization of “hidden gems” pertinent to the natural transformation system. Such strategies have recently been employed in a variety of different species. In S. mutans, for example, genome editing combined with the power of promoter mapping and RNA-Seq allowed for the identification of a link between the ComCDE and the ComRS systems, a ComR positive feedback loop mediated by SigX, and the XrpA peptide, encoded within sigX, which inhibits competence. In S. pneumoniae, a novel member of the competence regulon termed BriC was found to be directly under control of ComE and to promote biofilm formation and nasopharyngeal colonization but not competence. Together these new technologies enable us to discover new links and to revisit old pathways in the compelling study of natural genetic transformation.
The Mine of Natural Genetic Transformation in Streptococci
Natural transformation is the only bacterial horizontal gene transfer (HGT) mechanism in which the recipient cell has primary control over the transfer process. This is in contrast with conjugation and transduction, where the transfer relies primarily on genetic elements in the donor cells. Natural transformation allows microbes to better adapt to different stressors by assimilating divergent genetic information to ensure plasticity of their genomes, correct detrimental mutations, and spread important traits, such as resistance to antibiotics (Johnston et al., 2014; Fontaine et al., 2015; Straume et al., 2015). In addition, natural transformation has for decades been a valuable resource in microbiology as means to create mutant strains.
The discovery of natural transformation in bacteria was first reported in 1928 by Frederick Griffith with his studies on Streptococcus pneumoniae (Griffith, 1928). Over the ensuing 90 years, analysis of underlying mechanisms by classical genetic tools and monitoring of gene expression by transcriptional gene fusions and Northern blots gradually revealed intricate regulatory mechanisms that coordinate transient abilities to lyse nearby target cells and to take up and integrate genes released by that attack (reviewed by Fontaine et al., 2015; Straume et al., 2015; Shanker and Federle, 2017). Key elements include secreted “quorum sensing” peptide pheromone signals that coordinate competence development among cooperating cells (Håvarstein et al., 1995), and an alternative sigma factor (SigX) that responds to these signals to orchestrate expression of a large regulon of effector genes within cells (Lee and Morrison, 1999). Hybridization microarrays expanded the roster of genes apparently under control of these circuits (Rimini et al., 2000; Peterson et al., 2004; Vickerman et al., 2007), but their low resolution allowed some elements to be missed and others to be misinterpreted.
With the proliferation of high-throughput tools for parallel measurement of gene expression and the higher precision analysis offered by massive RNA sequencing methods, it has recently become possible to refine the understanding of competence regulation by identification of new actors and new linkages between known actors (Khan et al., 2016; Aprianto et al., 2018; Salvadori et al., 2018). Here we highlight several cases among streptococcal competence systems where new elements or new links are revealed or old interpretations corrected.
Mining the Hidden Gems
The distribution of competence among streptococcal species long remained obscure. After competence gene expression was found to depend on a peptide pheromone (CSP; Håvarstein et al., 1995) and a two-component signal transduction system (TCSTS) receptor for the peptide (ComDE; Håvarstein et al., 1996; Pestova et al., 1996), the restriction of orthologous quorum sensing (QS) circuits to the mitis and anginosus groups of streptococci raised doubts as to functional competence regulons in species of the other groups, among which reports of transformation were largely sporadic at best (Martin et al., 2006).
A completely different class of peptide-mediated QS regulation of competence genes was not discovered until the application of a new tool for high-throughput parallel determination of gene expression. Use of LC-MS quantitation of tryptic digests of hundreds of proteins to analyze gene expression changes in Streptococcus thermophilus when peptide import was blocked revealed an actively-regulated competence regulon in this species of the salivarius group, and led to discovery of control of sigX expression by an Rgg class receptor/peptide QS circuit mediated by a small peptide pheromone, named ComR/S (Fontaine et al., 2010; Fleuchot et al., 2011). Remarkably, all streptococcal species outside the mitis and anginosus groups regulate their SigX sigma factors through ComR/S circuits (Fontaine et al., 2013). Exceptionally, S. mutans engages both classes of QS circuits for competence regulation (Song et al., 2013; Shanker and Federle, 2017). Identification of characteristic consensus cis-acting regulatory site sequences for the three key competence regulators, ComE, SigX, and ComR, the “ComE-box” (aCAnTTcaG-12-aCAgTTgaG; Ween et al., 1999), the “SigX-box” (TACGAATA; Campbell et al., 1998), and the ComR-box (GACA-12-TGTC; Fontaine et al., 2013), facilitates the interpretation of high-resolution transcript maps.
After the discovery of the ComRS cell-to-cell signaling system in the salivarius group (Fontaine et al., 2010), an orthologous circuit was identified in S. mutans, where it regulates competence via XIP, a 7-amino-acid peptide pheromone derived from ComS (Mashburn-Warren et al., 2010; Khan et al., 2012). Under alternative conditions, competence in S. mutans can be stimulated either by an 18-aa CSP acting through a ComD receptor (Petersen et al., 2006; Hossain and Biswas, 2012) or by XIP, acting through a ComR receptor (Mashburn-Warren et al., 2010; Khan et al., 2012). A link between the two regulatory pathways remained elusive until 2015, when a binding site for SigX was identified upstream of comE, where it could link ComRS-regulated SigX to expression of ComED (aka BlpRH) pathway for control of bacteriocin production (Reck et al., 2015; Son et al., 2015; Khan et al., 2016). To uncover this new regulatory link, the authors initially combined the use of dual-reporter strains for assessment of population heterogeneity with RNA-seq data to determine gene expression in regulatory mutants (Reck et al., 2015). Additional studies arrived to the same conclusion through different approaches (Son et al., 2015; Khan et al., 2016).
In S. mutans, the presence of an intragenic SigX-box within the ORF SMU.60 came under scrutiny particularly because comR is immediately downstream of that gene and early microarray data had indicated an unexplained up-regulation of comR during competence (Perry et al., 2009; Okinaga et al., 2010; Dufour et al., 2011; Lemme et al., 2011). High-resolution RNA-Seq mapping of competence-associated transcripts at this locus revealed increased unidirectional RNA synthesis beginning not in the SMU.60-comR intergenic region but ~200 bp upstream, within the SMU.60 ORF and closely downstream of a candidate SigX-box. Nested deletions at the 3′ end of SMU.60 allowed the up-regulation of comR transcription only when this putative SigX-box remained intact. Three single-base mutations in the consensus SigX-box caused only silent mutations in SMU.60, but significantly decreased the induced comR transcription, showing that the presence of the SigX-box was essential for the up-regulation of comR at competence (Khan et al., 2017). This gem was discovered by coupling high-resolution RNA-Seq mapping to search for signature boxes upstream of the induced transcript, revealing an unexpected feedback loop for comR mediated by its regulatory target, SigX.
A series of studies of competence regulation in S. mutans culminated in identification of an open reading frame nested within the sigX gene that regulates genetic competence and oxidative stress tolerance in S. mutans (Kaspar et al., 2015). Investigating a link between oxidative stress tolerance, (p)ppGpp metabolism, and competence in the rcrRPQ operon, the authors observed that an rcrR polar (ΔrcrR-P) mutant was hypertransformable while a non-polar (ΔrcrR-NP) mutant did not transform. The expression levels of the sigX gene as a whole were 100-fold higher in both mutants compared to the WT (Seaton et al., 2011); however, the number of reads mapped to the 5′ region of sigX in the ΔrcrR-NP mutant were much less than in the ΔrcrR-P mutant. Conversely, in the central and 3′ regions of sigX, the expression pattern was reversed, revealing that a sense transcript located within the 3′ portion of sigX was highly expressed in the ΔrcrR-NP mutant. The level of full-length sigX transcript was actually reduced in the non-polar mutant, despite both transcripts being expressed from the same promoter. This internal transcript is translated from a second reading frame, yielding a 69-aa peptide product, termed sigX regulatory peptide A (xrpA). SigX production and transformation levels were restored when xrpA expression was disrupted by introduction of two silent mutations within sigX in the ΔrcrR-NP background. Furthermore, the target of XrpA has been traced directly to the binding of the ComR-XIP complex to the sigX promoter (Kaspar et al., 2018). The gem in this case was hidden in plain sight as a transcript internal to one of the most studied genetic components of natural transformation: SigX. However, it was revealed only when the power of RNA-seq quantitatively distinguished transcripts representing different regions of sigX (Kaspar et al., 2015).
The pneumococcal transcriptome was recently assessed by RNA-Seq in 22 different conditions relevant to infection, including competence for genetic transformation (Aprianto et al., 2018). Upon investigating genes with expression correlating with expression of comCDE, the authors identified a candidate that had not been noticed in two previous array-based studies of the competence regulons. Expression levels of this gene were strongly correlated with expression of comCDE across infection-relevant conditions, and directly repeated comE boxes in its promoter region indicated direct regulation by ComE. In addition, searches for double glycine leaders at the N-termini of pneumococcal proteins had identified 25 sequence-clusters within a predicted “secretome” including this same product (Cuevas et al., 2017). Interestingly, deletion of the candidate gene had no effect on transformation (Aprianto et al., 2018), but rather affected biofilm formation and nasopharyngeal colonization (Aggarwal et al., 2018; Aprianto et al., 2018); thus, it was termed Biofilm-regulating peptide induced by competence (BriC) and added to the list of genes under regulation in S. pneumoniae during competence (Cuevas et al., 2017; Aggarwal et al., 2018; Aprianto et al., 2018). This hidden gem was revealed by combining the power of large numbers of WGS for individual streptococcal species to identify conserved structures with the high-throughput of RNA-Seq to reveal correlated patterns of expression.
Avoiding the Fool's Gold
The early data on the transcriptional response during genetic competence were produced using strategies of microarray construction and analysis that were known to have technical limitations, including a lack of strand specificity, the use of probes restricted to annotated ORFs, the use of probes with low spatial resolution, and the use of cDNA preparation techniques that can result in false positives. In contrast, the current use of RNA-sequencing for transcriptome analyses enables improved genome coverage, a higher spatial resolution, strand specificity, analysis of intergenic or unannotated genomic regions, and provides improved mapping of candidate promoter sites (Vivancos et al., 2010; Mills et al., 2013; Zhao et al., 2015). During searches for hidden gems using early transcriptome approaches, it is not rare to encounter fool's gold. For instance, the gene SMU63c was reported as being upregulated during competence (Reck et al., 2015; Khan et al., 2016; Wenderska et al., 2017). However, when re-examined using directional RNA-seq (Khan et al., 2017), the apparent competence-associated transcription of SMU63.c was restricted to the anti-sense direction. In fact, transcription of SMU63c in the sense direction was actually down-regulated 35-fold.
Other false positives, arising through differential expression of genes located downstream of SigX-boxes but in the antisense direction (Khan et al., 2016), highlight the possibility of read-through transcripts without biological significance. Such findings result both from lack of strand-specific information, as detailed in the case above, and from pervasive transcription due to inefficient or missing transcription termination signals. Regarding the competence response particularly, pervasive transcription may be responsible for the upregulation of many so-called “non-core” genes of the competence response in various species (Khan et al., 2016), which comprise mostly genes with yet unknown functions. Although it represents a widespread phenomenon in bacteria (Wade and Grainger, 2014), we cannot rule out that some instances of pervasive transcription might also contribute to the optimization of the competent state. On the other hand, the lack of inter-species conservation of competence-specific pervasive transcripts in bacteria suggests that they lack such a functional role (Raghavan et al., 2012). In the case of competence operons, there may simply be little selective pressure favoring rigorous termination of transcripts that are perhaps made rarely and for a short time.
Finding the right conditions
Finding the right environmental and experimental conditions for competence has been the key to successful research throughout the history of discoveries on transformation, starting with numerous efforts to reproduce in vitro the observations by Griffith of transformation in mice (Griffith, 1928). Growth in the presence of anti-serum was one of the factors that seemed to do the trick (Bracco et al., 1957). However, higher and reproducible levels of competence were only achieved later on, with the discovery of the competence signaling peptide in streptococci of the mitis, anginosus, and mutans groups, and the possibility to use it in a synthetic form (Håvarstein et al., 1995; Li et al., 2001; Petersen et al., 2004).
A remarkable example of combining the identification of optimal growth conditions for competence with proteomics and transcriptomics to reveal a hidden gem came from studies with streptococci of the salivarius group. In this group, the use of a chemically defined medium without peptides, combined with proteomic (Gardan et al., 2009) and transcriptomic (Fontaine et al., 2010) approaches, revealed a new class of quorum sensing signaling that controls competence. The sequence for a Rgg regulator was located close to bacteriocin genes induced during competence, and it was followed by a putative pheromone gene. This was found to encode a competence signaling peptide recognized by the Rgg and internalized by an oligopeptide permease (Opp; Gardan et al., 2009; Fontaine et al., 2010, 2013). This discovery paved the way for the identification of similar systems in the pyogenic, bovis, suis, and mutans groups of streptococci (Mashburn-Warren et al., 2010, 2012; Fleuchot et al., 2011; Khan et al., 2012; Fontaine et al., 2013; Gardan et al., 2013; Morrison et al., 2013; Zaccaria et al., 2014). The reason why the function of this quorum sensing system was only apparent in peptide-free medium is not yet entirely clear, but peptides in the medium may compete with internalization of the competence signal by the oligopeptide permease.
Still, transformable strains and species can often show high differences in transformation levels even with the use of synthetic competence pheromones. This is illustrated by S. pneumoniae and S. mitis, which can show high levels of transformation efficiency in the laboratory (Hotchkiss, 1958; Ephrussi-Taylor et al., 1965; Lacks, 1966; Salvadori et al., 2016) in comparison to S. pyogenes, which rarely transforms despite seemingly possessing all the necessary genetic apparatus (Mashburn-Warren et al., 2012; Marks et al., 2014). Clues to these enigmas can most likely be found by studying the environmental cues surrounding each species. Indeed, a number of experimental conditions, e.g., temperature, pH, nutrient availability, are already known to be able to affect competence for natural transformation (Johnsborg and Håvarstein, 2009; Guo et al., 2014; Moreno-Gamez et al., 2017), and there are likely many more to be discovered. By mastering the use of powerful resources such as transcriptome mapping, we increase our chances to generate relevant and accurate data about these systems, which are thought to play an important role in bacterial adaptation and evolution.
Gems Awaiting Discovery
In the “mining” process to find real gems, important additional clues have been obtained by obtaining transcriptome maps that reflect different stages during competence development (Peterson et al., 2004; Vickerman et al., 2007; Lemme et al., 2011; Khan et al., 2016). Maps derived from samples with prolonged exposure to pheromone show that the expression of hundreds of genes can be affected in addition to the early and late classes of competence genes, but these are not conserved across the genus and do not seem to include any genes essential for transformation. On the other hand, reducing as much as possible the time point of evaluation following treatment with competence pheromones to obtain snapshots that reflect the sequential events following competence has been particularly revealing. This is illustrated by the fact that all genes identified as essential for transformation that are induced by the competence pheromones are either early or late genes. Comparison of transcriptome maps across streptococcal species using a S. mutans detailed map to filter away genes that apparently belong to “the fool's gold” has indeed led to the identification of a core set of 27 pan-streptococcal genes regulated by SigX two thirds of which have a defined function in transformation (Khan et al., 2016). The functions of the remaining third of the core regulon are not yet defined, but due to their conserved presence as part of the SigX regulon across species, these have a high potential to represent gems awaiting the right experimental conditions to reveal their roles.
Concluding Remarks and Future Perspectives
The contribution of these new “gems” to our understanding of natural genetic transformation in streptococci comes in different shapes and forms. Thus, the findings can highlight a novel ORF or gene that is found embedded in the competence cascade of activation or a novel pathway in the wiring of the competence system. Regardless, these insights together provide the possibility of a more accurate assemblage of the natural genetic transformation puzzle. In the case of S. mutans, these regulatory components and pathways recently revealed by high-resolution transcriptional profiling contributed significantly to our appreciation of how the system is wired (Figure 1).
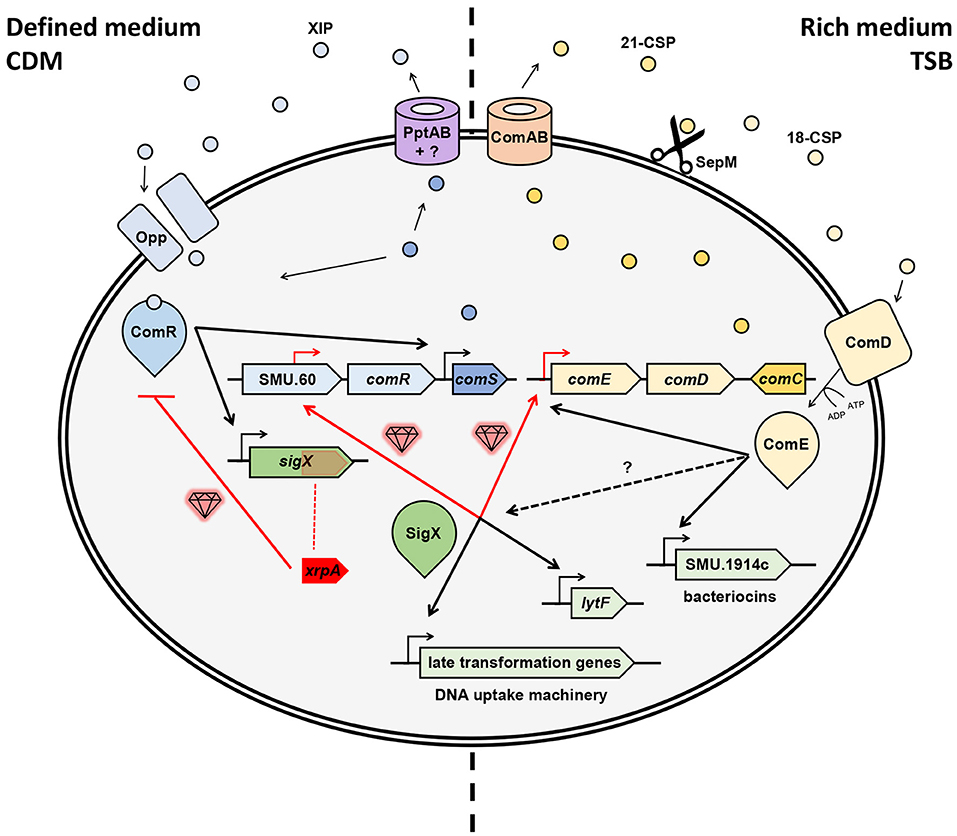
Figure 1. Hidden gems discovered in the regulation of natural genetic transformation in S. mutans. Competence can be triggered in chemically defined medium (CDM) by XIP or in rich medium (e.g., TSB) by CSP (aka MIP). Gems illustrated in red indicate novel pathways or elements. In CDM, ComS is exported by PptAB and another unknown element (Chang and Federle, 2016), after which the mature XIP is internalized the via Opp (Gardan et al., 2009), then binds to ComR to activates sigX and a comS positive feedback loop (Mashburn-Warren et al., 2010; Khan et al., 2012; Fontaine et al., 2013). A “hidden gem” was revealed here as XrpA, an internal peptide to sigX that hinders competence by inhibiting the ComR-XIP complex to bind the promoter region of sigX (Kaspar et al., 2015, 2018). Following, SigX, the master regulator of competence, activates transcription of late transformation genes involved in the DNA uptake machinery and also other genes such as lytF (Dufour and Levesque, 2013; Fontaine et al., 2013). Two new gems were found here as SigX recognizes SigX boxes upstream of ComED (Reck et al., 2015; aka BlpRH) and in the central part of SMU.60, upstream of ComR (Khan et al., 2016), and thus proximally regulates comED and comR. Recently, new evidence indicates that ComS can play a role as both an intercellular and an intracellular signal (Underhill et al., 2018). In rich medium, ComC (aka MIP) is exported by ComAB (aka NlmTE; Hui and Morrison, 1991; Hui et al., 1995; Hale et al., 2005) into 21-CSP, which is then cleaved by SepM (Hossain and Biswas, 2012) into its most active form 18-CSP (Petersen et al., 2006). Once it reaches the threshold, the mature pheromone activates ComD, a histidine kinase (HK) transmembrane receptor, which phosphorylates and, thus, activates the response regulator ComE (similarly as in S. pneumonia; Håvarstein et al., 1996; Pestova et al., 1996). Such activation up-regulates the early genes comABCDE, creating a positive feedback loop, and activates a variety of bacteriocin genes (Kreth et al., 2007). SigX can also be induced in this pathway through unknown mechanisms.
Building a more comprehensive overview of natural genetic transformation and its fine-tuning mechanisms in bacteria is highly warranted considering the relevance of this system in adaptation to different environments and stress response (reviewed by Johnston et al., 2014; Blokesch, 2016). An understanding of the conditions that facilitate or interrupt the development of transformation can aid to the overall comprehension of its role and evolution, in light of the tight evolutionary links between natural transformation and other adaptive responses seen in bacteria, such as the clustered regularly interspaced short palindromic repeats (CRISPRs; Jorth and Whiteley, 2012). We also gain a better insight how the process differs across species that are often inhabiting a similar (or even the same) niche, as it is the case with streptococci, and how this can play a role in their relationship with the host. This is of special relevance in light of the spread of virulence traits and antibiotic resistance demonstrated by a variety of emergent disease isolates that carry new segments of DNA that were likely acquired by HGT (Hiller et al., 2010; Frank et al., 2011; Gottig et al., 2015; Juhas, 2015; Cleary et al., 2016; Brynildsrud et al., 2018). More current than ever, the study of natural genetic transformation and other forms of HGT is a top priority in the global scenario.
Author Contributions
RJ, GS, TC, DM, and FP conceptualized the work, drafted, and critically revised the article. All authors approved the final version to be published.
Funding
This article was supported by the program international partnership for outstanding education, research, and innovation (INTPART) grant number 274867.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aggarwal, S. D., Eutsey, R., West-Roberts, J., Domenech, A., Xu, W., Abdullah, I. T., et al. (2018). Function of BriC peptide in the pneumococcal competence and virulence portfolio. PLoS Pathog. 14:e1007328. doi: 10.1371/journal.ppat.1007328
Aprianto, R., Slager, J., Holsappel, S., and Veening, J. W. (2018). High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res. 46, 9990–10006. doi: 10.1093/nar/gky750
Blokesch, M. (2016). Natural competence for transformation. Curr. Biol. 26, R1126–R1130. doi: 10.1016/j.cub.2016.08.058
Bracco, R. M., Krauss, M. R., Roe, A. S., and Macleod, C. M. (1957). Transformation reactions between Pneumococcus and three strains of Streptococci. J. Exp. Med. 106, 247–259. doi: 10.1084/jem.106.2.247
Brynildsrud, O. B., Eldholm, V., Bohlin, J., Uadiale, K., Obaro, S., and Caugant, D. A. (2018). Acquisition of virulence genes by a carrier strain gave rise to the ongoing epidemics of meningococcal disease in West Africa. Proc. Natl. Acad. Sci. U.S.A. 115, 5510–5515. doi: 10.1073/pnas.1802298115
Campbell, E. A., Choi, S. Y., and Masure, H. R. (1998). A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27, 929–939. doi: 10.1046/j.1365-2958.1998.00737.x
Chang, J. C., and Federle, M. J. (2016). PptAB exports Rgg quorum-sensing peptides in Streptococcus. PLoS ONE 11:e0168461. doi: 10.1371/journal.pone.0168461
Cleary, D. W., Devine, V. T., Jefferies, J. M., Webb, J. S., Bentley, S. D., Gladstone, R. A., et al. (2016). Comparative genomics of carriage and disease isolates of Streptococcus pneumoniae serotype 22F reveals lineage-specific divergence and niche adaptation. Genome Biol. Evol. 8, 1243–1251. doi: 10.1093/gbe/evw066
Cuevas, R. A., Eutsey, R., Kadam, A., West-Roberts, J. A., Woolford, C. A., Mitchell, A. P., et al. (2017). A novel streptococcal cell-cell communication peptide promotes pneumococcal virulence and biofilm formation. Mol. Microbiol. 105, 554–571. doi: 10.1111/mmi.13721
Dufour, D., Cordova, M., Cvitkovitch, D. G., and Levesque, C. M. (2011). Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J. Bacteriol. 193, 6552–6559. doi: 10.1128/JB.05968-11
Dufour, D., and Levesque, C. M. (2013). Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J. Bacteriol. 195, 105–114. doi: 10.1128/JB.00926-12
Ephrussi-Taylor, H., Sicard, A. M., and Kamen, R. (1965). Genetic recombination in DNA-induced transformation of Pneumococcus. I. the problem of relative efficiency of transforming factors. Genetics 51, 455–475.
Fleuchot, B., Gitton, C., Guillot, A., Vidic, J., Nicolas, P., Besset, C., et al. (2011). Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80, 1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x
Fontaine, L., Boutry, C., de Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2010). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Fontaine, L., Goffin, P., Dubout, H., Delplace, B., Baulard, A., Lecat-Guillet, N., et al. (2013). Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87, 1113–1132. doi: 10.1111/mmi.12157
Fontaine, L., Wahl, A., Flechard, M., Mignolet, J., and Hols, P. (2015). Regulation of competence for natural transformation in streptococci. Infect. Genet. Evol. 33, 343–360. doi: 10.1016/j.meegid.2014.09.010
Frank, C., Werber, D., Cramer, J. P., Askar, M., Faber, M., an der Heiden, M., et al. (2011). Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 365, 1771–1780. doi: 10.1056/NEJMoa1106483
Gardan, R., Besset, C., Gitton, C., Guillot, A., Fontaine, L., Hols, P., et al. (2013). Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J. Bacteriol. 195, 1845–1855. doi: 10.1128/JB.02196-12
Gardan, R., Besset, C., Guillot, A., Gitton, C., and Monnet, V. (2009). The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191, 4647–4655. doi: 10.1128/JB.00257-09
Gottig, S., Gruber, T. M., Stecher, B., Wichelhaus, T. A., and Kempf, V. A. (2015). In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin. Infect. Dis. 60, 1808–1815. doi: 10.1093/cid/civ191
Griffith, F. (1928). The significance of Pneumococcal types. J. Hyg. 27, 113–159. doi: 10.1017/S0022172400031879
Guo, Q., Ahn, S. J., Kaspar, J., Zhou, X., and Burne, R. A. (2014). Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J. Bacteriol. 196, 227–236. doi: 10.1128/JB.00995-13
Håvarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Håvarstein, L. S., Gaustad, P., Nes, I. F., and Morrison, D. A. (1996). Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21, 863–869. doi: 10.1046/j.1365-2958.1996.521416.x
Hale, J. D., Heng, N. C., Jack, R. W., and Tagg, J. R. (2005). Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J. Bacteriol. 187, 5036–5039. doi: 10.1128/JB.187.14.5036-5039.2005
Hiller, N. L., Ahmed, A., Powell, E., Martin, D. P., Eutsey, R., Earl, J., et al. (2010). Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6:e1001108. doi: 10.1371/journal.ppat.1001108
Hossain, M. S., and Biswas, I. (2012). An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J. Bacteriol. 194, 5886–5896. doi: 10.1128/JB.01381-12
Hotchkiss, R. D. (1958). Size limitations governing the incorporation of genetic material in the bacterial transformations and other non-reciprocal recombinations. Symp. Soc. Exp. Biol. 12, 49–59.
Hui, F. M., and Morrison, D. A. (1991). Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173, 372–381. doi: 10.1128/jb.173.1.372-381.1991
Hui, F. M., Zhou, L., and Morrison, D. A. (1995). Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153, 25–31. doi: 10.1016/0378-1119(94)00841-F
Johnsborg, O., and Håvarstein, L. S. (2009). Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 33, 627–642. doi: 10.1111/j.1574-6976.2009.00167.x
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Jorth, P., and Whiteley, M. (2012). An evolutionary link between natural transformation and CRISPR adaptive immunity. mBio 3:5. doi: 10.1128/mBio.00309-12
Juhas, M. (2015). Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 41, 101–108. doi: 10.3109/1040841X.2013.804031
Kaspar, J., Ahn, S. J., Palmer, S. R., Choi, S. C., Stanhope, M. J., and Burne, R. A. (2015). A unique open reading frame within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance. Mol. Microbiol. 96, 463–482. doi: 10.1111/mmi.12948
Kaspar, J., Shields, R. C., and Burne, R. A. (2018). Competence inhibition by the XrpA peptide encoded within the comX gene of Streptococcus mutans. Mol. Microbiol. 109, 345–364. doi: 10.1111/mmi.13989
Khan, R., Junges, R., Amdal, H. A., Chen, T., Morrison, D. A., and Petersen, F. C. (2017). A positive feedback loop mediated by Sigma X enhances expression of the streptococcal regulator ComR. Sci. Rep. 7:5984. doi: 10.1038/s41598-017-04768-5
Khan, R., Rukke, H. V., Hovik, H., Amdal, H. A., Chen, T., Morrison, D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:2. doi: 10.1128/mSystems.00038-15
Khan, R., Rukke, H. V., Ricomini Filho, A. P., Fimland, G., Arntzen, M. O., Thiede, B., et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194, 3781–3788. doi: 10.1128/JB.00624-12
Kreth, J., Hung, D. C., Merritt, J., Perry, J., Zhu, L., Goodman, S. D., et al. (2007). The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153(Pt 6), 1799–1807. doi: 10.1099/mic.0.2007/005975-0
Lacks, S. (1966). Integration efficiency and genetic recombination in pneumococcal transformation. Genetics 53, 207–235.
Lee, M. S., and Morrison, D. A. (1999). Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181, 5004–5016.
Lemme, A., Grobe, L., Reck, M., Tomasch, J., and Wagner-Dobler, I. (2011). Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193, 1863–1877. doi: 10.1128/JB.01363-10
Li, Y. H., Lau, P. C., Lee, J. H., Ellen, R. P., and Cvitkovitch, D. G. (2001). Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183, 897–908. doi: 10.1128/JB.183.3.897-908.2001
Marks, L. R., Mashburn-Warren, L., Federle, M. J., and Håkansson, A. P. (2014). Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J. Infect. Dis. 210, 25–34. doi: 10.1093/infdis/jiu058
Martin, B., Quentin, Y., Fichant, G., and Claverys, J. P. (2006). Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14, 339–345. doi: 10.1016/j.tim.2006.06.007
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78, 589–606. doi: 10.1111/j.1365-2958.2010.07361.x
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2012). The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 194, 4589–4600. doi: 10.1128/JB.00830-12
Mills, J. D., Kawahara, Y., and Janitz, M. (2013). Strand-specific RNA-seq provides greater resolution of transcriptome profiling. Curr. Genomics 14, 173–181. doi: 10.2174/1389202911314030003
Moreno-Gamez, S., Sorg, R. A., Domenech, A., Kjos, M., Weissing, F. J., van Doorn, G. S., et al. (2017). Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun. 8:854. doi: 10.1038/s41467-017-00903-y
Morrison, D. A., Guedon, E., and Renault, P. (2013). Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus. J. Bacteriol. 195, 2612–2620. doi: 10.1128/JB.00230-13
Okinaga, T., Xie, Z., Niu, G., Qi, F., and Merritt, J. (2010). Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol. Oral Microbiol. 25, 165–177. doi: 10.1111/j.2041-1014.2010.00574.x
Perry, J. A., Cvitkovitch, D. G., and Levesque, C. M. (2009). Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol. Lett. 299, 261–266. doi: 10.1111/j.1574-6968.2009.01758.x
Pestova, E. V., Håvarstein, L. S., and Morrison, D. A. (1996). Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21, 853–862. doi: 10.1046/j.1365-2958.1996.501417.x
Petersen, F. C., Fimland, G., and Scheie, A. A. (2006). Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 61, 1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x
Petersen, F. C., Pecharki, D., and Scheie, A. A. (2004). Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J. Bacteriol. 186, 6327–6331. doi: 10.1128/JB.186.18.6327-6331.2004
Peterson, S. N., Sung, C. K., Cline, R., Desai, B. V., Snesrud, E. C., Luo, P., et al. (2004). Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51, 1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x
Raghavan, R., Sloan, D. B., and Ochman, H. (2012). Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio 3:4. doi: 10.1128/mBio.00156-12
Reck, M., Tomasch, J., and Wagner-Dobler, I. (2015). The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet. 11:e1005353. doi: 10.1371/journal.pgen.1005353
Rimini, R., Jansson, B., Feger, G., Roberts, T. C., de Francesco, M., Gozzi, A., et al. (2000). Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36, 1279–1292. doi: 10.1046/j.1365-2958.2000.01931.x
Salvadori, G., Junges, R., Åmdal, H. A., Chen, T., Morrison, D. A., and Petersen, F. C. (2018). High-resolution profiles of the Streptococcus mitis CSP signaling pathway reveal core and strain-specific regulated genes. BMC Genomics 19:453. doi: 10.1186/s12864-018-4802-y
Salvadori, G., Junges, R., Morrison, D. A., and Petersen, F. C. (2016). Overcoming the barrier of low efficiency during genetic transformation of Streptococcus mitis. Front. Microbiol. 7:1009. doi: 10.3389/fmicb.2016.01009
Seaton, K., Ahn, S. J., Sagstetter, A. M., and Burne, R. A. (2011). A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J. Bacteriol. 193, 862–874. doi: 10.1128/JB.01257-10
Shanker, E., and Federle, M. J. (2017). Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 8:1. doi: 10.3390/genes8010015
Son, M., Shields, R. C., Ahn, S. J., Burne, R. A., and Hagen, S. J. (2015). Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol. Lett. 362:19. doi: 10.1093/femsle/fnv159
Song, L., Wang, W., Conrads, G., Rheinberg, A., Sztajer, H., Reck, M., et al. (2013). Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics 14:430. doi: 10.1186/1471-2164-14-430
Straume, D., Stamsås, G. A., and Håvarstein, L. S. (2015). Natural transformation and genome evolution in Streptococcus pneumoniae. Infect. Genet. Evol. 33, 371–380. doi: 10.1016/j.meegid.2014.10.020
Underhill, S. A. M., Shields, R. C., Kaspar, J., Haider, M., Burne, R. A., and Hagen, S. J. (2018). Intracellular signaling through the comRS system in Streptococcus mutans genetic competence. mSphere. doi: 10.1128/mSphere.00444-18
Vickerman, M. M., Iobst, S., Jesionowski, A. M., and Gill, S. R. (2007). Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 189, 7799–7807. doi: 10.1128/JB.01023-07
Vivancos, A. P., Guell, M., Dohm, J. C., Serrano, L., and Himmelbauer, H. (2010). Strand-specific deep sequencing of the transcriptome. Genome Res. 20, 989–999. doi: 10.1101/gr.094318.109
Wade, J. T., and Grainger, D. C. (2014). Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653. doi: 10.1038/nrmicro3316
Ween, O., Gaustad, P., and Håvarstein, L. S. (1999). Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33, 817–827. doi: 10.1046/j.1365-2958.1999.01528.x
Wenderska, I. B., Latos, A., Pruitt, B., Palmer, S., Spatafora, G., Senadheera, D. B., et al. (2017). Transcriptional profiling of the oral pathogen Streptococcus mutans in response to competence signaling peptide XIP. mSystems 2:1. doi: 10.1128/mSystems.00102-16
Zaccaria, E., van Baarlen, P., de Greeff, A., Morrison, D. A., Smith, H., and Wells, J. M. (2014). Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS ONE 9:e99394. doi: 10.1371/journal.pone.0099394
Keywords: pheromone, competence, natural transformation, mutagenesis, oral cavity, CSP, streptococcus
Citation: Junges R, Salvadori G, Chen T, Morrison DA and Petersen FC (2019) Hidden Gems in the Transcriptome Maps of Competent Streptococci. Front. Mol. Biosci. 5:116. doi: 10.3389/fmolb.2018.00116
Received: 28 October 2018; Accepted: 11 December 2018;
Published: 04 January 2019.
Edited by:
Kurt Henry Piepenbrink, University of Nebraska-Lincoln, United StatesReviewed by:
Adam Blanchard, Nottingham Trent University, United KingdomM. Gabriela Kramer, Centro de Investigaciones Biomédicas (CEINBIO), Uruguay
Copyright © 2019 Junges, Salvadori, Chen, Morrison and Petersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernanda C. Petersen, Zi5jLnBldGVyc2VuQG9kb250LnVpby5ubw==
 Roger Junges
Roger Junges Gabriela Salvadori
Gabriela Salvadori Tsute Chen
Tsute Chen Donald A. Morrison
Donald A. Morrison Fernanda C. Petersen
Fernanda C. Petersen