- 1Center for Disease Control and Prevention of People’s Liberation Army, Beijing, China
- 2Academy of Military Medical Sciences, Beijing, China
- 3College of Environmental and Chemical Engineering, Nanchang Hangkong University, Nanchang, China
- 4Department of Laboratory Diagnosis, Sun Yat-sen University Affiliated Zhongshan Hospital, Zhongshan, China
- 5The Sixth Medical Center of People’s Liberation Army General Hospital, Beijing, China
Background: Enterobacter cloacae is an opportunistic pathogen which is responsible for serious nosocomial infections. A gene which plays an important role in resistance to carbapenems is the New Delhi metallo-β-lactamase 1 (NDM-1). Currently, the spread of NDM-1-producing E. cloacae strains is a serious public threat.
Methods: A multidrug-resistant E. cloacae ssp. dissolvens strain CBG15936 was recovered in 2017 in Guangzhou, China. PCR, S1-pulsed-field gel electrophoresis, and Southern blotting were performed to locate the blaNDM–1 gene. Susceptibility testing and conjugation experiments were also performed. Illumina HiSeq and Nanopore sequencers were used to perform whole-genome sequencing.
Results: Strain CBG15936 belongs to ST932 and is resistant to carbapenems. The blaNDM–1 gene was found on a ∼62-kb plasmid, which has a conjugation frequency of 1.68 × 10–3 events per donor cell. Genome sequencing and analysis revealed that the NDM-1-carrying IncN1 plasmid contained a new transposon Tn6696, which consists of an intact qnrS1-carrying Tn6292 element, an inverted 8.3-kb Tn3000 remnant, ISkpn19, ΔtnpA, and IS26.
Conclusion: A new transposon, Tn6696, has been detected on a blaNDM–1-carrying plasmid recovered from multidrug-resistant E. cloacae ssp. dissolvens CBG15936 from China. This finding provides a new perspective regarding the potential for blaNDM–1 to undergo horizontal transfer among drug-resistant bacteria.
Introduction
Enterobacter cloacae is a gram-negative opportunistic pathogen which belongs to Enterobacteriaceae. Previous studies have reported that E. cloacae are ubiquitous in nature and can be isolated from clinical samples (Davin-Regli et al., 2019). With the extensive use of antibiotics, carbapenem-resistant E. cloacae have become an important nosocomial pathogen which can cause septicemia and lower respiratory tract infections (Mayhall et al., 1979; Daw et al., 1989). In recent years, carbapenem-resistant E. cloacae isolates have been reported worldwide (Davin-Regli and Pagès, 2015). New Delhi metallo-β-lactamase 1 (NDM-1) was first detected in a Swedish patient transferred from India, and it plays a major role in the appearance and dissemination of carbapenem-resistant gram-negative bacteria (Yong et al., 2009; Jamal et al., 2013). NDM-1 has become a burden on the health care system especially in intensive care units (Ahmad et al., 2018; Khalid et al., 2019). The detection rate of NDM-1 in E. cloacae has been increasing globally (Karlowsky et al., 2017). Prevalence of NDM-1-carrying E. cloacae was observed in France (Perry et al., 2011) and Mexico (Torres-Gonzalez et al., 2015). Genome sequencing and analysis also revealed an NDM-1 E. cloacae outbreak in a hospital in the UK (Fairley et al., 2019). In the southwest region of China, 132 carbapenem-resistant E. cloacae isolates were obtained from patients between 2012 and 2016. Twenty (15.2%) of these strains were identified as NDM-1 positive (Jia et al., 2018). It is important to monitor the spread of blaNDM–1 among E. cloacae stains.
Transposons are mobile genetic elements which are able to translocate chromosome or plasmids. Transposons have been shown to carry drug resistance genes and provide antibiotic resistance in pathogenic bacteria (Whittle et al., 2002). Tn6292 is a Tn-family unit transposon which belongs to the Tn3-family and has an IS26 at the right end. In addition, Tn6292 contains a quinolone resistance region qnrS1 (Feng et al., 2016). Multidrug-resistant (MDR) bacteria containing Tn6292 have been reported many times in China (Li et al., 2018). Tn3000 is bracketed by IS3000 at both ends and contains the blaNDM–1 gene. Tn3000 has been shown to be responsible for transmission of the blaNDM–1 gene in various parts of the world (Campos et al., 2015). Thus, it is important to elucidate the genetic features of transposons in order to explore possible mechanisms of bacterial resistance.
Therefore, in this study, we report a carbapenem-resistant E. cloacae ssp. dissolvens strain and the genetic features of the blaNDM–1-harboring plasmid it carries. In addition, we identify a new transposon, Tn6696, present in this plasmid. These findings provide a new perspective regarding possible mechanisms of gene transmission to mediate drug resistance.
Materials and Methods
Bacterial Identification
Strain CBG15936 was recovered from the sputum of a patient in Guangzhou, China, in 2017. The strain was identified by using the Vitek 2 Compact System (BioMérieux, France) and confirmed with 16S rRNA gene sequencing. The blaNDM–1 gene was detected by PCR amplification as previously described (Zhang et al., 2013). This isolate was collected through routine surveillance, and verbal consent was obtained as no personally identifiable data were included. The ethics of the study was reviewed and supervised by the Center for Disease Control and Prevention of PLA. All experiments were performed in the biosafety cabinet following the standard procedure.
S1-Pulsed-Field Gel Electrophoresis (PFGE) and Conjugation Experiments
Genomic DNA was prepared in agarose plugs and digested with the S1 endonuclease (Takara, Dalian, China). DNA fragments were electrophoresed on a CHEF-DR III system (Bio-Rad, Hercules, United States) for 15 h at 14°C with run conditions of 6 V/cm and pulse times from 0.22 to 26.29 s. The Salmonella enterica serotype Braenderup H9812 was used as the size marker. To determine the location of the blaNDM–1 gene, DNA was transferred to a positively charged nylon membrane (Roche) and then hybridized with digoxigenin-labeled blaNDM–1. Conjugation experiments were carried out in LB broth cultures. Azide-resistant Escherichia coli strain J53 [F(-) met pro Azi(r)] (recipient) and strain CBG15936 (donor) were mixed at a 1:3 ratio and then incubated at 37°C (Yi et al., 2012). After 18 h, transconjugants were selected for 12 h on MacConkey agar plates supplemented with meropenem (4 μg/ml) and sodium azide (150 μg/ml). Horizontal transferability of drug resistance was confirmed with antimicrobial susceptibility testing. Transconjugant-carrying plasmids were subsequently confirmed by PCR amplification and PFGE.
Susceptibility Testing
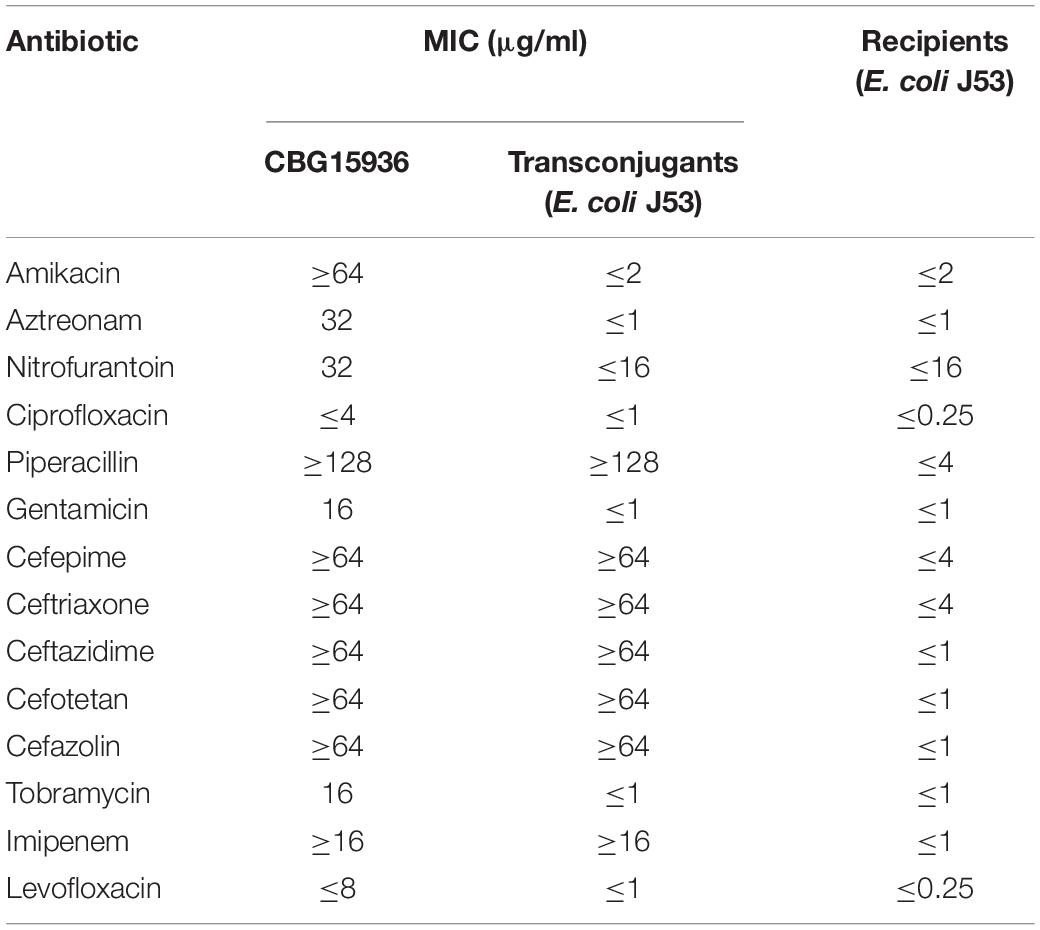
The minimum inhibitory concentrations (MICs) of amikacin, aztreonam, nitrofurantoin, ciprofloxacin, piperacillin, gentamicin, cefepime, ceftriaxone, ceftazidime, cefotetan, cefazolin, tobramycin, imipenem, and levofloxacin were determined with the Vitek 2 Compact System (Bobenchik et al., 2015). E. coli reference strain ATCC25922 was used as quality control. The MIC value of meropenem was determined using an E-test. The results were interpreted according to Clinical and Laboratory Standards Institute [CLSI] (2018) guidelines.
Whole-Genome Sequencing and Comparative Genome Analysis
Genomic DNA was extracted by using the High Pure PCR Template Preparation Kit (Qiagen, Inc., Valencia, CA, United States). Whole-genome sequencing was subsequently performed by using Illumina HiSeq according to the 350-bp paired-end protocol available from Novogene Company (Beijing, China) and MinION in our lab. The genome was assembled de novo by using a Unicycler (Li et al., 2010). RAST 2.0 was used to annotate the genome sequences obtained (Aziz et al., 2008). ResFinder v3.2 was used to identify acquired antibiotic resistance genes (Jia et al., 2016). Plasmid replicon type was analyzed by using PlasmidFinder (Carattoli et al., 2014). IS sequences were analyzed by using ISfinder (Siguier et al., 2006). Sequences of pNDM1-CBG and three similar plasmids were compared by using BLAST and Easyfig software (Sullivan et al., 2011). Whole-genome sequences of CBG15936 were used to determine multilocus sequence typing (MLST) based on detection of seven housekeeping genes (arcA, aspC, clpX, dnaG, fadD, lysP, and mdh) in pubMLST1 (Larsen et al., 2012). Twenty-nine sequences of the hsp60 gene of eight Enterobacter clusters were retrieved from NCBI and used for phylogenetic analysis to identify the species of the strain.
Nucleotide Sequence Accession Numbers
The entire sequence of strain, CBG15936, as well as plasmids, pTEM-CBG and pNDM1-CBG, have been deposited in GenBank under accession numbers CP046116, CP046117, and CP046118, respectively.
Results
Microbiological and Genetic Features of E. cloacae ssp. dissolvens CBG15936
Strain CBG15936 was identified as E. cloacae using Vitek and 16S rRNA and further classified as E. cloacae ssp. dissolvens based on hsp60 genotyping (Figure 1). Susceptibility testing showed that E. cloacae ssp. dissolvens strain CBG15936 exhibits resistance to amikacin, aztreonam, piperacillin, cefepime, ceftriaxone, ceftazidime, cefotetan, cefazolin, and imipenem (Table 1). The E-test showed that strain CBG15936 (MIC ≥ 16) and transconjugants (MIC > 32) both exhibit resistance to meropenem. The strain was found positive for the blaNDM–1 gene by PCR. S1-PFGE showed that this strain contains two different plasmids ∼60 and 75 kb in length (Figure 2). Southern blotting indicated that the blaNDM–1 gene is present in the ∼60-kb plasmid. S1-PFGE and conjugation assays revealed that transfer of the plasmid carrying the blaNDM–1, qnrS1, and dfrA14 genes from strain CBG15936 to E. coli J53 occurs at a frequency of 1.68 × 10–3 events per donor cell (Figure 2). And the transconjugants were observed to acquire resistance to piperacillin, cefepime, ceftriaxone, ceftazidime, cefotetan, cefazolin, and imipenem (Table 1). According to MLST, strain CBG15936 is Sequence Type 932. Unicycler assembly results showed that this strain contains two plasmids with lengths of 75,044 and 62,663 bp. The ∼75-kb plasmid was designated pTEM-CBG, and it contains four drug resistance genes: fosA3 (conferring fosfomycin resistance), rmtB (conferring aminoglycosides resistance), blaCTX–M–65 (conferring cephalosporin resistance), and blaTEM–1B (conferring penicillin resistance). The ∼62-kb plasmid was designated pNDM1-CBG, and it contains three resistance genes: qnrS1 (conferring quinolone resistance), dfrA14 (conferring trimethoprim resistance), and blaNDM–1 (conferring carbapenem resistance).
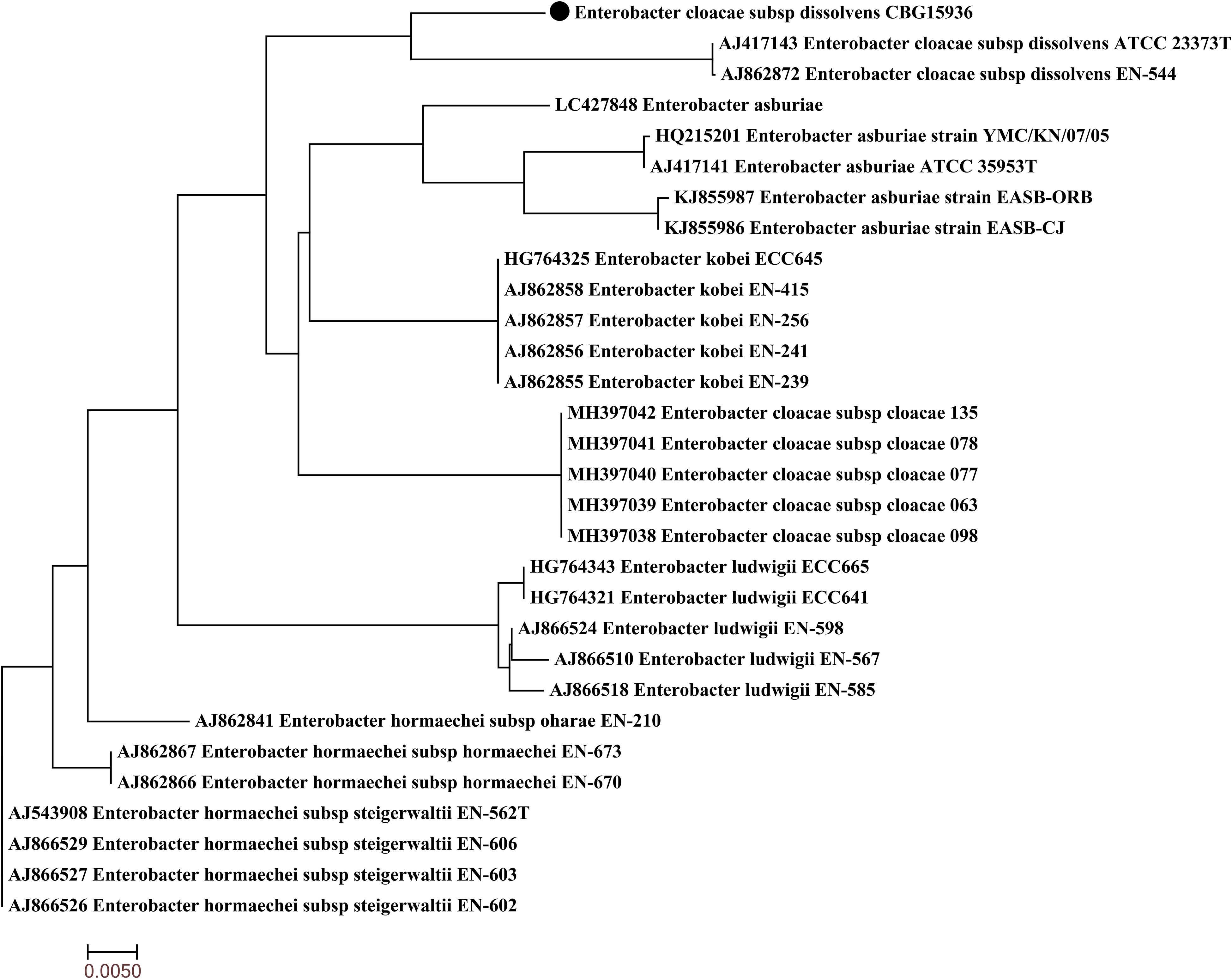
Figure 1. Phylogenetic tree of Enterobacter cloacae ssp. dissolvens strain CBG15936 with other 29 available E. cloacae complex hsp60 genes from GenBank. Strain CBG15936 is marked with the black dot.
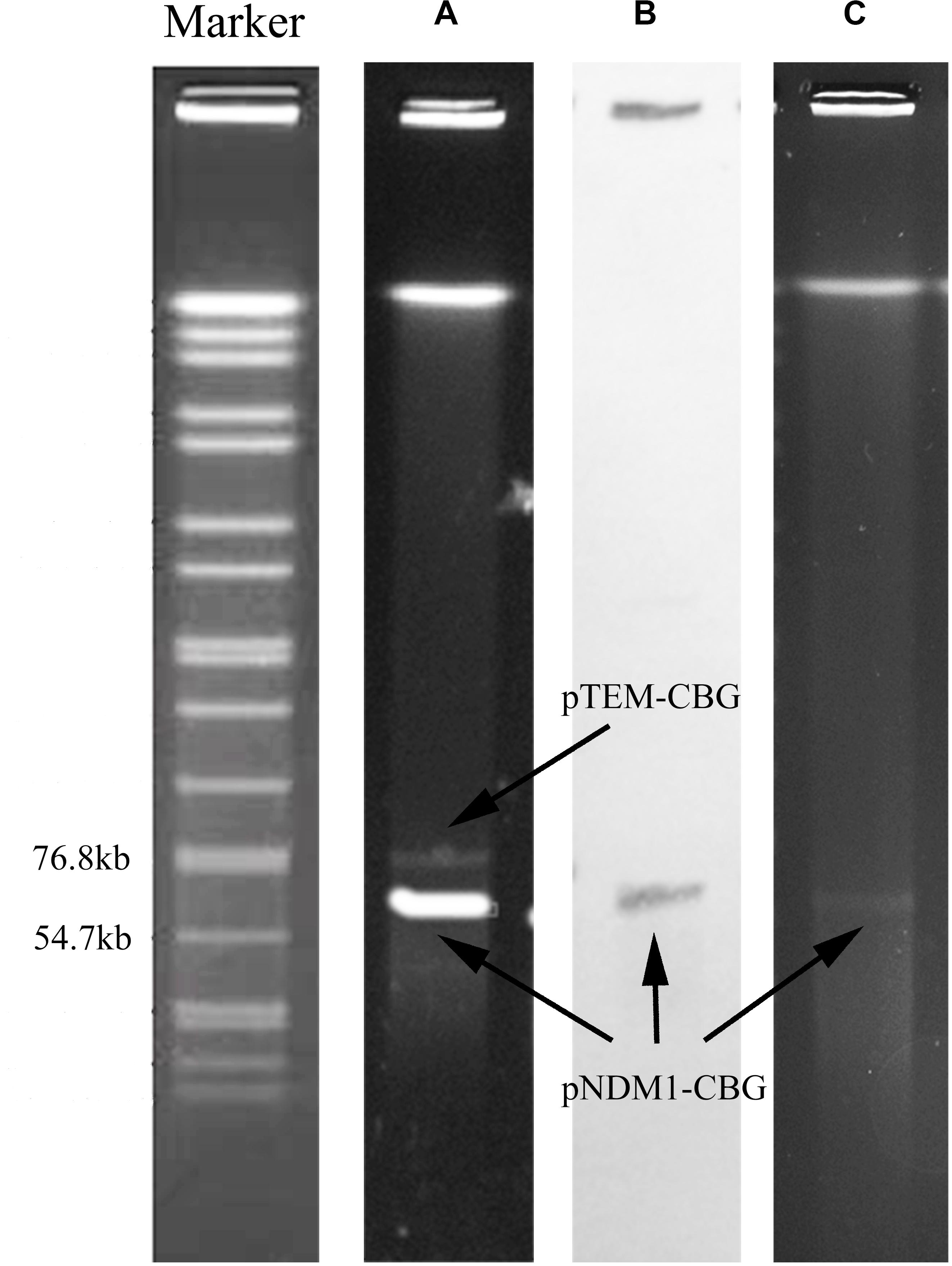
Figure 2. S1-PFGE pattern and Southern blotting for strain CBG15936 and E. coli J53 transconjugants. Lanes: Marker, Salmonella serotype Braenderup strain H9812 as a reference size standard. (A) PFGE result for S1-digested plasmid DNA of strain CBG15936. (B) Southern blot hybridization with probes specific to blaNDM–1; (C) PFGE result for S1-digested plasmid DNA of strain E. coli J53 transconjugants.
Genetic Features of the blaNDM–1-Carrying Plasmid
Plasmid pNDM1-CBG belongs to IncN1, has an average GC content of 52.34%, and has 91 open reading frames. Plasmid pNDM1-CBG also shares 99.95% identity and 100% coverage with plasmid pNDM-BTR, carried by E. coli BTR in Beijing, which was identified in 2013 (Zhao et al., 2017). Meanwhile, pNDM1-CBG shares 100% identity and 83% coverage with plasmid p378-IMP, carried by Pseudomonas aeruginosa in Chongqing, which was identified in 2013 (Feng et al., 2016). Both plasmids are composed of a similar ∼40-kb backbone and a variable multidrug resistance region. The backbone is composed of replication (repA), antirestriction (ardA and klcA), stability (stdB), conjugation (tra), and type IV secretion system (virB) genes. Compared with pNDM-BTR, pNDM1-CBG had an inversed ΔTn3000, an insertion of ISkpn19, a deletion of 144 bp upstream of the MDR region, and deletion of IS26 downstream of the MDR region. Compared with p378-IMP, pNDM1-CBG is missing ardA downstream of the antirestriction region, yet it is present before the stability region. Replacement of In823b (which contains blaIMP–4 downstream of the type IV secretion region) with In191 (which contains dfrA14) is observed in plasmid pNDM1-CBG (Figure 3A).
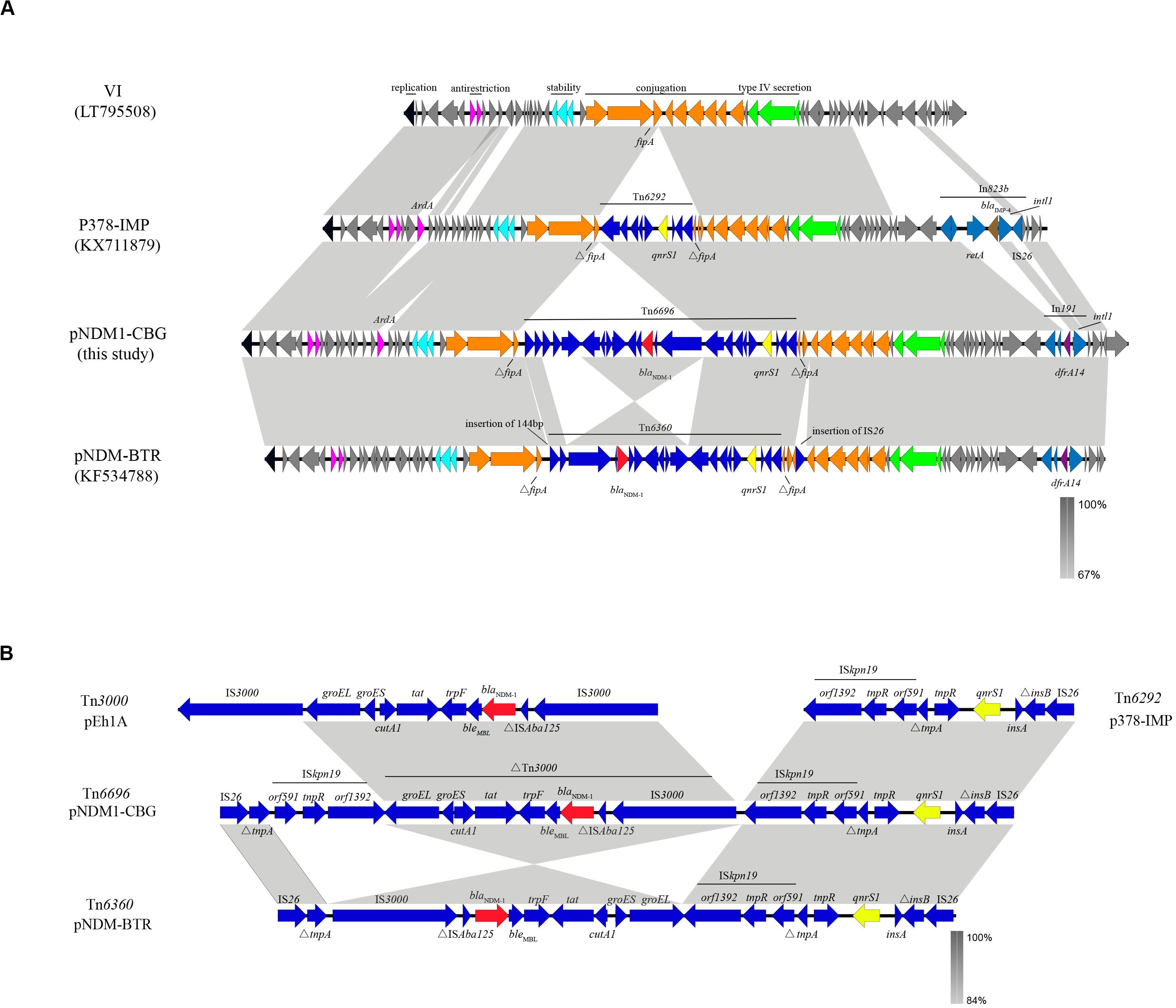
Figure 3. A comparative schematic diagram of (A) entire plasmids including VI (39,744 bp), p379-IMP (51,207 bp), pNDM1-CBG (62,663 bp), and pNDM-BTR (59,400 bp) and (B) transposons including Tn6292 in p37-IMP, Tn6696 in pNDM1-CBG, Tn6360 in pNDM-BTR, and Tn3000. Open reading frames are indicated with arrows. Homology regions among different plasmids are denoted with light gray coloring. Gene backbones are shown in dark gray. Black, carmine, cyan, orange, and green arrows represent genes association with replication, antirestriction, stability, conjugation, and the type IV secretion system, respectively. Accessory modules are shown in blue; gene backbones of integron are shown in royal blue; blaNDM–1 is shown in red; qnrS1 is shown in yellow; blaIMP–4 is shown in brown; dfrA14 is shown in purple.
The MDR region in pNDM1-CBG is composed of a novel transposon designated Tn6696. The latter consists of ΔTn3000, Tn6292, IS26, ISkpn19, and ΔtnpA. The blaNDM–1 gene is located in ΔTn3000 between a truncated ISAba125 and the ble gene. Compared with Tn3000, IS3000 is deleted in ΔTn3000, which is organized as IS3000-ΔISAba125-blaNDM–1-ble-trpF-tat-ΔcutA1-groES-groEL. Tn6292 was first identified in p378-IMP and organized as orf1393-tnpR-orf591-ΔtrpA-tnpR-qnrs1-insA-ΔinsB-IS26. Tn6696 is a combination of Tn6292 and a truncated Tn3000 and is similar to Tn6360, which was first identified in pNDM-BTR. Meanwhile, an inversed ΔTn3000 and deletion of ISkpn19 characterize Tn6360 (Figure 3B).
Discussion
E. cloacae have a broad range of hosts and have been isolated from wastewater, patients, and soil microcosms (Osborn et al., 2000; Chen et al., 2014; Zhang et al., 2014). Here, we report an NDM-1-producing MDR E. cloacae ssp. dissolvens strain, CBG15936, recovered from China. Our previous study showed that the expression of blaNDM–5 differed in plasmids and that the transconjugants with one plasmid exhibited higher-level carbapenem resistance than those with two plasmids or larger plasmids (Yang et al., 2020). Resistance to meropenem of transconjugants and strain CBG15936 may be affected by different NDM expressions and numbers of plasmids. This strain contains a novel transposon, Tn6696, carrying blaNDM–1 and located on the IncN1 plasmid pNDM1-CBG. Considering broad-host-range plasmids generally occurs at a variable frequency from 10–3 to 10–6 (Grohmann et al., 2003), pNDM1-CBG exhibited a relatively high transfer frequency. Our results support the potential for cross-species transmission to occur.
The novel transposon, Tn6696, identified in pNDM1-CBG contains an inverted Tn3000 remnant and an intact Tn6292. The latter is a Tn3-family transposon with an IS26 at the right end and has previously been found in a pP378-IMP plasmid obtained from P. aeruginosa recovered in China (Feng et al., 2016). Tn3000 was first located in a pEh1A plasmid from Enterobacter hormaechei E0083033-1 and in pEc2A from E. coli E0083033-2 recovered from Brazil (Campos et al., 2015). Therefore, a reorganization event involving fusion of Tn3000 and Tn6292 may have created Tn6696. The NDM-1 gene in Tn6696 is bracketed by two copies of the insertion sequence, ISkpn19. It is hypothesized that the resulting special structure has the potential to mobilize a drug resistance gene (Wu et al., 2016). However, Tn6696 had an additional copy of ISkpn19 and an inverted ΔTn3000 compared with Tn6360, suggesting that the former may originate from the latter through transposition and inversion of the ISkpn19-ΔTn3000 structure.
Plasmid pNDM1-CBG also shares high similarity, yet lower coverage, with plasmid VI from E. coli. The main difference between these plasmids is the quite variable MDR regions, which are all inserted in the fipA gene at the same position. In a previous study, fipA was shown to be interrupted into two fragments by MDRs in many different plasmids (Zhao et al., 2017; Yang et al., 2018). Thus, fipA may serve as a “hot spot” for integration of mobile genetic elements.
Conclusion
Here, we report a new transposon, Tn6696, in the IncN1 plasmid, pNDM1-CBG, from a ST932 MDR E. cloacae ssp. dissolvens strain recovered in China. We describe the structures of both pNDM1-CBG and Tn6696 in detail. Our work provides a new perspective regarding the potential for a novel horizontal transfer of NDM-1 via Tn6696 to occur among IncN1 plasmids. The latter finding is of great significance for future studies of the dissemination of blaNDM–1 in different species and its monitoring.
Data Availability Statement
The datasets generated for this study can be found in the CP046116, CP046117, and CP046118.
Author Contributions
PL, HM, and HS conceived and designed the experiments. QC, LL, YL, and KW performed the experiments. ZL, PHL, and LY analyzed the data. QC and PL wrote the manuscript. All authors read and approved the final manuscript.
Funding
The study was supported by the grants from the National Science and Technology Major Project (Grant No. 2018ZX10712001-002-002) and the Beijing Nova Program (Grant No. Z181100006218110).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Ahmad, N., Khalid, S., Ali, S. M., and Khan, A. U. (2018). Occurrence of blaNDM variants among Enterobacteriaceae from a neonatal intensive care unit in a northern india hospital. Front. Microbiol. 26:92.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., and Edwards, R. A. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bobenchik, A. M., Deak, E., Hindler, J. A., Charlton, C. L., and Humphries, R. M. (2015). Performance of vitek 2 for antimicrobial susceptibility testing of Enterobacteriaceae with vitek 2 (2009 FDA) and 2014 CLSI breakpoints. J. Clin. Microbiol. 53, 816–823. doi: 10.1128/JCM.02697-14
Campos, J. C., da Silva, M. J., dos Santos, P. R., Barros, E. M., and Pereira, M. D. O. (2015). Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco and India. Antimicrob. Agents Chemother. 59, 7387–7395. doi: 10.1128/AAC.01458-15
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Hu, H., Chavda, K. D., Zhao, S., Liu, R., Liang, H., et al. (2014). Complete sequence of a KPC-Producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob. Agents Chemother. 58, 2422–2425. doi: 10.1128/AAC.02587-13
Clinical and Laboratory Standards Institute [CLSI] (2018). M100 S28 Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical Laboratory Standards Institute.
Davin-Regli, A., Lavigne, J. P., and Pagès, J. M. (2019). Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 32:e00002-19. doi: 10.1128/CMR.00002-19
Davin-Regli, A., and Pagès, J. M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6:392. doi: 10.3389/fmicb.2015.00392
Daw, M. A., Munnelly, P., Mccann, S. R., Daly, P. A., and Keane, C. T. (1989). Value of surveillance cultures in the management of neutropenic patients. Eur. J. Clin. Microbiol. 7, 742–747. doi: 10.1007/BF01975040
Fairley, D., Protaschik, Y., Finn, R., Turton, J., and Doumith, M. (2019). Investigation of a hospital Enterobacter cloacae NDM-1 outbreak using whole genome sequencing. Microbiol. Soc. 1:1. doi: 10.1099/acmi.ac2019.po0391
Feng, W., Zhou, D., Wang, Q., Luo, W., Zhang, D., and Sun, Q. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Grohmann, E., Muth, G., and Espinosa, M. (2003). Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301. doi: 10.1128/mmbr.67.2.277-301.2003
Jamal, W., Rotimi, V. O., Albert, M. J., Khodakhast, F., Nordmann, P., and Poirel, L. (2013). High prevalence of VIM-4 and NDM-1 metallo-β-lactamase among carbapenem-resistant Enterobacteriaceae. J. Med. Microbiol. 62, 1239–1244. doi: 10.1099/jmm.0.059915-0
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2016). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jia, X., Dai, W., Ma, W., Yan, J., He, J., Li, S., et al. (2018). Carbapenem-Resistant E. cloacae in Southwest China: molecular analysis of resistance and risk factors for infections caused by NDM-1-producers. Front. Microbiol. 9:658. doi: 10.3389/fmicb.2018.00658
Karlowsky, J. A., Lob, S. H., Kazmierczak, K. M., Badal, R. E., Young, K., and Motyl, K. R. (2017). In vitro activity of imipenem against carbapenemase-positive Enterobacteriaceae isolates collected by the SMART global surveillance program from 2008 to 2014. J. Clin. Microbiol. 55, 1638–1649. doi: 10.1128/JCM.02316-16
Khalid, S., Ahmad, N., Ali, S. M., and Khan, A. U. (2019). Outbreak of efficiently transferred carbapenem-resistant blaNDM-producing gram-negative Bacilli isolated from neonatal intensive care unit of an indian hospital. Microb. Drug Resist. 26, 284–289. doi: 10.1089/mdr.2019.0092
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, B., Feng, J., Zhan, Z., Yin, Z., Jiang, Q., and Wei, P. (2018). Dissemination of KPC-2-encoding IncX6 plasmids among multiple Enterobacteriaceae species in a single chinese hospital. Front. Microbiol. 9:478. doi: 10.3389/fmicb.2018.00478
Li, R., Zhu, H., Ruan Qian, W., Fang, X., Shi, Z., Li, Y., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Mayhall, C. G., Lamb, V. A., Gayle, W. E., and Haynes, B. W. (1979). Enterobacter cloacae septicemia in a bum center: epidemiology and control of an outbreak. J. Infect. Dis. 139, 166–171. doi: 10.1093/infdis/139.2.166
Osborn, A. M., Pickup, R. W., and Saunders, J. R. (2000). Development and application of molecular tools in the study of IncN-related plasmids from lakewater sediments. FEMS Microbiol. Lett. 186, 203–208. doi: 10.1111/j.1574-6968.2000.tb09105.x
Perry, J. D., Naqvi, S. H., Mirza, I. A., Alizai, S. A., Hussain, A., Ghirardi, S., et al. (2011). Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J. Antimicrob. Chemother. 66, 2288–2294. doi: 10.1093/jac/dkr299
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(Suppl. 1), 32–36. doi: 10.1093/nar/gkj014
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Torres-Gonzalez, P., Bobadilla-Del Valle, M., Tovar-Calderon, E., Leal-Vega, F., Hernandez-Cruz, A., Martinez-Gamboa, A., et al. (2015). Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-β-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob. Agents Chemother. 59, 7080–7083. doi: 10.1128/AAC.00055-15
Whittle, G., Shoemaker, N. B., and Salyers, A. A. (2002). The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59, 2044–2054. doi: 10.1007/s000180200004
Wu, W., Espedido, B., Feng, Y., and Zong, Z. (2016). Citrobacter freundii carrying blaKPC-2 and blaNDM-1: characterization by whole genome sequencing. Sci. Rep. 6:30670. doi: 10.1038/srep30670
Yang, L., Li, P., Liang, B., Hu, X., Li, J., and Xie, J. (2018). Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci. Rep. 8:10653. doi: 10.1038/s41598-018-28879-9
Yang, L., Lin, J., Lu, L., Xue, M., Ma, H., and Guo, H. (2020). Coexistence of two blaNDM- 5 genes carried on IncX3 and IncFII plasmids in an Escherichia coli isolate revealed by illumina and nanopore sequencing. Front Microbiol. 11:195. doi: 10.3389/fmicb.2020.00195
Yi, H., Cho, Y.-J., Yong, D., and Chun, J. (2012). Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J. Bacteriol. 194, 3742–3743. doi: 10.1128/JB.00641-12
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a New Metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhang, G. Q., Yao, Y. H., Yu, X. L., and Niu, J. J. (2014). A survey of five broad-host-range plasmids in gram-negative bacilli isolated from patients. Plasmid 74, 9–14. doi: 10.1016/j.plasmid.2014.05.002
Zhang, X., Lou, D., Xu, Y., Shang, Y., Li, D., Huang, X., et al. (2013). First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol. 13:282. doi: 10.1186/1471-2180-13-282
Keywords: Enterobacter cloacae, carbapenem resistance, blaNDM1, Tn6696, whole genome sequencing
Citation: Chen Q, Lin Y, Li Z, Lu L, Li P, Wang K, Yang L, Ma H, Li P and Song H (2020) Characterization of a New Transposon, Tn6696, on a blaNDM–1-Carrying Plasmid From Multidrug-Resistant Enterobacter cloacae ssp. dissolvens in China. Front. Microbiol. 11:525479. doi: 10.3389/fmicb.2020.525479
Received: 09 January 2020; Accepted: 18 August 2020;
Published: 15 September 2020.
Edited by:
Constantinos Tsioutis, European University Cyprus, CyprusReviewed by:
Asad U. Khan, Aligarh Muslim University, IndiaSulagna Basu, National Institute of Cholera and Enteric Diseases (ICMR), India
Copyright © 2020 Chen, Lin, Li, Lu, Li Wang, Yang, Ma, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Ma, mahui-28@163.com; Peng Li, jiekenlee@126.com; Hongbin Song, hongbinsong@263.net
†These authors have contributed equally to this work
 Qichao Chen1†
Qichao Chen1† Yanfeng Lin
Yanfeng Lin Zhonghong Li
Zhonghong Li Kaiying Wang
Kaiying Wang Peng Li
Peng Li Hongbin Song
Hongbin Song