- Key Laboratory of Environmental Pollution and Microecology of Liaoning Province, Shenyang Medical College, Shenyang, China
A Gram-positive, α-hemolytic, catalase-negative, facultative anaerobic and non-motile coccus was isolated form throat swabs taken from the oropharynx of healthy children. The genome was shown to be 1.950,659 bp long and contained 42.03 mol% G + C bases with 1,942 protein-coding and 53 RNA coding genes. The results of 16S rRNA gene sequencing strongly suggested that the strain is a member of the Streptococcus genus, with 98.04, 98.11, and 97.34% similarities to Streptococcus australis ATCC 700641T, Streptococcus rubneri LMG 27207T and Streptococcus parasanguinis ATCC 15912T, respectively. A sodA gene comparison exhibited a sequence identity of 92.6% with the closest strain Streptococcus australis ATCC 700641T. In silico DNA-DNA hybridization showed a highest DNA similarity value of 52% with Streptococcus australis ATCC 700641T. Comparing 18 biochemical traits, the similarities of the Streptococcus strain E24 were 72% with Streptococcus rubneri LMG 27207T, 78% with Streptococcus australis ATCC 700641T and 44% with Streptococcus parasanguinis ATCC 15912T. We suggest that based on the genotypic and phenotypic results that the strain E24 is a novel species of the Streptococcus genus and propose the name Streptococcus xiaochunlingii sp. nov. E24.
Introduction
The microbiota of the upper respiratory tract has been denoted as gatekeepers to respiratory health, since it prevent pathogens from overgrowing and disseminating toward the lungs (Man et al., 2017; Tsang et al., 2019). In the first week after birth, the upper respiratory microbiota of infants develop from a mixed toward a Streptococcus viridans-predominated bacterial community, followed by niche differentiation to Staphylococcus aureus and other bacteriae frequently leading to a composition of Moraxella spp., Dolosigranulum spp., and Streptococcus spp. (Bosch et al., 2016). In a previous study, we isolated Streptococcus strain E24 as one of 4 antagonistic strains which had bacteriostatic effects on Klebsiella pneumoniae, Proteus vulgaris, Enterobacter cloacae, and Acinetobacter Baumannii (Li et al., 2019).
In recent decades, due to improved phenotypic and molecular identification methods and with the development of the new taxonogenomics concept (Fournier et al., 2015), the genus Streptococcus has been significantly expanded with the addition of many newly discovered species (Huch et al., 2013; Ricaboni et al., 2017); more than 110 species are now recognized within the genus Streptococcus1. With 16S rRNA gene sequence analyses revealed that Streptococcus species were clustered into six groups comprising S. anginosus, S. mitis, S. mutans, S. bovis, S. pyogenes, and S. salivarius (Kawamura et al., 1995). Here we present the results of taxonogenomics concept analyses of a novel strain termed Streptococcus strain E24.
Materials and Methods
Collection of Bacterial Samples
Bacterial colonies from throat swabs of healthy children were cultivated aerobically for 24 h at 37°C on agar plates enriched with sheep blood (5%) (Becton Dickinson, Heidelberg, Germany). During streptococci isolation a single, circular, unpigmented and α-hemolytic colony of 0.5–1.0 mm diameter emerged, which was purified by repeated streaking and finally stored at −80°C. The study was conducted in accordance with the “Declaration of Helsinki” guidelines and approved by the Ethics Committee of Shenyang Medical College (approval number: No. 2015052902). Written informed consent was obtained from the legal representatives of the participating children.
16S rRNA sodA Gene Analyses
The nucleic acids of the strain were extracted for gene amplification and cloning of the 16S rRNA gene was accomplished by using a commercial DNA Purification Kit (Wizard Genomic DNA Purification Kit, Promega, Madison, WI, United States) and the 16S rRNA universal primers 27F/1492R (5′-AGAGTTTGATCMTGGCTCAG-3′ and 5′-GGYTACCTTGTTACGACTT-3′, respectively). The sequence obtained by sequencing was aligned using Basic Local Alignment Search Tool (BLAST) of NCBI. We collected 16S rRNA gene sequences of all known Streptococcus species from the NCBI database (Pontigo et al., 2015). Phylogenetic trees were constructed with two different algorithms, namely neighbor-joining (Saitou and Nei, 1987) and maximum-likelihood (Guindon and Gascuel, 2003) by close-neighbor-interchange (CNI) (search level = 2, random additions = 100) using MEGA version 7.0 (Kumar et al., 2016) and the reliability of the nodes was estimated by bootstrap analysis (1000 replications) (Felsenstein, 1985). Additionally, we analyzed the sodA gene since it has been proposed for phylogenetic characterization especially for differentiating streptococci (Poyart et al., 1998) with the primers forward: 5′-CCITAYICITAYGAYGCIYTIGARCC-3′ and reverse: 5′-ARRTARTAIGCRTGYTCCCAIACRTC-3′ leading to a 430 bp fragment.
Biochemical and Phenotypic Characterizations
Isolate growths were examined in aerobic, anaerobic and microaerophilic atmospheres, with or without 5% CO2 and at 4, 15, 22, 30, 35, 37 and 42°C using nutrient agar containing sheep blood (5%, Solarbio, Beijing, China) (Facklam and Elliott, 1995). The nutrient agar consisted of peptone 10.0 g/L, beef powder 3.0 g/L sodium chloride 5.0 g/L, agar 15.0 g/L with pH of 7.3 ± 0.1 (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). Cell motility and chain building was evaluated with the aid of a Leica DM1000 light microscope (Leica Microsystems, France). Specimens were also examined after the classical Gram straining (Austrian, 1960). The susceptibility of each isolates to salt was tested by growing the strains in the presence of 2.5, 3.5, 4.5 or 6.5 g/L NaCl. Moreover, nine different pHs were tested: 4.0, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 9.0 (Supplementary Table 1).
Cell sizes were calculated from images captured on a scanning electron microscope. Bacteria were fixed in glutaraldehyde phosphate (2.5%) buffer overnight at 4°C. They were then washed twice with phosphate buffer (PBS), fixed for 30 min in a 1% osmium tetroxide solution (SPI supplies Inc., West Chester, PA, United States) and then washed 3 times with PBS before being dehydrated with 50, 70, 80, 90, 95 (twice), and 100% ethanol (three times), respectively (5 min per ethanol concentration). After natural drying, each sample was subjected to ion sputtering and then the gold was sprayed onto the surface and then viewed on a Hitachi 3400N scanning electron microscope (Hitachi High Technologies, Japan).
Sporulation was tested with Streptococcus strain E24 solutions (concentration: 1 × 106 cfu/mL) divided into two groups, from which one group was heated to 80–90°C for 20 min and the other served as control. Then 50 μl solutions of each group were applied on nutrient agar plates containing 5% sheep blood. The strains were incubated under normal conditions at 37°C overnight to observe the bacterial colony formation. No colony growth was observed on the plate of the heat treatment group on the next day.
Biochemical Assays
Isolates were characterized with Rapid ID32 Strep, API ZYM and API 50CH systems (bioMérieux, Marcy-l’Étoile, France) as well as catalase (Becton Dickinson, Franklin Lakes, NJ, United States) and oxidase (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China) assays were carried out separately.
Susceptibility to Antibiotics
The antibiotic susceptibility of the strain Streptococcus strain E24 was determined using disk diffusion (Le Page et al., 2015) on Mueller-Hinton E agar (bioMérieux, Marcy-l’Étoile, France) consisting of casamino acid 17.5 g/L, beef extract 2.0 g/L, soluble starch 1.5 g/L Agar 17.0 g/L at pH 7.2 ± 0.2 (25°C) (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). The antimicrobial agents tested were: vancomycin, ceftriaxone, cefepime, cefotaxime, Rina Thiazole Amine and chloramphenicol (all at 30 μg); ampicillin and penicillin (10 μg); and clindamycin (2 μg).
Analysis of Fatty Acids in Bacteria
Fatty acid methyl esters (FAMEs) were analyzed using a gas chromatography/mass spectrometry [see (Dione et al., 2016) and (Sasser, 1990)]. FAMEs were prepared from a sample consisting of 40 mg of bacterial biomass, separated using an MS column (Elite 5) and examined with a mass spectrometry (Clarus 500-SQ 8 S, PerkinElmer, France) followed by appropriate searches of spectral databases.
Extraction of DNA and Sequencing of the Genome
Genomic DNA was extracted using the SDS methodology (Lim et al., 2016). Agarose gel electrophoresis (HE-120, Tanon Science & Technology Inc., Shanghai, China) was used to detect the DNA, which was analyzed and quantified with a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States). 1 μg samples were used as preparations of DNA and sequence libraries were constructed using a NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs Inc., Ipswich, MA, United States), with appropriate index codes. Samples were sonicated to sizes of about 350 bp and fragments end-polished, A-tailed and ligated for subsequent amplification by PCR. The PCR products were purified and libraries evaluated to determine the distribution of sizes using real-time PCR (Agilent 2100 Bioanalyzer, Agilent Technologies, Palo Alto, CA, United States). Beijing Novogene Bioinformatics Technology Co., Ltd sequenced the genome (Illumina NovaSeq PE150, Illumina Inc., San Diego, United States).
Genome Sequencing Assembly and Annotation
After preprocessing, appropriate data were gathered using SOAP (version 2.04) (Li et al., 2008, 2010), SPAdes (Bankevich et al., 2012) and ABySS (Simpson et al., 2009) assembly software, and finally integrated using CISA software. The preliminary assembly results were optimized and holes filled with GapCloser (version 1.12) to obtain the final assembly result. Fragments below 500 bp were filtered out. Encoded genes were predicted using GeneMarkS software. The protein-coding sequences produced with BLAST were annotated by comparison with the NR, GO, KEGG, COG, PFAM, TCDB, and Swiss-Prot protein databases.
Genome Comparison
Average nucleotide identity (ANI) was used to assess genomic similarity and to define species with values of 95%. The ANI analysis was performed with OAT (Yoon et al., 2017) and DNA-DNA hybridization (DDH) was used to assess the genomic similarity and to define species with 70% hybridization values (Tindall et al., 2010). The Genome-to-Genome Distance Calculator (GGDC) (Meier-Kolthoff et al., 2013) was employed to conduct a digital DNA-DNA hybridization (dDDH) analysis. The adjacent to the Streptococcus strain E24 located strains were selected based on BLAST alignment and the phylogenetic tree constructed from 16S rRNA gene results data.
Results
Phylogenetic Analysis
The newly determined 16S rRNA (Figures 1A,B) isolate gene sequence was aligned with sequences of other Streptococcus species retrieved from GenBank. Strain Streptococcus strain E24 shared 98.04, 98.11, and 97.34 % 16S rRNA gene sequence similarity with S. australis ATCC 700641T, S. rubneri LMG 27207T and S. parasanguinis ATCC 15912T, respectively. The housekeeping gene sodA (Figures 1C,D) of Streptococcus strain E24 shared 94.94, 92.74, and 90.80% similarities with S. australis ATCC 700641T, S. timonensis Marseille-P2915T and S. infantis ATCC 700779T, respectively. Figures 1A,C represent phylogenetic trees constructed with the neighbor-joining and Figures 1B,D phylogenetic trees constructed with the maximum-likelihood method. Taken together, the phylogenetic analyses demonstrated that Streptococcus strain E24 represents a novel species within the genus Streptococcus. The 16S rRNA gene sequences of Streptococcus strain E24 is recorded in the NCBI database (accession no. MN592637).
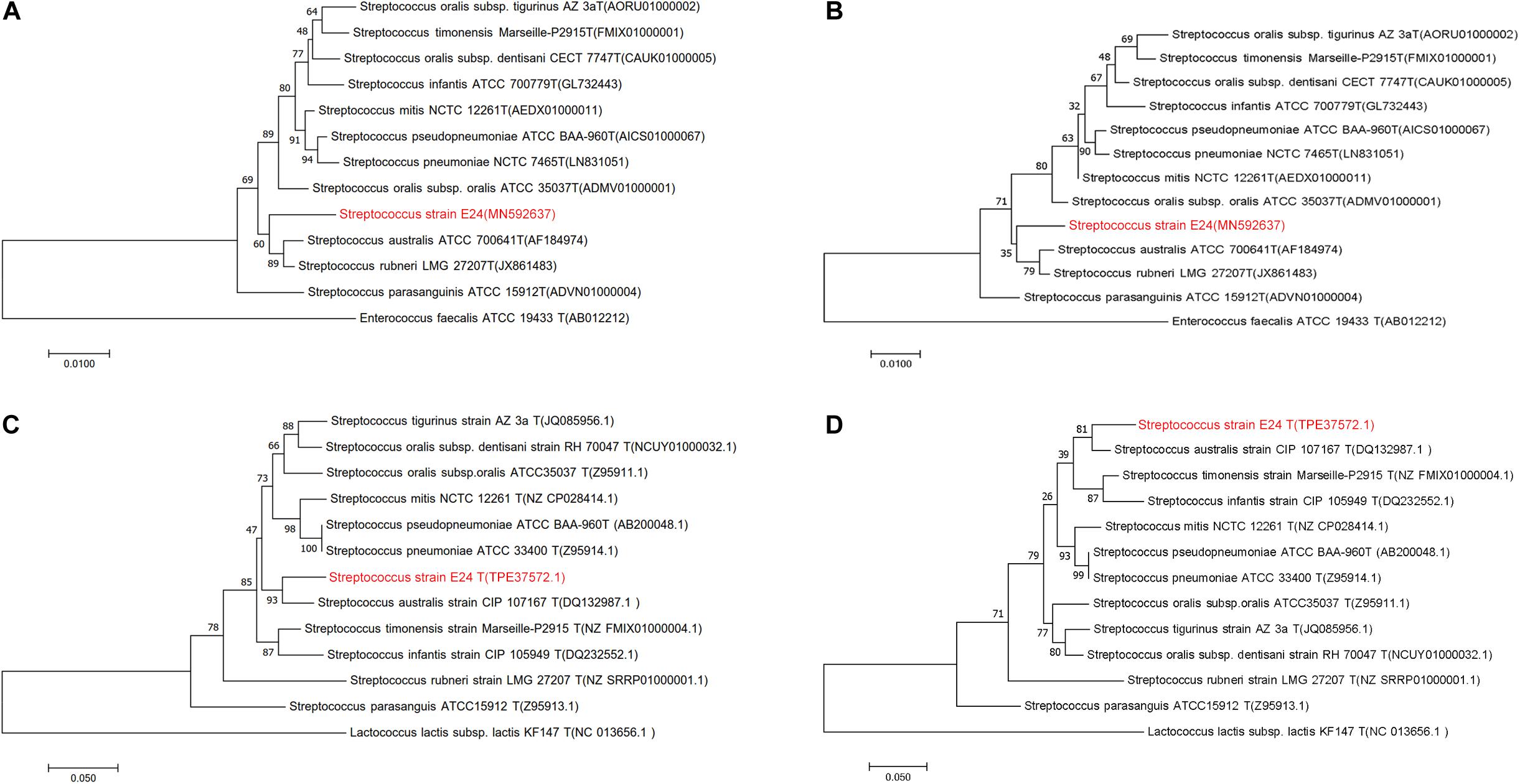
Figure 1. Phylogenetic trees of Streptococcus strain E24 and closely related members of the Streptococcal species groups. (A) Neighbor-joining and (B) maximum-likelihood 16S rRNA gene sequences comparisons, (C) neighbor-joining and (D) maximum-likelihood sodA gene sequences comparisons. Sequences of Enterococcus faecalis and Lactococcus lactis subsp. lactis were used as out-groups. Bootstrap values ≥ 50 are indicated and the bars indicate 0.01 and 0.05 substitutions per site.
Phenotypic Characteristics
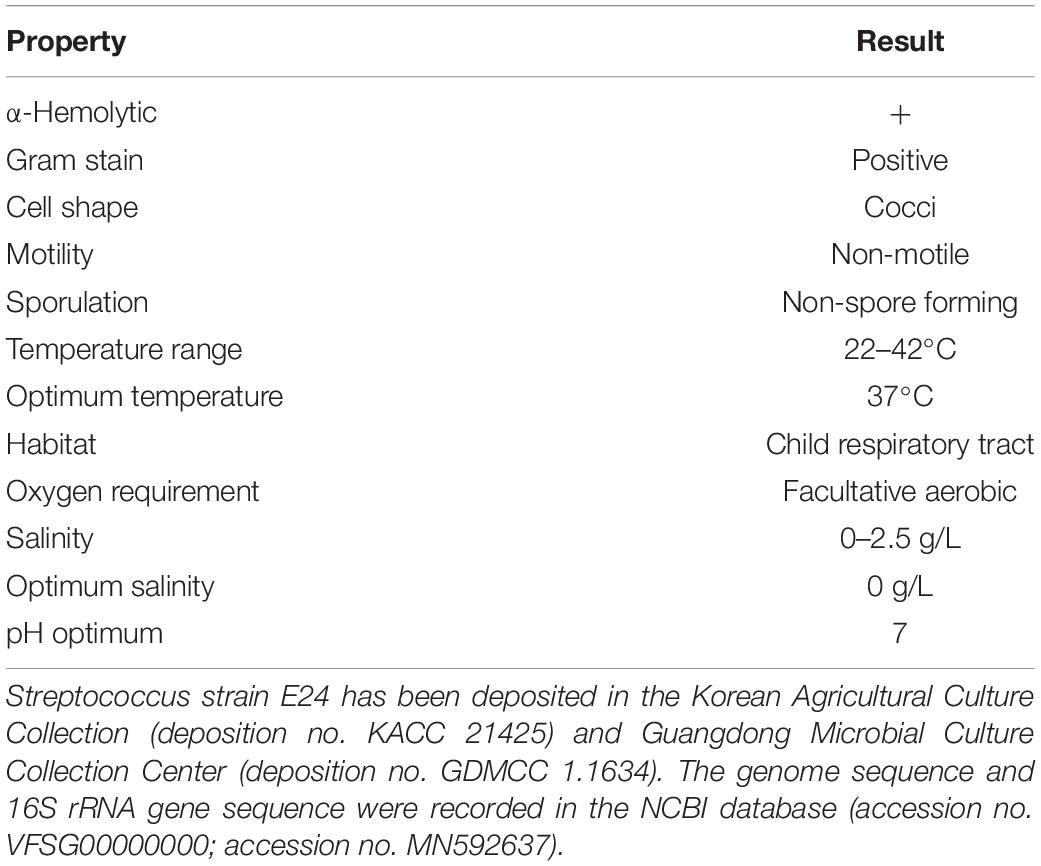
Morphological features were determined using cells cultured aerobically at 37°C on enriched nutrition agar (vide supra) for 24 h, when the bacteria became punctate, gray and formed α-hemolytic colonies having edges that were undulated. Gram staining was performed using the classical Gram stain procedure (Austrian, 1960) and light microscopy images showed Gram-positive cocci (Figure 2A). Cells had an average diameter of about 5 μm as viewed by an electron microscopy (Figure 2B). Salt tolerance was determined by the growth of the strains in the presence of 2.5 g/L. The isolate was found to be unable to grow at or below 22°C but could grow at up to 42°C, with 37°C being the optimal temperature. The cells were facultative anaerobe, and the pH for optimal growth was 7 (Table 1 and Supplementary Table 1).
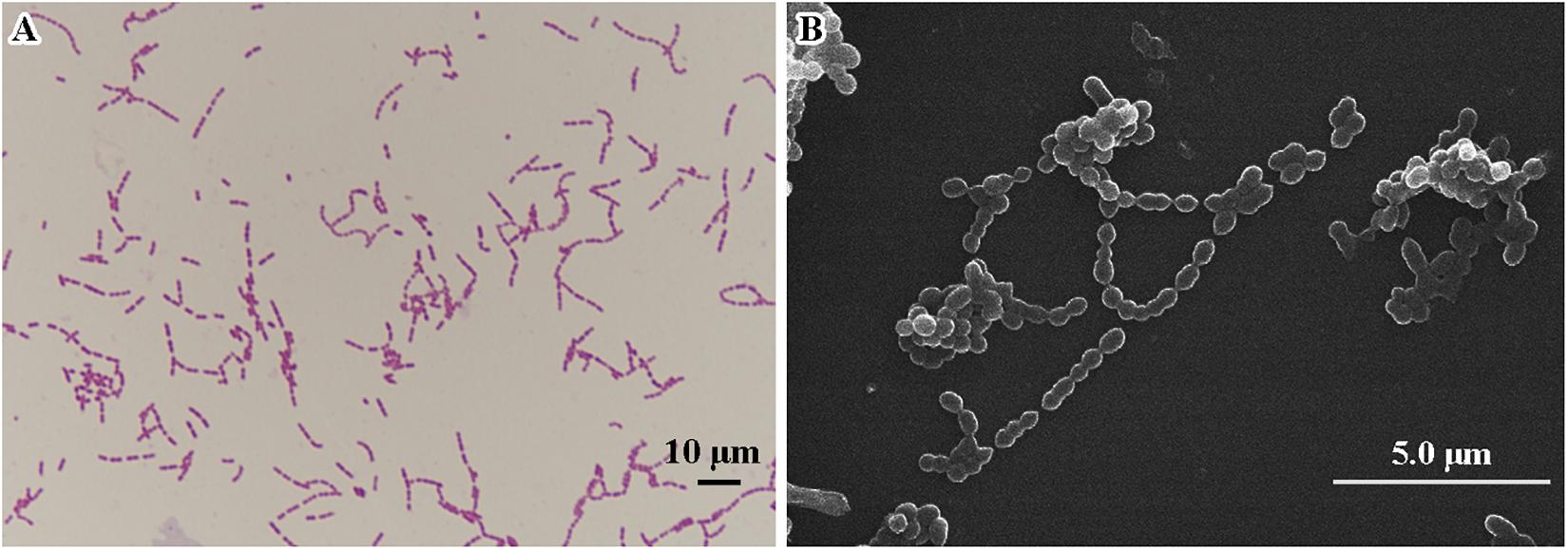
Figure 2. Microscopic imaging of Streptococcus strain E24 (A) Gram stained coccus under light microscope 10 × 100 magnification, scale bar = 10 μm. (B) Transmission electron microscope image (operating voltage of 30.0 kV), scale bar = 5.0 μm.
Biochemical Characterization and FAME Profile
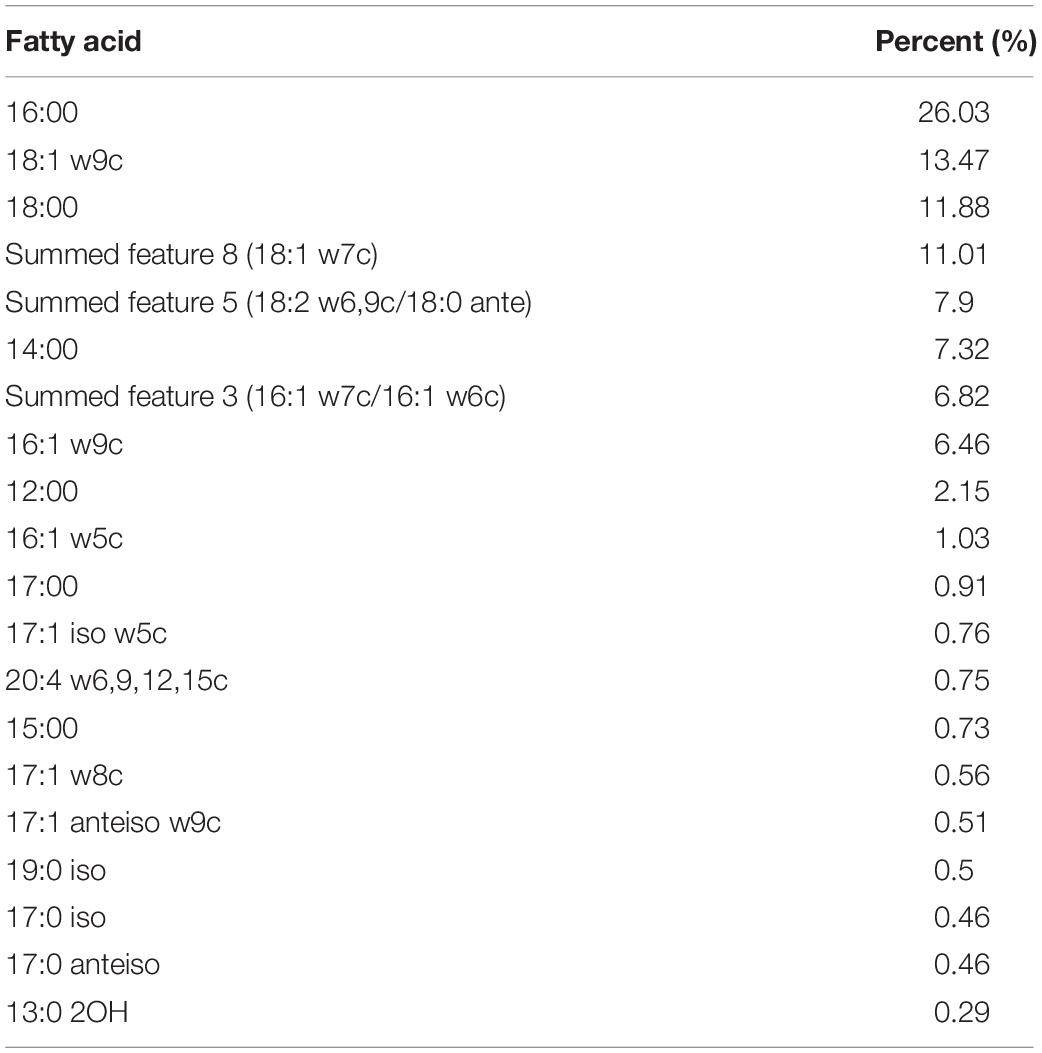
Catalase and oxidase tested negative and Streptococcus strain E24 did not hydrolyse arginine, aesculin or starch except hippurate and was negative for esterase, leucin arylamidase, valin arylamidase and β-D-galactosidase, but β-D-glucuronidase, β-D-glucosidase and alkaline phosphatase produced positive results. Streptococcus strain E24 does not produce acid from glycerol, erythritol, d- or l-arabinose, d-ribose, d- or l-xylose, D-adonitol, methyl β-D-xyloside, d-fructose-l-sorbose, l-rhamnose, dulcitol, d-mannitol, methyl α-D-mannopyranoside, methyl α-D-glucopyranoside, amygdalin, arbutin, aesculin, salicin, cellobiose, d-maltose, trehalose, inulin, melezitose, starch, glycogen, xylitol, gentiobiose, d-raffinose, d-melibiose, d-sorbitol, d-lyxose, d-tagatose, d- or l-fucose, d- or l-arabitol or 2- or 5-ketogluconate. Utilization of d-lactose is variable. Streptococcus strain E24 was capable of producing acid from D-galactose, D-glucose, D-mannose, inositol, N-acetylglucosamine and sucrose. Different biochemical characteristics between the novel Streptococcus strain E24 (strain number 1) and other members of the Streptococcus genus are shown in Table 2. Hexadecanoic acid (16:00), (9Z)-9-octadecenoic acid (18:1 w9c), octadecanoic acid (18:00) and (11Z)-11-octadecenoic acid (18:1 w7c) comprised 62% of all fatty acids (Table 3).
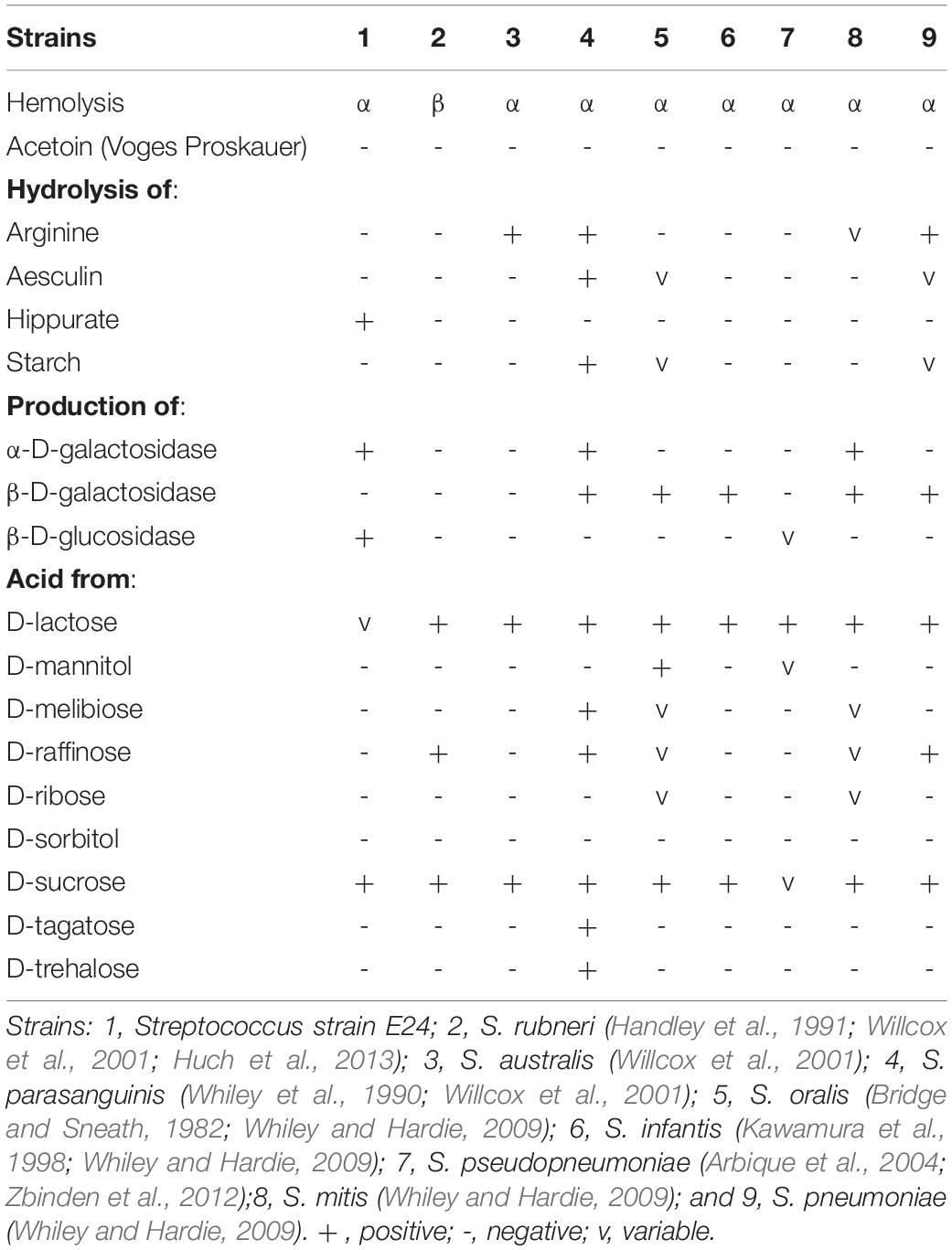
Table 2. Differential characteristics of Streptococcus strain E24 in comparison with other related organisms.
Antibiotic Susceptibility
Antibiotic susceptibility tests of Streptococcus strain E24 revealed susceptibility to vancomycin, cefepime, Rina thiazole amine, penicillin, chloramphenicol and clindamycin, but resistance to ceftriaxone, ampicillin, and cefotaxime.
Genome Properties
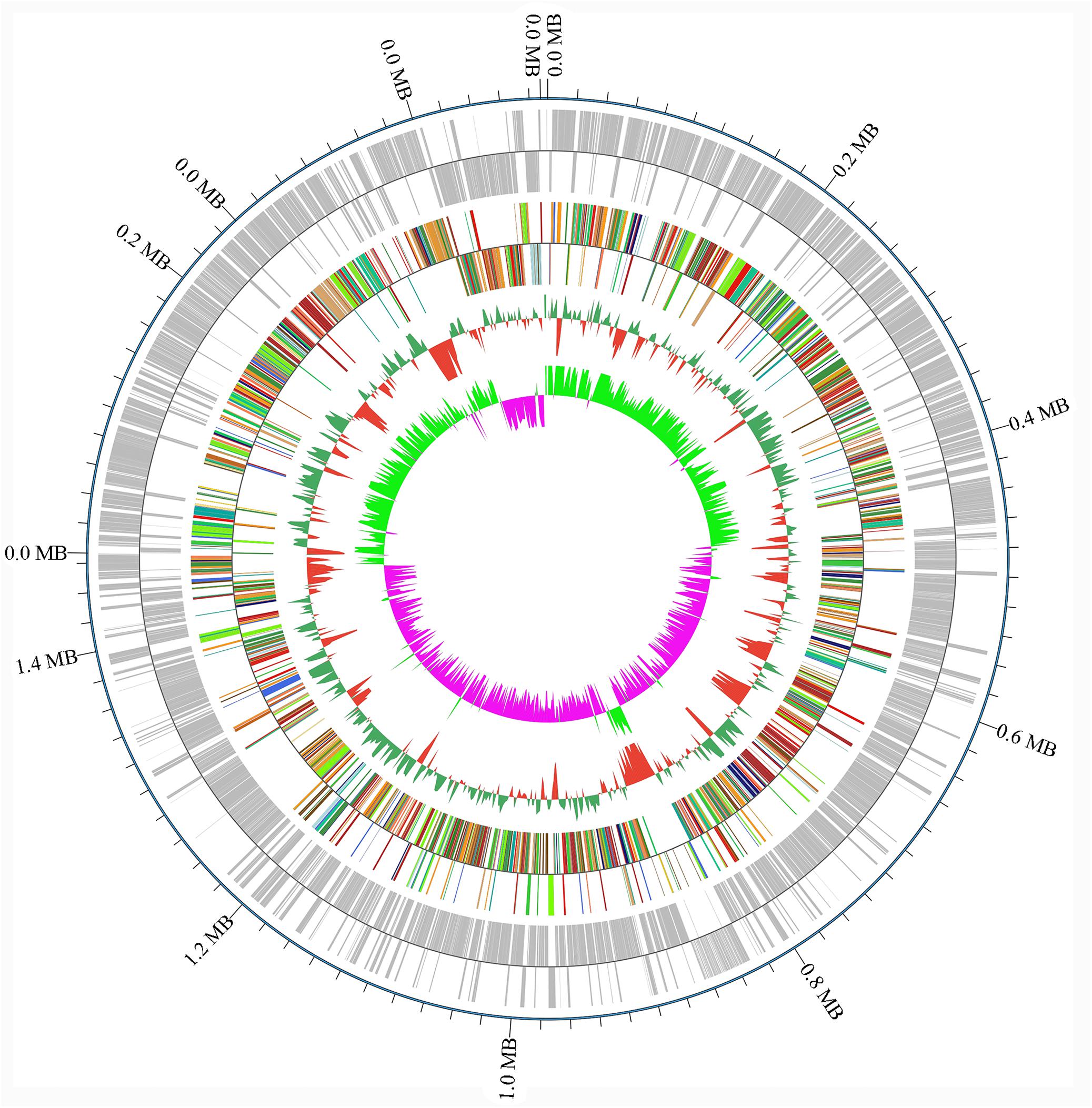
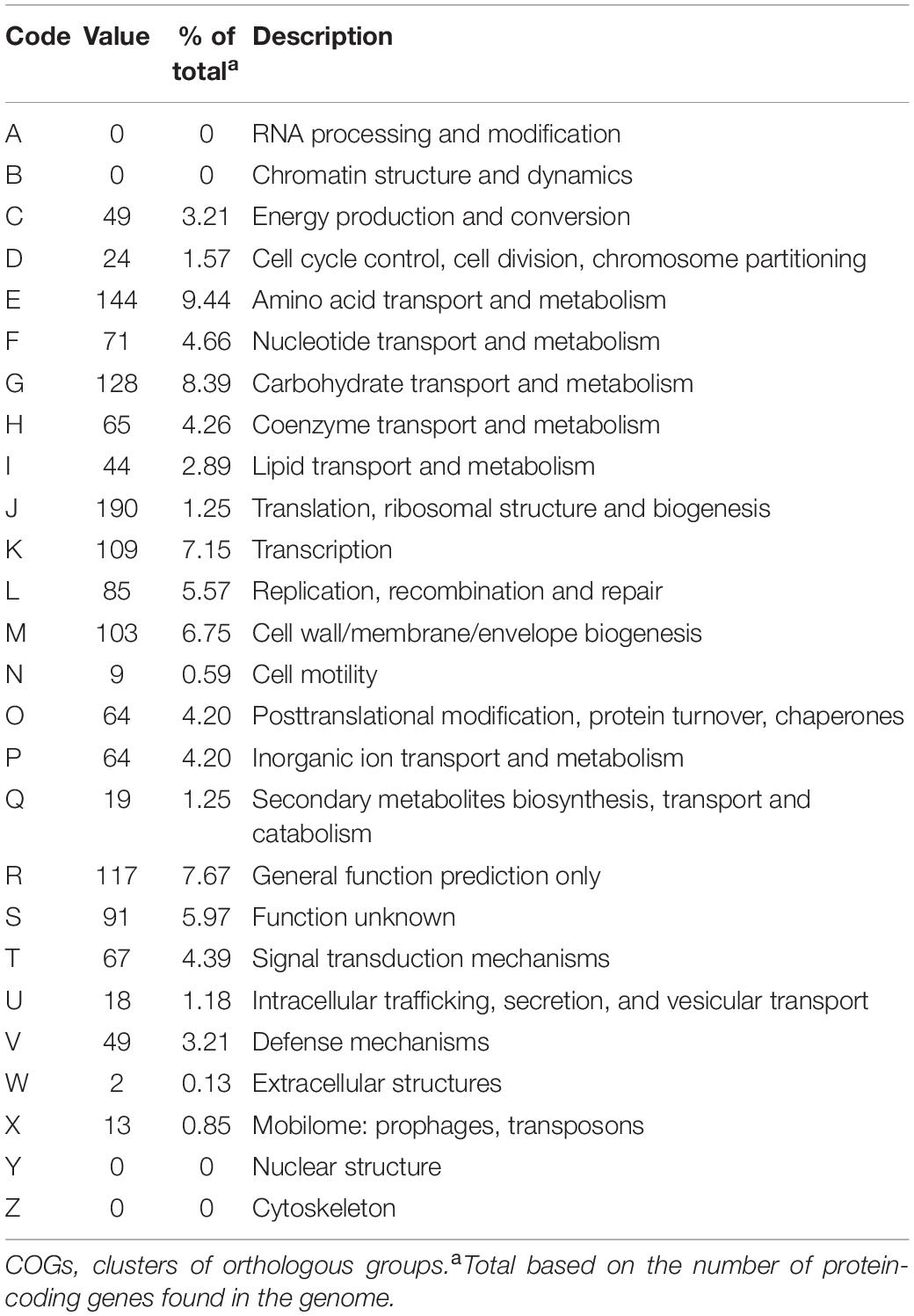
A draft of the genome of Streptococcus strain E24 revealed that it was comprised of 1,950,659 bp with a 42.03% G + C content (Figure 3). It is comprised of five scaffolds (five contigs). Among the 1,994 genes predicted, 1,942 encoded proteins and 52 RNAs (3 genes were 5S rRNA, 1 gene 16S rRNA, 1 gene 23S rRNA and 47 genes tRNA). Of these, 1,525 genes (78.52%) were specified with putative functions according to COG or NR BLAST analysis. Twenty-two genes were ORFs (1.13%) and the remaining genes were responsible for hypothetical proteins. Table 4 shows the COG functional categories of the sorted genes. The sequence of the genome was recorded in the NCBI database (accession no. VFSG00000000).
Comparisons of Genomes
The genome sequence length of Streptococcus strain E24 (1,950 Mb) was less than S. pseudopneumoniae ATCC BAA-960T (2,086 Mb), S. tigurinus AZ 3aT (2,185 Mb), S. pneumoniae NCTC 7465T (2,161 Mb), S. parasanguinis ATCC 15912T (2,154 Mb) and S. sanguinis ATCC 10556T (2,303 Mb), but greater than that of S. dentisani CECT 7747T (1,884 Mb), S. infantis ATCC 700779T (1,857 Mb), S. mitis NCTC 12261T (1,916 Mb) and S. oralis ATCC 35037T (1,914 Mb). The G + C contents of Streptococcus strain E24 (42.03%) was greater than that of S. pseudopneumoniae ATCC BAA-960T (39.8%), S. dentisani CECT 7747T (41.1%), S. infantis ATCC 700779T (38.9%), S. mitis NCTC 12261T (41.1%), S. oralis (41.4%), S. pneumoniae NCTC 7465T (39.7%), S. parasanguinis ATCC 15912T (41.7%) and S. tigurinus AZ 3aT (40.3%), but less than that of S. sanguinis ATCC 10556T (43.2%).
The distribution of genes into categories of COG was identical for all genomes investigated. To determine similarities between genomes between strains, we assigned two parameters, namely dDDH, which had a high correlation with DDH (Auch et al., 2010), and AGIOS, which was independent of DDH. The degree of nucleotide sequence similarity and numbers of orthologous genes between different genomes was also examined. Concerning dDDH values, it was found that the range varied between 23.5 and 57.8% within the 11 comparison strains, and 24 to 52% when Streptococcus strain E24 was matched with 11 comparison strains (Table 5).
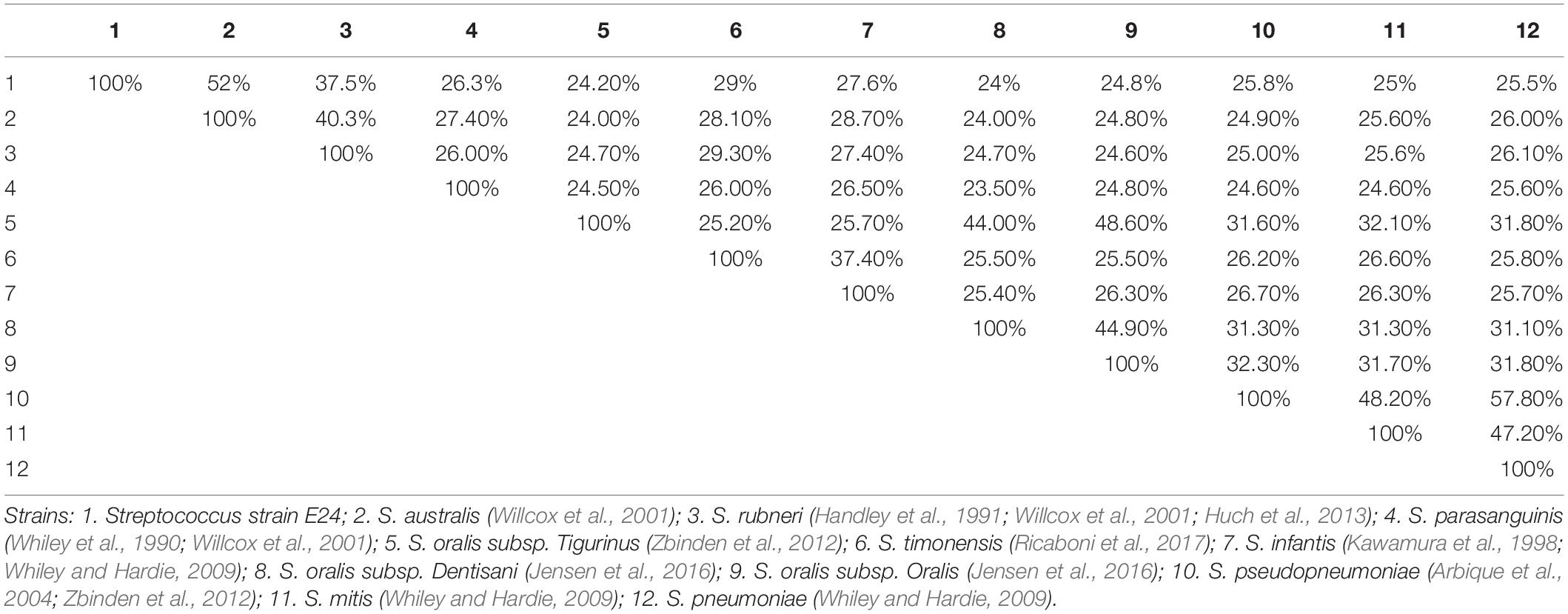
Table 5. Pairwise comparison of Streptococcus strain E24 with nine other Streptococcal species using genome-to-genome distances and formula 2 (DDH estimates based on identities/high-scoring segment pair length).
Discussion
The phenotypic analysis obtained from API strips (Table 2) revealed, that the Streptococcus strain E24 was the only one which hydrolyzed hippurate and there were essential differences between the nine analyzed streptococci, which has been described also for other Streptococcus strains in which the probability that a strain was correctly identified by nine phenotypic tests was 90–100% (Raemy et al., 2013). The similarities of the 16S rRNA gene sequences from Streptococcus strain E24 with the closest related streptococci were 98.04, 98.11, and 97.34% for Streptococcus australis ATCC 700641T, Streptococcus rubneri LMG 27207T and Streptococcus parasanguinis ATCC 15912T, whereas the sodA gene comparison exhibited a sequence identity of 92.6% with the closest strain Streptococcus australis ATCC 700641T. These similarities are in line with another newly identified Streptococcus strain exhibiting 98.7 and 92.6% similarities of 16S rRNA and rpoB gene sequences between the Streptococcus timonensis sp. nov. and the Streptococcus infantis strain JCM 10157T (Ricaboni et al., 2017). There were also essential differences of G + C contents and genome sequence lengths between Streptococcus strain E24 and eight other streptococci, but also within the 8 streptococci species, which has been described also for other Streptococcus strains (Thompson et al., 2013) and attributed to frequent occurrence of horizontal gene transfers (Bellanger et al., 2009; Harvey et al., 2011; Zhang et al., 2011). Most obviously, the dDDH analysis revealed 24 to 52% similarities between Streptococcus strain E24 and 11 comparison strains and the similarities between the 12 different streptococci strains was between 23.5 and 57.8%, which is less than the proposed limit of 70% similarity for discrimination of Streptococcus species (Auch et al., 2010; Thompson et al., 2013).
In summary, based on the genotypic, phylogenetic and phenotypic results, it is concluded that the Streptococcus strain E24 is a novel species of the genus Streptococcus, which is distinct from its closest phylogenetic neighbors Streptococcus australis ATCC 700641T, Streptococcus rubneri LMG 27207T and Streptococcus parasanguinis ATCC 15912T and for which the name Streptococcus xiaochunlingii sp. nov.E24 is proposed.
Description of Streptococcus xiaochunlingii sp. nov. E24
Streptococcus xiaochunlingii (xiao’chun’ lingi.i. N.L. gen masc. named after Professor Xiao Chunling, who is the leader of the laboratory in which the strain has been isolated).
Streptococcus strain E24 is a non-motile, non-spore-forming, facultative anaerobic and Gram-positive coccus, isolated from the oropharynx of healthy children. Growth is achieved under aerobic, microaerophilic and anaerobic atmospheres and had a temperature growth range of 22 to 42°C, with the optimum temperature being 37°C. After 48 h of aerobic incubation on 5% sheep’s blood–enriched nutrition agar, colonies are pinpoint, grayish and α-hemolytic, with undulated edges and with a diameter of 0.5 to 1 mm. Cells are roughly round with a 0.5 μm diameter. Cells did not hydrolyse arginine, aesculin or starch except hippurate and were negative in tests for esterase, leucin arylamidase, valin arylamidase and β-D-galactosidase. Positive for β-D-glucuronidase, β-D-glucosidase and alkaline phosphatase. Does not produce acid from glycerol, erythritol, d- or l-arabinose, d-ribose, d- or l-xylose, d-adonitol, methyl β-D-xyloside, d-fructose-l-sorbose, l-rhamnose, dulcitol, d-mannitol, methyl α-D-mannopyranoside, methyl α-D-glucopyranoside, amygdalin, arbutin, aesculin, salicin, cellobiose, d-maltose, trehalose, inulin, melezitose, starch, glycogen, xylitol, gentiobiose, d-lyxose, d-raffinose, d-tagatose, d- or l-fucose, d- or l-arabitol, d-sorbitol, d-melibiose or 2- or 5-ketogluconate. Utilization of d-lactose is variable. They were capable of producing acid from D-galactose, D-glucose, d-mannose, inositol, N-acetylglucosamine and sucrose. Catalase and oxidase tested negative. The genome was shown to be 1.950,659 bp long and contained 42.03 mol% G + C bases with 1,942 protein-coding and 53 RNA coding genes. The results of 16S rRNA gene sequencing strongly suggested that the strain was a member of the Streptococcus genus, with 98.04, 98.11, and 97.34% similarities to Streptococcus australis ATCC 700641T, Streptococcus rubneri LMG 27207T and Streptococcus parasanguinis ATCC 15912T, respectively. A sodA gene comparison exhibited a sequence identity of 92.6% with the closest strain Streptococcus australis ATCC 700641T. In silico DNA–DNA hybridization showed a closest DNA similarity value of 52% with Streptococcus australis ATCC 700641T. Streptococcus strain E24 has been deposited in the Korean Agricultural Culture Collection (deposition no. KACC 21425) and Guangdong Microbial Culture Collection Center (deposition no. GDMCC 1.1634). The genome and 16S rRNA gene sequences are recorded in the NCBI database (accession no. VFSG00000000; accession no. MN592637).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/, VFSG00000000; https://www.ncbi.nlm.nih.gov/, MN592637.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Shenyang Medical College (approval number: No. 2015052902). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
XL and YZ were responsible for the conception and design of the study, drafted, commented, and revised the manuscript. XL, YZ, YS, HQ, and DL were in charge of statistical analysis. All authors were responsible for data acquisition and analysis, read and approved the final version of this manuscript.
Funding
This research was supported by the Science and Technology Program of Liaoning Province (Grant No. 2017225076), the Scientific Research Program of Liaoning Educational Committee (Grant No. 2020), and the Natural Science Foundation of Liaoning Province (Leading Program) (Grant No. 20180551234). The funder had no role in the design of the study or the collection, analysis, and interpretation of data, or in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.563213/full#supplementary-material
Footnotes
References
Arbique, J. C., Poyart, C., Trieu-Cuot, P., Quesne, G., Carvalho Mda, G., Steigerwalt, A. G., et al. (2004). Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 42, 4686–4696. doi: 10.1128/jcm.42.10.4686-4696.2004
Auch, A. F., von Jan, M., Klenk, H. P., and Goker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic. Sci. 2, 117–134. doi: 10.4056/sigs.531120
Austrian, R. (1960). The Gram stain and the etiology of lobar pneumonia, an historical note. Bacteriol. Rev. 24, 261–265. doi: 10.1128/mmbr.24.3.261-265.1960
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bellanger, X., Roberts, A. P., Morel, C., Choulet, F., Pavlovic, G., Mullany, P., et al. (2009). Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J. Bacteriol. 191, 2764–2775. doi: 10.1128/jb.01412-08
Bosch, A., Levin, E., van Houten, M. A., Hasrat, R., Kalkman, G., Biesbroek, G., et al. (2016). Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9, 336–345. doi: 10.1016/j.ebiom.2016.05.031
Bridge, P. D., and Sneath, P. H. A. (1982). Streptococcus gallinarum sp. nov. and Streptococcus oralis sp. nov. Int. J. Syst. Evol. Micr. 32, 410–415. doi: 10.1099/00207713-32-4-410
Dione, N., Sankar, S. A., Lagier, J. C., Khelaifia, S., Michele, C., Armstrong, N., et al. (2016). Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 10, 66–76. doi: 10.1016/j.nmni.2016.01.002
Facklam, R., and Elliott, J. A. (1995). Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8, 479–495. doi: 10.1128/cmr.8.4.479
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fournier, P. E., Lagier, J. C., Dubourg, G., and Raoult, D. (2015). From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 36, 73–78. doi: 10.1016/j.anaerobe.2015.10.011
Guindon, S., and Gascuel, O. (2003). A simpl, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Handley, P., Coykendall, A., Beighton, D., Hardie, J. M., and Whiley, R. A. (1991). Streptococcus crista sp. nov., a viridans Streptococcus with tufted fibrils, isolated from the human oral cavity and throat. Int. J. Syst. Bacteriol. 41, 543–547. doi: 10.1099/00207713-41-4-543
Harvey, R. M., Stroeher, U. H., Ogunniyi, A. D., Smith-Vaughan, H. C., Leach, A. J., and Paton, J. C. (2011). A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence. PLoS One 6:e19650. doi: 10.1371/journal.pone.0019650
Huch, M., De Bruyne, K., Cleenwerck, I., Bub, A., Cho, G. S., Watzl, B., et al. (2013). Streptococcus rubneri sp. nov., isolated from the human throat. Int. J. Syst. Evol. Microbiol. 63(Pt 11), 4026–4032. doi: 10.1099/ijs.0.048538-0
Jensen, A., Scholz, C. F. P., and Kilian, M. (2016). Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov. Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int. J. Syst. Evol. Microbiol. 66, 4803–4820. doi: 10.1099/ijsem.0.001433
Kawamura, Y., Hou, X. G., Sultana, F., Miura, H., and Ezaki, T. (1995). Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45, 406–408. doi: 10.1099/00207713-45-2-406
Kawamura, Y., Hou, X. G., Todome, Y., Sultana, F., Hirose, K., Shu, S. E., et al. (1998). Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Int. J. Syst. Bacteriol. 48(Pt 3), 921–927. doi: 10.1099/00207713-48-3-921
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le Page, S., van Belkum, A., Fulchiron, C., Huguet, R., Raoult, D., and Rolain, J. M. (2015). Evaluation of the PREVI(R) Isola automated seeder system compared to reference manual inoculation for antibiotic susceptibility testing by the disk diffusion method. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1859–1869. doi: 10.1007/s10096-015-2424-8
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Li, X., Yang, B., Sun, Y., Li, S., Liu, D., Zou, Y., et al. (2019). Screening of antagonistic strains of respiratory origin and analysis of their bacteriostatic effects on pathogens. Microbiologyopen 8:e940. doi: 10.1002/mbo3.940
Lim, H. J., Lee, E. H., Yoon, Y., Chua, B., and Son, A. (2016). Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 120, 379–387. doi: 10.1111/jam.13011
Man, W. H., de Steenhuijsen Piters, W. A., and Bogaert, D. (2017). The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270. doi: 10.1038/nrmicro.2017.14
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Pontigo, F., Moraga, M., and Flores, S. V. (2015). Molecular phylogeny and a taxonomic proposal for the genus Streptococcus. Genet. Mol. Res. 14, 10905–10918. doi: 10.4238/2015.September.21.1
Poyart, C., Quesne, G., Coulon, S., Berche, P., and Trieu-Cuot, P. (1998). Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36, 41–47. doi: 10.1128/JCM.36.1.41-47.1998
Raemy, A., Meylan, M., Casati, S., Gaia, V., Berchtold, B., Boss, R., et al. (2013). Phenotypic and genotypic identification of streptococci and related bacteria isolated from bovine intramammary infections. Acta. Vet. Scand. 55:53. doi: 10.1186/1751-0147-55-53
Ricaboni, D., Mailhe, M., Lagier, J. C., Michelle, C., Armstrong, N., Bittar, F., et al. (2017). Noncontiguous finished genome sequence and description of Streptococcus timonensis sp. nov. isolated from the human stomach. New Microbes New Infect. 15, 77–88. doi: 10.1016/j.nmni.2016.11.013
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20, 1–6.
Simpson, J. T., Wong, K., Jackman, S. D., Schein, J. E., Jones, S. J., and Birol, I. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123. doi: 10.1101/gr.089532.108
Thompson, C. C., Emmel, V. E., Fonseca, E. L., Marin, M. A., and Vicente, A. C. P. (2013). Streptococcal taxonomy based on genome sequence analyses. F1000Research 2, 67–67. doi: 10.12688/f1000research.2-67.v1
Tindall, B. J., Rossello-Mora, R., Busse, H. J., Ludwig, W., and Kampfer, P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60(Pt 1), 249–266. doi: 10.1099/ijs.0.016949-0
Tsang, T. K., Lee, K. H., Foxman, B., Balmaseda, A., Gresh, L., Sanchez, N., et al. (2019). Association between the respiratory microbiome and susceptibility to influenza virus infection. Clin. Infect. Dis. 71, 1195–1203. doi: 10.1093/cid/ciz968
Whiley, R. A., Fraser, H. Y., Douglas, C. W., Hardie, J. M., Williams, A. M., and Collins, M. D. (1990). Streptococcus parasanguis sp. nov., an atypical viridans Streptococcus from human clinical specimens. FEMS Microbiol. Lett. 56, 115–121. doi: 10.1111/j.1574-6968.1990.tb04133.x
Whiley, R. A., and Hardie, J. M. (2009). Genus I. Streptococcus Rosenbach 1884, 22AL. New York, NY: Springer.
Willcox, M., Zhu, H., and Knox, K. (2001). Streptococcus australis sp. nov., a novel oral Streptococcus. Int. J. Syst. Evol. Micr. 51, 1277–1281. doi: 10.1099/00207713-51-4-1277
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zbinden, A., Mueller, N. J., Tarr, P. E., Sproer, C., Keller, P. M., and Bloemberg, G. V. (2012). Streptococcus tigurinus sp. nov., isolated from blood of patients with endocarditis, meningitis and spondylodiscitis. Int. J. Syst. Evol. Microbiol. 62(Pt 12), 2941–2945. doi: 10.1099/ijs.0.038299-0
Keywords: Streptococcus, phenotypic analyses, 16S rRNA, novel species, genome sequencing
Citation: Zou Y, Sun Y, Qi H, Liu D, Tian H, Wang N and Li X (2020) Streptococcus xiaochunlingii sp. nov. E24 Isolated From the Oropharynx of Healthy Chinese Children. Front. Microbiol. 11:563213. doi: 10.3389/fmicb.2020.563213
Received: 19 May 2020; Accepted: 08 September 2020;
Published: 29 September 2020.
Edited by:
Daniel Yero, Autonomous University of Barcelona, SpainReviewed by:
Maryam Tidjani Alou, Aix-Marseille Université, FranceGavin Humphreys, The University of Manchester, United Kingdom
Copyright © 2020 Zou, Sun, Qi, Liu, Tian, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinming Li, lixinming277@163.com
 Yang Zou
Yang Zou Ye Sun
Ye Sun He Qi
He Qi Defeng Liu
Defeng Liu Han Tian
Han Tian Nan Wang
Nan Wang Xinming Li
Xinming Li