
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 September 2020
Sec. Systems Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.568853
This article is part of the Research TopicInsect Microbiome: From Diversity To ApplicationsView all 35 articles
Bark beetles are destructive forest pests considering their remarkable contribution to forest depletion. Their association with fungi is useful against the challenges of survival on the noxious and nutritionally limited substrate, i.e., conifer tissues. Fungal symbionts help the beetles in nutrient acquisition and detoxification of toxic tree secondary metabolites. Although gut is the prime location for food digestion and detoxification, limited information is available on gut-mycobiome of bark beetles. The present study screened the gut-mycobiont from six bark beetles (five Ips and one non-Ips) from Scolytinae subfamily using high-throughput sequencing and explored their putative role in symbiosis with the host insect. Results revealed the predominance of four fungal classes- Sordariomycetes, Saccharomycetes, Eurothiomycetes, and Dothidomycetes in all bark beetles. Apart from these, Agaricomycetes, Leothiomycetes, Incertae sedis Basidiomycota, Tremellomycetes, Lecanoromycetes, and Microbotryomycetes were also documented in different beetles. Five Ips bark beetles share a consortium of core fungal communities in their gut tissues consisting of 47 operational taxonomic units (OTUs) belonging to 19 fungal genera. The majority of these core fungal genera belong to the phylum Ascomycota. LEfSe analysis revealed a set of species-specific fungal biomarkers in bark beetles. The present study identified the gut mycobiont assemblage in bark beetles and their putative ecological relevance. An enriched understanding of bark beetle-fungal symbiosis is not only filling the existing knowledge gap in the field but may also unleash an unforeseen potential for future bark beetle management.
The evolutionary success of insects is based on myriad associations with microorganisms having complementary potential that is otherwise lacking in insects and restricts them when inhabiting an ecologically challenging niche or invading new environments (Linnakoski et al., 2012; Sun et al., 2013; Douglas, 2015; García-Fraile, 2018). Hence, insects serve as an excellent model system to study such evolutionary associations (i.e., symbiosis) concerning animal-plant ecology. Bark beetle (Coleoptera: Curculionidae: Scolytinae) and fungi association is one of such model systems. The evolution of bark beetle (BB)-fungi mutualisms is based on reciprocal interactions such as nutritional mutualisms, protective mutualisms, and dispersal mutualism (Biedermann and Vega, 2020). In BB exploiting sapwood (xylem), fungal associates often alleviate the deficiency of such plant tissue that is one of the most nutritionally-limiting and recalcitrant organic substrates on earth to thrive on (Filipiak and Weiner, 2014, 2017). The high abundance of substances like lignin, hemicellulose, cellulose (i.e., bio-polymers with rich carbon content) in the wood is not accessible to beetles until prior degradation of microbes, i.e., fungi (Kirk and Cowling, 1984).
Fungi often benefit coleopteran insects including BB by detoxifying plant allelochemicals (Dowd and Shen, 1990; Wang et al., 2013; Tsui et al., 2014; Zhao et al., 2019). It is believed that fungi and BB together involved in the exhaustion of host defense during the mass attack and subsequent tree-killing (Krokene and Solheim, 1996; Paine et al., 1997). Hammerbacher et al. (2013) showed blue-staining ascomycete Ceratocystis polonica helping the spruce BB, Ips typographus to break down the stilbene (phenolic compound) defenses of Norway spruce. The similar multipartite relationship was observed between symbiotic fungi (Grosmannia clavigera, Leptographium longiclavatum, Ophiostoma montium) and mountain pine beetle (Dendroctonus ponderosae Hopkins), where the presence of such fungal symbionts likely benefited the beetles under a wide range of environmental conditions (Ojeda Alayon et al., 2017). However, results from many other studies raise an argument against the importance of fungi for beetle fitness and killing the host trees as the pathogenic fungi are also found to be associated rather regularly with parasitic beetles that are less aggressive and do not kill the host conifers (Harrington, 1993; Wingfield, 1995). Furthermore, symbiotic fungus (i.e., Candida nitratophila isolated from Ips typographus) is also reported to oxidize pheromone compound cis-verbenol to trans-verbenol or verbenone. In return to all those favors, fungi benefit from dispersal by insect hosts (dispersal mutualism).
Bark beetles (BB) are economically important insects (Huang et al., 2020). The BB subfamily Scolytinae of weevils (Curculionidae) is composed of approximately 6000 species (Kirkendall et al., 2015). Association with fungi such as Ophiostoma, Ceratocystiopsis, Ceratocystis, and Grosmannia is very common among Scolytinae (Six, 2003; Kirisits, 2004; Zipfel et al., 2006). Some BB also associated with Basidiomycetes fungi (Whitney et al., 1987; Hsiau and Harrington, 2003). All strongly mutualistic associations are upheld generation after generation by vertically transmitting fungi through dedicated structures called mycangia (Six, 2020). Non-mycangial beetles transmit fungi via gut or body surface (Six, 2003; Stefanini, 2018). Ayres et al. (2000) showed that the N concentration of phloem tissues with Dendroctonus frontalis (mycangial beetle) larvae are twice than non-infested tissues indicating the importance of fungi for the growth of D. frontalis larvae. The same study also reveals that mycangial beetles (D. frontalis) construct smaller feeding galleries than non-mycangial beetle (Ips grandicollis), which is understandable considering the nutritional availability in host plant tissues influencing high or low consumption strategy for livelihood (Ayres et al., 2000).
Insect gut provides a typical environment for microbial inhabitation, including fungal colonization (Engel and Moran, 2013; Stefanini, 2018). Besides, herbivore insect gut is the site where plant allelochemical digestion, detoxification, and nutritional exchange occurs (Linser and Dinglasan, 2014). Gut-associated fungi can facilitate some or all of those vital processes inside the gut of its host (Dowd and Shen, 1990; Genta et al., 2006; DiGuistini et al., 2011; Wang et al., 2013; Itoh et al., 2018). Despite the functional importance of the gut inhabiting fungi in the adaptive physiology of its hosts, very few studies on BB have investigated the gut-associated fungal community exclusively. The whole-body microbiome studies, common in BB mycobiome research area, often represent a mix of biota from the digestive tract, external biota that escapes surface sterilization (mostly from mouthparts and entomogenous fungi from exoskeleton), and hemocoel and gonad rudiments (presence of fungi unknown).
Furthermore, basic information about the core fungal community in BB gut (subfamily: Scolytinae; Ips species) and their ecological role, including the metabolic interaction with the host is also limited. Hence, it is necessary to formulate studies on tissue-specific BB gut fungal assemblages, which are critical to a comprehensive understanding of the adaptive physiology of BB in general and their fungal symbionts inside the gut. For non-mycangial BB where gut serves as an essential site for carrying symbiotic microbes, such studies are even more crucial. Moreover, classic culture-dependent study methods are frequently biased to some specific groups of the fungal communities depending on the culture condition and laboratory processes involved downstream. Recent culture-independent, high-throughput next-generation sequencing-based screening of fungal communities provides an opportunity for exclusive identification and facilitate studying the ecological relevance of fungal species inhabiting inside BB gut that are never detected before. Furthermore, the gut fungal community assemblages may differ in same beetle species due to geographical and climatic differences between sampling sites, other biotic and abiotic factors including tree tissue, season or time of sampling, methodological strategies, and even cross-contamination during molecular processing or sample handling as described clearly in the review by Linnakoski et al. (2012). Hence, more studies are needed in different geographic locations with the same or different BB species to obtain a quick, comprehensive overview of such multipartite BB-fungal associations, which may expose surprising opportunities in bark beetle management (Popa et al., 2012).
Taken together, the current understanding of BB-fungal association and the ecological relevance of the gut tissue-specific association is incomplete, although relevant from both eco-evolutionary and applied perspectives. In Czech Republic, Ips typographus (IT), I. duplicatus (ID), and Polygraphus poligraphus (PP) are common spruce feeding beetles. Ips typographus is one of the most damaging pests in Europe, causing landscape-level mortality of spruce (Lieutier et al., 2004). I. duplicatus attacks green standing trees only whereas Polygraphus poligraphus (PP, non-Ips) likes spruce trees growing under dense and shaded condition (Kolk et al., 1996). Among the pine feeding beetles in Czech Republic, Ips acuminatus (IAC) causes considerable damage on the top and branches of Scots pine (Davydenko et al., 2017). Being a secondary pest of pine, Ips sexdentatus (SX) attacks stressed and weekend tress (Kolk et al., 1996). Ips cembrae (IC), secondary pest of European larix population, prefers wind-blown and dying trees for colonization. However, during an outbreak, IC can colonize not only on larch but also on spruce (Kolk et al., 1996). All these beetles can attack green standing trees under drought conditions and thus possess an increasing threat to forests. Hence, in the present study, we attempt to study gut tissue-specific fungal community assemblages in six economically important bark beetles (BB) from Scolytinae subfamily [IT, ID, IC, SX, IAC, and PP] collected from forests in Czech Republic using culture-independent molecular ecological approaches based on internal transcribed spacer (ITS) sequencing. Subsequently, for the core fungal communities inside the gut irrespective of beetle species, sampling location or host plant are identified, and their potential function within the gut communities is explored. Such information fills the existing knowledge gap and provides a sound scientific basis for a future shotgun or meta-transcriptomics studies followed by downstream functional studies to unravel the fine-tuning of the metabolic exchange between gut inhabiting mycobionts and their insect host.
Emerged adult bark beetle (Coleoptera: Curculionidae: Scolytinae) were sampled from infested trees during May and June 2018 from local forests in the Czech Republic. Precisely, Ips typographus (L.) (IT), Ips cembrae (IC), Ips sexdentatus (SX), Ips duplicatus (ID), were collected from Rouchovany (49°04′08.0″N 16°06′15.4″E, under State Forest Enterprise); Polygraphus poligraphus (PP) was collected from Kostelec nad Černými lesy Kostelec nad Èernımi lesy (50°00′07.2″N 14°50′56.3″E, under Military Forest Enterprise) and Ips acuminatus (IAC) was collected from Libavá (49°40′18.8″N 17°31′44.1″E, under School Forest Enterprise) in the Czech Republic (Figure 1A). The ecology and population phase of the collected beetles were summarized in Figure 1B. More than 120 living and healthy beetles were collected and pooled from logs belonging to more than eight infested trees per locality to make six biological replicates. Bark beetle species identification was performed based on morphology and published classical taxonomic work (Pfeffer, 1955; Nunberg, 1981). Beetles were stored at 4°C until shock frozen under liquid nitrogen for future use. Due to the pooling of beetles during sampling, the individual colony specific variability of the beetle gut fungal diversity could not be explored. Alternatively, such an approach somewhat reduces the stochastic variations connected with the heterologous natural tissue material (i.e., bark beetles from different colonies) and enables the separation of transient OTUs using statistical methods.
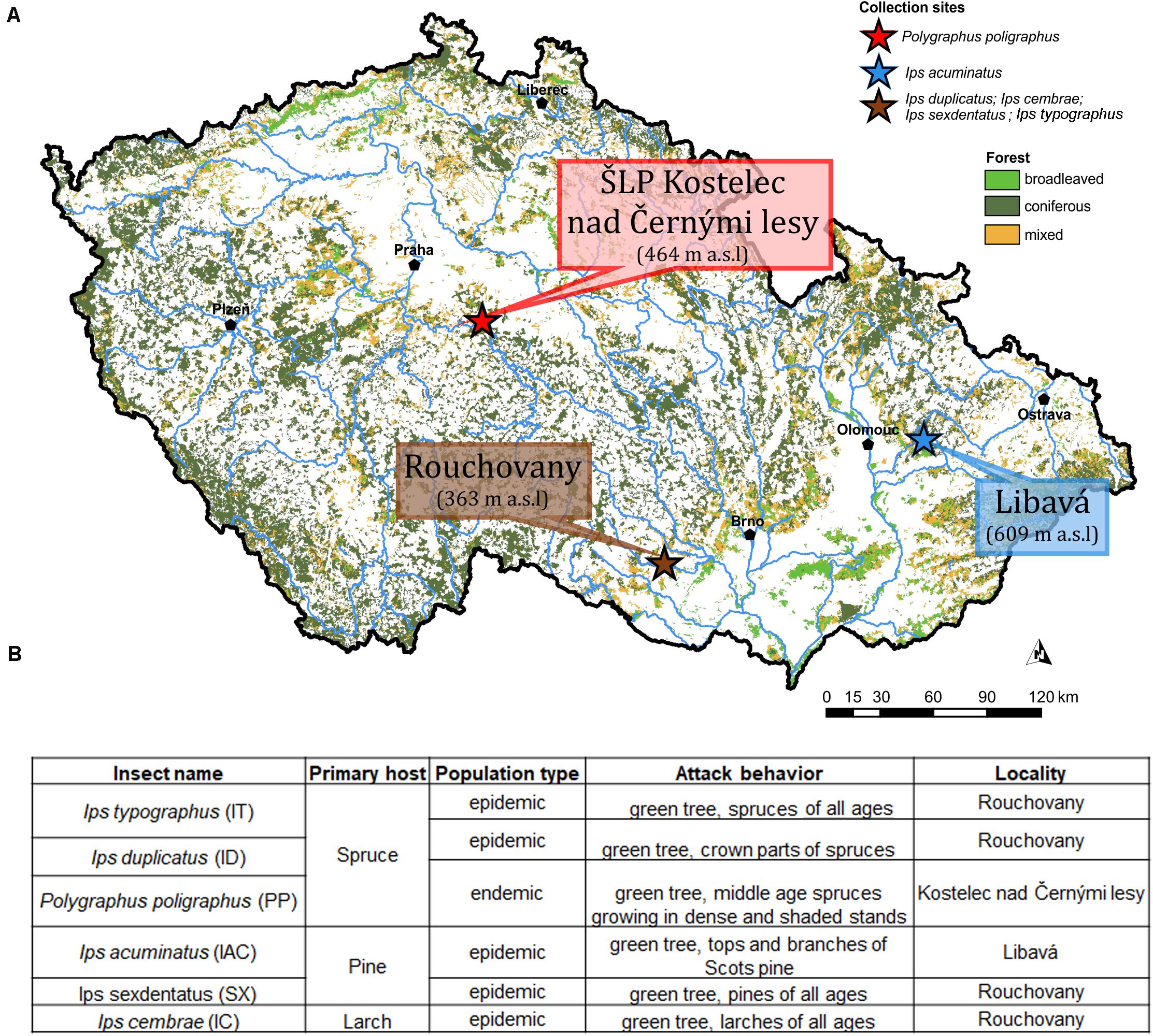
Figure 1. (A) Sampling site. The different sampling location for bark beetle collection. Ips duplicatus (ID), Ips typographus (IT), Ips sexdentatus (SX), and Ips cembrae (IC) are collected from Rochovany, Czech Republic. Polygraphus poligraphus (PP) is collected from Kostelec nad Černými lesy, and Ips acuminatus (IAC) is collected from Libavá, Czech Republic. (B) Ecology, population phase and attack behavior of the collected beetles (Lieutier et al., 2004).
Randomly selected bark beetles were surface sterilized following the standard protocol and dissected under sterile conditions in the biosafety cabinet under the microscope with a sterile entomological dissection kit. Beetle gut with apparent nematode infection was discarded. Beetle gut tissues (10 guts per replicate) were homogenized, and microbial DNA was extracted using PureLink Microbiome DNA Purification Kit from Invitrogen company. Beetle gut DNA was quantified using Qubit 2.0 Fluorometer (Thermo Scientific), and the integrity was assessed by 1% agarose gel electrophoresis. High-quality DNA from all beetle species (six biological replicates per species) was sent for high-throughput amplicon sequencing (Internal transcribed spacer, ITS) at Novogene Company, China.
Amplicon sequencing was done at the Novogene company, China, following the standard company protocol. Precisely, after diluting the DNA to 1 ng/μL in sterile water, fungal ITS genes of distinct regions (ITS2) were amplified using a specific set of primers (ITS3, ITS4; amplicon size 386 bp) (White et al., 1990) with unique barcodes. PCR reactions were performed using Phusion High-Fidelity PCR Master Mix (New England Biolabs). PCR products with amplification between 400 and 450 bp were selected and mixed in equidensity ratios for gel purification. No template control in PCR reaction did not show any amplification; hence it was not used for gel purification and further library preparation. After purification of PCR products using the Qiagen Gel Extraction Kit (Qiagen, Germany), sequencing libraries were created using NEBNext Ultra DNA Library Pre-Kit from Illumina, and index codes were ligated. The library quantity and quality were analyzed using Qubit 2.0 Fluorometer (Thermo Fisher Scientific), Q-PCR and in Agilent Bioanalyser 2100 system respectively. The libraries were sequenced to generate 250 bp paired-end reads using an Illumina platform.
After read assignment to different samples, barcode, and primer sequence removal, paired-end reads were assembled using FLASH (V1.2.7)1 (Magoč and Salzberg, 2011). Quality filtering was performed to obtain high-quality clean tags applying pre-set parameters (Bokulich et al., 2013) in QIIME (V1.7.0)2 (Caporaso et al., 2010). UCHIME algorithm3 (Edgar et al., 2011) was used to detect chimera sequences4 taking the UNITE database (Nilsson et al., 2019) as a reference, and subsequently, the chimera sequences were removed (Haas et al., 2011), and the effective Tags were finally collected.
Data analysis was conducted using UPARSE software (UPARSE v7.0.1001)5 (Edgar, 2013). After assigning all sequences with ≥ 97% similarity to the same OTU, obtained OTUs were blasted against the UNITE Database for fungal species annotation. Multiple sequence alignment (MSA) was performed to explore the phylogenetic relationship of different OTUs using MUSCLE software (Version 3.8.31)6 (Edgar, 2004). During the analysis, the singletons (n < 2) were discarded, and OTU abundance was normalized using the sequence number corresponding to the sample with the least sequences. The alpha and beta diversity analysis was subsequently performed based on the normalized OTU abundance data.
The complexity of fungal species diversity (Alpha diversity) was estimated for each sample using standard indices such as observed-species, sequence depth (Good’s coverage) (Chao et al., 1988), community richness (Chao1, ACE), diversity (Shannon, Simpson) (Magurran, 1988). These indices in all 36 tested samples were calculated in QIIME (Version 1.7.0) (Caporaso et al., 2010) and presented using R software (Version 2.15.3; R Core Team, 2013, Vienna, Austria) (R Core Team, 2013).
The differences in fungal species complexity in different beetle samples (Beta diversity) (Lozupone et al., 2007) was estimated using QIIME software (Caporaso et al., 2010) (Version 1.7.0). Weighted and unweighted UniFrac distance matrices were used to measure the dissimilarity coefficient between pairwise samples. Non-metric multidimensional scaling analysis (NMDS) (Oksanen et al., 2010) was performed to get the principal coordinates and visualize the complex, multidimensional data. Unweighted Pair-group Method with Arithmetic Means (UPGMA) Clustering (Knight et al., 2010) was done to interpret the distance matrix using average linkage in QIIME software (Version 1.7.0) (Caporaso et al., 2010). Variation analysis of fungal community structure between different bark beetle (BB) gut was assessed by standard statistical methodologies such as Analysis of Similarity (ANOSIM) (Clarke, 1993), Multi-response permutation procedure (MRPP) analysis (Cai, 2006) and ADONIS (Anderson, 2001) using R software (Vegan package) (Oksanen et al., 2007). Furthermore, Metastats (Paulson et al., 2011) was used to observe the variation in fungal species between the different BB sample groups. The significance of observed fungal species abundance differences among groups was further evaluated by p-value calculated by the method of permutation test and q-value calculated by the method of Benjamini and Hochberg False Discovery Rate (FDR) (White et al., 2009). Lastly, LEfSe [linear discriminant analysis (LDA) Effect Size] analysis was performed to detect key fungal species with a significant intra-group variation among beetle sample groups using LEfSe software (Segata et al., 2011).
Illumina paired-end sequencing of the gut tissue of six bark beetles from the Scolytinae subfamily yielded a total of 4,564,713 reads, of which 4,400,177 reads were obtained after quality control tests (sequences with Phred Quality score < 30 were discarded). The fungal ITS2 clean-reads observed per bark beetle species were used for further downstream bioinformatic data processing (Supplementary Excels 1, 2).
The comprehensiveness of sampling was represented by the Good’s coverage estimator (>99%), and the rarefaction curves that tend to attain a plateau indicating most of the gut fungal diversity in the bark beetles (BB) were sequenced (Supplementary Table 1 and Supplementary Figure 1). The number of observed species was significantly higher in PP (176.5 ± 6.84) compared to all other beetles (P < 0.01) (Supplementary Table 1). A total of 955 fungal OTU (operational taxonomic unit) clusters at 97% similarity were observed in the six different bark beetles (Supplementary Excel 3). The gut mycobiome represented using GraPhlAn revealed the predominance of four different classes- Sordariomycetes, Saccharomycetes, Eurothiomycetes, and Dothidomycetes that were present in all BB (Supplementary Figures 2,3). Apart from these classes, Agaricomycetes, Leothiomycetes, Incertae sedis Basidiomycota, Tremellomycetes, Lecanoromycetes, Microbotryomycetes were also observed in different BB (Supplementary Figures 2,3). However, the GraPhlAn display of IAC was represented at the phylum level due to the presence of an increased number of different classes compare to other beetles (Supplementary Figure 3). Considering the spruce feeding beetles, Saccharomycetes was most abundant in ID (90.3%) followed by IT (78.9%) and lowest in PP (19%) whereas Sordariomycetes showed similar abundance in PP (11.6%) and IT (11.8%) compare to ID (5.1%). Furthermore, PP showed a high relative abundance of Eurothiomycetes (12.1%) and Agaricomycetes (4.2%) compared to other BB (Figure 2A and Supplementary Excel 4). Nonetheless, comparing the pine feeding beetles, IAC showed a similar relative abundance of Saccharomycetes (27.7%) and Sordariomycetes (21.8%) in the gut. Likewise, SX also followed a similar trend (Saccharomycetes- 11.8% and Sordariomycetes- 11.7%) (Figure 2A and Supplementary Excel 4). Additionally, the larch feeding IC documented the high abundance of Saccharomycetes (83.8%) followed by Sordariomycetes (3.4%) and Eurothiomycetes (2.8%) (Figure 2A and Supplementary Excel 4).
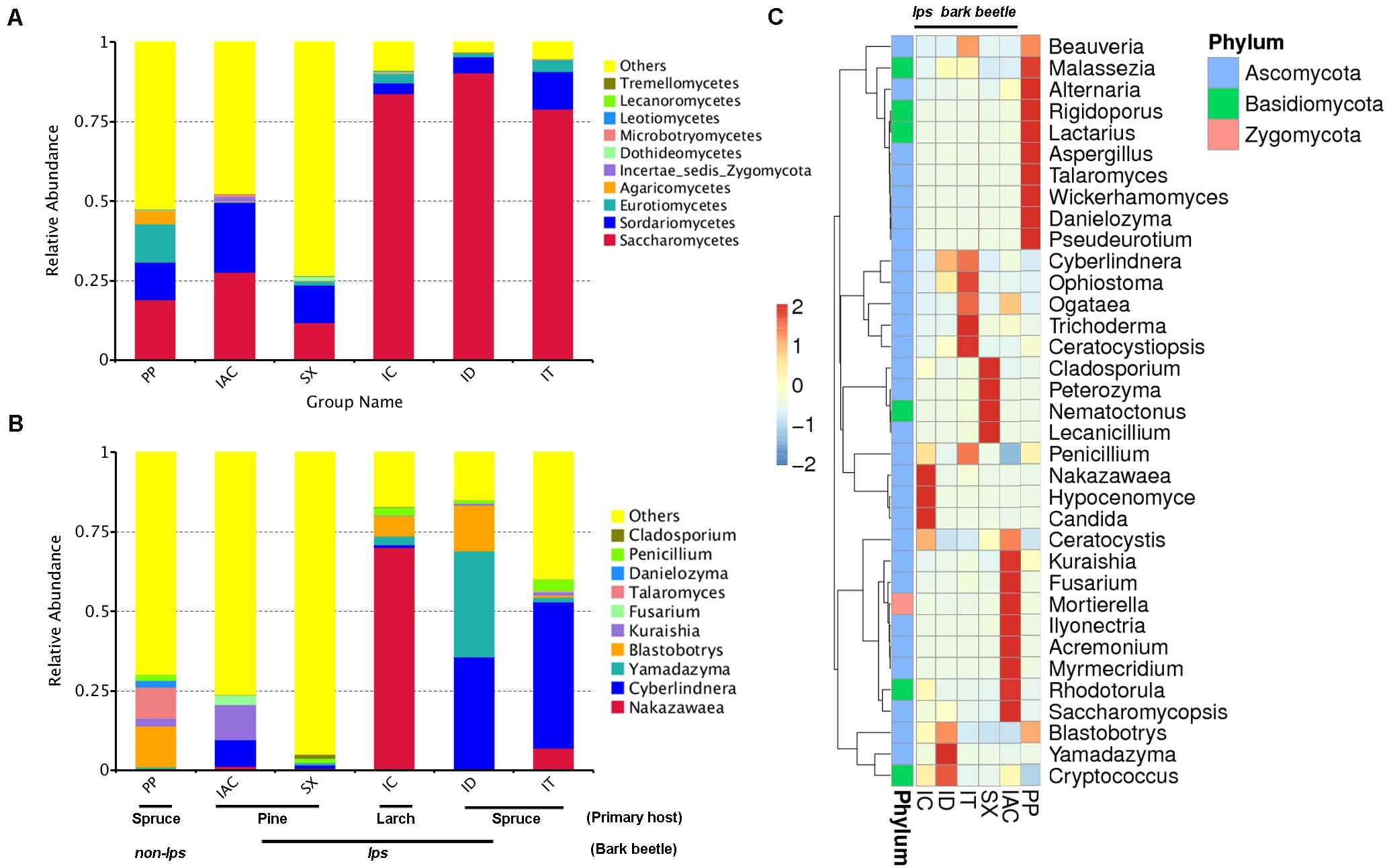
Figure 2. Fungal diversity observed in the gut of different bark beetles. (A) The bar plot illustrating the relative abundance of the gut mycobiome at the class level (top 10). “Others” signifies the total relative abundance of the rest of the class. (B) The histogram depicting the relative abundance of top 10 fungal genera in the bark beetle gut. Likewise, “Others” represents the total relative abundance of rest of the fungal communities in the beetle gut. (C) Heatmap displaying the abundance of 35 dominant fungal genera in the gut of the bark beetles. The color gradient shows the relative OTU abundance for each bark beetle where the darker color signifies higher abundance and lighter color indicates a low abundance of the particular fungal genera.
The evolutionary tree illustrated the relative abundance of top 100 fungal genera, and further, the top 20 fungal species identified in our study were represented in the taxonomic tree (Supplementary Figure 4). The relative abundance of Ascomycota was highest, followed by Basidiomycota and Zygomycota (Supplementary Excel 4). In particular, the presence of Cladosporium, Penicillium, Danielozyma, Talaromyces, Fusarium Kuraishia, Blastobotrys, Yamadazyma, Cyberlindnera, and Nakazawaea belonging to Ascomycota phylum were among the most abundant fungal genera present in the gut of BB (Figure 2B and Supplementary Excel 4). Other dominant genera include Beauveria, Malassezia, Alternaria, Aspergillus, Wickerhamomyces, Ophiostoma, Ogataea, Trichoderma, Ceratocystiopsis, Peterozyma, Lecanicillium, Candida, Ceratocystis, Mortierella, Rhodotorula, Saccharomycopsis, and Cryptococcus (Figure 2C and Supplementary Excel 4). Furthermore, the ternary plots representing the predominant fungal genera (top 10) among the spruce feeding BB revealed the predominance of Yamadazyma in ID (33.3%). In contrast, Nakazawaea was abundant in IT (6.9%) and Talaromyces in PP (9.6%). Additionally, Cyberlindnera was documented to be prevalent in both IT (46%) and ID (35.1%) whereas, Blastobotrys was dominant in ID (14.4%) and PP (12.8%) (Supplementary Figure 5 and Supplementary Excel 4). Furthermore, considering the pine and larch feeding beetles (IAC, SX, and IC), IC showed the predominance of Nakazawaea (70%), Blastrobotrys (6.2%), Yamadazyma (2.7%) and Penicillium (2.4%). Nevertheless, Cyberlindnera (8.4%), Kuraishia (10.9%), and Fusarium (3.17%) were most abundant in IAC whereas, Cladosporium was highly present in SX (1.2%) (Supplementary Figure 5 and Supplementary Excel 4).
The fungal community richness and diversity present within the gut tissue of the BB was indicated by the alpha diversity indices (Figure 3 and Supplementary Table 1). The Shannon and Simpson indices representing the community diversity in the gut tissue of spruce feeding BB revealed similar diversity in between Ips typographus (IT) (Shannon-2.67 ± 0.20 and Simpson-0.69 ± 0.05) and non-Ips BB, Polygraphus poligraphus (PP), (Shannon-2.64 ± 0.21 and Simpson- 0.67 ± 0.05) compared to Ips duplicatus (ID) (Shannon- 1.81 ± 0.28 and Simpson- 0.51 ± 0.08) (Figures 3C,D). Interestingly, the expected fungal richness (Chao1 and ACE; Wilcoxon signed-rank test) among these spruce feeding beetles documented higher richness in PP (Chao1- 210.72 ± 11.63 and ACE- 218.96 ± 13.20) compared to IT (Chao1- 128.19 ± 9.24 and ACE-128.75 ± 9.24) and ID (Chao1- 109.76 ± 6.76 and ACE- 109.76 ± 6.76) (Figures 3A,B). Furthermore, considering the pine feeders, Ips acuminatus (IAC) (Shannon- 2.84 ± 0.13 and Simpson- 0.77 ± 0.02) showed significantly higher gut fungal diversity than Ips sexdentatus (SX) (Shannon-1.46 ± 0.23 and Simpson-0.42 ± 0.08) (Figures 3C,D). However, no significant difference in fungal richness was documented between IAC (Chao1- 136.38 ± 16.99 and ACE- 139.74 ± 17.68) and SX (Chao1- 134.34 ± 6.15 and ACE- 140.90 ± 5.45) (Figures 3A,B). Additionally, the gut mycobionts of Ips beetles showed similar community richness and diversity among them compared to PP (Figure 3 and Supplementary Table 1).
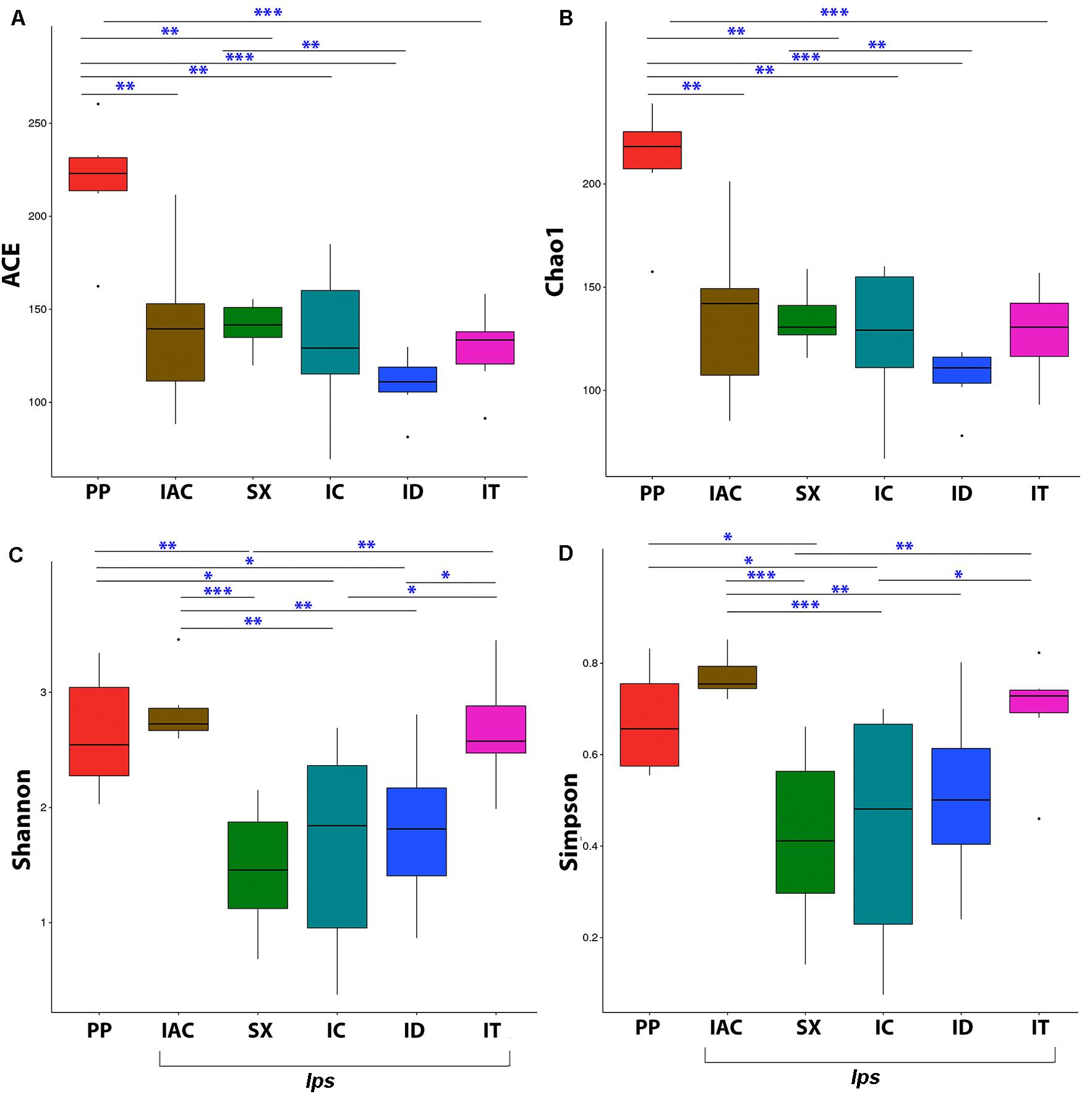
Figure 3. Alpha diversity indices representing the fungal community richness and diversity among the six different bark beetles. The expected richness estimated by (A) ACE analysis and (B) Chao1 analysis indicated significant variation between Polygraphus poligraphus (PP) and other spruce feeding beetles while no such difference in community richness was observed between pine feeding beetles (IAC and SX). The gut fungal diversity illustrated by (C) Shannon index and (D) Simpson index documented similar diversity measures between Polygraphus poligraphus (PP) and Ips typographus (IT) compared to Ips duplicatus (ID) whereas Ips acuminatus (IAC) represented higher diversity than Ips sexdentatus (SX). The statistical analysis for the significant differences between groups is done by Wilcoxon signed-rank test where “∗” denotes p < 0.05, “∗∗” designates p < 0.01 and “∗∗∗” resembles p < 0.001.
Our data suggest that the BB share a consortium of core fungal communities in their gut tissues (Figure 4). The core mycobionts present in the spruce feeding BB (PP, IT, and ID) consists of 76 OTUs belonging to 22 fungal genera (Figure 4A and Supplementary Excel 5). Similarly, 81 OTUs assigned to 25 genera were shared between the pine beetles (SX and IAC) (Figure 4B and Supplementary excel 6). Interestingly, comparing the five Ips bark beetles the occurrence of 47 core fungal OTUs were observed that were assigned to 19 fungal genera constituting Cyberlindnera, Rhodotorula, Saccharomycopsis, Yamadazyma, Blastobotrys, Talaomyces, Fusarium, Kuraishia, Penicillium, Cladosporium, Candida, Ogataea, Cryptococcus, Ceratocystis, Ophiostoma, Trichoderma, Malassezia, Cuniculitrema, and Beauveria (Figure 4C and Supplementary excel 7). Nevertheless, the core fungal consortium compared among all of the six BB (PP, IT, ID, IC, IAC, and SX) revealed the presence of 37 common OTUs predominantly belonging to 16 fungal genera (Supplementary Figure 6 and Supplementary Excel 8). Besides, the core mycobiome present in all the beetles, the occurrence of other fungal genera were also documented in this study. The existence of such fungal communities might be acquired from the environment as fungal spores or during feeding. Among these fungal communities, Peterozyma and Hypocenomyce were observed in the BB from Rouchavany (SX, IC, IT, and ID), whereas Wickerhamomyces, Myrmecridium, and Lactarius were documented in PP (Kostelec) and IAC (Libavá). Furthermore, Grapium was found in all the spruce feeding BB (PP, IT, and ID), and IIyonectria and Coprinellus were observed in pine (IAC, SX) and larch feeding (IC) bark beetles, respectively (Supplementary Excel 4).
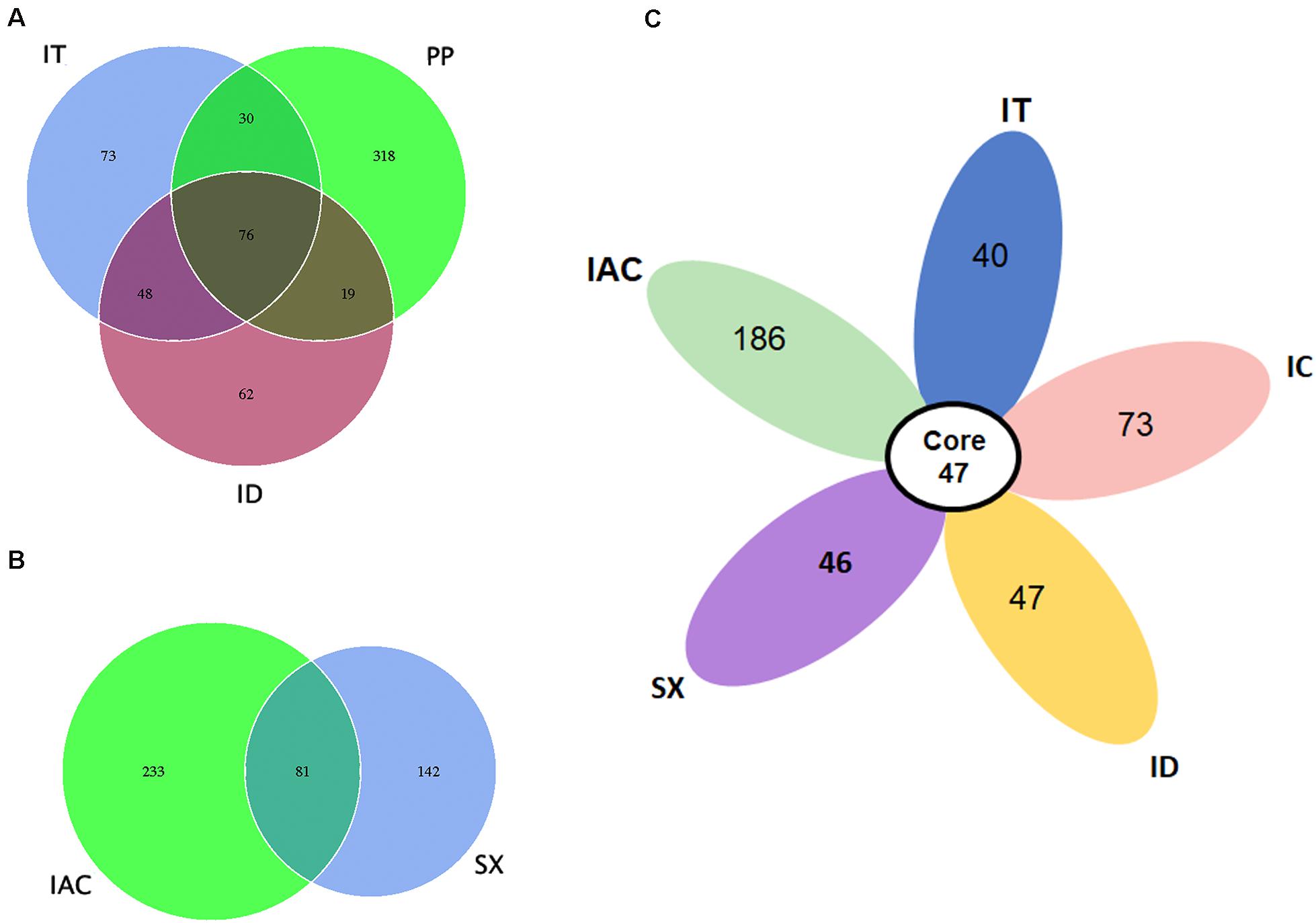
Figure 4. Core gut mycobiome. (A) Venn diagram showing the gut fungal OTU distribution between the spruce feeding bark beetles (IT, PP, and ID) where the number of OTUs (common or unique) shared among the beetles is denoted. (B) Venn diagram displaying the number of common and unique OTUs between the pine feeding beetles (IAC and SX). (C) Flower diagram showing the presence of 47 core OTUs shared among the five different Ips bark beetles.
The beta diversity calculated based on Weighted and Unweighted UniFrac distances reflects the differences in the gut mycobiome of six different BB (Figure 5 and Supplementary Figure 7). The box plot based on the Weighted UniFrac distances between the samples suggested that among the spruce feeding BB, the gut fungal communities in IT were significantly different from ID and PP (p < 0.01) (Figure 5A). However, no significant variation was observed between ID and PP. Furthermore, considering the larch feeding beetle, IC, showed a substantial difference in the gut mycobionts compare to ID (p < 0.01) and PP (p < 0.001). In contrast, no significant differences were documented between the pine feeding beetles (IAC and SX) (Figure 5A). However, there was considerable variation between PP and pine feeding BB (IAC, SX) (p < 0.05). Interestingly, the hierarchical clustering based on the Weighted Unifrac distances clustered all the Ips species together in one clade and placed PP (non- Ips species) in a different clade (Figure 5B) demonstrating the convergence of fungal association in Ips species. Furthermore, the influence of host tree feeding on the BB gut mycobiome was also observed where pine feeding (IAC and SX), spruce feeding (IT and ID), and larch feeding (IC) Ips beetles clusters together within the same clade (Figure 5B).
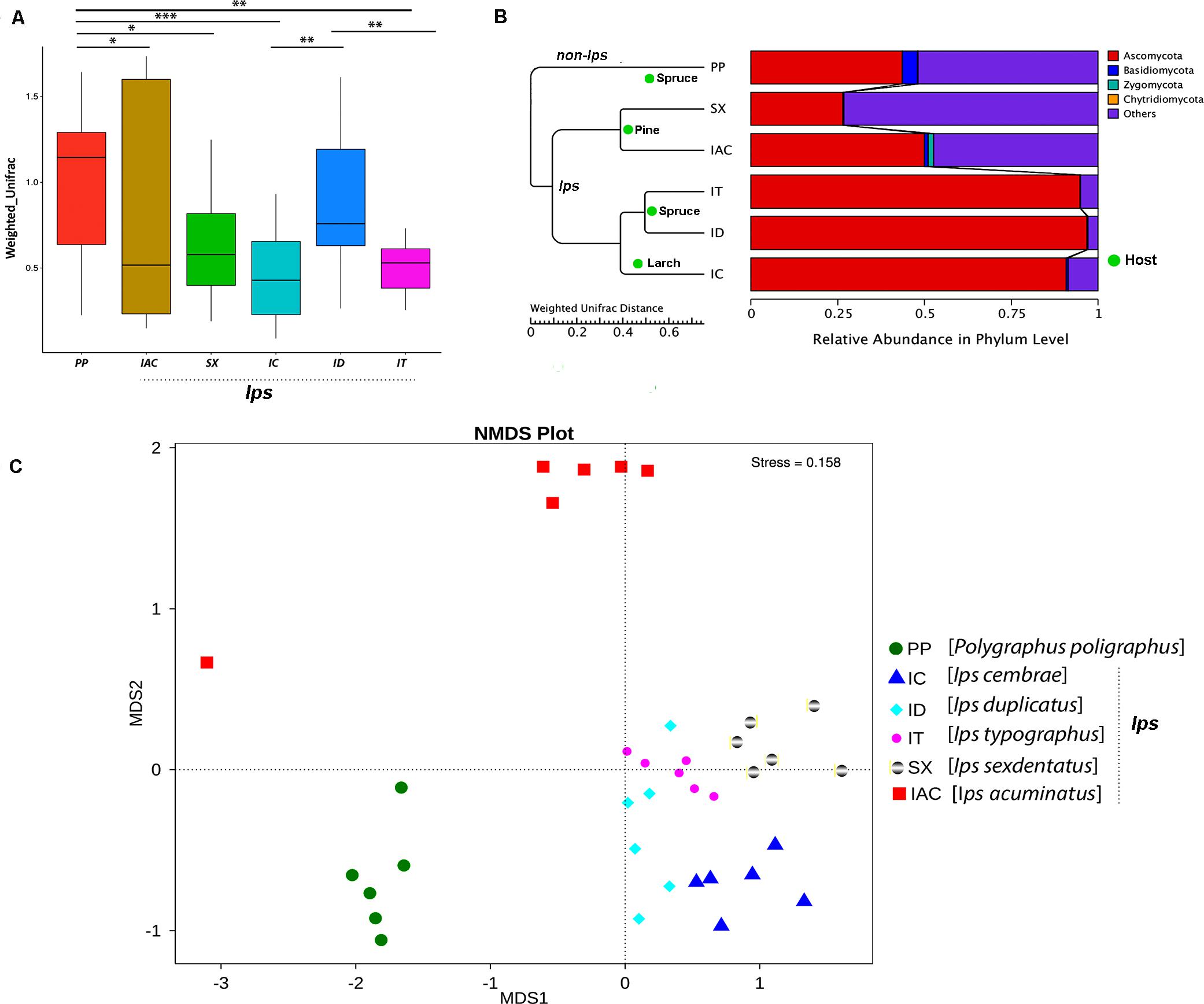
Figure 5. Beta diversity analysis. (A) The boxplot representing the diversity between the bark beetles based on the Weighted Unifrac distances showing significant differences between the PP and other bark beetles. The statistical analysis for significance is performed by Wilcoxon signed-rank test where the significance level at p < 0.05 is denoted by “∗,” “∗∗” designates p < 0.01 and “∗∗∗” signifies p < 0.001. (B) The WPGMA (Weighted pair group method with arithmetic mean) tree cluster based on Weighted Unifrac distance illustrates the similarity in gut fungal communities among the Ips species of bark beetles (IT, ID, IC, SX, and IAC) compare to non- Ips bark beetle PP. Additionally, bark beetles feeding on similar conifer host are grouped. The WPGMA tree displayed with the relative abundance of the gut mycobiome at phylum level. (C) Non-metric multidimensional scaling analysis (NMDS) representing the variation in the gut fungal communities between the six bark beetles. The data points in the same color denote the same bark beetle species. The different bark beetles are denoted by different symbols. The bark beetles collected from Rouchovany (IT, ID, IC, SX) are clustered together and are significantly different from the bark beetles sampled from other locations, Kostelec (PP) and Libavá (IAC).
Alternatively, Non-Metric Multidimensional Scaling (NMDS) analysis revealed the environmental influence on the beetle gut communities resulting from different sampling locations. The NMDS plot illustrated differences in the gut mycobionts of bark beetles (BB) from Rouchavany (IT, ID, IC, and SX), Kostelec (PP), and Libavá (IAC) representing three clusters (Figure 5C). Such observation explaining the influence of the site-specific environmental conditions on the gut communities need further experimental validation.
The fungal species-specific variation present in the gut of bark beetles represented by Metastat analysis revealed the preponderance of Cyberlindera amylophila, Yamadazyma mexicana, and Ophiostoma bicolor in the spruce feeding beetles from Rouchavany (IT and ID). In contrast, Talaromycetes rugulosus, Staphylotrichum coccosporum were significantly abundant in PP (Kostelec site) while, Fusarium solani was prevalent in both PP and IT. Furthermore, Ogateae neopine, Rhodotorula hylophila, and Cyberlindera mississippiensis were highly abundant in IAC (Figure 6). Statistical analysis (ANOSIM, MRPP, and ADONIS) illustrates significant variation between the fungal communities’ present in the six BB under study (Supplementary Tables 2,3). The gut mycobionts differed considerably between BB with different conifer feeding and were also influenced by the environment due to different collection site. Nevertheless, our data showed that the fungal communities associated with the BB were unique among different beetle species. Although the NMDS analysis documented the clustering of the bark beetles collected from the same site nearby, there were also noteworthy differences in the gut mycobiome between these beetles (Figure 5C and Supplementary Tables 2,3).
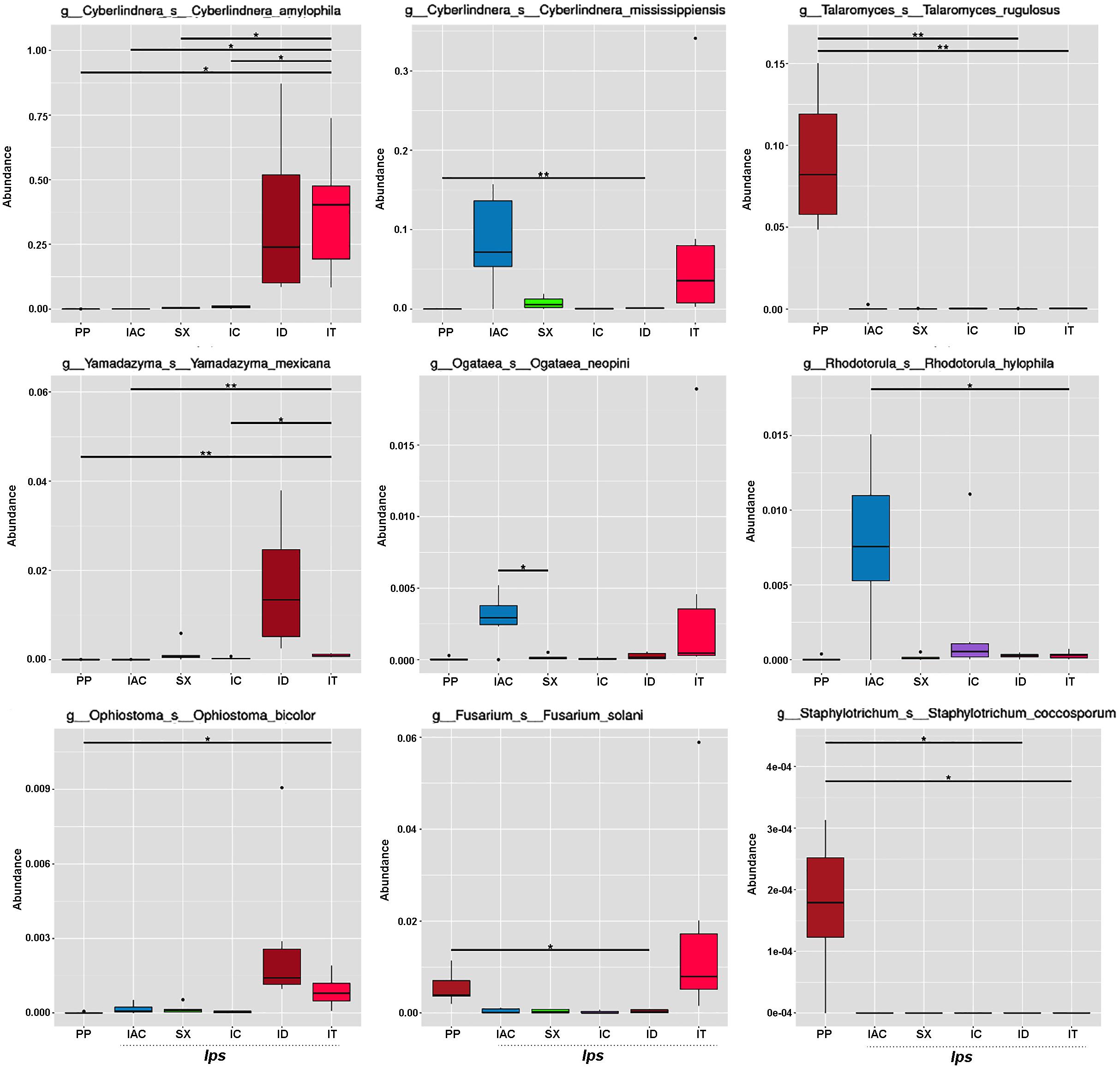
Figure 6. Metastats analysis. The barplot represents the fungal species-specific variation (top 9 species) in the gut of the bark beetles. The significant differences in species abundance among beetles are evaluated by the FDR test. The horizontal line designates the two groups with significant variation. “*” represents significant difference at q < 0.05 while “**” denotes significance at q < 0.01.
Moreover, the presence of significantly abundant fungal communities evaluated by Linear discriminant analysis effect size (LefSe) was described as important fungal biomarkers. The LDA (Linear discriminant analysis) scores [(log10) > 4 as threshold] represented by the histogram were used for the estimation of the biomarkers (Figure 7). The LDA score reflected the significant abundance of fungal classes that include- Eurotiomycetes, Agaricomycetes, Saccharomycetes along with fungal families belonging to Trichocomaceae, Peniophoraceae, Ceratocystidaceae, and Pichiaceae in the spruce feeding BB (IT, ID, and PP) (Figure 7A). Furthermore, the prevalence of fungal species belonging to Pichiaceae, Saccharomycetales, and Sccharomycetes was identified as biomarkers for ID. In contrast, Incertae sedis Saccharomycetales were abundant in IT. Additionally, the fungal communities belonging to Trichocomaceae, Eurotiomycetes, Hypocreales, Ceratocystidaceae, Microascales, Peniophoraceae, Russulales, and Agaricomycetes were observed as dominant markers in PP (Figure 7B). While in the pine feeding BB (IAC and SX), the histogram of the LDA scores illustrated the significant abundance of the fungal class belonging to Eurotiomycetes and Dothideomycetes and the families Trichocomaceae and Davidiellaceae in SX whereas fungal genera Rhodotorula and Nakazawaea in IAC (Figure 7C). Moreover, the cladogram indicated Davidiellaceae, Capnodiales, Dothideomycetes, Trichocomaceae, Eurotiales, and Eurotiomycetes as biomarkers in pine feeding beetle, SX (Figure 7D).
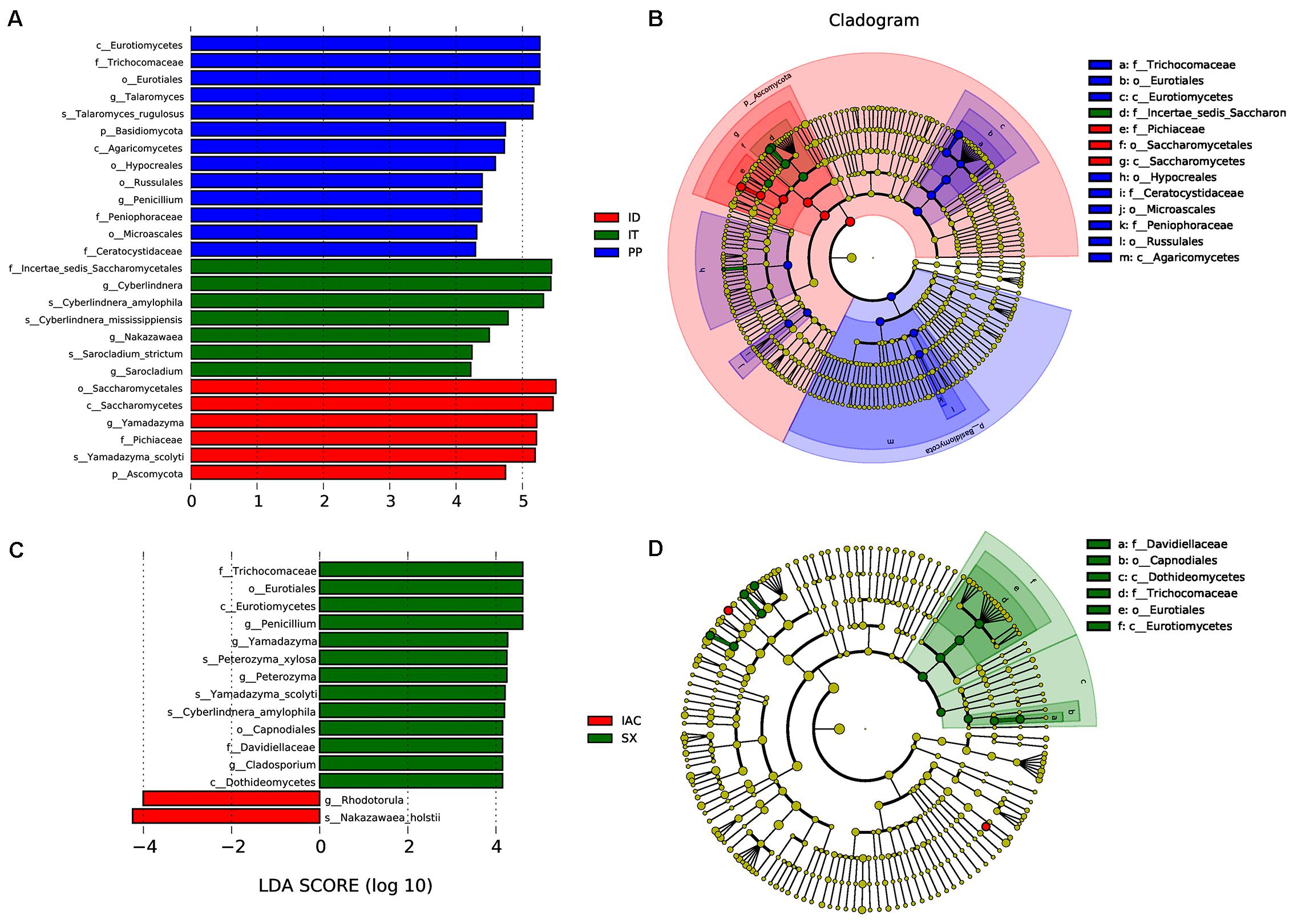
Figure 7. LEfSe analysis. (A) The histogram of the Linear discriminant analysis (LDA) scores illustrating the differentially abundant fungal communities in the spruce feeding bark beetles (IT, ID, and PP). The LDA score at log 10 > 4 is set as threshold and the length of each bin, i.e., LDA score represents the extent to which the fungal biomarker differs among the groups. (B) The cladogram illustrating the presence of fungal biomarkers that are significantly different between the spruce beetles. (C) The LDA score plot (log 10 > 4 as the threshold) representing the significantly abundant gut fungal communities in the pine feeding beetles (IAC and SX). (D) The cladogram describing the fungal biomarkers in pine beetles. The circles representing from inside to outside denotes different taxonomic level (phylum to genus). Each circle resembles a distinct taxon at the respective taxonomic level. The relative abundance of a particular taxon is proportional to the size of that circle. The color of the fungal biomarkers is according to the color of corresponding bark beetle while yellowish-green dots designate the non-significant fungal species. Red and green nodes denote that these fungi contribute highly to the group. Letters above the circles denote the fungal biomarker.
Taken together, the histogram representing all the six bark beetles (Supplementary Figure 8), illustrated the significant abundance of Penicillium and Peterozyma in SX while Microbotryomycetes, Sordariomycetes, Saccharomycetaceae, Ophiostomataceae, Nectriaceae, Plectosphaerellaceae, Fusarium, Rhodotorula, Nakazawaea, Incertae sedis Sordariomycetes in IAC. The fungal communities belonging to Eurotiomycetes, Agaricomycetes, Trichocomaceae, Trichomonascaceae, Ceratocystidaceae, Peniophoraceae were highly present in PP. Moreover, IT documented the significant abundance of Incertae sedis Saccharomycetales and Cyberlindera, whereas Yamadazyma, Microascaceae, Saccharomycetales were observed ID. The larch feeding BB (IC) showed the presence of Pichiaceae, Candida, and Nakazawaea in the histogram of LDA scores (Supplementary Figure 8A). Consequently, the overall cladogram representing the biomarkers from all the six bark beetles documented Incertae sedis Sordariomycetes, Ophiostomataceae, Plectosphaerellaceae, and Nectriaceae as markers in IAC while Pichiaceae in IC. The fungal order Saccharomycetales and family Microascaceae were the biomarkers in ID, whereas Incertae sedis Saccharomycetales was the dominant marker for IT. The non- Ips BB, Polygraghus poligraphus (PP) demonstrated Trichocomaceae, Trichomonascaceae, Ceratocystidaceae, Peniophoraceae as biomarkers (Supplementary Figure 8B).
Fungi are often associated with bark beetles (BB) as their symbionts where fungi are either carried in specialized mycangia or on their exoskeleton (Klepzig and Six, 2004; Six, 2012). Most of the previous BB-fungal symbiosis studies are conducted non-tissue specific manner, i.e., taking the whole bark beetles comprising endo and exo-mycobiome (Kirisits, 2007; Six, 2012). Hence, we have a limited understanding of the tissue-specific assemblage and interaction between BB and its fungal associates, which is an important aspect for having a better understanding of symbiosis. Not surprisingly, BB gut serves as a rich source of mycobionts assemblage. As insect gut is the prime site for digestion and detoxification of food (Linser and Dinglasan, 2014), often gut-associated mycobionts play a crucial role in those processes and decide the fate of host-derived compounds (Itoh et al., 2018). Hence, the present study is designed to screen the gut fungal diversity in five Ips and one non-Ips bark beetles (BB) collected from three different sites (Figure 1) in Czech Republic. Our data reported higher fungal diversity in the gut of Ips acuminatus (IAC) and Ips typographus (IT) compared to other tested Ips bark beetles and Polygraphus poligraphus (PP) (Supplementary Table 1). The pine feeding BB, Ips acuminatus (IAC) documented higher gut fungal diversity may be due to their fungus feeding behavior at the larval stage. However, there is no apparent difference in the gut mycobiome richness compare to other pine inhabitants, Ips sexdentatus (SX) (Figure 3). Congruently, Ips acuminatus (IAC) considered as less aggressive yet attacked healthier coniferous trees (Wermelinger et al., 2008) compare to the other secondary pest on pine, Ips sexdentatus (SX) (Pineau et al., 2017). Hence, the higher mycobiome diversity in IAC could thereby contribute to coping with the toxic defensive pine compounds (Davis et al., 2018) and nutritional acquisition during development (Villari et al., 2012). On the contrary, the aggressive secondary BB (Six, 2020), Ips typographus (IT), exhibiting mass attack to combat the tree defense showed higher fungal diversity among the spruce feeding BB in this study. It is worth to mention here that some fungi such as Endocondiophora polonica, Grosmannia penicillate, and Grosmannia europhioides associated with the aggressive beetles (e.g., Ips typographus) often utilize the tree defensive toxic compounds as carbon source and thus reduce their concentration in addition to lessen the competition between symbiotic partners for available carbohydrate like glucose (Zhou et al., 2016; Zhao et al., 2019). Hence, the fungal association may have a dual benefit for bark beetles (BB). However, such assumptions need further experimental corroborations.
In addition to tree colonization behavior, other factors such as sampling site, microenvironment of the habitat, nutrient availability and defensive host compounds influence the gut mycobiome communities in BB. Despite all these variabilities, our data suggest the presence of core fungal communities constituting of 19 genera that were conserved among all five Ips bark beetles (Figure 4). The majority of these core fungal genera belong to the phylum Ascomycota. Interestingly, some of the Ascomycota associates (Grosmannia, Ophiostoma, Endocondiophora, Ceratocystis, and Leptographium) in the BB (e.g., Dendroctonus ponderosae, Ips typographus) were reported to endure host tree defense mechanism and utilize these defensive secondary metabolites as carbon sources (Hammerbacher et al., 2013; Cale et al., 2016; Wadke et al., 2016; Zhou et al., 2016; Davis et al., 2018). The detoxification of defensive plant compounds and the ability to exploit these metabolites as carbon sources by the symbiotic fungi benefits the BB and boosts their fitness. Our study revealed the presence of ophiostomatoid fungi Ophiostoma, Ceratocystis belonging to Ophiostomataceae and Ceratocystidaceae family, as the core gut members in the BB from Scolytinae subfamily. Several studies have already documented ophiostomatoid fungi (e.g., Grosmannia penicillate, Ophiostoma bicolor, Ophiostoma piceae, Ceratocystis polonica, Leptographium chlamydatum) to be frequently associated with I. typographus (Viiri, 1997; Persson et al., 2009; Linnakoski et al., 2010). These ophiostomatoid fungi, commonly referred to as blue staining fungi, are serious tree pathogens (Wingfield et al., 1993; Kirisits, 2004), and their association with the BB has been formed over a long period (Hartig, 1878). The fungal spores of ophiostomatoid fungi are typically dispersed by BB (Masuya et al., 2009). As the BB invade the coniferous trees and the fungi get transported to the phloem and the bark where they function in nitrogen acquisition and maintains the nitrogen balance that is critical for beetle development (Ayres et al., 2000). Similarly, D. brevicomis beetle surviving on the outer bark largely depend on the two mutualistic fungi (Ceratocystiopsis brevicomi and Entomocorticium sp.) for maintaining the nitrogen and phosphorous ratios in their diet (Six and Elser, 2019). Furthermore, Ceratocystis sp. also contributes to the detoxification of phenolic compounds released as tree defensive response against beetle or fungal attack (Hammerbacher et al., 2013). Such association between the ophiostomatoid fungi (Ophiostoma ulmi) and the Scolytus spp. BB has been previously reported to kill numerous elm trees in Europe and North America (Hubbes, 1999; Santini and Faccoli, 2015). In contrast to Ophiostoma sp., the association of Ceratocystis sp. with the bark beetles are not very specific (Wingfield et al., 2016). The fungi Ceratocystis are often vectored by different insects and can infect host plants belonging to different orders (Roux et al., 2007; Lee et al., 2016). It is noteworthy that the presence of such fungal genus (Ceratocystis) in the core consortium could presumably pose an additional threat to forests, causing serious tree diseases such as Eucalyptus wilt (caused by C. fimbriata s.l.) or Ceratocystis wilt (caused by C. manginecans) (De Beer et al., 2014).
The additional presence of non-ophiostomatoid fungi belonging to the genera Fusarium, Penicillium, Trichoderma, Beauveria, and Cladosporium in the core communities in BB gut were also observed in this study. Similar to the ophiostomatoid fungi, Fusarium, an important plant pathogen, was documented to induce necrotic lesion on the phloem and disturb water conduction, ultimately killing the tree (Solheim, 1988; Yamaoka et al., 2015). Previous studies reported the association of Fusarium with Ips sexdentatus (SX) and detected their presence in SX galleries (Bezos et al., 2018). Moreover, studies showed mutualistic interaction between Fusarium with Hypothenemus hampei infesting coffee beans (Morales-Ramos et al., 2000). The occurrence of Penicillium and Trichoderma was also demonstrated in the previous study done by Krokene and Solheim (1996). It is interesting to note that Fusarium and Beauveria associated with the bark beetle core gut mycobiome in our study are entomopathogenic (Teetor-Barsch and Roberts, 1983; Zimmermann, 2007). The presence of such entomopathogenic fungus in the BB gut might occur through wounds or by sub-lethal feeding of the fungal spores by BB (Reay et al., 2005). Congruently, the presence of entomopathogenic fungus associated with bark beetles (BB) could eventually serve as a potential candidate for biocontrol strategies against forest pests.
Besides, the filamentous fungi, our results revealed the occurrence of yeasts in the BB gut microbiome. The existence of yeasts symbionts (Suh et al., 2003; Lee et al., 2006) was often attributed to nutrient acquisition, detoxification of defensive plant compounds, and production of volatile organic semiochemicals in BB (Davis et al., 2011). Several yeast genera present within the core mycobiome communities include Kusarishia, Ogataea, Yamadazyma, Candida, Cyberlindnera, and Cryptococcus. The presence of Kusarishia and Ogataea was also observed in the mountain pine beetle, D. ponderosae (Davis et al., 2011). Interestingly, these yeasts are capable of converting cis- and trans-verbenol to verbenone, anti-aggregation pheromone (Hunt and Borden, 1990). Similarly, Kusarishia and Candida enabled such interconversion in spruce feeding I. typographus (Leufvén et al., 1984). Besides, some Candida species have been reported to produce lipases that hydrolyze the long-chain triacylglycerols and play a significant role in metabolic processes (Suh and Blackwell, 2004). The presence of such fungal species in BB gut might serve a similar function. However, such possibilities demand further experimental corroborations.
Often symbionts interact with each other to fine-tune their co-existence within the host, i.e., Yamadazyma and Candida identified in the core consortium in the present study has been shown to influence the growth of other fungal symbionts O. montium in D. ponderosae (Adams et al., 2008). The previous studies suggested that yeast isolated from I. typographus outcompetes the fungus Ophiostoma in culture. Such influence of yeast on other BB fungal symbionts could compromise the beetle performance as this blue staining fungus may act in parallel to contribute to the death of the beetle-infested trees (Furniss et al., 1990). The yeast Cyberlindnera, earlier reported as a common symbiont of Dendroctonus beetles (Rivera et al., 2009), detected in our study, can degrade starch and lipids. It can also abide by the secondary metabolites in pine and detoxify them (Briones-Roblero et al., 2017). Additionally, the presence of Cryptococcus observed in this study was documented previously in Ips pini and Ips typographus, having a putative role in the production of semiochemicals (Leufvén and Nehls, 1986).
The persistence of core fungal consortium in the bark beetles (BB) in the present study suggested the involvement of gut mycobiome directly or indirectly in host physiology and performance. Nonetheless, the variation in the gut mycobiome illustrated by the β-diversity may be contributed by the factors such as different feeding habits, environmental impact due to different sampling locations and differences in BB species (Figure 5). The BB belonging to Ips and non-Ips species showed significant differences in their gut mycobiome. Interestingly, different conifer host feeding also shaped the gut fungal communities where Ips bark beetle feeding on spruce (IT and ID) were clustered in one clade similar to pine feeders, IAC and SX. It is worth to mention here that, in the present study, autochthonous mycobiota of the digestive tract of BB cannot be distinguished from the mycobiome that is associated with the diet of beetles. Furthermore, the NMDS analysis revealed that BB gut fungal diversity varied significantly based on their sampling locations and was clustered in three distinct groups. The non- Ips species PP collected from Kostelec clustered distantly similar to Ips acuminatus (IAC) collected from Libavá. The fungus feeding behavior and the influence of varying sampling location possibly placed apart IAC in a separate cluster than other Ips species after NMDS analysis. In contrast, separate clustering of PP may be due to the combined contribution of species and sampling location difference. Although bark beetles (BB) collected from Rouchovany were grouped, there was a distinct separation between different Ips species collected from the same site. This observation reflects the feeding behavior of BB on the different coniferous hosts and conserved species-specific mycobiont association within them. However, such conclusions need to be investigated further.
It is interesting to note that each of the BB in the present study represented a set of fungal biomarkers that play a crucial role in their physiology and the performance during and after tree infestation. For instance, the biomarker belonging to the fungal family Ceratocystidaceae was able to detoxify the defensive tree compounds released on a beetle or fungal attack (Hammerbacher et al., 2013). Moreover, the genera Ceratocytosis belonging to the family Ceratocystidaceae identified as the core member of the gut mycobiome have a broad tree host range, i.e., this fungus was pathogenic to a wide array of trees (Juzwik et al., 2008; Tsopelas et al., 2017) thereby delivers the BB with the opportunity to infest and survive on different tree hosts. Similarly, the fungal species belonging to Ophiostomataceae family was also considered as a potential biomarker contributing to nutrient acquisition through detoxification of secondary metabolites of host trees and thereby supporting in beetle development (Ayres et al., 2000; Cale et al., 2016; Wadke et al., 2016; Zhou et al., 2016). Other yeast biomarkers such as Pichiaceae and Saccharomycetaceae observed in the BB have been reported to impact the growth of other fungal symbionts in the bark beetles (Adams et al., 2008). Such yeast species were also considered to have a putative role in the biosynthesis of anti-aggregation pheromone in beetles (Hunt and Borden, 1990).
Taken together, the present study identified and explored the core mycobiome community in five Ips bark beetle (BB) species of economic importance. Present findings increase the current understanding of fungal composition and α-diversity in BB (five Ips and one non-Ips) gut. β-diversity analysis further pinpoints toward the importance of sampling location, host-feeding and species differences as critical contributors for shaping up the BB gut-mycobiont. Further screening the mycobiome within beetle food (phloem), decaying wood and wood from BB galleries may provide higher insight into beetle-fungus interaction. Nevertheless, the present study could be of considerable value and interest in forest entomology, providing insights into the gut-specific core fungal communities and offering a foundation for future follow ups on the functional role that core mycobiome community could have in the gut of Ips bark beetles.
The datasets generated during this study are available under NCBI Bio-project PRJNA632703 (Bio-sample accessions SAMN14917923–SAMN14917958, SRA accession PRJNA632703; https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA632703).
AC, FS, and AR planned the research. AC and AR conducted the experiments, data analysis, and biological interpretation of results. RM, MA, and JS collected the beetles. MA and JS dissected the beetles. AC, RM, FS, and AR prepared the manuscript. All authors approved the manuscript.
The project was financed by the Internal Grant Agency (IGA) from the Faculty of Forestry and Wood Sciences, Czech University of Life Sciences. The financial support for AC was provided by “EVA 4.0,” No. CZ.02.1.01/0.0/0.0/16_019/0000803 financed by OP RDE. Infrastructural support and salary for AR, RM, and FS is obtained from “EXTEMIT – K,” No. CZ.02.1.01/0.0/0.0/15_003/0000433 financed by OP RDE.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Novogene Sequencing Company, China, is acknowledged for its sequencing service and bioinformatic assistance. Jiri Trombik from the Faculty of Forestry and Wood Sciences is acknowledged for his contribution to the preparation of Figure 1.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.568853/full#supplementary-material
Adams, A. S., Six, D. L., Adams, S. M., and Holben, W. E. (2008). In vitro interactions between yeasts and bacteria and the fungal symbionts of the mountain pine beetle (Dendroctonus ponderosae). Microb. Ecol. 56, 460–466. doi: 10.1007/s00248-008-9364-0
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Ayres, M. P., Wilkens, R. T., Ruel, J. J., Lombardero, M. J., and Vallery, E. (2000). Nitrogen budgets of phloem-feeding bark beetles with and without symbiotic fungi. Ecology 81, 2198–2210. doi: 10.1890/0012-9658(2000)081[2198:nbopfb]2.0.co;2
Bezos, D., Martínez-Álvarez, P., Sanz-Ros, A. V., Martín-García, J., Fernandez, M. M., and Diez, J. J. (2018). Fungal communities associated with bark beetles in Pinus radiata plantations in Northern Spain affected by Pine Pitch Canker, with special focus on Fusarium species. Forests 9:698. doi: 10.3390/f9110698
Biedermann, P. H., and Vega, F. E. (2020). Ecology and evolution of insect–fungus mutualisms. Annu. Rev. Entomol. 65, 431–455. doi: 10.1146/annurev-ento-011019-024910
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Briones-Roblero, C. I., Rodríguez-Díaz, R., Santiago-Cruz, J. A., Zúñiga, G., and Rivera-Orduña, F. N. (2017). Degradation capacities of bacteria and yeasts isolated from the gut of Dendroctonus rhizophagus (Curculionidae: Scolytinae). Folia Microbiol. 62, 1–9. doi: 10.1007/s12223-016-0469-4
Cai, L. (2006). Multi-response permutation procedure as an alternative to the analysis of variance: an SPSS implementation. Behav. Res. Methods 38, 51–59. doi: 10.3758/bf03192749
Cale, J. A., Collignon, R. M., Klutsch, J. G., Kanekar, S. S., Hussain, A., and Erbilgin, N. (2016). Fungal volatiles can act as carbon sources and semiochemicals to mediate interspecific interactions among bark beetle-associated fungal symbionts. PLoS One 11:e0162197. doi: 10.1371/journal.pone.0162197
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chao, A., Lee, S.-M., and Chen, T.-C. (1988). A generalized Good’s nonparametric coverage estimator. Chin. J. Math. 16, 189–199.
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Davis, T. S., Hofstetter, R. W., Foster, J. T., Foote, N. E., and Keim, P. (2011). Interactions between the yeast Ogataea pini and filamentous fungi associated with the western pine beetle. Microb. Ecol. 61, 626–634. doi: 10.1007/s00248-010-9773-8
Davis, T. S., Horne, F. B., Yetter, J. C., and Stewart, J. E. (2018). Engelmann spruce chemotypes in Colorado and their effects on symbiotic fungi associated with the North American spruce beetle. J. Chem. Ecol. 44, 601–610. doi: 10.1007/s10886-018-0961-1
Davydenko, K., Vasaitis, R., and Menkis, A. (2017). Fungi associated with Ips acuminatus (Coleoptera: Curculionidae) in Ukraine with a special emphasis on pathogenicity of ophiostomatoid species. Eur. J. Entomol. 114, 77–85. doi: 10.14411/eje.2017.011
De Beer, Z. W., Duong, T., Barnes, I., Wingfield, B. D., and Wingfield, M. J. (2014). Redefining Ceratocystis and allied genera. Stud. Mycol. 79, 187–219. doi: 10.1016/j.simyco.2014.10.001
DiGuistini, S., Wang, Y., Liao, N. Y., Taylor, G., Tanguay, P., Feau, N., et al. (2011). Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. PNAS 108, 2504–2509. doi: 10.1073/pnas.1011289108
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Dowd, P. F., and Shen, S. K. (1990). The contribution of symbiotic yeast to toxin resistance of the cigarette beetle (Lasioderma serricorne). Entomol. Exp. Appl. 56, 241–248. doi: 10.1111/j.1570-7458.1990.tb01402.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Filipiak, M., and Weiner, J. (2014). How to make a beetle out of wood: multi-elemental stoichiometry of wood decay, xylophagy and fungivory. PLoS One 9:e115104. doi: 10.1371/journal.pone.0115104
Filipiak, M., and Weiner, J. (2017). Nutritional dynamics during the development of xylophagous beetles related to changes in the stoichiometry of 11 elements. Physiol. Entomol. 42, 73–84. doi: 10.1111/phen.12168
Furniss, M. M., Solheim, H., and Christiansen, E. (1990). Transmission of blue-stain fungi by Ips typographus (Coleoptera: Scolytidae) in Norway spruce. Ann. Entomol. Soc. Am. 83, 712–716. doi: 10.1093/aesa/83.4.712
García-Fraile, P. (2018). Roles of bacteria in the bark beetle holobiont–how do they shape this forest pest? Ann. Appl. Biol. 172, 111–125. doi: 10.1111/aab.12406
Genta, F. A., Dillon, R. J., Terra, W. R., and Ferreira, C. (2006). Potential role for gut microbiota in cell wall digestion and glucoside detoxification in Tenebrio molitor larvae. J. Insect Physiol. 52, 593–601. doi: 10.1016/j.jinsphys.2006.02.007
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hammerbacher, A., Schmidt, A., Wadke, N., Wright, L. P., Schneider, B., Bohlmann, J., et al. (2013). A common fungal associate of the spruce bark beetle metabolizes the stilbene defenses of Norway spruce. Plant Physiol. 162, 1324–1336. doi: 10.1104/pp.113.218610
Harrington, T. (1993). “Diseases of conifers caused by species of Ophiostoma and Leptographium,” in Ceratocystis and Ophiostoma: Taxonomy, Ecology, and Pathogenicity eds M. J. Wingfield, K. A. Seifert, and J. F. Webber (St. Paul: American Phytopathological Society Press) 161–172
Hartig, R. (1878). Die Zersetzungserscheinungen des Holzes der Nadelholzbäume und der Eiche in forstlicher, Botanischer und Chemischer Richtung. Springer: Berlin. doi: 10.5962/bhl.title.25641
Hsiau, P. T., and Harrington, T. C. (2003). Phylogenetics and adaptations of basidiomycetous fungi fed upon by bark beetles (Coleoptera: Scolytidae). Symb. Rehovot. 34, 111–132.
Huang, J., Kautz, M., Trowbridge, A. M., Hammerbacher, A., Raffa, K. F., Adams, H. D., et al. (2020). Tree defence and bark beetles in a drying world: carbon partitioning, functioning and modelling. New Phytol. 225, 26–36. doi: 10.1111/nph.16173
Hubbes, M. (1999). The American elm and Dutch elm disease. For. Chron. 75, 265–273. doi: 10.5558/tfc75265-2
Hunt, D., and Borden, J. (1990). Conversion of verbenols to verbenone by yeasts isolated fromDendroctonus ponderosae (Coleoptera: Scolytidae). J. Chem. Ecol. 16, 1385–1397. doi: 10.1007/bf01021034
Itoh, H., Tago, K., Hayatsu, M., and Kikuchi, Y. (2018). Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 35, 434–454. doi: 10.1039/c7np00051k
Juzwik, J., Harrington, T. C., MacDonald, W. L., and Appel, D. N. (2008). The origin of Ceratocystis fagacearum, the oak wilt fungus. Annu. Rev. Phytopathol. 46, 13–26. doi: 10.1146/annurev.phyto.45.062806.094406
Kirisits, T. (2004). “Fungal associates of European bark beetles with special emphasis on the ophiostomatoid fungi,” in Bark and Wood Boring Insects in Living Trees in Europe. A Synthesis eds F. Lieutier, K. R. Day, and A. Battisti J. C. Gregoire H. F. Evans (London: Kluwer Academic Publishers).
Kirisits, T. (2007). “Fungal associates of European bark beetles with special emphasis on the ophiostomatoid fungi,” in Bark and Wood Boring Insects in Living Trees in Europe, A Synthesis. Eds F. Lieutier, K.R. Day, A. Battisti, and JC. Grégoire H.F. Evans (Dordrecht: Springer) 181–236. doi: 10.1007/978-1-4020-2241-8_10
Kirk, T. K., and Cowling, E. B. (1984). “Biological decomposition of solid wood,” in Advances in Chemistry Series 207, ed. R.M. Rowell. (Washington, DC: American Chemical Society) 455–487. doi: 10.1021/ba-1984-0207.ch012
Kirkendall, L., Biedermann, P., and Jordal, B. (2015). “Evolution and diversity of bark and ambrosia beetles in Bark Beetles: Biology and Ecology of Native and Invasive Species eds F. E. Vega and R. W. Hofstetter (San Diego, CA: Academic Press).
Klepzig, K. D., and Six, D. (2004). Bark beetle-fungal symbiosis: context dependency in complex associations. Symbiosis 37, 189–205.
Knight, R., Lozupone, C., Lladser, M., Knights, D., and Stombaugh, J. (2010). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Kolk, A., Starzyk, J., Kinelski, S., and Dzwonkowski, R. (1996). Atlas of Forest Insect Pests. Warsaw: MULTICO Publishing House Ltd.
Krokene, P., and Solheim, H. (1996). Fungal associates of five bark beetle species colonizing Norway spruce. Can. J. For. Res. 26, 2115–2122. doi: 10.1139/x26-240
Lee, D.-H., Roux, J., Wingfield, B. D., Barnes, I., Mostert, L., and Wingfield, M. J. (2016). The genetic landscape of Ceratocystis albifundus populations in South Africa reveals a recent fungal introduction event. Fungal Biol. 120, 690–700. doi: 10.1016/j.funbio.2016.03.001
Lee, S., Kim, J.-J., and Breuil, C. (2006). Diversity of fungi associated with mountain pine beetle, Dendroctonus ponderosae, and infested lodgepole pines in British Columbia. Fungal Divers. 22, 91–105.
Leufvén, A., Bergström, G., and Falsen, E. (1984). Interconversion of verbenols and verbenone by identified yeasts isolated from the spruce bark beetleIps typographus. J. Chem. Ecol. 10, 1349–1361. doi: 10.1007/bf00988116
Leufvén, A., and Nehls, L. (1986). Quantification of different yeasts associated with the bark beetle, Ips typographus, during its attack on a spruce tree. Microb. Ecol. 12, 237–243. doi: 10.1007/bf02011208
Lieutier, F., Day, K. R., Battisti, A., Grégoire, J.-C., and Evans, H. F. (2004). Bark and Wood Boring Insects in Living Trees in Europe: A Synthesis. Berlin: Springer.
Linnakoski, R., De Beer, Z. W., Ahtiainen, J., Sidorov, E., Niemelä, P., Pappinen, A., et al. (2010). Ophiostoma spp. associated with pine-and spruce-infesting bark beetles in Finland and Russia. Persoonia 25, 72–93. doi: 10.3767/003158510x550845
Linnakoski, R., De Beer, Z. W., Niemelä, P., and Wingfield, M. J. (2012). Associations of conifer-infesting bark beetles and fungi in Fennoscandia. Insects 3, 200–227. doi: 10.3390/insects3010200
Linser, P. J., and Dinglasan, R. R. (2014). “Insect gut structure, function, development and target of biological toxins,” in Advances in Insect Physiology, eds T. S. Dhadialla and S. S. Gill (Academic Press Inc.: Cambridge) 1–37. doi: 10.1016/b978-0-12-800197-4.00001-4
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/aem.01996-06
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Magurran, A. E. (1988). Ecological Diversity and its Measurement. Princeton, NJ: Princeton university press.
Masuya, H., Kaneko, S., Yamaura, Y., and Yamaoka, Y. (2009). Ophiostomatoid fungi isolated from Japanese red pine and their relationships with bark beetles. Mycoscience 50, 212–223. doi: 10.1007/s10267-008-0474-9
Morales-Ramos, J. A., Rojas, M. G., Sittertz-Bhatkar, H., and Saldaña, G. (2000). Symbiotic relationship between Hypothenemus hampei (Coleoptera: Scolytidae) and Fusarium solani (Moniliales: Tuberculariaceae). Ann. Entomol. Soc. Am. 93, 541–547. doi: 10.1603/0013-8746(2000)093[0541:srbhhc]2.0.co;2
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264.
Nunberg, M. (1981). Klucze do rozpoznawania owadów Polski. Czȩść XIX. Chrzz̧szcze–Coleoptera, Korniki–Scolytidae, Wyrynniki–Platypodidae. Zeszyt 99–100.
Ojeda Alayon, D. I., Tsui, C. K., Feau, N., Capron, A., Dhillon, B., Zhang, Y., et al. (2017). Genetic and genomic evidence of niche partitioning and adaptive radiation in mountain pine beetle fungal symbionts. Mol. Ecol. 26, 2077–2091. doi: 10.1111/mec.14074
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., O’hara, R., Simpson, G.L., et al. (2010). Vegan: Community Ecology Package. R Package Version 1.17-4. Available online at: http://CRAN.R-project.org/package=vegan (accessed October 20, 2019).
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Stevens, M. H. H., Oksanen, M. J., et al. (2007). The vegan package. Commun. Ecol. Pack. 10:719.
Paine, T., Raffa, K., and Harrington, T. (1997). Interactions among scolytid bark beetles, their associated fungi, and live host conifers. Annu. Rev. Entomol. 42, 179–206. doi: 10.1146/annurev.ento.42.1.179
Paulson, J. N., Pop, M., and Bravo, H. C. (2011). Metastats: an improved statistical method for analysis of metagenomic data. Genome Biol. 12:17.
Persson, Y., Vasaitis, R., Långström, B., Öhrn, P., Ihrmark, K., and Stenlid, J. (2009). Fungi vectored by the bark beetle Ips typographus following hibernation under the bark of standing trees and in the forest litter. Microb. Ecol. 58, 651–659. doi: 10.1007/s00248-009-9520-1
Pfeffer, A. (1955). Kùrovci-Scolytoidea:(Rád: Brouci-Coleoptera). Prague: Československá akademie věd.
Pineau, X., Bourguignon, M., Jactel, H., Lieutier, F., and Sallé, A. (2017). Pyrrhic victory for bark beetles: successful standing tree colonization triggers strong intraspecific competition for offspring of Ips sexdentatus. For. Ecol. Manage. 399, 188–196. doi: 10.1016/j.foreco.2017.05.044
Popa, V., Déziel, E., Lavallée, R., Bauce, E., and Guertin, C. (2012). The complex symbiotic relationships of bark beetles with microorganisms: a potential practical approach for biological control in forestry. Pest Manage. Sci. 68, 963–975. doi: 10.1002/ps.3307
R Core Team (2013). R: A Language and Environment for Statistical Computing (Version 2.15. 3). R Foundation for Statistical Computing. Vienna.
Reay, S., Thwaites, J., and Farrell, R. (2005). A survey of Ophiostoma species vectored by Hylastes ater to pine seedlings in New Zealand. For. Pathol. 35, 105–113. doi: 10.1111/j.1439-0329.2004.00393.x
Rivera, F. N., Gonzalez, E., Gomez, Z., Lopez, N., Hernandez-Rodriguez, C., Berkov, A., et al. (2009). Gut-associated yeast in bark beetles of the genus Dendroctonus Erichson (Coleoptera: Curculionidae: Scolytinae). Biol. J. Linn. Soc. 98, 325–342. doi: 10.1111/j.1095-8312.2009.01289.x
Roux, J., Heath, R., Labuschagne, L., Nkuekam, G. K., and Wingfield, M. (2007). Occurrence of the wattle wilt pathogen, Ceratocystis albifundus on native South African trees. For. Pathol. 37, 292–302. doi: 10.1111/j.1439-0329.2007.00507.x
Santini, A., and Faccoli, M. (2015). Dutch elm disease and elm bark beetles: a century of association. Iforest 8:126. doi: 10.3832/ifor1231-008
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60.
Six, D. L. (2003). A comparison of mycangial and phoretic fungi of individual mountain pine beetles. Can. J. For. Res. 33, 1331–1334. doi: 10.1139/x03-047
Six, D. L. (2012). Ecological and evolutionary determinants of bark beetle—fungus symbioses. Insects 3, 339–366. doi: 10.3390/insects3010339
Six, D. L. (2020). A major symbiont shift supports a major niche shift in a clade of tree-killing bark beetles. Ecol. Entomol. 45, 190–201. doi: 10.1111/een.12786
Six, D. L., and Elser, J. J. (2019). Extreme ecological stoichiometry of a bark beetle–fungus mutualism. Ecol. Entomol. 44, 543–551. doi: 10.1111/een.12731
Solheim, H. (1988). Pathogenicity of some Ips typographus-associated blue-stain fungi to Norway spruce. Meddelelser fra Norsk Institut Skogforskning 40, 1–11.
Stefanini, I. (2018). Yeast-insect associations: it takes guts. Yeast 35, 315–330. doi: 10.1002/yea.3309
Suh, S.-O., and Blackwell, M. (2004). Three new beetle-associated yeast species in the Pichia guilliermondii clade. FEMS Yeast Res. 5, 87–95. doi: 10.1016/j.femsyr.2004.06.001
Suh, S. O., Marshall, C. J., Mchugh, J. V., and Blackwell, M. (2003). Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol. Ecol. 12, 3137–3145. doi: 10.1046/j.1365-294x.2003.01973.x
Sun, J., Lu, M., Gillette, N. E., and Wingfield, M. J. (2013). Red turpentine beetle: innocuous native becomes invasive tree killer in China. Annu. Rev. Entomol. 58, 293–311. doi: 10.1146/annurev-ento-120811-153624
Teetor-Barsch, G. H., and Roberts, D. W. (1983). Entomogenous Fusarium species. Mycopathologia 84, 3–16. doi: 10.1007/bf00436991
Tsopelas, P., Santini, A., Wingfield, M. J., and Wilhelm de Beer, Z. (2017). Canker stain: a lethal disease destroying iconic plane trees. Plant Dis. 101, 645–658. doi: 10.1094/pdis-09-16-1235-fe
Tsui, C. K.-M., Farfan, L., Roe, A. D., Rice, A. V., Cooke, J. E., El-Kassaby, Y. A., et al. (2014). Population structure of mountain pine beetle symbiont Leptographium longiclavatum and the implication on the multipartite beetle-fungi relationships. PLoS One 9:e105455. doi: 10.1371/journal.pone.0105455
Viiri, H. (1997). Fungal associates of the spruce bark beetle Ips typographus L.(Col. Scolytidae) in relation to different trapping methods. J. Appl. Entomol. 121, 529–533. doi: 10.1111/j.1439-0418.1997.tb01444.x
Villari, C., Battisti, A., Chakraborty, S., Michelozzi, M., Bonello, P., and Faccoli, M. (2012). Nutritional and pathogenic fungi associated with the pine engraver beetle trigger comparable defenses in Scots pine. Tree Physiol. 32, 867–879. doi: 10.1093/treephys/tps056
Wadke, N., Kandasamy, D., Vogel, H., Lah, L., Wingfield, B. D., Paetz, C., et al. (2016). The bark-beetle-associated fungus, Endoconidiophora polonica, utilizes the phenolic defense compounds of its host as a carbon source. Plant Physiol. 171, 914–931.
Wang, Y., Lim, L., DiGuistini, S., Robertson, G., Bohlmann, J., and Breuil, C. (2013). A specialized ABC efflux transporter G c ABC-G 1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol. 197, 886–898. doi: 10.1111/nph.12063
Wermelinger, B., Rigling, A., Schneider Mathis, D., and Dobbertin, M. (2008). Assessing the role of bark-and wood-boring insects in the decline of Scots pine (Pinus sylvestris) in the Swiss Rhone valley. Ecol. Entomol. 33, 239–249. doi: 10.1111/j.1365-2311.2007.00960.x
White, J. R., Nagarajan, N., and Pop, M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5:e1000352. doi: 10.1371/journal.pcbi.1000352
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press). 18, 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Whitney, H., Bandoni, R., and Oberwinkler, F. (1987). Entomocorticium dendroctoni gen. et sp. nov.(Basidiomycotina), a possible nutritional symbiote of the mountain pine beetle in lodgepole pine in British Columbia. Can. J. Bot. 65, 95–102. doi: 10.1139/b87-013
Wingfield, M. J., Garnas, J. R., Hajek, A., Hurley, B. P., de Beer, Z. W., and Taerum, S. J. (2016). Novel and co-evolved associations between insects and microorganisms as drivers of forest pestilence. Biol. Invas. 18, 1045–1056. doi: 10.1007/s10530-016-1084-7
Wingfield, M. J., Seifert, K. A., and Webber, J. F. (1993). Ceratocystis and Ophiostoma: Taxonomy, Ecology, and Pathogenicity. American Phytopathological Society: Saint Paul, MI
Wingfield, M. J. (1995). “Do conifer bark beetles require fungi to kill trees?,” in Barkbeetles, Blue-Stain Fungi, and Conifer Defence Systems ed E.; Christiansen (Norway: Aktuelt fra Skogforsk).
Yamaoka, Y., Kuroki, D., Matsutani, K., Aoyama, T., Masuya, H., and Kajimura, H. (2015). Pathogenicity of Fusarium solani associated with a bark beetle, Scolytogenes birosimensis, to Pittosporum tobira. J. For. Res. 20, 514–521. doi: 10.1007/s10310-015-0505-2
Zhao, T., Kandasamy, D., Krokene, P., Chen, J., Gershenzon, J., and Hammerbacher, A. (2019). Fungal associates of the tree-killing bark beetle, Ips typographus, vary in virulence, ability to degrade conifer phenolics and influence bark beetle tunneling behavior. Fungal Ecol. 38, 71–79. doi: 10.1016/j.funeco.2018.06.003
Zhou, F., Lou, Q., Wang, B., Xu, L., Cheng, C., Lu, M., et al. (2016). Altered carbohydrates allocation by associated bacteria-fungi interactions in a bark beetle-microbe symbiosis. Sci. Rep. 6:20135. doi: 10.1038/srep20135
Zimmermann, G. (2007). Review on safety of the entomopathogenic fungi Beauveria bassiana and Beauveria brongniartii. Biocontrol Sci. Technol. 17, 553–596. doi: 10.1080/09583150701309006
Keywords: OTUs, gut-mycobiont, fungal-biomarkers, core community, Scolytinae, symbiosis, bark beetles, Ips
Citation: Chakraborty A, Modlinger R, Ashraf MZ, Synek J, Schlyter F and Roy A (2020) Core Mycobiome and Their Ecological Relevance in the Gut of Five Ips Bark Beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 11:568853. doi: 10.3389/fmicb.2020.568853
Received: 02 June 2020; Accepted: 12 August 2020;
Published: 03 September 2020.
Edited by:
Adly M. M. Abdalla, International Atomic Energy Agency, AustriaReviewed by:
Kin-Ming (Clement) Tsui, Weill Cornell Medicine-Qatar, QatarCopyright © 2020 Chakraborty, Modlinger, Ashraf, Synek, Schlyter and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Roy, Um95QGZsZC5jenUuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.