
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 September 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.571903
This article is part of the Research Topic Bacteriocins and Other Ribosomally Synthesised and Post-translationally Modified Peptides (RiPPs) as Alternatives to Antibiotics View all 13 articles
 Takeshi Zendo1*
Takeshi Zendo1* Chihiro Ohashi1
Chihiro Ohashi1 Shintaro Maeno2
Shintaro Maeno2 Xingguo Piao1
Xingguo Piao1 Seppo Salminen3
Seppo Salminen3 Kenji Sonomoto1
Kenji Sonomoto1 Akihito Endo2
Akihito Endo2Apilactobacillus kunkeei FF30-6 isolated from healthy honey bees synthesizes the bacteriocin, which exhibits antimicrobial activity against Melissococcus plutonius. The bacteriocin, kunkecin A, was purified through three-step chromatography, and mass spectrometry revealed that its relative molecular mass was 4218.3. Edman degradation of purified kunkecin A showed only the N-terminal residue, isoleucine. Hence, alkaline alkylation made the subsequent amino acid residues accessible to Edman degradation, and 30 cycles were sequenced with 11 unidentified residues. Whole genome sequencing of A. kunkeei FF30-6, followed by Sanger sequencing, revealed that the genes encoding the proteins involved in lantibiotic biosynthesis were within the plasmid, pKUNFF30-6. Most of the identified proteins exhibited significant sequence similarities to the biosynthetic proteins of nisin A and its variants, such as subtilin. However, the kunkecin A gene cluster lacked the genes corresponding to nisI, nisR, and nisK of the nisin A biosynthetic gene cluster. A comparison of the gene products of kukA and nisA (kunkecin A and nisin A structural genes, respectively) suggested that they had similar post-translational modifications. Furthermore, the structure of kunkecin A was proposed based on a comparison of the observed and calculated relative molecular masses of kunkecin A. The structural analysis revealed that kunkecin A and nisin A had a similar mono-sulfide linkage pattern. Purified kunkecin A exhibited a narrow antibacterial spectrum, but high antibacterial activity against M. plutonius. Kunkecin A is the first bacteriocin to be characterized in fructophilic lactic acid bacteria and is the first nisin-type lantibiotic found in the family Lactobacillaceae.
Fructophilic lactic acid bacteria (FLAB) are only found in fructose-rich niches, such as flowers and fruits, and Fructobacillus spp. and Apilactobacillus kunkeei, formerly classified as Lactobacillus kunkeei (Zheng et al., 2020), are representatives of this group (Endo and Okada, 2008; Endo et al., 2012). Recent studies revealed that the genotypic and phenotypic characteristics of FLAB enable them to adapt to the fructose-rich niches (Endo et al., 2009, 2015, 2018). A. kunkeei was originally isolated from wine (Edwards et al., 1998) and was recently characterized as one of the major components of the gut microbiota in honey bee queens and larvae (Endo and Salminen, 2013; Vojvodic et al., 2013; Anderson et al., 2018). The species has been linked to the nectar and hive materials of honey bees (Anderson et al., 2013; Kwong and Moran, 2016). As A. kunkeei has a symbiotic relationship with the host insects, it has potential probiotic and paratransgenic applications to honey bees (Rangberg et al., 2015; Arredondo et al., 2018). A previous study reported that the culture supernatant from an A. kunkeei isolate exhibited anti-Melissococcus plutonius activity (Endo and Salminen, 2013), the causative agent of European foulbrood in honey bee larvae (Arai et al., 2012). Furthermore, this antibacterial activity was inhibited by a treatment with proteases (Endo and Salminen, 2013), which suggested the proteinaceous nature of this substance.
Bacteriocins are ribosomally synthesized antimicrobial peptides that exhibit bactericidal or bacteriostatic activity (Cotter et al., 2005b; Alvarez-Sieiro et al., 2016). Various bacterial species, including lactic acid bacteria (LAB), produce bacteriocins, whereas properties for bacteriocin production vary according to strains. The application of bacteriocins, particularly those derived from LAB, in food safety has been attracting increasing interest because most of them are active against several food spoilage bacteria and foodborne pathogens and are also easily degraded by gut proteases (Cleveland et al., 2001; Perez et al., 2014). Moreover, LAB bacteriocins have been evaluated as an alternative antibiotic agent for clinical applications (Heunis et al., 2013; Perez et al., 2014).
Nisin is the most-studied LAB bacteriocin. It is a class I bacteriocin that is also referred to as lantibiotics and is widely used as a safe food preservative (Delves-Broughton et al., 1996). Several natural variants of nisin are produced by strains belonging to Lactococcus lactis, such as nisin A (Gross and Morell, 1971), nisin Z (Mulders et al., 1991), nisin Q (Zendo et al., 2003; Fukao et al., 2008), and nisin F (de Kwaadsteniet et al., 2008), in addition to subtilin produced by strains of Bacillus subtilis (Banerjee and Hansen, 1988). Natural nisin variants produced by strains of other genera have recently been reported, such as nisin U (Wirawan et al., 2006), nisin H (O’Connor et al., 2015), nisin O (Hatziioanou et al., 2017), nisin J (O’Sullivan et al., 2020), and nisin P (Garcia-Gutierrez et al., 2020) by Blautia spp., Staphylococcus spp., and Streptococcus spp. All nisin variants share a basal structure consisting of five mono-sulfide bridges (lanthionine rings), and three dehydrated amino acid residues, which result from post-translational modifications but have some amino acid substitutions. The biosynthesis of nisin A requires eleven genes in three transcription units (Kuipers et al., 1995; de Ruyter et al., 1996). After the synthesis of NisA, the precursor of nisin A, NisB catalyzes the dehydration of Ser and Thr residues in NisA to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively. NisC then catalyzes the cyclization of the dehydrated residues with five Cys residues to form lanthionine (Lan) and 3-methyllanthionine (MeLan), respectively. Modified NisA containing five mono-sulfide bridges and three dehydrated residues is secreted from the producer cell through NisT [an ATP-binding cassette (ABC) transporter]. The 23-amino-acid-long leader sequence of NisA is then cleaved by NisP (protease) outside the cell, and the 34-amino acid mature nisin A is released. The two-component regulatory system, NisK (histidine kinase)/NisR (response regulator) upregulates transcription units to enhance the synthesis of nisin A. Mature nisin A can also serve as an autoinducer. NisI (membrane protein) and NisFEG (ABC transporter) are two independent self-immunity systems that protect producer cells against the antimicrobial activity of nisin A. The other nisin variants are synthesized by a similar mechanism employing similar biosynthetic proteins (Yoneyama et al., 2008).
The honey bee isolate A. kunkeei FF30-6 produces an antibacterial peptide that exhibits anti-M. plutonius activity (Endo and Salminen, 2013). In the present study, we purified and characterized the structure and activity of a novel bacteriocin, which we named kunkecin A. The gene cluster encoding the proteins involved in bacteriocin biosynthesis was identified through a whole genome analysis of A. kunkeei FF30-6. The results of our analysis revealed that kunkecin A is a variant of nisin A, which had not yet been reported in the family Lactobacillaceae. Kunkecin A had a narrow antimicrobial spectrum but exhibited high antimicrobial activity against a few bacteria originating from honey bees, including M. plutonius.
The bacteriocin produced by A. kunkeei FF30-6 was purified from the culture supernatant using a three-step chromatography procedure. The active fraction containing the purified bacteriocin was obtained in the final step of reverse-phase high-performance liquid chromatography (HPLC). The active fraction was used to characterize the structure and assess its antibacterial activity. Electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) revealed that the relative molecular mass of the purified bacteriocin was 4218.3 (Figure 1). It was considered to be a novel bacteriocin and termed kunkecin A, because, to the best of our knowledge, its relative molecular mass has not been previously reported for any other bacteriocins in the literature or databases dedicated to bacteriocins such as Bactibase (Hammami et al., 2010) and BAGEL4 (Van Heel et al., 2018).
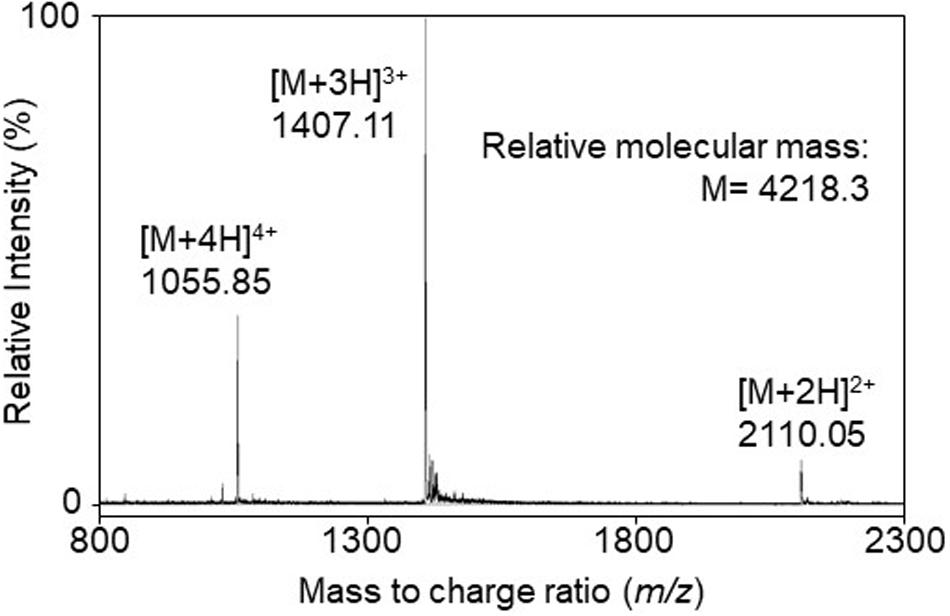
Figure 1. The electrospray ionization time-of-flight (ESI-TOF) mass spectrum of the purified bacteriocin, kunkecin A, produced by A. kunkeei FF30-6. Multiple charged molecular ions were detected and are indicated.
Edman degradation of purified kunkecin A detected only the N-terminal residue, isoleucine, but no other residues. This result suggested that there was a modified residue at the second position, such as a dehydrated amino acid residue, which inhibited Edman degradation. The purified peptide was further treated with alkaline 2-mercaptoethanol to enable the degradation reaction to access the modified residues in kunkecin A, following the methods described by Meyer et al. (1994). After 30 cycles of Edman degradation of the treated peptide, we obtained the following amino acid sequence: IXXYVLXXPG XIXGRLMGXN NKXKXXHXHS, where X indicates the cycle at which no amino acid was identified. This result strongly suggested that kunkecin A is a lantibiotic and that the second residue and subsequent X residues were post-translationally modified.
A previous study reported the draft genome sequence of A. kunkeei FF30-6 (NZ_BDDX00000000) with 25 contigs (Maeno et al., 2016). One of the contigs (contig no. 12) was suspected to be a putative plasmid sequence because it had a gene-encoding DNA replication initiator protein A. In the present study, we used Sanger sequencing to determine the possible plasmid structure. We obtained a circular plasmid, termed pKUNFF30-6, which consisted of 19,498 bp (Supplementary Figure S1 and Supplementary Table S1). The complete plasmid sequence was analyzed using the DDBJ Fast Annotation and Submission Tool (DFAST,1) (Tanizawa et al., 2016), which revealed that eight bacteriocin-related genes were located on the plasmid (accession number, AP019008). The amino acid sequences of the gene products exhibited significant similarities to those of the proteins involved in the biosynthesis of nisin A (Table 1). This putative gene cluster consisting of eight consecutive genes (orf10-17 of pKUNFF30-6) lacked the genes corresponding to nisI, nisR, and nisK in the nisin A biosynthetic cluster. Half of the genes in the cluster (orf10-13) were coded in the opposite direction to the other half, orf14-17. This is in contrast to nisin A biosynthetic genes, which were all coded in the same direction (Figure 2). Each gene in the kunkecin A biosynthetic gene cluster was named after the corresponding gene in the nisin A biosynthetic gene cluster (Table 1 and Figure 2).
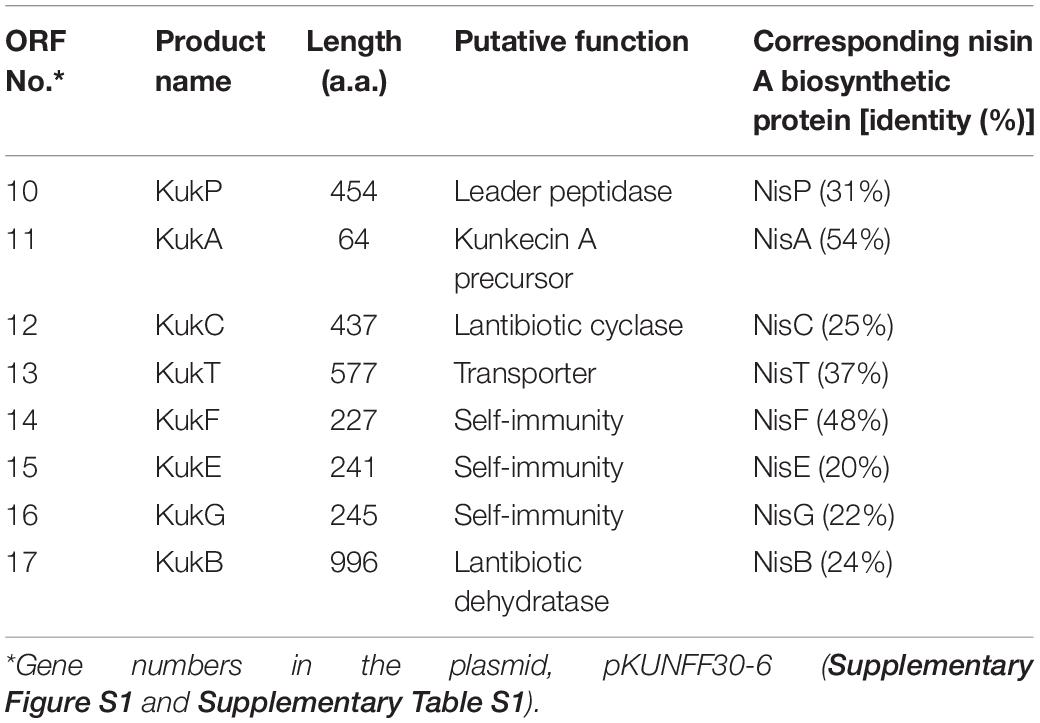
Table 1. Putative kunkecin A biosynthetic proteins encoded on genes identified in the plasmid, pKUNFF30-6, harbored by A. kunkeei FF30-6.
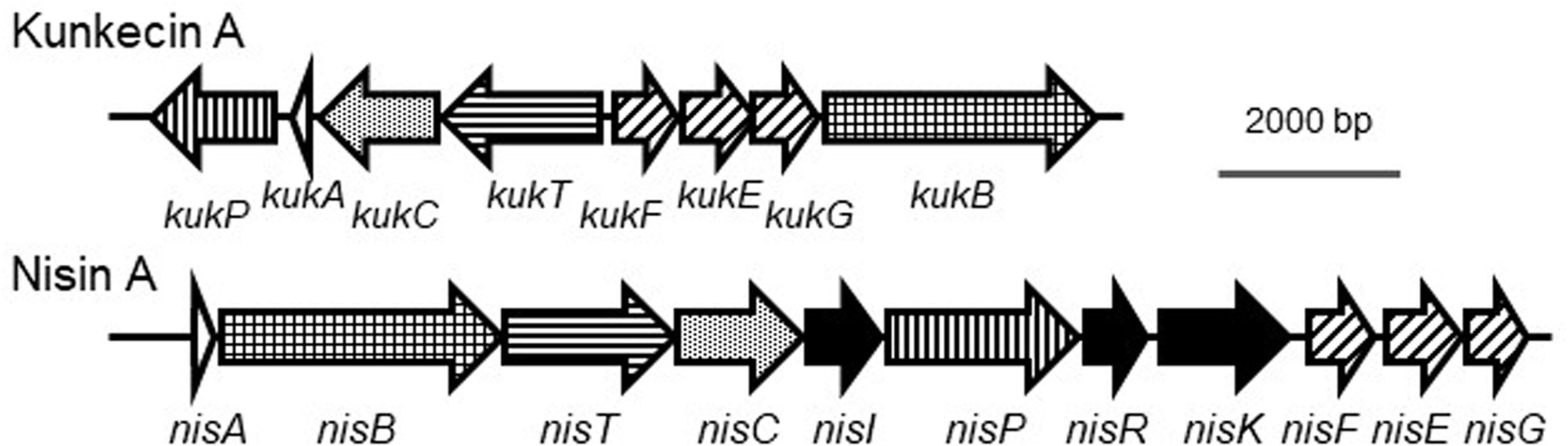
Figure 2. Comparison of biosynthetic gene clusters of kunkecin A and nisin A. The putative kunkecin A biosynthetic genes are labeled with the same pattern as each corresponding gene in the nisin A biosynthetic cluster. No genes corresponding to nisI, nisR, or nisK (filled with black) were found in the kunkecin biosynthetic gene cluster.
The precursor peptide of kunkecin A, KukA was similar to those of nisin A (NisA) and its natural variants (Figure 3). The cleavage site of the leader peptide of KukA was identified based on the alignment with NisA and the result of Edman degradation, as shown in Figure 3. The difference in the calculated (4327.0) and observed (4218.3) relative molecular masses of the KukA core peptide (Figure 1) indicated that six dehydrations occurred in the core peptide. Additionally, a comparative analysis of the structures of nisin A and kunkecin A revealed that modified residues were located at the position of X in the N-terminal sequence of kunkecin A obtained by Edman degradation. The positions of the modified residues were similar in nisin A, except for the 5th and 33rd residues, at which valine and histidine, respectively, were detected in kunkecin A both by Edman degradation and DNA sequence analysis. These modifications involved six dehydrations and concurred with the observed difference between the calculated and observed relative molecular masses of kunkecin A. Consequently, we proposed the structure of kunkecin A as shown in Figure 4. The structure of kunkecin A includes a dehydrobutyrine, lanthionine, and four 3-methyllanthionine residues with a similar ring pattern to that of nisin A. However, the C-terminal of kunkecin A was longer than that of nisin A.

Figure 3. Amino acid sequence alignment of precursor peptides of kunkecin A and nisin variants. Each precursor was aligned using the Clustal Omega program. The dotted line indicates the cleavage site of the leader peptides. Asterisks indicate fully conserved positions. Colons and periods indicate positions conserved by amino acid residues with strong and weak similarity, respectively.
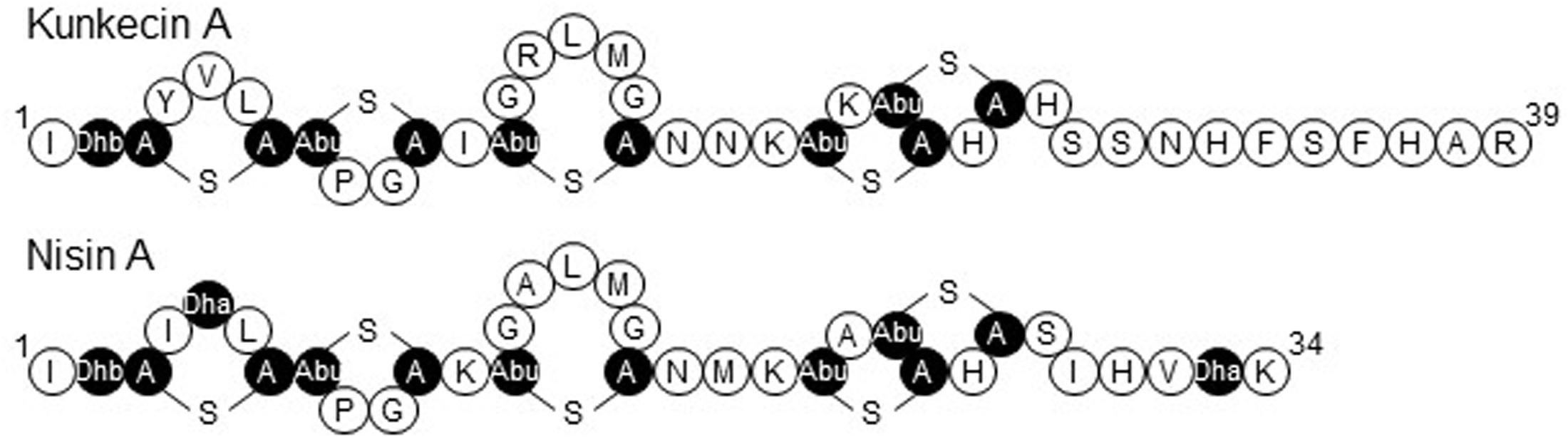
Figure 4. The proposed primary structure of kunkecin A and the primary structure of nisin A. Unusual amino acids generated by post-translational modifications are indicated in black. Dha and Dhb indicate the dehydrated amino acids, dehydroalanine, and dehydrobutyrine, respectively. A-S-A and Abu-S-A indicate lanthionine and 3-methyllanthionine, respectively.
The antimicrobial activity of purified kunkecin A was evaluated against a panel of bacterial indicator strains as the minimum concentrations, causing clear inhibition zones by the spot-on-lawn assay. We also compared the antibacterial activity of purified kunkecin A with that of nisin A (Table 2). The antibacterial activity of kunkecin A was two-fold higher than that of nisin A against M. plutonius. Honey bee commensals show different sensitivities to the bacteriocins at the species level. Lactobacillus apis, L. kullabergensis, Bombilactobacillus mellis, and Bifidobacterium asteroides exhibited markedly higher tolerance (>four-fold) to kunkecin A than to nisin A, whereas others, including Apilactobacillus apinorum, Bombilactobacillus mellifer, and Lactobacillus melliventiris, showed the opposite reactions. The kunkecin A-producing strain, A. kunkeei FF30-6, and the nisin A-producer strain, L. lactis subsp. lactis NCDO 497, were highly tolerant to kunkecin A and nisin A. The tolerance to the kunkecin A and nisin A treatments may be attributed to self-immunity against their own and homologous bacteriocins. The American foulbrood-causing species, Paenibacillus larvae PL-1, was tolerant to kunkecin A or nisin A. The minimum concentrations for the inhibition of kunkecin A against some of the indicator strains, including a Gram-negative strain, Escherichia coli, were higher than the concentration at which self-immunity was observed.
The self-immunity was further examined by the deferred antagonism assay. Against A. kunkeei FF30-6, a very tiny inhibition zone with a clear edge was caused by L. lactis subsp. lactis NCDO 497, but no inhibition zone was formed by itself. On the other hand, against L. lactis subsp. lactis NCDO 497, no inhibition zones were formed by the both strains.
We previously reported that a culture supernatant of one of the A. kunkeei isolates, strain FF30-6, originated from honey bees inhibited the growth of the type strain of M. plutonius (Endo and Salminen, 2013). Since A. kunkeei is a promising candidate for probiotic and paratransgenic in honey bees, it would be of interest to study its antagonistic activities in honey bee commensals.
The relative molecular mass of kunkecin A is unique among known bacteriocins and within the reported range for LAB bacteriocins and larger than those of most lantibiotics, including nisin A (Cotter et al., 2005a). However, further structural analyses on purified kunkecin A suggested that it shares amino acid sequences and positions of modified residues with nisin A and its variants. The conserved motif sequence (FNLD) of the leader peptide in nisin-group lantibiotics (Plat et al., 2011) was also detected in the kunkecin A leader peptide, except for the final 4th residue, which changed to glycine (Figure 3). The amino acid sequence of the N-terminal in kunkecin A begins with isoleucine and has a dehydrobutyrine at the second position, which is consistent with the results of Edman degradation. The comparative analysis of the observed and calculated relative molecular masses of kunkecin A revealed that the peptide underwent six dehydrations, including dehydrobutyrine at the second position. Furthermore, the comparison of kunkecin A and nisin A strongly suggested that kunkecin A contained totally five mono-sulfide linkages (one lanthionine and four 3-methyllanthionine), which are also found in nisin A. The amino acid sequences of the putative modification enzymes, KukB and KukC, also exhibited high similarities to those of the respective lantibiotic modification enzymes of nisin A and its variants. This result strongly indicated that the ring pattern of kunkecin A was similar to that of the nisin A-group lantibiotics.
In the putative kunkecin A biosynthetic gene cluster, eight genes on the plasmid pKUNFF30-6 encoded putative lantibiotic biosynthetic proteins and the kunkecin A precursor peptide. Putative kunkecin A biosynthetic proteins also exhibited significant sequence similarities to those involved in the biosynthesis of nisin A (Table 1). The function of these proteins was predicted to be similar to that of nisin A biosynthetic proteins. The kunkecin A precursor peptide, KukA, is dehydrated and cyclized by the lantibiotic modification enzymes, KukB and KukC, respectively. Modified KukA is secreted by the ABC transporter, KukT, and the leader peptide of KukA is cleaved by the cell-wall-anchored leader peptidase, KukP, outside the producer cell. The producer cell is protected by the self-immunity proteins, KukF, KukE, and KukG, which comprise another ABC transporter. In contrast to the nisin A biosynthetic gene cluster, the kunkecin A gene cluster lacks the genes corresponding to nisI, nisR, and nisK. NisI is responsible for self-immunity against nisin A (Kuipers et al., 1993), and NisR and NisK form a two-component regulatory system with nisin A as the autoinducer to regulate the biosynthesis of nisin A (Kuipers et al., 1995; de Ruyter et al., 1996). In the reported nisin-variant gene clusters, that of nisin H lacks the gene corresponding to nisI (O’Connor et al., 2015), while that of nisin J lacks the genes to nisI, nisR, and nisK (O’Sullivan et al., 2020).
NisI is a membrane-anchored lipoprotein that functions coordinately with the ABC transporter comprising NisF, NisE, and NisG for self-immunity in the producer cell (Stein et al., 2003). NisFEG or SpaFEG, the corresponding system to subtilin, alone is known to be sufficient to impart self-immunity against each cognate lantibiotic (Kuipers et al., 1993; Stein et al., 2003). This finding suggests that the KukFEG system was sufficient to protect producer cells against kunkecin A without the need for a protein corresponding to NisI or SpaI in nisin A or subtilin, respectively. The producer strain, A. kunkeei FF30-6 exhibited tolerance to both kunkecin A and nisin A (Table 2), while it was inhibited very weakly by nisin A-producing L. lactis NCDO 497 in the deferred antagonism assay. The less cross-immunity of A. kunkeei FF30-6 against the nisin A producer can be attributed to high production of nisin A and/or the lack of the NisI homologous protein. Although NisI and SpaI contributes less to total immunity than NisFEG and SpaFEG, respectively, previous studies reported that they protected producer cells from the pore formation activity of their respective cognate bacteriocins (Stein et al., 2005; AlKhatib et al., 2014). Additionally, SpaI has a very similar structure to NisI (Hacker et al., 2015) and interacts with the C-terminal of subtilin (Geiger et al., 2019). The lack of the NisI homologous protein in kunkecin A biosynthesis may be related to the C-terminal extension of kunkecin A and its resulting activity.
During nisin A biosynthesis, nisin A can activate the promoters located upstream of nisA and nisF via the two-component NisR/NisK regulatory system and trigger the synthesis of nisin A (Kuipers et al., 1995; de Ruyter et al., 1996). No regulatory genes were identified in the kunkecin A biosynthetic gene cluster. Additionally, gene organization in the kunkecin A gene cluster differed from that in the nisin A gene cluster. These two lines of evidence suggest that the regulation of kunkecin A synthesis differs from that of nisin A synthesis.
Kunkecin A exhibited higher antibacterial activity against M. plutonius than nisin A, even though these two lantibiotics share structural similarity. However, although they shared a mono-sulfide bridge pattern, kunkecin A lacked two dehydrated residues at positions five and 33 and possessed five extra amino acid residues in the C-terminal. These changes may be responsible for the differences observed in their antibacterial activities.
Melissococcus plutonius was one of the most sensitive strains to kunkecin A among the honey-bee-related microbes tested. Antibiotics generally kill pathogens and commensals, which may result in diarrhea and the development of drug-resistant bacterial strains in animals. The long-term antibiotic treatment of honey bee colonies in apiaries has led to the selection of genes that confer antibiotic-resistance to the gut microbiota of the honey bee (Tian et al., 2012). Consequently, the incidence of antibiotic-resistant foulbrood pathogens is high in countries, which use oxytetracycline in apiaries (Murray and Aronstein, 2006; Alippi et al., 2007; Murray et al., 2007; Tian et al., 2012). Furthermore, antibiotic treatments increase the mortality of honey bees (Raymann et al., 2017). The administration of a broad spectrum bacteriocin, such as nisin A, may also affect the gut microbiota in honey bees. The microbiota is crucial for the healthy development of honey bees (Zheng et al., 2017; Raymann et al., 2018). The population of commensal bacteria, such as Bifidobacterium is inversely associated with the population of M. plutonius in healthy honey bees. Further studies are needed to clarify whether kunkecin A is a useful tool for controlling pathogens in apiaries.
In the present study, we described a novel bacteriocin and lantibiotic, kunkecin A. This is the first bacteriocin reported from FLAB and is the first nisin-type lantibiotic found in the family Lactobacillaceae. A more detailed understanding of the regulatory mechanisms underlying kunkecin A biosynthesis may enhance the production of this novel lantibiotic for future applications.
The bacteriocin-producing strain, A. kunkeei FF30-6 isolated from healthy honey bees (Apis mellifera mellifera) (Endo and Salminen, 2013) was cultured in fMRS medium, Lactobacilli MRS broth (MRS; BD Difco, Sparks, MD) supplemented with 2% (w/v) D-fructose (Nacalai Tesque, Kyoto, Japan). Lapidilactobacillus dextrinicus JCM 5887T and M. plutonius ATCC 35311T, used as general indicator strains for bacteriocin activity, were cultured in MRS medium at 30°C and KSBHI medium at 37°C, respectively. KSBHI medium was composed of Brain Heart Infusion (BHI) medium (Oxoid, Hampshire, United Kingdom) supplemented with 20.4 g/L of KH2PO4 and 10 g/L of soluble starch (Arai et al., 2012). The other bacterial strains that were used as indicator strains for the bacteriocin assay (Table 2) were cultured for 18 h under the optimal conditions recommended by the respective culture collections. All bacterial cultures were stored at –80°C with 15% glycerol and were propagated in the respective media at the recommended temperatures for 18 h before use.
The antibacterial activity of bacteriocin was evaluated using the spot-on-lawn method (Ennahar et al., 2001), in which 10 μL of the bacteriocin preparation was spotted onto the bacterial lawn. Regarding general indicator strains, Lactobacilli Agar AOAC (BD Difco) inoculated with an overnight culture of the indicator strain at a density of 107 CFU/mL was overlaid on an MRS agar plate [MRS supplemented with 1.2% (w/v) agar]. KSBHI medium and BHI medium supplemented with 5% (v/v) horse blood and 1.5% (w/v) agar were used for M. plutonius and P. larvae, respectively, instead of the double layer of Lactobacilli Agar AOAC and MRS agar. After an overnight incubation at the respective temperatures recommended for the indicator strain, the bacterial lawn was checked for inhibition zones. Regarding purified bacteriocins, two-fold serial dilutions of the solution containing a fixed concentration of a bacteriocin were assayed, and the minimum concentrations that resulted in a clear zone of inhibition on each indicator lawn were recorded as the intensity of the antibacterial activity. Each assay was performed in triplicate to confirm reproducibility.
To study a self-immunity against the own bacteriocins, the deferred-antagonism assay was further conducted by using the bacteriocin producers, A. kunkeei FF30-6 and L. lactis subsp. lactis NCDO 497 as indicator strains. The bacteriocin producing strains were stabbed into fMRS agar plate [fMRS supplemented with 1.5% (w/v) agar] and grown at 30°C for 8 h. Then, fMRS agar inoculated with an indicator strain was overlaid on the agar plate. After an overnight incubation, the bacterial lawn was examined for inhibition zones.
The bacteriocin was purified using a three-step chromatography procedure that was previously described with minor modifications (Masuda et al., 2011). Purification was performed using four 250-mL cultures (total of 1 L) of A. kunkeei FF30-6 grown to the early stationary phase in fMRS medium at 30°C for 8 h with reciprocal shaking at 140 strokes/min. Cells were pelleted by centrifugation at 8000 g at 4°C for 20 min. Further, 20 g of activated Amberlite XAD-16 resin (Sigma-Aldrich, St. Louis, MO, United States) was added to the culture supernatant. The resin matrix was shaken slowly at 4°C for 4 h, and was then washed with 200 mL Milli-Q water, followed by 400 mL 50% (v/v) ethanol. The active fraction was eluted with 200 mL 70% (v/v) isopropanol containing 0.1% trifluoroacetic acid. The eluted active fraction was evaporated to 60 mL to remove isopropanol. The sample was then diluted with an equal volume of 50 mM sodium citrate buffer (pH 3.0, CB), and applied to an SP Sepharose Fast Flow cation-exchange chromatography column (internal diameter, 10 mm; length, 100 mm; GE Healthcare, Uppsala, Sweden) equilibrated with 50 mL CB. The column was washed with 50 mL CB and 100 mL of 0.3 M NaCl in CB. The active bacteriocin fraction was eluted with 40 mL of 0.6 M NaCl in CB. This active fraction was applied to a Capcell-Pak C18 MGII S5 column (internal diameter 4.6 mm; length 150 mm; Shiseido, Tokyo, Japan) in an LC-2000Plus HPLC system (JASCO, Tokyo, Japan). The active fractions were eluted at a flow rate of 1 mL/min with a linear gradient of 15–45% (v/v) of acetonitrile in the Milli-Q water-acetonitrile mobile phase containing 0.1% trifluoroacetic acid for 30 min. The main active fraction was further purified using reverse-phase HPLC under the same conditions. Purified active fractions were stored at –30°C. The antibacterial activities of the fractions obtained at each purification step were determined as described above using L. dextrinicus JCM 5887T as an indicator strain. Nisin A was purified from a commercial nisin A preparation (Sigma) by using cation-exchange chromatography and reverse-phase HPLC, as described previously (Fujita et al., 2007). Regarding structural characterization, the purified fractions were concentrated using a SpeedVac concentrator (Savant, Farmingdale, NY, United States). To assess antibacterial activity, the solvent was completely removed by lyophilization, and the purified bacteriocin was dissolved in 10% (v/v) dimethyl sulfoxide (DMSO). Peptide concentrations were determined using a Pierce® BCATM Protein Assay Kit (Takara Bio, Otsu, Japan).
The relative molecular masses of the purified fractions were analyzed by ESI-TOF MS using a JMS-T100LC mass spectrometer (JEOL, Tokyo, Japan). Amino acid sequences were determined based on Edman degradation using a PPSQ-31 protein sequencer (Shimadzu, Kyoto, Japan). In further analyses of the N-terminal sequences of peptides containing dehydrated residues and lanthionine, the purified peptide was treated with alkaline 2-mercaptoethanol, following the methods described by Meyer et al. (1994) and subjected to Edman degradation.
The draft genome sequence of A. kunkeei FF30-6 containing 25 contigs was obtained in the previous study with the accession number NZ_BDDX00000000 (Maeno et al., 2016). One of the contigs (contig no. 12), which exhibited plasmid-like genetic characteristics, was further sequenced using the following primers: FF306-c12-F (5′-AAAAGAATAGACAACCACCCA-3′) and FF306-c12-R (5′-CCTTTCTAAGAGGAATATGG-3′). The sequence of the plasmid termed pKUNFF30-6 was analyzed using DFAST2 to detect bacteriocin-related genes. Potential genes were further analyzed using the BLAST program of the National Center for Biotechnology Information database3. The DNA and amino acid sequences obtained were analyzed using GENETYX-WIN software (GENETYX, Tokyo, Japan). The amino acid sequence alignment was analyzed using Clustal Omega4.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, AP019008.
TZ and AE designed the project, analyzed the data, and wrote the manuscript. CO, SM, and XP performed the experiments and analyzed the data. SS and KS supervised the project. All authors contributed to the article and approved the submitted version.
This work was partially supported by JSPS KAKENHI grant numbers JP24380051, JP26850054, and JP17H03797.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Prof. M. Kimura and Asst. Prof. T. Nakashima of Kyushu University, Japan, for allowing us to access the automated protein sequencer. Computational analysis was performed on the NIG supercomputer at ROIS.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.571903/full#supplementary-material
Alippi, A. M., López, A. C., Reynaldi, F. J., Grasso, D. H., and Aguilar, O. M. (2007). Evidence for plasmid-mediated tetracycline resistance in Paenibacillus larvae, the causal agent of American Foulbrood (AFB) disease in honeybees. Vet. Microbiol. 125, 290–303. doi: 10.1016/j.vetmic.2007.05.018
AlKhatib, Z., Lagedroste, M., Fey, I., Kleinschrodt, D., Abts, A., and Smits, S. H. J. (2014). Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS One 9:e102246. doi: 10.1371/journal.pone.0102246
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Anderson, K. E., Ricigliano, V. A., Mott, B. M., Copeland, D. C., Floyd, A. S., and Maes, P. (2018). The queen’s gut refines with age: longevity phenotypes in a social insect model. Microbiome 6:108. doi: 10.1186/s40168-018-0489-1
Anderson, K. E., Sheehan, T. H., Mott, B. M., Maes, P., Snyder, L., Schwan, M. R., et al. (2013). Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS One 8:e83125. doi: 10.1371/journal.pone.0083125
Arai, R., Tominaga, K., Wu, M., Okura, M., Ito, K., Okamura, N., et al. (2012). Diversity of Melissococcus plutonius from honeybee larvae in Japan and experimental reproduction of European foulbrood with cultured atypical isolates. PLoS One 7:e33708. doi: 10.1371/journal.pone.0033708
Arredondo, D., Castelli, L., Porrini, M. P., Garrido, P. M., Eguaras, M. J., Zunino, P., et al. (2018). Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Benef. Microb. 9, 279–290. doi: 10.3920/BM2017.0075
Banerjee, S., and Hansen, J. N. (1988). Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J. Biol. Chem. 263, 9508–9514.
Cleveland, J., Montville, T. J., Nes, I. F., and Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/s0168-1605(01)00560-8
Cotter, P. D., Hill, C., and Ross, R. P. (2005a). Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci. 6, 61–75. doi: 10.2174/1389203053027584
Cotter, P. D., Hill, C., and Ross, R. P. (2005b). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273
de Kwaadsteniet, M., ten Doeschate, K., and Dicks, L. M. T. (2008). Characterization of the Structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 74, 547–549. doi: 10.1128/AEM.01862-07
de Ruyter, P. G., Kuipers, O. P., Beerthuyzen, M. M., van Alen-Boerrigter, I., and de Vos, W. M. (1996). Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178, 3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996
Delves-Broughton, J., Blackburn, P., Evans, R. J., and Hugenholtz, J. (1996). Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek 69, 193–202. doi: 10.1007/bf00399424
Edwards, C. G., Haag, K. M., Collins, M. D., and Hutson, R. A. (1998). Lactobacillus kunkeei sp. nov.: a spoilage organism associated with grape juice fermentations. J. Appl. Microbiol. 84, 698–702. doi: 10.1046/j.1365-2672.1998.00399.x
Endo, A., Futagawa-Endo, Y., and Dicks, L. M. T. (2009). Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst. Appl. Microbiol. 32, 593–600. doi: 10.1016/J.SYAPM.2009.08.002
Endo, A., Irisawa, T., Futagawa-Endo, Y., Takano, K., du Toit, M., Okada, S., et al. (2012). Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int. J. Syst. Evol. Microbiol. 62, 500–504. doi: 10.1099/ijs.0.031054-0
Endo, A., Maeno, S., Tanizawa, Y., Kneifel, W., Arita, M., Dicks, L., et al. (2018). Fructophilic lactic acid bacteria, a unique group of fructose-fermenting microbes. Appl. Environ. Microbiol. 84:e01290-18. doi: 10.1128/AEM.01290-18
Endo, A., and Okada, S. (2008). Reclassification of the genus Leuconostoc and proposals of Fructobacillus fructosus gen. nov., comb. nov., Fructobacillus durionis comb. nov., Fructobacillus ficulneus comb. nov. and Fructobacillus pseudoficulneus comb. nov. Int. J. Syst. Evol. Microbiol. 58, 2195–2205. doi: 10.1099/ijs.0.65609-0
Endo, A., and Salminen, S. (2013). Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 36, 444–448. doi: 10.1016/J.SYAPM.2013.06.002
Endo, A., Tanizawa, Y., Tanaka, N., Maeno, S., Kumar, H., Shiwa, Y., et al. (2015). Comparative genomics of Fructobacillus spp. and Leuconostoc spp. reveals niche-specific evolution of Fructobacillus spp. BMC Genomics 16:1117. doi: 10.1186/s12864-015-2339-x
Ennahar, S., Asou, Y., Zendo, T., Sonomoto, K., and Ishizaki, A. (2001). Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. Int. J. Food Microbiol. 70, 291–301. doi: 10.1016/S0168-1605(01)00565-7
Fujita, K., Ichimasa, S., Zendo, T., Koga, S., Yoneyama, F., Nakayama, J., et al. (2007). Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73, 2871–2877. doi: 10.1128/AEM.02286-06
Fukao, M., Obita, T., Yoneyama, F., Kohda, D., Zendo, T., Nakayama, J., et al. (2008). Complete covalent structure of nisin Q, new natural nisin variant, containing post-translationally modified amino acids. Biosci. Biotechnol. Biochem. 72, 1750–1755. doi: 10.1271/bbb.80066
Garcia-Gutierrez, E., O’Connor, P. M., Saalbach, G., Walsh, C. J., Hegarty, J. W., Guinane, C. M., et al. (2020). First evidence of production of the lantibiotic nisin P. Sci. Rep. 10:3738. doi: 10.1038/s41598-020-60623-0
Geiger, C., Korn, S. M., Häsler, M., Peetz, O., Martin, J., Kötter, P., et al. (2019). LanI-mediated lantibiotic immunity in Bacillus subtilis: functional analysis. Appl. Environ. Microbiol. 85:e00534-19. doi: 10.1128/AEM.00534-19
Hacker, C., Christ, N. A., Duchardt-Ferner, E., Korn, S., Göbl, C., Berninger, L., et al. (2015). The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J. Biol. Chem. 290, 28869–28886. doi: 10.1074/jbc.M115.679969
Hammami, R., Zouhir, A., Le Lay, C., Ben Hamida, J., and Fliss, I. (2010). BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol. 10:22. doi: 10.1186/1471-2180-10-22
Hatziioanou, D., Gherghisan-Filip, C., Saalbach, G., Horn, N., Wegmann, U., Duncan, S. H., et al. (2017). Discovery of a novel lantibiotic nisin O from Blautia obeum A2-162, isolated from the human gastrointestinal tract. Microbiology 163, 1292–1305. doi: 10.1099/mic.0.000515
Heunis, T. D. J., Smith, C., and Dicks, L. M. T. (2013). Evaluation of a nisin-eluting nanofiber scaffold to treat Staphylococcus aureus-induced skin infections in mice. Antimicrob. Agents Chemother. 57, 3928–3935. doi: 10.1128/AAC.00622-13
Kuipers, O. P., Beerthuyzen, M. M., de Ruyter, P. G., Luesink, E. J., and de Vos, W. M. (1995). Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304.
Kuipers, O. P., Beerthuyzen, M. M., Siezen, R. J., and de Vos, W. M. (1993). Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216, 281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x
Kwong, W. K., and Moran, N. A. (2016). Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384. doi: 10.1038/nrmicro.2016.43
Maeno, S., Tanizawa, Y., Kanesaki, Y., Kubota, E., Kumar, H., Dicks, L., et al. (2016). Genomic characterization of a fructophilic bee symbiont Lactobacillus kunkeei reveals its niche-specific adaptation. Syst. Appl. Microbiol. 39, 516–526. doi: 10.1016/J.SYAPM.2016.09.006
Masuda, Y., Ono, H., Kitagawa, H., Ito, H., Mu, F., Sawa, N., et al. (2011). Identification and characterization of leucocyclicin Q, a novel cyclic: bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 77:e06348-11. doi: 10.1128/AEM.06348-11
Meyer, H. E., Heber, M., Eisermann, B., Korte, H., Metzger, J. W., and Jung, G. (1994). Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal. Biochem. 223, 185–190. doi: 10.1006/ABIO.1994.1571
Mulders, J. W., Boerrigter, I. J., Rollema, H. S., Siezen, R. J., and de Vos, W. M. (1991). Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201, 581–584. doi: 10.1111/j.1432-1033.1991.tb16317.x
Murray, K. D., and Aronstein, K. A. (2006). Oxytetracycline-resistance in the honey bee pathogen Paenibacillus larvae is encoded on novel plasmid pMA67. J. Apic. Res. 45, 207–214. doi: 10.1080/00218839.2006.11101349
Murray, K. D., Aronstein, K. A., and de León, J. H. (2007). Analysis of pMA67, a predicted rolling-circle replicating, mobilizable, tetracycline-resistance plasmid from the honey bee pathogen, Paenibacillus larvae. Plasmid 58, 89–100. doi: 10.1016/j.plasmid.2007.02.001
O’Connor, P. M., O’Shea, E. F., Guinane, C. M., O’Sullivan, O., Cotter, P. D., Ross, R. P., et al. (2015). Nisin H is a new nisin variant produced by the gut-derived strain Streptococcus hyointestinalis DPC6484. Appl. Environ. Microbiol. 81, 3953–3960. doi: 10.1128/AEM.00212-15
O’Sullivan, J. N., O’Connor, P. M., Rea, M. C., O’Sullivan, O., Walsh, C. J., Healy, B., et al. (2020). Nisin J, a novel natural nisin variant, is produced by Staphylococcus capitis sourced from the human skin microbiota. J. Bacteriol. 202:e0639-19. doi: 10.1128/JB.00639-19
Perez, R. H., Zendo, T., and Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact 13:S3. doi: 10.1186/1475-2859-13-S1-S3
Plat, A., Kluskens, L. D., Kuipers, A., Rink, R., and Moll, G. N. (2011). Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl. Environ. Microbiol. 77, 604–611. doi: 10.1128/AEM.01503-10
Rangberg, A., Mathiesen, G., Amdam, G. V., and Diep, D. B. (2015). The paratransgenic potential of Lactobacillus kunkeei in the honey bee Apis mellifera. Benef. Microb. 6, 513–523. doi: 10.3920/BM2014.0115
Raymann, K., Bobay, L.-M., and Moran, N. A. (2018). Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol. 27, 2057–2066. doi: 10.1111/mec.14434
Raymann, K., Shaffer, Z., and Moran, N. A. (2017). Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15:e2001861. doi: 10.1371/journal.pbio.2001861
Stein, T., Heinzmann, S., Düsterhus, S., Borchert, S., and Entian, K.-D. (2005). Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187, 822–828. doi: 10.1128/JB.187.3.822-828.2005
Stein, T., Heinzmann, S., Solovieva, I., and Entian, K.-D. (2003). Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278, 89–94. doi: 10.1074/jbc.M207237200
Tanizawa, Y., Fujisawa, T., Kaminuma, E., Nakamura, Y., and Arita, M. (2016). DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 35, 173–184. doi: 10.12938/bmfh.16-003
Tian, B., Fadhil, N. H., Powell, J. E., Kwong, W. K., and Moran, N. A. (2012). Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3:e0377-12. doi: 10.1128/mBio.00377-12
Van Heel, A. J., De Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
Vojvodic, S., Rehan, S. M., and Anderson, K. E. (2013). Microbial gut diversity of Africanized and European honey bee larval instars. PLoS One 8:e72106. doi: 10.1371/journal.pone.0072106
Wirawan, R. E., Klesse, N. A., Jack, R. W., and Tagg, J. R. (2006). Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 72, 1148–1156. doi: 10.1128/AEM.72.2.1148-1156.2006
Yoneyama, F., Fukao, M., Zendo, T., Nakayama, J., and Sonomoto, K. (2008). Biosynthetic characterization and biochemical features of the third natural nisin variant, nisin Q, produced by Lactococcus lactis 61-14. J. Appl. Microbiol. 105, 1982–1990. doi: 10.1111/j.1365-2672.2008.03958.x
Zendo, T., Fukao, M., Ueda, K., Higuchi, T., Nakayama, J., and Sonomoto, K. (2003). Identification of the lantibiotic nisin Q, a new natural nisin variant produced by Lactococcus lactis 61-14 isolated from a river in Japan. Biosci. Biotechnol. Biochem. 67, 1616–1619. doi: 10.1271/bbb.67.1616
Zheng, H., Powell, J. E., Steele, M. I., Dietrich, C., and Moran, N. A. (2017). Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U.S.A. 114, 4775–4780. doi: 10.1073/pnas.1701819114
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: bacteriocins, lantibiotics, nisin, Apilactobacillus kunkeei, Melissococcus plutonius, fructophilic lactic acid bacteria
Citation: Zendo T, Ohashi C, Maeno S, Piao X, Salminen S, Sonomoto K and Endo A (2020) Kunkecin A, a New Nisin Variant Bacteriocin Produced by the Fructophilic Lactic Acid Bacterium, Apilactobacillus kunkeei FF30-6 Isolated From Honey Bees. Front. Microbiol. 11:571903. doi: 10.3389/fmicb.2020.571903
Received: 12 June 2020; Accepted: 12 August 2020;
Published: 16 September 2020.
Edited by:
Des Field, University College Cork, IrelandReviewed by:
Auke J. van Heel, University of Groningen, NetherlandsCopyright © 2020 Zendo, Ohashi, Maeno, Piao, Salminen, Sonomoto and Endo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeshi Zendo, emVuZG9AYWdyLmt5dXNodS11LmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.