- Department of Bionanoscience, Kavli Institute of Nanoscience, Delft University of Technology, Delft, Netherlands
The molecule guanosine tetraphophosphate (ppGpp) is most commonly considered an alarmone produced during acute stress. However, ppGpp is also present at low concentrations during steady-state growth. Whether ppGpp controls the same cellular targets at both low and high concentrations remains an open question and is vital for understanding growth rate regulation. It is widely assumed that basal ppGpp concentrations vary inversely with growth rate, and that the main function of basal ppGpp is to regulate transcription of ribosomal RNA in response to environmental conditions. Unfortunately, studies to confirm this relationship and to define regulatory targets of basal ppGpp are limited by difficulties in quantifying basal ppGpp. In this Perspective we compare reported concentrations of basal ppGpp in E. coli and quantify ppGpp within several strains using a recently developed analytical method. We find that although the inverse correlation between ppGpp and growth rate is robust across strains and analytical methods, absolute ppGpp concentrations do not absolutely determine RNA synthesis rates. In addition, we investigated the consequences of two separate RNA polymerase mutations that each individually reduce (but do not abolish) sensitivity to ppGpp and find that the relationship between ppGpp, growth rate, and RNA content of single-site mutants remains unaffected. Both literature and our new data suggest that environmental conditions may be communicated to RNA polymerase via an additional regulator. We conclude that basal ppGpp is one of potentially several agents controlling ribosome abundance and DNA replication initiation, but that evidence for additional roles in controlling macromolecular synthesis requires further study.
Introduction
How might a bacteria cell measure its own growth rate? In the model bacterium Escherichia coli, the small molecule guanosine tetraphosphate (ppGpp) is closely tied to growth rate control. However, due to the circumstances of its discovery, ppGpp is more familiar as a stress or starvation signal. ppGpp and guanosine pentaphosphate (pppGpp), collectively called (p)ppGpp, were first identified in E. coli as compounds produced in strains that inhibit stable RNA synthesis upon amino acid starvation, a phenomenon known as the stringent response (Cashel and Gallant, 1969). The source of (p)ppGpp during the stringent response is the enzyme RelA, which synthesizes (p)ppGpp in response to uncharged (non-aminoacyl) tRNA binding the acceptor site of an actively translating ribosome (Haseltine and Block, 1973). The high concentrations of (p)ppGpp observed during acute starvation (600–1000 pmol OD–1 for ppGpp) (Harshman and Yamazaki, 1971; Lazzarini et al., 1971) drive profound responses via both transcriptional and post-translational mechanisms (Traxler et al., 2008; Kanjee et al., 2012). The overall result of the stringent response is strong inhibition of all macromolecule synthesis (rRNA, DNA, proteins, phospholipids, and peptidoglycan), leading to growth arrest. (p)ppGpp is hydrolyzed by the essential enzyme SpoT. SpoT also carries an active (p)ppGpp synthase domain, although the specific biochemical trigger of (p)ppGpp synthesis by SpoT has not yet been identified. As a recent study suggests that ppGpp is more potent than pppGpp in mediating growth rate control in E. coli (Mechold et al., 2013) we focus exclusively upon ppGpp.
Comparatively less well-understood are the functions of “basal ppGpp”: the ppGpp concentrations observed during steady-state growth in E. coli in the absence of stress (between 10 and 90 pmol OD–1). When growth rate is varied by nutritional quality, basal ppGpp correlates inversely with growth rate. Basal ppGpp is essential in minimal media as it is required to activate transcription of amino acid pathways (Traxler et al., 2011). Just as ppGpp strongly inhibits stable RNA synthesis at high concentrations, basal ppGpp mildly inhibits transcriptional initiation from ribosomal RNA (rRNA) promoters, thus at least partly determining ribosome abundance during steady-state growth (Ryals et al., 1982). The inverse relationship between ppGpp and growth rate is thought to reflect the rheostat-like function of ppGpp as a regulator of ribosomal biosynthesis in response to nutrient availability. If ppGpp is artificially deviated from natural basal concentrations, growth is slowed, suggesting that ppGpp finds a growth-optimum level and adjusts rRNA expression accordingly (Zhu and Dai, 2019). The rate of DNA replication initiation also adjusts to small increases in basal ppGpp (Schreiber et al., 1995), and a strain entirely lacking ppGpp [relA– spoT–, or (p)ppGpp0] does not vary DNA replication initiation in response to growth rate, suggesting that ppGpp participates in regulating the DNA-biomass ratio (Fernández-Coll et al., 2020). As ribosome abundance is one of the factors determining the global protein synthesis rate, ppGpp may act as a growth rate-reporting signal that smoothly adjusts the steady-state rate of overall biomass synthesis (Hui et al., 2015). ppGpp should be thus considered a growth rate speedometer as well as a stress signal.
The observation that high ppGpp concentrations inhibit biomass synthesis suggests that basal ppGpp concentrations might also directly regulate all biomass synthesis pathways during steady-state growth, in addition to regulating stable RNA synthesis. This hypothetical layer of regulation would complement control of rRNA transcription, which determines the maximum rate of steady-state protein synthesis. For example, studies suggest that basal ppGpp might also regulate the instantaneous rate of protein synthesis by limiting purine synthesis (PurF, Wang et al., 2018) or translation cofactor activities (e.g., IF2, Dai et al., 2016) or the fraction of active ribosomes (Zhang et al., 2018). Basal ppGpp might also regulate cell envelope biosynthesis (Noga et al., 2020), as high ppGpp inhibits phospholipid synthesis, and inhibition of phospholipid synthesis arrests peptidoglycan synthesis (Ishiguro et al., 1980; Heath et al., 1994). In this extreme “orchestra conductor” model, ppGpp would not only regulate ribosome abundance, but would also tightly synchronize the synthesis rates of each macromolecule. This model expands the role of ppGpp beyond its better-established role, which is to inhibit general biomass synthesis at starvation-induced concentrations (>600 pmol OD–1) like an emergency brake.
Defining the targets of basal ppGpp and identifying how basal ppGpp is maintained are two goals essential to understand how E. coli controls growth. To further encourage the recent revival of interest in the mechanisms of steady-state growth regulation and homeostasis (Scott et al., 2010, 2014; Dai et al., 2016), we contribute this Perspective on basal ppGpp in E. coli to evaluate the widely assumed notion that ppGpp is always inversely proportional to growth rate.
Quantifying Basal ppGpp Is Difficult but Essential
Actual ppGpp measurements are essential for determining which cellular processes are watching the growth rate speedometer. The rarity of basal ppGpp measurements is understandable as basal ppGpp is difficult to accurately quantify. The main challenges in measuring basal ppGpp in vivo are (1) its low abundance; (2) its chemical instability; (3) the presence of environmentally sensitive enzymes that rapidly hydrolyze and synthesize ppGpp. This means the analytical method must both chemically stabilize ppGpp and immediately denature all enzymes that synthesize or hydrolyze ppGpp. Moreover, in order to study ppGpp dynamics relevant to the rapid ppGpp response (<5 s), the method must enable fast sampling.
Despite these difficulties, several (p)ppGpp measurement methods have been developed, including thin layer chromatography (TLC) (Bochners and Ames, 1982; Sarubbi et al., 1988; Fernández-Coll and Cashel, 2018), high performance liquid chromatography (HPLC) (Ryals et al., 1982; Buckstein et al., 2008; Bokinsky et al., 2013; Varik et al., 2017; Jin et al., 2018) and liquid chromatography mass spectrometry (LC-MS) (Ihara et al., 2015; Patacq et al., 2018). The advantages and disadvantages of each method have summarized in Supplementary Table 1. Whichever method is used, rapid quenching of metabolism during sampling (i.e., no centrifuging of live cells) is required.
ppGpp Is a Reliable Growth Rate Speedometer: Most Basal ppGpp Measurements Indicate That the Inverse Correlation Between ppGpp and Growth Rate Trends Is Robust
A survey of reported basal ppGpp concentrations combined with our own measurements (Figure 1) indicates that despite apparent variability in absolute concentrations, the inverse correlation with growth rate is robust. We do not include pppGpp measurements, which are less-often reported. All data can be found in the Supplementary Material.
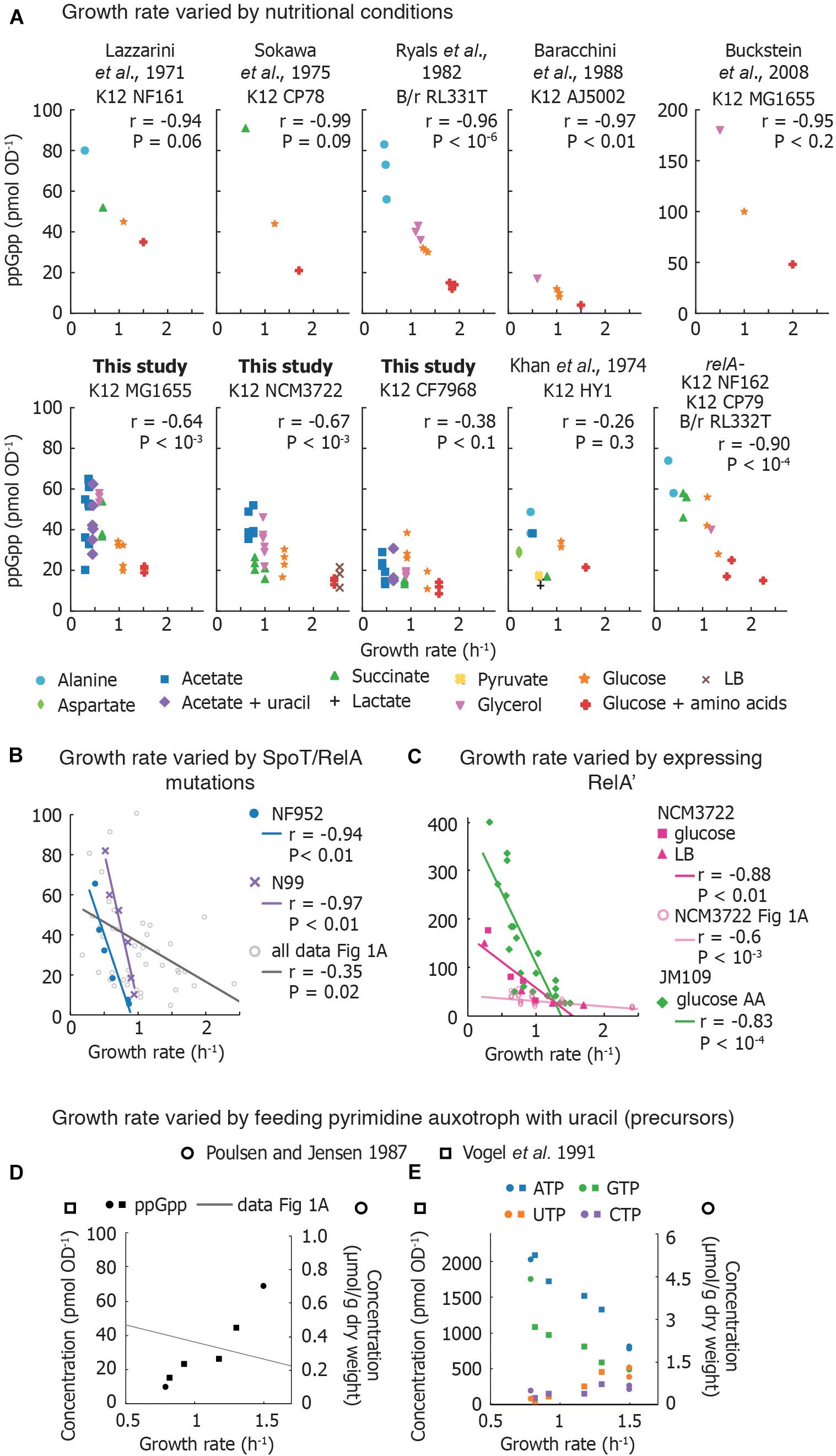
Figure 1. Compilation of measurements of basal ppGpp vs. growth rate. (A) Basal ppGpp concentrations measured in E. coli strains that generally follow an inverse correlation with growth rate. Both literature data and data obtained for this study are shown (Supplementary Section 3). For data from this study, three technical replicates per biological replicate are shown. For data taken from the literature, the number of biological replicates varies and can be found in Supplementary Material. (B) Trends obtained by several E. coli strains bearing various mutations in ppGpp synthase/hydrolase enzyme SpoT grown in glucose minimal medium. Each point represents values obtained from one strain. This is compared to compiled data from A. The genotype of each strain used can be found in Supplementary Material. (C) Basal ppGpp measured during overexpression of RelA in E. coli JM109 (in glucose amino acids without Gln, Glu) and in E. coli NCM3722 in LB and glucose minimal medium. (D,E) Ribonucleotide concentrations in E. coli strains defective in pyrimidine synthesis. (D) ppGpp concentrations as a function of growth rate, overlaid with the compiled ppGpp vs. growth rate data of (A), and (E) intracellular ATP, GTP, UTP, and CTP concentrations. References to data sets are provided in the Supplementary Material. For (A–D), the Pearson correlation coefficient r and significance figure P (from a two-tailed significance test) is shown.
ppGpp Is an Accurate Growth Rate Speedometer in Wild-Type E. coli When Growth Rate Is Varied by Nutrient Source
Previously Reported Measurements
Measurements in laboratory-adapted E. coli show an inverse correlation between growth rate and basal ppGpp (15–90 pmol OD–1) (Figure 1A). Early studies connected basal ppGpp, growth rate, and stable RNA abundance (Lazzarini et al., 1971; Sokawa et al., 1975; Ryals et al., 1982). Khan and Yamazaki (1974) measured basal ppGpp in an E. coli patient isolate and found several conditions in which ppGpp concentrations do not align smoothly with the expected trend (Figure 1A). However, RNA/DNA ratios in the outlier cultures followed the expected trend with growth rate (Khan and Yamazaki, 1974).
Reported ppGpp concentrations may differ perhaps due to differences in strains, turbidimeter calibration, or sampling method (Baracchini et al., 1988; Buckstein et al., 2008). Interestingly, even biological replicates show substantial variability (Ryals et al., 1982). Using an assay that exhibited less than 10% variation between individual measurements, Murray and Bremer (1996) report that ppGpp concentrations show 20% variations between biological replicates.
New Measurements (This Study)
We measured basal ppGpp concentrations in three E. coli K-12 strains using LC-MS (Figure 1A). Concentrations in MG1655 have been reported in Buckstein et al. (2008) for only three conditions (Figure 1A). NCM3722 is becoming increasingly popular as it lacks several genetic defects of MG1655 (Soupene et al., 2003; Brown and Jun, 2015). CF7968 (MG1655 rph+ but not isogenic with the MG1655 reported here) has been used to demonstrate correlation between the RNA/protein ratio and the growth rate (Potrykus et al., 2011). All three strains exhibit the expected inverse correlation between ppGpp and growth rate. We observed 20% variation in technical replicates from a single culture and 23% average variation between biological replicates, similar to previous reports (Murray and Bremer, 1996).
RelA Is Not Required to Establish the Correlation Between Basal ppGpp and Growth Rate
At least three studies have compared basal ppGpp concentrations of isogenic relA+ and relA– strains (Lazzarini et al., 1971; Sokawa et al., 1975; Ryals et al., 1982). Each study observed no significant difference in ppGpp concentrations and growth rates in relA+ and relA–, indicating that SpoT alone is able to establish an inverse correlation between ppGpp and growth rate (Figure 1A).
What Metabolic Process Sets Basal ppGpp Concentrations?
The observation that SpoT is able to maintain the ppGpp-growth rate correlation on its own does not necessarily indicate that RelA activity plays no role in maintaining basal ppGpp. As deacylated tRNA stimulates RelA activity (Haseltine and Block, 1973), one might presume that deacyl-tRNA abundance also correlates inversely with growth rate. However, the aminoacylated fraction of at least six tRNA species remains constant across growth rates. As total tRNA increases in parallel with rRNA, the absolute concentration of deacylated tRNA should also increase with growth rate (Dai et al., 2016). There are 43 different tRNA species in E. coli, and currently it is not known whether the acylation of each individual tRNA species varies. However, we surmise that RelA may contribute to basal ppGpp as relA– mutants exhibit different ribosome pausing behavior than wild-type (Li et al., 2018).
Identifying how SpoT maintains basal ppGpp is essential to understand the metabolic cues that lead to the inverse ppGpp-growth rate correlation. As hydrolysis and synthesis activities of a SpoT homolog are mutually exclusive (Hogg et al., 2004), basal ppGpp is likely set by a balance between the fraction of SpoT proteins engaged in either ppGpp synthesis or ppGpp hydrolysis. Environmental triggers that bias SpoT toward ppGpp synthesis were explored by Murray and Bremer (1996), and included carbon starvation, azide exposure, and simultaneous removal of all 20 amino acids. Although potential regulators have been identified [acyl carrier protein (Battesti and Bouveret, 2006), anti-sigma factor Rsd (Lee et al., 2018) and the small protein YtfK (Germain et al., 2019)], it is unclear how these regulate SpoT.
Tricking the Growth Rate Speedometer: Artificial Variations of ppGpp Within Fixed Nutritional Conditions
Mutation and overexpression of the relA and spoT genes enable different ppGpp concentrations in identical nutritional conditions. This also leads to an inverse relationship between ppGpp and growth rate.
Point Mutations in RelA and SpoT Change Basal ppGpp by Rebalancing Rates of ppGpp Synthesis and Hydrolysis
Non-disabling mutations in spoT and relA perturb the balance between ppGpp synthesis and hydrolysis, resulting in varied basal ppGpp levels while retaining viability (Sarubbi et al., 1988). Two sets of mutations in different E. coli strains (Supplementary Material) exhibited inverse relationships between ppGpp and growth rate that appear to be steeper than the majority of basal level trends observed in various media (Figure 1B). However, as the ppGpp-growth rate relationship was not determined in the parental strains from which these mutants were obtained, direct comparisons with wild-type behaviors are not possible. rRNA decreases as basal ppGpp increases in spoT mutant strains as expected (Sarubbi et al., 1988; Hernandez and Bremer, 1990).
Ectopic Overexpression of RelA Generates a Steep ppGpp-Growth Rate Trendline
Overexpressing the catalytic domain of RelA (referred to as RelA′ or RelA∗) artificially elevates ppGpp, inhibits rRNA synthesis and decreases growth rate. Data from ppGpp titrations using RelA′ are compared in Figure 1C. Schreiber et al. (1991) titrated ppGpp concentrations using RelA′ and obtained a ppGpp-growth rate trend that appears steeper than curves obtained for other strains. Two groups recently observed that RelA′ overexpression leads to higher basal ppGpp than expected for a given growth rate. RelA′ overexpression in E. coli NCM3722 in both LB medium and glucose minimal medium yields a ppGpp-growth rate curve steeper than the curve obtained in nutrient-limited NCM3722 (confirmed using a Chi-squared test, P < 10–6) (Zhu et al., 2019; Noga et al., 2020). However, the RNA/protein ratio vs. growth rate trends measured in ppGpp- and carbon-limited cultures closely overlap. This suggests that in the absence of stress, growth rate and RNA synthesis control can be decoupled from absolute ppGpp concentrations while still obeying an inverse relationship.
Breaking the Growth Rate Speedometer: When ppGpp Is Not Inversely Correlated With Growth Rate
Nucleotide Starvation Causes a Positive Correlation Between Growth Rate and ppGpp
The most dramatic departure from the canonical ppGpp-growth rate trend has been accomplished by disrupting nucleotide metabolism. When the growth rates of nucleotide auxotrophs are titrated by adding limiting nucleotides or nucleotide precursors, ppGpp increases in parallel with growth rate. Poulsen and Jensen (1987) first observed this phenomenon in E. coli mutants unable to synthesize specific nucleotides [carAB- guaB(ts)]. When growth rate was titrated with pyrimidine and purine sources, the authors inverted the correlation between ppGpp and growth rate (Figure 1D). Vogel et al. (1991) also found that ppGpp concentrations increased from 15 to 44 pmol OD–1 in parallel with growth rate and total RNA in a pyrimidine auxotroph (Figure 1D).
Why does ppGpp correlate positively with growth rate when growth is limited by pyrimidine (uracil) supply? First, uracil limitation does not activate ppGpp synthesis in wild-type strains (Cashel and Gallant, 1969), indicating that RelA and SpoT do not detect all forms of starvation. Second, these auxotrophs exhibit low concentrations of UTP and CTP (Figure 1E) suggesting that ribosome abundance – and thus translation rate – is controlled by substrate concentrations in these mutants (NTP) rather than by inhibitor concentrations (ppGpp). As NTP limitation is relieved, other metabolites likely become limiting for growth, triggering ppGpp synthesis.
Growth Rate and RNA Content Do Not Strictly Follow ppGpp Concentrations During Out-of-Steady-State Growth Transitions
Steady-state correlations such as the correlation between ppGpp and growth rate imply but do not establish regulatory connections. Hypotheses inspired by correlations must be tested by environmental perturbations. Baracchini and Bremer (1988) monitored growth and rRNA synthesis while adjusting basal ppGpp by adding pseudomonic acid to E. coli glucose cultures. Pseudomonic acid causes accumulation of uncharged tRNA and increases ppGpp. High concentrations of pseudomonic acid abruptly increased ppGpp and rapidly arrested growth, consistent with the stringent response. Low concentrations of pseudomonic acid also triggered ppGpp synthesis (up to 60–100 pmol OD–1) and an immediate but smaller decrease in rRNA synthesis. However, the instantaneous growth rate (monitored by optical density) was not perturbed in the short term by small increases in ppGpp concentrations.
These out-of-steady-state experiments reveal several important limitations of basal ppGpp regulation. First, the correlation between basal ppGpp and growth rate is broken when growth is out of steady state: were the relationship between growth rate and ppGpp to be as strict during growth transitions as during steady-state growth, a tripling in basal ppGpp would cause an immediate corresponding reduction in growth. Second, unlike the rapid protein synthesis inhibition caused by high ppGpp concentrations (Svitil et al., 1993), small increases of ppGpp (<100 pmol OD–1) do not seem to immediately inhibit biomass synthesis (with the exception of stable RNA). This undermines any notion that basal ppGpp directly regulates the instantaneous translation rate. Finally, two additional studies demonstrate that ppGpp and the rate of stable RNA synthesis can be transiently decoupled during nutritional upshifts, suggesting that additional signals may regulate rRNA synthesis (Friesen et al., 1975; Hansen et al., 1975).
Measurements of Basal ppGpp Reveal That Disruption of ppGpp Binding Sites on RNA Polymerase Does Not Abolish Correlation Between Basal ppGpp, RNA, and Growth Rate
To determine whether RNA polymerase (RNAP) retains regulation by basal ppGpp if its two ppGpp binding sites are disrupted, we measured basal ppGpp levels, growth rates and cellular RNA in E. coli strains expressing RNAP mutants (Ross et al., 2013, 2016). Although we did not test a strain bearing both mutations together, we reasoned that mutations in either individual binding site might nevertheless strongly affect RNA synthesis control by basal ppGpp and weaken the relationship between RNA and growth rate, as observed in a ppGpp0 strain by Potrykus et al. (2011).
We transferred mutations that disrupt ppGpp binding site 1 [rpoZ(wt) rpoC R362A R417A K615A; Ross et al., 2013] or that disrupt ppGpp binding site 2 (rpoC N680A K681A; Ross et al., 2016) from MG1655 to NCM3722. We confirmed that the stringent response does not arrest RNA synthesis in our mutant strains as strongly as in wild-type (Supplementary Figure 1), qualitatively consistent with results previously observed (Ross et al., 2016). We sampled cultures that had been grown directly from fresh colonies (i.e., without dilution from overnight cultures) to reduce the outgrowth of cells bearing additional RNAP mutations (Murphy and Cashel, 2003).
ppGpp concentrations remain inversely correlated with growth rate in both mutants. However, both mutants grow more slowly and have correspondingly higher ppGpp concentrations in most growth media than wild-type NCM3722 (Figures 2A–C). Furthermore, the RNA content of both mutants correlates positively with growth rate, as it does for the wild-type strain (Figure 2D), with exception of the lower RNA concentration for the rpoC2- mutant in LB medium. At first glance, this is consistent with the notion that the RNAP mutants are less sensitive to ppGpp, as apparent from the slopes of cellular RNA content vs. ppGpp (Figure 2E). A chi-squared goodness-of-fit test verified that the mutants do not fit the wild-type pattern (P < 10–6). In other words, higher ppGpp concentrations may be required to inhibit RNA synthesis in these strains. While it might be expected that the cultures expressing ppGpp-insensitive RNAP thus contain a higher RNA abundance than wild-type, we found that for every medium aside from MOPS/acetate, both mutant strains exhibit equivalent or even less RNA per OD unit than does the wild-type (Figure 2F). This is inconsistent with the abolition of growth rate control of RNA content observed in ppGpp0 strains (Potrykus et al., 2011).
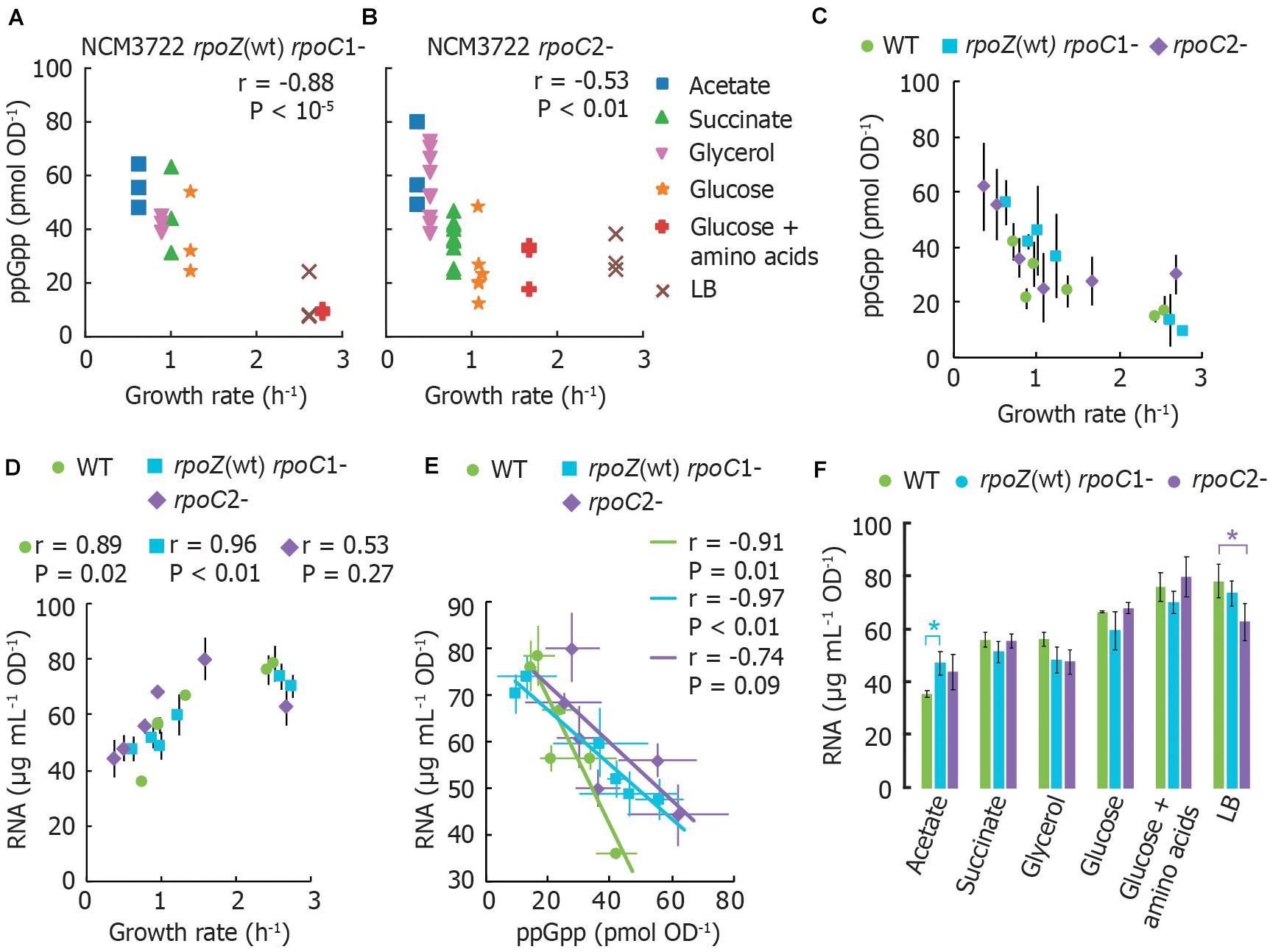
Figure 2. RNA polymerase mutants without ppGpp binding site 1 or 2 still exhibit the typical ppGpp and RNA vs. growth rate trends. (A) Basal ppGpp in NCM3722 rpoZ(WT) rpoC1- and in (B) NCM3722 rpoC2- (showing technical replicates, 3 per culture). (C) Averaged data of A and B overlapped with average NCM3722 wild type data from Figure 1A. (D–F) The total RNA concentration of NCM3722 wild type and rpoC mutants in various growth media. Error bars represent standard deviations. (D) Total RNA concentration plotted as a function of growth rate. (E) Data of D plotted as a function of ppGpp concentrations with a linear fit. The average or each condition is shown. See Supplementary Section 3 for materials and methods. (F) Data of D plotted to compare RNA content between strains grown in identical media. Bars represent the average of three technical replicates of one culture, with exception of rpoC2- in glycerol (2 cultures, 6 replicates) and succinate (3 cultures, 9 replicates). *P < 0.05 for a two-tailed student’s t-test. For (A,B,D,E), the Pearson correlation coefficient r and significance figure P (from a two-tailed significance test) is shown.
While our results indicate that neither ppGpp binding site on RNAP is individually sufficient to mediate ppGpp control over RNA content, we cannot exclude the possibility that the simultaneous removal of both ppGpp binding sites is required to fully eliminate the ppGpp-RNA content relationship. Other factors may also be implicated in RNA synthesis control in the NCM3722 strain. For instance, TraR, a transcription factor encoded on the F plasmid carried by NCM3722 is known to mimic the action of DksA and ppGpp (Gopalkrishnan et al., 2017).
Conclusion
We find that the inverse correlation between ppGpp and growth rate during steady-state growth is quite robust, even against removal of the ppGpp synthesis enzyme RelA. Deviations from the trend (e.g., RelA′ overexpression, pyrimidine starvation, and growth transitions) deserve fuller exploration as they likely hint at poorly understood facets of ppGpp biology. Disrupting either individual ppGpp binding site of RNAP did not eliminate the correlation between growth rate, RNA content, and basal ppGpp concentrations. Despite compelling evidence for basal ppGpp control of rRNA synthesis, incorporating ppGpp into quantitative models of cell behavior requires a better understanding of both transcriptional and post-translational targets. In order to advance this goal, we suggest several questions for the field:
1. What targets are responsive to basal ppGpp concentrations? As basal ppGpp varies with growth rate in parallel with all biosynthetic fluxes during balanced growth, it is tempting to overextend models of ppGpp control. Targets thought to be regulated during the stringent response may prove insensitive to basal ppGpp. Experiments that monitor ppGpp during growth transitions already suggest that small changes in basal ppGpp do not immediately affect instantaneous protein synthesis or total biomass production. Studies of basal ppGpp concentrations during growth transitions are essential for distinguishing what is influenced by ppGpp. We suggest experiments that monitor protein synthesis during small controlled variations in basal ppGpp (±50 pmol OD–1).
2. What establishes basal ppGpp concentrations? It is unknown which metabolic cues drive RelA and SpoT to generate basal ppGpp.
3. What regulates stable RNA synthesis during steady-state growth, aside from basal ppGpp? Our observation that RNA polymerase mutants lacking either one of the two ppGpp binding sites still exhibit an inverse relationship between RNA content and ppGpp concentrations implies that other factors also regulate RNA content, as has been suggested (Fernández-Coll and Cashel, 2018). However, experiments in a strain simultaneously bearing both RNAP mutations will be necessary to confirm this.
4. Does basal ppGpp vary inversely with growth rate when growth rate is varied by other nutrients than carbon source? Measuring basal ppGpp vs. growth rate in conditions that have not yet been tested (e.g., nitrogen source) would also determine whether basal ppGpp is an accurate growth rate speedometer.
5. Do all species with RSH proteins also maintain basal concentrations of ppGpp (or pppGpp) during steady-state growth? As RSH proteins are widely distributed (Atkinson et al., 2011), basal ppGpp may be a defining feature of bacterial growth.
We further recommend the use of common reference strains (preferably E. coli NCM3722) and defined media to enable comparisons between labs.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
NI and GB conceived and designed the experiments and wrote the manuscript. NI performed experiments and data analysis. NI, NB, and MN developed the LC-MS method. All authors contributed to the article and approved the submitted version.
Funding
NI was supported by a grant to GB from the Frontiers of Nanoscience program. MN was supported by a grant to GB from the Netherlands Organisation for Scientific Research (ALW Open 824.15.018). GB and NB were supported by startup funds provided by the Department of Bionanoscience of TU Delft.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Michael Cashel and Richard L. Gourse for providing strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.574872/full#supplementary-material
References
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT Homolog (RSH) superfamily: distribution and functional evolution of ppGpp Synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479
Baracchini, E., and Bremer, H. (1988). Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J. Biol. Chem. 263, 2597–2602.
Baracchini, E., Glass, R., and Bremer, H. (1988). Studies in vivo on Escherichia coli RNA polymerase mutants altered in the stringent response. Mol. Gen. Genet. 213, 379–387. doi: 10.1007/bf00339606
Battesti, A., and Bouveret, E. (2006). Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x
Bochners, B. R., and Ames, B. N. (1982). Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257, 9759–9769.
Bokinsky, G., Baidoo, E. E. K., Akella, S., Burd, H., Weaver, D., Alonso-Gutierrez, J., et al. (2013). HipA-triggered growth arrest and β-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195, 3173–3182. doi: 10.1128/JB.02210-12
Brown, S. D., and Jun, S. (2015). Complete genome sequence of Escherichia coli NCM3722. Genome Announc. 3:e00879-15. doi: 10.1128/genomea.00879-15
Buckstein, M. H., He, J., and Rubin, H. (2008). Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726. doi: 10.1128/jb.01020-07
Cashel, M., and Gallant, J. (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Dai, X. F., Zhu, M. L., Warren, M., Balakrishnan, R., Patsalo, V., Okano, H., et al. (2016). Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2:16231. doi: 10.1038/nmicrobiol.2016.231
Fernández-Coll, L., and Cashel, M. (2018). Contributions of SpoT hydrolase, SpoT synthetase, and RelA synthetase to carbon source diauxic growth transitions in Escherichia coli. Front. Microbiol. 9:1802. doi: 10.3389/fmicb.2018.01802
Fernández-Coll, L., Maciag-Dorszynska, M., Tailor, K., Vadia, S., Levin, P. A., Szalewska-Palasz, A., et al. (2020). The absence of (p)ppGpp renders initiation of Escherichia coli chromosomal DNA synthesis independent of growth rates. mBio 11:e03223-19. doi: 10.1128/mBio.03223-19
Friesen, J., Fiil, N., and von Meyenburg, K. (1975). Synthesis and turnover of basal level guanosine tetraphosphate in Escherichia coli. J. Biol. Chem. 250, 304–309.
Germain, E., Guiraud, P., Byrne, D., Douzi, B., Djendli, M., and Maisonneuve, E. (2019). YtfK activates the stringent response by triggering the alarmone synthetase SpoT in Escherichia coli. Nat. Commun. 10:5763. doi: 10.1038/s41467-019-13764-4
Gopalkrishnan, S., Ross, W., Chen, A. Y., and Gourse, R. L. (2017). TraR directly regulates transcription initiation by mimicking the combined effects of the global regulators DksA and ppGpp. Proc. Natl. Acad. Sci. U.S.A. 114, E5539–E5548. doi: 10.1073/pnas.1704105114
Hansen, M. T., Pato, M. L., Molin, S., and Fiil, N. P. (1975). Simple downshift and resulting lack of correlation between ppGpp pool size and ribonucleic acid accumulation. J. Bacteriol. 122, 585–591. doi: 10.1128/jb.122.2.585-591.1975
Harshman, R. B., and Yamazaki, H. (1971). Formation of ppGpp in a Relaxed and Stringent Strain of E. coli during diauxie lag. Biochemistry 10, 3980–3982. doi: 10.1021/bi00797a027
Haseltine, W. A., and Block, R. (1973). Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U.S.A. 70, 1564–1568. doi: 10.1073/pnas.70.5.1564
Heath, R. J., Jackowski, S., and Rock, C. O. (1994). Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB). J. Bacteriol. 269, 26584–26590.
Hernandez, V. J., and Bremer, H. (1990). Guanosine tetraphosphate (ppGpp) dependence of the growth rate control of rrnB P1 promoter activity in Escherichia coli. J. Biol. Chem. 265, 11605–11614.
Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004). Conformational antagonism between opposing active sites in a bifunctional Re1A/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117, 57–68. doi: 10.1016/S0092-8674(04)00260-0
Hui, S., Silverman, J. M., Chen, S. S., Erickson, D. W., Basan, M., Wang, J., et al. (2015). Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol. Syst. Biol. 11:784. doi: 10.15252/msb.20145697
Ihara, Y., Ohta, H., and Masuda, S. (2015). A highly sensitive quantification method for the accumulation of alarmone ppGpp in Arabidopsis thaliana using UPLC-ESI-qMS/MS. J. Plant Res. 128, 511–518. doi: 10.1007/s10265-015-0711-1
Ishiguro, E. E., Mirelman, D., and Harkness, R. E. (1980). Regulation of the terminal steps in peptidoglycan biosynthesis in ether-treated cells of Escherichia coli. FEBS Lett. 120, 175–178. doi: 10.1016/0014-5793(80)80291-2
Jin, H., Lao, Y. M., Zhou, J., Zhang, H. J., and Cai, Z. H. (2018). A rapid UHPLC-HILIC method for algal guanosine 5′-diphosphate 3′-diphosphate (ppGpp) and the potential separation mechanism. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1096, 143–153. doi: 10.1016/j.jchromb.2018.08.009
Kanjee, U., Ogata, K., and Houry, W. A. (2012). Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85, 1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x
Khan, S. R., and Yamazaki, H. (1974). Inapparent correlation between guanosine tetraphosphate levels and RNA contents in Escherichia coli. Biochem. Biophys. Res. Commun. 59, 125–132. doi: 10.1016/s0006-291x(74)80183-x
Lazzarini, R. A., Cashel, M., and Gallant, J. (1971). On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J. Biol. Chem. 246, 4381–4385.
Lee, J. W., Park, Y. H., and Seok, Y. J. (2018). Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, E6845–E6854. doi: 10.1073/pnas.1722514115
Li, S. H., Li, Z., Park, J. O., King, C. G., Rabinowitz, J. D., Wingreen, N. S., et al. (2018). Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947. doi: 10.1038/s41564-018-0199-2
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S., and Cashel, M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189. doi: 10.1093/nar/gkt302
Murphy, H., and Cashel, M. (2003). Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 371, 596–601. doi: 10.1016/s0076-6879(03)71044-1
Murray, K. D., and Bremer, H. (1996). Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 259, 41–57. doi: 10.1006/jmbi.1996.0300
Noga, M. J., Buke, F., van den Broek, N. J. F., Imholz, N., Scherer, N., and Bokinsky, G. (2020). Post-translational control is sufficient to coordinate membrane synthesis with growth in Escherichia coli. mBio 11:e02703-19. doi: 10.1101/728451
Patacq, C., Chaudet, N., and Létisse, F. (2018). Absolute quantification of ppGpp and pppGpp by double-spike isotope dilution ion chromatography-high-resolution mass spectrometry. Anal. Chem. 90, 10715–10723. doi: 10.1021/acs.analchem.8b00829
Potrykus, K., Murphy, H., Philippe, N., and Cashel, M. (2011). ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575. doi: 10.1111/j.1462-2920.2010.02357.x
Poulsen, P., and Jensen, K. F. (1987). Effect of UTP and GTP pools on attenuation at the pyrE gene of Escherichia coli. Mol. Gen. Genet. 208, 152–158. doi: 10.1007/bf00330436
Ross, W., Sanchez-Vazquez, P., Chen, A. Y., Lee, J. H., Burgos, H. L., and Gourse, R. L. (2016). ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62, 811–823. doi: 10.1016/j.molcel.2016.04.029
Ross, W., Vrentas, C. E., Sanchez-Vazquez, P., Gaal, T., and Gourse, R. L. (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429. doi: 10.1016/j.molcel.2013.03.021
Ryals, J., Little, R., and Bremer, H. (1982). Control of ribosomal-RNA and transfer-RNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 151, 1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982
Sarubbi, E., Rudd, K. E., and Cashel, M. (1988). Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. MGG Mol. Gen. Genet. 213, 214–222. doi: 10.1007/BF00339584
Schreiber, G., Metzger, S., Aizenman, E., Roza, S., Cashel, M., and Glaser, G. (1991). Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 266, 3760–3767.
Schreiber, G., Ron, E. Z., and Glaser, G. (1995). ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr. Microbiol. 30, 27–32. doi: 10.1007/BF00294520
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z., and Hwa, T. (2010). Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102. doi: 10.1126/science.1192588
Scott, M., Klumpp, S., Mateescu, E. M., and Hwa, T. (2014). Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 10:747. doi: 10.15252/msb.20145379
Sokawa, Y., Sokawa, J., and Kaziro, Y. (1975). Regulation of stable RNA synthesis and ppGpp levels in growing cells of Escherichia coli. Cell 5, 69–74. doi: 10.1016/0092-8674(75)90093-8
Soupene, E., Van Heeswijk, W. C., Plumbridge, J., Stewart, V., Bertenthal, D., Lee, H., et al. (2003). Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185, 5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003
Svitil, A. L., Cashel, M., and Zyskind, J. W. (1993). Guanosine tetraphosphate inhibits protein synthesis in vivo. J. Biol. Chem. 268, 2307–2311.
Traxler, M. F., Summers, S. M., Nguyen, H. T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Traxler, M. F., Zacharia, V. M., Marquardt, S., Summers, S. M., Nguyen, H. T., Stark, S. E., et al. (2011). Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the “feast to famine” gradient in Escherichia coli. Mol. Microbiol. 79, 830–845. doi: 10.1111/j.1365-2958.2010.07498.x
Varik, V., Oliveira, S. R. A., Hauryliuk, V., and Tenson, T. (2017). HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7:11022. doi: 10.1038/s41598-017-10988-6
Vogel, U., Pedersen, S., and Jensen, K. F. (1991). An unusual correlation between ppGpp pool size and rate of ribosome synthesis during partial pyrimidine starvation of Escherichia coli. J. Bacteriol. 173, 1168–1174. doi: 10.1128/jb.173.3.1168-1174.1991
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2018). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150. doi: 10.1038/s41589-018-0183-4
Zhang, Y., Zbornikova, E., Rejman, D., and Gerdes, K. (2018). Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9, 1–20. doi: 10.1128/mBio.02188-17
Zhu, M., and Dai, X. (2019). Growth suppression by altered (p)ppGpp levels results from non-optimal resource allocation in Escherichia coli. Nucleic Acids Res. 47, 4684–4693. doi: 10.1093/nar/gkz211
Keywords: ppGpp, growth rate, RNA polymerase, RNA synthesis, Escherichia coli
Citation: Imholz NCE, Noga MJ, van den Broek NJF and Bokinsky G (2020) Calibrating the Bacterial Growth Rate Speedometer: A Re-evaluation of the Relationship Between Basal ppGpp, Growth, and RNA Synthesis in Escherichia coli. Front. Microbiol. 11:574872. doi: 10.3389/fmicb.2020.574872
Received: 21 June 2020; Accepted: 25 August 2020;
Published: 17 September 2020.
Edited by:
Katarzyna Potrykus, University of Gdańsk, PolandReviewed by:
Johannes Geiselmann, Université Grenoble Alpes, FranceRegis Hallez, University of Namur, Belgium
Llorenc Fernandez-Coll, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), United States
Copyright © 2020 Imholz, Noga, van den Broek and Bokinsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole C. E. Imholz, Ti5DLkUuSW1ob2x6QHJ1Zy5ubA==; Gregory Bokinsky, Ry5FLkJva2luc2t5QHR1ZGVsZnQubmw=
†Present address: Nicole C. E. Imholz, Stratingh Institute for Chemistry, University of Groningen, Groningen, Netherlands; Marek J. Noga, Translational Metabolic Laboratory, Department of Laboratory Medicine, Radboudumc, Nijmegen, Netherlands; Niels J. F. van den Broek, Vaccine Process & Analytical Development, Janssen Vaccines & Prevention B.V., Leiden, Netherlands
 Nicole C. E. Imholz
Nicole C. E. Imholz Marek J. Noga
Marek J. Noga Niels J. F. van den Broek†
Niels J. F. van den Broek† Gregory Bokinsky
Gregory Bokinsky