
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 August 2020
Sec. Systems Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.02027
Steam explosion is an environment-friendly pretreatment method to improve the subsequent hydrolysis process of lignocellulosic biomass. Steam explosion pretreatment improved ruminal fermentation and changed fermentation pattern of corn stover during ruminal fermentation in vitro. The study gave a comprehensive insight into how stream explosion pretreatment shifted archaeal and bacterial community structure to change ruminal fermentation in vitro of corn stover. Results showed that steam explosion pretreatment dramatically improved the apparent disappearance of dry matter (DM), neutral detergent fiber (NDF), and acid detergent fiber (ADF). Steam explosion pretreatment significantly increased the molar proportion of propionate and decreased the ratio of acetate to propionate. At archaeal level, steam explosion pretreatment significantly increased the relative abundance of Methanobrevibacter, which can effectively remove metabolic hydrogen to keep the fermentation continuing. At bacterial level, the shift in fermentation was achieved by increasing the relative abundance of cellulolytic bacteria and propionate-related bacteria, including Spirochaetes, Elusimicrobia, Fibrobacteres, Prevotella, Treponema, Ruminococcus, and Fibrobacter.
Corn stover is an abundant agricultural residue that can be harvested as a ruminant feedstuff or used to produce biofuel. As a lignocellulosic material, corn stover is mainly composed of three types of polymers: cellulose, hemicellulose, and lignin. These three polymers form a complex network to resist the degradation by microorganisms and enzymes (Malinovsky et al., 2014). Several physical and chemical pretreatment methods have been applied to enhance the utilization of lignocellulosic materials, such as grinding, acid, alkali, enzyme, and steam explosion pretreatment (Schumacher et al., 2014; Schroyen et al., 2015; Bolado-Rodriguez et al., 2016). Although chemical pretreatment methods are useful strategies to improve the utilization of lignocellulosic materials, the chemicals used during processes must be removed and environmental pollution also follows these methods. Compared to chemical pretreatment methods, steam explosion has been recognized as an environment-friendly pretreatment and broadly used for numerous lignocellulosic biomass such as corn stover (Zhao et al., 2018), wheat straw (Ferreira et al., 2013), birch (Vivekanand et al., 2013), and Salix (Horn et al., 2011). Steam explosion is an explosion caused by violent boiling of water into steam, which heats the biomass at high temperature and rapidly drops pressure to disrupt the biomass fibers (Horn et al., 2011). Recent studies have shown that steam explosion can effectively improve digestibility of lignocellulosic biomass (Zhao et al., 2018) and increase the methane (CH4) production during anaerobic digestion (Lizasoain et al., 2017). Steam explosion mainly reduces cellulose degree of polymerization to enhance the digestibility of lignocellulosic biomass (Yu et al., 2015; Jin et al., 2016).
The rumen is a fermentation vat that is home to a vast array of bacteria, protozoa, fungi, and archaea. The structure of ruminal microbiomes is closely related to the process of rumen microbial fermentation. The microorganisms in rumen have been found to be able to effectively ferment lignocellulosic biomass into chemical compounds (Jami et al., 2013; Yue et al., 2013). Volatile fatty acids (VFA) and CH4 are the two major products in rumen microbial fermentation process. VFA will be subsequently absorbed by ruminants to convert into food products (Jami et al., 2013); While, CH4 will be released to atmosphere and presents a loss of gross energy intake (2–12%), depending on types of diets (Johnson and Johnson, 1995). In the typical ruminal microbial fermentation process, the formation of acetate and butyrate both result in the release of metabolic hydrogen which must then be re-oxidized to continue the fermentation process (Ungerfeld, 2015). Methanogenic archaea are the only producer of CH4 in rumen and methanogenesis is the main route to remove metabolic hydrogen which is transferred from bacteria, protozoa, and fungi to archaea (Wolin and Miller, 1997). Steam explosion pretreatment could lead to increased digestibility and metabolic hydrogen production, usually accompanied by CH4 production. Propionate formation is also considered as an alternative pathway to utilize metabolic hydrogen in rumen (Moss et al., 2000). The strong negative correlation between propionate formation and CH4 formation has been proved by Pinares-Patino et al. (2003) and Goopy et al. (2006). Ungerfeld (2015) reported that inhibition of methanogenesis increased the propionate production by redirecting the flow of metabolic hydrogen.
Zhao et al. (2018) reported that steam explosion pretreatment increased digestibility and fermentation of corn stover by improving ruminal microbial colonization. Du et al. (2019) reported that stream explosion pretreatment significantly altered the fermentation parameters in vitro of wheat straw and decreased the ratio of acetate to propionate by facilitating sugar production and microbial colonization. However, it is unknown whether stream explosion pretreatment shifts the structure of ruminal microbiomes to improve fermentation and change fermentation pattern. The objective of this study was to explore how steam explosion pretreatment shifts archaeal and bacterial community structure to enhance fermentation of corn stover and change ruminal fermentation pattern by building a model of corn stover and steam-exploded corn stover ruminal fermentation in vitro.
Corn stover used in present study was harvested in a cornland at Yanqin District Beijing, China. The stover was left and dried in the field after harvest for about 2 months. The full stalk was cut into 50 cm fragments to make it easy to carry. The stover was dried at 65°C for 24 h to obtain air-dried samples. The air-dried corn stover was manually chopped to 3–5 cm fragments by scissors. One 500 g of (dry matter, DM) chopped corn stover samples was milled to pass through a 1 mm sieve using a Wiley mill (Thomas Scientific, Swedesboro, NJ, United States) as the control group fermentation substrate. A 20 L steam explosion reactor connected to a steam generator (QB-200, Hebi Gentle Bioenergy Co. Ltd., China) was used to produce steam-exploded corn stover. Another 500 g of (DM) chopped corn stover samples was accurately weighed, and then atomized evenly 50 g of deionized water on the samples. The samples were sealed in a vacuum bag and infiltrated for about 12 h at room temperature. The steam explosion parameters selected in present study were at 1.5 MPa steam pressure and 10% moisture for 3 min. The steam-exploded corn stover was oven-dried at 65°C for 24 h and then used for the treatment group fermentation substrate. A completely randomized design was applied in an in vitro incubation, and there are two treatments in the current study: CON (the control group fermentation substrate, corn stover) and TRT (the treatment group fermentation substrate, steam-exploded corn stover).
The experimental procedures were approved by the Chinese Academy of Agricultural Sciences Animal Care and Use Committee (Beijing, China). Rumen fluid was collected 2 h before morning feeding from three cannulated lactating Holstein cows fed a total mixed ration (TMR) composed (% DM basis) of corn silage (25%), alfalfa hay (15%), steam-flaked corn (27%), soybean meal (8.5%), cottonseed meal (8.5%), beet pulp (5.5%), distillers dried grains with solubles (7.5%), minerals, and vitamins (3%). The donor cows were fed twice daily at 0500 and 1700 h. Ruminal fluid was filtered through four layers of cheesecloth and immediately brought to the laboratory. Rumen fluid sampled from each of the donor cows was combined on an equal volume basis and diluted with buffer solution (1:2 v/v) formulated as the method of Menke (1988), containing per liter 8.75 g NaHCO3, 1.00 g NH4HCO3, 1.43 g Na2HPO4, 1.55 g KH2PO4, 0.15 g MgSO4·7H2O, 0.52 g Na2S, 0.017 g CaCl·2H2O, 0.015 g MnCl2·4H2O, 0.002 g CoCl·6H2O, 0.012 g FeCl3·6H2O, and 1.25 mg resazurin. The preparation process of the mixed liquid is carried out under continuous flushing with carbon dioxide (CO2). The starting pH of the mixed liquid after equilibration under CO2 is 6.80. The mixed fluid was transferred into 120-ml serum bottles (75 ml/bottle) containing 500 mg of fermentation substrates (CON or TRT) under the continuous flow of CO2 to remove the air from headspace. In addition, an extra four bottles containing no substrate served as blank for correction of analytes and gases. The serum bottles were immediately sealed with butyl rubber stoppers plus aluminum caps and then connected with vacuumed air bags. The batch fermentation was incubated at 39°C for 72 h with horizontal shaking at 60 rpm. The in vitro batch fermentation was carried out in triplicates and four serum bottles per treatment were arranged in each batch.
The 100-ml calibrated glass syringes (Häberle Labortechnik, Lonsee-Ettlenschieß, Germany) were used to measure the total gas production of each air bag. The pH of incubations was measured using a SevenGo™ portable pH meter (Mettler Toledo, Switzerland). The pre-weighed nylon bags (8 cm × 12 cm, 42 μm) were used to filter the whole biomass material of each bottle. Then, the nylon bags were washed with cold running water until the effluent ran clear. Afterwards, the nylon bags were dried at 55°C for 48 h for analysis of the apparent disappearance of DM, neutral detergent fiber (NDF), and acid detergent fiber (ADF). The filtrate samples (2.5 ml) from each bottle were individually collected for determining VFA profiles, and another 2.5 ml of filtrate sample was collected for microbial analysis. The samples for VFA profiles were frozen at −20°C and the samples for microbial analysis were frozen immediately in liquid nitrogen. The ANKOM A200 fiber analyzer (ANKOM Technology, Macedon, NY, United States) was used to measure the contents of NDF and ADF according to the approach described by Van Soest et al. (1991). Sodium sulfite and α-amylase were used for the analysis of NDF. The concentrations of CH4 and hydrogen (H2) were determined using a gas chromatograph (7890B, Agilent Technologies, United States) fitted with a thermal conductivity detector and a packed column (Porapak Q, Agilent Technologies, United States). The VFA concentrations were analyzed by a gas chromatograph (7890B, Agilent Technologies, United States) equipped with a flame ionization detector and a capillary column (30 m × 0.250 mm × 0.25 μm; BD-FFAP, Agilent Technologies, United States). More detail methods were described by Wang et al. (2018).
Microbial DNA was extracted using an E.Z.N.ATM Mag-Bind Soil DNA kit (Omega, Norcross, Georgia, United States), in accordance with the manufacturer’s instructions. The quality and concentration of the extracted DNA were assessed by 1% agarose gel electrophoresis and a Qubit 3.0 spectrometer (Invitrogen, United States), respectively. This method gave DNA yields sufficient for analysis (averaging 35 ng/μl), but kit-based assays may yield non-representative taxonomic sampling (Henderson et al., 2013).
The primers 515F (5'-GTGCCAGCMGCCGCGGTAA-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') were selected for bacterial community analysis (Caporaso et al., 2011). The analysis of archaeal community was conducted by the nested PCR according to the lower abundance of archaeal community compared with bacterial community. The process of nested PCR was conducted using the primers Arch340F (5'-CCCTAYGGGGYGCASCAG-3')/Arch1000R (5'-GAGARGWRGTGCATGGCC-3') and Arch349F (5'-GYGCASCAGKCGMGAAW-3')/Arch806R (50-GGACTACVSGGGTATCTAAT-3'), as previously described by Wang et al. (2018). The 16S ribosomal RNA (rRNA) gene amplicons were pooled and sent to Shanghai Sangon Biotech Co., Ltd. for pair-end sequencing using the Illumina MiSeq platform (2 × 300 bp).
Sequencing reads were matched to different samples in accordance with the unique barcode of different samples, and then pair-end reads were merged using FLASH (Magoč and Salzberg, 2011). The quality of these merged reads was controlled by using PRINSEQ (Schmieder and Edwards, 2011). Then, the barcode and primers were removed, and UCHIME was used to filter out PCR chimeras to get clean sequences (Edgar et al., 2011). After filtration, the average length of all the clean reads was 416 and 379 bp, and the average sequencing depth was ca. 58,574 and 61,208 clean reads for bacterial and archaeal community analysis, respectively. After removal of singletons, operational taxonomic units (OTU) were clustered at 97% sequence identity using UPARSE (Edgar et al., 2011). The taxonomic classification of the sequences was carried out using the ribosomal database project (RDP) classifier at the bootstrap cutoff of 80%, as suggested by the RDP. The alpha diversity indices including Simpson, Shannon, Chao1, Coverage, and ACE were calculated through the QIIME 2 software package. The principal coordinate analysis (PCoA) was conducted by the weighted UniFrac distance (Lozupone et al., 2007), and the significant difference between treatments was assessed by an analysis of similarity (ANOSIM) in QIIME with 999 permutations (R Core Team, 2013). The extended error bar plot was performed to visualize the difference in relative abundance of bacteria and archaea by bioinformatics software (STAMP; Parks et al., 2014).
All the raw sequences were submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/Traces/sra/), under accession number SRP189861.
Data were analyzed using PROC MIXED of SAS (version 9.4; SAS Institute Inc., Cary, NC). Spearman’s rank correlations between the relative abundance of microbial genera and fermentation variables were analyzed using the PROC CORR procedure of SAS (version 9.4; SAS Institute Inc., Cary, NC). p < 0.05 was considered statistically significant.
Data of substrate apparent disappearance, the molar proportion of VFA, and CH4 and H2 production are presented in Table 1. Based on the previous studies on in vitro forage fiber digestion, we terminated the incubation process at 72 h (Cross et al., 1974; Zhao et al., 2018). Steam explosion pretreatment dramatically improved the apparent disappearance of DM, NDF, and ADF. This was similar to the previous study, which reported that the improvement in digestibility of the steam-exploded corn stover was consistent with the enhanced colonization of microorganism (Zhao et al., 2018). Some other studies also reported that steam explosion pretreatment can enhance the degradability of lignocellulosic biomass by increasing porosity (Dan et al., 2017) and reducing degree of polymerization (Jin et al., 2016). In response to the improved DM degradability, the concentration of total VFA was significantly increased, which was similar to the previous studies (Zhao et al., 2018; Du et al., 2019). Steam explosion pretreatment increased the release of sugars from corn stover to improve the production of VFA (Zhao et al., 2018; Du et al., 2019). Steam explosion pretreatment changed the fermentation pattern and promoted a shift in the fermentation pattern toward a higher molar proportion of propionate in the present study. Du et al. (2019) also reported that steam explosion pretreatment increased the molar proportion of propionate. Fermentation of nonstructural carbohydrates (i.e., sugars, starches, organic acids, and other reserve carbohydrates), compared to fermentation of structural carbohydrates (i.e., cellulose, hemicellulose, and lignin), increased amounts of propionate and decreased the ratio of acetate and propionate (Dijkstra, 1994). Therefore, the higher molar proportion of propionate should be attributed to more soluble nonstructural carbohydrates released from steam explosion pretreatment. Zhao et al. (2018) and Du et al. (2019) both reported that steam explosion pretreatment increased the release of soluble nonstructural carbohydrates. Corresponded with the increased total VFA concentration and the higher molar proportion of propionate, the pH of TRT was significantly decreased compared with CON. Previous studies also reported that increases in total VFA concentrations and the molar proportion of propionate were associated with the decrease in pH due to containing more soluble nonstructural carbohydrates in diets (Penner and Oba, 2009; Song et al., 2018). Total gas production and CH4 production from TRT were both higher than that from CON, but the increases were both statistically insignificant. The results are not consistent from previous studies (Lizasoain et al., 2017; Mulat et al., 2018), which reported that steam explosion pretreatment significantly increased the total gas production and CH4 production. Steam explosion pretreatment improved the digestibility of corn stover and more readily fermentable substrates should produce more gas. While the lack of significant increase in total gas production and CH4 production in the present study might be attributed to the limited number of substrates initially added in the in vitro incubation. In addition, the anaerobic digestion inoculum used in the present study is rumen fluid, which is home to a vast array of ciliate protozoa, fungi, bacteria, and archaea. The compositions and interactions of different microbes are more complex than those used in the previous studies (Lizasoain et al., 2017; Mulat et al., 2018). In the present study, the formation of propionate might be served as an alternative to H2 formation and then reduce the utilization of H2 in methanogenesis, shifting H2 flow from CH4 to propionate production. Zhang et al. (2016) reported that propionate accumulation led to lower CH4 production of microwave pretreatment food waste compared to microwave pretreatment sewage sludge. Therefore, the higher proportion of propionate or the changed fermentation pattern may limit the significant increase in CH4 production in the present study. In the present study, there was a 24% increase in DM disappearance from the TRT fermentation compared to the CON fermentation (from 61.9 to 76.7%), but only a 4.6% increase in VFA production (from 62.8 to 65.7 mM), and increases in gas production were not significant. Much more DM disappeared than was recovered in VFA production. The reason may be due to sugars being released, and not being fermented to VFA. Zhao et al. (2018) and Du et al. (2019) reported that the concentration of reducing sugar, xylose, and fructose in steam explosion group increased after the 72 h of incubation. The chemical composition of corn stover and steam-exploded corn stover was shown in Supplementary Material.
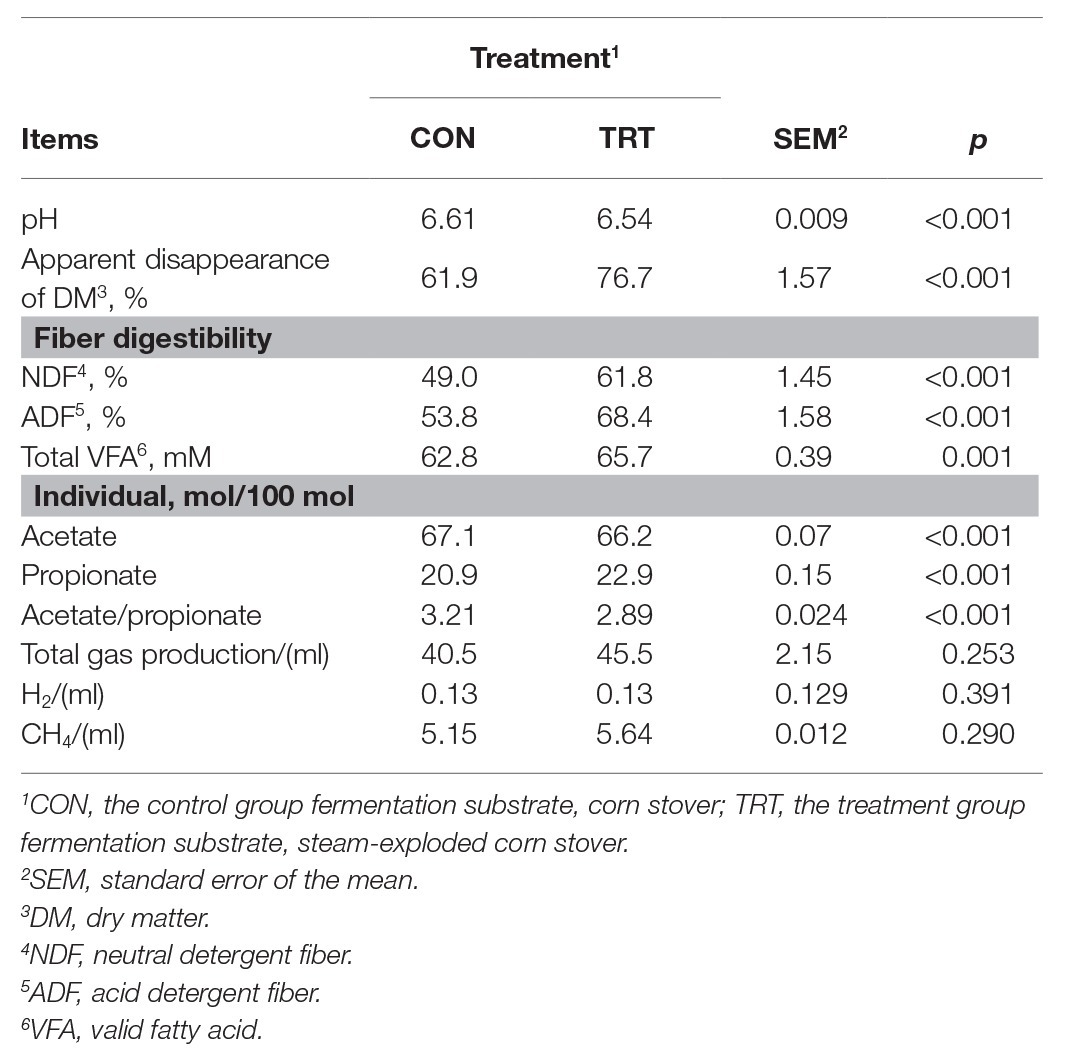
Table 1. Effects of steam explosion pretreatment on fermentation and gas production after 72 h of incubation (n = 12).
After merging and quality control, a total of 1,637,655 sequences from 24 samples were generated, and 1,572,671 high-quality sequences were acquired, with an average read length of 379 bp. After chimera removal, OTUs were obtained from the remaining 1,468,988 sequences, with 97% sequence similarity. After filtering, the remaining 24,035 OTUs were used for subsequent analysis. At archaeal phylum level, Euryarchaeota and Thaumarchaeota represented the archaeal community, where Euryarchaeota represented on average 99.99%. At archaeal genus level, Methanobrevibacter (38.53%), Methanomassiliicoccus (34.79%), Methanomicrobium (25.36%), and Methanosphaera (0.31%, Figure 1A) were the four predominant genera. Methanobrevibacter was recognized to be the most common genus in rumen (Janssen and Kirs, 2008; Danielsson et al., 2017). Methanomassiliicoccus has a high relative abundance similar to Methanobrevibacter in the present study.
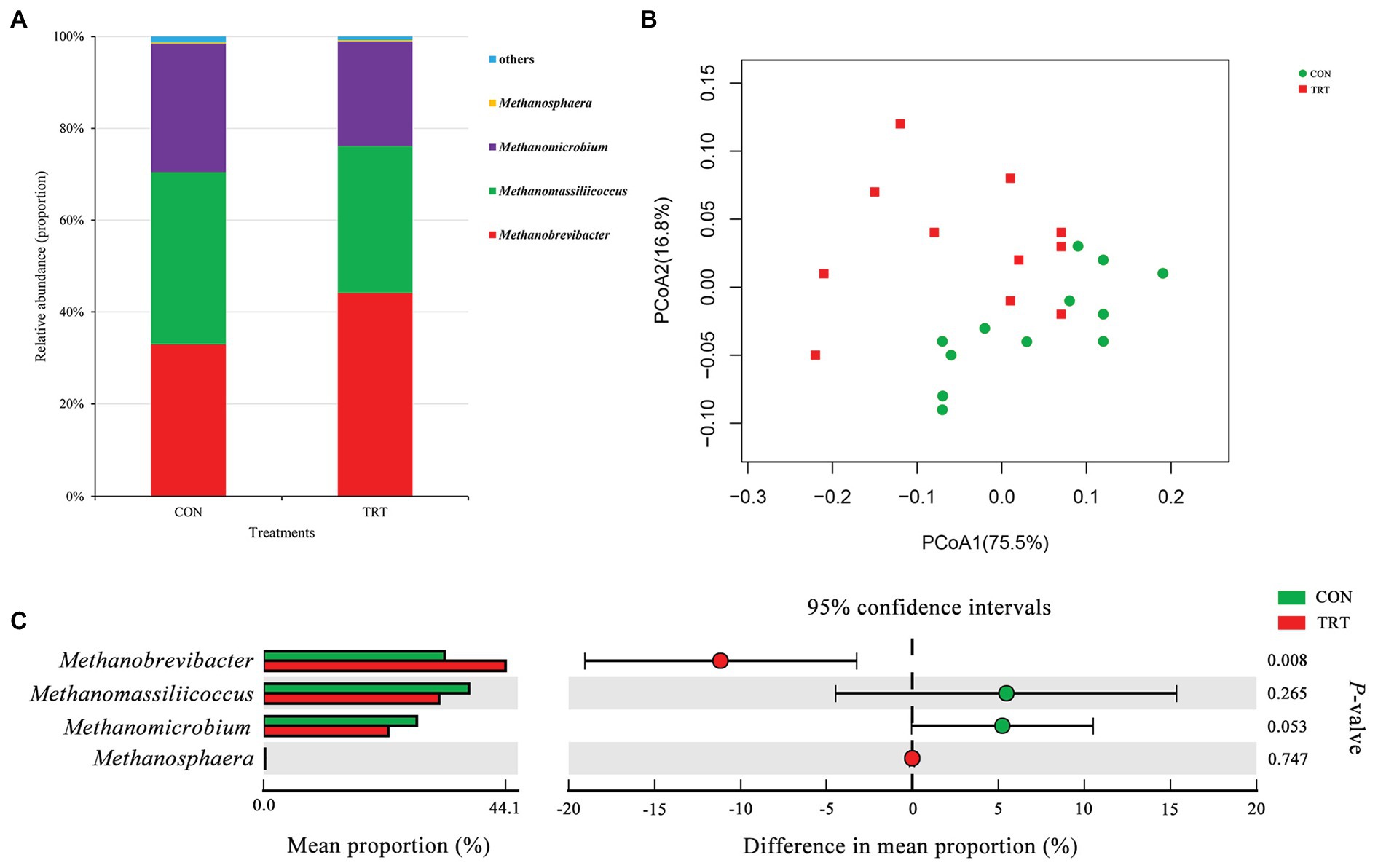
Figure 1. Changes in archaeal community structure. (A) The composition of the predominant archaeal genera. (B) Principal coordinate analysis (PCoA) of archaeal community structure. (C) Difference in the relative abundance of archaeal genera. CON, the control group fermentation substrate, corn stover; TRT, the treatment group fermentation substrate, steam-exploded corn stover (n = 12).
Alpha archaeal diversity was presented in Table 2. No significant differences were observed between treatments based on the alpha diversity indices of Chao1, ACE, Shannon, and Simpson, showing that the archaeal community richness and diversity were not affected by steam explosion pretreatment. The analysis of PCoA based on the weighted UniFrac metrics (Figure 1B) showed that TRT was not distinctly separated from CON. Principal coordinate 1 and 2 accounted for 75.5 and 16.8% of the total variation, respectively. The ANOSIM analysis revealed a significant difference between CON and TRT (R = 0.125, p = 0.034).
At archaeal genus level, steam explosion pretreatment significantly increased the relative abundance of Methanobrevibacter. While the relative abundance of Methanomassiliicoccus and Methanomicrobium from TRT was lower than that from CON, the decreases were not statistically significant (Figure 1C). Spearman’s rank correlations between the relative abundance of archaeal genera and fermentation parameters were analyzed using the PROC CORR procedure of SAS 9.4 (Figure 3A). Within the archaeal community, no significant correlations were observed between the relative abundance of archaeal genera and CH4 production. As the sole producers of CH4 in rumen, the number of methanogens was considered to be related to CH4 production (Wallace et al., 2014; Veneman et al., 2015). However, the efficiency of different methanogens was considered to be more important than the number of methanogens in methanogenesis (Shi et al., 2014). Previous studies reported that Methanobrevibacter had positive correlations with CH4 production (Zhou et al., 2011; Danielsson et al., 2012; Wallace et al., 2015). Cunha et al. (2017) found that the relative abundance of Methanosphaera had negative correlations with CH4 production. There are two common methanogenic pathways in rumen: hydrogenotrophic pathway and methylotrophic pathway. Most methanogenic archaea use H2 and CO2 as substrates to produce CH4, but some species can also metabolize formate, methanol, or acetate for methanogenesis. Methanobrevibacter is the common hydrogenotrophic archaea, which produces 1 mole of CH4 for each mole of CO2 by hydrogenotrophic pathway (Hook et al., 2010), while Methanosphaera requires 4 moles of methanol to produce 3 moles of CH4 by methylotrophic pathway (Fricke et al., 2006). Methanomassiliicoccus has ability to use methylamine substrates to generate CH4 by H2-dependent methylotrophic pathway (Moissl-Eichinger et al., 2018). Methanogenic archaea commonly acquired substrates from environment, while some species would improve efficiency by building contacts with protozoa, which generated large amounts of H2 using hydrogenosomes (Embley et al., 2003). In rumen, Methanobrevibacter is recognized as the most common protozoa-associated methanogens, and the contribution of Methanomicrobium to protozoa-associated methanogenic community is different among studies (Belanche et al., 2014). In the present study, the improvement of corn stover in fermentation efficiency may be related to changes in archaeal community structure. Steam explosion mainly reduces cellulose degree of polymerization to enhance the digestibility of lignocellulosic biomass (Yu et al., 2015; Jin et al., 2016). Steam explosion pretreatment also increased the release of soluble nonstructural carbohydrates to improve the fermentation efficiency of corn stover (Zhao et al., 2018; Du et al., 2019). During the incubation in vitro, cellulose, hemicellulose, and soluble nonstructural carbohydrates are hydrolyzed to glucose and other monosaccharides, which are further metabolized to VFAs, CO2, and metabolic hydrogen (Beauchemin et al., 2020). The improvement of fermentation efficiency produced more fermentation products, including more metabolic hydrogen. Methanogenesis is the main route to remove metabolic hydrogen in rumen and Methanobrevibacter is an efficient hydrogenotrophic methanogenic archaea through the reduction of CO2 via H2 to produce CH4 (Hook et al., 2010). To keep the fermentation continuing, metabolic hydrogen must be removed to keep the partial pressure H2 low (Moss et al., 2000). Therefore, steam explosion pretreatment increased the relative abundance of Methanobrevibacter to enhance fermentation efficiency of corn stover.
After merging and quality control, a total of 1,543,897 sequences from 24 samples were generated, and 14,926,333 high-quality sequences were acquired, with an average read length of 416 bp. After chimera removal, OTUs were obtained from the remaining 1,405,778 sequences, with 97% sequence similarity. After filtering, the remaining 85,546 OTUs were used for subsequent analysis. At bacterial phylum level, Firmicutes and Bacteroidetes were the dominant phyla, representing 34.07 and 30.64% of the total sequences, respectively. Verrucomicrobia, Lentisphaerae, Proteobacteria, Actinobacteria, Spirochaetes, Planctomycetes, Elusimicrobia, Euryarchaeota, and Fibrobacteres represented average percentages of 12.78, 5.30, 4.25, 3.76, 2.33, 0.62, 0.36, 0.27, and 0.18%, respectively (Figure 2A). At bacterial genus level, the 10 predominant genera were Olawnella (3.48%), Prevotella (3.39%), Oligosphaera (3.35%), Campylobacter (2.69%), Treponema (1.65%), Succiniclasticum (1.51%), Ruminococcus (1.51%), Sporobacter (1.41%), Paraprevotella (1.17%), and Acetobacteroides (1.04%; Figure 2B).
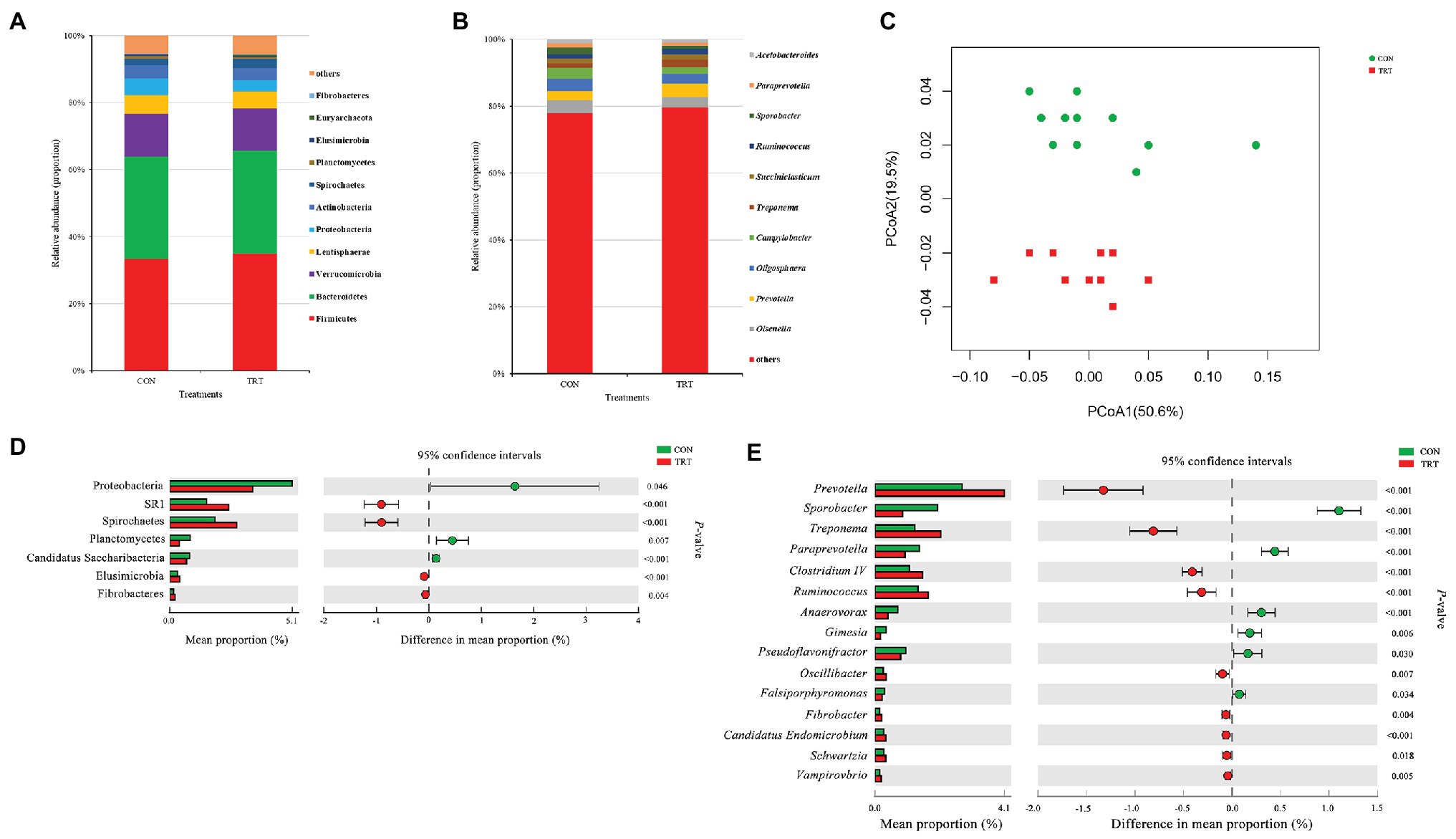
Figure 2. Changes in bacteria community structure. (A) The composition of the predominant bacterial phyla. (B) The composition of the predominant bacterial genera. (C) PCoA of bacterial community structure. (D) Difference in the relative abundance of bacterial phyla. (E) Difference in the relative abundance of bacterial genera. CON, the control group fermentation substrate, corn stover; TRT, the treatment group fermentation substrate, steam-exploded corn stover (n = 12).
Alpha bacterial diversity was presented in Table 2. No significant differences were observed between treatments based on Chao1 and ACE, showing that steam explosion pretreatment did not change the bacterial community richness. Changes of Shannon index and Simpson index indicated that steam explosion pretreatment significantly increased the diversity of bacterial community. The analysis of PCoA based on the weighted UniFrac metrics (Figure 2C) showed that TRT was significantly separated from CON. Principal coordinate 1 and 2 accounted for 50.6 and 19.5% of the total variation, respectively. The ANOSIM analysis revealed significant differences between treatments (R = 0.215, p = 0.001).
At bacterial phylum level, steam explosion pretreatment significantly increased the relative abundance of Spirochaetes, Elusimicrobia, and Fibrobacteres (Figure 2D). In rumen, Spirochetes can ferment xylan, pectin, and arabinogalactan, which are polymers commonly present in plant materials. Hydrolysis products of plant polymers (e.g., d-xylose, ʟ-arabinose, d-galacturonic acid, d-glucuronic acid, and cellobiose) also can be utilized as growth substrates by Spirochetes cultured from rumen fluid. Spirochetes fermented glucose to formate and acetate, some of them also produced succinate or ethanol (Paster and Canale-Parola, 1982). Succinate is not the end product of rumen fermentation and it will be further metabolized to propionate by other rumen species (Scheifinger and Wolin, 1973). Some important species belong to Spirochaetes, such as Treponema bryantii and Treponema saccharophilum, which were engaged primarily in the fermentation of soluble carbohydrates (Stanton and Canale-Parola, 1980). Steam explosion pretreatment destructed the integrity of cover stover and increased the release of sugars (Zhao et al., 2018), which could be served as fermentation substrates to produce succinate by Spirochaetes and finally converted to propionate by other rumen species. In the present study, the increased relative abundance of Spirochaetes may be related to the higher proportion of propionate in TRT group. Elusimicrobia is an enigmatic bacterial phylum in the hindgut of insects, as well as in rumen where it occurs as symbionts of various flagellated protists or as free-living bacteria (Méheust et al., 2019). Cultivation and genome-based studies revealed that some species belong to Elusimicrobia are capable of glucose fermentation (Geissinger et al., 2009; Zheng et al., 2016). In the present study, the increase in the relative abundance of Elusimicrobia may be attributed to the more release of sugars of corn stover resulted from steam explosion pretreatment. Fibrobacteres is a small bacterial phylum which currently comprises sole cultured representative genus, Fibrobacter, and Fibrobacter succinogenes and Fibrobacter intestinalis are the two cultured species, which were considered as major bacterial degraders of lignocellulosic material in rumen (Ransom-Jones et al., 2014). Steam explosion pretreatment broke the bonds between lignin and hemicellulose and cellulose, and damaged the surface structure of corn stover, so that fibrotic microbes had better interaction with lignocellulosic material (Zhao et al., 2018). More digestible lignocellulosic material may result in the increased relative abundance of Fibrobacteres.
At bacterial genus level, steam explosion pretreatment significantly increased the relative abundance of Prevotella, Treponema, Ruminococcus, Oscillibacter, Fibrobacter, and Schwartzia (Figure 2E). Spearman’s rank correlations indicated that the relative abundance of Prevotella, Treponema, and Ruminococcus has significantly positive correlations with propionate proportion (r > 0.55, p < 0.05) (Figure 3B). Prevotella is considered as the most abundant genus in rumen (de Menezes et al., 2011; Jami and Mizrahi, 2012) and one of the main propionate producers (Strobel, 1992). Different Prevotella species produced different fermentation end products, including that Prevotella albensis produced acetate, Prevotella brevis and Prevotella bryantii primarily produced propionate, and P. ruminicola produced propionate and acetate (Emerson and Weimer, 2017). Treponema is typically associated with the fermentation of soluble carbohydrates (Stanton and Canale-Parola, 1980). Treponema belongs to the phylum of Spirochaetes, which may be related to the higher proportion of propionate in the present study, as we talked about above. In addition, positive interactions have been demonstrated between T. bryantii and rumen cellulolytic bacteria (Kudo et al., 1987). Ruminococcus plays crucial roles in the fermentation of dietary polysaccharides in rumen (Leschine, 1995). Some Ruminococcus species are cellulolytic, including Ruminococcus flavefaciens and Ruminococcus albus, both of which were originally isolated from the bovine rumen (La Reau and Suen, 2018). Some Ruminococcus species are non-cellulolytic, including Ruminococcus callidus and Ruminococcus bicirculans. R. callidus can ferment saccharides and its major fermentation products are succinate and acetate, with a less amount of formate, lactate, and pyruvate (La Reau and Suen, 2018). Some of Ruminococcus can ferment polysaccharides to produce succinate, which will be further metabolized to propionate by other rumen species (Scheifinger and Wolin, 1973). At bacterial level, the improvement in fermentation efficiency and the shift in fermentation pattern may be achieved by increasing the relative abundance of cellulolytic bacteria and propionate-related bacteria. However, only filtrates were used for DNA isolation, so only the non-adherent (planktonic) community was analyzed in the present study. This community would be expected to be under-represented with respect to cellulolytic microbes when comparing the microbial data to fiber digestibility. Therefore, the potential bias associated with under-sampling the particle-associated community inevitably existed in the present study.
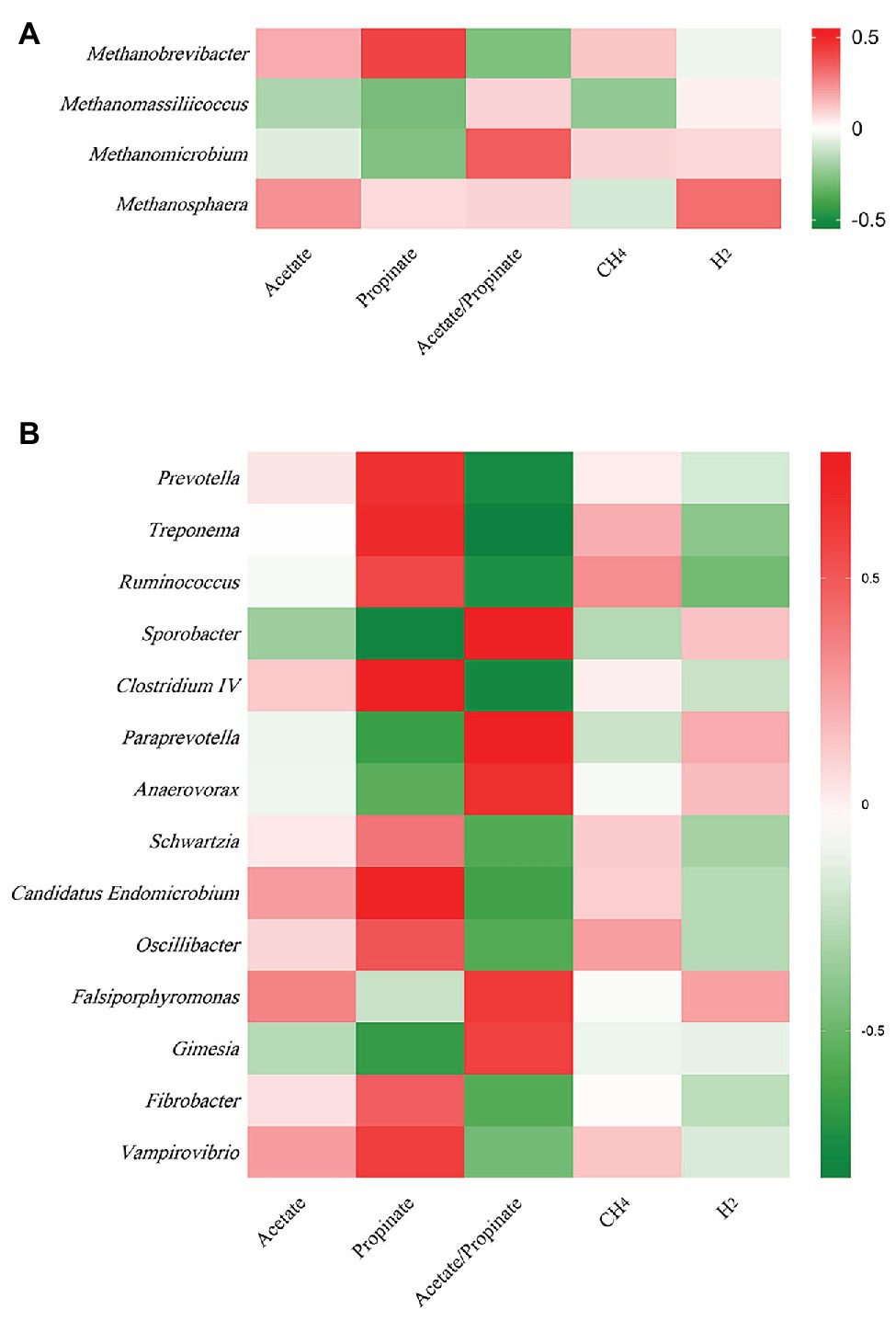
Figure 3. Spearman’s rank correlation between the relative abundances of archaeal and bacterial genera and fermentation parameters. (A) Between archaeal genera and fermentation parameters. (B) Between bacteria genera and fermentation parameters (n = 12).
Steam explosion pretreatment improved the fermentation efficiency and changed the fermentation pattern of corn stover during ruminal fermentation in vitro by shifting archaeal and bacterial community structure. Analysis of archaeal community structure revealed that steam explosion pretreatment significantly increased the relative abundance of Methanobrevibacter, which can effectively remove metabolic hydrogen to keep the fermentation continuing. At bacterial level, steam explosion pretreatment significantly increased the relative abundance of cellulolytic bacteria, including Fibrobacteres, Fibrobacter, and Ruminococcus. Steam explosion pretreatment significantly increased the relative abundance of propionate-related bacteria, including Spirochaetes, Elusimicrobia, Prevotella, Treponema, and Ruminococcus.
The datasets generated for this study can be found in the NCBI Sequence Read Archive/accession number SRP189861.
The experimental procedures were approved by the Chinese Academy of Agricultural Sciences Animal Care and Use Committee (Beijing, China).
KW, BX, and LJ designed the study. KW and XN conducted the experiment and analyzed the data. KW wrote the manuscript. JT, XN, GZ, LJ, and BX revised the paper. All authors contributed to the article and approved the submitted version.
The study was financially supported by the National Key Research and Development Plan (Grant No. 2016YFD0500507, 2016YFD0700205, and 2016YFD0700201).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmicb.2020.02027/full#supplementary-material
Beauchemin, K. A., Ungerfeld, E. M., Eckard, R. J., and Wang, M. (2020). Review: fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal 14, s2–s16. doi: 10.1017/S1751731119003100
Belanche, A., de la Fuente, G., and Newbold, C. J. (2014). Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 90, 663–677. doi: 10.1111/1574-6941.12423
Bolado-Rodriguez, S., Toquero, C., Martin-Juarez, J., Travaini, R., and Garcia-Encina, P. A. (2016). Effect of thermal, acid, alkaline and alkaline-peroxide pretreatments on the biochemical methane potential and kinetics of the anaerobic digestion of wheat straw and sugarcane bagasse. Bioresour. Technol. 201, 182–190. doi: 10.1016/j.biortech.2015.11.047
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Cross, H. H., Smith, L. W., and DeBarth, J. V. (1974). Rates of in vitro forage fiber digestion as influenced by chemical treatment. J. Anim. Sci. 39, 808–812. doi: 10.2527/jas1974.394808x
Cunha, C. S., Veloso, C. M., Marcondes, M. I., Mantovani, H. C., Tomich, T. R., Pereira, L. G. R., et al. (2017). Assessing the impact of rumen microbial communities on methane emissions and production traits in Holstein cows in a tropical climate. Syst. Appl. Microbiol. 40, 492–499. doi: 10.1016/j.syapm.2017.07.008
Dan, S., Alam, A., Tu, Y., Zhou, S., Wang, Y., Tao, X., et al. (2017). Steam-exploded biomass saccharification is predominately affected by lignocellulose porosity and largely enhanced by Tween-80 in Miscanthus. Bioresour. Technol. 239, 74–81. doi: 10.1016/j.biortech.2017.04.114
Danielsson, R., Dicksved, J., Sun, L., Gonda, H., Muller, B., Schnurer, A., et al. (2017). Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 8:226. doi: 10.3389/fmicb.2017.00226
Danielsson, R., Schnurer, A., Arthurson, V., and Bertilsson, J. (2012). Methanogenic population and CH4 production in Swedish dairy cows fed different levels of forage. Appl. Environ. Microbiol. 78, 6172–6179. doi: 10.1128/AEM.00675-12
de Menezes, A. B., Lewis, E., O’Donovan, M., O’Neill, B. F., Clipson, N., and Doyle, E. M. (2011). Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 78, 256–265. doi: 10.1111/j.1574-6941.2011.01151.x
Dijkstra, J. (1994). Production and absorption of volatile fatty acids in the rumen. Livest. Prod. Sci. 39, 61–69. doi: 10.1016/0301-6226(94)90154-6
Du, C., Nan, X., Wang, K., Zhao, Y., and Xiong, B. (2019). Evaluation of the digestibility of steam-exploded wheat straw by ruminal fermentation, sugar yield and microbial structure in vitro. RSC Adv. 9, 41775–41782. doi: 10.1039/C9RA08167D
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Embley, T. M., van der Giezen, M., Horner, D. S., Dyal, P. L., Bell, S., and Foster, P. G. (2003). Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life 55, 387–395. doi: 10.1080/15216540310001592834
Emerson, E. L., and Weimer, P. J. (2017). Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 101, 4269–4278. doi: 10.1007/s00253-017-8150-7
Ferreira, L. C., Donoso-Bravo, A., Nilsen, P. J., Fdz-Polanco, F., and Perez-Elvira, S. I. (2013). Influence of thermal pretreatment on the biochemical methane potential of wheat straw. Bioresour. Technol. 143, 251–257. doi: 10.1016/j.biortech.2013.05.065
Fricke, W. F., Seedorf, H., Henne, A., Kruer, M., Liesegang, H., Hedderich, R., et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 188, 642–658. doi: 10.1128/JB.188.2.642-658.2006
Geissinger, O., Herlemann, D. P., Mörschel, E., Maier, U. G., and Brune, A. (2009). The ultramicrobacterium “Elusimicrobium minutum” gen. nov., sp. nov., the first cultivated representative of the termite group 1 phylum. Appl. Environ. Microbiol. 75, 2831–2840. doi: 10.1128/AEM.02697-08
Goopy, J. P., Hegarty, R. S., and Dobos, R. C. (2006). The persistence over time of divergent methane production in lot fed cattle. Int. Congr. Ser. 1293, 111–114. doi: 10.1016/j.ics.2006.02.019
Henderson, G., Cox, F., Kittelmann, S., Miri, V. H., Zethof, M., Noel, S. J., et al. (2013). Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One 8:e74787. doi: 10.1371/journal.pone.0074787
Hook, S. E., Wright, A. -D. G., and McBride, B. W. (2010). Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010, 1–11. doi: 10.1155/2010/945785
Horn, S. J., Estevez, M. M., Nielsen, H. K., Linjordet, R., and Eijsink, V. G. H. (2011). Biogas production and saccharification of Salix pretreated at different steam explosion conditions. Bioresour. Technol. 102, 7932–7936. doi: 10.1016/j.biortech.2011.06.042
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jami, E., and Mizrahi, I. (2012). Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7:e33306. doi: 10.1371/journal.pone.0033306
Janssen, P. H., and Kirs, M. (2008). Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74, 3619–3625. doi: 10.1128/AEM.02812-07
Jin, W., Chen, L., Hu, M., Sun, D., Li, A., Li, Y., et al. (2016). Tween-80 is effective for enhancing steam-exploded biomass enzymatic saccharification and ethanol production by specifically lessening cellulase absorption with lignin in common reed. Appl. Energy 175, 82–90. doi: 10.1016/j.apenergy.2016.04.104
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Kudo, H., Cheng, K. -J., and Costerton, J. (1987). Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can. J. Microbiol. 33, 244–248. doi: 10.1139/m87-041
La Reau, A. J., and Suen, G. (2018). The ruminococci: key symbionts of the gut ecosystem. J. Microbiol. 56, 199–208. doi: 10.1007/s12275-018-8024-4
Leschine, S. B. (1995). Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 49, 399–426. doi: 10.1146/annurev.mi.49.100195.002151
Lizasoain, J., Trulea, A., Gittinger, J., Kral, I., Piringer, G., Schedl, A., et al. (2017). Corn stover for biogas production: effect of steam explosion pretreatment on the gas yields and on the biodegradation kinetics of the primary structural compounds. Bioresour. Technol. 244, 949–956. doi: 10.1016/j.biortech.2017.08.042
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Malinovsky, F. G., Fangel, J. U., and Willats, W. G. T. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5:178. doi: 10.3389/fpls.2014.00178
Méheust, R., Castelle, C. J., Carnevali, P. B. M., Farag, I. F., He, C., Chen, L. -X., et al. (2019). Aquatic Elusimicrobia are metabolically diverse compared to gut microbiome Elusimicrobia and some have novel nitrogenase-like gene clusters. bioRxiv [preprint]. doi: 10.1101/765248.
Menke, K. H. (1988). Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Dev. 28, 7–55.
Moissl-Eichinger, C., Pausan, M., Taffner, J., Berg, G., Bang, C., and Schmitz, R. A. (2018). Archaea are interactive components of complex microbiomes. Trends Microbiol. 26, 70–85. doi: 10.1016/j.tim.2017.07.004
Moss, A. R., Jouany, J. P., and Newbold, J. (2000). Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49, 231–253. doi: 10.1051/animres:2000119
Mulat, D. G., Huerta, S. G., Kalyani, D., and Horn, S. J. (2018). Enhancing methane production from lignocellulosic biomass by combined steam-explosion pretreatment and bioaugmentation with cellulolytic bacterium Caldicellulosiruptor bescii. Biotechnol. Biofuels 11:19. doi: 10.1186/s13068-018-1025-z
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Paster, B. J., and Canale-Parola, E. (1982). Physiological diversity of rumen spirochetes. Appl. Environ. Microbiol. 43, 686–693. doi: 10.1128/AEM.43.3.686-693.1982
Penner, G. B., and Oba, M. (2009). Increasing dietary sugar concentration may improve dry matter intake, ruminal fermentation, and productivity of dairy cows in the postpartum phase of the transition period. J. Dairy Sci. 92, 3341–3353. doi: 10.3168/jds.2008-1977
Pinares-Patino, C. S., Ulyatt, M. J., Waghorn, G. C., Lassey, K. R., Barry, T. N., Holmes, C. W., et al. (2003). Methane emission by alpaca and sheep fed on lucerne hay or grazed on pastures of perennial ryegrass/white clover or birdsfoot trefoil. J. Agric. Sci. 140, 215–226. doi: 10.1017/S002185960300306X
Ransom-Jones, E., Jones, D. L., Edwards, A., and McDonald, J. E. (2014). Distribution and diversity of members of the bacterial phylum fibrobacteres in environments where cellulose degradation occurs. Syst. Appl. Microbiol. 37, 502–509. doi: 10.1016/j.syapm.2014.06.001
R Core Team (2013). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available at: http://www.R-project.org/
Scheifinger, C. C., and Wolin, M. J. (1973). Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 26, 789–795. doi: 10.1128/AEM.26.5.789-795.1973
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Schroyen, M., Vervaeren, H., Vandepitte, H., Van Hulle, S. W. H., and Raes, K. (2015). Effect of enzymatic pretreatment of various lignocellulosic substrates on production of phenolic compounds and biomethane potential. Bioresour. Technol. 192, 696–702. doi: 10.1016/j.biortech.2015.06.051
Schumacher, B., Wedwitschka, H., Hofmann, J., Denysenko, V., Lorenz, H., and Liebetrau, J. (2014). Disintegration in the biogas sector--technologies and effects. Bioresour. Technol. 168, 2–6. doi: 10.1016/j.biortech.2014.02.027
Shi, W., Moon, C. D., Leahy, S. C., Kang, D., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Song, S. D., Chen, G. J., Guo, C. H., Rao, K. Q., Gao, Y. H., Peng, Z. L., et al. (2018). Effects of exogenous fibrolytic enzyme supplementation to diets with different NFC/NDF ratios on the growth performance, nutrient digestibility and ruminal fermentation in Chinese domesticated black goats. Anim. Feed Sci. Technol. 236, 170–177. doi: 10.1016/j.anifeedsci.2017.12.008
Stanton, T., and Canale-Parola, E. (1980). Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 127, 145–156. doi: 10.1007/BF00428018
Strobel, H. J. (1992). Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl. Environ. Microbiol. 58, 2331–2333. doi: 10.1128/AEM.58.7.2331-2333.1992
Ungerfeld, E. M. (2015). Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: a meta-analysis. Front. Microbiol. 6:37. doi: 10.3389/fmicb.2015.00037
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Veneman, J. B., Muetzel, S., Hart, K. J., Faulkner, C. L., Moorby, J. M., Perdok, H. B., et al. (2015). Does dietary mitigation of enteric methane production affect rumen function and animal productivity in dairy cows? PLoS One 10:e0140282. doi: 10.1371/journal.pone.0140282
Vivekanand, V., Olsen, E. F., Eijsink, V. G., and Horn, S. J. (2013). Effect of different steam explosion conditions on methane potential and enzymatic saccharification of birch. Bioresour. Technol. 127, 343–349. doi: 10.1016/j.biortech.2012.09.118
Wallace, R. J., Rooke, J. A., Duthie, C. A., Hyslop, J. J., Ross, D. W., McKain, N., et al. (2014). Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci. Rep. 4:5892. doi: 10.1038/srep05892
Wallace, R. J., Rooke, J. A., McKain, N., Duthie, C. A., Hyslop, J. J., Ross, D. W., et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0
Wang, K., Nan, X., Chu, K., Tong, J., Yang, L., Zheng, S., et al. (2018). Shifts of hydrogen metabolism from methanogenesis to propionate production in response to replacement of forage fiber with non-forage fiber sources in diets in vitro. Front. Microbiol. 9:2764. doi: 10.3389/fmicb.2018.02764
Wolin, M., and Miller, T. (1997). “Microbe–microbe interactions” in The rumen microbial ecosystem. eds. P. N. Hobson and C. S. Stewart (London: Chapman & Hall), 467–491.
Yu, H., Wei, X., Zhou, S., Liu, M., Tu, Y., Ao, L., et al. (2015). Steam explosion distinctively enhances biomass enzymatic saccharification of cotton stalks by largely reducing cellulose polymerization degree in G. barbadense and G. hirsutum. Bioresour. Technol. 181, 224–230. doi: 10.1016/j.biortech.2015.01.020
Yue, Z. -B., Li, W. -W., and Yu, H. -Q. (2013). Application of rumen microorganisms for anaerobic bioconversion of lignocellulosic biomass. Bioresour. Technol. 128, 738–744. doi: 10.1016/j.biortech.2012.11.073
Zhang, J., Lv, C., Tong, J., Liu, J., Liu, J., Yu, D., et al. (2016). Optimization and microbial community analysis of anaerobic co-digestion of food waste and sewage sludge based on microwave pretreatment. Bioresour. Technol. 200, 253–261. doi: 10.1016/j.biortech.2015.10.037
Zhao, S., Li, G., Zheng, N., Wang, J., and Yu, Z. (2018). Steam explosion enhances digestibility and fermentation of corn stover by facilitating ruminal microbial colonization. Bioresour. Technol. 253, 244–251. doi: 10.1016/j.biortech.2018.01.024
Zheng, H., Dietrich, C., Radek, R., and Brune, A. (2016). Endomicrobium proavitum, the first isolate of E ndomicrobia class. nov.(phylum Elusimicrobia)–an ultramicrobacterium with an unusual cell cycle that fixes nitrogen with a G roup IV nitrogenase. Environ. Microbiol. 18, 191–204. doi: 10.1111/1462-2920.12960
Zhou, M., Chung, Y. H., Beauchemin, K. A., Holtshausen, L., Oba, M., McAllister, T. A., et al. (2011). Relationship between rumen methanogens and methane production in dairy cows fed diets supplemented with a feed enzyme additive. J. Appl. Microbiol. 111, 1148–1158. doi: 10.1111/j.1365-2672.2011.05126.x
Keywords: steam explosion, corn stover, ruminal fermentation in vitro , archaeal community, bacterial community
Citation: Wang K, Nan X, Tong J, Zhao G, Jiang L and Xiong B (2020) Steam Explosion Pretreatment Changes Ruminal Fermentation in vitro of Corn Stover by Shifting Archaeal and Bacterial Community Structure. Front. Microbiol. 11:2027. doi: 10.3389/fmicb.2020.02027
Received: 18 January 2020; Accepted: 31 July 2020;
Published: 28 August 2020.
Edited by:
Antonio Faciola, University of Florida, United StatesReviewed by:
Todd Riley Callaway, University of Georgia, United StatesCopyright © 2020 Wang, Nan, Tong, Zhao, Jiang and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linshu Jiang, amxzQGJhYy5lZHUuY24=; Benhai Xiong, eGlvbmdiZW5oYWlAY2Fhcy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.