
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 18 August 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.02045
This article is part of the Research Topic Bioremediation of Chemical Pesticides Polluted Soil View all 8 articles
Acephate is an organophosphate pesticide that has been widely used to control insect pests in agricultural fields for decades. However, its use has been partially restricted in many countries due to its toxic intermediate product methamidophos. Long term exposure to acephate and methamidophos in non-target organisms results in severe poisonous effects, which has raised public concern and demand for the removal of these pollutants from the environment. In this paper, the toxicological effects of acephate and/or methamidophos on aquatic and land animals, including humans are reviewed, as these effects promote the necessity of removing acephate from the environment. Physicochemical degradation mechanisms of acephate and/or methamidophos are explored and explained, such as photo-Fenton, ultraviolet/titanium dioxide (UV/TiO2) photocatalysis, and ultrasonic ozonation. Compared with physicochemical methods, the microbial degradation of acephate and methamidophos is emerging as an eco-friendly method that can be used for large-scale treatment. In recent years, microorganisms capable of degrading methamidophos or acephate have been isolated, including Hyphomicrobium sp., Penicillium oxalicum, Luteibacter jiangsuensis, Pseudomonas aeruginosa, and Bacillus subtilis. Enzymes related to acephate and/or methamidophos biodegradation include phosphotriesterase, paraoxonase 1, and carboxylesterase. Furthermore, several genes encoding organophosphorus degrading enzymes have been identified, such as opd, mpd, and ophc2. However, few reviews have focused on the biochemical pathways and molecular mechanisms of acephate and methamidophos. In this review, the mechanisms and degradation pathways of acephate and methamidophos are summarized in order to provide a new way of thinking for the study of the degradation of acephate and methamidophos.
Organophosphate compounds (OPs) are one of the most widely used pesticides because of their broad spectrum, specificity, and high efficiency toward insects and pests (Mulla et al., 2020). Acephate and methamidophos are two of the most common and efficient OPs that are used for pest control in agriculture (Maia et al., 2011; Kumar et al., 2015; Pan et al., 2015).
Acephate [O, S-dimethyl-acetyl-phosphoramidothioate, molecular weight (MW) = 183.17] (Figure 1) is a systemic insecticide that effectively controls various pests on ornamental plants, cotton, beans, and head lettuce as well as parasites on mammalians. It is a good substitute because it is less toxic than methamidophos (Mahajna et al., 1997). Acephate is a class II “moderately hazardous” pesticide, but methamidophos is classified as a class IV “highly toxic” pesticide (World Health Organization [WHO], 2009). Acephate is highly water-soluble and can easily contaminate groundwater and soil, which is also easily absorbed by plants and accumulated in edible parts of plants (Mohapatra et al., 2011; Syed et al., 2014).
Methamidophos (O, S-dimethyl phosphoramidothioate, MW = 141.12) (Figure 1) is also a systemic insecticide effective against chewing and sucking insects and is one of the intermediate products of acephate (Farag et al., 2012; Araoud et al., 2016). Methamidophos mainly leads to enzyme inactivation by phosphorylation of the serine residues of acetylcholinesterase (AChE) and butyrylcholinesterase (BuChE) active site (Lugokenski et al., 2012). It exerts high toxicity not only through the persistent inhibition of AChE but also through the complex blocking action on neuronal nicotinic acetylcholine receptors (nAChRs), which is different from the inhibition of AChE (Di et al., 2004). In addition, methamidophos can inhibit the activity of carboxylase in cells (Mahajna et al., 1997). Because of its high efficiency, methamidophos was extensively used in large amounts in many parts of the world before 2013. Methamidophos has been restricted in China since 2013 but is still extensively used in many other developing countries (Araoud et al., 2016).
Acephate and methamidophos are used in enormous quantities because of their high efficiency in agriculture and other industrial purposes, which leads them to remain in various environments for many years (Konstantinou et al., 2006; Lugokenski et al., 2012; Dar et al., 2020). Acephate and methamidophos have both hydrophilic and hydrophobic properties that assist them easy movement between water and soil (He et al., 2017). Studies have shown that methamidophos residues may decrease the total microbial biomass carbon and fungal biomass (Li et al., 2008; Wang et al., 2008). In recent years, acephate and methamidophos have been detected in a variety of vegetables and fruits (Mohapatra et al., 2011; Syed et al., 2014; Andrade et al., 2015). At present, acephate and methamidophos have become a focus of public concern due to the harmful effects of pesticide exposure on invertebrates, mammals, and humans (Yao et al., 2018). The human body has limited metabolic capacity for acephate and methamidophos, and it may results in hyperglycemia, lipid metabolism disorder, DNA damage, increased oxidative stress, and risk of carcinogenesis after long-term exposure (Chang et al., 2009).
In recent years, many researchers have studied the different degradation processes for the removal of acephate and methamidophos from polluted environments. photo-Fenton processes, UV/TiO2, ultrasonic ozonation, and ionizing irradiation, etc. are widely studied for acephate or methamidophos (Dai et al., 2008; Wang et al., 2015; Yang et al., 2015; Zheng et al., 2016). Efficient and environmentally friendly microbial degradation has been considered as the most promising remediation method (Cycoń and Piotrowska-Seget, 2016; Bhatt et al., 2019; Pang et al., 2020; Feng et al., 2020; Zhang et al., 2020). Several degrading microorganisms with partial or complete degradation capacity of acephate or methamidophos have been isolated and characterized, such as Lysinibacillus fusiformis, Pseudomonas sp., Pseudomonas pseudo alcaligenes, Bacillus subtilis, Pseudomonas azotoformans, and Pseudomonas putida (Lin et al., 2016; Maddela and Venkateswarlu, 2017; Singh et al., 2017, 2020).
Furthermore, there are many studies on the degradation pathways of acephate and methamidophos as well as studies on related functional genes and enzymes (Li et al., 2007; Shen et al., 2010; Chino-Flores et al., 2012; Kumar et al., 2018). Microorganisms metabolize pesticides through their respective degradation pathways, and the metabolic capacity of microorganisms can be improved by recombination technology (Pinjari et al., 2013; Arora et al., 2017; Cycoń et al., 2017; Zhang et al., 2018; Lin et al., 2020). Microbial degradation of acephate and methamidophos is a potential tool for large-scale pollutant removal. Thus, biodegradation of acephate and methamidophos also should be concerned. We emphasize the role of physicochemical degradation and biodegradation for large-scale treatment of acephate and/or methamidophos contamination. However, few reviews have focused on the degradation mechanisms and biochemical pathways of acephate and methamidophos (Maqbool et al., 2016; Kumar et al., 2018; Jiang et al., 2019). We emphasize the role of biodegradation for large-scale treatment of acephate and/or methamidophos contamination.
This paper reviews (I) the toxicity of acephate and methamidophos, and their removal; (II) the different oxidation or reduction methods of physicochemical degradation; (III) the different types of natural degradation strains, or genetically engineered microorganisms; (IV) the organophosphorus degrading enzymes and their encoding genes.
The extensive use and long term exposure to acephate and methamidophos results in the cumulative effect of these compounds release into the environment, which consequently results in the direct or indirect poisoning of non-target organisms. We summarize the toxic effects of acephate and methamidophos on aquatic and land animals, including humans in Table 1.
Zebrafish is a commonly used model test organism due to its advantages of high genetic and organ system homology to humans, external fertilization, high fecundity, and transparency in early adulthood (Liu et al., 2018). It has reported that methamidophos exposure may affect neurodevelopmental genes and activate intracellular apoptosis, leading to early developmental neurodamage in zebrafish (He et al., 2017). The brain may be an important target of methamidophos toxicity in zebrafish, revealing the potential neurotoxicity of methamidophos to other aquatic species and humans (Rodríguez et al., 2012; Peng et al., 2015; He et al., 2017). Zebrafish embryo development retardation, larval deformities, and decreased chorionic surface tension were also induced by exposure to acephate (Liu et al., 2018). Since acephate and methamidophos are prone to accumulate in water, large quantities of residual pesticides are frequently exposed to and ingested by aquatic organisms. Water is the source of life, and pesticide residues in water can also pass to other living beings through the food chain.
Drosophila is also a model test organism that has amazing similarities in gene structure and function with higher vertebrates. Acephate contamination at higher concentrations than 5 g/mL can cause DNA damage and mortality in the fruit fly, and it may disrupt the balance of oxidase and antioxidant enzymes, such as catalase (CAT), glutathione-S-transferase (GST), cytochrome P450 (CYP450), AChE, and superoxide dismutase (SOD) (Rajak et al., 2017). Chronic toxicity of acephate to honeybees is characterized by weight loss and esterase inhibition (Yao et al., 2018). Earthworms exposed to soil containing acephate also experience oxidative stress, characterized by lipid peroxidation, protein oxidation, DNA damage, and changes in antioxidant enzyme status (Phugare et al., 2012). Acephate not only significantly reduces the antioxidant capacity of bird liver and kidney but also increases lipid peroxidation, interleukin and tumor necrosis factor in both organs and also affects the immune response (Tripathi et al., 2012; Farag et al., 2017). Methamidophos had a toxic effect on the number of viable counts, morphology, and histological changes of corpus luteal cells and progesterone production in bovines (Afzal et al., 2011).
Overuse of acephate and methamidophos can lead to an increase in high blood sugar, impaired metabolism, DNA damage, reproductive barriers, and cancer in rats (Maia et al., 2011; Araoud et al., 2016; Ribeiro et al., 2016). Acephate alters glucose metabolism in pregnant and lactating rats and predisposes their offspring to type 2 diabetes in adulthood (Ribeiro et al., 2016). Exposure to acephate may lead to metabolic disorders in mice, whereas changes in some endogenous metabolites lead to kidney damage and disrupt normal metabolic processes in rats, including glucose, nucleic acid, and protein metabolism (Hao et al., 2012). Methamidophos can cause lesions in the testis and epididymis, characterized by obstruction of spermatogenesis in the testis and severe edema in the epididymis, respectively (Farag et al., 2012). Methamidophos can cause DNA damage at different stages of spermatogenesis and reduce sperm quality in mice through acrosomal response and fertilization ability assessment (Urióstegui-Acosta et al., 2014). In addition, methamidophos also decreases the expression of zonula occludens protein 2 (ZO-2) in sperm cells of spermatic tubules, induces phosphorylation of ZO-2, and occludens in the testes. It also reduces the interactions between these proteins assessed by immunoprecipitation, which leads to reproductive toxicity in male mice (Ortega-Olvera et al., 2018).
The human body can be very vulnerable when exposed to methamidophos or acephate. Acephate could be a genotoxin, which would make it a severe threat to human health. It can cause chromosomal changes and DNA damage in human lymphocytes (Özkan et al., 2009). Moreover, it is suggested that acephate also exhibits cytotoxic and genotoxic effects on human sperm by disrupting sperm motility, cell membrane integrity, and sperm volume (Dhanushka and Peiris, 2017). In addition, methamidophos increases the generation of oxidative stress in human peripheral blood mononuclear cells (Ramirez-Vargas et al., 2017). The accumulation caused by the widespread use of acephate and methamidophos has led to toxic effects among many biological systems and has caused many health problems. Growing concern about the accumulation of pesticide residues in our food, soil, and wastewater has led to a great deal of interest in their removal.
Physical adsorption and chemical degradation methods were first proposed to remove acephate and methamidophos from the environment. The advantages of physical adsorption and chemical degradation methods lie in their broad spectrum of pollutants, their adaptability to the environment, and their effectiveness. Advanced oxidation processes (AOPs) are a more efficient composite technology that combines the advantages of different physicochemical degradation methods to more efficiently remove methamidophos and acephate; examples of these methods include photo-Fenton process, UV/TiO2, and ultrasonic ozonation (Dai et al., 2008; Wang et al., 2015; Zheng et al., 2016).
Physicochemical methods play an important role in the degradation of acephate. In the absence of surfactants, colloidal manganese dioxide has been shown to degrade acephate by oxidation (Qamruzzaman, and Nasar, 2014). Ultrasonic degradation is a physical degradation method with simple operation and few byproducts, whose main mechanisms are mechanical bond breaking and free radical reaction. When the ultrasonic power is high enough, the liquid produces instantaneous negative pressure, local high temperature, and a high-pressure environment, which finally cause water molecules to break up and become strong oxidants, such as hydrogen peroxide radicals (•OOH) and hydroxyl radicals (•OH) (Shriwas and Gogate, 2011; Golash and Gogate, 2012). Ultrasound and ozonization have synergistic effects in complex systems. The degradation efficiency of acephate ozonization can be increased from 60.6 to 87.6% in 60 min with 160 kHz ultrasound irradiation because it can improve the oxidizability of nitrogen atoms (Wang et al., 2015). Under visible light irradiation, nanocomposite Co3O4/McM-41 can completely remove acephate in 40 min (50 mg/L) or 60 min (100 mg/L) (AbuKhadra et al., 2020). Under the same conditions of Co3O4/McM-41, ZnFe2O4–TiO2 can remove 89.5% of acephate within 120 min, and the optimal value of the photocatalyst and H2O2 dosage of this degradation is 2.0 g/L and 8 mmol/L, respectively (Fu et al., 2012). TiO2 is often used as a catalyst due to its advantages of low cost, good chemical stability, and high catalytic activity. Compared to Sr/TiO2–PCFM, which is not modified, the modified Sr/TiO2–PCFM shows preferential degradation of acephate and can degrade target pollutants more quickly and efficiently (Liu et al., 2019). Furthermore, photocatalyst Fe3O4@SiO2@mTiO2 has a larger surface area and a stronger magnetic response, which can effectively degrade acephate, omethoate, and other organophosphorus pesticides (Zheng et al., 2016). The photocatalytic degradation rate of acephate (675 g.a.i./h) was shown to be 93.5% in a field trial after adding cerium-doped nano titanium dioxide (TiO2/Ce) (2400 g.a.i./h) for 20 h (Liu et al., 2015). TiO2 is a potential and efficient chemical catalyst for the removal of environmental pesticide residues, which deserves further study. In addition, a combination of multiple degradation methods may improve the degradation efficiency.
The degradation of acephate mainly involves the destruction of N-C, P-N, P-S, or P-O bonds and the generation of the main intermediate products O-methl-N-acetylphosphoramidate and the more toxic methamidophos (as shown in Figure 2). Previous researchers have used 60Co irradiation, which has led to many studies on the degradation efficiency, degradation pathways, and degradation dynamics (Sun et al., 2015; Yang et al., 2015; Huang et al., 2018). Chemical degradation mainly results in the generation of free radicals in the medium, such as and negative hydrogen ions (•H) and hydroxyl radicals (•OH) (Yang et al., 2015). Reducing and oxidizing free radicals play different roles in the degradation of acephate and methamidophos, so negative hydrogen ions (•H) and hydroxyl radicals (•OH) have different reaction kinetics from acephate, which leads to two different degradation pathways of acephate (Zhao et al., 2009). Negative hydrogen ions (•H) degrade acephate more quickly but produce fewer inorganic ions, while hydroxyl radicals (•OH) catalyze over more steps to produce more inorganic ions. As nucleophiles, negative hydrogen ions (•H) may first attack the P = O and C = O bonds, causing the electrons to delocalize and form temporary conjugated systems in the O-P-N-C-O bonds. When the P-N bond is disconnected, the acephate is subsequently formed with ammonium ions () and formic acid. At the same time, the C-S and C-O bonds of the product methamidophos are broken successively, resulting in phosphoric acid, sulfate radical, and a methyl group. In contrast, hydroxyl radicals (•OH) are strong electrophilic reagents that first attack the negatively charged S, O, and N atoms of acephate. As a result, the fracture of the P-N bond produces acetamide and phosphoric acid. The break between the P-N bond and the P-S bond produces acetaminophosphoric acid, which then produces acetamide, phosphoric acid, or ethanedioic acid. Whether in the presence of negative hydrogen ions (•H) or hydroxyl radicals (•OH), acephate will eventually be completely degraded to inorganic salts.
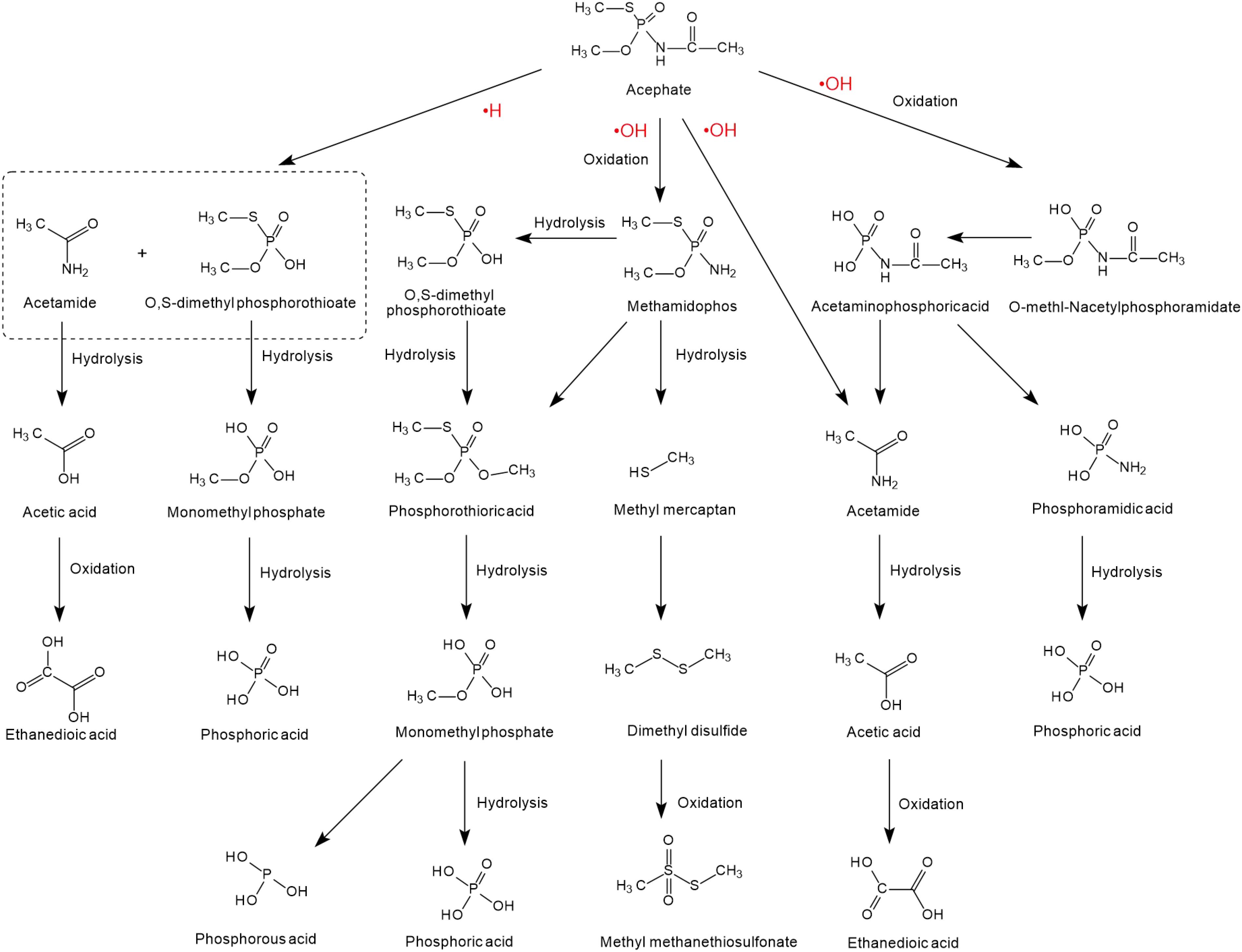
Figure 2. The degradation pathways of acephate reacted with •H and •OH (based on Liu et al., 2015; Wang et al., 2015; Huang et al., 2018; AbuKhadra et al., 2020).
Many physicochemical degradation methods have been developed for the degradation of methamidophos, which is very important for the removal of pesticide residues in the environment. Reductive hydrogen ions radicals (•H) can catalyze the degradation of acephate, but they cannot catalyze the degradation of methamidophos (Zhao et al., 2009). In contrast, oxidizing hydroxyl radicals (•OH) can catalyze not only the degradation of acephate but also the degradation of methamidophos (Zhao et al., 2009). Gamma irradiation can effectively degrade methamidophos into inorganic ions in aqueous solution by promoting the generation of hydroxyl radicals (•OH) in water (Zhao et al., 2009). Combined use of multiple degradation methods is a trend to improve degradation efficiency. The catalysis of methamidophos by UV and TiO2 has been studied. Under UV irradiation, nano TiO2 catalyzed 95% degradation of methamidophos within 4 h (Dai et al., 2008). In another study, the best degradation rate of methamidophos in water using nano TiO2 in the first 5 min reached 83.55%, which was significantly faster than the degradation reported by other studies (Zhang et al., 2006). It was found that the degradation efficiency increased with the increasing temperature and pH of the medium when the optimal amount of TiO2 was 12 g/L (Liu et al., 2009). It was also proved that the microwave degradation method is an effective method to remove methamidophos. In the presence of K2S2O8, over 96% of methamidophos can be degraded by microwave irradiation within 6 min, while the MW/K2S2O8 corresponding kinetic equations, half-life (t1/2) and the pseudofirst-order rate constants are lnC = −0.757t + 5.9, R2 = 0.9776; 0.916 min and 0.757/min, respectively (Zhang et al., 2009). Even in acidic environments, the electrolysis of the Pb/PbO2 electrode can better facilitate the removal of methamidophos (Martiìnez-Huitle et al., 2008). Electrode materials play a key role in electrolysis and influence the type of byproducts; thus, subsequent research on electrode materials is of great significance for electrochemical degradation. In addition, an adsorption material with good removal performance of methamidophos was obtained through modifying the structure and morphology of natural zeolite with 25 mmol/L hexadecyl-trimethyl-ammonium bromide (HDTMA) (Alvarez-García et al., 2020). Adsorption is considered to be a low-cost and simple water separation process, but it still needs to improve its material structure to enhance its adsorption capacity.
The possible photodegradation pathways of methamidophos catalyzed by TiO2 is shown in Figure 3 and first involves the cracking of P-S, P-N, and P-O bonds. When the P-S and P-O bonds of methamidophos are broken, P atoms react with methyl radicals to form [amino (methylsulfanyl) phosphoryl] methane and [amino (methoxy) phosphoryl] methane, which are further oxidized to P-formylphosphonamiddothioate and phosphine carboxylic acid, respectively. When the P-N bond of methamidophos is broken, an O, S-dimethyl phosphonothioate dimer is generated, which is more prone to further degradation. The intermediate products 1-aminoethanol and trimethoxymethane form the products acetamide and dimethoxymethanol, respectively, resulting in complete mineralization. Phosphoric acid is the main final product.
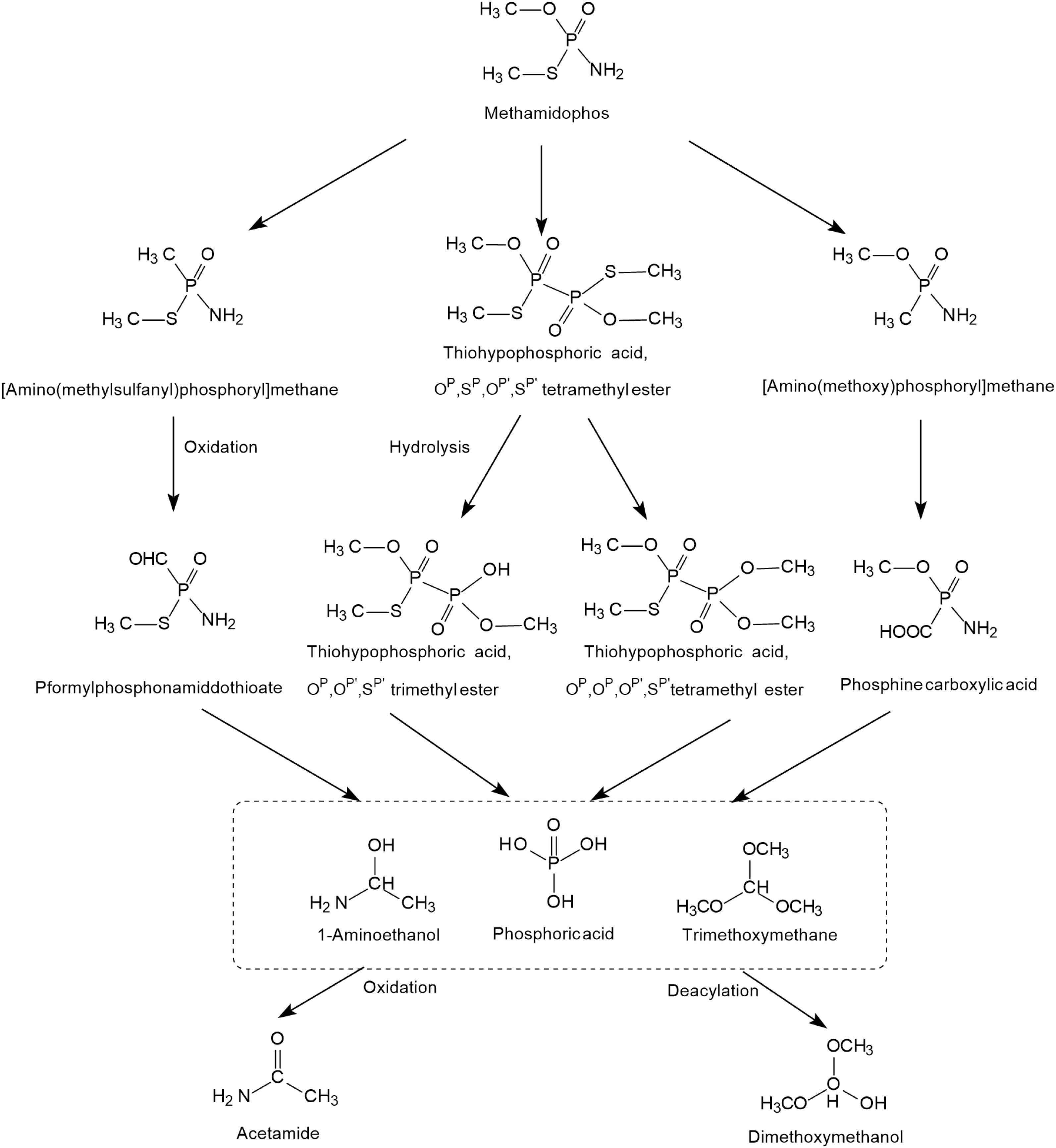
Figure 3. The photodegradation pathways of methamidophos catalyzed by TiO2 (based on Dai et al., 2008; Martiìnez-Huitle et al., 2008).
There is growing interest in microbiological degradation technologies because of the risk to life of pesticide residues, which provide cheap and efficient acephate and methamidophos detoxification to complement expensive chemical methods. Microbial degradation is a potential way to decontaminate pesticide-contaminated sites (Chen et al., 2015; Yang et al., 2018; Zhan et al., 2018; Huang et al., 2019; Bhatt et al., 2020a). Biodegradation microorganisms including bacteria, fungi, actinomycetes, yeasts, and algae can be obtained by enrichment culture and recombination technology (Pinjari et al., 2013; Chen et al., 2014; Birolli et al., 2019; Bhatt et al., 2020b). At present, many researchers are looking for effective acephate or methamidophos degrading microorganisms through enrichment culture, including sewage treatment systems, organophosphorus contaminated areas, industries, and agricultural fields (Chen et al., 2012; Gao et al., 2012; Li et al., 2014; Mohan and Naveena, 2015; Lin et al., 2016). To date, only a small number of microorganisms have been isolated and identified that can fully mineralize or degrade acephate or methamidophos, as shown in Table 2.
Acephate is degraded in two main ways, including through methamidophos or O-methyl-N-acetylphosphoramidate. Pseudomonas sp. Ind01 was able to degrade acephate by promoting the first step of acephate mineralization but could not utilize the methamidophos, which is involved in the process of O-methyl-N-acetylphosphoramidate (Pinjari et al., 2012). Pseudomonas pseudoalcaligenes PS-5 also completely transforms acephate into O-methyl-N-acetylphosphoramidate, even in the presence of heavy metal ions Cu2+, Fe3+, and humic acid (Singh et al., 2017). Pseudomonas aeruginosa Is-6, a biodegradable strain isolated from soil, can use acephate as a sole source of carbon, phosphorus, and energy and showed 92% degradation of acephate (1000 mg/L) in 7 days. This strain not only degrades acephate but is also able to efficiently degrade other pesticides like dimethoate, parathion, methyl parathion, chlorpyrifos, and malathion. Thus, P. aeruginosa strain Is-6 shows high degradation ability in contaminated soil, indicating its potential in environmental remediation (Ramu and Seetharaman, 2014). Mohan and Naveena (2015) isolated five bacterial strains, which are identified as Bacillus cereus ADI-10, Nibacillus fusiformis ADI-01, Pseudomonas pseudoalcaligenes ADI-03, Pseudomonas sp. ADI-04, and Pseudomonas pseudoalcaligenes ADI-06. These strains could efficiently grow and degrade acephate at 500 mg/L without any additional carbon source and were further utilized to analyze the mechanism of acephate degradation. In comparison to single bacterial cultures, the use of a combined culture of multiple strains in bioremediation was found to be more effective in terms of better metabolic and pollutant removal capabilities (Bhatt et al., 2020c; Li et al., 2020; Zhan et al., 2020). Phugare et al. (2012) reported the degradation of acephate by a bacterial consortium composed of Exiguobacterium sp. BCH4 and Rhodococcus sp. BCH2, which was capable of degrading 75.85% of 50 mg/L acephate. Microbial fixation is beneficial to the improvement of microbial utilization and degradation efficiency (Zang et al., 2013). In another study, recombinant Escherichia coli containing organophosphorus hydrolase (OPH) encoding plasmid was immobilized at a low temperature in polyvinyl alcohol (PVA) cryogel to form a biocatalyst with high activity, stability, and mechanical strength (Hong and Raushel, 1999). Fungi can be potential degrading microorganisms due to their extensive mycelium networks and low specificity of degrading enzymes, but only Mucor sp. has been found to be able to degrade methamidophos and acephate (Zang et al., 2013; Maqbool et al., 2016). However, many degrading microorganisms have not yet been tested in the field, which is much needed for social progress.
As shown in Figure 4, there is a diversity of acephate degradation pathways in microorganisms. Under the catalysis of carboxylesterase enzymes, acephate first releases the acetate residue to form the main product methamidophos (Pinjari et al., 2012). The product methamidophos is then hydrolyzed to produce methyl dihydrogen phosphate, S-methyl dihydrogen thiophosphate, or O-methylphosphoramidate with the catalysis of phosphotriesterase (PTE), and eventually phosphoric acid is generated. In the process of biodegradation, PTE plays a primary role in catalyzing the first step of acephate degradation. The breaking of the P-S and P-O bonds of acephate and its intermediates is mainly dependent on the hydrolysis of PTE (Chae et al., 1994; Lai et al., 1995). With the catalysis of PTE, acephate is hydrolyzed to O-methyl-N-acetylphosphoramidate or S-methyl phosphoramidate. The amino group is hydrolyzed by the phosphoamide hydrolase enzyme and releases ions (Ramu and Seetharaman, 2014). S-methyl phosphoramidate is eventually catalyzed to produce phosphorous acid, phosphenic amides, or phosphorous acid of lower molecular mass.
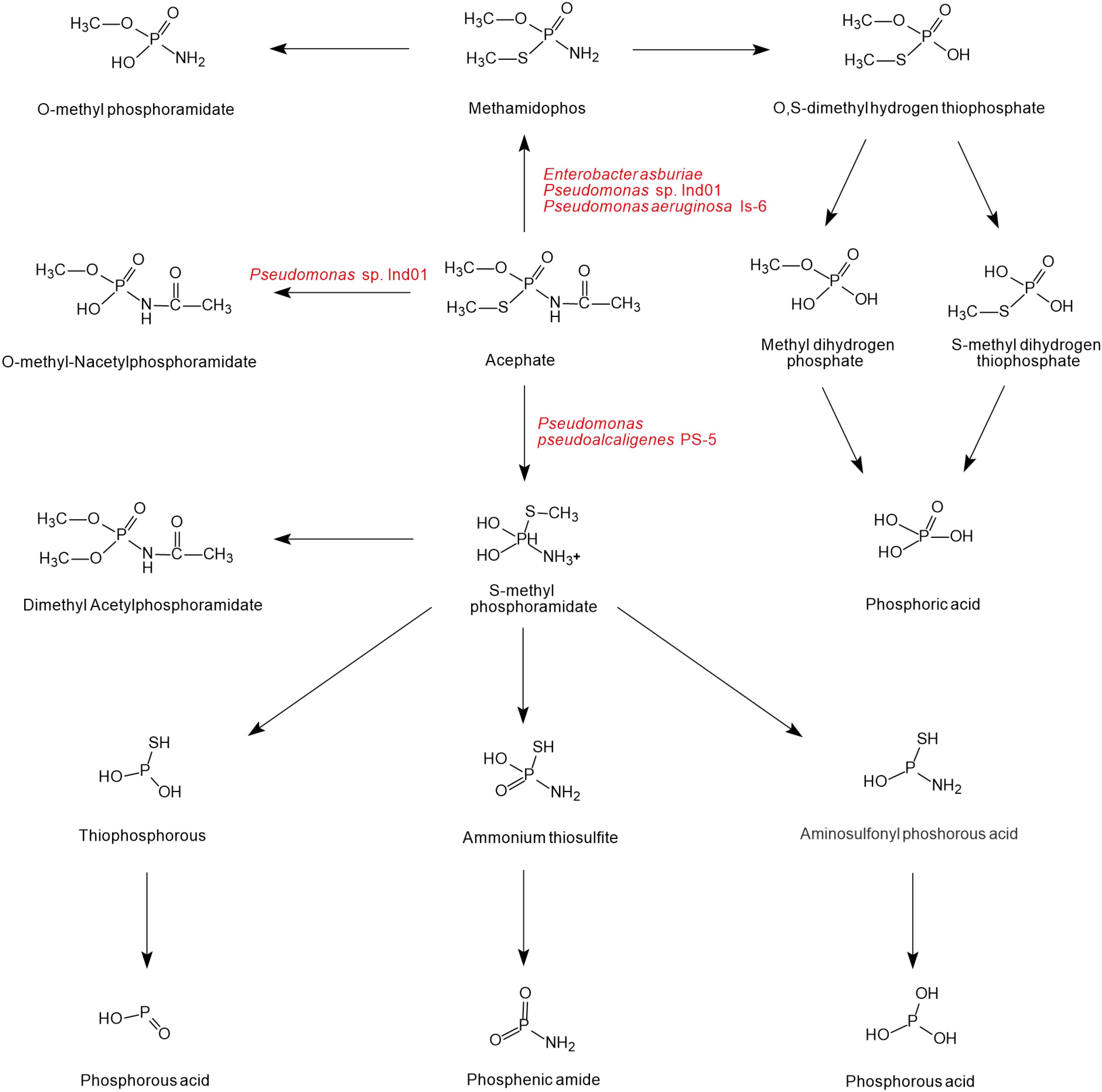
Figure 4. The microbial degradation pathways of acephate (based on Phugare et al., 2012; Pinjari et al., 2012; Ramu and Seetharaman, 2014; Singh et al., 2017).
Hyphomicrobium sp. MAP-1 can be grown using methamidophos as its sole source of carbon, nitrogen, and phosphorus, which can completely degrade 3000 mg/L of methamidophos within 84 h under optimum conditions (pH 7.0 and temperature 30°C) (Wang et al., 2010). Penicillium oxalicum ZHJ6 can degrade 99.9% of 1000 mg/L methamidophos within 12 days by co-metabolizing ethanol, glucose, fructose, sucrose, lactose, starch, and dextrin as carbon sources (Zhao et al., 2010). Acinetobacter sp., Pseudomonas stutzeri, and Pseudomonas aeruginosa can degrade 80% of 500 mg/L of methamidophos in 3 days under optimal conditions (pH 7.0 and temperature 30–35°C), which can not only use glucose, fructose, ethanol, and gal as carbon sources and energy but can also degrade various organophosphorus (Li et al., 2014). Isolated Luteibacter jiangsuensis sp. nov. can effectively use methamidophos as the sole carbon source (Wang et al., 2011). The intracellular degrading enzymes of methamidophos have been preliminarily discovered, and the optimum conditions have been explored using the response surface method (Wang et al., 2012; Zhang et al., 2013). Interestingly, some bacteria can degrade a series of organophosphorus pesticides including methamidophos, such as Aspergillus sp., Enterobacter ludwigi M2, and Brevundimonas faecalis MA-B12 (Zhao et al., 2014; Adelowo et al., 2015; Jiang et al., 2019). In particular, Sphingobium sp. K22212 and Sphingobium sp. Cam5-1 could only degrade organophosphorus insecticides with P-S bonds (Ahn et al., 2018). Few microorganisms can degrade acephate or methamidophos, or even completely mineralize them. Some bacteria only have partial degradation abilities, in which case, the combination of multiple strains can have a better degradation effect. Therefore, more strains capable of adapting to the environment and of high degradation need to be discovered.
As shown in Figure 5, only one pathway of methamidophos degradation has been identified in microorganisms. Like acephate degradation, PTE and phosphoamide hydrolase are involved in the intracellular degradation. The P-N bond of methamidophos is first hydrolyzed by phosphoamide hydrolase. With the hydrolysis of PTE, the products O, S-dimethyl hydrogen thiophosphate generate phosphoric acid, methyl mercaptan, and methanol with the breaking of P-O and P-S bonds. In particular, methyl mercaptan in methamidophos degradation bacteria generates further dimethyl sulfide and dimethyl disulfide, which are not found in the acephate biodegradation pathway.
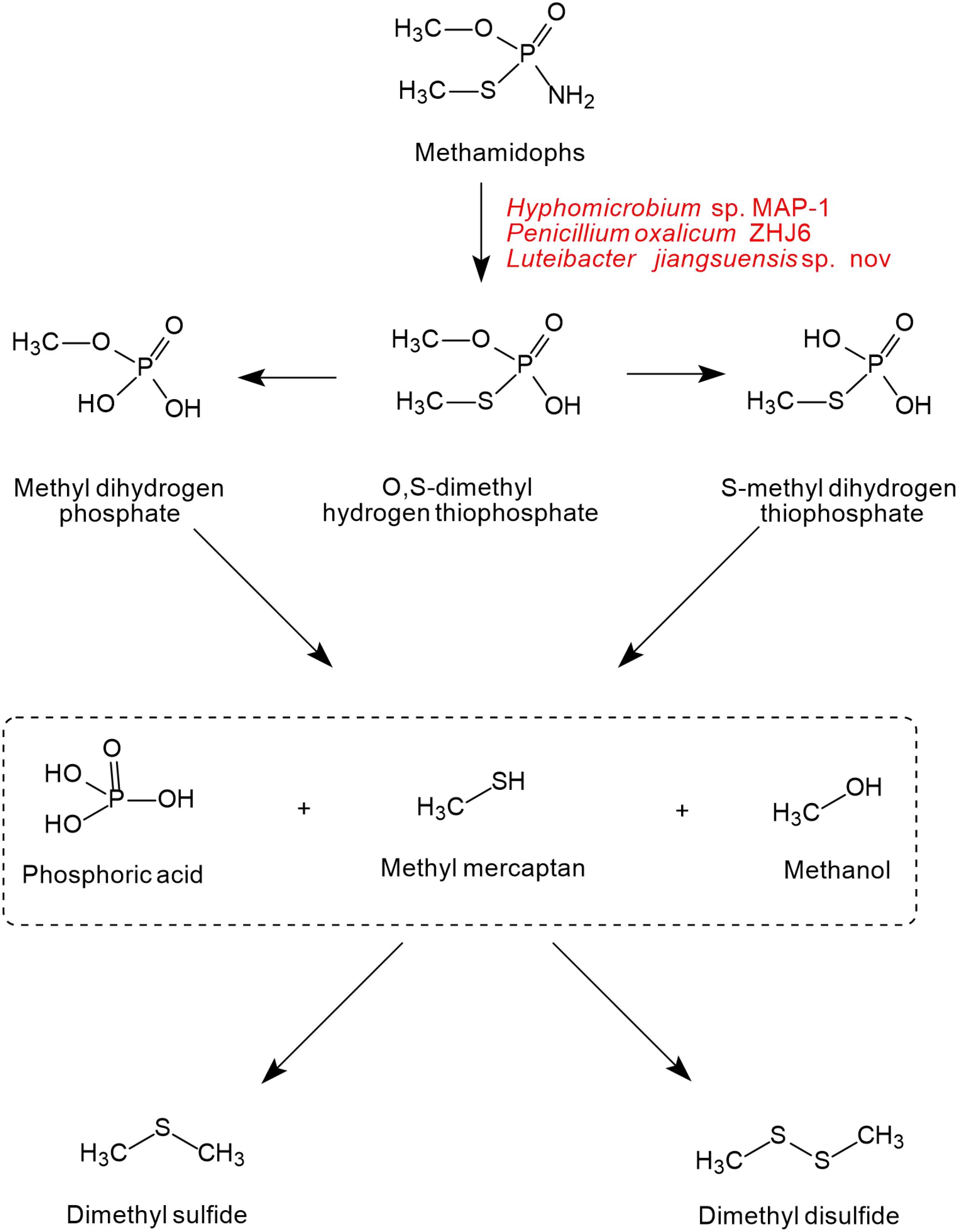
Figure 5. The microbial degradation pathway of methamidophos (based on Wang et al., 2010).
Acephate and methamidophos are highly toxic organophosphate pesticides for long term use, and their microbial degradation (including molecular and genetic mechanism) has attracted extensive attention due to its ecofriendly effectiveness (Scott et al., 2008). In addition, many biodegradable bacteria are effective in laboratory liquid media, but have poor degradation ability in field soil (Chen et al., 2013; Huang et al., 2020; Mishra et al., 2020). Therefore, more powerful enzymatic and genetic engineering techniques need to be applied. In the past, many researchers have focused on extracting PTE from microbial cells, most of which come from microorganisms and a few from other animals (Liu et al., 2017).
Microorganisms play an important role in promoting the degradation of these organophosphorus pesticides, and PTE (EC 3.1.8.1) is an effective tool to degrade organophosphorus pesticides. PTE has the stereoselectivity to degrade organophosphorus pesticides and tends to degrade (Sp)-isomers of acephate and methamidophos (Chae et al., 1994; Hong and Raushel, 1999). Most studies of bacterial PTE have shown extensive pH activity, and PTE purified from two bacterial strains Pseudomonas aeruginosa F10B and Clavibacter michiganensis subsp. insidiosum SBL11 reached peak activity at the pH value of 9.0 (Das and Singh, 2006). Another form of PTE was isolated from Pseudomonas monteilii, and phosphotriestercoroxon was the only phosphorus source (Horne et al., 2002). PTE isolated from the soil bacterium Pseudomonas diminuta significantly promoted the hydrolysis of organophosphate triester bonds (Ghanem and Raushel, 2005). Phosphotriesterase OPHC2, isolated from Pseudomonas pseudoalcaligenes and belonging to the metallo-β-lactamase superfamily, had unusual heat resistance and some OP degradation ability (Gotthard et al., 2013).
The production of degrading enzymes is responsible for the resistance in many animals, and the enzymes or genes involved in organophosphorus degradation and resistance are summarized in Table 3. The evolutionary relationships among the functional enzymes involved in the degradation of organophosphate pesticides are shown in Figure 6. In addition to phosphotriesterase, other enzymes that detoxify organophosphorus pesticides are also being isolated, such as paraoxonase 1 (PON1), parathion hydrolase, and somanase (Kumar et al., 2018). As an A-esterase, PON1 can detoxify organophosphate pesticides in the phase-I metabolism of the liver. The PON1-L55M and PON1-Q192R genotypes, the two most important coding region polymorphisms of PON1, are related to the occurrence of Parkinson’s disease, Alzheimer’s disease, and amyotrophic lateral sclerosis (Androutsopoulos et al., 2011). Through quantitative polymerase chain reaction (qPCR) analysis, acetylcholinesterase 2 (AChE2) and esterase 1 (EST1) were found to be metabolic enzymes involved in the process of allogenic biological detoxification of organophosphate pesticides; increased synthesis of these enzymes can distinguish between organophosphorus-resistant and -susceptible populations (Brito et al., 2017). Acetylcholinesterase (AChE, 3.1.1.7), isolated from Schizaphis graminum (Rondani), is tolerant to organophosphate pesticides, and it can significantly improve the resistance ability of Schizaphis graminum (Gao and Zhu, 2002). A high biotechnology potential phosphotriesterase-like lactonase (PLL) has been isolated, encoded by Vmut 2255 in the hyperthermoacidophilic crenarchaeon Vulcanisaeta moutnovskia (VmutPLL) (Kallnik et al., 2014). A Phe362Tyr mutation in the AChE gene of Lepeophtheirus salmonis protects from pesticide toxicity (Helgesen et al., 2019). Cattle ticks, also known as Boophilus microplus, generally have resistance to the main organophosphate pesticides. Research that isolated acetyl cholinesterase cDNA from Boophilus microplus indicates that it can encode a 62 kDa protein. After translation and modification, acetylcholinesterase can develop resistance to organophosphate pesticides (Baxter and Barker, 1998). Rumen ecosystems are made up of a variety of microorganisms and protozoa that produce enzymes that help to detoxify toxic compounds in the rumen, such as organophosphate pesticides. The cow rumen bacterial esterase gene (est5S) is 1098 bp long, encodes a protein with 366 amino acid residues, and has a molecular weight of 40 kDa. The enzyme has great potential in cleaning up contaminated pesticides (Kambiranda et al., 2009). Enzymes isolated from insects can also be used for the biodegradation of organophosphate pesticides.
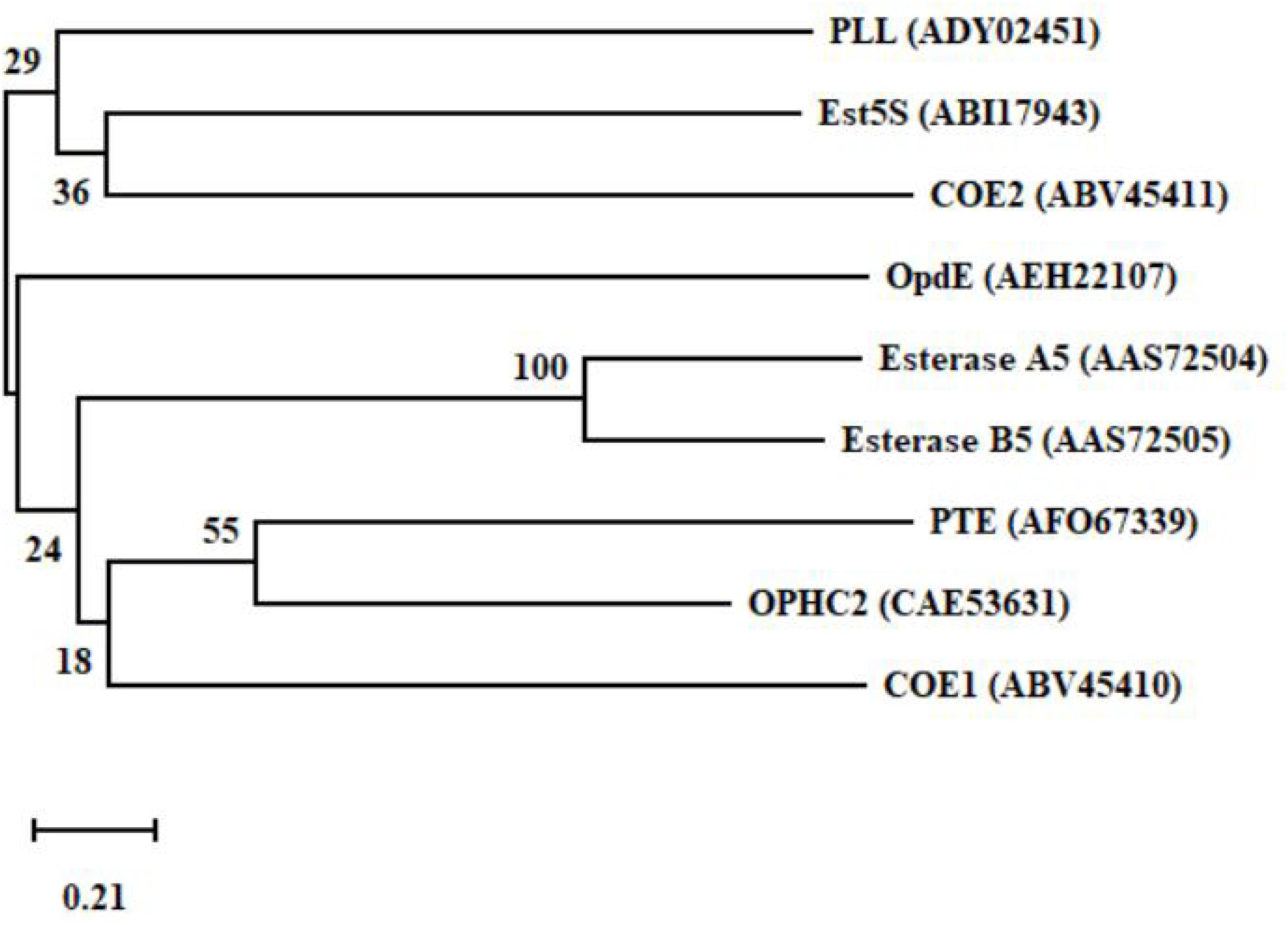
Figure 6. A phylogenetic tree of the key functional enzymes involved in the degradation of organophosphate pesticides. PTE was isolated from Pseudomonas monteilii (Horne et al., 2002). OPHC2 was isolated from Pseudomonas pseudoalcaligenes (Gotthard et al., 2013). PLL was isolated from Vulcanisaeta moutnovskia (Kallnik et al., 2014). Est5S was isolated from cow rumen bacteria (Kambiranda et al., 2009). OpdE was isolated from Enterobacter sp. (Chino-Flores et al., 2012). COE1 was isolated from Bemisia tabaci (Alon et al., 2008). COE2 was isolated from Bemisia tabaci (Alon et al., 2008). Esterase A5 was isolated from Culex pipiens (Buss and Callaghan, 2004). Esterase B5 was isolated from Culex pipiens (Buss and Callaghan, 2004).
Enzymes that are fixed can generally be used to improve efficiency. In the presence of appropriate immobilization, the degradation effect of the degrading enzyme is slightly better than that of the free enzyme, with a higher reuse rate and easier preservation (Jesionowski et al., 2014; Doraiswamy et al., 2019). After a series of tests, it was determined that Wax Encapsulated Organophosphate Hydrolase (WEOH) can be retained for 1 year under normal temperature, with an activity reduction of 10% (Muthyala and Gundala, 2019). More methods to improve the degradation efficiency and reuse efficiency of enzymes need to be developed.
Biodegradability is determined by genes, and genetic engineering technology may be used to carry out gene recombination and to enhance biodegradability. Therefore, it is very important to extract resistance genes. To date, many genes encoding organophosphorus degrading enzymes have been found, which will be of great support for the promotion of organophosphorus biodegradation, such as opd, mpd, and ophc2.
The gene opd is a widely studied gene. The opdE gene has been isolated from Enterobacter sp. cons002, which has a wide range of degrading activities against organophosphorus pesticides (Chino-Flores et al., 2012). The gene opdE consists of 753 bp and encodes a protein of 25 kDa (Chino-Flores et al., 2012). The sequential mutation and screening of OPH encoding genes in specific codons by saturation mutagenesis can improve the degradation ability of OPH mutated enzymes on the P-S bonds of organophosphorus pesticides (Schofield and DiNovo, 2010). Through genetic engineering technology, the opd gene cloned from Brevundimonas mututa was isogenously expressed in the acephate-mineralizing strain Pseudomonas sp. Ind01, suggesting that the engineered strain could degrade a variety of OP insecticides (Pinjari et al., 2013).
The mpd and ophc2 genes are also very important in degradation. The gene mpd has a low similarity with the opd gene, and the mpd gene encoded enzyme can degrade organophosphorus pesticides such as parathion and fenitrothion (Cui et al., 2001). The mpd gene was first cloned from Plesiomonas sp. strain M6 and then cloned from Sphingomonas sp. strain dsp-2 (Cui et al., 2001; Li et al., 2007). The gene ophc2 has a size of 975 bp, and its homology is less than 50% compared with other organophosphorus hydrolase genes in GenBank (Wu et al., 2004). The gene ophc2 was successfully heterogeneously expressed in Pichia pastoris and Escherichia coli by gene cloning (Chu et al., 2006; Shen et al., 2010). The emergence of metagenomics technology will rapidly promote the discovery of degrading microorganisms at the gene level and gradually replace the arduous enrichment and culture method (Jeffries et al., 2018). With the increase of people’s knowledge of the application of enzymes, more and more new biocatalysts with functional properties have been studied.
Due to the frequent use of acephate/methamidophos in agriculture over a long period of time, insect resistance to insecticides is on the rise. There are two molecular mechanisms for Plutella xylostella resistance, including the reduction in target sensitivity and the increase in detoxification. The acephate resistance of resistant varieties of diamondback moth Plutella xylostella was 47 times higher than that of sensitive varieties because of the metabolic detoxification mediated by GST and sensitivity reduction of AChEs (Sonoda and Igaki, 2010). Further research found that A298S and G324A mutations in AChE1 of Plutella xylostella also reduced the sensitivity to methamidophos (Sonoda and Igaki, 2010). In addition, CYP450 can directly detoxify OP compounds by catalyzing oxidation/reduction reactions, and esterase can hydrolyze OP pesticides. In a laboratory study, the increase of esterase activity was an important reason for acephate resistance enhancement in Indian brown planthoppers (Malathi et al., 2017).
There is a significant correlation between mutation frequency and resistance level in natural populations. The first two carboxylesterase genes (coe1 and coe2) were isolated from Bemisia tabaci, in which coe1 was overexpressed (about 4-fold) in the organophosphate-resistant strain. Sufficient data and biochemical evidence support that increased coe1 and coe2 activity is involved in Bemisia tabaci resistance to organophosphate insecticides (Alon et al., 2008). The resistance of organophosphate in biotype B is closely related to the point mutation in ace1 and ace2 acetylcholinesterase. By comparison of nucleic acids and derived amino acid sequences, there are only silent nucleotide polymorphisms in the ace2 strain and only one mutation in the ace1 strain, namely, Phe392Trp (Phe331 in Torpedo californica) (Alon et al., 2008). Under pressure from organophosphate pesticides, G137D mutations in the carboxylesterase E3 gene in the New World screwworm are associated with a general form of resistance to acetylcholinesterase through the metabolic detoxification of pesticides (Da Silva et al., 2011). Carboxylesterase gene amplification can increase the organophosphate resistance of Culex pipiens. The amplified carboxylesterase gene of Culex pipiens resistance was originally collected in Cyprus. Genomic DNA fragments containing two loci encoding the carboxylesterase alleles A5 and B5 have been cloned and sequenced; the gene spacer is 3.7 Kb in length in the A5-B5 amplituter (Buss and Callaghan, 2004). In the case of continuous use of organophosphate pesticides, some organisms will become more and more resistant to them.
In addition, insect resistance can also be induced under artificial conditions. One researcher screened 25 generations of mutant Laodelphax striatellus to obtain insect cross-resistance to acephate, deltamethrin, imidacloprid, methomyl, carbosulfan, and diazinon (Xu et al., 2014). In addition, the cross resistance of Laodelphax striatellus may also be related to the participation of CYP450s and esterases, and the overexpression of CYP6AY3v2, CYP306A2v2, CYP353D1v2, and LSCE36 genes (Xu et al., 2014). Insecticide-detoxifying carboxylesterase (CE) was shown to metabolize acephate and is controlled by the gene CpCE-1 of wild Cydia pomonella (CP) (Yang et al., 2014). Earthworms promote the detoxification of methamidophos mainly through their own biosorption and syntropy, especially the biodegradation of enzymes released in the soil (Zhou et al., 2008). The emergence of insect resistance is the result of organisms gradually adapting to acephate and methamidophos, which is beneficial to the extraction of resistance genes and the screening of new biodegradation agents.
In modern agricultural production, acephate and methamidophos play important roles in controlling pests and increasing the crop yield. However, improper use of acephate and methamidophos can lead to critical environmental pollution and life-threatening health problems due to the high toxicity of methamidophos to non-target organisms. Many physicochemical methods have been developed and applied for the remediation of acephate and methamidophos, but they are thought to be too expensive to use on a large scale. Therefore, microbial degradation is considered to be a better and effective degradation method for acephate and methamidophos. Microorganisms capable of metabolizing acephate and methamidophos have been isolated, but further studies are needed, such as the continued development and utilization of degrading enzymes, the development of engineered degrading microorganisms, and overcoming environmental complexity. Microbial degradation kinetics, degradation pathways, and related enzymes and functional genes need to be further studied, as this is conducive to the in situ repair of methamidophos and methamidophos. Therefore, in order to understand the genetic analysis of acephate and its related catabolic genes, we need to use advanced molecular technologies such as metagenomics, proteomics, and transcriptome analysis to reveal the missing links and evolutionary mechanism and metabolic pathway involved in the process of bio-degradation. In the future, metagenomics will serve as a useful tool to predict microbial degradation in polluted habitats and may facilitate the practical application of acephate and methamidophos degrading microorganisms from various contaminated sites.
SC conceived of the presented idea. ZL contributed to the writing and prepared the figures and tables. SP, WZ, SM, PB, and SC participated in revising the manuscript. All the authors approved it for publication.
We acknowledge the grants from the Key-Area Research and Development Program of Guangdong Province (2018B020206001), the National Natural Science Foundation of China (31401763), and the Guangdong Special Branch Plan for Young Talent with Scientific and Technological Innovation (2017TQ04N026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AbuKhadra, M. R., Mohamed, A. S., El-Sherbeeny, A. M., and Elmeligy, M. A. (2020). Enhanced photocatalytic degradation of acephate pesticide over MCM-41/Co3O4 nanocomposite synthesized from rice husk silica gel and Peach leaves. J. Hazard. Mater. 389:122129. doi: 10.1016/j.jhazmat.2020.122129
Adelowo, F. E., Amuda, O. S., Giwa, A. A., Onawumi, O., and Andfalana, O. O. E. O. (2015). Biodegradation of organophosphonates by Aspergillus specie. Orient. J. Chem. 31, 165–171. doi: 10.13005/ojc/31
Afzal, G. S., Rizvi, F., Zargham, K. M., and Khan, A. (2011). Toxic effects of cypermethrin and methamidophos on bovine corpus luteal cells and progesterone production. Exp. Toxicol. Pathol. 63, 131–135. doi: 10.1016/j.etp.2009.10.007
Ahn, J., Lee, S., Kim, S., You, J., Han, B., Weon, H., et al. (2018). Biodegradation of organophosphorus insecticides with P-S bonds by two Sphingobium sp. strains. Int. Biodeter. Biodegr. 132, 59–65. doi: 10.1016/j.ibiod.2018.05.006
Alon, M., Alon, F., Nauen, R., and Morin, S. (2008). Organophosphates’ resistance in the B-biotype of Bemisia tabaci (Hemiptera: Aleyrodidae) is associated with a point mutation in an ace1-type acetylcholinesterase and overexpression of carboxylesterase. Insect Biochem. Mol. 38, 940–949. doi: 10.1016/j.ibmb.2008.07.007
Alvarez-García, S., Ramírez-García, J. J., Granados-Correa, F., Granados-Correa, F., and Sánchez-Meza, J. C. (2020). Structural and textural influences of surfactant-modified zeolitic materials over the methamidophos adsorption behavior. Sci. Technol. 55, 619–634. doi: 10.1080/01496395.2019.1568476
Andrade, G. C. R. M., Monteiro, S. H., Francisco, J. G., Figueiredo, L. A., Botelho, R. G., and Tornisielo, V. L. (2015). Liquid chromatography-electrospray ionization tandem mass spectrometry and dynamic multiple reaction monitoring method for determining multiple pesticide residues in tomato. Food Chem. 175, 57–65. doi: 10.1016/j.foodchem.2014.11.105
Androutsopoulos, V. P., Kanavouras, K., and Tsatsakis, A. M. (2011). Role of paraoxonase 1 (PON1) in organophosphate metabolism: implications in neurodegenerative diseases. Toxicol. Appl. Pharm. 256, 418–424. doi: 10.1016/j.taap.2011.08.009
Araoud, M., Neffeti, F., Douki, W., Khaled, L., Najjar, M. F., Kenani, A., et al. (2016). Toxic effects of methamidophos on paraoxonase 1 activity and on rat kidney and liver and ameliorating effects of alpha-tocopherol. Environ. Toxicol. 31, 842–854. doi: 10.1002/tox.22095
Arora, P. K., Srivastava, A., Garg, S. K., and Singh, V. P. (2017). Recent advances in degradation of chloronitrophenols. Bioresour. Technol. 250, 902–909. doi: 10.1016/j.biortech.2017.12.007
Baxter, G. D., and Barker, S. C. (1998). Acetylcholinesterase cDNA of the cattle tick, Boophilus microplus: characterisation and role in organophosphate resistance. Insect Biochem. Mol. 28, 581–589. doi: 10.1016/S0965-1748(98)00034-34
Bhatt, P., Bhatt, K., Huang, Y., Lin, Z., and Chen, S. (2020a). Esterase is a powerful tool for the biodegradation of pyrethroid insecticides. Chemosphere 244:125507. doi: 10.1016/j.chemosphere.2019.125507
Bhatt, P., Huang, Y., Rene, E. R., Kumar, A. J., and Chen, S. (2020b). Mechanism of allethrin biodegradation by a newly isolated Sphingomonas trueperi strain CW3 from wastewater sludge. Bioresour. Technol. 305:123074. doi: 10.1016/j.biortech.2020.123074
Bhatt, P., Rene, E. R., Kumar, A. J., Zhang, W., and Chen, S. (2020c). Binding interaction of allethrin with esterase: bioremediation potential and mechanism. Bioresour. Technol. 310:123845. doi: 10.1016/j.biortech.2020.123845
Bhatt, P., Huang, Y., Zhan, H., and Chen, S. (2019). Insight into microbial applications for the biodegradation of pyrethroid insecticides. Front. Microbiol. 10:1778. doi: 10.3389/fmicb.2019.01778
Birolli, W. G., Lima, R. N., and Porto, A. L. M. (2019). Applications of marine-derived microorganisms and their enzymes in biocatalysis and biotransformation, the underexplored potentials. Front. Microbiol. 10:1453. doi: 10.3389/fmicb.2019.01453
Brito, L. G., de Oliveira Nery, L., da Silva Barbieri, F., Huacca, M. E. F., dos Santos Pereira, S., da Silva, R. R., et al. (2017). Molecular quantitative assay for esterase-mediated organophosphate resistance in Rhipicephalus microplus. Ticks Tick Borne Dis. 8, 725–732. doi: 10.1016/j.ttbdis.2017.05.006
Buss, D. S., and Callaghan, A. (2004). Molecular comparisons of the Culex pipiens (L.) complex esterase gene amplicons. Insect Biochem. Mol. 34, 433–441. doi: 10.1016/j.ibmb.2004.02.002
Carvalho, R. K., Rodrigues, T. C., Júnior, W. D., Mota, G. M. P., Andersen, M. L., and Mazaro-Costa, R. (2020). Short- and long-term exposure to methamidophos impairs spermatogenesis in mice. Reprod. Biol. doi: 10.1016/j.repbio.2020.05.003 [Epub ahead of print].
Chae, M. Y., Postula, J. F., and Raushel, F. M. (1994). Stereospecific enzymatic hydrolysis of phosphorus-sulfur bonds in chiral organphosphatetriesters. Bioorg. Med. Chem. Lett. 12, 1473–1478. doi: 10.1016/S0960-894X(01)80516-3
Chang, A. S., Montesano, M. A., Barr, D. B., Thomas, J., and Geller, R. J. (2009). Urinary elimination kinetics of acephate and its metabolite, methamidophos, in urine after acute ingestion. J. Med. Toxicol. 5, 68–72. doi: 10.1007/BF03161090
Chen, S., Chang, C., Deng, Y., An, S., Dong, Y. H., Zhou, J., et al. (2014). Fenpropathrin biodegradation pathway in Bacillus sp. DG-02 and its potential for bioremediation of pyrethroid-contaminated soils. J. Agric. Food Chem. 62, 2147–2157. doi: 10.1021/jf404908j
Chen, S., Deng, Y., Chang, C., Lee, J., Cheng, Y., Cui, Z., et al. (2015). Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci. Rep. 5:8784. doi: 10.1038/srep08784
Chen, S., Dong, Y. H., Chang, C., Deng, Y., Xi, F. Z., Zhong, G., et al. (2013). Characterization of a novel cyfluthrin-degrading bacterial strain Brevibacterium aureum and its biochemical degradation pathway. Bioresour. Technol. 132, 16–23. doi: 10.1016/j.biortech.2013.01.002
Chen, S., Liu, C., Peng, C., Liu, H., Hu, M., and Zhong, G. (2012). Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by a new fungal strain Cladosporium cladosporioides Hu-01. PLoS One 7:e47205. doi: 10.1371/journal.pone.0047205
Chino-Flores, C., Dantán-González, E., Vázquez-Ramos, A., Tinoco-Valencia, R., Díaz-Méndez, R., Sánchez-Salinas, E., et al. (2012). Isolation of the opdE gene that encodes for a new hydrolase of Enterobacter sp. capable of degrading organophosphorus pesticides. Biodegradation 23, 387–397. doi: 10.1007/s10532-011-9517-6
Chu, X. Y., Wu, N. F., Deng, M. J., Tian, J., Yao, B., and Fan, Y. L. (2006). Expression of organophosphorus hydrolase OPHC2 in Pichia pastoris: purification and characterization. Protein Expres. Purif. 49, 9–14. doi: 10.1016/j.pep.2006.03.013
Cui, Z. L., Li, S. P., and Fu, G. P. (2001). Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 10, 4922–4925. doi: 10.1128/AEM.67.10.4922-4925.2001
Cycoń, M., Mrozik, A., and Piotrowska-Seget, Z. (2017). Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: a review. Chemosphere 172, 52–71. doi: 10.1016/j.chemosphere.2016.12.129
Cycoń, M., and Piotrowska-Seget, Z. (2016). Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Front. Microbiol. 7:1463. doi: 10.3389/fmicb.2016.01463
Da Silva, N. M., de Carvalho, R. A., and de Azeredo-Espin, A. M. L. (2011). Acetylcholinesterase cDNA sequencing and identification of mutations associated with organophosphate resistance in Cochliomyia hominivorax (Diptera: Calliphoridae). Vet. Parasitol. 177, 190–195. doi: 10.1016/j.vetpar.2010.11.017
Dai, K., Peng, T. Y., Chen, H., Zhang, R. X., and Zhang, Y. X. (2008). Photocatalytic degradation and mineralization of commercial methamidophos in aqueous titania suspension. Environ. Sci. Technol. 42, 1505–1510. doi: 10.1021/es702268p
Dar, M. A., Kaushik, G., Francisco, J., and Chiu, J. F. (2020). “Pollution status and biodegradation of organophosphate pesticides in the environment,” in Abatement of Environmental Pollutants, eds S. Pardeep, K. Ajay, and B. Anwesha, (Amsterdam: Elsevier), 25–66. doi: 10.1016/b978-0-12-818095-2.00002-3
Das, S., and Singh, D. K. (2006). Purification and characterization of phosphotriesterases from Pseudomonas aeruginosa F10B and Clavibactermichiganense subsp. insidiosum SBL11. Can. J. Microbiol. 52, 157–168. doi: 10.1139/w05-113
Dhanushka, M. A. T., and Peiris, L. D. C. (2017). Cytotoxic and genotoxic effects of acephate on human sperm. J. Toxicol. 2017, 1–6. doi: 10.1155/2017/3874817
Di, A. S., Bernardi, G., and Mercuri, N. B. (2004). Methamidophos transiently inhibits neuronal nicotinic receptors of rat substantia nigra dopaminergic neurons via open channel block. Neurosci. Lett. 369, 208–213. doi: 10.1016/j.neulet.2004.07.074
Doraiswamy, N., Sarathi, M., and Pennathur, G. (2019). Cross-linked esterase aggregates (CLEAs) using nanoparticles as immobilization matrix. Prep. Biochem. Biotechnol. 49, 270–278. doi: 10.1080/10826068.2018.1536993
Farag, A. T., Radwan, A. H., Eweidah, M. H., Elmazoudy, R. H., and El-Sebae, A. (2012). Evaluation of male-mediated reproductive toxic effects of methamidophos in the mouse. Andrologia 44, 116–124. doi: 10.1111/j.1439-0272.2010.01113.x
Farag, M. R., Alagawany, M., and Dhama, K. (2017). Flaxseed oil alleviates toxic effects of subacute exposure to acephate on liver and kidney of broiler chicks. Asian J. Anim. Vet. Adv. 12, 61–70. doi: 10.3923/ajava.2017.61.70
Feng, Y., Huang, Y., Zhan, H., Bhatt, H., and Chen, S. (2020). An overview of strobilurin fungicide degradation: current status and future perspective. Front. Microbiol. 10:389. doi: 10.3389/fmicb.2020.00389
Fu, W. N., Wang, Y. H., He, C., and Zhao, J. L. (2012). Photocatalytic degradation of acephate on ZnFe2O4-TiO2 photocatalyst under visible-light irradiation. J. Adv. Oxid. Technol. 1, 177–182. doi: 10.1515/jaots-2012-0120
Gao, J. R., and Zhu, K. Y. (2002). Increased expression of an acetylcholinesterase gene may confer organophosphate resistance in the greenbug, Schizaphis graminum (Homoptera: Aphididae). Pestic. Biochem. Phys. 73, 164–173. doi: 10.1016/S0048-3575(02)00105-0
Gao, Y., Chen, S., Hu, M., Hu, Q., Luo, J., and Li, Y. (2012). Purification and characterization of a novel chlorpyrifos hydrolase from Cladosporium cladosporioides Hu-01. PLoS One 7:e38137. doi: 10.1371/journal.pone.0038137
Ghanem, E., and Raushel, F. (2005). Detoxification of organophosphate nerve agents by bacterial phosphotriesterase. Toxicol. Appl. Pharm. 207, 459–470. doi: 10.1016/j.taap.2005.02.025
Golash, N., and Gogate, P. R. (2012). Degradation of dichlorvos containing wastewaters using sonochemical reactors. Ultrason. Sonochem. 19, 1051–1060. doi: 10.1016/j.ultsonch.2012.02.011
Gotthard, G., Hiblot, J., Gonzalez, D., Elias, M., and Chabriere, E. (2013). Structural and enzymatic characterization of the phosphotriesterase OPHC2 from Pseudomonas pseudoalcaligenes. PLoS One 8:e77995. doi: 10.1371/journal.pone.0077995
Hao, D. F., Xu, W., Wang, H., Du, L. F., Yang, J. D., Zhao, X. J., et al. (2012). Metabolomic analysis of the toxic effect of chronic low-dose exposure to acephate on rats using ultra-performance liquid chromatography/mass spectrometry. Ecotoxixol. Environ. Saf. 83, 25–33. doi: 10.1016/j.ecoenv.2012.06.006
He, X. W., Gao, J. W., Dong, T. Y., Chen, M. J., Zhou, K., Chang, C. X., et al. (2017). Developmental neurotoxicity of methamidophos in the embryo-larval stages of zebrafish. Int. J. Environ. Res. Pub. Health 14:23. doi: 10.3390/ijerph14010023
Helgesen, K. O., Røyset, K., Aspehaug, V., and Jansen, P. A. (2019). The protective effect of the Phe362Tyr mutation in salmon lice’ (Lepeophtheirus salmonis) AChE when exposed to full-scale azamethiphos bath treatments. Aquaculture 505, 517–522. doi: 10.1016/j.aquaculture.2019.03.016
Hong, S., and Raushel, F. M. (1999). Stereochemical preferences for chiral substrates by the bacterial phosphotriesterase. Chem. Biol. Interact. 119–120, 225–234. doi: 10.1016/S0009-2797(99)00031-9
Horne, I., Harcourt, R. L., Sutherland, T. D., Russell, R. J., and Oakeshott, J. G. (2002). Isolation of a Pseudomonas monteilli strain with a novel phosphotriesterase. FEMS Microbiol. Lett. 206, 51–55. doi: 10.1016/S0378-1097(01)00518-3
Huang, Y., Lin, Z., Zhang, W., Pang, S., Bhatt, P., Rene, E. R., et al. (2020). New insights into the microbial degradation of D-cyphenothrin in contaminated water/soil environments. Microorganisms 8:473. doi: 10.3390/microorganisms8040473
Huang, Y., Zhan, H., Bhatt, P., and Chen, S. (2019). Paraquat degradation from contaminated environments: current achievements and perspectives. Front. Microbiol. 10:1754. doi: 10.3389/fmicb.2019.01754
Huang, Y. Y., Zhao, R. B., Hung, Y. C., Gao, H. Y., Zhang, P. H., Wang, Y., et al. (2018). The mechanisms and process of acephate degradation by hydroxyl radical and hydrated electron. Saudi J. Biol. Sci. 25, 226–233. doi: 10.1016/j.sjbs.2017.10.022
Jeffries, T. C., Rayu, S., Nielsen, U. N., Lai, K., Ijaz, A., Nazaries, L., et al. (2018). Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils. Front. Microbiol. 9:147. doi: 10.3389/fmicb.2018.00147
Jesionowski, T., Zdarta, J., and Krajewska, B. (2014). Enzyme immobilization by adsorption: a review. Adsorption. 20, 801–821. doi: 10.1007/s10450-014-9623-y
Jiang, B., Zhang, N. N., Xing, Y., Lian, L. N., Chen, Y. T., Zhang, D. Y., et al. (2019). Microbial degradation of organophosphorus pesticides: novel degraders, kinetics, functional genes, and genotoxicity assessment. Environ. Sci. Pollut. Res. 26, 21668–21681. doi: 10.1007/s11356-019-05135-9
Kallnik, V., Bunescu, A., Sayer, C., Bräsen, C., Wohlgemuth, R., Littlechild, J., et al. (2014). Characterization of a phosphotriesterase-like lactonase from the hyperthermoacidophilic crenarchaeon Vulcanisaetamoutnovskia. J. Biotechnol. 190, 11–17. doi: 10.1016/j.jbiotec.2014.04.026
Kambiranda, D. M., Asraful-Islam, S. M., Cho, K. M., Math, R. K., Lee, Y. H., Kim, H., et al. (2009). Expression of esterase gene in yeast for organophosphates biodegradation. Pestic. Biochem. Phys. 94, 15–20. doi: 10.1016/j.pestbp.2009.02.006
Konstantinou, I. K., Hela, D. G., and Albanis, T. A. (2006). The status of pesticide pollution in surface waters (rivers and lakes) of Greece. Part I. Review on occurrence and levels. Environ. Pollut. 141, 555–570. doi: 10.1016/j.envpol.2005.07.024
Kumar, S., Kaushik, G., Dar, M. A., Nimesh, S., López-chuken, U. J., and Villarreal-chiu, J. F. (2018). Microbial degradation of organophosphate pesticides: a review. Pedosphere 28, 190–208. doi: 10.1016/S1002-0160(18)60017-7
Kumar, V., Upadhyay, N., Kumara, V., and Sharma, S. (2015). A review on sample preparation and chromatographic determination of acephate and methamidophos in different samples. Arab. J. Chem. 8, 624–631. doi: 10.1016/j.arabjc.2014.12.007
Lai, K., Stolowich, N. J., and Wild, J. R. (1995). Characterization of P-S bond hydrolysis in organophosphorothioate pesticides by organophosphorus hydrolase. Arch. Biochem. Biophys. 318, 59–64. doi: 10.1006/abbi.1995.1204
Li, D. D., Yuan, Y. Z., Chen, Y. L., Li, N., Li, J. L., Zheng, Y. L., et al. (2014). Isolation and characterization of three bacterium strains for biodegradation of methamidophos. Adv. Mater. Res. 955–959, 235–238. doi: 10.4028/www.scientific.net/amr.955-959.235
Li, J., Wu, C., Chen, S. H., Lu, Q., Shim, H., Huang, X., et al. (2020). Enriching indigenous microbial consortia as a promising strategy for xenobiotics’ cleanup. J. Clean. Prod. 261:121234. doi: 10.1016/j.jclepro.2020.121234
Li, X. H., He, J., and Li, S. P. (2007). Isolation of a chlorpyrifos-degrading bacterium, Sphingomonas sp. strain Dsp-2, and cloning of the mpd gene. Res. Microbiol. 158, 143–149. doi: 10.1016/j.resmic.2006.11.007
Li, X. Y., Zhang, H. W., Wu, M. N., Zhang, Y., and Zhang, C. G. (2008). Effect of methamidophos on soil fungi community in microcosms by plate count, DGGE and clone library analysis. J. Environ. Sci. 20, 619–625. doi: 10.1016/s1001-0742(08)62103-8
Lin, W. T., Huang, Z., Li, X. Z., Liu, M. H., and Cheng, Y. J. (2016). Bio-remediation of acephate-Pb(II) compound contaminants by Bacillus subtilis FZUL-33. J. Environ. Sci. China 45, 94–99. doi: 10.1016/j.jes.2015.12.010
Lin, Z., Zhang, W., Pang, S., Huang, Y., Mishra, S., Bhatt, P., et al. (2020). Current approaches to and future perspectives on methomyl degradation in contaminated soil/water environments. Molecules 25:738. doi: 10.3390/molecules25030738
Liu, W., Chen, S. F., Zhao, W., and Zhang, S. J. (2009). Titanium dioxide mediated photocatalytic degradation of methamidophos in aqueous phase. J. Hazard. Mater. 164, 154–160. doi: 10.1016/j.jhazmat.2008.07.140
Liu, X. L., Liang, M. J., Liu, Y. H., and Fan, X. J. (2017). Directed evolution and secretory expression of a pyrethroid-hydrolyzing esterase with enhanced catalytic activity and thermostability. Microb. Cell Fact 16:81. doi: 10.1186/s12934-017-0698-5
Liu, X. Y., Wang, L. F., Zhou, X. M., Liu, K. L., Bai, L. Y., and Zhou, X. G. (2015). Photocatalytic degradation of acephate in pakchoi, Brassica chinensis, with Ce-doped TiO2. J. Environ. Sci. Health B 50, 331–337. doi: 10.1080/03601234.2015.1000177
Liu, X. Y., Zhang, Q. P., Li, S. B., Mi, P., Chen, D. Y., Zhao, X., et al. (2018). Developmental toxicity and neurotoxicity of synthetic organic insecticides in zebrafish (Danio rerio): a comparative study of deltamethrin, acephate, and thiamethoxam. Chemosphere 199, 16–25. doi: 10.1016/j.chemosphere.2018.01.176
Liu, Z. S., Zhang, Y., Kong, L. W., Liu, L., Luo, J., Liu, B. Y., et al. (2019). Preparation and preferential photocatalytic degradation of acephate by using the composite photocatalyst Sr/TiO2-PCFM. Chem. Eng. J. 374, 852–862. doi: 10.1016/j.cej.2019.06.013
Lugokenski, T. H., Gubert, P., Bueno, D. C., Nogara, P. A., Saraiva, R. D. A., Barcelos, R. P., et al. (2012). Effect of different oximes on rat and human cholinesterases inhibited by methamidophos: a comparativein vitro andin silico study. Basic Clin. Pharmacol. 111, 362–370. doi: 10.1111/j.1742-7843.2012.00912.x
Maddela, N. R., and Venkateswarlu, K. (2017). “Bacterial utilization of Acephate and Buprofezin,” in Insecticides-Soil Microbiota Interactions, eds N. R. Maddela, and K. Venkateswarlu, (Cham: Springer), 87–101. doi: 10.1007/978-3-319-66589-4_10
Mahajna, M., Quistad, G. B., and Casida, J. E. (1997). Acephate insecticide toxicity: safety conferred by inhibition of the bioactivating carboxyamidase by the metabolite methamidophos. Chem. Res. Toxicol. 10, 64–69. doi: 10.1021/tx9601420
Maia, L. O., Júnior, W. D., Carvalho, L. S., Jesus, L. R., Paiva, G. D., Araujo, P., et al. (2011). Association of methamidophos and sleep loss on reproductive toxicity of male mice. Environ. Toxicol. Phar. 32, 155–161. doi: 10.1016/j.etap.2011.04.007
Malathi, V. M., Jalali, S. K., Gowda, D. K. S., Mohan, M., and Venkatesan, T. (2017). Establishing the role of detoxifying enzymes in field-evolved resistance to various insecticides in the brown planthopper (Nilaparvatalugens) in South India. Insect Sci. 24, 35–46. doi: 10.1111/1744-7917.12254
Maqbool, Z., Hussain, S., Imran, M., Mahmood, F., Shahzad, T., Ahmed, Z., et al. (2016). Perspectives of using fungi as bioresource for bioremediation of pesticides in the environment: a critical review. Environ. Sci. Pollut. Res. 23, 16904–16925. doi: 10.1007/s11356-016-7003-8
Martiìnez-Huitle, C. A., De Battisti, A., Ferro, S., Reyna, S., Cerro-Loìpez, M., and Quiro, M. A. (2008). Removal of the pesticide methamidophos from aqueous solutions by electrooxidation using Pb/PbO2, Ti/SnO2, and Si/BDD Electrodes. Environ. Sci. Technol. 42, 6929–6935. doi: 10.1021/es8008419
Mishra, S., Zhang, W., Lin, Z., Pang, S., Huang, Y., Bhatt, P., et al. (2020). Carbofuran toxicity and its microbial degradation in contaminated environments. Chemosphere 259:127419. doi: 10.1016/j.chemosphere.2020.127419
Mohan, N., and Naveena, L. (2015). Isolation and determination of efficacy of acephate degrading bacteria from agricultural soil. J. Environ. Sci. Toxicol. Food Technol. 9, 10–20. doi: 10.9790/2402-09321020
Mohapatra, S., Ahuja, A. K., Deepa, M., and Sharma, D. (2011). Residues of acephate and its metabolite methamidophos in/on mango fruit (Mangiferaindica L.). B. Environ. Contam. Tox. 86, 101–104. doi: 10.1007/s00128-010-0154-2
Mulla, S. I., Ameen, F., Talwar, M. P., Eqani, S. A. M. A. S., Saxena, R. N. B. G., Tallur, P. N., et al. (2020). “Organophosphate pesticides: impact on environment, toxicity, and their degradation,” in Bioremediation of Industrial Waste for Environmental Safety, eds R. N. Bharagava, and S. Saxena, (Cham: Springer), doi: 10.1007/978-981-13-1891-7_13
Muthyala, V. R., and Gundala, P. H. (2019). The wax encapsulated organophosphate hydrolase (WEO) displays prolonged shelf-life and effective in decontamination of methyl parathion and sarin. Proc. Natl. Acad. Sci. India B 89, 631–637. doi: 10.1007/s40011-018-0977-x
Ortega-Olvera, J. M., Winkler, R., Quintanilla-Vega, B., Shibayama, M., Chávez-Munguía, B., Martín-Tapia, D., et al. (2018). The organophosphate pesticide methamidophos opens the blood-testis barrier and covalently binds to ZO-2 in mice. Toxicol. Appl. Pharm. 360, 257–272. doi: 10.1016/j.taap.2018.10.003
Özkan, D., Yüzbaşıoǧlu, D., Ünal, F., Yılmaz, S., and Aksoy, H. (2009). Evaluation of the cytogenetic damage induced by the organophosphorous insecticide acephate. Cytotechnology 59, 73–80. doi: 10.1007/s10616-009-9195-y
Pan, R., Chen, H., Wang, C., Wang, Q., and Jiang, Y. (2015). Enantioselective dissipation of acephate and its metabolite, methamidophos, during tea cultivation, manufacturing, and infusion. J. Agric. Food Chem. 63, 1300–1308. doi: 10.1021/jf504916b
Pang, S., Lin, Z., Zhang, W., Mishra, S., Bhatt, P., and Chen, S. (2020). Insights into the microbial degradation and biochemical mechanisms of neonicotinoids. Front. Microbiol. 11:868. doi: 10.3389/fmicb.2020.00868
Peng, H. F., Bao, X. D., Zhang, Y., Huang, L., and Huang, H. Q. (2015). Identification of differentially expressed proteins of brain tissue in response to methamidophos in flounder (Paralichthysolivaceus). Fish Shellf. Immun. 44, 555–565. doi: 10.1016/j.fsi.2015.03.028
Phugare, S. S., Gaikwad, Y. B., and Jadhav, J. P. (2012). Biodegradation of acephate using a developed bacterial consortium and toxicological analysis using earthworms (Lumbricusterrestris) as a model animal. Int. Biodeter. Biodegr. 69, 1–9. doi: 10.1016/j.ibiod.2011.11.013
Pinjari, A. B., Novikov, B., Rezenom, Y. H., Russell, D. H., Wales, M. E., and Siddavattam, D. (2012). Mineralization of acephate, a recalcitrant organophosphate insecticide is initiated by a Pseudomonad in environmental samples. PLoS One 7:e31963. doi: 10.1371/journal.pone.0031963
Pinjari, A. B., Pandey, J. P., Kamireddy, S., and Siddavattam, D. (2013). Expression and subcellular localization of organophosphate hydrolase in acephate-degrading Pseudomonas sp. strain Ind01 and its use as a potential biocatalyst for elimination of organophosphate insecticides. Lett. Appl. Microbiol. 57, 63–68. doi: 10.1111/lam.12080
Qamruzzaman, Nasar, A. (2014). Degradation of acephate by colloidal manganese dioxide in the absence and presence of surfactants. Desalin. Water Treat. 8, 2155–2164. doi: 10.1080/19443994.2014.937752
Rajak, P., Dutta, M., Khatun, S., Mandi, M., and Roy, S. (2017). Exploring hazards of acute exposure of Acephate in Drosophila melanogaster and search for l-ascorbic acid mediated defense in it. J. Hazard Mater. 321, 690–702. doi: 10.1016/j.jhazmat.2016.09.067
Ramirez-Vargas, M. A., Huerta-Beristain, G., Guzman-Guzman, I. P., Alarcon-Romero, L. D. C., Flores-Alfaro, E., Rojas-Garcia, A. E., et al. (2017). Methamidophos induces cytotoxicity and oxidative stress in human peripheral blood mononuclear cells. Environ. Toxicol. 32, 147–155. doi: 10.1002/tox.22220
Ramu, S., and Seetharaman, B. (2014). Biodegradation of acephate and methamidophos by a soil bacterium Pseudomonas aeruginosa strain Is-6. J. Environ. Sci. Health B 49, 23–34. doi: 10.1080/03601234.2013.836868
Ribeiro, T. A., Prates, K. V., Pavanello, A., Malta, A., Tófolo, L. P., Martins, I. P., et al. (2016). Acephate exposure during a perinatal life program to type 2 diabetes. Toxicology 372, 12–21. doi: 10.1016/j.tox.2016.10.010
Rodríguez, T., de Joode, B. V. W., Lindh, C. H., Rojas, M., Lundberg, I., and Wesseling, C. (2012). Assessment of long-term and recent pesticide exposure among rural school children in Nicaragua. Occup. Environ. Med. 69, 119–125. doi: 10.1136/oem.2010.062539
Schofield, D. A., and DiNovo, A. A. (2010). Generation of a mutagenized organophosphorus hydrolase for the biodegradation of the organophosphate pesticides malathion and demeton-S. J. Appl. Microbiol. 109, 548–557. doi: 10.1111/j.1365-2672.2010.04672.x
Scott, C., Pandey, G., Hartley, C. J., Jackson, C. J., Cheesman, M. J., Taylor, M. C., et al. (2008). The enzymatic basis for pesticide bioremediation. Indian J. Microbiol. 48, 65–79. doi: 10.1007/s12088-008-0007-4
Shen, Y., Lu, P., Mei, H., Yu, H., Hong, Q., and Li, S. (2010). Isolation of a methyl parathion-degrading strain Stenotrophomonas sp. SMSP-1 and cloning of the ophc2 gene. Biodegradation 21, 785–792. doi: 10.1007/s10532-010-9343-9342
Shriwas, A. K., and Gogate, P. R. (2011). Ultrasonic degradation of methyl parathion in aqueous solutions: intensification using additives and scale up aspects. Sep. Purif. Technol. 79, 1–7. doi: 10.1016/j.seppur.2011.02.034
Singh, S., Kumar, V., Singla, S., Sharma, M., Singh, D. P., Prasad, R., et al. (2020). Kinetic study of the biodegradation of acephate by indigenous soil bacterial isolates in the presence of humic acid and metal ions. Biomolecular 10:433. doi: 10.3390/biom10030433
Singh, S., Kumar, V., Upadhyay, N., Singh, J., Singla, S., and Datta, S. (2017). Efficient biodegradation of acephate by pseudomonas pseudoalcaligenes PS-5 in the presence and absence of heavy metal ions [Cu(II) and Fe(III)], and humic acid. 3 Biotech. 7:262. doi: 10.1007/s13205-017-0900-9
Sonoda, S., and Igaki, C. (2010). Characterization of acephate resistance in the diamondback moth plutellaxylostella. Pestic. Biochem. Phys. 98, 121–127. doi: 10.1016/j.pestbp.2010.05.010
Sun, W., Chen, L., Zhang, Y., and Wang, J. (2015). Synergistic effect of ozonation and ionizing radiation for PVA decomposition. J. Environ. Sci. China 34, 63–67. doi: 10.1016/j.jes.2015.01.020
Syed, J. H., Alamdar, A., Mohammad, A., Ahad, K., Shabir, Z., Ahmed, H., et al. (2014). Pesticide residues in fruits and vegetables from Pakistan: a review of the occurrence and associated human health risks. Environ. Sci. Pollut. Res. 21, 13367–13393. doi: 10.1007/s11356-014-3117-z
Takayasu, T., Yamamoto, H., Ishida, Y., Nosakab, M., Kuninakab, Y., Shimadab, E., et al. (2019). Postmortem distribution of acephate and its metabolite methamidophos in body fluids and organ tissues of an intoxication case. Forensic. Sci. Int. 300, e38–e43. doi: 10.1016/j.forsciint.2019.02.015
Tripathi, S. M., Thaker, A. M., Joshi, C. G., and Sankhala, L. N. (2012). Acephateimmunotoxicity in White Leghorn cockerel chicks upon experimental exposure. Environ. Toxicol. Phar. 34, 192–199. doi: 10.1016/j.etap.2012.04.002
Urióstegui-Acosta, M., Hernández-Ochoa, I., Sánchez-Gutiérrez, M., Piña-Guzmán, B., Rafael-Vázquez, L., Solís-Heredia, M. J., et al. (2014). Methamidophos alters sperm function and DNA at different stages of spermatogenesis in mice. Toxicol. Appl. Pharm. 279, 391–400. doi: 10.1016/j.taap.2014.06.017
Wang, B., Zhu, C. P., Gong, R. H., Zhu, J., Huang, B., Xu, F., et al. (2015). Degradation of acephate using combined ultrasonic and ozonation method. Water Sci. Eng. 8, 233–238. doi: 10.1016/j.wse.2015.03.002
Wang, L., Wang, G. L., Li, S. P., and Jiang, J. D. (2011). Luteibacter jiangsuensis sp. nov.: a methamidophos-degrading bacterium isolated from a methamidophos-manufacturing factory. Curr. Microbiol. 62, 289–295. doi: 10.1007/s00284-010-9707-1
Wang, L., Wen, Y., Guo, X. Q., Wang, G. L., Li, S. P., and Jiang, J. D. (2010). Degradation of methamidophos by Hyphomicrobium species MAP-1 and the biochemical degradation pathway. Biodegradation 21, 513–523. doi: 10.1007/s10532-009-9320-9
Wang, M. C., Liu, Y. H., Wang, Q., Gong, M., Hua, X. M., Pang, Y. J., et al. (2008). Impacts of methamidophos on the biochemical, catabolic, and genetic characteristics of soil microbial communities. Soil Biol. Biochem. 40, 778–788. doi: 10.1016/j.soilbio.2007.10.012
Wang, S. H., Zhang, C., and Yan, Y. C. (2012). Biodegradation of methyl parathion and p-nitrophenol by a newly isolated Agrobacterium sp. strain Yw12. Biodegradation 23, 107–116. doi: 10.1007/s10532-011-9490-0
World Health Organization [WHO] (2009). The WHO Recommended Classification of Pesticides by Hazard and Guidelines to Classification. Geneva: WHO.
Wu, N. F., Deng, M. J., Liang, G. Y., Chu, X. Y., Yao, B., and Fan, Y. L. (2004). Cloning and expression of ophc2, a new organphosphorus hydrolase gene. Chin. Sci. Bull. 49, 1245–1249. doi: 10.1360/04wc0146
Xu, L., Wu, M., and Han, Z. J. (2014). Biochemical and molecular characterisation and cross-resistance in field and laboratory chlorpyrifos-resistant strains of Laodelphaxstriatellus (Hemiptera: Delphacidae) from eastern China. Pest Manag. Sci. 70, 1118–1129. doi: 10.1002/ps.3657
Yang, J., Feng, Y., Zhan, H., Liu, J., Yang, F., Zhang, K., et al. (2018). Characterization of a pyrethroid-degrading Pseudomonas fulva strain P31 and biochemical degradation pathway of D-phenothrin. Front. Microbiol. 9:1003. doi: 10.3389/fmicb.2018.01003
Yang, J., Gao, Y. P., Liu, W. H., Ge, W., and Zhao, R. B. (2015). The degradation and mineralization of acephate by ionization irradiation. Environ. Prog. Sustain. 34, 324–332. doi: 10.1002/ep.11983
Yang, X. Q., Liu, J. Y., Li, X. C., Chen, M. H., and Zhang, Y. L. (2014). Key amino acid associated with acephate detoxification by Cydiapomonella carboxylesterase based on molecular dynamics with alanine scanning and site-directed mutagenesis. J. Chem. Inf. Model. 54, 1356–1370. doi: 10.1021/ci500159q
Yao, J., Zhu, Y. C., Adamczyk, J., and Luttrell, R. (2018). Influences of acephate and mixtures with other commonly used pesticides on honey bee (Apis mellifera) survival and detoxification enzyme activities. Comp. Biochem. Phys. C 209, 9–17. doi: 10.1016/j.cbpc.2018.03.005
Zang, S. Y., Yin, L. L., Ma, Y., Wang, T. T., and Wu, H. X. (2013). Removal of acephate and Hg2+ in compounding wastewater with immobilized microorganism. Adv. Mater. Res. 838–841, 2400–2403. doi: 10.4028/www.scientific.net/AMR.838-841.2400
Zhan, H., Huang, Y., Lin, Z., Bhatt, P., and Chen, S. (2020). New insights into the microbial degradation and catalytic mechanism of synthetic pyrethroids. Environ. Res. 182:109138. doi: 10.1016/j.envres.2020.109138
Zhan, H., Wang, H., Liao, L., Feng, Y., Fan, X., Zhang, L. H., et al. (2018). Kinetics and novel degradation pathway of permethrin in Acinetobacter baumannii ZH-14. Front. Microbiol. 9:98. doi: 10.3389/fmicb.2018.00098
Zhang, J., Lu, L., Chen, F., Chen, L., Yin, J., and Huang, X. (2018). Detoxification of diphenyl ether herbicide lactofen by Bacillus sp. Za and enantioselective characteristics of an esterase gene lacE. J. Hazard Mater. 341, 336–345. doi: 10.1016/j.jhazmat.2017.07.064
Zhang, L., Yan, F., Shu, M. M., Li, Q., and Zhao, Z. Y. (2009). Investigation of the degradation behaviour of methamidophos under microwave irradiation. Desalination 247, 396–402. doi: 10.1016/j.desal.2008.12.037
Zhang, L., Yan, F., Wang, Y. N., Guo, X. J., and Zhang, P. (2006). Photocatalytic degradation of methamidophos by UV irradiation in the presence of Nano-TiO2. Inorgan. Mater. 42, 1379–1387. doi: 10.1134/S002016850612017X
Zhang, N., Guo, Q. Q., Liu, X. F., Song, K., Wang, S., Wang, M. X., et al. (2013). Optimization of liquid culture conditions of methamidophos degrading enzyme from Staphylococcus rosehbach. Appl. Mech. Mater. 303–306, 2683–2686. doi: 10.4028/www.scientific.net/AMM.303-306.2683
Zhang, W., Lin, Z., Pang, S., Bhatt, P., and Chen, S. (2020). Insights into the biodegradation of lindane (γ-hexacholocyclohexane) using a microbial system. Front. Microbiol. 11:522. doi: 10.3389/fmicb.2020.00522
Zhao, R. B., Bao, H. Y., and Liu, Y. X. (2010). Isolation and characterization of Penicillium oxalicum ZHJ6 for biodegradation of methamidophos. Agr. Sci. China 9, 695–703. doi: 10.1016/S1671-2927(09)60145-0
Zhao, R. B., Bao, H. Y., and Xia, L. Y. (2009). γ -Irradiation degradation of methamidophos. Chin. J. Chem. 9, 1749–1754. doi: 10.1002/cjoc.200990294
Zhao, Y., Zhao, P., Wang, Y., and Qi, W. J. (2014). Isolation, identification, and characterization of an organophosphorous pesticide degrading bacterium, Enterobacter ludwigii M2. Adv. Mater. Res. 1051, 398–403. doi: 10.4028/www.scientific.net/AMR.1051.398
Zheng, L. L., Pi, F. W., Wang, Y. F., Xu, H., Zhang, Y. Z., and Sun, X. L. (2016). Photocatalytic degradation of acephate, omethoate, and methyl parathion by Fe3O4@SiO2@mTiO2nanomicrospheres. J. Hazard Mater. 315, 11–22. doi: 10.1016/j.jhazmat.2016.04.064
Keywords: toxicology, physicochemical degradation, microbial degradation, degradation pathways, degradation mechanisms, gene
Citation: Lin Z, Pang S, Zhang W, Mishra S, Bhatt P and Chen S (2020) Degradation of Acephate and Its Intermediate Methamidophos: Mechanisms and Biochemical Pathways. Front. Microbiol. 11:2045. doi: 10.3389/fmicb.2020.02045
Received: 23 May 2020; Accepted: 03 August 2020;
Published: 18 August 2020.
Edited by:
Xing Huang, Nanjing Agricultural University, ChinaReviewed by:
Jun wei Huang, Anhui Agricultural University, ChinaCopyright © 2020 Lin, Pang, Zhang, Mishra, Bhatt and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaohua Chen, c2hjaGVuQHNjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.