
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 15 September 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.569282
Arsenic (As) is a metalloid that occurs widely in the environment. The biological oxidation of arsenite [As(III)] to arsenate [As(V)] is considered a strategy to reduce arsenic toxicity and provide energy. In recent years, research interests in microbial As(III) oxidation have been growing, and related new achievements have been revealed. This review focuses on the highlighting of the novel regulatory mechanisms of bacterial As(III) oxidation, the physiological relevance of different arsenic sensing systems and functional relationship between microbial As(III) oxidation and those of chemotaxis, phosphate uptake, carbon metabolism and energy generation. The implication to environmental bioremediation applications of As(III)-oxidizing strains, the knowledge gaps and perspectives are also discussed.
Arsenic (As) is a ubiquitous metalloid in the environment, and it belongs to group 15 in the periodic table, positioned directly below phosphorus (Strawn, 2018). Arsenic exists in various forms, such as inorganic trivalent arsenite [As(III)], inorganic pentavalent arsenate [As(V)], trivalent organoarsenicals, pentavalent organoarsenicals and thioarsenicals (Chen J. et al., 2015; McDermott et al., 2020). Of which, As(III) and As(V) are the most prevalent forms (Zhu et al., 2014). The toxicity of arsenic makes it to be a class I human carcinogen on the list announced by the International Agency of Research on Cancer (IARC) (Li et al., 2014; Minatel et al., 2018). Arsenic is released into the environment by both natural processes (such as volcanic eruption and parent material weathering) and the consumption of arsenic-containing products (such as chemotherapeutic drugs, insecticides and wood preservatives) (Zhu et al., 2014; Bhowmick et al., 2018). Over 200 million people are currently affected by high groundwater arsenic pollution in several areas, including Bangladesh, Brazil, Canada, China, Hungary, India, Indonesia, Mexico, Pakistan, United States and Vietnam (Chakraborti et al., 2009; Rodriguez-Lado et al., 2013; Bhowmick et al., 2018; de Souza et al., 2018). Chronic exposure to arsenic has been documented to lead to cardiovascular, diabetic, reproductive, hematological, hepatic, neurological and respiratory diseases as well as to cause bladder, lung and skin cancers (Rahman et al., 2009; Carlin et al., 2015).
Microorganisms play an important role in arsenic bioavailability, mobility and speciation in nature (Zhu et al., 2014). Microbial As(III) oxidation is considered environmental bioremediation because As(V) is less toxic and mobile than As(III) (Xiong et al., 2010; Wang et al., 2015b). To date, different genera of As(III)-oxidizing bacteria and archaea have been reported, including α-,β-,γ-Proteobacteria, Thermus, green sulfur bacteria, filamentous green non-sulfur bacteria, Crenarchaeota and Euryarchaeota (Stolz et al., 2006; Li X. et al., 2013). Furthermore, some As(III)-oxidizing bacteria use As(III) oxidation as an energy source (Oremland and Stolz, 2003; Stolz et al., 2006; Cai et al., 2009a). Chemolithoautotrophic As(III)-oxidizing bacteria, such as Rhizobium sp. NT-26, use As(III) as the electron donor, oxygen as the electron acceptor, and carbon dioxide-bicarbonate as the carbon source to maintain bacterial growth (Santini et al., 2000). Moreover, heterotrophic As(III)-oxidizing bacteria, such as Hydrogenophaga sp. N14 and Agrobacterium tumefaciens GW4, employ As(III) oxidation as an energy resource (vanden Hoven and Santini, 2004; Wang et al., 2015a). In recent years, the detailed regulatory mechanisms of bacterial As(III) oxidation have been revealed by our group and other researchers.
Microbial As(III) oxidation is catalyzed by As(III) oxidase, AioBA (Anderson et al., 1992), which is widely distributed in heterotrophic and chemolithoautotrophic microorganisms (Yan et al., 2019). AioBA was first purified from the Alcaligenes faecalis bacterium in 1992 (Anderson et al., 1992), and additional characteristics were identified in different As(III) oxidizers, such as Herminiimonas arsenicoxydans ULPAs1 and Rhizobium sp. NT-26 (Santini et al., 2000; Muller et al., 2003; Warelow et al., 2017; Watson et al., 2017). To date, homologs of genes encoding AioBA have been found in phylogenetically diverse strains including members of α-,β-,γ-Proteobacteria, Actinobacteria, Aquificae, Bacteroidetes, Chlorobi, Chloroflexi, Crenarchaeota, Deinococcus-Thermus, Firmicutes and Nitrospira (Yamamura and Amachi, 2014).
AioBA is a heterotetramer consisting of a large catalytic subunit (AioA) and a small subunit (AioB) (Silver and Phung, 2005). AioA is divided into four domains, in which, domain I contains a bound [3Fe-4S] cluster and a Rieske subunit and domains II-IV contain bound molybdenum (Ellis et al., 2001). AioB contains a Rieske [2Fe-2S] domain, which is homologous to cytochrome bc1, cytochrome b6f and naphthalene 1,2-dioxygenase (Ellis et al., 2001). A recent study has shown that AioBA also acts as an antimonite [Sb(III)] oxidase, indicating the complex function of AioBA (Wang et al., 2015b). Additionally, another type of As(III) oxidase, ArxA, has also been identified in a limited number of bacteria, predominantly in γ-Proteobacteria isolated from lakes characterized by high pH and salinity (Zargar et al., 2010). ArxA represents a distinct phylogenetic clade of As(III) oxidases and has bifunctional activity for both As(III) oxidation and As(V) reduction in vitro (Zargar et al., 2012; Ospino et al., 2019). Besides, trivalent forms of organoarsenicals, such as methylarsenate [MAs(V)] and roxarsone [Rox(V)], are more toxic than inorganic arsenicals, (Chen J. et al., 2015; Shi et al., 2018a). A member of the NADPH-dependent FMN oxidoreductase superfamily, ArsH, is found in many bacterial arsenic resistance operons and oxidizes trivalent organoarsenicals (Chen J. et al., 2015).
A two-component bacterial transcriptional regulatory system was identified by Kashyap et al. (2006) in A. tumefaciens 5A, and additional details of AioSR have been investigated in Rhizobium sp. NT-26 (Sardiwal et al., 2010). The sensor kinase AioS and its cognate response regulator AioR form the two-component system together (Kashyap et al., 2006). AioS contains transmembrane helices and autokinase domain, while AioR has a helix-turn-helix domain to bind DNA (Sardiwal et al., 2010). Moreover, the phosphorylation assays in Rhizobium sp. NT-26 reveal that His273 is the autophosphorylation site in AioS, while Asp58, Asp13 and Asp53 compose the conserved transphosphorylation site in AioR (Sardiwal et al., 2010).
Afterward, a periplasmic binding protein gene (aioX) is found in the upstream of bacterial aioSR genes, and its expression is induced by As(III) (Figure 1; Liu et al., 2012). Disruption of aioX inhibited the upregulation of aioBA genes, and consequently, As(III) oxidation (Liu et al., 2012). Without expression of a TAT-type signal peptide, purified AioX is exclusively associated with the cytoplasmic membrane, and its association constant with As(III) is 2.4 μM (Liu et al., 2012). However, site-directed mutation of Cys108 to either alanine or serine resulted in disruption of As(III) binding and aioBA induction, consequently causing loss of the As(III) oxidation phenotype (Liu et al., 2012). Taken together, the characterization of AioX suggests that a novel As(III)-sensing mechanism is present in a range of bacteria, which further modifies our understanding of the regulatory mechanism of As(III) oxidation from a two-component signal transduction system to a three-component signal transduction system (AioXSR). In the presence of As(III), AioX senses As(III) signals in the periplasm, thereby inducing the autophosphorylation of AioS (Kashyap et al., 2006). The phosphoryl group is then transferred from AioS to AioR, creating an active positive regulator to initiate the expression of aioBA genes (Kashyap et al., 2006).
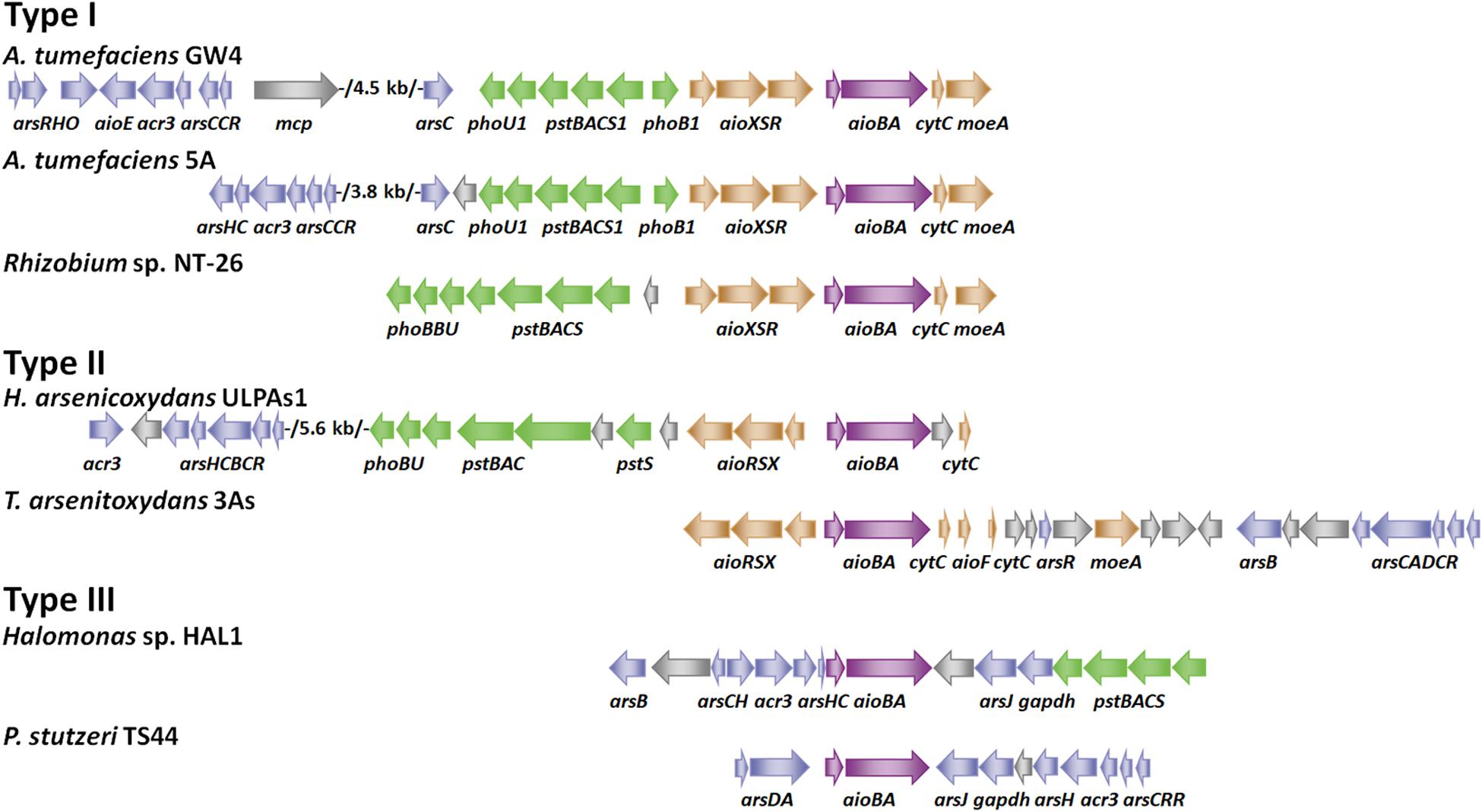
Figure 1. Gene arrangements of arsenic gene clusters. Arrows of different colors represent the following genes: blue for ars operon, green for pst/pho system, brown for aioXSR, purple for aioBA and gray for other genes. GenBank accession numbers are as follows: A. tumefaciens GW4 (NZ_AWGV00000000.1), A. tumefaciens 5A (NZ_AGVZ00000000.1), Rhizobium sp. NT-26 (NZ_FO082820.1, NZ_FO082821.1 and NZ_FO082822.1), H. arsenicoxydans ULPAs1 (NC_009138.1), T. arsenitoxydans 3As (NC_014145.1 and NC_014144.1), Halomonas sp. HAL1 (NZ_AGIB00000000.1) and Pseudomonas stutzeri TS44 (NZ_AJXE00000000.1).
According to the gene arrangements of aioXSR and aioBA, we divide As(III)-oxidizing bacteria into three types (Figure 1). Type I (aioXSR) is adjacent to aioBA with the same transcriptional direction. The three-component signal transduction system, AioXSR, is the main regulatory mechanism of As(III) oxidation in Type I typical bacteria. Type II (aioXSR) is adjacent to aioBA but has the opposite transcriptional direction. Type III has no aioXSR system in the bacterial genome (Cai et al., 2009b; Li et al., 2012; Li H. et al., 2013; Moinier et al., 2014; Chen F. et al., 2015; Wang et al., 2015a).
In Thiomonas arsenitoxydans 3As strain, a typical Type II gene arrangement was found (Figure 1), the ArsR/SmtB family regulator AioF is encoded in the aioBA operon, revealing that AioF is likely involved in the regulation of aioBA (Moinier et al., 2014). An electrophoretic mobility shift assay (EMSA) has demonstrated that AioF binds two distinct regions upstream from aioB (Moinier et al., 2014). Unlike other ArsR family regulators, which bind to the genetic regulatory region to repress expression without metal(loid) and release in the presence of metal(loid) (Shi et al., 2018a), AioF specifically binds the aioB regulatory region in the presence of arsenic, but the interaction between AioF and DNA disappears in the absence of arsenic (Moinier et al., 2014). Additionally, the promoters of aioB and aioX are convergent, and the regulatory region of aioB has a putative AioR-binding site and two AioF-binding sites (Moinier et al., 2014). Thus, the regulatory mechanism in T. arsenitoxydans 3As strain, which consists of both AioXSR and AioF, should be a rigorous regulatory system leading to the transcription of aioBA in two steps. In the absence of As(III), the transcriptional aioBA operon is completely silent, while aioXSR is expressed. In the presence of As(III), As(III) is detected by AioXS, and AioR is phosphorylated, leading to transcription of aioBA and aioF at low levels (Moinier et al., 2014). In the presence of both AioR and AioF, the aioBA operon expression is optimal (Moinier et al., 2014).
AioSR belongs to the Sigma54 family and RpoN is the main Sigma factor for binding of the Sigma54 family regulator with the promoter (Kang et al., 2012a). Several putative RpoN-binding sites have been identified within promoters throughout the genome of A. tumefaciens 5A, and one putative binding site has been found in aioBA promoter (Kang et al., 2012a). Disruption of the RpoN-binding site in aioBA promoter reduced the expression of aioBA and bacterial As(III) oxidation (Kang et al., 2012a). In A. tumefaciens 5A and H. arsenicoxydans ULPAs1, the presence of As(III) induced the expression of aioX, aioS and aioR (Cleiss-Arnold et al., 2010; Liu et al., 2012). However, there is no effect on the expression of aioX, aioS or aioR when Rhizobium sp. NT-26 or T. arsenitoxydans 3As was grown with As(III) (Slyemi et al., 2013; Corsini et al., 2018). Interestingly, the aioXSR operon was upregulated in stationary phase, and the growth phase-dependent promoter RpoE2 has been reported to influence the expression of the aioXSR operon (Corsini et al., 2018).
Many bacteria have two phosphate (Pi) transporters, a low affinity, high velocity Pi transporter (Pit system) and a high affinity, lower velocity Pi-specific transporter (Pst/Pho system) (Hsieh and Wanner, 2010). The Pit system serves as the primary Pi transporter when Pi is in excess, while the Pst/Pho system is activated only under low Pi concentrations (≤250 μM) (Hsieh and Wanner, 2010; Pontes and Groisman, 2018). Interestingly, genomic surveys have revealed that the two pst/pho systems often occur in As(III)-oxidizing bacteria (Figure 1; Li H. et al., 2013). The pst1/pho1 system is adjacent to aioBA genes (Figure 1), and the pst2/pho2 system is located elsewhere in the genome (Kang et al., 2012b; Li H. et al., 2013; Chen F. et al., 2015). The Pst/Pho system, including a Pi transport complex PstSABC and a two-component system PhoR/PhoB, is involved in Pi uptake under low Pi concentration (Qiao et al., 2019). The expression of phoB increased dramatically only under low Pi concentrations (≤250 μM) (Pontes and Groisman, 2018). In nature, other As(III) oxidizing bacteria were also isolated under low Pi conditions, and the efficiency of As(III) oxidation was reduced or interrupted under medium (1 mM) or high Pi conditions (>10 mM) (Kang et al., 2012b; Wang et al., 2015a, 2018). In A. tumefaciens strains, the disruption of phoB1 or pstS1 significantly decreased the transcription of aioB, and the efficiency of As(III) oxidation was decreased in the phoB1 mutant (Kang et al., 2012b; Qiao et al., 2019). An EMSA assay demonstrated that PhoB1 interacted with aioXSR promoter to regulate bacterial As(III) oxidation (Qiao et al., 2019). Moreover, the AioSR and PhoRB signal transduction systems were capable of transphosphorylation cross-talk in A. tumefaciens 5A, closely integrating bacterial As(III) oxidation with the phosphate starvation response (Wang et al., 2018).
In nature, there are another type of As(III) oxidation strains (Type III) which have no aioXSR regulation system in the genomes (Figure 1). The bacterial As(III) oxidation regulatory mechanism in Type III strains is unique compared to those of Type I and Type II (Lin et al., 2012; Chen F. et al., 2015). In the Type III As(III)-oxidizing Halomonas sp. HAL1 strain, transposon mutagenesis and gene knockout assays have both shown that PhoBR affect As(III)-oxidation phenotypes in only low Pi conditions (0.1 mM, Chen F. et al., 2015). A putative Pho box was found in the aioBA regulatory region, and PhoB could bind the Pho box in vivo and in vitro (Chen F. et al., 2015). In conclusion, PhoBR is a regulator of some As(III)-oxidizing bacteria, implying the co-regulation of As(III) oxidation and Pi uptake under low Pi conditions.
Generally, elements in the same group have similar chemical properties, such as silicon vs. carbon (Brzezinski, 2008) and selenium vs. sulfur (Stadtman, 1991), and substitution between these chemical analogs is common (Stadtman, 1991; Brzezinski, 2008; Rosen et al., 2011). Phosphorus is an essential nutrient for microbial biological processes. However, phosphorus is typically scarce and normally present at nanomolar levels in water and soil environments (Vieira et al., 2008). Thus, it is not surprising that As(V) can be incorporated into arsenolipids as a methylated pentavalent As-C bond and to form several types of arsenolipids in marine organisms (Edmonds and Francesconi, 1987; Dembitsky and Levitsky, 2004).
Recently, several studies have revealed that As(V) can substitute for its chemical analog Pi in bacteria (Wang et al., 2015a, 2018). Though nucleic acid and ATP may not be formed due to the instability of arsenal easter (Rosen et al., 2011), As(V) can incorporate into arsenosugars and arsenolipids with a methylated As(V)-C bond. This bond is very stable and well documented in fungi, plants, lichens and marine organisms (Dembitsky and Levitsky, 2004). More interestingly, our previous studies showed evidences of As(V) substitution of Pi in lipids in A. tumefaciens (Wang et al., 2015a, 2018): (1) Approximately half of As(V) was incorporated into cellular lipids in the 5A and GW4 strains under Pi-starvation and As-enriched conditions (Wang et al., 2018); (2) Compared to non-As treatment, the contents of five lipids were different with the addition of As(III), only one lipid type was increased with As(III), and this lipid contains 2-octyl-cyclopropaneoctanoic acid tail structure (Wang et al., 2015a). (3) The addition of As(V) was beneficial to the growth of As(III)-oxidizing strains 5A and GW4 in media composed of As(V):Pi ratios of 10:1, which means As(V) (1 mM for strain GW4 and 0.5 mM for strain 5A) is not toxic but beneficial to the bacteria (Wang et al., 2018).
Given that interconnection between As and Pi, it is counterintuitive that As(III)-oxidizing bacteria contain a complete and systematic regulatory link between arsenic regulation systems and the Pi signal transduction system. (1) When As(III) is present and environmental Pi is depleted, the arsenic-resistant negative regulator ArsR1 binds with As(III) and separates from the promoters of phoB1 and pstS1 (Garbinski et al., 2019); (2) PhoB1 is phosphorylated by the constitutively expressed histidine kinases PhoR and activates the expression of the pst/pho system and aioXSR system (Shi et al., 2018b; Rawle et al., 2019); (3) AioX senses As(III) in the environment, and self-phosphorylated AioS phosphorylates AioR to activate the expression of aioBA; (4) Meanwhile, phosphorylated PhoR and AioS both phosphorylate PhoB1 and AioR, which activate the pst/pho and aio systems (Wang et al., 2018; Rawle et al., 2019); (5) Expressed AioBA oxidize As(III) to As(V), PstS1 binds As(V), and the Pst1 transporter system transfers As(V) into bacterial cells. All of those evidences indicate that arsenic and Pi regulatory systems are deeply integrated and function in Pi-starvation response together (Wang et al., 2018; Rawle et al., 2019). Under Pi-starvation and arsenic-enrichment conditions, the integrated coregulation of arsenic and phosphorus metabolism is beneficial for bacteria to generate energy and use the Pi chemical analog, As(V). The consequent generation of arsenolipids may spare Pi to be recycled for nucleic acid synthesis, thereby enhancing bacterial survival potential.
Because As(III) is a toxic metalloid, it is recognized as a repellent to some microorganisms, such as Bradyrhizobium japonicum E10 and Azospirillum brasilense Az39 (Talano et al., 2013; Armendariz et al., 2015). Interestingly, As(III) is an attractant to some As(III)-oxidizing bacteria at low concentrations, such as A. tumefaciens GW4, H. arsenicoxydans ULPAs1 and Rhizobium sp. NT-26 (Muller et al., 2005; Andres et al., 2013; Shi et al., 2017). The As(III) chemoreceptor was first identified in A. tumefaciens GW4 bacterium in 2017 (Shi et al., 2017). On the genome of A. tumefaciens GW4, the mcp gene is located adjacent to the aioXSRBA operon, while the expression of the mcp, cheA and cheY2 is induced by As(III) (Shi et al., 2017). Disruption of mcp abolished bacterial As(III) chemotaxis ability, and the purified sensing domain of Mcp specifically bound to As(III), but not to As(V) or Pi (Shi et al., 2017). Moreover, a putative AioR-binding site has been found in the regulatory region of the mcp gene in A. tumefaciens GW4, H. arsenicoxydans ULPAs1 and Rhizobium sp. NT-26 (Shi et al., 2017). The aioR mutant interrupted the expression of mcp, consequently destroying As(III) chemotaxis in A. tumefaciens GW4 (Shi et al., 2017). Moreover, the As(III) oxidation regulator AioR has been shown to regulate the As(III)-oxidizing bacterial chemotactic response toward As(III) (Figure 2; Shi et al., 2017).
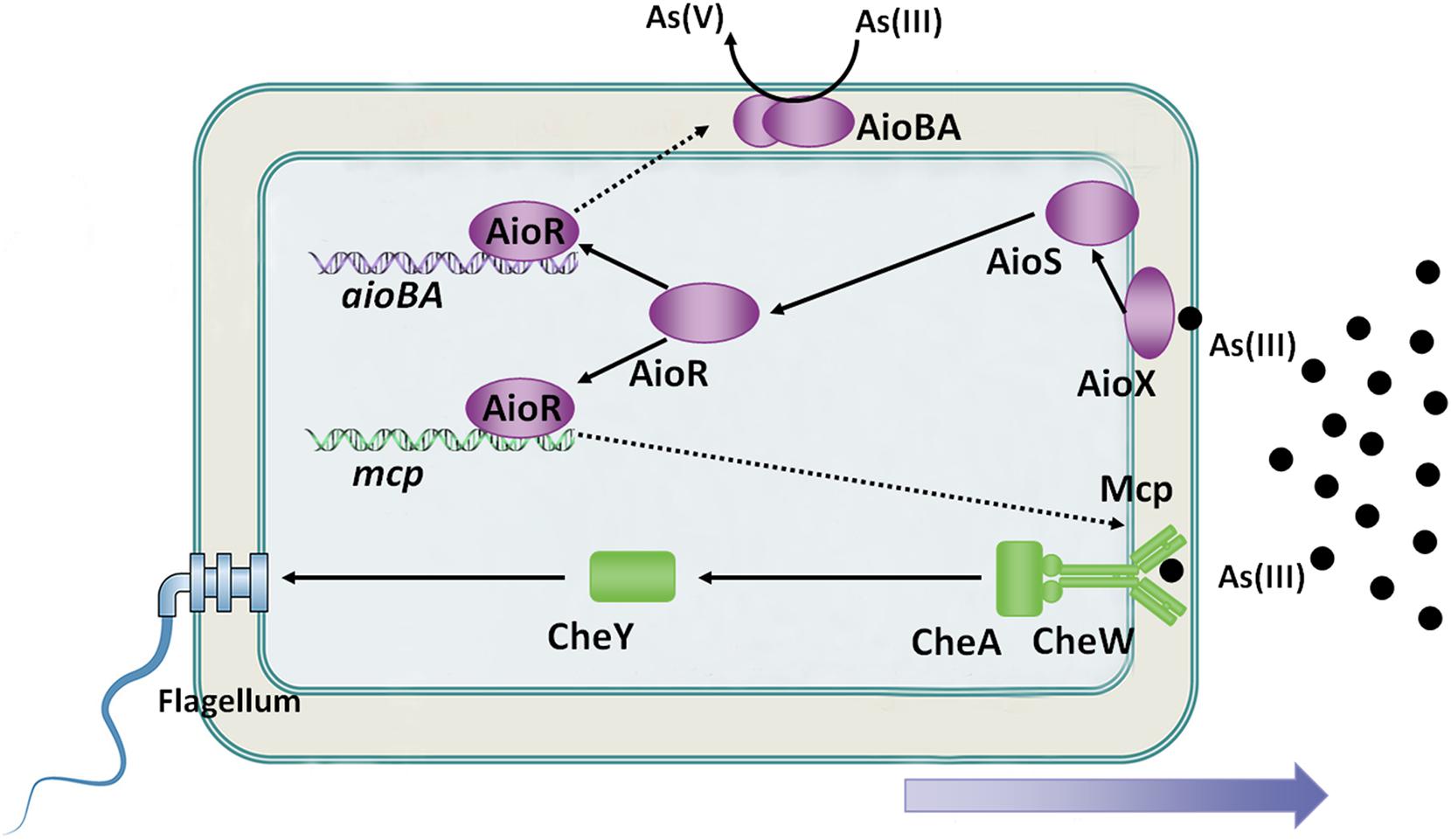
Figure 2. Overview of bacterial As(III) oxidation and chemotaxis. AioX binds As(III) and delivers the signal to the histidine kinase, AioS, simulating autophosphorylation of AioS. Autophosphorylated AioS then phosphorylates its cognate response regulator AioR, and subsequently AioR binds to the promoter of As(III) oxidase, which encodes aioBA genes, to switch on AioBA expression. Furthermore, AioR regulates expression of the As(III) chemoreceptor-encoding gene, mcp. Mcp detects the As(III) concentration gradient and then transports the signal to the flagellum via the chemotaxis system, CheWAY. Ultimately, the GW4 strain exhibits chemotaxis toward As(III).
An interesting observation derived from these studies suggests that bacterial As(III) oxidation is the essential condition of As(III) positive chemotaxis. (1) The deletion of As(III)-oxidizing genes resulted in disruption of bacterial As(III) positive chemotaxis phenotypes were observed in strains NT-26, ULPAs1 and GW4 (Koechler et al., 2010; Andres et al., 2013; Shi et al., 2017). (2) Both Mcp (association constant of 11.0 μM) and AioX (association constant of 2.4 μM) bind As(III) in the periplasm, but AioX is more sensitive to As(III) than Mcp (Liu et al., 2012; Shi et al., 2017). It indicates that AioXSR system serves as the initial As(III) response and Mcp serves as the downstream response, and such result is in agreement that bacterial As(III) chemotaxis is ultimately controlled by AioR (Figure 2; Shi et al., 2017). (3) In H. arsenicoxydans ULPAs1, dnaJ is essential for the expression of the flagellar master operon in Escherichia coli, and a disruption of dnaJ affected in both the motility and As(III) oxidation, suggesting that bacterial motility and As(III) oxidation may be co-regulated (Koechler et al., 2010). (4) Even though no clear connection was observed between AioR binding sites and motility-genes in Rhizobium sp. NT-26, TEM observations of the aioR mutant showed the presence of flagella in the early log phase of growth, suggesting that AioR may be involved in the repression of motility (Andres et al., 2013). Taken together, there is a strong correlation between bacterial As(III) oxidation and As(III) chemotaxis, and As(III) positive chemotaxis helps bacteria move toward As(III) to obtain more energy and transform As(III) to As(V), thereby enhancing bacterial survival (Shi et al., 2017).
In A. tumefaciens strains, there are different systems that recognize arsenic (Figure 3). Aio or Mcp system sense As(III) in the periplasmic space, Pst/Pho system senses As(V) and Pi in the periplasmic space, and ArsR senses As(III) in the cytosol. The different arsenic sensing systems relate to each other via the regulatory network. Excessive accumulation of As(III) in the cytosol is harmful to cells, and the central premise of bacterial As(III) oxidation is to enable bacteria growth in As(III)-contained environment. ArsR recognizes As(III) in the cytosol and then activates the expression of the ars operon to excrete As(III) out of cells. Furthermore, bacterial As(III) oxidation occurs in the periplasmic space, which means that As(V) production also accumulates in the periplasmic space. Thus, it is not surprising that Aio and Mcp systems sense As(III) and Pst/Pho system senses As(V) in the periplasmic space, which is more conducive for oxidization of As(III) and acquisition of As(V). These arsenic sensing systems promote each other and enhance cell survival (Figure 3). These findings contribute significantly to our current understanding of the metabolic impact and genetic circuitry involved during As(III) exposure in microorganisms. This indicates arsenic will impact and tightly link microbial activities in global biogeochemical cycles.
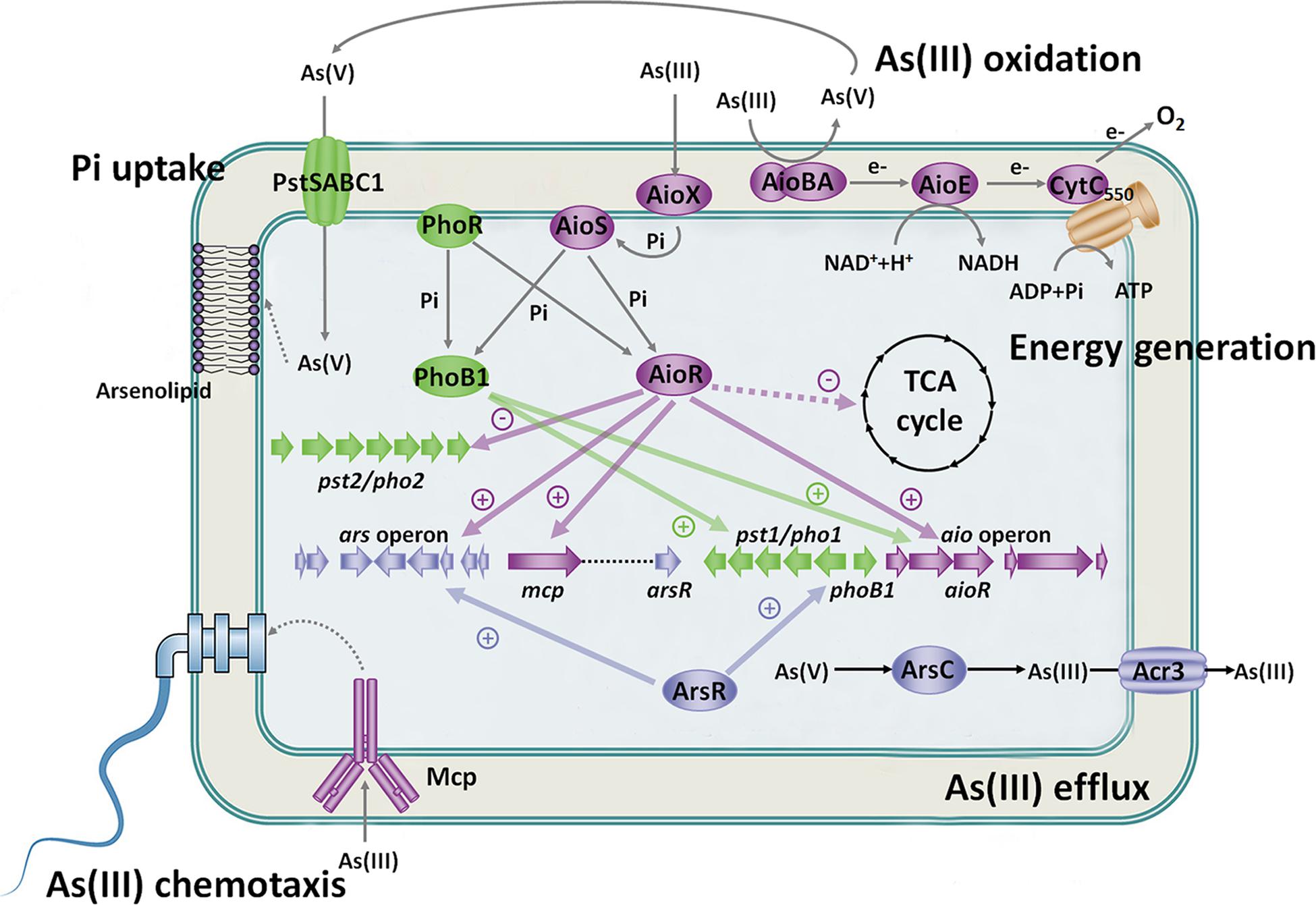
Figure 3. Regulatory and metabolic network related to As(III) oxidation. Under Pi-starvation and arsenic-enriched conditions, AioX and PstS sense the As(III) and As(V)/Pi signal first, and then AioS and PhoR are autophosphorylated. AioR and PhoB1 are phosphorylated by both AioS and PhoR. AioR positively regulates the expression of the As(III) oxidase AioBA and suppresses the TCA cycle so that As(III) oxidation is the main energy resource. Moreover, AioR increases the expression of the ars operon to enhance bacterial As(III) efflux. Moreover, ArsR activates the pst1/pho1 system, while AioR restrains the pst2/pho2 system to transport Pi and As(V). PhoB1 also increases the expression of the aio gene cluster and pst1/pho1 system. The connected regulatory network between the Aio, Ars and Pst/Pho systems indicates the close relationship between As(III) oxidation, As(III) efflux and Pi uptake. Furthermore, AioR regulates the expression of mcp and helps bacteria move toward As(III) to produce more energy and As(V). The synthesis of arsenolipids containing As(V) may spare Pi for nucleic acid synthesis, thereby enhancing bacterial survival potential. These findings imply that As(III) oxidation is biologically part of the Pi-starvation response.
For some As(III) oxidizers, As(III) oxidation is not only a detoxification mechanism but is also related to energy generation (Santini et al., 2000; Wang et al., 2015a). This may explain why As(III)-oxidizing bacteria exhibit positive chemotaxis to As(III) (Muller et al., 2005; Andres et al., 2013; Shi et al., 2017). Some autotrophic As(III) oxidizing strains use NO3– or O2 as the final electron acceptor to oxidize As(III), assisting in generation of energy for bacterial growth (Santini et al., 2000; Oremland et al., 2002). A photosynthetic As(III)-oxidizing bacterium has been reported to use As(III) as its sole photosynthetic electron donor (Kulp et al., 2008). Additionally, some heterotrophic As(III)-oxidizing bacteria, such as Hydrogenophaga sp. NT-14 and A. tumefaciens GW4, have also been reported to generate energy from As(III) oxidation (vanden Hoven and Santini, 2004; Wang et al., 2015a). Of which, A. tumefaciens GW4 was effective in improving the generation of ATP and NADH by As(III) oxidation (Wang et al., 2015a).
Bacterial As(III) oxidation is essentially a process of electron transport, and the electrochemical disequilibrium between reducing and oxidizing substrates is associated with energy generation (Mitchell, 1977; Ducluzeau et al., 2009). The c-type cytochrome CytC550 has been identified as an As(III) oxidation electron transporter in Rhizobium sp. NT-26 (Santini et al., 2007; Kalimuthu et al., 2014). However, the disruption of cytC550 did not completely block bacterial As(III) oxidation in A. tumefaciens GW4 (Wang et al., 2017). A further study identified a novel As(III) oxidation electron transporter, AioE, which belongs to the TrkA family, corresponding the generation of NADH and H+ (Belenky et al., 2007; Wang et al., 2009). Deletion of aioE resulted in a null As(III) oxidation phenotype and decreased the production of NADH, indicating that AioE is involved in As(III) oxidation electron transport (Wang et al., 2017). Furthermore, the redox potential gradient during the process of bacterial As(III) oxidation is AioBA > AioE > CytC550 (Wang et al., 2017). The decreased cellular concentration of ATP or NADH occurs in cytC550 or aioE mutants, respectively, revealing that electrons may be transferred from AioBA to AioE and finally to CytC550 with the generation of NADH or ATP, respectively (Wang et al., 2017).
Bacterial As(III) oxidation is associated with carbon metabolism. In the autotrophic As(III)-oxidizing strains Thiomonas arsenivorans, proteins involved in carbon fixation (ribulose-1,5-biphosphate carboxylase and fructose-1,6-biphosphatase) were upregulated in the presence of As(III) and the ability of carbon fixation and As(III) oxidation were both enhanced (Bryan et al., 2009). Additionally, transcriptomics and proteomics revealed that several genes and proteins involved in carbon fixation via the Calvin cycle were upregulated in Rhizobium sp. NT-26 grown with As(III) (Andres et al., 2013). It indicates that autotrophic As(III)-oxidizing strains may improve their capacity to fix carbon when As(III) is present (Bryan et al., 2009; Andres et al., 2013).
In the heterotrophic As(III)-oxidizing strain H. arsenicoxydans ULPAs1, the expression of proteins involved in the TCA cycle was upregulated in the presence of As(III) (Carapito et al., 2006; Weiss et al., 2009). An upregulated TCA cycle may be beneficial to counter the toxicity of As(III) (Carapito et al., 2006; Weiss et al., 2009). However, in A. tumefaciens GW4 and 5A, proteins related to the TCA cycle were downregulated, but proteins related to fermentation and As(III) oxidation processes were upregulated by As(III) (Tokmina-Lukaszewska et al., 2017; Shi et al., 2018b). Furthermore, AioR has been shown to negatively regulate the 5-dehydro-4-deoxyglucarate dehydratase encoding kdgD, and kdgD uses 5-dehydro-4-deoxy-D-glucarate as a substrate to produce 2,5-dioxopentanoate, which enters the TCA cycle (Jeffcoat et al., 1969; Shi et al., 2018b). These findings indicate that As(III) oxidation and fermentation are the main energy resources that directly contribute to energy generation in some heterotrophic As(III)-oxidizing bacteria (Shi et al., 2018b). In conclusion, the integrated relationship between As(III) oxidation and carbon metabolism may enhance bacterial survival potential under an oxygen-deficient, Pi-starvation and As-enriched environment.
As(III)-oxidizing microorganisms are potential candidates for bioremediation of arsenic contaminated environments due to the ability to oxidize As(III) to As(V) (Marinho et al., 2019). Apart from directly changing the chemical forms of arsenic compounds by microbial As(III) oxidation process, As(III)-oxidizing bacteria can also enhance the arsenic uptake in arsenic accumulating plants. For example, the presence of Ensifer sp. M14 strain in soil contributed to the increase of both growth and arsenic-uptake efficiency in alfalfa (Medicago sativa L., TANGO type) (Debiec-Andrzejewska et al., 2020). Compared to non-bioaugmented soil, the biomass of plants increased by about 60%, while the arsenic accumulation by alfalfa increased more than twice after 40 days (Debiec-Andrzejewska et al., 2020).
Microorganisms promote the interaction of arsenic and iron. In nature, the presence, enrichment and migration of arsenic is often associated with iron-containing minerals and As(III)-oxidizing microorganisms are used as biological reagents in combination with iron for the treatment of arsenic contamination (Karn et al., 2017; Yang et al., 2017). The combination of an As(III)-oxidizing bacterium Brevibacterium sp. YZ-1 and biogenic schwertmannite could immobilize arsenic in the highly arsenic contaminated soil (Yang et al., 2017). Combining an As(III)-oxidizing bacterium XS4 with FeCl3 also significantly stabilized arsenic in soil (Karn et al., 2017). Simultaneous removal of As(III) and was achieved by the immobilized Enterobacter strain in combination with FeCl3 and Ca(OH)2 (Shi et al., 2020). In addition, iron-oxidizing bacteria are also important in the oxidation of As(III). For instance, the inoculation of three arsenic resistant iron-oxidizing bacteria (Bacillus sp. T2, Pseudomonas sp. Yangling I4 and Bacillus sp. TF1-3) enhanced the formation of iron plaque and then decreased arsenic amount in brown rice (Xiao et al., 2020). Taken together, combining As(III)-oxidizing bacteria with chemical treatments are promising strategies for the removal or immobilization of arsenic from contaminated environment.
The concentration of Pi is usually low in the environment, and microorganisms can oxidize As(III) in low Pi concentration environments. Therefore, it is not unusual that relationships between bacterial As(III) oxidation, chemotaxis, Pi transport and energy metabolism appear under Pi-starvation conditions. Here, we highlight microbial survival strategies under Pi-limited and As(III)-enriched conditions (Figure 3). As(III)-oxidizing bacteria utilize As(III) oxidation to generate energy and detoxify As(III). At a certain level of As(III), bacteria activate the As(III) positive chemotaxis system to acquire As(III) and perform As(III) oxidation. Meanwhile, the pst1/pho1 system is upregulated to simultaneously acquire Pi and the As(III) oxidation product As(V), and As(V) may be incorporated into specific biomolecules, such as arsenolipids and arsenoproteins. The coregulation of bacterial arsenic and phosphorus metabolism benefits bacteria to recycle Pi for nucleic acid synthesis. Furthermore, the As(III) oxidation regulator AioR controls multiple As(III) resistance mechanisms to endow bacteria with strong As(III) resistance. Bacterial As(III) oxidation, chemotaxis, Pi transport and energy metabolisms synergistically benefit the survival of As(III)-oxidizing bacteria under Pi-limited and arsenic-riched conditions.
Recent literatures and our published works provide great insights for the understanding of microbial arsenite oxidation mechanisms. However, knowledge gaps still exist and more questions need to be resolved in the future. (1) Arsenic is found in bacterial lipids under Pi-limited conditions. However, the understanding of As(V) uptake and whether As(V) is incorporated in arsenolipids or replaces Pi in arsenolipids remain unknown. (2) In marine organisms, the As(III) S-adenosylmethionine methyltransferase ArsM has been reported to catalyze As(V) and other molecules to form arsenolipids (Qin et al., 2006). However, no recognizable arsM homolog has been identified in the A. tumefaciens GW4 and 5A strains, implying that arsenolipids originate from a different biosynthesis strategy in bacteria. (3) Genomic surveys have shown that genes related to the phosphonate transport system (phn) are often adjacent to aioBA genes in some As(III)-oxidizing bacteria (Manav et al., 2018). However, it remains unclear whether the Phn system is involved in the metabolism of arsenolipids. (4) As(III) oxidation regulators not only regulate As(III) oxidation but also affect multiple bacterial metabolic pathways. However, the regulatory network requires further study to clarify the biological significance of bacterial As(III) oxidation. Additionally, microbial As(III) oxidation is mainly investigated in bacteria and archaea. However, no As(III) oxidase demonstrates conserved homology in fungi (Li et al., 2014), suggesting that As(III) oxidation in fungi still needs to be investigated. (5) The application of As(III)-oxidizing microorganisms is only in the preliminary stage and the interactions among microbes, soils and plants need to be further investigated.
GW performed the study conceptualization and supervision. KS performed the literature search and wrote the original draft. KS, QW, and GW carried out the critical discussion, writing, reviewing, and editing. All authors read and approved the final manuscript.
This work is financially supported by the National Key Research and Development Program of China (2016YFD0800702) and the National Natural Science Foundation of China (31900078 and 31870086).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anderson, G. L., Williams, J., and Hille, R. (1992). The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J. Biol. Chem. 267, 23674–23682.
Andres, J., Arsene-Ploetze, F., Barbe, V., Brochier-Armanet, C., Cleiss-Arnold, J., Coppee, J. Y., et al. (2013). Life in an arsenic-containing gold mine, genome and physiology of the autotrophic arsenite-oxidizing bacterium Rhizobium sp. NT-26. Genome Biol. Evol. 5, 934–953. doi: 10.1093/gbe/evt061
Armendariz, A. L., Talano, M. A., Wevar Oller, A. L., Medina, M. I., and Agostini, E. (2015). Effect of arsenic on tolerance mechanisms of two plant growth-promoting bacteria used as biological inoculants. J. Environ. Sci. 33, 203–210. doi: 10.1016/j.jes.2014.12.024
Belenky, P., Bogan, K. L., and Brenner, C. (2007). NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19. doi: 10.1016/j.tibs.2006.11.006
Bhowmick, S., Pramanik, S., Singh, P., Mondal, P., Chatterjee, D., and Nriagu, J. (2018). Arsenic in groundwater of West Bengal, India: a review of human health risks and assessment of possible intervention options. Sci. Total. Environ. 612, 148–169. doi: 10.1016/j.scitotenv.2017.08.216
Bryan, C. G., Marchal, M., Battaglia-Brunet, F., Kugler, V., Lemaitre-Guillier, C., Lièvremont, D., et al. (2009). Carbon and arsenic metabolism in Thiomonas strains: differences revealed diverse adaptation processes. BMC Microbiol. 9:127. doi: 10.1186/1471-2180-9-127
Brzezinski, M. A. (2008). Mining the diatom genome for the mechanism of biosilicification. Proc. Natl. Acad. Sci. U.S.A. 105, 1391–1392. doi: 10.1073/pnas.0711994105
Cai, L., Liu, G., Rensing, C., and Wang, G. (2009a). Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9:4. doi: 10.1186/1471-2180-9-4
Cai, L., Rensing, C., Li, X., and Wang, G. (2009b). Novel gene clusters involved in arsenite oxidation and resistance in two arsenite oxidizers: Achromobacter sp. SY8 and Pseudomonas sp. TS44. Appl. Microbiol. Biotechnol. 83, 715–725. doi: 10.1007/s00253-009-1929-4
Carapito, C., Muller, D., Turlin, E., Koechler, S., Danchin, A., Van Dorsselaer, A., et al. (2006). Identification of genes and proteins involved in the pleiotropic response to arsenic stress in Caenibacter arsenoxydans, a metalloresistant beta-proteobacterium with an unsequenced genome. Biochimie 88, 595–606. doi: 10.1016/j.biochi.2005.11.004
Carlin, D. J., Naujokas, M. F., Bradham, K. D., Cowden, J., Heacock, M., Henry, H. F., et al. (2015). Arsenic and environmental health: state of the science and future research opportunities. Environ. Health Perspect. 124, 890–899. doi: 10.1289/ehp.1510209
Chakraborti, D., Das, B., Rahman, M. M., Chowdhury, U. K., Biswas, B., Goswami, A. B., et al. (2009). Status of groundwater arsenic contamination in the state of West Bengal, India: a 20-year study report. Mol. Nutr. Food Res. 53, 542–551. doi: 10.1002/mnfr.200700517
Chen, F., Cao, Y., Wei, S., Li, Y., Li, X., Wang, Q., et al. (2015). Regulation of arsenite oxidation by the phosphate two-component system PhoBR in Halomonas sp. HAL1. Front. Microbiol. 6:923. doi: 10.3389/fmicb.2015.00923
Chen, J., Bhattacharjee, H., and Rosen, B. P. (2015). ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 96, 1042–1052. doi: 10.1111/mmi.12988
Cleiss-Arnold, J., Koechler, S., Proux, C., Fardeau, M. L., Dillies, M. A., Coppee, J. Y., et al. (2010). Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709. doi: 10.1186/1471-2164-11-709
Corsini, P. M., Walker, K. T., and Santini, J. M. (2018). Expression of the arsenite oxidation regulatory operon in Rhizobium sp. str. NT-26 is under the control of two promoters that respond to different environmental cues. Microbiologyopen 7:e00567. doi: 10.1002/mbo3.567
de Souza, A. C. M., de Almeida, M. G., Pestana, I. A., and de Souza, C. M. M. (2018). Arsenic exposure and effects in humans: a mini-review in Brazil. Arch. Environ. Contam. Toxicol. 76, 357–365. doi: 10.1007/s00244-018-00586-6
Debiec-Andrzejewska, K., Krucon, T., Piatkowska, K., and Drewniak, L. (2020). Enhancing the plants growth and arsenic uptake from soil using arsenite-oxidizing bacteria. Environ. Pollut. 264:114692. doi: 10.1016/j.envpol.2020.114692
Dembitsky, V. M., and Levitsky, D. O. (2004). Arsenolipids. Prog. Lipid Res. 43, 403–448. doi: 10.1016/j.plipres.2004.07.001
Ducluzeau, A. L., van Lis, R., Duval, S., Schoepp-Cothenet, B., Russell, M. J., and Nitschke, W. (2009). Was nitric oxide the first deep electron sink? Trends Biochem. Sci. 34, 9–15. doi: 10.1016/j.tibs.2008.10.005
Edmonds, J. S., and Francesconi, K. A. (1987). Transformations of arsenic in the marine environment. Experientia 43, 553–557. doi: 10.1007/bf02143584
Ellis, P. J., Conrads, T., Hille, R., and Kuhn, P. (2001). Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A. Structure 9, 125–132. doi: 10.1016/s0969-2126(01)00566-4
Garbinski, L. D., Rosen, B. P., and Chen, J. (2019). Pathways of arsenic uptake and efflux. Environ. Int. 126, 585–597. doi: 10.1016/j.envint.2019.02.058
Hsieh, Y. J., and Wanner, B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203. doi: 10.1016/j.mib.2010.01.014
Jeffcoat, R., Hassall, H., and Dagley, S. (1969). Purification and properties of D-4-deoxy-5-oxoglucarate hydro-lyase (decarboxylating). Biochem. J. 115, 977–983. doi: 10.1042/bj1150977
Kalimuthu, P., Heath, M. D., Santini, J. M., Kappler, U., and Bernhardt, P. V. (2014). Electrochemically driven catalysis of Rhizobium sp. NT-26 arsenite oxidase with its native electron acceptor cytochrome c552. Biochim. Biophys. Acta 1837, 112–120. doi: 10.1016/j.bbabio.2013.07.010
Kang, Y. S., Bothner, B., Rensing, C., and McDermott, T. R. (2012a). Involvement of RpoN in regulating bacterial arsenite oxidation. Appl. Environ. Microbiol. 78, 5638–5645. doi: 10.1128/aem.00238-12
Kang, Y. S., Heinemann, J., Bothner, B., Rensing, C., and McDermott, T. R. (2012b). Integrated co-regulation of bacterial arsenic and phosphorus metabolisms. Environ. Microbiol. 14, 3097–3109. doi: 10.1111/j.1462-2920.2012.02881.x
Karn, S. K., Pan, X., and Jenkinson, I. R. (2017). Bio-transformation and stabilization of arsenic (As) in contaminated soil using arsenic oxidizing bacteria and FeCl3 amendment. 3 Biotech 7:50.
Kashyap, D. R., Botero, L. M., Franck, W. L., Hassett, D. J., and McDermott, T. R. (2006). Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188, 1081–1088. doi: 10.1128/jb.188.3.1081-1088.2006
Koechler, S., Cleiss-Arnold, J., Proux, C., Sismeiro, O., Dillies, M. A., Goulhen-Chollet, F., et al. (2010). Multiple controls affect arsenite oxidase gene expression in Herminiimonas arsenicoxydans. BMC Microbiol. 10:53. doi: 10.1186/1471-2180-10-53
Kulp, T. R., Hoeft, S. E., Asao, M., Madigan, M. T., Hollibaugh, J. T., Fisher, J. C., et al. (2008). Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321, 967–970. doi: 10.1126/science.1160799
Li, H., Li, M., Huang, Y., Rensing, C., and Wang, G. (2013). In silico analysis of bacterial arsenic islands reveals remarkable synteny and functional relatedness between arsenate and phosphate. Front. Microbiol. 4:347. doi: 10.3389/fmicb.2013.00347
Li, X., Gong, J., Hu, Y., Cai, L., Johnstone, L., Grass, G., et al. (2012). Genome sequence of the moderately halotolerant, arsenite-oxidizing bacterium Pseudomonas stutzeri TS44. J. Bacteriol. 194, 4473–4474. doi: 10.1128/jb.00907-12
Li, X., Hu, Y., Gong, J., Zhang, L., and Wang, G. (2013). Comparative genome characterization of Achromobacter members reveals potential genetic determinants facilitating the adaptation to a pathogenic lifestyle. Appl. Microbiol. Biotechnol. 97, 6413–6425. doi: 10.1007/s00253-013-5018-3
Li, X., Zhang, L., and Wang, G. (2014). Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PLoS One 9:e92236. doi: 10.1371/journal.pone.0092236
Lin, Y., Fan, H., Hao, X., Johnstone, L., Hu, Y., Wei, G., et al. (2012). Draft genome sequence of Halomonas sp. strain HAL1, a moderately halophilic arsenite-oxidizing bacterium isolated from gold-mine soil. J. Bacteriol. 194, 199–200. doi: 10.1128/jb.06359-11
Liu, G., Liu, M., Kim, E. H., Maaty, W. S., Bothner, B., Lei, B., et al. (2012). A periplasmic arsenite-binding protein involved in regulating arsenite oxidation. Environ. Microbiol. 14, 1624–1634. doi: 10.1111/j.1462-2920.2011.02672.x
Manav, M. C., Sofos, N., Hove-Jensen, B., and Brodersen, D. E. (2018). The abc of phosphonate breakdown: a mechanism for bacterial survival. Bioessays 40:e1800091.
Marinho, B. A., Cristóvão, R. O., Boaventura, R. A., and Vilar, V. J. (2019). As(III) and Cr(VI) oxyanion removal from water by advanced oxidation/reduction processes—a review. Environ. Sci. Pollut. Res. Int. 26, 2203–2227. doi: 10.1007/s11356-018-3595-5
McDermott, T. R., Stolz, J. F., Ronald, S., and Oremland, R. S. (2020). Arsenic and the gastrointestinal tract microbiome. Environ. Microbiol. Rep. 12, 136–159. doi: 10.1111/1758-2229.12814
Minatel, B. C., Sage, A. P., Anderson, C., Hubaux, R., Marshall, E. A., Lam, W. L., et al. (2018). Environmental arsenic exposure: from genetic susceptibility to pathogenesis. Environ. Int. 112, 183–197. doi: 10.1016/j.envint.2017.12.017
Mitchell, P. (1977). Vectorial chemiosmotic processes. Annu. Rev. Biochem. 46, 996–1005. doi: 10.1146/annurev.bi.46.070177.005024
Moinier, D., Slyemi, D., Byrne, D., Lignon, S., Lebrun, R., Talla, E., et al. (2014). An ArsR/SmtB family member is involved in the regulation by arsenic of the arsenite oxidase operon in Thiomonas arsenitoxydans. Appl. Environ. Microbiol. 80, 6413–6426. doi: 10.1128/aem.01771-14
Muller, D., Lievremont, D., Simeonova, D. D., Hubert, J. C., and Lett, M. C. (2003). Arsenite oxidase aox genes from a metal-resistant beta-proteobacterium. J. Bacteriol. 185, 135–141. doi: 10.1128/jb.185.1.135-141.2003
Muller, D., Médigue, C., Koechler, S., Barbe, V., Barakat, M., Talla, E., et al. (2005). A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 3:e53. doi: 10.1371/journal.pgen.0030053
Oremland, R. S., Hoeft, S. E., Santini, J. M., Bano, N., Hollibaugh, R. A., and Hollibaugh, J. T. (2002). Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68, 4795–4802. doi: 10.1128/aem.68.10.4795-4802.2002
Oremland, R. S., and Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944. doi: 10.1126/science.1081903
Ospino, M. C., Kojima, H., and Fukui, M. (2019). Arsenite oxidation by a newly isolated betaproteobacterium possessing arx genes and diversity of the arx gene cluster in bacterial genomes. Front. Microbiol. 10:1210. doi: 10.3389/fmicb.2019.01210
Pontes, M. H., and Groisman, E. A. (2018). Protein synthesis controls phosphate homeostasis. Genes Dev. 32, 79–92. doi: 10.1101/gad.309245.117
Qiao, Z., Huang, J., Cao, Y., Shi, K., and Wang, G. (2019). Genetics and proteomics analyses reveal the roles of PhoB1 and PhoB2 regulators in bacterial responses to arsenite and phosphate. Res. Microbiol. 170, 263–271. doi: 10.1016/j.resmic.2019.06.006
Qin, J., Rosen, B. P., Zhang, Y., Wang, G., Franke, S., and Rensing, C. (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 2075–2080. doi: 10.1073/pnas.0506836103
Rahman, M. M., Ng, J. C., and Naidu, R. (2009). Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ. Geochem. Health 31(Suppl. 1), 189–200. doi: 10.1007/s10653-008-9235-0
Rawle, R. A., Kang, Y. S., Bothner, B., Wang, G., and McDermott, T. R. (2019). Transcriptomics analysis defines global cellular response of Agrobacterium tumefaciens 5A to arsenite exposure regulated through the histidine kinases PhoR and AioS. Environ. Microbiol. 21, 2659–2676. doi: 10.1111/1462-2920.14577
Rodriguez-Lado, L., Sun, G., Berg, M., Zhang, Q., Xue, H., Zheng, Q., et al. (2013). Groundwater arsenic contamination throughout China. Science 341, 866–868. doi: 10.1126/science.1237484
Rosen, B. P., Ajees, A. A., and McDermott, T. R. (2011). Life and death with arsenic. Arsenic life: an analysis of the recent report “A bacterium that can grow by using arsenic instead of phosphorus”. Bioessays 33, 350–357. doi: 10.1002/bies.201100012
Santini, J. M., Kappler, U., Ward, S. A., Honeychurch, M. J., vanden Hoven, R. N., and Bernhardt, P. V. (2007). The NT-26 cytochrome c552 and its role in arsenite oxidation. Biochim. Biophys. Acta 1767, 189–196. doi: 10.1016/j.bbabio.2007.01.009
Santini, J. M., Sly, L. I., Schnagl, R. D., and Macy, J. M. (2000). A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: phylogenetic, physiological, and preliminary biochemical studies. Appl. Environ. Microbiol. 66, 92–97. doi: 10.1128/aem.66.1.92-97.2000
Sardiwal, S., Santini, J. M., Osborne, T. H., and Djordjevic, S. (2010). Characterization of a two-component signal transduction system that controls arsenite oxidation in the chemolithoautotroph NT-26. FEMS Microbiol. Lett. 313, 20–28. doi: 10.1111/j.1574-6968.2010.02121.x
Shi, K., Dai, X., Fan, X., Zhang, Y., Chen, Z., and Wang, G. (2020). Simultaneous removal of chromate and arsenite by the immobilized Enterobacter bacterium in combination with chemical reagents. Chemosphere 259:127428. doi: 10.1016/j.chemosphere.2020.127428
Shi, K., Fan, X., Qiao, Z., Han, Y., McDermott, T. R., Wang, Q., et al. (2017). Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4. Sci. Rep. 7:43252.
Shi, K., Li, C., Rensing, C., Dai, X., Fan, X., and Wang, G. (2018a). Efflux transporter ArsK is responsible for bacterial resistance to arsenite, antimonite, trivalent roxarsone, and methylarsenite. Appl. Environ. Microbiol. 84:e01842-18.
Shi, K., Wang, Q., Fan, X., and Wang, G. (2018b). Proteomics and genetic analyses reveal the effects of arsenite oxidation on metabolic pathways and the roles of AioR in Agrobacterium tumefaciens GW4. Environ. Pollut. 235, 700–709.
Silver, S., and Phung, L. T. (2005). Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71, 599–608.
Slyemi, D., Moinier, D., Talla, E., and Bonnefoy, V. (2013). Organization and regulation of the arsenite oxidase operon of the moderately acidophilic and facultative chemoautotrophic Thiomonas arsenitoxydans. Extremophiles 17, 911–920.
Stadtman, T. C. (1991). Biosynthesis and function of selenocysteine-containing enzymes. J. Biol. Chem. 266, 16257–16260.
Stolz, J. F., Basu, P., Santini, J. M., and Oremland, R. S. (2006). Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130.
Strawn, D. G. (2018). Review of interactions between phosphorus and arsenic in soils from four case studies. Geochem. Trans. 19:10.
Talano, M. A., Cejas, R. B., Gonzalez, P. S., and Agostini, E. (2013). Arsenic effect on the model crop symbiosis Bradyrhizobium-soybean. Plant Physiol. Biochem. 63, 8–14.
Tokmina-Lukaszewska, M., Shi, Z., Tripet, B., McDermott, T. R., Copie, V., Bothner, B., et al. (2017). Metabolic response of Agrobacterium tumefaciens 5A to arsenite. Environ. Microbiol. 19, 710–721.
vanden Hoven, R. N., and Santini, J. M. (2004). Arsenite oxidation by the heterotroph Hydrogenophaga sp. str. NT-14: the arsenite oxidase and its physiological electron acceptor. Biochim. Biophys. Acta 1656, 148–155.
Vieira, R. P., Gonzalez, A. M., Cardoso, A. M., Oliveira, D. N., Albano, R. M., Clementino, M. M., et al. (2008). Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environ. Microbiol. 10, 189–199.
Wang, Q., Han, Y., Shi, K., Fan, X., Wang, L., Li, M., et al. (2017). An oxidoreductase AioE is responsible for bacterial arsenite oxidation and resistance. Sci. Rep. 7:41536.
Wang, Q., Kang, Y. S., Alowaifeer, A., Shi, K., Fan, X., Wang, L., et al. (2018). Phosphate starvation response controls genes required to synthesize the phosphate analog arsenate. Environ. Microbiol. 20, 1782–1793.
Wang, Q., Qin, D., Zhang, S., Wang, L., Li, J., Rensing, C., et al. (2015a). Fate of arsenate following arsenite oxidation in Agrobacterium tumefaciens GW4. Environ. Microbiol. 17, 1926–1940.
Wang, Q., Warelow, T. P., Kang, Y. S., Romano, C., Osborne, T. H., Lehr, C. R., et al. (2015b). Arsenite oxidase also functions as an antimonite oxidase. Appl. Environ. Microbiol. 81, 1959–1965.
Wang, Z., Wang, Z., Shen, J., Wang, G., Zhu, X., and Lu, H. (2009). Identification of Phytophthora sojae genes involved in asexual sporogenesis. J. Genet. 88, 141–148.
Warelow, T. P., Pushie, M. J., Cotelesage, J. J. H., Santini, J. M., and George, G. N. (2017). The active site structure and catalytic mechanism of arsenite oxidase. Sci. Rep. 7:1757.
Watson, C., Niks, D., Hille, R., Vieira, M., Schoepp-Cothenet, B., Marques, A. T., et al. (2017). Electron transfer through arsenite oxidase: insights into Rieske interaction with cytochrome c. Biochim. Biophys. Acta 1858, 865–872.
Weiss, S., Carapito, C., Cleiss, J., Koechler, S., Turlin, E., Coppee, J. Y., et al. (2009). Enhanced structural and functional genome elucidation of the arsenite-oxidizing strain Herminiimonas arsenicoxydans by proteomics data. Biochimie 91, 192–203.
Xiao, A., Li, W. C., and Ye, Z. (2020). Effects of Fe-oxidizing bacteria (FeOB) on iron plaque formation, As concentrations and speciation in rice (Oryza sativa L.). Ecotoxicol. Environ. Saf. 190:110136.
Xiong, J., Wu, L., Tu, S., Van Nostrand, J. D., He, Z., Zhou, J., et al. (2010). Microbial communities and functional genes associated with soil arsenic contamination and the rhizosphere of the arsenic-hyperaccumulating plant Pteris vittata L. Appl. Environ. Microbiol. 76, 7277–7284.
Yamamura, S., and Amachi, S. (2014). Microbiology of inorganic arsenic: from metabolism to bioremediation. J. Biosci. Bioeng. 118, 1–9. doi: 10.1016/j.jbiosc.2013.12.011
Yan, G., Chen, X., Du, S., Deng, Z., Wang, L., and Chen, S. (2019). Genetic mechanisms of arsenic detoxification and metabolism in bacteria. Curr. Genet. 65, 329–338. doi: 10.1007/s00294-018-0894-9
Yang, Z., Wu, Z., Liao, Y., Liao, Q., Yang, W., and Chai, L. (2017). Combination of microbial oxidation and biogenic schwertmannite immobilization: a potential remediation for highly arsenic-contaminated soil. Chemosphere 181, 1–8. doi: 10.1016/j.chemosphere.2017.04.041
Zargar, K., Conrad, A., Bernick, D. L., Lowe, T. M., Stolc, V., Hoeft, S., et al. (2012). ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ. Microbiol. 14, 1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x
Zargar, K., Hoeft, S., Oremland, R., and Saltikov, C. W. (2010). Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J. Bacteriol. 192, 3755–3762. doi: 10.1128/JB.00244-10
Keywords: microbial arsenite oxidation, transcriptional regulation, chemotaxis, phosphate uptake, energy generation
Citation: Shi K, Wang Q and Wang G (2020) Microbial Oxidation of Arsenite: Regulation, Chemotaxis, Phosphate Metabolism and Energy Generation. Front. Microbiol. 11:569282. doi: 10.3389/fmicb.2020.569282
Received: 03 June 2020; Accepted: 21 August 2020;
Published: 15 September 2020.
Edited by:
Davide Zannoni, University of Bologna, ItalyReviewed by:
Lucia Cavalca, University of Milan, ItalyCopyright © 2020 Shi, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gejiao Wang, Z2VqaWFvQG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.