
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 16 September 2020
Sec. Systems Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.537783
 Valentina Mazzurco Miritana1
Valentina Mazzurco Miritana1 Giulia Massini1,2*
Giulia Massini1,2* Andrea Visca1
Andrea Visca1 Paola Grenni1
Paola Grenni1 Luisa Patrolecco3
Luisa Patrolecco3 Francesca Spataro3
Francesca Spataro3 Jasmin Rauseo3
Jasmin Rauseo3 Gian Luigi Garbini1
Gian Luigi Garbini1 Antonella Signorini2
Antonella Signorini2 Silvia Rosa2
Silvia Rosa2 Anna Barra Caracciolo1
Anna Barra Caracciolo1Anaerobic digestion (AD) treatment of cattle manure and slurry makes it possible to produce biogas, a renewable and storable biofuel, as well as digestate, a residual organic matter that can be used to replace chemical fertilizers. On the other hand, the intense use of antibiotics (e.g., sulfamethoxazole) in animal husbandry practices is showing increasing negative impacts resulting from the release of still metabolically active molecules into agroecosystems. In the present study, cattle manure collected from an AD plant-feeding tank was used as feedstock for AD experiments in which some batches were spiked with 5 mg L–1 of sulfamethoxazole (SMX). Adding the antibiotic affected the microbial community dynamic; in particular, the efficiency of the acidogenic and acetogenic phases of the process corresponded to higher CH4 and H2 production than in the control. SMX was also degraded, and at the end of the experiment (69 days), just 20% of its initial concentration was found. The relative abundance (ARG/16S) of resistance genes sul1, sul2, and the proxy intI1 initially found in the ingestate decreased during the AD in both the spiked and control batches, suggesting that this process lowers the likelihood of antibiotic resistance genes spreading.
Anaerobic digestion (AD) is an efficient process that transforms organic waste, e.g., agriculture and breeding residues, into renewable energy (biogas) and digestate (Guo et al., 2015; Sui et al., 2017; European Biogas Association, 2018; Styles et al., 2018). The latter, containing considerable amounts of plant-essential macro- and micro-nutrients, is a good alternative to chemical fertilizers (Bustamante et al., 2012; Barra Caracciolo et al., 2015; Bharathiraja et al., 2018).
Other advantages of AD are the significant reduction of pathogens in the digestate, a complete recycling of organic waste, and mitigation of global CO2 emissions, in line with the circular economy approach (Pampillón-González et al., 2017). In Italy, digestate is commonly used as a fertilizer, and its application in agriculture is regulated by the Inter-Ministerial Decree No. 5046/2016, which lists some parameters (e.g., total nitrogen and phosphorous, Pb, Cd, Ni, Zn, Cu, Hg, and Cr) to be checked. Recently, the EU Fertilizing Products Regulation (EU 2019/1009) has been proposed for promoting digestate use over all Europe. However, the possible presence of emerging contaminants, such as antibiotic residues, has not been considered so far. In fact, the environmentally friendly application of digestate on agricultural soil does not exclude the likelihood of introducing antibiotics, such as sulfamethoxazole (SMX); the latter is commonly used in livestock production worldwide (Rauseo et al., 2019).
Antibiotic residues are found in animal waste (Widyasari-Mehta et al., 2016), and consequently, antibiotic-resistant bacteria and antibiotic resistance genes (ARGs) can enter farm digesters, where their effect and fate are not well known. The AD process relies on a complex microbial community, in which different functional groups cooperate in sequential metabolic steps with a final production of methane-rich biogas (Madigan et al., 2014). Antibiotics are reported to reduce microbial activities or influence organic matter degradation during biogas production (Ji et al., 2013; Grenni et al., 2018). Among the functional microbial groups acting in AD, methanogens, which have the slowest growth rates, are the most sensitive to any changes in process conditions (Bharathiraja et al., 2018). They control the last and critical step of AD, and the proportion of Bacteria/Archaea is related to the process efficiency (Ferraro et al., 2019). Some research evaluated the effect of SMX on AD performance: a concentration of 500 mg L–1 produced a complete inhibition of methane production (Cetecioglu et al., 2012), and lower amounts (45–50 mg L–1) caused high accumulation of volatile fatty acids (VFAs) and a consequent lowering of the pH, thus altering the AD process efficiency (Aydin et al., 2015). Lower antibiotic concentrations (ranging from 1 to 10 mg L–1) only partially impacted the microbial community and AD process (Cetecioglu et al., 2012).
Other research demonstrates that SMX can be degraded during AD, depending on the specific experimental conditions and its initial concentration (Mohring et al., 2009; Cetecioglu et al., 2015; Cheng et al., 2018). Feng et al. (2017) report that antibiotic residues (e.g., SMX 1 μg L–1) were effectively removed during the AD of sludge and manure.
The selection of sulfonamide-resistant bacteria and spread of ARGs (Sköld, 2000; Enne et al., 2002) (sul1, sul2, sul3) have been demonstrated in various research on effluents, reclaimed water, and manure (Baquero et al., 2008; Martinez, 2009; Mckinney et al., 2018; Rauseo et al., 2019). However, there has been little published research on digestate, and the results have been quite different (Tasho and Cho, 2016; Grenni et al., 2018). Some authors find that most ARGs are diminished during the AD process except sul2 (Polesel et al., 2016; Zhang et al., 2018). On the other hand, other authors report that mesophilic AD was able to remove ARGs, including sul1 and sul2, while other ARGs increased (Ma et al., 2011). In this work, the possible effects of SMX on the dynamics of the AD process and H2 and CH4 production were investigated in batch mode by testing manure samples collected from an AD plant-feeding tank and adding the antibiotic SMX at a final concentration of 5 mg L–1. For this purpose, cattle manure was used testing the AD process as both the substrate and inoculum in the AD experiment. Changes in the microbial community structure [fluorescence in situ hybridization (FISH) and next-generation sequence (NGS) techniques]; occurrence of sul1, sul2, and intI1 genes; and antibiotic degradation were also investigated.
The organic input for the AD plants consists of cattle manure partially digested in the herbivorous ruminal ecosystem, and for this, it can be named “ingestate.”
The ingestate was collected from the feed pipe of a full-scale continuous stirred-tank reactor plant located on a beef and dairy cattle farm (Lazio, Italy). It was mainly composed of cow manure with a minor fraction (8–10%) of food waste (manager’s communication). The ingestate was mixed, and glass bottles of 2 L each (three replicates) were filled and transported to the laboratory where the batch experiment was immediately set-up. At the same time, some ingestate aliquots (three replicates) were used for its characterization in terms of total and volatile solids (g L–1), pH, organic carbon, total nitrogen, water content (%), and microbiological abundance (N. cells mL–1). These data are reported in Table 1. Moreover, the initial SMX concentration was determined.
The experiment was set-up in batch mode using six Pyrex reactors of 600 mL each (three replicates for each experimental condition) for evaluating the AD performance of the ingestate collected from an AD plant-feeding tank and of the latter with SMX. Each reactor was filled with 300 mL of the ingestate (corresponding to 31 g VS) without pretreatment (e.g., no filtration, no dilution) in order to mimic a full-scale biogas plant condition. The antibiotic SMX was added to three batches (SMX batches). For this purpose, we dissolved SMX (SMX purity 99%, Sigma-Aldrich S7507) powder in a methanol/MilliQ solution (stock solution); then, we further diluted the stock solution in ultrapure water. The final amount of methanol in each batch was very low (0.18 in 300 mL of working volume).
Aliquots of the SMX solution were added to each batch in order to reach a final concentration of 5 mg L–1. This concentration was used because, in other works (Cetecioglu et al., 2012), it did not completely inhibit the AD, and it was also close to a concentration found in cattle manure in another previous research (Rauseo et al., 2019).
The other three batch replicates were considered controls. All the batches were sealed with rubber stoppers and metal rings and then flushed with N2 (10′) to establish an anoxic environment. The experiment was carried out in mesophilic (37–38°C) and orbital shaking (100 rpm) conditions. Biogas and organic acid and microbial abundance analyses were performed over the experimental time (69 days). The sampling frequency of the gas and the liquid medium varied in line with the evolution in the AD process. During the first 6 days, the biogas measurements were performed daily. For the next 2 weeks, the measurements were performed every 3 days and then weekly. The experiment ended when no more CH4 had been produced for 2 weeks. At selected times (5, 10, 15, 21, and 69 days), slurry aliquots were sampled for SMX determinations and evaluation of the antibiotic effect on the structure of the microbial community. The latter was characterized using both the fluorescence in situ hybridization and next-generation sequencing methods. Moreover, the 16S rRNA gene, the resistance genes sul1 and sul2, and the class 1 integron-integrase gene intl1 were also quantified by quantitative PCR (qPCR).
The total biogas volume was measured using a water displacement system (Martínez-Sibaja et al., 2011). The biogas composition in the headspace of the reactors was analyzed using a gas chromatograph (Focus GC, by Thermo Fisher Scientific, United States) equipped with a thermal conductivity detector (TCD) and a 3-m stainless steel column packed with Hayesep Q (800/100 mesh). Nitrogen gas was used as a carrier at a flow rate of 35 mL min–1. The temperature of the column and of the injector was 120°C and that of the TCD was 200°C. The cumulative H2 and CH4 productions were calculated using the Logan equation (Logan et al., 2002). The metabolic products of fermentation, such as acetic, butyric, propionic, lactic, succinic, and formic acids, were analyzed with a high-performance liquid chromatograph (Thermo Spectra System, United States) equipped with both a UV detector (l1/4 210 nm) and a refractive index detector. The column, a 300 mm × 7.8 mm Rezex ROA-Organic Acid Hþ (8%) (Phenomenex, United States) with a 4 × 30 mm security guard cartridge Carbo-H (Phenomenex, United States), was operated at 75°C, using a solution of 5 mN H2SO4 as the mobile phase (flow rate, 0.5 mL min–1). The liquid samples were diluted 1:10 in H2SO4 5 mN and filtered with a 0.22-μm membrane before injection into the HPLC.
Sulfamethoxazole was extracted by pressurized liquid extraction (PLE) using the Thermo Scientific Dionex, United States, ASETM 150 (accelerated solvent extractor) system as described in Rauseo et al. (2019). Briefly, each sample (2 g) was mixed and homogenized with inert diatomaceous earth (Thermo 062819) to completely fill the PLE cell. The extraction solvent was a mixture of water: methanol (50:50, v/v). The final extract (about 20 mL volume) was then diluted with pure water to reduce the methanol content to below 5% and was purified following the solid phase extraction (SPE) procedure as reported in detail in a previous work (Rauseo et al., 2019). SMX was eluted from the SPE cartridges (Waters Oasis hydrophilic–lipophilic balance, HLB cartridges, 6 mL, 1 g) using 2 × 1.5 mL methanol-ethyl acetate (1:1 v/v) and 2 × 1.5 mL methanol containing 1% ammonia. The eluates were evaporated under a gentle nitrogen stream at room temperature to reach a final volume of about 50 μL and then reconstituted with the mobile phase used for the chromatographic analysis.
Sulfamethoxazole in the purified extracts was analyzed by using high-performance liquid chromatography (HPLC, column oven mod. LC-100 and micro Pump Series 200, Perkin Elmer, United States) equipped with a 7125 Rheodyne injector valve with a 20-μL loop and interfaced to a triple quadrupole mass spectrometer detector connected with an electrospray ionization detector (MS-MS, mod. API 3000, AB Sciex, Germany). The chromatographic Gemini column (150 × 4.6 mm, 5 μm RP C 18, Phenomenex, France), preceded by a guard column packed with the same stationary phase, was maintained at 25°C. The elution was at a 1.0 mL min–1 flow rate. The mobile phase was composed of acidified MeOH (0.1% formic acid–phase A), acidified ultrapure water (0.1% formic acid–phase B), and acetonitrile (ACN–phase C) of an HPLC grade. The elution was in an isocratic mode (phases A, B, C: 33, 33, 34%, respectively). The MS/MS detector was set in a positive ESI mode, and the source temperature was set at 400°C. The ionspray voltage was +5 kV, and the nebulizer and curtain gas were at 14 and 12 units, respectively. Nitrogen was used as both a collision and drying gas. Declustering, focusing, and entrance potential were set at 31.2, 75, and 9 V, respectively. Multiple reaction monitoring detection was based on the precursor (254.2 m/z) and product ion transitions (92.0, 108.1, and 156.1 m/z) and on the comparison of the retention time (2.23 min.).
The calibration curve for the target compound was obtained by analyzing working standard solutions at the following concentrations: (0.1, 0.25, 0.5, 1.0, 2.5, 5.0, and 7.5 mg L–1). The linearity was confirmed with an R2 ≥ 0.98 for the concentration range investigated. Deuterated SMX (SMX–d4, Clearsynth) was used as the internal standard in the calibration procedure and to compensate for possible matrix effects.
In order to check the performance of both the extraction/purification and analytical procedures, SMX recoveries from the ingestate matrix, artificially spiked at different concentrations of SMX (1.0, 5.0, and 7.5 mg L–1), were determined in triplicate. For each final concentration, triplicated spiked samples were prepared. SMX detection was performed by subtracting the blank level of the ingestate non-spiked sample. SMX recoveries ranged between 85 and 95%, and the overall repeatability (n = 3) of the extraction procedure was satisfactory (relative standard deviation value ranging from 1.5 to 7.8%). The limit of detection (LOD) was calculated in accordance with the IUPAC method (Thompson et al., 2002) and was 1.5 μg L–1. The quantification limits were set at three times the LOD.
Total microbial abundance (N. cells mL–1) was measured with the epifluorescence direct count method, using DAPI (4′,6-diamidino-2-phenylindole) as a DNA fluorescent intercalant. Slurry samples (1 g for three replicates) were collected and transferred to a test tube containing 9 mL of a fixing solution (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, 2% formaldehyde (v/v), 0.5% Tween 20 (v/v), and 100 mM sodium pyrophosphate) and processed as described in detail in a previous work (Barra Caracciolo et al., 2005a). The microscope used was a Zeiss epifluorescence microscope AXIOSKOP 40 (Carl Zeiss, Germany) equipped with a ZEISS HXP 120v light source and 1000× magnification.
Samples were collected from each experimental reactor and fixed with a formalin solution (4% w/v, 6 h at 4°C) as reported in Pernthaler et al. (2001). After being rinsed, the pellets were resuspended in an ethanol (96%) and PBS (1×) solution and stored until use (−20°C).
Before the FISH analysis, a cell-extraction procedure was performed to detach and separate cells from inorganic particles as described in a previous work (Barra Caracciolo et al., 2005a).
The FISH analysis was carried out as described in detail in Amann et al. (1995) and Barra Caracciolo et al. (2005b). In particular, each sample (three replicates for each condition) was collected onto a membrane filter (white polycarbonate filters, pore size 0.22 μm, diameter 25 mm, Millipore, United States). Each filter was cut into sections, and at least two of them were hybridized with the same probe. Hybridization buffers (5M NaCl, 1M Tris/HCl, 100 g L–1 sodium dodecyl sulfate and a specific amount of formamide) with each specific probe were prepared. Oligonucleotide probes for the Bacteria (EUB338 I, II, III) and Archaea (ARC915) domains were used (Greuter et al., 2016). Moreover, the LGC354 II, III, SRB385, MB1174 probes were also used for the detection of Firmicutes, Sulfate-Reducing Bacteria (SRB) (class Deltaproteobacteria) and methanogens (Methanobacteriales), respectively. These are key microbial taxa involved in the AD process. All oligonucleotide probes (50 ng μL–1), labeled with carboxyfluorescein (FAM) or indocarbocyanine (Cy3) dye at the 5′-end, were purchased from MWG AG Biotech, Germany. The filters were mounted on microscope slides and kept in a hybridization chamber for 90 min at 46°C. The filters were then washed in a buffer at 48°C for 15 min, rinsed with distilled water and air-dried. Finally, the slides were mounted with a few drops of VectaShield (Vector Laboratories, United States) and DAPI in order to estimate the proportion of cells targeted by each specific probe out of the total cells. The slides were viewed using the abovementioned Zeiss epifluorescence microscope. The micrographs were obtained using the AxioVision Release 4.6.3 program. The results are expressed as the percentage of DAPI cells that hybridized with the fluorescent probe (% positive cells vs. DAPI).
Total DNA was extracted from each slurry sample using the DNeasy PowerSoil kit (Qiagen, United States) in accordance with the manufacturer’s recommendations. A non-template sample (DNA-free water) was included as a negative control during the whole workflow. The extraction yield and quality of the DNA were assessed with spectrophotometry (Multiskan Sky Microplate Spectrophotometer, Thermo Fisher Scientific, United States). All extractions were stored at −20°C until use. The extracted DNA was used for both the qPCR and NGS analyses.
Quantitative PCR was used to quantify two SMX-resistance genes (sul1 and sul2), and the class 1 integron-integrase gene (intl1). In addition, the 16S rRNA gene copy numbers were determined to assess the overall microbial load and to calculate the relative abundance of the resistance genes targeted in the samples collected. All qPCR assays were performed in the CFX96 real-time PCR detection system (Bio-Rad, United States) using SYBR green detection. The primers used for the ARGs, 16S, and intl1 gene quantification are listed in Supplementary Table 2. Each reaction was carried out in a total volume of 20 μL containing 10 μL SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, United States), 0.5 μL of each primer (10 μM), and 15 ng of DNA template. For all genes analyzed, the thermal cycling conditions were as follows: 95°C for 3 min, 45 cycles at 95°C for 15 s, annealing temperature (Tm) specific for each gene and primer pair for 30 s, and 72°C for 30 s. The fluorescence signal was read after each elongation step.
Amplicon specificity for each gene was verified by performing a melting curve starting at 55°C and increasing by 0.5°C until 95°C. Moreover, preliminary PCR was conducted to test the efficiency and the quality of primers (and amplicons), running gel electrophoresis of amplified DNA, and matching DNA bands with the expected fragment size.
Each assay was run in triplicate including the no template controls. Any possible qPCR inhibition was assessed by conducting an inhibition test using samples diluted from 10- to 100-fold, and no inhibition was observed. The quantitative PCR data were expressed as the ratio of ARG or intI1 gene copy number per 16S copy number to evaluate the relative proportion of each target gene in the bacterial community.
Aliquots of the DNA extracted (15 ng μL–1) from each replicate were used for NGS. The V3–V4 region of 16S rRNA genes was amplified with the Pro341F and Pro805R primers reported in Supplementary Table 3. These primers were selected in order to ensure the simultaneous identification of Bacteria and Archaea (Takahashi et al., 2014), both of which are involved in AD processes.
The size of the amplicons was checked with an agarose gel (1%), together with sample quality. The amplicons were sequenced with a MiSeq instrument (Illumina, Chesterford, United Kingdom) using the MiSeq Reagent Kit v3 for 600 cycles. The reads were imported and demultiplexed using QIIME 2 v2019.11 and denoised with the DADA2 (Callahan et al., 2016) plugin. The primers were removed using the “trim-left-f” (forward) and “trim-left-r” (reverse) primer DADA2 functions. These functions remove the sequences from the beginning to a specific position. The exact length of the primers was 17 nucleotides for the forward and 21 nucleotides for reverse one. The amplicon sequencing variants obtained were compared to the 97% identical clustered Ribosomal Database Project (RDP release 11) using a naive Bayes classifier trained on the amplified region with 80% confidence. The principal coordinate analysis based on the Bray–Curtis distance and PERMANOVA as a statistical test was performed using the online tool Microbiome Analyst2 in order to evaluate the composition of the bacterial community during the AD process.
The sequenced data obtained were filtered with a minimum count of four and prevalence in samples of 20%. All samples were rarefied to an even sequence depth based on the sample having the lowest sequencing depth. The principal coordinate analysis based on the Bray–Curtis distance was performed using Past 3.25 (Hammer et al., 2001) in order to evaluate the composition of the bacterial community in different conditions. The comparison between conditions was performed using STAMP 2.1.3 (Parks et al., 2014) with the two-way ANOVA statistical test and Tukey-Kramer post hoc test. The t-test between two groups was performed using MS Excel 2013.
H2 and CH4 were produced in both SMX and control batches. In both cases, the production of H2 started at day 3 and continued until days 10–15, when the CH4 production also started (Figure 1).
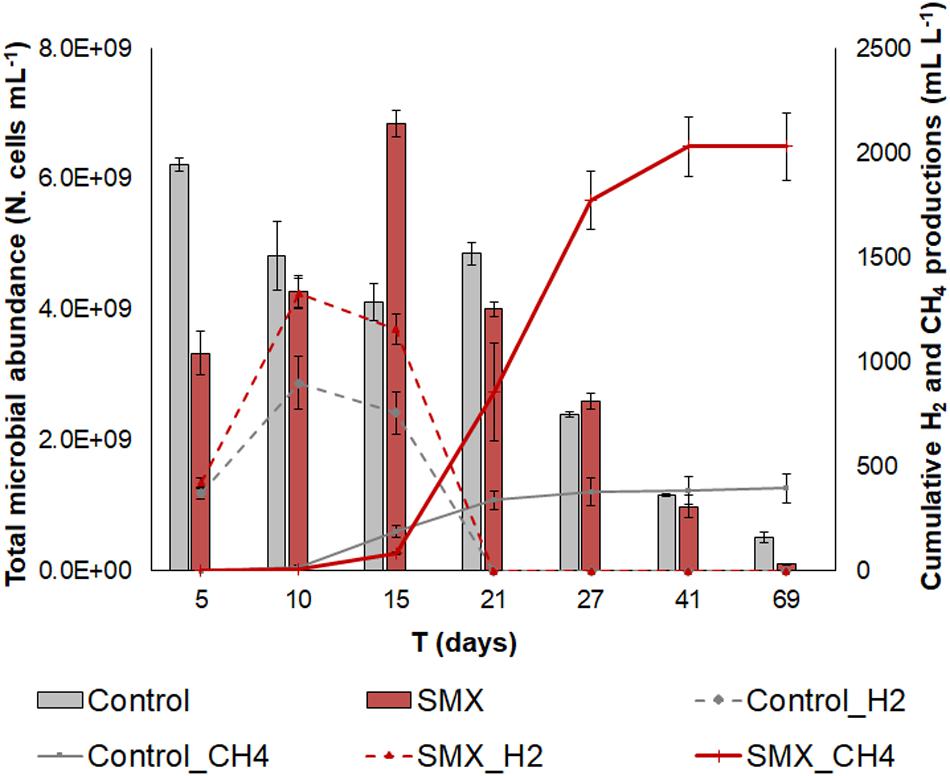
Figure 1. Total microbial abundance (N. cells mL– 1) in SMX-spiked (SMX) and control batches (histograms); cumulative biogas (CH4 and H2) production (lines) over the experimental time.
However, a lower production (p < 0.01) of hydrogen as well as of methane was observed in the controls than in the SMX batches (Figure 1). In particular, the cumulative CH4 production was about five times higher in the SMX condition than the control one (2030.6 ± 143.3 mL L–1 vs. 386.8 ± 65.4 mL L–1). Furthermore, the maximum methane concentration was observed (71% from day 27) in SMX, and in control it never exceeded 31%. The highest H2 productions of 1239.6 ± 72.3 mL L–1 for SMX and 899.6 ± 83.3 mL L–1 for control, respectively, were reached at day 10. Subsequently, the cumulative H2 values decreased, and they were never found from day 21 (Figure 1). Contrary to these results, other authors (Cetecioglu et al., 2012) reported that biogas (CH4 and H2) production was not affected by adding SMX (Cetecioglu et al., 2012) at concentrations of 1 and 10 mg L–1 in an anaerobic system. However, an increase in hydrogen production was also observed in the presence of the antimicrobial triclocarban (Wang et al., 2019). To the best of our knowledge, the results of the experiment reported here are the first finding an increase in methane production when SMX was present at a 5 mg L–1. The presence of the SMX antibiotic favored the production of H2, which, in turn, presumably favored the production of CH4. The stimulation of H2 production due to bioaugmentation with a bacterial pool of hydrogen producers also led to an increase in methane production in another previous work (Ferraro et al., 2018). High hydrogen production is an indicator of the imbalance between the hydrolysis/acidogenesis and methanogenesis phases during the AD process. The results of this work can be ascribed to a high substrate/microorganisms ratio of the inoculum, which presumably promoted an enrichment of the hydrogen-producing bacteria, as is also found in another work (Yuan et al., 2019).
The initial microbial cell abundance (day 5) was higher in the control than in the SMX condition, showing an initial negative effect by the antibiotic on the microbial community (Figure 1). However, an increasing trend was observed in the SMX batches with a peak at day 15 when methane production started. At the end of the experiment, no differences in cell numbers were observed between the SMX and control conditions.
The process intermediate compositions, which include VFAs, succinic and lactic acids, and ethanol, are reported in Figure 2. In the ingestate, high concentrations of acetic, propionic, and lactic acid (3.7 g L–1, 2.5 g L–1, and 1.6 g L–1, respectively) and ethanol (4.3 g L–1) were measured. The pH value (pH 6.4) at the start of the experiment was higher than the acids’ dissociation constant (≤4.86 at 25°C); this result suggests that the VFAs were mainly in their dissociated forms and, therefore, unable to permeate inside cells and inhibit the microbial activity (Wainaina et al., 2019). Although the high concentration of soluble metabolites suggests that the methanogenesis and acetogenesis started quickly, a high hydrogen production was also observed. Hydrogen production occurred immediately in both experimental conditions, and the acetic, butyric, formic, succinic, and valeric acids were detected, indicating a mixed acid fermentation. Moreover, a simultaneous consumption of lactic acid and ethanol for both experimental conditions was detected, indicating an early acetogenic pathway. The oxidation of lactate and ethanol is inhibited when hydrogen partial pressure is approaching 1 ATM, as evaluated by thermodynamic calculations (Harper and Pohland, 1986). The highest calculated values of H2 partial pressure were 0.47 and 0.70 ATM in the control and SMX conditions, respectively (day 6). During hydrogen production, the pH dropped (day 3) to values of 4.8 and 5.1 in control and SMX, respectively. In SMX, it increased to 6.3 (day 21) and gradually decreased to 5.4 at the end of experiment. In the control condition, the highest pH value was 6.0 (day 15), but it quickly decreased to 5.5 (day 21). During methane production, a high production of ethanol (2.6 g L–1) and a further acetic acid increase were observed from 15 to 21 days in the SMX condition. Subsequently, the ethanol was promptly consumed, and the methane production reached its maximum value; in this case, a late fermentative acetate–ethanol pathway followed by an ethanol acetogenic phase can be hypothesized. Another relevant difference between the two experimental conditions was the total VFA concentration over time: In control batches, a value of 17 g L–1 was reached at day 15, decreasing to 9.92 g L–1 at the end of AD (day 69); in the SMX condition, a lower value than 15.0 g L–1 was measured over the AD process. The latter results are in accordance with a recent experimental study on codigestion of food waste and pig manure, which reports that the threshold inhibition concentration of VFAs ranged from 16.5 to 18.0 g L–1 (Jiang et al., 2018). However, in the present experiment, hydrogenotrophic metabolism was the active pathway for methane production, particularly for SMX. In this condition, the hydrogen production resulted mainly from the acidogenic phase as well as from lactic acid and ethanol acetogenesis. These results are in line with the fact that methane production from acetic acid is inhibited by H2 partial pressure above 10–4 ATM and that propionic acid oxidation to acetate is favorable only at H2 partial pressure below 10–4 ATM. Moreover, butyric acid oxidation is favorable at H2 partial pressures of 10–3 ATM or below. For these reasons, the production of methane from H2/CO2 is favored compared to acetate cleavage when H2 partial pressure is above 10–4 ATM. In the present work, the threshold H2 partial pressure of 10–4 ATM was reached at day 41, when an oxidation of VFAs and a further accumulation of acetic acid were observed, suggesting that the acetogenesis phase was coupled with homoacetogenesis, as demonstrated by the absence of methane and hydrogen production.
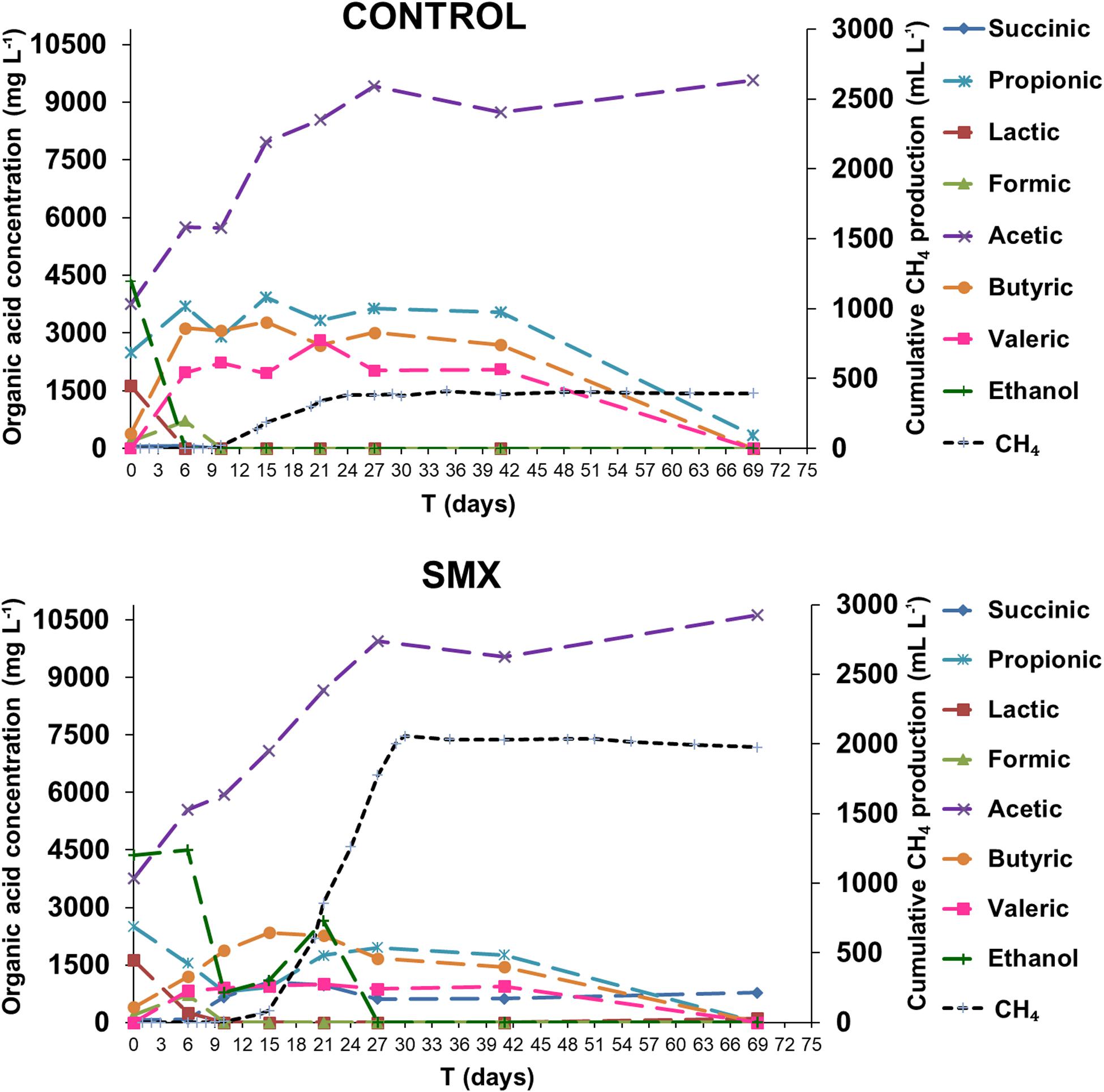
Figure 2. Organic acid concentrations and ethanol during the AD process (the standard deviations range from 7.6 to 13.4%).
The FISH analysis of the ingestate used to start the experiment showed a predominance of Bacteria (92.2%) corresponding to 1.74 109 cells mL–1 and a presence of Archaea (0.6%) corresponding to 1.13 107 cells mL–1. The NGS sequencing of the ingestate supported this FISH analysis, showing similar initial percentages of 99% of Bacteria and 0.8% of Archaea, respectively. Among Bacteria, the main groups found were Clostridia (38%) Bacteroidia (26%), and Bacilli (14%) followed by Gamma-Proteobacteria (11%) and Erysipelotrichia (5%). Minor groups were Actinobacteria (4%), Synergista (1%), and Verrucomicrobiae (1%).
In the batch experiment, the FISH analysis showed an initial detrimental antibiotic effect (t-test, p < 0.01) both for Bacteria and Archaea, as shown by the significantly lower number of cells (N. cells mL–1) in SMX than in control at 5 days (Figures 3A,B). These results are in line with those of the total microbial abundance and show how SMX, at the initial concentration used in this experiment, negatively affected some microbial populations. However, this effect was transient, and an increase in Bacteria was observable until day 15 (at the end of H2 production) and subsequently in Archaea (Figures 3A,B). In particular, an increase in Archaea abundance was detected from days 15 and 21 for SMX and control, respectively (Figure 3B) conferring a more balanced structure to the microbial community in line with the highest rate of methane production and hydrogen consumption. The FISH analysis made it possible to visualize and quantify at day 15, a significant increase in the abundance of Firmicutes and in the functional group of SRB in the SMX condition compared to control (Figure 3C). Firmicutes is one of the most abundant populations found to compose the bacterial populations of the AD community, and their increase can be related to an improvement in the process performance (Chen et al., 2016).
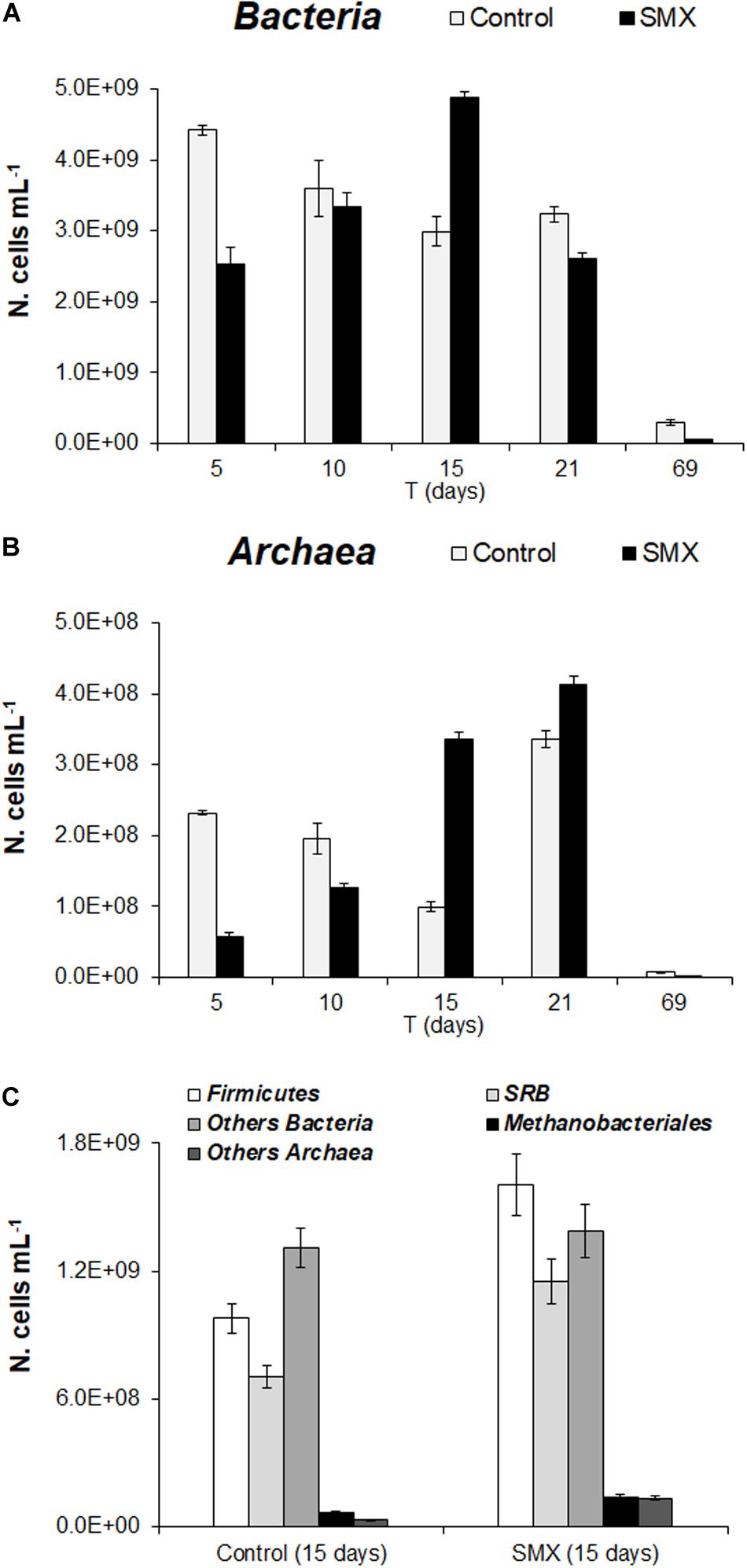
Figure 3. (A) N. cells mL– 1 of Bacteria and (B) Archaea in control and SMX-spiked (SMX) batches over the experimental time evaluated by FISH. (C) N. cell mL– 1 of Firmicutes, SRB, and Methanobacteriales detected at day 15.
Sulfate-reducing bacteria form a functional group able to use H2 for producing H2S, competing with methanogenic bacteria in the metabolic pathway of H2/CO2; on the contrary, in this work, SRB activity presumably helped to consume H2, contrasting the lowering of pH and, therefore, promoting the performance of the process.
The NGS results (Figures 4A,B, 5A–D) provide a deeper insight into the microbial community and its dynamic during the AD process and made it possible to show its shifts under the experimental conditions at different phylogenetic levels. The Chao index calculated for estimating the number of species in a community (Jiwon et al., 2017) was significantly (t-test, p < 0.01) different between the ingestate and the control and SMX conditions (Table 2). A higher value (Two-way ANOVA, p < 0.01) of this index was observed in the control than in the SMX over the experimental time. These results show a direct effect of the antibiotic on the microbial community.
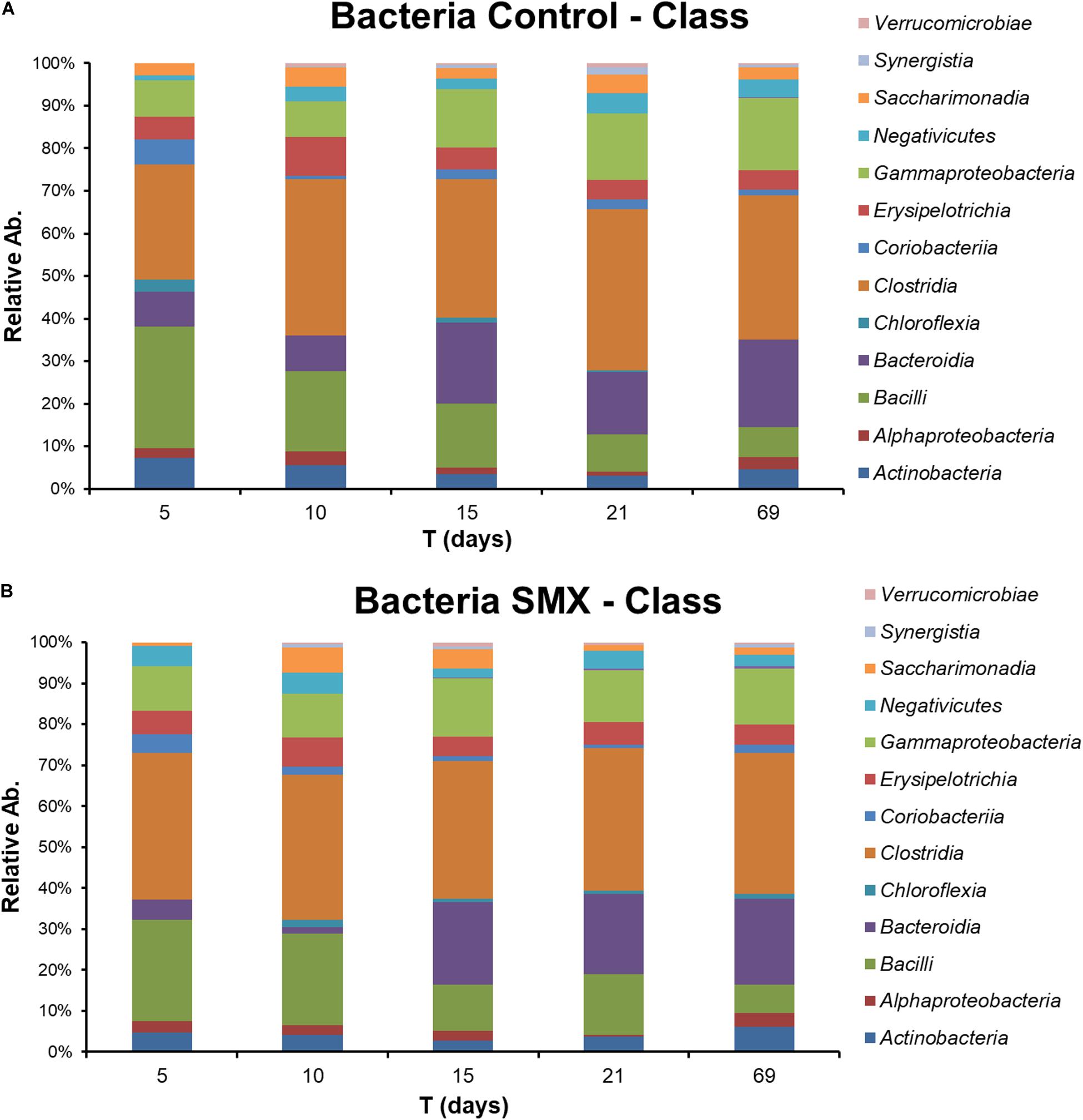
Figure 4. Relative abundance of Bacteria at different experimental times evaluated by NGS in Control (A) and (B) SMX batches.
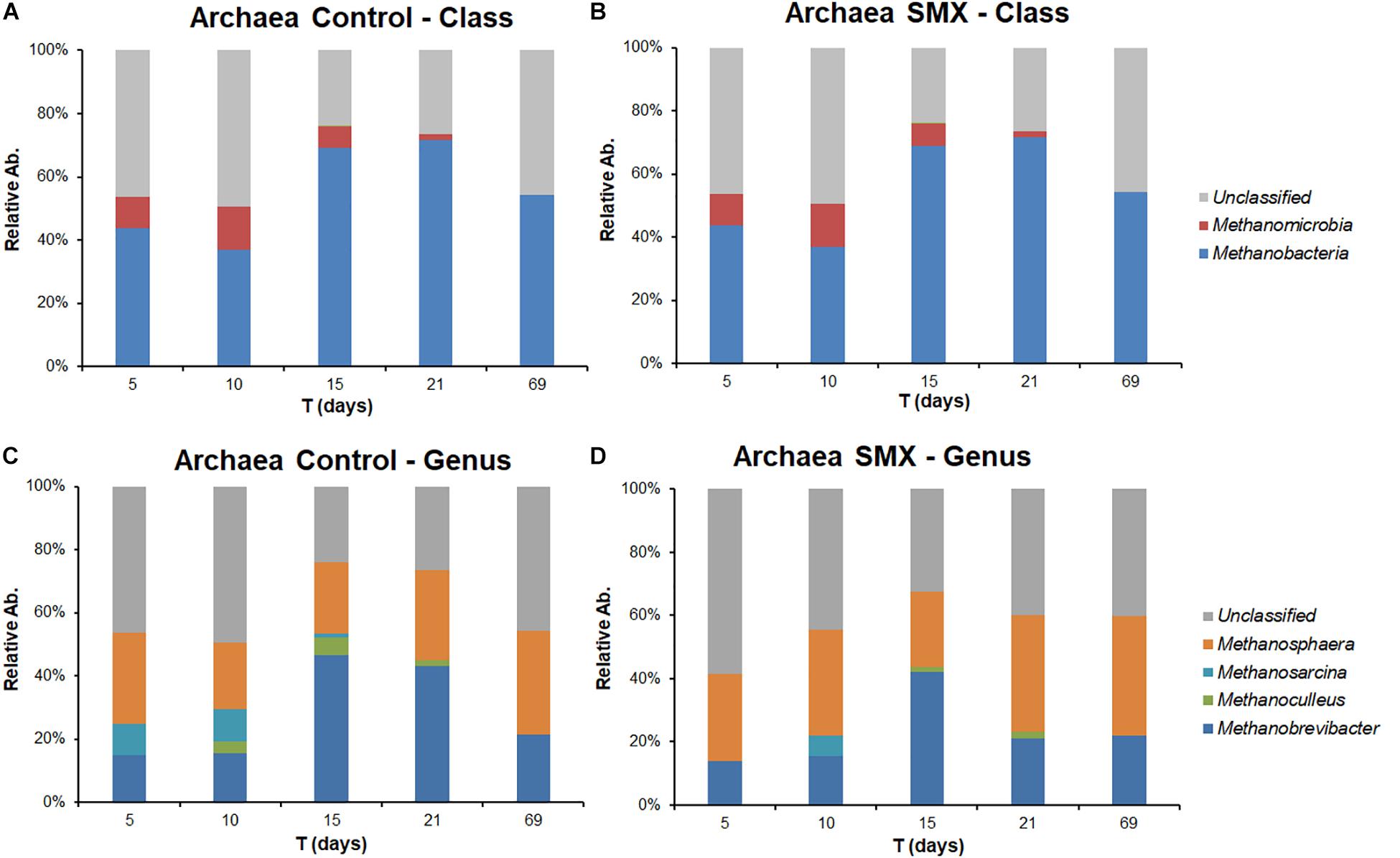
Figure 5. Relative abundance of Archaea in control and SMX conditions evaluated by NGS at class (A,B) and genus level (C,D), respectively.
Moreover, the Bray–Curtis principal coordinate analysis shows significant differences (Permanova, p < 0.001) between the ingestate, the first experimental phase of H2 production (days 5–10), and the second phase of CH4 evolution (days 15–69), demonstrating how the microbial community changed during the AD process (Figure 6).
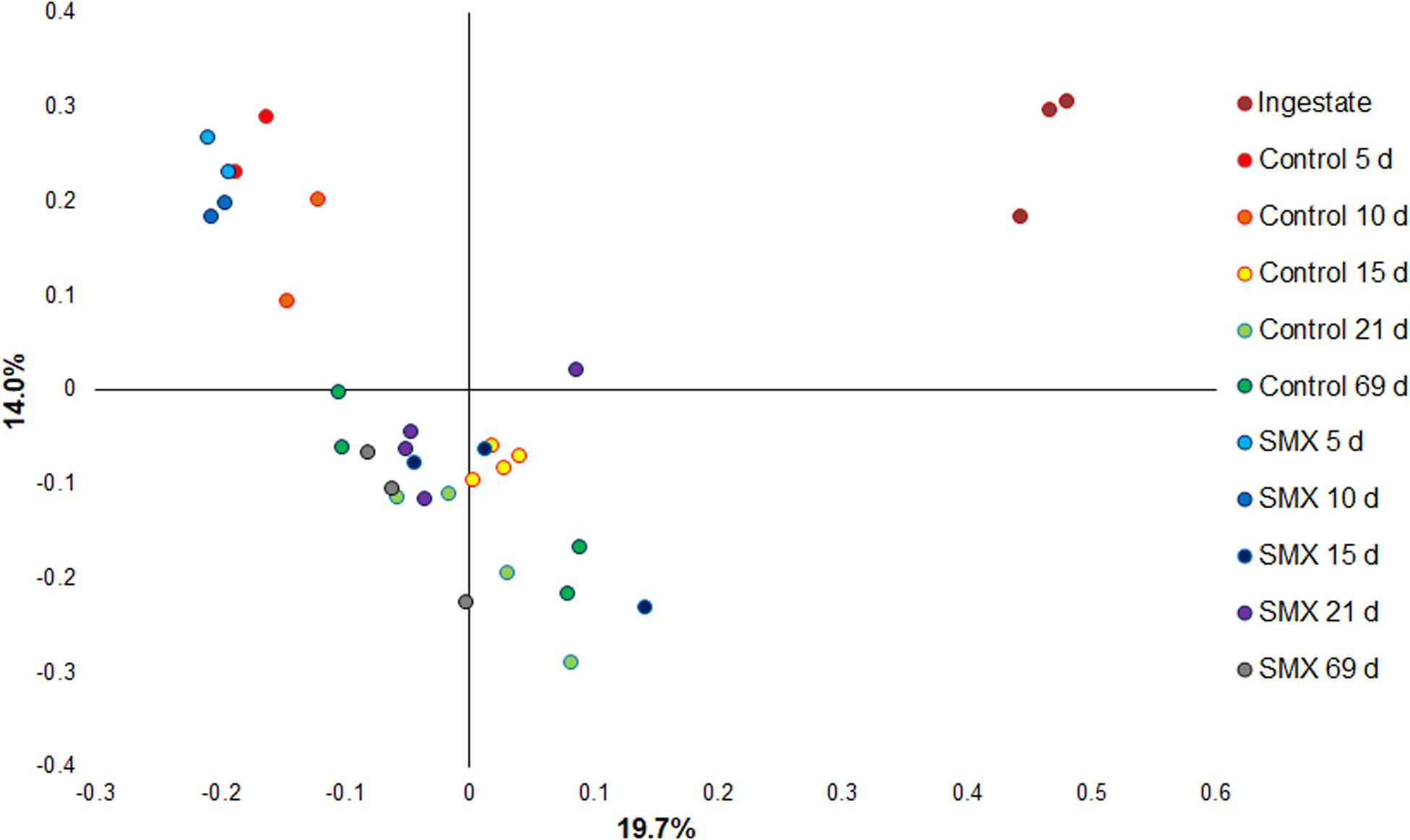
Figure 6. Principal coordinate analysis based on Bray–Curtis (Permanova p < 0.001) for evaluating the difference in composition of the bacterial community under the different conditions.
Significant differences were found inside some archaeal and bacterial groups between the control and SMX batches at day 5. In particular, significantly lower percentages (p < 0.01) of Methanomicrobia (10% control vs. 0% SMX), Bacilli (28% control vs. 25% SMX) and Bacteroidia (8% control vs. 5% SMX) were found in the SMX batches.
The archaeal fraction was mainly composed of two classes: Methanomicrobia and Methanobacteria. At genus level, two genera predominated, Methanobrevibacter (26%) and Methanosphaera (17%), which are known to thrive in a range of pH values between 5.0 and 7.5 (Madigan et al., 2014) in line with the values recorded in this experiment (Figure 5A–D). In particular, Methanobrevibacter is acid-tolerant and can grow from pH 5.0 to 7.5 (Savant et al., 2002). The methane production involves mainly acetotrophic (e.g., Methanosarcinales) and hydrogenotrophic archaeal groups, the former producing methane using acetic acid and the latter using H2/CO2 (Castellano-Hinojosa et al., 2018); the results of the present work show that, in the control condition, there was a higher percentage (p < 0.01) of Methanomicrobia than in the SMX one (Figure 5). Inside the class Methanomicrobia, the genus Methanosarcina, able to use acetic acid for producing methane, was detected in the control condition at days 5, 10, and 15 (9.3, 9.9, and 1.3% respectively), and in the SMX one only at day 10 (6.0%). The overall microbial community characterization results compared with the VFA production trend (Figure 2) clearly indicate that hydrogenotrophic metabolism was the active pathway for methane production, especially for the SMX condition. The acetoclastic pathway was presumably inactive as can be deduced from the accumulation of acetic acid as well as the absence or low amount of Methanosarcina in SMX.
The relative abundance (ARG/16S) of sul1, sul2, and intI1 genes in the ingestate and in the batch experiment are reported in Table 3. The most abundant genes detected in the ingestate were sul1 and the proxy intI1, and the ARG sul2 was detected in a lower concentration. The overall abundance of ARGs decreased in the digestate anaerobic process in both SMX and control conditions with significantly lower values (p < 0.01) at the end of the experiment. However, in the control the residual concentration of SMX (ca. 0.30 mg L–1) found in the ingestate still persisted (Table 3).
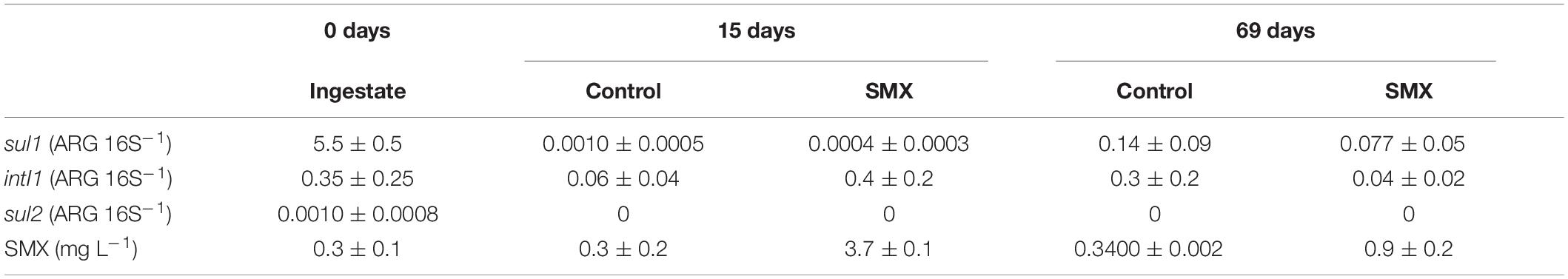
Table 3. Relative abundance (ARG 16S–1 ± standard error) of sul1, sul2, and intI1 and quantification of SMX (mg L–1 ± standard error) in the ingestate (0 days) and batch experiments at 15 and 69 days.
The antibiotic concentration decreased over time in the SMX-spiked batches and in line with the increase in microbial abundance; at the end of the experiment, about 80% was degraded. This result shows that SMX can also be degraded in anaerobic conditions; SMX biodegradation has been found in previous works in effluents (Patrolecco et al., 2018) and digestate-treated soil in aerobic conditions (Rauseo et al., 2019). Recent works (Wang et al., 2016; Gurmessa et al., 2020) report SMX to be removed mainly by biodegradation and that adsorption processes were negligible. Moreover, in AD experiments, in which the substrate was pig manure, SMX was removed with variable elimination rates from 0 to 100%, depending on specific process parameters, such as temperature and hydraulic retention time (Gurmessa et al., 2020).
The results show a selective effect of SMX on the microbial community, which not only did not hamper AD, but also stimulated some microbial populations involved in biogas production and in the antibiotic degradation.
The AD process promoted an overall lowering of the ARG load inside the microbial community in both control and SMX conditions. Interestingly, the decrease in the relative abundance of ARGs was not directly related to the reduction in antibiotic concentration. This supports the hypothesis that whatever the initial antibiotic concentration, the AD process seems to discourage ARG maintenance among the microbial community, owing presumably to its fitness costs. Further studies are in progress for evaluating in real biogas plants if the AD process, with manure as feedstock, is able to decrease the antibiotic residues and ARGs. The latter aspect is particularly important because AD might lower the risk of ARG spread and make digestate more suitable than manure for replacing chemical fertilizers.
The datasets generated for this study are available at: https://www.ebi.ac.uk/ena/browser/view/PRJEB37044.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was funded by Regione Lazio – Lazio Innova (Italy) – Project name: AZeRO Antibiotics N. 85-2017-15065 – CUP B56C18000730002 (Progetti Gruppi di Ricerca – Programma strategico regionale per la ricerca, innovazione ed il trasferimento tecnologico 2017–2020 – Conoscenza e Cooperazione per un nuovo modello di sviluppo L.R. 13/2008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the CIB-Italian Biogas Consortium for supporting the project (www.consorziobiogas.it/en/). We thank Domenico Mastroianni and Francesca Falconi for their analyses of metals and organic carbon. We are grateful to Flavia Piccinini for having participated in all the experimental work for her master’s thesis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.537783/full#supplementary-material
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169. doi: 10.1128/mmbr.59.1.143-169.1995
Aydin, S., Ince, B., Cetecioglu, Z., Arikan, O., Ozbayram, E. G., Shahi, A., et al. (2015). Combined effect of erythromycin, tetracycline and sulfamethoxazole on performance of anaerobic sequencing batch reactors. Bioresour. Technol. 186, 207–214. doi: 10.1016/j.biortech.2015.03.043
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Barra Caracciolo, A., Giuliano, G., Grenni, P., Cremisini, C., Ciccoli, R., and Ubaldi, C. (2005a). Effect of urea on degradation of terbuthylazine in soil. Environ. Toxicol. Chem. 24, 1035–1040. doi: 10.1897/04-253R.1
Barra Caracciolo, A., Grenni, P., Cupo, C., and Rossetti, S. (2005b). In situ analysis of native microbial communities in complex samples with high particulate loads. FEMS Microbiol. Lett. 253, 55–58. doi: 10.1016/j.femsle.2005.09.018
Barra Caracciolo, A., Topp, E., and Grenni, P. (2015). Pharmaceuticals in the environment: biodegradation and effects on natural microbial communities. A review. J. Pharm. Biomed. Anal. 106, 25–36. doi: 10.1016/j.jpba.2014.11.040
Bharathiraja, B., Sudharsana, T., Jayamuthunagai, J., Praveenkumar, R., Chozhavendhan, S., and Iyyappan, J. (2018). Biogas production – A review on composition, fuel properties, feed stock and principles of anaerobic digestion. Renew. Sustain. Energy Rev. 90, 570–582. doi: 10.1016/j.rser.2018.03.093
Bustamante, M. A., Alburquerque, J. A., Restrepo, A. P., de la Fuente, C., Paredes, C., Moral, R., et al. (2012). Co-composting of the solid fraction of anaerobic digestates, to obtain added-value materials for use in agriculture. Biomass Bioenergy 43, 26–35. doi: 10.1016/j.biombioe.2012.04.010
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Castellano-Hinojosa, A., Armato, C., Pozo, C., González-Martínez, A., and González-López, J. (2018). New concepts in anaerobic digestion processes: recent advances and biological aspects. Appl. Microbiol. Biotechnol. 102, 5065–5076. doi: 10.1007/s00253-018-9039-9
Cetecioglu, Z., Ince, B., Gros, M., Rodriguez-Mozaz, S., Barceló, D., Ince, O., et al. (2015). Biodegradation and reversible inhibitory impact of sulfamethoxazole on the utilization of volatile fatty acids during anaerobic treatment of pharmaceutical industry wastewater. Sci. Total Environ. 536, 667–674. doi: 10.1016/J.scitotenv.2015.07.139
Cetecioglu, Z., Ince, B., Orhon, D., and Ince, O. (2012). Acute inhibitory impact of antimicrobials on acetoclastic methanogenic activity. Bioresour. Technol. 114, 109–116. doi: 10.1016/j.biortech.2012.03.020
Chen, S., Cheng, H., Wyckoff, K. N., and He, Q. (2016). Linkages of Firmicutes and Bacteroidetes populations to methanogenic process performance. J. Ind. Microbiol. Biotechnol. 43, 71–81. doi: 10.1007/s10295-016-1760-8
Cheng, D. L., Ngo, H. H., Guo, W. S., Liu, Y. W., Zhou, J. L., Chang, S. W., et al. (2018). Bioprocessing for elimination antibiotics and hormones from swine wastewater. Sci. Total Environ. 621, 1664–1682. doi: 10.1016/j.scitotenv.2017.10.059
Enne, V. I., King, A., Livermore, D. M., and Hall, L. M. C. (2002). Sulfonamide resistance in Haemophilus influenzae mediated by acquisition of sul2 or a short insertion in chromosomal folP. Antimicrob. Agents Chemother. 46, 1934–1939. doi: 10.1128/AAC.46.6.1934-1939.2002
European Biogas Association (2018). EBA Statistical Report. Available online at: http://european-biogas.eu/2017/12/14/eba-statistical-report-2017-published-soon/ (accessed January 2020).
Feng, L., Casas, M. E., Ottosen, L. D. M., Møller, H. B., and Bester, K. (2017). Removal of antibiotics during the anaerobic digestion of pig manure. Sci. Total Environ. 603–604, 219–225. doi: 10.1016/J.scitotenv.2017.05.280
Ferraro, A., Dottorini, G., Massini, G., Mazzurco Miritana, V., Signorini, A., Lembo, G., et al. (2018). Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour. Technol. 260, 364–373. doi: 10.1016/j.biortech.2018.03.128
Ferraro, A., Massini, G., Mazzurco Miritana, V., Signorini, A., Race, M., and Fabbricino, M. (2019). A simplified model to simulate bioaugmented anaerobic digestion of lignocellulosic biomass: biogas production efficiency related to microbiological data. Sci. Total Environ. 691, 885–895. doi: 10.1016/j.scitotenv.2019.07.051
Grenni, P., Ancona, V., and Barra Caracciolo, A. (2018). Ecological effects of antibiotics on natural ecosystems: a review. Microchem. J. 136, 25–39. doi: 10.1016/j.microc.2017.02.006
Greuter, D., Loy, A., Horn, M., and Rattei, T. (2016). ProbeBase-an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 44, D586–D589. doi: 10.1093/nar/gkv1232
Guo, M., Song, W., and Buhain, J. (2015). Bioenergy and biofuels: history, status, and perspective. Renew. Sustain. Energy Rev. 42, 712–725. doi: 10.1016/j.rser.2014.10.013
Gurmessa, B., Pedretti, E. F., Cocco, S., Cardelli, V., and Corti, G. (2020). Manure anaerobic digestion effects and the role of pre- and post-treatments on veterinary antibiotics and antibiotic resistance genes removal efficiency. Sci. Total Environ. 721;137532. doi: 10.1016/j.scitotenv.2020.137532
Hammer, Ø, Harper, D. A. T., and Ryan, P. D. (2001). Past: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Harper, S. R., and Pohland, F. G. (1986). Recent developments in hydrogen management during anaerobic biological wastewater treatment. Biotechnol. Bioeng. 28, 585–602. doi: 10.1002/bit.260280416
Ji, J. Y., Xing, Y. J., Ma, Z. T., Zhang, M., and Zheng, P. (2013). Acute toxicity of pharmaceutical wastewaters containing antibiotics to anaerobic digestion treatment. Chemosphere 91, 1094–1098. doi: 10.1016/j.chemosphere.2013.01.009
Jiang, Y., Dennehy, C., Lawlor, P. G., Hu, Z., McCabe, M., Cormican, P., et al. (2018). Inhibition of volatile fatty acids on methane production kinetics during dry co-digestion of food waste and pig manure. Waste Manag. 79, 302–311. doi: 10.1016/j.wasman.2018.07.049
Jiwon, S., Guevarra, R., Lee, J., and Kim, H. B. (2017). Deciphering diversity indices for better understanding of the microbial communities. J Microbiol Biotechnol. 27, 2089–2093. doi: 10.4014/jmb.1709.09027
Logan, B. E., Oh, S. E., Kim, I. S., and Van Ginkel, S. (2002). Biological hydrogen production measured in batch anaerobic respirometers. Environ. Sci. Technol. 37:1055. doi: 10.1021/es015783i
Ma, Y., Wilson, C. A., Novak, J. T., Riffat, R., Aynur, S., Murthy, S., et al. (2011). Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class i integrons. Environ. Sci. Technol. 45, 7855–7861. doi: 10.1021/es200827t
Madigan, M. T., Martinko, J. M., Bender, K. S., Buckley, D. H., and Stahl, D. A. (2014). Brock Biology of Microorganisms -, 14th Edn. London: Pearson.
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
Martínez-Sibaja, A., Alvarado-Lassman, A., Astorga-Zaragoza, C. M., Adam-Medina, M., Posada-Gómez, R., and Rodríguez-Jarquin, J. P. (2011). Volumetric gas meter for laboratory-scale anaerobic bioreactors. Meas. J. Int. Meas. Confed. 44, 1801–1805. doi: 10.1016/j.measurement.2011.08.018
Mckinney, C. W., Dungan, R. S., Moore, A., and Leytem, A. B. (2018). Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 94:fiy010. doi: 10.1093/femsec/fiy010
Mohring, S. A. I., Strzysch, I., Fernandes, M. R., Kiffmeyer, T. K., Tuerk, J., and Hamscher, G. (2009). Degradation and elimination of various sulfonamides during anaerobic fermentation: a promising step on the way to sustainable pharmacy? Environ. Sci. Technol. 43, 2569–2574. doi: 10.1021/es802042d
Pampillón-González, L., Luna-Guido, M., Ruíz-Valdiviezo, V. M., Franco-Hernández, O., Fernández-Luqueño, F., Paredes-López, O., et al. (2017). Greenhouse gas emissions and growth of wheat cultivated in soil amended with digestate from biogas production. Pedosphere 27, 318–327. doi: 10.1016/S1002-0160(17)60319-9
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Patrolecco, L., Rauseo, J., Ademollo, N., Grenni, P., Cardoni, M., Levantesi, C., et al. (2018). Persistence of the antibiotic sulfamethoxazole in river water alone or in the co-presence of ciprofloxacin. Sci. Total Environ. 640–641, 1438–1446. doi: 10.1016/j.scitotenv.2018.06.025
Pernthaler, J., Glöckner, F. O., Schönhuber, W., and Amann, R. (2001). Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30, 207–226. doi: 10.2105/ajph.36.12.1446-a
Polesel, F., Andersen, H. R., Trapp, S., and Plósz, B. G. (2016). Removal of Antibiotics in Biological Wastewater Treatment Systems - A critical assessment using the activated sludge modeling framework for xenobiotics (ASM-X). Environ. Sci. Technol. 50, 10316–10334. doi: 10.1021/acs.est.6b01899
Rauseo, J., Barra Caracciolo, A., Ademollo, N., Cardoni, M., Di Lenola, M., Gaze, W. H., et al. (2019). Dissipation of the antibiotic sulfamethoxazole in a soil amended with anaerobically digested cattle manure. J. Hazard. Mater. 378:120769. doi: 10.1016/j.jhazmat.2019.120769
Savant, D. V., Shouche, Y. S., Prakash, S., and Ranade, D. R. (2002). Methanobrevibacter acididurans sp. nov., a novel methanogen from a sour anaerobic digester. Int. J. Syst. Evol. Microbiol. 52, 1081–1087. doi: 10.1099/ijs.0.01903-0
Sköld, O. (2000). Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3, 155–160. doi: 10.1054/drup.2000.0146
Styles, D., Adams, P., Thelin, G., Vaneeckhaute, C., Chadwick, D., and Withers, P. J. A. (2018). Life cycle assessment of biofertilizer production and use compared with conventional liquid digestate management. Environ. Sci. Technol. 52, 7468–7476. doi: 10.1021/acs.est.8b01619
Sui, Q., Zhang, J., Tong, J., Chen, M., and Wei, Y. (2017). Seasonal variation and removal efficiency of antibiotic resistance genes during wastewater treatment of swine farms. Environ. Sci. Pollut. Res. 24, 9048–9057. doi: 10.1007/s11356-015-5891-7
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T., and Nishijima, M. (2014). development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592
Tasho, R. P., and Cho, J. Y. (2016). Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: a review. Sci. Total Environ. 563–564, 366–376. doi: 10.1016/j.scitotenv.2016.04.140
Thompson, M., Ellison, S. L. R., and Wood, R. (2002). Harmonized guidelines for single-laboratory validation of methods of analysis (IUPAC technical report). Pure Appl. Chem. 74, 835–855. doi: 10.1351/pac200274050835
Wainaina, S., Lukitawesa, K., Awasthi, M., and Taherzadeh, M. J. (2019). Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production: a critical review. Bioengineered 10, 437–458. doi: 10.1080/21655979.2019.1673937
Wang, J., Lin, H., Sun, W., Xia, Y., Ma, J., Fu, J., et al. (2016). Variations in the fate and biological effects of sulfamethoxazole, norfloxacin and doxycycline in different vegetable-soil systems following manure application. J. Hazard. Mater. 304, 49–57. doi: 10.1016/j.jhazmat.2015.10.038
Wang, Y., Wang, D., Chen, F., Yang, Q., Li, Y., Li, X., et al. (2019). Effect of triclocarban on hydrogen production from dark fermentation of waste activated sludge. Bioresour. Technol. 279, 307–316. doi: 10.1016/j.biortech.2019.02.016
Widyasari-Mehta, A., Hartung, S., and Kreuzig, R. (2016). From the application of antibiotics to antibiotic residues in liquid manures and digestates: a screening study in one European center of conventional pig husbandry. J. Environ. Manage. 177, 129–137. doi: 10.1016/j.jenvman.2016.04.012
Yuan, T., Bian, S., Ko, J. H., Wu, H., and Xu, Q. (2019). Enhancement of hydrogen production using untreated inoculum in two-stage food waste digestion. Bioresour. Technol. 282, 189–196. doi: 10.1016/j.biortech.2019.03.020
Zhang, J., Sui, Q., Zhong, H., Meng, X., Wang, Z., Wang, Y., et al. (2018). Impacts of zero valent iron, natural zeolite and Dnase on the fate of antibiotic resistance genes during thermophilic and mesophilic anaerobic digestion of swine manure. Bioresour. Technol. 258, 135–141. doi: 10.1016/j.biortech.2018.03.005
Keywords: cattle manure, sulfamethoxazole, biogas, anaerobic digestion, microbial community, antibiotic degradation, ARGs
Citation: Mazzurco Miritana V, Massini G, Visca A, Grenni P, Patrolecco L, Spataro F, Rauseo J, Garbini GL, Signorini A, Rosa S and Barra Caracciolo A (2020) Effects of Sulfamethoxazole on the Microbial Community Dynamics During the Anaerobic Digestion Process. Front. Microbiol. 11:537783. doi: 10.3389/fmicb.2020.537783
Received: 25 February 2020; Accepted: 20 August 2020;
Published: 16 September 2020.
Edited by:
Dimitrios Georgios Karpouzas, University of Thessaly, GreeceReviewed by:
Eldon R. Rene, IHE Delft Institute for Water Education, NetherlandsCopyright © 2020 Mazzurco Miritana, Massini, Visca, Grenni, Patrolecco, Spataro, Rauseo, Garbini, Signorini, Rosa and Barra Caracciolo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulia Massini, Z2l1bGlhLm1hc3NpbmlAZW5lYS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.