- 1LISM, Institut de Microbiologie de la Méditerranée, CNRS, Aix-Marseille University, Marseille, France
- 2SAMe Unit, Microbiology Department, Pasteur Institute, Paris, France
The SlyA transcriptional regulator controls the expression of genes involved in virulence and production of surface components in S. Typhimurium and E. coli. Its mode of action is mainly explained by its antagonism with the H-NS repressor for the same DNA binding regions. Interestingly, it has been reported that the alarmone ppGpp promotes SlyA dimerization and DNA binding at the promoter of pagC, enhancing the expression of this gene in Salmonella. A recurring problem in the field of stringent response has been to find a way of following ppGpp levels in vivo in real time. We thought that SlyA, as a ppGpp responsive ligand, was a perfect candidate for the development of a specific ppGpp biosensor. Therefore, we decided to characterize in depth this SlyA control by ppGpp. However, using various genes whose expression is activated by SlyA, as reporters, we showed that ppGpp does not affect SlyA regulation in vivo. In addition, modulating ppGpp levels did not affect SlyA dimerization in vivo, and did not impact its binding to DNA in vitro. We finally showed that ppGpp is required for the expression of hlyE in E. coli, a gene also activated by SlyA, and propose that both regulators are independently required for hlyE expression. The initial report of ppGpp action on SlyA might be explained by a similar action of SlyA and ppGpp on pagC expression, and the complexity of promoters controlled by several global regulators, such as the promoters of pagC in Salmonella or hlyE in E. coli.
Introduction
SlyA is a transcriptional regulator that belongs to the MarR superfamily of regulators (Will and Fang, 2020). Since its discovery as an inducer of hemolytic activity (Libby et al., 1994), several genes have been shown to be regulated by SlyA in Salmonella enterica and in Escherichia coli, however their regulons are different in these two bacteria. In Salmonella, SlyA controls the expression of genes required for virulence (Navarre et al., 2005; Ellison and Miller, 2006). A slyA mutant is impaired for growth within macrophages and is hyper susceptible to oxidative stress (Ellison and Miller, 2006). In E. coli, SlyA activates the expression of the cryptic hemolysin hlyE (clyA) (Wyborn et al., 2004; Lithgow et al., 2007), of Type1 fimbriae (McVicker et al., 2011), of pagP involved in lipid A palmitoylation in biofilm (Chalabaev et al., 2014), and of K5 capsule gene cluster (Corbett et al., 2007). In addition to these reports on specific genes, a global study has recently expanded the proposed repertoire of the SlyA regulon in E. coli, with cryptic genes coding for potential fimbrial-like adhesins that contribute to biofilm formation (Curran et al., 2017). Furthermore, this latter study permitted the refinement of a SlyA binding motif in E. coli.
SlyA binds DNA as a dimer. It functions mainly as counter-silencer by antagonizing and displacing the H-NS repressor (Stoebel et al., 2008; Will et al., 2015). Interestingly, in E. coli, slyA expression is positively autoregulated, independently of H-NS (Corbett et al., 2007). However, the majority of SlyA targets reported so far are known or predicted to be repressed by H-NS (Curran et al., 2017). The condition of induction of slyA itself or the potential ligand molecule of SlyA have not been elucidated. SlyA has been crystalized with a bound salicylate molecule and it was shown in vitro that this binding inhibited SlyA binding to DNA (Dolan et al., 2011; Will et al., 2019).
It has been reported that ppGpp nucleotide promotes SlyA dimerization and binding to its target promoters in Salmonella, and that ppGpp is required for SlyA activity in vivo (Zhao et al., 2008). ppGpp is an important nucleotide acting as a secondary messenger of the stringent response (Potrykus and Cashel, 2008). This global stress response plays a central role in the physiology of bacteria, and its main role is to slow down ribosome biosynthesis and activity while promoting survival programs. This response has been the subject of a strong and renewed interest in the last years when its importance in pathogenicity and resistance to antibiotics has been (re)discovered (Dalebroux et al., 2010; Hobbs and Boraston, 2019). There are two main modes of action of ppGpp, whose relative importances depend on bacteria:in E. coli and closely related bacteria, ppGpp binds RNAP in conjunction with DksA, influencing globally the transcriptome landscape of the cell (Gourse et al., 2018). In addition, ppGpp inhibits enzymes of the guanosine synthesis pathway and ribosomal GTPases (Bennison et al., 2019). The possible allosteric regulation of SlyA by ppGpp triggered high interest at the time, as shown by its highlight in an important review discussing the role of ppGpp in virulence (Dalebroux et al., 2010). If validated, this behavior might have provided a good base for the design of direct ppGpp biosensors that are still missing in the field for live detection and/or imaging of ppGpp in bacteria. However, there has been no further mention of this result or follow-up in the literature. It was only mentioned in a discussion that ppGpp was not required for fimB activation by SlyA in E. coli (McVicker et al., 2011).
Therefore, we decided to study and characterize clearly this proposed role of ppGpp in controlling SlyA mechanism. The results presented here, based on a combination of genetics, molecular, and biochemical approaches, show that ppGpp is not directly involved in the molecular mechanism of SlyA dimerization and DNA binding. However, for some SlyA regulated genes (like hlyE in E. coli or pagCD in Salmonella), complex regulation network involving H-NS and other global regulators might explain indirect ppGpp effects.
Materials and Methods
Plasmid Constructions
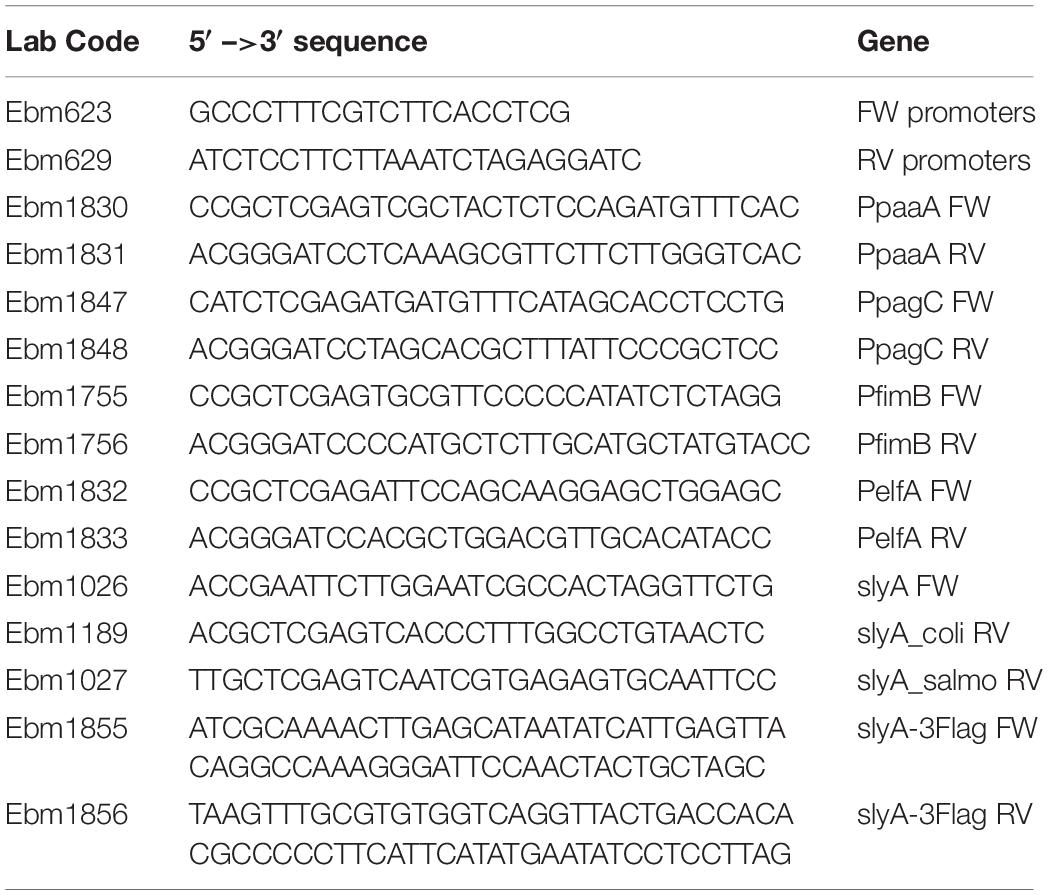
Plasmid constructions are described succinctly in Table 1. The slyA ORFs from E. coli or Salmonella enterica s. Typhimurium 12023 were amplified by PCR on genomic DNA template using the indicated oligonucleotides and cloned in the pBAD24 (pEB227) and pET-6His-Tev vectors (pEB1188).
Transcriptional fusions with GFP were constructed in the pUA66 (pEB898) or pUA139 (pEB987) vector backbone (Zaslaver et al., 2006). When available, transcriptional fusions were retrieved from the Zaslaver collection (Zaslaver et al., 2006), or else the promoter regions were PCR amplified using the oligonucleotides listed in Table 2 and cloned between XhoI and BamHI restriction sites. The Ecocyc website (Karp et al., 2018) was used for sequence retrieval.
Strain Constructions
The construction of the various strains is described succinctly in Table 3. Insertion of the 3Flag sequence in fusion with the slyA ORF on the chromosome was done by direct recombination of a PCR fragment amplified with oligonucleotides ebm1855/1856 and pJL148 plasmid (Zeghouf et al., 2004) as template, following the Datsenko and Wanner procedure (Datsenko and Wanner, 2000). Deletion mutant alleles obtained from the Keio collection (Baba et al., 2006) or tagged alleles obtained by recombination were transduced from one genetic background to another by generalized transduction with phage P1. The kanamycin resistance cassette was removed by transformation with the pCP20 plasmid (Cherepanov and Wackernagel, 1995).
Measure of Expression Using Transcriptional Fusions With GFP
Escherichia coli strains were transformed by the plasmids carrying the GFP transcriptional fusions, with or without pBAD plasmids producing SlyA proteins, and the selection plates were incubated at 37°C for 16 h. 600 μl of LB medium supplemented with the required antibiotics, and 0.05% arabinose for pBAD induction, were inoculated (three to six replicates for each assay) and grown for 16 h at 30°C in 96-well polypropylene plates of 2.2-ml wells under aeration and agitation. Fluorescent intensity measurement was performed in a Tecan infinite M200 plate reader. 150 μl of each well was transferred into a black Greiner 96-well plate for reading optical density at 600 nm (OD600) and fluorescence (excitation, 485 nm; emission, 530 nm). The expression levels were calculated by dividing the intensity of fluorescence by the OD600. After mean values were calculated, values from the control vector were subtracted. The results are given in arbitrary units, because the intensity of fluorescence is acquired with an optimal and variable gain; hence, the absolute values cannot be compared between different panels. The error bars on the figures show the standard error of the mean (SEM).
Purification of SlyA Proteins
BL21(DE3)pLysS strain was transformed with plasmids pET6HisTev-slyA_stm (pEB1885) or pET6HisTev-slyA_ecoli (pEB2004). The strains were grown in 500 ml LB at 30°C. At OD600nm = 0.9, 1 mM IPTG was added and the cultures incubated during 6 h at 23°C. The proteins were then purified following the procedure described previously (Wahl et al., 2011).
Electrophoretic Mobility Shift Assay
DNA fragments containing the pagC_stm, hlyE, or slyA promoters were obtained by PCR using the corresponding transcriptional fusion plasmids as matrices, and the ebm623 and ebm629 primers that hybridize at the limit of the cloning sites. The PCR fragments were then purified using Macherey Nagel PCR purification kit. 20 nM PCR fragments were incubated with purified SlyA and ppGpp (TriLink Biotechnologies) (see legends of Figure 4 and Supplementary Figure S3 for the concentrations), in a 20 μl final reaction buffer containing 25 mM Tris–HCl (pH 7.2), 10 mM MgCl2, 1 mM CaCl2, 0.5 mM EDTA, 50 mM KCl, and 5% glycerol. The mix was incubated for 30 min at 20°C. The reactions were then analyzed by native PAGE (Acrylamide 10% 29:1). DNA was stained with GelRed (Fluo-Probes).
In vivo Crosslinking With Formaldehyde
Cells were pelleted and washed once with 10 mM potassium phosphate buffer, pH 6.8, and resuspended in the same volume, with (+F) or without (-F) formaldehyde 1%. Samples were incubated for 15 min at room temperature. The cells were then pelleted and washed again before solubilization in Laemmli loading buffer (volume normalized according to the OD600 of the initial cultures). Before loading on SDS-PAGE, the samples were either heated 10 min at 37°C to maintain the cross-links, or heated 20 min at 96°C to destroy them. SDS-PAGE, electrotransfer onto nitrocellulose membranes, and Western blot analyses were performed as previously described (Bouveret et al., 1995). The monoclonal anti-M2 Flag antibody used for 3Flag tag detection was purchased from Sigma.
Results
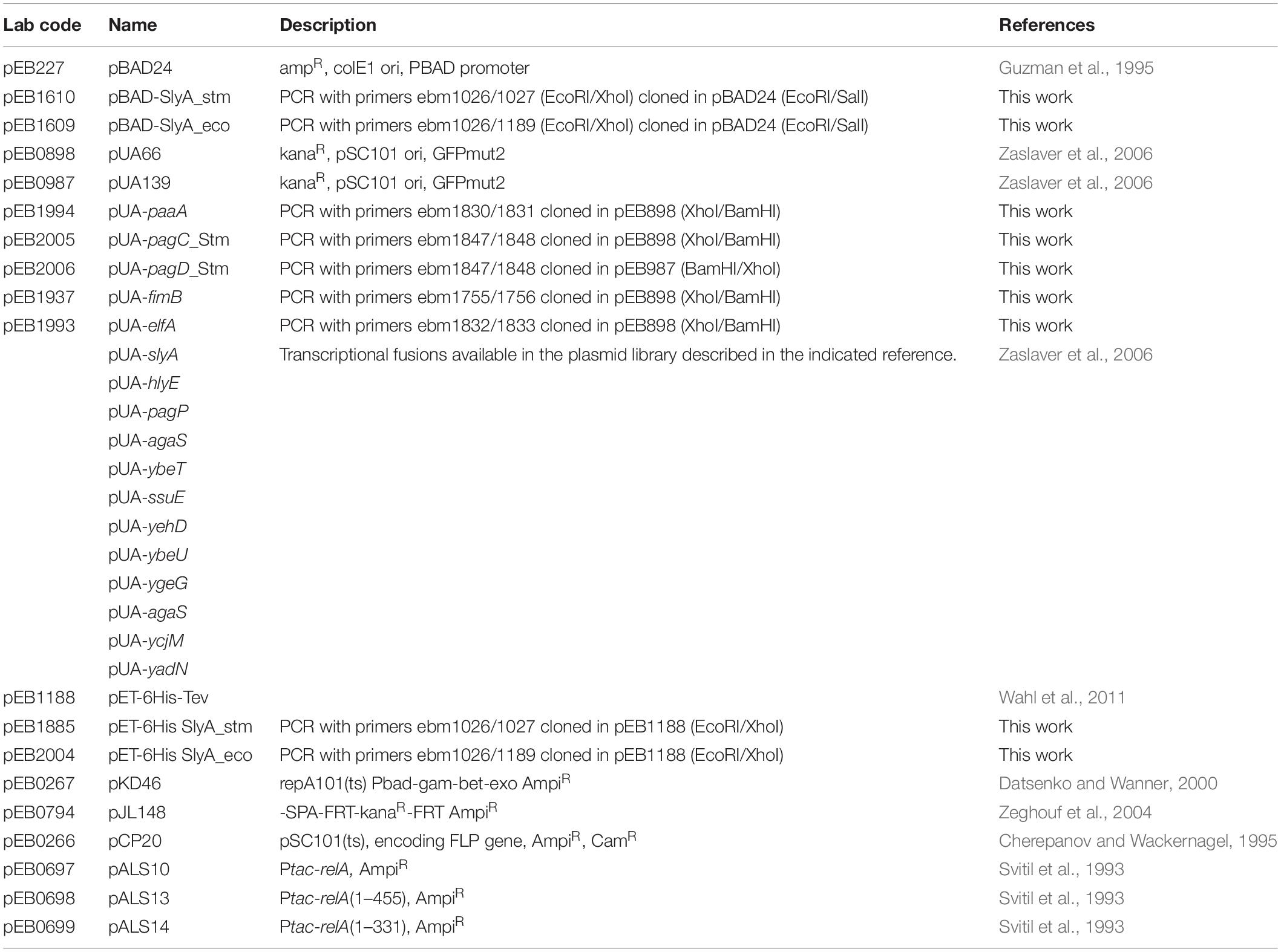
We wanted to study the effect of ppGpp in the activation of gene expression by SlyA in E. coli. In addition to the known SlyA targets hlyE, fimB, and slyA itself, it was reported that SlyA might influence the expression of many genes when overexpressed (Curran et al., 2017). Based on this study, we tested a set of transcriptional fusions to select the ones that will allow us to follow the activity of SlyA. We used transcriptional fusions with GFP already available in a published E. coli promoter library (Zaslaver et al., 2006):pagP, slyA, hlyE, agaS, ybeT, ssuE, yehD, ybeU, ygeG, agaS, ycjM, and yadN. We completed this set by constructing transcriptional fusions missing in the library with the promoters of elfA, fimB, and paaA of E. coli, and also with promoters of Salmonella pagC and pagD, which are regulated by the SlyA/H-NS antagonism and reported to be affected by ppGpp (Zhao et al., 2008). We then measured the expression of all these transcriptional fusions in wild type and ΔslyA strains (Figure 1A and Supplementary Figure S1A), as well as in the ΔslyA strain overproducing or not SlyA from a pBAD inducible plasmid (Figure 1B and Supplementary Figure S1B). From this, we selected 4 reporters that responded robustly to SlyA: slyA itself, paaA, hlyE, and pagC_Stm (Figure 1 and Supplementary Figure S1).
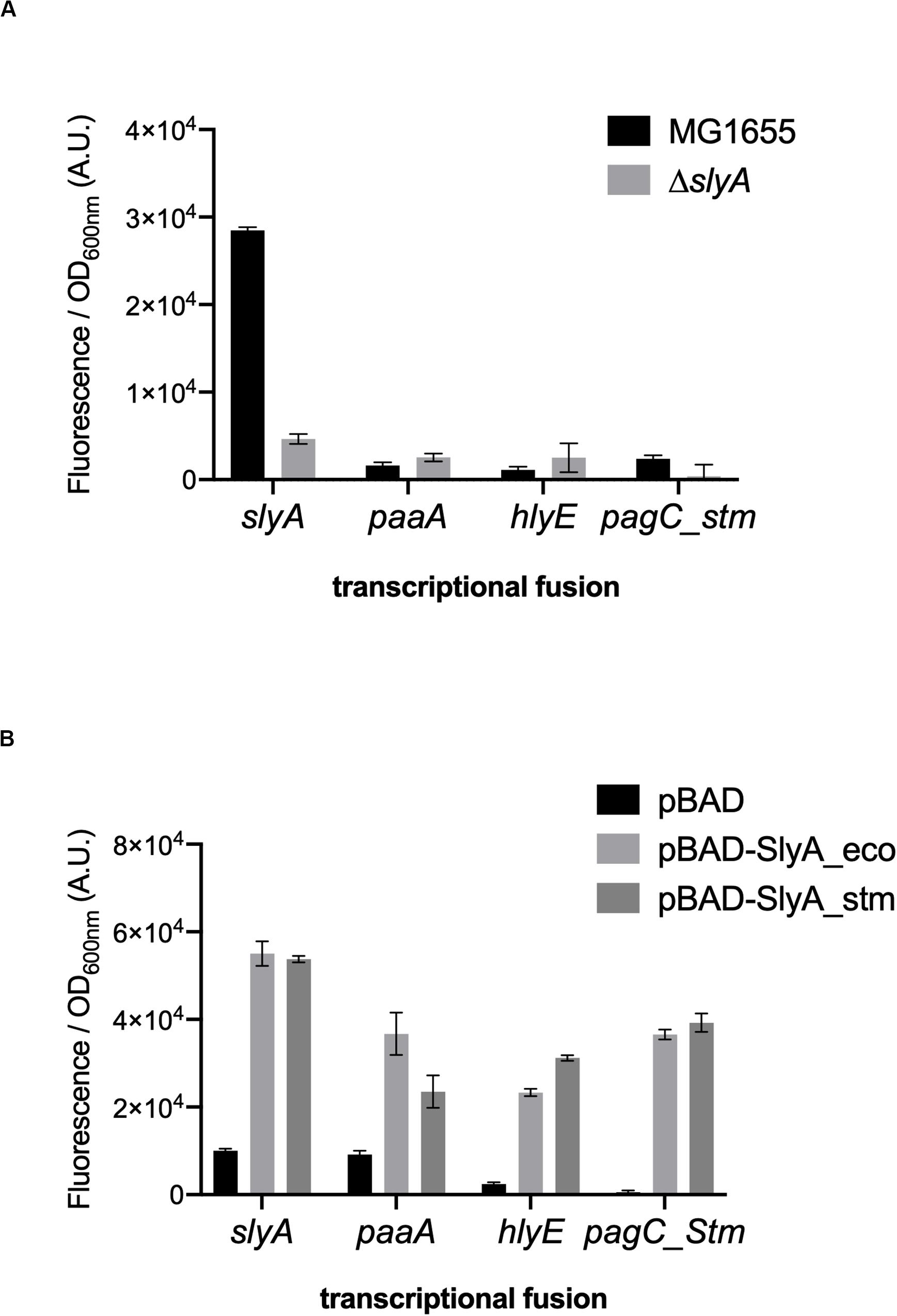
Figure 1. slyA, paaA, hlyE, and pagC_stm promoters are induced by SlyA. (A) Comparison of transcriptional fusion activity in wild type MG1655 and in the slyA mutant EB1073 strains grown overnight at 30°C in LB. (B) Transcriptional fusion activity when SlyA protein is overproduced. MG1655 strains transformed by the indicated transcriptional fusions and the pBAD24 (pEB227), pBAD-slyA_ecoli (pEB1609), or pBAD_slyA_stm (pEB1610) plasmids were incubated overnight at 30°C in LB supplemented with 0.05% arabinose. The activities correspond to the ratio between GFP fluorescence and OD600nm of 6 replicates, given in arbitrary units (A.U.). The error bars show the SEM.
The slyA transcriptional fusion was the only one to show a strong expression level in the wild type strain (Figure 1A). We therefore compared its expression in strains devoid of ppGpp (strains deleted of the relA and spoT genes). The absence of ppGpp did not modify the expression of slyA (Figure 2A). We then compared the expression of the four transcriptional fusions selected above in ΔslyA strains overproducing SlyA, in the presence or in the absence of ppGpp (Figure 2B). For the slyA, paaA, and pagC_Stm fusions, the absence of ppGpp did not prevent the induction by SlyA. These results indicate that in vivo, ppGpp is not required for the mechanism of transcription activation by SlyA.
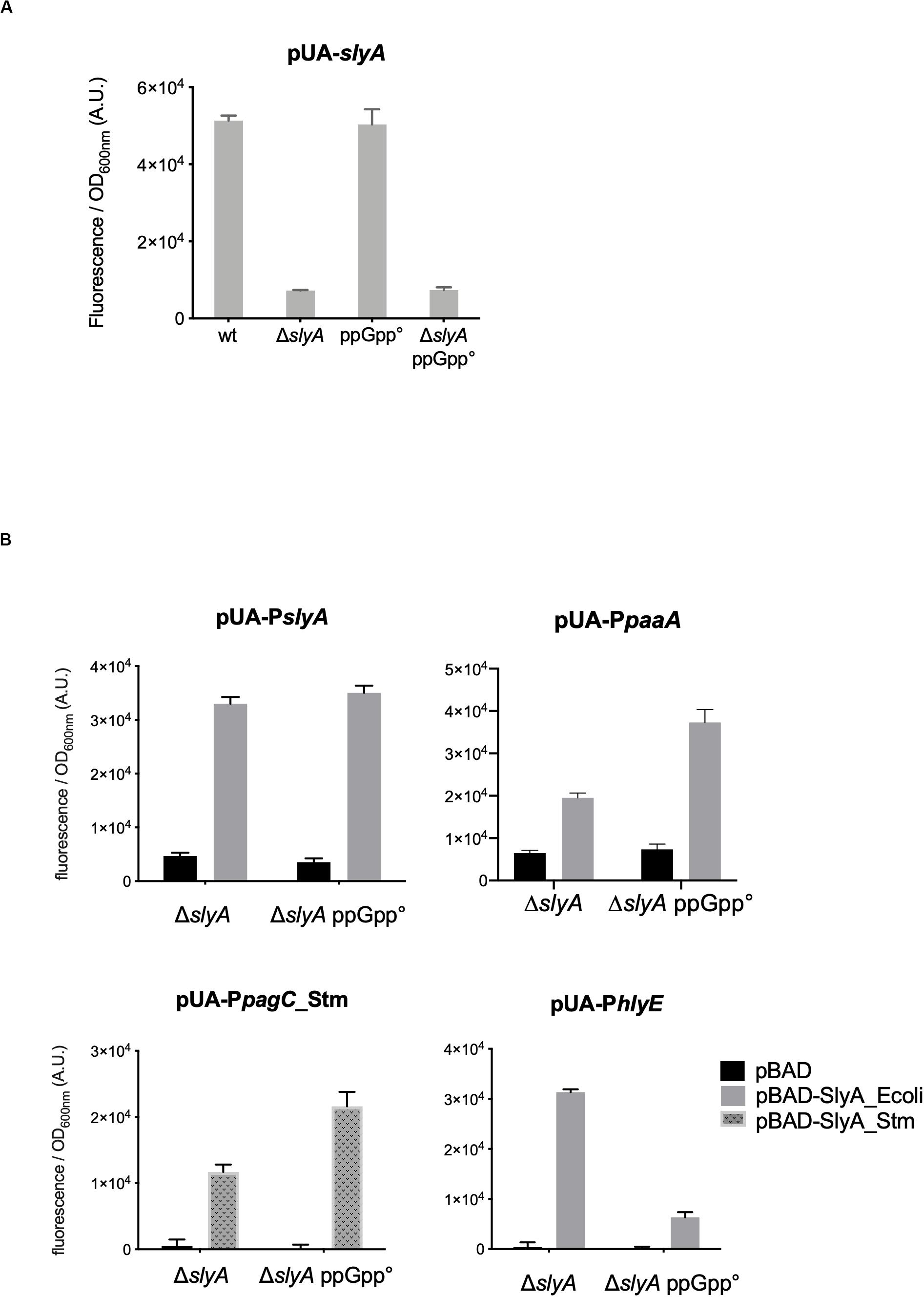
Figure 2. In vivo induction by SlyA does not require ppGpp. (A) Comparison of slyA transcriptional fusion activity in wild type MG1655, ΔslyA (EB1073), ppGpp° (EB425), and ΔslyA_ppGpp° (EB1077) strains grown overnight at 30°C in LB. (B) Effect of pBAD-SlyA overproduction in ΔslyA or ΔslyA_ppGpp° strains on the expression of slyA, paaA, pagC_Stm, and hlyE transcriptional fusions, in the same conditions as in Figure 1B. The activities correspond to the ratio between GFP fluorescence and OD600nm of six (A) or four (B) replicates, given in arbitrary units (A.U.). The error bars show the SEM.
However, the induction of the hlyE fusion by SlyA was strongly decreased in the absence of ppGpp (Figure 2B). To characterize better the specific effect of ppGpp on hlyE, we tested its induction by SlyA in different mutants for global regulatory factors. First, we tested the action of SlyA in the ΔdksA mutant. DksA is a cofactor of the RNA polymerase, required for the regulation of RNAP by ppGpp (Gourse et al., 2018). While ppGpp is still present in this mutant, dksA deletion mimics the global effects of a ppGpp° mutant on gene transcription due to the action of ppGpp on RNA polymerase. While SlyA still activated the expression of the slyA transcriptional fusion in the ΔdksA mutant, as in the ppGpp° mutant (Supplementary Figure S2), SlyA induction of hlyE was strongly decreased in the ΔdksA mutant, similarly to what was observed in the ppGpp° mutant (Figure 3A). This result suggests that the effect of ppGpp on hlyE expression is due to its role in controlling expression through RNAP regulation (at hlyE promoter or others), and not to a direct control of SlyA activity. For full activation of its expression, hlyE would therefore need both SlyA overproduction (or activation by unknown conditions), and the presence of ppGpp.
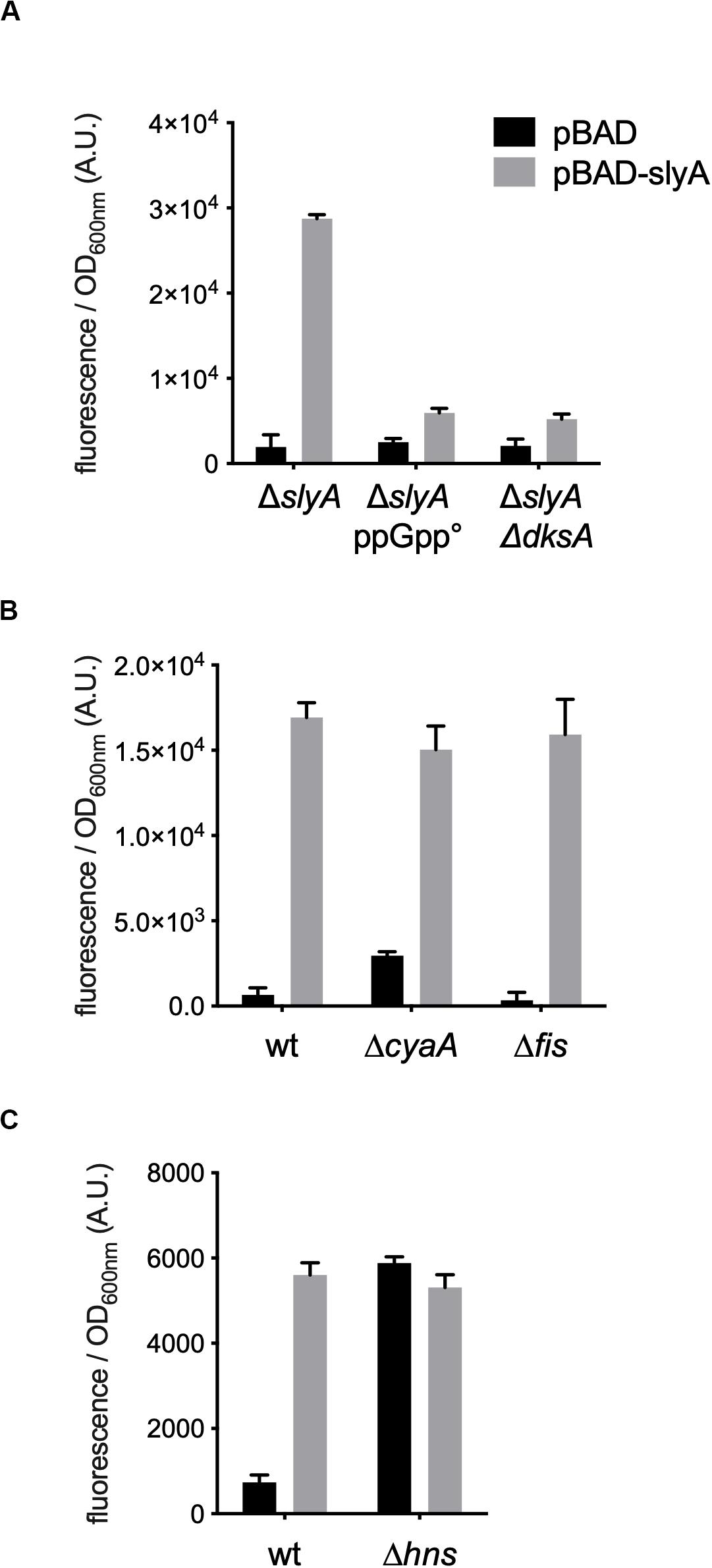
Figure 3. hlyE activation by SlyA in global regulatory mutants. Induction of the hlyE transcriptional fusion by pBAD-slyA_ecoli was tested in the same conditions as in Figure 1B, in the indicated mutant strains: wt (MG1655), ΔslyA (EB1073), ΔslyA_ppGpp (EB1077), ΔslyAΔdksA (EB1100), ΔcyaA (EB781), Δfis (EB743), and Δhns (EB951). The activities correspond to the ratio between GFP fluorescence and OD600nm of 4 (A,C) or 3 (B) replicates, given in arbitrary units (A.U.). The error bars show the SEM.
In addition to SlyA, hlyE expression is controlled by a network of global regulators, such as H-NS (Wyborn et al., 2004; Lithgow et al., 2007), CRP-cAMP and FNR (Westermark et al., 2000), and it was also reported that it is negatively regulated by Fis (Bradley et al., 2007). ppGpp is also a member of this complex network controlling bacterial physiology (Travers and Muskhelishvili, 2005). Therefore the ppGpp/DksA effect observed on the expression of hlyE might be indirect through one or several of these global regulators. We tested hlyE induction by SlyA in hns, fis, and cyaA mutants. SlyA was still able to induce hlyE expression in fis and cyaA mutants (Figure 3B). As expected, the expression of hlyE was de-repressed in the Δhns mutant, and not further induced by the presence of SlyA (Figure 3C). This confirmed that SlyA activation of hlyE expression is due to the displacement of H-NS. This set of experiments suggests that ppGpp role in hlyE expression is not due to an indirect effect through CRP-cAMP or Fis regulators, but probably through the regulation of RNAP at the hlyE promoter in synergy with DksA.
Our results obtained in vivo suggested that ppGpp had no role in SlyA function, contrary to what was reported before (Zhao et al., 2008). Therefore, it was necessary to also test the effect of ppGpp on SlyA DNA binding in vitro. Using gel shift assays, we were able to detect a robust binding of SlyA on the promoter regions tested:hlyE, slyA, and pagC_Stm (Supplementary Figure S3A). We then choose for each binding assay, SlyA/DNA ratios that were just sufficient to detect a shift in order to test the effect of adding ppGpp. With addition of 50 μM or 100 μM ppGpp [the same concentrations used by Zhao et al. (2008)], the shifts were not affected (Figure 4). Because we used purified SlyA proteins with a 6his tag fused at the N-terminal, we also performed the same experiments after removing the tag by TEV cleavage. Also, ppGpp might have been trapped with SlyA during the purification, therefore we performed the purifications in a ppGpp null strain and obtained the same negative result (Supplementary Figure S3B).
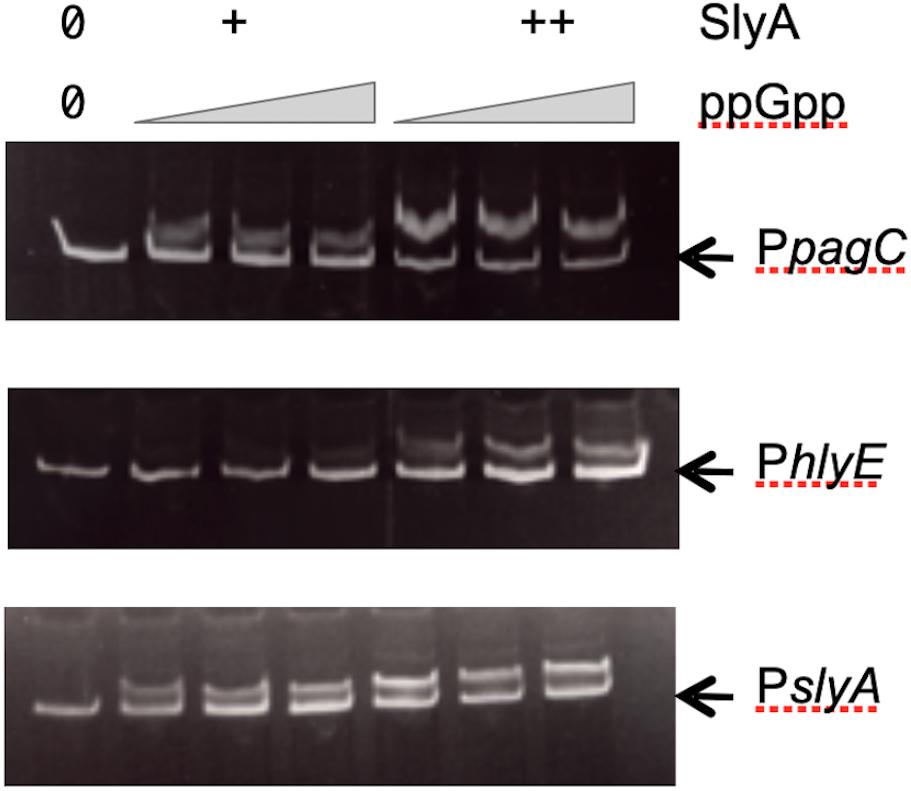
Figure 4. Effect of ppGpp on SlyA binding to DNA. Electrophoretic mobility shift assays were performed using purified SlyA from E. coli [at 50 (+) or 100 (++) nM] or purified SlyA from Salmonella for the pagC experiment [at 200 (+) or 400 (++) nM]. For each SlyA concentration, 0, 50, or 100 μM ppGpp were added.
The last reported effect of ppGpp on SlyA, was that it enhanced its dimerization, as shown by in vivo cross-linking experiments (Zhao et al., 2008). In order to detect SlyA by Western blot, we constructed wild type and ppGpp° strains producing a SlyA-3Flag tagged protein expressed from its endogenous locus. SlyA-3Flag was readily detected in the two genetic backgrounds, at the expected size of approximately 20 kDa (Figure 5A). To test the dimerization, we used whole cell cross-linking with formaldehyde. This cross-linker produces covalent bonds that can be destroyed by heating at 96°C. In the wild type background, dimerization of SlyA was clearly detected by cross-linking with formaldehyde (Figure 5A). The dimerization was identical in the ppGpp° background (Figure 5A). In reverse, we decided to test if an excess of ppGpp might affect SlyA dimerization, by overproducing the RelA ppGpp synthase. Plasmids pALS10, pALS13, and pALS14 code, respectively, for a full RelA protein, a constitutively active truncated RelA protein, and an inactive RelA protein (Svitil et al., 1993). We performed the cross-linking experiment in MG1655 strain transformed by these three plasmids and with induction of RelA variants expression. In the samples with induced ppGpp production (pALS10 and pALS13), SlyA dimerization was not affected, or even slightly diminished (Figure 5B). In conclusion, we were not able to see any positive effect of ppGpp on SlyA dimerization.
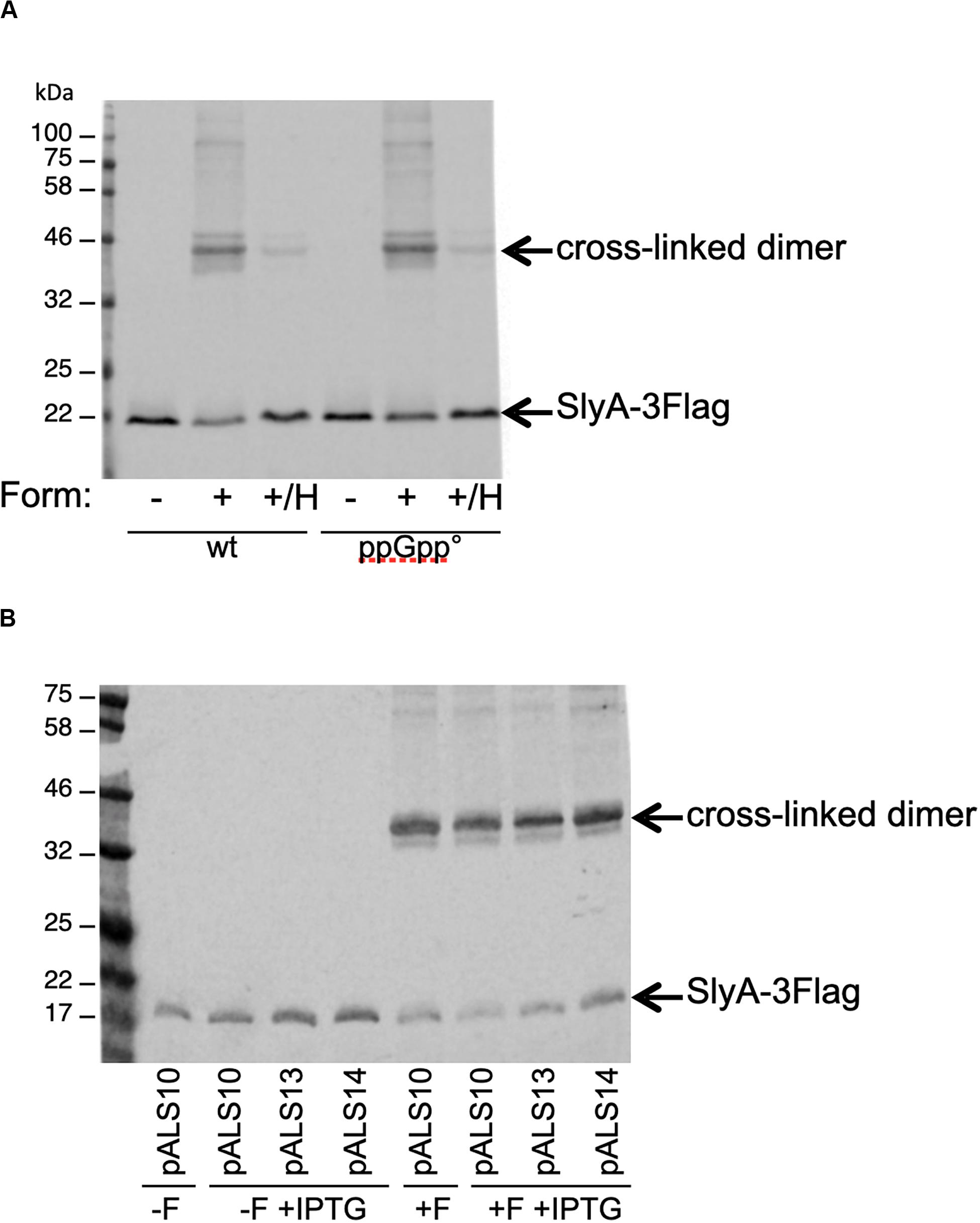
Figure 5. Effect of ppGpp on SlyA dimerization in vivo. (A) MG1655_SlyA-3Flag (EB1106) and ppGpp°_SlyA-3Flag (EB1110) strains were grown to OD600 = 1.3. (B) Strain MG1655_SlyA-3Flag (EB1106) was transformed by plasmids pALS10, pALS13, and pALS14 (pEB0697, pEB0698, and pEB0699, respectively) (Svitil et al., 1993). These transformed strains were grown to OD600 = 1.5 and then relA expression was induced with 1 mM IPTG for 30 min. Then, for both panels, the cells were cross-linked with formaldehyde as described in section “Materials and Methods.” +F, with formaldehyde; –F, without formaldehyde; +/H, with formaldehyde and then heated at 96°C. After SDS-PAGE and Western blot, the SlyA-3Flag tag was detected with monoclonal anti-Flag M2.
Discussion
In this study, we showed that ppGpp is not required for SlyA function in E. coli. The expression of several reporter genes was still induced by SlyA overproduction in the absence of ppGpp in vivo, SlyA binding to DNA was not improved by adding ppGpp in vitro, and finally SlyA dimerization was not affected by ppGpp absence or increased levels in vivo. Even if the initial report of ppGpp effect on SlyA is now more than 10 years old (Zhao et al., 2008), we think this information is important and of public good for the community of researchers working on ppGpp. Indeed, we are aware of several groups that were interested in developing ppGpp sensors based on this observation, including ourselves. To our knowledge, no confirmation or disproof of ppGpp effect on SlyA was reported since then, apart from a brief mention that ppGpp had no effect on SlyA control of the fimB promoter in E. coli (McVicker et al., 2011). Furthermore, SlyA was not spotted in two independent global studies aiming at identifying ppGpp binding proteins (Zhang et al., 2018; Wang et al., 2019). It is therefore still unclear what molecule can regulate SlyA activity. However, recent work provided strong evidence of SlyA control by Salicylate, which fits with SlyA belonging to the MarR family containing proteins known to respond to small aromatic carboxylate compounds (Dolan et al., 2011; Will et al., 2019).
The obvious difference that could explain the discrepancy between our work and the one reported in Zhao et al., 2008 is that we have studied the activity of SlyA in E. coli, while the previous study was done in Salmonella (Zhao et al., 2008), and that we have studied different reporter genes controlled by SlyA. In particular, the expression of slyA itself was a very useful reporter of SlyA action, since it is not dependent on H-NS, and permitted to observe that it was not affected in the absence of ppGpp (Figure 2A). Furthermore, we do not contradict the fact that pagCD promoters are shut down in a ppGpp° strain in Salmonella as shown in Zhao et al., 2008. We propose that it is in fact very similar to what we observed for the expression of hlyE in E. coli, for which SlyA overproduction can partially counteract the negative effect of ppGpp absence (Figure 2B), as it was observed for pagCD in Salmonella (Zhao et al., 2008). Production of SlyA_eco or SlyA_stm had identical effects when produced in E. coli (Figure 1B). Inversely, it was shown that production of SlyA_eco in Salmonella is able to counter silence the expression of pagC, similarly, to SlyA_stm (Will et al., 2019). Therefore, we think the molecular mechanism of SlyA is identical in the two bacteria. However, it is clear that the regulons and the physiological role of SlyA are very different in the two bacteria. This difference does not come from the SlyA protein itself, but from the variations in intergenic and regulatory regions of the target genes. A striking difference is for example that SlyA represses its own expression in Salmonella (Stapleton et al., 2002; Will et al., 2019), whereas it auto-activates its expression in E. coli as we showed here (Figure 1) and as it was demonstrated before (Corbett et al., 2007). The expression level of slyA might also play a role, as it has been suggested that slyA expression is much lower in E. coli than in Salmonella (Will et al., 2019). However, in our experiments, the PslyA transcriptional fusion was one of the few to display a robust basal expression level, and we were able to detect the SlyA-3Flag tagged protein expression in E. coli (Figure 5). Still, only SlyA overproduction using pBAD-SlyA plasmid permitted to detect expression of paaA, hlyE, and pagC_Stm, suggesting a strong excess of SlyA is necessary to overcome H-NS repression on these genes.
Concerning the effect of ppGpp on SlyA binding to DNA in vitro, and the dimerization of SlyA in vivo, the discrepancy between our results and the previous ones (Zhao et al., 2008) is more difficult to understand. Indeed, the SlyA proteins of E. coli and Salmonella are highly similar (91% identical and 95% similar over 142 amino acids), and we performed in vitro binding experiments with SlyA proteins purified from both E. coli and Salmonella, including a binding experiment on a similar Salmonella pagCD intergenic region as the one used previously (Figure 4). As described in the result section, we performed several control experiments to rule out any effect of the tag or the purification procedure of SlyA proteins (Supplementary Figure S3B). For the in vivo dimerization detected by cross-linking with formaldehyde, we performed the experiment in an E. coli strain producing a SlyA-tagged protein expressed from its endogenous locus. Zhao et al. performed this experiment in Salmonella, with a SlyA-tagged protein expressed from a plasmid. In this case, an indirect effect of ppGpp on slyA expression might explain the different results.
The interpretation of the experiments performed in strains mutated for global regulators (such as ppGpp) is complicated by the mode of action of SlyA, which is not a direct and classical activator, but acts mainly as a counter silencer of H-NS. It has been shown that ppGpp physiological effects are intermixed with global regulators such as Fis, CRP, or H-NS, and even DNA supercoiling state (Johansson et al., 2000; Travers and Muskhelishvili, 2005). Therefore, it is to be expected that any tinkering of ppGpp concentrations in vivo will affect a complex network of global regulations. Particular promoters such as pagC in Salmonella or hlyE in E. coli are controlled by an especially high numbers of specific and global factors, not only H-NS and SlyA, but also PhoPQ and EmrR in the case of pagC in Salmonella (Zhao et al., 2008; Yang et al., 2019) or CRP and FNR for hlyE in E. coli (Westermark et al., 2000; Bradley et al., 2007). Obviously, these complex regulatory networks might be affected by ppGpp levels, together with a possible direct effect of ppGpp on the RNAP depending on the nature of the promoter itself (Gourse et al., 2018), as it might be the case for hlyE in our study or pagC in Salmonella (Zhao et al., 2008). More generally, because ppGpp impacts global regulatory networks central to the physiology of bacteria, our study should be taken as a warning of caution in the interpretation of in vivo effects triggered by the modification of ppGpp levels.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
EB and JV designed the study. EB and JB designed and performed the experiments. EB, JV, and JB discussed the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Centre National de la Recherche Scientifique (CNRS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Laetitia My for collaborating with exploratory experiments on SlyA, preliminary to this study, and Frédéric Barras for helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01856/full#supplementary-material
Abbreviations
E. coli, Escherichia coli; GFP, green fluorescent protein; ppGpp, guanosine tetraphosphate.
References
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008.
Bennison, D. J., Irving, S. E., and Corrigan, R. M. (2019). The impact of the stringent response on TRAFAC GTPases and prokaryotic ribosome assembly. Cells 8:1313. doi: 10.3390/cells8111313
Bouveret, E., Derouiche, R., Rigal, A., Lloubès, R., Lazdunski, C., and Bénédetti, H. (1995). Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J. Biol. Chem. 270, 11071–11077. doi: 10.1074/jbc.270.19.11071
Bradley, M. D., Beach, M. B., de Koning, A. P. J., Pratt, T. S., and Osuna, R. (2007). Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 153, 2922–2940. doi: 10.1099/mic.0.2007/008565-0
Chalabaev, S., Chauhan, A., Novikov, A., Iyer, P., Szczesny, M., Beloin, C., et al. (2014). Biofilms formed by gram-negative bacteria undergo increased lipid a palmitoylation, enhancing in vivo survival. mBio 5:e01116-14. doi: 10.1128/mBio.01116-14
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-a
Corbett, D., Bennett, H. J., Askar, H., Green, J., and Roberts, I. S. (2007). SlyA and H-NS regulate transcription of the Escherichia coli K5 capsule gene cluster, and expression of slyA in Escherichia coli is temperature-dependent, positively autoregulated, and independent of H-NS. J. Biol. Chem. 282, 33326–33335. doi: 10.1074/jbc.m703465200
Curran, T. D., Abacha, F., Hibberd, S. P., Rolfe, M. D., Lacey, M. M., and Green, J. (2017). Identification of new members of the Escherichia coli K-12 MG1655 SlyA regulon. Microbiology 163, 400–409. doi: 10.1099/mic.0.000423
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C., and Swanson, M. S. (2010). ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199. doi: 10.1128/mmbr.00046-09
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dolan, K. T., Duguid, E. M., and He, C. (2011). Crystal structures of SlyA protein, a master virulence regulator of Salmonella, in free and DNA-bound states. J. Biol. Chem. 286, 22178–22185. doi: 10.1074/jbc.m111.245258
Ellison, D. W., and Miller, V. L. (2006). Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9, 153–159. doi: 10.1016/j.mib.2006.02.003
Gourse, R. L., Chen, A. Y., Gopalkrishnan, S., Sanchez-Vazquez, P., Myers, A., and Ross, W. (2018). Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184. doi: 10.1146/annurev-micro-090817-062444
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Hobbs, J. K., and Boraston, A. B. (2019). (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect. Dis. 5, 1505–1517. doi: 10.1021/acsinfecdis.9b00204
Johansson, J., Balsalobre, C., Wang, S. Y., Urbonaviciene, J., Jin, D. J., Sondén, B., et al. (2000). Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell 102, 475–485. doi: 10.1016/s0092-8674(00)00052-0
Karp, P. D., Ong, W. K., Paley, S., Billington, R., Caspi, R., Fulcher, C., et al. (2018). The EcoCyc database. EcoSal Plus 8:10.1128/ecosalplus.ESP-0009-2013. doi: 10.1128/ecosalplus.ESP-0009-2013
Libby, S. J., Goebel, W., Ludwig, A., Buchmeier, N., Bowe, F., Fang, F. C., et al. (1994). A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. U.S.A. 91, 489–493. doi: 10.1073/pnas.91.2.489
Lithgow, J. K., Haider, F., Roberts, I. S., and Green, J. (2007). Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol. Microbiol. 66, 685–698. doi: 10.1111/j.1365-2958.2007.05950.x
McVicker, G., Sun, L., Sohanpal, B. K., Gashi, K., Williamson, R. A., Plumbridge, J., et al. (2011). SlyA protein activates fimB gene expression and type 1 fimbriation in Escherichia coli K-12. J. Biol. Chem. 286, 32026–32035. doi: 10.1074/jbc.m111.266619
Navarre, W. W., Halsey, T. A., Walthers, D., Frye, J., McClelland, M., Potter, J. L., et al. (2005). Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56, 492–508. doi: 10.1111/j.1365-2958.2005.04553.x
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical. Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Stapleton, M. R., Norte, V. A., Read, R. C., and Green, J. (2002). Interaction of the Salmonella Typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J. Biol. Chem. 277, 17630–17637. doi: 10.1074/jbc.m110178200
Stoebel, D. M., Free, A., and Dorman, C. J. (2008). Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154, 2533–2545. doi: 10.1099/mic.0.2008/020693-0
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi: 10.1016/0022-2836(86)90385-2
Svitil, A. L., Cashel, M., and Zyskind, J. W. (1993). Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 268, 2307–2311.
Travers, A., and Muskhelishvili, G. (2005). DNA supercoiling - a global transcriptional regulator for enterobacterial growth. Nat. Rev. Microbiol. 3, 157–169. doi: 10.1038/nrmicro1088
Wahl, A., My, L., Dumoulin, R., Sturgis, J. N., and Bouveret, E. (2011). Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol. Microbiol. 80, 1260–1275. doi: 10.1111/j.1365-2958.2011.07641.x
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150. doi: 10.1038/s41589-018-0183-4
Westermark, M., Oscarsson, J., Mizunoe, Y., Urbonaviciene, J., and Uhlin, B. E. (2000). Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182, 6347–6357. doi: 10.1128/jb.182.22.6347-6357.2000
Will, W. R., Brzovic, P., Le Trong, I., Stenkamp, R. E., Lawrenz, M. B., Karlinsey, J. E., et al. (2019). The evolution of SlyA/RovA transcription factors from repressors to countersilencers in Enterobacteriaceae. mBio 10:e00009-e19. doi: 10.1128/mBio.00009-19
Will, W. R., and Fang, F. C. (2020). The evolution of MarR family transcription factors as counter-silencers in regulatory networks. Curr. Opin. Microbiol. 55, 1–8. doi: 10.1016/j.mib.2020.01.002
Will, W. R., Navarre, W. W., and Fang, F. C. (2015). Integrated circuits: how transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr. Opin. Microbiol. 23, 8–13. doi: 10.1016/j.mib.2014.10.005
Wyborn, N. R., Stapleton, M. R., Norte, V. A., Roberts, R. E., Grafton, J., and Green, J. (2004). Regulation of Escherichia coli hemolysin E expression by H-NS and Salmonella SlyA. J. Bacteriol. 186, 1620–1628. doi: 10.1128/jb.186.6.1620-1628.2004
Yang, D., Kong, Y., Sun, W., Kong, W., and Shi, Y. (2019). A dopamine-responsive signal transduction controls transcription of Salmonella enterica serovar typhimurium virulence genes. mBio 10:e02772-18. doi: 10.1128/mBio.02772-18
Zaslaver, A., Bren, A., Ronen, M., Itzkovitz, S., Kikoin, I., Shavit, S., et al. (2006). A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628. doi: 10.1038/nmeth895
Zeghouf, M., Li, J., Butland, G., Borkowska, A., Canadien, V., Richards, D., et al. (2004). Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3, 463–468. doi: 10.1021/pr034084x
Zhang, Y., Zborníková, E., Rejman, D., and Gerdes, K. (2018). Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9:e02188-17. doi: 10.1128/mBio.02188-17
Keywords: SlyA, ppGpp, Escherichia coli, stress response, hlyE
Citation: Bartoli J, Viala JP and Bouveret E (2020) SlyA Transcriptional Regulator Is Not Directly Affected by ppGpp Levels. Front. Microbiol. 11:1856. doi: 10.3389/fmicb.2020.01856
Received: 24 April 2020; Accepted: 15 July 2020;
Published: 04 August 2020.
Edited by:
Michael Cashel, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), United StatesReviewed by:
William Ryan Will, University of Washington, United StatesCarlos Balsalobre, University of Barcelona, Spain
Gert Bange, University of Marburg, Germany
Copyright © 2020 Bartoli, Viala and Bouveret. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emmanuelle Bouveret, ZW1tYW51ZWxsZS5ib3V2ZXJldEBwYXN0ZXVyLmZy
 Julia Bartoli1
Julia Bartoli1 Emmanuelle Bouveret
Emmanuelle Bouveret