- 1Department of Microbiology, Tokushima University Graduate School of Medical Science, Tokushima, Japan
- 2Department of Microbiology, Kansai Medical University, Osaka, Japan
Human coronaviruses (HCoVs) are of zoonotic origins, and seven distinct HCoVs are currently known to infect humans. While the four seasonal HCoVs appear to be mildly pathogenic and circulate among human populations, the other three designated SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause severe diseases in some cases. The newly identified SARS-CoV-2, a causative virus of COVID-19 that can be deadly, is now spreading worldwide much more efficiently than the other two pathogenic viruses. Despite evident differences in these properties, all HCoVs commonly have an exceptionally large genomic RNA with a rather peculiar gene organization and have the potential to readily alter their biological properties. CoVs are characterized by their biological diversifications, high recombination, and efficient adaptive evolution. We are particularly concerned about the high replication and transmission nature of SARS-CoV-2, which may lead to the emergence of more transmissible and/or pathogenic viruses than ever before. Furthermore, novel variant viruses may appear at any time from the CoV pools actively circulating or persistently being maintained in the animal reservoirs, and from the CoVs in infected human individuals. In this review, we describe knowns of the CoVs and then mention their unknowns to clarify the major issues to be addressed. Genome organizations and sequences of numerous CoVs have been determined, and the viruses are presently classified into separate phylogenetic groups. Functional roles in the viral replication cycle in vitro of non-structural and structural proteins are also quite well understood or suggested. In contrast, those in the in vitro and in vivo replication for various accessory proteins encoded by the variable 3′ one-third portion of the CoV genome mostly remain to be determined. Importantly, the genomic sequences/structures closely linked to the high CoV recombination are poorly investigated and elucidated. Also, determinants for adaptation and pathogenicity have not been systematically investigated. We summarize here these research situations. Among conceivable projects, we are especially interested in the underlying molecular mechanism by which the observed CoV diversification is generated. Finally, as virologists, we discuss how we handle the present difficulties and propose possible research directions in the medium or long term.
Introduction
People around the world now have been seeing a global devastating outbreak of COVID-19, caused by a new human coronavirus (HCoV) designated severe acute respiratory syndrome CoV 2 (SARS-CoV-2) (Lu et al., 2020; Wu A. et al., 2020; Zhu et al., 2020). Various CoVs were isolated from mammals and birds, and were long considered to be weakly pathogenic until the identification of SARS-CoV (Drosten et al., 2003; Fouchier et al., 2003; Ksiazek et al., 2003; Zhong et al., 2003) followed by the Middle East respiratory syndrome virus MERS-CoV (Zaki et al., 2012) as a causative virus for serious human infectious disease. Before the three outbreaks, a number of human coronaviruses were discovered and found to be responsible for a seasonally prevalent viral disease with mild symptoms such as the common cold and/or diarrhea (Forni et al., 2017; de Wilde et al., 2018; Cui et al., 2019; Tse et al., 2020; Wang N. et al., 2020; Ye et al., 2020). These include HCoV-NL63 (van der Hoek et al., 2004), HCoV-229E (Hamre and Procknow, 1966), HCoV-OC43 (McIntosh et al., 1967), and HCoV-HKU1 (Woo et al., 2005) in Figure 1. The principal scientific question for virologists and the investigators of other research fields is what makes each CoV or group of CoVs behave so distinctively from the others. Although a large number of excellent articles on the clinical outcomes of COVID-19 and relevant host immune responses have been published very recently (Biswas et al., 2020; Bost et al., 2020; Broggi et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Davies et al., 2020; Giamarellos-Bourboulis et al., 2020; Gordon et al., 2020; Grifoni et al., 2020; Hansen et al., 2020; Ju et al., 2020; Kadkhoda, 2020; Kim D. et al., 2020; Long et al., 2020; McKechnie and Blish, 2020; Oberfeld et al., 2020; Ong et al., 2020; Polycarpou et al., 2020; Robbiani et al., 2020; Shi R. et al., 2020; Subbarao and Mahanty, 2020; Tang D. et al., 2020; Tay et al., 2020; Vabret et al., 2020; Wilk et al., 2020; Xu et al., 2020; Ye et al., 2020; Zhang et al., 2020; Zhou P. et al., 2020; Zhou Z. et al., 2020; Ziegler et al., 2020; Zohar and Alter, 2020), fundamental studies aimed at the above issue have been poorly carried out. Needless to mention, biological and molecular bases for the observed CoV divergence should be elucidated urgently for basic science and clinical applications in the future. As for the origin and evolution of the seven HCoVs (Figure 1) described above, researchers have sufficiently clarified this particular subject by their extensive efforts through field and in silico analyses (Su et al., 2016; Forni et al., 2017; Cui et al., 2019; Adachi et al., 2020; Biswas et al., 2020; Tang D. et al., 2020; Ye et al., 2020). However, mechanistic bases for the adaptive mutations to generate distinct virus groups/lineages/clades are insufficiently elucidated as yet. In summary, we have focused on the baseline studies on the HCoV diversification in this review article by picking up on relevant biological and molecular biological issues from previously published reports. The selected subjects should be experimentally and conclusively analyzed by molecular genetic methods of the day to obtain definitive answers.
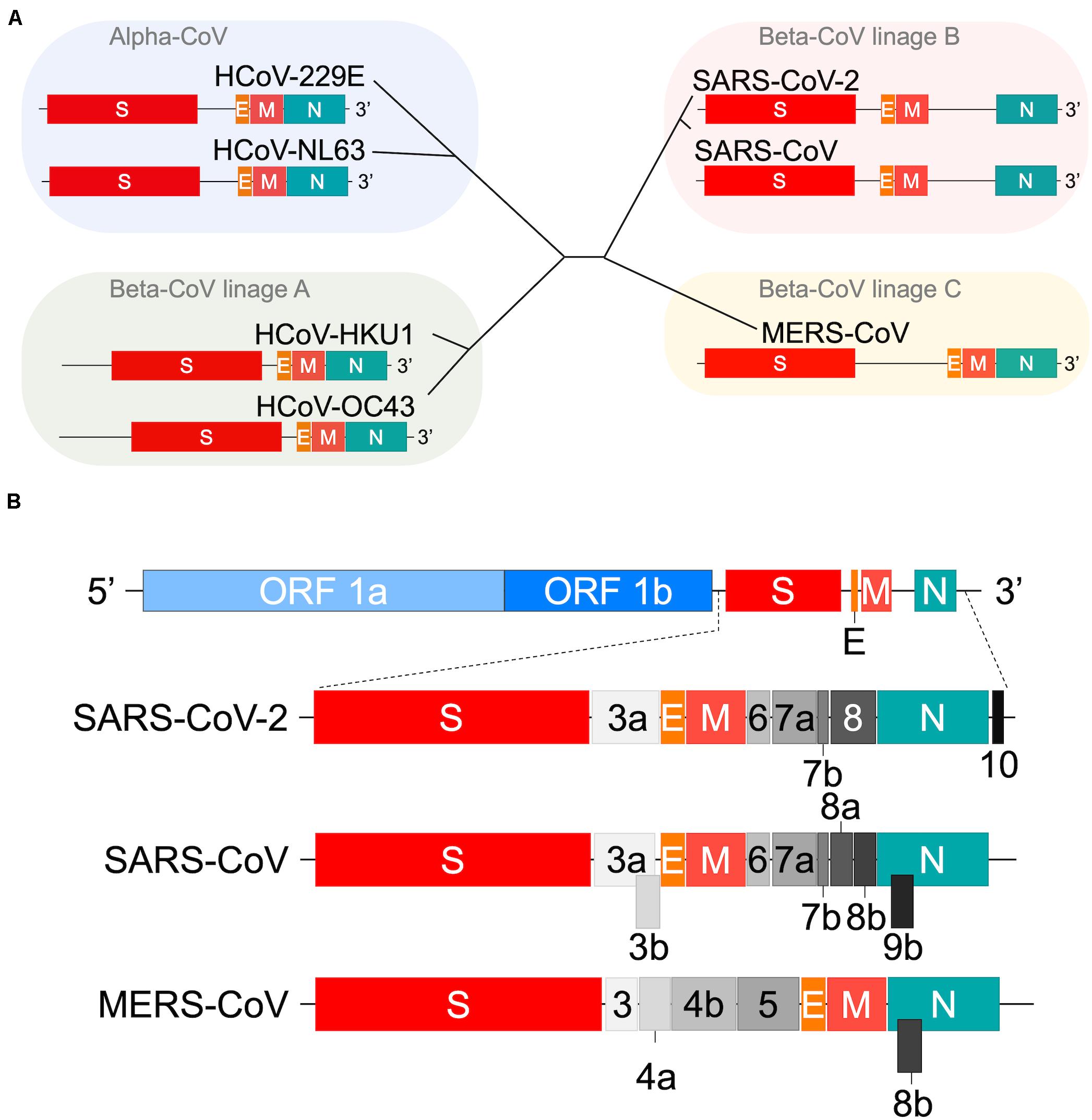
Figure 1. Genome organization of human coronaviruses. (A) Schematic representation of the variable 3′ genomic region. The genome organization of viruses in the four viral groups (Su et al., 2016; Forni et al., 2017) including SARS-CoV-2 (Tse et al., 2020; Wang N. et al., 2020) are shown. Those for various accessory proteins are omitted. The HE orf sequences of HCoV-HKU1 and HCoV-OC43 (Table 3 footnote) are also omitted. The beta-CoV lineage D group that contains some bat viruses only is not presented in this panel. (B) Comparison of SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. All orf sequences in the full-length viral genome are shown (Totura and Baric, 2012; Su et al., 2016; Forni et al., 2017; Cui et al., 2019; Biswas et al., 2020; Hou et al., 2020; Thao et al., 2020; Wang N. et al., 2020; Wu A. et al., 2020; Xie et al., 2020). It has been reported that SARS-CoV-2 contains orf 9b (Cagliani et al., 2020), suggesting a more similar genome structure between SARS-CoV and SARS-CoV-2 than shown in panel B.
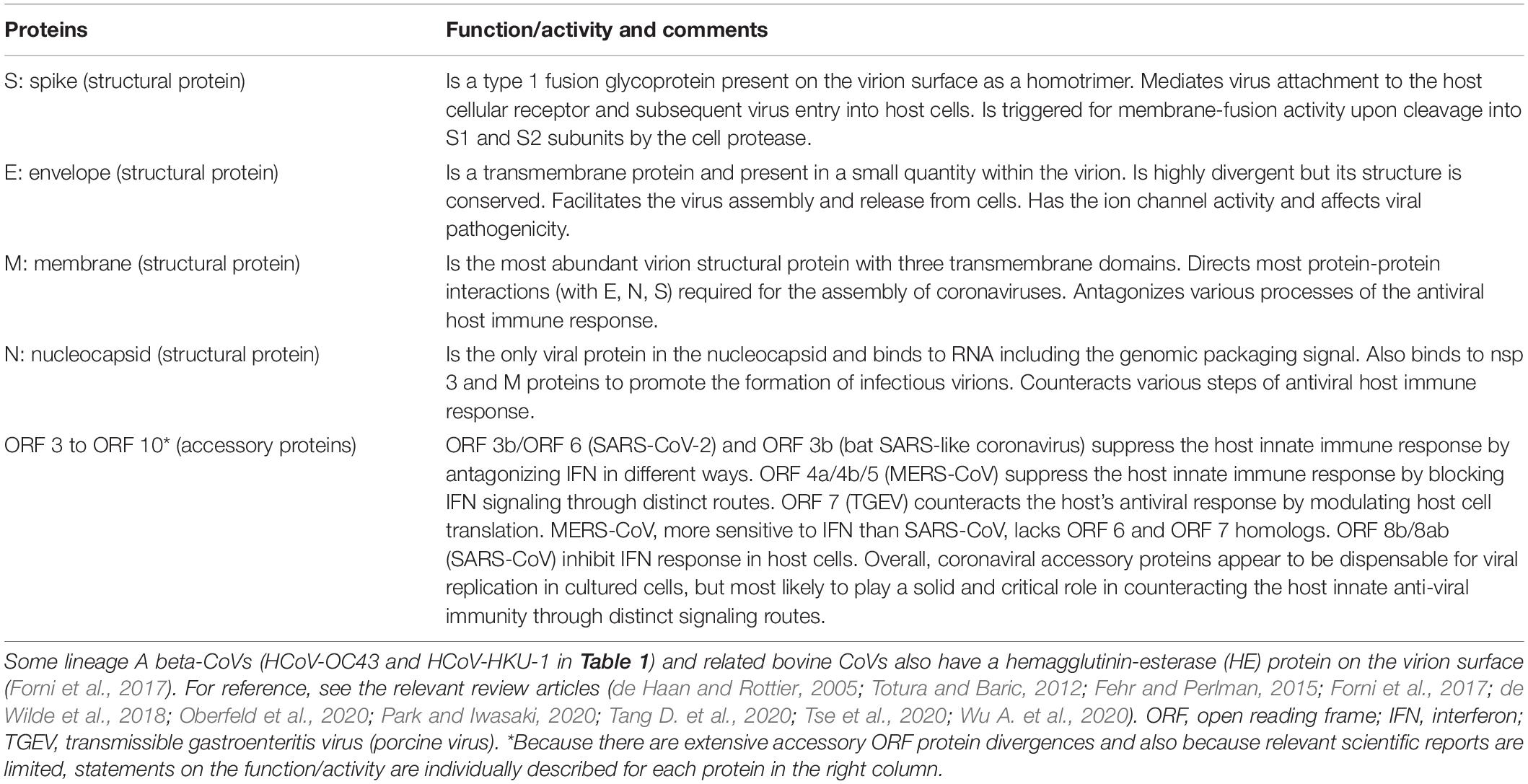
The most prominent feature of CoVs is their exceptionally large genome RNA (∼30 kb) (Fehr and Perlman, 2015; Forni et al., 2017; Cui et al., 2019; Han et al., 2019; Hou et al., 2020; Thao et al., 2020; Wu A. et al., 2020; Xie et al., 2020). Furthermore, it is single-stranded, non-segmented, and polycistronic. While the conserved 5′ two-thirds of the genome encodes a series of non-structural proteins for the replicase-transcriptase complex, the variable 3′ one-third encodes a variety of structural and accessory proteins (Figure 1). Thus, the regulation for well-timed CoV gene expressions should be quite complicated. Another outstanding characteristic of CoVs is their ability for extremely high genomic recombination (Lai, 1992; Nagy and Simon, 1997; Rowe et al., 1997; Lauring et al., 2013; Su et al., 2016; Forni et al., 2017; Cui et al., 2019; Adachi et al., 2020). Among numerous animal virus species, CoVs are known to be first-class for their recombination frequency (Lai, 1992). Probably consistent with this observation, considerably many cis-acting sequences/structures critical for RNA replication and transcription have been identified throughout the coronavirus genome (Fehr and Perlman, 2015). The gene recombination is considered to have dual evolutionary consequences (Simon-Loriere and Holmes, 2011). While it can increase the rate of adaptive evolution by creating advantageous genetic variations, it also can stabilize genomic RNA by generating a functional genome through removing deleterious mutations/deletions. The observed high genomic recombination rates thus confer the plasticity to the CoV genome. Finally, the CoV genome encodes diverse accessory proteins at the variable 3′ one-third portion (Figure 1). These accessory proteins differ in the number and sequence even among the CoVs of the same viral lineage (Forni et al., 2017). They are believed to play a role in suppressing host innate immunity (Cruz et al., 2011; Totura and Baric, 2012; Forni et al., 2017; de Wilde et al., 2018; Cui et al., 2019; Park and Iwasaki, 2020), thus promoting viral adaptation to some specific host species and individuals. Obviously, a precisely organized description of knowns and unknowns about the general picture of HCoV would certainly generate new and significant insights into the corona-virology and shed light on the CoV research today.
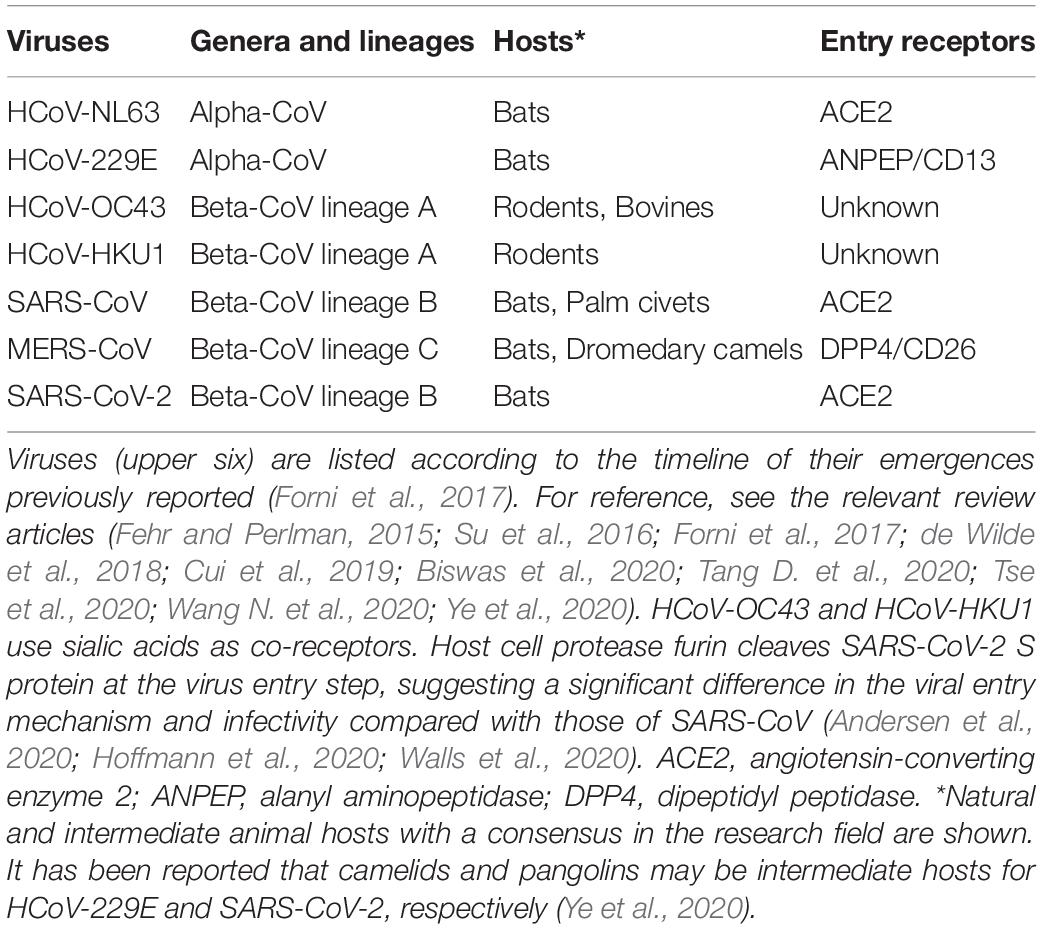
The transmission of CoVs between host species (species tropism) and individuals is a major issue to be addressed. The tissue and cell tropism of the viruses within individuals is critically important as well. In general, viral tropism is determined at the surface of target cells by direct binding of the virus and cellular receptor molecule(s) and/or at the post-entry intracellularly replication step(s) (Nomaguchi et al., 2012a, b). As for pathogenic HCoVs, the primary cellular receptors have been identified as angiotensin-converting enzyme 2 (ACE2) for SARS-CoV (Li et al., 2003), dipeptidyl peptidase 4 (DPP4) for MERS-CoV (Raj et al., 2013), and ACE2 for SARS-CoV-2 (Letko et al., 2020a; Lu et al., 2020; Walls et al., 2020; Wu A. et al., 2020; Zhou P. et al., 2020; Zhu et al., 2020). It has been well-established that ACE2 and DPP4 work for the coronaviral receptors and determinants of the coronavirus tropism (Table 1; Fehr and Perlman, 2015; Forni et al., 2017; de Wilde et al., 2018; Biswas et al., 2020; Letko et al., 2020a; Oberfeld et al., 2020; Tang D. et al., 2020; Tse et al., 2020; Wang N. et al., 2020; Zhou P. et al., 2020). It is unclear as yet on the biological and mechanistic bases by which HCoV-NL63 and SARS-CoV/SARS-CoV-2 of two distinct phylogenetic groups use the same receptor ACE2. Many CoVs utilize peptidases as the cellular receptor, despite that their enzymatic domains are not required for the viral entry process (Fehr and Perlman, 2015). It has been reported that the receptor-specific clustering for viral receptor-binding proteins from the family Coronaviridae is absent (Ng et al., 2020), and that there exist many HCoV receptors other than ACE2 (Fehr and Perlman, 2015; de Wilde et al., 2018; Ye et al., 2020). These reports suggest a complicated evolutionary pathway for HCoVs, which may include the switch to the same receptor on multiple occasions (Li F. et al., 2005; Wu et al., 2009; Song et al., 2018). Further study is necessary to elucidate this biologically important issue. Notably, some co-factors such as cellular proteases and sialic acids are required for efficient CoV entry into cell cytosols for subsequent viral replication (Fehr and Perlman, 2015; Forni et al., 2017; de Wilde et al., 2018; Oberfeld et al., 2020; Tang D. et al., 2020). Whether there is another/other receptor(s) for CoVs remains elusive (Letko et al., 2020a). Also, whether some unknown cell factor(s) restricts the CoV intracellular replication needs to be determined. These cellular factors may influence viral tropism, replication, transmission, pathogenicity, and thus viral ecology.
Based on the above described considerations, in this review article, we describe and discuss: (i) the integrative virology of HCoVs, (ii) reverse genetics systems for human and animal CoVs, and (iii) conclusion: future studies in a demonstrative and perspective manner. In this challenging time, we, as experimental virologists, need to initiate basic HCoV studies to counteract SARS-CoV-2. While focusing on studies on human and simian retroviruses for a long time, we also have significant research experience in many other viruses. Coronaviruses and retroviruses are virologically distinct, but it is quite clear that the principal purpose, main concept, and major research strategy for current virology are commonly shared among basic researchers. We have summarized important scientific issues from the viewpoint of our own. Here, we aim to concentrate on studies in the medium or long term. First, we outline basic factual matters such as grouping viruses based on their ecology/evolution/pathogenicity, genome organization, replication cycle, and functional aspects of individual viral proteins. We then summarize the applications of the reverse genetics system, a powerful tool regularly used in current virology, to CoVs with an extremely large RNA genome to demonstratively analyze all kinds of viral properties. Finally, as a whole, we present basic research directions against coronaviruses severely pathogenic for humans, which would also lead to the establishment of effective anti-viral strategies against possible re-emerging and emerging viruses of various viral species.
Integrative Virology of HCoVs
Classification, Genome Organization, and Basic Properties
Coronavirus is a positive-sense RNA virus [RNA (+) virus] and a member of the family Coronaviridae. All coronaviruses have a highly conserved total genome organization and commonly have a specific open reading frame (ORF) structure (Figure 1). Based on extensive sequence comparisons, coronaviruses are divided into four genera, i.e., alpha-CoV, beta-CoV, gamma-CoV, and delta-CoV (Su et al., 2016; Cui et al., 2019; Wang N. et al., 2020; Ye et al., 2020). HCoVs belong to the alpha-CoV genus, or to the beta-CoV genus constituting a major large phylogenetic group. Beta-CoVs are further classified into lineages A, B, C, and D. As clearly observed in Figure 1A, each virus lineage is readily distinguished by its ORF structure. Close examination of the ORF structure of the viral genome has revealed that each virus has its own organization at the 3′ variable genomic region, whereas no variations are found for the 5′ conserved genomic region (Figure 1B). Also, the ORF structure at the 3′ genomic region appears to vary from strain to strain within a viral group (Forni et al., 2017; Cui et al., 2019). Thus, HCoVs have a unique set of genes of their own.
Currently, seven different HCoVs are known as representatives of each distinctive virus group that infects humans as described above in “Introduction” section. Table 1 lists these HCoVs with some virological information. While certain seasonal HCoVs (HCoV-NL63 and HCoV-229E) belong to alpha-CoV and are of bat origin, others (HCoV-OC43 and HCoV-HKU1) belong to beta-CoV and are of rodent origin. In general, these four viruses appear to be well-adapted to humans and broadly circulate among human populations in some countries in specific seasons (Su et al., 2016). As a cellular receptor, while HCoV-NL63 utilizes ACE2 like SARS-CoV and SARS-CoV-2, HCoV-229E uses alanyl aminopeptidase (ANPEP). Pathogenic SARS-CoV, MERS-CoV, and SARS-CoV-2 are grouped into the lineage B or C, and of bat origin. These three viruses can cause severe diseases in humans and furthermore, COVID-19 by SARS-CoV-2 is prevalent worldwide. Its high transmission rate and incidence are notably evident among the three diseases. However, the fatality of individuals infected with MERS-CoV is significantly higher relative to that of those with SARS-CoV or SARS-CoV-2. Biological and molecular bases for the observed difference between the seasonal and pathogenic HCoVs, and also those among the pathogenic HCoVs must be determined as soon as possible.
Replication in Cells
CoVs utilize numerous proteins encoded by their corresponding genes (Figure 1) for replication. Accordingly, CoVs have a conserved genome structure with a high protein-coding capacity. There are 16 nsp (at most), encoded by ORF 1a and ORF 1b and generated from precursor proteins pp1a and pp1ab, for the viral RNA replication and transcription events (Figure 2). Engagement of the remarkably many proteins in the processes is probably to maintain the replication fidelity. This seems somewhat paradoxical with the highly diverse viral phenotypes observed. However, this mechanism should be essential for CoVs to survive in hostile environments. It may connote a built-in viral strategy to generate a variety of structural and accessory proteins encoded by the 3′ genomic region (Figure 1). CoVs are known to possess a unique proof-reading mechanism by the RNA-dependent RNA polymerase (RdRp) to maintain the integrity of long genomic RNA (Denison et al., 2011; Smith et al., 2013). Indeed, ongoing researches show that the mutation rate of SARS-CoV-2 is not significantly different from those of the other CoVs (Tang X. et al., 2020). The mutation rates of HCoV genomes are estimated to be moderate among those of the other single-stranded RNA virus genomes (Zhao et al., 2004; Pyrc et al., 2006; Cotton et al., 2014; Ren et al., 2015; Su et al., 2016). Therefore, it is not unreasonable to assume that the high recombination capacity of HCoVs is a major cause of the observed HCoV diversification. It deserves noting that hot spots of the high genomic recombination for SARS-CoVs are, in the higher order, S, orf 8, and orf 3 genes (Cui et al., 2019). Remarkably, a novel accessory gene designated orf x has been recently identified between the orf 6 and orf 7 genes in the bat SARS-like CoV genomes (Ge et al., 2013; Yang et al., 2015; Hu et al., 2017; Cui et al., 2019).
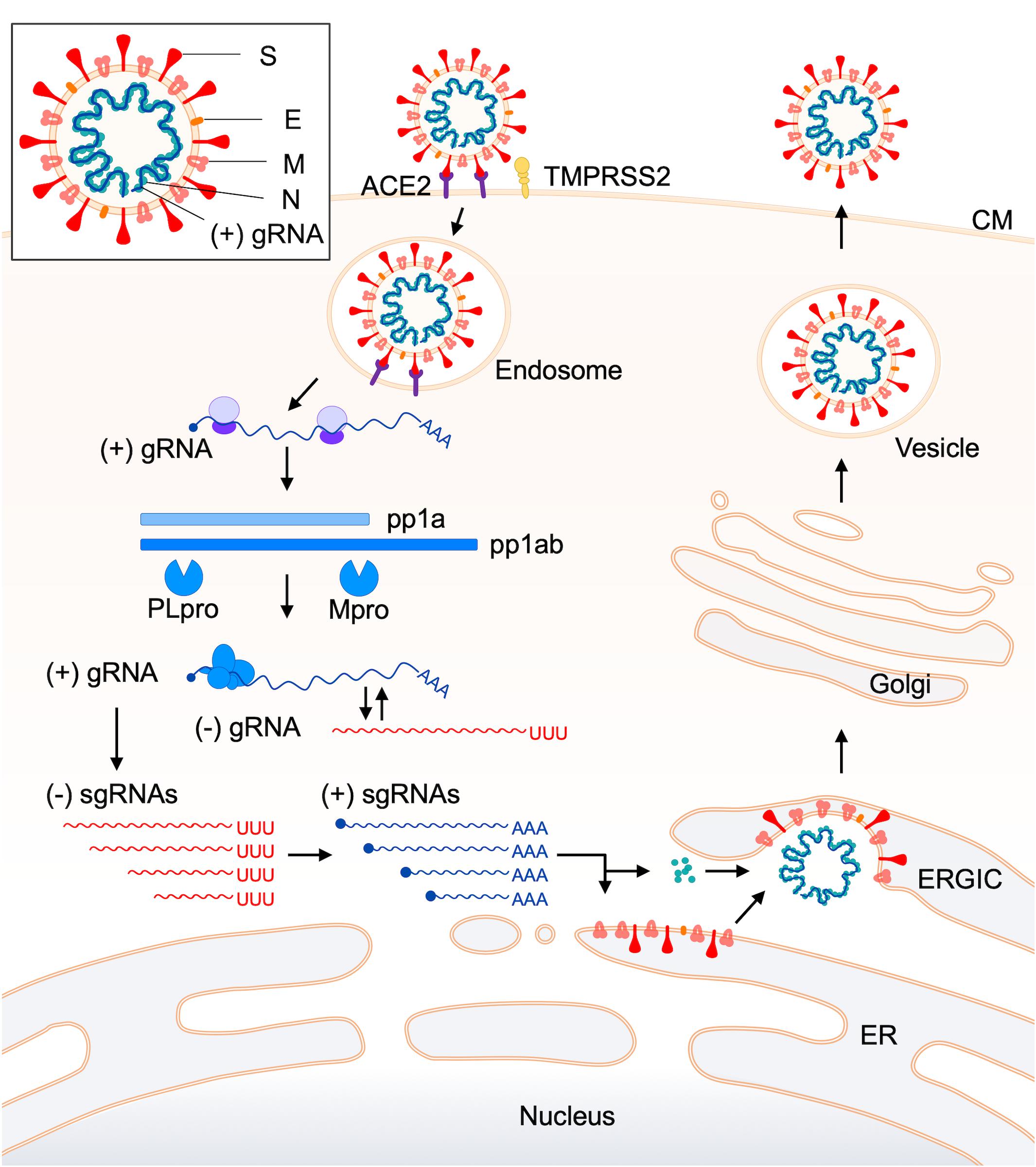
Figure 2. Replication cycle of coronaviruses. The replication process of coronaviruses is schematically shown from the virus attachment to target cells up to the virus release from infected cells (de Haan and Rottier, 2005; Fehr and Perlman, 2015; de Wilde et al., 2018; Oberfeld et al., 2020; Tse et al., 2020). An entry receptor and a co-factor for efficient viral entry (ACE2 and TMPRSS2, respectively, in this case) are indicated. For details of the replication steps, see the text. A schema of the coronavirus virion is also shown in a box at the top-left. Abbreviations (Fehr and Perlman, 2015; de Wilde et al., 2018): ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease, serine 2; CM, cell membrane; gRNA, genomic RNA; pp, polyprotein; PLpro, papain-like protease; Mpro, main protease; sgRNA, subgenomic RNA; ERGIC, endoplasmic reticulum-Golgi intermediate compartment; ER, endoplasmic reticulum.
The viral replication cycle in cells starts with the binding of virions to specific cellular receptors (Table 1) and ends with the release of infectious virions to the extracellular environments (Figure 2). For clarity, here, reported functions and/or activities associated with viral replication in vitro or in vivo are summarized in Table 2 for the non-structural protein (nsp) group (nsp 1–nsp 16) and in Table 3 for structural (S, E, M, and N) and accessory (ORF 3-ORF 10) proteins. CoV replication in cells is schematically outlined in Figure 2 (de Haan and Rottier, 2005; Fehr and Perlman, 2015; de Wilde et al., 2018). At the initial virus-entry step, viral S protein plays a major role in attaching to cells via the receptor. Regarding SARS-CoV, the entry step is as follows. Following virus-receptor binding, S is cleaved into two subunits S1 (receptor-binding domain, RBD) and S2 (fusion domain) by a protease such as TMPRRS2 for efficient virus entry into the cytoplasm. After endocytosis, S is further cleaved by lysosomal proteases for exposing the fusion peptide and leads to the fusion of virus envelope and endosome membrane, finally resulting in the viral RNA (+) spouting into the cytosol. Some lineage A beta-CoVs (HCoV-OC43 and HCoV-HKU1) carry another structural protein, hemagglutinin-esterase (HE), for the binding activity to sialic acids (Tables 1, 3). However, its function was being lost by the accumulation of adaptive mutations, suggesting its non-essential role in the virus entry process (Bakkers et al., 2017). The next major step is the translation and modulation of a series of non-structural proteins (Table 2), followed by the synthesis of viral genomic and various sub-genomic RNAs (van Hemert et al., 2008; Hillen et al., 2020; Romano et al., 2020; Wang Q. et al., 2020). Each sub-genomic RNA serves as mRNA for structural (S, E, M, and N) and accessory (ORF 3–ORF 10) proteins in Table 3. Notably, numerous cis-acting sequences/structures have been recognized in the genome (Fehr and Perlman, 2015). These contain a 5′ leader sequence, untranslated region, transcriptional regulatory sequences at the beginning of each structural and accessory gene, and 3′ untranslated region (Fehr and Perlman, 2015). Next to the viral RNA replication and RNA syntheses, viral structural and accessory proteins are produced (Figure 2). Structural proteins are inserted into the endoplasmic reticulum (ER) and move to the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), where they form mature virions with viral genomic RNA. Subsequently, progeny virions in vesicles are transported to the cell surface and released to the outside environments. Although a general picture of the CoV replication steps is acquired, biological and molecular biological studies in more detail are still required, especially for the accessory proteins (Table 3) and for the functional interactions of various viral proteins. Generally, while the pathogenic CoVs (SARS-CoV, MERS-CoV, and SARS-CoV-2 in Table 1) grow well in cultured cell lines, the seasonal CoVs (HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1 in Table 1) propagate very poorly or negligibly. Also, systemic and comparative studies on the cellular tropism of various HCoVs have not yet been performed. As such, numerous projects at the cellular level remain to be carried out to understand biological and molecular bases for the HCoV virology.
Host Responses and Viral Adaptations
While scientifically confirmed knowledge on viral replication and related issues in vitro underpins the understanding of the complicated nature of pathogenic CoVs, knowing various responses of hosts to the virus infection may be critically important as well to solve the present scientific issues in the laboratories. Because the virus infection process sharply reflects the halfway and final results of viral conflict or interaction with hosts, extensive studies at the levels of the cell, individual, and population are essential. In this regard, a number of articles regarding SARS/MERS (Totura and Baric, 2012; Channappanavar et al., 2016; de Wit et al., 2016; Su et al., 2016; Park and Iwasaki, 2020) and also regarding COVID-19 (Biswas et al., 2020; Bost et al., 2020; Broggi et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Giamarellos-Bourboulis et al., 2020; Gordon et al., 2020; Grifoni et al., 2020; Hansen et al., 2020; Ju et al., 2020; Kadkhoda, 2020; Kim D. et al., 2020; McKechnie and Blish, 2020; Oberfeld et al., 2020; Ong et al., 2020; Polycarpou et al., 2020; Robbiani et al., 2020; Shi R. et al., 2020; Sun J. et al., 2020; Tang D. et al., 2020; Tay et al., 2020; Vabret et al., 2020; Wilk et al., 2020; Xu et al., 2020; Ye et al., 2020; Zhang et al., 2020; Zhou P. et al., 2020; Zhou Z. et al., 2020; Ziegler et al., 2020; Zohar and Alter, 2020) have been published and have provided detailed information on the epidemiology of the diseases, pathophysiological nature of the disease, clinical features of the patients, viral and host factors associated with the infection, the clinical symptoms, host innate immune responses, neutralizing antibodies against viruses, and so on. Scientific dealing with these huge amounts of information is surely a next, important, and challenging step for experimental virologists currently working on SARS-CoV-2.
With the unique genome organization and the genomic sequence characteristics described above, CoVs have a highly flexible potential to mutate in fluxing environments. Generally, adaptive mutations can occur in an amino acid-dependent and/or in a nucleotide-dependent manner to a high degree for RNA viruses. Moreover, drastic alterations of the genome organization such as the gain and loss of genes, which are frequently observed for CoVs (Forni et al., 2017), would give the concerned virus the potential to fiercely change its biological properties. Given the diverse receptor usage (Table 1; Fehr and Perlman, 2015; Forni et al., 2017; de Wilde et al., 2018; Tang D. et al., 2020), high replicative ability (Yount et al., 2000, 2002, 2003; Scobey et al., 2013; Thao et al., 2020; Xie et al., 2020), broad host tropism (Su et al., 2016; Forni et al., 2017; Cui et al., 2019; Tang D. et al., 2020), ongoing inter- and intra-species transmission among animals (Lau et al., 2005; Li et al., 2005a; Woo et al., 2012; Huang et al., 2013; Brook and Dobson, 2015; Zhou et al., 2018; Han et al., 2019; Wang and Anderson, 2019; Letko et al., 2020b), and occasional zoonotic transmission to humans (Table 1; Drosten et al., 2003; Fouchier et al., 2003; Ksiazek et al., 2003; Zhong et al., 2003; Zaki et al., 2012; Lu et al., 2020; Wu A. et al., 2020; Zhu et al., 2020), CoVs are tractable and outstanding targets for experimental studies on the recombination, mutation, adaptation, and evolution. One clue that may be attributed to the adaptations and diversifications of HCoVs is the possible switching of receptor usages. As shown in Table 1, there are a variety of receptors for HCoVs: ACE2, ANPEP/CD13, DPP4/CD26, and maybe some more. Therefore, these types of receptor switchings occur frequently during the evolution of CoVs. Interestingly, S protein, the spike protein on the virion surface that primarily and directly interacts with host cell receptors, contains a recombination hotspot (Hon et al., 2008; Wu et al., 2016; Hu et al., 2017; Cui et al., 2019), which has been positively selected (Lau et al., 2015; Forni et al., 2017). Thus, the high frequency of RNA recombination observed in CoVs (Lai et al., 1985; Keck et al., 1987; Lai, 1992; Nagy and Simon, 1997), possibly through the secondary structure and replication stalling of RNA and the non-processive replicase-driven template switching mechanism (Lai, 1992; Rowe et al., 1997), supports that the recombination at this hotspot is important for adaptations to new host types, and the diversification/evolution of CoVs. Indeed, there is evidence for in vivo recombination between animal and human CoVs (Wu et al., 2003). Also, there are a number of reports that demonstrate the adaptive recombination/mutations in the S gene of HCoV-229E, SARS-CoV, MERS-CoV, and SARS-CoV-2 (Li et al., 2005b; Wong et al., 2017; Letko et al., 2018, 2020a; Gussow et al., 2020), as described above for the HE gene of lineage A beta-CoVs (HCoV-OC43 and HCoV-HKU1). Of note, a growth-enhancing mutation has been found in the S protein of SARS-CoV-2 during this pandemic (Korber et al., 2020). CoV S protein is a key viral factor that primarily shoulders the host cell and species tropism, one of the critical viral properties, and thus, its biologically significant variations would influence much the viral phenotype. Of note here, the structure-based phylogenetic analysis of the receptor-binding S1 domain (Forni et al., 2017; Ng et al., 2020) and also the extensive systemic study on the receptor-usage and infection ability to cell types of different species (Letko et al., 2020a) have suggested the presence of an unknown receptor(s) for HCoVs. The current seven species of HCoVs are most likely to have appeared through evolution via multiple complicated receptor switching events. Totally and historically, CoV adaptive determinants for tropism, replication ability and/or pathogenicity have not yet been systemically analyzed except for S protein. In particular, the possible involvement of the HCoV accessory proteins in these fundamental virus properties is poorly studied in a demonstrative manner so far. Extensive studies in this direction need to be urgently carried out.
Reverse Genetics Technology for Animal and Human Coronaviruses
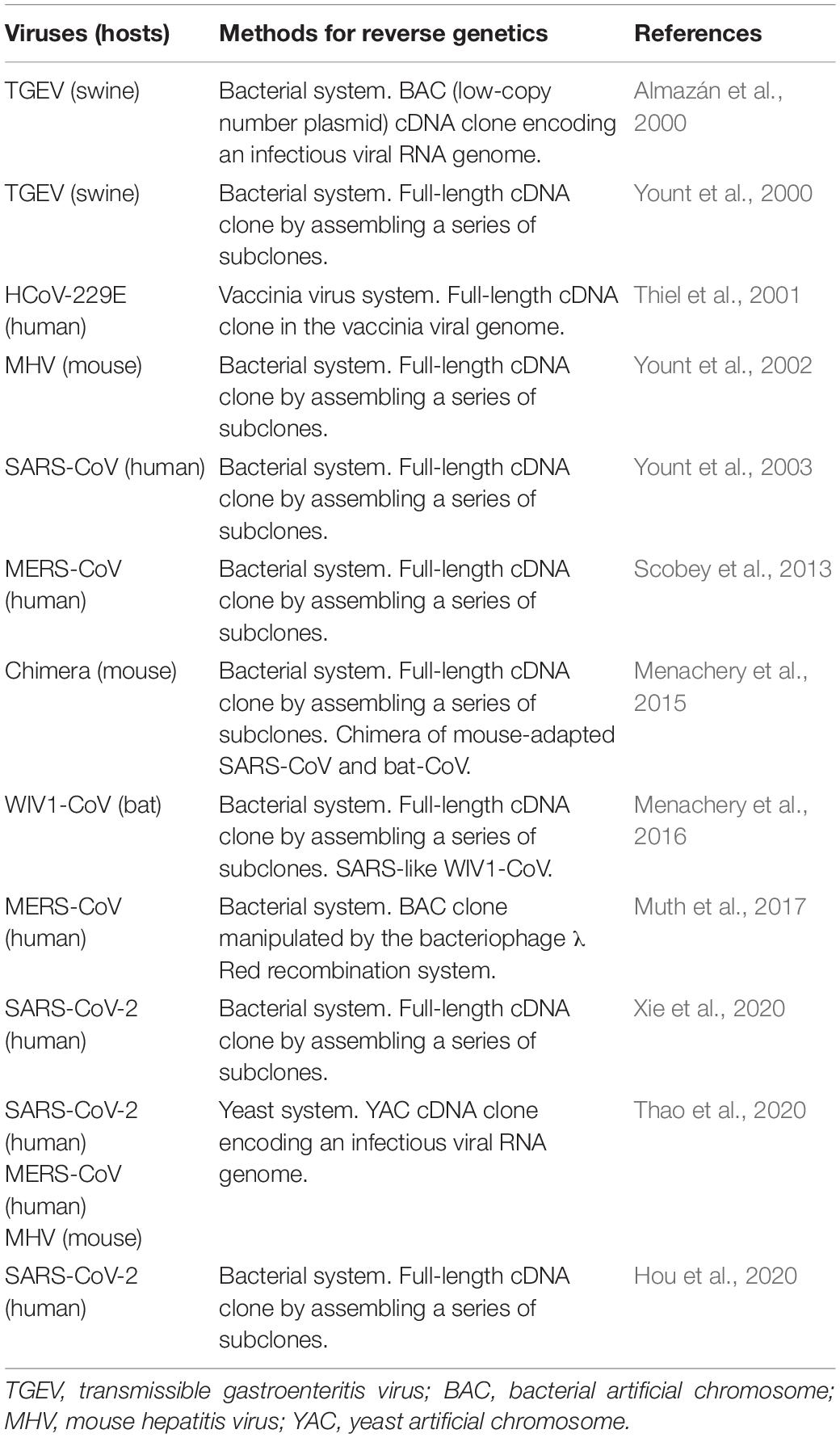
The most frequently and widely utilized experimental system for analytical studies on human/animal viruses would be the reverse genetics. To study biology and molecular biology of viruses, reverse genetic systems are almost prerequisite methods in the current virology. With the aid of the genetic system, we can readily perform a series of mutational functional studies on any coding or non-coding regions of any genes, expecting to have reproducible experimental data on a solid basis. We can apply it, other than the orthodox functional studies, to a wide variety of research projects such as those on the effects of spontaneously occurring natural variations, adaptive mutations in relation to virus evolution, interactions of multiple viruses, prediction of viral drug/vaccine resistance, and so on. The reverse genetics is the most powerful and superior method in today’s virology. However, mostly due to their extraordinarily large RNA genomes, it was quite difficult to establish valid reverse genetics systems for CoVs. It is hard to stably maintain such a long genome in the DNA vectors in the microbes for genetics, and sometimes the cloned DNAs contain some toxic sequences to the microbes concerned. In addition, in most cases, cloned DNAs need to be transcribed in vitro into RNAs for RNA (+) viruses like CoVs before experimental use (for RNA transfection). Researchers thus have come up with various resources to make the methodology easy to use (Almazán et al., 2014). As listed in Table 4, the reverse genetics methods for various CoVs are technically divided into four types: BAC (bacterial artificial chromosome), YAC (yeast artificial chromosome), a large DNA virus vaccinia, and plasmid assembly systems. Outlines of the methods to produce genetically engineered CoVs by RNA transfection are schematically presented in Figure 3.
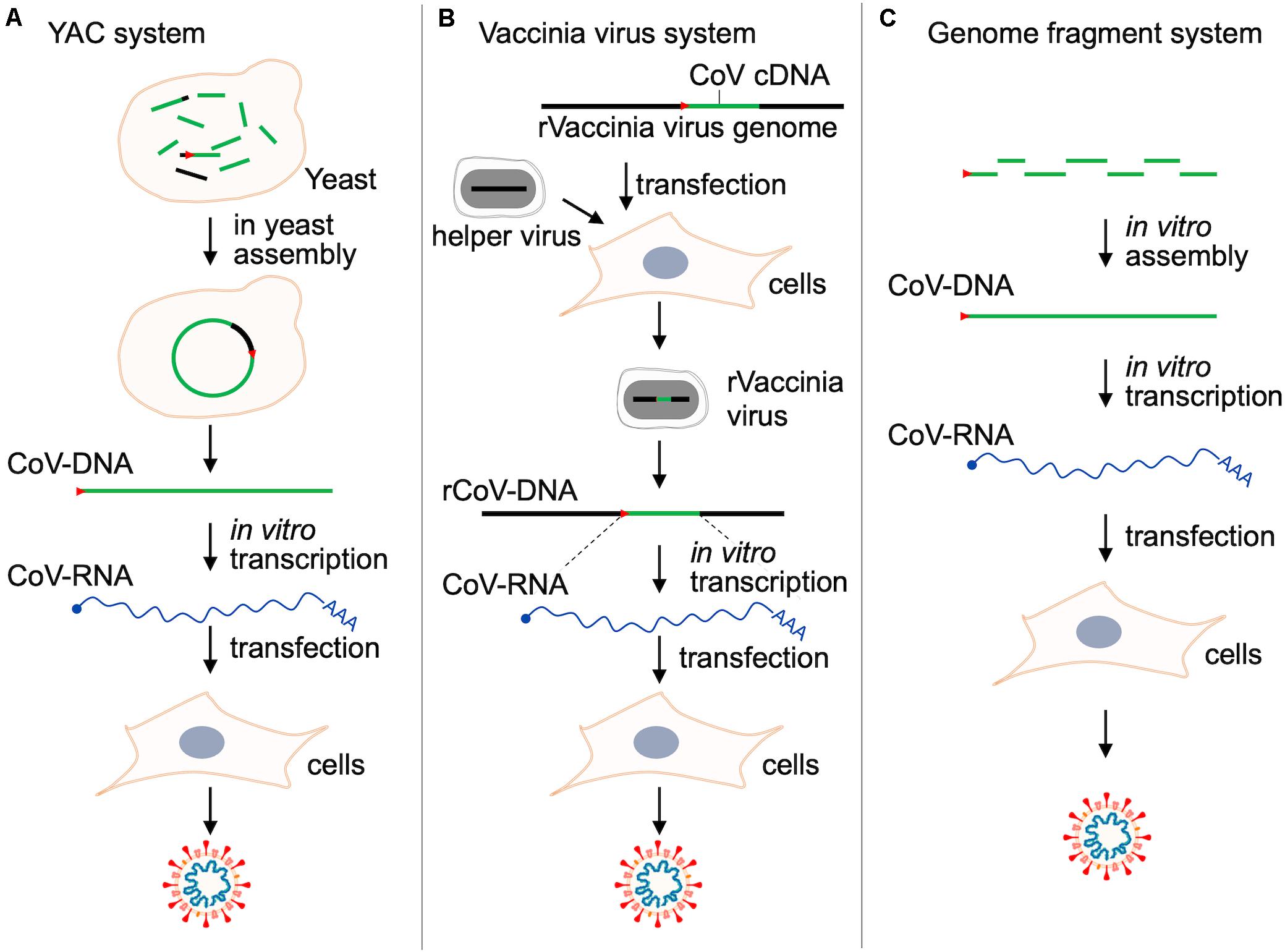
Figure 3. Reverse genetics systems for studies on CoVs. Outlines of the three major methods to produce CoVs by RNA transfection are shown. For details, see Thao et al. (2020) for panel (A), Thiel et al. (2001) for panel (B), and Hou et al. (2020); Xie et al. (2020) for panel (C). The BAC system (Table 4) is essentially quite similar with that in panel (A), but a complete full-length CoV-DNA must be constructed in vitro as BAC before transformation into bacteria and the following preparation of plasmid DNA (Almazán et al., 2000). Red arrow heads in this figure indicate the T7 promoter sequence. r, recombinant.
We have summarized these systems chronologically in Table 4. In a pioneer study, a low-copy number BAC cDNA clone encoding a full-length viral RNA genome was used to generate the infectious transmissible gastroenteritis virus (TGEV) (Almazán et al., 2000). This paper successfully demonstrated that viral tropism and virulence can be modified in vitro. Then, Baric and his team generated an infectious TGEV construct (Yount et al., 2000). They initiated and developed a novel technology by which a complete clone was made through assembling several subclones in order. They successively and successfully generated full-length infectious clones of mouse hepatitis virus (MHV) (Yount et al., 2002), SARS-CoV (Yount et al., 2003), MERS-CoV (Scobey et al., 2013), mouse-adapted SARS-CoV (Menachery et al., 2015), bat-derived WIV1-CoV (Menachery et al., 2016), and finally SARS-CoV-2 (Hou et al., 2020). Furthermore, the methods other than those described above were proposed for genetic studies on CoVs. These are a vaccinia-based and a new BAC system for HCoV-229E (Thiel et al., 2001) and MERS-CoV (Muth et al., 2017), respectively. In 2020, complying with researchers’ expectations and strong requests, two reverse genetics methodologies for SARS-CoV-2, i.e., the assembly (Hou et al., 2020; Xie et al., 2020) and the YAC (Thao et al., 2020) systems, have been reported. The authors of the three articles (Table 4) have demonstrated that their systems are useful for various functional analyses on SARS-CoV-2. Of note, both systems can be or were applied to the other CoVs such as MHV, SARS-CoV, MERS-CoV, HCoV-229E, and HCoV-HKU1. Also, full-length constructs carrying a marker reporter gene have been constructed for MHV, MERS-CoV, SARS-CoV, and SARS-CoV-2 (Hou et al., 2020; Xie et al., 2020; Thao et al., 2020). Importantly and notably, it has been successfully demonstrated that the specific infectivity of SARS-CoV-2 for the respiratory tract region is determined in an ACE2-dependent manner (Hou et al., 2020). Furthermore, it has been shown that ciliated airway cells and AT-2 cells (type II pneumocytes, constituents of adult alveolar epithelium) are the primary targets for SARS-CoV-2 (Hou et al., 2020). Researchers can follow one of these or all of them as a powerful analyzing tool for SARS-CoV-2 biology and molecular biology, depending on their scientific preference and/or experience. By using the experimental systems described above (Figure 3), researchers will be able to perform a systemic analysis on SARS-CoV-2 and related issues in a demonstrative manner. Besides studies on highly pathogenic HCoVs, comparative functional analyses by the reverse genetics system using seasonal HCoVs and animal CoVs closely related to HCoVs may be important to systematically understand the biology of HCoVs. The comparative molecular virology is a legitimate approach of experimental virology today. Coupled with in vitro laboratory and in vivo animal experiments, the reverse genetics system (Figure 3) would prove a real worth (Totura and Baric, 2012; Almazán et al., 2014; Bakkers et al., 2017; Wong et al., 2017; Letko et al., 2018, 2020a; Bao et al., 2020; Chandrashekar et al., 2020; de Wit et al., 2020; Hou et al., 2020; Jiang et al., 2020; Lakdawala and Menachery, 2020; Shi J. et al., 2020; Sun S.-H. et al., 2020; Thao et al., 2020; Williamson et al., 2020; Xie et al., 2020; Yu et al., 2020).
Conclusion: Future Studies in a Demonstrative and Perspective Manner
In the present context that so many review articles on CoVs are being published, we emphasize our review as containing new concepts and viewpoints regarding the corona-virology based on the CoV unique and critical properties so far reported. We do care about the diversification of coronaviruses and care for the underlying molecular mechanisms by which the highly diverse phenotypes of CoVs are created. Needless to mention, the principal cause lies in the CoV genome itself. Its unusually long RNA genome and complicatedly regulated expression system certainly constitute a foundation for ever-changing appearance of CoVs. In addition, plenty of circumstantial evidence fully indicates that the environmental factors strongly assist its diversifying potential. Thus, we need to eagerly engage or be interested in both of the laboratory/clinical research and the fieldwork (Adachi et al., 2020).
On the basis of the fundamental studies consistently continued by the CoV investigators and the stimulating and thought-provoking experience in the recent three CoV outbreaks, researchers must prepare for the future in the medium or long term. We long have been involved in molecular genetic studies of human and simian immunodeficiency viruses (HIV/SIVs) as described above. Despite considerable differences in their biological and molecular biological properties, research concepts, strategies, and tools are common between the two virus species, CoV and HIV/SIV (Adachi, 2020). More than anything, we have to handle the global disease-causing nature of the two viruses, SARS-CoV-2 and HIV-1. We, as experimental virologists, analyze the CoVs in a solidly demonstrative and perspective manner by utilizing the reverse genetics methodology and related technology (Table 4 and Figure 3) as exactly is the case for studies on HIV/SIVs, and use the various animal model systems available (Bao et al., 2020; Chandrashekar et al., 2020; de Wit et al., 2020; Jiang et al., 2020; Lakdawala and Menachery, 2020; Shi J. et al., 2020; Sun S.-H. et al., 2020; Williamson et al., 2020; Yu et al., 2020) when necessary as a part of experimental virology. Having a bird’s-eye view is essential for studies on viruses of this kind. Finally, because numerous papers on COVID-19/SARS-CoV-2 have been published on a day-by-day basis in 2020, we sort out the latest publications (original, review, and other types of articles so far published in 2020, as of June), and cite them here in parentheses according to the four categories below. These articles are generally important for CoV research and worth referencing from our standpoint. Although drugs, neutralizing antibodies, and vaccines against SARS-CoV-2 and related viruses are urgently important, of course, in this challenging time, we do not take up the issue in this review. Refer to the articles below, instead.
Drugs, Neutralizing Antibodies and Vaccines
(Baum et al., 2020; Brouwer et al., 2020; Burton and Walker, 2020; Dai et al., 2020; de Wit et al., 2020; Diamond and Pierson, 2020; Gordon et al., 2020; Hansen et al., 2020; Hassan et al., 2020; Jin et al., 2020a, b; Kim E. et al., 2020; Rogers et al., 2020; Tang D. et al., 2020; Tse et al., 2020; Wang N. et al., 2020; Wec et al., 2020; Williamson et al., 2020; Wu Y. et al., 2020; Yu et al., 2020; Zhou and Zhao, 2020).
Host Responses to Infection and Clinical Outcomes
(Biswas et al., 2020; Bost et al., 2020; Broggi et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Davies et al., 2020; Giamarellos-Bourboulis et al., 2020; Gordon et al., 2020; Grifoni et al., 2020; Hansen et al., 2020; Ju et al., 2020; Kadkhoda, 2020; Kim D. et al., 2020; Long et al., 2020; McKechnie and Blish, 2020; Oberfeld et al., 2020; Ong et al., 2020; Park and Iwasaki, 2020; Polycarpou et al., 2020; Robbiani et al., 2020; Shi R. et al., 2020; Subbarao and Mahanty, 2020; Tang D. et al., 2020; Tay et al., 2020; Vabret et al., 2020; Wilk et al., 2020; Xu et al., 2020; Ye et al., 2020; Zhang et al., 2020; Zhou P. et al., 2020; Zhou Z. et al., 2020; Ziegler et al., 2020; Zohar and Alter, 2020).
Viral Basic Properties, Adaptations, and Diversifications
(Andersen et al., 2020; Baum et al., 2020; Biswas et al., 2020; Chan et al., 2020; Gussow et al., 2020; Hillen et al., 2020; Hoffmann et al., 2020; Hou et al., 2020; Jaimes et al., 2020; Kim D. et al., 2020; Letko et al., 2020a, b; Li et al., 2020; Matsuyama et al., 2020; Ng et al., 2020; Oberfeld et al., 2020; Prather et al., 2020; Qi et al., 2020; Rogers et al., 2020; Romano et al., 2020; Shang et al., 2020; Su et al., 2016; Sun J. et al., 2020; Tang D. et al., 2020; Thao et al., 2020; Walls et al., 2020; Wang Q. et al., 2020; Wec et al., 2020; Wrapp et al., 2020; Wu A. et al., 2020; Xie et al., 2020; Ye et al., 2020; Ziegler et al., 2020).
Host Animals and Animal Experiments
(Bao et al., 2020; Chandrashekar et al., 2020; de Wit et al., 2020; Hansen et al., 2020; Hassan et al., 2020; Jiang et al., 2020; Lakdawala and Menachery, 2020; Letko et al., 2020b; Rogers et al., 2020; Shi J. et al., 2020; Sun S.-H. et al., 2020; Williamson et al., 2020; Yu et al., 2020; Zhou and Zhao, 2020).
Author Contributions
TK, AA, and MN conceived the idea. TK depicted the figures. AA and SA made a draft. TK, ND, and MN reviewed it and discussed its content. AA and MN wrote a final manuscript. All authors approved its submission.
Funding
This research was supported by Japan Agency for Medical Research and Development (AMED) under Grant Number JP20he0822006 and by the FY 2020 supplementary budget from Tokushima Prefecture, Japan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms. Kazuko Yoshida (Tokushima University, Tokushima, Japan) and Ms. Fumie Nishina (Kansai Medical University, Osaka, Japan) for their excellent editorial assistance.
References
Adachi, A. (2020). Grand challenge in human/animal virology: unseen, smallest replicative entities shape the whole globe. Front. Microbiol. 11:431. doi: 10.3389/fmicb.2020.00431
Adachi, S., Koma, T., Doi, N., Nomaguchi, M., and Adachi, A. (2020). Commentary: origin and evolution of pathogenic coronaviruses. Front. Immunol. 11:811. doi: 10.3389/fimmu.2020.00811
Almazán, F., González, J. M., Pénzes, Z., Izeta, A., Calvo, E., Plana-Durán, J., et al. (2000). Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 97, 5516–5521. doi: 10.1073/pnas.97.10.5516
Almazán, F., Sola, I., Zuñiga, S., Marquez-Jurado, S., Morales, L., Becares, M., et al. (2014). Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 189, 262–270. doi: 10.1016/j.virusres.2014.05.02
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., and Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452. doi: 10.1038/s41591-020-0820-9
Bakkers, M. J., Lang, Y., Feitsma, L. J., Hulswit, R. J., de Poot, S. A., van Vliet, A. L., et al. (2017). Betacoronavirus adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe 21, 356–366. doi: 10.1016/j.chom.2017.02.008
Bao, L., Deng, W., Huang, B., Gao, H., Liu, J., Ren, L., et al. (2020). The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:405. doi: 10.1038/s41586-020-2312-y
Baum, A., Fulton, B. O., Wloga, E., Copin, R., Pascal, K. E., Russo, V., et al. (2020). Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. doi: 10.1126/science.abd0831 [Epub ahead of print].
Biswas, A., Bhattacharjee, U., Chakrabarti, A. K., Tewari, D. N., Banu, H., and Dutta, S. (2020). Emergence of novel coronavirus and COVID-19: Whether to stay or die out? Crit. Rev. Microbiol. 46, 182–193. doi: 10.1080/1040841X.2020.1739001
Bost, P., Giladi, A., Liu, Y., Bendjelal, Y., Xu, G., David, E., et al. (2020). Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 181, 1475–1488.e12. doi: 10.1016/j.cell.2020.05.006
Broggi, A., Ghosh, S., Sposito, B., Spreafico, R., Balzarini, F., Antonino Lo Cascio, A. L., et al. (2020). Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. doi: 10.1126/science.abc3545 [Epub ahead of print].
Brook, C. E., and Dobson, A. P. (2015). Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 23, 172–180. doi: 10.1016/j.tim.2014.12.004
Brouwer, P. J. M., Caniels, T. G., van der Straten, K., Snitselaar, J. L., Aldon, Y., Bangaru, S., et al. (2020). Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. doi: 10.1126/science.abc5902 [Epub ahead of print]
Burton, D. R., and Walker, L. M. (2020). Rational vaccine design in the time of COVID-19. Cell Host Microbe 27, 695–698. doi: 10.1016/j.chom.2020.04.022
Cagliani, R., Forni, D., Clerici, M., and Sironi, M. (2020). Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 83:104353. doi: 10.1016/j.meegid.2020.104353
Cao, Y., Su, B., Guo, X., Sun, W., Deng, Y., and Bao, L. (2020). Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. doi: 10.1016/j.cell.2020.05.025 [Epub ahead of print].
Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., et al. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9, 221–236. doi: 10.1080/22221751.2020.1719902
Chandrashekar, A., Liu, J., Martinot, A. J., McMahan, K., Mercado, N. B., Peter, L., et al. (2020). SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science eabc4776. doi: 10.1126/science.abc4776 [Epub ahead of print].
Channappanavar, R., Fehr, A. R., Vijay, R., Mack, M., Zhao, J., Meyerholz, D. K., et al. (2016). Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193. doi: 10.1016/j.chom.2016.01.007
Cotton, M., Watson, S. J., Zumla, A. I., Makhdoom, H. Q., Palser, A. L., Ong, S. H., et al. (2014). Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5:e1062-13. doi: 10.1128/mBio.01062-13
Cruz, J. L., Sola, I., Becares, M., Alberca, B., Plana, J., Enjuanes, L., et al. (2011). Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 7:e1002090. doi: 10.1371/journal.ppat.1002090
Cui, J., Li, F., and Shi, Z.-L. (2019). Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192. doi: 10.1038/s41579-018-0118-9
Dai, W., Zhang, B., Su, H., Li, J., Zhao, Y., Xie, X., et al. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335. doi: 10.1126/science.abb4489
Davies, N. G., Klepac, P., Liu, Y., Prem, K., Jit, M., Eggo, R. M., et al. (2020). Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. doi: 10.1038/s41591-020-0962-9 [Epub ahead of print].
de Haan, C. A., and Rottier, P. J. (2005). Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64, 165–230. doi: 10.1016/S0065-3527(05)64006-7
de Wilde, A. H., Snijder, E. J., Kikkert, M., and van Hemert, M. J. (2018). Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 419, 1–42. doi: 10.1007/82_2017_25
de Wit, E., Feldmann, F., Cronin, J., Jordan, R., Okumura, A., Thomas, T., et al. (2020). Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 117, 6771–6776. doi: 10.1073/pnas.1922083117
de Wit, E., van Doremalen, N., Falzarano, D., and Munster, V. J. (2016). SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534. doi: 10.1038/nrmicro.2016.81
Denison, M. R., Graham, R. L., Donaldson, E. F., Eckerle, L. D., and Baric, R. S. (2011). Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 8, 270–279. doi: 10.4161/rna.8.2.15013
Diamond, M. S., and Pierson, T. C. (2020). The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe 27, 699–703. doi: 10.1016/j.chom.2020.04.021
Drosten, C., Günther, S., Preiser, W., van der Werf, S., Brodt, H. R., Becker, S., et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976. doi: 10.1056/NEJMoa030747
Fehr, A. R., and Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23. doi: 10.1007/978-1-4939-2438-7_1
Forni, D., Cagliani, R., Clerici, M., and Sironi, M. (2017). Molecular evolution of human coronavirus genomes. Trends Microbiol. 25, 35–48. doi: 10.1016/j.tim.2016.09.001
Fouchier, R. A., Kuiken, T., Schutten, M., van Amerongen, G., van Doornum, G. J., van den Hoogen, B. G., et al. (2003). Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 423:240. doi: 10.1038/423240a
Ge, X. Y., Li, J. L., Yang, X. L., Chmura, A. A., Zhu, G., Epstein, J. H., et al. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538. doi: 10.1038/nature12711
Giamarellos-Bourboulis, E. J., Netea, M. G., Rovina, N., Akinosoglou, K., Antoniadou, A., Antonakos, N., et al. (2020). Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992–1000.e3. doi: 10.1016/j.chom.2020.04.009
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Grifoni, A., Weiskopf, D., Ramirez, S. I., Mateus, J., Dan, J. M., Moderbacher, C. R., et al. (2020). Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15. doi: 10.1016/j.cell.2020.05.015
Gussow, A. B., Auslander, N., Faure, G., Wolf, Y. I., Zhang, F., and Koonin, E. V. (2020). Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 177, 15193–15199. doi: 10.1073/pnas.2008176117
Hamre, D., and Procknow, J. J. (1966). A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121, 190–193. doi: 10.3181/00379727-121-30734
Han, Y., Du, J., Su, H., Zhang, J., Zhu, G., Zhang, S., et al. (2019). Identification of diverse bat alphacoronaviruses and betacoronaviruses in China provides new insights into the evolution and origin of coronavirus-related diseases. Front. Microbiol. 10:1900. doi: 10.3389/fmicb.2019.01900
Hansen, J., Baum, A., Pascal, K. E., Russo, V., Giordano, S., Wloga, E., et al. (2020). Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science eabd0827. doi: 10.1126/science.abd0827
Hassan, A. O., Case, J. B., Winkler, E. S., Thackray, L. B., Kafai, N. M., Bailey, A. L., et al. (2020). A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. doi: 10.1016/j.cell.2020.06.011 [Epub ahead of print].
Hillen, H. S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D., and Cramer, P. (2020). Structure of replicating SARS-CoV-2 polymerase. Nature 548, 154–156. doi: 10.1038/s41586-020-2368-8
Hoffmann, M., Kleine-Weber, H., and Pöhlmann, S. (2020). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5. doi: 10.1016/j.molcel.2020.04.022
Hon, C. C., Lam, T.-Y., Shi, Z.-L., Drummond, A. J., Yip, C.-W., Zeng, F., et al. (2008). Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 82, 1819–1826. doi: 10.1128/JVI.01926-07
Hou, Y. J., Okuda, K., Edwards, C. E., Martinez, D. R., Asakura, T., Dinnon, K. H. III, et al. (2020). SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446.e14. doi: 10.1016/j.cell.2020.05.042
Hu, B., Zeng, L. P., Yang, X. L., Ge, X. Y., Zhang, W., Li, B., et al. (2017). Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13:e1006698. doi: 10.1371/journal.ppat.1006698
Huang, Y. W., Dickerman, A. W., Piñeyro, P., Li, L., Fang, L., Kiehne, R., et al. (2013). Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4:e00737-13. doi: 10.1128/mBio.00737-13
Jaimes, J. A., Millet, J. K., and Whittaker, G. R. (2020). Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 23:101212. doi: 10.1016/j.isci.2020.101212
Jiang, R.-D., Liu, M.-Q., Chen, Y., Shan, C., Zhou, Y.-W., Shen, X.-R., et al. (2020). Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182, 50-58.e8. doi: 10.1016/j.cell.2020.05.027
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020a). Structure of M (pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. doi: 10.1038/s41586-020-2223-y
Jin, Z., Zhao, Y., Sun, Y., Zhang, B., Wang, H., Wu, Y., et al. (2020b). Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 27, 529–532. doi: 10.1038/s41594-020-0440-6
Ju, B., Zhang, Q., Ge, J., Wang, R., Sun, J., Ge, X., et al. (2020). Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119. doi: 10.1038/s41586-020-2380-z
Kadkhoda, K. (2020). COVID-19: an immunopathological view. mSphere 5:e344-20. doi: 10.1128/mSphere.00344-20
Keck, J. G., Makino, S., Soe, L. H., Fleming, J. O., Stohlman, S. A., and Lai, M. M. C. (1987). RNA recombination of coronavirus. Adv. Exp. Med. Biol. 218, 99–107. doi: 10.1007/978-1-4684-1280-2_11
Kim, D., Lee, J. Y., Yang, J. S., Kim, J. W., Kim, V. N., and Chang, H. (2020). The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e10. doi: 10.1016/j.cell.2020.04.011
Kim, E., Erdos, G., Huang, S., Kenniston, T. W., Balmert, S. C., Carey, C. D., et al. (2020). Microneedle array delivered recombinant coronavirus vaccines: immunogenicity and rapid translational development. EBioMedicine 55:102743. doi: 10.1016/j.ebiom.2020.102743
Korber, B., Fischer, W., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., et al. (2020). Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID19 virus. Cell. doi: 10.1016/j.cell.2020.06.043 [Epub ahead of print].
Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966. doi: 10.1056/NEJMoa030781
Lai, M. M. C. (1992). RNA recombination in animal and plant viruses. Microbiol. Rev. 56, 61–79. doi: 10.1128/mmbr.56.1.61-79.1992
Lai, M. M. C., Baric, R. S., Makino, S., Keck, J. G., Egbert, J., Leibowitz, J. L., et al. (1985). Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 56, 449–456. doi: 10.1128/jvi.56.2.449-456.1985
Lakdawala, S. S., and Menachery, V. D. (2020). The search for a COVID-19 animal model. Science 368, 942–943. doi: 10.1126/science.abc6141
Lau, S. K., Woo, P. C., Li, K. S., Huang, Y., Tsoi, H. W., Wong, B. H., et al. (2005). Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 102, 14040–14045. doi: 10.1073/pnas.0506735102
Lau, S. K. P., Feng, Y., Chen, H., Luk, H. K. H., Yang, W.-H., Li, K. S. M., et al. (2015). Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 89, 10532–10547. doi: 10.1128/jvi.01048-15
Lauring, A. S., Frydman, J., and Andino, R. (2013). The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 11, 327–336. doi: 10.1038/nrmicro3003
Letko, M., Marzi, A., and Munster, V. (2020a). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569. doi: 10.1038/s41564-020-0688-y
Letko, M., Miazgowicz, K., McMinn, R., Seifert, S. N., Sola, I., Enjuanes, L., et al. (2018). Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 24, 1730–1737. doi: 10.1016/j.celrep.2018.07.045
Letko, M., Seifert, S. N., Olival, K. J., Plowright, R. K., and Munster, V. J. (2020b). Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 18, 461–471. doi: 10.1038/s41579-020-0394-z
Li, F., Li, W., Farzan, M., and Harrison, S. C. (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868. doi: 10.1126/science.1116480
Li, M.-Y., Li, L., Zhang, Y., and Wang, X.-S. (2020). Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 9:45. doi: 10.1186/s40249-020-00662-x
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi: 10.1038/nature02145
Li, W., Shi, Z., Yu, M., Ren, W., Smith, C., Epstein, J. H., et al. (2005a). Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679. doi: 10.1126/science.1118391
Li, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., et al. (2005b). Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24, 1634–1643. doi: 10.1038/sj.emboj.7600640
Long, Q. X., Tang, X. J., Shi, Q. L., Li, Q., Deng, H. J., Yuan, J., et al. (2020). Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. doi: 10.1038/s41591-020-0965-6 [Epub ahead of print].
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Matsuyama, S., Nao, N., Shirato, K., Kawase, M., Saito, S., Takayama, I., et al. (2020). Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 117, 7001–7003. doi: 10.1073/pnas.2002589117
McIntosh, K., Becker, W. B., and Chanock, R. M. (1967). Growth in suckling mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U.S.A. 58, 2268–2273. doi: 10.1073/pnas.58.6.2268
McKechnie, J. L., and Blish, C. A. (2020). The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe 27, 863–869. doi: 10.1016/j.chom.2020.05.009
Menachery, V. D., Yount, B. L. Jr., Debbink, K., Agnihothram, S., Gralinski, L. E., Plante, J. A., et al. (2015). A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513. doi: 10.1038/nm.3985
Menachery, V. D., Yount, B. L. Jr., Sims, A. C., Debbink, K., Agnihothram, S. S., Gralinski, L. E., et al. (2016). SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U.S.A. 113, 3048–3053. doi: 10.1073/pnas.1517719113
Muth, D., Meyer, B., Niemeyer, D., Schroeder, S., Osterrieder, N., Müller, M. A., et al. (2017). Transgene expression in the genome of Middle East respiratory syndrome coronavirus based on a novel reverse genetics system utilizing Red-mediated recombination cloning. J. Gen. Virol. 98, 2461–2469. doi: 10.1099/jgv.0.000919
Nagy, P. D., and Simon, A. E. (1997). New insights into the mechanisms of RNA recombination. Virology 235, 1–9. doi: 10.1006/viro.1997.8681
Ng, W. M., Stelfox, A. J., and Bowden, T. A. (2020). Unraveling virus relationships by structure-based phylogenetic classification. Virus Evol. 6:veaa003. doi: 10.1093/ve/veaa003
Nomaguchi, M., Doi, N., Matsumoto, Y., Sakai, Y., Fujiwara, S., and Adachi, A. (2012a). Species tropism of HIV-1 modulated by viral accessory proteins. Front. Microbiol. 3:267. doi: 10.3389/fmicb.2012.00267
Nomaguchi, M., Fujita, M., Miyazaki, Y., and Adachi, A. (2012b). Viral tropism. Front. Microbiol. 3:281. doi: 10.3389/fmicb.2012.00281
Oberfeld, B., Achanta, A., Carpenter, K., Chen, P., Gilette, N. M., Langat, P., et al. (2020). SnapShot: COVID-19. Cell 181, 954–954.e1. doi: 10.1016/j.cell.2020.04.013
Ong, E. Z., Chan, Y. F. Z., Leong, W. Y., Lee, N. M. Y., Kalimuddin, S., Haja Mohideen, S. M., et al. (2020). A dynamic immune response shapes COVID-19 progression. Cell Host Microbe 27, 879–882.e2. doi: 10.1016/j.chom.2020.03.021
Park, A., and Iwasaki, A. (2020). Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 27, 870–878. doi: 10.1016/j.chom.2020.05.008
Polycarpou, A., Howard, M., Farrar, C. A., Greenlaw, R., Fanelli, G., Wallis, R., et al. (2020). Rationale for targeting complement in COVID-19. EMBO Mol. Med. e202012642. doi: 10.15252/emmm.202012642 [Epub ahead of print].
Prather, K. A., Wang, C. C., and Schooley, R. T. (2020). Reducing transmission of SARS-CoV-2. Science 368, 1422–1424. doi: 10.1126/science.abc6197
Pyrc, K., Dijkman, R., Deng, L., Jebbink, M. F., Ross, H. A., Berkhout, B., et al. (2006). Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 364, 964–973. doi: 10.1016/j.jmb.2006.09.074
Qi, F., Shen Qian, S., Zhang, S., and Zheng Zhang, Z. (2020). Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 526, 135–140. doi: 10.1016/j.bbrc.2020.03.044
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254. doi: 10.1038/nature12005
Ren, L., Zhang, Y., Li, J., Xiao, Y., Zhang, J., Wang, Y., et al. (2015). Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 5:11451. doi: 10.1038/srep11451
Robbiani, D. F., Gaebler, C., Muecksch, F., Lorenzi, J. C. C., Wang, Z., Cho, A., et al. (2020). Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. doi: 10.1038/s41586-020-2456-9 [Epub ahead of print].
Rogers, T. F., Zhao, F., Huang, D., Beutler, N., Burns, A., He, W. T., et al. (2020). Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science eabc7520. doi: 10.1126/science.abc7520 [Epub ahead of print].
Romano, M., Ruggiero, A., Squeglia, F., Maga, G., and Berisio, R. (2020). A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 9:E1267. doi: 10.3390/cells9051267
Rowe, C. L., Fleming, J. O., Nathan, M. J., Sgro, J. Y., Palmenberg, A. C., and Baker, S. C. (1997). Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol. 71, 6183–6190. doi: 10.1128/jvi.71.8.6183-6190.1997
Scobey, T., Yount, B. L., Sims, A. C., Donaldson, E. F., Agnihothram, S. S., Menachery, V. D., et al. (2013). Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 110, 16157–16162. doi: 10.1073/pnas.131154211
Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., et al. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224. doi: 10.1038/s41586-020-2179-y
Shi, J., Wen, Z., Zhong, G., Yang, H., Wang, C., Huang, B., et al. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368, 1016–1020. doi: 10.1126/science.abb7015
Shi, R., Shan, C., Duan, X., Chen, Z., Liu, P., Song, J., et al. (2020). A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. doi: 10.1038/s41586-020-2381-y [Epub ahead of print].
Simon-Loriere, E., and Holmes, E. C. (2011). Why do RNA viruses recombine? Nat. Rev. Microbiol. 9, 617–626. doi: 10.1038/nrmicro2614
Smith, E. C., Blanc, H., Surdel, M. C., Vignuzzi, M., and Denison, M. R. (2013). Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9:e1003565. doi: 10.1371/journal.ppat.1003565
Song, W., Gui, M., Wang, X., and Xiang, Y. (2018). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 14:e1007236. doi: 10.1371/journal.ppat.1007236
Su, S., Wong, G., Shi, W., Liu, J., Lai, A. C. K., Zhou, J., et al. (2016). Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24, 490–502. doi: 10.1016/j.tim.2016.03.003
Subbarao, K., and Mahanty, S. (2020). Respiratory virus infections: understanding COVID-19. Immunity 52, 905–909. doi: 10.1016/j.immuni.2020.05.004
Sun, J., He, W. T., Wang, L., Lai, A., Ji, X., Zhai, X., et al. (2020). COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 26, 483–495. doi: 10.1016/j.molmed.2020.02.008
Sun, S.-H., Chen, Q., Gu, H. J., Yang, G., Wang, Y. X., Huang, X. Y., et al. (2020). A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. doi: 10.1016/j.chom.2020.05.020 [Epub ahead of print].
Tang, D., Comish, P., and Kang, R. (2020). The hallmarks of COVID-19 disease. PLoS Pathog. 16:e1008536. doi: 10.1371/journal.ppat.1008536
Tang, X., Wu, C., Li, X., Song, Y., Yao, X., Xinkai Wu, X., et al. (2020). On the origin and continuing evolution of SARS-CoV-2. Nat. Sci. Rev. 7, 1012–1023. doi: 10.1093/nsr/nwaa036
Tay, M. Z., Poh, C. M., Rénia, L., MacAry, P. A., and Ng, L. F. P. (2020). The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374. doi: 10.1038/s41577-020-0311-8
Thao, T. T. N., Labroussaa, F., Ebert, N., V’kovski, P., Stalder, H., Portmann, J., et al. (2020). Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582, 561–565. doi: 10.1038/s41586-020-2294-9
Thiel, V., Herold, J., Schelle, B., and Siddell, S. G. (2001). Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82, 1273–1281. doi: 10.1099/0022-1317-82-6-1273
Totura, A. L., and Baric, R. S. (2012). SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2, 264–275. doi: 10.1016/j.coviro.2012.04.004
Tse, L. V., Meganck, R. M., Graham, R. L., and Baric, R. S. (2020). The current and future state of vaccines, antivirals and gene therapies against emerging coronaviruses. Front. Microbiol. 11:658. doi: 10.3389/fmicb.2020.00658
Vabret, N., Britton, G. J., Gruber, C., Hegde, S., Kim, J., Kuksin, M., et al. (2020). Immunology of COVID-19: current state of the science. Immunity 52, 910–941. doi: 10.1016/j.immuni.2020.05.002
van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J., Wolthers, K. C., et al. (2004). Identification of a new human coronavirus. Nat. Med. 10, 368–373. doi: 10.1038/nm1024
van Hemert, M. J., van den Worm, S. H., Knoops, K., Mommaas, A. M., Gorbalenya, A. E., and Snijder, E. J. (2008). SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. doi: 10.1371/journal.ppat.1000054
Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., and Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6. doi: 10.1016/j.cell.2020.02.058
Wang, L. F., and Anderson, D. E. (2019). Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 34, 79–89. doi: 10.1016/j.coviro.2018.12.007
Wang, N., Shang, J., Jiang, S., and Du, L. (2020). Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 11:298. doi: 10.3389/fmicb.2020.00298
Wang, Q., Wu, J., Wang, H., Gao, Y., Liu, Q., Mu, A., et al. (2020). Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 182, 417–428.e13. doi: 10.1016/j.cell.2020.05.034
Wec, A. Z., Wrapp, D., Herbert, A. S., Maurer, D. P., Haslwanter, D., Sakharkar, M., et al. (2020). Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science eabc7424. doi: 10.1126/science.abc7424 [Epub ahead of print].
Wilk, A. J., Rustagi, A., Zhao, N. Q., Roque, J., Martínez-Colón, G. J., McKechnie, J. L., et al. (2020). A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. doi: 10.1038/s41591-020-0944-y [Epub ahead of print].
Williamson, B. N., Feldmann, F., Schwarz, B., Meade-White, K., Porter, D. P., Schulz, J., et al. (2020). Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. doi: 10.1038/s41586-020-2423-5 [Epub ahead of print].
Wong, A. H. M., Tomlinson, A. C. A., Zhou, D., Satkunarajah, M., Chen, K., Sharon, C., et al. (2017). Receptor-binding loops in alphacoronavirus adaptation and evolution. Nat. Commun. 8:1735. doi: 10.1038/s41467-017-01706-x
Woo, P. C., Lau, S. K., Chu, C. M., Chan, K. H., Tsoi, H. W., Huang, Y., et al. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895. doi: 10.1128/JVI.79.2.884-895.2005
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/JVI.06540-11
Wrapp, D., De Vlieger, D., Corbett, K. S., Torres, G. M., Wang, N., and Van Breedam, W. (2020). Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1436–1441. doi: 10.1016/j.cell.2020.05.047
Wu, A., Peng, Y., Huang, B., Ding, X., Wang, X., Niu, P., et al. (2020). Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328. doi: 10.1016/j.chom.2020.02.001
Wu, H.-Y., Guy, J. S., Yoo, D., Vlasak, R., Urbach, E., and Brian, D. A. (2003). Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology 315, 174–183. doi: 10.1016/S0042-6822(03)00511-7
Wu, K., Li, W., Peng, G., and Li, F. (2009). Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U.S.A. 106, 19970–19974. doi: 10.1073/pnas.0908837106
Wu, Y., Li, C., Xia, S., Tian, X., Kong, Y., Wang, Z., et al. (2020). Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe 27, 891–898.e5. doi: 10.1016/j.chom.2020.04.023
Wu, Z., Yang, L., Ren, X., Zhang, J., Yang, F., Zhang, S., et al. (2016). ORF8-related genetic evidence for chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 213, 579–583. doi: 10.1093/infdis/jiv476
Xie, X., Muruato, A., Lokugamage, K. G., Narayanan, K., Zhang, X., Zou, J., et al. (2020). An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e3. doi: 10.1016/j.chom.2020.04.004
Xu, X., Sun, J., Nie, S., Li, H., Kong, Y., Liang, M., et al. (2020). Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. doi: 10.1038/s41591-020-0949-6 [Epub ahead of print].
Yang, X. L., Hu, B., Wang, B., Wang, M. N., Zhang, Q., Zhang, W., et al. (2015). Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 90, 3253–3256. doi: 10.1128/JVI.02582-15
Ye, Z.-W., Yuan, S., Yuen, K.-S., Fung, S.-Y., Chan, C.-P., and Dong-Yan Jin, D.-Y. (2020). Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 16, 1686–1697. doi: 10.7150/ijbs.45472
Yount, B., Curtis, K. M., and Baric, R. S. (2000). Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74, 10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000
Yount, B., Curtis, K. M., Fritz, E. A., Hensley, L. E., Jahrling, P. B., Prentice, E., et al. (2003). Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 100, 12995–13000. doi: 10.1073/pnas.1735582100
Yount, B., Denison, M. R., Weiss, S. R., and Baric, R. S. (2002). Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76, 11065–11078. doi: 10.1128/jvi.76.21.11065-11078.2002
Yu, J., Tostanoski, L. H., Peter, L., Mercado, N. B., McMahan, K., Mahrokhian, S. H., et al. (2020). DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science eabc6284. doi: 10.1126/science.abc6284
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., and Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. doi: 10.1056/NEJMoa1211721
Zhang, X., Tan, Y., Ling, Y., Lu, G., Liu, F., Yi, Z., et al. (2020). Viral and host factors related to the clinical outcome of COVID-19. Nature 583, 437–440. doi: 10.1038/s41586-020-2355-0
Zhao, Z., Li, H., Wu, X., Zhong, Y., Zhang, K., Zhang, Y. P., et al. (2004). Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 4:21. doi: 10.1186/1471-2148-4-21
Zhong, N. S., Zheng, B. J., Li, Y. M., Poon, L. L. M., Xie, Z. H., Chan, K. H., et al. (2003). Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362, 1353–1358. doi: 10.1016/s0140-6736(03)14630-2
Zhou, G., and Zhao, Q. (2020). Perspectives on therapeutic neutralizing antibodies against the novel coronavirus SARS-CoV-2. Int. J. Biol. Sci. 16, 1718–1723. doi: 10.7150/ijbs.45123
Zhou, P., Fan, H., Lan, T., Yang, X. L., Shi, W. F., Zhang, W., et al. (2018). Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556, 255–258. doi: 10.1038/s41586-018-0010-9
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhou, Z., Ren, L., Zhang, L., Zhong, J., Xiao, Y., Jia, Z., et al. (2020). Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27, 883–890.e2. doi: 10.1016/j.chom.2020.04.017
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Ziegler, C. G. K., Allon, S. J., Nyquist, S. K., Mbano, I. M., Miao, V. N., Tzouanas, C. N., et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19. doi: 10.1016/j.cell.2020.04.035
Keywords: COVID-19, SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV, biological diversification, recombination, adaptive evolution
Citation: Koma T, Adachi S, Doi N, Adachi A and Nomaguchi M (2020) Toward Understanding Molecular Bases for Biological Diversification of Human Coronaviruses: Present Status and Future Perspectives. Front. Microbiol. 11:2016. doi: 10.3389/fmicb.2020.02016
Received: 22 June 2020; Accepted: 30 July 2020;
Published: 25 August 2020.
Edited by:
Yasuko Tsunetsugu Yokota, Tokyo University of Technology, JapanReviewed by:
Tetsuo Tsukamoto, Kindai University, JapanMinato Hirano, The University of Texas Medical Branch at Galveston, United States
Yohei Kurosaki, Nagasaki University, Japan
Copyright © 2020 Koma, Adachi, Doi, Adachi and Nomaguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akio Adachi, YWRhY2hpQHRva3VzaGltYS11LmFjLmpw; YWRhY2hpYWtAaGlyYWthdGEua211LmFjLmpw; Masako Nomaguchi, bm9tYWd1Y2hpQHRva3VzaGltYS11LmFjLmpw
 Takaaki Koma
Takaaki Koma Shun Adachi
Shun Adachi Naoya Doi
Naoya Doi Akio Adachi
Akio Adachi Masako Nomaguchi
Masako Nomaguchi