
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 08 September 2020
Sec. Microbe and Virus Interactions with Plants
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.02095
This article is part of the Research TopicForests and Their Interactions with the EnvironmentView all 21 articles
Biological diversity plays an important role in the stability of ecosystems. The Mu Us Desert (MUD), located in Northern China, is an aeolian desert. Although it has been governed by a series of ecological restoration programs, the MUD still has limited biological diversity. Populus euphratica (P. euphratica), a xerophytic plant, has great potential to improve the biological diversity of the MUD. However, the survival rate of P. euphratica in the MUD has been very low. The current study tried to explore the mechanism of the high death rate of P. euphratica in the microbiome perspective. The correlation study between soil community composition and soil properties showed that water-filled pore space (WFPS), pH, EC, AP, NO3–, and NH4+ possess higher potential to change the bacterial community (18%) than the fungal community (9%). Principal coordinate analysis indicated that the composition of both bacteria (Proteobacteria and Bacteroidetes) and fungi (Ascomycota) in the root soil can be increased by P. euphratica. By systematically comparing between the fungal diversity in the root soil around P. euphratica and the pathogenic fungus extract from the pathogenic site of P. euphratica, we found that the high death rate of P. euphratica was associated with specific pathogenic fungus Alternaria alternate and Didymella glomerata. In addition, the microbiome composition analysis indicated that P. euphratica planting could also influence the portions of bacteria community, which also has great potential to lead to future infection. However, as the extraction and separation of bacteria from plants is challenging, the correlation between pathogenic bacteria and the high death rate of P. euphratica was not studied here and could be explored in future work.
- Soil microbiome can be influenced significantly by Populus euphratica.
- A. alternate and D. glomerata in soil can lead to the high death rate of Populus euphratica.
- Plant-pathogen interactions within bacteria and Populus euphratica death was observed.
The Mu Us Desert (MUD) was once covered by 81 km2 of sand dunes. Fortunately, after the efforts of several generations, its ecological environment has been greatly improved. Since 1978, the desert region in the MUD has been moved 400 km to the north (Feng et al., 2013; Li et al., 2013; Lai et al., 2016). The sand retreat was a great achievement and victory that made the MUD one of the most famous regional vegetation restorations (Li et al., 2019; Sun et al., 2019). However, the achievement of environmental management in the MUD is mainly attributed to the introduction of two kinds of plants: conifer pine and shrub. It is difficult for other conventional green plants to survive in the MUD due to the natural climate, and as a result, most of the restored areas were dominated by these two kinds of plants. Although proliferation of these species temporarily controlled the spread of the local desert and improved the local vegetation coverage, the limited biodiversity in the region renders the ecological environment fragile. The resulting risk of secondary desertification has always been an urgent problem for the ecological stability of the MUD region (Shu et al., 2018; Li et al., 2019).
YuLin (Yuyang District), located in the interior of the MUD, possesses 118.27 km2 of grassland reclamation. YuLin is one of most typical vegetated areas of the MUD and has great potential as a pilot site due to its ecological stability (Li et al., 2019). Populus euphratica, a perennial woody plant with high salinity and aridity tolerance, is widely spread in Western China and adjacent Central Asian countries (Keram et al., 2019). Considering the growth habit of P. euphratica, the climatic and environmental conditions of the Yuyang District can be suitable for its growth (Shu et al., 2018). However, after several years of trying, the success of P. euphratica, which should be able to improve the ecological development in the MUD, has stagnated due to its high mortality rate. At the same time, some factors governing the success of P. euphratica have been discovered. After these P. euphratica trees were planted in the nursery garden, the survival rate was close to 100% within the first year. Once the P. euphratica trees were transferred outside, almost 50% of the saplings died within the second year. During the third year, less than 5% of these P. euphratica trees survived. Only 5% of these tested trees grew healthy after a probationary period of 3 years. By analyzing the dead plants, some typical symptoms, like stem cankers and rust disease, etc., were found. The symptoms identified in almost all of the dead P. euphratica were consistent. These fatal diseases seem to be a reliable reason for the high death rate of P. euphratica. At the same time, why was there such a large difference in the death rate among different growth periods? Where or how were the P. euphratica trees infected?
Previous studies indicated that, the plant-associated microbial communities could be affected by the tissue age of the plant, environmental conditions, and agronomical practices (Vorholt, 2012; Leff et al., 2015; Arrigoni et al., 2018). In this plant–pathogen interaction system, microbiome plays a vital role for the health of the plant. The microbiome transplanted by the soil could always predetermine future plant health (Wei et al., 2019). This study is focused on the bacterial and fungal community in the soil around P. euphratica and the endophyte pathogenic to P. euphratica. The biological diversity in different places at different periods was studied by high-throughput sequencing. In addition, the endophytes pathogenic to P. euphratica were extracted, isolated, and authenticated by DNA sequencing. We analyzed the correlations between microorganisms in soils and endophytes in the pathogenetic P. euphratica.
According to the tree age, four different sampling sites were arranged: Sampling Site 1 (half year), the new development area of Yulin where P. euphratica had not been transplanted (XKWY, 38°9′37.7″N, 109°41′9.2″E) in the MUD, Yu Yang District, Northwest China; Sampling Site 2 (1 year), the new development area of Yunlin where P. euphratica were transplanted (XKY, 38°26′38.0″N, 109°35′38.2″E) in the MUD, Yu Yang District, Northwest China; Sampling Site 3 (2 years), the afforestation land (ZLD, 38°25′58.2″N, 109°37′18.7″E) in the MUD, Yu Yang District, Northwest China; Sampling Site 4 (3 years), Airport Road study site (JCL, 38°23′11.2″N, 109°37′11.78026″E) in the MUD, Yu Yang District, Northwest China. At the JCL study site, three types of different soils were investigated: JCLH, the soil around the surviving P. euphratica after 3 years of planting in JCL; JCLS, the soil around the dead P. euphratica after 3 years of planting in JCL; and JCLYLT, the soil in JCL without P. euphratica planting.
Six replications were conducted at all sampling sites of the selected soil samples. Soil samples were collected surrounding the soil stem, approximately 20 cm in diameter, of P. euphratica at depths from 10 to 50 cm. After thoroughly mixing, 100 g of prepared soil samples was stored at −80°C for sequencing. Corresponding air-dried soil samples were sieved by a 2-mm mesh and stored at 4°C for the analysis of soil physicochemical properties. The diseased parts of P. euphratica at different study sites were imaged and collected by cutting off the target stem.
Twenty-four pathogenetic P. euphratica samples were collected from XKY, ZLD, and JCL. The collected samples were disinfected by 75% ethanol for 1 min and 10% NaClO for 5 min (surface treatment). After then, the disinfected samples were rinsed thoroughly by sterile water and then dried in aseptic fume hood. Finally, the sterilized symptomatic stems were cultured by potato dextrose agar (PDA) culture medium, with antibiotic supplements (50 μg/ml of ampicillin and streptomycin sulfate), at 25°C. The characteristics of cultured mycelium on PDA were observed daily and isolated. The microscopic morphological characteristics of isolated colony were observed and recorded (Olympus, Tokyo, Japan). All the valuable biological materials have been protected, controlled, and accounted according the laboratory biosecurity guidance (WHO/CDS/EPR/2006.6) of World Health Organization (WHO).
Details of DNA extraction are shown in Supplementary Text S1. In brief, total DNA extraction of 36 soil samples was conducted (six groups with six repeats). DNA extraction of each sample possesses two technical replicates. The quality and concentration of extracted DNA were assessed. Samples were stored at −20°C before use. Details of bacteria 16S rRNA and fungi ITS gene amplicon sequencing are shown in Supplementary Text S2. Amplicon sequences were analyzed using the “DADA2” package in the R environment (version 3.6.1). Corresponding details are shown in Supplementary Text S3.
The bacterial community composition differences among treatments were tested by PERMANOVA (adonis, transformed data by Bray–Curtis, permutation = 999), implemented in R version 3.6.1. The DESeq function of the “DESeq2” package (version 1.18.1) was employed to test for differentially abundant ASVs among treatments. Statistical significance was based on a value of p < 0.05 (with FDR < 5% under the Benjamini–Hochberg correction).
The correlation between the community composition and soil properties was statistically analyzed based on six different types of soil. These parameters, including water-filled pore space (WFPS), pH, EC, AP, NO3–, and NH4+, are shown in Supplementary Table S1 and Figure 1. For the bacteria, the influence degree of the abovementioned parameters ranged as follows: pH > EC > WFPS > NO3– > AP > NH4+ (Figure 1A). These soil properties contributed to 18% of the change in the bacterial community (Figure 1A). According to the results of the correlation analysis, the influence of pH mainly affected Massilia and Rhodococcus, EC had a higher influence on Devosia; and NO3– on Candidatus nitrososphaera. For the fungi, the degree of influence of the abovementioned parameters ranged as follows: EC > pH > NO3– > WFPS > AP > NH4+ (Figure 1C). These soil properties contributed to 9% of the change in the fungal community (Figure 1C). In addition, the influence of EC mainly focused on Psathyrella, Coprinellus, Mallocybe, Peziza, and Arcopilus; NO3– mainly influenced Psathyrella, Coprinellus, Mallocybe, and Arcopilus; and WFPS mainly influenced Psathyrella.
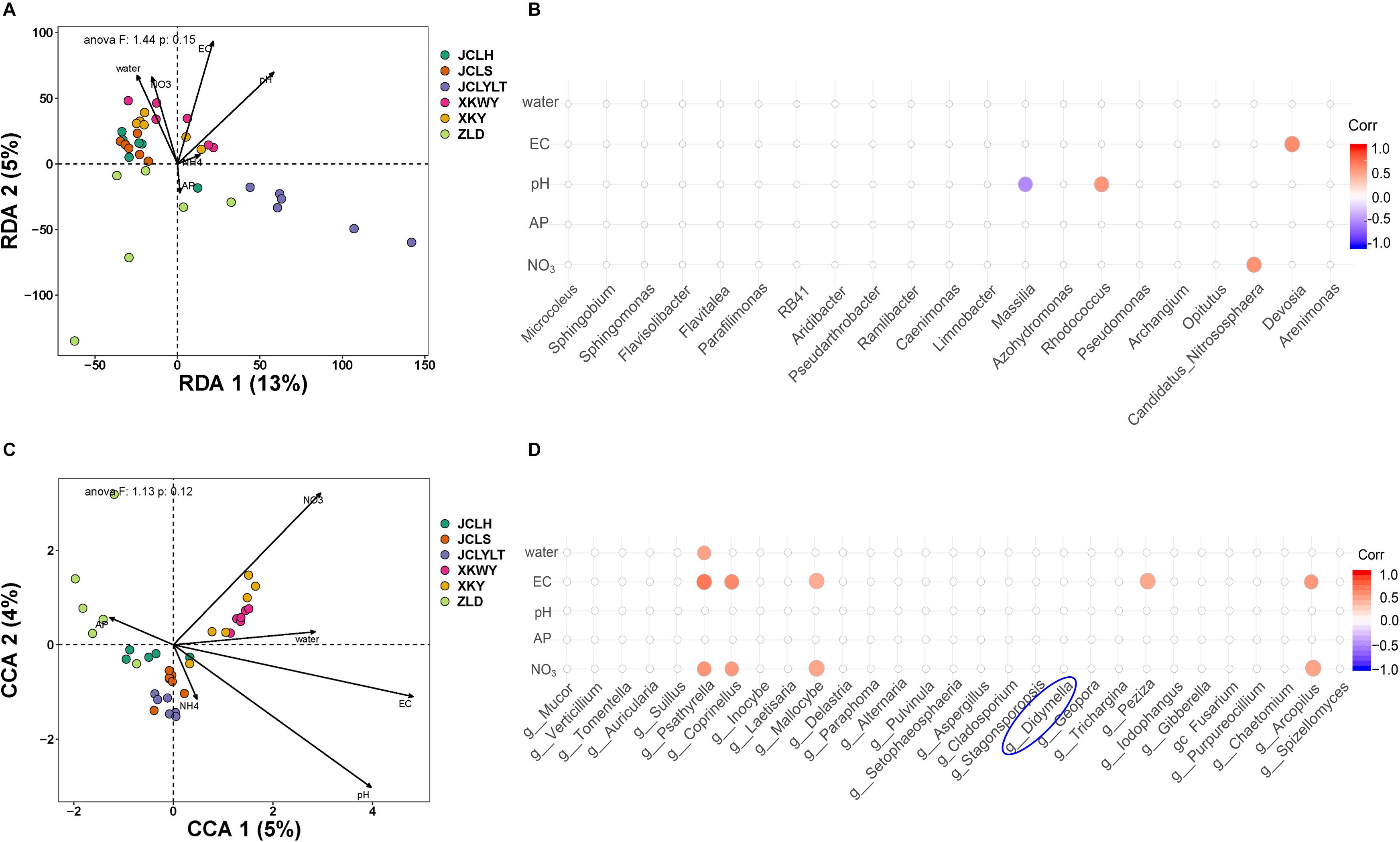
Figure 1. The influence degree of soil properties to the (A) bacterial community, (C) fungal community, and corresponding correlations between soil properties and (B) bacteria and (D) fungi.
To explore the correlations between stem pathogen infection and microbiome assemblage in the root soil, the endophytic fungi in the symptomatic segments were isolated and identified (Figure 2). A–D in Figure 2 is four typical symptoms of diseased P. euphratica in XKY, ZLD, and JCL. Finally, a total of 10 types of endophytic fungi were isolated and identified. Among these fungi, Epicoccum nigrum can produce some colored pigments, which are antifungal agents against other pathogenic fungi (Brown et al., 1987; Sun et al., 2011; Radić and Štrukelj, 2012), and Alternaria species (Alternaria alternate, Alternaria tenuissima), whose killing power release depends on a high-humidity environment (Moral et al., 2018). Phoma glomerata, which has an excellent ecological plasticity, can be permitted by 90 different kinds of plants and normally can cause leaf spot (Dörr et al., 2011). Trichoderma is a widespread fungus that is considered as an opportunistic avirulent plant symbiont (Harman et al., 2004; Bae et al., 2011). Previously, reports of Cladosporium oxysporum indicated that it has pathogenic effects on tomato and some vegetables, while it normally causes leaf spot (Wilingham et al., 2002; Baiswar et al., 2011; Zheng et al., 2014). Didymella glomerata is within the Didymellaceae family, which can cause stem lesions or cankers (Chen et al., 2015; Basim et al., 2016; Yao et al., 2016; Donati et al., 2018). The Valsa genus (Valsasordida, Valsanivea, Valsamalicola, and Valsamali) belongs to the family of Valsaceae, which can cause trunk diseases. Additionally, trunk diseases possess great power to kill young poplar trees 2 or 3 years after infection (Yi and Chi, 2011; Ma et al., 2016; Aghdam et al., 2017; Liu et al., 2018; Yu et al., 2018).
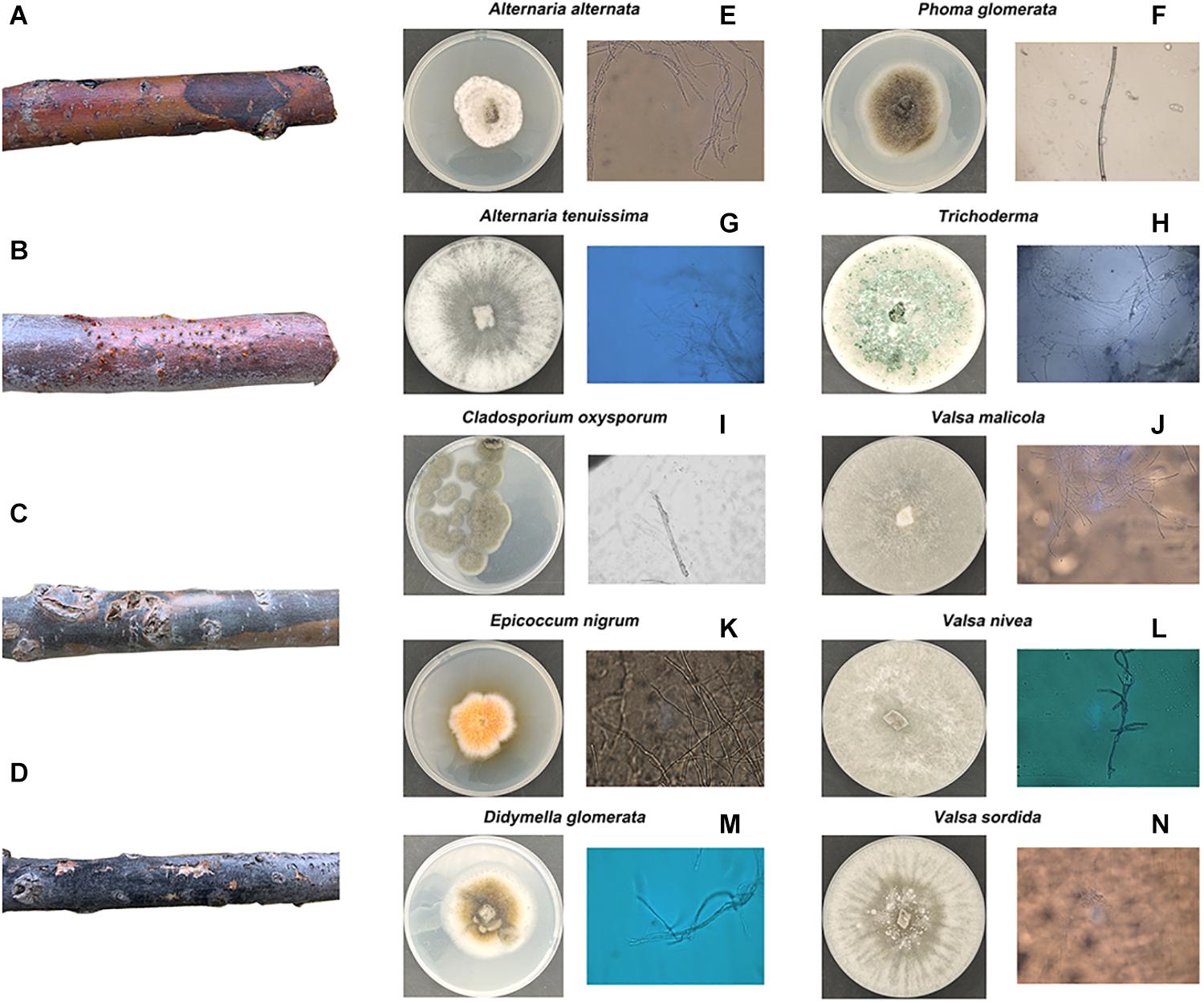
Figure 2. Four kinds of common morphological characteristics of diseased Populus euphrasae in XKY, ZLD, and JCL are shown in (A–D). The endophytic fungi isolated and identified from (A–D) are shown in (E–N).
According to the results of relative abundance analysis, for the bacteria, the most common phyla in the six types of soils were Proteobacteria, Bacteroidetes, Acidobacteria, Actinobacteria, and Thaumarchaeota. After P. euphratica planting, the relative abundance of Proteobacteria (dominant OTUs: OTU_12049, OTU_12644) (Supplementary Figure S1) and Bacteroidetes (dominant OTUs: OTU_13655, OTU_13821) (Supplementary Figure S1) increased (Figure 3A). However, the portions of Actinobacteria (dominant OTUs: OTU_22374) (Supplementary Figure S1) and Thaumarchaeota (dominant OTUs: OTU_5953, OTU_5954) (Supplementary Figure S1) were decreased (Figure 3A). At the same time, the fungi Ascomycota (dominant OTUs: OTU_3086, OTU_3548 and OTU_3550) (Figure 6) and Basidiomycota (dominant OTUs: OTU_2456) (Figure 6) were two of the most common fungal phyla. After P. euphratica planting, compared with the soil without P. euphratica, the relative abundance of Ascomycota in all the selected soils, except XKY, was increased (Figure 3B). However, the relative abundance of Basidiomycota was decreased in JCLH, JCLS, XKWY, and ZLD. Interestingly, XKY was the only exception (Figure 3B). By comparing the results of the relative abundance changing between the phyla level and the dominant OTUs level, we found that the relative abundance changing of similar trend dominant OTUs could meet with the phyla level well (Figures 3, 6 and Supplementary Figure S1).
The results of the observed α-diversity are shown in Figures 4A, 5A, which indicated that P. euphratica planting could influence the community diversity of both bacteria and fungi significantly. Overall, the number of OTUs in the same soil sample was higher for bacteria than for fungi. For the bacteria, the community richness (Figure 4A, eveness Simpson) was increased after P. euphratica planting compared with JCLYLT. As for the species diversity (Figure 4A, Shannon), XKWY, XKY, and ZLD have higher species diversity than JCL. After P. euphratica planting, the species diversity difference of JCLH, JCLS, and JCLYL was not obvious. For the fungi, the community richness in JCLS was higher than that in other experimental sites (Figure 5A, eveness Simpson). Additionally, a similar tendency existed in the community diversity (Figure 5A, Shannon).
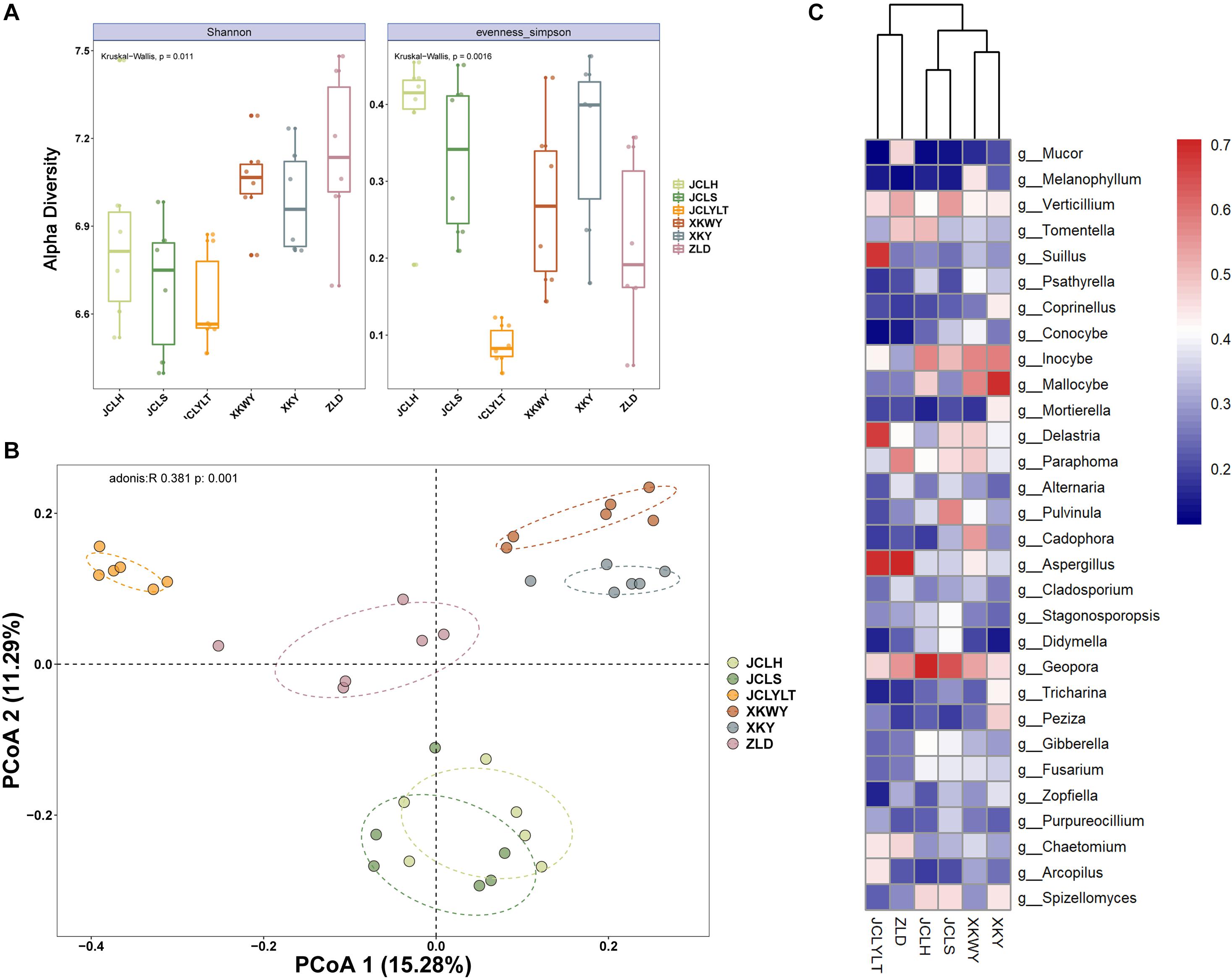
Figure 4. (A) Alpha diversity analysis based on the OTUs account for the species richness using eveness Simpson index and species diversity using Shannon index. (B) The Bray–Curtis dissimilarity principal coordinate analysis (PCoA) of bacterial community structures in different soils. (C) Characteristic bacterial populations with significant community differences in different soils.
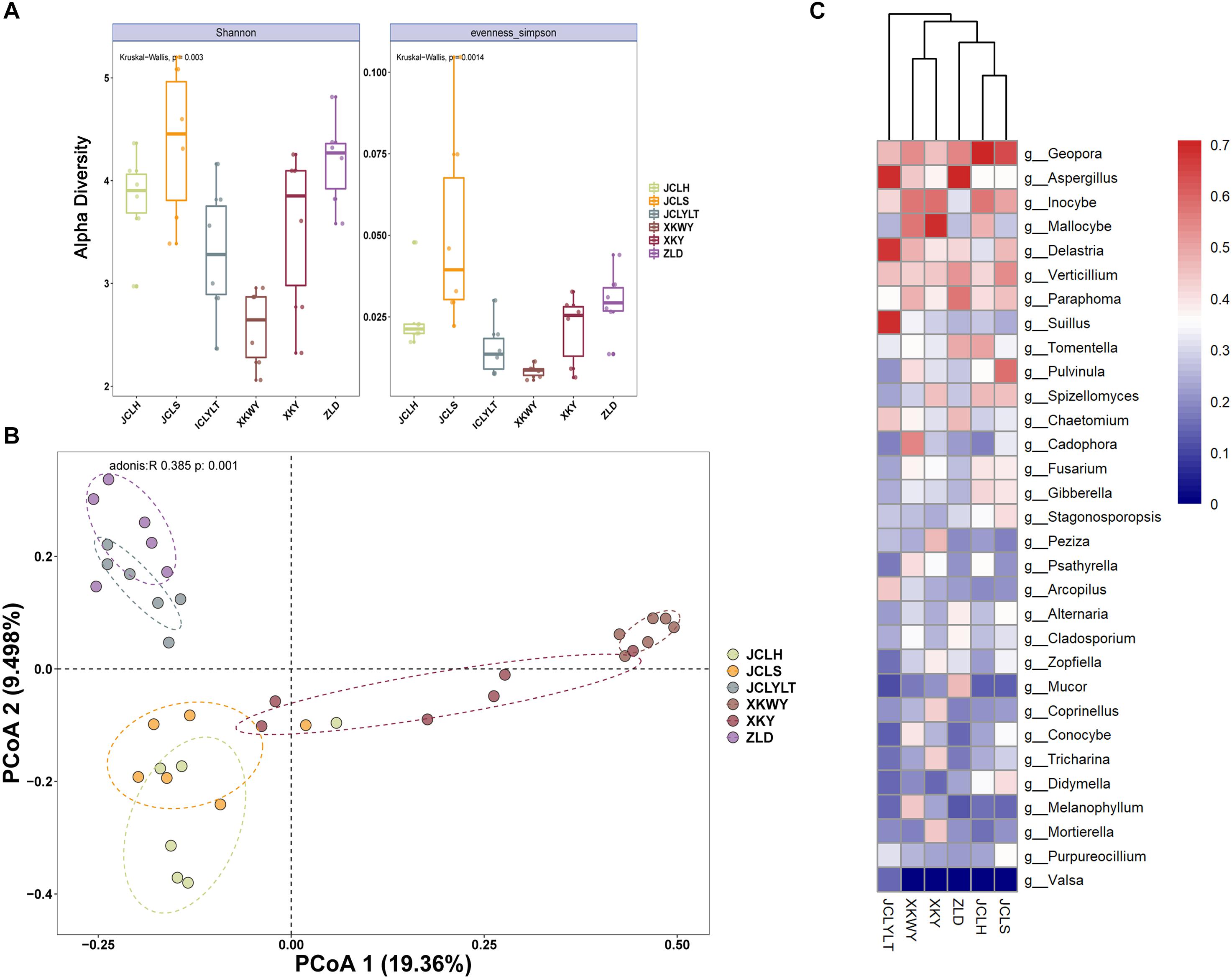
Figure 5. (A) Alpha diversity analysis based on the OTUs account for the species richness using eveness Simpson index and species diversity using Shannon index. (B) Bray–Curtis dissimilarity principal coordinate analysis (PCoA) of fungal community structures in different soils. (C) Characteristic fungal populations with significant community differences in different soils.
For the bacterial community, the Bray–Curtis dissimilarity principal coordinate analysis (PCoA) results showed significant differences in the soils of JCL, XKWY, XKY, and ZLD (Figure 4B). In the JCL soils, after P. euphratica planting, both JCLS and JCLH showed pronounced differences compared to JCLYLT. The bacterial community between JCLS and JCLH was not completely separated. For the fungal community, the PCoA results indicated that the fungal community in the selected soil showed significant differences (Figure 5B). In the JCL soils, after P. euphratica planting, both JCLS and JCLH showed pronounced differences compared to JCLYLT. Similar to the bacterial community, the fungal community between JCLS and JCLH was not completely separated.
The differential abundance of bacteria and fungi of six selected soil sites are shown in Figures 4C, 5C. The phyla with significant community differences from all the selected OTUs were presented.
The differential abundances of bacteria and fungi of six selected soil sites are shown in Figures 4C, 5C. The phyla with significant community differences based on all the selected OTUs were presented.
The composition of soil microbiomes can be influenced by multiple environmental factors, and further interactions between pathogens and plants face the same situation (Xu et al., 2015; Wei et al., 2019). The results of this study indicated that soil physicochemical properties changed the composition of bacterial and fungal communities. Different parameters always possess, to some extent, a preference for specific phyla. For the bacteria, four different phyla were identified after taking the bacterial composition of all the sequenced soil samples into consideration. These affected phyla included Massilia, Rhodococcus, Devosia, and Candidatus Nitrososphaera. Previous studies have shown that most Rhodococcus species are benign (Mcleod et al., 2006). The genus Massilia possesses bacteria that are able to suppress pathogens for healthy plants. Studies of Devosia and Candidatus Nitrososphaera are scarce, and the pathogenicity of these genera remains unclear (Scola et al., 1998; Vannini et al., 2004; Zhalnina et al., 2014). For the fungi, EC, NO3–, and WFPS contributed some effect to five phyla, independent or synergistic. All the phyla had not been isolated and identified in symptomatic segments. Bacterial isolation and identification from infected segments were not studied here due to the large complexity.
Figures 4B, 5B show that the microbial community structure in the soils around the surviving P. euphratica is markedly different (P < 0.001). These results show the foundation difference of soil microbiome composition between different study sites. For the JCL sites, after P. euphratica planting, the microbiome community structures in both JCLH and JCLS were separated completely from JCLYLT (P < 0.001) (Figures 4B, 5B). At the same time, the soil bacterial and fungal community structures between JCLH and JCLS almost clustered into two distinct groups but were not thoroughly separated (P < 0.001, accounting for 26.57% of the bacterial community and 28.86% of the fungal community). Early reports showed that variations in soil microbiome community structures and functions can affect target plants and determine whether the plants survived or succumbed to disease (Wei et al., 2019). In addition to composition, microbiome diversity level is also an important predictor for the soil health and plant growth (Saleem et al., 2019). Therefore, significant soil microbiome community structures have a great potential to lead to the high death rate of P. euphratica.
In this study, 10 types of endophytic fungi were isolated and identified. After systematic literature review, we found that Alternaria (A. alternate, A. tenuissima), D. glomerata, and the Valsa genus (Valsa sordida, Valsa nivea, Valsa malicola, Valsa mali) have great potential to participate in plant–pathogen interactions. Results showed that Ascomycota was the major fungal component in the selected soils with or without P. euphratica planting. Interestingly, Alternaria, Didymella, and Valsa all belong to the division of Ascomycota. By analyzing several characteristic fungal populations (the abundance of specific phyla in the studied soils that possess dramatic differences except Valsa, p < 0.05), we found that for the Alternaria, the relative abundance of Alternaria (OTU_3086, on behalf of Alternaria alternate) in JCLS was the highest within six selected soils (Figure 6A). The abundance of Alternaria in different studied sites followed an interesting rank: JCLS > ZLD > JCLH > XKWY ≈ XKY ≈ JCLYLT. The relative abundance of Didymella in JCLS was the highest. The abundance of Didymella (3,550 OTUs, on behalf of D. glomerata) in different studied sites followed an interesting rank: JCLS > JCLH > ZLD > XKWY ≈ XKY ≈ JCLYLT (Figure 6B). For the genus Valsa, there were almost no differences in the relative abundance among the selected soils. In this respect, the high death rate of Populus may be caused by Alternaria alternate and D. glomerata.
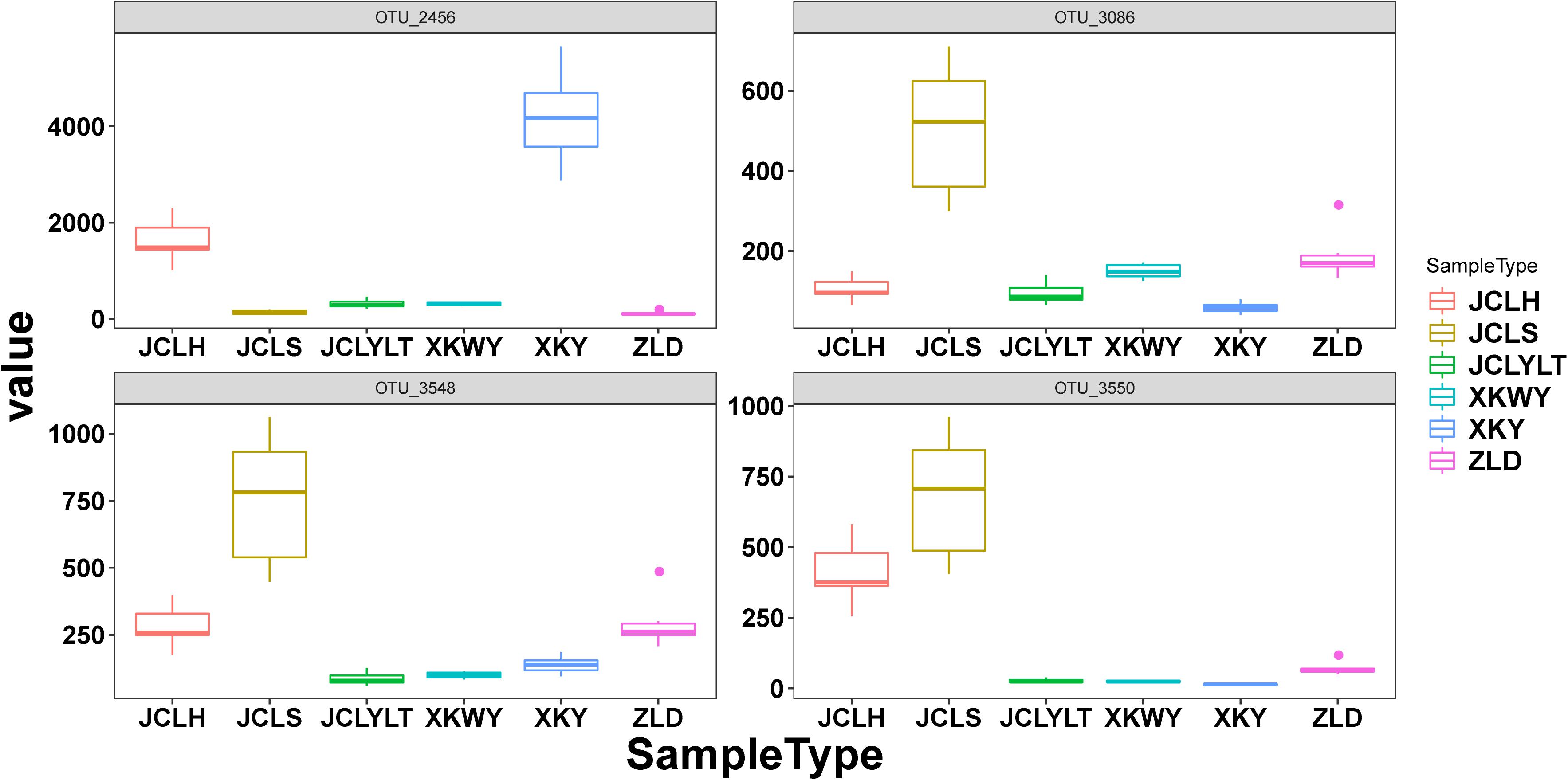
Figure 6. The relative abundance of OTUs 2456, OTUs 3086, OTUs 3548, and OTUs 3550 in the studied soils.
According to the plant–pathogen interactions discovered in the fungi perspective, we found that higher relative abundance of the target fungi in the P. euphratica-planted soil poses a high risk of infection. When we do a reasonable extrapolation of this result in the bacteria view, several candidate bacteria (OTU_12049 and OTU_12644, on behalf of JG34-KF-161 and Sphingomonadaceae Sphingomonas, respectively, within the Proteobacteria phyla) could be listed (Supplementary Figure S1). In addition, we should also pay attention to the relative abundance decrease of Actinobacteria (dominant OTUs: OTU_22374) (Supplementary Figure S1) and Thaumarchaeota (dominant OTUs: OTU_5953, OTU_5954) (Supplementary Figure S1) after P. euphratica planting. Thaumarchaeota has been regarded as the key player within the global nitrogen cycle (Shao et al., 2019). The phylogeny and the function of the class Actinobacteria remains controversial up to now (Ludwig et al., 2012; Sen et al., 2014). Its function within the plant–pathogen interaction system should be explored thoroughly. So, to do a further confirmation about this prediction will be a challenging, but meaningful, object in our future studies.
In conclusion, the results of the present study revealed deeper interactions between plant and pathogen. Increased attention on the presence of pathogens during tree planting will be a crucial factor in plant survival rates. Massart and Mark point out that in order to achieve a better biological control, we must combine as much potential factors together (e.g., host plants, host genotype, integrate microbial community, pathogen, biocontrol agents, molecules, etc.) (Massart et al., 2015; Mazzola and Gu, 2002). In addition, previous studies have indicated that the tolerance of plants to pathogens increases with increasing tree age and growth state (Mansfeld et al., 2017; Berendsen et al., 2018). Plant–pathogen interactions as a complicated dynamic equilibrium process warrant further study.
The data can be found in the NCBI database, with accession number – PRJNA597436.
YT puts forward the whole idea of this manuscript and implemented all the tasks. ZD had given several constructive instructions to this manuscript. XW and FT had given site instructions and integtate theory with practice. BG helped ZD with some interdisciplinary theories. CZ works as a assistant and made many contributions to this manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Mr. Kailin Tian for the isolation of plant endophytes and Dr. Tao Wen for the DNA extraction and sequencing. We are also grateful to the American Journal Experts (T5LH6SVF) for their helpful efforts on the language edit of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02095/full#supplementary-material
Aghdam, A. R., Asadollah, B. A., Karimi, K., and Cryptosphaeria, M. A. (2017). Canker of populusnigra caused by cryptosphaeriapullmanensis, a new threat to poplar industry in Iran. J. Phytopathol. 165, 387–396. doi: 10.1111/jph.12572
Arrigoni, E., Antonielli, L., Pindo, M., Pertot, I., and Perazzolli, M. (2018). Tissue age and plant genotype affect the microbiota of apple and pear bark. Microbiol. Res. 211, 57–68. doi: 10.1016/j.micres.2018.04.002
Bae, H., Roberts, D. P., Lim, H. S., Strem, M. D., and Park, S. C. (2011). EndophyticTrichoderma isolates from tropical environments delay disease onset and induce resistance against Phytophthora capsici in hot pepper using multiple mechanisms. Mol. Plant Microbe Interact. 24, 336–351. doi: 10.1094/mpmi-09-10-0221
Baiswar, P., Chandra, S., Bag, T. K., Patel, R. K., and Ngachan, S. V. (2011). Cladosporiumoxysporum on Prunusnepalensis in India. Australas. Plant Dis. Notes 6, 3–6.
Basim, E., Basım, H., Abdulai, M., Baki, D., and Öztürka, N. (2016). Dentification and characterization of Didymellabryoniaecausing gummy stem blight disease of watermelon (Citrullus lanatus) in Turkey. Crop. Prot. 90, 150–156. doi: 10.1016/j.cropro.2016.08.026
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., and Jonge, R. D. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Brown, A. E., Finlay, R., and Ward, J. S. (1987). Antifungal compounds produced by Epicoccumpurpurascens against soil-borne plant pathogenic fungi. Soil Biol. Biochem. 19, 657–664. doi: 10.1016/0038-0717(87)90044-7
Chen, Q., Jiang, J. R., Zhang, G. Z., Cai, L., and Crous, P. W. (2015). Resolving the Phoma enigma. Stud. Myco. 82, 137–217. doi: 10.1016/j.simyco.2015.10.003
Donati, I., Cellini, A., Buriani, G., Mauri, S., and Kay, C. (2018). Pathways of flower infection and pollen-mediated dispersion of Pseudomonas syringaepv. actinidiae, the causal agent of kiwifruit bacterial canker. Hortic. Res. 5:56.
Dörr, A. J. M., Rodolfi, M., Scalici, M., Eliaa, A. C., and Garzoli, L. (2011). Phomaglomerata, a potential new threat to Italian inland waters. J. Nat. Conserve. 19, 370–373. doi: 10.1016/j.jnc.2011.06.006
Feng, X., Fu, B., Lu, N., Zeng, Y., and Wu, B. (2013). How ecological restoration alters ecosystem services: an analysis of carbon sequestration in China’s loess Plateau. Sci. Rep. 3:2846.
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species—opportunistic avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Keram, A., Halik, Ü, Keyimu, M., Aishan, T., Mamat, Z., and Rouzi, A. (2019). Gap dynamics of natural Populuseuphratica floodplain forests affected by hydrological alteration along the Tarim River: implications for restoration of the riparian forests. For. Ecol. Manage. 438, 103–113. doi: 10.1016/j.foreco.2019.02.009
Lai, Z., Zhang, Y., Liu, J., Wu, B., Qin, S., and Fa, K. (2016). Fine-root distribution, production, decomposition, and effect on soil organic carbon of three revegetation shrub species in northwest China. For. Ecol. Manage. 359, 381–388. doi: 10.1016/j.foreco.2015.04.025
Leff, J. W., Del Tredici, P., Friedman, W. E., and Fierer, N. (2015). Spatial structuring of bacterial communities within individual Ginkgo biloba trees. Environ. Microbiol. 17, 2352–2361. doi: 10.1111/1462-2920.12695
Li, S., Yan, C., Wang, T., and Du, H. Q. (2019). Monitoring grassland reclamation in the Mu us desert using remote sensing from 2010 to 2015. Environ. Earth Sci. 78:311.
Li, X. R., Zhang, Z. S., Huang, L., and Wang, X. (2013). Review of the ecohydrological processes and feedback mechanisms controlling sand-binding vegetation systems in sandy desert regions of China. Chin. Sci. Bull. 58, 1483–1496. doi: 10.1007/s11434-012-5662-5
Liu, P., Shi, Y. Y., and Zhu, L. W. (2018). Genetic variation in resistance to valsa canker is related to arbutin and gallic acid content in Pyrus bretschneideri. Hortic. Plant J. 4, 233–238. doi: 10.1016/j.hpj.2018.09.002
Ludwig, W., Euzéby, J., and Whitman, W. B. (2012). Taxonomic Outline of the Phylum Actinobacteria. In Bergey’s Manual§of Systematic Bacteriology. New York, NY: Springer, 29–31.
Ma, R., Zhu, Y. F., Fan, X. L., and Tian, C. M. (2016). Canker disease of willow and poplar caused by Cryptosphaeriapullmanensis recorded in China. Forest Pathol. 46, 327–335. doi: 10.1111/efp.12261
Mansfeld, B. N., Colle, M., Kang, Y., Jones, A. D., and Grumet, R. (2017). Transcriptomic and metabolomic analyses of cucumber fruit peels reveal a developmental increase in terpenoid glycosides associated with age-related resistance to Phytophthora.capsici. Hortic. Res. 4:17022.
Massart, S., Martinez-Medina, M., and Jijakli, M. H. (2015). Biological control in the microbiome era: challenges and opportunities. Biol. Control 89, 98–108. doi: 10.1016/j.biocontrol.2015.06.003
Mazzola, M., and Gu, Y. H. (2002). Wheat genotype-specific induction of soil microbial communities suppressive to disease incited by Rhizoctonia solani anastomosis group (AG)-5 and AG-8. Phytopathology 92, 1300–1307. doi: 10.1094/phyto.2002.92.12.1300
McLeod, M. P., Warren, R. L., Hsiao, W. W., Araki, N., Myhre, M., Fernandes, C., et al. (2006). The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U.S.A. 103, 15582–15587. doi: 10.1073/pnas.0607048103
Moral, J., Lichtemberg, P. S. F., Papagelis, A., Sherman, J., and Michailides, T. J. (2018). Didymellaglomeratacausing leaf blight on pistachio. Eur. J. of Plant Pathol. 151, 1095–1099. doi: 10.1007/s10658-018-1422-y
Radić, N., and Štrukelj, B. (2012). Endophytic fungi—The treasure chest of antibacterial substances. Phytomedicine 19, 1270–1284. doi: 10.1016/j.phymed.2012.09.007
Saleem, M., Hu, J., and Jousset, A. (2019). More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annu. Rev. Ecol. Evol. Syst. 50, 145–168. doi: 10.1146/annurev-ecolsys-110617-062605
Scola, L. B., Birtles, R. J., Mallet, M. N., and Raoult, D. (1998). Massiliatimonae gen. nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J. Clin. Microbiol. 36, 2847–2852. doi: 10.1128/jcm.36.10.2847-2852.1998
Sen, A., Daubin, V., Abrouk, D., Gifford, I., Berry, A. M., and Normand, P. (2014). Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders ‘Frankiales’ and Micrococcales should be split into coherent entities: proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov. and Nakamurellales ord. nov. Int. J. Syst. Evol. Micr. 64, 3821–3832. doi: 10.1099/ijs.0.063966-0
Shao, K., Jiang, X., Hu, Y., Tang, X., and Gao, G. (2019). Thaumarchaeota affiliated with soil crenarchaeotic group are prevalent in the alkaline soil of an alpine grassland in northwestern China. Ann. Microbiol. 69, 867–870. doi: 10.1007/s13213-019-01492-5
Shu, P. X., Li, B. S., Wang, H., Qiu, Y. H., and Niu, D. F. (2018). Geochemical characteristics of surface dune sand in the Mu us desert, inner mongolia, and implications for reconstructing the paleoenvironment. Quatern. Int. 479, 106–116. doi: 10.1016/j.quaint.2017.05.053
Sun, H. H., Mao, W. J., Jiao, J. Y., Xu, J. C., and Li, H. Y. (2011). Structural characterization of extracellular polysaccharides produced by the marine fungus Epicoccum nigrum JJY-40 and their antioxidant activities. Mar. Biotechnol. 13, 1048–1055. doi: 10.1007/s10126-011-9368-5
Sun, Y., Zhang, Y., Feng, W., Qin, S. G., and Liu, Z. (2019). Revegetated shrub species recruit different soil fungal assemblages in a desert ecosystem. Plant Soil 435, 81–93. doi: 10.1007/s11104-018-3877-1
Vannini, C., Rosati, G., Verni, F., and Petroni, G. (2004). Identification of the bacterial endosymbionts of the marine ciliate Euplotesmagnicirratus (Ciliophora, Hypotrichia) and proposal of ‘CandidatusDevosiaeuplotis’. Int. J. Syst. Evol. Microbiol. 54, 1151–1156. doi: 10.1099/ijs.0.02759-0
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wei, Z., Gu, Y. A., Friman, V. P., Kowalchuk, G. A., and Xu, Y. C. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5:eaaw0759. doi: 10.1126/sciadv.aaw0759
Wilingham, S. L., Pegg, K. G., Langdon, P. W. B., Cooke, A. W., and Peasley, D. (2002). Combinations of strobilurin fungicides and acibenzolar (Bion) to reduce scab on passionfruit caused by Cladosporium oxysporum. Australas. Plant Path. 31, 333–336. doi: 10.1071/ap02036
Xu, X., Passey, T., Wei, F., Saville, R., and Harrison, R. J. (2015). Amplicon-based metagenomics identified candidate organisms in soils that caused yield decline in strawberry. Hortic. Res. 2:15022.
Yao, X. F., Li, P. F., Xu, J. H., Zhang, M., and Ren, R. S. (2016). Rapid and sensitive detection of Didymella bryoniae by visual loop-mediated isothermal amplification assay. Front. Microbiol. 7:1372. doi: 10.3389/fmicb.2016.01372
Yi, H. W., and Chi, Y. J. (2011). Biocontrol of Cytospora canker of poplar in north-east China with Trichodermalongibrachiatum. Forest Pathol. 41, 299–307. doi: 10.1111/j.1439-0329.2010.00704.x
Yu, C., Li, T., Shi, X., Saleem, M., Li, B., Liang, W., et al. (2018). Deletion of endo-β-1, 4-xylanase VmXyl1 impacts the virulence of Valsamali in apple tree. Front. Plant Sci. 9:663. doi: 10.3389/fpls.2018.00663
Zhalnina, K. V., Dias, R., Leonard, M. T., Quadros, P. D. D., and Camargo, F. A. O. (2014). Genome Sequence of candidatus nitrososphaera evergladensis from group I.1b enriched from everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS One 9:e101648. doi: 10.1371/journal.pone.0101648
Keywords: Populus euphratica, Mu Us Desert, biological diversity, microbial communities, pathogen
Citation: Tuo Y, Dong Z, Wang X, Gao B, Zhu C and Tuo F (2020) Metagenomics Reveal Correlations Between Microbial Organisms in Soils and the Health of Populus euphratica. Front. Microbiol. 11:2095. doi: 10.3389/fmicb.2020.02095
Received: 17 December 2019; Accepted: 10 August 2020;
Published: 08 September 2020.
Edited by:
Amy Brunner, Virginia Tech, United StatesReviewed by:
Muhammad Saleem, Alabama State University, United StatesCopyright © 2020 Tuo, Dong, Wang, Gao, Zhu and Tuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Tuo, eXR1b0Bzbm51LmVkdS5jbg==; Zhibao Dong, emJkb25nQHNubnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.