
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 12 August 2020
Sec. Evolutionary and Genomic Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01937
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) are diverse immune systems found in many prokaryotic genomes that target invading foreign DNA such as bacteriophages and plasmids. There are multiple types of CRISPR with arguably the most enigmatic being Type IV. During an investigation of CRISPR carriage in clinical, multi-drug resistant, Klebsiella pneumoniae, a Type IV-A3 CRISPR-Cas system was detected on plasmids from two K. pneumoniae isolates from Egypt (isolated in 2002–2003) and a single K. pneumoniae isolate from the United Kingdom (isolated in 2017). Sequence analysis of all other genomes available in GenBank revealed that this CRISPR-Cas system was present on 28 other plasmids from various Enterobacteriaceae hosts and was never found on a bacterial chromosome. This system is exclusively located on IncHI1B/IncFIB plasmids and is associated with multiple putative transposable elements. Expression of the cas loci was confirmed in the available clinical isolates by RT-PCR. In all cases, the CRISPR-Cas system has a single CRISPR array (CRISPR1) upstream of the cas loci which has several, conserved, spacers which, amongst things, match regions within conjugal transfer genes of IncFIIK/IncFIB(K) plasmids. Our results reveal a Type IV-A3 CRISPR-Cas system exclusively located on IncHI1B/IncFIB plasmids in Enterobacteriaceae that is likely to be able to target IncFIIK/IncFIB(K) plasmids presumably facilitating intracellular, inter-plasmid competition.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-Cas) are widespread, adaptive, RNA-mediated, immune systems found in the genomes of prokaryotic organisms (bacteria and archaea) that target invading foreign DNA such as bacteriophages and conjugative plasmids (Barrangou et al., 2007; Marraffini and Sontheimer, 2010a). CRISPR functions through a three-stage process: adaptation involving the acquisition of foreign DNA molecules as spacers, expression and maturation of the short CRISPR RNAs (crRNAs), and the interference with a cognate invading foreign DNA molecule (Rath et al., 2015). The classification of CRISPR-Cas systems is continuously updated to include newly identified subtypes. To date, CRISPR-Cas systems are classified into two classes, six Types (I–VI), and ∼ 33 subtypes (Koonin et al., 2017; Makarova et al., 2018, 2020). There is ongoing discovery of multiple, novel class 2 CRISPR-Cas systems (Makarova et al., 2020). The two classes differ according to the effector module; class 1 utilizes multi-protein effector Cas complexes, while class 2 utilizes a single-protein effector [Type II contains Cas9; Type V contains Cas 12a (previously known as Cpf1), Cas12b (previously known as C2c1), Cas12c (previously known as C2c3), Cas12d (previously known as CasY), and Cas12e (previously known as CasX); and Type VI contains Cas13a (previously known as C2c2), Cas13b, and Cas13c] (Makarova and Koonin, 2015; Makarova et al., 2015, 2017; Pyzocha and Chen, 2018). CRISPR-Cas systems are confirmed, or expected, to provide immunity against viruses and other mobile genetic elements (MGEs), except for transposon-encoded CRISPR-Cas systems that lack the interference module and therefore are predicted to perform functions distinct from adaptive immunity (Makarova et al., 2020). Most of the CRISPR types target DNA, some types specifically target RNA such as Type VI, while Type III CRISPR systems are unique because they exhibit both RNA interference and DNA interference in vivo to protect their microbial hosts (Marraffini and Sontheimer, 2008, 2010b; Hale et al., 2009; Deng et al., 2013; Manica et al., 2013; Goldberg et al., 2014; Tamulaitis et al., 2014; Zebec et al., 2014; Peng et al., 2015; Samai et al., 2015; Elmore et al., 2016; Estrella et al., 2016; Kazlauskiene et al., 2016; Zhang and Ye, 2017; Ozcan et al., 2019; Lin et al., 2020).
Type IV was previously called the Unknown Type (Type U), due to its rare occurrence and lack of the adaptation module, until an updated classification in 2015 (Makarova et al., 2013; Zhang and Ye, 2017). It was then named Type IV (putative) after its identification in Acidithiobacillus ferrooxidans presenting a different genetic arrangement of Type U cas genes (Makarova et al., 2015). In 2017, Type IV classification was updated, after its identification in Thioalkalivibrio sp. K90mix (TK90_2699-TK90_2703), to show an associated repeat-spacer array for a cas loci that have csf4 (dinG), csf5 (cas6-Like), csf1 (cas8-Like), csf2 (cas7), and csf3 (cas5) genetic arrangement, respectively, and was then assigned as Type IV-A (Koonin et al., 2017). In 2018, a variant of Type IV that lacks a repeat-spacer array from Rhodococcus jostii RHA1 (RHA1_ro10069-RHA1_ro10072), was assigned as Type IV-B (Figure 1; Makarova et al., 2018). In 2019, the Type IV-C CRISPR-Cas system was formally classified as a distinct subtype after its identification in nine contigs; mostly from thermophilic microorganisms (Makarova et al., 2020). Other papers have also proposed the classification of Type IV-D, Type IV-E, and subgroups of Type IV-A(1-4) (Crowley et al., 2019; Pinilla-Redondo et al., 2019), however, the suggested subgroups did not have a unified genetic arrangement corresponding for each of the named Type IV-A(1-4) variants. Type IV CRISPR-Cas systems were shown to employ crRNA-guided effector complexes (Ozcan et al., 2019). Type IV is the only type to possesses csf4 (dinG) in its CRISPR-Cas loci (Dwarakanath et al., 2015; Koonin et al., 2017), and it was recognized initially as the signature proteins for Type IV systems, (Makarova and Koonin, 2015; Crawley et al., 2018) although recently subtype IV-D has been shown to carry a helicase of the RecD family in place of the archetypal DinG (Pinilla-Redondo et al., 2019). To date, Type IV variants (IV-A, IV-B, and IV-C) described above show different genetic arrangements and orientation of cas loci, and they all lack the adaptation module. Also, all Type IV CRISPR-Cas systems are encoded by bacterial plasmids, prophages or other, uncharacterized integrated elements (Faure et al., 2019b). Thus, it has been hypothesized that Type IV is similar to an ancestral innate immune system that gained adaptive ability by associating with a transposon-like element containing cas1 and cas2 (Rath et al., 2015).
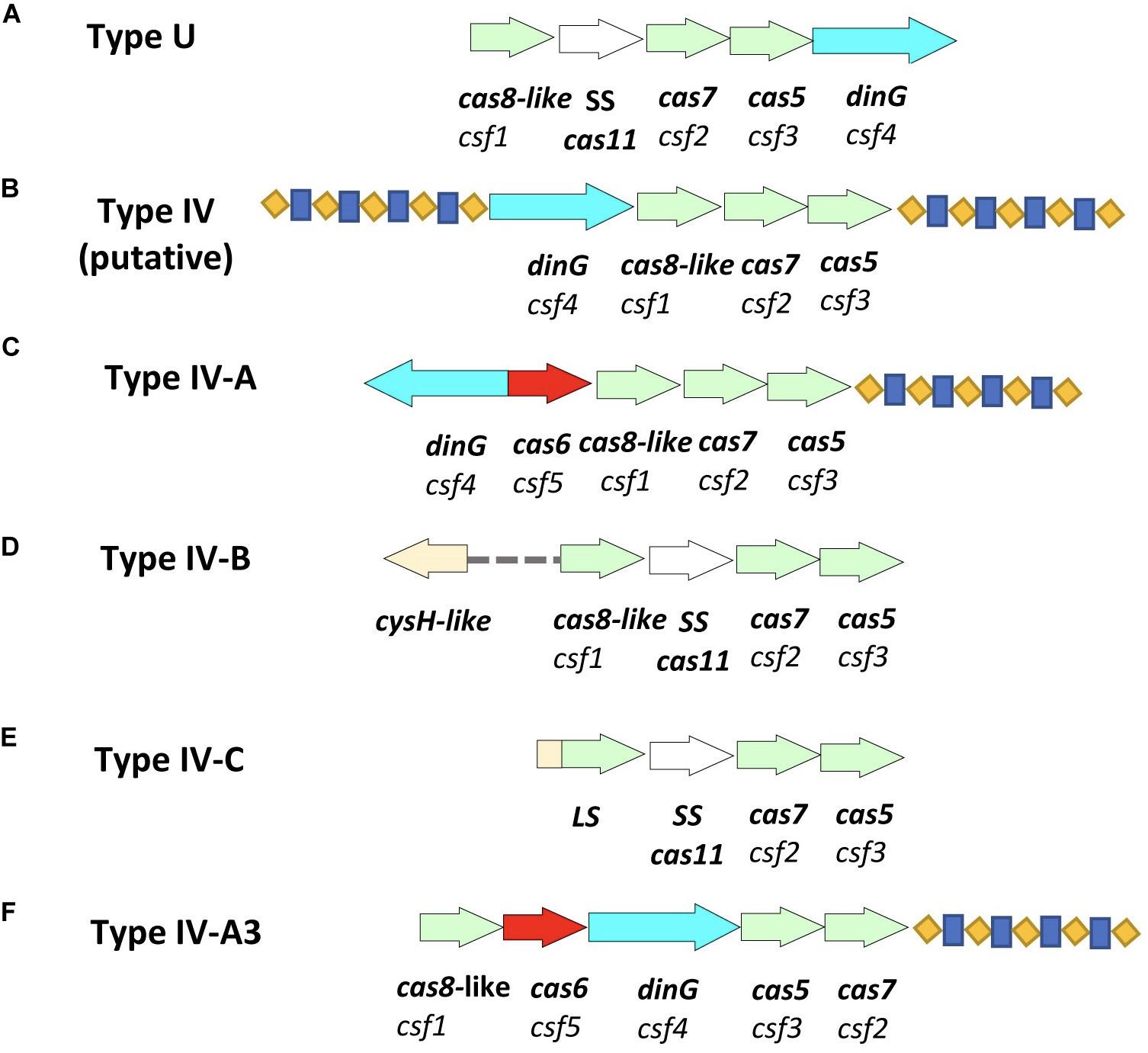
Figure 1. Schematic representation of Type IV CRISPR-Cas systems and the newly identified variant of Type IV-A. (A) Type U (unknown) as identified in 2013 (Makarova et al., 2013). (B) Type IV (putative) as identified in Acidithiobacillus ferrooxidans in 2015 (Makarova et al., 2015), however, the two associated repeat-spacer arrays were identified in this study. (C) Type IV-A identified in Thioalkalivibrio sp. K90mix (TK90_2699-TK90_2703) in 2017 (Makarova et al., 2018). (D) Type IV-B identified in Rhodococcus jostii RHA1 (RHA1_ro10069-RHA1_ro10072) in 2017 (Makarova et al., 2018). (E) Type IV-C identified in Thermoflexia bacterium D6793_05715-D6793_05700 (Makarova et al., 2020). (F) Type IV-A3 as detected in Enterobacteriaceae isolates and genomes in this study. Arrows in different colors represent genes; red represents cas6; bright blue represents dinG; light green represents other essential genes of the system; cas8-like/LS, cas7 and cas5; white represents cas11; blue-yellow pattern represents the direct repeat-spacer loci.
Three clinical isolates were investigated; Klebsiella pneumoniae-53 and K. pneumoniae-65 were isolated from Egyptian university teaching hospitals (2002–2003), and K. pneumoniae-CR5 from University College London Hospital in the United Kingdom (2017). The bacterial genomic DNA sequencing was conducted at MicrobesNG (Birmingham, United Kingdom). Isolates were sequenced using an Illumina HiSeq 2500 and an Illumina MiSeq instruments, to boost coverage, with a 2 × 250 bp paired end sequencing using Nextera XT library prep.
DNA sequences were analyzed using CRISPRFinder, CRISPRCasFinder, CRISPRTarget, and Snapgene (GSL Biotech) (Grissa et al., 2007; Lundgren et al., 2015; Couvin et al., 2018; GSL, 2020). The Cas domain analyses were performed by HHpred (Sensitive protein homology detection, function, and structure prediction based on HMM–HMM comparison) at MPI bioinformatics Toolkit1 (Zimmermann et al., 2018). HHpred was performed using NCBI_Conserved_Domains(CD)_v3.16 and TIGRFAMs_v15.1 databases and Bac_Escherichia_coli_K12_07_Mar_2017 proteome settings. Multi-Locus Sequence Typing, resistance genes and plasmids were identified using MLST, ResFinder, and PlasmidFinder, respectively (Carattoli et al., 2014). Spacer analysis was performed by BLAST and Geneious (Kearse et al., 2012). A phylogenetic UPGMA-based tree was constructed for CRISPR arrays and Cas proteins using MEGA X 10.1 (Kumar et al., 2016; Shen et al., 2017). The alignment of the regions containing protospacers (including 10 bp flanking the protospacer) associated with CRISPR1 repeats were investigated to identify putative protospacer adjacent motif (PAM) signature, as described in Pinilla-Redondo et al. (2019). PAMs were identified based on compared alignments of nucleotides immediately preceding each detected protospacer in all or up to ten unique protospacers of all the investigated sequences. The leader sequence was identified by sequence alignment. Direct repeats and PAM conservation were assessed using WebLogo, RNA secondary structure was predicted using RNAfold (Hofacker, 2003; Crooks et al., 2004; Mathews, 2005; Mojica et al., 2009). The GC skew plots were generated using the GenSkew online analysis tool2. Presence in other GenBank sequences was investigated by BLASTn3.
The CRISPR-Cas loci expression was tested. RT-PCR was performed using LightCycler® RNA Amplification Kit SYBR Green (Roche Diagnostics Ltd., United Kingdom). The primers were designed for fully characterized genes (csf2-fw:AAAATGCGGTCTCAACTTCCG; csf2-rev:TGACGAAGAG TTCCCCGAATG), (dinG-fw:GAGTCTGCCGGATTGTCGTTA; dinG-rev:GTACCAGATAGCCCAGCGTTT), and (cas6-fw:AAT GCGTTTCGGTTGCGTATC; cas6-rev:GAGTACGGCAGCTT CTCTCC).
Type IV-A-3 CRISPR-Cas, based on the gene composition and genetic architecture of the IV-A variants detected in K. pneumoniae as described in Pinilla-Redondo et al. (2019), was detected on a total of thirty-one (three clinical isolates and twenty eight sequences from GenBank) IncHI1B/IncFIB(Mar) plasmid sequences within Enterobacteriaceae (Figure 1 and Supplementary Table S1). The IncH1B/IncFIB plasmids are large, low copy number, conjugative plasmids with narrow-host-ranges, which are found in multiple genera of Enterobacteriaceae (Zhong et al., 2005; Suzuki et al., 2010; Faure et al., 2019a). An important feature of IncH1B/IncFIB plasmid biology is the entry exclusion by which the cells that contain an IncF/IncH plasmid become poor recipients in additional conjugation rounds (Garcillan-Barcia and de la Cruz, 2008; Ravenhall et al., 2015); which frees a resident plasmid from competition with related plasmids at segregation during bacterial division but may contribute to limiting plasmid dissemination among potential hosts.
This Type IV-A3 CRISPR-Cas is characterized by the presence of a cas loci containing dinG, which is a distinct feature of Type IV-A CRISPR-Cas system that was shown to be a requirement for the system functional activity in Pseudomonas aeruginosa (Crowley et al., 2019; Pinilla-Redondo et al., 2019; Makarova et al., 2020), a conserved leader sequence and a CRISPR array in all the detected sequences and they all show homology to each other. These Type IV-A3 CRISPR-Cas systems were initially detected by BLAST that confirmed the presence of three genes; cas7, dinG, and cas6, and further HHpred analysis of other associated ORFs revealed the presence of two more genes; cas5 and cas8-like.
We also identified partial related Type IV-A3 systems either (cas8-like, cas6, and dinG) or (cas7 and a CRISPR array) occurring on other IncHI1B/IncFIB(Mar) plasmids (Supplementary Table S1). Partial and complete Type IV-A3 system characterization showed occurrence of a range of different IS elements and retrotransposons (group II introns) (Supplementary Table S1). The average GC content of this Type IV-A3 CRISPR-Cas loci (47.7 ± 0.01%) was found to be closer to that of the IncHI1B/IncFIB(Mar) plasmids on which they reside (46.2 ± 0.01%), compared to the chromosomal sequences of the bacterial host (57 ± 0.02%), (Table 1).
A single CRISPR array (CRISPR1) was identified upstream of all cas loci. The repeats have a predicted stem-loop secondary-structure (Figures 2A,B) and is likely involved in a pre-crRNA Cas6-mediated process. The alignment of the regions around and containing the protospacer, particularly the last six positions preceding the protospacer, associated with CRISPR1 repeats revealed the conservation of the putative PAM signature (AAG) adjacent to the end of the protospacers (Figure 2C). A highly conserved 65 bp leader sequence occurring between the CRISPR-Cas loci and the CRISPR array was observed in all the sequences (Figure 2D). The minor variations in the leader sequence only occurred in two sequences (C in position −63 is A in CP014776.1 Pluralibacter gergoviae, and G in position −41 is A, and C in position −39 is T in K. pneumoniae-CR5 ST-392). The high conservation of the leader sequence is unlike that presented in Pinilla-Redondo et al. (2019). RT-PCR of the confirmed cas loci (cas7, dinG, and cas6) demonstrated that they are expressed in all three of the available clinical isolates (Table 2).
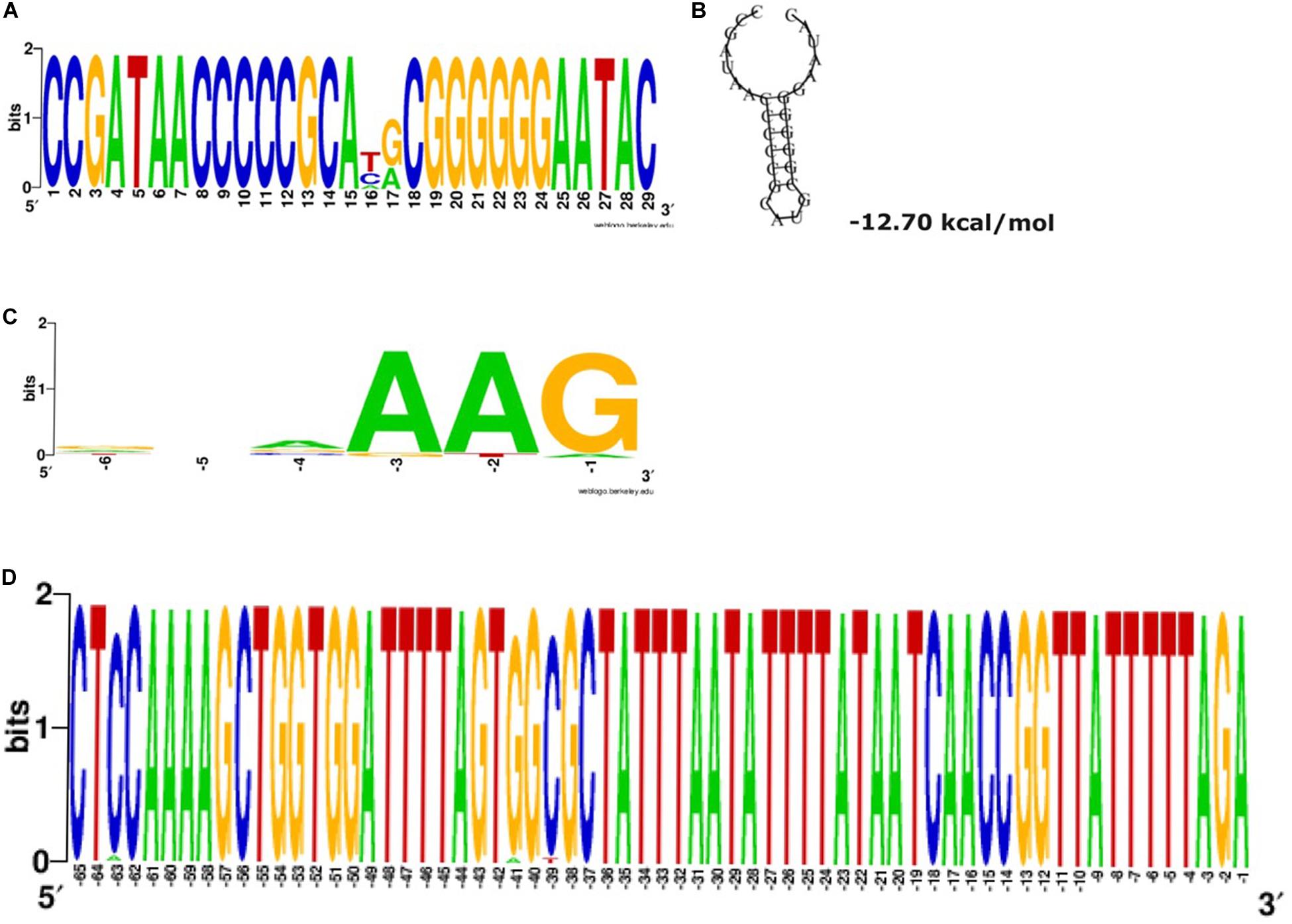
Figure 2. Type IV-A3 conserved repeats and the predicted stable stem-loop secondary structure, putative PAM and leader sequence. (A) Type IV-A-variant conserved repeats. The height of the letters in the sequence logo shows the relative frequency of their recurrence at that position. Wobbles at positions 16 and 17 are within the loop of the predicted stem-loop structure and are therefore tolerated in the structural prediction shown in (B). (B) The predicted secondary structure of direct repeats and the associated Minimum Free Energy (MFE) estimated in (kcal/mol) shown underneath the structure. This structure is predicted to be involved in the mechanism of pre-crRNA processing. (C) Type IV-A-variant conserved putative protospacer adjacent motifs (PAMs). The alignment of the regions containing protospacers shows the conservation of putative PAM signature (AAG), position –3 to –1, adjacent to the end of the protospacers, using WebLogo. The analysis was performed on all the detected (467 spacers) of the 31 CRISPR1 arrays analyzed, specifically, spacer matching 9% (42/467) to bacteriophages and 25.5% (119/467) to plasmid sequences. Searches for other subtypes/variants were unsuccessful, likely due to the low number of spacer–protospacer matches (D) Conserved Type IV-A-variant CRISPR leader. The WebLogo shows a highly conserved 65 bp occurring between the CRISPR-Cas loci an the CRISPR array among the sequences investigated in this study.
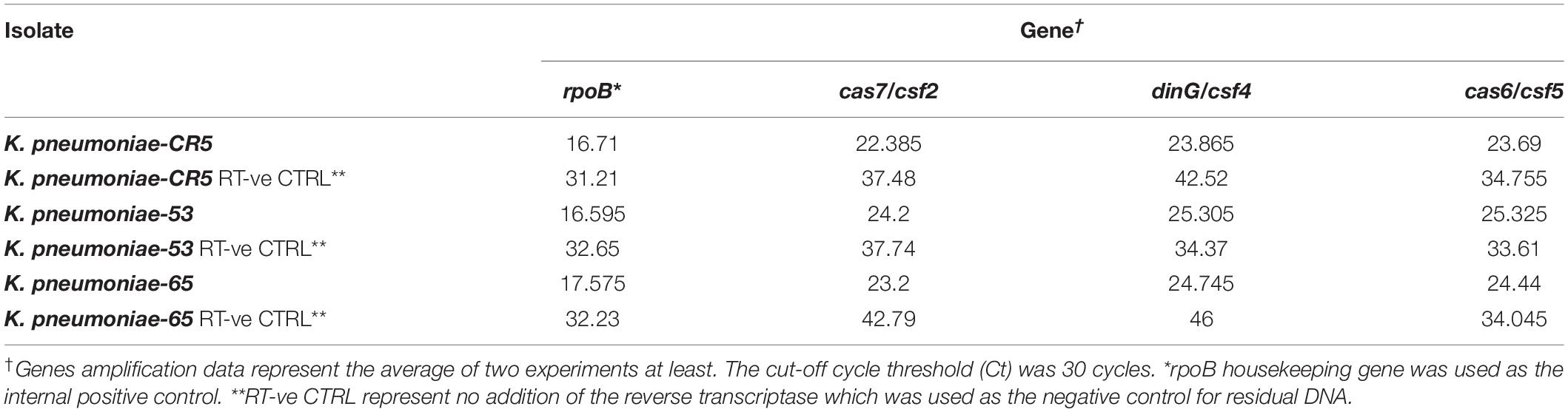
Table 2. RT-PCR data of the confirmed cas loci (cas7, dinG, and cas6) in the three clinical isolates.
We have detected a total of 467 spacers in the 31 CRISPR1 arrays analyzed, out of which 9% (42/467) match to bacteriophages and 25.5% (119/467) match to plasmid sequences. The majority of spacer sequences are present in more than one spacer array and some are present more than once within the same array (Figure 3). Plasmid targeting spacers appeared in every example of this Type IV-A3 associated CRISPR array analyzed. Sequence analysis revealed that spacers correspond to IncFIIK conjugal transfer genes; traN and traL (Figure 3). Limited conservation within the order of the spacer arrays showed that the arrays cluster into two distinct groups which share geographical associations and suggest persistence within the plasmid pool in isolates from certain countries over time (Figure 4).
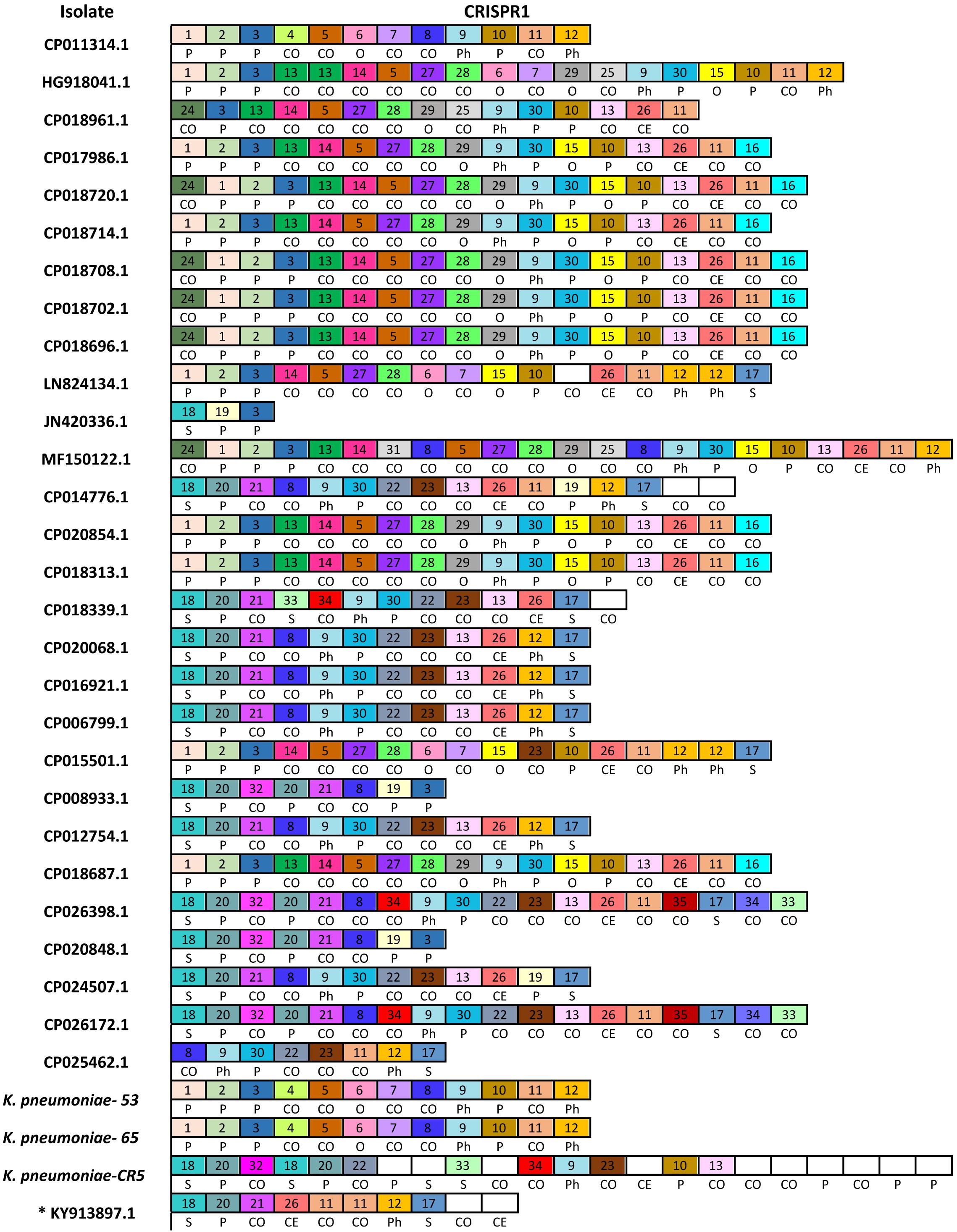
Figure 3. Newly described Type IV-A3 CRISPR spacer polymorphism. The spacers map. Only spacers are represented by boxes, and no repeats are included. Identical spacers are represented by the same number and color, while unique spacers are represented by white color and no number is associated with the box. Self-targeting spacers are indicated by letter (S) and show 100% identity to host DNA, plasmid-targeting spacers are indicated by letter (P), phage targeting spacers are indicated by letters (Ph), other Enterobacteriaceae targeting spacers (100% identity) are indicated by letter (O), cryptic spacers with similarity to other bacterial DNA are indicated by letters (CO), and those with similarity to Eukaryotic DNA are indicated by letters (CE) that are positioned underneath the relevant spacer. CE spacers showed at least 57% identity to eukaryotic DNA. CE spacers were confirmed by multiple sequences alignments. * KY913897.1 is the isolate that only has a CRISPR array and a cas7 (csf2) (not a complete Type IV system) and therefore is not included in the total analysis in Figure 4.
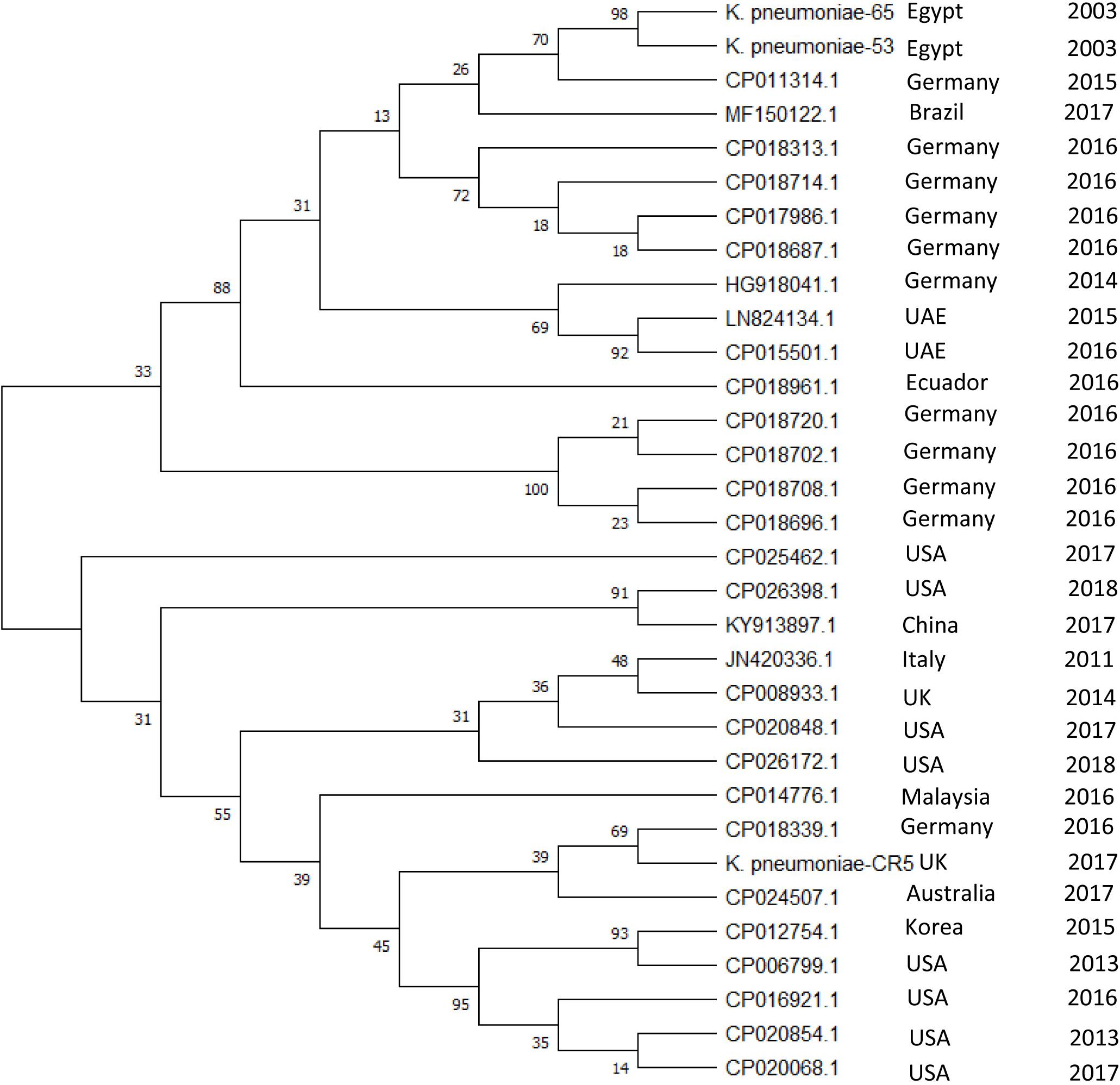
Figure 4. Evolutionary relationships of Type IV-A3 CRISPR spacer. The phylogenetic tree illustrating the evolutionary relationships of Type IV-A3 CRISPR array nucleotide sequences. Phylogenetic UPGMA tree was constructed using the MUSCLE algorithm of MEGA7. The evolutionary distances were computed using the Maximum Likelihood method and Tamura-Nei model, bootstrap test (1000 replicates), and the rate variation among sites was modeled with a gamma distribution (shape parameter = 2). The percentage of trees in which the associated taxa clustered together is shown next to the branches. The year and geographical origin of the isolate are listed to the right of the branch ends.
The CRISPR system described here is always found associated with IncH1B/IncFIB plasmids in Enterobacteriaceae, has dinG and cas7 (involved in interference), and cas6, cas5, and cas8-like (involved in expression and maturation of short crRNAs) (Cass et al., 2015; Dwarakanath et al., 2015; Koonin and Krupovic, 2015; Lundgren et al., 2015; Crowley et al., 2019; Faure et al., 2019b). The detection of a csf1/cas8-like in the Type IV-A-variant described here updates the initial (Newire et al., 2019) and subsequent (Kamruzzaman and Iredell, 2019) reports of this system. Additionally, our results agree with other reports suggesting a need for Type IV-A variant classification (Crowley et al., 2019; Pinilla-Redondo et al., 2019).
Notably, some of the previously described Type IV systems do not possess a dinG or cas8-like (e.g., Type IV-C); however, cas7 genes are consistently found in all the previously and presently described Type IV sequences. Also, Cas7 is the most conserved protein among members of the Type IV CRISPR family (Pinilla-Redondo et al., 2019). This highlights the role of cas7 in Type IV identification.
This Type IV-A3 described here has a variable CRISPR array and a conserved leader sequence. The conserved leader sequence occurrence in a wide variety of K. pneumoniae sequence types may reflect their narrow association with IncH1B/IncFIB plasmids. Conserved leader sequences in other types (Type I-E) were shown to increase acquisition efficiency by presumably stabilizing the Cas1–2-leader-repeat interaction (Kieper et al., 2019). The order of spacers demonstrated conservation with some polymorphism, and they cluster into two main groups (Figure 4) matching DNA from a variety of geographical sources. Expression of this Type IV-A cas genes suggests immunity to incoming DNA matching the spacers. Crowley et al. (2019) posit that interference is mediated, similar to type I and type III systems, through multi-subunit complexes composed of Csf proteins and the use of crRNA as a guide to bind complementary nucleic acid forming R-loops. In this case, it was hypothesized that DinG is then recruited to these R-loops, where it either acts directly to destroy the foreign DNA (e.g., a plasmid) or recruits an endogenous nuclease to mediate RNA-guided interference (Crowley et al., 2019); however, this needs to be tested. Also, we note that the adaptation module is missing, thus adding new spacers will require cas1 and cas2 from other CRISPR-Cas systems. Like other Type IV systems that cannot function as independent adaptive immune systems (Koonin and Krupovic, 2015), we suspect that the Type IV-A3 CRISPR-Cas described here is likely to co-operate with other cas loci, whenever they exist within the Enterobacteriaceae host genomes, for spacer acquisition. Those CRISPR-Cas loci could belong to those CRISPR systems that are known to be frequently associated with an Enterobacteriaceae host, such as Type I-E/I-E∗ or Type I-F (Aydin et al., 2017). The association between Type IV-A and cas6e and cas6f (cas6 sequences observed in subtypes I-E and I-F, respectively) was previously reported in other bacterial families, suggesting functional links (Pinilla-Redondo et al., 2019; Taylor et al., 2019). These functional links were inferred based on the similarities in the leader and the repeat sequences of Type I-E and Type IV-A3 (Pinilla-Redondo et al., 2019). Another evidence that supports possible functional co-operation is the presence of Cas6 that shows 99%+ identity to Type I-E Cas6 in Enterobacteriaceae in the Type IV-A3 described here. Furthermore, unlike other Cas proteins associated with Type IV-A3 described here, Cas6 were highly conserved sequences, showing no particular association with interrupting IS elements, which may further support the recruitment of cas6 is form Type I-E. These possible adaptation functional links appear to be a feature that can be switched on/off, which requires the presence of the IncHI1B/IncFIB(Mar) plasmids (that carry this Type IV-A-variant) inside a bacterial host that has a functional CRISPR-Cas system in its genome. Although a previous report suggested that Type IV CRISPR-Cas system-positive plasmids were only found in Enterobacteriaceae with chromosomal Type I-E/I-E∗ CRISPR-Cas (Kamruzzaman and Iredell, 2019), we could not identify Type I-E/I-E∗ in all the isolate genomes that have Type IV-A3. For example, K. pneumoniae-53, CP011314.1, HG918041.1, JN420336.1, MF150122.1, CP014776.1, CP018339.1, CP026398.1, CP020848.1, CP024507.1, CP026172.1, and CP025462.1 did not have Type I-E/I-E∗ CRISPR-Cas systems. Thus, we assume there is no conditional connection between the presence of Type IV-A3 and Type I-E/I-E∗ CRISPR-Cas systems in Enterobacteriaceae.
Type IV-A3 CRISPR system reported here is exclusively located on IncH1B/IncFIB plasmids. We have also spotted an imperfect spacer target in traN of an IncFIIK plasmid in K. pneumoniae-53 which suggests this plasmid may be able to evade plasmid mediated CRISPR interaction within this strain (Jiang et al., 2013). Therefore, these spacers are likely to be involved in plasmid competition; protecting the resident Type IV-A CRISPR-Cas carrying plasmid in Enterobacteriaceae as previously suggested (Newire et al., 2019; Pinilla-Redondo et al., 2019). Recently, some Type IV-A system variants that are associated with P. aeruginosa were shown to target invasive plasmids, which strengthens the involvement of Type IV-A CRISPR-Cas systems in plasmid competition (Crowley et al., 2019).
Type IV CRISPR-Cas systems demonstrate a notable diversity of molecular organization (Figure 1) and some appear to have taken on roles in addition to adaptive cellular immunity (Faure et al., 2019a). For example, some of the Type IV CRISPR–Cas loci were previously predicted to encode bacterial toxins that together with the Cas proteins of the Type IV systems may contribute to plasmid stabilization (Faure et al., 2019b). The Type IV-A3 system described here demonstrates a complex evolutionary connection with MGEs in terms of parasitism and immunity (Koonin and Makarova, 2017). The association between this Type IV-A3 system and multiple MGEs, plus the identification of partial cas loci genes with and without the CRISPR array on other IncHI1B/IncFIB(Mar) plasmids (Figure 1 and Supplementary Table S1), plus the identification of similar arrays in different plasmids in the same host from the same country (Figures 3, 4), indicates that dynamic, MGE mediated movement and rearrangement of this CRISPR-Cas Type IV-A system is ongoing. The similarity in the GC content between this Type IV-A and the IncHI1B/IncFIB plasmids in contrast with the higher chromosomal GC content supports the observations that the system is exclusively plasmid associated, both in this study and in others (Ravenhall et al., 2015). Because reporting standard deviations from comparisons of element-wide GC contents across different genomes could be misleading, since the strains are closely related that statistical observations are not independent, we have investigated the GC skew in a reasonably sized sliding window (1000 bp) across the length of the element (Type IV-A3 system and plasmid DNA sequences) in a single genome (K. pneumoniae-65), which also confirmed that the system is exclusively plasmid associated. This demonstrates unique evolutionarily juxtaposed connections between CRISPR-Cas and MGEs which is worthy of further investigation. To our knowledge, this is the first identification of a CRISPR-Cas system exclusively associated with IncHI1B/IncFIB plasmids that demonstrates an evolutionary association with MGEs and is likely to be involved in plasmid competition.
This manuscript has been released as a pre-print at BioRxiv (Newire et al., 2019).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, SGOJ00000000; https://www.ncbi.nlm.nih.gov/genbank/, SGOK00000000; and https://www.ncbi.nlm.nih.gov/genbank/, SGOL00000000.
EN discovered the CRISPR system within the genomes of her Egyptian isolate collection, analyzed the sequence data, and wrote the first draft of the manuscript. SJ, AA, and VE designed and carried out the experiments to test cas loci expression. AR analyzed the data and wrote the manuscript. All authors critically reviewed and approved the manuscript.
EN was supported by a grant from the Schlumberger Foundation’s Faculty for the Future Program (2012–2016).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01937/full#supplementary-material
Aydin, S., Personne, Y., Newire, E., Laverick, R., Russell, O., Roberts, A. P., et al. (2017). Presence of Type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J. Antimicrob. Chemother. 72, 2213–2218. doi: 10.1093/jac/dkx137
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Cass, S. D., Haas, K. A., Stoll, B., Alkhnbashi, O. S., Sharma, K., Urlaub, H., et al. (2015). The role of Cas8 in type I CRISPR interference. Biosci. Rep. 35:e00197.
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Neron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251.
Crawley, A. B., Henriksen, J. R., and Barrangou, R. (2018). CRISPRdisco: an automated pipeline for the discovery and analysis of CRISPR-Cas systems. CRISPR J. 1, 171–181. doi: 10.1089/crispr.2017.0022
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Crowley, V. M., Catching, A., Taylor, H. N., Borges, A. L., Metcalf, J., Bondy-Denomy, J., et al. (2019). A Type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference In Vivo. CRISPR J. 2, 434–440. doi: 10.1089/crispr.2019.0048
Deng, L., Garrett, R. A., Shah, S. A., Peng, X., and She, Q. (2013). A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 87, 1088–1099. doi: 10.1111/mmi.12152
Dwarakanath, S., Brenzinger, S., Gleditzsch, D., Plagens, A., Klingl, A., Thormann, K., et al. (2015). Interference activity of a minimal Type I CRISPR-Cas system from Shewanella putrefaciens. Nucleic Acids Res. 43, 8913–8923. doi: 10.1093/nar/gkv882
Elmore, J. R., Sheppard, N. F., Ramia, N., Deighan, T., Li, H., Terns, R. M., et al. (2016). Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 30, 447–459. doi: 10.1101/gad.272153.115
Estrella, M. A., Kuo, F. T., and Bailey, S. (2016). RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 30, 460–470. doi: 10.1101/gad.273722.115
Faure, G., Makarova, K. S., and Koonin, E. V. (2019a). CRISPR-Cas: complex functional networks and multiple roles beyond adaptive immunity. J. Mol. Biol. 431, 3–20. doi: 10.1016/j.jmb.2018.08.030
Faure, G., Shmakov, S. A., Yan, W. X., Cheng, D. R., Scott, D. A., Peters, J. E., et al. (2019b). CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nature Rev. Microbiol. 17, 513–525. doi: 10.1038/s41579-019-0204-7
Garcillan-Barcia, M. P., and de la Cruz, F. (2008). Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60, 1–18. doi: 10.1016/j.plasmid.2008.03.002
Goldberg, G. W., Jiang, W., Bikard, D., and Marraffini, L. A. (2014). Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637. doi: 10.1038/nature13637
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57.
GSL (2020). SnapGene Software. (Biotech Chicago, IL). (from GSL Biotech). Available online at: snapgene.com
Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., et al. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956. doi: 10.1016/j.cell.2009.07.040
Hofacker, I. L. (2003). Vienna RNA secondary structure server. Nucleic Acids Res. 31, 3429–3431. doi: 10.1093/nar/gkg599
Jiang, W., Maniv, I., Arain, F., Wang, Y., Levin, B. R., and Marraffini, L. A. (2013). Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 9:e1003844. doi: 10.1371/journal.pgen.1003844
Kamruzzaman, M., and Iredell, J. R. (2019). CRISPR-cas system in antibiotic resistance plasmids in Klebsiella pneumoniae. Front. Microbiol. 10:2934. doi: 10.3389/fmicb.2019.02934
Kazlauskiene, M., Tamulaitis, G., Kostiuk, G., Venclovas, È, and Siksnys, V. (2016). Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306. doi: 10.1016/j.molcel.2016.03.024
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kieper, S. N., Almendros, C., and Brouns, S. J. J. (2019). Conserved motifs in the CRISPR leader sequence control spacer acquisition levels in Type I-D CRISPR-Cas systems. FEMS Microbiol. Lett. 366:fnz129.
Koonin, E. V., and Krupovic, M. (2015). Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat. Rev. Genet. 16, 184–192. doi: 10.1038/nrg3859
Koonin, E. V., and Makarova, K. S. (2017). Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol. Evol. 9, 2812–2825. doi: 10.1093/gbe/evx192
Koonin, E. V., Makarova, K. S., and Zhang, F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lin, J., Feng, M., Zhang, H., and She, Q. (2020). Characterization of a novel type III CRISPR-Cas effector provides new insights into the allosteric activation and suppression of the Cas10 DNase. Cell Discov. 6:29.
Lundgren, M., Charpentier, E., and Fineran, P. C. (2015). CRISPR Methods and Protocols. New York, NY: Springer Science Business Media New York.
Makarova, K. S., and Koonin, E. V. (2015). Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 1311, 47–75. doi: 10.1007/978-1-4939-2687-9_4
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nature Rev. Microbiol. 13, 722–736.
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2013). The basic building blocks and evolution of CRISPR-CAS systems. Biochem. Soc. Trans. 41, 1392–1400. doi: 10.1042/bst20130038
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2018). Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1, 325–336. doi: 10.1089/crispr.2018.0033
Makarova, K. S., Zhang, F., and Koonin, E. V. (2017). SnapShot: class 1 CRISPR-Cas systems. Cell 168, 946–946.e1. doi: 10.1016/j.cell.2017.02.018
Manica, A., Zebec, Z., Steinkellner, J., and Schleper, C. (2013). Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 41, 10509–10517. doi: 10.1093/nar/gkt767
Marraffini, L. A., and Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. doi: 10.1126/science.1165771
Marraffini, L. A., and Sontheimer, E. J. (2010a). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Rev. Genet. 11, 181–190. doi: 10.1038/nrg2749
Marraffini, L. A., and Sontheimer, E. J. (2010b). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. doi: 10.1038/nature08703
Mathews, D. H. (2005). Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics 21, 2246–2253. doi: 10.1093/bioinformatics/bti349
Mojica, F. J. M., Diez-Villasenor, C., Garcia-Martinez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155(Pt 3), 733–740. doi: 10.1099/mic.0.023960-0
Newire, E., Aydin, A., Juma, S., Enne, V. I., and Roberts, A. P. (2019). Identification of a Type IV CRISPR-Cas system located exclusively on IncHI1B/IncFIB plasmids in Enterobacteriaceae. bioRxiv [Preprint]. doi: 10.1101/536375
Ozcan, A., Pausch, P., Linden, A., Wulf, A., Schuhle, K., Heider, J., et al. (2019). Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat. Microbiol. 4, 89–96. doi: 10.1038/s41564-018-0274-8
Peng, W., Feng, M., Feng, X., Liang, Y. X., and She, Q. (2015). An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 43, 406–417. doi: 10.1093/nar/gku1302
Pinilla-Redondo, R., Mayo-Muñoz, D., Russel, J., Garrett, R. A., Randau, L., Sørensen, S. J., et al. (2019). Type IV CRISPR–Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 48, 2000–2012. doi: 10.1093/nar/gkz1197
Pyzocha, N. K., and Chen, S. (2018). Diverse class 2 CRISPR-Cas effector proteins for genome engineering applications. ACS Chem. Biol. 13, 347–356. doi: 10.1021/acschembio.7b00800
Rath, D., Amlinger, L., Rath, A., and Lundgren, M. (2015). The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117, 119–128. doi: 10.1016/j.biochi.2015.03.025
Ravenhall, M., Skunca, N., Lassalle, F., and Dessimoz, C. (2015). Inferring horizontal gene transfer. PLoS Comput. Biol. 11:e1004095. doi: 10.1371/journal.pcbi.1004095
Samai, P., Pyenson, N., Jiang, W., Goldberg, G. W., Hatoum-Aslan, A., and Marraffini, L. A. (2015). Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174. doi: 10.1016/j.cell.2015.04.027
Shen, J., Lv, L., Wang, X., Xiu, Z., and Chen, G. (2017). Comparative analysis of CRISPR-Cas systems in Klebsiella genomes. J. Basic Microbiol. 57, 325–336.
Suzuki, H., Yano, H., Brown, C. J., and Top, E. M. (2010). Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192, 6045–6055. doi: 10.1128/jb.00277-10
Tamulaitis, G., Kazlauskiene, M., Manakova, E., Venclovas, È, Nwokeoji, A. O., Dickman, M. J., et al. (2014). Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell 56, 506–517. doi: 10.1016/j.molcel.2014.09.027
Taylor, H. N., Warner, E. E., Armbrust, M. J., Crowley, V. M., Olsen, K. J., and Jackson, R. N. (2019). Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease. RNA Biol. 16, 1438–1447. doi: 10.1080/15476286.2019.1634965
Zebec, Z., Manica, A., Zhang, J., White, M. F., and Schleper, C. (2014). CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 42, 5280–5288. doi: 10.1093/nar/gku161
Zhang, Q., and Ye, Y. (2017). Not all predicted CRISPR-Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinform. 18:92. doi: 10.1186/s12859-017-1512-4
Zhong, Z., Helinski, D., and Toukdarian, A. (2005). Plasmid host-range: restrictions to F replication in Pseudomonas. Plasmid 54, 48–56. doi: 10.1016/j.plasmid.2004.11.001
Keywords: Type IV, IncFIIK, IncFIB(K), inter-plasmid competition, mobile genetic element
Citation: Newire E, Aydin A, Juma S, Enne VI and Roberts AP (2020) Identification of a Type IV-A CRISPR-Cas System Located Exclusively on IncHI1B/IncFIB Plasmids in Enterobacteriaceae. Front. Microbiol. 11:1937. doi: 10.3389/fmicb.2020.01937
Received: 04 May 2020; Accepted: 22 July 2020;
Published: 12 August 2020.
Edited by:
Kira Makarova, National Center for Biotechnology Information (NLM), United StatesReviewed by:
Qunxin She, Shandong University, ChinaCopyright © 2020 Newire, Aydin, Juma, Enne and Roberts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam P. Roberts, YWRhbS5yb2JlcnRzQGxzdG1lZC5hYy51aw==
†Present address: Enas Newire, Institute of Systems, Molecular & Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, United Kingdom Alp Aydin, Quadram Institute, Norwich, United Kingdom
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.