- 1Proteomics and Spectroscopy Unit (ZBS 6), Centre for Biological Threats and Special Pathogens, Robert Koch Institute, Berlin, Germany
- 2Food Chemistry and Molecular Sensory Science, Technical University of Munich, Munich, Germany
- 3Functional Microbiology, Institute of Microbiology, Department of Pathobiology, University of Veterinary Medicine Vienna, Vienna, Austria
Bacillus cereus plays an often unrecognized role in food borne diseases. Food poisoning caused by this pathogen is manifested by either diarrhea or emesis. Due to the relatively high prevalence of emetic toxin cereulide associated food poisoning, methods for simple and reliable detection of cereulide producing strains are of utmost importance. Recently, two different studies reported on the application of MALDI-ToF MS for either the differentiation of emetic and non-emetic strains of B. cereus or for direct detection of cereulide from bacterial colony smears. However, for implementation of cereulide detection using MALDI-ToF MS in routine microbiological diagnostics additional investigations on the sensitivity and specificity as well as on the fitting into common workflows for bacterial identification are needed. These aspects prompted us to investigate open issues and to test sample preparation methods, commonly used for microbial identification for their suitability to detect the emetic toxin from bacteria. Based on our experimental findings we propose a workflow that allows identification of B. cereus and sensitive detection of cereulide in parallel, using linear-mode MALDI-ToF MS equipment. The protocol was validated in a blinded study and is based on the well-established ethanol/formic acid extraction method. Cereulide is detected in the ethanol wash solution of samples identified as B. cereus as peaks at m/z 1175 and 1191. Peak position difference of 16 Th (Thomson) indicates detection of the sodium and potassium adducts of cereulide. This sample treatment offers possibilities for further characterization by more sophisticated LC-MS-based methods. In summary, the ease of use and the achieved level of analytical sensitivity as well as the wide-spread availability of MALDI-ToF MS equipment in clinical microbiological laboratories provides a promising tool to improve and to facilitate routine diagnostics of B. cereus associated food intoxications.
Introduction
Bacillus cereus is ubiquitously distributed in nature and commonly isolated from soil and food. It is a well-recognized causative agent of two different types of gastrointestinal diseases mediated by toxins but is also increasingly reported to be linked to non-gastrointestinal diseases, such as nosocomial or eye infections. The diarrheal form of food borne diseases is presumably caused by enterotoxin production of B. cereus in the intestine while the emetic syndrome is caused by the depsipeptide toxin cereulide, preformed in foods contaminated with emetic strains of B. cereus (Ehling-Schulz et al., 2019). Intoxication caused by cereulide leads to nausea and vomiting after 0.5–6 h incubation and lasts up to 24 h (Ehling-Schulz et al., 2004). The emetic symptoms are usually self-limiting, however severe cases, such as liver failure and acute encephalopathy, have been reported (Rouzeau-Szynalski et al., 2020). Using a pig model, it was shown recently that cereulide is taken up by the body, efficiently translocated and accumulate in certain host organs and tissues (Bauer et al., 2018).
Cereulide is synthesized by a NRPS, which is encoded by the cereulide synthetase (ces) gene (Ehling-Schulz et al., 2005b) located on a megaplasmid (Ehling-Schulz et al., 2006). This heat- and acid-stable cyclic dodecadepsipeptide toxin has a molecular mass of 1152.5 Da and contains three repeats of the amino acids L-O-Val-L-Val-D-O-Leu-D-Ala. A number of variants, such as isocereulides A-G (Marxen et al., 2015a) or homocereulide (Pitchayawasin et al., 2004; Naka et al., 2019) have been described in detail, which vary in their ionophoric properties and toxicity, thereby demonstrating the remarkably high level of chemodiversity of this potent toxin.
Bacillus cereus can be frequently found as a microbiological contaminant in a wide range of different foods. As an endospore forming bacterium it is difficult to eliminate from the food chain and therefore is considered a major problem in food production processes (Andersson et al., 1995; Rouzeau-Szynalski et al., 2020). Most of the B. cereus strains from randomly collected food samples are able to produce at least one of the diarrheal toxins (>90%) while only a small portion is able to produce the emetic toxin cereulide (1–10%) (Hoton et al., 2009; Messelhausser et al., 2014). Although the prevalence of emetic strains is generally rather low, up to 30% of certain food classes linked to foodborne outbreaks contained emetic B. cereus strains (Hoton et al., 2009; Messelhausser et al., 2014). It is thought that the incidence of foodborne B. cereus intoxications is currently underestimated, due to short duration of illness, missing awareness in the clinical field and the fact that reporting of B. cereus intoxications to authorities is obligatory in many countries only in case of outbreaks (EFSA, 2016). Furthermore, detection of the cereulide toxin itself is tedious and rather challenging due to the comparably low toxic dose and the lack of enrichment strategies.
A simple, reliable and sensitive direct detection method for cereulide in frame of routine microbiology diagnostics would therefore be desirable. Polymerase chain reaction (PCR) is used to screen bacterial isolates for the ces gene, which is, however, not a verification of the actual toxin production (Fricker et al., 2007) and recently a machine learning based Fourier Transform Infrared spectroscopy method for discrimination of emetic and non-emetic strains has been described (Bagcioglu et al., 2019). Currently, liquid-chromatography coupled to mass spectrometry (LC-MS) is the gold standard for cereulide detection and quantification (Bauer et al., 2010; Biesta-Peters et al., 2010; Stark et al., 2013). Based on a SIDA assay (Bauer et al., 2010), an ISO method (EN-ISO, 18465) for accurate quantitation of cereulide in complex matrices, such as foods, has been established lately. However, the latter technique is complex and is available mostly in specialized analytical laboratories. Within the last decade MALDI-ToF MS has revolutionized the way pathogens are identified (Seng et al., 2009; Nomura, 2015; Schubert and Kostrzewa, 2017; Welker et al., 2019). Today, MALDI-ToF MS is used worldwide in laboratories for routine identification of clinically relevant pathogenic microorganisms.
Thus, it was evident that the potential of standard MALDI-ToF MS-based biotyping for detection of emetic strains and the cereulide toxin was tested. The suitability of MALDI-ToF MS for differentiation between emetic and non-emetic B. cereus strains was shown by statistical analyses of the linear-mode low-mass MALDI-ToF MS data (Ulrich et al., 2019). However, the specific biomarkers used for differentiation were found at m/z positions at 1171 and 1187 and would give an inexplicably large difference of approx. 4000 ppm, provided that the observed biomarkers represent the sodium and potassium adducts of the cereulide. As the molecular identity of the biomarker peaks was obviously not a subject of the aforementioned study, this inconsistency remains to be investigated. It was also reported that reflectron-mode MALDI-ToF MS permits detection of the sodium and potassium adducts of cereulide at m/z 1175.6 ([M + Na]+) and m/z 1191.6 ([M + K]+), respectively (Ducrest et al., 2019). The same study proposed matrix-free detection by laser desorption/ionization (LDI) MS, which was reported to result in a 1000-fold improvement of LOD (Ducrest et al., 2019). However, neither spectra or calibration curves were provided supporting the claimed analytic sensitivity for the emetic toxin.
Furthermore, both recently published studies (Ducrest et al., 2019; Ulrich et al., 2019) did not provide method evaluation or optimization of the experimental protocol for cereulide detection. The inconsistencies of the data and the limitations in the applicability of the proposed experimental protocols for simple, rapid and sensitive cereulide detection in a routine clinical laboratory have motivated us to systematically investigate the potential and limits of MALDI-ToF MS for cereulide detection in routine diagnostics.
To this end, we evaluated three different sample preparation methods commonly used in routine MS-based workflows for pathogen identification and tested different organic solvents for their ability to extract the emetic toxin from B. cereus materials. The experimental cereulide detection protocol was evaluated under blinded conditions and the LOD of cereulide detection from MALDI- and LDI-ToF MS measurements was determined. Furthermore, fragment analysis was performed for unambiguous identification of cereulide by means of MALDI-ToF/ToF MS. Our study demonstrates that a routine sample preparation method for MALDI-ToF MS biotyping allows both, identification of B. cereus and parallel detection of cereulide with sufficiently high analytical sensitivity.
Materials and Methods
Bacillus cereus Strains
The B. cereus emetic reference strain F4810/72 (Ehling-Schulz et al., 2005a) was used for protocol optimization experiments, and a test panel of strains, derived from the B. cereus strain collection of the Institute of Microbiology at the Vetmeduni Vienna, was compiled for evaluation of the optimized protocol in frame of a blinded study (see Table 1). The test panel, which comprised high, medium and low emetic toxin producing B. cereus strains as well as a so-called emetic-like [non-cereulide producer, but genetically closely related to emetic strain (Ehling-Schulz et al., 2005a)], and the non-emetic B. cereus type strain was sent blinded by Vetmeduni to the Robert-Koch Institute (RKI) Berlin. Classification of strains as high, medium or low toxin producers was carried out using UPLC-TOF MS (Stark et al., 2013).
Cultivation of B. cereus for MALDI-ToF MS Analysis
Bacteria were cultured on different media used in bacterial diagnostic labs under aerobic conditions for 24 h at 37°C before toxin extraction. The tested cultivation media included Casein-Soy-Peptone (Caso, Merck KGaA, Darmstadt, Germany), Luria-Bertani (LB, produced in house) or blood agar produced in house from Columbia blood agar base (CM0331, Oxoid, Wesel, Germany) and sheep blood [5% (v/v)].
Sample Preparation for Cereulide Detection in Bacterial Biotyping MALDI-ToF MS Workflows
Three sample preparation methods for bacterial biotyping by MALDI-ToF MS, (i) direct transfer, (ii) ethanol/FA and (iii) TFA extraction were compared for their suitability to detect the emetic toxin. Direct transfer of bacterial cells from blood, LB or Caso agar plates onto the MALDI target was performed according to the manufacturer’s instructions of the MALDI Biotyper system (MALDI Biotyper 2.0 User Manual, 2008, Revision 2, Bruker Daltonik, Bremen, Germany). The spots were overlaid with 1 μL of saturated matrix solution [12 mg/mL HCCA, in 70% (v/v) ACN, and 3% (v/v) TFA] and allowed to dry.
TFA and ethanol/FA extractions of the different B. cereus cultures were carried out as previously published (Lasch et al., 2008, 2016; Freiwald and Sauer, 2009). In case of TFA extraction 1 μL of the 1:10 diluted TFA extract was mixed on target with 1 μL of the saturated HCCA matrix solution. For ethanol/FA extraction B. cereus cells were brought in 300 μL double-distilled water before 900 μL ethanol were added; cf. (Freiwald and Sauer, 2009). Volumes of 1 μL of the 75% (v/v) ethanol fractions were mixed on target with 1 μL of the above-mentioned saturated matrix solution. The remaining volume (∼1200 μL) of the ethanol solution was dried and resolubilized in a volume of 20 μL pure ACN. From the concentrated ACN extract a volume of 1 μL was spotted onto a ground steel target (Bruker) and mixed with 1 μL of saturated matrix solution before drying. 1 μL of the FA sample solution was dried on target, overlaid by 1 μL of the above-mentioned matrix solution and dried again. ACN extraction of the cereulide from B. cereus cells was carried out under identical conditions, except that water and ethanol were replaced by 1200 μL ACN.
Cereulide Extraction From B. cereus Colony Material by Solvents
Colony material from blood cultures of B. cereus F4810/72 (24 h, 37°C) equivalent to three inoculation loops was suspended in 500 μL organic solvent (methanol, n-pentane, ethanol, ACN and acetone) by vortexing. The samples were sonicated for 5 min at room temperature in an ultra-sonication bath before insoluble material was removed by centrifugation for 5 min at 4000 × g. The supernatants were evaporated to dryness in a vacuum concentrator (SpeedVac, Uniequip, Martinsried, Germany). Dried samples were resolubilized in 20 μL ACN; resolubilized samples are in the following termed concentrated extracts. For MALDI-ToF MS measurements, 1 μL of the concentrated extracts were mixed on target with an equal volume of saturated HCCA matrix solution (see above) and allowed to air-dry before MS measurements.
Cereulide Detection and Characterization Using MALDI-ToF and LIFT-ToF/ToF MS
Dried samples were analyzed using an Autoflex Speed MALDI-ToF/ToF mass spectrometer (Bruker) in positive ion reflectron mode. The instrument was controlled by the FlexControl Software (Version 3.3.108) supplied by Bruker. External calibration was carried out using the peptide calibration standard II (Bruker). Full scan mass spectra were acquired in the m/z range of 700–3500 at 1 kHz repetition rate of the Smartbeam II Nd-YAG laser (excitation wavelength λ = 355 nm). Each recorded spectrum was summed from eight profiles, which were accumulated from 1000 laser shots each. Peaks were picked after smoothing and baseline subtraction using the SNAP algorithm implemented in the FlexAnalysis software (v. 3.3.80, Bruker). Cereulide was further characterized by LIFT-ToF/ToF MS analysis (Suckau et al., 2003). For this purpose, fragmentation spectra were accumulated at 200 Hz laser repetition rate and elevated laser energies from 1000 shots in parent acquisition and 1000 shots in fragment acquisition mode. The precursor ion selection range was 0.85% of the precursor mass.
Linear MALDI-ToF spectra were collected in the positive ion mode by means of an Autoflex I mass spectrometer (Bruker). The instrument was controlled by the FlexControl software and equipped with a nitrogen laser (λ = 337 nm) operating at a pulse rate of 10 Hz. Pulse ion extraction time was 10 ns and the sampling rate was set to 2.0 gigasamples per second (GS/s). MS measurements were carried out using an acceleration voltage of 20.00 (ion source 1) and 18.45 (ion source 2) kV. Lens voltage was 6.70 kV. Spectra were stored in the range between m/z 700 and 3500. Bruker’s peptide calibration standard II was employed for external calibration.
Blinded Validation of the MALDI-ToF MS Protocol for Detection of Cereulide Producing B. cereus Strains
All B. cereus strains belonging to the test panel, provided blinded by Vetmeduni Vienna to RKI (for details of strains see Table 1), were grown on Caso agar for 24 h at 37°C. Colony material was then processed by the ethanol/FA method (see above), which involved transfer into 300 μL of double-distilled water and adding of 900 μL ethanol. From the 75% (v/v) ethanol fractions a volume of 1 μL was mixed on target with 1 μL of saturated HCCA solution.
In addition, concentrated extracts were prepared by evaporating the remaining supernatants to dryness in a vacuum concentrator (SpeedVac) followed by resolubilization in 20 μL ACN (100%). Volumes of 1 μL of the concentrated extracts were mixed on target with the HCCA matrix solution (see above) before the measurements. MALDI-ToF mass spectra were obtained in positive reflectron mode using the Autoflex Speed instrument from Bruker. Peaks were detected using the SNAP algorithm implemented in FlexAnalysis (Bruker) with the TopHat baseline substitution. A minimum SNR of 3 was chosen in order to allow comparison with a recently published study (Ducrest et al., 2019). MALDI-ToF MS test results were then transmitted to the Vienna group for unblinding and for comparison with cereulide toxin data from UPLC-TOF MS profiling (Stark et al., 2013).
Determination of Detection Limits by MALDI- and Matrix-Free Laser Desorption Ionization (LDI) -ToF MS
Cereulide standard (50 μg/mL in ACN, product ID CX20422) was purchased from Chiralix (Nijmegen, Netherlands) and used to prepare two dilution series. In the first series, the cereulide standard was diluted by pure ACN to obtain concentrations between 0.5 ng/mL and 1 μg/mL; from these solutions a volume of 1 μL was either directly deposited onto a ground steel target (LDI) and allowed to air dry, or mixed before sample spotting with an equal volume of a saturated solution of HCCA (MALDI). For MALDI-ToF MS 5, or 6 target position spots were prepared while one mass spectrum was collected per spot. Samples for LDI-ToF MS measurements were prepared twice so at least 11, sometimes 12 LDI spectra were collected and analyzed. In the second dilution series, appropriate amounts of the cereulide standard were spiked into 75% (v/v) ethanol washing solutions of B. cereus ATCC 10987 cells (emetic-like strain without the ces gene, grow conditions see above) to adjust cereulide concentrations between 2 ng/mL and 1 μg/mL. Again, a volume of 1 μL was spotted onto a steel target (LDI), or mixed with 1 μL of saturated HCCA solution (MALDI).
Mass spectra from both cereulide concentration series were collected in reflector mode as described above, except that each recorded spectrum was obtained by summing up 500 laser shots fired consecutively at each of 20 predefined shot step positions (large spiral geometry, altogether 10,000 laser shots per target position spot). The objective of this highly standardized procedure was to allow reproducible determination of the cereulide amount.
Ultraperformance Liquid Chromatography – Mass Spectrometry (UPLC-MS/MS) Analysis of Cereulide and Isocereulides
Mass spectrometric analyses of ethanol washing solutions was performed on a Waters Xevo TQ-S mass spectrometer (Waters, Manchester, United Kingdom) coupled to an Acquity UPLC i-class core system (Waters, Milford, MA, United States) consisting of a binary solvent manager, sample manager, and column oven. Aliquots (2 μL) of prepared ethanolic extracts were injected into the UPLC/MS-MS system equipped with a 2.1 × 150 mm, 1.7 μm, UPLC CSH C18 column (Waters, Manchester, United Kingdom) and analyzed according to Marxen et al. (2015b).
Results
Direct Cereulide Detection From Colony Materials Prepared for Bacterial Typing
The suitability of different MALDI-ToF MS biotyping workflows, such as direct transfer, ethanol/FA and TFA extraction, for detection of the emetic toxin was investigated in B. cereus F4810/72 grown on different solid agars, blood, LB and Caso. The obtained reflectron MALDI-ToF mass spectra are shown in Figure 1.
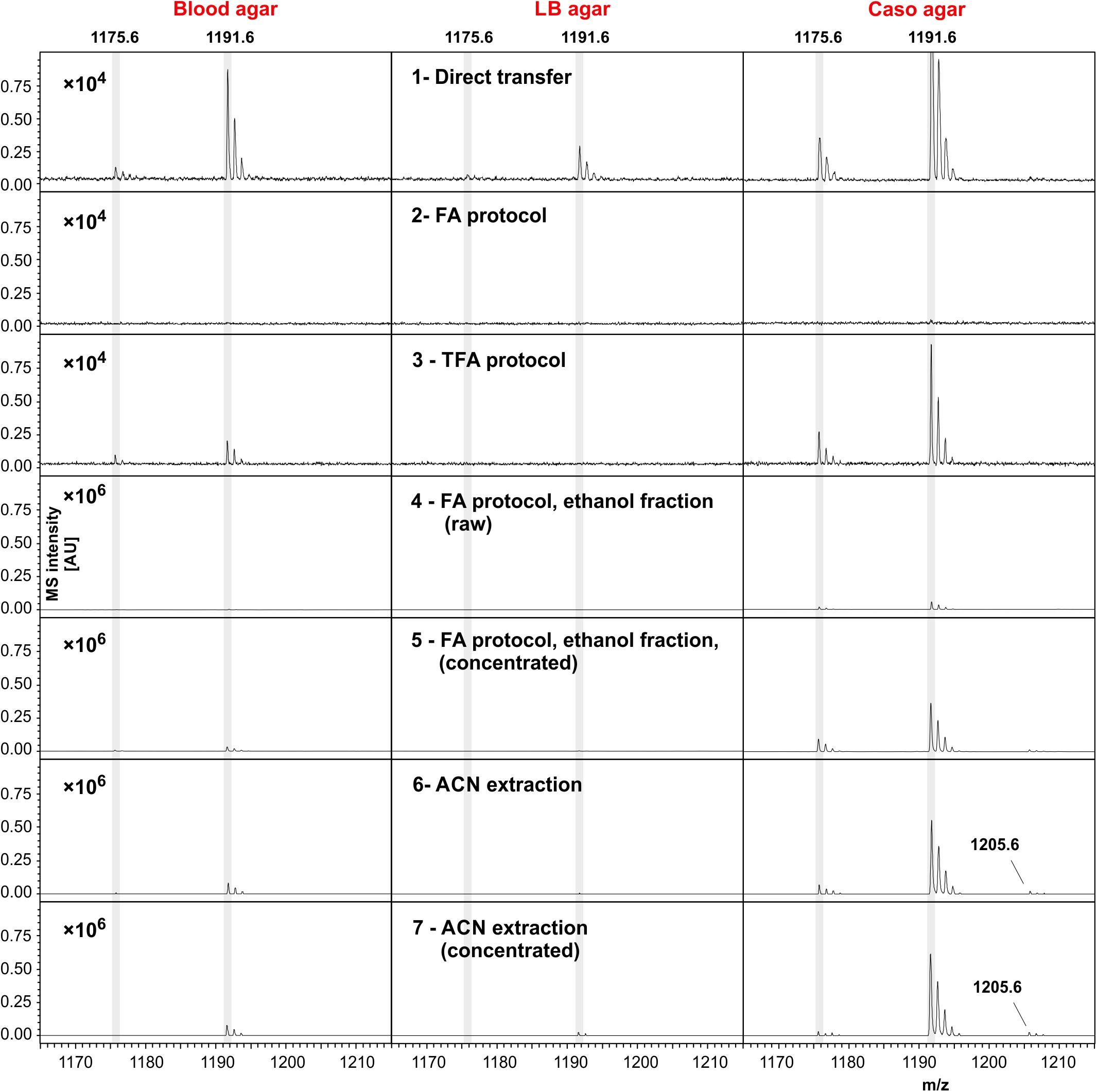
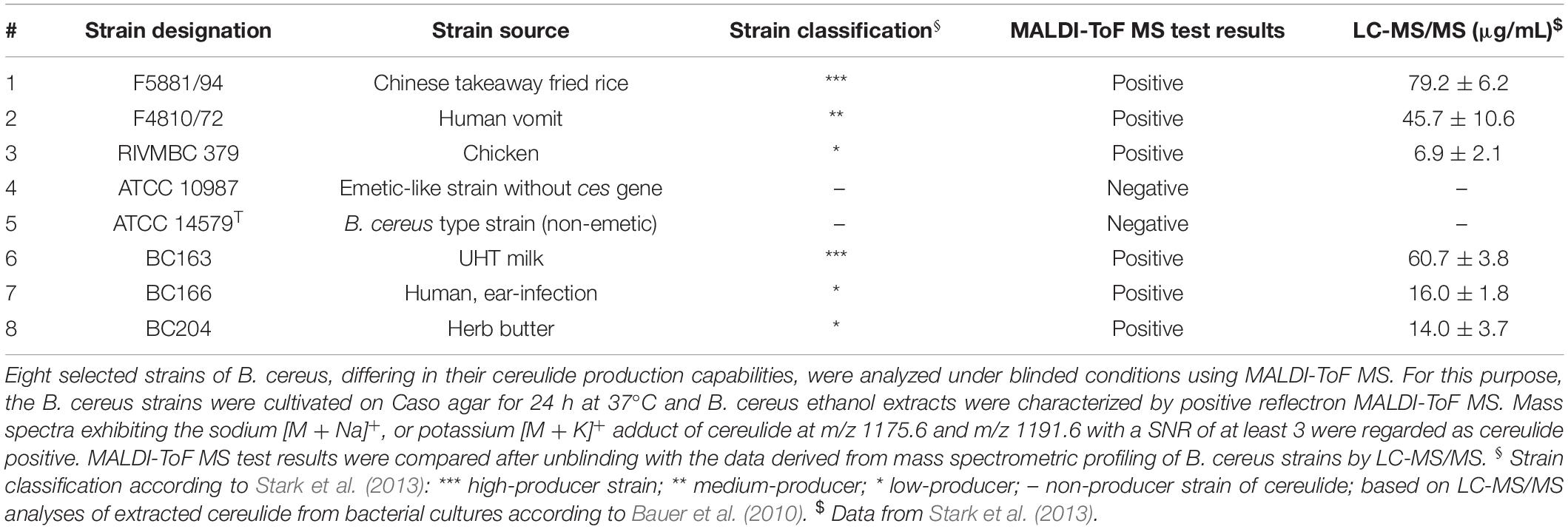
Figure 1. Cereulide detection in B. cereus samples cultivated using different cultivation media and different sample preparation, or cereulide extraction methods. Three commonly applied sample preparation techniques for bacterial biotyping workflows, namely the direct transfer (colony-smear) method and ethanol-formic acid (FA), or trifluoroacetic acid (TFA) extraction were utilized (cf. first three rows). In these cases mass spectra were recorded from B. cereus F4810/72 samples, or extracts thereof, grown on blood (left column), LB (middle column) or Caso agar (right column) using the positive reflectron mode. Rows 4 and 5 display mass spectra from the ethanol washing solution (raw, or concentrated) obtained when applying the standard ethanol/FA extraction technique. To obtain spectra of row 5, ethanol washing solutions were completely dried and solubilized afterward in a volume of 20 μL ACN. Spectra from these solutions were recorded after drying 1 μL and adding matrix on target steel plates. Row 6 and 7: Mass spectra acquired after extracting the cereulide by ACN (again as raw and concentrated extracts, respectively). Cereulide was detected as sodium [M + Na]+ adduct at m/z 1175.6 and as potassium [M + K]+ adduct at m/z 1191.6 (cf. shaded areas). Isocereulide A and/or isocereulide F was detected as potassium [M + K]+ adduct at m/z 1205.6 of B. cereus samples cultivated on Caso agar (visible only in rows 5–7). Note the different scaling of the intensity axes in rows 1–3 (factor 104) and lines 4–7 (×106). Sample preparation and MS measurements were carried under highly standardized conditions: same matrix, same instrument, 6000 laser shots co-added per sample and highly similar laser intensity.
In Figure 1 mass spectra obtained from B. cereus F4810/72 cultures grown on solid blood agar are depicted in the left column whereas the middle and right columns display spectra from LB and Caso agar cultures. Rows 1–3 illustrate reflector mode MALDI-ToF spectra from samples produced by the different preparation methods for MALDI biotyping in the m/z region between 1165 and 1215. Rows 4–5 depict spectra from the washing fraction used in the widely used ethanol/FA extraction protocol, namely the unprocessed (raw) ethanol washing fraction containing 25% (v/v) water and 75% (v/v) ethanol (row 4) and the concentrated ethanol fraction (row 5). Note that the intensity scaling in these panels differs from the scaling in rows 1–3 by a factor of 100. Cereulide was detected as sodium [M + Na]+ and potassium [M + K]+ adduct at m/z 1175.6 and 1191.6, respectively (see shaded areas). The highest intensities of emetic toxin peaks were generally found in preparations, or extracts of Caso agar cultures. Moderate or even low intensities of the peaks at m/z 1175.6 and 1191.6 were typically present in spectra from B. cereus cultures grown on blood or LB agar, respectively. Additionally, isocereulide A and/or isocereulide F was detected as potassium [M + K] + adduct at m/z 1205.6 in both measurements of ACN extracts.
The absence of cereulide signals in samples processed by the ethanol/FA method might be explained by the wash steps with 75% (v/v) ethanol solution (row 2 in Figure 1). Indeed, spectra from the raw and concentrated ethanol fractions exhibit mass signals of the cereulide (rows 4–5 in Figure 1). Thus, cereulide detection from cell extracts obtained by the widely used ethanol/FA extraction method cannot be recommended. Instead, the ethanol washing solution should be used to test for the presence of cereulide.
The effectiveness of cereulide extraction from the microbial samples was improved by replacing of the water/ethanol washing solution by an equal volume of pure ACN (rows 6–7 in Figure 1.) Under the given experimental conditions (B. cereus F4810/72, same growth, measurement and sample treatment conditions, cultivation on Caso or blood agar), direct detection of cereulide was possible from microbial cells washed by pure ACN.
Extraction of the Toxin From B. cereus Colony Material by Different Solvents
Five different organic solvents were tested for their efficacy to extract cereulide from B. cereus cultures. For this purpose, equal amounts of cell material from B. cereus F4810/72 cultivated on blood agar were used for systematically comparing the intensities of MALDI-ToF MS cereulide signals (Figure 2). In order to eliminate effects of the solvents on the matrix crystallization, all extracts were dried completely and resolubilized in ACN before spotting onto the MALDI target plates. The sodium [M + Na]+ and potassium [M + K]+ adducts of cereulide were detected at m/z 1175.6 and m/z 1191.6 in all samples with high signal-to-noise (SNR) ratios, however at different signal intensities. The protonated form of the cereulide at 1153.6 [M + H]+ could not be detected in any of the spectra. The systematic comparison of different solvents for cereulide extraction revealed, that direct acetone extraction of the bacterial cultures resulted in the highest and n-pentane in the lowest cereulide signal intensities. Nevertheless, ACN and ethanol, resulting in the second and third highest intensity, respectively, were chosen as extraction solvents for further experiments because detection of cereulide was possible in both cases by raw extracts, i.e., without further concentration by vacuum centrifugation. In contrast to ACN and ethanol, acetone is generally less suitable for MALDI-TOF MS because it may trigger the formation of large matrix crystals leading to limited reproducibility. ACN results in a more suitable crystallization whereas ethanol is used as a washing solution in a well-established sample processing method for microbial biotyping.
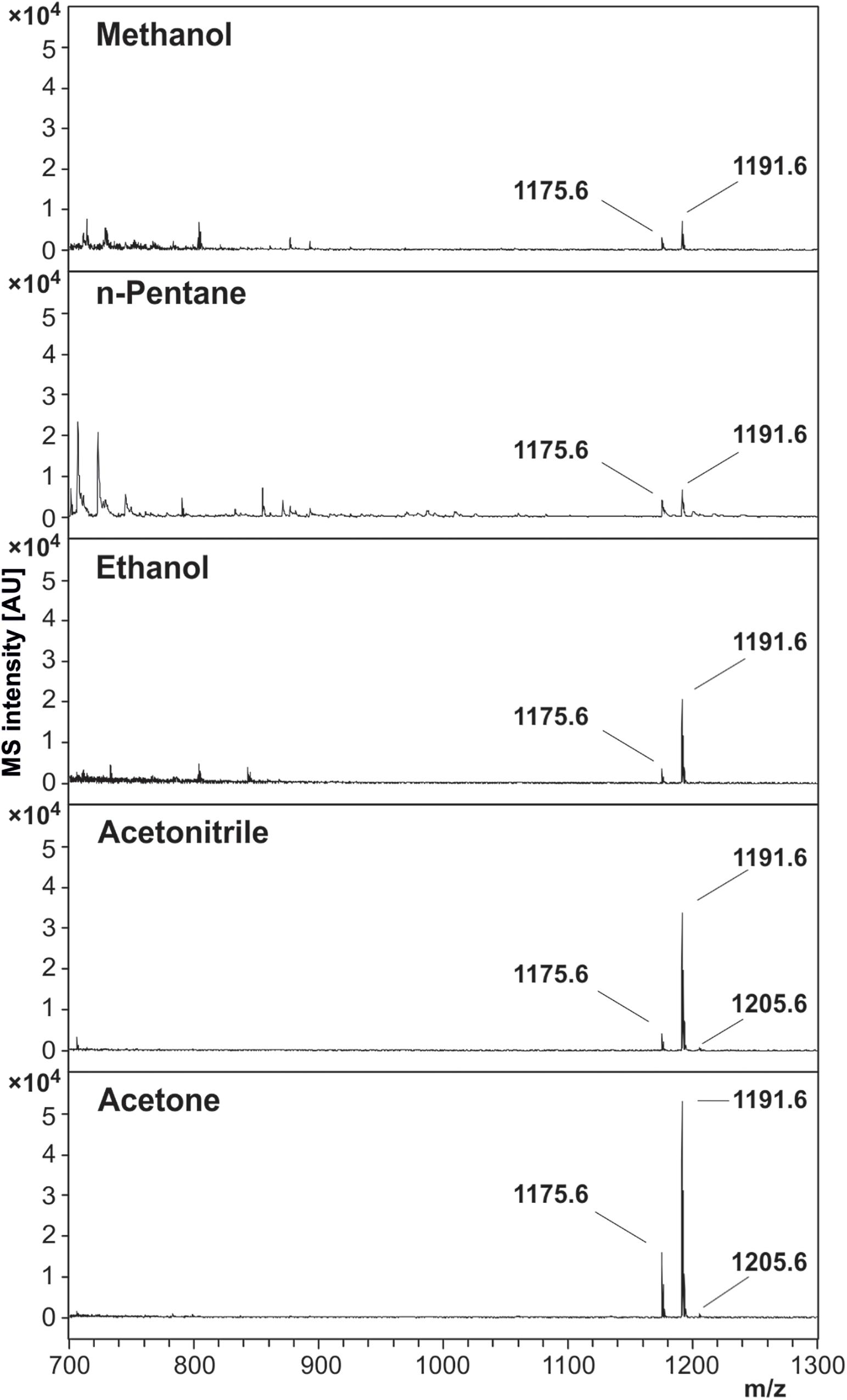
Figure 2. Effectivity of cereulide extraction by different solvents from B. cereus F4810/72 colony material. Five solvents, methanol, n-pentane, ethanol, ACN and acetone, were compared for their efficacy to extract the emetic toxin from cells of B. cereus. Equal amounts B. cereus F4810/72 colony material were used for solvent extraction. MALDI-ToF mass spectra were acquired from concentrated solvent extracts by accumulating altogether 8000 laser shots in the m/z range of 700–3500 (reflector mode, 1 μL extract + 1 μL matrix solution). Spectra are equally scaled. Cereulide peaks of each spectrum are labeled with their corresponding m/z value, whereas 1175.6 is the sodium [M + Na]+ and 1191.6 the potassium [M + K]+ adduct of cereulide. The peak at position 1205.6 is the potassium [M + K]+ adduct of isocereulide A and/or isocereulide F. The protonated form [M + H]+ of cereulide at m/z 1153.6 could not be detected in any of the mass spectra.
MALDI LIFT-ToF/ToF MS Spectrum of Cereulide
In order to confirm the molecular identity of peaks assigned in the literature as sodium ([M + Na]+) and potassium ([M + K]+) adducts of the cereulide, the peak at m/z 1191.6 was sequenced by MALDI LIFT ToF/ToF MS. For this purpose, 1 μL of the concentrated ACN extract of B. cereus F4810/72 was spotted onto a ground steel target. LIFT-ToF/ToF MS measurements revealed the formation of sodiated and potassiated fragment series of the cereulide. Both fragment series unambiguously confirmed the molecular identity of the peak at m/z 1191.6 as the potassium adduct ion peak of cereulide (Figure 3).
Figure 3. MALDI LIFT-ToF/ToF MS spectrum of cereulide. The potassium [M + K]+ adduct of cereulide at m/z 1191.6 was fragmented using laser induced dissociation (LID). Fragmentation resulted in observation of two series of sodiated and potassiated fragment ion adducts in the MALDI LIFT-ToF mass spectrum. The experimental m/z peak positions of these signals enabled assignment to a variety of fragment ions specific for cereulide and thus allowed unambiguous molecular assignment of the toxin.
Blinded Validation of MALDI-ToF MS for Cereulide Production of B. cereus Strains
Cereulide was qualitatively determined by MALDI-ToF MS under blinded conditions from the ethanol washing solutions of a panel of B. cereus strains, provided blindly by the Vetmeduni Vienna group to the RKI partner institution (Table 1). B. cereus strains were selected from the strain collection of the Vetmeduni group to test whether MALDI-ToF MS is suitable for detecting the emetic toxin over a broad concentration range. The test panel comprised a total of eight B. cereus strains, including two high-producer, one medium-producer, three low-producer and two non-producing strains. Cereulide production capacity of the strains was previously determined by means of SIDA and HPLC–MS/MS (Stark et al., 2013). The non-emetic B. cereus type strain ATCC 14579 and the emetic-like strain ATCC 10987, which is genetically closely related to the emetic B. cereus reference strain F4810/72 but is lacking the ces genes, were included as negative controls. Spectra were first obtained from raw ethanol washing solutions. In case of negative test results concentrated extracts were prepared and characterized afterward. All cereulide producing strains were correctly identified using MALDI-ToF MS, as described in the Materials and Methods section. Both cereulide negative strains were also correctly classified. Ethanol wash solutions were also analyzed by LC-MS to show that sample preparation is compatible with this technique, which could be used for further cereulide characterization (Supplementary Figure SI-01). An overview of the results is provided in Table 1.
Determination of the Limit of Detection (LOD) of Cereulide by MALDI- and LDI-ToF MS
It has been stated recently, that the sensitivity of cereulide detection can be improved by a factor of 1000 using matrix-free laser-desorption/ionization (LDI) MS (Ducrest et al., 2019). However, neither spectra, nor other LDI-derived data such as calibration curves were presented. Therefore, one of the goals of the present study was to investigate the applicability of LDI-ToF MS and to systematically compare the LODs of MALDI- and LDI-ToF MS-based cereulide detection. To this end, a dilution series of a commercial cereulide standard within the range of 0.5 ng/mL to 1 μg/mL was analyzed by performing 5 (MALDI) or 11 (LDI) replicate measurements using standardized MS parameters for data acquisition. The number of replicate measurements was increased for LDI in order to address the larger signal intensity variation compared to MALDI. The results of these reflectron-mode investigations are illustrated in Figure 4. The mean, minimum and maximum intensity values of the potassium peak of cereulide (m/z 1191.6) are plotted against the amount of cereulide per sample spot for MALDI and LDI-ToF MS measurements, respectively. The double-logarithmic representation of Figure 4A illustrates successful detection of an amount of 0.25 ng cereulide per sample spot in 3 of 5 MALDI-ToF MS measurements. These data suggest that the LOD under the given experimental conditions (MALDI-ToF MS, reflectron mode measurements, preparations of the pure toxin, Autoflex Speed mass spectrometer) equals approximately 0.25 ng (Figure 4). Furthermore, despite our large efforts to standardize data acquisition, application of LDI-TOF MS resulted in substantial variations of cereulide signal intensities, which significantly hampered accurate determination of the LOD. The intensity variations of the cereulide’s potassium adduct peak in MALDI- and LDI-ToF MS measurements are illustrated by the blue shaded areas in the Figures 4A,B. In the LDI case, a cereulide amount of 0.05 ng per spot was sufficient in 11 out of 11 cases whereas [M + K]+ peaks of 0.01 ng cereulide were found in 3 out of 11 spectra. Selected MALDI- and LDI-ToF MS spectra of the dilution series are presented in the Supporting Information (Supplementary Figure SI-02). The experimental data of the cereulide standard dilution series suggest that LDI-ToF MS improves the detection limit for cereulide by a factor of about 5–25. The improvement of the LOD by such a factor is remarkable, but clearly deviates from the claimed 1000-fold improvement by LDI (Ducrest et al., 2019). As the complex background of bacterial cells extracts could potentially alter the LOD, we repeated LOD determination in a second dilution series by spiking cereulide into the ethanolic wash fractions of the emetic-like strain B. cereus ATCC 10987. The results of these investigations were highly similar to data obtained by measurements of pure cereulide with LODs of 0.25 ng for MALDI- and 0.1 ng per spot for LDI-ToF MS (Supplementary Figure SI-03).
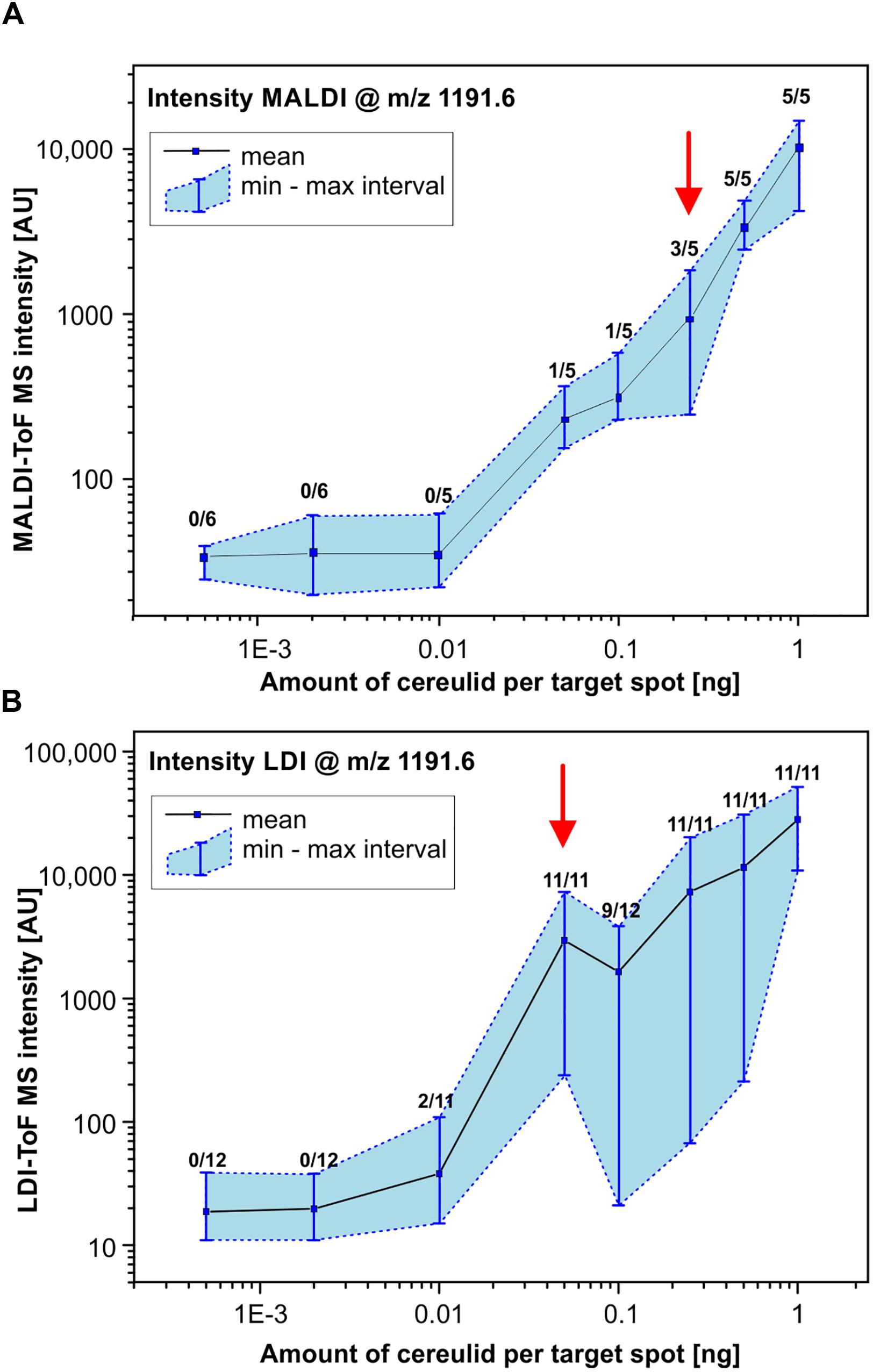
Figure 4. Determination of the limit of detection (LOD) of cereulide by MALDI- and LDI-ToF MS. MALDI-ToF MS (A) and matrix-free LDI-ToF MS (B) were used to determine the LOD of cereulide. For this purpose, a commercial cereulide standard was obtained and used to prepare a dilution series. Mass spectra were collected in reflector mode under strictly standardized conditions. Criteria for a positive test result are (i) a mass peak of the potassium adduct [M + K]+ of cereulide at m/z of 1191.6 and (ii) a SNR threshold of at least 3. Data points represent the mean and the min/max MS intensity values from 5 (6) (MALDI) or 11 (12) (LDI) measurements at each point of the dilution series. Furthermore, numbers of positive test results in relation to the total number of replicate measurements are indicated for each point of the curves. Note the double logarithmic scaling (see also text for details).
Detection of Cereulide in the Linear Mode of MALDI- and LDI-ToF MS
Bacterial identification is usually performed in an automated fashion using specialized MALDI-ToF MS instruments with dedicated and simplified hardware compared to the one’s utilized for cereulide detection by us at RKI and others (Ducrest et al., 2019; Ulrich et al., 2019). MS instruments for microbial identification often allow m/z determination in linear mode only and offer moderate levels of resolution and mass accuracy compared to reflector-based MALDI-ToF/ToF instruments. Therefore, we performed the analysis in the linear mode also by an instrument designed in the 1990s that is equipped with a low-cost N2 laser (Autoflex I) to examine if the detection of both cereulide adduct ions is possible by instrumentation similar to mass spectrometers used for microbial biotyping. Selected linear-mode spectra for MALDI- and LDI-ToF MS are opposed to the respective reflector-based spectra in Supplementary Figure SI-04. It was found, that cereulide detection is possible in linear mode and that LDI-type measurements improve the peak resolution compared to MALDI. However, as expected, spectral resolution and mass accuracy were lower in linear-mode mass spectra and particularly low in spectra obtained by the Autoflex I instrument.
Discussion
Bacillus cereus is a major concern in food production processes because of its high prevalence in food samples related to poisoning (Messelhausser et al., 2014). Recently, it has been proposed that MALDI-ToF MS might represent a suitable method for identifying cereulide producing strains of B. cereus (Ulrich et al., 2019) and for cereulide detection directly from bacterial cultures (Ducrest et al., 2019; Ulrich et al., 2019). Both studies use the colony-smear method, also known as direct transfer, for sample preparation of B. cereus cultures and rather advanced MALDI-ToF MS instrumentation compared to the equipment usually used for characterizing bacterial isolates. However, with regard to analytical sensitivity, specificity and applicability of cereulide detection in routine microbiological diagnostics, these studies left a number of questions unanswered. Therefore, this study aimed at identifying optimal sample preparation conditions to maximize the analytical sensitivity and to evaluate the fitting of the MALDI-ToF MS cereulide detection method into current diagnostic workflows. Our results of the comparison of common sample preparation methods for bacterial biotyping and the efficiency tests of different organic solvents for cereulide detection open up new possibilities for cereulide detection based on the well-accepted ethanol/FA sample preparation protocol proposed by Bruker in the commercial MALDI Biotyper workflow (Freiwald and Sauer, 2009). We propose a workflow for B. cereus identification and cereulide detection, which is illustrated in Figure 5. Bacterial cells are prepared using standard ethanol/FA extraction without discarding the wash fraction. Microbial species are identified using standard spectral database matching of experimental MALDI-ToF MS spectra in the m/z range between 2 and 20 kDa and, if B. cereus is identified, the wash fraction can be further analyzed by linear mode MALDI-ToF MS measurements in the m/z region between 700 and 2000 Da. If necessary, the sensitivity of this workflow can be increased concentrating the ethanol wash solution. If peaks at m/z 1175 and 1191 are detected in samples identified as B. cereus and if the peak position difference equals 16 Th (Thomson), these peaks are the sodium and potassium adducts of cereulide and therefore the cells must be emetic B. cereus strains. The workflow based on the ethanol/FA protocol was successfully evaluated in a blinded test, involving eight B. cereus strains with varying cereulide production capacities. These experiments revealed that all non-concentrated ethanol extracts tested contained sufficient amounts of the emetic toxin, which demonstrates the high analytical sensitivity of the technique. Furthermore, as revealed by our current study, the cereulide containing wash solution can be directly subjected to LC-MS/MS, which offers the possibility for unambiguous cereulide identification, isoform discrimination and accurate quantification (Supplementary Figure SI-01).
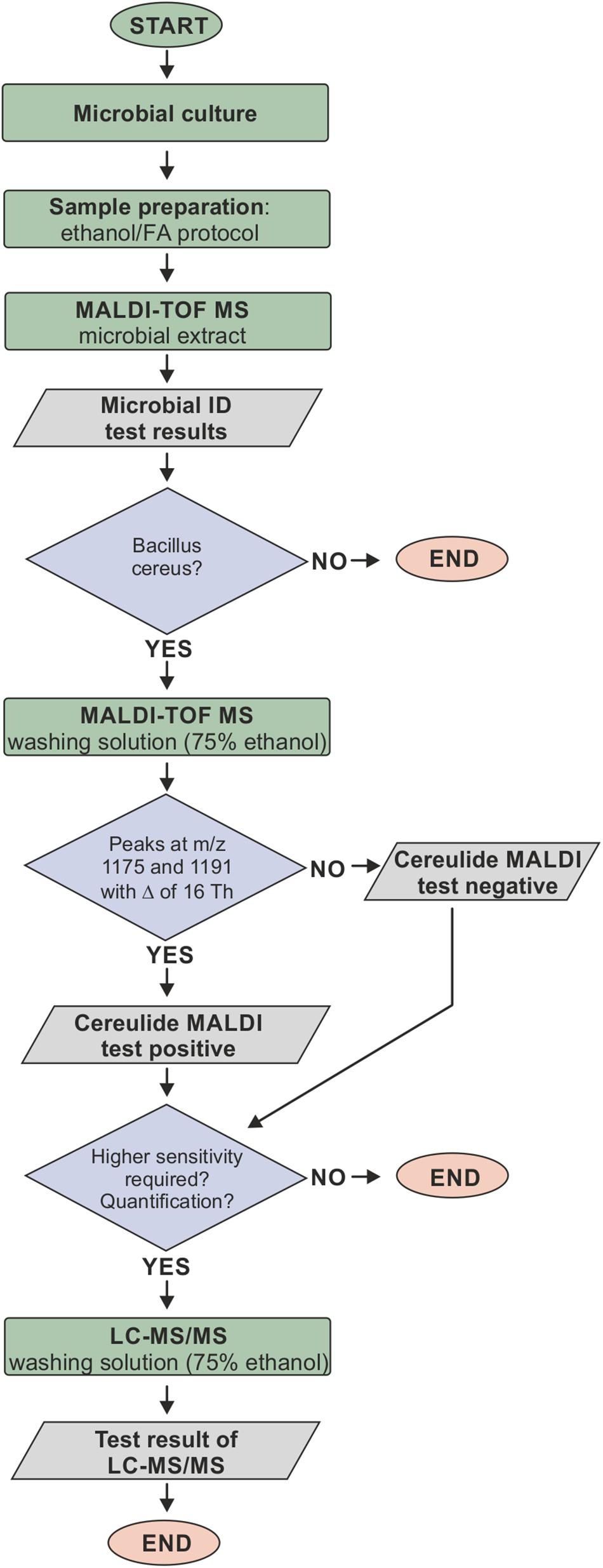
Figure 5. Proposed workflow for cereulide detection based on the ethanol/formic acid (FA) extraction protocol. Bacterial cells are prepared using the ethanol/FA extraction protocol without discarding the wash fraction. Microbial species are identified using standard spectral database matching of experimental MALDI-ToF mass spectra in the m/z range between 2 and 20 kDa. If B. cereus is identified, the wash fraction is analyzed in the m/z region between 700 and 2000 Da. The detection of sodium and potassium adduct peaks at m/z 1175 and 1191 with a m/z difference of 16 Th in samples identified as B. cereus indicates that these cells are producing cereulide. The wash fraction can furthermore be directly subjected to LC-MS/MS, which offers the possibility for unambiguous cereulide identification, isoform discrimination and accurate quantification.
Selectivity in mass spectrometry is largely dependent on mass accuracy and resolution. However, MALDI biotyping does not require particularly high mass resolution or high mass accuracy. Therefore, instruments specially designed for routine microbial biotyping, such as Microflex mass spectrometers from Bruker, are fairly widespread on the one hand but offer only limited options to achieve high mass accuracy and resolution. Consequently, hitherto rather advanced MS instruments were used for establishing cereulide detection methods (Ducrest et al., 2019; Ulrich et al., 2019). However, since the proposed method should ideally be widely employed to meet the needs of routine diagnostics, we aimed to establish a method for cereulide detection by utilizing entry-level mass spectrometers (linear mode, N2 laser, limited mass resolution, etc.).
Ulrich et al. (2019) claimed to have identified two specific biomarkers for cereulide production at m/z 1171 and 1187, which were initially identified by t-tests but later assigned to cereulide itself, because these mass peaks were “similar” to a cereulide standard. Obviously, the biomarker peaks at m/z 1171 and 1187 in Ulrich et al. (2019) correspond to the sodium and potassium adduct of cereulide, which were, however, detected in linear positive mode with mass errors of roughly 4000 ppm. The difficulty in the correct peak assignment by Ulrich et al. (2019) illustrates the specificity problem of MALDI-ToF MS if the molecule identification is solely based on the precursor mass, especially when using linear mode mass spectra of low mass accuracy and resolution. This issue becomes even more apparent when the complexity of the background increases, e.g., if complex samples such as food products are analyzed. Our work and the work of others (Ducrest et al., 2019) showed that fragmentation of cereulide from bacterial isolates using MALDI LIFT-TOF/TOF MS provides sequence information that enables unambiguous toxin detection and enhances selectivity, compared to the assignment of the precursor mass alone (Figure 3). However, fragment analysis requires rather advanced MALDI-ToF/ToF MS equipment, usually not available in routine diagnostic laboratories. Nevertheless, the detection of the emetic toxin is possible in linear mode with sufficient certainty (despite the lower resolution and the increased mass error of standard biotyping equipment), as at least two adduct ion peaks are usually detected that must occur at a mass difference of 16 Th. The distance between the sodium and potassium peaks of the cereulide is almost independent from calibration inaccuracies. In conjunction with bacterial species identification, this approach should enable molecular assignment of mass signals as cereulide at sufficiently high specificity.
Analytical sensitivity is another important aspect of cereulide detection. In the literature, the LOD for cereulide by MALDI-ToF MS was controversially reported as 1 μg/mL [cereulide standard dilution series, linear mode, Autoflex Speed (Ulrich et al., 2019)] and 30 ng/mL [cereulide standard, reflectron mode (Ducrest et al., 2019)] or even 30 pg/mL in case of LDI-ToF MS measurements (Ducrest et al., 2019). Of note, the latter data was unfortunately reported without providing any supporting information, such as spectra, calibration curves, etc. The claimed sensitivity for LDI-ToF MS-based detection of cereulide would correspond to approximately 25 amol cereulide per target spot.
Under our experimental conditions we were unable to confirm the reported massive sensitivity increase by LDI-ToF MS. For reflectron MALDI-ToF MS of the cereulide standard we determined a LOD of 0.25 ng per sample spot which falls in the range between the two reported values. The LOD of LDI-ToF MS was determined to vary between values of 10–50 pg per spot. This increase of sensitivity was, however, achieved at the costs of technical difficulties linked to LDI measurements and a largely reduced reproducibility (Figure 4). As an illustration, visual assessment of matrix crystals enables an experienced operator to identify suitable measurement positions (so called sweet spots) and to select positions with homogeneous crystal structures for subsequent data acquisition. In case of LDI-ToF MS measurements, however, matrix and matrix crystals are absent so that the quality of spectra acquired in the operator-guided mode may vary greatly. In frame of this study, MALDI- and LDI-ToF MS measurements were thus carried out by means of a pre-defined geometry of laser shot step positions. The results of these measurements clearly indicate low levels of repeatability and reproducibility, which may considered limiting factors for the applicability of LDI-ToF MS in a routine environment.
Conclusion
The ease of use and the wide-spread availability of MALDI-ToF MS equipment should be helpful to identify cereulide producing B. cereus strains in a routine setup and to diagnose food-associated cereulide intoxications by a simple, rapid and cost-effective technology. The proposed workflow is based on the well-established ethanol/FA extraction protocol and can be used without any modification of a sample preparation procedure routinely used for bacterial identification. It can be carried out with routine linear mode MALDI-ToF MS instrumentation and represents a suitable high-throughput technique to screen B. cereus strains for their actual emetic toxin production.
Data Availability Statement
The mass spectrometry data have been deposited at Zenodo with the dataset identifier 4020648 (https://zenodo.org/record/4020648).
Author Contributions
PL, ME-S, AS, and JD contributed to the conception and design of the study. AS and TS collected the data and performed the experiments. PL, ME-S, TS, and JD analyzed and interpreted the data. JD wrote the first draft of the manuscript. All authors wrote sections of the manuscript, contributed to manuscript revision, and read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank D. Jacob (RKI, Highly Pathogenic Microorganisms, ZBS 2) for providing emetic and non-emetic test strains of B. cereus. Furthermore, the authors are grateful to J. Rau (Chemisches und Veterinäruntersuchungsamt Stuttgart, Germany) for fruitful discussions and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.511674/full#supplementary-material
Abbreviations
ACN, acetonitrile; FA, formic acid; HCCA, α-cyano -4-hydroxycinnamic acid; LC, liquid-chromatography; LDI, laser desorption/ionization; LOD, limit of detection; MALDI-ToF, matrix-assisted laser desorption/ionization - time-of-flight; MS, mass spectrometry; NRPS, non-ribosomal peptide synthetase; SIDA, stable isotope dilution analysis; SNR, signal-to-noise ratio; Th, Thomson; TFA, trifluoroacetic acid.
References
Andersson, A., Rönner, U., and Granum, P. E. (1995). What problems does the food industry have with the spore-forming pathogens Bacillus cereus and Clostridium perfringens? Int. J. Food Microbiol. 28, 145–155. doi: 10.1016/0168-1605(95)00053-4
Bagcioglu, M., Fricker, M., Johler, S., and Ehling-Schulz, M. (2019). Detection and identification of Bacillus cereus, Bacillus cytotoxicus, Bacillus thuringiensis, Bacillus mycoides and Bacillus weihenstephanensis via machine learning based FTIR spectroscopy. Front. Microbiol. 10:902.
Bauer, T., Sipos, W., Stark, T. D., Kaser, T., Knecht, C., Brunthaler, R., et al. (2018). First insights into within host translocation of the bacillus cereus toxin cereulide using a porcine model. Front. Microbiol. 9:2652.
Bauer, T., Stark, T., Hofmann, T., and Ehling-Schulz, M. (2010). Development of a stable isotope dilution analysis for the quantification of the Bacillus cereus toxin cereulide in foods. J. Agric Food Chem. 58, 1420–1428. doi: 10.1021/jf9033046
Biesta-Peters, E. G., Reij, M. W., Blaauw, R. H., In ’T Veld, P. H., Rajkovic, A., Ehling-Schulz, M., et al. (2010). Quantification of the emetic toxin cereulide in food products by liquid chromatography-mass spectrometry using synthetic cereulide as a standard. Appl. Environ. Microbiol. 76, 7466–7472. doi: 10.1128/aem.01659-10
Ducrest, P. J., Pfammatter, S., Stephan, D., Vogel, G., Thibault, P., and Schnyder, B. (2019). Rapid detection of Bacillus ionophore cereulide in food products. Sci. Rep. 9:5814.
EFSA (2016). Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J. 14:e04524.
Ehling-Schulz, M., Fricker, M., Grallert, H., Rieck, P., Wagner, M., and Scherer, S. (2006). Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20.
Ehling-Schulz, M., Fricker, M., and Scherer, S. (2004). Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 48, 479–487. doi: 10.1002/mnfr.200400055
Ehling-Schulz, M., Lereclus, D., and Koehler, T. M. (2019). The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol. Spectr. 7:32.
Ehling-Schulz, M., Svensson, B., Guinebretiere, M. H., Lindback, T., Andersson, M., Schulz, A., et al. (2005a). Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151, 183–197. doi: 10.1099/mic.0.27607-0
Ehling-Schulz, M., Vukov, N., Schulz, A., Shaheen, R., Andersson, M., Martlbauer, E., et al. (2005b). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71, 105–113. doi: 10.1128/aem.71.1.105-113.2005
Freiwald, A., and Sauer, S. (2009). Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 4, 732–742. doi: 10.1038/nprot.2009.37
Fricker, M., Messelhausser, U., Busch, U., Scherer, S., and Ehling-Schulz, M. (2007). Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl. Environ. Microbiol. 73, 1892–1898. doi: 10.1128/aem.02219-06
Hoton, F. M., Fornelos, N., N’guessan, E., Hu, X., Swiecicka, I., Dierick, K., et al. (2009). Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ. Microbiol. Rep. 1, 177–183.
Lasch, P., Nattermann, H., Erhard, M., Stammler, M., Grunow, R., Bannert, N., et al. (2008). MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal. Chem. 80, 2026–2034. doi: 10.1021/ac701822j
Lasch, P., Stämmler, M., and Schneider, A. (2016). A MALDI-TOF mass spectrometry database for identification and classification of highly pathogenic microorganisms from the robert koch-institute (RKI). Zenodo 27:2017.
Marxen, S., Stark, T. D., Frenzel, E., Rutschle, A., Lucking, G., Purstinger, G., et al. (2015a). Chemodiversity of cereulide, the emetic toxin of Bacillus cereus. Anal. Bioanal. Chem. 407, 2439–2453. doi: 10.1007/s00216-015-8511-y
Marxen, S., Stark, T. D., Rutschle, A., Lucking, G., Frenzel, E., Scherer, S., et al. (2015b). Multiparametric quantitation of the Bacillus cereus toxins cereulide and isocereulides A-G in foods. J. Agric. Food Chem. 63, 8307–8313. doi: 10.1021/acs.jafc.5b03705
Messelhausser, U., Frenzel, E., Blochinger, C., Zucker, R., Kampf, P., and Ehling-Schulz, M. (2014). Emetic Bacillus cereus are more volatile than thought: recent foodborne outbreaks and prevalence studies in Bavaria (2007-2013). Biomed. Res. Int. 2014:465603.
Naka, T., Hattori, Y., Takenaka, H., Ohta, Y., Kirihata, M., and Tanimori, S. (2019). Synthesis of the reported structure of homocereulide and its vacuolation assay. Bioorg. Med. Chem. Lett. 29, 734–739. doi: 10.1016/j.bmcl.2019.01.007
Nomura, F. (2015). Proteome-based bacterial identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS): a revolutionary shift in clinical diagnostic microbiology. Biochim. Biophys. Acta 1854, 528–537. doi: 10.1016/j.bbapap.2014.10.022
Pitchayawasin, S., Isobe, M., Kuse, M., Franz, T., Agata, N., and Ohta, M. (2004). Molecular diversity of cereulide detected by means of nano-HPLC-ESI-Q-TOF-MS. Int. J. Mass Spectrom. 235, 123–129. doi: 10.1016/j.ijms.2004.04.007
Rouzeau-Szynalski, K., Stollewerk, K., Messelhausser, U., and Ehling-Schulz, M. (2020). Why be serious about emetic Bacillus cereus: cereulide production and industrial challenges. Food Microbiol. 85:103279. doi: 10.1016/j.fm.2019.103279
Schubert, S., and Kostrzewa, M. (2017). MALDI-TOF MS in the microbiology laboratory: current trends. Curr. Issues Mol. Biol. 23, 17–20. doi: 10.21775/cimb.023.017
Seng, P., Drancourt, M., Gouriet, F., La Scola, B., Fournier, P. E., Rolain, J. M., et al. (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49, 543–551. doi: 10.1086/600885
Stark, T., Marxen, S., Rutschle, A., Lucking, G., Scherer, S., Ehling-Schulz, M., et al. (2013). Mass spectrometric profiling of Bacillus cereus strains and quantitation of the emetic toxin cereulide by means of stable isotope dilution analysis and HEp-2 bioassay. Anal. Bioanal. Chem. 405, 191–201.
Suckau, D., Resemann, A., Schuerenberg, M., Hufnagel, P., Franzen, J., and Holle, A. (2003). A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 376, 952–965. doi: 10.1007/s00216-003-2057-0
Ulrich, S., Gottschalk, C., Dietrich, R., Martlbauer, E., and Gareis, M. (2019). Identification of cereulide producing Bacillus cereus by MALDI-TOF MS. Food Microbiol. 82, 75–81. doi: 10.1016/j.fm.2019.01.012
Keywords: cereulide, B. cereus, MALDI-ToF, mass spectrometry, toxin detection
Citation: Doellinger J, Schneider A, Stark TD, Ehling-Schulz M and Lasch P (2020) Evaluation of MALDI-ToF Mass Spectrometry for Rapid Detection of Cereulide From Bacillus cereus Cultures. Front. Microbiol. 11:511674. doi: 10.3389/fmicb.2020.511674
Received: 15 November 2019; Accepted: 16 September 2020;
Published: 06 October 2020.
Edited by:
Aldo Corsetti, University of Teramo, ItalyReviewed by:
Sean Doyle, Maynooth University, IrelandLaura Sanchez, University of Illinois at Chicago, United States
Copyright © 2020 Doellinger, Schneider, Stark, Ehling-Schulz and Lasch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joerg Doellinger, doellingerj@rki.de
 Joerg Doellinger
Joerg Doellinger Andy Schneider1
Andy Schneider1 Monika Ehling-Schulz
Monika Ehling-Schulz Peter Lasch
Peter Lasch