- 1School of Life Sciences, Lanzhou University, Lanzhou, China
- 2Life Science and Engineering College of Northwest University for Nationalities, Lanzhou, China
- 3College of Agronomy, Gansu Agricultural University, Lanzhou, China
Actinomycetes in extreme alpine habitat have attracted much attention due to their unique physiological activities and functions. However, little is known about their ecological distribution and diversity. Here, we explored the phylogenetic relationship and physiological heterogeneity of cultivable actinomycetes from near-root soils of different plant communities in the Laohu Ditch (2200 – 4200 m) and Gaize County area (5018 – 5130 m) on the Qinghai-Tibetan Plateau. A total of 128 actinomycete isolates were obtained, 16S rDNA-sequenced and examined for antimicrobial activities and organic acid, H2S, diffusible pigments, various extracellular enzymes production. Seventy three isolates of the total seventy eight isolates from the Laohu Ditch, frequently isolated from 2200 to 4200 m, were closely related to Streptomyces spp. according to the 16S rDNA sequencing, while four isolates within the genus Nocardia spp. were found at 2200, 2800, and 3800 m. In addition, one potential novel isolate with 92% sequence similarity to its nearest match Micromonospora saelicesensis from the GenBank database, was obtained at 2200 m. From the Gaize County area, fifty Streptomyces isolates varied in diversity at different sites from 5018 to 5130 m. The investigation of phenotypic properties of 128 isolates showed that 94.5, 78.9, 68, 64.8, 53, 51.6, 50, 36.7, 31.2, and 22.7% of the total isolates produced catalase, lipase 2, urease, protease, H2S, lipase 3, amylase, lipase 1, diffusible pigment and organic acid, respectively. The antimicrobial assays of the total isolates revealed that 5, 28, 19, and 2 isolates from Streptomyces spp. exhibited antimicrobial activity against Escherichia coli, Staphylococcus aureus, Candida albicans, and Pseudomonas aeruginosa, respectively. This study intends to bring helpful insights in the exploitation and utilization of alpine actinomycetes for novel bioactive compounds discovery.
Introduction
Actinomyces had served as the sources of novel antibiotic and bioactive molecule candidates with application in many fields (Ramesh and Mathivanan, 2009; Genilloud et al., 2011). Today, the exploitation of actinomycetes with bioactive metabolites from unexplored or extreme ecosystems may be an efficient way to satisfy the everlasting demand for novel natural products, which have antimicrobial and therapeutic properties to combat human and plant pathogens (Intra et al., 2011; Ouyang et al., 2011). During the last decades, the research of actinomycetes in several alpine habitats has gained some remarkable results. For example, the study in the high Arctic permafrost of Spitsbergen has shown that there is a high diversity of actinobacterial communities in this alpine soil and many of the phylotypes identified may represent novel, uncultured species, which might be the sources of genetic diversity and ultimately novel bioactive compounds (Hansen et al., 2007). Ivanova et al. (2009) presented that a large amount of trehalose and glycerol as cryopreservation additives produced by Streptomyces spp. from the permafrost in Spitsbergen could be a reason for the strains to survive in cold and dry conditions. Further studies revealed that production of a range of bioactive metabolites including antibiotics, trehalose, lipase and pigment by actinobacteria from alpine cold habitats, could be a strategy for the strains in response to harsh environmental conditions (Fong et al., 2001; Dillon et al., 2003; Zhang et al., 2008; Babalola et al., 2009; Ivanova et al., 2009; Sivalingam et al., 2019). Malviya et al. (2009) documented that some Streptomyces isolates from alpine zones of Pindari glacier region in Indian Himalaya exhibited strong antifungal properties. These findings provide evidence that a wide diversity of Actinobacteria can survive in alpine environments, and most of which could yield bioactive compounds. In addition, as pivotal participants in the biogeochemical cycles, actinobacteria were known to possess diverse physiology and metabolic flexibility to survive in unfavorable environments (Shivlata and Satyanarayana, 2015). Thus exploring the phylogenetic and physiological diversity patterns of alpine actinomycetes may provide an opportunity for selecting strains that are under environmental pressures, which may drive adaptions that produce unique biosynthetic or hydrolytic capabilities.
The Qinghai-Tibetan Plateau, as the largest and highest plateau on Earth, possesses unique climate characteristics: alpine hypoxia, less precipitation, low humidity, osmotic stress and high incident radiation, which involves the plants and soil on the plateau in extreme harsh environmental conditions such as low temperature, anoxygenous, prolonged UV radiation and oligotrophic (Zhang et al., 2007a). Accordingly, the actinomycetes living in it may represent novel species and develop unique physiological adaption mechanism, ultimately yield bioactive compounds. However, the cultivable soil actinomycetes in this unique habitat remain relatively unexplored except for the preliminary survey on the Potentilla fruticosa L. alpine meadow and reports of novel actinomycetes (Wang et al., 2004; Chen et al., 2011; Zhang et al., 2016a, 2019). Thus, we had isolated and characterized actinomycetes from near-root soils of different plant communities, which distributed in two geographically diverse alpine habitats along an altitudinal gradient (2200 – 4200 m at the Laohu Ditch site; 5018 – 5130 m at the Gaize County area) on the Qinghai-Tibetan Plateau, aiming to explore the phylogenetic and physiological diversity of the cultivable actinomycetes.
Materials and Methods
Site Description and Sample Collection
The Laohu Ditch, located in the northeast of the Qinghai-Tibetan Plateau, is characterized by distinct vertical distribution of plant communities, from desert and desertification grassland (2200 m), mountain grassland (2800 m), alpine bushwood (3350 m), alpine meadow (3800 m), to alpine cold-desert (4200 m). The mean annual precipitation (MAP) ranged from 73.3 mm (2200 m) to 279.4 mm (4200 m), and the mean annual temperature (MAT) was from 8.1°C (2200 m) to – 5.3°C (4200 m) (offered by Qilian Mountains Station, State Key Laboratory of Cryospheric Sciences). The area of Gaize County underlain by permafrost in the central Tibetan Plateau, is distinguished by its high elevation (>5000 m) and unique plant communities adapting to this low temperature, anoxia and strong UV habitats. The MAT in the area of Gaize sampling sites was approximately 0°C with monthly mean temperature - 12.1°C in January and 12.8°C in July, and the MAP was approximately 150 mm year–1 (Qiao et al., 2015). The detailed information was depicted in Table 1.
Near-root soils were sampled from fifteen dominant plants distributing along the altitude gradients in the Laohu Ditch and five distinct dominant plants in the Gaize County area in July. The samples were aseptically processed according to the previous described (Xu et al., 1996; Zhang et al., 2007a,b; Mingma et al., 2014). Briefly, five soil cores, which were 4.5 cm in diameter and 10 cm deep, were collected radially up to ∼10 cm from the base of the plant with ∼20 cm distances, mixed to form one composite soil sample. All the samples were immediately placed in sterilized ice coolers and transported to the laboratory within 24 h.
Isolation of Actinomycetes
Actinomycetes were isolated by spreading dilutions of soil samples on petri dishes using Gao’s No. 1 medium (20 g of soluble starch, 1 g of KNO3, 0.5 g of K2HPO4, 0.5 g of MgSO4⋅7H2O, 0.5 g of NaCl, 0.01 g of FeSO4⋅7H2O, 20 g of agar, pH 7.2 – 7.4). The media also contained 25 μg/mL potassium dichromate to minimize bacterial and fungal contamination (Xu et al., 1996). All the plates were incubated at 20°C ± 1 for 30 days.
The growth and appearance of actinomycetes were observed every day on the medium plates and the colonies were recognized by their characteristics such as leathery or powdery appearance with concave, convex, crumpled or flate surface etc. Representative isolates of 128 that formed colonies with visually different morphologies were selected from 200 initially recovered colonies and subcultured to obtain pure colonies for further studies.
DNA Extraction, 16S rDNA Amplification, and Sequencing
Total DNA were extracted from subcultures as described by Orsini and Romano-Spica (2001) with minor adjustment. Briefly, fresh biomass (around 50 mg) was suspended in 1 mL washing solution [50 mM Tris–HCl, pH 7.7, 25 mM EDTA, 0.1% sodium dodecyl sulfate (SDS), 0.1% polyvinylpyrrolidone (PVP)]. After centrifuging at 12,000 × g for 2 min, the biomass was resuspended in 100 μl lysis solution (50 mM Tris–HCl, pH 8.0, 25 mM EDTA, 3% SDS, 1.2% PVP) and heated in a microwave oven at 700 W for 45 s, then added 400 μl preheated (65°C) extraction solution (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.3 M sodium acetate, 1.2% PVP). The DNA pellet was phenol−chloroform extracted, precipitated in isopropyl alcohol, washed with 70% ethanol, air-dried at room temperature then resuspended in deionized distilled water for use. The universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1504R (5′-TTAAGGATGGTGATGCCGCA-3′) were used for amplification of 16S rDNA sequences, and the PCR amplication was performed as follows: 5 min at 95°C, followed by 30 cycles of 1 min at 95°C, 1 min at 56°C for annealing and 2 min at 72°C for extension, and a final extension for 8 min at 72°C. The PCR products were confirmed by electrophoresis in a 1% (w/v) agarose gel, stained with ethidium bromide in TAE buffer, then sent to purify and cycle sequencing using an ABI3100 automated sequencer at Beijing Sangon Biotech (Beijing, China).
Nucleotide Sequence Accession Numbers
The 16S rDNA sequences of 128 isolates reported in this study have been submitted to the GenBank nucleotide sequence databases under accession nos. JQ812058-JQ812111 and JQ838073-JQ838150 available at https://www.ncbi.nlm.nih.gov/nucleotide.
Phylogenetic Analysis
For further phylogenetic analysis, the sequenced 16S rDNA of the 128 isolates were matched with those in a public database using the EzBioCloud tool, and the nearest representative gene sequences of related type strains were downloaded then aligned with the isolated sequences using Clustal W program. Phylogenetic trees of the isolates were constructed by using the Maximum Likelihood method and Tamura-Nei model (Tamura and Nei, 1993) with bootstrap analysis of 1,000 replicates (Felsenstein, 1985) performed in the MEGA X package, and then the trees were edited by Evolview1.
Screening of Soil Actinomycetes for Organic Acid, H2S and Extracellular Enzymes Production
All the 128 soil actinomycetes were examined qualitatively for the production of organic acid, H2S and extracellular enzymes including lipase 1, lipase 2, and lipase 3, amylase, protease, urease, catalase. Each isolate was inoculated on the center of the respective substrates such as tween 20 (lipase 1), tween 40 (lipase 2), tween 80 (lipase 3), starch (amylase), gelatin (protease) amended agar plates separately and incubated for 7–14 days at room temperature, the plates directly detected by clearing zones around the colonies were regarded as positive for enzyme activity (Sanchez-Porro et al., 2003; Ramesh and Mathivanan, 2009). The assays of urease, catalase, organic acid and H2S production were performed as described by Shirling and Gottlieb (1966). In addition, the diffusible pigments were also documented. Each test was conducted in triplicate, and plates or tubes with the same medium but without actinomycete isolates were maintained for controls.
Screening of Soil Actinomycetes for Antimicrobial Activity
All the 128 soil actinomycete isolates were screened for antimicrobial activity by agar overlay method as described by Anand et al. (2006) with minor adjustment. Spore suspensions of actinomycetes were inoculated on Gao’s No. 1 medium and cultured at 20°C ± 1 for 7 days, then overlaid with 5 ml of soft nutrient agar (0.6% agar) containing 500 μL of overnight growing culture of the tested microorganisms, including Escherichia coli ATCC25922 representing Gram-negative bacteria, Staphylococcus aureus ATCC25923 representing Gram-positive bacteria, Candida albicans ATCC66415 representing yeast-like fungi and a clinical isolated Pseudomonas aeruginosa strain representing freshly pathogenic multi-resistant bacterial strain. The overlaid plates were then incubated at 28°C for 24 h and the clear inhibition zone around each isolate was recorded as positive for antimicrobial activity (Ramesh and Mathivanan, 2009). Plates with the same medium simultaneously inoculated with the tested human pathogens but without actinomycete isolates were maintained for controls.
Results
16S rDNA-Based Phylogenetic Diversity of the Cultivable Actinomycetes
Seventy eight actinomycetes with representative phenotypes were recovered from the Laohu Ditch, and the 16S rDNA sequencing revealed that the predominant genus was Streptomyces (73 isolates), followed by Nocardia (4 isolates), and one isolate with only 92% sequence similarity to its nearest match Micromonospora saelicesensis from the GenBank database, indicating that it may be a potential novel isolate at genus level. The phylogenetic tree demonstrating the relationship between the genera Streptomyces spp. (randomly selected representative sequences of Streptomyces spp.), Nocardia spp., and Micromonospora spp. was presented on Figure 1.

Figure 1. Maximum Likelihood tree based on nearly complete 16S rDNA sequences showing relationship between the isolates from the Laohu Ditch and the genera Streptomyces spp., Nocardia spp., and Micromonospora spp.
As the dominant isolates, a phylogenetic tree displaying the relationships between 73 Streptomyces isolates in this study, and between them and their nearest relatives from GenBank, was also constructed as shown in Figure 2. This allowed the sorting of the sequences into two disparate groups, designed as Groups I and II. In Group I, the single isolate QLS81 from near-root soil of Potentilla saudersiana Royle at 2800 m, a putatively new species of Streptomyces spp. to be furtherly confirmed by DNA-DNA pairing value, formed a distinct branch with the highest 16S rDNA sequence similarity of 99% to the type strain of Streptomyces specialis. Strikingly, it can produce substantial diffusible black pigment which biological significances needed to be furtherly explored (Supplementary Figure S1). In Group II, 52 out of the 72 Streptomyces isolates formed relatively distinct phyletic lines with their 16S rDNA sequences showing 99 to 100% similarities to their type strains obtained from the GenBank database. Interestingly, 42 isolates amongst the 52 isolates were sorted into ten clusters, with most of the isolates originating from the same plant assigned to the same cluster. Cluster 1 consisted of 10 isolates showing 99–100% similarity to the sequence of Streptomyces cavourensis. One isolate was obtained from Ceratoides latens Reveal & N.H. Holmgren at 2200 m, four (QLS40, QLS67, QLS84, and QLS87) from Astragalus licentianus Hand.-Mazz. at 2800 m, the other two (QLS02 and QLS09) from A. licentianus Hand.-Mazz. at 4200 m, one (QLS85) from Androsace umbellata (Lour.) Merr. at 3800 m, and two (QLS18 and QLS22) from Leontopodium leontopodoides Beauverd and Potentilla fruticosa L. at 3350 m, respectively. Four isolates in Cluster 2 showing 99–100% identity to Streptomyces anulatus, were found in Salsola collina Pall at 2200 m (QLS04), A. licentianus Hand.-Mazz. at 2800 m (QLS31), P. fruticosa L. at 3350 m (QLS13) and A. licentianus Hand.-Mazz. at 4200 m (QLS29). Cluster 3 consisting of 5 isolates displaying 99–100% identity to Streptomyces microflavus could be divided into two sub-clusters. Two isolates, QLS03 and QLS08 from A. licentianus Hand.-Mazz. at 4200 m, with brown pigmented aerial mycelium and brown diffusible pigment, respectively, formed a distinct sub-clade; While isolates QLS16 from Arenaria kansuensis Maxim at 3800 m, QLS17 from Saussurea japonica Kuntze at 3350 m, QLS19 from Poa annua L. at 3800 m producing pale yellow, deep yellow and labile yellow diffusible pigments, respectively, formed another sub-clade. Cluster 4 contained 3 isolates from Saussurea japonica Kuntze at 3350 m showing 99–100% similarity to Streptomyces cyaneofuscatus. And 3 isolates in Cluster 5 showing 99% identity to Streptomyces aureus were isolated from Rhodiola quadrifida Fisch. & C.A. Mey at 3350 m. Cluster 6 containing three isolates showing 99–100% similarity to Streptomyces chryseus were obtained from R. quadrifida Fisch. & C.A. Mey at 3350 m. Two isolates in Cluster 7, QLS62, and QLS68 from A. licentianus Hand.-Mazz. at 4200, 2800 m, respectively, demonstrated 99% similarity to Streptomyces enissocaesilis. Cluster 8 consisted of 4 isolates (QLS10, QLS82, QLS74, and QLS24), obtained from Ceratoides latens Reveal & N.H. Holmgren at 2200 m, A. licentianus Hand.-Mazz. at 2800 m, R. quadrifida Fisch. & C.A. Mey at 3350 m and A. licentianus Hand.-Mazz. at 4200 m, respectively, showing 99–100% identity to Streptomyces agglomeratus. And Cluster 9 consisted of 5 isolates showing 99–100% identity to Streptomyces durmitorensis, isolated from P. saudersiana Royle (QLS33) and A. licentianus Hand.-Mazz. (QLS45, QLS50, QLS76, and QLS77) at 2800 m. Cluster 10 contained three isolates from Ceratoides latens Reveal & N.H. Holmgren (QLS11, QLS80) and Stipa glareosa PA Smirn (QLS37) at 2200 m.
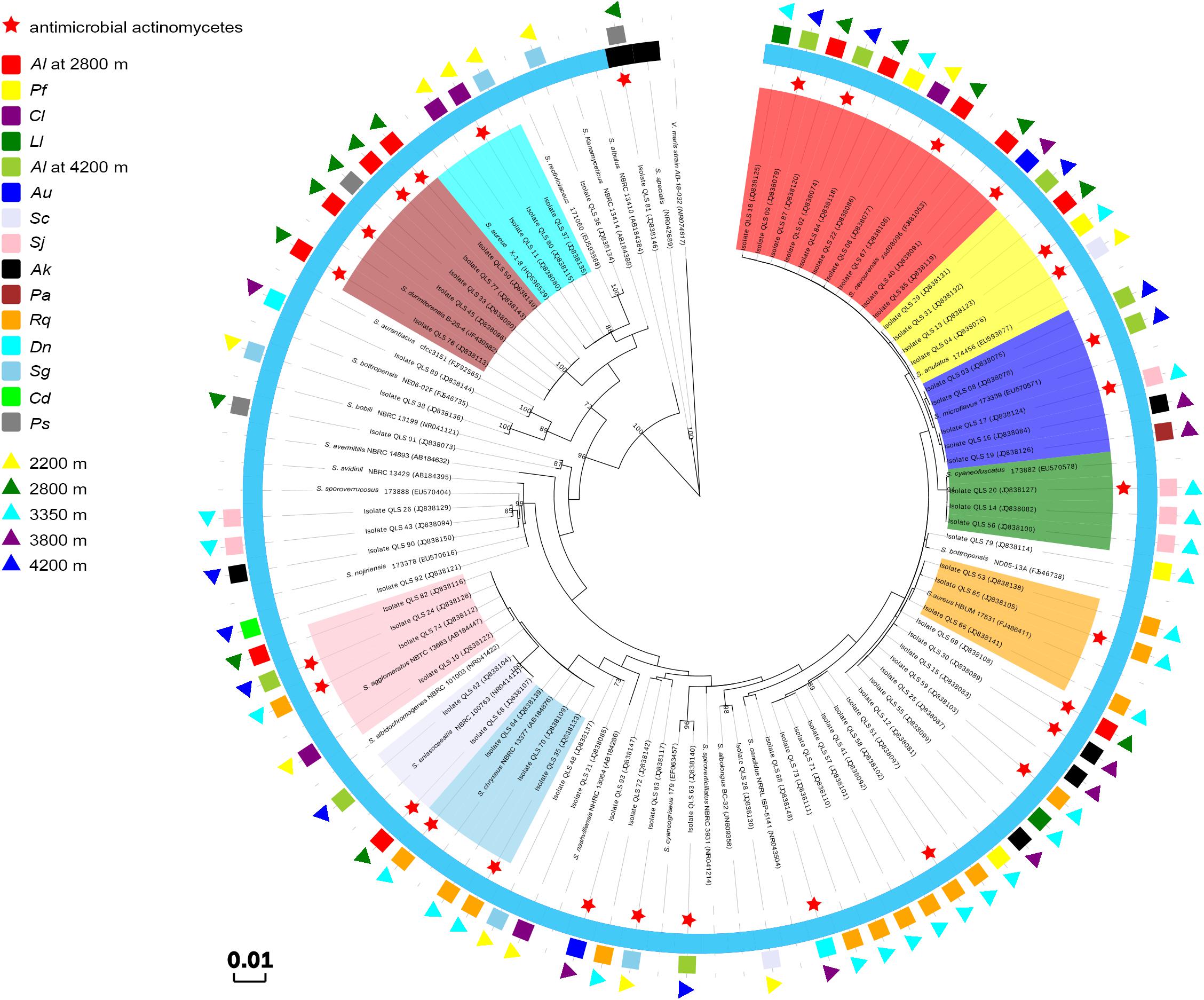
Figure 2. Phylogenetic tree of the Streptomyces isolates from the Laohu Ditch and their nearest relatives based on 16S rDNA sequences. The inner ring indicates the relationship of the Streptomyces spp., Cluster 1 in shade of red, Cluster 2 in shade of yellow, Cluster 3 in shade of blue, Cluster 4 in shade of green, Cluster 5 in shade of orange, Cluster 6 in shade of lavender, Cluster 7 in shade of skyblue, Cluster 8 in shade of pink, Cluster 9 in shade of brown, Cluster 10 in shade of cyan; the black semi-ring and the light blue semi-ring represent Group I and GroupII, respectively; the middle ring indicates the distribution of the 73 isolates in near-root soils of 15 plant communities. Al, Astragalus licentianus Hand.-Mazz.; Ll, Leontopodium leontopodoides Beauverd; Pf, Potentilla fruticosa L.; Cl, Ceratoides latens Reveal & N.H. Holmgren; Au, Androsace umbellata (Lour.) Merr.; Sc, Salsola collina Pall; Sj, Saussurea japonica Kuntze; Ak, Arenaria kansuensis Maxim; Pa, Poa annua L.; Rq, Rhodiola quadrifida Fisch. & C.A. Mey; Dn, Draba nemorosa L.; Sg, Stipa glareosa PA Smirn; Cd, Cancrinia discoidea Poljakov ex Tzvelev; Ps, P. saudersiana Royle; the outer ring indicates the distribution of the 73 isolates in diverse altitudes.
From the Gaize County area, fifty isolates with typical colony morphologies were obtained and they all pertained to Streptomyces spp. according to the 16S rDNA sequencing. The phylogenetic tree exhibiting the relationships between the fifty isolates, and between them and their closest relatives, was presented in Figure 3. As shown in Figure 3, isolate QZGYFj1 from Stipa capillata L. at 5018 m, solely formed a distinct phyletic branch, although it demonstrated 98% sequence similarity to its nearest match Streptomyces rimosus, indicating that it may be a potential new isolate. Isolate QZGYEb4 from Stipa purpurea Griseb. at 5020 m, showing 99% identity to Streptomyces rectiviolaceus, formed relatively phylogenetically distinct clade. While isolate QZGYEd3 solely formed relatively distinct phyletic line. The 47 remaining isolates were assigned to seven clusters. Cluster I contained 16 isolates distributing in S. capillata L. at 5018 m, Carex moocroftii Falc. ex Boott at 5056 m, Kobresia pygmaea C.B. Clarke at 5100 m and Dracocephalum heterophyllum Benth at 5130 m, exhibiting 99–100% similarity to Streptomyces griseus. Cluster II containing four isolates at 5056 m demonstrated 99–100% identity to S. cyaneofuscatus. Cluster III consisted of 5 isolates distributing at 5020, 5018, 5100, and 5130 m, showing 99–100% identity to Streptomyces bottropensis. Cluster IV consisted of three isolates QZGYEb5, QZGYEb2 and QZGYEc1, with isolates QZGYEc1 at 5130 m and QZGYEb2 at 5018 m forming a subclade. And three isolates in Cluster V displayed 99% similarity to Streptomyces subrutilus, with QZGYFe2 and QZGYEf1 at 5056 m and QZGYFc8 at 5130 m. Cluster VI contained eight isolates, with four isolates, QZGYFd1 and QZGYFc5 at 5130 m, QZGYFa1 and QZGYFb3 at 5020 m forming a sub-cluster, while another four isolates clustering together showing 99–100% similarity to Streptomyces phaeochromogenes. And eight isolates in Cluster VII distributed at 5020, 5056, and 5130 m, with 3 isolates forming a sub-cluster and the other 5 clustering together demonstrating 99–100% identity to S. chryseus.
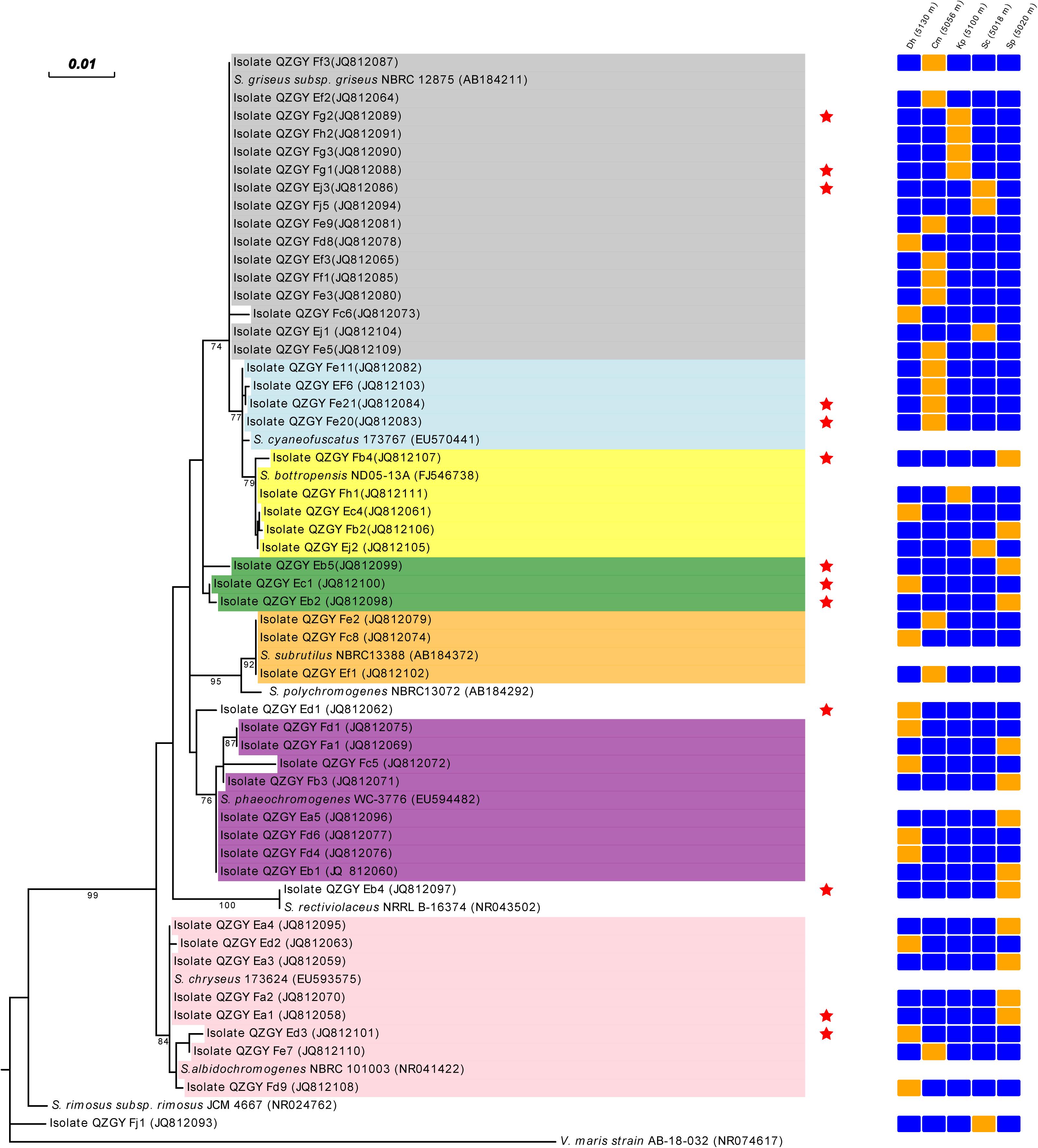
Figure 3. Phylogenetic tree displaying relationships of the Streptomyces isolates from the Gaize County area and their nearest relatives based on the 16S rDNA sequences (⋆ indicates antimicrobial actinomycetes). Dh, Dracocephalum heterophyllum Benth; Cm, Carex moocroftii Falc. ex Boott; Kp, Kobresia pygmaea C.B. Clarke; Sp, Stipa purpurea Griseb.; Sc, Stipa capillata L.; orange cell, detected; blue cell, not detected; Cluster I in shade of dark gray, Cluster II in shade of light blue; Cluster III in shade of yellow; Cluster IV in shade of green; Cluster V in shade of orange; Cluster VI in shade of purple; Cluster VII in shade of pink.
As shown in Figures 2–4, isolates with 99–100% 16S rDNA sequence similarity to S. cavourensis widely distributed in different plant communities across five altitudes on the Laohu Ditch. While isolates with 99–100% 16S rDNA sequence identity to S. griseus predominated among the isolates on the Gaize County area, constituting 32% of the total isolates. In addition, isolates with 99–100% 16S rDNA sequence identities to S. rectiviolaceus, S. chryseus, S. cyaneofuscatus and S. bottropensis occurred both in the Laohu Ditch and Gaize County area. However, variances of Streptomyces isolates in the Laohu Ditch and Gaize County area were observed. For example, isolates with 99–100% 16S rDNA sequence identities to S. specialis, Streptomyces goshikiensis, Streptomyces aurantiacus, Streptomyces nojiriensis, Streptomyces purpureus only occurred at 2800, 3350, 3800, 4200, and 5130 m, respectively.
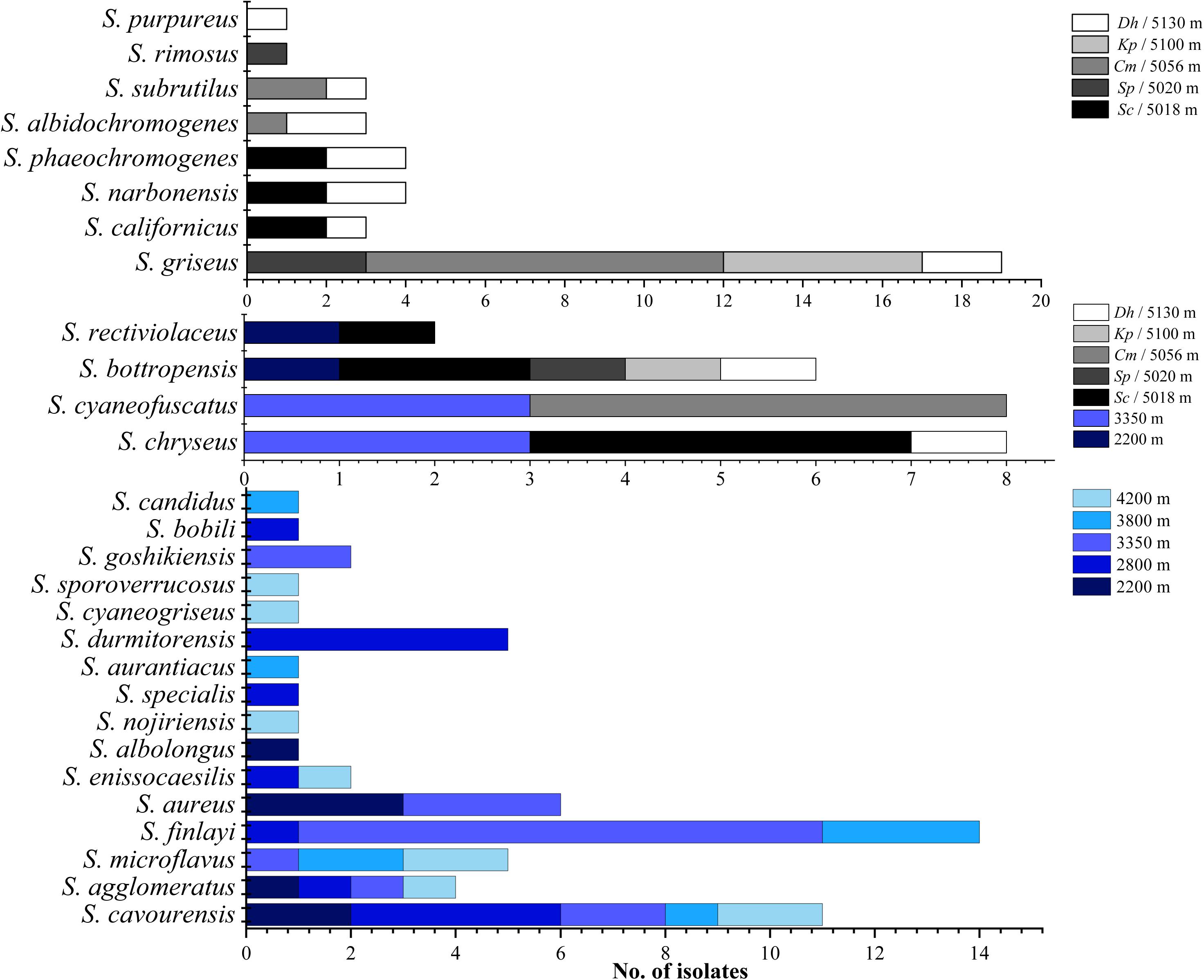
Figure 4. Distribution of isolates related to different Streptomyces species on diverse altitudes of the Laohu Ditch and Gaize County area.
Phenotypic Characteristics of the Cultivable Actinomycetes
The phenotypic properties of the isolates were shown in Figures 5, 6. From the Laohu Ditch, more than half of the 78 isolates produced catalase, lipase 2, urease, gelatinase, lipase 3 amylase. H2S, lipase 1, pigment and organic acid producing isolates accounted for 48.7, 44.9, 33.3, and 17.9% of the total isolates, respectively. As shown in Figure 5, H2S, organic acid, diffusible pigment and various extracellular enzymes producing isolates were widely distributed in the five altitudes, with diffusible pigment, urease, H2S, lipase 1, lipase 3 and protease producing isolates dominating at 2800 m, amylase and lipase 1 producing isolates predominant at 3350 m, organic acid producing isolates dominating at 4200 m. However, as displayed in Figure 6 the proportion of H2S, organic acid, diffusible pigment and extracellular enzymes producing isolates demonstrated great discrepancy in near-root soils of different plant communities. Catalase and urease producing isolates were distributed across 15 plant communities; lipase 2, protease producing isolates occurred in 14 plants; amylase, H2S, diffusible pigment producing isolates occurred in 13 plants, and lipase 1, lipase 3 in 12 plants, organic acid producing isolates only occurred in 10 plants. In addition, H2S, organic acid, diffusible pigment and various enzymes producing isolates frequently occurred in two medicinal plants, A. licentianus Hand.-Mazz. and R. quadrifida Fisch. & C.A. Mey.
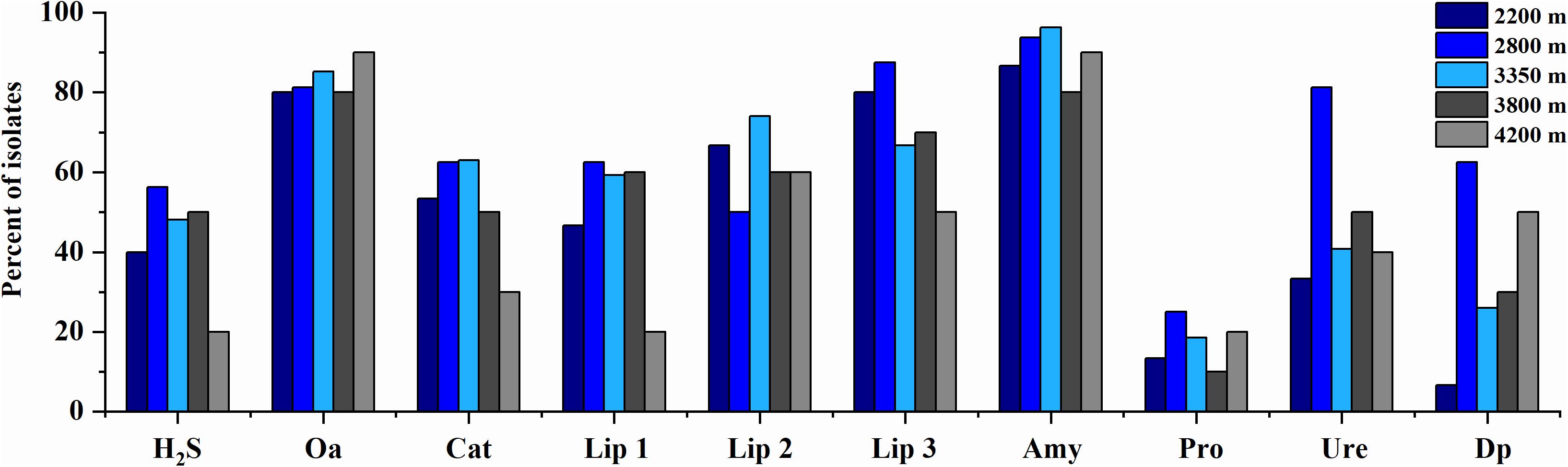
Figure 5. Distribution of H2S, organic acid, diffusible pigments and various enzymes producing isolates at the increasing altitudes of the Laohu Ditch. Cat, catalase; Lip 2, lipase 2; Ure, urease; Pro., protease; Lip 3, lipase 3; Amy, amylase; Lip 1, lipase 1; Dp, diffusible pigment; Oa, organic acid.
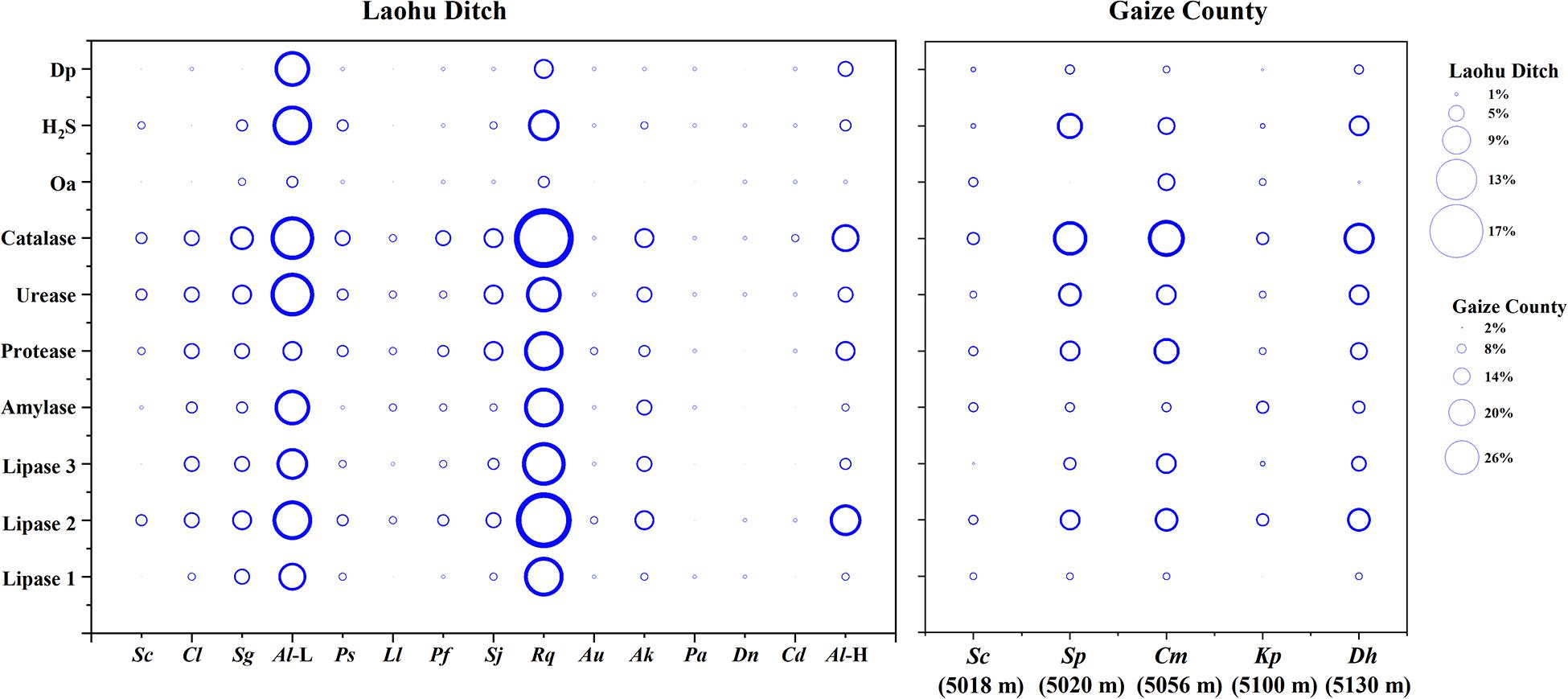
Figure 6. Distribution of H2S, organic acid, diffusible pigment and various enzymes producing isolates in near-root soils of twenty plant communities on the Tibetan Plateau. (The size of the bubble indicates the proportion of organic acid, H2S, diffusible pigment and various enzymes producing isolates to the total isolates, respectively). Al (L), Astragalus licentianus Hand.-Mazz. at 2800 m, Al (H), Astragalus licentianus Hand.-Mazz. at 4200.
Among 50 Streptomyces isolates from the Gaize County area, more than half of the isolates produced catalase, lipase 2, urease, protease, H2S. Amylase, lipase 3, lipase 1, pigment and organic acid producing isolates accounted for 46, 46, 24, 28, and 30% of the total isolates, respectively. As shown in Figure 6 organic acid and lipase 1 producing isolates occurred across four plant communities except for Stipa purpurea Griseb. at 5020 m and Kobresia pygmaea C.B. Clarke at 5100 m, respectively. The other eight were found in all of the five plant communities, with discrepancy in proportion and variety.
Antimicrobial Activity of the Cultivable Actinomycetes
The antimicrobial activity varied among the actinomycetes as shown in Table 2. Among 78 actinomycete isolates from the Laohu Ditch, 29 isolates (37.2%) exhibited antimicrobial activity against the human pathogens. Of which, 2, 19, 14, 1 isolates demonstrated antimicrobial activity against E. coli, S. aureus, C. albicans, and P. aeruginosa, respectively. In addition, 7 isolates showed antimicrobial activity against two of the four human pathogens. And the 29 antagonistic actinomycetes were all screened from Streptomyces spp. As shown in Figure 2, they were widely spread in distinct phylogenetic clusters from 12 plant communities at different altitudes. Noticeably, 51.4% of the tested isolates (35 in total) from near-root soils of two medicinal plants (A. licentianus Hand.-Mazz. and R. quadrifida Fisch. & C.A. Mey) demonstrated antimicrobial activity, which comprised 62% of the total antagonistic isolates.
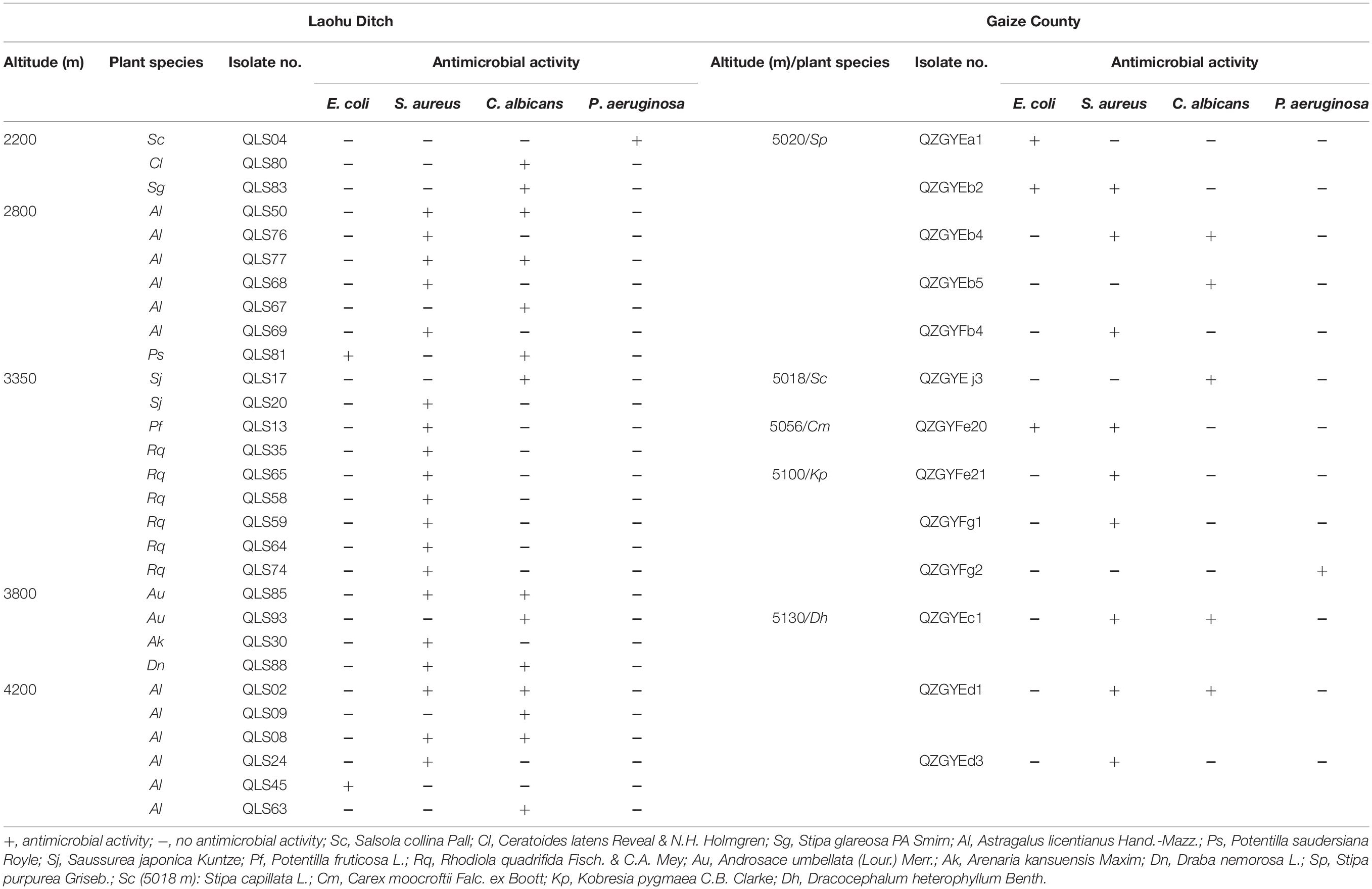
Table 2. Antimicrobial activity of the actinomycetes from the alpine habitats on the Tibetan Plateau.
While from the Gaize County area, thirteen of the fifty actinomycete isolates (26.0%) showed growth inhibitory activity against the human pathogens. Among them, 3, 9, 5, 1 isolates displayed antimicrobial activity against E. coli, S. aureus, C. albicans, and P. aeruginosa, respectively. Moreover, five isolates showed a broad antimicrobial spectrum against two of the four human pathogens. As shown in Table 2, antagonistic actinomycetes were scattered around five plant communities, with the most occurring in Stipa purpurea Griseb. at 5020 m.
In this study, the distinct phenotypic characters and antimicrobial activities were detected among isolates with identical 16S rDNA sequences as shown in Table 3. Though apparent division of the isolates to different clusters in accordance with the 16S rDNA sequences, there was just as much physiological variation among the isolates within the same clusters obtained from the same area, as among the isolates with identical 16S rDNA sequences but in disparate area, such as isolates with 99–100% 16S rDNA sequence identities to S. cavourensis, S. griseus, and S. rectiviolaceus (Table 3), S. chryseus, S. cyaneofuscatus, and S. bottropensis (Supplementary Tables 1–3).
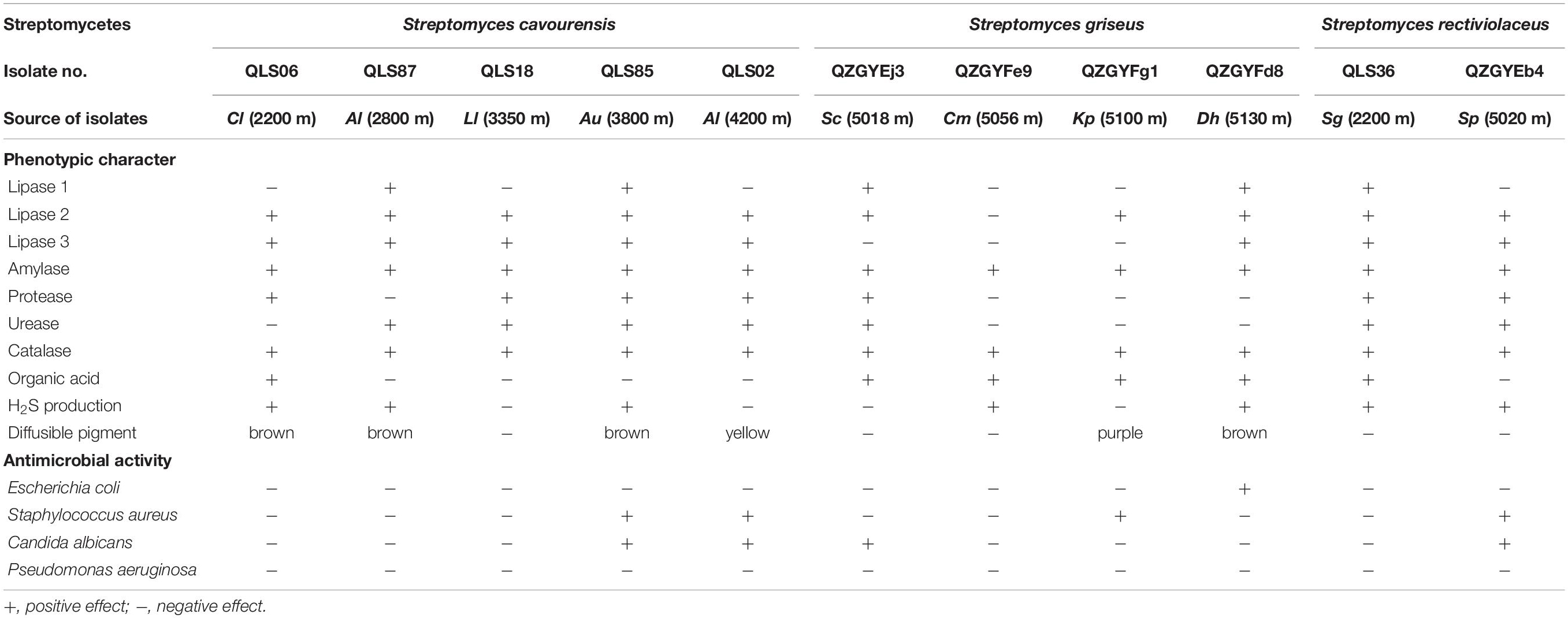
Table 3. Physiological heterogeneity between different isolates of the same clusters in the same area or disparate area.
Discussion
Actinomycetes remain a mainstream source of antibiotics against constantly emerging multidrug resistant pathogenic microorganisms (Tiwari and Gupta, 2013; Shivlata and Satyanarayana, 2015; Barka et al., 2016). To raise the screening efficiency of valuable strains, knowledge about the diversity, physiological activity and ecological distribution of unexploited actinomycete flora is urgently needed (Tiwari and Gupta, 2013). Our study added to this field by displaying the phylogenetic diversity and physiological heterogeneity of the cultivable actinomycetes from near-root soils of different plant communities at the increasing altitudes on the Qinghai-Tibetan Plateau.
It is notable that the 16S rDNA sequence as the sole marker for phylogenetic and taxonomic analysis is limited. In fact, it has been reported that Streptomyces isolates with 97% 16S rDNA sequence identities can vary by as much as 30% in core genome divergence with average nucleotide identities ranging from 100–78.3% (Chevrette et al., 2019). However, the 16S rDNA sequence is widely used as an indispensable gene marker in the bacterial taxonomic analysis (Martina et al., 2019). In this study, the 128 isolates were assigned to Streptomyce spp., Nocardia spp. and one potential new genus based on the 16S rDNA sequences. Moreover, the 97% 16S rDNA sequence identity threshold has been extensively used as a boundary for bacterial species delineation (Meier-Kolthoff et al., 2013), however, this threshold value has been raised to 99% for actinomycetes (Stach et al., 2003; Guo et al., 2015) based on comparative studies between 16S rDNA sequence identities, average nucleotide identity (ANI) values of whole genomes and DNA–DNA hybridization (DDH) (Guo et al., 2015). Accordingly, we empolyed a threshold of 99–100% to assign Streptomyces isolates to different clusters. Surely, based on this metric, the isolates identified as potential novel isolates according to the 16S rDNA sequences deserved to be furtherly confirmed by polyphasic taxonomic approach (Zhang et al., 2019).
Streptomyces spp. possess high adaptive capability for surviving in many extreme environmental conditions and their incidence had been documented in diverse extreme habitats including frozen soils, deserts, oceans, Arctic and Antarctic regions (Maldonado et al., 2008; Le Roes-Hill et al., 2009; Malviya et al., 2009; Okoro et al., 2009; Ramesh and Mathivanan, 2009; Verma et al., 2009; Ivanova et al., 2010; Zothanpuia et al., 2018). Studies in frozen soils, Arctic and Antarctic regions had revealed a great diversity of cultivable Streptomyces species (Le Roes-Hill et al., 2009; Ivanova et al., 2010; Li et al., 2011; Zhang et al., 2016b; Kamjam et al., 2019), while in marine and deserts, Streptomyces spp. had been reported as one of the dominant culturable genera (Okoro et al., 2009; Prieto-Davo et al., 2016). In this study, Streptomyces isolates were isolated from near-root soils of the studied plant communities from an altitude of 2200 to 5130 m on the Qinghai-Tibetan Plateau, with variances in diversity on different sites. By contrast, four Nocardia isolates and one potential novel genus just detected in Salsola collina Pall of 2200 m (QLS27, QLS44), Stipa glareosa PA Smirn of 2200 m (QLS96), P. saudersiana Royle of 2800 m (QLS42) and Arenaria kansuensis Maxim of 3800 m (QLS54) from the Laohu Ditch. This may be attributed to high dependency of the rare actinomycetes (Nocardia spp. and the potential new genus) upon their living conditions, with a relatively narrow ecological niche (Horner-Devine et al., 2004). Additionally, differences in the basic nutrients such as energy, carbon and nitrogen sources necessary for actinomycetes supplied by vegetation litter and/or plant root exudates, could cause variation in actinobacterial composition (Rasche et al., 2011; Zhang et al., 2013). For example, isolates with 98–100% 16S rDNA sequence identities to 28 species of Streptomyces spp. were detected in 20 plant communities along different altitudes on the Tibetan Plateau. Compared to the frequent occurrence of members related to S. cavourensis and S. griseus in different plant communities at the increasing altitudes, quite a few Streptomyces isolates specifically occurred in near-root soils of diverse plant communities at varying altitudes. This finding implied that a high degree of Streptomyces spp. diversity could adapt to the extreme alpine habitats and different plants may be selective to their root-associated Streptomyces isolates (Adil et al., 2017; Naylor et al., 2017). Noticeably, isolates with 99–100% 16S rDNA sequence identities to S. chryseus, S. rectiviolaceus, S. bottropensis, and S. cyaneofuscatus were detected in both of the studied geographically diverse cold environments. Furthermore, based on the same taxonomy according to the 16S rDNA sequences, the occurrence of some Streptomyces isolates in the studied extreme alpine habitats was observed in other extreme environments as well. For example, members related to S. griseus and S. rimosus had been isolated even from thermophilic environment (Shivlata and Satyanarayana, 2015). Isolate ISP 5300 affiliated to S. cavourensis had been reported as an alkaliphilic Streptomyces species by Mikami et al. (1982). And two isolates M-169 and M-157 related to S. cyaneofuscatus explored in deep-sea were found to produce novel antimicrobial and antitumor compounds (Ortiz-López et al., 2018; Rodríguez et al., 2018).
In this study, almost all of the Streptomyces isolates from the alpine habitats produced catalase, which was consistent with the catalase-positive of Streptomyces spp. described in Bergey’s Manual of Systematic Bacteriology (Ruan, 2013). Colony pigment or diffusible pigment produced by bacteria is a physiological strategy of adaption to low temperature and of resistant to environmental stress (Dillon et al., 2003; Ponder et al., 2005; Zhang et al., 2008). For example, microorganisms in diverse cold habitats would increase carotenoid production to keep membrane stabilization at lower temperature and secrete dark pigment to absorb UV light (Fong et al., 2001; Dillon et al., 2003; Zhang et al., 2008). Marine actinomycetes producing diffusible pigments were able to survive longer than those with no diffusible pigment production (Ramesh and Mathivanan, 2009). In our study, forty Streptomyces isolates obtained from the two alpine habitats produced diffusible pigments, which may be of significance for the isolates adapting to the alpine habitats on the Tibetan Plateau. In addition, as saprophytic inhabitants, soil actinomycetes thrive on decomposing organic materials such as lignin, chitin, cellulose, sulfocompounds etc., which is enabled by means of producing diverse extracellular hydrolytic enzymes (Bhatnagar and Kim, 2010; Suela Silva et al., 2013). The result of biochemical assays for the obtained actinomycetes showed that lipase 2, urease, protease, amylase and H2S producing strains accounted for 82.1, 70.5, 62.8, 52.6, and 48.7% of the total isolates from the Laohu Ditch, while 72, 62, 66, 46, and 60% of the total isolates from the Gaize County area, respectively. This demonstrated that most of the Streptomyces isolates possessed the capability of hydrolyzing activity on polyesters, urea, gelatin, amylum and sulfur-containing amino acids, reflecting that those isolates may associate with the cyclings of carbon, nitrogen and sulfur on the Tibetan Plateau (Shivlata and Satyanarayana, 2015; Barka et al., 2016). However, variances in proportion of H2S, diffusible pigment, organic acid and enzymes producing isolates in each plant community at increasing altitudes on the Tibetan Plateau were detected, implying the differences in soil ingredient of diverse plant root habitats closely relate to distinct plant litter and/or root exudates (Goodfellow and Williams, 1983; Bais et al., 2006). For example, the enrichment of H2S, diffusible pigment, organic acid and enzymes producing isolates in near-root soils of two medicinal plants, A. licentianus Hand.-Mazz. and R. quadrifida Fisch. & C.A. Mey, was observed. Studies have shown that medicinal plant roots are rich in bioactive compounds that could affect the physiological activities of their root-associated actinomycetes (Li et al., 2008; Zhao et al., 2012; Patrycja et al., 2015; Martina et al., 2019).
Moreover, the physiological heterogeneity of some Streptomyces isolates was detected on the Tibetan Plateau. For example, 13.7% of the total Streptomyces isolates related to S. cavourensis frequently occurring at different plant communities across five altitudes on the Laohu Ditch, and 32% of the total isolates in the Gaize County area affiliated with S. griseus across four sites, exhibited distinct physiological capabilities. Most notably, with the same taxonomy according to the 16S rDNA sequences, isolate QZGYEb4 from 5020 m on the Gaize County area showed diverse antimicrobial activity and phenotypic traits from its counterpart QLS36 from 2200 m on the Laohu Ditch. The study of marine actinomycetes also found that though nearly or fully identical 16S rDNA sequences to known terrestrial organisms, the actinomycetes still have signs of adaption to their marine environment (Bredholt et al., 2008). Zhang et al. (2008) revealed that physiological variation between strains with close evolutionary relationships suggested the differences in the ecological conditions of bacteria survival habitats.
It has been demonstrated that actinomycetes in extreme environments are capable of yielding antagonistic bioactive compounds (Shivlata and Satyanarayana, 2015; Sivalingam et al., 2019). Twenty nine actinomycetes from the Laohu Ditch and thirteen from the Gaize County area, respectively, exhibiting antimicrobial activity against four kinds of pathogenic microorganisms, were all screened from Streptomyces spp., which were widely spread at the increasing altitudes of the studied areas on the Tibetan Plateau. Interestingly, the antagonistic isolates still slightly enriched in the near-root soils of two medicinal plants, A. licentianus Hand.-Mazz. (28.6% of the total antagonistic streptomycetes) and R. quadrifida Fisch. & C.A. Mey (14.3% of the total antagonistic streptomycetes). Barakate et al. (2002) presented that 14 and 68% of the 131 streptomycetes from plant rhizospheric soils showed antimicrobial activity against Escherichia coli and Staphylococcus aureus, respectively. Several researchers had reported antimicrobial activity of actinomycetes against various pathogenic microorganisms from other unique habitats. Thirty nine actinomycetes recovered from the Antarctic soils, of which, fifteen displayed antagonistic activity against clinical Gram-positive and Gram-negative bacteria (Lee et al., 2012). Ramesh and Mathivanan (2009) obtained 208 marine actinomycetes, of which, 14.9, 8.7, and 13.5% exhibited antimicrobial activity against E. coli, P. aeruginosa and C. albicans, respectively. These results revealed that the unexplored unique habitats have the potential to discover actinomycetes with antimicrobial activities as well as various bioactive compounds.
To the best of our knowledge, this study offers for the first time a prelude about the unexplored culturable soil actinomycetes diversity associated with the typical alpine habitats on the Qinghai-Tibetan Plateau and their bioactive capabilities. However, more in-depth investigations on extraction, purification of the bioactive compounds produced by the actinomycetes, as well as the cold adaption mechanism of actinomycetes in alpine habitats will strengthen the development and utilization of those actinomycete isolates.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SX, XZ, and CZ planned and designed the research. KJ, JLL, MDW, and YW provided the help in sampling. MMW and JHL analyzed the data. AM conducted the experiments and wrote the manuscript. All authors were involved in revising the manuscript critically.
Funding
This study was funded jointly by grants from the National Science Foundation of China (nos. 31870470 and 31570393).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Editor and reviewers for helpful comments that improved this manuscript and to Cryosphere Research Station on the Qinghai-Tibet Plateau, Chinese Academy of Sciences for sampling the Gaize soils.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.555351/full#supplementary-material
Footnotes
References
Adil, E., Nicholas, L., Kistler, H. C., and Kinkel, L. L. (2017). Plant community richness mediates inhibitory interactions and resource competition between Streptomyces and Fusarium Populations in the Rhizosphere. Microbiol. Ecol. 74, 157–167. doi: 10.1007/s00248-016-0907-5
Anand, T. P., Bhat, A. W., Shouche, Y. S., Roy, U., Siddharth, J., and Sarma, S. P. (2006). Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of south east india. Microbiol. Res. 161, 252–262. doi: 10.1016/j.micres.2005.09.002
Babalola, O. O., Kirby, B. M., Le Roes-Hill, M., Cook, A. E., Cary, S. C., Burton, S. G., et al. (2009). Phylogenetic analysis of actinobacterial populations associated with Antarctic Dry Valley mineral soils. Environ. Microbiol. 11, 566–576. doi: 10.1111/j.1462-2920.2008.01809.x
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Ann. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Barakate, M., Ouhdouch, Y., Oufdou, K., and Beaulieu, C. (2002). Characterization of rhizospheric soil streptomycetes from Moroccan habitats and their antimicrobial activities. World J. Microbiol. Biotechnol. 18, 49–54. doi: 10.1023/A:1013966407890
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/mmbr.00019-15
Bhatnagar, I., and Kim, S. K. (2010). Immense essence of excellence: marine microbial bioactive compounds. Mar. Drugs 8, 2673–2701. doi: 10.3390/md8102673
Bredholt, H., Fjaervik, E., Johnsen, G., and Zotchev, S. B. (2008). Actinomycetes from sediments in the trondheim fjord, norway: diversity and biological activity. Mar. Drugs 6, 12–24. doi: 10.3390/md6010012
Chen, J., Chen, H. M., Zhang, Y. Q., Wei, Y. Z., Li, Q. P., Liu, H. Y., et al. (2011). Agromyces flavus sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 61, 1705–1709. doi: 10.1099/ijs.0.023242-0
Chevrette, M. G., Carlos-Shanley, C., Louie, K. B., Bowen, B. P., and Currie, C. R. (2019). Taxonomic and metabolic incongruence in the ancient genus Streptomyces. Front. Microbiol. 10:2170. doi: 10.3389/fmicb.2019.02170
Dillon, J. G., Miller, S. R., and Castenholz, R. W. (2003). UV-acclimation responses in natural populations of cyanobacteria (Calothrix sp.). Environ. Microbiol. 5, 473–483. doi: 10.1046/j.1462-2920.2003.00435.x
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fong, N. J., Burgess, M. L., Barrow, K. D., and Glenn, D. R. (2001). Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 56, 750–756. doi: 10.1007/s002530100739
Genilloud, O., Gonzalez, I., Salazar, O., Martin, J., Tormo, J. R., and Vicente, F. (2011). Current approaches to exploit actinomycetes as a source of novel natural products. J. Ind. Microbiol. Biotechnol. 38, 375–389. doi: 10.1007/s10295-010-0882-7
Goodfellow, M., and Williams, S. T. (1983). Ecology of actinomycetes. Ann. Rev. Microbiol. 37, 189–216. doi: 10.1146/annurev.mi.37.100183.001201
Guo, X., Liu, N., Li, X., Ding, Y., Fei, S., Gao, Y., et al. (2015). Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol. 81, 3086–3103. doi: 10.1128/AEM.03859-14
Hansen, A. A., Herbert, R. A., Mikkelsen, K., Jensen, L. L., Kristoffersen, T., Tiedje, J. M., et al. (2007). Viability, diversity and composition of the bacterial community in a high arctic permafrost soil from spitsbergen, northern norway. Environ. Microbiol. 9, 2870–2884. doi: 10.1111/j.1462-2920.2007.01403.x
Horner-Devine, M. C., Lage, M., Hughes, J. B., and Bohannan, B. J. (2004). A taxa-area relationship for bacteria. Nature 432, 750–753. doi: 10.1038/nature03073
Intra, B., Mungsuntisuk, I., Nihira, T., Igarashi, Y., and Panbangred, W. (2011). Identification of actinomycetes from plant rhizospheric soils with inhibitory activity against Colletotrichum spp., the causative agent of anthracnose disease. BMC Res. Notes 4:98. doi: 10.1186/1756-0500-4-98
Ivanova, V., Lyutskanova, D., Kolarova, M., Aleksieva, K., and Stoilova-Disheva, M. (2010). Structural Elucidation of a Bioactive Metabolites Produced by Streptomyces avidinii SB9 strain, isolated from permafrost soil in Spitsbergen, Arctic. Biotechnol. Biotechnol. Equipment 24, 2092–2095. doi: 10.2478/V10133-010-0080-9
Ivanova, V., Lyutskanova, D., Stoilova-Disheva, M., Kolarova, M., Aleksieva, K., Raykovska, V., et al. (2009). Isolation and identification of alpha,alpha-trehalose and glycerol from an arctic psychrotolerant Streptomyces sp. SB9 and their possible role in the strain’s survival. Prep. Biochem. Biotechnol. 39, 46–56. doi: 10.1080/10826060802589585
Kamjam, M., Nopnakorn, P., Zhang, L., Peng, F., Deng, Z., and Hong, K. (2019). Streptomyces polaris sp. nov. and Streptomyces septentrionalis sp. nov., isolated from frozen soil. Antonie Van Leeuwenhoek 112, 375–387. doi: 10.1007/s10482-018-1166-x
Le Roes-Hill, M., Rohland, J., Meyers, P. R., Cowan, D. A., and Burton, S. G. (2009). Streptomyces hypolithicus sp. nov., isolated from an Antarctic hypolith community. Int. J. Syst. Evol. Microbiol. 59, 2032–2035. doi: 10.1099/ijs.0.007971-0
Lee, L. H., Cheah, Y. K., Mohd Sidik, S., Ab Mutalib, N. S., Tang, Y. L., Lin, H. P., et al. (2012). Molecular characterization of antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotechnol. 28, 2125–2137. doi: 10.1007/s11274-012-1018-1
Li, J., Tian, X. P., Zhu, T. J., Yang, L. L., and Li, W. J. (2011). Streptomyces fildesensis sp. nov., a novel streptomycete isolated from antarctic soil. Antonie Van Leeuwenhoek 100, 537–543. doi: 10.1007/s10482-011-9609-7
Li, J., Zhao, G.-Z., Chen, H.-H., Wang, H.-B., Qin, S., Zhu, W.-Y., et al. (2008). Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Lett. Appl. Microbiol. 47, 574–580. doi: 10.1111/j.1472-765X.2008.02470.x
Maldonado, L., Fragoso-Yáñez, D., Pérez-García, A., Rosellón-Druker, J., and Quintana, E. (2008). Actinobacterial diversity from marine sediments collected in Mexico. Antonie van Leeuwenhoek 95, 111–120. doi: 10.1007/s10482-008-9294-3
Malviya, M. K., Pandey, A., Trivedi, P., Gupta, G., and Kumar, B. (2009). Chitinolytic activity of cold tolerant antagonistic species of Streptomyces isolated from glacial sites of Indian himalaya. Curr. Microbiol. 59, 502–508. doi: 10.1007/s00284-009-9466-z
Martina, O., Jaqueline, H., Marlene, L., Florian, G., Thomas, R., Christoph, W., et al. (2019). Exploring actinobacteria associated with rhizosphere and endosphere of the native alpine medicinal plant leontopodium nivale subspecies alpinum. Front. Microbiol. 10:2531. doi: 10.3389/fmicb.2019.02531
Meier-Kolthoff, J. P., Göker, M., Spröer, C., and Klenk, H. P. (2013). When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 195, 413–418. doi: 10.1007/s00203-013-0888-4
Mikami, Y., Miyashita, K., and Arai, T. (1982). Diaminopimelic acid profiles of alkalophilic and alkaline-resistant strains of Actinomycetes. J. Gen. Microbiol. 128, 1709–1712. doi: 10.1099/00221287-128-8-1709
Mingma, R., Pathom-aree, W., Trakulnaleamsai, S., Thamchaipenet, A., and Duangmal, K. (2014). Isolation of rhizospheric and roots endophytic actinomycetes from leguminosae plant and their activities to inhibit soybean pathogen, xanthomonas campestris pv. glycine. World J. Microbiol. Biotechnol. 30, 271–280. doi: 10.1007/s11274-013-1451-9
Naylor, D., DeGraaf, S., Purdom, E., and Coleman-Derr, D. (2017). Drought and host selection influence bacterial community dynamics in the grass root microbiome. Isme J. 11, 2691–2704. doi: 10.1038/ismej.2017.118
Okoro, C. K., Brown, R., Jones, A. L., Andrews, B. A., Asenjo, J. A., Goodfellow, M., et al. (2009). Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert. Chile. Antonie Van Leeuwenhoek 95, 121–133. doi: 10.1007/s10482-008-9295-2
Orsini, M., and Romano-Spica, V. (2001). A microwave-based method for nucleic acid isolation from environmental samples. Lett. Appl. Microbiol. 33, 17–20. doi: 10.1046/j.1472-765X.2001.00938.x
Ortiz-López, F. J., Alcalde, E., Sarmiento-Vizcaíno, A., Díaz, C., Cautain, B., García, L. A., et al. (2018). New 3-Hydroxyquinaldic acid derivatives from cultures of the marine derived actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs 16:371. doi: 10.3390/md16100371
Ouyang, Y., Wu, H., Xie, L., Wang, G., Dai, S., Chen, M., et al. (2011). A method to type the potential angucycline producers in actinomycetes isolated from marine sponges. Antonie Van Leeuwenhoek 99, 807–815. doi: 10.1007/s10482-011-9554-5
Patrycja, G., Magdalena, W., Gauravi, A., Dnyaneshwar, R., Hanna, D., and Mahendra, R. (2015). Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Antonie van Leeuwenhoek 108, 267–289. doi: 10.1007/s10482-015-0502-7
Ponder, M. A., Gilmour, S. J., Bergholz, P. W., Mindock, C. A., Hollingsworth, R., Thomashow, M. F., et al. (2005). Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol. Ecol. 53, 103–115. doi: 10.1016/j.femsec.2004.12.003
Prieto-Davo, A., Dias, T., Gomes, S. E., Rodrigues, S., Parera-Valadez, Y., Borralho, P. M., et al. (2016). The madeira archipelago as a significant source of marine-derived actinomycete diversity with anticancer and antimicrobial potential. Front. Microbiol. 7:1594. doi: 10.3389/fmicb.2016.01594
Qiao, Y., Zhao, L., Pang, Q., Chen, J., Zou, D., and Gao, Z. (2015). [Characteristics of permafrost in gerze county on the Tibetan Plateau]. J. Glaciol. Geocryol. 37, 1453–1460. doi: 10.7522/j.isnn.1000-0240.2015.0161
Ramesh, S., and Mathivanan, N. (2009). Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J. Microbiol. Biotechnol. 25, 2103–2111. doi: 10.1007/s11274-009-0113-4
Rasche, F., Knapp, D., Kaiser, C., Koranda, M., Kitzler, B., Zechmeister-Boltenstern, S., et al. (2011). Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. Isme J. 5, 389–402. doi: 10.1038/ismej.2010.138
Rodríguez, V., Martín, J., Sarmiento-Vizcaíno, A., de la Cruz, M., García, L. A., Blanco, G., et al. (2018). Anthracimycin B, a potent antibiotic against gram-positive bacteria isolated from cultures of the deep-sea actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs 16:406. doi: 10.3390/md16110406
Ruan, J. (2013). [Bergey’s manual of systematic bacteriology (second edition) volume 5 and the study of actinomycetes systematic in China]. Acta Microbiol. Sinica 53, 521–530.
Sanchez-Porro, C., Martin, S., Mellado, E., and Ventosa, A. (2003). Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. J. Appl. Microbiol. 94, 295–300. doi: 10.1046/j.1365-2672.2003.01834.x
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Bacteriol. 16, 312–340. doi: 10.1099/00207713-16-3-313
Shivlata, L., and Satyanarayana, T. (2015). Thermophilic and alkaliphilic Actinobacteria: biology and potential applications. Front. Microbiol. 6:1014. doi: 10.3389/fmicb.2015.01014
Sivalingam, P., Hong, K., Pote, J., and Prabakar, K. (2019). Extreme environment streptomyces: potential sources for new antibacterial and anticancer drug leads? Int. J. Microbiol. 2019:5283948. doi: 10.1155/2019/5283948
Stach, J. E. M., Maldonado, L. A., Masson, D. G., Ward, A. C., Goodfellow, M., and Bull, A. T. (2003). Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 69, 6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003
Suela Silva, M., Naves Sales, A., Teixeira Magalhaes-Guedes, K., Ribeiro Dias, D., and Schwan, R. F. (2013). Brazilian cerrado soil actinobacteria ecology. Biomed. Res. Int. 2013:503805. doi: 10.1155/2013/503805
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Tiwari, K., and Gupta, R. K. (2013). Diversity and isolation of rare actinomycetes: an overview. Crit. Rev. Microbiol. 39, 256–294. doi: 10.3109/1040841x.2012.709819
Verma, V. C., Gond, S. K., Kumar, A., Mishra, A., Kharwar, R. N., and Gange, A. C. (2009). Endophytic actinomycetes from Azadirachta indica A. Juss.: isolation, diversity, and anti-microbial activity. Microb. Ecol. 57, 749–756. doi: 10.1007/s00248-008-9450-3
Wang, Q. L., Cao, G. M., Jiang, W. B., and Zhang, Y. S. (2004). [Study on actinomycetes population of alpine meadow soil in Qinghai]. Wei Sheng Wu Xue Bao 44, 733–736. doi: 10.1161/01.ATV.0000116865.98067.31
Xu, L., Li, Q., and Jiang, C. (1996). Diversity of soil actinomycetes in yunnan, china. Appl. Environ. Microbiol. 62, 244–248. doi: 10.1021/bk-1996-0647.ch017
Zhang, B., Tang, S., Chen, X., Zhang, L., Zhang, G., Zhang, W., et al. (2016a). Streptomyces lacrimifluminis sp. nov., a novel actinobacterium that produces antibacterial compounds, isolated from soil. Int. J. Syst. Evol. Microbiol. 66, 4981–4986. doi: 10.1099/ijsem.0.001456
Zhang, L., Ruan, C., Peng, F., Deng, Z., and Hong, K. (2016b). Streptomyces arcticus sp. nov., isolated from frozen soil. Int. J. Syst. Evol. Microbiol. 66, 1482–1487. doi: 10.1099/ijsem.0.000907
Zhang, B., Tang, S., Yang, R., Chen, X., Zhang, D., Zhang, W., et al. (2019). Streptomyces dangxiongensis sp. nov., isolated from soil of Qinghai-Tibet Plateau. Int. J. Syst. Evol. Microbiol. 69, 2729–2734. doi: 10.1099/ijsem.0.003550
Zhang, G., Ma, X., Niu, F., Dong, M., Feng, H., An, L., et al. (2007a). Diversity and distribution of alkaliphilic psychrotolerant bacteria in the Qinghai-Tibet Plateau permafrost region. Extremophiles 11, 415–424. doi: 10.1007/s00792-006-0055-9
Zhang, G., Niu, F., Ma, X., Liu, W., Dong, M., Feng, H., et al. (2007b). Phylogenetic diversity of bacteria isolates from the Qinghai-Tibet Plateau permafrost region. Can. J. Microbiol. 53, 1000–1010. doi: 10.1139/w07-031
Zhang, X. F., Yao, T. D., Tian, L. D., Xu, S. J., and An, L. Z. (2008). Phylogenetic and physiological diversity of bacteria isolated from Puruogangri ice core. Microb. Ecol. 55, 476–488. doi: 10.1007/s00248-007-9293-3
Zhang, X. F., Zhao, L., Xu, S. J. Jr., Liu, Y. Z., Liu, H. Y., et al. (2013). Soil moisture effect on bacterial and fungal community in Beilu River (Tibetan Plateau) permafrost soils with different vegetation types. J. Appl. Microbiol. 114, 1054–1065. doi: 10.1111/jam.12106
Zhao, K., Penttinen, P., Chen, Q., Guan, T., and Lindström, K. (2012). The rhizospheres of traditional medicinal plants in Panxi, China, host a diverse selection of actinobacteria with antimicrobial properties. Appl. Microbiol. Biotechnol. 94, 1321–1335. doi: 10.1007/s00253-011-3862-6
Keywords: actinomycetes, phylogenetic diversity, physiological activities, alpine habitat, Qinghai-Tibetan Plateau
Citation: Ma A, Zhang X, Jiang K, Zhao C, Liu J, Wu M, Wang Y, Wang M, Li J and Xu S (2020) Phylogenetic and Physiological Diversity of Cultivable Actinomycetes Isolated From Alpine Habitats on the Qinghai-Tibetan Plateau. Front. Microbiol. 11:555351. doi: 10.3389/fmicb.2020.555351
Received: 24 April 2020; Accepted: 20 August 2020;
Published: 02 October 2020.
Edited by:
Laura Zucconi, University of Tuscia, ItalyReviewed by:
Reed M. Stubbendieck, University of Wisconsin-Madison, United StatesGina Lewin, Georgia Institute of Technology, United States
Copyright © 2020 Ma, Zhang, Jiang, Zhao, Liu, Wu, Wang, Wang, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijian Xu, xushijian@lzu.edu.cn
 Aiai Ma
Aiai Ma Xinfang Zhang1
Xinfang Zhang1 Changming Zhao
Changming Zhao