- 1Grupo de Inmunología Celular e Inmunogenética, Facultad de Medicina, Universidad de Antioquia, Medellín, Colombia
- 2Grupo de Productos Naturales Marinos, Facultad de Ciencias Farmacéuticas y Alimentarias, Universidad de Antioquia, Medellín, Colombia
- 3Grupo de Parasitología, Facultad de Medicina, Universidad de Antioquia, Medellín, Colombia
- 4Centro Nacional de Secuenciación Genómica, Facultad de Medicina, Universidad de Antioquia, Medellín, Colombia
The upsurge and persistence of drug resistant strains of Mycobacterium tuberculosis (Mtb) is an important limitant to the battery of drugs available for the elimination of tuberculosis (TB). To avoid future scarcity of antibiotics against Mtb, it is important to discover new effective anti-mycobacterial agents. In this study, we present data from a series of experiments to determine in vitro and in vivo anti-mycobacterial activity of a library of epidioxy-sterol analogs. We test 15 compounds for their ability to reduce the viability of Mtb. We found that one compound called T5 epidioxy-sterol-ANB display significant potency against Mtb in vitro specifically inside macrophages but without effectivity in axenic cultures. A viability assay confirms that this T5 compound is less toxic for macrophages in vitro as compared to the current Mtb drug Rifampicin at higher concentrations. We use a transcriptomic analysis of Mtb inside macrophages after T5 epidioxy-sterol-ANB treatment, and we found a significant down-regulation of enzymes involved in the cholesterol and folic acid pathways. In vivo, significant differences were found in the lungs and spleen CFUs of Mtb infected mice treated with the T5 epidioxy-sterol-ANB as compared with the untreated control group, which provides additional evidence of the effectivity of the T5 compound. Altogether these results confirm the potential of this T5 epidioxy-sterol-ANB compound against Mtb.
Introduction
Tuberculosis (TB), produced by Mycobacterium tuberculosis (Mtb), is the primary cause of death worldwide due to an infectious agent (McShane, 2019). In 2018, TB killed 1.5 million people, and 10 million were infected in the same year (World Health Organization [WHO], 2018). The worldwide prevalence of TB is sustained by the continuing HIV-AIDS pandemic, widespread antibiotic multi-drug-resistant (MDR) strains, and extensively drug-resistant (XDR) strains and poverty. Also, more than half of a million cases of rifampicin resistant TB were reported annually, with nearly 78% of those cases being MDR-TB. In addition, WHO statistics reported that about 1 in 3 deaths of TB are due to antimicrobial resistance (World Health Organization [WHO], 2018).
TB requires treatment with a combination of drugs; four anti-TB drugs taken for 6 months are required for the most drug-sensitive forms of TB (Lange et al., 2019). Treatment for MDR-TB and XDR-TB has been demonstrated to be lengthy and complicated; usually for XDR-TB patients the treatment consists of a combination of at least eight drugs, in some cases involving daily injections, for 18 months or even longer (Pontali et al., 2019). In 2019, the US Food and Drug Administration (FDA) approved the new BPaL regimen containing the drugs pretomanid, bedaquiline, and linezolid (Burki, 2019). In the Nix-TB trial in South Africa, this drug combination cured within just 6 months up to 90% of XDR tuberculosis with 5 pills a day (Nunn et al., 2019).
The development of new antibiotics by the pharmaceutical industry had mostly been stuck due to economic and regulatory hurdles (Spellberg, 2014; Anonymous., 2019). When new drugs against infectious diseases are used, the emergence of resistance is almost inevitable. Though bacterial evolution is unpredictable, the timeline for the development of resistance is uncertain. Different mechanisms have been associated with antibiotic resistance that includes mutations in genes targeted by antibiotics, the degradation or modification of antibiotics by the bacteria, the overexpression of efflux pumps to reduce the uptake of antibiotics, and alterations of the cell wall by osmoregulation in the phagosome (Larrouy-Maumus et al., 2016; Gygli et al., 2017; Laws et al., 2019). Intracellular persistence within macrophages is an essential feature of Mtb pathogenesis (Baena and Porcelli, 2009; Queval et al., 2017). When Mtb is inside macrophages it normally replicates in phagosomes, which are believed to be a restricted and stressful environment (Ehrt and Schnappinger, 2009; Bussi and Gutierrez, 2019). To be able to replicate in this closed environment, Mtb utilizes particular metabolic pathways to obtain host-derived nutrients (Lopez-Agudelo et al., 2017; Baena et al., 2019; Mashabela et al., 2019). A variety of transcriptional profiling studies in macrophages have indicated that lipid metabolism and cholesterol are important for Mtb survival (Aguilar-Ayala et al., 2017; Zimmermann et al., 2017; Baena et al., 2019; Lee et al., 2019). Additionally, genes involved in cholesterol utilization, gluconeogenesis, or the methyl citrate cycle (MCC) are required for full Mtb virulence during infection; mutants in some genes along these metabolic pathways fail to establish infection in macrophages (Joshi et al., 2006; Pandey and Sassetti, 2008; Miner et al., 2009; Russell, 2011; Griffin et al., 2012; Lopez-Agudelo et al., 2017; Fieweger et al., 2019). Based on this information, the central carbon metabolic pathways of Mtb are thought to be potential targets for TB drug discovery. An important study showed a structurally diverse set of compounds that target the Mtb cholesterol pathway that causes growth restriction of the bacteria inside macrophages but not in axenic cultures in the absence of cholesterol (VanderVen et al., 2015).
In summary, it is vital to unveil new drugs to treat TB that inhibit new biological targets and pathways. Identifying small molecules that are capable of inhibiting specific enzymatic targets in Mtb using target-based screens is still challenging. In this report we used a chemical library screen to identify new compounds that inhibit Mtb replication during infection inside macrophages, which allow us to identify the T5 epidioxy-sterol-ANB compound with a good anti-mycobacterial activity. The T5 compound is the result of the conjugation of the 4-nitrobenzoic acid (ANB) and the 5α,8α-epidioxy-3β-cholesterol. Although there are a few reports in the literature showing some activity of these molecules, none of them have been used in vivo in animal models for tuberculosis (Cateni et al., 2007; Giampaglia et al., 2007). Here we show in vivo activity and a metabolic effect of this T5 compound that suggest that it may inhibit simultaneously the cholesterol degradation and folic acid synthesis pathways in Mtb.
Materials and Methods
Bacteria and Plasmid
H37Rv was grown at 120 rpm in a shaker at 37°C in a square bottle containing 10 ml of 7H9 (Difco, Sparks, MD. United States) supplemented with 10% of oleic acid-albumin-dextrose-catalase (OADC) (Becton Dickinson Microbiology Systems, NJ, United States), 0.5% Glycerol (Sigma, Saint Louis, MO, United States), and 0.05% Tyloxapol (Sigma, Saint Louis, MO, United States), to an optical density of 0.5 at OD600 nm. H37Rv-pMV261.kan-GFP was grown in the presence of 50 μg/ml of kanamycin. The pMV261.kan-GFP is a multicopy plasmid under the control of the hsp60 that expresses GFP.
Compound Library Synthesis
Syntheses of the 5 epidioxy steryl esters were done in two steps. The first step is the synthesis of the compound 1, 5α,8α-epidioxy-3β-cholesterol, by the photochemical reaction of 7-dehydrocholesterol with oxygen mediated by eosine like photosensitizer (Feng et al., 2007). In this reaction the endoperoxide group was formed in positions C-5 and C-8 in a [4 + 2] cycloaddition reaction. The aromatic esters were done by the Steglich methodology with compound 1 using DCC and DMAP in CHCl3 with different aromatic acids (Farshori et al., 2010) (Supplementary Figure S1).
NMR of Compound T5
The NMR experiments were obtained in CDCl3 using a Bruker spectrometer AVANCE III NMR operating at 600 MHz for 1H and 150 MHz for 13C.
1H NMR (600 MHz, Chloroform-d) δ 9.22 (d, J = 2.1 Hz, 1H), 9.14 (d, J = 2.2 Hz, 2H), 6.56 (d, J = 8.5 Hz, 1H), 6.29 (d, J = 8.5 Hz, 1H), 5.36 (q, J = 11.2, 5.1 Hz, 1H), 2.31 (ddd, J = 13.6, 5.6, 1.7 Hz, 1H), 2.26 (dd, J = 13.6, 11.6 Hz, 1H), 2.16 – 2.07 (m, 2H), 2.05 – 1.98 (m, 1H), 1.98 – 1.89 (m, 1H), 1.85 – 1.74 (m, 2H), 1.65 (m, J = 11.6, 9.0, 8.1, 2.5 Hz, 1H), 1.61 – 1.49 (m, 4H), 1.49 – 1.39 (m, 1H), 1.41 – 1.36 (m, 1H), 1.39 – 1.30 (m, 2H), 1.30 – 1.17 (m, 3H), 1.14 (s, 2H), 1.19 – 1.07 (m, 2H), 1.06 – 0.98 (m, 1H), 0.98 (s, 3H), 0.91 (d, J = 6.5 Hz, 3H), 0.87 (dd, J = 6.7, 2.9 Hz, 7H), 0.85 (s, 3H), 0.82 (s, 3H).
13C NMR (151 MHz, Chloroform-d) δ 148.61, 134.70, 131.24, 129.45, 122.29, 81.75, 79.62, 72.74, 56.39, 51.47, 50.97, 44.77, 39.42, 39.35, 36.99, 35.93, 35.22, 34.27, 33.13, 28.24, 27.99, 26.30, 23.78, 23.42, 22.81, 22.55, 20.60, 18.57, 18.12, 12.65.
Cell Viability Assay
The AMJ2-C8 macrophages were infected at an MOI of 10:1. Cell viability was determined by fluorescence microscopy. After 48 hrs of treatment with the compound library, the treated cells were stained with Acridine Orange (AO) and Propidium Iodide (PI) in PBS (4 mg/ml). AO is excited at wavelengths near 502 nanometers (nm) when intercalated with dsDNA, emitting a green fluorescence with wavelengths of approximately 525 nm. AO stains acidic compartments such as lysosomes, where it becomes sequestered and protonated in live cells. Within this low lysosomal pH, the vesicles emit red fluorescence when loaded with the dye. When cells have an affected plasma membrane integrity, the PI is permeable and emits red fluorescence in the nucleus once it is intercalated with the DNA, which is excited at wavelengths of approximately 535 nm and fluoresces in the spectrum of 617 nm. Thus, cell viability could be assessed by the differential uptake of both dyes. The method for the cell viability quantification is showed in Supplementary Figure S4.
MTT Assay
The MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay is founded on the conversion of MTT into formazan crystals by viable bacteria, which determines succinate dehydrogenase activity. The MTT assay was carried out as described before (Mshana et al., 1998). Briefly, in a 96 well plate we put 5 × 107 bacteria (H37Rv) with 40 μL of culture medium into each well, and then each compound was added at different concentrations diluted with DMSO. We performed three independent experiments for each compound concentration. The plates were incubated at 37°C for 48 h. The compound MTT (Sigma, St. Louis, MO, United States) was dissolved in PBS (pH 7.2) to get a final concentration of 5 mg/ml. Following, 10 μL of the MTT solution was added to each well in the plate and incubated for 4 h at 37°C. Finally, 50 μL of a lysing buffer (20% sodium dodecyl sulfate in 50% N1N-dimethylformamide [pH 4.7]) was added to each well, and the plates were incubated overnight. The absorbance was measured with a spectrophotometer at a wavelength of 570 nm. The H37Rv bacteria was grown as described above in the bacteria and plasmid section. CFU counts of H37Rv axenic culture after 48 h of treatment with the T5 and Rifampicin compounds were determined in 7H10 agar plates.
Macrophage Cell Lines and Mtb Infection
The AMJ2-C8 murine alveolar macrophage cell line was obtained as a gift from Steven A. Porcelli at the Albert Einstein College of Medicine. The cells were grown in DMEM supplemented with 10% FBS, 1X Pen/strep, 0.05 mM of 2-mercaptoethanol, and 1X NEAA.
The THP-1 cell line was obtained from ATCC (TIB-202) and maintained in sterile RPMI-1640 medium supplemented with 10% FBS, 1X Pen/strep, 0.05 mM of 2-mercaptoethanol, and 1X NEAA. For differentiation 2 × 106 THP-1 cells were seeded per well in a 6-well plate and differentiated to macrophages for 48 h, in the presence of 100 ng/ml PMA (162 nM). After a resting period of 24 h, macrophages were then infected with Mtb H37Rv for 4 h at a multiplicity of infection (MOI) of 10:1, and incubated for 4 and 20 h (RNA-seq experiments) and also for 48 h (CFUs experiments in the presence of the T5 and rifampicin compounds), at 37°C with 5% CO2. To measure the survival of the cell-associated Mtb-H37Rv, THP-1 cells were lysed in 1.5 mL of distilled water with 1% SDS to collect the intracellular bacteria. The lysates were serially diluted in 7H9 broth, plated on 7H10 agar plates, and incubated for 3 weeks at 37°C. Colony counting was then performed in triplicate.
In vivo Experiment With the T5 Compound
Female wild type Balb/C mice (6–8 weeks of age) were used in the experiment and maintained under a sterile BSL3 facility. Four mice per group were infected with 2 × 105 bacteria intra peritoneal (i.p) in 200 μL of PBS with 0.05% Tween 80. The amount of bacteria during the challenge was verified by plating the inoculum on 7H10 plates. After 4 days of infection, the mice were treated with the T5 compound or the vehicle. The mice received 550 μg/200 μL of T5 compound daily by i.p injection (25 mg/Kg). On day 32, the mice were sacrificed, and the lungs and spleens were smashed and plated in 7H10 media plates. After 24 days, the CFUs were estimated. All experiments were performed in accordance with the Ethics Committee for Animal Experiments (CEEA) of the University of Antioquia.
RNA Extraction
The Mtb infected THP-1 cells were lysed at 4°C by adding 1 ml of RLT buffer (RNeasy Plus mini kit, QIAGEN, Hilden, Germany). We maintained the lysates frozen in liquid nitrogen for 15 s and then the lysates were homogenized for 20 s with a tissue tearor homogenizer (Biospec, Bartlesville, OK, United States; model 985–370 at 5,000 rpm). The homogenization procedure was repeated two times with an intermediate incubation of 1 min on ice. Next, the lysates were transferred to an Eppendorf 1.5-ml tube where they were disrupted using bead beating (Bead beater Instrument; applying six cycles of 30 s at maximum speed with cooling on ice between the different cycles) using high impact zirconium-silica beads (BenchmarkScientific, Bartlesville, OK, United States). Following this, the samples were centrifuged at 10,000 rpm for 10 min at 4°C. Finally, we used the aqueous phase to extract the total RNA with the Qiagen RNeasy plus mini kit, which includes the DNA retention column according to the manufacturer’s instructions. We performed three independent experiments for each condition, and the extracted RNA was pooled for its related replicates. The RNA Integrity Number (RIN) was determined by a Bioanalyzer 2000 instrument; values for all the samples were between 8.1 and 8.6.
RNA-seq
RNA-seq libraries were sequenced using an Illumina platform (Macrogen, South Korea). The libraries for the RNA-seq were generated using the TruSeq®RNA-Sample-Preparation Kitv2 following the manufacturer instructions (Illumina, Inc., San Diego, CA, United States). Briefly, the sequencing library is prepared by random fragmentation of the RNA sample, followed by 5′ and 3′ adapter ligation. The libraries started with double-stranded cDNA synthesized from RNA with insert sizes from 300–500 bp that are used for paired-end sequencing. Blunt-end DNA fragments are produced using a combination of fill-in reactions and exonuclease activity. An ‘A’- base is then added to the blunt ends of each strand, preparing them for ligation to the sequencing adapters. Each adapter contains a ‘T’-base overhang on 3′-end providing a complementary overhang for ligating the adapter to the A-tailed fragmented DNA. These newly redesigned adapters contain the full complement of sequencing primer hybridization sites for single, paired-end, and multiplexed reads. rRNA from macrophages and bacteria was depleted (Illumina Ribo-Zero Gold rRNA Removal Kit -Epidemiology), and sequencing was performed (TrueSeq, stranded, paired-end reads of 100 bp). The “clean” read dataset (clean reads refer to reads that passes the process of adapter removal, low quality bases removal at both ends [<Q30] and that after this two process the reads retain a minimum length of 70 bases) was used in order to map to the reference genome of the H37Rv strain of Mtb (AL123456_update130713), using the software BOWTIE2 with the default settings. We converted the SAM file to BAM, and then we sorted and indexed the reads using SAMTOOLS. Reads counts assigned to each gene present in the reference Mtb strain were done by means of the HTSEQ script with the stranded option. Next, we used the R package EDGER for the differential expression analysis, following the directions of the software authors for the different RNA-seq experiments. We stablished the coefficient of biological variation to 0.3, due to the lack of library replicates. The results of the EDGER analysis were printed as graphs and tables. The RNA-seq transcriptomic data was deposited in https://www.ncbi.nlm.nih.gov/bioproject: DMSO 4 h, BaenaRNA1_1.fastq.gz (SRR11195990); DMSO 20 hrs, BaenaRNA3_1.fastq.gz (SRR11195989); T5 4 h, BaenaRNA2_1.fastq.gz (SRR11195988); and T5 20 h, BaenaRNA4_1.fastq.gz (SRR11195987).
Results
The T5 Compound Has Anti-mycobacterial Activity
Previous reports have shown that epidioxy-sterols-like compounds have activity against Mtb (Saludes et al., 2002; Cateni et al., 2007; Duarte et al., 2007; Zhou et al., 2015). Thus, we decided to evaluate the activity of a unique library of 15 epidioxy-sterol analog conjugated compounds against this bacterium (Figure 1). For this purpose, we implemented an assay using H37Rv containing a plasmid that expresses GFP under a constitutive promoter. In this assay, Mtb replicates in the THP-1 macrophages and constitutively produces GFP. In the presence of an anti-mycobacterial compound GFP fluorescent signal is quenched in a concentration-dependent manner, as we observed with the positive control drug Rifampicin (Figures 2A,B). A similar strategy was previously used in the discovery of novel inhibitors of cholesterol degradation pathway in Mtb (VanderVen et al., 2015). The only compound that showed good activity in the infected THP-1 macrophages was the T5 compound, evidenced in the low IC50 of 0.116 μg/mL (0.2 μM) although 6.7 times higher than rifampicin that showed an IC50 of 0.024 μg/mL (0.03 μM) (Figures 2C,D). As we mentioned above, this T5 compound is the result of conjugation of the compound T1 and ANB. In contrast to T5, the compound T1 and ANB showed very low activity against Mtb (Figures 2C,D). Similar results were obtained by using the murine alveolar macrophage cell line AMJ2-C8 (data not showed). To confirm this result, we evaluate the Mtb viability by lysing the macrophages and plating the recovered bacteria and counted CFUs. The data shows undetectable CFUs at as low as 0.1 μg/ml, which is comparable to the treatment of Rifampicin (Figure 2E and Supplementary Figure S2). Thus, both the sterol ring and the ANB together in the T5 compound are required for effective anti-mycobacterial activity. We next analyzed the effectivity of the library compounds against an axenic Mtb culture. While the ANB compound has an anti-mycobacterial activity, the T5 compound did not show any significant activity under this experimental condition (Figures 3A,B). We confirmed this result with the CFU measurement in which rifampicin is able to reduce the bacterial numbers significantly but the result with the T5 compound is similar to the negative control (Figure 3C). The ANB also known as 4-nitrobenzoic acid was previously shown to have anti-mycobacterial activity against Mtb in axenic cultures in the diagnostic setting, but there is no data showing if the compound is effective inside macrophages (Giampaglia et al., 2007). Thus, we have a T5 compound with effective anti-mycobacterial activity inside macrophages but with no activity against extracellular bacteria. This could suggest that a particular processing condition inside the macrophage (e.g., activating enzymes) may be required to transform the T5 compound in order to work against Mtb.
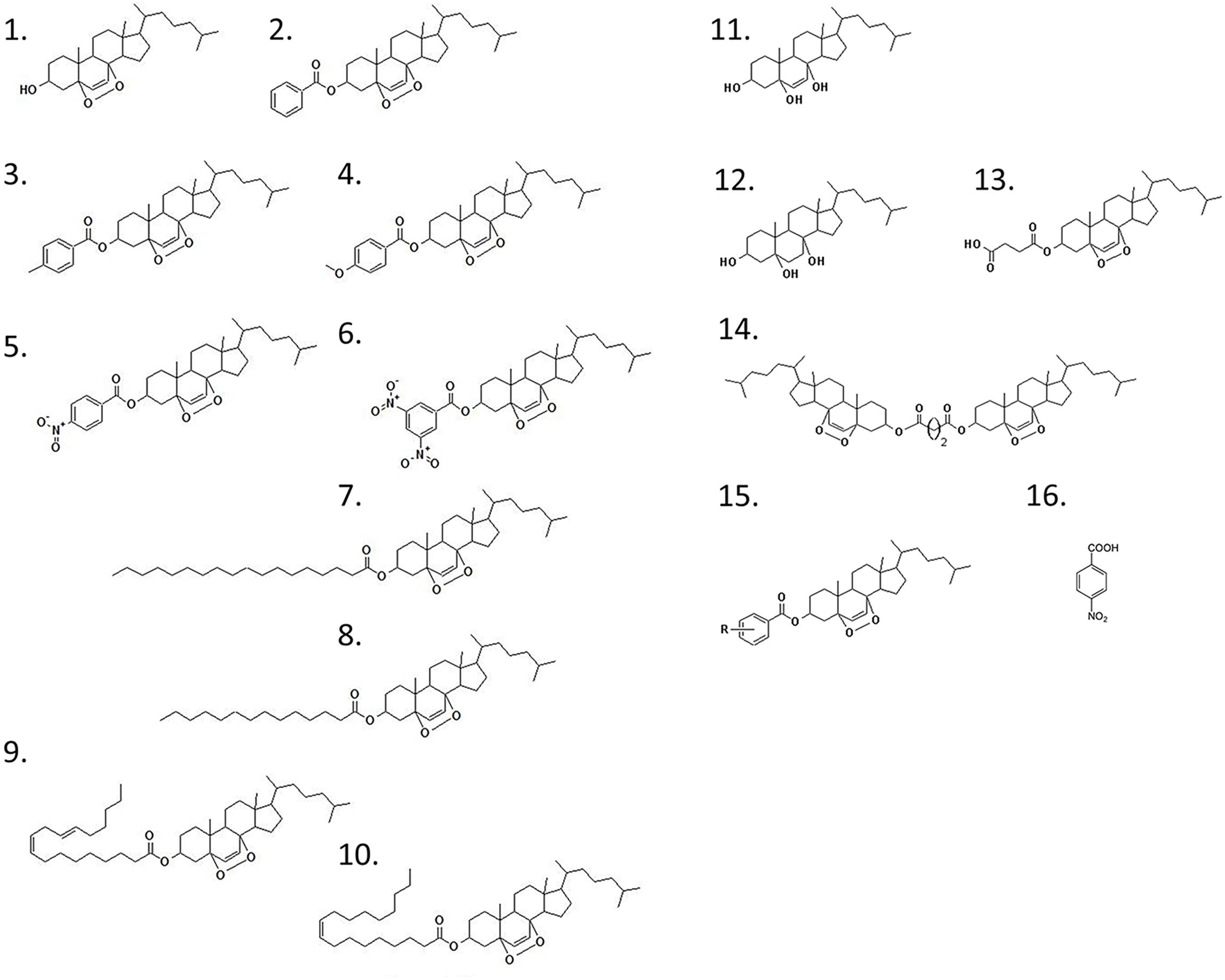
Figure 1. Chemical structure of the epidioxy-sterol analogs. This compound library started with the compound 1 (T1) that corresponded to a 5α,8α-epidioxy- 3β-cholesterol. The other compounds are called after the addition of a second (R) group from which we have generated the compounds 1–15 that in this paper we called T1–T15. The compound 16 is the 4-nitrobenzoic acid (ANB).
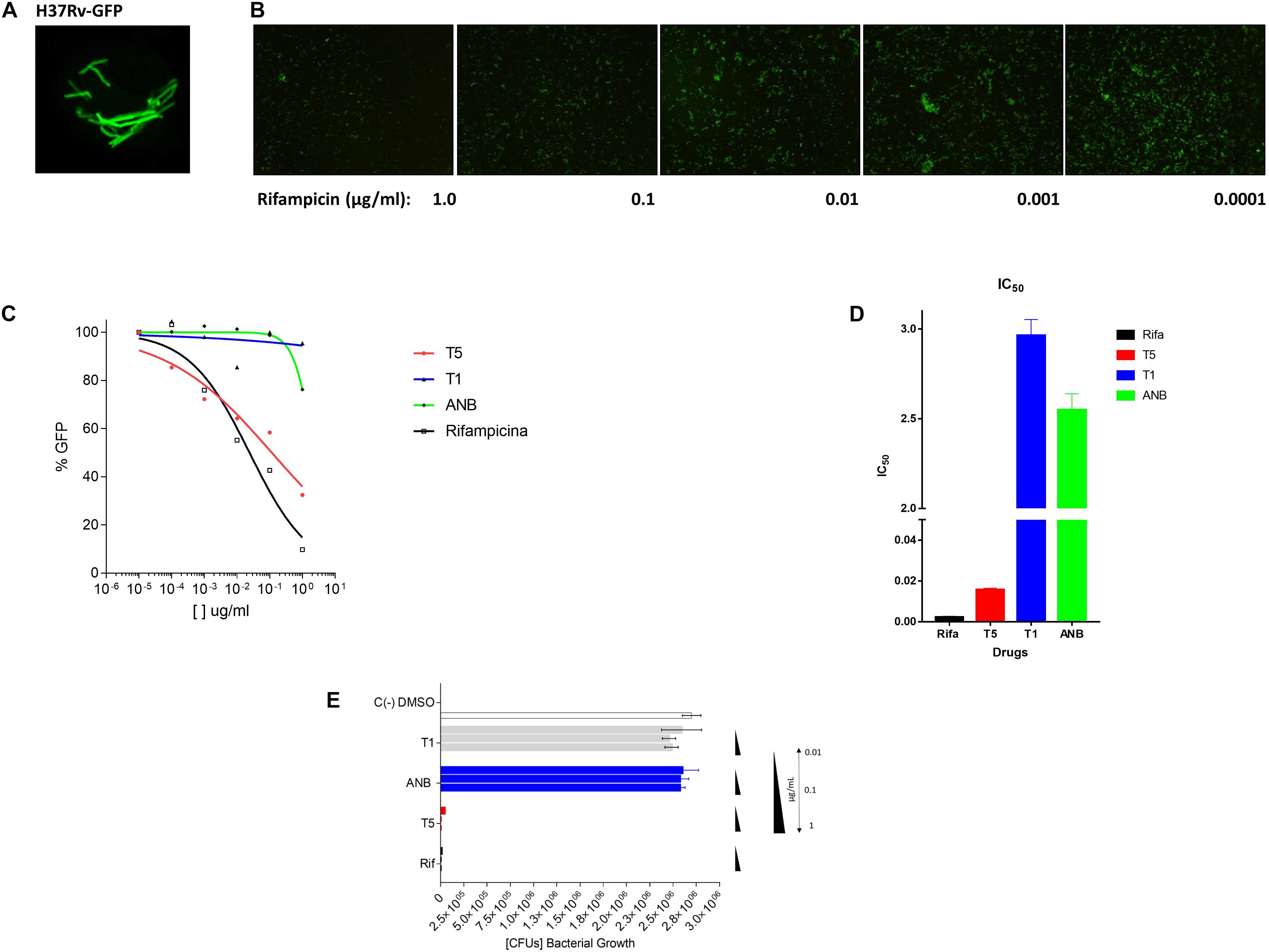
Figure 2. Inhibitory assay for the selection of compound analogs with activity against Mtb. (A) The Mtb bacteria H37Rv expressing the plasmid pMV261-GFP. (B) Inhibitory effect of Rifampicin (48 h) in the expression of GFP after the Mtb infection (4 h) of the THP-1 macrophage cell line. The panel shows 5 concentrations of the rifampicin compound. (C) Inhibitory GFP curves for the compound T5 relative to the compound T1, ANB, and the control compound rifampicin after 48 h of treatment post-infection. (D) Inhibitory concentration 50 (IC50) calculated based on the inhibitory curves. (E) The colony forming units (CFUs) that resulted after the lysis of the infected THP-1 cell line. The molecular weight (g/mol) of the compounds is as follow: T1 (416.6), T5 (565.7), ANB (167.11), and rifampicin (822.94). The equivalent molar concentration to the μg/mL used in the assays (1, 0.1, 0.01, 0.001, and 0.0001) for the different compound are: T1 (2.4, 0.24 μM, 24, 2.4, and 0.24 nM), T5 (1.7, 0.17 μM, 17, 1.7, and 0.17 nM), ANB (6, 0.6 μM, 60, 6, and 0.6 nM), and for rifampicin (1.2, 0.12 μM, 12, 1.2, and 0.12 nM).
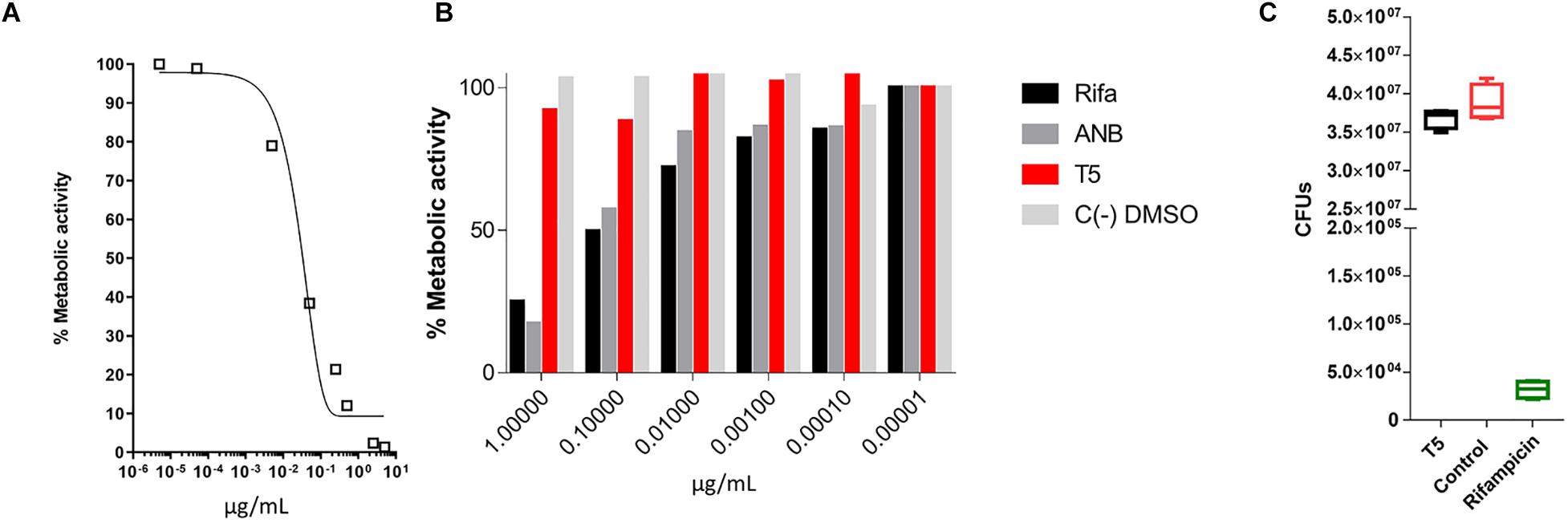
Figure 3. MTT assay to evaluate the inhibitory capacity of the T5 compound against an axenic culture of Mtb. (A) Evaluation of the% metabolic activity of the control compound rifampicin by the MTT assay in vitro for 48 h, against an axenic culture of Mtb. (B) Determination of the % metabolic activity based on the MTT assay of the T5 compound relative to the control compounds ANB and rifampicin at different concentrations. (C) CFU counts of H37Rv axenic culture after 48 h of treatment with the T5 and Rifampicin compounds. The optical density values (OD) obtained in the MTT assay were converted to percentage of metabolic activity relative to the DMSO control treatment which represent 100% of metabolic activity.
The T5 Compound Has Low Cell Toxicity at Higher Concentrations
Cytotoxicity of the T5 compound was tested using the AMJ2-C8 macrophage cell line by staining the cells with AO and PI (Figure 4A). We used a range of concentrations (0.0001, 0.001, 0.01, 0.1, and 1 μg/ml) of the T5 as well as for T1 and ANB compounds, Rifampicin and DMSO, as the positive negative controls respectively. We found that while the T1 and Rifampicin are highly toxic at the higher concentration of 1 μg/mL, the T5 compound showed low cytotoxicity at the same concentration (Figure 4B). This is an interesting result because it is expected that any new candidate compound has low toxicity, which could be associated with fewer side effects for future experiments such as the experimentation in animal models.
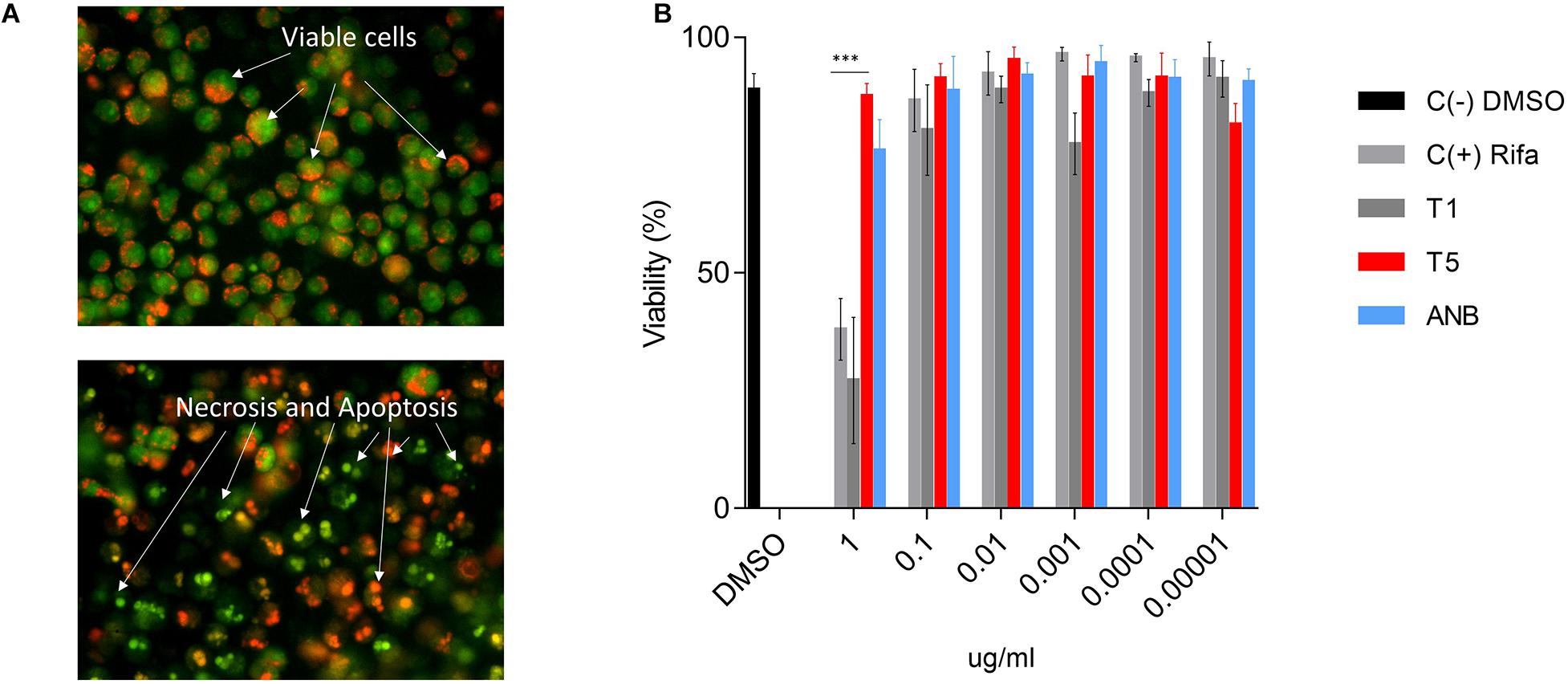
Figure 4. Cytotoxicity assay for T5 compound in the AMJ2-C8 cell line. (A) A representation of healthy and viable cells (no treatment) and cells treated with a high concentration of rifampicin (1 μg/mL). The cells were stained with acridine orange (AO) and propidium iodide (PI). (B) Viability (%) of the AM cell line after 48 h treatment with the T5 compound relative to the control compounds T1, ANB, and Rifampicin. ∗∗∗p-value < 0.0001. We are showing data of a representative experiment from four independent ones.
The Transcriptome of Infected THP-1 Cells Treated With the T5 Compound
In order to precisely define the genes in Mtb and in the THP-1 macrophages, which are affected by the T5 compound, we performed a dual transcriptome by RNA-seq. The macrophages were infected for 4 and 20 h and then the total RNA was extracted and sequenced (methods section, Supplementary Figure S3). Using log2FC > 2.0, log2FC < -2.0, and a p-value < 0.05 as selection criteria for differentially expressed genes, we found that macrophages expressed very few genes up-regulated and down-regulated in response to the T5 compound either at 4 or 20 h post-treatment (Figures 5A,B). The most noticeable up-regulated gene at 4 h was miR-3648 with a log2FC > 10 (Figure 5A). This gene was shown to be involved in the negative regulation of the adenomatous polyposis coli 2 (APC2) that is expressed in different cell lines and is a tumor suppressor gene (Rashid et al., 2017). Thus, the reduced levels of APC2 may result in an increased expression of the Wnt/β-catenin signaling pathway genes, and promotes cell proliferation. Another interesting up-regulated gene is the BCO2 that encodes for a β-carotene-9′, 10′-oxygenase 2 (Figure 5A). The increased expression of BCO2 may confer and increase survival since its knockdown causes an increment in apoptosis due to its protective role to oxidative stress (Ref: Lei woo, Exp Biol Med [Maywood], 2016). Also, we have the up-regulation of the EFNA1 gene that codes for the Ephrin A1 protein, which is a member of the A-type ephrin family and has been implicated as a negative inductor of apoptosis (Ref: Spencer Alford, Cancer Cell Int. 2010). The up-regulation of the genes miR-3648, BCO2, and EFNA1 by the T5 compound could be associated with its lower cytotoxicity.
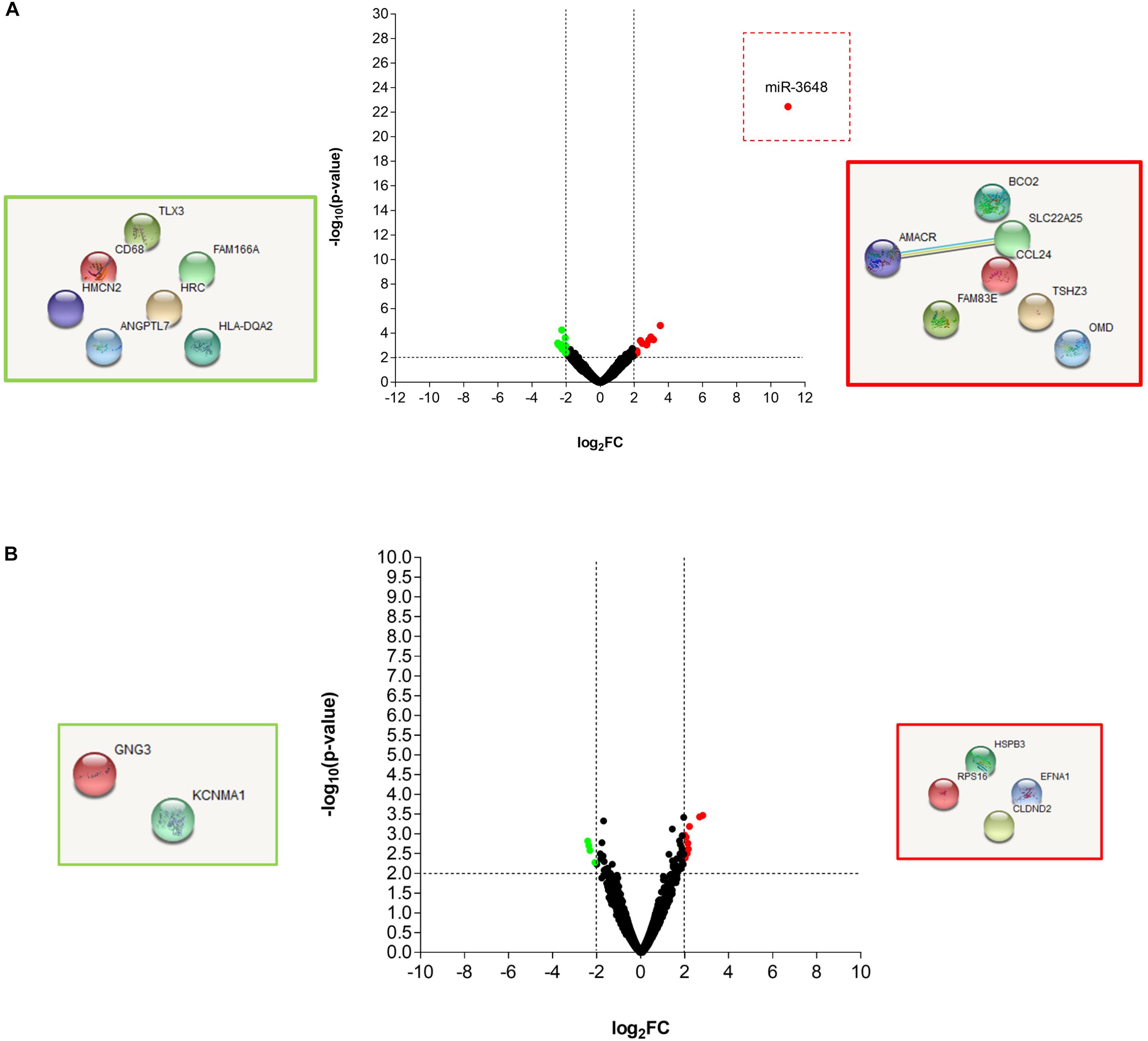
Figure 5. RNA-seq analysis of the THP-1 macrophages Mtb infected, and treated with the T5 compound. (A) Volcano plot and STRING analysis of the up-regulated and down-regulated genes at 4 h with a log2FC > 2.0 or log2FC < 2.0 and a p-value < 0.05. (B) Volcano plot and STRING analysis of the up-regulated and down-regulated genes at 20 h with a log2FC > 2.0 or log2FC < 2.0 and a p-value < 0.05.
Transcriptome of Mtb Treated With the T5 Compound Affects the Cholesterol and Folic Acid Pathways
While the transcriptome of the macrophage remained mostly unaffected by the treatment of the T5 compound, the Mtb transcriptome showed a significant number of up-regulated and down-regulated genes (Figures 6A,B). To find out which were the most affected pathways in Mtb under the treatment of the T5 compound, we used the Mtb pathway/genome from the BioCyc database collection1 (version 23.1). Using this approach, we found a significant down-regulation of enzymes involved in the cholesterol and folic acid pathways like the para-aminobenzoate synthase (pabB) (Figures 7A,B). In addition, we found a significant up-regulation of the ammonium-transport integral protein (Amt), which is important for detoxification of the bacteria from the accumulation of ammonia (Figure 7C).
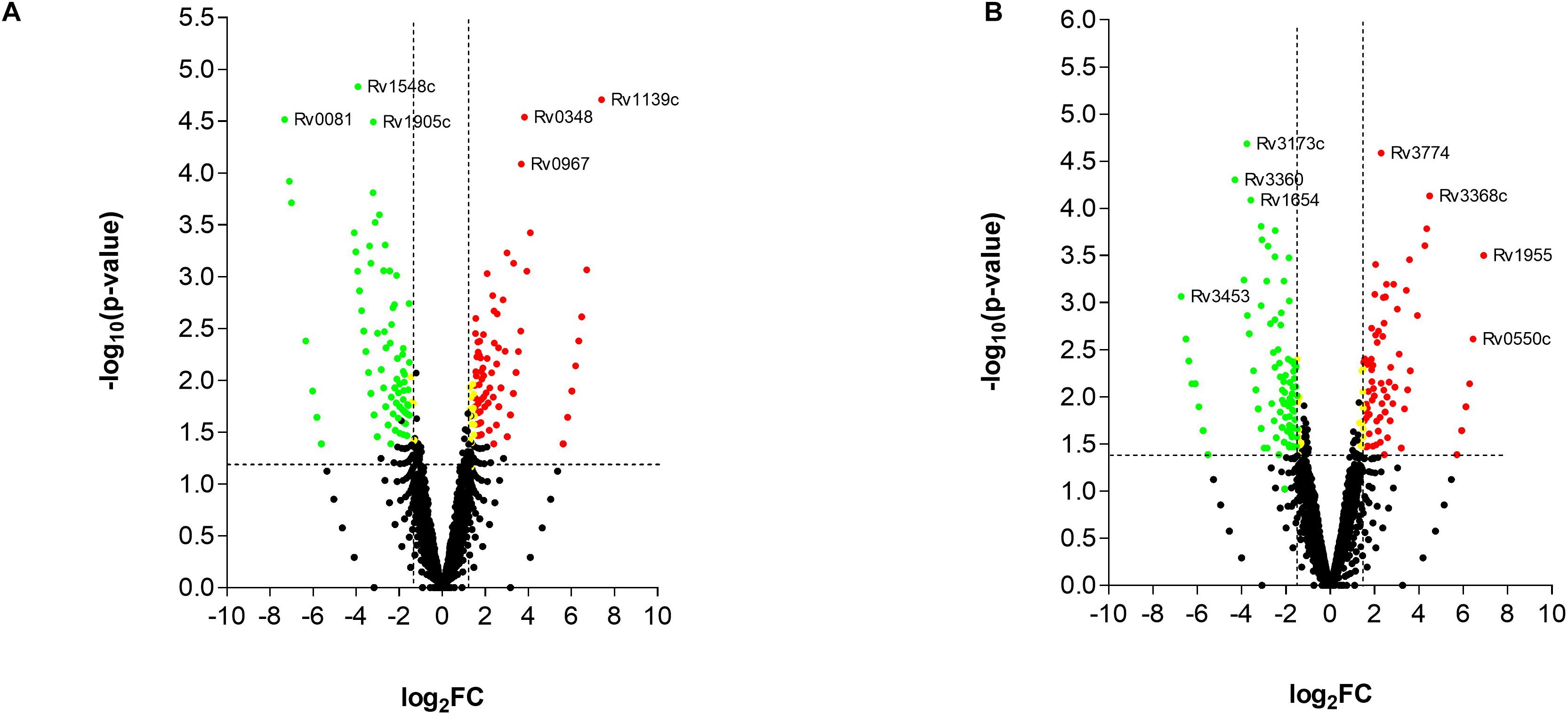
Figure 6. RNA-seq analysis of Mtb after the treatment with the T5 compound. (A) Volcano plot analysis of the up-regulated and down-regulated genes at 4 h with a log2FC > 2.0 or log2FC < 2.0 and a p-value < 0.05. (B) Volcano plot analysis of the up-regulated and down-regulated genes at 20 h with a log2FC > 2.0 or log2FC < 2.0 and a p-value < 0.05.

Figure 7. T5 compound affect the cholesterol and folic acid pathways in Mtb. (A) Shows the main enzymes and reactions of the cholesterol degradation pathway in Mtb that were significantly affected by the treatment of the T5 compound using the BioCyc database Collection (https://biocyc.org/organism-summary? object=MTBH37RV, version 23.1). (B) Shows the main enzymes and reactions of the folic acid synthesis pathway that were significantly affected by the treatment of the T5 compound using the BioCyc database. (C) Shows the up-regulation of the ammonium-transport integral protein (Amt) in response to the treatment with the T5 compound using the BioCyc database. The data in (A,B) are from the 4 h and in (C) from 20 h transcriptome.
The Compound T5 Reduces the Mtb CFUs in the Mouse Model
To validate the anti-mycobacterial activity of the T5 compound, we did a pilot in vivo experiment using Balb/C mice. In this experiment we observed that the T5 compound cause a significant 2–3 fold reduction of the Mtb CFUs in the lungs as well as in the spleen of the treated mice, as compared to the control vehicle treated group (Figures 8A,B). We did not observe significant differences in weight of the animals at the end of the treatment and 15 days post-infection between the T5 treated mice and the control group (Figure 8C). In summary, these findings suggest that this T5 compound is not only active in vitro but also in vivo.
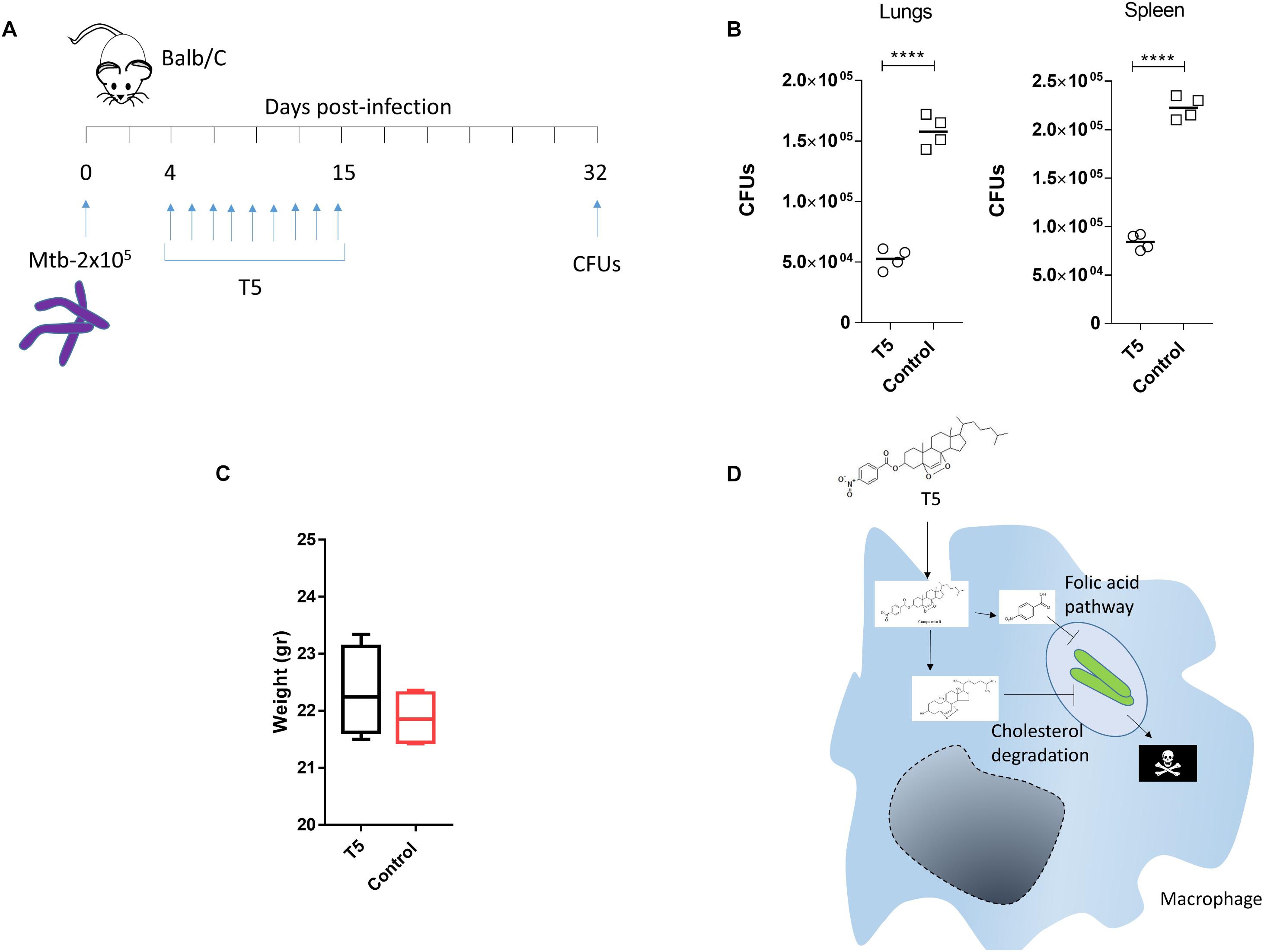
Figure 8. The T5 compound has anti-mycobacteral activity in vivo. (A) The diagram shows the treatment scheme for the T5 compound and the CFU determination time points. Four mice per group were infected with 2 × 105 bacteria intra peritoneal (i.p) in 200 μL of PBS with 0.05% Tween 80. (B) Quantification of the CFUs in spleen and lungs of the T5 treated Balb/C mice after 32 days. (C) Body weight of Balb/C mice at the end of drug treatment 15 days post-infection. (D) Diagram showing a model of the possible mechanism of action of the T5 compound against Mtb. Unpaired t-test ****p-value < 0.0001. Representative of two independent experiments.
Discussion
One difficulty in TB drug discovery arises from a limited knowledge of the physiological environment and growth conditions that Mtb experienced during infection. A rising in drug-resistant TB is a major impediment to successfully deal with this disease. The WHO recognizes antimicrobial resistance as one of the most demanding global threats to cure TB. Drug resistance is probably a consequence of overuse of antibiotics in the treatment of humans or lack of adherence to the treatment. When a new antibiotic is introduced, it can have great results for a period of time, but then the bacteria gradually adapt and the antibiotic becomes less effective. This is why we need to maintain a constant search for new effective antibiotics that target different pathways in an organism like Mtb.
Our results define the activity of a new compound called T5, which has anti-mycobacterial activity against Mtb, specifically when the bacteria locate inside the macrophages, but surprisingly not in the extracellular environment. We hypothesize that this T5 compound may have a combinatorial effect in which the conjugation may help their entry into cells, likely due to the requirement of Mtb to utilize cholesterol. Once inside of the bacteria, an enzymatic reaction, probably mediated by a macrophage specific esterase, could release the ANB part from the T1 compound, and then the ANB may interfere with the enzymes of the folic acid pathway. In addition, the released T1 part of the T5 compound may simultaneously affect the enzymes of the cholesterol degradation pathways (Figure 8D).
Even though anti-folate drugs have been very successful as anticancer and antimicrobial agents, they are poorly effective in the TB therapy (Minato et al., 2015a; Hajian et al., 2019). The ineffectiveness of these type of drugs is probably due to the lack of permeability to them in Mtb (Nixon et al., 2014). Moreover, anti-folates have a past history of use in TB therapy especially through the use of the drug para-aminosalicylic-acid (PAS) that worked as a bacteriostatic anti-tubercular agent. Finding a compound that is both permeable and effective to inhibit this folate pathway would be highly desirable. On the other hand, the cholesterol catabolic pathway is an important therapeutic target in Mtb in which many of its genes are involved in pathogenesis. This has been demonstrated by the mutation or deletion of these genes and the attenuation of the bacteria. The cholesterol catabolic pathway in Mtb is accomplished in two major lanes, first by the degradation of the aliphatic side chain and second by the degradation of the sterol A-D rings. A compound that could disrupt the two major steps in the degradation of cholesterol in Mtb could be a great finding.
It is important to mention that spontaneous resistance to the ANB part of the T5 compound, which is similar to the PAS compound, could emerge via multiple mechanisms that include limited bio-activation within the folate synthesis pathway, efflux pumps, specific mutations like in the pabB gene, and an insufficient accumulation within the bacilli (Minato et al., 2015a). We do not have information about spontaneous drug-resistant mutation for the epidioxy-sterol-T1 part of the molecule. However, it is interesting that a compound like the T5 compound that acts over different metabolic pathways is less likely to encounter resistance in the same bacteria, which could be advantageous for this compound in the fight against Mtb.
The up-regulation of the ammonium transporter could be a consequence of the nitro group present in the ANB part of the T5 compound. This ANB could behave similarly to a nitroimidazole compound releasing nitric oxide (NO) and converted into ammonium by the bacteria, which has to be shuttled out to avoid toxicity. The activity that we observed for the T5 compound in vivo is significant but not as effective if we compared the results with drugs like Rifampicin; however, we know that the T5 compound is highly hydrophobic and may require a better vehicle in order to obtain more effective results in future animal experiments.
Conclusion
We found a new compound called T5 with a good anti-mycobacterial activity in vivo and in vitro which may be functioning as a simultaneous inhibitor of the folate and cholesterol pathways.
Data Availability Statement
The RNA-seq transcriptomic data was deposited in https://www.ncbi.nlm.nih.gov/bioproject: DMSO 4 h, BaenaRNA1_1.fastq.gz (SRR11195990); DMSO 20 h, BaenaRNA3_1.fastq.gz (SRR11195989); T5 4 h, BaenaRNA2_1.fastq.gz (SRR11195988); and T5 20 h, BaenaRNA4_1.fastq.gz (SRR11195987).
Ethics Statement
The animal study was reviewed and approved by the animal ethics committee of the Universidad de Antioquia.
Author Contributions
AB, JA, AM, and LB conceived and designed the experiments. AB, EV, and MP performed the experiments. AB, JA, MP, and EV analyzed the data. AB, LB, MP, JA, and AM wrote and revised the manuscript. All the authors read and approved the manuscript.
Funding
We want to acknowledge our funding source CODI Universidad de Antioquia 2015-6726.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge “Estrategia de Sostenibilidad” Universidad de Antioquia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.537935/full#supplementary-material
FIGURE S1 | Compound library and T5 NMR. (A) Reaction to produce the T5 compound. (B) 1H NMR spectrum of compound T5. (C) 13C NMR spectrum of compound T5. (D) 1H- 13C HMBC 2D NMR correlation spectroscopy of compound T5. (E) 1H- 13C HMBC 2D NMR correlation spectroscopy of compound T5. (F) Inhibitory GFP curves for the compounds T2–T4 and T6–T15 after 48 h of treatment post-infection.
FIGURE S2 | CFUs of the Mtb after the THP-1 macrophage lysis. The THP-1 macrophages were lysed and 5 μl of bacteria were plated for each dilution in 7H10 media for 23 days. The plates were divided in 4 quadrants and labeled clockwise with the dilution (Undiluted [1], 10–1, 10–2, 10–3), starting with upper left quadrant.
FIGURE S3 | RNA quality of the samples used for the RNA-seq. (A) Agarose gel showing the integrity of the ribosomal RNA bands. (B) RNA integrity number (RIN) obtained for each sample. (C) Showing the bio-analyzer electropherograms for each sample.
FIGURE S4 | Cell viability assay quantification. (A) The cell death phenotypes encountered after the treatment with the different compounds. (B) Example of the cell death quantification procedure that we have used to generate the viability data in Figure 4B.
TABLE S1 | THP-1 Macrophage transcriptome.
TABLE S2 | Mtb-H37Rv transcriptome.
Footnotes
References
Aguilar-Ayala, D. A., Tilleman, L., Van Nieuwerburgh, F., Deforce, D., Palomino, J. C., Vandamme, P., et al. (2017). The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 7:17665.
Anonymous. (2019). Antibiotic susceptibility diagnostics for the future. Nat. Microbiol. 4:1603. doi: 10.1038/s41564-019-0577-4
Baena, A., Cabarcas, F., Alvarez-Eraso, K. L. F., Isaza, J. P., Alzate, J. F., Barrera, L. F., et al. (2019). Differential determinants of virulence in two Mycobacterium tuberculosis colombian clinical isolates of the LAM09 family. Virulence 10, 695–710. doi: 10.1080/21505594.2019.1642045
Baena, A., and Porcelli, S. A. (2009). Evasion and subversion of antigen presentation by Mycobacterium tuberculosis. Tissue Antigens 74, 189–204. doi: 10.1111/j.1399-0039.2009.01301.x
Burki, T. (2019). BPaL approved for multidrug-resistant tuberculosis. Lancet Infect. Dis. 19, 1063–1064. doi: 10.1016/s1473-3099(19)30489-x
Bussi, C., and Gutierrez, M. G. (2019). Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol. Rev. 43, 341–361. doi: 10.1093/femsre/fuz006
Cateni, F., Doljak, B., Zacchigna, M., Anderluh, M., Piltaver, A., Scialino, G., et al. (2007). New biologically active epidioxysterols from Stereum hirsutum. Bioorg. Med. Chem. Lett. 17, 6330–6334. doi: 10.1016/j.bmcl.2007.08.072
Duarte, N., Ferreira, M. J., Martins, M., Viveiros, M., and Amaral, L. (2007). Antibacterial activity of ergosterol peroxide against Mycobacterium tuberculosis: dependence upon system and medium employed. Phytother. Res. 21, 601–604. doi: 10.1002/ptr.2119
Ehrt, S., and Schnappinger, D. (2009). Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell Microbiol. 11, 1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x
Farshori, N. N., Banday, M. R., Zahoor, Z., and Rauf, A. (2010). DCC/DMAP mediated esterification of hydroxy and non-hydroxy olefinic fatty acids with beta-sitosterol: in vitro antimicrobial activity. Chin. Chem. Lett. 21, 646–650. doi: 10.1016/j.cclet.2010.01.003
Feng, K., Wu, L. Z., Zhang, L. P., and Tung, C. H. (2007). IRA-200 resin-supported platinum(II) complex for photooxidation of olefins. Tetrahedron 63, 4907–4911. doi: 10.1016/j.tet.2007.03.148
Fieweger, R. A., Wilburn, K. M., and VanderVen, B. C. (2019). Comparing the metabolic capabilities of bacteria in the Mycobacterium tuberculosis complex. Microorganisms 7:177. doi: 10.3390/microorganisms7060177
Giampaglia, C. M., Martins, M. C., Chimara, E., Oliveira, R. S., de Oliveira Vieira, G. B., Marsico, A. G., et al. (2007). Differentiation of Mycobacterium tuberculosis from other mycobacteria with rho-nitrobenzoic acid using MGIT960. Int. J. Tuberc. Lung. Dis. 11, 803–807.
Griffin, J. E., Pandey, A. K., Gilmore, S. A., Mizrahi, V., McKinney, J. D., Bertozzi, C. R., et al. (2012). Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227. doi: 10.1016/j.chembiol.2011.12.016
Gygli, S. M., Borrell, S., Trauner, A., and Gagneux, S. (2017). Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 41, 354–373. doi: 10.1093/femsre/fux011
Hajian, B., Scocchera, E., Shoen, C., Krucinska, J., Viswanathan, K., G-Dayanandan, N., et al. (2019). Drugging the folate pathway in Mycobacterium tuberculosis: the role of multi-targeting agents. Cell Chem. Biol. 26, 781.e6–791.e6.
Joshi, S. M., Pandey, A. K., Capite, N., Fortune, S. M., Rubin, E. J., Sassetti, C. M., et al. (2006). Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl. Acad. Sci. U.S.A. 103, 11760–11765. doi: 10.1073/pnas.0603179103
Lange, C., Dheda, K., Chesov, D., Mandalakas, A. M., Udwadia, Z., Horsburgh, C. R., et al. (2019). Management of drug-resistant tuberculosis. Lancet 394, 953–966.
Larrouy-Maumus, G., Marino, L. B., Madduri, A. V. R., Ragan, T. J., Hunt, D. M., Bassano, L., et al. (2016). Cell-envelope remodeling as a determinant of phenotypic antibacterial tolerance in Mycobacterium tuberculosis. ACS Infect. Dis. 2, 352–360. doi: 10.1021/acsinfecdis.5b00148
Laws, M., Shaaban, A., and Rahman, K. M. (2019). Antibiotic resistance breakers: current approaches and future directions. FEMS Microbiol. Rev. 43, 490–516. doi: 10.1093/femsre/fuz014
Lee, J., Lee, S., Kim, K. K., Lim, Y., Choi, J., Cho, S., et al. (2019). Characterisation of genes differentially expressed in macrophages by virulent and attenuated Mycobacterium tuberculosis through RNA-Seq analysis. Sci. Rep. 9:4027.
Lopez-Agudelo, V. A., Baena, A., Ramirez-Malule, H., Ochoa, S., Barrera, L. F., Ríos-Estepa, R., et al. (2017). Metabolic adaptation of two in silico mutants of Mycobacterium tuberculosis during infection. BMC Syst. Biol. 11:107. doi: 10.1186/s12918-017-0496-z
Mashabela, G. T., de Wet, T. J., and Warner, D. F. (2019). Mycobacterium tuberculosis metabolism. Microbiol. Spectr. 7:a021121.
McShane, H. (2019). Insights and challenges in tuberculosis vaccine development. Lancet Respir. Med. 7, 810–819. doi: 10.1016/s2213-2600(19)30274-7
Minato, Y., Thiede, J. M., Kordus, S. L., McKlveen, E. J., Turman, B. J., Baughn, A. D., et al. (2015a). Mycobacterium tuberculosis folate metabolism and the mechanistic basis for para-aminosalicylic acid susceptibility and resistance. Antimicrob. Agents Ch. 59, 5097–5106. doi: 10.1128/aac.00647-15
Miner, M. D., Chang, J. C., Pandey, A. K., Sassetti, C. M., and Sherman, D. R. (2009). Role of cholesterol in Mycobacterium tuberculosis infection. Indian J. Exp. Biol. 47, 407–411. doi: 10.1007/978-981-32-9413-4_22
Mshana, R. N., Tadesse, G., Abate, G., and Miorner, H. (1998). Use of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide for rapid detection of rifampin-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 36, 1214–1219. doi: 10.1128/jcm.36.5.1214-1219.1998
Nixon, M. R., Saionz, K. W., Koo, M. S., Szymonifka, M. J., Jung, H., Roberts, J. P., et al. (2014). Folate pathway disruption leads to critical disruption of methionine derivatives in Mycobacterium tuberculosis. Chem. Biol. 21, 819–830. doi: 10.1016/j.chembiol.2014.04.009
Nunn, A. J., Phillips, P. P. J., Meredith, S. K., Chiang, C., Conradie, F., Dalai, D., et al. (2019). A trial of a shorter regimen for rifampin-resistant tuberculosis. N. Engl. J. Med. 380, 1201–1213.
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Pontali, E., Raviglione, M. C., and Migliori, G. B. the writing group members of the Global Tb Network Clinical Trials Committee (2019). Regimens to treat multidrug-resistant tuberculosis: past, present and future perspectives. Eur. Respir. Rev. 28:190035. doi: 10.1183/16000617.0035-2019
Queval, C. J., Brosch, R., and Simeone, R. (2017). The macrophage: a disputed fortress in the battle against Mycobacterium tuberculosis. Front. Microbiol. 8:2284. doi: 10.3389/fmicb.2017.02284
Rashid, F., Awan, H. M., Shah, A., Chen, L., and Shan, G. (2017). Induction of miR-3648 Upon ER stress and its regulatory role in cell proliferation. Int. J. Mol. Sci. 18:1375. doi: 10.3390/ijms18071375
Russell, D. G. (2011). Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol. Rev. 240, 252–268. doi: 10.1111/j.1600-065x.2010.00984.x
Saludes, J. P., Garson, M. J., Franzblau, S. G., and Aguinaldo, A. M. (2002). Antitubercular constituents from the hexane fraction of Morinda citrifolia Linn. (Rubiaceae). Phytother. Res. 16, 683–685. doi: 10.1002/ptr.1003
VanderVen, B. C., Fahey, R. J., Lee, W., Liu, Y., Abramovitch, R. B., Memmott, C., et al. (2015). Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog. 11:e1004679. doi: 10.1371/journal.ppat.1004679
Zhou, H., Zhao, L., Li, W., Yang, Y., Xu, L., Ding, Z., et al. (2015). Anti-Mycobacterium tuberculosis active metabolites from an endophytic Streptomyces sp YIM65484. Rec. Nat. Prod. 9, 196–200.
Keywords: antimicobacterial, macrophages, cholesterol pathway, folic acid pathway, Mycobacterium tuberculosis
Citation: Baena A, Vasco E, Pastrana M, Alzate JF, Barrera LF and Martínez A (2020) New Conjugated Compound T5 Epidioxy-Sterol-ANB Inhibits the Growth of Mycobacterium tuberculosis Affecting the Cholesterol and Folate Pathways. Front. Microbiol. 11:537935. doi: 10.3389/fmicb.2020.537935
Received: 25 February 2020; Accepted: 13 August 2020;
Published: 10 September 2020.
Edited by:
Maria Rosalia Pasca, University of Pavia, ItalyReviewed by:
Vikram Saini, All India Institute of Medical Sciences, IndiaEvgeniya V. Nazarova, Genentech, United States
Copyright © 2020 Baena, Vasco, Pastrana, Alzate, Barrera and Martínez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andres Baena, YW5kcmVzLmJhZW5hZ0B1ZGVhLmVkdS5jbw==
 Andres Baena
Andres Baena Emanuel Vasco
Emanuel Vasco Manuel Pastrana2
Manuel Pastrana2 Juan F. Alzate
Juan F. Alzate Luis F. Barrera
Luis F. Barrera Alejandro Martínez
Alejandro Martínez