- 1American University of Beirut, Center for Infectious Diseases Research (CIDR) and WHO Collaborating Center for Reference and Research on Bacterial Pathogens, Beirut, Lebanon
- 2Department of Pathology and Laboratory Medicine, American University of Beirut Medical Center, Beirut, Lebanon
- 3Department of Microbiology, Faculty of Medicine and University Hospital in Plzen, Charles University, Plzeň, Czechia
- 4Department of Pediatrics and Adolescent Medicine, Faculty of Medicine, American University of Beirut, Beirut, Lebanon
Background: The globally emerging Candida auris pathogens poses heavy burden to the healthcare system. Their molecular analyses assist in understanding their epidemiology, dissemination, treatment, and control. This study was warranted to describe the genomic features and drug resistance profiles using whole genome sequencing (WGS) among C. auris isolates from Lebanon.
Methods: A total of 28 C. auris clinical isolates, from different hospital units, were phenotypically identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and tested for antifungal resistance using Vitek-2 system and E test. The complete genomes were determined by WGS using long reads sequencing (PacBio) to reveal the clade distribution and antifungal resistance genes.
Results: Candida auris revealed uniform resistance to fluconazole and amphotericin B, with full susceptibility to echinocandins. Among key resistance genes studied, only two mutations were detected: Y132F in ERG11 gene and a novel mutation, D709E, found in CDR1 gene encoding for an ABC efflux pump. Phylogenetically, C. auris genomes belonged to South Asian clade I and showed limited genetic diversity, suggesting person to person transmission.
Conclusion: This characterization of C. auris isolates from Lebanon revealed the exclusivity of clade I lineage together with uniform resistance to fluconazole and amphotericin B. The control of such highly resistant pathogen necessitates an appropriate and rapid recovery and identification to contain spread and outbreaks.
Introduction
Candida auris has been an emerging fungal infection, characterized by high transmissibility, multidrug resistance, and poor outcomes. As such, it is posing serious nosocomial health concerns globally (Forsberg et al., 2019). Its potency in colonizing patients’ skin enables patient-to-patient spread, causing outbreaks in healthcare settings (Schelenz et al., 2016; Lockhart et al., 2017).
The recognition and identification of C. auris is challenging, as the isolates of this yeast can be misidentified using commonly phenotypic laboratory methods. However, its speciation can be determined by automated systems such as the matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and Vitek System (Rychert et al., 2018; Patel, 2019). Molecular methods based on sequencing of the D1–D2 region of the 28s rDNA or internal transcribed spacer region provide reliable confirmation of species identification (Schoch et al., 2012; Cernakova et al., 2021). Moreover, whole-genome sequencing (WGS)-based methods have increasingly been used to detect and characterize phylogeographic types and transmission dynamics for this emerging pathogen (Lockhart et al., 2017). Since its first recognition in Japan in 2009, genetically divergent lineages have been globally identified and stratified geographically into four main clades: clade I (Southern Asia), clade II (Eastern Asia), clade III (Africa), and clade IV (South America) (Chow et al., 2020). A potentially clade V has been identified in an isolate from a patient in Iran showing a difference of >200,000 single-nucleotide polymorphisms from the other clades (Chow et al., 2019).
Several studies have revealed the genetic profiles of recovered C. auris isolates from different countries worldwide (Lockhart et al., 2017; Forsberg et al., 2019; Chow et al., 2020), including few reports from different countries in our MENA region (Al Maani et al., 2019; Alfouzan et al., 2019, 2020; Almaghrabi et al., 2020; Salah et al., 2021). In Lebanon, however, only two clinical studies were reported from the American University of Beirut Medical Center (AUBMC): the first addressed the profile of C. auris infection among 14 infected cases (Allaw et al., 2021). The second was a case report on C. duobushaemulonii associated with coronavirus disease 2019 (COVID-19) disease (Awada et al., 2021). Thus, in the absence of any characterization of C. auris isolates in Lebanon, this study was warranted to describe the genomic features, genetic relationships, and drug resistance profiles using WGS among C. auris isolates recovered at a major tertiary care center in this country.
Materials and Methods
Candida auris Isolates Collection
Candida auris isolates analyzed in this study were those recovered from patient specimens submitted for fungal investigation (prior to commencing patients’ therapy) at AUBMC Clinical Microbiology Laboratory (CML), accredited by the College of American Pathologists since 2004.
A total of 28 isolates were retrieved from 21 patients by plating the specimens on Sabouraud dextrose agar (SAB) medium and incubated them at 37°C. The specimen source of these isolates were as follows: 4 isolates from peripheral blood, 1 from central line catheter, 12 from deep tracheal aspirate (DTA), 8 from urine, 2 from skin screen, and 1 from bronchioalveolar lavage (BAL).
In six patients, isolates were simultaneously recovered from different specimen sources of the same patient: blood and central line catheter from 1 patient; DTA and urine from 2 patients; DTA and skin screening from 1 patient; DTA, skin screening, and urine from 1 patient; and DTA and blood from 1 patient.
Identification and Speciation of Candida auris Isolates
The recovered Candida species on SAB medium was submitted directly for identification or subcultured on chocolate agar medium prior to identification. The colonies of these isolates were identified by MALDI-TOF system (Bruker Daltonik, GmbH, Bremen, Germany) and by the Vitek 2 system (BioMérieux, Marcy L’Etoile, France). Species identities were confirmed by PCR following by sequencing using ITS1F/ITS4R primers for the variable internal transcribed spacers ITS1 and ITS2 regions, located between universally conserved genes 18S, 5.8S, and 28S and NL1F/NL2R primers used to detect the D1–D2 region located at the 5′ end of the gene 28S encoding for the large nuclear ribosomal subunit (Schoch et al., 2012).
Antifungal Susceptibility Testing
The Vitek 2 system, employing the antifungal susceptibility cards (AST-YS 08), was used to determine the minimum inhibitory concentrations (MICs) of the following antifungal agents: fluconazole, voriconazole, caspofungin, micafungin, amphotericin B, and flucytosine. The E-test (AB Biodisk, Solna, Sweden) was used to determine the MICs (μg/ml) of itraconazole (strip concentration range, 0.002–32 μg/ml), using Roswell Park Memorial Institute (RPMI) 1640 media (Sigma, St. Louis, MO, United States), according to what was reported earlier from our laboratory (Araj et al., 2015).
The interpretation of the minimum inhibitory concentrations (MICs) susceptibility breakpoints (μg/ml) for C. auris were based on CDC1 (accessed July 7, 2021) and Clinical and Laboratory Standards Institute (CLSI) guidelines, essentially defined based on those established for closely related Candida species (Candida haemuloni) and on expert opinion. In this context, the designated resistant breakpoints are as follows: fluconazole, ≥32 μg/ml; anidulafungin, ≥4 μg/ml; caspofungin, ≥2 μg/ml; micafungin, ≥4 μg/ml; amphotericin B, ≥2 μg/ml. Voriconazole susceptibility breakpoints are not applicable and recommended to consider using fluconazole susceptibility as a surrogate susceptibility assessment. There is no published guidance about flucytosine’s breakpoint susceptibility.
Quality Control Isolates
The quality of test performance was controlled by including the reference strains C. albicans (ATCC 10231), C. parapsilosis (ATCC 22019), and C. kruseii (ATCC 6258).
Whole Genome Sequencing
The genomic DNA of 29 isolates (28 C. auris isolates and one C. haemuloni as outgroup) was extracted using the NucleoSpin microbial DNA kit (Macherey-Nagel, Duren, Germany). Subsequently, the extracted DNA was sheared to obtain 15-kb fragments using Hydropore long on the Megaruptor 2 (Diagnode). Express kit 2.0 (Pacific Biosciences, Menlo Park, CA, United States) was used for library preparation using the microbial multiplexing protocol according to the manufacturer’s recommendations. Library size selection was applied using the AMPure PB beads (Pacific Biosciences, Menlo Park, CA, United States) to select for fragments above 3 kb. Using the SMRT Link v9.0, HGAP4 and microbial assembly pipelines were used to assemble the sequences with a minimum seed coverage of 30, 40, and 50 depending on the coverage. The assemblies of CA3LBN and CA7LBN were annotated; gene prediction was achieved using BRAKER2 v2.1.6 pipeline on fungus mode, which combines GeneMarK-ES v4.65 and AUGUSTUS 3.4.0 for fungal gene prediction and identifying gene locations with the corresponding CDS and messenger RNA (mRNA) qualifiers (Altschul et al., 1990; Stanke et al., 2006, 2008; Ter-Hovhannisyan et al., 2008; Camacho et al., 2009; Hoff et al., 2016, 2019; Bruna et al., 2021). Interproscan 5.50-84.0 (Jones et al., 2014) was used on the COG database to create the xml file to further incorporate in the functional annotation pipeline created by the Funannotate 1.8.7 (Blachowicz et al., 2019; Vasquez-Gross et al., 2020; Smith, 2021). The pipeline starts by running HMMscan (HMMer v3.3) (hmmer.org) with default parameters on the PFAM database, then using emapper 2.1.2 based on eggnog orthology data (Huerta-Cepas et al., 2017, 2019). Sequence searches were performed using Diamond Blastp (Buchfink et al., 2015) on UniProt DB version 2021_02 and MEROPS v12.0; the resulting annotations were combined using Gene2Product v1.69, and later, Signalp 5.0 was used to predict secreted proteins (Almagro Armenteros et al., 2019). Furthermore, transfer RNA (tRNA) identification was done using ARAGORN v1.2.41 (Laslett and Canback, 2004); tRNAs identified and found to be overlapping with any CDS sequences were removed. The ribosomal RNA (rRNA) identification was done by downloading C. auris rRNA sequences from the National Center for Biotechnology Information (NCBI) and then by using BLAST + v2.11.0 (Camacho et al., 2009). Assemblies and annotations were assessed using BUSCO V5.2.2 (Manni et al., 2021).
Phylogeny
The corresponding phylogenies of the 29 genomes of this study, along with all of the 80 C. auris sequences found in the NCBI assembly database, were performed. Briefly, the alignment of the core genome, detection of recombination events, and single nucleotide polymorphisms (SNPs) detection were performed using Parsnp v1.2, available in the Harvest suite (Treangen et al., 2014) using CA7LBN (index case), a reference genome for clustering and using CA3LBN (identified as C. haemuloni) as an outgroup as described elsewhere (Prakash et al., 2016). SNPs identified in local collinear blocks were subsequently used for reconstructing an approximate maximum-likelihood tree using FastTree2 (Price et al., 2010) while including the general time reversible (GTR) model of nucleotide substitution. The Shimodaira–Hasegawa test was used to assess the support for significant clustering in the observed phylogeny. The interactive tree of life or iTOL (Letunic and Bork, 2019) was used to annotate, modify, and edit the resulting phylogeny.
Single Nucleotide Polymorphisms Detection
In this study, SNPs of the 27 C. auris genomes were compared to the SNPs of the first C. auris (CA7LBN detected in October 2020) by using snippy multicommand (snippy-base application v4.5.0) (Seemann, 2015) that generates a core genome multiple alignment against a common reference. The CA7LBN was used as a reference, since it was the index case. The pipeline detects the variants and generates single file for each isolate listing the different variations. The results were compared to detect any possible microevolution events among the genomes with respect to the time of detection.
Molecular Detection of Antifungal Resistance Genes Mutations
The sequences of CDR1, CDR2, ERG1, ERG2, ERG3, ERG5, ERG6, ERG11, ERG24, MDR1, MRR1, TAC1, and UPC2 were extracted from the genome of C. auris B11221 and were used as reference. C. auris B11221 was chosen, since it is susceptible against fluconazole and amphotericin B. Then, the reference genes were blasted against the generated assemblies from the study, and the corresponding genes were detected using BLAST + v2.11.0 (Camacho et al., 2009). Protein products were then compared, and the corresponding amino acid substitutions were detected. All mutations have been confirmed by visually inspecting the alignment of the CCS reads to the assembly.
Data Availability
All genome assemblies have been deposited at GenBank under the following accession numbers: CP077052–CP077058, CP077045–CP077051, CP076661–CP076667, CP077038–CP07 7044, CP077031–CP077037, CP077024–CP077030, CP076749–CP076755, CP077017–CP077023, CP077010–CP077016, CP07 7003–CP077009, CP076996–CP077002, CP076989–CP076995, CP076982–CP076988, CP076975–CP076981, CP076968–CP07 6974, CP076961–CP076967, CP076954–CP076960, CP076947–CP076953, CP076940–CP076946, CP076933–CP076939, CP07 6926–CP076932, CP076919–CP076925, CP076912–CP076918, CP076905–CP076911, CP076898–CP076904, CP076891–CP07 6897, CP076884–CP076890, CP076877–CP076883, and CP076 870–CP076876. We annotated two isolates (the index and the outgroup) with the following ascension codes CP076661–CP076667 and CP076749–CP076755 corresponding to CA3LBN and CA7LBN, respectively (Supplementary Table 1).
Results
Patients Demographics
The gender distribution among the 21 patients showed 12 male (57%) and 9 female (43%). The average and range of age for the male and female patients were 71 years (range, 57–80 years) and 63 years (range, 34–82 years), respectively. The distribution of patients based on their clinical hospital location were nine in intensive care unit (ICU), two in respiratory care unit (RCU), four in coronary care unit (CCU), one in neuro ICU (NICU), one in emergency department (ED), and four in medical floor.
Antifungal Susceptibility Results of Candida auris
The MIC50/MIC90 and range of MICs (μg/ml) for each of the tested antifungal agents against the C. auris isolates are shown in Table 1 as follows: itraconazole, 0.25/1 (range, 0.19–1); fluconazole, ≥32/≥32 (16–32); voriconazole, 0.25/0.25 (range, 0.12–4); caspofungin, 0.25/0.25 (range, 0.25–0.25); micafungin 0.12/0.12 (range, 0.064–0.12); amphotericin B, 8/8 (range, 2–16); flucytosine 1/1 (range, 1–1).
Among the triazole class drugs, C. auris was considered to be uniformly resistant to fluconazole, although only 54% of the isolates showed clear resistance to fluconazole (MICs ≥ 32 μg/ml), while 46% of these isolates revealed a level close to the resistance values, an MIC of 16 μg/ml. The voriconazole rates of susceptible, intermediate, and resistant C. auris were 36, 61, and 3%, respectively, and that against itraconazole (n = 8 isolates tested) were 0, 75, and 25%, respectively.
Concerning the polyene class, the specified breakpoints of resistance (MICs ≥ 2 μg/ml) for amphotericin B was detected in 100% of the tested isolates. Extrapolating the breakpoint susceptibility reported for C. auris isolates by CDC, our results indicated uniform susceptibility (100%, MICs ≤ 4 μg/ml) of these isolates against the tested echinocandin class drugs (micafungin and caspofungin) (Table 1).
Mutations of Drug-Resistance-Associated Genes
All strains expressed fluconazole and amphotericin B resistant phenotypes. Corresponding mutations in drug-resistance-associated genes, namely, CDR1, CDR2, ERG1, ERG2, ERG3, ERG5, ERG6, ERG11, ERG24, MDR1, MRR1, TAC1, and UPC2, were investigated in comparison with the reference genes in B11221 strain. Two mutations were detected: the first one in the lanosterol 14-α-demethylase-encoding gene ERG11 (Y132F) particularly in the first “hot-spot” region located between amino-acids (105–165) and another novel mutation D709E, found in CDR1 encoding for an ABC efflux pump.
Phylogenetic Studies and Single Nucleotide Polymorphism Analysis
The 29 genomes generated in our study, with CA3LBN (C. Haemulonii isolate) being the outgroup, were compared to all C. auris genomes available in the NCBI database (80 genomes). The results indicated that our isolates belonged to clade I (South Asian) among the five known global clades and closely clustered with the branch length showing zero differences between our isolates and the ones from United States, Germany, and India and located in close proximity to C. auris isolates recovered from Saudi Arabia, United Arab Emirates, and Hong Kong (Figure 1).
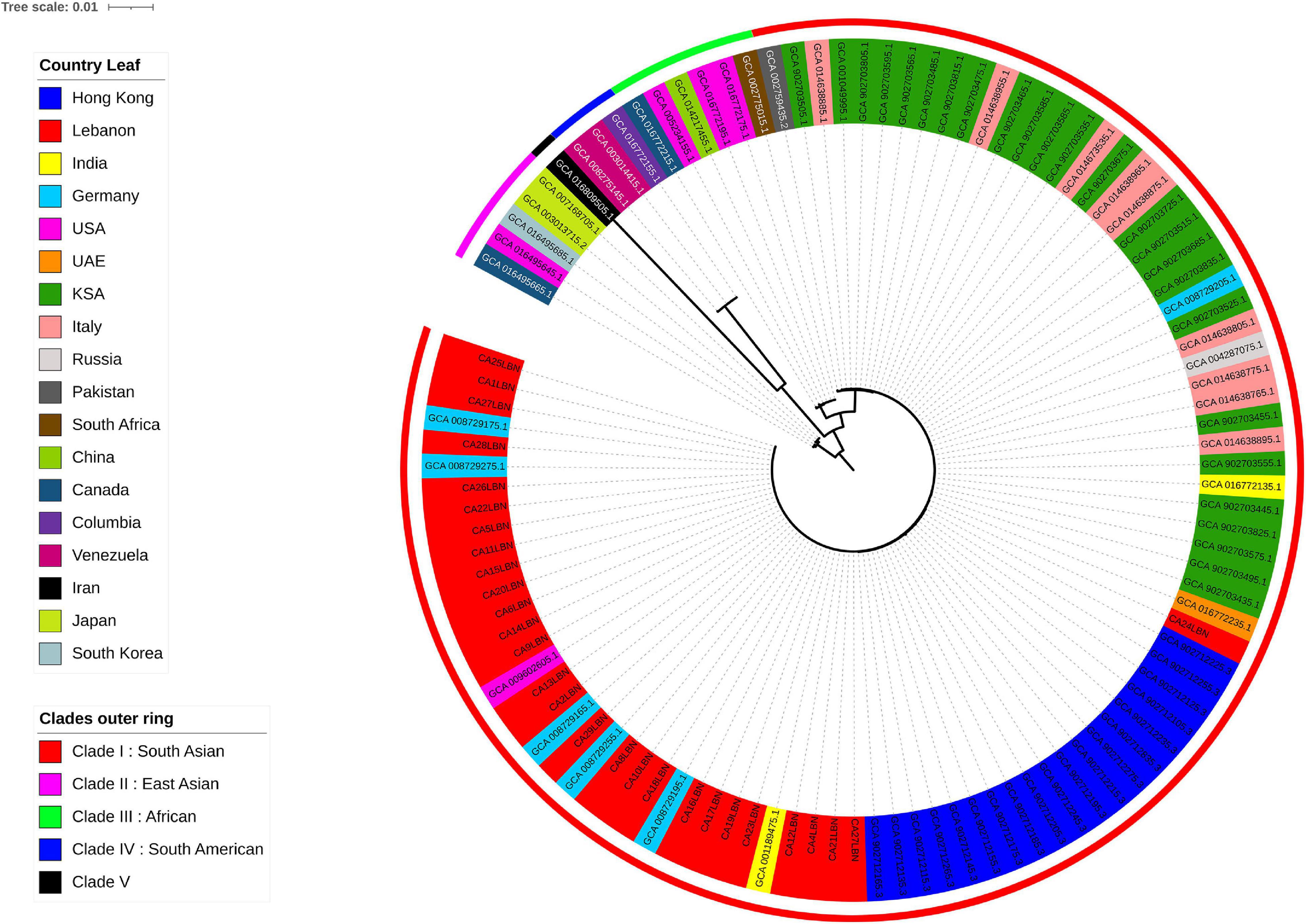
Figure 1. Phylogenetic tree of C. auris genomes, including 28 from the present study and 80 publicly available from the NCBI database. The outer most perimeter represents the five clades geographic distribution: clade I (red), clade II (purple), clade III (green), clade IV (electric blue), and clade V (black). The leaf labels indicate the strains’ ID and colored according to the geographic origin of each strain. Our 28 isolates from Lebanon fall in clade I.
The genetic variations and microevolution among the 28 C. auris isolates were analyzed and compared to the first isolate detected in October 2020 (CA7LBN). Results revealed a range of one to six SNPs with one SNP in three isolates, two SNPs in five isolates, three SNPs in seven, four SNPs in four, five SNPs in six, and six SNPs in two isolates (Supplementary Table 1). SNPs were detected in coding and non-coding regions in the studied isolates. All isolates showed a mutation in THR1, a gene encoding for trihydroxynaphthalene reductase, with an amino acid substitution L188G. In addition, five other mutations were detected in different isolates as shown in Supplementary Table 2. The rest of the mutations were either in genes coding for uncharacterized proteins (hypothetical proteins) or in non-coding regions.
Discussion
Candida auris infection has been tolling the healthcare systems at major hospitals globally due to its serious threatening impact and its resistance to multiple antifungal agents that limit treatment options. In Lebanon, this is the first study addressing the antifungal profile and molecular features of C. auris isolates using the long reads sequencing technique that generated complete genome sequences of these pathogens.
Identification of C. auris is generally a challenging experience. Interestingly, the first suspected encounter of C. auris happened when yeast colonies were recovered from the blood culture of a 34-year-old Lebanese man who works and lives in Africa (Sierra Leone) and was transferred to AUBMC for management of confirmed COVID-19 severe infection. Identification (ID) by Vitek 2 revealed C. auris (96% probability, Bionumber 4110145251301771). Repeat identification on Vitek 2 (August 12, 2020) revealed low discrimination organism (C. auris and C. duobushaemuloins; Bionumber, 4110145255321771). Unfortunately, our MALDI-TOF was affected by severe tremors of the devastating explosion of the port of Beirut (on August 4, 2020) and could not reveal any identification. Thus, discrimination between these species warranted further testing (molecular, MALDI-TOF, WGS), which was kindly extended by colleagues from Lebanon, United States, and Canada (thanked under acknowledgment), whereby all of them revealed the identification as C. duobushaemulonii (Awada et al., 2021). This reflected the difficulties in the proper identification of C. auris (Iguchi et al., 2019). Subsequently, the recovered C. auris under study were identified by the MALDI-TOF and WGS.
Concerning the phenotypic and genotypic antifungal susceptibility of C. auris, it also has its challenges, and it is worth to compare our findings with those reported from different parts of the world. With regard to the antifungal agents mostly used for the treatment of infection due to this pathogen, a couple of agents are relied upon including caspofungin, micafungin, fluconazole, voriconazole, and amphotericin B. Our study generally showed comparable uniform susceptible findings to those reported regionally concerning echinocandins (caspofungin and micafungin) (Emara et al., 2015; Khan et al., 2018; Al Maani et al., 2019; Almaghrabi et al., 2020). However, a sporadic resistance to echinocandins was reported from the United States (1%) (Forsberg et al., 2020), and from India and South Africa (7%) (Lockhart et al., 2017). Moreover, the uniform resistance rates to fluconazole in our study was also comparable to those reported regionally (Emara et al., 2015; Al-Siyabi et al., 2017; Mohsin et al., 2017; Salah et al., 2021) except that of one study in Oman, which reported a lower resistant rate (58%) (Al Maani et al., 2019). Globally, 93% of C. aris isolates were resistant to fluconazole (Lockhart et al., 2017). Concerning voriconazole, our results revealed a range of susceptibility against C. auris, namely, 36% S, 61% I, and 3% R, while resistant data from our region and other parts of the world showed a range of 8–73.6% (Lockhart et al., 2017; Almaghrabi et al., 2020). Nevertheless, among these triazoles, fluconazole susceptibility has been suggested by CDC to be used as a surrogate marker for second generation triazole (e.g., voriconazole) susceptibility assessment. Still, isolates that are resistant to fluconazole may respond to other triazoles occasionally (see text footnote 1).
As for amphotericin B, the resistance rate against C. auris in our study was uniform (100%). This resistant rate is higher than those reported rates (0–62%) from different countries in our region such as Saudi Arabia (62%) (Almaghrabi et al., 2020), Oman (33–50%) (Al-Siyabi et al., 2017; Mohsin et al., 2017; Al Maani et al., 2019), Kuwait (23%) (Emara et al., 2015), United Arab Emirates (0%) (Alatoom et al., 2018), and other countries including Pakistan and India (35%) (Lockhart et al., 2017). In the United States, the reported amphotericin B resistant rate was 33% (Forsberg et al., 2020). Fortunately, the pan- and echinocandin-resistant C. auris strains were not detected in Lebanon, as was recently reported from Texas and District of Columbia (DC) in United States (Lyman et al., 2021).
The molecular characterization of the resistance genes in our study indicated the presence of Y132F mutations in Erg11 gene in all isolates, reflecting the resistance to azoles. These Y132F substitutions were commonly detected among South Asian isolates particularly Indian and Pakistani strains and considered clade-I-specific markers of resistance against fluconazole (Lockhart et al., 2017). Moreover, Y132F substitution in ERG11 gene appears to be combined with a novel mutation in the CDR1 gene, an ATP-binding cassette (ABC)-type efflux pump-encoding gene, which has previously been shown to substantially contribute to azole resistance in C. auris (Chowdhary et al., 2018). However, the genetic determinants promoting the increased expression of efflux pump-encoding genes in C. auris remain unidentified (Kim et al., 2019; Rybak et al., 2019). Rybak et al. study demonstrates that fluconazole-resistant clinical isolates of C. auris exhibit elevated levels of CDR1 expression and contribute significantly to clinical resistance against the entire class of triazole antifungals. The deletion of CDR1 in this fluconazole-resistant clinical isolate was sufficient to restore triazole resistance and increase the susceptibility of resistant strains from 64- to 128-fold. Several mechanisms have been proposed for the increased gene expression such as higher levels of mRNA stability, gene amplification, or deregulation because of point mutations in the promoter region.
Unfortunately, there is little or no information about CDR1 hotspot mutations in C. auris in the literature.
However, sequence analysis of the Candida albicans CDR1 gene showed several point mutations located near the promoter of the resistant strains. These point mutations could be distributed within the recognition sequence for the binding of trans-acting transcription factors, and hence, changes to the nucleotide sequence could cause either less efficient binding of transcriptional repressors or increase in the affinity of activators to the promoter region, therefore upregulating CDR1 (Looi et al., 2005).
For amphotericin B, the 100% resistance among our isolates was detected despite the absence of main resistance drivers related to ERG2, ERG3, and ERG6 gene mutation (Frias-De-Leon et al., 2020). Such finding indicates that the mechanism of amphotericin resistance remains to be fully elucidated.
All isolates showed a mutation in THR1 (L188G), a gene encoding a homoserine kinase involved in the biosynthesis of threonine. THR1 is considered a potential molecular target for antifungal chemotherapy, since THR1 genes are essential for growth and are required for virulence of C. albicans and C. neoformans cells (Kingsbury and McCusker, 2008). C. albicans cells lacking THR1 accumulate the toxic biosynthetic intermediate homoserine and are attenuated in terms of virulence and die rapidly upon threonine starvation and serum incubation (Kingsbury and McCusker, 2010a,b). Moreover, C. albicans THR1-depleted mutants exhibited increased sensitivity to oxidative and osmotic stress (Lee et al., 2018). Regarding the phylogenetic analysis, different findings have been reported globally. In our study, all C. auris genomes belonged to clade I showing a limited genetic diversity with SNP difference of ≤6, regardless of the recovered source, site of specimen, or time span between isolations, thereby highly reflecting an outbreak due to hospital-associated transmission and confirming what was reported earlier from the same medical center (Allaw et al., 2021). This clade I finding was similar to that reported in Qatar, Saudi Arabia, Oman, Pakistan, and India (Alfouzan et al., 2019). Other studies from different parts of the world reported other clades, for instance, clade III (Africa) among Australian isolates (Biswas et al., 2020), clade IV (South America) among isolates from Chicago (Roberts et al., 2021) and Venzuela (Lockhart et al., 2017), clade II (East Asia) from Japan, and clade V from Iran (Chow et al., 2019).
Conclusion
This first molecular characterization of C. auris from Lebanon revealed the exclusivity of clade I lineage among the studied isolates. This clade I together with its uniform resistance to fluconazole and amphotericin B are similar to what was reported from different countries in our region. The control of such highly resistant pathogen necessitates an appropriate and rapid recovery and identification to contain spread and outbreaks.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
GA designed the study. GA and RE collected the Candida samples. IB, LR, MF, and RE conducted the experiments. GA, IB, LR, MF, and GD analyzed the data and wrote the manuscript. All authors revised and approved the final draft.
Funding
This study was supported by the Charles University Research Fund PROGRES (project number Q39) and by project CZ.02.1. 01/0.0/0.0/16_019/0000787 “Fighting Infectious Diseases,” provided by the Ministry of Education Youth and Sports of the Czech Republic.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Appreciation and thanks are extended to our colleagues and staff noted below for their kind assistance in analyzing the first recovered isolates: Nancy L. Wengenack, Director, Mycology and Mycobacteriology Laboratories, Mayo College of Medicine, Minnesota, United States; Philippe Dufresne, Director of Laboratoire de Santé Publique du Québec, Canada; Shawn R. Lockhart, Senior Clinical Laboratory Advisor, Mycotic Diseases Branch, Centers for Disease Control and Prevention, Atlanta, GA, United States; and Samia Naccache, Technical Director Microbiology and Molecular Department, LabCorp/Dynacare NW for Swedish Medical Center, Seattle, United States; and to the Clinical Microbiology staff at AUBMC and Fata Akl at CIDR for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.770635/full#supplementary-material
Supplementary Table 1 | Candida auris isolates accession numbers.
Supplementary Table 2 | The number and location of SNPs among Candida auris isolates.
Footnotes
References
Al Maani, A., Paul, H., Al-Rashdi, A., Wahaibi, A. A., Al-Jardani, A., Al Abri, A. M. A., et al. (2019). Ongoing challenges with healthcare-associated Candida auris outbreaks in Oman. J. Fungi 5:101. doi: 10.3390/jof5040101
Alatoom, A., Sartawi, M., Lawlor, K., AbdelWareth, L., Thomsen, J., Nusair, A., et al. (2018). Persistent candidemia despite appropriate fungal therapy: first case of Candida auris from the United Arab emirates. Int. J. Infect. Dis. 70, 36–37. doi: 10.1016/j.ijid.2018.02.005
Alfouzan, W., Ahmad, S., Dhar, R., Asadzadeh, M., Almerdasi, N., Abdo, N. M., et al. (2020). Molecular epidemiology of Candida auris outbreak in a major secondary-care hospital in Kuwait. J. Fungi 6:307. doi: 10.3390/jof6040307
Alfouzan, W., Dhar, R., Albarrag, A., and Al-Abdely, H. (2019). The emerging pathogen Candida auris: a focus on the middle-eastern countries. J. Infect. Public Health 12, 451–459. doi: 10.1016/j.jiph.2019.03.009
Allaw, F., Kara Zahreddine, N., Ibrahim, A., Tannous, J., Taleb, H., Bizri, A. R., et al. (2021). First Candida auris outbreak during a COVID-19 pandemic in a tertiary-care center in Lebanon. Pathogens 10:157. doi: 10.3390/pathogens10020157
Almaghrabi, R. S., Albalawi, R., Mutabagani, M., Atienza, E., Aljumaah, S., Gade, L., et al. (2020). Molecular characterisation and clinical outcomes of Candida auris infection: single-centre experience in Saudi Arabia. Mycoses 63, 452–460. doi: 10.1111/myc.13065
Almagro Armenteros, J. J., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Al-Siyabi, T., Al Busaidi, I., Balkhair, A., Al-Muharrmi, Z., Al-Salti, M., and Al’Adawi, B. (2017). First report of Candida auris in Oman: clinical and microbiological description of five candidemia cases. J. Infect. 75, 373–376. doi: 10.1016/j.jinf.2017.05.016
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Araj, G. F., Asmar, R. G., and Avedissian, A. Z. (2015). Candida profiles and antifungal resistance evolution over a decade in Lebanon. J. Infect. Dev. Ctries 9, 997–1003. doi: 10.3855/jidc.6550
Awada, B., Alam, W., Chalfoun, M., Araj, G., and Bizri, A. R. (2021). COVID-19 and Candida duobushaemulonii superinfection: a case report. J. Mycol. Med. 31:101168. doi: 10.1016/j.mycmed.2021.101168
Biswas, C., Wang, Q., van Hal, S. J., Eyre, D. W., Hudson, B., Halliday, C. L., et al. (2020). Genetic heterogeneity of Australian Candida auris isolates: insights from a nonoutbreak setting using whole-genome sequencing. Open Forum Infect. Dis. 7:ofaa158. doi: 10.1093/ofid/ofaa158
Blachowicz, A., Chiang, A. J., Elsaesser, A., Kalkum, M., Ehrenfreund, P., Stajich, J. E., et al. (2019). Proteomic and metabolomic characteristics of extremophilic fungi under simulated mars conditions. Front. Microbiol. 10:1013. doi: 10.3389/fmicb.2019.01013
Bruna, T., Hoff, K. J., Lomsadze, A., Stanke, M., and Borodovsky, M. (2021). BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 3:lqaa108. doi: 10.1093/nargab/lqaa108
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using diamond. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Cernakova, L., Roudbary, M., Bras, S., Tafaj, S., and Rodrigues, C. F. (2021). Candida auris: a quick review on identification, current treatments, and challenges. Int. J. Mol. Sci. 22:4470. doi: 10.3390/ijms22094470
Chow, N. A., de Groot, T., Badali, H., Abastabar, M., Chiller, T. M., and Meis, J. F. (2019). Potential fifth clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 25, 1780–1781. doi: 10.3201/eid2509.190686
Chow, N. A., Munoz, J. F., Gade, L., Berkow, E. L., Li, X., Welsh, R. M., et al. (2020). Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. doi: 10.1128/mBio.03364-19
Chowdhary, A., Prakash, A., Sharma, C., Kordalewska, M., Kumar, A., Sarma, S., et al. (2018). A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73, 891–899. doi: 10.1093/jac/dkx480
Emara, M., Ahmad, S., Khan, Z., Joseph, L., Al-Obaid, I., Purohit, P., et al. (2015). Candida auris candidemia in Kuwait, 2014. Emerg. Infect. Dis. 21, 1091–1092. doi: 10.3201/eid2106.150270
Forsberg, K., Lyman, M., Chaturvedi, S., and Schneider, E. C. (2020). Public health action-based system for tracking and responding to U.S. Candida drug resistance: AR Lab Network, 2016-2019. Open Forum Infect. Dis. 7(Suppl. 1), S206–S207. doi: 10.1093/ofid/ofaa439.465
Forsberg, K., Woodworth, K., Walters, M., Berkow, E. L., Jackson, B., Chiller, T., et al. (2019). Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 57, 1–12. doi: 10.1093/mmy/myy054
Frias-De-Leon, M. G., Hernandez-Castro, R., Vite-Garin, T., Arenas, R., Bonifaz, A., Castanon-Olivares, L., et al. (2020). Antifungal resistance in Candida auris: molecular determinants. Antibiotics 9:568. doi: 10.3390/antibiotics9090568
Hoff, K. J., Lange, S., Lomsadze, A., Borodovsky, M., and Stanke, M. (2016). BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769. doi: 10.1093/bioinformatics/btv661
Hoff, K. J., Lomsadze, A., Borodovsky, M., and Stanke, M. (2019). Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962, 65–95. doi: 10.1007/978-1-4939-9173-0_5
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Iguchi, S., Itakura, Y., Yoshida, A., Kamada, K., Mizushima, R., Arai, Y., et al. (2019). Candida auris: a pathogen difficult to identify, treat, and eradicate and its characteristics in Japanese strains. J. Infect. Chemother. 25, 743–749. doi: 10.1016/j.jiac.2019.05.034
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Khan, Z., Ahmad, S., Al-Sweih, N., Joseph, L., Alfouzan, W., and Asadzadeh, M. (2018). Increasing prevalence, molecular characterization and antifungal drug susceptibility of serial Candida auris isolates in Kuwait. PLoS One 13:e0195743. doi: 10.1371/journal.pone.0195743
Kim, S. H., Iyer, K. R., Pardeshi, L., Munoz, J. F., Robbins, N., Cuomo, C. A., et al. (2019). Erratum for Kim et al., “genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance”. mBio 10:e00346-19. doi: 10.1128/mBio.00346-19
Kingsbury, J. M., and McCusker, J. H. (2008). Threonine biosynthetic genes are essential in Cryptococcus neoformans. Microbiology 154(Pt. 9), 2767–2775. doi: 10.1099/mic.0.2008/019729-0
Kingsbury, J. M., and McCusker, J. H. (2010a). Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2Δ) mutants is influenced by the carbon source and rapamycin. Microbiology 156, 929–939. doi: 10.1099/mic.0.034348-0
Kingsbury, J. M., and McCusker, J. H. (2010b). Fungal homoserine kinase (thr1Δ) mutants are attenuated in virulence and die rapidly upon threonine starvation and serum incubation. Eukaryot. cell 9, 729–737. doi: 10.1128/EC.00045-10
Laslett, D., and Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Lee, Y. T., Fang, Y. Y., Sun, Y. W., Hsu, H. C., Weng, S. M., Tseng, T. L., et al. (2018). THR1 mediates GCN4 and CDC4 to link morphogenesis with nutrient sensing and the stress response in Candida albicans. Int. J. Mol. Med. 42, 3193–3208. doi: 10.3892/ijmm.2018.3930
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Lockhart, S. R., Etienne, K. A., Vallabhaneni, S., Farooqi, J., Chowdhary, A., Govender, N. P., et al. (2017). Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64, 134–140. doi: 10.1093/cid/ciw691
Looi, C. Y., Ec, D. S., Seow, H. F., Rosli, R., Ng, K. P., and Chong, P. P. (2005). Increased expression and hotspot mutations of the multidrug efflux transporter, CDR1 in azole-resistant Candida albicans isolates from vaginitis patients. FEMS Microbiol. Lett. 249, 283–289. doi: 10.1016/j.femsle.2005.06.036
Lyman, M., Forsberg, K., Reuben, J., Dang, T., Free, R., Seagle, E. E., et al. (2021). Notes from the field: transmission of pan-resistant and echinocandin-resistant Candida auris in health care facilities - Texas and the District of Columbia, January-April 2021. MMWR Morb. Mortal. Wkly Rep. 70, 1022–1023. doi: 10.15585/mmwr.mm7029a2
Manni, M., Berkeley, M. R., Seppey, M., and Zdobnov, E. M. (2021). BUSCO: assessing genomic data quality and beyond. Curr. Protoc. 1:e323. doi: 10.1002/cpz1.323
Mohsin, J., Hagen, F., Al-Balushi, Z. A. M., de Hoog, G. S., Chowdhary, A., Meis, J. F., et al. (2017). The first cases of Candida auris candidaemia in Oman. Mycoses 60, 569–575. doi: 10.1111/myc.12647
Patel, R. (2019). A moldy application of MALDI: MALDI-ToF Mass Spectrometry for Fungal Identification. J Fungi 5:4. doi: 10.3390/jof5010004
Prakash, A., Sharma, C., Singh, A., Kumar Singh, P., Kumar, A., Hagen, F., et al. (2016). Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin. Microbiol. Infect. 22, 277.e1–9. doi: 10.1016/j.cmi.2015.10.022
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Roberts, S. C., Zembower, T. R., Ozer, E. A., and Qi, C. (2021). Genetic evaluation of nosocomial Candida auris transmission. J. Clin. Microbiol. 59, e02252–20. doi: 10.1128/JCM.02252-20
Rybak, J. M., Doorley, L. A., Nishimoto, A. T., Barker, K. S., Palmer, G. E., and Rogers, P. D. (2019). Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob. Agents Chemother. 63, e00057–19. doi: 10.1128/AAC.00057-19
Rychert, J., Slechta, E. S., Barker, A. P., Miranda, E., Babady, N. E., Tang, Y. W., et al. (2018). Multicenter evaluation of the Vitek MS v3.0 system for the identification of filamentous fungi. J. Clin. Microbiol. 56, e01353–17. doi: 10.1128/JCM.01353-17
Salah, H., Sundararaju, S., Dalil, L., Salameh, S., Al-Wali, W., Tang, P., et al. (2021). Genomic epidemiology of Candida auris in qatar reveals hospital transmission dynamics and a South Asian origin. J. Fungi 7:240. doi: 10.3390/jof7030240
Schelenz, S., Hagen, F., Rhodes, J. L., Abdolrasouli, A., Chowdhary, A., Hall, A., et al. (2016). First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 5:35. doi: 10.1186/s13756-016-0132-5
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Seemann, T. (2015). Snippy: Fast Bacterial Variant Calling From NGS Reads [Internet]. San Francisco, CA: github.
Smith, C. A. (2021). Macrosynteny analysis between lentinula edodes and lentinula novae-zelandiae reveals signals of domestication in lentinula edodes. Sci. Rep. 11:9845. doi: 10.1038/s41598-021-89146-y
Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. doi: 10.1093/bioinformatics/btn013
Stanke, M., Schoffmann, O., Morgenstern, B., and Waack, S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 7:62. doi: 10.1186/1471-2105-7-62
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y. O., and Borodovsky, M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18, 1979–1990. doi: 10.1101/gr.081612.108
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Keywords: Candida auris, whole-genome sequencing, antifungal resistance, South Asian clade, Lebanon
Citation: Reslan L, Araj GF, Finianos M, El Asmar R, Hrabak J, Dbaibo G and Bitar I (2022) Molecular Characterization of Candida auris Isolates at a Major Tertiary Care Center in Lebanon. Front. Microbiol. 12:770635. doi: 10.3389/fmicb.2021.770635
Received: 04 September 2021; Accepted: 23 November 2021;
Published: 25 January 2022.
Edited by:
Annamari Heikinheimo, University of Helsinki, FinlandReviewed by:
Kin-Ming (Clement) Tsui, University of British Columbia, CanadaJoão Nobrega De Almeida Júnior, Universidade de São Paulo, Brazil
Copyright © 2022 Reslan, Araj, Finianos, El Asmar, Hrabak, Dbaibo and Bitar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George F. Araj, garaj@aub.edu.lb; Ibrahim Bitar, ibrahimbitar5@gmail.com
 Lina Reslan
Lina Reslan George F. Araj
George F. Araj Marc Finianos
Marc Finianos Rima El Asmar
Rima El Asmar Jaroslav Hrabak
Jaroslav Hrabak Ghassan Dbaibo
Ghassan Dbaibo Ibrahim Bitar
Ibrahim Bitar