- Department of Microbiology, Institute for Water and Wetland Research, Faculty of Science, Radboud University Nijmegen, Nijmegen, Netherlands
Anaerobic ammonium-oxidizing (anammox) bacteria, members of the “Candidatus Brocadiaceae” family, play an important role in the nitrogen cycle and are estimated to be responsible for about half of the oceanic nitrogen loss to the atmosphere. Anammox bacteria combine ammonium with nitrite and produce dinitrogen gas via the intermediates nitric oxide and hydrazine (anammox reaction) while nitrate is formed as a by-product. These reactions take place in a specialized, membrane-enclosed compartment called the anammoxosome. Therefore, the substrates ammonium, nitrite and product nitrate have to cross the outer-, cytoplasmic-, and anammoxosome membranes to enter or exit the anammoxosome. The genomes of all anammox species harbor multiple copies of ammonium-, nitrite-, and nitrate transporter genes. Here we investigated how the distinct genes for ammonium-, nitrite-, and nitrate- transport were expressed during substrate limitation in membrane bioreactors. Transcriptome analysis of Kuenenia stuttgartiensis planktonic cells showed that four of the seven ammonium transporter homologs and two of the nine nitrite transporter homologs were significantly upregulated during ammonium-limited growth, while another ammonium transporter- and four nitrite transporter homologs were upregulated in nitrite limited growth conditions. The two nitrate transporters were expressed to similar levels in both conditions. In addition, genes encoding enzymes involved in the anammox reaction were differentially expressed, with those using nitrite as a substrate being upregulated under nitrite limited growth and those using ammonium as a substrate being upregulated during ammonium limitation. Taken together, these results give a first insight in the potential role of the multiple nutrient transporters in regulating transport of substrates and products in and out of the compartmentalized anammox cell.
Introduction
Anaerobic ammonium-oxidizing (anammox) bacteria play a major role in the biological nitrogen cycle where they contribute significantly to oceanic nitrogen loss and are estimated to be responsible for up to 50% of the dinitrogen gas release to the atmosphere (Francis et al., 2007; Lam and Kuypers, 2011). In addition, they are applied in wastewater treatment for cost-effective and environment-friendly nitrogen removal (Kartal et al., 2010; Lackner et al., 2014). Anammox bacteria belong to the family Brocadiaceae and order Brocadiales and so far five different genera have been described: “Ca. Kuenenia,” “Ca. Brocadia,” “Ca. Anammoxoglobus,” “Ca. Jettenia,” and “Ca. Scalindua” (Jetten et al., 2009). Anammox bacteria are not genetically tractable and grow slowly, with reported doubling times ranging from days to weeks (Lotti et al., 2015; Zhang and Okabe, 2020). They are cultivated as enrichment cultures of up to 95% purity in bioreactor systems. Although anammox cells grow generally in larger cell aggregates, the “type strain” Kuenenia stuttgartiensis and Brocadia sinica can be cultivated as a planktonic enrichment culture (van der Star et al., 2008; Zhang and Okabe, 2017).
Anammox bacteria have a unique cell organelle, the anammoxosome, which occupies a major part of the cell volume (van Niftrik and Jetten, 2012). The anammox reaction takes place in the anammoxosome and is postulated to be coupled to energy conservation over the anammoxosome membrane (van Niftrik et al., 2008a, b; Neumann et al., 2014). The anammox reaction proceeds via three consecutive redox reactions (for a review see Kartal and Keltjens, 2016). First, a nitrite reductase performs the one-electron reduction of nitrite to nitric oxide. It is proposed that this step is carried out by distinct enzymes in different anammox species, more specifically the iron cytochrome cd1 NirS (K. stuttgartiensis and marine Scalindua species), the octaheme hydroxylamine oxidoreductase kustc0458 (K. stuttgartiensis RU1), or the copper containing NirK (Jettenia caeni) (Strous et al., 2006; Kartal et al., 2011b, 2013; Hira et al., 2012; van de Vossenberg et al., 2013; de Almeida et al., 2016; Speth et al., 2016; Hu et al., 2019).
The next step in the anammox reaction is the condensation of nitric oxide and ammonium with four electrons to produce hydrazine performed by the hydrazine synthase (HZS) (Dietl et al., 2015). Finally, hydrazine is oxidized to dinitrogen gas by the hydrazine dehydrogenase (HDH) (Maalcke et al., 2016; Akram et al., 2019a). The oxidation of hydrazine releases four low potential electrons that are proposed to be shuttled via soluble cytochromes to an electron transport chain in the anammoxosome membrane and ultimately back to the nitrite reductase and hydrazine synthase (Strous et al., 2006; Kartal et al., 2013; Kartal and Keltjens, 2016). The electrons would flow via an electron transport chain consisting of Rieske-heme b complexes (bc1 complexes) and a quinone pool in order to establish a proton motive force (and subsequent ATP synthesis) across the anammoxosome membrane. Some electrons are “lost” from the cycle to provide the reducing power for carbon fixation and assimilation, and these lost electrons are thought to be supplemented by the oxidation of nitrite to nitrate by the nitrite oxidoreductase Nxr. This produces the nitrate that is observed as an additional product when anammox bacteria are grown on nitrite and ammonium (van de Graaf et al., 1997; Kartal and Keltjens, 2016).
As the anammox reaction takes place in the anammoxosome, the substrates and products need to cross both the outer- and cytoplasmic membrane as well as the anammoxosome membrane. In other prokaryotes, ammonium-, nitrite- and nitrate transport is facilitated by dedicated transporter proteins that reside in the cytoplasmic membrane. Ammonium transport is mediated by the AmtB/Mep/Rh family of proteins that are found in all domains of life (Andrade and Einsle, 2007; McDonald and Ward, 2016). The ammonium transporter consists of a homotrimer and each monomer contains an ammonium channel (Zheng et al., 2004). In different members of the AmtB/Mep/Rh family, the substrate for transport may vary: NH3 for Rh family transporters and NH4+ or H+/NH3 for Amt and Mep transporters (Neuhauser et al., 2014; Wacker et al., 2014; Baday et al., 2015). GlnK PII regulators modulate ammonium transporters by binding near the exit port and thereby blocking the transport channel when ammonium is present in high levels (Conroy et al., 2007; Gruswitz et al., 2007). These GlnK PII regulators and AmtB transporters are conserved gene pairs in prokaryotes (Thomas et al., 2000; Coutts et al., 2002).
Nitrite transport is mediated via the formate/nitrite transporter (FNT) family, containing FocA, NirC and HSC transporters that are associated with formate, nitrite and hydrosulphide transport, respectively (Lu et al., 2013; Mukherjee et al., 2017). They all transport anions and different weak acids and transport is driven via proton binding, resulting in transfer of neutral substrate through the pore, similar to the mechanism proposed for AmtB transporters (Atkovska and Hub, 2017; Wiechert and Beitz, 2017a, b). Nitrate is transported via the NarK family transporters which are members of the Major Facilitator Superfamily (MFS) of transporters (Andrade and Einsle, 2013; Noinaj and Buchanan, 2014). Two NarK subtypes have been identified: the NarK1 type transporters are associated with nitrate assimilation and transport is thought to occur via nitrate/proton symport. The NarK2 type transporters are generally used during dissimilatory nitrate reduction and are thought to act as nitrate:nitrite antiporters. This would give them a double function of nitrate import and removal of the toxic nitrite produced by nitrate reduction (see Goddard et al., 2017 for review).
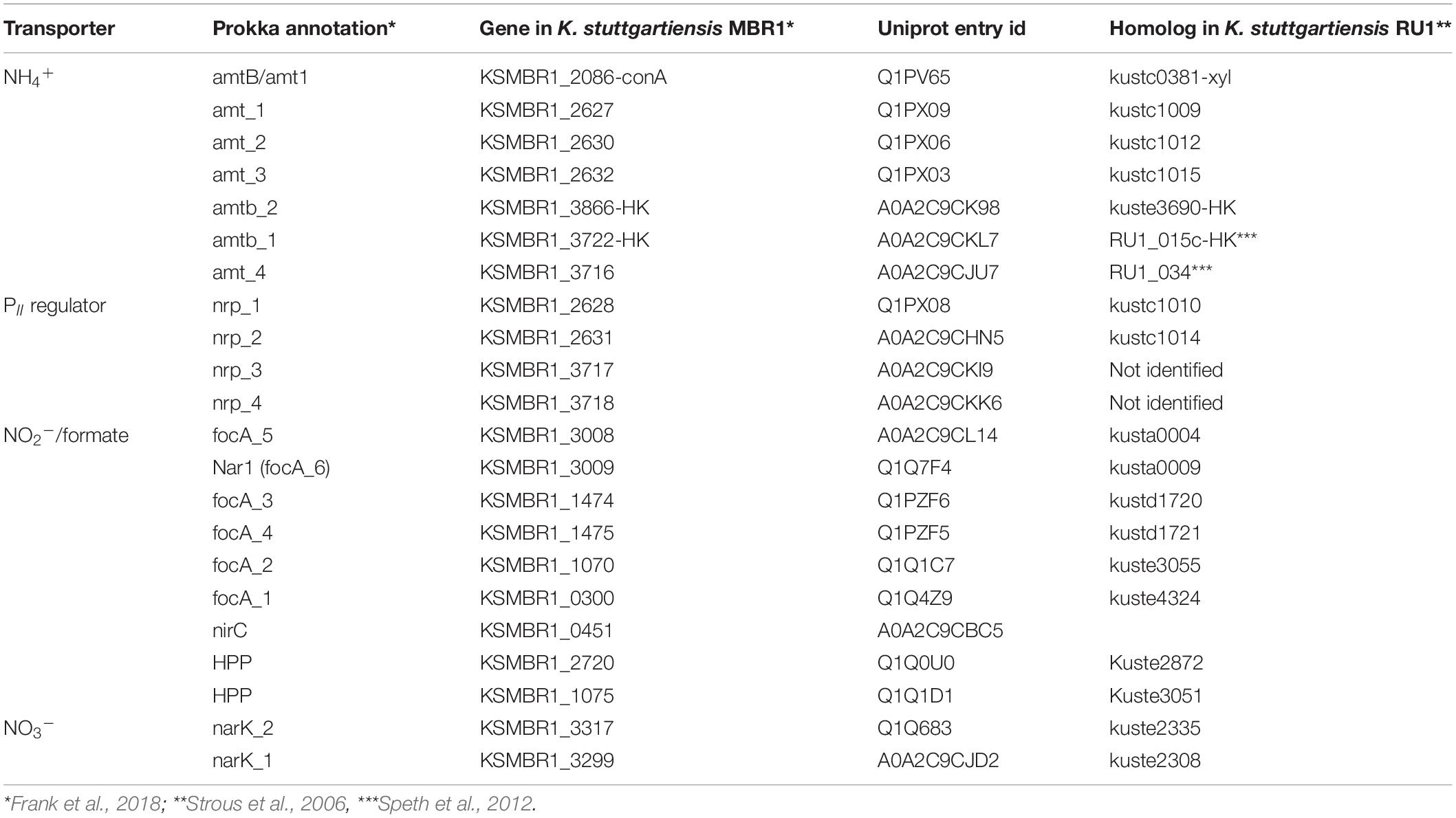
All published anammox (meta)genomes harbor multiple copies of ammonium, nitrite and nitrate transporters that could facilitate transport into- or out of the anammoxosome crossing several membrane bilayers (Strous et al., 2006; Speth et al., 2012; van de Vossenberg et al., 2013; Oshiki et al., 2015; Speth et al., 2015; Frank et al., 2018). The closed genome of the type strain Kuenenia stuttgartiensis MBR1 (Frank et al., 2018) encodes no less than seven AmtB-type ammonium transporter homologs, four PII ammonium transport regulators (GlnK) and seven FNT-type transporters that are proposed to transport nitrite. There is also one annotated NirC-type nitrite transporter (Table 1). To transport nitrate out of the cell, two narK genes encoding nitrate transporters have been identified, both of which are of the NarK2-type and annotated as nitrate:nitrite transporters (Frank et al., 2018). In anammox bacteria, these may usually function in the opposite direction compared with other bacteria: exporting the by-product nitrate while simultaneously importing some additional nitrite as substrate for the anammox reaction. However the direction of transport will depend on the concentration gradients of nitrate and nitrite: Li et al. (2016) found that addition of nitrate to nitrite-stressed Brocadia cells alleviated nitrite toxicity, presumably via nitrate uptake coupled to nitrite export from the cell (Li et al., 2016).
The reason for the high number of homologous nutrient transporters in anammox bacteria is not yet known. We hypothesized that distinct transporter proteins are expressed under different growth conditions, i.e., the availability of the substrates ammonium and nitrite. Transporters with higher affinity would be upregulated upon low or limiting nutrient conditions. Nearly all previous transcriptome work in K. stuttgartiensis and in other anammox species has been performed under nitrite limitation with an excess of ammonium to prevent nitrite toxicity (de Almeida et al., 2011; Hu et al., 2013, 2019; Kartal et al., 2013; Neumann et al., 2014). The past body of work is therefore not fit to determine the influence of different nutrient limitations on the expression levels of transporters. In this paper, we tested the effect of ammonium- versus nitrite limitation on the expression level of the various genes encoding for nutrient transporters. Based on the results we discuss the redundancy in nutrient transport proteins in anammox bacterium K. stuttgartiensis MBR1.
Materials and Methods
Bacterial Growth
Kuenenia stuttgartiensis MBR1 was cultivated in a single cell membrane bioreactor (MBR) Applikon B.V. Schiedam, Netherlands) as described previously (Kartal et al., 2011a). The growth medium consisted of (per litre): 1.25 g KHCO3, 0.025 g NaH2PO4, 0.5 ml 1.2 M HCl, 0.15 g CaCl2⋅2H2O, 0.1 g MgSO4⋅7H2O and 6.25 mg FeSO4⋅7 H2O. The reactor volume was 2 L. The pH was maintained at 7.3 using 100 g/L KHCO3 buffer solution. To ensure an anaerobic system both the reactor and the main medium bottle were flushed with a mixture of Argon/CO2 (95%/5%) at 10 ml/min. Excess biomass was removed at 0.138 L per day, resulting in a dilution rate of 0.07 (doubling time of 10 days). For nitrite limited conditions, the reactor received 15 mmol/day NaNO2 and 7.5 mmol/day (NH4)2SO4, resulting in 15 mmol/day NO2– and 15 mmol/day NH4+. For ammonium limited conditions, the NaNO2 load of the reactor was increased in two steps to 18 and finally to 21 mmol/day NaNO2, without changing the (NH4)2SO4 load. A secondary medium bottle containing all medium salts as described above but without NaNO2, only 5 mM (NH4)2SO4 and with reduced FeSO4⋅7 H2O (3.13 mg/L) to prevent precipitation, was used to add approximately 0.45 mmol (NH4)2SO4 per day. This allowed for fine tuning of the ammonium:nitrite ratio and enabled the nitrite levels to be kept below toxic levels, but between 2 and 20 mg/L (40 and 400 μM).
Ammonium and Nitrite Determination
Nitrite and nitrate concentrations in the reactor were determined daily using MQuantTM 10–500 mg/l nitrate detector strips (Merck KGaA, Darmstadt, Germany) during the operation of the bioreactor. Supernatants of reactor samples were used for accurate determination of nitrite and ammonium concentrations in the reactor. Nitrite concentrations were determined using the colorimetric Griess-reaction (Feigl and Amaral, 1958): 5 μl supernatant was mixed with 45 μl milliQ, 50 μl 1% sulfanilic acid in 1 M HCL and 50 μl 0.1% naphtylethylene diaminedihydrochloride in water. The solution was then incubated for 10 min at room temperature, and the absorbance was analyzed at a wavelength of 540 nm. MilliQ Water was used as a blank control.
Ammonium concentrations were determined using ortho-phtaldialdehyde (OPA), either by the colorimetric assay or the more sensitive fluorescence method (Taylor et al., 1974). For the colorimetric ammonium assay, 50 μl supernatant was mixed with 750 μl OPA reagent [0.54% (w/v) ortho-phthaldialdehyde, 0.05% (v/v) β-mercaptoethanol, and 10% (v/v) ethanol in 400 mM potassium phosphate buffer (pH 7.3)], incubated for 20 min at room temperature and absorbance at 420 nm was read using a SPEKTRAmaxTM plate reader (Molecular Devices, San Jose, California).
For the sensitive ammonium assay, 10 or 20 μl supernatant was mixed with 200 μl OPA reagent that had been diluted 10-times in 400 mM potassium phosphate buffer (pH 7.3). Samples were incubated in the dark for 20 min and fluorescence was measured using a SparkTM plate reader (Biocompare, South San Francisco, California). Samples were shaken 5 s pre-measurement, with excitation and emission wavelengths at 411 and 482 nm, respectively.
Sampling for Transcriptome Analysis
When the bioreactor had been growing in steady state for 8–25 days in either ammonium- or nitrite limited conditions, 2 ml samples were taken on three separate days and immediately centrifuged at room temperature for 2 min at 16,100 × g. The supernatant was removed, and the pellets snap-frozen in liquid nitrogen and stored at −80°C.
RNA Isolation, Library Preparation, and Sequencing
From each of the 6 samples, total RNA was isolated using TRIzolTM Reagent (Thermo Fisher Scientific, Waltham, United States) according to the manufacturer’s instructions with the exception that 100-fold higher cell numbers were used, since the recommended cell number resulted in very low to no RNA yields. The isolated RNA was treated with DNAse 1, as described and provided by the RiboPureTM kit (Thermo Fisher Scientific, Waltham, United States). RNA was eluted in DEPC treated water in volumes between 20 and 25 μl of which 1–2 μl was used to quantify the isolated RNA. Concentrations ranged between 40 and 112 ng/μl and RNA Integrity Numbers (RIN) were between 8.9 and 9.4. Ribosomal RNA was partly removed using the MICROBExpress kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Although this kit removes most of the 23S rRNA, it does not remove the 16S rRNa of K. stuttgartiensis efficiently. RNA was fragmented to 260 bp fragments, cDNA was synthesized, adapters (including barcodes) were ligated and cDNA was amplified using the TruSeq stranded mRNA Library Prep Kit v2, according to the protocol supplied by the manufacturer (Illumina, San Diego, CA, United States). The 6 barcoded cDNA libraries were pooled prior to sequencing, which was performed with the Illumina MiSeq® sequencer (Illumina, San Diego, CA, United States) using the 150 bp sequence chemistry and the Miseq Reagent Kit v3 (Illumina, San Diego, CA, United States).
RNA and cDNA Quantification
RNA and cDNA were quantified using the QubitTM RNA HS Assay Kit and dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, United States), respectively. The quality and size distribution of the RNA before and after rRNA removal, and of the cDNA libraries was determined using the Agilent 2100 Bioanalyser with the Agilent RNA 6000 Nano Kit and the high sensitivity DNA kit (Agilent technologies, Santa Clara, CA, United States), respectively. The cDNA fragments in the libraries peaked around the recommended 260 bp.
Sequence Reads Analysis
The number of sequencing reads per sample varied between 1.4 and 4.8 million with an average read length of 137 bp. CLC Genomics Workbench 11.0.1 (Qiagen bioinformatics, Denmark) was used to trim reads using the following settings: quality limit was set to 0.01, with a maximum of 2 ambiguous nucleotides, no adapter trimming, and removal of 10 nucleotides at the 5′ and 5 nucleotides at the 3′ end. Reads smaller than 50 bp were discarded. The number of reads left after trimming varied between 1.3 and 4.5 million with an average read length of 118 bp. Non-mRNA sequences were removed by mapping the reads to all non-coding RNAs obtained from the K. stuttgartiensis MBR1 genome (Frank et al., 2018). The non-mapped reads, consisting of the mRNA reads, were subsequently mapped to the annotated K. stuttgartiensis MBR1 genome and mapping results were expressed in RPKM values (Reads Per Kilobase gene per Million mapped reads). Average RPKM values were calculated for the triplicate samples per condition. For differential expression analysis a Wald test was performed using DEseq2 in R and adjusted p-values were calculated, taking into account the false discovery rate (Love et al., 2014). Genes with significant changes in expression between ammonium and nitrite limited growth conditions were selected using the following three criteria: (1) mean of normalized counts (basemean values) > 4, (2) log2foldchange > 0.58 or < -0.58 (i.e., a difference in expression of more than 1.5-fold), and (3) adjusted p < 0.05. Assignment of K. stuttgartiensis MBR1 genes to Clusters of orthologous groups (COG) was performed at NCBI using the Conserved Domain Search Service (Marchler-Bauer and Bryant, 2004).
Nucleotide Sequence Accession Number
The transcriptome sequences were deposited at NCBI in the Gene Expression Omnibus (GEO) under accession number GSE148825.
Results and Discussion
Nitrite and Ammonium Limited Growth of K. stuttgartiensis MBR1
The expression of the multiple ammonium, nitrite and nitrate transporters encoded in the genome of K. stuttgartiensis MBR1 was studied under ammonium- and nitrite limited growth conditions. Hereto, we set up and optimized K. stuttgartiensis cultivation in a dedicated bioreactor system. The experiment was started with cultivation of planktonic K. stuttgartiensis cells in a membrane reactor with 15 mM nitrite and ammonium, resulting in nitrite limited conditions, a steady state cell density of OD600 0.6 and a doubling time of 10 days. To obtain ammonium limited conditions, we increased the nitrite concentration in the medium over 2 days in two steps, via 18–21 mM nitrite, while maintaining the doubling time at 10 days and maintaining the cell density at OD600 0.6. Reaching a stable steady state under ammonium limited conditions is more challenging, as the excess nitrite left in the medium can cause cell stress and toxicity and the onset of biofilm formation. We found that reducing cell densities in the reactor helped to avoid biofilm formation. Therefore, for this experiment we cultivated K. stuttgartiensis at a lower cell density than we usually do in our maintenance bioreactor that is run at OD600 1.2. To avoid toxicity and biofilm formation, the reactor was carefully monitored for nitrite build up. A separate medium, containing 5 mM ammonium but no nitrite, was pumped into the reactor to maintain the concentration of nitrite in the reactor between 40 and 400 μM while keeping the ammonium concentration at 0 mM. Despite careful fine-tuning, it proved difficult to maintain the cell density at a steady state for more than 1 month. Several times the reactor lost biomass with concomitant increase in ciliates in the reactor, presumably consuming the K. stuttgartiensis cells. Also, the cells in the reactor occasionally still started producing biofilm resulting in loss of optical density.
Transcriptomes of K. stuttgartiensis MBR1 Under Nitrite and Ammonium Limited Conditions
We used transcriptomics to investigate and compare differential gene expression under the two growth conditions. Gene expression of cells grown under nitrite- versus ammonium limited continuous culture conditions was compared using triplicate samples for each condition (Figure 1A). Before analysis the reads mapping to 53 structural RNA (rRNA, sRNA, and tRNA) genes were removed from the data set. Of the total of 4095 (minus the 53 structural RNA genes) annotated K. stuttgartiensis MBR1 genes, transcripts of 654 genes were not detected in either ammonium or in nitrite limited growth conditions. Over half of these genes (474) were annotated as hypothetical protein or putative ORF and 36 genes were either insertion elements or annotated as (putative) transposases. Of the genes that were expressed, 3179 were expressed in both growth conditions; 58 genes were only detected during ammonium limited growth; and 151 were only detected during nitrite limited growth (Figure 1B). The 58 genes that were expressed only during ammonium limitation showed very low to medium expression levels (average 12 and a maximum expression of 58 RPKM). Only seven of these genes were annotated, of which two genes involved in flagellum biosynthesis, a gene containing a DNA polymerase 3 finger domain, and rubredoxin gene rub_1, had expression levels above 4 RPKM (Supplementary File 1). The 151 genes that were expressed only during nitrite limited conditions also had low to medium expression levels (average 12 and a maximum of 79 RPKM). Only 23 of these genes were annotated and 18 had an expression of > 4 RPKM or more (Supplementary File 1). These included four transposon-like elements, a flagellar biosynthesis gene, and rubredoxin gene rub_2. Rubredoxins are iron containing electron carriers. They were recently found to be expressed in Brocadia sinica when grown using an electrode instead of nitrite as terminal electron acceptor and they were proposed to shuttle electrons from the anammoxosome to the exterior (Shaw et al., 2020). K. stuttgartiensis encodes three rubredoxin genes and the third gene, KSMBR1_2919, is expressed at around 300 RPKM in both nitrite and ammonium limited conditions so this gene may encode the dominant rubredoxin present in the cell. It is unclear at present what the role of the rubredoxins is in ammonium- and nitrite-limited growth conditions.
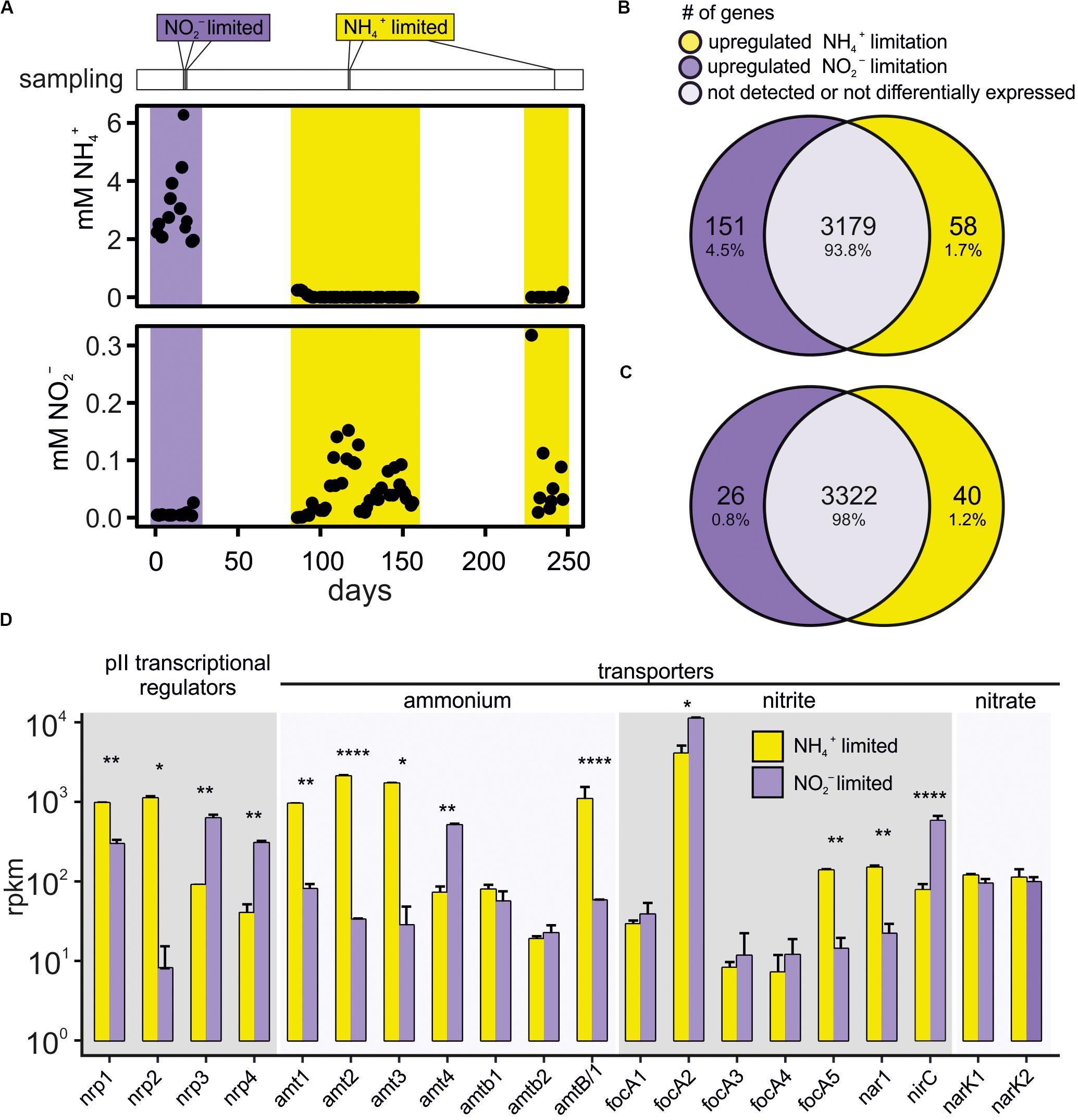
Figure 1. (A) Measured ammonium and nitrite levels in the K. stuttgartiensis MBR1 bioreactor and sampling times for transcriptomic analyses, indicated with vertical lines above the graph. (B) Venn diagram showing the number of genes that were exclusively expressed during either ammonium- (yellow) or nitrite-limitation (purple), and the genes that were expressed in both conditions. (C) Venn diagram showing only the genes significantly upregulated > 1.5-fold in either ammonium- (yellow) or nitrite-limitation (purple). The overlap includes all genes with either (1) very low expression levels (basemean < 4), or (2) expression levels that were within 1.5-fold change between ammonium- and nitrite limited conditions, or (3) with changes in expression levels that were below the significance level (padj > 0.05). Note that the 707 genes not expressed in either condition are not included in the Venn diagrams. (D) Bar graph of PII regulator genes (nrp1-4), ammonium transporter genes (amt1-4, amtb1-2, and amtB/1), nitrite transporter genes (focA1-5 and nar1), nitrate transporter genes (narK1-2) and their expression levels under both ammonium- (yellow) and nitrite (purple) limited growth. Statistical significance ns: *p = 0.05, **p = 0.01, ****p = 0.0001. RPKM values corresponding to each bar are shown in Supplementary File 2.
Of the genes that were expressed in both conditions, we found 40 genes significantly upregulated during ammonium limited growth versus 26 genes during nitrite limited growth (Figure 1C and Supplementary File 2). When assigned to categories of orthologous groups, these genes mainly cluster in C (energy production and conversion), P (inorganic metabolism, including the nutrient transport genes) and T (signal transduction) (Supplementary Figure 1). Most genes (2950) were not assigned to any orthologous group, similar to reports for other anammox species (van de Vossenberg et al., 2013; Bagchi et al., 2016).
Many of the differentially expressed genes were organized in operon-like structures (Figure 2). Of the putative operons that were significantly upregulated in nitrite limited conditions, one contained nitrite transporter gene focA_2 and another ammonium transporter gene Amt_4 (Figures 2BII,V) and three code for enzymes involved in nitrite metabolism (Figures 2BI,III,IV). Putative operons that were significantly upregulated in ammonium limited conditions, contained nutrient transporter genes (Figures 2BVI,VIII,X), one contained nuo genes that encode NADH dehydrogenase subunits (Figure 2BVII, see also below), and two contained in total all three copies of the hydrazine synthase cluster (Figures 2BIX,XI, see also below). A seventh putative operon contained a gene encoding Hsp20/alpha crystallin family protein, a Yfdx protein with unknown function and a Do/DeqQ family serine endopeptidase.
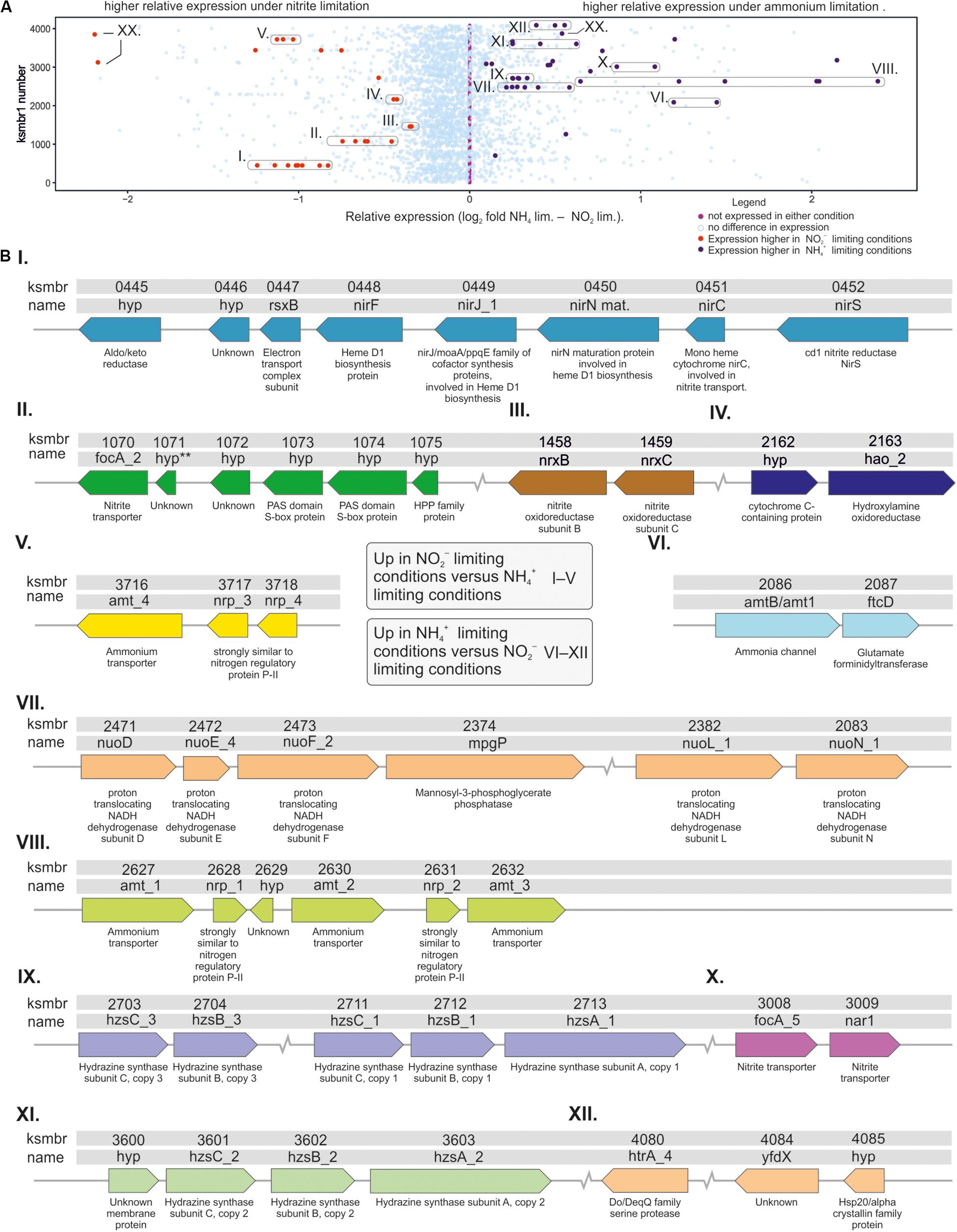
Figure 2. Schematic representation of K. stuttgartiensis MBR1 genes that are either up- or downregulated in response to ammonium- or nitrite-limiting bioreactor growth conditions. (A) graph showing all genes as dots on vertical line, with the relative expression on the y-axis. Genes indicated in translucent blue were either not significantly expressed, or not significantly different between the two conditions. Genes indicated in purple were expressed at a significantly higher level in response to ammonium limitation compared to nitrite limitation. Genes indicated as red dots were expressed at a significantly higher level in response to nitrite limitation when compared to ammonium limitation. Circles were drawn around genes that are in very close genomic proximity. XX = KSMBR1_3177, KSMBR1_3122, and KSMBR1_3846. (B) Synteny and description of genes that were expressed at a significantly different level between nitrite- and ammonium-limiting conditions. Roman numerals I–V indicate genes that were expressed relatively more during nitrite-limiting conditions, and roman numerals VI–XII indicate genes expressed relatively more during ammonium-limiting conditions.
Ammonium Transporters
Of all seven K. stuttgartiensis MBR1 ammonium transporters the amt_1 (KSMBR1_2627), amt_2 (KSMBR1_2630) and amt_3 (KSMBR1_2632) cluster (Figure 2BVIII), and amtB/amt1 (KSMBR1_2086, Figure 2BVI) were significantly upregulated in response to ammonium limitation. The amtB/amt1 gene has an additional N-terminal concanavalin A-like domain as found in β-xylosidases (Jordan and Wagschal, 2010), that is transcribed with the amtB part of the gene as one transcript (data not shown). Concanavalin A is a lectin, but it is unclear at present whether its role in the AmtB/amt1 protein is indeed carbohydrate binding. The amtB/amt1 homolog was previously identified as one of the rare prokaryotic Rh members of the AmtB/Mep/Rh transporter family. In the ammonia oxidizer Nitrosomonas europaea, which lacks AmtB transporters, this Rh homolog is used to transport ammonium into the cell (Lupo et al., 2007; Weidinger et al., 2007; Matassi, 2017). However, the function of the AmtB/amt1 in K. stuttgartiensis remains to be elucidated.
In response to nitrite limitation the ammonium transporter amt_4 (KSMBR1_3716, Figure 2BV) was significantly upregulated (Figure 1D), reflecting results obtained for ammonium limited Scalindua brodae (Russ et al., 2019). The two ammonium transporters with histidine kinase domains, amtb_1 (KSMBR1_3722) and amtb_2 (KSMBR1_3866), were expressed at a low level (<100 RPKM) and expression did not differ between the tested conditions (Figure 1D). The amtb2 gene product (named KsAmt-5 by the authors) has been shown to function as an ammonium sensor in vivo, but does not transport ammonium across the membrane (Pfluger et al., 2018). In fungal species, multiple Mep-type ammonium transporters are present as well and some of these ammonium “transceptors” have a signaling function (van den Berg et al., 2019). So it is likely that in K. stuttgartiensis more than one ammonium transporter homologs has a function other than transport.
GlnK PII regulators regulate ammonium transport by binding near the exit port and thereby blocking the transport channel under high ammonium conditions (Conroy et al., 2007; Gruswitz et al., 2007). These GlnK PII regulators and AmtB transporters are well known to be conserved gene pairs in prokaryotes (Thomas et al., 2000; Coutts et al., 2002). Four GlnK PII regulators are present in the genome of K. stuttgartiensis MBR1. Regulators nrp1 (KSMBR1_2628) and nrp2 (KSMBR1_2631) were upregulated in response to ammonium limitation (Figure 1D), and are located next to amt_1, amt_2, and amt_3. These genes likely form one operon as the transcriptome reads overlap in the whole region between amt_1 and amt_3 (Figure 2BVIII and data not shown). Regulators nrp3 (KSMBR1_3717) and nrp4 (KSMBR1_3718) were upregulated in response to nitrite limitation (Figure 1D) and overlapping transcriptome reads indicated these form an operon with amt_4 (Figure 2BV and data not shown).
Although anammox bacteria contain a strikingly large number of ammonium transporters, the presence of more than one amtB copy in one organism is not unusual. In fact, although model organism Escherichia coli only has one amtB gene this appears to be an exception, as multiple homologs of amtB genes and of the other members in the AMT/MEP/Rh ammonium transporter family have been observed in organisms from all three domains of life (Merrick et al., 2006; Andrade and Einsle, 2007; Tremblay and Hallenbeck, 2009; McDonald and Ward, 2016). In plants and yeast with multiple transporters, some have been identified to differ in their affinity to ammonium (Loqué and von Wirén, 2004; Qin et al., 2018). Our results indicate that this might also be the case in K. stuttgartiensis, with amt_4 representing a low affinity ammonium transporter expressed when ammonium concentrations are relatively high, and amt_1, 2, 3 and amtB/amt1 potential high affinity transporters. However, the amtB/amt1 with the concanavalin A-like domain may have a function other than ammonium transport. It is most homologous to the Rh members of the AMT/MEP/Rh transporter family, which have rather low affinity for ammonium (Lupo et al., 2007; Baday et al., 2015) and have also been associated with CO2 transport (Peng and Huang, 2006).
In addition to the observed differential expression of potential high and low affinity transporters, and the indication that some of the transporters may have alternative substrates, it is also possible that different transporters are located in the different membranes of the anammox cell. The anammox reaction takes place in the anammoxosome meaning that both substrates ammonium and nitrite and by-product nitrate need to cross three double membranes: the outer-, cytoplasmic- and anammoxosome membranes. The three membranes could each have separate dedicated transporters. Indeed, Medema et al. (2010) predicted Amt_1 to be located in the cytoplasmic membrane and Amt_2 and -3 in the anammoxosome membrane. At present it is unclear how membrane proteins would be specifically targeted to the different membranes, as signal sequences have not been identified. It has therefore been suggested that chaperones may be required (Medema et al., 2010). Location of Amt transporters on the anammoxosome membrane poses the intriguing question of how these transporters could be regulated by their PII regulators. These bind at the exit of the Amt pore in the cytoplasm, blocking ammonium transport (Conroy et al., 2007; Gruswitz et al., 2007). The method of regulation has been well studied for ammonium uptake in assimilatory processes by the GS/GOGAT cycle in E. coli: a drop in internal ammonium concentration results in accumulation of 2-oxoglutarate in the cytoplasm, which binds to GlnK (the PII regulator) together with UDP and ATP. This causes release of GlnK from AmtB and subsequent transport of ammonium into the cell (Kim et al., 2012; Huergo et al., 2013). Regulation of Amt transporters in the anammoxosome membrane of K. stuttgartiensis by the same mechanism would require the GS/GOGAT system as well as a large part of the central carbon metabolism present inside the anammoxosome, which is unlikely as biosynthetic processes most probably take place in the cytoplasm. Whether ammonium import is differently regulated when ammonium is used for dissimilation rather than only for assimilatory processes is not known. A parallel with aerobic ammonium oxidizers cannot be drawn here as these organisms are thought to oxidize ammonium in the periplasm and they do therefore not have to import ammonium for dissimilation into the cell (Lehtovirta-Morley, 2018). Not all Amt transporters appear to be regulated through a PII regulator (Strösser et al., 2004; Tremblay et al., 2007), and this could be the case for potential Amt transporters in the anammoxosome membrane as well. Alternatively, it is possible that in anammox cells, high affinity ammonium transporters located on the cytoplasmic membrane concentrate ammonium/ammonia in the cytoplasm enough to allow sufficient diffusion of ammonia into the anammoxosome without additional dedicated transporters, or with help of a low affinity transporter. Since hydrazine can diffuse freely through dense ladderane membranes that are characteristic for anammox cells (Moss et al., 2018), the smaller ammonia will also be able to diffuse through, with or without a facilitator.
Nitrite and Nitrate Transporters
The genome of K. stuttgartiensis MBR1 encodes for six nitrite focA transporters. Multiple copies of nitrite transporters in one organism have been described for other organisms, but they are not usually all of the focA type (Clegg et al., 2002). Of the 6 focA transporters, focA_2 (KSMBR1_1070, Figure 2BII) was highly expressed in both conditions and further upregulated in response to nitrite limitation, and focA_5 (KSMBR1_3008) had low expression but was upregulated under ammonium limiting conditions (Figure 1D). The transporters focA_1 (KSMBR1_0300), focA_3 (KSMBR1_1474) and focA_4 (KSMBR1_1475) were barely expressed under either condition (∼38, 12, and 12 RPKM, respectively). In addition to the five annotated focA genes, a sixth focA homolog is annotated as the nitrate oxidoreductase nar1 (KSMBR1_3009, Table 1). This gene is located adjacent to focA_5 (Figure 2BX) and is upregulated (from 22 to 127 RPKM) upon ammonium limitation. Although PROKKA annotates it a nar1, BlastP analyses and information reported in the genome browser MaGe (Vallenet et al., 2013) suggest that this transporter belongs to the formate/nitrite transporter family instead. Previously, the homologous gene in strain K. stuttgartiensis RU1 was manually annotated as a focA formate/nitrite transporter (kusta0009) (Strous et al., 2006). We therefore conclude that nar1 is a nitrite transporter and suggest renaming it focA_6 (Table 1). We identified a further nitrite transporter homolog, nirC (KSMBR1_0451) that showed upregulation (from 83 to 623 RPKM) during nitrite limited growth.
The hypothetical genes KSMBR1_2720 (Supplementary Table 1) and KSMBR1_1075 (Figure 2BII) were upregulated in response to nitrite limitation and are predicted to be part of the HPP protein family. This recently described protein family consists of membrane proteins that have four trans-membrane domains and a conserved HPP motif. In cyanobacteria and chloroplasts, a HPP protein was shown to play a role in nitrite transport (Maeda et al., 2014). In addition, KSMBR_1075 is located in close proximity to the most highly expressed nitrite transporter focA_2. This suggests that KSMBR1_2720 and KSMBR_1075 may also function as nitrite transporters in K. stuttgartiensis and brings the total nitrite transporter homologs in the genome to nine.
In a study where K. stuttgartiensis RU1 was cultivated on nitric oxide and ammonium (without nitrite), the expression of the focA_2 homolog kuste3055 and the nirC homolog kuste4137 was downregulated 36- and 27-fold respectively (Hu et al., 2019). FocA/NirC transporters are relatively non-selective and can also transport other small anions (Lu et al., 2012; Lu et al., 2013). Therefore it is possible that FocA_2 and perhaps also NirC constitute the main nitrite transporters and the other FocA homologs may have a physiological function other than nitrite transport in K. stuttgartiensis. As anammox bacteria are able to use formate as an alternative electron donor to ammonium (Kartal et al., 2007, 2008; van de Vossenberg et al., 2008), transport of formate is a likely function for one or more of the other FocA transporters.
The genome also encodes for two nitrate/nitrite transporters, both of the NarK2-type, named narK1 (KSMBR1_3299), and narK2 (KSMBR1_3317), which were constitutively expressed at around 100 RPKM and did not respond to the investigated nutrient limitations. The NarK transporters are thought to export the by-product nitrate, which is produced from nitrite, probably by the nitrite oxidoreductase Nxr complex, thereby supplementing the electrons lost from the electron transport chain to carbon fixation (Reimann et al., 2015; Kartal and Keltjens, 2016). As the growth rates in both the ammonium- and nitrite limited conditions was kept the same, the rate of electrons lost to carbon fixation would be similar in both conditions and therefore the rate of nitrate production by Nxr is not expected to vary significantly between the two conditions.
Genes Involved in Energy Metabolism
Several genes whose products are involved in the anammox reaction were differentially regulated in ammonium- versus nitrite limited conditions (Figures 2B, 3). The genes significantly upregulated during nitrite limitation included those whose gene products require nitrite as a substrate: (1) the nirS gene (KSMBR1_0452) encoding a nitrite reductase, a candidate for catalyzing the conversion of nitrite to nitric oxide in the anammox reaction (Kartal et al., 2013). This gene was expressed at a low level (100 RPKM), but was 10-fold upregulated in response to nitrite limitation. The nirS gene is situated adjacent to the nirC nitrite transporter homolog (KSMBR1_0451) described above. (2) The nitrite oxidoreductase genes nrxCB (Figure 2BIII). Please note there is a typing error in the text of the KSMBR1 annotation here: nrx instead of nxr. These are the homologs of the nxrCB genes kustd1704 and 1703 in K. stuttgartiensis RU1 (Strous et al., 2006; Kartal and Keltjens, 2016) and are part of a larger gene cluster that encodes the nitrite oxidoreductase complex Nxr (see above). The Nxr complex in K. stuttgartiensis consists of a catalytic α subunit (NxrA), a β subunit thought to function as an electron channel (NxrB) and a γ subunit (NxrC) that has a heme b binding domain and is homologous to NxrC subunits from other organisms that function as reductases or dehydrogenases (Reimann et al., 2015). All nxr subunits (A: KSMBR1_1455, B: 1458, and C: 1459), were highly expressed and subunit B and C were upregulated in response to nitrite limitation. (3) The hydroxylamine oxidoreductase HAO_2 (KSMBR1_2163), which is one of ten paralogous HAO type genes present in K. stuttgartiensis MBR1, was expressed at a high level (2000 RPKM), and was also 2.5-fold upregulated in response to nitrite limitation.
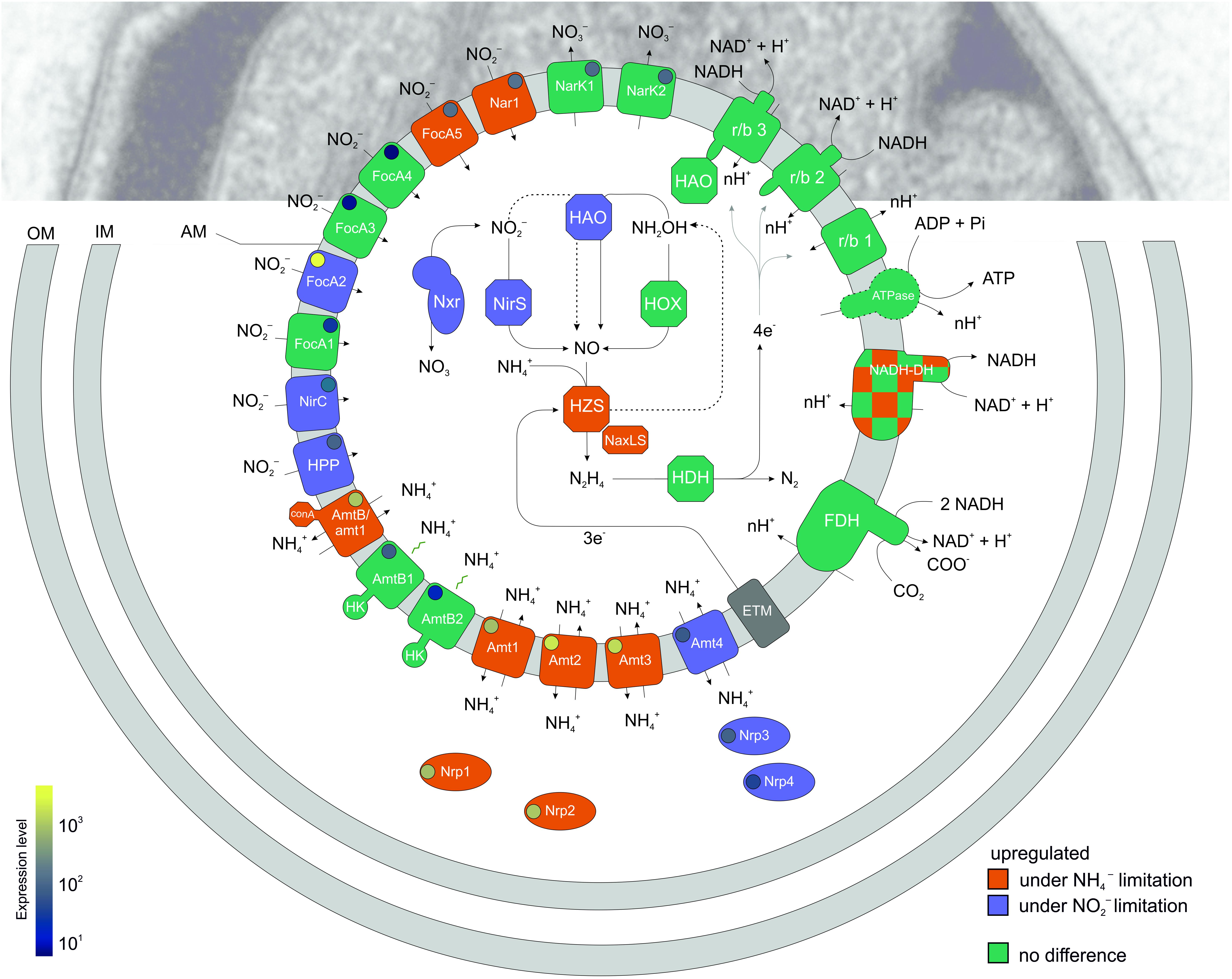
Figure 3. A schematic overview of a K. stuttgartiensis cell and the differential expression of gene products related to metabolism and nutrient transport under nitrite- versus ammonium limited bioreactor growth conditions. Transporters have been drawn on the anammoxosome membrane for comparison but their localization on either of the three anammox membranes awaits experimental validation. Genes up-regulated under nitrite- relative to ammonium limiting conditions are indicated in purple and genes upregulated under ammonium- relative to nitrite-limiting conditions are indicated in orange. Genes that are not affected when grown under either nitrite- or ammonium-limiting conditions are indicated in green. A circle within each transport related gene indicates relative expression level. OM, outer membrane; IM, inner (cytoplasmic) membrane; AM, anammoxosome membrane; HK, histidine kinase domain. Proteins depicted in this image: Ammonium transporters: AmtB/amt1 (KSMBR1_2086), Amt_1 (KSMBR1_2627), Amt_2 (KSMBR1_2630), Amt_3 (KSMBR1_2632), Amtb_2 (KSMBR1_3866), Amtb_1 (KSMBR1_3722), Amt_4 (KSMBR1_3716); Ammonium transport PII regulators: Nrp1 (KSMBR1_2628), Nrp2 (KSMBR1_2631), Nrp3 (KSMBR1_3717), Nrp4 (KSMBR1_3718); Nitrite transporters: FocA_5 (KSMBR1_3008), Nar1 (focA_6) (KSMBR1_3009), FocA_3 (KSMBR1_1474), FocA_4 (KSMBR1_1475), FocA_2 (KSMBR1_1070), FocA_1 (KSMBR1_0300), HPP (Putative nitrite transporter, KSMBR_1075); Nitrate transporters: NarK1 (KSMBR1_3317), NarK2 (KSMBR1_3299); Anammox key metabolism: NirS (KSMBR1_0452), HZS (hzsA: KSMBR1_2705, 2713, 3603), hzsB: (KSMBR1_2704, 2712 and 3602), and hzsC: (KSMBR1_2703, _2711, and 3601), NXR (A: KSMBR1_1455, B: 1458, and C: 1459), HAO (KSMBR1_2163), HAO r/b3 (KSMBR1_3792), HOX (KSMBR1_2670); Electron transport chain: FDH (KSMBR1_2469, 2470, 2471, 2479), r/b 1 (KSMBR1_1020 – 1021), r/b 3 (KSMBR1_ 3792 – 3797), r/b 2 (KSMBR1_1275 – 1283), ATPase (KSMBR1_0120 – 0130), NADH-DH (KSMBR1_3978 – 3990), ETM (unknown electron transport module).
The upregulation of the three genes nxrBC, nirS, and hao2 when growth was limited by nitrite may indicate that the affinities of the enzymes are too low to sustain enzyme activity and maintain the growth rate as the level of nitrite provided drops. This low affinity allows the organism to regulate the metabolic flux by modifying the amount of enzyme in the cell. Interestingly, both nirS and hao2 were upregulated. Both these enzymes have been proposed to be the nitrite reductase responsible for the conversion of nitrite to nitric oxide: In K. stuttgartiensis RU1, the nirS gene is expressed at lower levels compared to other genes involved in the energy metabolism and therefore the hydroxylamine oxidoreductase HAO_2 homolog kustc0458 is hypothesized to encode the enzyme that reduces nitrite to nitric oxide instead (Kartal et al., 2013). Our results show that both are expressed and upregulated under nitrite limitation, with hao2 showing 5-fold higher expression than nirS. Interestingly, Hu et al. (2019) reported that in K. stuttgartiensis RU1 cells cultivated with nitric oxide and ammonium rather than nitrite and ammonium, the nitrite reductase gene cluster (containing nirS and nirC), as well as the hao2 homolog kustc0458 were amongst the most downregulated genes in the transcriptome (Hu et al., 2019). So although nirS and hao2 are upregulated when nitrite concentrations are low, they shut down when nitrite is absent. This suggests that the expression of nirS and hao2 is regulated by a mechanism that does not respond linearly to the concentration of nitrite. It remains unclear whether one or both of these genes encode(s) the physiological nitrite reductase that produces the nitric oxide used by HZS – an important topic for future research.
In the next step in the anammox reaction, nitric oxide and ammonium are converted to hydrazine by the unique HZS complex, which consists of three subunits and two copies of each subunit (Dietl et al., 2015). In the genome of K. stuttgartiensis MBR1, there are three copies of the hzs operon present, all of which were very highly expressed. For comparison, hzsC_1 (KSMBR1_2711) is expressed at 12475 RPKM and hzsA_1 (KSMBR1_2713) at 28462 RPKM compared to genes involved in carbon fixation formate dehydrogenase which is expressed at 1584 RPKM and CO dehydrogenase (KSMBR1_3598)/acetyl-CoA synthase at 652 RPKM under nitrite-limited conditions. All copies of both hzsB (KSMBR1_3601, 2704, and 2712) and hzsC (KSMBR1_2703, 2711, and 3601) were upregulated in response to ammonium limitation, while only one copy of hzsA (hzsA_1, KSMBR1_2713) was upregulated (Figures 2BIX,XI, 3). However, as the three copies of the hzs cluster have nearly 100% identical gene sequences, most transcripts could map to all three copies. The algorithm maps each read only once so we could not determine from our data whether one or all hzs operons were highly transcribed. HZS is a slow enzyme when tested in vitro, but whether this is due to loss of activity during purification is not yet clear (Maalcke et al., 2014; Dietl et al., 2015). In all transcriptome studies of anammox species to date, the hzs genes were among the most highly expressed genes in the genome (Kartal et al., 2011b; Hu et al., 2013; van de Vossenberg et al., 2013; Bagchi et al., 2016; Russ et al., 2019). Also, in proteomics studies the HZS complex was shown to be a major part of the total protein complement (Kartal et al., 2011b; Hu et al., 2019) and maintained at these high levels even after 60 days of starvation (Bagchi et al., 2016; Ma and Wang, 2018). Even though HZS forms such a large part of total protein content of the cell, the hzs genes are still upregulated in response to ammonium limitation to ensure sufficient hydrazine production capacity. Russ et al. (2019) observed similar upregulation of the hzs genes of the marine anammox species Scalindua brodae during short term ammonium limitation (Russ et al., 2019). The large amount of HZS present in the cell is apparently not enough to maintain the level of hydrazine synthesis when ammonium levels are low, i.e., the affinity to ammonium is insufficient to maintain the rate of hydrazine synthesis.
Some of the genes predicted to be part of the electron transport chain were also upregulated in response to ammonium limitation, such as the complex I NADH-quinone oxidoreductase subunits D, E, F, G, L, and N (KSMBR 2471—2474, 2482, and 2485). The other subunits of this complex had higher, but not significantly higher, expression levels in ammonium- versus nitrite limited growth (Figures 2BVII, 3). The heterodimeric complex NaxLS with low redox potential (c-type cytochrome KSMBR1_3082 and KMSBR_3083) was highly expressed in both nitrite- and ammonium limited conditions (around 8,200 versus 10,000 RPKM, respectively) and the upregulation in response to ammonium limitation was significant (Supplementary Table 1). This complex binds to the HZS complex, and is hypothesized to serve one of two roles: to retain the volatile anammox intermediate nitric oxide inside the anammox cell and to shuttle it to hydrazine synthase or to shuttle electrons from an electron donor to hydrazine synthase (Ukita et al., 2010; Akram et al., 2019b). Akram et al. (2019b) hypothesize that if this complex is involved in retaining and shuttling nitric oxide, it should be upregulated in response to nitrite limitation to retain the precious nitric oxide within the cell. Indeed Hu et al. (2019) cultured K. stuttgartiensis on non-limiting amounts of nitric oxide instead of nitrite and observed 20- and 10-fold downregulation of the naxLS genes, respectively. However, we found upregulation during ammonium limitation when nitrite was not limiting, suggesting increased need when nitrite is not the limiting growth nutrient. As we also found upregulation of the hzs genes in ammonium limited conditions, it is possible that higher copy numbers of NaxLS are required to make sure all the HZS complexes in the anammoxosome are provided with sufficient NO.
Genes Involved in Ammonium Assimilation
During ammonium limitation, the dissimilatory enzymes have to compete with the assimilatory ammonium pathways for their substrate. Ammonium assimilation can occur via the high affinity GS/GOGAT system where glutamine synthase forms glutamine from glutamate and ammonium. Glutamate synthase subsequently transfers the amide group from glutamine to 2-oxoglutate (α-ketoglurate) to produce two glutamate molecules. Alternatively, ammonium can be assimilated by the low affinity glutamate dehydrogenase (GDH) which produces glutamate from 2-oxoglutarate and ammonium (Yan, 2007). K. stuttgartiensis MBR1 possesses two glutamine synthase genes, glnA_1 (KSMBR1_0034) and glnA_2 (KSMBR1_0678), and two hypothetical proteins with homology to glutamate synthase (KSMBR1_1164 and KSMBR1_3563). It also has two copies of the low affinity ammonium assimilation GDH gene gdh_1 (KSMBR1_1085) and gdh_2 (KSMBR1_2236). The GS/GOGAT genes were more highly expressed than the GDH genes but none showed upregulation during ammonium limited growth (Supplementary File 2), indicating that K. stuttgartiensis cells were able to maintain the intracellular concentration of ammonium during ammonium limited growth conditions. This is in contrast with the marine anammox species S. brodae, which did induce the GS/GOGAT genes when grown under ammonium limitation (Russ et al., 2019).
Other Genes
Three putative transposases showed significant differential expression: KSMBR_3038, an IS66 family transposase, was upregulated in response to ammonium limitation, and both KSMBR1_3122, an IS1634 family transposase, and KSMBR1_3846, an ISAs1 family transposase, were upregulated in response to nitrite limitation (Figure 2A and Supplementary Table 1).
The hypothetical gene KSMBR1_3177 was highly upregulated in response to ammonium limitation (Figure 2A). This gene has a large beta-barrel porin-2 (BBP2) domain, specific to outer membrane porins. This hypothetical porin may be upregulated to allow for a higher influx of ammonium from outside the cell. The glutamate formimidoyltransferase gene ftcD (KSMBR1_2087) is located just downstream of amtB/amt1 (KSMBR1_2086) on the genome and was upregulated in response to ammonium limitation (Figure 2BVII). Glutamate formimidoyltransferases are involved in histidine degradation to glutamate. There was some but not much transcript overlap between amtB/amt1 and ftcD and the expression of ftcD was much lower than amtB/amt1 so we conclude the genes are probably not forming an operon.
Conclusion
In summary, we have shown differential gene expression for several of the ammonium and nitrite transporter homologs in K. stuttgartiensis cells and this may partly explain the high copy number of these transporter genes in the genome. It is also clear that at least one of the ammonium transporters has a different function as an ammonium sensor rather than transporter (Pfluger et al., 2018). In addition, it is possible that specific transporters are located in different membranes in the cell. Our next step will be to investigate these other possible hypotheses for the high number of nutrient transporters in the K. stuttgartiensis genome. Although transcriptome analysis is a powerfool tool to study differential expression of genes, RNA levels do not necessarily reflect the levels of active protein in the cell. We therefore aim to follow up this work with proteome and activity studies to obtain more complete understanding of the physiology of K. stuttgartiensis in different growth conditions. The environmentally- and industrially- relevant anammox bacteria are food for thought with their compartmentalized energy metabolism. Investigating nutrient transport in a “complex” bacterial cell will ultimately help us understand how the cell regulates transport of nutrients and expression of transporters and how it responds to, and deals with, changing nutrient conditions in its environment.
Data Availability Statement
The datasets generated for this study can be found at NCBI in the Gene Expression Omnibus (GEO) under accession number GSE148825.
Author Contributions
MS, HC, and LN conceived parts of the project. MS and LN designed and discussed the experimental set-up. MS, DB, and GN designed, set-up, and maintained the bioreactors. MS, TA, and DB performed the experiments. MS, DB, SP, and LN analyzed the data. MS and SP prepared the figures. SP, MS, and LN wrote the manuscript with input from all authors. All authors were involved in research discussions.
Funding
SP and LN were supported by the Netherlands Organisation for Scientific Research (NWO) grant ALWOP.308 and VI.Vidi.192.001, respectively. MJ and MS were supported by the ERC AG Anammox 232970; MJ was also supported by the ERC AG Ecomom 339880; and MJ and GN were supported by the SIAM 024002002. HC was supported by the ERC Advanced Grant VOLCANO (669371).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Katinka van de Pas-Schoonen for her knowledge and help in operating the bioreactor. We thank Tom Bloembergen and Dorothee Junker for their work on projects that generated preliminary data, and Susana Andrade and Boran Kartal for helpful discussions. For invaluable bioinformatics advice, we are grateful to Chris Lawson, Geert Cremers, and Jeroen Frank. We would like to thank the reviewers for the valuable discussion on our text.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01959/full#supplementary-material
References
Akram, M., Dietl, A., Mersdorf, U., Prinz, S., Maalcke, W., Keltjens, J., et al. (2019a). A 192-heme electron transfer network in the hydrazine dehydrogenase complex. Sci. Adv. 5:eaav4310. doi: 10.1126/sciadv.aav4310
Akram, M., Reimann, J., Dietl, A., Menzel, A., Versantvoort, W., Kartal, B., et al. (2019b). A nitric oxide binding heterodimeric cytochrome c complex from the anammox bacterium Kuenenia stuttgartiensis binds to hydrazine synthase. J. Biol. Chem. 294, 16712–16728. doi: 10.1074/jbc.RA119.008788
Andrade, S. L. A., and Einsle, O. (2007). The Amt/Mep/Rh family of ammonium transport proteins. Mol. Membr. Biol. 24, 357–365. doi: 10.1080/09687680701388423
Andrade, S. L. A., and Einsle, O. (2013). The tricky task of nitrate/nitrite antiport. Angewandte Chemie Int. Ed. 52, 10422–10424. doi: 10.1002/anie.201305421
Atkovska, K., and Hub, J. S. (2017). Energetics and mechanism of anion permeation across formate-nitrite transporters. Sci. Rep. 7:12027. doi: 10.1038/s41598-017-11437-0
Baday, S., Orabi, E. A., Wang, S., Lamoureux, G., and Berneche, S. (2015). Mechanism of NH4+ recruitment and NH3 transport in Rh proteins. Structure 23, 1550–1557. doi: 10.1016/j.str.2015.06.010
Bagchi, S., Lamendella, R., Strutt, S., Van Loosdrecht, M. C. M., and Saikaly, P. E. (2016). Metatranscriptomics reveals the molecular mechanism of large granule formation in granular anammox reactor. Sci. Rep. 6:28327. doi: 10.1038/srep28327
Clegg, S., Yu, F., Griffiths, L., and Cole, J. A. (2002). The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol. Microbiol. 44, 143–155. doi: 10.1046/j.1365-2958.2002.02858.x
Conroy, M. J., Durand, A., Lupo, D., Li, X.-D., Bullough, P. A., Winkler, F. K., et al. (2007). The crystal structure of the Escherichia coli AmtB–GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. U.S.A. 104:1213. doi: 10.1073/pnas.0610348104
Coutts, G., Thomas, G., Blakey, D., and Merrick, M. (2002). Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21, 536–545. doi: 10.1093/emboj/21.4.536
de Almeida, N. M., Maalcke, W. J., Keltjens, J. T., Jetten, M. S. M., and Kartal, B. (2011). Proteins and protein complexes involved in the biochemical reactions of anaerobic ammonium-oxidizing bacteria. Biochem. Soc. Trans. 39, 303–308. doi: 10.1042/bst0390303
de Almeida, N. M., Wessels, H., De Graaf, R. M., Ferousi, C., Jetten, M. S. M., Keltjens, J. T., et al. (2016). Membrane-bound electron transport systems of an anammox bacterium: a complexome analysis. Biochim. Biophys. Acta Bioenerget. 1857, 1694–1704. doi: 10.1016/j.bbabio.2016.07.006
Dietl, A., Ferousi, C., Maalcke, W. J., Menzel, A., De Vries, S., Keltjens, J. T., et al. (2015). The inner workings of the hydrazine synthase multiprotein complex. Nature 527, 394–397. doi: 10.1038/nature15517
Feigl, F., and Amaral, J. R. (1958). Specific detection of nitrogen through pyrolytic oxidation in organic spot test analysis. Anal. Chem. 30, 1148–1150. doi: 10.1021/ac60138a046
Francis, C. A., Beman, J. M., and Kuypers, M. M. M. (2007). New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1, 19–27. doi: 10.1038/ismej.2007.8
Frank, J., Lucker, S., Vossen, R., Jetten, M. S. M., and Hall, R. J. (2018). Resolving the complete genome of Kuenenia stuttgartiensis from a membrane bioreactor enrichment using single-molecule real-time sequencing. Sci. Rep. 8:4580. doi: 10.1038/s41598-018-23053-7
Goddard, A. D., Bali, S., Mavridou, D. A. I, Luque-Almagro, V. M., Gates, A. J., Roldan, M. D., et al. (2017). The Paracoccus denitrificans NarK-like nitrate and nitrite transporters-probing nitrate uptake and nitrate/nitrite exchange mechanisms. Mol. Microbiol. 103, 117–133. doi: 10.1111/mmi.13546
Gruswitz, F., Connell, J., and Stroud, R. M. (2007). Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc. Natl. Acad. Sci. U.S.A. 104, 42–47. doi: 10.1073/pnas.0609796104
Hira, D., Toh, H., Migita, C. T., Okubo, H., Nishiyama, T., Hattori, M., et al. (2012). Anammox organism KSU-1 expresses a NirK-type copper-containing nitrite reductase instead of a NirS-type with cytochrome cd(1). FEBS Lett. 586, 1658–1663. doi: 10.1016/j.febslet.2012.04.041
Hu, Z., Van Alen, T., Jetten, M. S. M., and Kartal, B. (2013). Lysozyme and penicillin inhibit the growth of anaerobic ammonium-oxidizing Planctomycetes. Appl. Environ. Microbiol. 79, 7763–7769. doi: 10.1128/aem.02467-13
Hu, Z., Wessels, H. J. C. T., Van Alen, T., Jetten, M. S. M., and Kartal, B. (2019). Nitric oxide-dependent anaerobic ammonium oxidation. Nat. Commun. 10:1244. doi: 10.1038/s41467-019-09268-w
Huergo, L. F., Chandra, G., and Merrick, M. (2013). PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. 37, 251–283. doi: 10.1111/j.1574-6976.2012.00351.x
Jetten, M. S. M., Van Niftrik, L., Strous, M., Kartal, B., Keltjens, J. T., and Op Den Camp, H. J. M. (2009). Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 44, 65–84. doi: 10.1109/cleoe-eqec.2009.5193615
Jordan, D. B., and Wagschal, K. (2010). Properties and applications of microbial beta-D-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl. Microbiol. Biotechnol. 86, 1647–1658. doi: 10.1007/s00253-010-2538-y
Kartal, B., De Almeida, N. M., Maalcke, W. J., Op Den Camp, H. J. M., Jetten, M. S. M., and Keltjens, J. T. (2013). How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 37, 428–461. doi: 10.1111/1574-6976.12014
Kartal, B., Geerts, W., and Jetten, M. S. M. (2011a). Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Methods Enzymol. 486, 89–108. doi: 10.1016/b978-0-12-381294-0.00004-3
Kartal, B., and Keltjens, J. T. (2016). Anammox biochemistry: a tale of heme c proteins. Trends in Biochemical. Sciences. 41, 998–1011. doi: 10.1016/j.tibs.2016.08.015
Kartal, B., Kuypers, M. M. M., Lavik, G., Schalk, J., Den Camp, H., Jetten, M. S. M., et al. (2007). Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environmental. Microbiology. 9, 635–642. doi: 10.1016/j.syapm.2006.03.004
Kartal, B., Kuenen, J. G., and Van Loosdrecht, M. C. M. (2010). Sewage treatment with anammox. Science 328, 702–703. doi: 10.1126/science.1185941
Kartal, B., Maalcke, W. J., De Almeida, N. M., Cirpus, I., Gloerich, J., Geerts, W., et al. (2011b). Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–U159. doi: 10.1038/nature10453
Kartal, B., Van Niftrik, L., Rattray, J., De Vossenberg, J., Schmid, M. C., Damste, P. S., et al. (2008). Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol. 63, 46–55. doi: 10.1111/j.1574-6941.2007.00408.x
Kim, M., Zhang, Z. G., Okano, H., Yan, D. L., Groisman, A., and Hwa, T. (2012). Need-based activation of ammonium uptake in Escherichia coli. Mol. Syst. Biol. 8:616. doi: 10.1038/msb.2012.46
Lackner, S., Gilbert, E. M., Vlaeminck, S. E., Joss, A., Horn, H., and Van Loosdrecht, M. C. M. (2014). Full-scale partial nitritation/anammox experiences - an application survey. Water Res. 55, 292–303. doi: 10.1016/j.watres.2014.02.032
Lam, P., and Kuypers, M. M. M. (2011). Microbial nitrogen cycling processes in oxygen minimum zones. Annu. Rev. Mar. Sci. 3, 317–345. doi: 10.1146/annurev-marine-120709-142814
Lehtovirta-Morley, L. E. (2018). Ammonia oxidation: ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 365:fny058. doi: 10.1093/femsle/fny058
Li, G. B., Vilcherrez, D., Carvajal-Arroyo, J. M., Sierra-Alvarez, R., and Field, J. A. (2016). Exogenous nitrate attenuates nitrite toxicity to anaerobic ammonium oxidizing (anammox) bacteria. Chemosphere 144, 2360–2367. doi: 10.1016/j.chemosphere.2015.11.013
Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Lotti, T., Kleerebezern, R., Abelleira-Pereira, J. M., Abbas, B., and Van Loosdrecht, M. C. M. (2015). Faster through training: the anammox case. Water Res. 81, 261–268. doi: 10.1080/09593330.2014.982722
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, W., Du, J., Schwarzer, N. J., Gerbig-Smentek, E., Einsle, O., and Andrade, S. L. A. (2012). The formate channel FocA exports the products of mixed-acid fermentation. Proc. Natl. Acad. Sci. U.S.A. 109, 13254–13259. doi: 10.1073/pnas.1204201109
Lu, W., Schwarzer, N. J., Du, J., Wacker, T., Gerbig-Smentek, E., Einsle, O., et al. (2013). The Formate/Nitrite transporter family of anion channels. Eur. Biophys. J. Biophys. Lett. 42:S82. doi: 10.1515/hsz-2012-0339
Lupo, D., Li, X. D., Durand, A., Tomizaki, T., Cherif-Zahar, B., Matassi, G., et al. (2007). The 1.3-angstrom resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 19303–19308. doi: 10.1073/pnas.0706563104
Ma, X., and Wang, Y. Y. (2018). Anammox bacteria exhibit capacity to withstand long-term starvation stress: a proteomic-based investigation of survival mechanisms. Chemosphere 211, 952–961. doi: 10.1016/j.chemosphere.2018.07.185
Maalcke, W. J., Dietl, A., Marritt, S. J., Butt, J. N., Jetten, M. S. M., Keltjens, J. T., et al. (2014). Structural basis of biological NO generation by octaheme oxidoreductases. J. Biol. Chem. 289, 1228–1242. doi: 10.1074/jbc.M113.525147
Maalcke, W. J., Reimann, J., De Vries, S., Butt, J. N., Dietl, A., Kip, N., et al. (2016). Characterization of anammox hydrazine dehydrogenase, a key N2-producing enzyme in the global nitrogen cycle. J. Biol. Chem. 291, 17077–17092. doi: 10.1074/jbc.M116.735530
Maeda, S., Konishi, M., Yanagisawa, S., and Omata, T. (2014). Nitrite transport activity of a novel HPP family protein conserved in cyanobacteria and chloroplasts. Plant Cell Physiol. 55, 1311–1324. doi: 10.1093/pcp/pcz112
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Matassi, G. (2017). Horizontal gene transfer drives the evolution of Rh50 permeases in prokaryotes. BMC Evol. Biol. 17:2. doi: 10.1186/s12862-016-0850-6
McDonald, T. R., and Ward, J. M. (2016). Evolution of Electrogenic Ammonium Transporters (AMTs). Front. Plant Sci. 7:352. doi: 10.3389/fpls.2016.00352
Medema, M. H., Zhou, M., Van Hijum, S. A. F. T., Gloerich, J., Wessels, H. J. C. T., Siezen, R. J., et al. (2010). A predicted physicochemically distinct sub-proteome associated with the intracellular organelle of the anammox bacterium Kuenenia stuttgartiensis. BMC Genomics 11:299. doi: 10.1186/1471-2164-11-299
Merrick, M., Javelle, A., Durand, A., Severi, E., Thornton, J., Avent, N. D., et al. (2006). The Escherichia coli AmtB protein as a model system for understanding ammonium transport by Amt and Rh proteins. Transf. Clin. Biol. 13, 97–102. doi: 10.1016/j.tracli.2006.02.015
Moss, F. R. III, Shukena, S. R., Mercera, J. A. M., Cohena, C. M., Weissb, T. M., Boxera, S. G., et al. (2018). Ladderane phospholipids form a densely packed membrane with normal hydrazine and anomalously low proton/hydroxide permeability. Proc. Natl. Acad. Sci. U.S.A. 115, 9098–9103. doi: 10.1073/pnas.1810706115
Mukherjee, M., Vajpai, M., and Sankararamakrishnan, R. (2017). Anion-selective Formate/nitrite transporters: taxonomic distribution, phylogenetic analysis and subfamily-specific conservation pattern in prokaryotes. BMC Genomics 18:560. doi: 10.1186/s12864-017-3947-4
Neuhauser, B., Dynowski, M., and Ludewig, U. (2014). Switching substrate specificity of AMT/MEP/Rh proteins. Channels 8, 496–502. doi: 10.4161/19336950.2014.967618
Neumann, S., Wessels, H. J. C. T., Rijpstra, W. I. C., Damste, J. S. S., Kartal, B., Jetten, M. S. M., et al. (2014). Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol. Microbiol. 94, 794–802. doi: 10.1111/mmi.12816
Noinaj, N., and Buchanan, S. K. (2014). Structural insights into the transport of small molecules across membranes. Curr. Opin. Struct. Biol. 27, 8–15. doi: 10.1016/j.sbi.2014.02.007
Oshiki, M., Shinyako-Hata, K., Satoh, H., and Okabe, S. (2015). Draft genome sequence of an anaerobic ammonium-oxidizing bacterium, “Candidatus Brocadia sinica”. Microbiol. Resour. Announc. 3:e00267-15. doi: 10.1128/genomeA.00267-15
Peng, J., and Huang, C. H. (2006). Rh proteins vs Amt proteins: an organismal and phylogenetic perspective on CO2 and NH3 gas channels. Transf. Clin. Biol. 13, 85–94. doi: 10.1016/j.tracli.2006.02.006
Pfluger, T., Hernandez, C. F., Lewe, P., Frank, F., Mertens, H., Svergun, D., et al. (2018). Signaling ammonium across membranes through an ammonium sensor histidine kinase. Nat. Commun. 9:164. doi: 10.1038/s41467-017-02637-3
Qin, W., Amin, S. A., Lundeen, R. A., Heal, K. R., Martens-Habbena, W., Turkarslan, S., et al. (2018). Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 12, 508–519. doi: 10.1038/ismej.2017.186
Reimann, J., Jetten, M. S. M., and Keltjens, J. T. (2015). “Metal enzymes in “impossible” microorganisms catalyzing the anaerobic oxidation of ammonium and methane,” in Sustaining Life on Planet Earth: Metalloenzymes Mastering Dioxygen and Other Chewy Gases. Metal Ions in Life Sciences, Vol. 15, eds P. Kroneck and M. Sosa Torres (Cham: Springer).
Russ, L., Van Alen, T. A., Jetten, M. S. M., Op Den Camp, H. J. M., and Kartal, B. (2019). Interactions of anaerobic ammonium oxidizers and sulfide-oxidizing bacteria in a substrate-limited model system mimicking the marine environment. FEMS Microbiol. Ecol. 95:fiz137. doi: 10.1093/femsec/fiz137
Shaw, D. R., Ali, M., Katuri, K. P., Gralnick, J. A., Reimann, J., Mesman, R., et al. (2020). Extracellular electron transfer-dependent anaerobic oxidation of ammonium by anammox bacteria. BioRxiv [Preprint]. doi: 10.1101/855817
Speth, D. R., Hu, B., Bosch, N., Keltjens, J. T., Stunnenberg, H. G., and Jetten, M. S. M. (2012). Comparative genomics of two independently enriched “Candidatus Kuenenia stuttgartiensis” anammox bacteria. Front. Microbiol. 3:307. doi: 10.3389/fmicb.2012.00307
Speth, D. R., In’T Zandt, M. H., Guerrero-Cruz, S., Dutilh, B. E., and Jetten, M. S. M. (2016). Genome-based microbial ecology of anammox granules in a full-scale wastewater treatment system. Nat. Commun. 7:11172. doi: 10.1038/ncomms11172
Speth, D. R., Russ, L., Kartal, B., Op Den Camp, H. J. M., Dutilh, B. E., and Jetten, M. S. M. (2015). Draft genome sequence of anammox bacterium “Candidatus Scalindua brodae,” obtained using differential coverage binning of sequencing data from two reactor enrichments. Genome Announc. 3:e01415-14. doi: 10.1128/genomeA.01415-14
Strösser, J., Lüdke, A., Schaffer, S., Krämer, R., and Burkovski, A. (2004). Regulation of GlnK activity: modification, membrane sequestration and proteolysis as regulatory principles in the network of nitrogen control in Corynebacterium glutamicum. Mol. Microbiol. 54, 132–147. doi: 10.1111/j.1365-2958.2004.04247.x
Strous, M., Pelletier, E., Mangenot, S., Rattei, T., Lehner, A., Taylor, M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794. doi: 10.1038/nature04647
Taylor, S., Ninjoor, V., Dowd, D. M., and Tappel, A. L. (1974). Cathepsin b2 measurement by sensitive fluorometric ammonia analysis. Anal. Biochem. 60, 153–162. doi: 10.1016/0003-2697(74)90140-7
Thomas, G., Coutts, G., and Merrick, M. (2000). The glnKamtB operon: a conserved gene pair in prokaryotes. Trends Genet. 16, 11–14. doi: 10.1046/j.1365-2958.2000.01994.x
Tremblay, P. L., Drepper, T., Masepohl, B., and Hallenbeck, P. C. (2007). Membrane sequestration of PII proteins and nitrogenase regulation in the photosynthetic bacterium Rhodobacter capsulatus. J. Bacteriol. 189, 5850–5859. doi: 10.1128/JB.00680-07
Tremblay, P. L., and Hallenbeck, P. C. (2009). Of blood, brains and bacteria, the Amt/Rh transporter family: emerging role of Amt as a unique microbial sensor. Mol. Microbiol. 71, 12–22. doi: 10.1111/j.1365-2958.2008.06514.x
Ukita, S., Fujii, T., Hira, D., Nishiyama, T., Kawase, T., Migita, C. T., et al. (2010). A heterodimeric cytochrome c complex with a very low redox potential from an anaerobic ammonium-oxidizing enrichment culture. FEMS Microbiol. Lett. 313, 61–67. doi: 10.1111/j.1574-6968.2010.02122.x
Vallenet, D., Belda, E., Calteau, A., Cruveiller, S., Engelen, S., Lajus, A., et al. (2013). MicroScope-an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41, E636–E647. doi: 10.1093/nar/gks1194
van de Graaf, A. A., Debruijn, P., Robertson, L. A., Jetten, M. S. M., and Kuenen, J. G. (1997). Metabolic pathway of anaerobic ammonium oxidation on the basis of 15N studies in a fluidized bed reactor. Microbiology 143, 2415–2421. doi: 10.1099/00221287-143-7-2415
van de Vossenberg, J., Rattray, J. E., Geerts, W., Kartal, B., Van Niftrik, L., Van Donselaar, E. G., et al. (2008). Enrichment and characterization of marine anammox bacteria associated with global nitrogen gas production. Environ. Microbiol. 10, 3120–3129. doi: 10.1111/j.1462-2920.2008.01643.x
van de Vossenberg, J., Woebken, D., Maalcke, W. J., Wessels, H. J. C. T., Dutilh, B. E., Kartal, B., et al. (2013). The metagenome of the marine anammox bacterium ‘Candidatus Scalindua profunda’ illustrates the versatility of this globally important nitrogen cycle bacterium. Environ. Microbiol. 15, 1275–1289. doi: 10.1111/j.1462-2920.2012.02774.x
van den Berg, B., Lister, S., and Rutherford, J. C. (2019). Ammonium transceptors: novel regulators of fungal development. PLoS Pathog. 15:e1008059. doi: 10.1371/journal.ppat.1008059
van der Star, W. R. L., Miclea, A. I., Van Dongen, U., Muyzer, G., Picioreanu, C., and Van Loosdrecht, M. C. M. (2008). The membrane bioreactor: a novel tool to grow anammox bacteria as free cells. Biotechnol. Bioeng. 101, 286–294. doi: 10.1002/bit.21891
van Niftrik, L., Geerts, W. J. C., Van Donselaar, E. G., Humbel, B. M., Webb, R. I., Fuerst, J. A., et al. (2008a). Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 190, 708–717. doi: 10.1128/jb.01449-07
van Niftrik, L., Geerts, W. J. C., Van Donselaar, E. G., Humbel, B. M., Yakushevska, A., Verkleij, A. J., et al. (2008b). Combined structural and chemical analysis of the anammoxosome: a membrane-bounded intracytoplasmic compartment in anammox bacteria. J. Struct. Biol. 161, 401–410. doi: 10.1016/j.jsb.2007.05.005
van Niftrik, L., and Jetten, M. S. M. (2012). Anaerobic ammonium-oxidizing bacteria: unique microorganisms with exceptional properties. Microbiol. Mol. Biol. Rev. 76, 585–596. doi: 10.1128/mmbr.05025-11
Wacker, T., Garcia-Celma, J. J., Lewe, P., and Andrade, S. L. A. (2014). Direct observation of electrogenic NH4+ transport in ammonium transport (Amt) proteins. Proc. Natl. Acad. Sci. U.S.A. 111, 9995–10000. doi: 10.1073/pnas.1406409111
Weidinger, K., Neuhauser, B., Gilch, S., Ludewig, U., Meyer, O., and Schmidt, I. (2007). Functional and physiological evidence for a Rhesus-type ammonia transporter in Nitrosomonas europaea. FEMS Microbiol. Lett. 273, 260–267. doi: 10.1111/j.1574-6968.2007.00805.x
Wiechert, M., and Beitz, E. (2017a). Formate-nitrite transporters: monoacids ride the dielectric slide. Channels 11, 365–367. doi: 10.1080/19336950.2017.1329999
Wiechert, M., and Beitz, E. (2017b). Mechanism of formate-nitrite transporters by dielectric shift of substrate acidity. EMBO J. 36, 949–958. doi: 10.15252/embj.201695776
Yan, D. L. (2007). Protection of the glutamate pool concentration in enteric bacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 9475–9480. doi: 10.1073/pnas.0703360104
Zhang, L., and Okabe, S. (2017). Rapid cultivation of free-living planktonic anammox cells. Water Res. 127, 204–210. doi: 10.1016/j.watres.2017.10.029
Zhang, L., and Okabe, S. (2020). Ecological niche differentiation among anammox bacteria. Water Res. 171:115468. doi: 10.1016/j.watres.2020.115468
Keywords: anammox, planctomycete, anammoxosome, amtB, focA, narK, transcriptomics, nutrient limitation
Citation: Smeulders MJ, Peeters SH, van Alen T, de Bruijckere D, Nuijten GHL, op den Camp HJM, Jetten MSM and van Niftrik L (2020) Nutrient Limitation Causes Differential Expression of Transport- and Metabolism Genes in the Compartmentalized Anammox Bacterium Kuenenia stuttgartiensis. Front. Microbiol. 11:1959. doi: 10.3389/fmicb.2020.01959
Received: 20 April 2020; Accepted: 24 July 2020;
Published: 13 August 2020.
Edited by:
Martin Koenneke, University of Bremen, GermanyReviewed by:
Graeme W. Nicol, Université de Lyon, FrancePierre Offre, Royal Netherlands Institute for Sea Research (NIOZ), Netherlands
Willm Martens-Habbena, University of Florida, United States
Copyright © 2020 Smeulders, Peeters, van Alen, de Bruijckere, Nuijten, op den Camp, Jetten and van Niftrik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura van Niftrik, bC52YW5uaWZ0cmlrQHNjaWVuY2UucnUubmw=
†These authors have contributed equally to this work
 Marjan J. Smeulders
Marjan J. Smeulders Stijn H. Peeters
Stijn H. Peeters Theo van Alen
Theo van Alen Daan de Bruijckere
Daan de Bruijckere Guylaine H. L. Nuijten
Guylaine H. L. Nuijten Huub J. M. op den Camp
Huub J. M. op den Camp Mike S. M. Jetten
Mike S. M. Jetten Laura van Niftrik
Laura van Niftrik