
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 09 October 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.585404
This article is part of the Research TopicMultilateral Interactions In The RhizosphereView all 13 articles
 Ben Niu1,2*
Ben Niu1,2* Weixiong Wang2
Weixiong Wang2 Zhibo Yuan2
Zhibo Yuan2 Ronald R. Sederoff3
Ronald R. Sederoff3 Heike Sederoff4
Heike Sederoff4 Vincent L. Chiang1,3
Vincent L. Chiang1,3 Rainer Borriss5,6*
Rainer Borriss5,6*Major losses of crop yield and quality caused by soil-borne plant diseases have long threatened the ecology and economy of agriculture and forestry. Biological control using beneficial microorganisms has become more popular for management of soil-borne pathogens as an environmentally friendly method for protecting plants. Two major barriers limiting the disease-suppressive functions of biocontrol microbes are inadequate colonization of hosts and inefficient inhibition of soil-borne pathogen growth, due to biotic and abiotic factors acting in complex rhizosphere environments. Use of a consortium of microbial strains with disease inhibitory activity may improve the biocontrol efficacy of the disease-inhibiting microbes. The mechanisms of biological control are not fully understood. In this review, we focus on bacterial and fungal biocontrol agents to summarize the current state of the use of single strain and multi-strain biological control consortia in the management of soil-borne diseases. We discuss potential mechanisms used by microbial components to improve the disease suppressing efficacy. We emphasize the interaction-related factors to be considered when constructing multiple-strain biological control consortia and propose a workflow for assembling them by applying a reductionist synthetic community approach.
The interest in control of plant diseases by beneficial microbes, has increased recently due to the global need for environmentally friendly alternatives to chemical pesticides and fertilizers (Handelsman and Stabb, 1996; Fira et al., 2018; Syed Ab Rahman et al., 2018). A large number of bacterial and fungal strains, as well as viruses, nematodes, and insects have been employed as biological control agents (BCAs) in the management of soil-borne crop pathogens for decades. BCAs have become a crucial component of sustainable agriculture and forestry (Cazorla and Mercado-Blanco, 2016; Alvarez and Biosca, 2017; Busby et al., 2017; Umesha et al., 2018). Although numerous beneficial microbial strains performed well against pathogens under controlled conditions in the laboratory or the greenhouse, examples of successful BCA application in commercial field-based crop production are rare (Xu et al., 2011; Mazzola and Freilich, 2017). This is mainly due to inadequate colonization of host rhizosphere connected with inefficient inhibition of soil-borne pathogen growth (Sarma et al., 2015; Mazzola and Freilich, 2017).
Different BCA consortia, consisting of two or more microbial strains [multi-strain biological control agents (MSBCAs)], are assembled to improve the stability and efficiency of disease-inhibition (Sarma et al., 2015; Mazzola and Freilich, 2017; Vorholt et al., 2017; Woo and Pepe, 2018). The biotechnological potential of microbial consortia was reviewed recently and examples for their possible applications in areas of biopolymers, bioenergy, biochemicals, and bioremediation have been presented (Bhatia et al., 2018). Here, we focus on the application of MSBCAs in sustainable agriculture. In several cases, superior disease suppression exerted by MSBCAs has been reported (Table 1). Diverse modes of action were proposed: (i) diversity in biocontrol mechanisms offered by each microbial component (Pierson and Weller, 1994; Sarma et al., 2015), (ii) occupation of distinctive niches by probiotic microorganisms resulting in more stable communities (Pierson and Weller, 1994; Pliego et al., 2008; Thomloudi et al., 2019), (iii) enhanced modulation of genetic elements acting in the community (Lutz et al., 2004), and (iv) a broader spectrum of targeted phytopathogens (Sarma et al., 2015; Thomloudi et al., 2019) may contribute to greater biocontrol activity in communities. However, our understanding of the mechanisms underlying the reinforcement of their disease-inhibitory effects by MSBCAs is still very limited.
Interactions in communities of plant-associated microbes are essential for plant health (Whipps, 2001; Frey-Klett et al., 2011; Kemen, 2014; Hassani et al., 2018). A well-known example is disease suppressive soil. They are defined by their ability to suppress plant diseases such as “take-all” disease in wheat caused by the fungal pathogen Gaeumannomyces graminis. The suppressive effect is due to the presence of 2,4-diacetylphloroglucinol produced by a group of soil-borne Pseudomonas spp. (Kwak and Weller, 2013).
The interplay among the members of MSBCAs might be relevant to their elevated disease-suppressing effect. It is necessary to pay attention to the microbe−microbe interplay-related elements when constructing MSBCAs because microbial interactions within the plant microbiome are important selective forces forming complex microbial assemblages (Hassani et al., 2018). In general, two different methods can be distinguished when BCA consortia are prepared: (i) mixing existing single-strain biological control agents (SSBCAs) according to empirical experience or (ii) preparing MSBCAs as a reductionist synthetic community (RSC) (Liu et al., 2019). In the RSC approach, defined synthetic communities (SynCom) are assembled using a limited number of isolates from the natural microbiome. In the following we prefer to use the term “SynCom” given that synthetic communities contain usually a limited number of isolates.
In this review, we provide a brief overview of the current state of the use of MSBCAs in the management of soil-borne diseases and describe potential mechanisms used by their microbial components to improve disease-suppression. We describe interaction-related factors to be considered when constructing MSBCAs and propose a workflow for assembling them as a reductionist synthetic community (Vorholt et al., 2017; Liu et al., 2019).
Selection of novel biocontrol microbial strains via isolation and screening is a permanent approach to improve the disease-controlling efficiency of BCAs. Although novel disease-suppressive strains might overcome inadequate colonization of the host rhizosphere and inefficient inhibition of soil-borne pathogen growth, the discovery of taxonomically novel isolates possessing biological disease control activity becomes more difficult over time even after extensive searches. Another promising approach, exploiting genetically modified microbial strains with improved antagonistic function has been restricted or prohibited worldwide (Migheli, 2001; Stemke, 2004). When applying BCAs in natural settings, BCAs do not act independent of their environment but interact with many indigenous microbes to become components of local microbial communities. The members of such consortia may evolve niche−specific microbial interactions to influence plant health (Whipps, 2001). There is growing interest in the use of disease-suppressing microbial communities, specifically MSBCAs, for controlling soil-borne pathogens.
Multi-strain biological control agents have successfully controlled soil-borne diseases of valuable crops caused by fungi, oomycetes, bacteria and nematodes (Table 1). Several microbial combinations are possible, such as fungus to fungus, fungus to bacterium, and bacterium to bacterium. Similar to the single-strain biological control agents (SSBCAs), MSBCAs employ diverse modes of action for control, e.g., competition for resources and niches (McKellar and Nelson, 2003; Wei et al., 2015; Hu et al., 2016), production of antimicrobial compounds (Thakkar and Saraf, 2014; Santhanam et al., 2019), induction of systemic resistance (Sarma et al., 2015; Solanki et al., 2019), and regulation of microbial communities (Zhang L.-N. et al., 2019). MSBCAs appear to have higher efficiency for control of soil-borne disease than SSBCAs (Figure 1A).
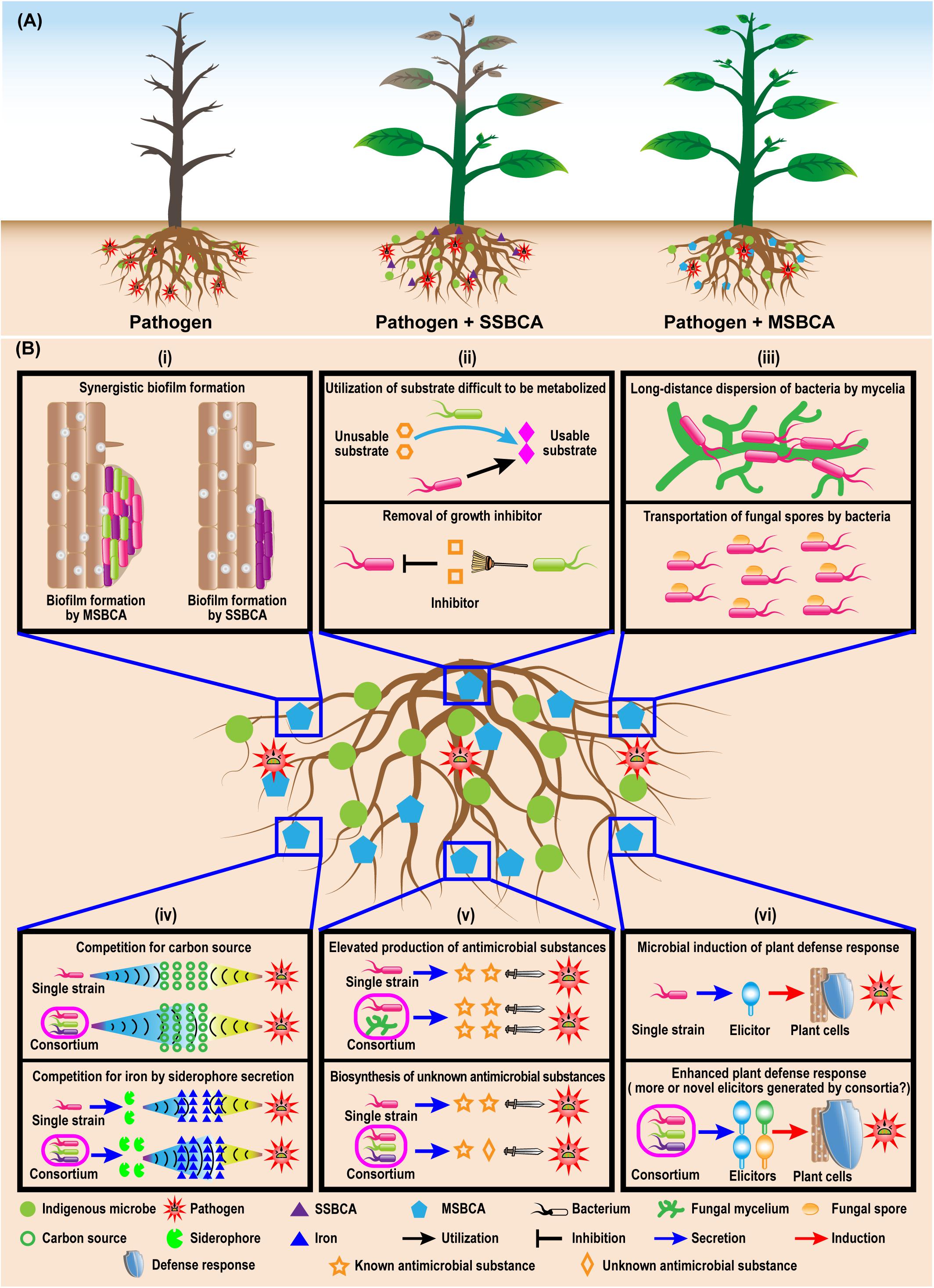
Figure 1. Enhanced biocontrol effects of multiple-strain biological control agents (MSBCA) against soil-borne pathogen (A) and the potential mechanisms underlying the elevated disease-suppressive efficacy (B). (i) enhanced biofilm formation, (ii) syntrophic microbial growth promotion, (iii) facilitated migration, (iv) boosted competition for resources, (v) stimulated antimicrobial substance biosynthesis, and (vi) elevated plant defense response induction.
Synergistic and/or additive effects exerted by carefully selected microbial consortia might explain their superior efficacy compared to single SSBCAs. In simple cases, MSBCAs consist of only two strains, e.g., a fungus and a bacterium where one or both have biocontrol activities. A consortium consisting of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens ACCC1111060 was found to be more efficient against infection by Botrytis cinerea (the agent of gray mold disease) than the individual strains (Wu et al., 2018). Similarly, when Trichoderma virens GI006 was combined with Bacillus velezensis Bs006, efficiency against Fusarium wilt of cape gooseberry was enhanced (Izquierdo-García et al., 2020). A bacterial consortium of Chitionophaga sp. 94 and Flavobacterium sp. 98 conferred more consistent protection against the infection of root rot, the infection of sugar beets by Rhizoctonia solani than the individual community members (Carrión et al., 2019). Thus, MSBCAs are capable of providing more effective protection of the hosts than inoculation with single-strains. A model resident bacterial community composed of five non-virulent Ralstonia spp. strains was more efficient at reducing the spread of the bacterial wilt of tomato caused by Ralstonia solanacearum than the single non-virulent strains (Wei et al., 2015). Further examples documenting the superior action of MSBCAs against soil-borne plant pathogens are listed in Table 1.
The enhancement of disease inhibition by MSBCAs is widely thought to be due to the addition of different features for control (Pierson and Weller, 1994; Sarma et al., 2015). Occupation of distinct niches in the rhizosphere may avoid competition among probiotic microorganisms, resulting in more stable rhizosphere communities (Pierson and Weller, 1994; Pliego et al., 2008; Thomloudi et al., 2019). Modulation of genetic elements (Lutz et al., 2004) and suppression of a broader range of phytopathogens (Pierson and Weller, 1994; Thomloudi et al., 2019) may account for the elevated biocontrol activity in microbial communities. In addition, some key features related to the disease-controlling effect of BCAs, including rhizosphere colonization and suppression of pathogen growth, can be promoted in consortia via a complex network of microbe−microbe interactions. This interplay might serve as the selective force building plant-associated microbial communities (Hassani et al., 2018). Members of the MSBCAs apply interspecies communication as a strategy to improve their control of soil-borne diseases.
Efficient colonization of the rhizosphere is the first and fundamental step to protect plants from soil-borne pathogens by BCAs. Insufficient rhizosphere colonization can impair the beneficial effects of the biocontrol microbial strains, resulting in reduction or failure of disease control. Inoculation with MSBCAs may enhance the colonization of the rhizosphere by biocontrol microbes. The rhizoplane colonization ability of a five-strain bacterial consortium suppressing a sudden wilt disease of Nicotiana attenuata was enhanced compared to that of each single community member (Santhanam et al., 2019). Survival of Pseudomonas species communities inhibiting bacterial wilt disease of tomato increased with increasing diversity (Hu et al., 2016). In addition, the total bacterial abundance on bean root tips rose when a two-membered biocontrol Pseudomonas species consortium for anthracnose was added (Bardas et al., 2009). Thus, using disease-controlling microorganisms as multi-strain consortia can indeed promote rhizosphere colonization by BCAs (Figure 1B). Such positive effects on microbial colonization may be due to positive regulation of some colonization-related biological processes, such as biofilm formation, growth and migration, by the interactions among microorganisms within consortia.
A promising example of successful use of empirical mixtures of BCA is the combination of the fungus Trichoderma spp. and the biocontrol bacterium Bacillus velezensis. According to in vitro observations the microbes appear to be incompatible. B. velezensis FZB42 produces an arsenal of antifungal compounds. The lipopeptides bacillomycin D and fengycin act antagonistically against filamentous fungi (Chowdhury et al., 2015) and it is to be expected that the bacilli might inhibit growth of Trichoderma when applied together. However, supernatants of B. velezensis stimulated growth of Trichoderma virens under in vitro conditions. Vice versa, addition of Trichoderma conidia did not affect viability of B. velezensis suggesting high compatibility of both microbes. Adhesion of B. velezensis spores to the conidia of T. virens without affecting their morphology was observed, supporting compatibility of both soil inhabitants (Izquierdo-García et al., 2020).
Germination of fungal conidiospores and Bacillus endospores, is a critical step in successful colonization of BCAs. The contact of Trichoderma conidia with Bacillus biofilms did not impair the ability of fungal spores to germinate and establish the fungus in soil (Izquierdo-García et al., 2020). The mycelia of Trichoderma can serve as a supporting layer for formation of bacterial biofilms and can aid bacterial migration in the soil. Growth of bacteria was supported by nutrients present in the fungal exudates (Warmink et al., 2011; Triveni et al., 2012).
Microbial colonization of plant roots can be promoted by the formation of biofilms (Fan et al., 2011; Beauregard et al., 2013). Biofilms are communities of surface-associated microorganisms encased in a self-produced extracellular matrix composed of exopolysaccharides, proteins and sometimes DNA (Vlamakis et al., 2013). Beside the well-studied single-species biofilms, rhizosphere microorganisms belonging to multiple taxa are able to form multi-species biofilms, of which the formation might be elevated by the microbe−microbe interactions within biofilm communities (Figure 1B). In a previous study, a consortium of five native bacterial isolates was found to be able to colonize the roots of N. attenuata by forming multiple-taxa biofilms on the root surfaces. Furthermore, under both in vitro and in vivo conditions, the amount of biofilm produced by each individual strain was significantly less than the biofilms formed by the five-membered bacterial community, which indicating a synergistic biofilm formation by the consortium (Santhanam et al., 2019). Similarly, a three-species biocontrol community composed of Xanthomonas sp. WCS2014-23, Stenotrophomonas sp. WCS2014-113 and Microbacterium sp. WCS2014-259 showed synergy, as the combination of three formed more biofilm than the single strains. Moreover, colonization of host roots by this community was stimulated by enhanced biofilm formation (Berendsen et al., 2018). Although the mechanisms of such positive effects on biofilm production are unclear, the improved efficacy can be attributed to the cooperative microbial interactions in consortia, triggering increased extracellular matrix deposition and cell-to-cell signaling (Santhanam et al., 2019).
Colonization of the rhizosphere requires robust microbial growth, which can be greatly improved by syntrophy, a nutritional situation in which multiple microorganisms combine their metabolic abilities to catabolize a substrate that cannot be degraded by either one of them alone (Morris et al., 2013; Mee et al., 2014). For example, Azospirillum brasilense, a well-known plant growth-promoting rhizobacterium (PGPR), is not able to use certain sugars and polysaccharides as carbon sources for growth in vitro. However, it is capable of associating with sugar- or polysaccharide-degrading bacteria, establishing a metabolic association where the sugar- and/or polysaccharide-degrading bacteria degrade the metabolites to products, which can be utilized as carbon source by A. brasilense. In turn A. brasilense provides the sugar- or polysaccharide-degrading bacteria with nitrogen by fixing the atmospheric N2 (Bashan and Holguin, 1997; Bashan, 1998). Such a synergistic catabolic effect on sugars and polysaccharides may boost the survival of A. brasilense in the rhizosphere, because plant roots release 5−21% of their photosynthetically fixed carbon as root exudates (Bais et al., 2006; Huang et al., 2014), including sugars and polysaccharides, which are used by the rhizosphere microbial communities. Beside nutrients, there are always microbial growth-inhibiting materials in the rhizosphere. While methanol can suppress the growth of methanotrophs in the rhizosphere, the methanotrophs are able to survive by coexisting with Hyphomicrobium spp. to build a rhizospheric microbial association, in which the Hyphomicrobium spp. is capable of removing methanol (Liechty et al., 2020). Therefore, the promotion of the growth of rhizosphere microbes can be achieved by syntrophic interactions leading to effective nutrient utilization and removal of harmful substances (Figure 1B).
Another crucial microbial trait for rhizosphere colonization is motility, defined as the ability of microorganisms to move or to perform mechanical work at the expense of metabolic energy (Harshey, 2003). There are six different categories of surface motility including swimming, swarming, gliding, twitching, sliding and darting (Harshey, 2003). According to Allard-Massicotte et al. (2016), motility is required for early root colonization by BCAs. The migration of microorganisms can be enhanced by the interactions among community members. For example, fungal hyphae are capable of serving as vectors for the dispersion of bacteria in the rhizosphere, which is known as a “fungal highway” (Kohlmeier et al., 2005; Warmink et al., 2011; Figure 1B). In a recent study, Zhang et al. (2020) showed that rhizobia use mycelia of Phomopsis liquidambaris as dispersal networks to migrate into legume rhizospheres and to trigger nodulation. Extraradical mycelium formed by the mycorrhiza fungus Glomus formosanum CNPAB020 can facilitate the translocation of Bradyrhizobium diazoefficiens USDA 110 in the rhizosphere (de Novais et al., 2020) in addition to its main activity in nutrient transfer. Prokaryotic cells are able to facilitate dispersal of non-motile asexual fungal spores as well (Figure 1B). Conidia of Aspergillus fumigatus, a non-motile rhizosphere fungus, can be transported by the rhizobacterium Paenibacillus vortex from niches of adverse growth conditions. Fungal mycelia may act as bridges to allow P. vortex to cross air gaps, which can be mutually facilitated dispersal, benefiting the life cycles of both of these very different rhizosphere inhabitants (Ingham et al., 2011). The enhanced dispersal may also occur between distinct bacterial species. An ampicillin-sensitive P. vortex strain was capable of swarming and colonizing on ampicillin plates using non-motile ampicillin-resistant Escherichia coli as cargo, dispersing both bacteria (Finkelshtein et al., 2015; Venieraki et al., 2016). Co-swarming or transporting other bacterial species may expand the abilities of the partners in occupying and exploiting ecological niches in diverse environments including the rhizosphere (Venieraki et al., 2016). Hence, interactions among the microbial components of a given community may bring about facilitated microbial migration, essential for efficient rhizosphere colonization.
In brief, microbe−microbe interactions can play a positive role in promoting rhizosphere colonization by beneficial microorganisms through boosting biofilm formation, microbial growth, migration inside of the microbiome, and interacting with plant roots. Thus, utilization of MSBCAs performing active interactions among their members may improve survival of disease-suppressing microbes, and their adaption to complex and changeable environmental conditions. In consequence, they may be able to stabilize their beneficial effects for the inhibition of soil-borne diseases.
Multi-strain biological control agents are able to exhibit stronger suppressive efficacy on the growth of soil-borne pathogens than SSBCAs. For instance, a bacterial strain mixture involving Bacillus subtilis S2BC-1 and GIBC-Jamog showed greater anti-fungal activity against the tomato vascular wilt pathogen, Fusarium oxysporum f.sp. lycopersici, than each of the individual strains (Shanmugam and Kanoujia, 2011). Similarly, Pseudomonas fluorescens T5 showed no inhibition against Rhizoctonia solani in vitro. However, when it was applied together with four non-antagonistic bacterial strains isolated from the rhizosphere of Tamarindus, this five-species bacterial community exhibited strong suppression of growth of R. solani (Kannan and Sureendar, 2009). Although the understanding of the enhanced pathogen-inhibiting effect of biocontrol consortia is limited, changes in resource competition and secretion of antimicrobial compounds triggered by microbial interactions may contribute to the enhanced suppression (Figure 1B).
Resource competition is a basic mechanism by which BCAs may protect plants from pathogens, implying that the beneficial microorganisms are able to rapidly and efficiently utilize the limited resources in the vicinity of the plant hosts to restrict or suppress the growth of phytopathogens. Plant exudates on root surfaces and in their surrounding rhizosphere, are the primary sources of nutrients for the rhizosphere microbiome. Successful suppression depends on the competition for nutrients in root exudates by biocontrol microbes and soil-borne pathogens. This contest can be elevated by the microbial interplay inside MSBCAs (Figure 1B). Two biocontrol consortia for tomato bacterial wilt caused by R. solanacearum, consisting of eight Pseudomonas and five non-virulent Ralstonia strains, exhibited much stronger inhibiting effects on the population density of R. solanacearum than each individual strain. The enhanced inhibition is caused by an increase of niche overlaps exerted by these consortia with R. solanacearum. Niche overlaps may be defined as ‘likeness’ between the communities and R. solanacearum in the catabolism of 48 different single-carbon resources found in tomato root exudates (Wei et al., 2015; Hu et al., 2016). The more diverse soil bacterial communities are, the better they are able to acquire many of the 31 individual carbon sources typical for soil, than the pathogen E. coli O157:H7 (van Elsas et al., 2012). Limited assimilatable iron resources remain in the rhizosphere, following the competition between disease-suppressing microorganisms and soil-borne pathogens (Gu et al., 2020). Many soil microbes scavenge iron by secreting siderophores, a chemically diverse group of secondary metabolites with a high affinity for iron, because iron predominantly occurs in soil in its insoluble ferric Fe (III) form (Traxler et al., 2013; Traxler and Kolter, 2015). The siderophores can both, to facilitate and suppress competitors, depending on whether the competitors possess the transporters or channels for siderophore uptake. The production of siderophores can be positively regulated by interspecies interactions among soil microbes. The interplay of Streptomyces coelicolor with five other soil actinobacteria increased the diversity of siderophores. Production of desferrioxamines by S. coelicolor, was triggered by siderophores from neighboring strains (Traxler et al., 2013). Therefore, the disease-inhibiting microorganisms in the rhizosphere may acquire elevated capability to utilize resources through microbial associations (Figure 1B).
Microorganisms are able to synthesize a multitude of compounds with antimicrobial activity, which is an important mode of action for direct inhibition or lethality on the microbial opponents in environments. So far, there have been a large number of reports of the antimicrobials produced by BCAs exhibiting suppressing effects on the growth of phytopathogens. These studies mainly focus on the biocontrol strains from the genera Bacillus, Pseudomonas, and Trichoderma, well known for the production of antibiotics including lipopeptides, polyketides, bacteriocins, phenazines, 2,4-diacetylphloroglucinol (DAPG) and chitinase (Ghisalberti and Sivasithamparam, 1991; Haas and Keel, 2003; Chen et al., 2007). Some metabolites with inhibitory functions are found in low concentration or are not expressed in pure culture but may be upregulated in a community (Nützmann et al., 2011; Brakhage, 2013; Pishchany et al., 2018). Lutz et al. (2004) examined the molecular interactions between bacterial and fungal BCAs, the DAPG-producing P. fluorescens and chitinase-producing Trichoderma atroviride P1. DAPG enhanced the expression of the nag1 chitinase gene, indicating that the positive regulation of key biocontrol genes may take place while mixing antagonists. Co-culturing the endophytic fungus Fusarium tricinctum with Bacillus subtilis, resulted in as much as a 78-fold increase in the accumulation of secondary metabolites including compounds with antimicrobial efficacy (Ola et al., 2013). Therefore, specific interactions among microorganisms belonging to different domains may enhance production of antimicrobial compounds. Not only are the microbial interactions able to upregulate the production of known antimicrobial compounds, but interactions may also activate the biosynthesis of hitherto unknown compounds with antimicrobial activity (Figure 1B). A novel antibiotic named amycomicin has been recently described (Pishchany et al., 2018). The production of this compound is dependent on the interaction between two soil-dwelling actinobacteria, Amycolatopsis sp. AA4 is the producer strain and Streptomyces coelicolor M145 is an inducer. According to these examples the synthesis of antimicrobial compounds can be stimulated or activated through both, inter- and intra-domain microbial interactions.
Therefore, the modulating effect of microbial interactions on resource competition and production of antimicrobial compounds may contribute to strengthening the inhibition of growth of pathogens (Figure 1B). Thus, applying BCAs as multi-strain mixtures can elevate the ability of biocontrol microorganisms to compete for the resources needed for rhizosphere survival with soil-borne pathogens and to stimulate the production of compounds toxic to specific pathogens. The increased niche overlaps and biosynthesis of novel antimicrobial compounds induced by microbe−microbe interactions may facilitate the BCAs to suppress a broader range of phytopathogens. The positive impact of interactions within MSBCAs may result in more efficient growth suppression of soil-borne pathogens, and improve the efficiency of soil-borne disease control by disease-inhibiting microbes.
Plants rely on rhizosphere microbiota to facilitate nutrient acquisition, in exchange for carbon-rich root exudates for bacterial nutrition. In addition, the rhizosphere microbiome is important for plant health and fitness (van der Heijden et al., 2008). The plant root microbiome consists of prokaryotic bacteria, eukaryotic filamentous fungi, and oomycetes. Besides a core microbiome ubiquitous in a multitude of hosts and geographical regions, a variable part of the microbiome is shaped by secretion of species-dependent plant secondary metabolites, which belong to diverse classes, such as coumarins, benzoxazinoids, phytoalexins and triterpenes (Jacoby et al., 2020). Consequently, diversity of species along the bulk-soil to root microbiota was found gradually decreasing. Positive correlations dominate within each of the three kingdoms. Reconstitution experiments performed with synthetic mono- or multi-kingdom microbial consortia and germ-free Arabidopsis plantlets revealed that the bacterial microbiota protects plants against potentially pathogenic fungi and oomycetes by mainly negative factors exerted against filamentous fungi (Durán et al., 2018). Widely distributed members of the core microbiota such as Variovorax, a gram-negative beta-proteobacterium, and Pseudomonas appeared to be important for plant protection but individual members of other bacterial taxa could overtake their function in biocontrol. Therefore, addition of an SSBCA or MSBCA might have positive effects in complex systems of agriculture and forestry.
The plant immune system also affects the composition of the microbiota in the vicinity of plant roots. The root-microbiome may expand plant immunity and acts as an additional layer of defense against plant pathogens (Teixeira et al., 2019). Interaction of beneficial microbes with plant roots can result in systemic host resistance to pathogens, which may be due to the activation of induced systemic resistance (ISR) (Sarma et al., 2015). In addition to promoting rhizosphere colonization and suppressing soil-borne pathogen growth, inducing enhanced plant defense responses to pathogens has been described in many studies as another important feature employed by the MSBCAs for their elevated disease-controlling effect. The additive or synergistic efficacy of the biocontrol consortia on the induction of elevated host immunities to plant pathogens is directed by activating several distinctive metabolic and signaling pathways against a given pathogen (Jain et al., 2012; Alizadeh et al., 2013; Sarma et al., 2015). However, how interactions among the members of MSBCAs can effectively boost specific systemic resistance to soil-borne pathogens remains to be better illustrated. One possible hypothesis is that the microbe-microbe interplay within the biocontrol consortia might lead to the production of larger amounts of specific elicitors and potent compounds capable of more efficiently eliciting ISR (Figure 1B).
Many root-associated gram-positive and gram-negative bacteria are able to produce plant growth hormones, such as indole-3-acetic acid (IAA) and thus promoting plant root growth, when auxin production does not exceed a critical level (Vessey, 2003; Lugtenberg and Kamilova, 2009). In case of some pathogenic bacteria, IAA production exceeds the critical threshold needed for plant growth and may negatively affect plant health (Spaepen et al., 2007; Subramoni et al., 2014; Segev et al., 2016). Some beneficial root-associated microbes such as Variovorax possess the IAA catabolic gene cluster and can reverse root growth inhibition occurring at high IAA concentrations by degrading IAA (Fitzpatrick et al., 2020).
Soil not only supports plant and animal life, but also hosts myriad microorganisms inside, referred to collectively as the soil microbiome (Berg and Smalla, 2009; Fierer, 2017; Jansson and Hofmockel, 2018; Thakur and Geisen, 2019), which governs biogeochemical cycling of macronutrients, micronutrients and other elements vital for the growth of plants and animals (Jansson and Hofmockel, 2020). The interactions between microbes and soil have always drawn the attention of microbiologists and ecologists. It has been widely accepted that microbial communities inhabiting soil are capable of alternating its physicochemical properties by organic litter deposition and metabolic activities (Jacoby et al., 2017; Jansson and Hofmockel, 2020), for example, by improving water retention (Naylor and Coleman-Derr, 2017), increasing carbon storage (Jansson et al., 2018) and mineral nutrition contents (van der Heijden et al., 2008; Jacoby et al., 2017). Vice versa, the variability in soil traits may impact the composition and function of soil microbial communities (Peiffer et al., 2013; Yang et al., 2019; Chen et al., 2020; Wang et al., 2020). Our increasing awareness of the influences of soil-feature changes on the microbiome has resulted in an emerging urgency to elevate the suppressing effect of soil microbiota against phytopathogens by managing the soil properties. Wang et al. (2020) demonstrated that the addition of biochar to the soil not only raises the pH and the available nutrient content, but also augments fungal richness and diversity, especially the abundance of potential biocontrol fungi, which led to the inhibition of Phytophthora blight of pepper. Similarly, biochar amendment controlled bacterial wilt through changing soil chemistry and the composition of the microbial community. The application of biochar specifically enriched beneficial bacteria and decreased pathogen abundance (Chen et al., 2020). Furthermore, Yang et al. (2019) showed that wheat straw return significantly increased soil nitrogen and reduced the relative abundance of pathogenic fungal genera in the soil microbial community, indicating a potential for disease control. Thus, promoting the biocontrol effects of the soil microbial community against soil-dwelling pathogens by manipulating soil features is a promising strategy for soil-borne disease management. Moreover, understanding the interplay between the soil and its associated microbiota will expand our knowledge about the impact of abiotic factors on biological soil-borne pathogen control.
Application of BCAs in the community context as MSBCAs can increase the ability to control soil-borne diseases of crops through interaction-mediated promotion of rhizosphere colonization outcompeting soil-borne pathogens. Thus, construction and utilization of MSBCAs could augment soil-borne disease control in sustainable agriculture and forestry. So far, there are at least two strategies for preparing effective MSBCAs, (i) mixing the existing SSBCAs according to empirical criteria, and (ii) assembling MSBCAs by applying the reductionist SynCom approach, also named RSC (Liu et al., 2019). Using either one of the two strategies, microbe-microbe interactions need to be taken into account.
Combining beneficial microbial isolates that may enhance the effect achieved by single isolates dates back to the discovery of PGPR (Kloepper et al., 1980). Selecting proper strains is critical. We noted that microorganisms used for developing biocontrol consortia were often selected according to their individual disease suppressive capacity. However, except for this property, no precise selection standards have been adopted to choose microbial components (Sarma et al., 2015; Thomloudi et al., 2019). This approach often results in equal or even lower efficacy of the multi-strain mixtures compared to the individual strains (Sarma et al., 2015). Thus, it is necessary to carefully evaluate the compatibility and interactions of the candidate strains before the MSBCA consortium is established. We propose, in addition to the disease-inhibiting activity of individual strains, to consider two interaction-related properties, (i) compatibility, and (ii) diversity.
The members of a probiotic consortium are considered to be compatible when they do not inhibit growth of each other during their in vitro co-culture and/or in rhizosphere colonization competition assays (Liu et al., 2018; Thomloudi et al., 2019). Co-inoculation with incompatible isolates might hinder one or more microbial agents from reaching the appropriate population threshold for plant disease control (Haas and Defago, 2005). The results of the in vitro co-culture compatibility tests often represent the interactions occurring among the members of the consortium. However, variation in media used to test in vitro compatibility (Lyons and Kolter, 2017), the colonization of different ecological niches on roots (Pliego et al., 2008), and interference among mechanisms for disease control (Stockwell et al., 2011) can lead to inconsistent compatibility assays. Thus, compatibility among members of a synthetic microbial community should be considered as a prerequisite in the engineering of MSBCAs applied to plants, and should be verified by further assays.
In addition, the degree of microbial diversity affects the assembly, survival, and functionality of BCAs in the rhizosphere and their ability to inhibit soil-borne diseases (Hu et al., 2016). First, a high level of species diversity can increase the resources that microbial species can collectively use as a community (the niche breadth), and enable microorganisms to survive in the rhizosphere more efficiently (Wei et al., 2015). Second, the amount and number of secondary metabolites that suppress pathogen growth increase with increasing taxonomic diversity in MSBCAs (Raaijmakers and Weller, 1998; Jousset et al., 2014). A combination of different secondary metabolites produced jointly by diverse microbes may strengthen the antagonistic effect against pathogens (Loper et al., 2012). Therefore, MSBCAs of high diversity could be more adaptive to the pressure of rhizosphere environments and act more efficiently against soil-borne plant diseases.
In summary, compatibility and diversity are two interaction relevant factors (Figure 2A) that may determine the success of MSBCAs. Some additional traits, such as microbial colonization of the rhizosphere, mode of action for disease control, safety to humans and the environment, ease of application and convenience of management systems need to be considered, when establishing the biocontrol microbial communities for soil-borne diseases (Raupach and Kloepper, 1998; Sikora et al., 2010; Bashan et al., 2013; Grosskopf and Soyer, 2014; Ahkami et al., 2017).
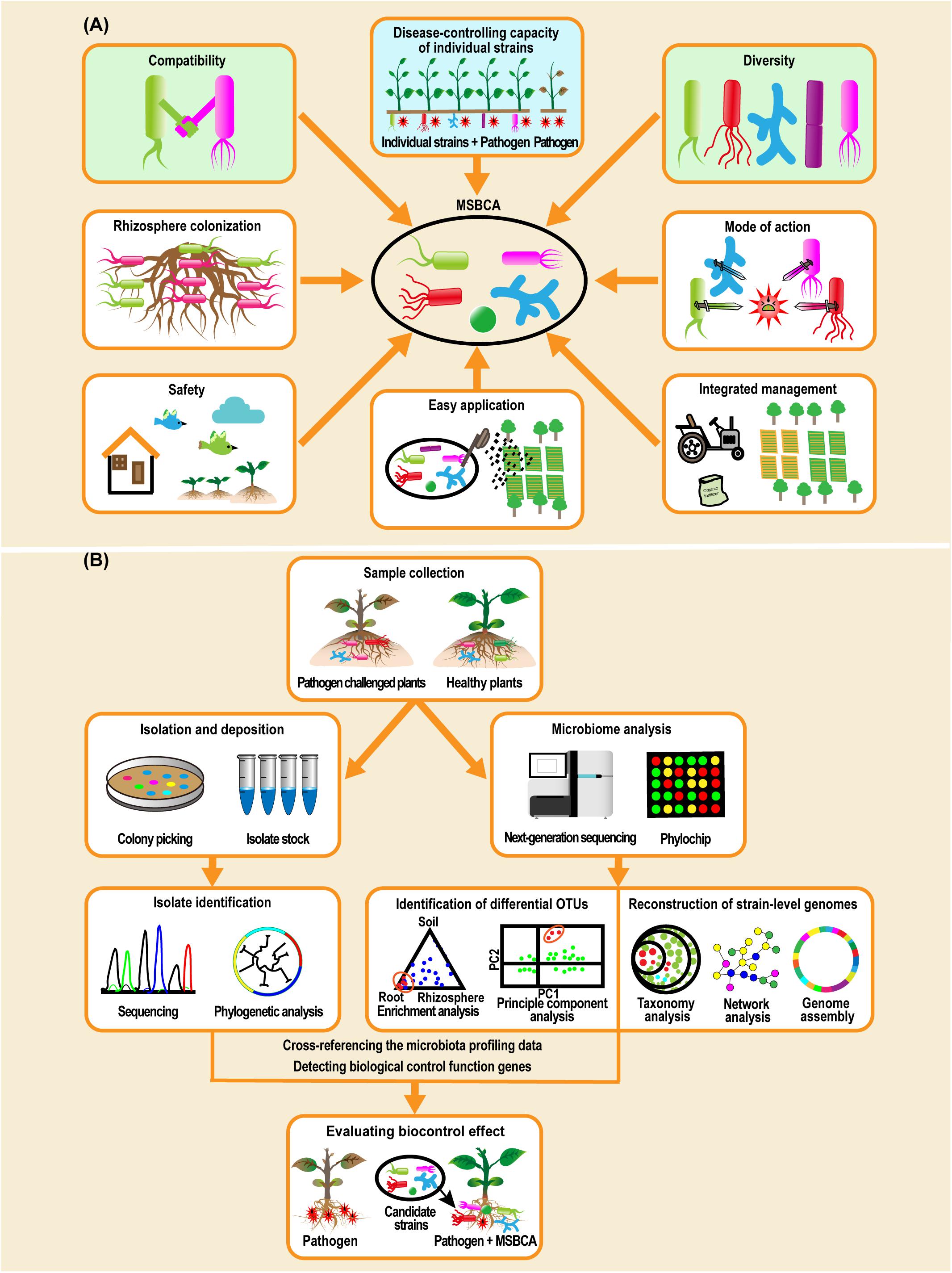
Figure 2. Biological features need to be considered when constructing multiple-strain biological control agents (MSBCAs) and workflow of assembling MSBCAs using a reductionist synthetic community (Vorholt et al., 2017; Liu et al., 2019). (A) Biological features involving disease-suppressive effects of each individual strain, compatibility, diversity, microbial colonization of rhizosphere, mode of action for disease control, safety to humans and the environment, easy application and convenience to be incorporated into an existing management system (Raupach and Kloepper, 1998; Sikora et al., 2010; Bashan et al., 2013; Grosskopf and Soyer, 2014; Ahkami et al., 2017), need to be taken into account when establishing the MSBCAs for soil-borne diseases. (B) In general, methodology of constructing MSBCAs by a reductionist synthetic community approach is built on the conception of host-mediated selection of plant-associated microbiota (Mueller and Sachs, 2015). Microbiome analysis by 16S rRNA gene amplicon sequencing or metagenomics sequencing, or by PhyloChip analysis, in parallel to the extended microbial strain isolation to achieve as much diversity as possible, is an early step to pick the potential disease-suppressing species by identifying the differential OTUs between the microbiome of the samples collected from pathogen challenged and control plants (Berendsen et al., 2018), or by reconstructing strain-level genomes based on functional diversity (Carrión et al., 2019). Then, after cross-referencing the microbiota profiling data with the taxonomic identities of the isolates in comprehensive culture collections (Niu et al., 2017; Berendsen et al., 2018), or by detecting the genes encoding the functions of biological control in the genomes of cultivated isolates (Carrión et al., 2019), the candidate strains will be characterized and selected for the multi-strain community, of which the disease-reducing effects will be further evaluated.
Although empirically combining existing microbial isolates with biocontrol activity is useful, it is nearly impossible to predict efficiency of such consortia in suppressing plant disease and strengthening plant growth in the context of the whole plant microbiome. In contrast, utilizing a reduced number of representative members of the target host microbiota to build SynCom (Vorholt et al., 2017; Liu et al., 2019) will likely simplify handling and production of such MSBCAs. SynCom analysis performed in gnotobiotic systems allows us to study the effect of the plant microbiota on host fitness under different environmental circumstances. It also allows us to investigate microbe−microbe interactions and microbial gene functions (Carlström et al., 2019; Liu et al., 2019), and to construct novel MSBCAs.
Several microbial communities able to suppress plant diseases have been assembled via the reductionist SynCom approach based on microbiome analysis and comprehensive culture collections (Liu et al., 2019). A synthetic bacterial consortium was constructed, able to reduce the severity of the maize seedling blight caused by Fusarium verticillioides (Niu et al., 2017). The biocontrol effect of the synthetic community against F. verticillioides was stronger than that of each individual strain. To prepare this synthetic community, Niu et al. (2017) started from microbiota established by maize roots, which were identified by 16S rRNA gene amplicon sequencing and additional strain cultivating methods. A greatly simplified SynCom was obtained, consisting of seven strains, Enterobacter ludwigii, Stenotrophomonas maltophilia, Ochrobactrum pituitosum, Herbaspirillum frisingense, Pseudomonas putida, Curtobacterium pusillum, and Chryseobacterium indologenes, representing three of the four most dominant phyla found in maize roots.
A three-membered bacterial community able to induce systemic resistance in Arabidopsis thaliana against Hyaloperonospora arabidopsidis (downy mildew) was constructed (Berendsen et al., 2018) via host-mediated microbiome selection (Mueller and Sachs, 2015). Carrión et al. (2019) showed that infection of sugar beets by a fungal pathogen, Rhizoctonia solani, is hindered by an endosymbiotic community of bacteria living inside plant roots. This endophytic community was enriched for Chitinophagaceae and Flavobacteriaceae harboring chitinase genes and biosynthetic gene clusters encoding non-ribosomal peptide synthetases and polyketide synthases. A MSBCA consortium of Chitinophaga and Flavobacterium strains was established, which consistently suppressed fungal root disease. Carrión et al. (2019) concluded that endophytic root microbiomes may harbor many functional traits that can protect synergistically their host plants (Carrión et al., 2019).
Building MSBCAs by a reductionist SynCom approach (Figure 2B) offers the chance to accurately and rapidly pick out the microbial strains qualified for establishing the MSBCA from thousands of isolates found in the natural host microbiome. In this way, the crucial disease control-interactions present in the plant microbiome (Hassani et al., 2018) can be mirrored in the few selected strains used for the MSBCA. Establishing SynComs should be the method of choice. SynComs represent a helpful complement to pesticides, and might be combined in future application with effective empirical mixtures and/or single representatives of existing SSBCAs.
Utilization of selected beneficial microorganisms in the community is an effective approach to improve the efficiency of BCA (Figure 1A; Sarma et al., 2015; Mazzola and Freilich, 2017; Vorholt et al., 2017; Woo and Pepe, 2018). A necessary precondition for its success is the analysis of the microbial interactions among the members and the effect exerted by the MSBCA on plant health (Figure 1). When designing a MSBCA, two crucial interaction-related factors, compatibility and diversity, need to be considered (Figure 2A). Constructing MSBCA by combining microbes with great taxonomic distance appears desirable. We recommend a reductionist SynCom approach based on the principle of host-mediated microbiome selection (Mueller and Sachs, 2015), and selection of representative microbes to form efficient biocontrol consortia. This allows us to assemble customized MSBCAs depending on the specific requirements of disease management in different crops and environments. This strategy will result in protecting against distinct pathogens and might be comparable to the concept of “precision medicine” for human health (Berg et al., 2020), that advocates treatments of patients on a personalized level (Collins and Varmus, 2015) based on the patient’s genome sequence and their specific genome-environment interaction. Beside the practical use of MSBCAs as biopesticides, they may also serve as useful tools for investigating how microbial interspecies interactions affect plant microbiome assembly (Niu et al., 2017), and how evolutionary processes act on the plant holobiont (integrating the plant, the microbiome and the environment) (Hassani et al., 2018).
In this review, we summarize the potential mechanisms deployed by microbial components of communities to improve their disease-suppressing functions. Our understanding of these processes at the level of molecular mechanisms is rudimentary, especially the mechanisms of the initiation of rhizosphere colonization and the resulting elevated host immunity. Next, the technology of functional genomics, transcriptomics, proteomics and metabolomics will need to be applied to elucidate the genetic basis of enhanced biofilm formation, syntrophic microbial growth promotion and migration, and enhanced ISR. Although a 16S rRNA gene amplicon sequencing-based reductionist SynCom approach is useful to characterize MSBCAs, the relatively short reads may not achieve the taxonomic resolution needed to distinguish related strains (Edgar, 2018; Fuks et al., 2018). Thus, beside the high cost of a metagenomics approach, the utilization of modified 16S rRNA gene amplicon sequencing-based methods with improved resolution, such as full-length 16S rRNA gene amplicon sequencing (Callahan et al., 2019), may be expanded in the future when constructing the SynComs with biocontrol activity. In addition to the disease-suppressing function of the SynComs, their plant growth-promoting effects are worth further investigation (Zhang J. et al., 2019; Zhuang et al., 2020). So far, most MSBCAs have been applied in agriculture (Table 1), Using BCAs in forestry for plant disease control should be recommended. Finally, as agrochemical companies such as BASF, Syngenta and Bayer have developed and launched several MSBCA-based commercialized products for sustainable management of soil-borne pathogens, application of MSBCAs should bring more efficient control of soil-borne diseases in agriculture, horticulture, and forestry.
BN and RB conceived the idea, designed the outlines of the review, and wrote the manuscript. WW and ZY prepared the figures and the table. VC, RS, and HS revised the manuscript. All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the National Science Foundation of China Grant No. 31801785 (to BN), Northeast Forestry University International Forest Biotech Center Project Grant (to BN), Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team to VC), Heilongjiang Provincial Science Foundation of China Grant No. YQ2019C002 (to BN), Fundamental Research Funds for the Central Universities Grants Nos. 2572020DY24 and 2572018BD05 (to BN), and ENDOBICA-Novel antimicrobial compounds and endophytic bacteria for developing improved biocontrol agents by the German Ministry for Education and Research Grant 031B0582A (to RB). RB acknowledges support by the German Research Foundation (DFG) and the Open Access Publication Fund of Humboldt-Universität zu Berlin.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank members of the BN laboratory for valuable advice. RB acknowledges support by the German Research Foundation (DFG) and the Open Access Publication Fund of Humboldt-Universität zu Berlin.
Ahkami, A. H., Allen White, R., Handakumbura, P. P., and Jansson, C. (2017). Rhizosphere engineering: enhancing sustainable plant ecosystem productivity. Rhizosphere 3, 233–243. doi: 10.1016/j.rhisph.2017.04.012
Alizadeh, H., Behboudi, K., Ahmadzadeh, M., Javan-Nikkhah, M., Zamioudis, C., Pieterse, C. M. J., et al. (2013). Induced systemic resistance in cucumber and Arabidopsis thaliana by the combination of Trichoderma harzianum Tr6 and Pseudomonas sp. Ps14. Biol. Control 65, 14–23. doi: 10.1016/j.biocontrol.2013.01.009
Allard-Massicotte, R., Tessier, L., Lecuyer, F., Lakshmanan, V., Lucier, J. F., Garneau, D., et al. (2016). Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 7:e01664-16. doi: 10.1128/mBio.01664-16
Alvarez, B., and Biosca, E. G. (2017). Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front. Plant Sci. 8:1218. doi: 10.3389/fpls.2017.01218
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bardas, G. A., Lagopodi, A. L., Kadoglidou, K., and Tzavella-Klonari, K. (2009). Biological control of three Colletotrichum lindemuthianum races using Pseudomonas chlororaphis PCL1391 and Pseudomonas fluorescens WCS365. Biol. Control 49, 139–145. doi: 10.1016/j.biocontrol.2009.01.012
Bashan, Y. (1998). Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 16, 729–770. doi: 10.1016/S0734-9750(98)00003-2
Bashan, Y., de-Bashan, L. E., Prabhu, S. R., and Hernandez, J.-P. (2013). Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998-2013). Plant Soil 378, 1–33. doi: 10.1007/s11104-013-1956-x
Bashan, Y., and Holguin, G. (1997). Azospirillum-plant relationships: environmental and physiological advances (1990-1996). Can. J. Microbiol. 43, 103–121. doi: 10.1139/m97-015
Beauregard, P. B., Chai, Y., Vlamakis, H., Losick, R., and Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 110, E1621–E1630. doi: 10.1073/pnas.1218984110
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M.-C. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13.
Bhatia, S. K., Bhatia, R. K., Choi, Y.-K., Kan, E., Kim, Y.-G., and Yang, Y.-H. (2018). Biotechnological potential of microbial consortia and future perspectives. Crit. Rev. Biotechnol. 38, 1209–1229. doi: 10.1080/07388551.2018.1471445
Bolton, A. T. (1980). Control of Pythium aphanidermatum in poinsettia in a soilless culture by Trichoderma viride and a Streptomyces sp. Can. J. Plant Pathol. 2, 93–95. doi: 10.1080/07060668009501446
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Busby, P. E., Soman, C., Wagner, M. R., Friesen, M. L., Kremer, J., Bennett, A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15:e2001793. doi: 10.1371/journal.pbio.2001793
Callahan, B. J., Wong, J., Heiner, C., Oh, S., Theriot, C. M., Gulati, A. S., et al. (2019). High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 47:e103. doi: 10.1093/nar/gkz569
Carlström, C. I., Field, C. M., Bortfeld-Miller, M., Müller, B., Sunagawa, S., and Vorholt, J. A. (2019). Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat Ecol Evol. 3, 1445–1454. doi: 10.1038/s41559-019-0994-z
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., de Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Cazorla, F. M., and Mercado-Blanco, J. (2016). Biological control of tree and woody plant diseases: an impossible task? BioControl 61, 233–242. doi: 10.1007/s10526-016-9737-0
Chen, S., Qi, G., Ma, G., and Zhao, X. (2020). Biochar amendment controlled bacterial wilt through changing soil chemical properties and microbial community. Microbiol. Res. 231, 1–9. doi: 10.1016/j.micres.2019.126373
Chen, X. H., Koumoutsi, A., Scholz, R., Eisenreich, A., Schneider, K., Heinemeyer, I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. doi: 10.1038/nbt1325
Choure, K., Dubey, R. C., and Maheswar, D. K. (2012). Development of plant growth promoting microbial consortium based on interaction studies to reduce wilt incidence in Cajanus cajan L var. Manak. MiddleEast J. Sci. Res. 12, 1459–1470. doi: 10.5829/idosi.mejsr.2012.12.11.7
Chowdhury, S. P., Hartmann, A., Gao, X. W., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front. Microbiol. 6:780. doi: 10.3389/fmicb.2015.00780
Collins, F. S., and Varmus, H. (2015). A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795. doi: 10.1056/NEJMp1500523
de Novais, C. B., Sbrana, C., da Conceicao Jesus, E., Rouws, L. F. M., Giovannetti, M., Avio, L., et al. (2020). Mycorrhizal networks facilitate the colonization of legume roots by a symbiotic nitrogen-fixing bacterium. Mycorrhiza 30, 389–396. doi: 10.1007/s00572-020-00948-w
Domenech, J., Reddy, M. S., Kloepper, J. W., Ramos, B., and Gutierrez-Mañero, J. (2006). Combined application of the biological product LS213 with Bacillus, Pseudomonas or Chryseobacterium for growth promotion and biological control of soil-borne diseases in pepper and tomato. Biocontrol 51, 245–258. doi: 10.1007/s10526-005-2940-z
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983. doi: 10.1016/j.cell.2018.10.020
Edgar, R. C. (2018). Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375. doi: 10.1093/bioinformatics/bty113
Estevez de Jensen, C., Percich, J. A., and Graham, P. H. (2002). Integrated management strategies of bean root rot with Bacillus subtilis and Rhizobium in Minnesota. Field Crops. Res. 74, 107–115. doi: 10.1016/S0378-4290(01)00200-3
Ezziyyani, M., Requena, M. E., Egea-Gilabert, C., and Candela, M. E. (2007). Biological control of Phytophthora root rot of pepper using Trichoderma harzianum and Streptomyces rochei in combination. J. Phytopathol. 155, 342–349. doi: 10.1111/j.1439-0434.2007.01237.x
Fan, B., Chen, X. H., Budiharjo, A., Bleiss, W., Vater, J., and Borriss, R. (2011). Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J. Biotechnol. 151, 303–311. doi: 10.1016/j.jbiotec.2010.12.022
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Finkelshtein, A., Roth, D., Ben Jacob, E., and Ingham, C. J. (2015). Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. mBio 6:e00074-15. doi: 10.1128/mBio.00074-15
Fira, D., Dimkic, I., Beric, T., Lozo, J., and Stankovic, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Fitzpatrick, C. R., Salas-González, I., Conway, J. M., Finkel, O. M., Gilbert, S., Russ, D., et al. (2020). The plant microbiome: from ecology to reductionism and beyond. Annu. Rev. Microbiol. 74, 81–100. doi: 10.1146/annurev-micro-022620-014327
Frey-Klett, P., Burlinson, P., Deveau, A., Barret, M., Tarkka, M., and Sarniguet, A. (2011). Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 75, 583–609. doi: 10.1128/MMBR.00020-11
Fuks, G., Elgart, M., Amir, A., Zeisel, A., Turnbaugh, P. J., Soen, Y., et al. (2018). Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 6:17. doi: 10.1186/s40168-017-0396-x
Ghisalberti, E. L., and Sivasithamparam, K. (1991). Antifungal antibiotics produced by Trichoderma spp. Soil Biol. Biochem. 23, 1011–1020. doi: 10.1016/0038-0717(91)90036-J
Grosskopf, T., and Soyer, O. S. (2014). Synthetic microbial communities. Curr. Opin. Microbiol. 18, 72–77. doi: 10.1016/j.mib.2014.02.002
Gu, S., Wei, Z., Shao, Z., Friman, V.-P., Cao, K., Yang, T., et al. (2020). Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002–1010. doi: 10.1038/s41564-020-0719-8
Haas, D., and Defago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Haas, D., and Keel, C. (2003). Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41, 117–153. doi: 10.1146/annurev.phyto.41.052002.095656
Handelsman, J., and Stabb, E. V. (1996). Biocontrol of soilborne plant pathogens. Plant Cell 8, 1855–1869. doi: 10.1105/tpc.8.10.1855
Harshey, R. M. (2003). Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. doi: 10.1146/annurev.micro.57.030502.091014
Hassani, M. A., Duran, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
Hu, J., Wei, Z., Friman, V.-P., Gu, S.-H., Wang, X.-F., Eisenhauer, N., et al. (2016). Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 7:e01790-16. doi: 10.1128/mBio.01790-16
Huang, X.-F., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q., and Vivanco, J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275. doi: 10.1139/cjb-2013-0225
Ingham, C. J., Kalisman, O., Finkelshtein, A., and Ben-Jacob, E. (2011). Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex. Proc. Natl. Acad. Sci. U.S.A. 108, 19731–19736. doi: 10.1073/pnas.1102097108
Izquierdo-García, L. F., González-Almario, A., Cotes, A. M., and Moreno-Velandia, C. A. (2020). Trichoderma virens Gl006 and Bacillus velezensis Bs006: a compatible interaction controlling Fusarium wilt of cape gooseberry. Sci. Rep. 10:6857. doi: 10.1038/s41598-020-63689-y
Jacoby, R., Peukert, M., Succurro, A., Koprivova, A., and Kopriva, S. (2017). The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant Sci. 8:1617. doi: 10.3389/fpls.2017.01617
Jacoby, R. P., Chen, L., Schwier, M., Koprivova, A., and Kopriva, S. (2020). Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Res. 9:151. doi: 10.12688/f1000research.21796.1
Jain, A., Singh, A., Singh, S., and Singh, H. B. (2015). Biological management of Sclerotinia sclerotiorum in pea using plant growth promoting microbial consortium. J. Basic Microbiol. 55, 961–972. doi: 10.1002/jobm.201400628
Jain, A., Singh, S., Sarma, B. K., and Singh, H. B. (2012). Microbial consortium-mediated reprogramming of defence network in pea to enhance tolerance against Sclerotinia sclerotiorum. J. Appl. Microbiol. 112, 537–550. doi: 10.1111/j.1365-2672.2011.05220.x
Jansson, C., Vogel, J., Hazen, S., Brutnell, T., and Mockler, T. (2018). Climate-smart crops with enhanced photosynthesis. J. Exp. Bot. 69, 3801–3809. doi: 10.1093/jxb/ery213
Jansson, J. K., and Hofmockel, K. S. (2018). The soil microbiome-from metagenomics to metaphenomics. Curr. Opin. Microbiol. 43, 162–168. doi: 10.1016/j.mib.2018.01.013
Jansson, J. K., and Hofmockel, K. S. (2020). Soil microbiomes and climate change. Nat. Rev. Microbiol. 18, 35–46. doi: 10.1038/s41579-019-0265-7
Jousset, A., Becker, J., Chatterjee, S., Karlovsky, P., Scheu, S., and Eisenhauer, N. (2014). Biodiversity and species indentity shape the antifungal activity of bacterial communities. Ecology 95, 1184–1190. doi: 10.1890/13-1215.1
Kannan, V., and Sureendar, R. (2009). Synergistic effect of beneficial rhizosphere microflora in biocontrol and plant growth promotion. J. Basic Microbiol. 49, 158–164. doi: 10.1002/jobm.200800011
Kavino, M., and Manoranjitham, S. K. (2017). In vitro bacterization of banana (Musa spp.) with native endophytic and rhizospheric bacterial isolates: novel ways to combat Fusarium wilt. Eur. J. Plant Pathol. 151, 371–387. doi: 10.1007/s10658-017-1379-2
Kemen, E. (2014). Microbe-microbe interactions determine oomycete and fungal host colonization. Curr. Opin. Plant Biol. 20, 75–81. doi: 10.1016/j.pbi.2014.04.005
Kloepper, J. W., Leong, J., Teintze, M., and Schroth, M. N. (1980). Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286, 885–886. doi: 10.1038/286885a0
Kohlmeier, S., Smits, T. H. M., Ford, R. M., Keel, C., Harms, H., and Wick, L. Y. (2005). Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39, 4640–4646. doi: 10.1021/es047979z
Kwak, M.-J., Kong, H. G., Choi, K., Kwon, S.-K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Kwak, Y.-S., and Weller, D. M. (2013). Take-all of wheat and natural disease suppression: a review. Plant Pathol. J. 29, 125–135. doi: 10.5423/PPJ.SI.07.2012.0112
Lian, J., Wang, Z., Cao, L., Tan, H., Patrik, I., Jiang, Z., et al. (2009). Artificial inoculation of banana tissue culture plantlets with indigenous endophytes originally derived from native banana plants. Biol. Control 51, 427–434. doi: 10.1016/j.biocontrol.2009.08.002
Liechty, Z., Santos-Medellin, C., Edwards, J., Nguyen, B., Mikhail, D., Eason, S., et al. (2020). Comparative analysis of root microbiomes of rice cultivars with high and low methane emissions reveals differences in abundance of methanogenic archaea and putative upstream fermenters. mSystems 5:e00897-19. doi: 10.1128/mSystems.00897-19
Liu, K., McInroy, J. A., Hu, C.-H., and Kloepper, J. W. (2018). Mixtures of plant-growth-promoting rhizobacteria enhance biological control of multiple plant diseases and plant-growth promotion in the presence of pathogens. Plant Dis. 102, 67–72. doi: 10.1094/PDIS-04-17-0478-RE
Liu, Y.-X., Qin, Y., and Bai, Y. (2019). Reductionist synthetic community approaches in root microbiome research. Curr. Opin. Microbiol. 49, 97–102. doi: 10.1016/j.mib.2019.10.010
Loper, J. E., Hassan, K. A., Mavrodi, D. V., Davis, E. W., Lim, C. K., Shaffer, B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. doi: 10.1371/journal.pgen.1002784
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Lutz, M. P., Wenger, S., Maurhofer, M., Defago, G., and Duffy, B. (2004). Signaling between bacterial and fungal biocontrol agents in a strain mixture. FEMS Microbiol. Ecol. 48, 447–455. doi: 10.1016/j.femsec.2004.03.002
Lyons, N. A., and Kolter, R. (2017). Bacillus subtilis protects public goods by extending kin discrimination to closely related species. mBio 8:e00723-17. doi: 10.1128/mBio.00723-17
Maciag, T., Krzyzanowska, D. M., Jafra, S., Siwinska, J., and Czajkowski, R. (2020). The great five-an artificial bacterial consortium with antagonistic activity towards Pectobacterium spp. and Dickeya spp.: formulation, shelf life, and the ability to prevent soft rot of potato in storage. Appl. Microbiol. Biotechnol. 104, 4547–4561. doi: 10.1007/s00253-020-10550-x
Mathiyazhagan, K., Karuppannan, M., Balamohan, T., Kumar, N., Khartiba, L., and Ramasamy, S. (2014). Enhancement of growth and panama wilt resistance by in vitro co-culturing of banana plantlets with PGPR and endophytes. Acta Horticult. 1024, 277–282. doi: 10.17660/ActaHortic.2014.1024.37
Mazzola, M., and Freilich, S. (2017). Prospects for biological soilborne disease control: application of indigenous versus synthetic microbiomes. Phytopathology 107, 256–263. doi: 10.1094/PHYTO-09-16-0330-RVW
McKellar, M. E., and Nelson, E. B. (2003). Compost-induced suppression of Pythium damping-off is mediated by fatty-acid-metabolizing seed-colonizing microbial communities. Appl. Environ. Microbiol. 69, 452–460. doi: 10.1128/aem.69.1.452-460.2003
Mee, M. T., Collins, J. J., Church, G. M., and Wang, H. H. (2014). Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. U.S.A. 111, E2149–E2156. doi: 10.1073/pnas.1405641111
Migheli, Q. (2001). Genetically modified biocontrol agents: environmental impact and risk analysis. J. Plant Pathol. 83, 47–56.
Mohandas, S., Manjula, R., Rawal, R. D., Lakshmikantha, H. C., Chakraborty, S., and Ramachandra, Y. L. (2010). Evaluation of arbuscular mycorrhiza and other biocontrol agents in managing Fusarium oxysporum f. sp. Cubense infection in banana cv. Neypoovan. Biocontrol. Sci. Technol. 20, 165–181. doi: 10.1080/09583150903438439
Mohd Fishal, E. M., Meon, S., and Yun, W. M. (2010). Induction of tolerance to Fusarium wilt and defense-related mechanisms in the plantlets of susceptible berangan banana pre-inoculated with Pseudomonas sp. (UPMP3) and Burkholderia sp. (UPMB3). Agric. Sci. China 9, 1140–1149. doi: 10.1016/s1671-2927(09)60201-7
Morris, B. E., Henneberger, R., Huber, H., and Moissl-Eichinger, C. (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. doi: 10.1111/1574-6976.12019
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Naylor, D., and Coleman-Derr, D. (2017). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8:2223. doi: 10.3389/fpls.2017.02223
Niu, B., Paulson, J. N., Zheng, X., and Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A. 114, E2450–E2459. doi: 10.1073/pnas.1616148114
Nützmann, H.-W., Reyes-Dominguez, Y., Scherlach, K., Schroeckh, V., Horn, F., Gacek, A., et al. (2011). Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. U.S.A 108, 14282–14287. doi: 10.1073/pnas.1103523108
Ola, A. R., Thomy, D., Lai, D., Brotz-Oesterhelt, H., and Proksch, P. (2013). Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 76, 2094–2099. doi: 10.1021/np400589h
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Pierson, E. A., and Weller, D. M. (1994). Use of mixtures of fluorescent pseudomonads to suppress take-all and improve the growth of wheat. Phytopathology 84, 940–947. doi: 10.1094/Phyto-84-940
Pishchany, G., Mevers, E., Ndousse-Fetter, S., Horvath, D. J. Jr., Paludo, C. R., Silva-Junior, E. A., et al. (2018). Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc. Natl. Acad. Sci. U.S.A. 115, 10124–10129. doi: 10.1073/pnas.1807613115
Pliego, C., de Weert, S., Lamers, G., de Vicente, A., Bloemberg, G., Cazorla, F. M., et al. (2008). Two similar enhanced root-colonizing Pseudomonas strains differ largely in their colonization strategies of avocado roots and Rosellinia necatrix hyphae. Environ. Microbiol. 10, 3295–3304. doi: 10.1111/j.1462-2920.2008.01721.x
Raaijmakers, J. M., and Weller, D. M. (1998). Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 11, 144–152. doi: 10.1094/MPMI.1998.11.2.144
Raupach, G. S., and Kloepper, J. (1998). Mixtures of plant growth-promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology 88, 1158–1164. doi: 10.1094/PHYTO.1998.88.11.1158
Santhanam, R., Menezes, R. C., Grabe, V., Li, D., Baldwin, I. T., and Groten, K. (2019). A suite of complementary biocontrol traits allows a native consortium of root-associated bacteria to protect their host plant from a fungal sudden-wilt disease. Mol. Ecol. 28, 1154–1169. doi: 10.1111/mec.15012
Sarma, B. K., Yadav, S. K., Singh, S., and Singh, H. B. (2015). Microbial consortium-mediated plant defense against phytopathogens: readdressing for enhancing efficacy. Soil Biol. Biochem. 87, 25–33. doi: 10.1016/j.soilbio.2015.04.001
Segev, E., Wyche, T. P., Kim, K. H., Petersen, J., Ellebrandt, C., Vlamakis, H., et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLife 5:e17473. doi: 10.7554/eLife.17473
Shanmugam, V., and Kanoujia, N. (2011). Biological management of vascular wilt of tomato caused by Fusarium oxysporum f.sp. lycospersici by plant growth-promoting rhizobacterial mixture. Biol. Control 57, 85–93. doi: 10.1016/j.biocontrol.2011.02.001
Sikora, R. A., Zum Felde, A., Mendoza, A., Menjivar, R., and Pocasangre, L. (2010). “In planta suppressiveness to nematodes and long term root health stability through biological enhancement-Do we need a cocktail?,” in IV International Symposium on Banana: International Conference on Banana and Plantain in Africa: Harnessing International, Mombasa, 879. doi: 10.17660/ActaHortic.2010.879.60
Solanki, M. K., Yandigeri, M. S., Kumar, S., Singh, R. K., and Srivastava, A. K. (2019). Co-inoculation of different antagonists can enhance the biocontrol activity against Rhizoctonia solani in tomato. Antonie Van Leeuwenhoek 112, 1633–1644. doi: 10.1007/s10482-019-01290-8
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Srivastava, R., Khalid, A., Singh, U. S., and Sharma, A. K. (2010). Evaluation of arbuscular mycorrhizal fungus, fluorescent Pseudomonas and Trichoderma harzianum formulation against Fusarium oxysporum f. sp. lycopersici for the management of tomato wilt. Biol. Control 53, 24–31. doi: 10.1016/j.biocontrol.2009.11.012
Stemke, D. J. (2004). “Genetically modified microorganisms,” in The GMO Handbook: Genetically Modified Animals, Microbes, and Plants in Biotechnology, ed. S. R. Parekh, (Totowa, NJ: Humana Press), 85–130.
Stockwell, V. O., Johnson, K. B., Sugar, D., and Loper, J. E. (2011). Mechanistically compatible mixtures of bacterial antagonists improve biological control of fire blight of pear. Phytopathology 101, 113–123. doi: 10.1094/PHYTO-03-10-0098
Subramoni, S., Nathoo, N., Klimov, E., and Yuan, Z.-C. (2014). Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front. Plant Sci. 5:322. doi: 10.3389/fpls.2014.00322
Syed Ab Rahman, S. F., Singh, E., Pieterse, C. M. J., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Teixeira, P. J. P., Colaianni, N. R., Fitzpatrick, C. R., and Dangl, J. L. (2019). Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17. doi: 10.1016/j.mib.2019.08.003
Thakkar, A., and Saraf, M. (2014). Development of microbial consortia as a biocontrol agent for effective management of fungal diseases in Glycine max L. Arch. Phytopathol. Plant Prot. 48, 459–474. doi: 10.1080/03235408.2014.893638
Thakur, M. P., and Geisen, S. (2019). Trophic regulations of the soil microbiome. Trends Microbiol. 27, 771–780. doi: 10.1016/j.tim.2019.04.008
Thangavelu, R., and Gopi, M. (2015a). Combined application of native Trichoderma isolates possessing multiple functions for the control of Fusarium wilt disease in banana cv. Grand Naine. Biocontrol. Sci. Technol. 25, 1147–1164. doi: 10.1080/09583157.2015.1036727
Thangavelu, R., and Gopi, M. (2015b). Field suppression of Fusarium wilt disease in banana by the combined application of native endophytic and rhizospheric bacterial isolates possessing multiple functions. Phytopathol. Mediterr. 54, 241–252. doi: 10.14601/Phytopathol_Mediterr-15160
Thomloudi, E.-E., Tsalgatidou, P. C., Douka, D., Spantidos, T.-N., Dimou, M., Venieraki, A., et al. (2019). Multistrain versus single-strain plant growth promoting microbial inoculants - The compatibility issue. Hell. Plant Prot. J. 12, 61–77. doi: 10.2478/hppj-2019-0007
Traxler, M. F., and Kolter, R. (2015). Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970. doi: 10.1039/c5np00013k
Traxler, M. F., Watrous, J. D., Alexandrov, T., Dorrestein, P. C., and Kolter, R. (2013). Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4:e00459-13. doi: 10.1128/mBio.00459-13
Triveni, S., Prasanna, R., and Saxena, A. K. (2012). Optimization of conditions for in vitro development of Trichoderma viride-based biofilms as potential inoculants. Folia Microbiol. 57, 431–437. doi: 10.1007/s12223-012-0154-1
Umesha, S., Singh, P. K., and Singh, R. P. (2018). “Microbial Biotechnology and Sustainable Agriculture,” in Biotechnology for Sustainable Agriculture, eds R. L. Singh and S. Mondal, (Amsterdam: Elsevier), 185–205.
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., Elhottova, D., Kristufek, V., and Salles, J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.1109326109
Venieraki, A., Tsalgatidou, P. C., Georgakopoulos, D. G., Dimou, M., and Katinakis, P. (2016). Swarming motility in plant-associated bacteria. Hell. Plant Prot. J. 9, 16–27. doi: 10.1515/hppj-2016-0002
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R., and Kolter, R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Vorholt, J. A., Vogel, C., Carlström, C. I., and Muller, D. B. (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155. doi: 10.1016/j.chom.2017.07.004
Wang, G., Ma, Y., Chenia, H. Y., Govinden, R., Luo, J., and Ren, G. (2020). Biochar-mediated control of phytophthora blight of pepper is closely related to the improvement of the rhizosphere fungal community. Front. Microbiol. 11:1427. doi: 10.3389/fmicb.2020.01427
Warmink, J. A., Nazir, R., Corten, B., and van Elsas, J. D. (2011). Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol. Biochem. 43, 760–765. doi: 10.1016/j.soilbio.2010.12.009
Wei, Z., Yang, T., Friman, V.-P., Xu, Y., Shen, Q., and Jousset, A. (2015). Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6:8413. doi: 10.1038/ncomms9413
Whipps, J. M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. doi: 10.1093/jexbot/52.suppl_1.487
Wong, C. K. F., Saidi, N. B., Vadamalai, G., Teh, C. Y., and Zulperi, D. (2019). Effect of bioformulations on the biocontrol efficacy, microbial viability and storage stability of a consortium of biocontrol agents against Fusarium wilt of banana. J. Appl. Microbiol. 127, 544–555. doi: 10.1111/jam.14310
Woo, S. L., and Pepe, O. (2018). Microbial consortia: promising probiotics as plant biostimulants for sustainable agriculture. Front. Plant Sci. 9:1801. doi: 10.3389/fpls.2018.01801
Wu, Q., Ni, M., Dou, K., Tang, J., Ren, J., Yu, C., et al. (2018). Co-culture of Bacillus amyloliquefaciens ACCC11060 and Trichoderma asperellum GDFS1009 enhanced pathogen-inhibition and amino acid yield. Microb. Cell Fact. 17:155. doi: 10.1186/s12934-018-1004-x
Xu, X.-M., Jeffries, P., Pautasso, M., and Jeger, M. J. (2011). Combined use of biocontrol agents to manage plant diseases in theory and practice. Phytopathology 101, 1024–1031. doi: 10.1094/PHYTO-08-10-0216
Yang, H., Ma, J., Rong, Z., Zeng, D., Wang, Y., Hu, S., et al. (2019). Wheat straw return influences nitrogen-cycling and pathogen associated soil microbiota in a wheat-soybean rotation system. Front. Microbiol. 10:811. doi: 10.3389/fmicb.2019.01811
Yang, W., Zheng, L., Liu, H.-X., Wang, K.-B., Yu, Y.-Y., Luo, Y.-M., et al. (2014). Evaluation of the effectiveness of a consortium of three plant-growth promoting rhizobacteria for biocontrol of cotton Verticillium wilt. Biocontrol. Sci. Technol. 24, 489–502. doi: 10.1080/09583157.2013.873389
Zhang, J., Liu, Y.-X., Zhang, N., Hu, B., Tao, J., Xu, H., et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684. doi: 10.1038/s41587-019-0104-4
Zhang, L.-N., Wang, D.-C., Hu, Q., Dai, X.-Q., Xie, Y.-S., Li, Q., et al. (2019). Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 10:1668. doi: 10.3389/fmicb.2019.01668
Zhang, W., Li, X.-G., Sun, K., Tang, M.-J., Xu, F.-J., Zhang, M., et al. (2020). Mycelial network-mediated rhizobial dispersal enhances legume nodulation. ISME J. 14, 1015–1029. doi: 10.1038/s41396-020-0587-5
Zhou, D. M., Feng, H., Schuelke, T., De Santiago, A., Zhang, Q. M., Zhang, J. F., et al. (2019). Rhizosphere microbiomes from root knot nematode non-infested plants suppress nematode infection. Microb. Ecol. 78, 470–481. doi: 10.1007/s00248-019-0319
Keywords: microbial interaction, biological control agents, soil-borne disease, consortia, microbiome and community
Citation: Niu B, Wang W, Yuan Z, Sederoff RR, Sederoff H, Chiang VL and Borriss R (2020) Microbial Interactions Within Multiple-Strain Biological Control Agents Impact Soil-Borne Plant Disease. Front. Microbiol. 11:585404. doi: 10.3389/fmicb.2020.585404
Received: 20 July 2020; Accepted: 14 September 2020;
Published: 09 October 2020.
Edited by:
Soumitra Paul Chowdhury, Helmholtz Zentrum München, GermanyReviewed by:
Xiaoyulong Chen, Guizhou University, ChinaCopyright © 2020 Niu, Wang, Yuan, Sederoff, Sederoff, Chiang and Borriss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben Niu, YmVuX25pdUBuZWZ1LmVkdS5jbg==; Rainer Borriss, cmFpbmVyLmJvcnJpc3NAcnouaHUtYmVybGluLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.