- State Key Laboratory of Veterinary Etiological Biology, OIE/National Foot and Mouth Disease Reference Laboratory, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
Foot-and-mouth disease (FMD) is a highly contagious disease that affects cloven-hoofed animals such as pigs, cattle, and sheep. The disease is caused by the foot-and-mouth disease virus (FMDV) which has a non-enveloped virion with icosahedral symmetry that encapsulates a positive-sense, single-stranded RNA genome of ∼8.4 kb. FMDV infection causes obvious immunosuppressive effects on the host. In recent years, studies on the immunosuppressive mechanism of FMDV have become a popular topic. In addition, studies have shown that many FMDV proteins are involved in the regulation of host innate immunity and have revealed mechanisms by which FMDV proteins mediate host innate immunity. In this review, advances in studies on the mechanisms of interaction between FMDV proteins and host innate immunity are summarized to provide a comprehensive understanding of FMDV pathogenesis and the theoretical basis for FMD prevention and control.
Introduction
Foot-and-mouth disease (FMD) is an acute, highly contagious livestock disease that affects cloven-hoofed animals such as pigs, cattle, and sheep, thereby causing severe economic loss. Typical clinical symptoms of FMD include high fever and numerous blisters on the oral mucosa, hoof, and breast, with the disease being both diverse and fast (Thomson et al., 2003). There have been several serious outbreaks of FMD in some countries, across Europe, the Middle East, Africa and Asia, and Taiwan, etc., which have hindered the development of livestock breeding and caused huge losses to the global economy (Sangare et al., 2001; Grubman and Baxt, 2004). The Foot-and-mouth disease virus (FMDV) is the pathogen that causes FMD, belonging to the Aphthovirus genus of Picornaviridae family. There are seven FMDV serotypes in the world, namely, A, O, C, South Africa 1, South Africa 2, South Africa 3, and Asia 1. Each serotype includes multiple subtypes and does not have an antigenic cross-protection reaction (Saiz et al., 2002). FMDV is a single-stranded and positive-sense RNA virus surrounded by an icosahedral capsid. Its genome, comprising of approximately 8,400 nucleotides, has a single open reading frame (ORF) that is translated into a polyprotein, which is then processed by the three viral proteases Lpro, 2A, and 3Cpro into the polypeptide products P1 (VP1 to VP4), P2 (2A, 2B, and 2C), and P3 (3A, 3B, 3Cpro, and 3Dpol) and subsequently generated four mature structural proteins (VP4, VP2, VP3, and VP1) and eight non-structural proteins (Lpro, 2A, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol) (Grubman and Baxt, 2004) (Figure 1).
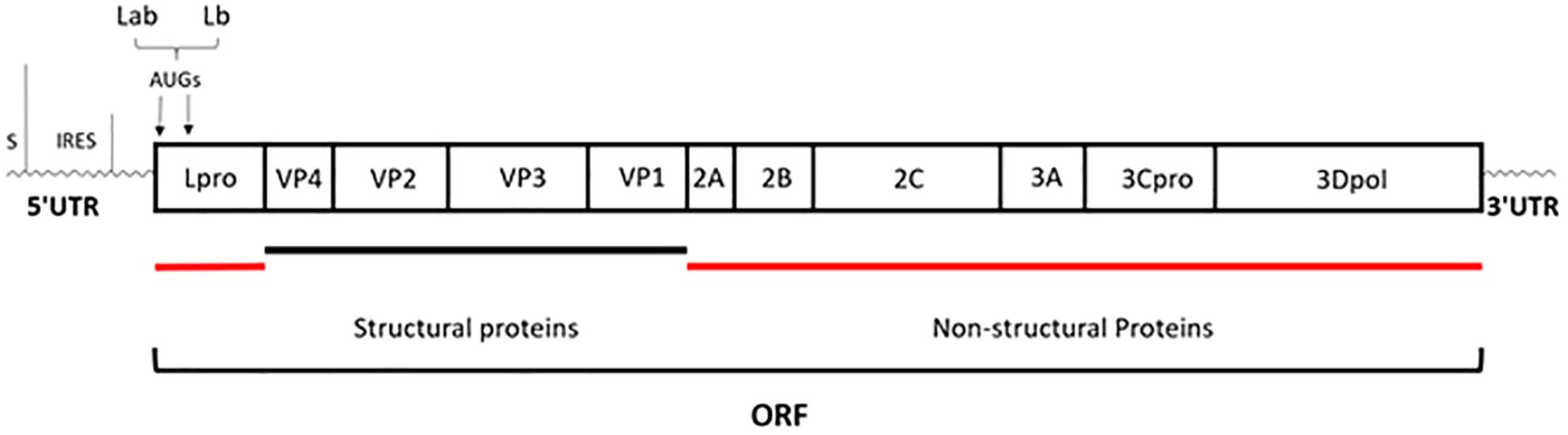
Figure 1. Schematic representation of genome organization. The genome of approximately 8,400 nucleotides has a single open reading frame (ORF); then is processed by the three viral proteases: Lpro, 2A, and 3Cpro; eventually generated four mature structural proteins (VP4, VP2, VP3, and VP1) and eight non-structural proteins (Lpro, 2A, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol) (Grubman and Baxt, 2004).
After the host is infected by microbial pathogens, pathogen recognition receptors (PRRs) in the host, recognize conserved molecular structures (i.e., pathogen-associated molecular patterns, PAMPs) from pathogens (Kumar et al., 2011). At present, depending on their different protein domain homologies, PRRs are classified four major families: Toll-like receptors (TLRs), RIG-I (retinoic acid-inducible-gene-I)-like receptors (RLRs), Nod-like receptors (NLRs), and C-type lectin receptors (CLRs) (Kumar et al., 2009). Once PRRs recognize PAMPs in the host cell, various anti-microbial immune responses are rapidly triggered via the induction of inflammatory cytokines, chemokines, and type I interferons (Zhong et al., 2010) (Figure 2). The innate immune system is the first line of defense against pathogens and plays a crucial role in controlling pathogen infection (Yoneyama and Fujita, 2010). Interferon (IFN) production plays a pivotal role in the host antiviral innate immune response, which can suppress viral replication especially in the early stage of the immune response. IFNs can be divided into three types according to their specific membrane-bound receptors, namely, type I, II, and III IFN (Randall and Goodbourn, 2008). Type I IFN (mainly as IFN-α and IFN-β) are more important than other interferons (such as type II, and III IFNs) in regulating the innate immune system. For example, the FMDV RNA is recognized by MDA5 and induces type I IFN production in the host to inhibit FMDV replication (Stetson and Medzhitov, 2006; Ma et al., 2018). Moreover, FMDV Lpro can inhibit IFN-α/β production by cleaving eIF4G and NF-κB (Hato et al., 2010). Recently, study has shown that a novel member of type I IFN, IFN-αω also has anti-FMDV activity (Li et al., 2019d). IFN-γ, the only member of the type II IFNs, can also regulate innate immunity responses (Deretic and Levine, 2018). Study has found that IFN-γ can potentially be used as a rapid method to detect the immune response of FMDV (Parida et al., 2006). Type III IFN (IFN-λ1, IFN-λ2, and IFN-λ3) are also involved in the regulation of FMDV infection. In addition to having pathogen defense functionality that is as good as that of the type I IFNs, type III IFNs can also increase adaptive immune responses in the respiratory mucosa (Ye et al., 2019). It has been demonstrated that poIFN-λ1 (novel recombinant porcine interferon lambda 1) can inhibit FMDV replication in IBRS-2 cells, and that FMDV Lpro has the ability to counteract this inhibition (Wang et al., 2011b). In addition, innate immune cells also play important roles in innate immune response processes, which can induce the production of IFNs in many ways to achieve antiviral effects (Golde et al., 2008). FMDV and its hosts are in a constant arms race, such that the virus has evolved multiple strategies to escape the host’s immune surveillance and defense system, which eventually destroys the balance between FMDV replication and host antiviral response (Table 1). In particular, FMDV proteins can directly or indirectly regulate the host’s innate immune response to survive and replicate in the host (Rodriguez Pulido and Saiz, 2017).
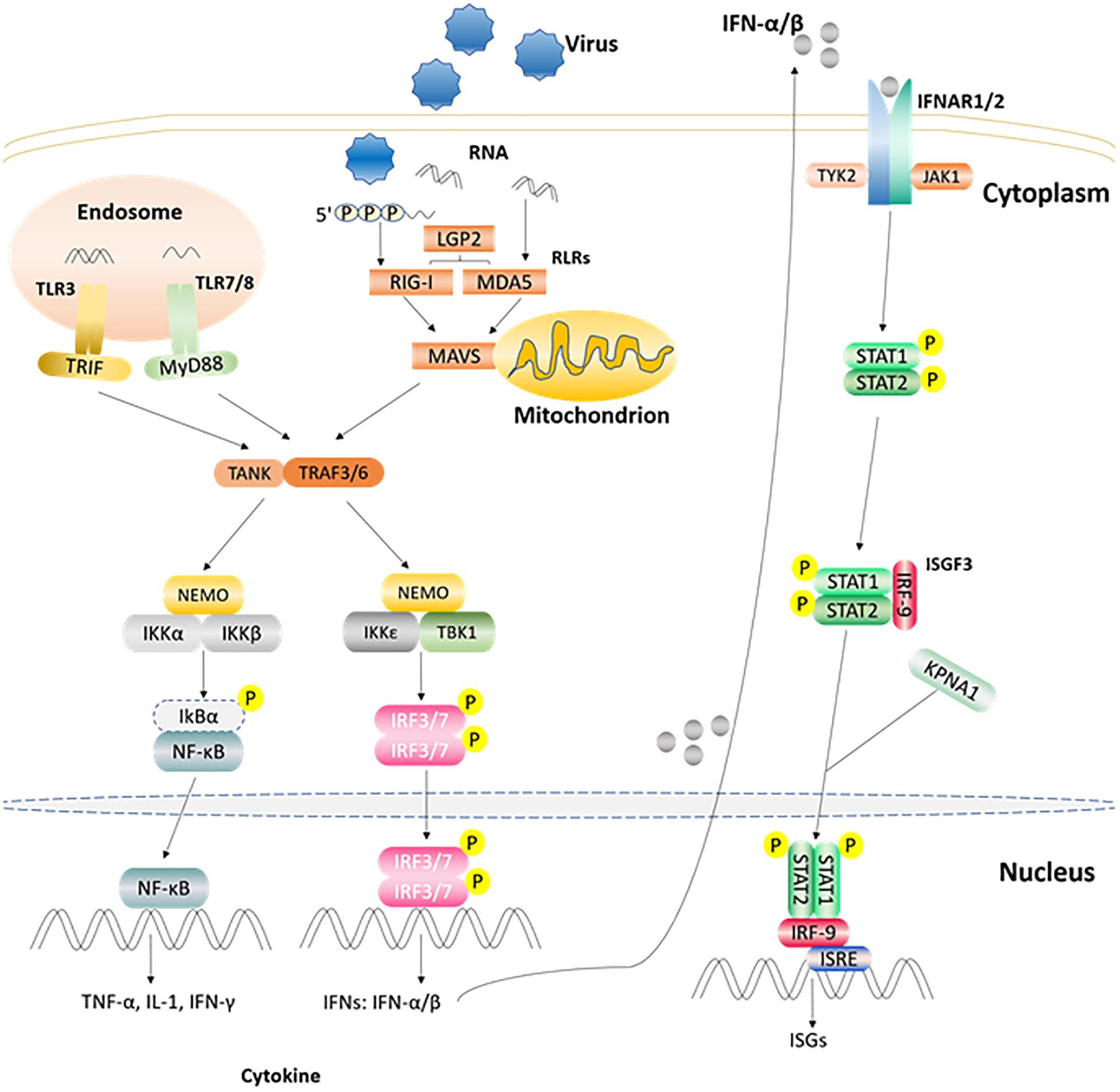
Figure 2. Part of the innate immune signaling pathways triggered after the viral RNA is recognized by TLRs and RLRs. After TLR3 and TLR7/8 recognize the ligand in endosome, they recruit and activate TRIF and MyD88, respectively. RLRs member recognizes corresponding ligand and activates the CARD region of MAVS. Activated TRIF, MyD88, MAVS induce TANK, and TRAF3/6. Then one signal pathway activates IKKα/β and NEMO complex, and the other activates TBKI and IKKε, NEMO complex. Subsequently, the IKKα/β and NEMO complex activates NF-κB, while TBKI and IKKε, NEMO complex phosphorylates and activates IRF3/7 to induce the expression of IFN α/β gene. The produced IFN α/β can bind to receptor IFNAR1/2, recruit STAT1/2 through the JAK1/TYK2 adaptor molecule, and recruit IRF9 through phosphorylated STAT1/2 to form the complex ISGF3 and enter the nucleus, then bind ISRE, and stimulate the expression of ISGs (Rodriguez Pulido and Saiz, 2017; Medina et al., 2018).

Table 1. FMDV proteins regulate and/or interact with the host’s innate immunity (Medina et al., 2018).
In the past two decades, thanks to the improved understanding of FMDV structure, many studies have confirmed that FMDV proteins can inhibit the production of IFNs. Therefore, this article reviews the relationship between FMDV proteins and the host’s innate immune-related proteins and also provides ideas for further studies regarding FMDV inhibiting the host’s innate immune response and regarding effective FMDV vaccines development.
FMDV Proteins Regulate Host Innate Immunity
The Role of FMDV Structural Proteins in the Regulation of Host Innate Immunity
Four structural proteins (VP4, VP2, VP3, and VP1) encoded by the P1 region of FMDV, mainly form the icosahedral capsid of virus particles: VP1-3 cooperate to form the capsid surface, while VP4 forms the internal structure of the virus particles. The four structural proteins differ in terms of conservation: VP1 is highly variable, VP2 and VP3 are relatively conserved, and VP4 is highly conserved among all serotypes. VP1 and VP3 play important roles in inhibiting the production of host interferons to weaken the host antiviral response. On the other hand, VP2 is involved in the induction of autophagy (Feng et al., 2018; Li et al., 2019b; Liu et al., 2020).
Advances in the Suppression of Innate Immunity by FMDV VP1
The FMDV structural proteins VP1, VP2, and VP3 of FMDV are folded into an eight-stranded wedge-shaped-barrel, and compose a major part of capsids; however, most of the antigen sites corresponding to the immune response are mainly found on the G-H loop of VP1 (Abubakar et al., 2018). There is a hypervariable region on the G–H loop which may lead to the high variability of VP1 (Fernandez-Sainz et al., 2019). VP1 plays crucial roles in FMDV infection, such as in inducing neutralizing antibodies, mediating cellular and humoral immunity, inducing host cell apoptosis, and promoting FMDV replication. It has been found that the FMDV structural protein VP1 can interact with soluble resistance-related calcium binding protein (sorcin) to inhibit type I IFN production induced by SeV or TNF-α, and the interaction between FMDV VP1 and sorcin can also disrupt the signal transduction of NF-κB, leading to persistent FMDV infection of the host (Li et al., 2013); however, further study is still required to determine its specific mechanism. Recently, the interaction of the host cell protein DnaJ heat shock protein family (Hsp40) member A3 (DNAJA3) with VP1 was identified through a yeast two-hybrid system. Studies has shown that DNAJA3 can significantly reduce the inhibitory effect of VP1 on SeV-induced IRF3 phosphorylation, dimerization, and nuclear localization through the lysosomal pathway, thus reducing the antagonism on the IFN-β signal pathway and inhibiting FMDV replication (Zhang et al., 2019). In addition, VP1 can also act on ribosomal protein SA (RPSA) to weaken its inhibitory effect on the mitogen-activated protein kinase (MAPK) pathway, which is conducive to the FMDV replication (Zhu et al., 2020). VP2 may indirectly inhibit the host’s type I IFN response pathway by interacting with the host protein heat shock protein family B [small] member 1 (HSPB1) to enhance viral replication, but the mechanism needs to be further elucidated (Sun et al., 2018).
Advances in the Suppression of Innate Immunity by FMDV VP3
Though VP3 is more conservative than VP1, it still plays an important role in the process of viral assembly and in suppressing the host innate immunity. For example, study has confirmed that arginine 56 in VP3 is relevant to FMDV virulence (Borca et al., 2012). Subsequent study found that VP3 also plays a vital role in escaping the host’s innate immune response, for example, FMDV VP3 can interact with janus kinase 1 (JAK1) and degrade it via the lysosomal pathway, thereby inhibiting IFN-γ-induced phosphorylation, signal transducer activator of transcription 1 (STAT1) dimerization and nuclear accumulation of phosphorylated STAT1, which ultimately leads to the inhibition of the type II IFN signaling pathway to evade the host innate immunity (Li et al., 2016b). Moreover, VP3 depends on its C-terminal (111–220 amino acids) to interact with virus-induced signaling adapter (VISA) and inhibits the expression of VISA by reducing VISA mRNA synthesis, finally inhibiting IFN-β production (Li et al., 2016c). Besides, FMDV VP0 protein interacts with Poly (rC) binding protein 2 (PCBP2) to degrade VISA via apoptotic pathway, thereby increasing FMDV replication (Li et al., 2019c). The latest study has found that the TANK-binding kinase 1 (TBK1) protein can interact with VP3 and degrade VP3 by its kinase and E3 ubiquitin ligase activity to resist FMDV infection (Li et al., 2019b). In addition, microRNA-1307 could enhance the host immune response by promoting VP3 degradation through the proteasome pathway in PK-15 cells to inhibit FMDV replication (Qi et al., 2019). Another structural protein, VP4, interacts with nucleoside diphosphate kinase 1 (NME1) to inhibit p53-induced activation of the IFN pathway (Feng et al., 2018).
The Role of FMDV Non-structural Proteins in the Regulation of Host Innate Immunity
Foot-and-mouth disease virus regions P2 and P3 encode partial precursors and eight mature non-structural proteins (Lpro, 2A, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol). Moreover, Lpro, 3Cpro, 2B, 2C, and 3A also regulate host antiviral innate immune responses.
Advances in the Suppression of Innate Immunity by FMDV Lpro
The RNA translation initiation site of FMDV has two AUGs separated by 84 nucleotides. As such, there are two different forms of Lpro after translation, namely, Lab and Lb. It is generally believed that Lb is more powerful and more effective than Lab (Grubman and Baxt, 2004; Liu et al., 2015). Lpro is related to viral virulence and is obtained by self-cleavage from the C-terminus of polypeptide chain (Gys-Gly site), and is the first mature viral protein after translation (Mason et al., 2003a). Study has shown that Lpro can specifically cleave eukaryotic initiation factor (eIF4G) to inhibit the translation of host cap-dependent mRNA, so FMDV can destroy synthesis of the host proteins, but it does not affect FMDV mRNA translation (Guan and Belsham, 2017). In addition, Lpro can also cleave NF-κB, which is related to IFN-α/β production, so as to facilitate viral replication (de Los Santos et al., 2006, 2007; Hato et al., 2010). Lpro can affect the translation and transcription of IFNs in various ways in order to interrupt antiviral activity mediated by IFN-α/β (de Los Santos et al., 2006). Lpro blocks nuclear translocation of the NF-κB subunit heterodimer p65/RelA to inhibit the IFN-β expression (de Los Santos et al., 2007). Lpro can also reduce the protein level of IRF3/7 through its catalytic activity to inhibit the transcription of IFN-β mRNA and MDA5-mediated type I IFN (Wang et al., 2010; Medina et al., 2018). Lpro also inhibits poly (I:C)-induced type III IFN (IFN-λ1) promoter activity through its enzyme activity and the conserved protein domain SAP region, thereby promoting the nuclear localization of Lpro, which is related to FMDV virulence and pathogenicity (Wang et al., 2011b; Segundo et al., 2012). As a novel type of viral DUB (deUbiquitinase), Lpro negatively regulates the type I IFN pathway by inhibiting the ubiquitination of innate immunity signaling molecules such as as retinoic acid-inducible gene I (RIG-I), tank-binding kinase 1 (TBK1), and TNF receptor-associated factor 3/6 (TRAF3/6) (Wang et al., 2011a). On the contrary, although Lpro also acts as a deISGylase, it is not important for inhibiting type I IFN, and this deISGylation activity is impaired after mutate Lpro W105 (the only conserved aromatic residue in all FMDV serotypes and can interact with ISG15), then leading to viral attenuation both in tissue culture and in vivo in mice (Medina et al., 2020). Recent study has shown that Lpro can inhibit the expression of IFNs and interferon-stimulated genes (ISGs) by interacting with activity dependent neuroprotective protein, a transcription factor that targets the IFN-α promoter region during early FMDV infection to increase viral replication (Medina et al., 2017); however, its mechanism still requires further study. Lpro can also target the innate immune molecule laboratory of genetics and physiology 2 (LGP2) to reduce the level of IFNs in infected cells, which then promotes the replication of FMDV (Rodriguez Pulido et al., 2018). In addition, Lpro cuts the SG scaffold proteins Ras GTPase SH3 domain binding protein 1 (G3BP1) and G3BP2 through its catalytic activity to inhibit SG formation, which may be relevant to the inhibition of the type I IFN response pathway (Visser et al., 2019). However, there are some issues about the mechanisms that are still unclear, such as the location of the cleavage site of G3BP1 and G3BP2 and the relationship between G3BP1/2 and Lpro inhibiting type I IFN signaling. In general, Lpro mainly regulates innate immunity via the aforementioned ways, so as to survive in the host and cause disease.
Advances in the Suppression of Innate Immunity by FMDV 3Cpro
3Cpro is a proteolytic enzyme that belongs to the chymotrypsin-like cysteine protease family and plays an important role in processing viral multiprotein precursors and in viral replication (Curry et al., 2007). 3Cpro is highly conservative and cleaves the viral polyprotein ten times (Mason et al., 2003b). Once the mutation occurs at 3Cpro, the original cleavage function may be inhibited, therefore, 3Cpro is thought to be a potential target for anti-FMDV drugs (Birtley et al., 2005; Curry et al., 2007). Furthermore, 3Cpro can also cleave many host proteins in infected cells to affect antiviral innate immunity. Early study has shown that 3Cpro is the sole viral protein that can cleave the 20 amino acids at the N-terminal of the histone H3. This cleavage may has an effect on the transcription level of cells infected with FMDV, as H3 is related to the regulatory domain of the transcriptional activity of eukaryotes, eventually almost completely destroying host cell functions (Falk et al., 1990). Based on earlier study, 3Cpro also has the ability to cleave eIF4G, although the cleavage site is different from the Lpro cleavage site, and the effect of 3Cpro is weaker than Lpro. Moreover, 3Cpro also cleaves eIF4A in the late stage of FMDV infection, thus inhibiting the synthesis of host cell proteins (Belsham et al., 2000). Moreover, 3Cpro can specifically cut NF-κB essential modulator (NEMO) at the Gln 383 site due to its proteolytic activity, which inhibits the activity of IRFs and NF-κB to abrogate NEMO-mediated type I IFN signaling and increase FMDV replication (Wang et al., 2012a). The 3Cpro proteolytic enzyme activity is essential to induce karyopherin α1 (KPNA1, a nuclear transport receptor of phosphorylated STAT1) degradation, and disrupt the nuclear transport of STAT1/STAT2 to block the JAK-STAT pathway, which then inhibits IFN signaling (Du et al., 2014). Autophagy contributes to the antiviral effect of type I and type II IFN, whereas the virus can evade innate immunity by manipulating the autophagy pathway (Richetta and Faure, 2013). Furthermore, 3Cpro can inhibit autophagy by degrading ATG5-ATG12, which could positively regulate type I IFN production, in order to enhance FMDV infection (Fan et al., 2017). Previous study has shown that 3Cpro cleaves the C-terminal nuclear nucleotide signal of src-associated protein in mitosis (Sam68), resulting in its redistribution in the cytoplasm, and then binds to the FMDV internal ribosome entry site (IRES), thereby affecting the life cycle of FMDV (Lawrence et al., 2012). Subsequent further study has found that 3Cpro can interact with Sam68 to regulate FMDV replication in vitro (Rai et al., 2015).
Advances in FMDV 2B Suppressing Innate Immunity
Studies have shown that the FMDV non-structural protein precursor 2BC can block endoplasmic reticulum-golgi apparatus transport, and 2B has been found to be closely associated with the endoplasmic reticulum and found to be involved in membrane rearrangement (Moffat et al., 2005, 2007). Further research has proven that 2B is an ion channel protein composed of 154 aa and crosses the endoplasmic reticulum membrane, whereas its C-terminus and N-terminus are exposed to the cytoplasm (Ao et al., 2015). In addition, 2B has the function of increasing the host cell membrane permeability and disrupting Ca2+ balance in the host cell, thereby inducing autophagy (Campanella et al., 2004; de Jong et al., 2008). Moreover, 2B amino acid residues (28–147) are related to FMDV replication (Ao et al., 2015; Gladue et al., 2018). During FMDV infection, 2B can regulate the LGP2 protein expression level to increase FMDV replication (Zhu et al., 2017). Moreover, 2B can decrease the expression levels of MDA5 and RIG-I followed by disrupting the phosphorylation of key factors TBK1 and IRF3, thereby inhibiting the IFN-β production in the RLRs antiviral signaling pathway (Zhu et al., 2016; Li et al., 2018). Recent study has demonstrated that 2B affects the NF-κB and IFN-β signaling pathway by downregulating nucleotide-binding oligomerization domain 2 (NOD2) expression to enhance FMDV replication (Liu et al., 2019). In addition, interaction of 2B with the host Cyclophilin A (CypA) reduced CypA-mediated degradation of Lpro to promote FMDV replication (Liu et al., 2018). Study has shown that CypA is involved in regulating the HCV-induced type I IFN pathway (Bobardt et al., 2013); however, whether CypA is involved in the FMDV-induced type I IFN pathway is not clear. Additionally, the transmembrane region of 2B promotes IL-1β production by activating NLRP3 inflammasomes to inhibit FMDV replication (Zhi et al., 2020).
Advances in the Suppression of Innate Immunity by FMDV 3A
Foot-and-mouth disease virus non-structural protein 3A is a conserved protein consisting of 153 amino acids and is much longer than other picornavirus 3A proteins (Medina et al., 2018). FMDV 3A plays a crucial role in viral replication, distinguishing the host range of infection, and virulence. For example, the deletion of 3A residues 87–106 reduces the replication and virulence of FMDV in cattle, but not in pigs (Pacheco et al., 2013). A study has shown that 3A (1–51 amino acids) can interact with RIG-I/MDA5/VISA and inhibit the TLR-mediated IFN-β signaling pathway by downregulating their mRNA expression level, thereby escaping the host innate immunity (Li et al., 2016a). Furthermore, the interaction between FMDV 3A and DEAD-box family protein (DDX56) inhibits type I IFN signaling pathway by reducing IRF3 phosphorylation to increase FMDV replication (Fu et al., 2019; Li et al., 2019a). In addition, recent study has shown that 3A can degrade G3BP1 by upregulating the expression of autophagy-related protein leucine rich repeat-containing 25 (LRRC25) to reduce IFN-β production, thereby facilitating the replication and growth of FMDV (Yang et al., 2020).
The Role of Other FMDV Proteins in the Regulation of Host Innate Immunity
Except for the aforementioned viral proteins that have been widely studied, there are other FMDV proteins involved in the process of regulating the host innate immune response. The non-structural FMDV protein 2C can interact with host protein Nmi (N-myc and STAT interactor), which may be involved in the FMDV 2C-induced apoptosis (Wang et al., 2012b). Further study has indicated that 2C and Nmi induce a type I IFN response, and expression of FMDV 2C or Nmi significantly suppresses VSV replication (Zheng et al., 2014). In addition, 3Dpol interacts with DEAD-box RNA helicase 1 (DDX1), which has been identified to inhibit FMDV replication due to its ATPase or helicase activity and induce the production of host IFN-β protein to enhance antiviral innate immunity (Xue et al., 2019). In addition, the non-coding regions (i.e., 5′Grubman and Baxt, 2004). For example, study has shown that the S fragment deletion mutants of the FMDV 5′UTR can increase mRNA levels of IFN-β and ISGs, and weaken FMDV virulence (Kloc et al., 2017). Both 3Dpol and ISES can also interact with Sam68 to regulate viral RNA translation (Rai et al., 2015). The host protein G3BP1 can directly interact with the IRES of FMDV 5′afterward, G3BP1 is cleaved by Lpro and 3Cpro, upon which both products, namely, Ct–G3BP1 and Nt–G3BP1, can inhibit FMDV cap– and IRES–dependent translation (Galan et al., 2017). In terms of its function in enhancing IRES activity and determining virulence, the FMDV 3′UTR can bind to S fragment and IRES, and form a 5′–3′ bridge (IRES–3′UTR or S–3′UTR) (Serrano et al., 2006; Gao et al., 2016).
Concluding Remarks and Further Perspectives
In this review, we mainly list the studies on the interaction of FMDV proteins with host proteins to regulate innate immunity. FMDV proteins can act on host proteins through a variety of ways to directly or indirectly block the innate immune signaling pathway in order to enhance its replication ability. For example, FMDV cleaves host proteins by its self-enzyme activity, and FMDV interacts with key factors in the innate immune pathway to increase its replication (Medina et al., 2018). These mechanisms have shown that FMDV disrupts the dynamic balance between virus and host, thereby providing the direction and theoretical basis for further FMDV research. Given that FMDV has a worldwide spread and seriously affects global economy and trade, FMD is listed as a class A infectious disease by the World Organization for Animal Health (OIE). Therefore, the aforementioned studies provide a solid theoretical foundation for FMDV vaccine development and technical guidance for FMD control (Aghaei et al., 2019; Dhanesh et al., 2020).
Due to the complexity of the FMDV structure and mutation-prone nature of the FMDV genome, the detailed mechanism of FMDV-mediated host’s innate immune response needs to be further investigated. Several preventive measures are not effective, leading to cross-infection of multiple serotypes, thereby making the diagnosis and prevention of FMDV more difficult. Consequently, there are still some questions that need to be explored: (1) Do the FMDV proteins have other sites to interact with host factors that suppress immune responses? (2) Are there interactions between FMDV proteins that promote FMDV replication? (3) What is the relationship between the inflammatory response and clinical symptoms caused by FMDV infection?
In summary, FMDV proteins interact with host proteins or cytokines to regulate the host’s innate and adaptive immune responses, which subsequently weakens the immunity of the animal and promotes FMDV replication as well as persistent infection. This review provided a theoretical basis for the development of FMDV vaccines and anti-FMDV drugs, as well as novel ideas regarding the diagnosis and control of FMD.
Author Contributions
JP and DL conceived and designed the study. JP, JY, WY, JR, YW, HZ, and DL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Gansu Provincial Science and Technology Department of China Grant 17JR5RA323.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the other members of the HZ lab for their constructive comments.
References
Abubakar, M., Manzoor, S., and Ahmed, A. (2018). Interplay of foot and mouth disease virus with cell-mediated and humoral immunity of host. Rev. Med. Virol. 28:e1966. doi: 10.1002/rmv.1966
Aghaei, A., Moghbeli, M., Kargar, M., Nazarian, S., and Kafilzadeh, F. (2019). Cloning and expression of a novel synthetic gene containing VP1 and 3A in Bacillus subtilis as a vaccine candidate against foot-and-mouth disease virus. Biologicals 60, 55–59. doi: 10.1016/j.biologicals.2019.05.003
Ao, D., Guo, H. C., Sun, S. Q., Sun, D. H., Fung, T. S., Wei, Y. Q., et al. (2015). Viroporin activity of the foot-and-mouth disease virus non-structural 2B protein. PLoS One 10:e0125828. doi: 10.1371/journal.pone.0125828
Belsham, G. J., McInerney, G. M., and Ross-Smith, N. (2000). Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J. Virol. 74, 272–280. doi: 10.1128/jvi.74.1.272-280.2000
Birtley, J. R., Knox, S. R., Jaulent, A. M., Brick, P., Leatherbarrow, R. J., and Curry, S. (2005). Crystal structure of foot-and-mouth disease virus 3C protease. new insights into catalytic mechanism and cleavage specificity. J. Biol. Chem. 280, 11520–11527. doi: 10.1074/jbc.m413254200
Bobardt, M., Hopkins, S., Baugh, J., Chatterji, U., Hernandez, F., Hiscott, J., et al. (2013). HCV NS5A and IRF9 compete for CypA binding. J. Hepatol. 58, 16–23. doi: 10.1016/j.jhep.2012.08.007
Borca, M. V., Pacheco, J. M., Holinka, L. G., Carrillo, C., Hartwig, E., Garriga, D., et al. (2012). Role of arginine-56 within the structural protein VP3 of foot-and-mouth disease virus (FMDV) O1 Campos in virus virulence. Virology 422, 37–45. doi: 10.1016/j.virol.2011.09.031
Campanella, M., de Jong, A. S., Lanke, K. W., Melchers, W. J., Willems, P. H., Pinton, P., et al. (2004). The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J. Biol. Chem. 279, 18440–18450. doi: 10.1074/jbc.m309494200
Curry, S., Roque-Rosell, N., Zunszain, P. A., and Leatherbarrow, R. J. (2007). Foot-and-mouth disease virus 3C protease: recent structural and functional insights into an antiviral target. Int. J. Biochem. Cell Biol. 39, 1–6. doi: 10.1016/j.biocel.2006.07.006
de Jong, A. S., de Mattia, F., Van Dommelen, M. M., Lanke, K., Melchers, W. J., Willems, P. H., et al. (2008). Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J. Virol. 82, 3782–3790. doi: 10.1128/jvi.02076-07
de Los Santos, T., de Avila Botton, S., Weiblen, R., and Grubman, M. J. (2006). The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 80, 1906–1914. doi: 10.1128/jvi.80.4.1906-1914.2006
de Los Santos, T., Diaz-San Segundo, F., and Grubman, M. J. (2007). Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J. Virol. 81, 12803–12815. doi: 10.1128/jvi.01467-07
Deretic, V., and Levine, B. (2018). Autophagy balances inflammation in innate immunity. Autophagy 14, 243–251. doi: 10.1080/15548627.2017.1402992
Dhanesh, V. V., Hosamani, M., Basagoudanavar, S. H., Saravanan, P., Biswal, J. K., Tamil Selvan, R. P., et al. (2020). Immunogenicity and protective efficacy of 3A truncated negative marker foot-and-mouth disease virus serotype A vaccine. Appl. Microbiol. Biotechnol. 104, 2589–2602. doi: 10.1007/s00253-020-10370-z
Du, Y., Bi, J., Liu, J., Liu, X., Wu, X., Jiang, P., et al. (2014). 3Cpro of foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 nuclear translocation. J. Virol. 88, 4908–4920. doi: 10.1128/jvi.03668-13
Falk, M. M., Grigera, P. R., Bergmann, I. E., Zibert, A., Multhaup, G., and Beck, E. (1990). Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J. Virol. 64, 748–756. doi: 10.1128/jvi.64.2.748-756.1990
Fan, X., Han, S., Yan, D., Gao, Y., Wei, Y., Liu, X., et al. (2017). Foot-and-mouth disease virus infection suppresses autophagy and NF-small ka, CyrillicB antiviral responses via degradation of ATG5-ATG12 by 3C(pro). Cell Death Dis. 8:e2561. doi: 10.1038/cddis.2016.489
Feng, H. H., Zhu, Z. X., Cao, W. J., Yang, F., Zhang, X. L., Du, X. L., et al. (2018). Foot-and-mouth disease virus induces lysosomal degradation of NME1 to impair p53-regulated interferon-inducible antiviral genes expression. Cell Death Dis. 9:885.
Fernandez-Sainz, I., Gavitt, T. D., Koster, M., Ramirez-Medina, E., Rodriguez, Y. Y., Wu, P., et al. (2019). The VP1 G-H loop hypervariable epitope contributes to protective immunity against foot and mouth disease virus in swine. Vaccine 37, 3435–3442. doi: 10.1016/j.vaccine.2019.05.019
Fu, S. Z., Yang, W. P., Ru, Y., Zhang, K. S., Wang, Y., Liu, X. T., et al. (2019). DDX56 cooperates with FMDV 3A to enhance FMDV replication by inhibiting the phosphorylation of IRF3. Cell Signal 64:109393. doi: 10.1016/j.cellsig.2019.109393
Galan, A., Lozano, G., Pineiro, D., and Martinez-Salas, E. (2017). G3BP1 interacts directly with the FMDV IRES and negatively regulates translation. FEBS J. 284, 3202–3217. doi: 10.1111/febs.14184
Gao, Y., Sun, S. Q., and Guo, H. C. (2016). Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol. J. 13:107.
Gladue, D. P., Largo, E., de la Arada, I., Aguilella, V. M., Alcaraz, A., Arrondo, J. L. R., et al. (2018). Molecular characterization of the viroporin function of foot-and-mouth disease virus nonstructural protein 2B. J. Virol. 92:e01360–18. doi: 10.1128/JVI.01360-18
Golde, W. T., Nfon, C. K., and Toka, F. N. (2008). Immune evasion during foot-and-mouth disease virus infection of swine. Immunol. Rev. 225, 85–95. doi: 10.1111/j.1600-065x.2008.00672.x
Guan, S. H., and Belsham, G. J. (2017). Separation of foot-and-mouth disease virus leader protein activities; identification of mutants that retain efficient self-processing activity but poorly induce eIF4G cleavage. J. Gen. Virol. 98, 671–680. doi: 10.1099/jgv.0.000747
Hato, S. V., Sorgeloos, F., Ricour, C., Zoll, J., Melchers, W. J., Michiels, T., et al. (2010). Differential IFN-alpha/beta production suppressing capacities of the leader proteins of mengovirus and foot-and-mouth disease virus. Cell Microbiol. 12, 310–317. doi: 10.1111/j.1462-5822.2009.01395.x
Kloc, A., Diaz-San Segundo, F., Schafer, E. A., Rai, D. K., Kenney, M., de Los Santos, T., et al. (2017). Foot-and-mouth disease virus 5′-terminal S fragment is required for replication and modulation of the innate immune response in host cells. Virology 512, 132–143. doi: 10.1016/j.virol.2017.08.036
Kumar, H., Kawai, T., and Akira, S. (2009). Pathogen recognition in the innate immune response. Biochem. J. 420, 1–16. doi: 10.1042/bj20090272
Kumar, H., Kawai, T., and Akira, S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. doi: 10.3109/08830185.2010.529976
Lawrence, P., Schafer, E. A., and Rieder, E. (2012). The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 425, 40–52. doi: 10.1016/j.virol.2011.12.019
Li, D., Fu, S., Wu, Z., Yang, W., Ru, Y., Shu, H., et al. (2019a). DDX56 inhibits type I interferon by disrupting assembly of IRF3-IPO5 to inhibit IRF3 nucleus import. J. Cell Sci. 133:jcs230409. doi: 10.1242/jcs.230409
Li, D., Yang, W., Ren, J., Ru, Y., Zhang, K., Fu, S., et al. (2019b). The e3 ubiquitin ligase TBK1 mediates the degradation of multiple picornavirus VP3 proteins by phosphorylation and ubiquitination. J. Virol. 93:e01438–19. doi: 10.1128/JVI.01438-19
Li, D., Zhang, J., Yang, W., He, Y., Ru, Y., Fu, S., et al. (2019c). Poly (rC) binding protein 2 interacts with VP0 and increases the replication of the foot-and-mouth disease virus. Cell Death Dis. 10:516.
Li, S. F., Gong, M. J., Xie, Y. L., Shao, J. J., Zhao, F. R., Zhang, Y. G., et al. (2019d). A novel type I interferon, interferon alphaomega, shows antiviral activity against foot-and-mouth disease virus in vitro. Microb. Pathog. 127, 79–84. doi: 10.1016/j.micpath.2018.11.040
Li, D., Lei, C., Xu, Z., Yang, F., Liu, H., Zhu, Z., et al. (2016a). Foot-and-mouth disease virus non-structural protein 3A inhibits the interferon-beta signaling pathway. Sci. Rep. 6:21888.
Li, D., Wei, J., Yang, F., Liu, H. N., Zhu, Z. X., Cao, W. J., et al. (2016b). Foot-and-mouth disease virus structural protein VP3 degrades Janus kinase 1 to inhibit IFN-gamma signal transduction pathways. Cell Cycle 15, 850–860. doi: 10.1080/15384101.2016.1151584
Li, D., Yang, W., Yang, F., Liu, H., Zhu, Z., Lian, K., et al. (2016c). The VP3 structural protein of foot-and-mouth disease virus inhibits the IFN-beta signaling pathway. FASEB J. 30, 1757–1766. doi: 10.1096/fj.15-281410
Li, M., Xin, T., Gao, X., Wu, J., Wang, X., Fang, L., et al. (2018). Foot-and-mouth disease virus non-structural protein 2B negatively regulates the RLR-mediated IFN-beta induction. Biochem. Biophys. Res. Commun. 504, 238–244. doi: 10.1016/j.bbrc.2018.08.161
Li, X., Wang, J., Liu, J., Li, Z., Wang, Y., Xue, Y., et al. (2013). Engagement of soluble resistance-related calcium binding protein (sorcin) with foot-and-mouth disease virus (FMDV) VP1 inhibits type I interferon response in cells. Vet. Microbiol. 166, 35–46. doi: 10.1016/j.vetmic.2013.04.028
Liu, C., Feng, H., Liu, Y., Chen, Y., Yang, S., Deng, R., et al. (2020). Soluble FMDV VP1 proteins fused with calreticulin expressed in Escherichia coli under the assist of trigger factor16 (Tf16) formed into high immunogenic polymers. Int. J. Biol. Macromol. 155, 1532–1540. doi: 10.1016/j.ijbiomac.2019.11.130
Liu, H., Xue, Q., Cao, W., Yang, F., Ma, L., Liu, W., et al. (2018). Foot-and-mouth disease virus nonstructural protein 2B interacts with cyclophilin a, modulating virus replication. FASEB J. 32, 6706–6723. doi: 10.1096/fj.201701351
Liu, Y., Zhu, Z., Zhang, M., and Zheng, H. (2015). Multifunctional roles of leader protein of foot-and-mouth disease viruses in suppressing host antiviral responses. Vet. Res. 46:127.
Liu, H., Zhu, Z., Xue, Q., Yang, F., Cao, W., Zhang, K., et al. (2019). Foot-and-mouth disease virus antagonizes NOD2-Mediated antiviral effects by inhibiting NOD2 protein expression. J. Virol. 93:e00124-19. doi: 10.1128/JVI.00124-19
Ma, X. X., Ma, L. N., Chang, Q. Y., Ma, P., Li, L. J., Wang, Y. Y., et al. (2018). Type I interferon induced and antagonized by foot-and-mouth disease virus. Front. Microbiol. 9:1862. doi: 10.3389/fmicb.2018.01862
Mason, P. W., Grubman, M. J., and Baxt, B. (2003a). Molecular basis of pathogenesis of FMDV. Virus Res. 91, 9–32. doi: 10.1016/s0168-1702(02)00257-5
Mason, P. W., Marvin, J., Grubman, and Baxt, B. (2003b). Molecular basis of pathogenesis of FMDV. Virus Res. 91, 9–32.
Medina, G. N., Knudsen, G. M., Greninger, A. L., Kloc, A., Diaz-San Segundo, F., Rieder, E., et al. (2017). Interaction between FMDV L(pro) and transcription factor ADNP is required for optimal viral replication. Virology 505, 12–22. doi: 10.1016/j.virol.2017.02.010
Medina, G. N., Segundo, F. D., Stenfeldt, C., Arzt, J., and de Los Santos, T. (2018). The different tactics of foot-and-mouth disease virus to evade innate immunity. Front. Microbiol. 9:2644. doi: 10.3389/fmicb.2018.02644
Medina, G. N., Azzinaro, P., Ramirez-Medina, E., Gutkoska, J., Fang, Y., Diaz-San Segundo, F., et al. (2020). Impairment of the DeISGylation activity of foot-and-mouth disease virus lpro causes attenuation in vitro and in vivo. J. Virol. 94:e00341-20. doi: 10.1128/JVI.00341-20
Moffat, K., Howell, G., Knox, C., Belsham, G. J., Monaghan, P., Ryan, M. D., et al. (2005). Effects of foot-and-mouth disease virus nonstructural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J. Virol. 79, 4382–4395. doi: 10.1128/jvi.79.7.4382-4395.2005
Moffat, K., Knox, C., Howell, G., Clark, S. J., Yang, H., Belsham, G. J., et al. (2007). Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J. Virol. 81, 1129–1139. doi: 10.1128/jvi.00393-06
Pacheco, J. M., Gladue, D. P., Holinka, L. G., Arzt, J., Bishop, E., Smoliga, G., et al. (2013). A partial deletion in non-structural protein 3A can attenuate foot-and-mouth disease virus in cattle. Virology 446, 260–267. doi: 10.1016/j.virol.2013.08.003
Parida, S., Oh, Y., Reid, S. M., Cox, S. J., Statham, R. J., Mahapatra, M., et al. (2006). Interferon-gamma production in vitro from whole blood of foot-and-mouth disease virus (FMDV) vaccinated and infected cattle after incubation with inactivated FMDV. Vaccine 24, 964–969. doi: 10.1016/j.vaccine.2005.08.108
Qi, L., Wang, K., Chen, H., Liu, X., Lv, J., Hou, S., et al. (2019). Host microRNA miR-1307 suppresses foot-and-mouth disease virus replication by promoting VP3 degradation and enhancing innate immune response. Virology 535, 162–170. doi: 10.1016/j.virol.2019.07.009
Rai, D. K., Lawrence, P., Kloc, A., Schafer, E., and Rieder, E. (2015). Analysis of the interaction between host factor Sam68 and viral elements during foot-and-mouth disease virus infections. Virol. J. 12:224.
Randall, R. E., and Goodbourn, S. (2008). Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47. doi: 10.1099/vir.0.83391-0
Richetta, C., and Faure, M. (2013). Autophagy in antiviral innate immunity. Cell Microbiol. 15, 368–376. doi: 10.1111/cmi.12043
Rodriguez Pulido, M., and Saiz, M. (2017). Molecular mechanisms of foot-and-mouth disease virus targeting the host antiviral response. Front. Cell Infect. Microbiol. 7:252. doi: 10.3389/fcimb.2017.00252
Rodriguez Pulido, M., Sanchez-Aparicio, M. T., Martinez-Salas, E., Garcia-Sastre, A., Sobrino, F., and Saiz, M. (2018). Innate immune sensor LGP2 is cleaved by the Leader protease of foot-and-mouth disease virus. PLoS Pathog 14:e1007135. doi: 10.1371/journal.ppat.1007135
Saiz, M., Nunez, J. I., Jimenez-Clavero, M. A., Baranowski, E., and Sobrino, F. (2002). Foot-and-mouth disease virus: biology and prospects for disease control. Microbes Infect. 4, 1183–1192. doi: 10.1016/s1286-4579(02)01644-1
Sangare, O., Bastos, A. D., Marquardt, O., Venter, E. H., Vosloo, W., and Thomson, G. R. (2001). Molecular epidemiology of serotype O foot-and-mouth disease virus with emphasis on West and South Africa. Virus Genes 22, 345–351.
Segundo, F. D., Weiss, M., Perez-Martin, E., Dias, C. C., Grubman, M. J., and Santos Tde, L. (2012). Inoculation of swine with foot-and-mouth disease SAP-mutant virus induces early protection against disease. J. Virol. 86, 1316–1327. doi: 10.1128/jvi.05941-11
Serrano, P., Pulido, M. R., Saiz, M., and Martinez-Salas, E. (2006). The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 87, 3013–3022. doi: 10.1099/vir.0.82059-0
Stetson, D. B., and Medzhitov, R. (2006). Type I interferons in host defense. Immunity 25, 373–381. doi: 10.1016/j.immuni.2006.08.007
Sun, P., Zhang, S., Qin, X., Chang, X., Cui, X., Li, H., et al. (2018). Foot-and-mouth disease virus capsid protein VP2 activates the cellular EIF2S1-ATF4 pathway and induces autophagy via HSPB1. Autophagy 14, 336–346. doi: 10.1080/15548627.2017.1405187
Thomson, G. R., Vosloo, W., and Bastos, A. D. (2003). Foot and mouth disease in wildlife. Virus Res. 91, 145–161. doi: 10.1016/s0168-1702(02)00263-0
Visser, L. J., Medina, G. N., Rabouw, H. H., de Groot, R. J., Langereis, M. A., de Los Santos, T., et al. (2019). Foot-and-Mouth disease virus leader protease cleaves G3BP1 and G3BP2 and inhibits stress granule formation. J. Virol. 93:e00922–18. doi: 10.1128/JVI.00922-18
Wang, D., Fang, L., Li, K., Zhong, H., Fan, J., Ouyang, C., et al. (2012a). Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 86, 9311–9322. doi: 10.1128/jvi.00722-12
Wang, J., Wang, Y., Liu, J., Ding, L., Zhang, Q., Li, X., et al. (2012b). A critical role of N-myc and STAT interactor (Nmi) in foot-and-mouth disease virus (FMDV) 2C-induced apoptosis. Virus Res. 170, 59–65. doi: 10.1016/j.virusres.2012.08.018
Wang, D., Fang, L., Li, P., Sun, L., Fan, J., Zhang, Q., et al. (2011a). The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 85, 3758–3766. doi: 10.1128/jvi.02589-10
Wang, D., Fang, L., Liu, L., Zhong, H., Chen, Q., Luo, R., et al. (2011b). Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-lambda1 pathway. Mol. Immunol. 49, 407–412. doi: 10.1016/j.molimm.2011.09.009
Wang, D., Fang, L., Luo, R., Ye, R., Fang, Y., Xie, L., et al. (2010). Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem. Biophys. Res. Commun. 399, 72–78. doi: 10.1016/j.bbrc.2010.07.044
Xue, Q., Liu, H., Zeng, Q., Zheng, H., Xue, Q., and Cai, X. (2019). The DEAD-Box RNA helicase DDX1 interacts with the viral protein 3D and inhibits foot-and-mouth disease virus replication. Virol. Sin. 34, 610–617. doi: 10.1007/s12250-019-00148-7
Yang, W., Li, D., Ru, Y., Bai, J., Ren, J., Zhang, J., et al. (2020). Foot-and-mouth disease virus 3A protein causes upregulation of autophagy-related protein LRRC25 To inhibit the G3BP1-mediated rig-like helicase-signaling pathway. J. Virol. 94:e02086–19. doi: 10.1128/JVI.02086-19
Ye, L., Schnepf, D., and Staeheli, P. (2019). Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 19, 614–625. doi: 10.1038/s41577-019-0182-z
Yoneyama, M., and Fujita, T. (2010). Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20, 4–22. doi: 10.1002/rmv.633
Zhang, W., Yang, F., Zhu, Z., Yang, Y., Wang, Z., Cao, W., et al. (2019). Cellular DNAJA3, a novel VP1-Interacting protein, inhibits foot-and-mouth disease virus replication by inducing lysosomal degradation of VP1 and attenuating its antagonistic role in the beta interferon signaling pathway. J. Virol. 93:e00588-19. doi: 10.1128/JVI.00588-19
Zheng, W., Li, X., Wang, J., Li, X., Cao, H., Wang, Y., et al. (2014). A critical role of interferon-induced protein IFP35 in the type I interferon response in cells induced by foot-and-mouth disease virus (FMDV) protein 2C. Arch. Virol. 159, 2925–2935. doi: 10.1007/s00705-014-2147-7
Zhi, X., Zhang, Y., Sun, S., Zhang, Z., Dong, H., Luo, X., et al. (2020). NLRP3 inflammasome activation by Foot-and-mouth disease virus infection mainly induced by viral RNA and non-structural protein 2B. RNA Biol. 17, 335–349. doi: 10.1080/15476286.2019.1700058
Zhong, B., Wang, Y. Y., and Shu, H. B. (2010). Regulation of virus-triggered type I interferon signaling by cellular and viral proteins. Front. Biol. (Beijing) 5:12–31. doi: 10.1007/s11515-010-0013-x
Zhu, Z., Li, C., Du, X., Wang, G., Cao, W., Yang, F., et al. (2017). Foot-and-mouth disease virus infection inhibits LGP2 protein expression to exaggerate inflammatory response and promote viral replication. Cell Death Dis. 8:e2747. doi: 10.1038/cddis.2017.170
Zhu, Z., Wang, G., Yang, F., Cao, W., Mao, R., Du, X., et al. (2016). Foot-and-Mouth Disease Virus Viroporin 2B Antagonizes RIG-I-Mediated Antiviral Effects by Inhibition of Its Protein Expression. J. Virol. 90, 11106–11121. doi: 10.1128/jvi.01310-16
Keywords: innate immunity, interferon, immunosuppression, foot-and-mouth disease virus, virus-host interactions
Citation: Peng J, Yi J, Yang W, Ren J, Wen Y, Zheng H and Li D (2020) Advances in Foot-and-Mouth Disease Virus Proteins Regulating Host Innate Immunity. Front. Microbiol. 11:2046. doi: 10.3389/fmicb.2020.02046
Received: 12 May 2020; Accepted: 04 August 2020;
Published: 09 October 2020.
Edited by:
Douglas Paul Gladue, United States Department of Agriculture, United StatesReviewed by:
Fayna Diaz San Segundo, United States Department of Agriculture, United StatesTakashi Irie, Hiroshima University, Japan
Copyright © 2020 Peng, Yi, Yang, Ren, Wen, Zheng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haixue Zheng, aGFpeHVlemhlbmdAMTYzLmNvbQ==; Dan Li, bGlkYW5AY2Fhcy5jbg==
 Jiangling Peng
Jiangling Peng