
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 August 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01918
Antimicrobial resistance is an ancient bacterial defense mechanism that has rapidly spread due to the frequent use of antibiotics for disease treatment and livestock growth promotion. We are becoming increasingly aware that pathogens, such as members of the genus Acinetobacter, are precipitously evolving drug resistances through multiple mechanisms, including the acquisition of antibiotic resistance genes. In this study, we isolated three multidrug resistant Acinetobacter species from birds on a free-range farm. Acinetobacter radioresistens, Acinetobacter lwoffii, and Acinetobacter johnsonii were isolated from hens, turkeys and ducks and were resistant to 14 clinically relevant antibiotics, including several listed by the World Health Organization as essential medicines. Co-culturing any of the three Acinetobacter species with Acinetobacter baumannii resulted in contact-dependent release of intact resistance determinants. We also isolated several lytic bacteriophages and selected two of these phages to be included in this study based on differences in plaquing characteristics, nucleic acid content and viral morphology. Both phages released host DNA, including antibiotic resistance genes during cell lysis and we demonstrated that these resistance determinants were transferable to a naïve strain of Escherichia coli. This study demonstrates that contact-dependent competition between bacterial species can readily contribute to DNA release into the environment, including antibiotic resistance determinants. We also highlight that the constant lysis and turnover of bacterial populations during the natural lifecycle of a lytic bacteriophage is an underappreciated mechanism for the liberation of DNA and subsequent genetic exchange.
In recent years, multidrug resistant (MDR) bacteria have become a serious concern for healthcare providers worldwide (Poirel et al., 2011; Bengtsson-Palme et al., 2017). Preeminent among these bacteria is Acinetobacter baumannii, a common nosocomial MDR pathogen resistant to desiccation and readily able to acquire MDR genes in both hospital and environmental settings (Poirel et al., 2008; Al Atrouni et al., 2016). In 2013, the CDC ranked carbapenem-resistant A. baumannii as the most concerning MDR pathogen requiring new antibiotics (Sievert et al., 2013) and the WHO published a similar report in 2017 (Lawe-Davies and Bennett, 2017). At the origin of this problem is the propensity of A. baumannii to become resistant to antibiotic treatment through acquisition of resistance genes (Wilharm et al., 2013). Although the extreme resistances of clinical isolates of A. baumannii are well documented (Valentine et al., 2008; Yang et al., 2015; Fernando et al., 2016), the prevalence of MDR isolates in agricultural settings is only recently being explored (Maboni et al., 2020). Furthermore, there has been limited characterization of A. baumannii interactions with reservoir species, such as Acinetobacter radioresistens, which can serve as a source of carbapenem resistance in hospital settings (Poirel et al., 2008). Thus, identifying MDR gene reservoirs and modes of gene release to naïve pathogens will be critical to understanding and combating the spread of antibiotic resistance.
In addition to encoding a number of antibiotic resistance determinants, A. baumannii is well equipped to outcompete neighboring bacteria in a number of approaches. The genus Acinetobacter has been confirmed to carry type 1 (T1SS), type 2 (T2SS), type 4 (T4SS), and type 6 (T6SS) secretion systems, as well as >40 contact-dependent inhibition (CDI) systems, all of which are virulence factors that give Acinetobacter species a competitive advantage (Weber et al., 2013; Hayes et al., 2014; Souza et al., 2015; Harding et al., 2017; Kinsella et al., 2017; De Gregorio et al., 2019; Sgro et al., 2019). T6SS and CDI systems are two mechanisms of direct bacterial competition that Acinetobacter species encode (Harding et al., 2018). Briefly, the T6SS is a complex multi-component system anchored in the inner cell membrane that builds a spike, which, upon contact, injects toxins into a prey cell (Weber et al., 2013). CDI is a two-component system anchored in the outer membrane, which consists of a large transporter (CdiB) and toxin (CdiA) that is released during contact with the prey cell (De Gregorio et al., 2018; Roussin et al., 2019). These virulence factors greatly increase the fitness of the acinetobacters that express them and enables the bacteria to thrive in diverse and competitive environments.
In recent years, there has been an effort to isolate more bacteriophages that use ESKAPE pathogens, such as A. baumannii, as a host. Bacteriophages have been and still are an under-appreciated distributor of antimicrobial resistance elements. These microbial viruses, which have been reported to infect ∼1024 bacterial cells per second globally (Deresinski, 2009), are the most abundant biological entity on earth (Suttle, 2005; Paez-Espino et al., 2016). Host gene transduction, including MDR genes, has been described for some bacteriophages and phage-derived elements (Penades et al., 2015). Although the mechanisms are poorly understood, it was generally accepted that upon infection, most non-transducing lytic bacteriophages will degrade host DNA to inhibit cellular activities or to repurpose nucleotides for viral use (Warren and Bose, 1968). However, there have been recent observations that some lytic phages release intact plasmid DNA (Keen et al., 2017). Keen et al. (2017) have designated these phages as “superspreaders” and demonstrated that drug resistance genes released by these phages can be acquired and expressed by unrelated bacteria found in the same environment. Although over 100 Acinetobacter-infecting bacteriophages have been isolated, this phenomenon has not been reported for this genus (Turner et al., 2017). A greater understanding of Acinetobacter phage impact on MDR dissemination is required before designing phage therapies to treat antibiotic resistant infections, except in the most desperate cases.
Understanding how Acinetobacter species are exposed to exogenous genetic material is crucial to further understanding MDR spread. In this report, we describe MDR Acinetobacter species isolated from a free-range farm (Rothrock et al., 2016). Based on their susceptibility to phage predation and killing when co-cultured with A. baumannii, we demonstrate that antibiotic resistance genes can be released by contact- and phage-lysed cells. In addition to identifying a potential reservoir of MDR-carrying Acinetobacter species in food animals, these findings highlight two underappreciated modes of DNA release that can partially account for the unchecked spread of antibiotic resistance within this genus.
Strains were isolated from fecal samples collected from layer hens, turkeys, and ducks on a free-range pastured poultry farm (Rothrock et al., 2016). Fecal samples were diluted 2 mL/g in sterile phosphate-buffered saline and plated on Brilliance Campycount agar (Remel) under microaerobic conditions (10% CO2, 5% O2, 85% N2) at 37°C for 18 h. Isolated red colonies were re-cultured on Campy-Line selective agar (Line, 2001).
Cell suspensions from agar plates were adjusted to an OD600 = 0.5. Suspensions were floated on formvar-coated copper grids for 1 h and 5% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, United States) was added to fix the samples before imaging. Transmission electron microscopy (TEM) was performed using a JEOL JEM1011 microscope (JEOL Inc., Peabody, MA, United States). MALDI-TOF VITEKTM MS (BioMérieux, Durham, NC, United States), and, when necessary, full 16S rRNA and rpoB sequencing (La Scola et al., 2006) confirmed the isolate identities with 99.9% confidence.
After positive Acinetobacter identification, the three Acinetobacter species we isolated: A. radioresistens, A. johnsonii, and A. lwoffii, along with the laboratory strains of A. baumannii, were incubated aerobically using Luria-Bertani (LB) medium (Becton, Dickinson and Company) at 30°C and under agitation at 200 RPM for liquid cultures.
Antibiotic MIC values for all Acinetobacter isolates was assessed using the Vitek 2XL (BioMérieux) and Sensititre (Trek Diagnostic Systems, Cleveland, OH, United States) platforms. The cards/plates used where GN65, GN69 (BioMérieux) and TrekCOMPGN1F, TrekGN4GF (Trek Diagnostic Systems, West Sussex, United Kingdom), following the instructions of the manufacturers and the CLSI guidelines (CLSI M07-A10). Two different MIC systems were used since some isolates did not grow in the Vitek2 card system. Antibiotic break points for both MIC methods were determined by CLSI M100 or EUCAST1 when no CLSI break points were published. Isolates were considered MDR if they were resistant to ≥3 antibiotic classes (Manchanda et al., 2010).
Imipenem resistance for A. radioresistens was also assessed using the imipenem ETEST® strip (BioMérieux). Briefly, a culture of A. radioresistens LH6 was adjusted to an OD600 = 0.7 in Mueller-Hinton (MH) broth and 150 μL was spread onto MH agar plates. The E-test strip was placed on top of the plate and incubated at 37°C overnight prior to imaging on the Chemidoc XRS+ imager (Bio-Rad).
The pBAV1K-T5-gfp plasmid (Bryksin and Matsumura, 2010) was introduced into LH2, LH6, and D16 by washing overnight liquid cultures with ice-cold 10% glycerol 3X. Then, 50 μL of the cell preparation was mixed with 500 ng of plasmid DNA in an electrocuvette and electroporation was done using the Micropulser Electroporation Apparatus (Bio-Rad) on the Ec2 (2.5 kV, 100 Ω and 25 μF) bacterial setting. The sample was mixed with 100 μL SOC medium [2% tryptone (w/v), 0.5% yeast extract (w/v), 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 20 mM glucose] and plated on LB-kanamycin (50 mg/mL) selective plates to isolate plasmid-containing colonies.
The contact-dependent killing assay was adapted from a previously described bacterial competition assay (Weber et al., 2015). Briefly, strains were grown in liquid culture and adjusted to OD600 = 1.0. Strains were washed in LB one time to remove any antibiotic, mixed in a 1:1 ratio and 5 μL was spotted onto LB agar. After 4 h, the agar containing the spot was excised and resuspended into 1 mL of LB broth. Ten-fold serial dilutions were made and spotted onto LB plates supplemented with 50 μg/mL kanamycin (GoldBio) to select for the prey strain and incubated overnight at 30°C to enumerate the surviving strain. When assessing contact-dependent inhibition, a 0.45 μm nitrocellulose membrane (Bio-Rad) separated the strains by spotting the prey strain on LB agar, placing the membrane on the prey strain, and spotting the predator strain on top of the nitrocellulose. This allows the possible passage of small molecules between the predator and prey during the 4-h incubation, but prevents migration of the predator (results not shown). After the incubation time, the membrane and predator strain were removed, and the agar spot was excised and resuspended in PBS for extracellular DNA isolation.
Following the co-culture experiments described above, cells were resuspended in phosphate-buffered saline. For all extracellular DNA isolations, cell suspensions and released DNA were filtered through a 0.22 μm membrane. The nucleic acids in the filtrate were isolated via phenol/chloroform (Thermo Fisher Scientific) extraction, precipitated with isopropanol and sodium acetate (Thermo Fisher Scientific), and resuspended in nuclease-free water. Care was taken to ensure that an equal fraction of the aqueous phase was taken from each sample so that the isolated DNA quantities would be proportional to each other.
Isolated extracellular DNA was PCR amplified using the kanamycin resistance gene primers: KanF-(CGCAGAAGGCAATGTCATAC) and KanR-(CACTTTGAACGGCATGATGG). Taq polymerase (Thermo Fisher Scientific) was used according to the manufacturer’s instructions, with a Tm of 55°C.
Bacteriophage isolation was performed as described (Gencay et al., 2017). All Acinetobacter strains were tested and only LH6 was capable of being infected by all phages. Thus, LH6 was used as the propagating strain and was grown overnight at 30°C with shaking at 200 RPM, the culture was then adjusted to OD600 = 0.4, and infected with bacteriophages at a multiplicity of infection (MOI) of 0.0001. The infected culture was incubated at 30°C with shaking at 200 RPM overnight. Afterward, the culture was centrifuged at 4255 × g for 15 min, the resulting supernatant was put through a 0.22 μm filter, and the phage-containing filtrate was collected.
The gfp-plasmid containing strain LH6g (LH6 with gfp) was grown in LB liquid culture at 30°C with shaking at 200 RPM overnight. The culture was adjusted to OD600 = 1.0, infected with phages at MOI = 0.001, and incubated at 30°C with shaking at 200 RPM for 18 h. DNA isolation was performed as described above.
Transformation of chemically competent TOP10 E. coli cells (Invitrogen) was done according to the manufacturer’s instructions. Briefly, 10 μL of isolated released DNA was incubated with 40 μL of competent cells on ice for 15 min. The cells were then heat shocked at 42°C for 45 s and placed back on ice for 2 min. Cells were mixed with 400 μL of LB medium and incubated at 37°C, 200 RPM for 45 min. This was followed by plating the cells on LB plates supplemented with 50 μg/mL kanamycin and incubation overnight at 37°C before counting the transformants.
While isolating campylobacters for our routine studies, colonies typically indicative of Campylobacter jejuni or Campylobacter coli from BrillianceTM agar followed by sub-culturing on Campy-Line agar were examined by TEM. These isolates did not show the characteristic curved rod morphology associated with C. jejuni and C. coli (results not shown). To obtain species identification with 99.9% confidence on our isolates, we compared each isolate by MALDI-TOF mass spectrometry, entire 16S rRNA sequencing, and rpoB sequencing when necessary (see Supplementary Sequencing Data). We also examined the antibiotic MIC values for these strains since we predicted these isolates would possess multiple resistances allowing them to be cultured on C. jejuni/C. coli selective plates. Table 1 summarizes the species identification and MIC results. Three Acinetobacter species were isolated: A. radioresistens, A. johnsonii, and A. lwoffii. In total, the isolates were resistant to 14 antibiotics, seven of which are on the WHO list of essential medicines (ampicillin, cefazolin, ceftazidime, chloramphenicol, nitrofurantoin, rifampicin, and tetracycline) (WHO, 2017). Additionally, 8/10 isolates were MDR (Manchanda et al., 2010) and were obtained from all bird hosts sampled (laying hen, turkey, and duck). Nine MICs were found to be exclusive to one bird type (laying hen- ceftriaxone, chloramphenicol, rifampicin; turkey- ertapenem, ceftazidime, aztreonam, tetracycline, and cefalexin ticarcillin/clavulanic acid), indicated in gray Table 1. These groupings suggested that drug resistances could be mobile within the respective laying hen and turkey populations, leading us to investigate drug resistance gene mobility further.
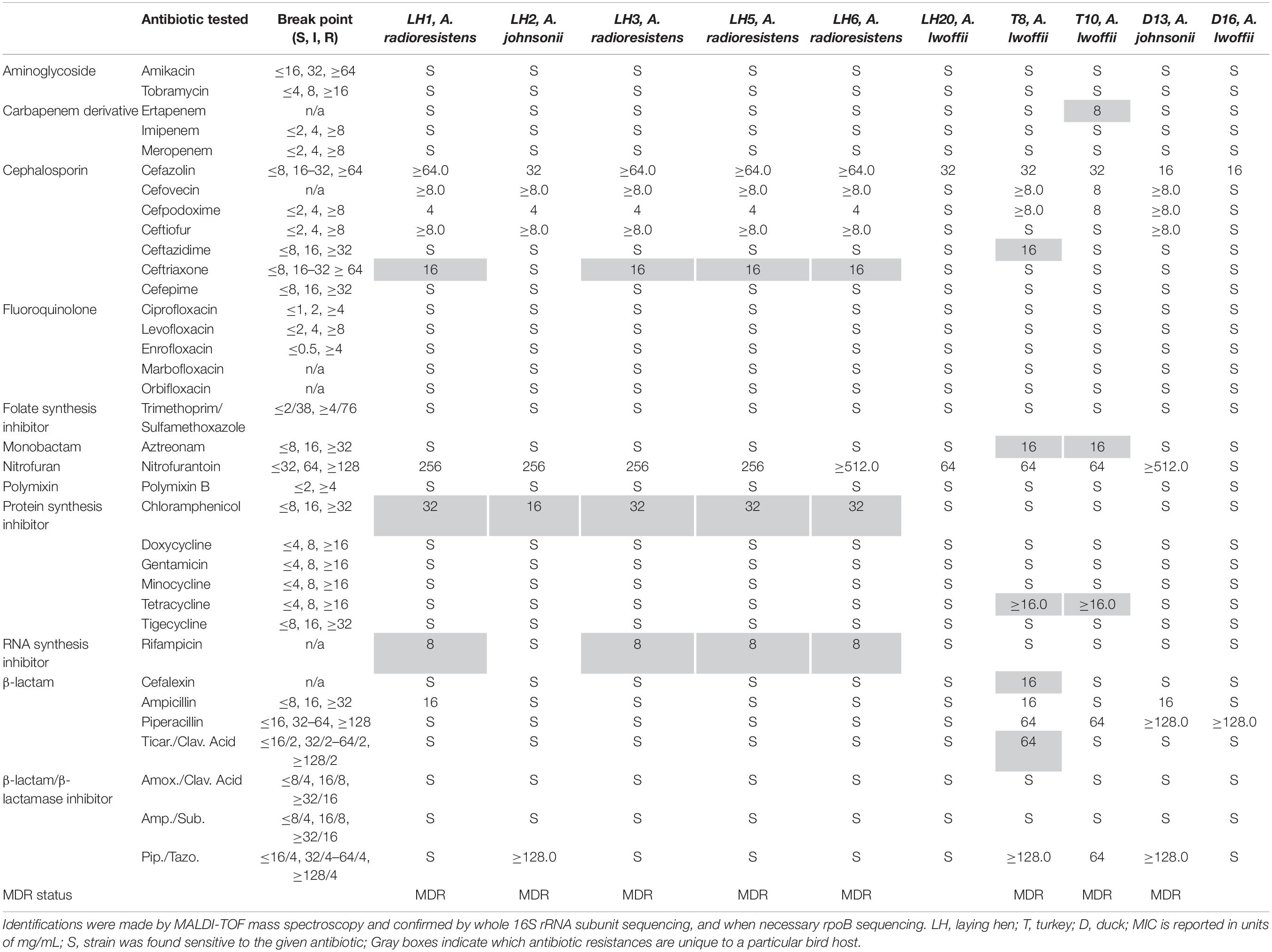
Table 1. Strain identification and minimum inhibitory concentrations of antibiotics for each Acinetobacter species isolated from avian fecal samples.
We hypothesized that contact-dependent killing mechanisms present in Acinetobacter species could assist in obtaining DNA from neighboring organisms by accelerating the death and lysis of those cells. Therefore, we performed bacterial competition assays to determine whether the isolated Acinetobacter species showed a reduction in cell numbers after incubation with A. baumannii. The clinically relevant pathogen A. baumannii ATCC 19606, which has a constitutively active T6SS, was used as the predator (Weber et al., 2015). After introducing the gfp-containing plasmid, pBAV1K-T5-gfp, into our Acinetobacter prey strains (A. johnsonii LH2, A. radioresistens LH6, and A. lwoffii D16), competition assays were performed to observe the susceptibility of these strains to A. baumannii 19606. Killing was observed for all strains, with LH2g (g denotes gfp-expressing variant) and D16g showing the most dramatic effect (Figure 1A). Incubating A. baumannii 19606 with D16g reduced the prey strains to the limit of detection, while LH2g and LH6g were reduced by ∼100-fold. In an effort to determine if the killing was through general CDI or T6SS, we obtained cdi1 and cdi2 deletion mutants in strain 19606 and an hcp (hemolysin co-regulated protein) mutant in A. baumannii ATCC 17978 (Weber et al., 2015; Harding et al., 2017) and compared their killing relative to the respective WT strains. The hcp mutation renders the strain unable to form the extracellular “needle” portion of the T6SS complex, causing the predator to be unable to interact with prey in a T6SS-dependent manner (Ho et al., 2014). We used both strains because we could not generate an hcp mutant in strain 19606 and bioinformatic analysis did not identify CDI systems in strain 17978. We observed minimal effect of CDI knockouts in LH2g and LH6g, compared to WT, while the deletion of cdi1 resulted in greater survival in prey strain D16g, compared to WT and the cdi2 deletion mutant (Figure 1A). When testing the hcp mutant, there was a contribution to killing by the T6SS with LH6g and D16g, and minimal killing of LH2g (Figure 1B). From these results, we concluded that LH2 has minimal susceptibility to CDI and T6SS, LH6 has minimal susceptibility to CDI, but is susceptible to T6SS, and D16 is susceptible to both CDI and T6SS.
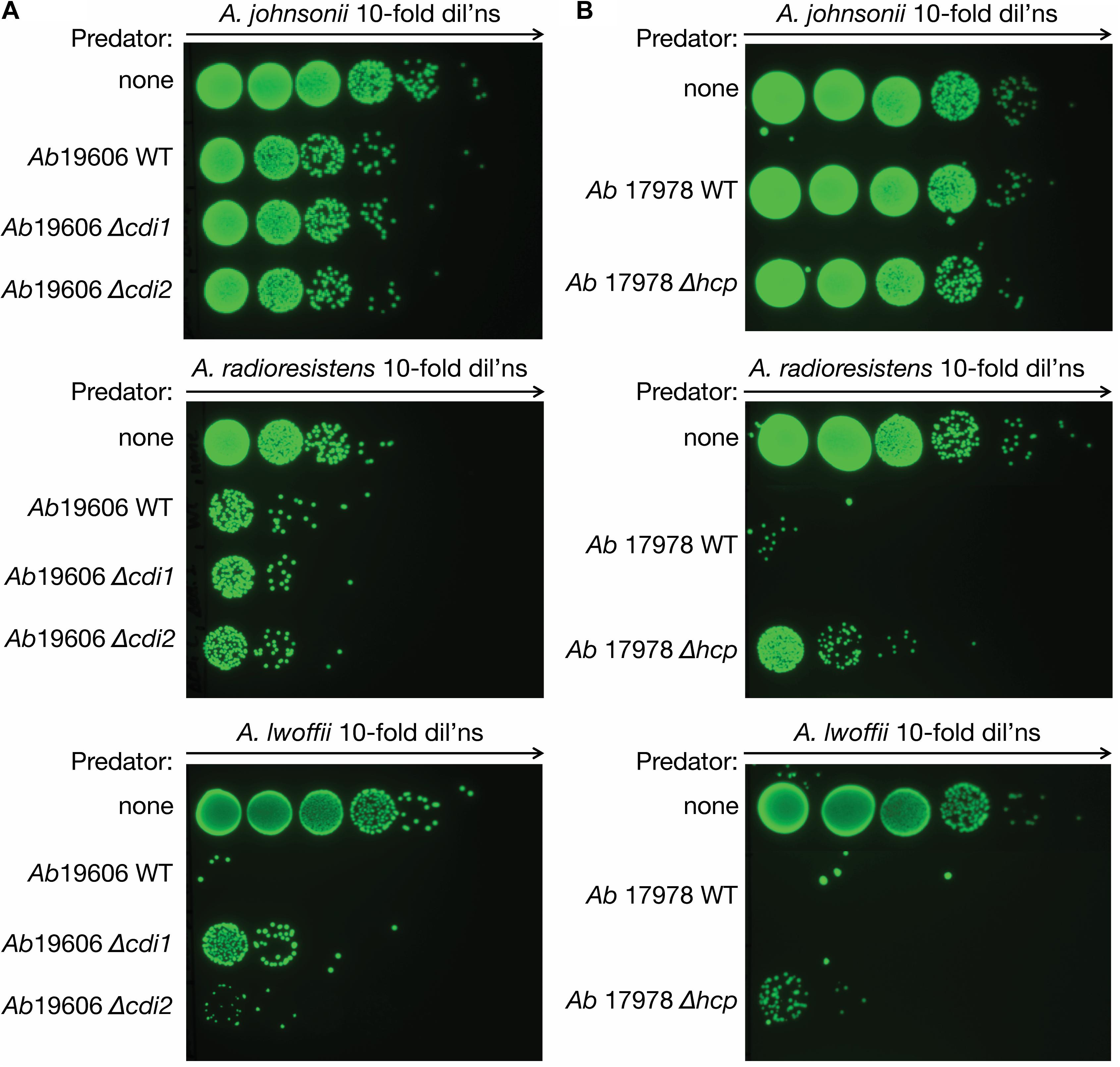
Figure 1. Bacterial killing assay. Acinetobacter contact-dependent killing. (A) A. baumannii kills all isolated Acinetobacter species (LH2, LH6, and D16). The surviving isolates are shown after 4 h of contact. A gfp-encoding plasmid was introduced into the prey species to distinguish them from A. baumannii strain 19606. The A. baumannii ATCC 19606 cdi1 and cdi2 mutant strains were compared to WT and no competition controls to assess the activity of CDI on the isolated strains. (B) The role of T6SS-dependent killing was tested for the isolated strains (LH2, LH6, and D16). The A. baumannii strain 17978 hcp mutant strain, deficient in T6SS activity, was compared to WT and no competition. The surviving isolates are shown after 4 h of contact. All assays were performed at least in triplicate.
We next determined if, after co-incubation with A. baumannii, the prey strains can release intact DNA into the environment through contact-dependent killing (schematic in Figure 2A). The A. baumannii 19606 competition assays were therefore repeated with and without a nitrocellulose membrane between the two strains (to inhibit cell contact, while facilitating the exchange of other molecules) and the DNA was isolated after co-culture (Figure 2B). To test whether or not the released DNA contains intact antibiotic resistance genes, the kanamycin resistance (KmR) gene on the gfp-plasmid was probed by PCR. Figure 2C and Supplementary Figure S1 show that the KmR gene was present in all recovered DNA samples from co-cultures, but not in cultures where contact was inhibited by the membrane or in control incubations. The relative intensities of the PCR products in each lane were measured by densitometry, and significantly more plasmid was released by LH2g and D16g during contact than when inhibited by a membrane (Figure 2D). This demonstrates that contact is necessary for cell killing and that an intact KmR gene is released from these cells.
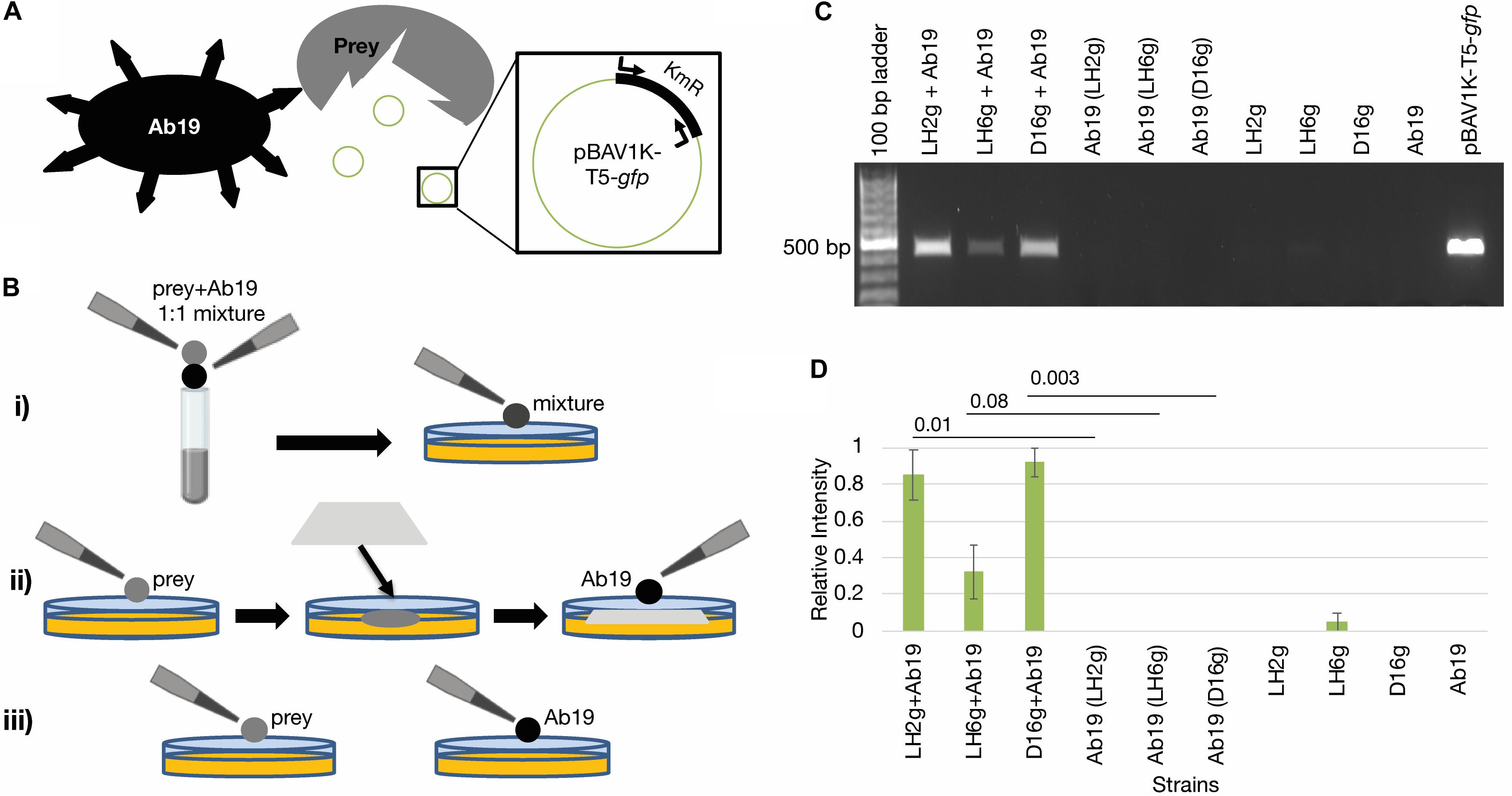
Figure 2. Contact-dependent DNA release. (A) Schematic showing assay concept. The predator strain (Ab19) liberates the pBAV1K-T5-gfp plasmid from the prey strains being tested. Then the presence of the plasmid is detected by amplification of the KmR gene on the plasmid. (B) Schematic detailing the different incubation conditions being used. (i) Prey strain and Ab19 are mixed 1:1 and spotted directly onto the agar plate; (ii) prey strain and Ab19 are separated by a membrane; (iii) controls include incubations of each strain on the plates individually. (C) A 1.0% agarose gel depicting the PCR-based KmR gene (450 bp) detection of released DNA mediated by contact dependent killing representative of triplicate experiments shown in Supplementary Figure S1. Co-cultures separated by a membrane to inhibit T6SS are denoted with parentheses. (D) Relative intensities of PCR products in each lane from Supplementary Figure S1 were measured by densitometry using Image LabTM. The band produced by amplification of pBAV1K-T5-gfp was used as the standard (Rel. Intensity = 1.0). The averages are represented, and error bars represent the standard error of the mean. Student’s paired T-tests were performed for each co-incubation compared to its corresponding membrane-separated control incubation. The p-values for the LH2g, LH6g, and D16g datasets are indicated.
During initial strain isolation, we also isolated several bacteriophages capable of propagating on A. radioresistens strain LH6 with two differing phenotypes: a typical “pinprick” and a “halo” plaque morphology. One representative phage from each phenotypic group (CAP1 and CAP3, respectively) was selected to determine if intact KmR DNA is released after phage infection and cell lysis. The CAP1 phage is a DNA phage likely belonging to the podoviridae and the CAP3 phage is an RNA phage belonging to the cystoviridae. The characterization of the isolated phages will be described elsewhere (Crippen et al., in preparation). To test the ability of phages CAP1 and CAP3 to release host DNA that contains MDR genes, we performed a phage-mediated DNA release assay. The gfp-expressing A. radioresistens LH6g was infected with either CAP1 or CAP3 overnight until a clear culture was obtained and the released host DNA was recovered along with encapsidated phage nucleic acids (Figures 3A,B). Bands at approximately 1 and 1.7 kb were determined to be cell RNA (Supplementary Figure S2). PCR amplification with KmR gene primers indicated that both CAP1 and CAP3 released more KmR DNA than was released during cell growth (Figure 3C and Supplementary Figure S3). This shows that these phages exhibit the potential to accelerate the spread of antibiotic resistance genes through the release of intact resistance genes. We further wished to test the transferability of the released DNA by transforming chemically competent E. coli cells with equal amounts of released DNA and plating for gfp-expressing colonies on Km selective media (Figure 3D). We found that released DNA did contain intact plasmids, which was transformed into E. coli cells. Together, these results show that phage-released DNA can be source of resistance determinants for competent cells.
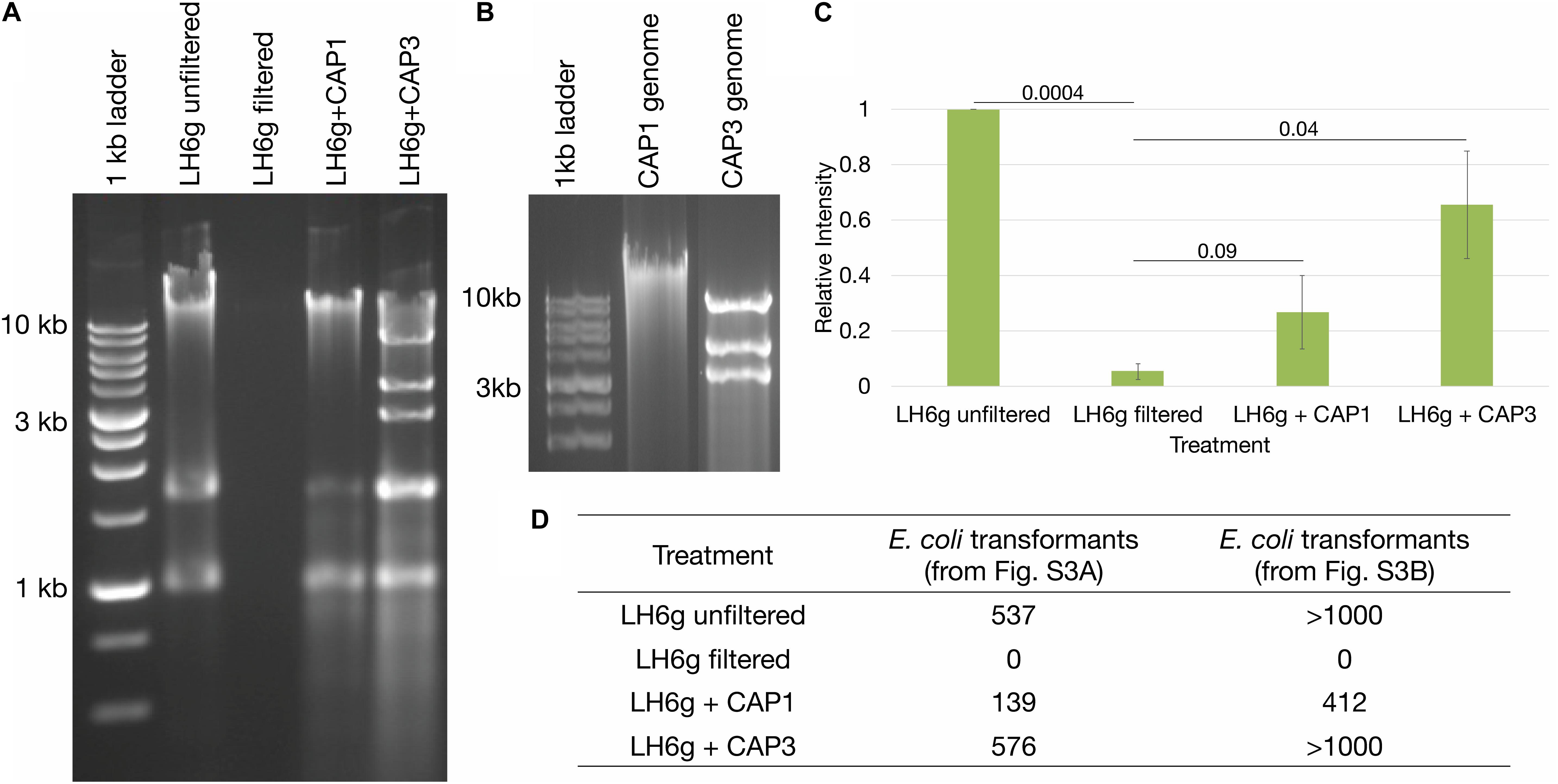
Figure 3. Bacteriophage-mediated DNA release. (A) Analyses of extracellular DNA from cultures with and without phage infection. Agarose gel electrophoresis showing intact DNA release from phage-infected cultures, including cellular RNA at approximately 1 and 1.7 kb (see Supplementary Figure S2). (B) Agarose gel electrophoresis of CAP1 and CAP3 genomes. (C) Relative intensities of PCR products from Supplementary Figure S3 were averaged for each treatment and are represented from triplicate experiments. Standard error of the mean is represented by the error bars, and Student’s paired T-tests were performed with p-values indicated. (D) Chemically competent E. coli were transformed with equal amounts of isolated DNA from sample sets from Supplementary Figures S3A,B and the number of kanamycin resistant colonies are indicated.
In this study, we unexpectedly isolated three Acinetobacter species on C. jejuni/C. coli selective media from laying hen, turkey and duck fecal samples obtained from a free-range pasture fed farm. Species such as C. jejuni are ubiquitous on poultry farms and have been shown to develop resistances, even in the absence of a selective pressure (Luo et al., 2005; Luangtongkum et al., 2009). Previously, a high incidence of MDR E. coli and Listeria spp. were also detected in bird fecal samples from this farm, however, Acinetobacter species were not examined in that study (Rothrock et al., 2016). The species that we isolated were resistant to 14/31 clinically relevant antibiotics that were tested. Each of these Acinetobacter species has been previously reported to carry carbapenemase genes on the blaNDM or blaOXA containing plasmids, the latter predicted to have actually originated from A. radioresistens (Poirel et al., 2008; Zong and Zhang, 2013; Yang et al., 2015). Given that those plasmids are found in divergent Acinetobacter isolates, it is unlikely that the genes are subject to exclusive vertical transmission. In this study, 8/10 of our reservoir isolates were MDR, with one A. lwoffii MDR isolate showing an additional resistance to the last resort carbapenem, ertapenem, with an MIC of 8.0 mg/mL. Given the identification of MDR bacteria on an antibiotic-free farm, it is important to understand the dynamics of gene exchange between naïve pathogens and non-pathogenic MDR reservoir species in the absence of selective antibiotic pressure. Comparison of our drug resistance profiles to the previous study indicates a potential exchange of resistances across bacterial barriers. Most notably, tetracycline resistance was found in all the previous genera tested, including Campylobacter, Salmonella, Listeria, and Escherichia and in both of our turkey-derived A. lwoffii isolates. Resistance to tetracycline is most commonly conferred by the activity of a drug efflux pump, which are typically found on mobile elements (Espinal et al., 2011). It will be interesting to sequence the genomes of the A. lwoffii and A. johnsonii isolates to compare the resistance profiles determined in this study with the genetic composition and extra chromosomal elements that these strains may possess. Additionally, the resistance to β-lactam derivatives was observed in the previous study, while resistance to cephalosporins, which was widespread in our study, was not (Rothrock et al., 2016). These resistance phenotypes can be effected by the activity of AmpC, a metallo-β-lactamase (MBL), or an oxacillinase, all of which have been reported to be associated with mobile genetic elements (Pfeifer et al., 2010; Evans and Amyes, 2014). We previously sequenced strain A. radioresistens LH6 described in this study. LH6 lacks plasmids, but encodes several putative MBLs, which can account for its resistance profile (Crippen et al., 2018). Interestingly, LH6 also encodes blaOXA–23 with 100% amino acid homology to the A. baumannii ASM74664v1 homolog, but no broad carbapenem, cephalosporin or β-lactamase resistance phenotype was observed (Table 1 and Supplementary Figure S4), which is normally associated with the presence of this gene (Pfeifer et al., 2010). This could be due to the lack of IMP-1, OXA-58, and ISAcra1 in the LH6 genome, any of which are required for the trademark carbapenemase activity associated with the OXA-23 carbapenemase (Poirel et al., 2008; Higgins et al., 2013). Intermediate levels of nitrofurantoin resistance were found in Listeria spp. isolated in the Rothrock et al. (2016) study, while 9/10 of our isolates were resistant to nitrofurantoin, explained by the intrinsic nitrofurantoin resistance inherent to Acinetobacter spp. (Giske, 2015). This could be mediated by point mutations in nsfA or nsfB, which metabolize nitrofurantoin into the reactive intermediates that interfere with ribosomal subunits, or by oxqAB in some Acinetobacter spp., but the LH6 sequenced strain does not possess these efflux genes (Sekyere, 2018; Gardiner et al., 2019). Previously, a high incidence of fluoroquinolone resistance was also found among the Listeria spp., while aminoglycoside resistance was found in E. coli and Salmonella spp. (Rothrock et al., 2016). All of our isolates were susceptible to quinolone and aminoglycoside derivatives, indicating a barrier to genetic mobility that was potentially not present for tetracycline or β-lactam resistance elements. To further explore this phenomenon, we used a gfp-labeled resistance marker to demonstrate that plasmid DNA can be released through both contact-dependent killing by A. baumannii and by bacteriophage-mediated lysis.
In our study, we performed bacterial competition assays with two strains of A. baumannii and tested CDI and T6SS deletion mutants. Co-incubation of the isolated strains with A. baumannii strain 19606 and its isogenic cdi1 and cdi2 mutants resulted in extensive killing of A. lwoffii, and less with A. radioresistens and A. johnsonii. We observed that deletion of cdi1 (Harding et al., 2017) did not increase survival of A. johnsonii and A. radioresistens, but did show less killing of A. lwoffii. In this study, cdi2 mutation did not increase survival for any of the isolated strains. These results indicate that CDI systems have a minimal impact on Acinetobacter interspecies competition, except in the case of CDI1 and A. lwoffii strain D16. When using the A. baumannii 17978 strain and its isogenic hcp mutant, we observed minimal killing of A. johnsonii, while A. radioresistens and A. lwoffii were extensively killed and the T6SS could only account for part of this reduction. This killing could be due to an undescribed CDI system that does not resemble known CDI systems in that strain or other combat mechanisms expressed by the 17978 strain (Le et al., 2020). These recent findings are particularly relevant to consider in environments that are under constant selective pressure, such as the poultry gut (Singer and Hofacre, 2006). Rapid expansion of MDR has also been described in low-resource urban areas and rural farming areas where waste management is underdeveloped (Pehrsson et al., 2016). These observations make it important to understand the transfer of resistance determinants, prompting our subsequent experiments tracking an exogenously added resistance marker. We confirmed that release of the KmR gene was proportional to the amount of cell killing observed, consistent with observing the greatest reduction in A. lwoffii growth concomitant with the highest levels of KmR gene detection/release. Therefore, bacterial prey susceptibility to attack will likely impact the availability of new MDR genes for A. baumannii to acquire. Due to these combinations of selective pressures and diversity of bacterial inhabitants, reservoir species accumulate MDR genes that can be released by related pathogens such as A. baumannii, which has also been isolated from livestock in earlier studies (Fernando et al., 2016), and then incorporated into their own genomes.
Bacteriophages have also been described as vehicles for MDR spread through the transduction of genes or mobile elements that contain MDR genes (Brown-Jaque et al., 2015). If we consider the quantity of DNA that can be released when approximately half of the earth’s bacterial population is killed by bacteriophages every 2 days (Deresinski, 2009), then the exchange of MDR genes across species through phage release of DNA not only becomes likely, but inevitable. Evidence for this gene exchange was first demonstrated in the case of the E. coli “superspreader” bacteriophages (Keen et al., 2017). In addition to the findings of Keen et al. (2017), we have demonstrated the ability of the novel A. radioresistens bacteriophages CAP1 and CAP3 to release an intact KmR gene that could then potentially be incorporated by certain naturally competent bacteria that share the same environment, including A. baumannii. We further demonstrated this phenomenon by transforming the recovered DNA from phage-lysed cells into chemically competent E. coli. The probability that these two phage isolates, from different taxonomic families, are unique in their ability to release intact MDR genes is highly unlikely. Thus, great consideration must be given to all lytic phages when designing phage therapy treatments, because of the potential accelerated risk for horizontal gene transfer. These findings provide evidence for alternate methods for rapid MDR gene spread in bacterial species, especially in food animals. Additionally, these types of studies should be replicated for phage-derived elements such as endolysins, which are also being considered as potential therapeutics along with intact phages (Fischetti et al., 2006).
Because phage transduction can also be a mechanism for AMR determinant spread, we investigated the genomes of the isolated phages and their propagating strain, A. radioresistens LH6. Interestingly, 5/7 of the isolated phages were segmented RNA phages that are known to undergo recombination at high rates (Simon-Loriere and Holmes, 2011). In addition, genomic sequencing of A. radioresistens LH6 (Crippen et al., 2018) identified two possible chromosomal prophages, including one Mu-like, that may also be involved in AMR gene transduction (Braid et al., 2004). One of these prophages was indeed isolated after induction with mitomycin C and is being described in a separate manuscript (Crippen et al., in preparation). The presence of this active prophage and the regularity of prophages within the genus Acinetobacter indicates that prophages are also likely to participate in the dissemination of resistance determinants (Touchon et al., 2014; Costa et al., 2018).
Microbial antibiotic resistance is an ancient defense mechanism that is developing into a major crisis for healthcare providers due our great dependence on antibiotics for most medical procedures. The prevalence and mechanisms associated with antibiotic resistance are better characterized, but the mechanisms of MDR gene transfer in the environment are less understood. It is important to continue studying antibiotic resistance gene transfer, so that innovative solutions to the current antibiotic resistance crisis can be found (Bush et al., 2011; Culp et al., 2020).
All datasets generated for this study are included in the article/Supplementary Material.
CC performed all the experiments, with the exception of the strain identification and resistance profiles, and drafted the manuscript. MR provided access to the farm and the fecal samples that the bacterial strains and bacteriophages were isolated from. SS facilitated the strain identification and determination of resistance profiles. CS coordinated all the experiments. All authors edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Ryan DeMone whose science fair project motivated this study, Stephanie Guerra for her contributions with the contact-dependent killing assays, and Corey Mealer for analysis of the phage nucleic acid content. We would also like to thank Laura Lee-Rutherford from the Agricultural Research Service at the United States Department of Agriculture for her assistance with sample collection, the Bacteriology and Molecular Team at the Athens Veterinary Diagnostic Laboratory for strain identification, and the University of Georgia Electron Microscopy Core for their assistance. We also thank Stephen Trent for supplying A. baumannii strain 19606 used in this study, Mario Feldman for providing A. baumannii ATCC 19606 Δcdi1 and Δcdi2 mutant strains, A. baumannii ATCC 17978 WT lacking the pAB04 plasmid and the isogenic Δhcp mutant also lacking pAB04 plasmid, and William Miller for consultation on sequencing results. We would also like to thank Mario Feldman, Brent Weber, and Gisela Di Venanzio for helpful discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01918/full#supplementary-material
Al Atrouni, A., Joly-Guillou, M. L., Hamze, M., and Kempf, M. (2016). Reservoirs of non-baumannii Acinetobacter species. Front. Microbiol. 7:49. doi: 10.3389/fmicb.2016.00049
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2017). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:fux053. doi: 10.1093/femsre/fux053
Braid, M. D., Silhavy, J. L., Kitts, C. L., Cano, R. J., and Howe, M. M. (2004). Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 186, 6560–6574. doi: 10.1128/JB.186.19.6560-6574.2004
Brown-Jaque, M., Calero-Caceres, W., and Muniesa, M. (2015). Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7. doi: 10.1016/j.plasmid.2015.01.001
Bryksin, A. V., and Matsumura, I. (2010). Rational design of a plasmid origin that replicates efficiently in both gram-positive and gram-negative bacteria. PLoS One 5:e13244. doi: 10.1371/journal.pone.0013244
Bush, K., Courvalin, P., Dantas, G., Davies, J., Eisenstein, B., Huovinen, P., et al. (2011). Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896. doi: 10.1038/nrmicro2693
Costa, A. R., Monteiro, R., and Azeredo, J. (2018). Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci. Rep. 8:15346. doi: 10.1038/s41598-018-33800-5
Crippen, C. S., Huynh, S., Miller, W. G., Parker, C. T., and Szymanski, C. M. (2018). Complete genome sequence of Acinetobacter radioresistens strain LH6, a multidrug-resistant bacteriophage-propagating strain. Microbiol Resour Announc 7:e00929-18. doi: 10.1128/MRA.00929-18
Culp, E. J., Waglechner, N., Wang, W., Fiebig-Comyn, A. A., Hsu, Y. P., Koteva, K., et al. (2020). Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 578, 582–587. doi: 10.1038/s41586-020-1990-9
De Gregorio, E., Esposito, E. P., Zarrilli, R., and Di Nocera, P. P. (2018). Contact-dependent growth inhibition proteins in Acinetobacter baylyi ADP1. Curr. Microbiol. 75, 1434–1440. doi: 10.1007/s00284-018-1540-y
De Gregorio, E., Zarrilli, R., and Di Nocera, P. P. (2019). Contact-dependent growth inhibition systems in Acinetobacter. Sci. Rep. 9:154. doi: 10.1038/s41598-018-36427-8
Deresinski, S. (2009). Bacteriophage therapy: exploiting smaller fleas. Clin. Infect. Dis. 48, 1096–1101. doi: 10.1086/597405
Espinal, P., Roca, I., and Vila, J. (2011). Clinical impact and molecular basis of antimicrobial resistance in non-baumannii Acinetobacter. Future Microbiol. 6, 495–511. doi: 10.2217/fmb.11.30
Evans, B. A., and Amyes, S. G. (2014). OXA beta-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Fernando, D. M., Khan, I. U., Patidar, R., Lapen, D. R., Talbot, G., Topp, E., et al. (2016). Isolation and characterization of Acinetobacter baumannii recovered from Campylobacter selective medium. Front. Microbiol. 7:1871. doi: 10.3389/fmicb.2016.01871
Fischetti, V. A., Nelson, D., and Schuch, R. (2006). Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24, 1508–1511. doi: 10.1038/nbt1206-1508
Gardiner, B. J., Stewardson, A. J., Abbott, I. J., and Peleg, A. Y. (2019). Nitrofurantoin and fosfomycin for resistant urinary tract infections: old drugs for emerging problems. Aust. Prescriber 42, 14–19.
Gencay, Y. E., Birk, T., Sorensen, M. C., and Brondsted, L. (2017). Methods for isolation, purification, and propagation of bacteriophages of Campylobacter jejuni. Methods Mol. Biol. 1512, 19–28. doi: 10.1007/978-1-4939-6536-6_3
Giske, C. G. (2015). Contemporary resistance trends and mechanisms for the old antibiotics colistin, temocillin, fosfomycin, mecillinam and nitrofurantoin. Clin. Microbiol. Infect. 21, 899–905. doi: 10.1016/j.cmi.2015.05.022
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Harding, C. M., Pulido, M. R., Di Venanzio, G., Kinsella, R. L., Webb, A. I., Scott, N. E., et al. (2017). Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J. Biol. Chem. 292, 9075–9087. doi: 10.1074/jbc.M117.781575
Hayes, C. S., Koskiniemi, S., Ruhe, Z. C., Poole, S. J., and Low, D. A. (2014). Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb. Perspect. Med. 4:a010025. doi: 10.1101/cshperspect.a010025
Higgins, P. G., Zander, E., and Seifert, H. (2013). Identification of a novel insertion sequence element associated with carbapenem resistance and the development of fluoroquinolone resistance in Acinetobacter radioresistens. J. Antimicrob. Chemother. 68, 720–722. doi: 10.1093/jac/dks446
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Keen, E. C., Bliskovsky, V. V., Malagon, F., Baker, J. D., Prince, J. S., Klaus, J. S., et al. (2017). Novel “Superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio 8:e02115-16. doi: 10.1128/mBio.02115-16
Kinsella, R. L., Lopez, J., Palmer, L. D., Salinas, N. D., Skaar, E. P., Tolia, N. H., et al. (2017). Defining the interaction of the protease CpaA with its type II secretion chaperone CpaB and its contribution to virulence in Acinetobacter species. J. Biol. Chem. 292, 19628–19638. doi: 10.1074/jbc.M117.808394
La Scola, B., Gundi, V. A., Khamis, A., and Raoult, D. (2006). Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44, 827–832. doi: 10.1128/JCM.44.3.827-832.2006
Lawe-Davies, O., and Bennett, S. (2017). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Geneva: WHO.
Le, N. H., Peters, K., Espaillat, A., Sheldon, J. R., Gray, J., Di Venanzio, G., et al. (2020). Peptidoglycan editing provides immunity to Acinetobacter baumannii during bacterial warfare. Sci. Adv. 6:eabb5614. doi: 10.1126/sciadv.abb5614
Line, J. E. (2001). Development of a selective differential agar for isolation and enumeration of Campylobacter spp. J. Food Protect. 64, 1711–1715. doi: 10.4315/0362-028x-64.11.1711
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Luo, N., Pereira, S., Sahin, O., Lin, J., Huang, S., Michel, L., et al. (2005). Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U.S.A. 102, 541–546. doi: 10.1073/pnas.0408966102
Maboni, G., Seguel, M., Lorton, A., and Sanchez, S. (2020). Antimicrobial resistance patterns of Acinetobacter spp. of animal origin reveal high rate of multidrug resistance. Vet. Microbiol. 245:108702. doi: 10.1016/j.vetmic.2020.108702
Manchanda, V., Sanchaita, S., and Singh, N. P. (2010). Multidrug resistant Acinetobacter. J. Glob. Infect. Dis. 2, 291–304.
Paez-Espino, D., Eloe-Fadrosh, E. A., Pavlopoulos, G. A., Thomas, A. D., Huntemann, M., Mikhailova, N., et al. (2016). Uncovering earth’s virome. Nature 536, 425–430. doi: 10.1038/nature19094
Pehrsson, E. C., Tsukayama, P., Patel, S., Mejia-Bautista, M., Sosa-Soto, G., Navarrete, K. M., et al. (2016). Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216. doi: 10.1038/nature17672
Penades, J. R., Chen, J., Quiles-Puchalt, N., Carpena, N., and Novick, R. P. (2015). Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 23, 171–178. doi: 10.1016/j.mib.2014.11.019
Pfeifer, Y., Cullik, A., and Witte, W. (2010). Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 300, 371–379. doi: 10.1016/j.ijmm.2010.04.005
Poirel, L., Bonnin, R. A., and Nordmann, P. (2011). Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 63, 1061–1067. doi: 10.1002/iub.532
Poirel, L., Figueiredo, S., Cattoir, V., Carattoli, A., and Nordmann, P. (2008). Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52, 1252–1256. doi: 10.1128/AAC.01304-07
Rothrock, M. J., Hiett, K. L., Guard, J. Y., and Jackson, C. R. (2016). Antibiotic resistance patterns of major zoonotic pathogens from all-natural, antibiotic-free, pasture-raised broiler flocks in the Southeastern United States. J. Environ. Qual. 45, 593–603. doi: 10.2134/jeq2015.07.0366
Roussin, M., Rabarioelina, S., Cluzeau, L., Cayron, J., Lesterlin, C., Salcedo, S. P., et al. (2019). Identification of a contact-dependent growth inhibition (CDI) system that reduces biofilm formation and host cell adhesion of Acinetobacter baumannii DSM30011 strain. Front. Microbiol. 10:2450. doi: 10.3389/fmicb.2019.02450
Sekyere, J. O. (2018). Genomic insights into nitrofurantoin resistance mechanisms and epidemiology in clinical Enterobacteriaceae. Future Sci. OA 4:FSO293.
Sgro, G. G., Oka, G. U., Souza, D. P., Cenens, W., Bayer-Santos, E., Matsuyama, B. Y., et al. (2019). Bacteria-killing type IV secretion systems. Front. Microbiol. 10:1078. doi: 10.3389/fmicb.2019.01078
Sievert, D., Ricks, P., Edwards, J., Schneider, A., Patel, J., Srinivasan, A., et al. (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 34, 1–14. doi: 10.1086/668770
Simon-Loriere, E., and Holmes, E. C. (2011). Why do RNA viruses recombine? Nat. Rev. Microbiol. 9, 617–626. doi: 10.1038/nrmicro2614
Singer, R. S., and Hofacre, C. L. (2006). Potential impacts of antibiotic use in poultry production. Avian Dis. 50, 161–172. doi: 10.1637/7569-033106R.1
Souza, D. P., Oka, G. U., Alvarez-Martinez, C. E., Bisson-Filho, A. W., Dunger, G., Hobeika, L., et al. (2015). Bacterial killing via a type IV secretion system. Nat. Commun. 6:6453. doi: 10.1038/ncomms7453
Touchon, M., Cury, J., Yoon, E. J., Krizova, L., Cerqueira, G. C., Murphy, C., et al. (2014). The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol. Evol. 6, 2866–2882. doi: 10.1093/gbe/evu225
Turner, D., Wand, M. E., Briers, Y., Lavigne, R., Sutton, J. M., and Reynolds, D. M. (2017). Characterisation and genome sequence of the lytic Acinetobacter baumannii bacteriophage vB_AbaS_Loki. PLoS One 12:e0172303. doi: 10.1371/journal.pone.0172303
Valentine, S. C., Contreras, D., Tan, S., Real, L. J., Chu, S., and Xu, H. H. (2008). Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J. Clin. Microbiol. 46, 2499–2507. doi: 10.1128/JCM.00367-08
Warren, J., and Bose, S. (1968). Bacteriophage-induced inhibiton of host functions. J. Virol. 2, 327–334. doi: 10.1128/jvi.2.4.327-334.1968
Weber, B. S., Ly, P. M., Irwin, J. N., Pukatzki, S., and Feldman, M. F. (2015). A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 112, 9442–9447. doi: 10.1073/pnas.1502966112
Weber, B. S., Miyata, S. T., Iwashkiw, J. A., Mortensen, B. L., Skaar, E. P., Pukatzki, S., et al. (2013). Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142
Wilharm, G., Piesker, J., Laue, M., and Skiebe, E. (2013). DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J. Bacteriol. 195, 4146–4153. doi: 10.1128/JB.00754-13
Yang, D. K., Liang, H. J., Gao, H. L., Wang, X. W., and Wang, Y. (2015). Analysis of drug-resistant gene detection of blaOXA-like genes from Acinetobacter baumannii. Genet. Mol. Res. 14, 18999–19004. doi: 10.4238/2015.December.29.7
Keywords: Acinetobacter, environmental isolates, multidrug resistance, contact-dependent killing, bacteriophages, gene transfer
Citation: Crippen CS, Rothrock MJ Jr, Sanchez S and Szymanski CM (2020) Multidrug Resistant Acinetobacter Isolates Release Resistance Determinants Through Contact-Dependent Killing and Bacteriophage Lysis. Front. Microbiol. 11:1918. doi: 10.3389/fmicb.2020.01918
Received: 17 October 2019; Accepted: 21 July 2020;
Published: 14 August 2020.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Maria Soledad Ramirez, California State University, Fullerton, United StatesCopyright © 2020 Crippen, Jr., Sanchez and Szymanski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine M. Szymanski, Y3N6eW1hbnNAdWdhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.