- 1Biomedical Research Centre, Qatar University, Doha, Qatar
- 2Laboratory of Medical Microbiology, Vaccine and Infectious Disease Institute, University of Antwerp, Antwerp, Belgium
- 3Laboratory Medicine and Pathology, Hamad General Hospital, Doha, Qatar
- 4Infectious Disease Department, Hamad General Hospital, Doha, Qatar
This study was performed to investigate the genotypic causes of colistin resistance in 18 colistin-resistant Klebsiella pneumoniae (n = 13), Escherichia coli (n = 3) and Pseudomonas aeruginosa (n = 2) isolates from patients at the Hamad General Hospital, Qatar. MIC testing for colistin was performed using Phoenix (BD Biosciences, Heidelberg, Germany) and then verified with SensiTest Colistin (Liofilchem, Zona Ind. le, Italy). Strains determined to be resistant (MIC > 4-16 μg/mL) were then whole-genome sequenced (MiSeq, Illumina, Inc.). Sequences were processed and analysed using BacPipe v1.2.6, a bacterial whole genome sequencing analysis pipeline. Known chromosomal modifications were determined using CLC Genomics Workbench v.9.5.3 (CLCbio, Denmark). Two K. pneumoniae isolates (KPN-15 and KPN-19) harboured mcr-8.1 on the IncFII(K) plasmids, pqKPN-15 and pqKPN-19, and belonged to ST383 and ST716, respectively. One E. coli isolate harboured mcr-1.1 on the IncI2 plasmid pEC-12. The other 15 isolates harboured known chromosomal mutations linked to colistin resistance in the PhoPQ two-component system. Also, three K. pneumoniae strains (KPN-9, KPN-10 and KPN-15) showed disruptions due to IS elements in mgrB. To our knowledge, this marks the first description of mcr-8.1 in K. pneumoniae of human origin in Qatar. Currently, more research is necessary to trace the source of mcr-8.1 and its variants in humans in this region.
Introduction
Resistance to colistin, a last-resort antibiotic, has been identified in humans, livestock animals, and the environment (Forde et al., 2018). Colistin (polymyxin E) is a cationic polypeptide synthesised from Paenibacillus polymyxa subspecies Colistinus (Poirel et al., 2017). Colistin was first introduced for the treatment of GNB (Gram-negative bacteria) infections in 1952 and was used until the 1980s; however, its use in human medicine was limited due to neuro and nephrotoxicity (Akajagbor et al., 2013). Unfortunately, the emergence of multi and extensive drug resistance pathogens has necessitated colistin’s reintroduction to clinical practice (Catchpole et al., 1997). Colistin resistance is typically caused by chromosomally mediated modifications to its target site, the LPS (lipopolysaccharide) in the outer membrane of GNB, thereby reducing its binding affinity (Poirel et al., 2015; Aghapour et al., 2019). Colistin resistance was thought to have only a vertical spread, but recently horizontally transmitted plasmid-mediated resistance has emerged.
Colistin is a cationic molecule that interactions with the negatively charged lipopolysaccharide present on the outer membrane. This increases membrane permeability, eventually leading to bacterial death (Poirel et al., 2015). Interestingly, colistin resistance has been linked to higher levels of susceptibility to other antimicrobial agents, due to the outer membrane disruption enabling better passage of these into the cell. This appears to be particularly true for hydrophobic antibiotics (Li et al., 2020). Conversely, colistin resistance has also been implicated in resistance to cationic antimicrobial peptides present in the host. Studies have documented that mcr-1.1 can confer resistance to the cationic lysozyme as well as other cationic host antimicrobials (Napier et al., 2013; Sherman et al., 2016). This presents a significant concern due to the potential of mcr genes to be maintained due to the advantage they provide in surviving host-microbial (Napier et al., 2013; Sherman et al., 2016).
Transferable resistance was first reported in China in 2015 as the mcr-1.1 gene, a phosphoethanolamine (pEtN) transferase which adds pEtN to the lipid A moiety of the LPS, preventing colistin binding through the reduction of the negative charge on the LPS (Liu et al., 2016; Aghapour et al., 2019). Homologous pEtN transferases were identified in Gram-negative bacteria, such as mcr-2.1 from Belgian porcine E. coli (Xavier et al., 2016). Since then, nine different mcr genes and several variants have been identified across the globe (Carroll et al., 2019). In Qatar, mcr-1.1 was first reported in 2018 in E. coli isolated from broiler chickens and MDR E. coli from a patient with subarachnoid haemorrhage (Eltai et al., 2017; Forde et al., 2018). Additionally, studies have described mcr-1.1 from clinical isolates in Jordan and mcr-5.1 present in human isolates in Saudi Arabia (Redhwan et al., 2019). Mcr-8.1 has been previously reported in animal, and human Klebsiella pneumoniae isolates from China and Laos, and recently, Algeria, Morocco, France, Bangladesh, and Saudi Arabia (Wang et al., 2018, 2019; Hadjadj et al., 2019; Bonnin et al., 2020; Farzana et al., 2020; Hala et al., 2020; Nabti et al., 2020). A recent review on colistin resistance in the Middle East reported the presence of mcr genes in six countries in the region in K. pneumoniae isolates (Aris et al., 2020). Interestingly, it is reported that the majority of resistance is mediated by mgrB mutations with few appearing to harbour plasmid-mediated resistance. However, the proportion of resistance due to mcr gene has been increasing since their discovery in 2015. This increase in resistance has been suggested the use of antibiotics in the region (Aris et al., 2020).
While primarily present in animal associated isolates mcr genes are a typical example of the importance and necessity for a One Health approach, with recent reports showing carriage of mcr genes in both hospital and community-associated human bacterial pathogens such as K. pneumoniae and Acinetobacter spp. (Agaba et al., 2017). Therefore, surveillance of antimicrobial resistance mandates the determination of the prevalence of mcr in human bacterial pathogens and an assessment of their impact on clinical outcome. In this study, we analysed 18 isolates obtained from patients admitted to the Hamad General Hospital (HGH) in Qatar in order to determine the prevalence and identify the underlying mechanisms leading to their colistin resistance.
Materials and Methods
In total, 18 isolates belonging to K. pneumoniae (n = 13), E. coli (n = 3) and Pseudomonas aeruginosa (n = 2) were analysed in this study. Strain characteristics of the 18 isolates are described in Table 1 and Supplementary Table S1. Species confirmation was performed by MALDI-TOF (Bruker Daltonik GmbH, Leipzig, Germany). Ethical approval (MRC-01-17-198) was obtained from the Medical Research Centre (MRC), Hamad Medical Corporation (HMC), Doha, Qatar. Initial antimicrobial susceptibility testing was performed by Phoenix using the NMIC/ID-5 panel (BD Biosciences, Heidelberg, Germany) according to the manufacturer’s recommendations. Briefly, panels were inoculated with 0.5 McFarland pure culture, placed into the instrument and incubated at 35°C. The instrument tests panel every 20 min up to 16 h if necessary. MIC values of each antimicrobial agent automatically read as susceptible, intermediate, or resistant (SIR). Isolates showing colistin resistance by Phoenix were consequently confirmed using the SensiTest colistin kit (Liofilchem, Zona Ind. le, Italy), following the manufacturer’s instructions. Briefly, a suspension of 0.5 McFarland standard was prepared from the tested organism, and then diluted 1:20 in saline. 0.4 ml of this diluted suspension was added to a tube of Muller Hinton broth provided in the kit (solution B). 100 μl of solution B was dispensed to the seven wells in a row. Then the plate was incubated at 36 ± 2°C for 16-20 h. The lowest concentration of antibiotic that inhibits visible growth was recorded and interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines.
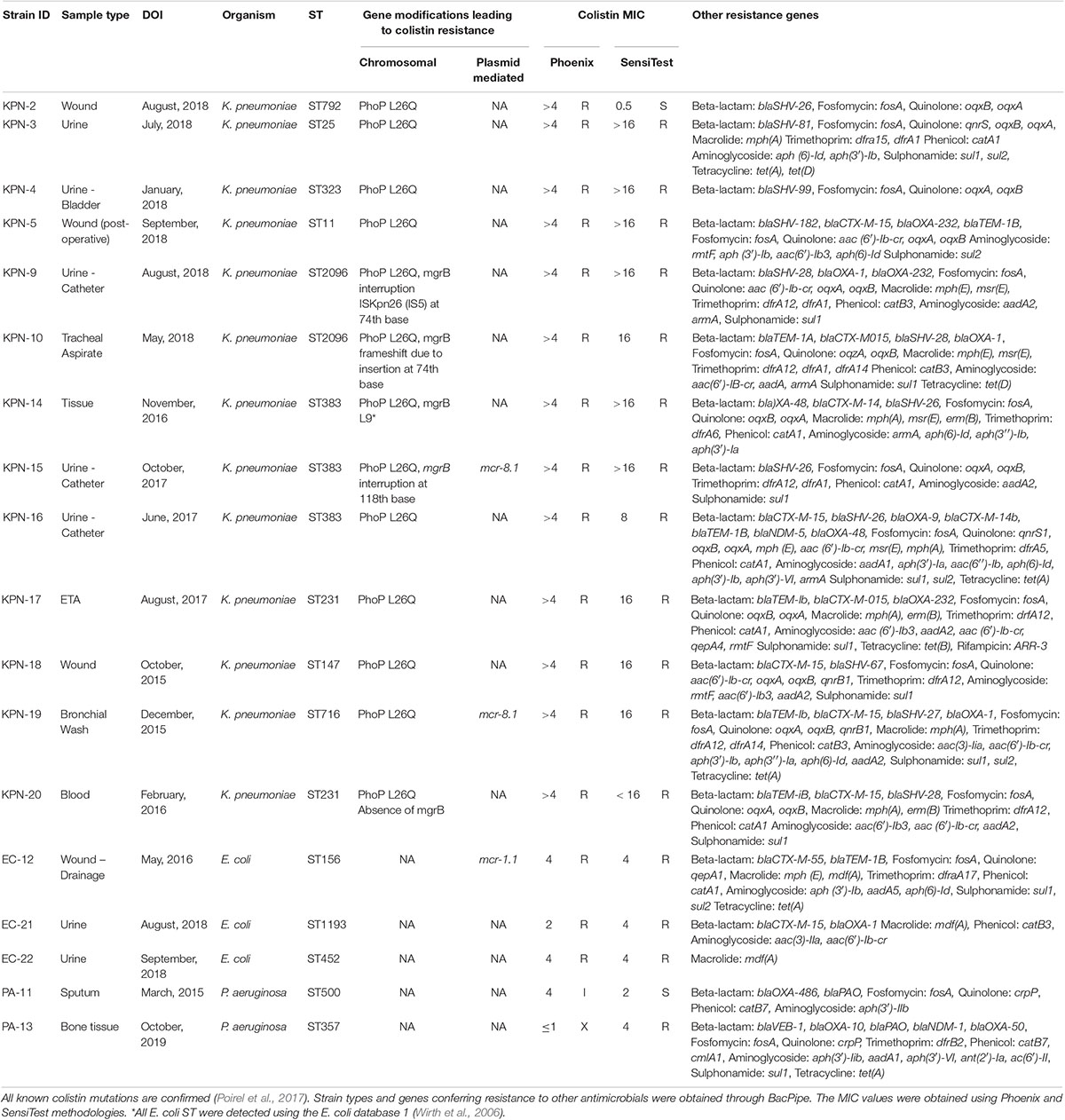
Table 1. Mutations present in isolates (n = 18) of K. pneumoniae (n = 13), E. coli (n = 3), and P. aeruginosa (n = 2) conferring resistance to colistin and other antimicrobials.
DNA Extraction and Sequencing
Genomic DNA was extracted from all isolates using a QIAamp® UCP Pathogen mini kit (Qiagen, Düsseldorf, Germany) according to the manufacturer’s protocol. Briefly, from each isolate’s genomic DNA was purified, and later quantified using a Qubit dsDNA high sensitivity assay (Thermo Fisher, Waltham, United States). Libraries were prepared using Nextera XT and sequenced via (2 bp × 250 bp) v2 500 cycle, MiSeq (Illumina, Inc., United States). The raw sequences were subjected to a quality check and analysis using our in-house pipeline, BacPipe v.1.2.6 (Xavier et al., 2020). Briefly, the reads were quality assessed, trimmed, followed by de novo assembly (SPAdes), typing (MLST), genome annotation (Prokka), and resistance genes (ResFinder and CARD). The scaffolds were run through Plasmid Finder (Carattoli et al., 2014) to determine the incompatibility type of the plasmids. Insertion element (IS) families were analysed through IS Finder (Siguier, 2006). Genetic characterisation, chromosomal gene modifications and mcr gene based phylogenetic analysis were done using CLC Genomics workbench v9.5.3 (CLCbio, Denmark).
Results
All isolates (n = 18) showed resistance to colistin with MICs > 4 μg/mL in both Phoenix and SensiTest results, except KPN-2 and EC-21 (Table 1). The clonal diversity among the isolates was determined; the K. pneumoniae isolates had nine different strain types (STs) (ST11, ST383, ST792, ST25, ST323, ST231, ST147, ST716, and ST2096, the E. coli isolates had two unique types (ST19, ST53), and one novel ST (Wirth et al., 2006), and P. aeruginosa had two types (ST500, ST357) (Table 1).
Two K. pneumoniae isolates (KPN-15 and KPN-19) harboured mcr-8.1 (Figure 1), and one E. coli (EC-12) carried mcr-1.1. For the mcr-8.1 harbouring strains, KPN-15 was found to have a MIC of > 16 μg/mL and KPN-19 a value of 16 μg/mL when evaluated using SensiTest. All other K. pneumoniae isolates had MIC values of 16 or > 16 μg/mL (Table 1). EC-12 harboured mcr-1.1 and was observed to have a MIC value by SensiTest of 4 μg/mL.
The contigs containing the mcr genes were analysed using BLAST to determine their origin and genetic context (Figure 1). Contig from isolate KPN-15 gave the top BLAST search hit to plasmid pLH94-8 (CP035204) with an 81% coverage and 99.7% sequence identity. Similarly, contig from isolate KPN-19, a top BLAST search hit was plasmid pLH94-8, with a 77% query coverage and 99.3% nucleotide identity. Isolates KPN-15 and KPN-19 harboured plasmid types (IncFII(Yp) and IncL) and IncFII(K), respectively. E. coli isolate EC-12 underwent the same analysis procedure as the K. pneumoniae strains. Isolate EC-12 contained mcr-1.1 with the top BLAST search hit being E. coli p1540-1 (CP019052) with 100% query coverage and 99.9% sequence identity. PlasmidFinder revealed the mcr-1.1 was harboured on plasmid type IncI2. KPN-15 and KPN-19 containing mcr-8.1 as well as the reference pLH94-8 were also examined for additional resistance genes to other antimicrobial agents and genetic similarity. The contigs containing the mcr-8.1 gene, harbours no other resistance genes were identified for either KPN-15 or KPN-19. However, it was found that all isolates harbours other resistance genes such as beta-lactams, aminoglycoside, macrolides and tetracycline (Table 1).
While only three isolates (KPN-15, KPN-19, and EC-12) carried mcr genes, other genetic mutations that have been previously associated with colistin resistance were observed and are reported in Table 1. In particular, mutations in the negative PhoPQ regulator mgrB, as well as in phoP, were the main modifications in study isolates linked to colistin resistance. K. pneumoniae isolates (KPN-9, KPN-10, KPN-14, and KPN-15) had modifications in the mgrB gene. Three of these isolates (KPN-9, KPN-10, and KPN-15) had MIC values of > 16 μg/mL while KPN-14 had a value of 16 μg/mL. KPN-9 and KPN-10 had the gene disruption at position 74th nucleotide, while KPN-15 had it at position 118th nucleotide. For KPN-9 it was determined that ISKpn-26 mediated the disruption. No insertion element could be found for KPN-10, and the change appeared to be due to a frameshift mutation. For KPN-15, while the disruption was mediated by IS1 family elements, the specific element could not be determined. Isolate KPN-20 appeared to be lacking the mgrB gene entirely, with only 10 bp remnants corresponding to the gene being found. While there were multiple mutations observed in the PhoPQ and PmrAB two-component systems in our isolates, only the phoP L26Q mutation was linked to colistin resistance. This mutation was present in all the K. pneumoniae isolates (n = 13). The evolutionary relationship of mcr-8.1 were compared with all other known mcr genes (1-10) and its variants. The mcr-8.1 gene is closely related to mcr-4 and mcr-5 genes (Supplementary Figure S1).
Discussion
Clonal Diversity and Plasmid Types
The clonal diversity of the isolates showed that the majority belonged to known clones. In particular, the ST for K. pneumoniae mostly belonged to known epidemic clones (ST147, ST11, ST231, ST792, ST383). For the isolates that harboured mcr genes (KPN15, KPN-19, EC-12), the ST type was examined to see if it had previously been linked to other antimicrobial-resistant mechanisms. KPN-15 belonged to ST383; a well-characterised ST known to harbour genes conferring carbapenem resistance (Sabirova et al., 2016), while KPN-19 was ST716, a strain type that has been previously linked to an outbreak of New-Delhi metallo-beta-lactamase resistant K. pneumoniae (Heinrichs et al., 2019). EC-12 belonged to ST156, reported previously in mcr-1.1 harbouring E. coli in Brazil, and a carbapenemase carrying isolate in China (Rossi et al., 2017).
The incompatibility types of the plasmids found in these isolates harbouring mcr-8.1 or mcr-1.1 were searched in the literature to look for previous reports of mcr carriage. The presence of IncFII plasmids in KPN-15 and KPN-19 potential source of origin harbouring mcr-8.1 gene variants has been reported in previous studies (Wang et al., 2018, 2019; Hadjadj et al., 2019; Wang et al., 2019). Similarly, the presence of IncI2 in EC-12 is unsurprising as it is the first Inc type described with mcr-1.1 and has been widely reported since then (Zurfluh et al., 2017).
mcr Genes
Studies have shown that mcr-1.1 confers low to moderate level resistance (Cao et al., 2018), as seen in isolate EC-12 which had a MIC value of 4 μg/mL. Previous studies on mcr-8.1 have reported MIC values of 16 μg/mL (Wang et al., 2018; Bonnin et al., 2020), however, for our study, we are unable to conclude if our isolates demonstrate higher than 16 μg/mL as the SensiTest method can only delineate colistin MICs up to 16 μg/mL. Nonetheless, all of the resistant K. pneumoniae strains, whether or not they carried mcr-8.1, had values of 16 μg/mL or > 16 μg/mL (Table 1). Additionally, mutations to the PhoPQ system, such as the L26Q phoP mutation seen in all K. pneumoniae strains, confer low to moderate level (8-64 μg/mL) polymyxin resistance (Miller et al., 2011).
A recent paper by Farzana et al. (2020) on isolates from Bangladesh describes the presence of mcr-8.1 on plasmid IncFIB(pQil) and is associated with a fitness cost. The study proposed that mcr-8.1 inserted at the same site as carbapenem resistance genes in IncFIB(pQil) (Farzana et al., 2020). This paper also noted that there was evidence suggesting that IS903B elements mediated the initial insertion of mcr-8.1 into the IncFIB(pQil) plasmid as these were seen to bracket all antimicrobial resistance elements in the studied strains (Farzana et al., 2020). IS903B has also been observed in other previously conducted studies on the mcr-8.1 carriage in K. pneumoniae (Hadjadj et al., 2019). Interestingly, isolates KPN-15 and KPN-19, harbour IS903B. This suggests that IS5 family elements may have some role in resistance gene transfer but is inconclusive regarding mcr-8.1 transfer specifically. For both our isolates and those in the Farzana et al. (2020) paper, the sequences flanking the mcr-8.1 gene were examined for similarities. It was found that the genetic organisation of the contigs containing the mcr-8.1 gene for our strains as well as the Farzana strains were highly similar. KPN-15/19 and the three Farzana strains share the same genetic context in the immediate flanking regions of mcr-8.1.
In contrast, the Farzana strains have approximately the entire length of the mcr-8.1 harbouring plasmid in common and KPN-15 and KNP-19 share approximately 31740 bp in the mcr-8.1 containing contigs (data not shown). Due to the pattern of similarities in the genetic organisation, it is hypothesised that all of the strains share a common mcr-8.1 ancestor but are not evidence of clonal spread between Qatar and Bangladesh. Some of the key genes observed flanking mcr-8.1 in all the strains were copR and sasA. In previous studies the presence of copR was also observed on a K. pneumoniae pK19 plasmid from pig faeces and a K. pneumoniae pLH94-8 IncFII plasmid from a human sample (Wang et al., 2018; Hadjadj et al., 2019).
A recent study described in conference proceedings documented the first known case of mcr-8.1 of human origin in Saudi Arabia and concluded that the resistance phenotype was due to not only mcr-8.1 but also the presence of mcr-1.1 (Hala et al., 2020). This corresponds to our findings that mcr-8.1 did not appear to be the sole source of colistin resistance and worked in collaboration with mgrB modifications. Due to our findings of mcr-8.1 and mcr-1.1 in human isolates in Qatar, the Saudi Arabia findings are of concern as they indicate a rise in prevalence of these resistance mechanisms in the region. Due to this, it is imperative that wider screening be implemented in order to improve the detection of mcr-8.1 and other variants in the region.
Chromosomal Modifications Linked to Colistin Resistance
In particular, mutations in the negative PhoPQ regulator mgrB, as well as in phoP, were the main modifications in our isolates linked to colistin resistance. Mutations, deletions, disruptions, etc. in the mgrB gene have been linked to high levels of colistin resistance, particularly in K. pneumoniae. While multiple mutations were observed in the PhoPQ and PmrAB two-component systems in our isolates, only the phoP L26Q mutation has been implicated in colistin resistance (Uz Zaman et al., 2018). Interestingly, the phoP L26Q mutation was also present in previously reported mcr-8.1 harbouring K. pneumoniae (Wang et al., 2018; Bonnin et al., 2020; Nabti et al., 2020). This question that role of this mutation in conferring resistance.
In a 2014 study, it was reported that mgrB genes are thought to play a significant role in the resistance phenotype (Cannatelli et al., 2014). A 2018 study by Pitt el al noted that mgrB disruptions at position 74 were the most common (Pitt et al., 2018). This disruption is also seen in our isolates KPN-9 and KPN-10. In particular, the disruption in KPN-9 was mediated by ISKpn26; the same IS element previously identified as the most common driver of mutation in mgrB in K. pneumoniae (Pitt et al., 2018). For isolate KPN-15 the mgrB gene was split at nucleotide 118 due to an insertion element of the IS1 family. Due to the use of short-read sequencing, the specific type of IS1 element responsible for this disruption could not be determined. The lack of mgrB in isolate KPN-20 suggests the complete disruption of the gene.
Conclusion
In this study, we show that different genetic factors play a role in colistin resistance among isolates of K. pneumoniae from various clinical samples in a hospital in Qatar. The primary mechanism of colistin resistance remains via chromosomal mutations. However, we have identified mcr-8.1 in K. pneumoniae and mcr-1.1 in E. coli that harbour both mcr genes and known chromosomal mutations, more specifically a single amino acid change in phoP, the net contribution of which needs to be studied further. This data is complementary to previous studies indicating the emergence of mcr-8.1 in humans, both globally and in the Middle East. This suggests a more widespread distribution of the gene and the necessity for wider screening to detect its presence in clinical centres.
Data Availability Statement
All datasets associated with this study are included in the article/Supplementary Material. The sequences of all 18 isolates described in this study submitted to NCBI GenBank under the BioProject: PRJNA609881.
Ethics Statement
Ethical approval (MRC-01-17-198) was obtained from the Medical Research Centre (MRC), Hamad Medical Corporation (HMC), Doha, Qatar.
Author Contributions
SM-K conceptualised the study. SM-K, BX, and NE designed the study. NE, EI, HA-M, HY, MA, CL, and AA carried out the experimental work. BK and BX performed the sequencing and analysis. BK, BX, NE, and SM-K drafted the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by grant MRC-01-17-198 from the Hamad General Hospital, Qatar.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01954/full#supplementary-material
FIGURE S1 | Evolutionary relationship of mcr genes: the complete protein sequences of mcr variants from the GenBank were aligned (clustalx) and used for building a maximum likelihood tree. Branch support value, in percent, indicates Bayesian posterior probability, followed by maximum likelihood bootstrap value (>70). iTOL (https://itol.embl.de/) was used to visualise the tree.
TABLE S1 | Strain and clinical characteristics of the isolates described in this study.
References
Agaba, P., Tumukunde, J., Tindimwebwa, J. V. B., and Kwizera, A. (2017). Nosocomial bacterial infections and their antimicrobial susceptibility patterns among patients in Ugandan intensive care units: a cross sectional study. BMC Res. Notes 10:349. doi: 10.1186/s13104-017-2695-2695
Aghapour, Z., Gholizadeh, P., Ganbarov, K., Bialvaei, A. Z., Mahmood, S. S., Tanomand, A., et al. (2019). Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 12, 965–975. doi: 10.2147/IDR.S199844
Aghapour, Z., Gholizadeh, P., Ganbarov, K., Zahedi Bialvaei, A., Saad Mahmood, S., Tanomand, A., et al. (2019). Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 12, 965–975. doi: 10.2147/idr.s199844
Akajagbor, D. S., Wilson, S. L., Shere-Wolfe, K. D., Dakum, P., Charurat, M. E., and Gilliam, B. L. (2013). Higher incidence of acute kidney injury with intravenous Colistimethate sodium compared with Polymyxin B in critically Ill patients at a tertiary care medical center. Clin. Infect. Dis. 57, 1300–1303. doi: 10.1093/cid/cit453
Aris, P., Robatjazi, S., Nikkhahi, F., and Marashi, M. A. (2020). Molecular mechanisms and prevalence of colistin resistance of Klebsiella pneumoniae in the Middle East region: a review over five last years. J. Glob. Antimicrob. Resist. 22, 625–630. doi: 10.1016/j.jgar.2020.06.009
Bonnin, R. A., Bernabeu, S., Jaureguy, F., Naas, T., and Dortet, L. (2020). MCR-8 mediated colistin resistance in a carbapenem-resistant Klebsiella pneumoniae isolated from a repatriated patient from Morocco. Int. J. Antimicrob. Agents 55:105920. doi: 10.1016/j.ijantimicag.2020.105920
Cannatelli, A., Giani, T., D’Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC.03110-3114
Cao, L., Li, X., Xu, Y., and Shen, J. (2018). Prevalence and molecular characteristics of mcr-1 colistin resistance in Escherichia coli: isolates of clinical infection from a Chinese University Hospital. Infect. Drug Resist. 11, 1597–1603. doi: 10.2147/IDR.S166726
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19
Catchpole, C. R., Andrews, J. M., Brenwald, N., and Wise, R. (1997). A reassessment of the in-vitro activity of colistin sulphomethate sodium. J. Antimicrob. Chemother. 39, 255–260. doi: 10.1093/jac/39.2.255
Eltai, N. O., Abdfarag, E. A., Al-Romaihi, H., Wehedy, E., Mahmoud, M. H., Alawad, O. K., et al. (2017). Antibiotic resistance profile of commensal Escherichia coli isolated from broiler chickens in qatar. J. Food Prot. 81, 302–307. doi: 10.4315/0362-028X.JFP-17-191
Farzana, R., Jones, L. S., Barratt, A., Rahman, M. A., Sands, K., Portal, E., et al. (2020). Emergence of mobile colistin resistance (mcr-8) in a Highly Successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere 5:e0023-20. doi: 10.1128/mSphere.00023-20
Forde, B. M., Zowawi, H. M., Harris, P. N. A., Roberts, L., Ibrahim, E., Shaikh, N., et al. (2018). Discovery of mcr-1 -mediated colistin resistance in a highly virulent Escherichia coli Lineage. mSphere 3:e00486-18. doi: 10.1128/mSphere.00486-18
Hadjadj, L., Baron, S. A., Olaitan, A. O., Morand, S., and Rolain, J.-M. (2019). Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 10:2720. doi: 10.3389/fmicb.2019.02720
Hala, S., Paul Antony, C., Alshehri, M., Alsaedi, A., Al Thaqafi, A., Momin, A., et al. (2020). Emergence of mobile colistin resistance genes mcr-1 and mcr-8 in Saudi Arabia in Abstracts of GCCMID 2019. J. Infect. Public Health 13:329. doi: 10.1016/j.jiph.2020.01.064
Heinrichs, A., Argudín, M. A., De Mendonça, R., Deplano, A., Roisin, S., Dodémont, M., et al. (2019). An outpatient clinic as a potential site of transmission for an outbreak of New Delhi Metallo-β-Lactamase-producing Klebsiella pneumoniae sequence type 716: a study using whole-genome sequencing. Clin. Infect. Dis. 68, 993–1000. doi: 10.1093/cid/ciy581
Li, B., Yin, F., Zhao, X., Guo, Y., Wang, W., Wang, P., et al. (2020). Colistin resistance gene mcr-1 Mediates cell permeability and resistance to hydrophobic antibiotics. Front. Microbiol. 10:3015. doi: 10.3389/fmicb.2019.03015
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-427
Miller, A. K., Brannon, M. K., Stevens, L., Johansen, H. K., Selgrade, S. E., Miller, S. I., et al. (2011). PhoQ mutations promote lipid a modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 55, 5761–5769. doi: 10.1128/AAC.05391-5311
Nabti, L. Z., Sahli, F., Ngaiganam, E. P., Radji, N., Mezaghcha, W., Lupande-Mwenebitu, D., et al. (2020). Development of real-time PCR assay allowed describing the first clinical Klebsiella pneumoniae isolate harboring plasmid-mediated colistin resistance mcr-8 gene in Algeria. J. Glob. Antimicrob. Resist. 20, 266–271. doi: 10.1016/j.jgar.2019.08.018
Napier, B. A., Burd, E. M., Satola, S. W., Cagle, S. M., Ray, S. M., McGann, P., et al. (2013). Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. mBio 4:e0021-13. doi: 10.1128/mBio.00021-13
Pitt, M., Elliott, A. G., Cao, M. D., Ganesamoorthy, D., Karaiskos, I., Giamarellou, H., et al. (2018). Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microb. Genomics 4:e000158.
Poirel, L., Jayol, A., Bontron, S., Villegas, M.-V., Ozdamar, M., Turkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Redhwan, A., Choudhury, M., Al Harbi, B., Kutbi, A., Alfaresi, M., AlJindan, R., et al. (2019). A Snapshot about the mobile colistin resistance (mcr) in the middle east and North Africa Region. J. Infect. Public Health 12, 149–150. doi: 10.1016/j.jiph.2018.10.125
Rossi, F., Girardello, R., Morais, C., Cury, A., Martins, L., Silva, A., et al. (2017). Plasmid-mediated mcr-1 in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics 72, 642–644. doi: 10.6061/clinics/2017(10)09
Sabirova, J. S., Xavier, B. B., Coppens, J., Zarkotou, O., Lammens, C., Janssens, L., et al. (2016). Whole-genome typing and characterisation of bla VIM19 -harbouring ST383 Klebsiella pneumoniae by PFGE, whole-genome mapping and WGS. J. Antimicrob. Chemother. 71, 1501–1509. doi: 10.1093/jac/dkw003
Sherman, E. X., Hufnagel, D. A., and Weiss, D. S. (2016). MCR -1 confers cross-resistance to lysozyme. Lancet Infect. Dis. 16, 1226–1227. doi: 10.1016/S1473-3099(16)30395-30394
Siguier, P. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Uz Zaman, T., Albladi, M., Siddique, M. I., Al Johani, S. M., and Balkhy, H. (2018). Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect. Drug Resist. 11, 1183–1187. doi: 10.2147/IDR.S161146
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infect. 7, 1–9. doi: 10.1038/s41426-018-0124-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front. Microbiol. 10:228. doi: 10.3389/fmicb.2019.00228
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Xavier, B. B., Mysara, M., Bolzan, M., Ribeiro-Gonçalves, B., Alako, B. T. F., Harrison, P., et al. (2020). BacPipe: a rapid, user-friendly whole-genome sequencing pipeline for clinical diagnostic bacteriology. iScience 23:100769. doi: 10.1016/j.isci.2019.100769
Keywords: mcr-8.1, mcr-1.1, colistin, Qatar, K. pneumoniae, E. coli, P. aeruginosa
Citation: Eltai NO, Kelly B, Al-Mana HA, Ibrahim EB, Yassine HM, Al Thani A, Al Maslmani M, Lammens C, Xavier BB and Malhotra-Kumar S (2020) Identification of mcr-8 in Clinical Isolates From Qatar and Evaluation of Their Antimicrobial Profiles. Front. Microbiol. 11:1954. doi: 10.3389/fmicb.2020.01954
Received: 29 April 2020; Accepted: 24 July 2020;
Published: 24 August 2020.
Edited by:
Patrick Rik Butaye, Ross University School of Veterinary Medicine, Saint Kitts and NevisReviewed by:
Youjun Feng, Zhejiang University, ChinaChengming Wang, Auburn University, United States
Copyright © 2020 Eltai, Kelly, Al-Mana, Ibrahim, Yassine, Al Thani, Al Maslmani, Lammens, Xavier and Malhotra-Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Surbhi Malhotra-Kumar, c3VyYmhpLm1hbGhvdHJhQHVhbnR3ZXJwZW4uYmU=
 Nahla O. Eltai
Nahla O. Eltai Brianna Kelly
Brianna Kelly Hassan A. Al-Mana
Hassan A. Al-Mana Emad B. Ibrahim3
Emad B. Ibrahim3 Hadi M. Yassine
Hadi M. Yassine Christine Lammens
Christine Lammens Basil B. Xavier
Basil B. Xavier Surbhi Malhotra-Kumar
Surbhi Malhotra-Kumar