
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 September 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.02099
This article is part of the Research Topic Establishment of the Microbiota in the Context of DOHAD: Long-Term Health Consequences and Possible Management View all 9 articles
 Gyungcheon Kim1,2†
Gyungcheon Kim1,2† Jaewoong Bae1†
Jaewoong Bae1† Mi Jin Kim3†
Mi Jin Kim3† Hyeji Kwon1
Hyeji Kwon1 Gwoncheol Park2
Gwoncheol Park2 Seok-Jin Kim1
Seok-Jin Kim1 Yon Ho Choe3
Yon Ho Choe3 Jisook Kim4
Jisook Kim4 Sook-Hyun Park4
Sook-Hyun Park4 Byung-Ho Choe4
Byung-Ho Choe4 Hakdong Shin2*
Hakdong Shin2* Ben Kang4*
Ben Kang4*The maternal vaginal microbiome is an important source for infant gut microbiome development. However, infants delivered by Cesarean section (CS) do not contact the maternal vaginal microbiome and this delivery method may perturb the early establishment and development of the gut microbiome. The aim of this study was to investigate the early gut microbiota of Korean newborns receiving the same postpartum care services for two weeks after birth by delivery mode using fecal samples collected at days 3, 7, and 14. Early gut microbiota development patterns were examined using 16S rRNA gene-based sequencing from 132 infants either born vaginally (VD, n = 64) or via Cesarean section (CS, n = 68). VD-born neonates showed increased alpha diversity in infant fecal samples collated at days 7 and 14 compared to those from day 3, while those of CS infants did not differ (p < 0.015). Bacterial structures of infants from both groups separated at day 7 (p < 0.001) and day 14 (p < 0.01). The bacterial structure of VD infants gradually changed over time (day 3 vs. day 7, p < 0.012; day 3 vs. day 14, p < 0.001). Day 14 samples of CS infants differed from day 3 and 7 samples (day 3 vs. day 14, p < 0.001). VD infant relative abundance of Bifidobacterium (days 7, 14), Bacteroides (days 7, 14), and Lachnospiraceae (day 7) significantly increased compared to CS infants, with a lower abundance of Enterobacteriaceae (found in all periods of the CS group) (LDA > 3.0). Relative abundances of Bifidobacterium, Lactobacillus, and Staphylococcus were significantly increased in both VD and CS groups at day 14 (LDA > 3.0). Predicted functional analysis showed that VD infants had overrepresented starch/sucrose, amino acid and nucleotide metabolism in gut microbiota with depleted lipopolysaccharide biosynthesis until day 14 compared to CS infants. This study confirmed that delivery mode is the major determinant of neonatal intestinal microbiome establishment and provides a profile of microbiota perturbations in CS infants. Our findings provide preliminary insight for establishing recovery methods to supply the specific microbes missing in CS infants.
The establishment and development of the early gut microbiome in infancy is important for immune maturation and metabolic programming (Lozupone et al., 2012). Delivery mode is a major determinant of the early gut microbial composition in infants. Infants delivered by Cesarean section (CS) miss contact with the maternal vaginal microbiome compared to infants born by vaginal delivery (VD), suggesting that CS delivery perturbs the early establishment and development of the infant gut microbiome (Dominguez-Bello et al., 2010). This immature gut microbiome in CS infants has been associated with adverse later-life outcomes such as immune and metabolic disorders (Renz et al., 2017; Stinson et al., 2018). Recently, several longitudinal studies have shown the impact of CS delivery on the establishment of early gut microbiome with subsequent health outcomes including the colonization of antimicrobial-resistance opportunistic pathogens in the gut (Shao et al., 2019), losing immune system priming (Wampach et al., 2018), and increased susceptibility to infectious outcomes (Reyman et al., 2019). The identification of missing microbes in CS infants during the early development of the gut microbiome will provide key information to develop recovery methods for the immature gut microbiome in CS infants.
Geographical location and postpartum care conditions can also affect the early life gut microbiome (Shin et al., 2015; Martin et al., 2016). Studies on the gut microbiome of Korean infants have been conducted with a limited number of subjects (less than 10 infants) or are missing early microbiome samples (Lee et al., 2015, 2016). In this study, we compared the composition, structure, and predicted metabolic function of microbiota in fecal samples from 132 healthy Korean neonates (64 from VD neonates; 68 from CS neonates) who received the same postpartum care services for 2 weeks after birth. We identified the microbial composition and predicted functions lost in the early gut microbiome of CS infants by comparing microbiota development in CS and VD infants. This study will provide sequential information on early gut microbiome establishment.
This study was approved by the Institutional Review Board (IRB) of Kyungpook National University Chilgok Hospital (IRB no. 2017-06-009), and written informed consent was obtained from the mothers. Stool samples were obtained from healthy infants born between a gestational age of 37 to 41 weeks. Infants born from mothers who had GBS infection and chronic diseases such as diabetes, hypertensive disorders, or autoimmune disease were excluded. Infants born from mothers in whom oral antibiotics were administered during the third trimester of pregnancy were also excluded. For vaginal births, only infants born within 12 h of amniotic sac breakage were included. Stool samples from the infants were collected three times during the study period at 3, 7, and 14 days after birth. Clinical data regarding the mother’s gravidity and parity history, mother’s medical history, antibiotic use during pregnancy, mode of delivery, and infant’s gestational age, sex, and birth weight were recorded.
Newborn fecal samples were collected by nurses from diapers using sterile swabs at 3, 7, and 14 days after birth. These fresh fecal samples were immediately transferred to sterile cryogenic tubes and stored at −20°C until delivery to the laboratory. The samples were then stored at −80°C until DNA extraction.
Total genomic DNA extraction was performed from 200 mg of stool sample using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) with additional bead beating following the manufacturer’s instructions. We measured the DNA concentration using a UV-vis spectrophotometer (Nanodrop 2000c, United States) and stored DNA samples at −20°C for subsequent experimentation.
The newborn microbiota composition was analyzed by 16S rRNA amplicon sequencing using Illumina MiSeq (Illumina, Inc., United States). For sequencing, the V3-V4 region of the bacterial 16S rRNA gene was amplified using primer set F319 (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACG GGNGGCWGCAG-3′) and R806 (5′-GTCTCGTGGGCTCG GAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAAT CC-3′). DNA templates (12.5 ng/μL) were amplified using a KAPA HiFi Hotstart PCR Kit (Kapa Biosystems, United States) with 5 μM of primers. Reaction conditions were as follows: 95°C for 3 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. After PCR clean-up, a secondary amplification to attach Illumina Nextera barcodes was performed using i5 forward primer and i7 reverse primer. The DNA was amplified according to the manufacturer’s protocol. PCR products were purified using an Agencourt AMpure XP PCR Purification Kit (Beckman Coulter, United Kingdom) according to the manufacturer’s protocol. The purified products were quantified using a QuantiFluor® ONE dsDNA System (Promega, United States). The product size and quality were evaluated on a Bioanalyzer 2100 (Agilent, United States). The pooled libraries were sequenced using an Illumina MiSeq instrument with a MiSeq v3 Reagent Kit (Illumina, Inc., United States). All amplicon sequence data and metadata have been uploaded through the EMP data portal (qiita.microbio.me/emp; Study ID: 13215).
Analysis of the 16S rRNA sequences was performed using the QIIME (v.1.9.1) bioinformatics pipeline (Caporaso et al., 2010). Using qualified sequences (paired-end, Phred ≥ Q20), the operational taxonomic units (OTUs) were identified based on an open-reference picking method using 97% identity to entries in the Greengenes database (v13_8) (DeSantis et al., 2006) using UCLUST (Edgar, 2010). Samples were rarefied to 3 116 reads per sample and OTU biome information of all samples used in this study is provided in Supplementary Table S1. The chimeric sequences were removed using usearch61. Sample alpha diversity was calculated using the phylogenetic distance and the number of observed OTUs. For beta diversity comparison between groups, weighted/unweighted UniFrac distances were evaluated (Lozupone et al., 2011). Permutational multivariate analysis of variance (PERMANOVA) and Adonis were used to determine the significant differences in bacterial structures (Anderson, 2001). To test statistical differences, a non-parametric t-test with 10 000 Monte Carlo permutations was used. We carried out linear discriminant analysis effect size (LEfSe) analysis (Segata et al., 2011) to detect significant differences in bacterial taxonomies (LDA score > 3.0). To verify the number of OTUs detected per samples, OTU picking was performed and analyzed using QIIME2 (v.2020.2) (Bolyen et al., 2019) based on DADA2 workflow (Callahan et al., 2016).
Core microbiota were analyzed to identify the stability of the infant gut microbiota for each individual at days 3 and 7 based on the day 14 values (defined as the normalized OTUs present in all samples) (Mortensen et al., 2018). OTUs that are present in at least 90% of the samples at each group were selected as core components in each group. To determine the overlapped OTUs between sampling points, SourceTracker (v2) with a Bayesian approach (Knights et al., 2011) and FEAST (Shenhav et al., 2019) were used.
Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) was used to predict the metabolic function of the metagenomes from the 16S rRNA gene dataset (Langille et al., 2013) using Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog classification.
A total of 132 (64 VD and 68 CS) infants were included in this study. Clinical and anthropometric infant characteristics are presented in Table 1. There were no significant differences in the clinical characteristics of mothers and infants according to the modes of delivery except maternal age (VD = 30.9 ± 3.3, CS = 34.1 ± 4.5; p < 0.001). All neonates were mainly breastfed (>50%) for 2 weeks after birth.
A total of 17 550 289 sequences (paired-end, Phred ≥ Q20) were obtained from infant fecal samples. The average sequence number per sample was 44 319 and these sequences were binned into 140 383 types of OTUs (Supplementary Table S1).
The alpha diversity of VD infant fecal samples increased over time but that of CS infants did not (non-parametric t-test, p < 0.015; Figure 1 and Supplementary Figure S1). While the alpha diversity of VD infant fecal samples tended to be higher in relation to CS infants, the statistical power to make comparisons was low (Supplementary Figure S1).
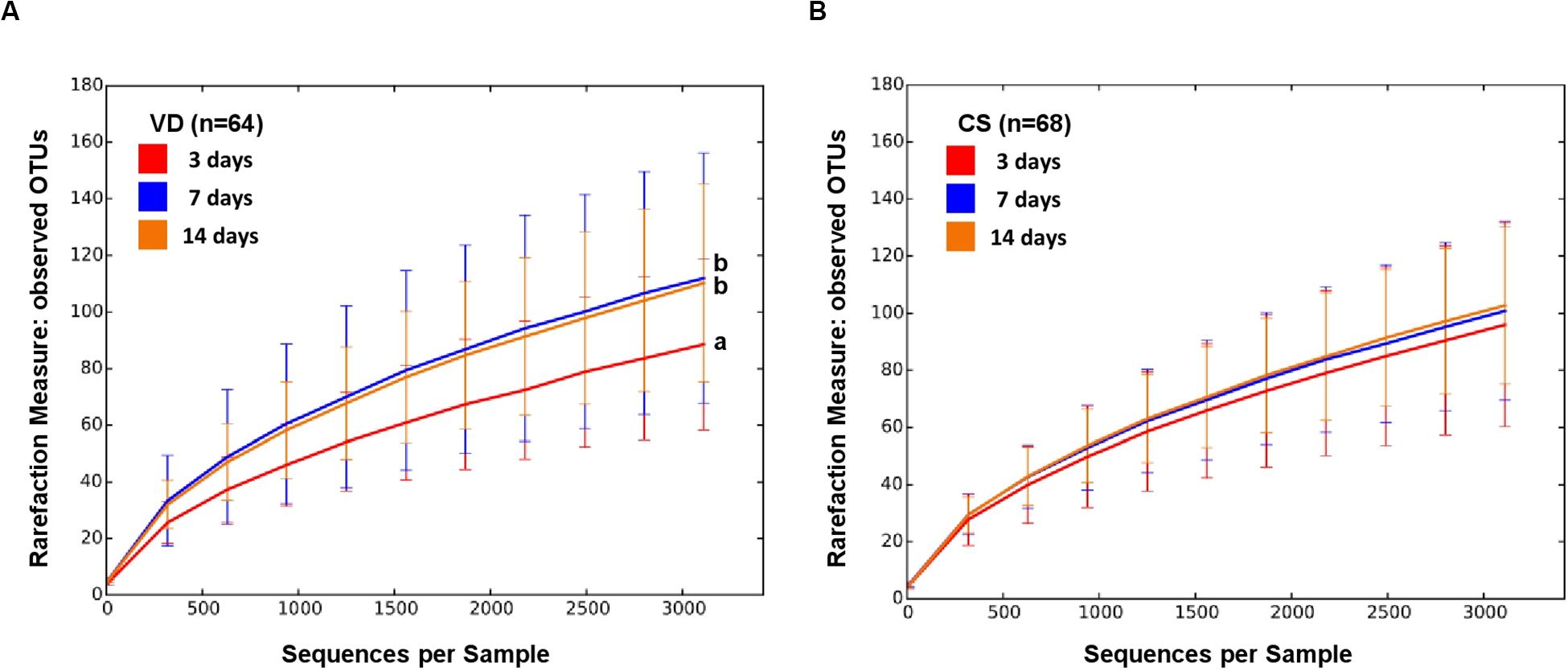
Figure 1. Alpha diversity of early gut microbiota according to delivery mode over time. Observed species metrics were used to plot bacterial diversity of VD (A) and CS (B). Different letters indicate significant differences between groups (p < 0.05).
Bacterial structure (beta diversity) of fecal microbiota differed significantly over time in VD infants (Figure 2A; Weighted UniFrac; Adonis, p < 0.012). However, the fecal microbiota of CS infants did not differ in beta diversity between day 3 and 7 samples (Figure 2B; Adonis, p < 0.260). This was supported by the UniFrac inter group distances between sampling days (Figures 2A,B and Supplementary Figures S2A,B).
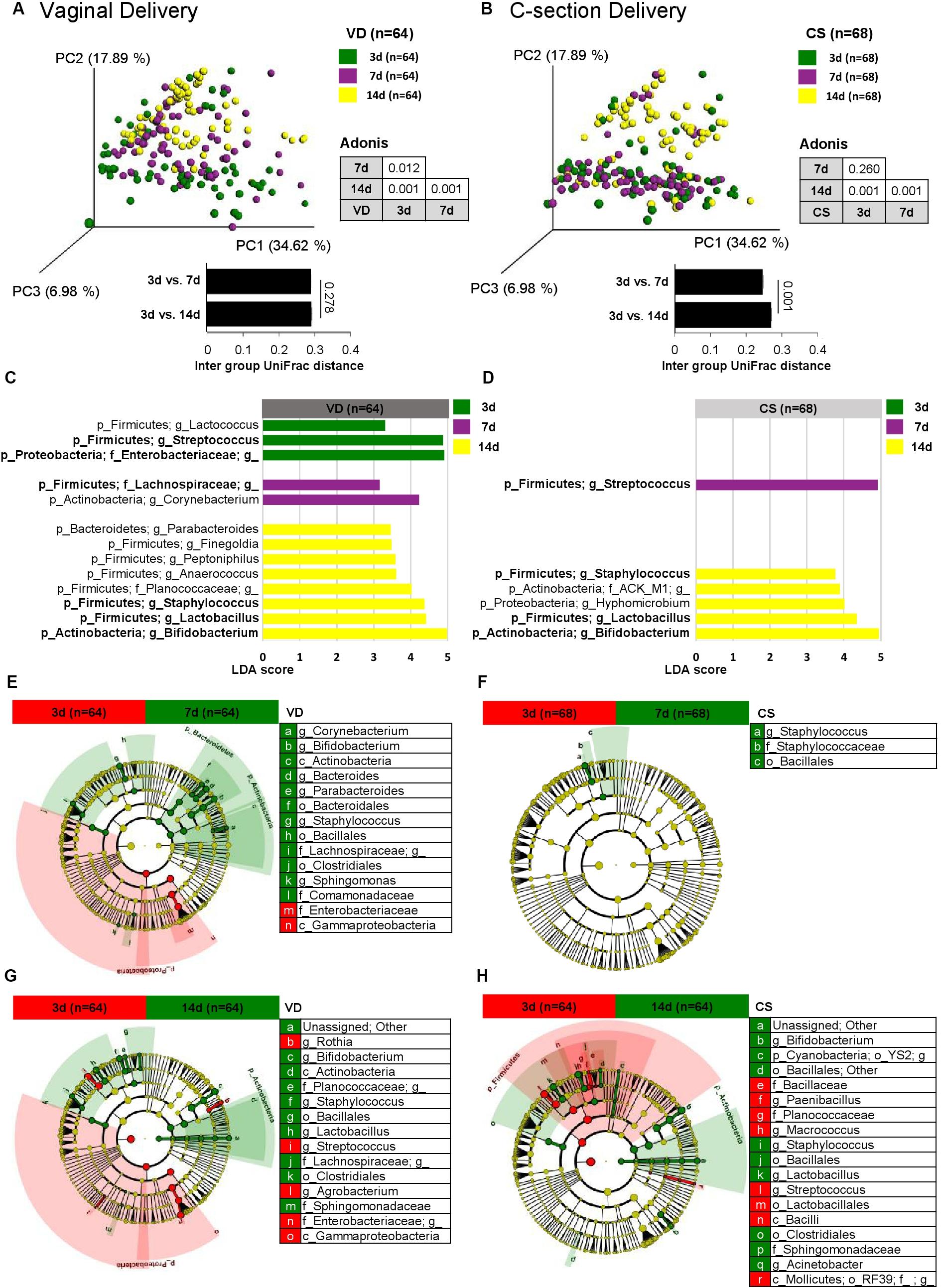
Figure 2. Bacterial structure differences in early gut microbiota according to delivery mode over time. Weighted UniFrac distances were used to evaluate the beta diversity of VD (A) and CS (B) infant gut microbiomes using PCoA and intra group distance. Adonis was used to test dissimilarity. LEfSe analysis (LDA > 3.0) was used to identify biomarkers between sampling points in VD (C) and CS infants (D). Cladograms were plotted to examine the phylogenetic differences (LDA > 3.0) between sampling points in VD (E,G) and CS infants (F,H).
Linear discrimination analysis (LDA) used to detect overrepresented bacterial taxonomies between sampling days showed that while six major bacterial taxonomies were overrepresented in VD infants (day 3: Streptococcus and Enterobacteriaceae; day 7: Lachnospiraceae; day 14: Staphylococcus, Lactobacillus, and Bifidobacterium), CS infants showed fewer overrepresented bacterial taxonomies (day 7: Streptococcus; day 14: Staphylococcus, Lactobacillus, and Bifidobacterium) (Figures 2C,D). Fecal microbiota on day 7 and 14 from VD infants showed a consistent profile of overrepresented bacterial taxa compared to day 3, but not in fecal samples from CS infants (Figures 2E–H).
Fecal samples from VD infants gradually shifted to the PC2 axis on the PCoA plot over time, but CS infant fecal samples moved abruptly only after day 14, suggesting the delayed establishment of early gut microbial structure in CS infants (Figures 3A-C and Supplementary Figures S2C–G). Core microbiota ratio and source tracking results based on OTUs detected from day 3 fecal samples also supported this observation of delayed early gut microbiota establishment in CS infants (Supplementary Figure S3). Enterococcaceae and Enterobacteriaceae families were overrepresented in CS infant fecal samples over the 2 week period compared to VD infants (LDA > 3.0 of bacterial OTUs in proportions > 1%; Figures 3D,E). VD infants had a higher relative abundance of Bifidobacterium, Bacteroides, Lactobacillus, and Lachnospiraceae in fecal samples at day 7 after birth compared to CS infants (LDA > 3.0 of bacterial OTUs in proportions > 1%; Figures 3D,E). The reproducibility of dataset in this study was verified based on the latest-generation tool for sequence quality control (DADA2), demonstrated that an equivalent tendency was derived from diversity and structure of infant fecal microbiota according to the mode of delivery as the results of 97% OTU clustering methods (Supplementary Figure S4). We also could find an increased dynamic tendency in bacterial structures during the establishment of early gut microbiota (Supplementary Figure S5).
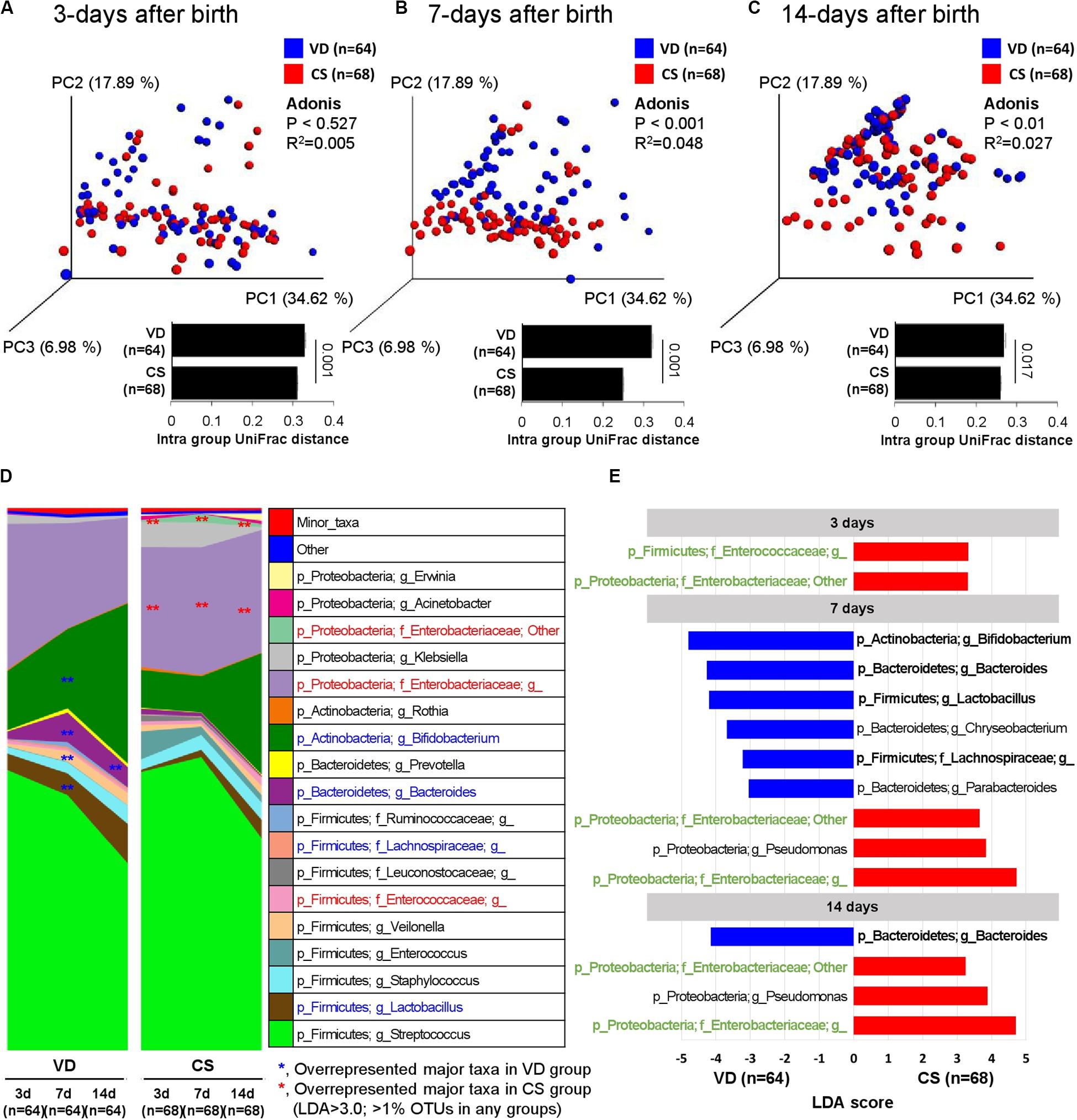
Figure 3. Development of early gut microbiota according to delivery mode. (A–C) Weighted UniFrac distances were used to evaluate beta diversity in early gut microbiota between delivery modes. Adonis was used to test dissimilarity. (D) Bacterial taxa plot between delivery modes. Different colors indicate each taxon at the genus level. (E) Differing abundance of bacterial communities between delivery modes using LEfSe; **Indicates overrepresented taxa (using LDA > 3.0) in delivery mode comparisons within sample type. Green indicates that infant gut microbiota maintenance from days 3–14 and bolded items represent the major taxa.
PICRUSt was used to predict KEGG functional profiling from our 16s rRNA dataset. Fecal microbiota on day 7 after birth showed higher discriminant predicted functions according to delivery modes than other samples (Table 2). Fecal microbiota of VD infants at day 7 after birth had a higher proportion of bacterial genes related to starch/sucrose metabolism, pyrimidine/purine metabolism and amino acid related enzymes (LDA > 3.0), and a lower proportion of genes related to lipopolysaccharide (LPS) biosynthesis proteins (LDA > 3.0; Table 2). Various types of genes (heptosyltransferase, rhamnosyltransferase, and glycosyltransferase) related to LPS synthesis were overrepresented from day 3 and remained to day 7 after birth in CS infants (Figure 4), suggesting premature exposure to foreign microbial antigens.
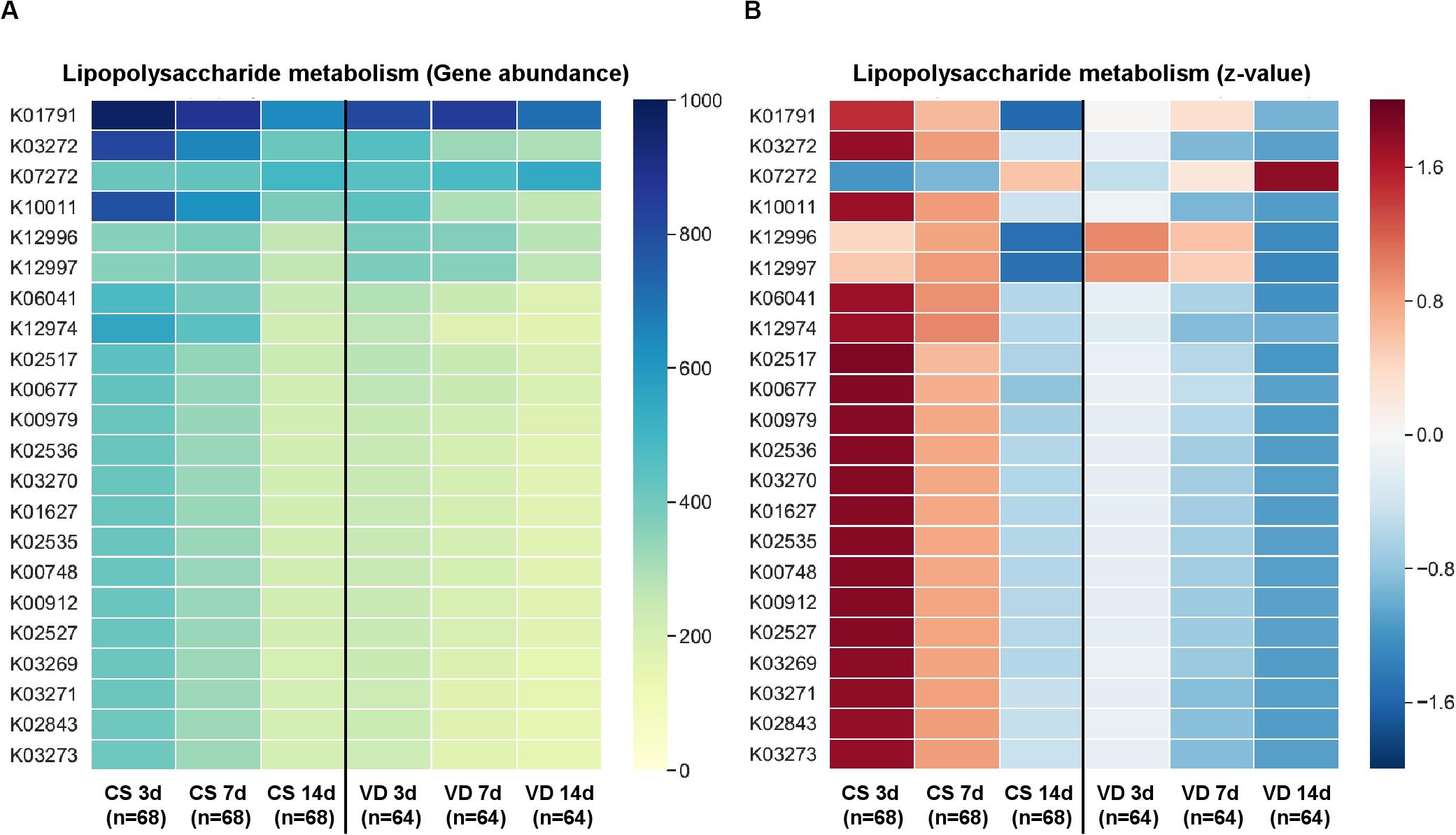
Figure 4. Predictive KEGG functional profiling for LPS metabolism according to delivery mode and sampling time. Gene abundance (A) based on PICRUSt analysis and z-score (B) were used to identify significant differences between groups.
This cohort study included 132 infants that received the same postpartum care services for two weeks after birth to evaluate the development patterns of early intestinal microbiota according to delivery mode. We confirmed previous reports that delivery mode is the major determinant of neonatal intestinal microbiome establishment (Dominguez-Bello et al., 2010; Dogra et al., 2015; Mueller et al., 2016). We also identified depleted bacterial composition associated with starch/sucrose, amino acid, and nucleotide metabolism in the early gut microbiome of CS infants compared to VD infants. These intestinal microbes missing in CS infants could play an important role in postnatal immune and metabolic system development (Cox et al., 2014; Jakobsson et al., 2014). This study provides a profile of the missing microbes in infants delivered by Cesarean section and this preliminary microbial insight should be examined for its association with subsequent health outcomes.
The developmental differences of infant gut microbiota are reflected in a chronological pattern. The increased microbiota diversity observed immediately after birth may be affected by the transmission of maternal gut microbiota, including non-colonizing microbiota, in vaginally delivered infants (Lee et al., 2016). Cesarean delivery is associated with the perturbation and delayed maturation of gut microbiota in early life, which in turn has been associated with increased risk of childhood obesity (Blustein et al., 2013), asthma (Kolokotroni et al., 2012) immune diseases (Sevelsted et al., 2015), and infectious outcomes (Reyman et al., 2019). Shao et al. conducted the largest known longitudinal study including 596 infants born in United Kingdom, revealed that maternal Bacteroides strains are the major missing microbes in the immature gut microbiome of CS infants with colonization by hospital environment-related species including Enterococcus, Enterobacter, and Klebsiella (Shao et al., 2019). These modifications of early gut microbiome development by the mode of delivery is matched with our results, which showed CS infant gut microbiota enriched in Enterobacteriaceae and Enterococcaceae families with Bacteroides depletion.
Evaluation of the temporal pattern of the changes in intestinal microbiota diversity and composition according to the delivery mode in Korean infants during the first two weeks showed that the bacterial diversity in VD neonate stools increased from day 3 onward. However, this was not the case CS infants. These results suggested that the early gut microbiota of VD infants had an ecological niche space for early gut microbiota development. We also found that the bacterial structure of infant gut microbiota changed gradually over time in VD infants, but hierarchically (without intermediate status) in CS infants, suggesting possible constrained changes in early gut bacterial structures of CS infants. The separation in bacterial structures over time was more pronounced when the weighted UniFrac measure was used, in relation to the unweighted. These results indicated that the dominant bacterial taxa played a major role in the change of bacterial structures over time. Our findings extend the potential recovery methods for missing microbes in CS infants by specifying an appropriate environmental niche for the establishment of early gut microbiota. Dominguez-Bello et al. demonstrated that transferring the vaginal microbiome of mothers to the neonates (vaginal seeding) partially restored the microbiome of CS neonates (Dominguez-Bello et al., 2016; Mueller et al., 2019). While this procedure should be studied in a large number of well-designed trials to identify the effects on microbiome and health benefits, trials to restore the gut microbiota niche in CS neonates could minimize the risk of microbiome-related diseases. Our study excluded mothers who received antibiotics before birth, suggesting that the difference in bacterial diversity between delivery modes may depend on interactions with other factors.
In this study, the major bacterium which formed infant gastrointestinal microbiota belonged to the Firmicutes and Proteobacteria phyla (Nagpal et al., 2016; Dong et al., 2018; Nagpal and Yamashiro, 2018). Specifically, the colonization levels of several genera belonging to Firmicutes, such as Enterococcoceae, Lachnospiraceae, and Lactobacillus were significantly influenced by delivery mode (Gronlund et al., 1999; Rutayisire et al., 2016). Other previous studies also showed that higher Enterobacteriaceae and Pseudomonas levels were observed in CS compared to VD infants (Adlerberth et al., 2006; Dominguez-Bello et al., 2010; Hesla et al., 2014; Dogra et al., 2015). The possible sources of these overrepresented microbes in CS infants should be examined to guide postnatal conditions for the establishment of early gut microbiota (Fryklund et al., 1992; Shin et al., 2015). Biomarker analysis using LEfSe also showed a higher relative abundance of Lactobacillus, Bifidobacterium, and Bacteroides in the stools of VD infants compared to those of CS infants; these findings are supported by previous studies (Mitsou et al., 2008; Kabeerdoss et al., 2013). The Bacteroides genus is a typical adult intestinal bacterium and several studies suggest that this genus may be transmitted from the maternal gut to neonates during delivery (Adlerberth et al., 2007; Vaishampayan et al., 2010; Rutayisire et al., 2016). The delayed establishment of Bacteroides in the gut microbiota of CS neonates could be related to health outcomes (Adlerberth et al., 2006; Penders et al., 2006). Some Bacteroides species (B. fragilis) are known to activate T cell-dependent immune responses that can affect both the development and homeostasis of the host immune system (Troy and Kasper, 2010). The predicted functional profiles related to LPS and xenobiotic metabolism were overrepresented in the early gut microbiome of CS infants at day 7 after birth (compared to VD infants). These differences are primarily contributed by enriched Enterobacteriaceae populations producing several types of LPS. Several studies show that lower LPS biosynthesis is related to developing a protective effect against asthma (Lotz et al., 2006; Stiemsma and Turvey, 2017), suggesting that finely timed exposure to foreign microbial antigens may help neonate immune system development. Interrogation of these predicted functional differences needs to be confirmed in future cohorts using true shotgun metagenomics analyses or metabolomics.
This study showed that the ratio of core gut microbiota from CS infants partially recovered by day 14 after birth compared to those of VD infants. This partial restoration of early gut microbiota from CS infants occurred relatively earlier than previously reported (Palmer et al., 2007; Eggesbo et al., 2011; Liu et al., 2019). While overrepresented Enterobacteriaceae and depleted Bacteroides were maintained at day 14 in CS infants, Bifidobacterium and Lactobacillus were restored to a level similar to those of VD infants. This reveals that a strategy for restoring microbiota is needed, excluding remedial supplementation with probiotic microbes. Also, further studies using Korean infants will help to identify factors causing the relatively rapid restoration of early gut microbiota in CS infants, such as Korean postpartum services (Kyung Kim et al., 2017).
A strength of this study is geographic specificity with a large sample size and density of sampling at early time points. However, limitations of this study included a lack of maternal samples (stools, skin/vaginal swabs) and differences in maternal and gestational ages (Table 1). The previous investigation on the impact of exclusive breastfeeding on gut microbiota in CS infants showed partial restoration of early gut microbiota establishment, in relation to VD infants (Liu et al., 2019). While we only documented whether the infants were majoritarily breastfed for the type of feeding, this study was limited to screen the exclusively breastfed group. As such, we are unable to suggest a direct link between other important cofactors (maternal microbiota and breastfeeding status) and infant early gut microbiota. Additionally, other limitations are the lack of follow-up measures to identify health outcomes and the rough resolution reachable by 16S rDNA. Further well-designed studies with multi-omics approaches are needed to increase our knowledge on the impact of mother-to-newborn microbiome transfer.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ebi.ac.uk/ena/browser/view/PRJEB39707.
The studies involving human participants were reviewed and approved by Kyungpook National University Chilgok Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
S-JK, YHC, HS, and BK conceived this study. YHC, JK, S-HP, B-HC, and BK designed the study and collected the microbiome samples. GK, JB, MJK, HK, GP, and HS analyzed the data. GK, JB, MJK, S-JK, HS, and BK wrote the manuscript. All authors contributed with valuable discussions and edition, approving the final version of manuscript.
HS was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2017R1D1A1B03029911) and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agricultural Microbiome R&D Program funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (No. 918022043CG000). BK was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (No. 2017R1C1B5076980).
GK, JB, HK, and S-JK were employed by BioEleven Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02099/full#supplementary-material
FIGURE S1 | Alpha diversity of early gut microbiota according to delivery mode over time. (A) Observed species metric. (B) Phylogenetic diversity. Non-parametric p-value was calculated using 10,000 Monte Carlo permutations.
FIGURE S2 | Bacterial structure differences of early gut microbiota according to delivery mode over time. Unweighted UniFrac distances were used to evaluate beta diversity of VD infants (A) and CS infants (B) using PCoA and intra group distance. Adonis was used to test dissimilarity. (C–E) Unweighted UniFrac distances were used to evaluate beta diversity between delivery modes. Cladograms were plotted to examine phylogenetic differences (LDA > 3.0) between sampling points in VD infants (F) and CS infants (G).
FIGURE S3 | The ratio of core microbiota differences of early gut microbiota according to delivery mode over time. The normalized OTUs present in all samples were used to calculate the ratio of core microbiota in stool samples from VD infants (A) and CS infants (B). Source tracking analyses were performed to identify the ratio of overlapped OTUs between sampling points using Source Tracker2 (C) and FEAST (D).
FIGURE S4 | Bacterial alpha and beta diversity in early gut microbiota according to delivery mode over time based on QIIME2 pipeline. Observed species metrics were used to plot bacterial diversity of VD (A) and CS (B). Weighted UniFrac distances were used to evaluate the beta diversity of VD (C) and CS (D) infant gut microbiomes using PCoA and intra group distance. PERMANOVA was used to test dissimilarity. (E) Bacterial taxa plot between delivery modes. Different colors indicate each taxon at the genus level; ∗∗Indicates overrepresented taxa (using LDA > 3.0) in delivery mode comparisons within sample type.
FIGURE S5 | Increased dynamics of bacterial structures during the establishment of early gut microbiota. Shannon diversity metrics were used to plot bacterial diversity of all samples (A), VD group (B), and CS group (C). Non-parametric p-value was calculated using 10,000 Monte Carlo permutations. Individual bacterial taxa plots at the genus level of VD group (D) and CS group (E). Different colors indicate each taxon at the genus level.
TABLE S1 | OTU biome information of all samples used in this study.
Adlerberth, I., Lindberg, E., Aberg, N., Hesselmar, B., Saalman, R., Strannegard, I. L., et al. (2006). Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr. Res. 59, 96–101. doi: 10.1203/01.pdr.0000191137.12774.b2
Adlerberth, I., Strachan, D. P., Matricardi, P. M., Ahrne, S., Orfei, L., Aberg, N., et al. (2007). Gut microbiota and development of atopic eczema in 3 European birth cohorts. J. Allergy Clin. Immunol. 120, 343–350. doi: 10.1016/j.jaci.2007.05.018
Anderson, M. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070
Blustein, J., Attina, T., Liu, M., Ryan, A. M., Cox, L. M., Blaser, M. J., et al. (2013). Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int. J. Obes 37, 900–906. doi: 10.1038/ijo.2013.49
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cox, L. M., Yamanishi, S., Sohn, J., Alekseyenko, A. V., Leung, J. M., Cho, I., et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721. doi: 10.1016/j.cell.2014.05.052
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dogra, S., Sakwinska, O., Soh, S. E., Ngom-Bru, C., Bruck, W. M., Berger, B., et al. (2015). Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 6:e02419-14. doi: 10.1128/mBio.02419-14
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Dominguez-Bello, M. G., De Jesus-Laboy, K. M., Shen, N., Cox, L. M., Amir, A., Gonzalez, A., et al. (2016). Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253. doi: 10.1038/nm.4039
Dong, T., Chen, T., White, R. A. III, Wang, X., Hu, W., Liang, Y., et al. (2018). Meconium microbiome associates with the development of neonatal jaundice. Clin. Transl. Gastroenterol. 9:182. doi: 10.1038/s41424-018-0048-x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eggesbo, M., Moen, B., Peddada, S., Baird, D., Rugtveit, J., Midtvedt, T., et al. (2011). Development of gut microbiota in infants not exposed to medical interventions. APMIS 119, 17–35. doi: 10.1111/j.1600-0463.2010.02688.x
Fryklund, B., Tullus, K., Berglund, B., and Burman, L. G. (1992). Importance of the environment and the faecal flora of infants, nursing staff and parents as sources of gram-negative bacteria colonizing newborns in three neonatal wards. Infection 20, 253–257. doi: 10.1007/bf01710789
Gronlund, M. M., Lehtonen, O. P., Eerola, E., and Kero, P. (1999). Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 28, 19–25. doi: 10.1097/00005176-199901000-00007
Hesla, H. M., Stenius, F., Jaderlund, L., Nelson, R., Engstrand, L., Alm, J., et al. (2014). Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol. Ecol. 90, 791–801. doi: 10.1111/1574-6941.12434
Jakobsson, H. E., Abrahamsson, T. R., Jenmalm, M. C., Harris, K., Quince, C., Jernberg, C., et al. (2014). Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63, 559–566. doi: 10.1136/gutjnl-2012-303249
Kabeerdoss, J., Ferdous, S., Balamurugan, R., Mechenro, J., Vidya, R., Santhanam, S., et al. (2013). Development of the gut microbiota in southern Indian infants from birth to 6 months: a molecular analysis. J. Nutr. Sci. 2:e18. doi: 10.1017/jns.2013.6
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763. doi: 10.1038/nmeth.1650
Kolokotroni, O., Middleton, N., Gavatha, M., Lamnisos, D., Priftis, K. N., and Yiallouros, P. K. (2012). Asthma and atopy in children born by caesarean section: effect modification by family history of allergies - a population based cross-sectional study. BMC Pediatr. 12:179. doi: 10.1186/1471-2431-12-179
Kyung Kim, S., Park, S., and Ahn, S. (2017). Effectiveness of psychosocial and educational prenatal and postnatal care interventions for married immigrant women in Korea: systematic review and meta-analysis. Asia Pac. J. Public Health 29, 351–366. doi: 10.1177/1010539517717364
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lee, E., Kim, B. J., Kang, M. J., Choi, K. Y., Cho, H. J., Kim, Y., et al. (2016). Dynamics of gut microbiota according to the delivery mode in healthy Korean infants. Allergy Asthma Immunol. Res. 8, 471–477. doi: 10.4168/aair.2016.8.5.471
Lee, E. K., Ahn, Y. T., Huh, C. S., Soo Kim, H., Kim, E., Chun, Y. H., et al. (2015). The early intestinal microbiota of healthy Korean newborns. Iran J. Pediatr. 25:e2079. doi: 10.5812/ijp.2079
Liu, Y., Qin, S., Song, Y., Feng, Y., Lv, N., Xue, Y., et al. (2019). The perturbation of infant gut microbiota caused by cesarean delivery is partially restored by exclusive breastfeeding. Front. Microbiol. 10:598. doi: 10.3389/fmicb.2019.00598
Lotz, M., Gutle, D., Walther, S., Menard, S., Bogdan, C., and Hornef, M. W. (2006). Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203, 973–984. doi: 10.1084/jem.20050625
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Martin, R., Makino, H., Cetinyurek Yavuz, A., Ben-Amor, K., Roelofs, M., Ishikawa, E., et al. (2016). Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498
Mitsou, E. K., Kirtzalidou, E., Oikonomou, I., Liosis, G., and Kyriacou, A. (2008). Fecal microflora of Greek healthy neonates. Anaerobe 14, 94–101. doi: 10.1016/j.anaerobe.2007.11.002
Mortensen, M. S., Hebbelstrup Jensen, B., Williams, J., Brejnrod, A. D., O’Brien Andersen, L., Roser, D., et al. (2018). Stability and resilience of the intestinal microbiota in children in daycare - a 12 month cohort study. BMC Microbiol. 18:223. doi: 10.1186/s12866-018-1367-5
Mueller, N. T., Dominguez-Bello, M. G., Appel, L. J., and Hourigan, S. K. (2019). ‘Vaginal seeding’ after a caesarean section provides benefits to newborn children: for: does exposing caesarean-delivered newborns to the vaginal microbiome affect their chronic disease risk? The critical need for trials of ‘vaginal seeding’ during caesarean section. Bjog 127:301. doi: 10.1111/1471-0528.15979
Mueller, N. T., Shin, H., Pizoni, A., Werlang, I. C., Matte, U., Goldani, M. Z., et al. (2016). Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci. Rep. 6:23133. doi: 10.1038/srep23133
Nagpal, R., Tsuji, H., Takahashi, T., Kawashima, K., Nagata, S., Nomoto, K., et al. (2016). Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front. Microbiol. 7:1997. doi: 10.3389/fmicb.2016.01997
Nagpal, R., and Yamashiro, Y. (2018). Gut microbiota composition in healthy japanese infants and young adults born by C-section. Ann. Nutr. Metab. 73, (Suppl. 3), 4–11. doi: 10.1159/000490841
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A., and Brown, P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. doi: 10.1371/journal.pbio.0050177
Penders, J., Thijs, C., Vink, C., Stelma, F. F., Snijders, B., Kummeling, I., et al. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521. doi: 10.1542/peds.2005-2824
Renz, H., Holt, P. G., Inouye, M., Logan, A. C., Prescott, S. L., and Sly, P. D. (2017). An exposome perspective: early-life events and immune development in a changing world. J. Allergy Clin. Immunol. 140, 24–40. doi: 10.1016/j.jaci.2017.05.015
Reyman, M., van Houten, M. A., van Baarle, D., Bosch, A., Man, W. H., Chu, M., et al. (2019). Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 10:4997. doi: 10.1038/s41467-019-13014-7
Rutayisire, E., Huang, K., Liu, Y., and Tao, F. (2016). The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 16:86. doi: 10.1186/s12876-016-0498-0
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sevelsted, A., Stokholm, J., Bonnelykke, K., and Bisgaard, H. (2015). Cesarean section and chronic immune disorders. Pediatrics 135, e92–e98. doi: 10.1542/peds.2014-0596
Shao, Y., Forster, S. C., Tsaliki, E., Vervier, K., Strang, A., Simpson, N., et al. (2019). Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121. doi: 10.1038/s41586-019-1560-1
Shenhav, L., Thompson, M., Joseph, T. A., Briscoe, L., Furman, O., Bogumil, D., et al. (2019). FEAST: fast expectation-maximization for microbial source tracking. Nat. Methods 16, 627–632. doi: 10.1038/s41592-019-0431-x
Shin, H., Pei, Z., Martinez, K. A. II, Rivera-Vinas, J. I., Mendez, K., Cavallin, H., et al. (2015). The first microbial environment of infants born by C-section: the operating room microbes. Microbiome 3:59. doi: 10.1186/s40168-015-0126-1
Stiemsma, L. T., and Turvey, S. E. (2017). Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin. Immunol. 13:3. doi: 10.1186/s13223-016-0173-6
Stinson, L. F., Payne, M. S., and Keelan, J. A. (2018). A critical review of the bacterial baptism hypothesis and the impact of cesarean delivery on the infant microbiome. Front. Med. 5:135. doi: 10.3389/fmed.2018.00135
Troy, E. B., and Kasper, D. L. (2010). Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front. Biosci. 15:25–34. doi: 10.2741/3603
Vaishampayan, P. A., Kuehl, J. V., Froula, J. L., Morgan, J. L., Ochman, H., and Francino, M. P. (2010). Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol. Evol. 2, 53–66. doi: 10.1093/gbe/evp057
Keywords: newborn, gut microbiome establishment, delivery mode, neonate microbiome, available microbiome niche
Citation: Kim G, Bae J, Kim MJ, Kwon H, Park G, Kim S-J, Choe YH, Kim J, Park S-H, Choe B-H, Shin H and Kang B (2020) Delayed Establishment of Gut Microbiota in Infants Delivered by Cesarean Section. Front. Microbiol. 11:2099. doi: 10.3389/fmicb.2020.02099
Received: 25 March 2020; Accepted: 10 August 2020;
Published: 11 September 2020.
Edited by:
Rebeca Martín, INRA Centre de Jouy-en-Josas, FranceReviewed by:
Léa Siegwald, Nestlé Research Center, SwitzerlandCopyright © 2020 Kim, Bae, Kim, Kwon, Park, Kim, Choe, Kim, Park, Choe, Shin and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hakdong Shin, aHNoaW5Ac2Vqb25nLmFjLmty; Ben Kang, YmVua2FuZ0BrbnUuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.